Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rapid detection of Candida albicans in urine by an Electrochemical Impedance Spectroscopy (EIS)-based biosensor†
Tina
D'Aponte
 a,
Maria
De Luca
a,
Maria
De Luca
 a,
Nikola
Sakač
b,
Martina
Schibeci
a,
Nikola
Sakač
b,
Martina
Schibeci
 c,
Angela
Arciello
c,
Emanuela
Roscetto
d,
Maria Rosaria
Catania
d,
Vincenzo
Iannotti
c,
Angela
Arciello
c,
Emanuela
Roscetto
d,
Maria Rosaria
Catania
d,
Vincenzo
Iannotti
 ae,
Raffaele
Velotta
ae,
Raffaele
Velotta
 a and
Bartolomeo
Della Ventura
a and
Bartolomeo
Della Ventura
 *a
*a
aDepartment of Physics “Ettore Pancini”, University of Naples “Federico II”, 80126 Naples, Italy. E-mail: bartolomeo.dellaventura@unina.it
bFaculty of Geotechnical Engineering, University of Zagreb, 42000 Varaždin, Croatia
cDepartment of Chemical Sciences, University of Naples “Federico II”, 80126 Naples, Italy
dDepartment of Molecular Medicine and Medical Biotechnology, University of Naples “Federico II”, 80131 Naples, Italy
eCNR – SPIN (Institute for Superconductors, Oxides and other Innovative Materials and Devices), Piazzale V. Tecchio 80, 80125 Naples, Italy
First published on 10th October 2023
Abstract
Candida albicans is a fungal organism commonly found in the human body, including the genitourinary tract. Overgrowth of this yeast can lead to candiduria, the abnormal presence of C. albicans in urine. The detection of candiduria relies on various methods that offer sensitivity and specificity for accurate diagnosis, but none of them is rapid and cost-effective. Biosensors may offer an answer; in particular, impedance-based biosensors have shown promise in addressing the detection of C. albicans since they offer high sensitivity, simplicity of fabrication, excellent selectivity, real-time detection, and cost-effectiveness. However, variations in the working surface of commercial screen-printed electrodes as well as manual functionalization can impact robustness and reproducibility. In this paper, we describe significant advances that we introduced to perform electrochemical impedance spectroscopy for biosensing purposes. Specifically, we designed a microfluidic cell for standardizing the electrode functionalization and target detection, ensuring sensor reproducibility and robustness. By optimizing several parameters like the flow rate and the density of antibodies on the electrode surface, we could achieve a limit of detection of 10 CFU mL−1 in urine with a measurement that lasted for less than 90 minutes. The modularity of the device and the measurement procedure makes the described biosensor extendable for conducting high-throughput analyses.
Introduction
Candida albicans is a fungal organism that commonly inhabits the human body, including the genitourinary tract.1,2 However, when this yeast overgrows, it can lead to a condition called candiduria, which involves the presence of C. albicans in urine. Candiduria is a significant issue that affects individuals of all ages and genders,3 particularly those with weakened immune systems or specific predisposing factors.4 Understanding the causes, symptoms, diagnosis, and treatment of candiduria is crucial for the effective management and prevention of complications.The gold standard method for diagnosing candiduria is urine culture. It involves inoculating urine samples onto appropriate culture media, such as Sabouraud dextrose agar or chromogenic agar, which selectively promote the growth of Candida species.5,6 Colonies are then identified through morphological characteristics, biochemical tests, or automated systems.7 This method provides both qualitative and quantitative information, but it is time-consuming since approximately 48 hours are required to obtain a response.
Polymerase chain reaction (PCR)8,9 and other molecular techniques have emerged as valuable tools for detecting Candida DNA in urine samples. These methods target specific Candida genes or regions and offer high sensitivity and specificity. PCR can differentiate between various Candida species and even detect low fungal loads. However, it is worth evidencing that PCR assays for Candida detection in urine typically do not provide quantitative information regarding the fungal load. This limitation may hinder the ability to determine the severity of infection or monitor treatment responses accurately.10
Enzyme-linked immunosorbent assays (ELISAs) are already utilized for Candida detection and demonstrate a good limit of detection (LOD). However, they are still associated with relatively long waiting times for obtaining results.11
In the last year, Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry MALDI-TOF MS has revolutionized microbial identification, including Candida species, since this technique analyses the protein profiles of microbial colonies and allows for rapid and accurate identification, even at the species level. Moreover, it can be applied to colonies obtained from urine culture. However, each of these methods has its own set of advantages and limitations, including factors like availability, cost, laboratory expertise, and the clinical context in which the test is being performed.12,13
Biosensors with enhanced sensitivity, selectivity, and ease-of-use would be crucial for improving the diagnosis and management of candiduria. In fact, a variety of biosensors have been proposed for the detection of Candida, offering different advantages and performance characteristics. Optical biosensing techniques,14 such as surface plasmon resonance and fluorescence-based assays,15,16 utilize the interaction of light with Candida-specific biomarkers for detection. Although they provide high sensitivity and real-time monitoring capabilities,17 these biosensors often require complex instrumentation, labelling processes, and expert personnel.
