Open Access Article
Open Access ArticleChromogenic enzyme substrates based on [2-(nitroaryl)ethenyl]pyridinium and quinolinium derivatives for the detection of nitroreductase activity in clinically important microorganisms†
Marie
Cellier-Rastit
a,
Valérie
Chalansonnet
a,
Arthur L.
James‡
b,
Annette
Johnston
b,
Sylvain
Orenga
a,
John D.
Perry
c,
Celine
Roger-Dalbert
a,
Vindhya L.
Salwatura
b,
Stephen P.
Stanforth
 *b,
Hannah E.
Sykes
b,
Viet T.
Truong
b and
Graeme
Turnbull
*b,
Hannah E.
Sykes
b,
Viet T.
Truong
b and
Graeme
Turnbull
 b
b
aResearch & Development Microbiology, bioMérieux SA, 3 route de Port Michaud, La-Balme-les-Grottes 38 390, France
bDepartment of Applied Sciences, Northumbria University, Newcastle upon Tyne, NE1 8ST, UK. E-mail: steven.stanforth@northumbria.ac.uk
cDepartment of Microbiology, Freeman Hospital, Newcastle upon Tyne NE7 7DN, UK
First published on 7th June 2023
Abstract
A series of [2-(nitroaryl)ethenyl]pyridinium and quinolinium derivatives have been synthesised as potential indicators of microbial nitroreductase activity. When assessed against a selection of 20 clinically important pathogenic microorganisms, microbial colonies of various colours (yellow, green, red, brown, black) were produced and attributed to nitroreductase activity. Most substrates elicited colour responses with Gram-negative microorganisms. In contrast, the growth of several species of Gram-positive microorganisms and yeasts was often inhibited by the substrates and hence coloured responses were not seen.
Introduction
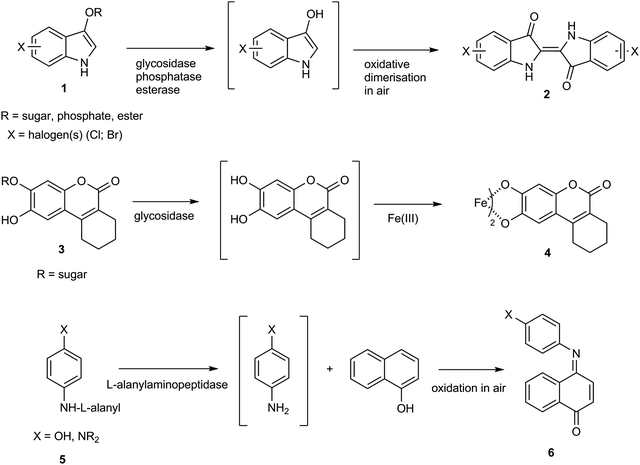
The detection and identification of pathogenic microorganisms using chromogenic culture media has emerged as an important diagnostic tool in clinical microbiology.1–4 Favourable attributes which have encouraged the extensive use of chromogenic culture media include ease of use, low cost and wide commercial availability. By incorporating a weakly coloured enzyme substrate into a culture medium, a colour change can be observed when a suitable microbial enzyme transforms the substrate into a highly coloured product. Many commercially available media incorporate halogenated indoxylic substrates 1 which, in the presence of a microbial hydrolase, produce strongly coloured indigo dyes 2 (Scheme 1).1–4 Such substrates generally show low toxicity towards microorganisms and a wide range of indoxylic substrates is commercially available for detection of a range of glucosidase and esterase enzymes. An important limitation is that the presence of oxygen is required for generation of the indigo dye 2 and they are therefore not suitable for detection of strictly anaerobic bacteria. No indoxylic substrates have been described for detection of nitroreductase activity.Glycosides of catechol derivatives, as exemplified by coumarins 3,5–8 have also been assessed for applications in chromogenic culture media. Hydrolysis of these substrates by a microbial glycosidase liberates the catechol which in the presence of an Fe(III) salt forms a black coloured chelate 4. More recently, the hydrolysis of derivatives of substrates 5 by an L-alanylaminopeptidase in the presence of 1-naphthol or a substituted 1-naphthol derivative has been shown to generate the either blue- or maroon-coloured dyes 6.9
Nitroreductases are widely distributed across microorganisms and are capable of reducing nitroaromatics to their corresponding arylamines (or arylhydroxylamines).10 A review describing the applications of small molecules as probes for nitroreductase activity has recently been published.11 In spite of ongoing interest in this area, relatively few applications of chromogenic nitroreductase substrates/probes in the microbial diagnostics arena have been reported. There are multiple potential uses of such substrates. A common test in diagnostic microbiology is the measurement of a total viable count to assess the overall microbial burden of water samples or foodstuffs or to confirm the sterility of pharmaceutical