Electrochemical biosensors are emerging as a powerful tool for detecting pathogens in biological fluids such as blood, sweat, saliva, tears, and urine.18,19 This technology is also suitable for portable and wearable devices, allowing continuous monitoring and enabling early management in diagnostics.20,21 In particular, electrochemical biosensors offer an alternative approach for detecting Candida cells by leveraging their electrochemical properties.22 For instance, amperometric23 and potentiometric20 biosensors utilize the measurement of current or potential changes, respectively, upon Candida interaction. These biosensors demonstrate good sensitivity, simplicity, and cost-effectiveness, but they may suffer from limited selectivity due to non-specific interactions.
Impedance-based biosensors,24,25 which measure changes in electrical impedance resulting from Candida-cell interactions,26,27 have shown promise in addressing the limitations of previous biosensors. They provide high sensitivity, simplicity of fabrication, and excellent selectivity for Candida detection; in fact, sometimes they can even discriminate between different species of Candida with good sensitivity.28,29 Further, impedance-based biosensors offer real-time and label-free detection, enabling rapid analysis of samples without additional reagents. In this scenario, they have the potential to improve early diagnosis and accuracy in detecting candiduria due to all their advantages, while overcoming the disadvantages of the currently most widely used techniques (such as time and cost).
The use of screen-printed electrodes (SPEs) makes them cost-effective and reliable. The increasingly widespread use of SPEs in recent years could lead to acceleration in the development of SPE-based electrochemical sensors in various fields, including health monitoring.30 However, several limitations can arise, including robustness and reproducibility, due to variations in the working surface of each electrode, even if minimal, and the different conditions of manual drop-by-drop functionalization.31,32
In fact, electrochemical measurements are often conducted by incubating SPEs in various solutions. This serves two primary purposes: i) functionalizing the surface of the working electrode with antibodies, and ii) facilitating interaction with the analyte to be detected. The actual measurement is then performed in an electrolyte solution. However, this procedure leads to poor measurement reproducibility. The main issue arises because in addition to binding to the working electrode, both the antibodies and the analytes may also bind to the other two electrodes (auxiliary and counter electrodes). To address this challenge, we developed a microfluidic system that allows solutions to interact exclusively with the working electrode before SPEs are manually introduced into the electrochemical cell. It's worth noting that the use of microfluidics inherently reduces the operator dependence. With this approach, we have not only improved the performance of the biosensor but also minimized operator-related variability.
Experimental
Chemicals and materials
Potassium hexacyanoferrate(II) trihydrate (K4Fe(CN)6·3H2O), potassium hexacyanoferrate(III) (K3Fe(CN)6), bovine serum albumin (BSA) and sulphuric acid (H2SO4 98%) were purchased from Sigma-Aldrich (Milano, Italy). Phosphate buffered saline (PBS, 0.01 M, pH 7.4) was prepared by dissolving PBS tablets (from Gold Bio, St Louis, MO, USA) in Milli-Q water; each tablet prepares 100 mL of a 0.01 M PBS solution. Polyclonal antibodies (5.0 mg mL−1) against C. albicans were purchased from Fitzgerald Industries. All water-based solutions were prepared in ultrapure water (24 MΩ cm) using a Milli-Q system.Gold screen printed electrodes (AuSPEs) were purchased from BVT Technologies (Stražek, Czech Republic). They consist of a gold disk-shaped working electrode (d = 1 mm), a silver/silver chloride reference electrode and a gold counter electrode. These three electrodes were printed onto a corundum ceramic base measuring 0.7 × 2.5 cm2. All potential values were referred to the silver/silver chloride reference electrode.