products. The potential to highlight all microbial colonies, which can sometimes be translucent, with a specific coloured reaction can facilitate visualization and counting by manual or automated methods. Although it is most useful to highlight bacterial pathogens using a specific hydrolase, such specific enzymes are often absent and it is therefore necessary to highlight them using a non-specific substrate and to rely on complementary hydrolase substrates to differentiate any competing flora that is able to grow despite the inclusion of targeted inhibitors. Finally, the deletion or modification of genes for nitroreductase can lead to resistance to antibiotics such as nitrofurantoin and metronidazole that require reduction by bacteria in order to exert their antibacterial effect.12,13 Such substrates may be useful for screening for mutants that lack nitroreductase activity.
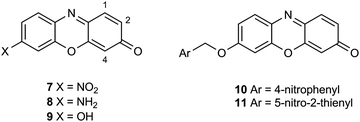
A series of halogenated nitrophenoxazinone derivatives 7 were examined as potential chromogenic substrates for the detection of pathogenic bacteria and several of these nitroaromatic compounds resulted in the formation of coloured bacterial colonies attributed to the formation of the amines 8 (Fig. 1).14 The 1,2,4-trifluoro derivative of heterocycle 7 produced the most encouraging results with Gram-negative bacteria where 9 out of a panel of 10 reduced this substrate thereby producing red-brown colonies. Most of the 8 Gram-positive bacteria studied were either unable to reduce this substrate or their growth was inhibited by this substrate and hence coloured colonies were not observed. Resorufin 9 is strongly fluorescent but it is also coloured and this heterocycle has been evaluated for a plethora of fluorogenic/chromogenic applications.15 The nitroreductase probes 10 and 11 which possess a self-immolative nitrobenzyl spacer have been evaluated as fluorogenic substrates for bacterial detection; reduction to the amine followed by fragmentation of the resulting aminobenzyl moiety liberates Resorufin.16,17 A fluorogenic probe designed around a cyanine dye possessing a pendant nitroimidazole has been described for the detection of ‘ESKAPE’ pathogens.18 The nitro-group quenches the fluorescence of the cyanine dye but after reduction of the nitro-group, the fluorescence of the cyanine dye is restored. Presumably a colour change also occurs following the reduction of the nitro-group, but this was not reported.
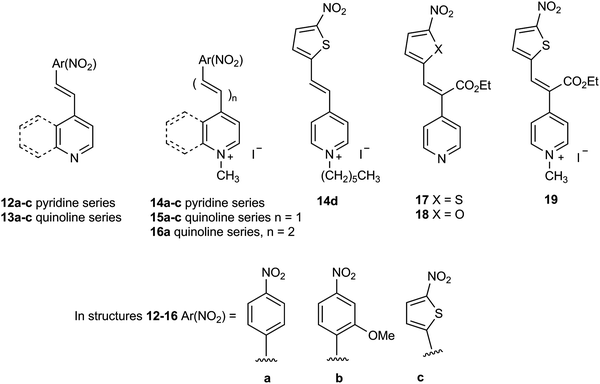
The widespread distribution of nitroreductase enzymes in microorganisms noted above combined with the relative sparsity of chromogenic culture media for detecting nitroreductase activity offers the potential to develop this area of microbial diagnostics. In this paper we describe the synthesis of a series of nitroreductase substrates 14–19 and their evaluation in agar media against clinically relevant, pathogenic bacteria (Fig. 2). It was anticipated that microbial reduction of the nitro-group in the quaternised structures 14–16 and 19 would generate a coloured ‘push–pull’ system in which the resulting amine (or hydroxylamine) lone-pair of electrons would be mesomerically associated with the pyridinium/quinolinium moiety thus demonstrating the capacity of microorganisms to reduce the substrate. Preliminary disclosures of some of these substrates have previously been made in the patent literature.19,20 In structures 17 and 18, the pyridine ring is not quaternised, but this is compensated to some extent by the presence of the electron-withdrawing ester substituent and hence these compounds were also included in this study. The agar media containing the nitroreductase substrates are only weakly coloured at the concentrations employed, but strongly coloured ‘push–pull’ systems are produced as a result of microbial nitroreductase activity. This dramatic change in colour enables visualisation of the microbial colonies and hence the detection of nitroreductase activity.
Results and discussion
(a) Synthesis of nitroreductase substrates
Synthetic procedures together with supporting characterisation data are given in the ESI.† The nitroreductase substrates 14a–c and 15a–c (76–98% yield) were prepared by quaternisation of the corresponding compounds 12a–c and 13a–c with methyl iodide respectively (Fig. 2). Heterocycle 14d was similarly prepared by quaternisation of compound 12c with n-hexyl iodide (53% yield). Heterocycle 16a was produced by the condensation of 1,4-dimethylquinolinium iodide with 4-nitrocinnamaldehyde under basic conditions (66% yield). The condensation of ethyl 4-pyridylacetate with either 5-nitrothiophene-2-carboxaldehyde or 5-nitrofuran-2-carboxaldehyde in the presence of acetic anhydride under basic conditions afforded compounds 17 (81% yield) and 18 (49% yield) respectively. Heterocycle 17 was subsequently quaternised with methyl iodide producing heterocycle 19 (59% yield).(b) Evaluation of substrates
A representative panel of 20 clinically important microorganisms (10 Gram-negative bacteria, 8 Gram-positive bacteria and 2 yeasts) were inoculated (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 colony forming units per spot) onto a single Columbia agar plate containing the substrate of interest (see tables for substrate concentration). All strains had been previously shown to reduce 7-nitrocoumarin-3-carboxylic acid using the method previously described by James et al.21
000 colony forming units per spot) onto a single Columbia agar plate containing the substrate of interest (see tables for substrate concentration). All strains had been previously shown to reduce 7-nitrocoumarin-3-carboxylic acid using the method previously described by James et al.21
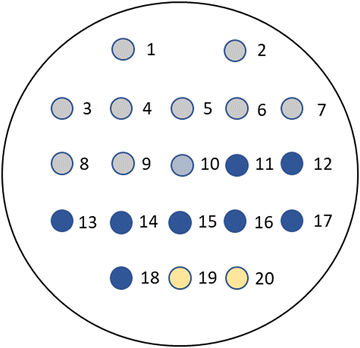
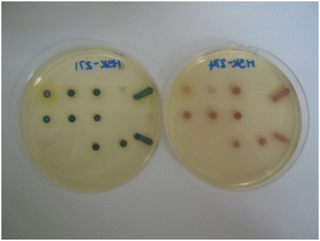
The arrangement of the microorganisms on each plate is depicted in Fig. 3 and the numbering (1–20) corresponds with the sequence of microorganisms shown in the evaluation tables. The growth of the microorganisms was compared with a control plate in which no substrate was present, but the corresponding volume of organic solvent required for dissolution of the substrate was added. Both the substrate-containing and control plates were then incubated at 37 °C in air for 18–20 hours.