C. albicans in GYP
C. albicans ATCC® 10231 cells were cultured in glucose–yeast extract–peptone (GYP) medium (glucose 20 g L−1; yeast extract 10 g L−1; peptone 20 g L−1) at 37 °C until exponential growth was reached. Afterwards, the optical density of the culture was measured, and the cells were diluted in 1/16 GYP medium at 2 × 106 CFU mL−1 before performing the analyses.Bacterial strain E. coli ATCC 25922 was grown in Muller Hinton broth (MHB, Becton Dickinson Difco, Franklin Lakes, NJ, USA) and on tryptic soy agar (TSA; Oxoid Ltd., Hampshire, UK). In all the experiments, bacteria were inoculated and grown overnight in MHB at 37 °C. The next day, bacteria were centrifuged and solubilized in 1× PBS at the desired cell densities (101–106 CFU mL−1). By colony counting assays, it was verified that bacterial growth was negligible in 1× PBS with respect to MHB through a time interval of 3 hours at room temperature, whereas bacterial death was not observed.
C. albicans in urine
Waste aliquots of urine samples used for quality controls were processed on dipslides (Urotube – Liofilchem) containing three different culture media (CLED agar, MacConkey agar, Sabouraud dextrose agar) and incubated for 48 hours at 37 °C to verify the absence of bacterial and fungal growth. Sterile urine aliquots were used for subsequent studies. Suspensions of the reference strain ATCC 10231 C. albicans were prepared at different concentrations (10n CFU mL−1 with n = 0, 1…7) in sterile urine aliquots. A control test was performed in sterile urine with null C. albicans concentration. The urine samples were real, anonymous urine samples, intended for disposal after being used as internal controls to test clinical biochemistry instrumentation. Before they were discarded, we used them for our experiments.Electrochemical cell
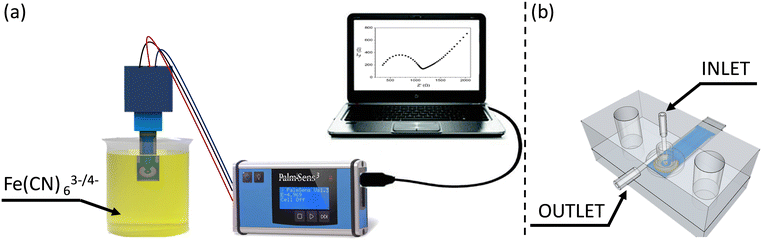
Electrochemical Impedance Spectroscopy (EIS) and cyclic voltammetry (CV) measurements were carried out by dipping the electrode into a beaker containing 3 mL of redox probe solution [Fe(CN)63−/Fe(CN)64− (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 40 mM)] at room temperature. The potentiostat and impedance analyser was a PALMSENS (model PalmSens3, Utrecht, The Netherlands) controlled using a computer through the PSTRACE version 5.5 software (Fig. 1a).
1, 40 mM)] at room temperature. The potentiostat and impedance analyser was a PALMSENS (model PalmSens3, Utrecht, The Netherlands) controlled using a computer through the PSTRACE version 5.5 software (Fig. 1a).
EIS measurements were performed at frequencies that ranged from 0.5 Hz to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Hz at a formal potential of 0.16 V and using an amplitude perturbation of 10 mV.
000 Hz at a formal potential of 0.16 V and using an amplitude perturbation of 10 mV.
The interaction of the Candida cells with the working electrode of the SPE was realized using a customized fluidic cell featuring an inlet that exclusively targets the working surface while keeping the auxiliary and reference electrodes isolated from the solutions. This cell is shown in Fig. 1b, in which the microfluidic cell is depicted in X-ray view, providing a clear visualization of the specific geometry utilized to selectively wet the working electrode without affecting the adjacent two electrodes.
In fact, microfluidics plays a crucial role in enhancing the capabilities of EIS and its integration with EIS offers several advantages for impedance-based biosensors. Firstly, microfluidic devices enable precise control and manipulation of fluid flow, allowing for a uniform distribution of samples and reagents across the sensing area. This uniformity ensures consistent and reliable measurements, minimizing variations that may arise from manual handling. Secondly, microfluidic channels can be designed to optimize the interaction between Candida cells and the electrode surface.
In this regard, by controlling the flow rate, cell concentration, and flowing time, the detection sensitivity can be optimized. The confined microscale environment also facilitates efficient mass transport, ensuring enhanced sensitivity and faster response times.
SPE cleaning procedure
The gold screen-printed electrodes were electrochemically cleaned using a 50 mM sulfuric acid (H2SO4).33 The potential was cycled from −0.4 V to 1.4 V, with a scan rate of 0.1 V s−1, until the cyclic voltammogram became stable (typically it took approximately 12 cycles). The electrode was then rinsed thoroughly with PBS and immersed in the electrolyte solution.Fig. S1a† shows the CV scans obtained before and after the cleaning procedure. The CV signal is that obtained after six scans in the range −0.6 V to +0.6 V, with a scan rate of 0.1 V. The CV scan after cleaning shows an increase in current, indicating a cleaner surface. Moreover, this procedure allowed us to measure the redox potential of the Fe(CN)63−/Fe(CN)64− solution, ultimately obtaining the formal potential of the redox couple for the subsequent EIS measurements. It is worth noticing that after cleaning, a reduction in impedance is also observed (as illustrated in Fig. S1b†); in particular, a lower RCT is revealing of a cleaner surface.