The extent of microbial growth and the colour intensities produced as a consequence of nitroreductase activity in the presence of the pyridinium series of substrates 14a–d are shown in Table 1. The inclusion of parent nitroreductase substrate 14a in the media resulted in the generation of pale-yellow colonies when nitroreductase activity was present but there was not a particularly strong colour contrast with the agar background. All the Gram-negative bacteria grew satisfactorily in the presence of this substrate except for E. coli for which growth was inhibited. The growth of the Gram-positive bacteria S. pyogenes and S. aureus (MRSA) was also inhibited but the other Gram-positive bacteria did grow and reduced this substrate thereby producing a coloured response. Neither of the two yeast species reduced this substrate and hence the formation of coloured colonies was not observed.
| Substrate | 14a | 14b | 14c | 14d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Colony colour | Pale yellow | Gold | Maroon | Deep red | |||||||||
| Substrate conc. (mg L−1) | 100 | 100 | 100 | 50 | 25 | 50 | |||||||
| Colony growth (G)a and colour intensity (CI)b | G | CI | G | CI | G | CI | G | CI | G | CI | G | CI | |
| a + good growth, +/− weak growth, Tr. trace of growth, − no growth. Growth on control plate was + (Gram-negative species) and + (Gram-positive species and yeasts). b + strong colour, +/− weak colour, Tr. trace of colour, − no colour. c NCTC: National Collection of Type Cultures; ATCC: American Type Culture Collection; NCPF: National Collection of Pathogenic Fungi. d Formerly named Klebsiella pneumoniae. e Formerly named Bacillus subtilis. | |||||||||||||
| Microorganism/referencec | |||||||||||||
| Gram-negative microorganisms | |||||||||||||
| 1 | Escherichia coli | Tr. | +/− | − | − | − | − | − | − | + | + | − | − |
| NCTC 10418 | |||||||||||||
| 2 | Raoultella planticola | + | + | Tr. | − | − | − | − | − | 1 col. | + | + | + |
| NCTC 9528 | |||||||||||||
| 3 | Providencia rettgeri | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 7475 | |||||||||||||
| 4 | Enterobacter cloacae | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 11936 | |||||||||||||
| 5 | Serratia marcescens | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 10211 | |||||||||||||
| 6 | Salmonella typhimurium | + | + | +/− | − | − | − | − | − | + | + | 2 col. | + |
| NCTC 74 | |||||||||||||
| 7 | Pseudomonas aeruginosa | + | + | + | + | + | + | + | + | + | + | 12 col | + |
| NCTC 10662 | |||||||||||||
| 8 | Yersinia enterocolitica | + | + | + | + | +/− | +/− | + | + | + | + | + | + |
| NCTC 11176 | |||||||||||||
| 9 | Burkholderia cepacia | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 10743 | |||||||||||||
| 10 | Acinetobacter baumannii | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 12156 | |||||||||||||
| Gram-positive microorganisms | |||||||||||||
| 11 | Streptococcus pyogenes | − | − | − | − | − | − | + | + | + | +/− | − | − |
| NCTC 8306 | |||||||||||||
| 12 | Staphylococcus aureus (MRSA) | − | − | − | − | +/− | +/− | + | + | + | + | − | − |
| NCTC 11939 | |||||||||||||
| 13 | Staphylococcus aureus (MSSA) | + | + | − | − | + | + | + | + | + | + | − | − |
| NCTC 6571 | |||||||||||||
| 14 | Staphylococcus epidermidis | + | + | Tr. | − | − | − | +/− | +/− | + | +/− | − | − |
| NCTC 11047 | |||||||||||||
| 15 | Listeria monocytogenes | + | +/− | + | + | Tr. | Tr. | + | + | + | +/− | − | − |
| NCTC 11994 | |||||||||||||
| 16 | Enterococcus faecium | + | +/− | + | + | + | + | + | + | + | +/− | − | − |
| NCTC 7171 | |||||||||||||
| 17 | Enterococcus faecalis | + | + | + | + | + | + | + | + | + | +/− | − | − |
| NCTC 775 | |||||||||||||
| 18 | Bacillus atrophaeus | + | + | − | − | Tr. | Tr. | + | + | + | Tr. | − | − |
| ATCC 9372 | |||||||||||||
| Yeasts | |||||||||||||
| 19 | Candida albicans | +/− | − | + | − | + | − | + | − | + | − | +/− | − |
| ATCC 90028 | |||||||||||||
| 20 | Candida glabrata | − | − | − | − | − | − | − | − | Tr. | − | − | − |
| NCPF 3943 | |||||||||||||
A bifurcated approach was adopted to shift the colour of reduced substrate towards the red region of the visible spectrum in order to improve the colour contrast between the microbial colonies and the agar media. The first approach was to replace the 4-nitrophenyl group in substrate 14a with a group of greater electron-releasing capacity. The incorporation of a methoxy group was therefore examined and the resulting substrate 14b was reduced, leading to the generation of gold-coloured colonies which exhibited good contrast against the background (Table 1 and Fig. 4). There was minimal diffusion of the reduced substrate into the medium which is essential for accurately identifying individual bacterial colonies when several microorganisms from clinical samples might be present on the culture medium. Three of the Gram-negative bacteria (E. coli, R. planticola and S. typhimurium) showed either poor growth or were inhibited by this substrate thereby failing to generate coloured microbial colonies. Three of the Gram-positive bacteria displayed moderate growth and hence produced microbial colonies that were less intensely coloured than those associated with their Gram-negative counterparts. Coloured colonies were not observed for either of the two yeast species with this substrate.