Electrode functionalization
The functionalization of the gold working electrode was accomplished using the Photochemical Immobilization Technique (PIT),34,35 which allows for the covalent attachment of IgG antibodies specific to C. albicans. This technique ensures that the fragment antigen binding site (Fab) of the antibodies is adequately exposed to the solution. To achieve this, antibodies in a solution (25 μg mL−1) were activated by UV-C light from a low-pressure mercury lamp, delivering approximately 0.3 W cm−2. The irradiation lasted for 30 seconds, after which the solution was introduced into the microfluidic cell and flowed over the working electrode of the AuSPE for 15 minutes. This duration was chosen as a trade-off between the short time required by PIT (the produced thiol is effective for approximately 10 minutes)34 and the extended time needed to conserve Abs (as a consumable material). Subsequently, a washing step with PBS was carried out to eliminate non-bonded Abs. The sequence of Abs irradiation, solution flow, and washing was repeated several times to optimize the surface coverage (see the section Results and discussion).EIS data fitting
In EIS, the data from impedance measurement are usually represented by a Nyquist plot, in which each point represents the response of the electrochemical cell at different frequencies of input voltage (Fig. S2a†). The simplest equivalent circuit to model the behaviour of the cell is the Randles circuit (Fig. S2b†).36 This circuit is composed of the resistance of the electrolyte solution (RS), the double-layer capacitance (CDL), the Warburg impedance (W) and the charge transfer resistance RCT. RS and W represent the bulk properties of the electrolyte solution and the diffusion features of the redox probe. These values are not influenced by modification on the electrode surface and are not modified by the antibodies–analyte interaction. In contrast, RCT depends critically on the dielectric and insulating features at the electrode–electrolyte interface; thus, since it is very sensitive to electrode modifications, it was used as a biosensing parameter.Frequently, the ideal capacitor CDL is replaced by a constant phase element CPE (Fig. S2c†) to account for inhomogeneity and defect areas of the biological layer. This element can fit the imperfect behaviour of the double-layer capacitance due to the porosity or the roughness of the electrode area.
A version of the Randles circuit reported in Fig. S2c† was used to find RCT from the Nyquist plots at the various target concentrations. To this aim, we relied on a tool from the PSTrace 5.5 software that used the Levenberg–Marquardt algorithm.
Results and discussion
Optimization of the functionalization procedure
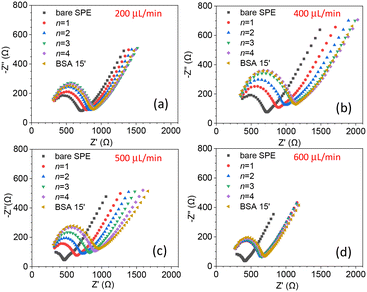
The biosensor described in this paper has several parameters that need to be optimized. In particular, both the flow rate, which affects the streamline distribution and the velocity of the particles, and the degree of surface coverage are crucial for sensitivity. We started by measuring the kinetics of the surface coverage by (UV-activated) antibodies at different flow rates. The results are shown in Fig. 2(a)–(d), in which n stands for the number of times the sequence of steps – UV activation of Abs, flowing of Abs solution and washing (see the Electrode functionalization section) – was carried out. The surface blocking was realized by conveying a solution of BSA [50 μg mL−1] to the cell for 15 minutes. As Fig. 2 shows, after each stage (n = 1, 4), while the impedance increased as a result of the presence of Abs on the surface, no significant change of impedance was produced by BSA. This means that for all the investigated flow rates, four functionalization stages – each including 15 minutes of UV-activated Abs flowing into the cell – were enough to fully cover the electrode surface.As is evident from Fig. 2, even the bare SPEs show different behavior (Nyquist plot and Rct) after the cleaning procedure – a feature that is unavoidable with typical commercial electrodes. To take into account the electrode variability, we normalized the change of the charge transfer resistance to its initial value Rct bare
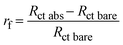 | (1) |
As a further check of the binding kinetics of Abs to the surface, we fixed the flow rate at 450 μL min−1 and measured rf after several functionalization stages. The results are reported in Fig. S4,† from which we deduced that n = 4 stages are sufficient to achieve saturation, i.e., an electrode surface fully covered by Abs.
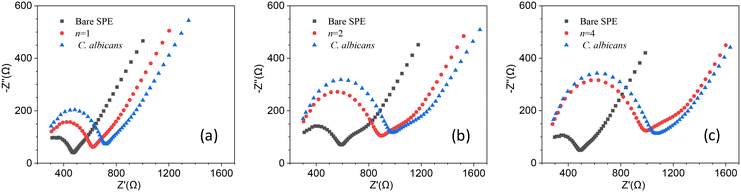
Since the degree of surface coverage by antibodies plays a crucial role in the charge transfer impedance, we wondered whether n ≥ 4 (surface fully covered by Abs) led to an optimal response for EIS, or rather, a surface with a lower degree of coverage could give rise to a stronger variation of Rct. The reason behind this question is that when the surface is fully covered, it may restrict the current variations resulting from target recognition, ultimately hampering the resistance change. To determine the optimal degree of surface coverage, we measured the change in Rct induced by a relatively low concentration of C. albicans (102 CFU mL−1) using electrodes that underwent different stages of functionalization (n = 1, 2 and 4). The results are shown in Fig. 3 from which one can immediately see that the change of the charge transfer resistance – i.e., the difference of Rct measured after and before the target is conveyed to the cell – is larger for n = 1 [Fig. 3(a)]. In fact, Rct increases as n goes from 1 to 4 as a result of more Abs on the electrode, but this condition is not beneficial for the sensitivity.
With the same arguments used to define rf, we can define the ratio rs, which is due to the target recognition, as follows:
 | (2) |
Detection of C. albicans in GYP
A volume of 1 mL of GYP containing 106 cells was serially diluted in GYP to have samples (1 mL) with C. albicans in the range of 101–106 cells per mL. The samples flowed in the circuit for 1 hour at room temperature, allowing for the capture of C. albicans by the immobilized antibodies on the electrode surface. Subsequently, the electrode was rinsed with 0.01 M PBS for 15 minutes to remove any non-specifically or weakly bound C. albicans cells. The charge transfer resistance was then measured by dipping the electrode in the electrolytic solution.The results are shown in Fig. S5† in which the errors arise from measurements carried out in duplicate or triplicate, whereas the experimental data were fitted by a logistic function. The shaded area represents the no-signal region when the 3SD criterion is adopted. In this case, the signal threshold is set above the control by 3 standard deviations. The relatively high uncertainty in the signal measured for the control can be attributed to fluctuations in the protein content of GYP, which is relatively high. It is evident that since the electrode surface was not fully covered, these proteins are capable of binding to the electrode in a non-specific manner. Nevertheless, even under these unfavorable conditions, a limit of detection of 102 CFU mL−1 can be safely deduced from Fig. S5.†
Detection of C. albicans in urine
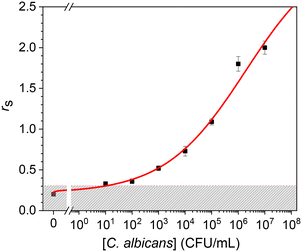
In an effort to develop a biosensor for medical diagnostic applications, we tested our procedure for detecting C. albicans in urine. We spiked an initial concentration of 107 CFU mL−1 of cells grown in GYP, and the dose–response curve is reported in Fig. 4. The experimental data resulted from measurements carried out in triplicate and were fitted by a logistic curve. All the measurements were performed by using different commercial gold screen printed electrodes, which are the main source of variability in this kind of biosensor as witnessed from the EIS spectra shown in Fig. 2. Thus, the tiny deviations of the experimental points from the logistic curve in Fig. 4 demonstrate the effectiveness of the normalization procedure described by eqn (1) and (2).A comparison of the data obtained in urine with those measured in GYP (Fig. S5†) reveals a lower control value – as well as a lower uncertainty – in urine than in GYP. This significant difference has a notable influence on the limit of detection because, by applying the 3SD criterion (shaded area in Fig. 4), we can reach a value of 10 CFU mL−1. As previously observed, this can be attributed to the high protein content in GYP. In fact, despite being a real sample, urine typically has a much lower concentration of analytes that could potentially interfere with the impedance measurement.
The presence of proteins in GYP can also explain the larger dynamic range exhibited by the detection of cells in urine. The dose–response curve suggests an asymptotic value for rs of approximately 3, whereas in GYP, a saturation value of approximately 0.8 is observed. In fact, the presence of analytes (proteins) capable of binding to the partially covered electrode surface inherently reduces the effective area available for detection, thereby limiting the maximum detectable concentration.