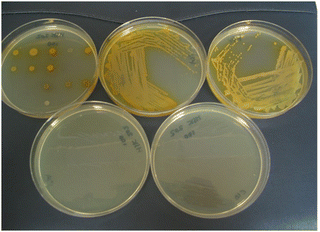 | ||
| Fig. 4 Evaluation of substrate 14b in Columbia agar (substrate concentration 100 mg L−1). Top left; 20 microorganisms (see Fig. 3 and Table 1 for the arrangement of microorganisms on the plate), top middle; Pseudomonas aeruginosa, top right; Acinetobacter baumannii, bottom left; Escherichia coli (inhibited); bottom right; Staphylococcus aureus (MRSA) (inhibited). | ||
By substituting the nitrophenyl-ring in substrates 14a with the more electron-rich nitrothiophene-ring, the thienyl-substrate 14c resulted in the production of maroon-coloured microbial colonies with excellent colour contrast against the background and minimal diffusion into the medium (Table 1 and Fig. 5). At a substrate concentration of 100 mg L−1, the growth of the Gram-negative bacteria E. coli, R. planticola and S. typhimurium was inhibited by this substrate and hence no coloured colonies were visible. The remaining Gram-negative bacteria exhibited good growth (apart from Y. enterocolitica for which growth was weak) and hence coloured colonies were generated. Two of the Gram-positive bacteria (S. pyogenes and S. epidermidis) were inhibited by this substrate, but the other Gram-positive bacteria all reduced this substrate thereby producing colonies with various colour intensities. Of the two yeast species, only C. albicans grew but nitroreductase activity was not detected since a coloured response was not observed. In order to alleviate growth inhibition by the substrate, its concentration was reduced to both 50 mg L−1 and 25 mg L−1. The substrate was less inhibitory to the growth of Y. enterocolitica, S. aureus (MRSA), S. epidermidis (previously inhibited) and L. monocytogenes at a concentration of 50 mg L−1 and a concomitant increase in colour intensity was observed. At a substrate concentration of 25 mg L−1, most of the Gram-negative and Gram-positive bacteria grew satisfactorily (although R. planticola produced only one colony, which is not clearly visible in Fig. 5), and coloured colonies could be seen. No coloured colonies were observed for either of the yeast species. The intensity of colour associated with some Gram-positive microorganisms decreased when the substrate concentration was reduced from 50 mg L−1 to 25 mg L−1, presumably due to lower concentrations of the resulting chromophore. The n-hexyl derivative 14d displayed a similar profile to its N-methylated analogue 14c with the Gram-negative microorganisms (substrate concentration 50 mg L−1) and deep, red-coloured colonies were observed. The growth of all Gram-positive bacteria was inhibited by compound 14d and hence no coloured microbial colonies were visible.