Specificity test
To assess the specificity of the developed immunosensor for C. albicans, we conducted a test to evaluate the response of the functionalized immunosensor to another non-specific microorganism, Escherichia coli, which is known to cause a significant proportion of urinary tract infections.37 Since E. coli can potentially be present in urine samples, it is a suitable candidate for the assessment of the lack of cross-reactivity. We carried out the test on different urine samples received in the laboratory for biochemical investigations and used as internal controls in bacteriology before being discarded.An aliquot of urine of 1 mL was incubated with E. coli at a concentration of 106 CFU mL−1 and made to flow through the circuit for one hour at room temperature.
Subsequently, a washing step with PBS (0.01 M) was performed for 15 minutes to remove any non-specific bonds or contaminants. Fig. 5, which illustrates the changes in Rct, demonstrates that the charge transfer resistance is only slightly higher than that of the negative control (1 mL of urine), whereas it is significantly lower than the value measured in C. albicans at the same concentration. This outcome was somewhat expected since we intentionally employed a surface that was not fully saturated with antibodies to enhance the visibility of the target microorganism. However, due to their smaller size, the bacteria may exhibit some interference. Nevertheless, in view of its application, we can confidently conclude that the specificity of the biosensor described here is more than satisfactory.
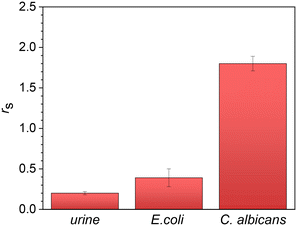 | ||
| Fig. 5 Specificity test. At the same concentration of 106 CFU mL−1, the signal from C. albicans is approximately four times larger than that from E. coli. | ||
Test of the biosensor
To verify the reliability of our biosensor, we spiked C. albicans in 1.1 mL urine to have a concentration of 104 CFU mL−1. Part of this volume (<100 μL) was used to yield four samples with different concentrations (104 CFU mL−1 and dilutions 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, and 1
100, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
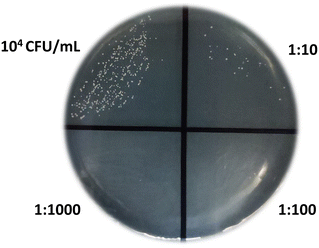
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000). A volume of 10 μL of each concentration was added to a Petri dish specifically designed for cultivating C. albicans, whereas the remaining part (≈1 mL) was used for biosensing measurements. Fig. 6 visually presents the presence and growth (24 hours) of fungal colonies at the spiked concentration and various dilutions. The colony count in the undiluted sample (10 μL) was ≈200 (197 colonies were actually recognized) and scaled with the dilutions accordingly. Thus the concentration measured by the cell growth method was 2 × 104 CFU mL−1.
1000). A volume of 10 μL of each concentration was added to a Petri dish specifically designed for cultivating C. albicans, whereas the remaining part (≈1 mL) was used for biosensing measurements. Fig. 6 visually presents the presence and growth (24 hours) of fungal colonies at the spiked concentration and various dilutions. The colony count in the undiluted sample (10 μL) was ≈200 (197 colonies were actually recognized) and scaled with the dilutions accordingly. Thus the concentration measured by the cell growth method was 2 × 104 CFU mL−1.
The measurement with the biosensor provided the value rs = 0.75 ± 0.10 that corresponds (see Fig. 4) to a cell concentration in the range 1–2 (×104) CFU mL−1, which is in satisfactory agreement with the value achieved from the cell culture.
Conclusions
C. albicans is the most common cause of superficial and invasive fungal infections, and urine samples are obviously suitable for diagnosing urinary tract infections. The microbiological diagnosis of urinary infections is based on a cut-off that is generally set at 104–105 CFU mL−1, although the detected concentration value is provided to a clinician for appropriate interpretation, even if it is lower than the cut-off. The need to have a good sensitivity at lower concentrations is especially critical in the case of newborns or suprapubic aspiration.38 Moreover, diagnosing Candida infections typically relies on culture examination, which can take 48–72 hours for fungal isolation and identification. Therefore, there is a strong need for a rapid and reliable test to detect this infection. To meet this demand, we have developed an electrochemical impedance immunosensor that utilizes commercial screen-printed electrodes for the fast and sensitive detection of C. albicans in urine.The gold surface of the electrode was functionalized with antibodies using a UV-activation technique known as the Photochemical Immobilization Technique (PIT)34 – a procedure recently recognized for its remarkable effectiveness in tethering Abs to gold surfaces.39 The change in charge transfer resistance (Rct) served as the indicator of the presence of C. albicans.
Interestingly, we found that for targets as large as fungi, partial coverage of the surface resulted in a stronger signal that allowed us to achieve a limit of detection of 10 CFU mL−1 in urine. While this limit can generally be reached or exceeded (at least in simple matrices), it is noteworthy that our biosensor utilizes commercial screen-printed electrodes, which we incorporated into a fluidic design to optimize the interaction between the target and the surface. As a result, we were able to measure the concentration of C. albicans in urine in less than 90 minutes with a procedure that is virtually independent of the operator.
Given the simplicity of the equipment required to carry out the measurements, our biosensor lends itself as a point-of-care device. Moreover, both the cell and the fluidic components can be easily upgraded to create a multiplexing device. Overall, the combination of microfluidics and electrochemical impedance spectroscopy described here offers significant advantages only in terms of improved sensitivity, enhanced throughput, and precise control of fluidic parameters.
Author contributions
Tina D'Aponte: investigation, data curation and writing – original draft. Maria De Luca: data curation and software. Nikola Sakač: formal analysis and writing – original draft. Martina Schibeci: resources and validation. Angela Arciello: resources and writing – review & editing. Emanuela Roscetto: resources and validation. Maria Rosaria Catania: resources and validation. Vincenzo Iannotti: methodology and writing – review & editing. Raffaele Velotta: methodology, formal analysis, writing – review & editing and supervision. Bartolomeo Della Ventura: conceptualization, methodology, data analysis, funding acquisition, and writing – review & editing.Conflicts of interest
“There are no conflicts to declare”.Acknowledgements
This study was carried out within the Agritech National Research Center and received funding from the European Union Next-GenerationEU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR) – MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4 – D.D. 1032 17/06/2022, CN00000022).References
- J. F. Fisher, Clin. Infect. Dis., 2011, 52, S429–S432 CrossRef PubMed.
- W. A. Alfouzan and R. Dhar, J. Mycol. Med., 2017, 27, 293–302 CrossRef CAS PubMed.
- P. Datta, M. Kaur, S. Gombar and J. Chander, Indian J. Crit. Care Med., 2018, 22, 56–57 CrossRef CAS PubMed.
- M. Gajdács, I. Dóczi, M. Ábrók, A. Lázár and K. Burián, Cent. Eur. J. Urol., 2019, 72, 209–214 Search PubMed.
- T. T. Özer, S. Durmaz and E. Yula, J. Infect. Chemother., 2016, 22, 629–632 CrossRef PubMed.
- P. Behzadi, E. Behzadi and R. Ranjbar, Cent. Eur. J. Urol., 2015, 68, 96–101 Search PubMed.
- L. Toner, N. Papa, S. H. Aliyu, H. Dev, N. Lawrentschuk and S. Al-Hayek, QJM, 2016, 109, 325–329 CrossRef CAS PubMed.
- E. E. Nejad, P. G. N. Almani, M. A. Mohammadi and S. Salari, J. Clin. Lab. Anal., 2020, 34, 1–8 Search PubMed.
- P. L. White, J. S. Price, A. Cordey and M. Backx, Curr. Fungal Infect. Rep., 2021, 15, 67–80 CrossRef PubMed.
- E. K. Dennis, S. Chaturvedi and V. Chaturvedi, Front. Microbiol., 2021, 12, 757835 CrossRef PubMed.
- K. Wang, Y. Luo, W. Zhang, S. Xie, P. Yan, Y. Liu, Y. Li, X. Ma, K. Xiao, H. Fu, J. Cai and L. Xie, Mycoses, 2020, 63, 181–188 CrossRef CAS PubMed.
- C. Rizzato, L. Lombardi, M. Zoppo, A. Lupetti and A. Tavanti, J. Fungi, 2015, 1, 367–383 CrossRef PubMed.
- E. Svetličić, L. Dončević, L. Ozdanovac, A. Janeš, T. Tustonić, A. Štajduhar, A. L. Brkić, M. Čeprnja and M. Cindrić, Molecules, 2022, 2717, 54–61 Search PubMed.
- K. K. Hussain, D. Malavia, E. M. Johnson, J. Littlechild, C. P. Winlove, F. Vollmer and N. A. R. Gow, J. Fungi, 2020, 6, 1–26 Search PubMed.
- L. Mendive-Tapia, D. Mendive-Tapia, C. Zhao, D. Gordon, S. Benson, M. J. Bromley, W. Wang, J. Wu, A. Kopp, L. Ackermann and M. Vendrell, Angew. Chem., 2022, 61, e202117218 CrossRef CAS PubMed.
- D. Yu, L. Wang, H. Zhou, X. Zhang, L. Wang and N. Qiao, Bioconjugate Chem., 2019, 30, 966–973 CrossRef CAS PubMed.