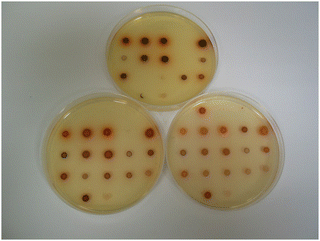 | ||
| Fig. 5 Evaluation of substrate 14c in columbia agar at various substrate concentrations. See Fig. 3 and Table 1 for the arrangement of microorganisms on the plates. Top image, 100 mg L−1; bottom left, 50 mg L−1; bottom right, 25 mg L−1. | ||
The second approach to shifting the pale-yellow colour of substrate 14a towards the red region of the visible spectrum was to increase the overall conjugation within the substrate by replacing the pyridinium ring in substrate 14a with a quinolinium ring. This change was beneficial and substrate 15a resulted in the generation of orange-coloured colonies when nitroreductase activity was present (Table 2). Most of the Gram-negative bacteria exhibited good growth when the substrate concentration was 100 mg L−1. Only E. coli was completely inhibited at this concentration and the growth of R. planticola was weak. Broadly similar results were observed at the lower substrate concentrations and although S. typhimurium and Y. enterocolitica both grew at 25 mg L−1, neither were associated with coloured colonies. Four of the Gram-positive bacteria were inhibited by this substrate at a concentration of 100 mg L−1. The inhibition of Gram-positive bacteria in Columbia agar media has previously been noted with halogenated derivatives of the nitrophenoxazinones 7.14 Reducing the substrate concentration to 25 mg L−1 did allow the detection of two additional Gram-positive bacteria (S. epidermidis and S. pyogenes), but their associated colour responses were weak at best. Of the two yeast species, only C. albicans colonies produced a trace of colour at low (25 mg L−1) substrate concentration. The profile of substrate 15b was similar to that of substrate 15a with the formation of red/orange-coloured colonies when nitroreductase activity occurred.
| Substrate | 15a | 15b | 15c | 16a | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Colony colour | Orange | Red or orangef | Black | Brown | |||||||||||||
| Substrate conc. (mg L−1) | 100 | 50 | 25 | 50 | 100 | 50 | 25 | 100 | |||||||||
| Colony growth (G)a and colour intensity (CI)b | G | CI | G | CI | G | CI | G | CI | G | CI | G | CI | G | CI | G | CI | |
| a + good growth, +/− weak growth, Tr. trace of growth, − no growth. Growth on control plate was + (Gram-negative species) and + (Gram-positive species and yeasts). b + strong colour, +/− weak colour, Tr. trace of colour, − no colour. c NCTC: National Collection of Type Cultures; ATCC: American Type Culture Collection; NCPF: National Collection of Pathogenic Fungi. d Formerly named Klebsiella pneumoniae. e Formerly named Bacillus subtilis. f Red (Gram-negative), orange (Gram-positive and yeasts). | |||||||||||||||||
| Microorganism/referencec | |||||||||||||||||
| Gram-negative microorganisms | |||||||||||||||||
| 1 | Escherichia coli | − | − | − | − | Tr. | − | Tr. | Tr. | − | − | − | − | − | − | − | − |
| NCTC 10418 | |||||||||||||||||
| 2 | Raoultella planticola | +/− | +/− | +/− | +/− | +/− | Tr. | + | + | − | − | − | − | − | − | − | − |
| NCTC 9528 | |||||||||||||||||
| 3 | Providencia rettgeri | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 7475 | |||||||||||||||||
| 4 | Enterobacter cloacae | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 11936 | |||||||||||||||||
| 5 | Serratia marcescens | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 10211 | |||||||||||||||||