- Y. Zhang, H. Duan, Y. Liu, Y. Li and J. Lin, Comput. Electron. Agric., 2023, 206, 107702 CrossRef.
- E. Cesewski and B. N. Johnson, Biosens. Bioelectron., 2020, 159, 112214 CrossRef CAS PubMed.
- M. Sarabaegi and M. Roushani, Anal. Methods, 2019, 11, 5591–5597 RSC.
- J. Wu, H. Liu, W. Chen, B. Ma and H. Ju, Nat. Rev. Bioeng., 2023, 1, 1–15 CrossRef.
- L. De Brito Ayres, J. Brooks, K. Whitehead and C. D. Garcia, Anal. Chem., 2022, 94, 16847–16854 CrossRef CAS PubMed.
- P. Dutta, Y. J. Lu, H. Y. Hsieh, T. Y. Lee, Y. T. Lee, C. M. Cheng and Y. J. Fan, Micromachines, 2021, 12, 166 CrossRef PubMed.
- S. Tvorynska, J. Barek and B. Josypčuk, Sens. Actuators, B, 2021, 13, 2–52 Search PubMed.
- M. Cimafonte, A. Fulgione, R. Gaglione, M. Papaianni, R. Capparelli, A. Arciello, S. B. Censi, G. Borriello, R. Velotta and B. Della Ventura, Sensors, 2020, 20, 274 CrossRef CAS PubMed.
- C. Zhang, Y. Su, S. Hu, K. Jin, Y. Jie, W. Li, A. Nathan and H. Ma, ACS Omega, 2020, 5, 5098–5104 CrossRef CAS PubMed.
- D. Kwasny, S. E. Tehrani, C. Almeida, I. Schjødt, M. Dimaki and W. E. Svendsen, Sensors, 2018, 18, 1–9 CrossRef PubMed.
- A. G. da Silva-Junio, I. A. M. Frias, R. G. Lima-Neto, L. Migliolo, P. S. e Silva, M. D. L. Oliveira and C. A. S. Andrade, J. Pharm. Biomed. Anal., 2022, 216, 114788 CrossRef CAS PubMed.
- K. L. Ribeiro, I. A. M. Frías, A. G. Silva, R. G. Lima-Neto, S. R. Sá, O. L. Franco, M. D. L. Oliveira and C. A. S. Andrade, Biochem. Eng. J., 2021, 167, 107918 CrossRef CAS.
- S. R. Sá, A. G. Silva Jr, R. G. Lima-Neto, C. A. S. Andrade and M. D. L. Oliveira, Talanta, 2020, 220, 121375 CrossRef PubMed.
- R. D. Crapnell, A. Garcia-Miranda Ferrari, N. C. Dempsey and C. E. Banks, Sens. Diagn., 2022, 1, 405–428 RSC.
- J. L. de Miranda, M. D. L. Oliveira, I. S. Oliveira, I. A. M. Frias, O. L. Franco and C. A. S. Andrade, Biochem. Eng. J., 2017, 124, 108–114 CrossRef CAS.
- L. A. Layqah and S. Eissa, Microchim. Acta, 2019, 186, 224 CrossRef PubMed.
- L. M. Fischer, M. Tenje, A. R. Heiskanen, N. Masuda, J. Castillo, A. Bentien, J. Émneus, M. H. Jakobsen and A. Boisen, Microelectron. Eng., 2009, 86, 1282–1285 CrossRef CAS.
- B. Della Ventura, M. Banchelli, R. Funari, A. Illiano, M. De Angelis, P. Taroni, A. Amoresano, P. Matteini and R. Velotta, Analyst, 2019, 144, 6871–6880 RSC.
- R. Funari, B. Della Ventura, C. Altucci, A. Offenhäusser, D. Mayer and R. Velotta, Langmuir, 2016, 32, 8084–8091 CrossRef CAS PubMed.
- T. Bertok, L. Lorencova, E. Chocholova, E. Jane, A. Vikartovska, P. Kasak and J. Tkac, ChemElectroChem, 2019, 6, 989–1003 CrossRef CAS.
- R. D. Klein and S. J. Hultgren, Nat. Rev. Microbiol., 2020, 18, 211–226 CrossRef CAS PubMed.
- M. G. Coulthard, Pediatr. Nephrol., 2019, 34, 1639–1649 CrossRef PubMed.
- M. Conrad, G. Proll, E. Builes-Münden, A. Dietzel, S. Wagner and G. Gauglitz, Microchim. Acta, 2023, 190, 62 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00209h |
| This journal is © The Royal Society of Chemistry 2023 |