| 6 | Salmonella typhimurium | + | +/− | + | +/− | + | − | + | + | − | − | − | − | − | − | + | +/− |
| NCTC 74 | |||||||||||||||||
| 7 | Pseudomonas aeruginosa | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 10662 | |||||||||||||||||
| 8 | Yersinia enterocolitica | + | +/− | + | Tr. | + | − | + | + | + | + | + | + | + | + | + | + |
| NCTC 11176 | |||||||||||||||||
| 9 | Burkholderia cepacia | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NCTC 10743 | |||||||||||||||||
| 10 | Acinetobacter baumannii | + | + | + | + | + | + | + | + | − | − | Tr. | +/− | Tr. | + | + | + |
| NCTC 12156 | |||||||||||||||||
| Gram-positive microorganisms | |||||||||||||||||
| 11 | Streptococcus pyogenes | − | − | − | − | + | Tr. | − | − | − | − | Tr. | +/− | + | +/− | − | − |
| NCTC 8306 | |||||||||||||||||
| 12 | Staphylococcus aureus (MRSA) | − | − | − | − | − | − | − | − | − | − | + | + | + | + | − | − |
| NCTC 11939 | |||||||||||||||||
| 13 | Staphylococcus aureus (MSSA) | − | − | − | − | − | − | +/− | +/− | +/− | + | + | + | +/− | + | − | − |
| NCTC 6571 | |||||||||||||||||
| 14 | Staphylococcus epidermidis | − | − | +/− | Tr. | +/− | +/− | − | − | − | − | − | − | − | − | − | − |
| NCTC 11047 | |||||||||||||||||
| 15 | Listeria monocytogenes | + | +/− | + | +/− | + | +/− | + | + | +/− | + | +/− | + | +/− | + | + | + |
| NCTC 11994 | |||||||||||||||||
| 16 | Enterococcus faecium | + | +/− | + | +/− | + | +/− | + | + | +/− | + | +/− | + | +/− | + | + | + |
| NCTC 7171 | |||||||||||||||||
| 17 | Enterococcus faecalis | + | +/− | + | +/− | + | +/− | + | + | Tr. | +/− | +/− | +/− | +/− | +/− | + | + |
| NCTC 775 | |||||||||||||||||
| 18 | Bacillus atrophaeus | +/− | +/− | +/− | +/− | +/− | +/− | Tr. | − | − | − | − | − | − | − | − | − |
| ATCC 9372 | |||||||||||||||||
| Yeasts | |||||||||||||||||
| 19 | Candida albicans | +/− | − | +/− | − | +/− | Tr. | + | +/− | Tr. | +/− | +/− | +/− | +/− | +/− | + | +/− |
| ATCC 90028 | |||||||||||||||||
| 20 | Candida glabrata | − | − | − | − | − | − | − | − | Tr. | − | Tr. | − | Tr. | − | Tr. | − |
| NCPF 3943 | |||||||||||||||||
The dramatic change in colour response going from substrate 14a (pale-yellow) to substrate 14c (maroon) suggested that the nitrothiophene derivative 15c should be investigated. Where nitroreductase activity was present, this substrate resulted in the formation of black colonies with exceptional colour contrast against the background (Table 2 and Fig. 6, 7). Most of the Gram-negative bacteria grew satisfactorily at all substrate concentrations (100, 50 and 25 mg L−1) except for E. coli and R. planticola which were both inhibited. The growth of A. baumannii was also inhibited at a substrate concentration of 100 mg L−1 but it did show some growth at the lower concentrations.
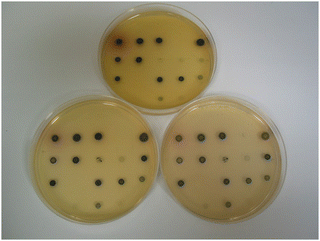 | ||
| Fig. 6 Evaluation of substrate 15c in columbia agar at various substrate concentrations. See Fig. 3 and Table 2 for the arrangement of microorganisms on the plates. Top image, 100 mg L−1; bottom left, 50 mg L−1; bottom right, 25 mg L−1. | ||
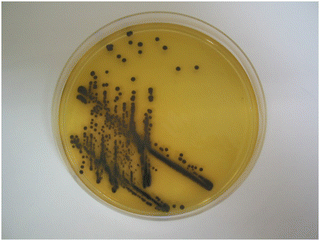 | ||
| Fig. 7 Columbia agar plate showing Pseudomonas aeruginosa in the presence of substrate 15c (100 mg L−1). | ||
Substrate 16a possessing the 1,3-butadienyl component has also been prepared and compared with its alkenyl counterpart, compound 15a (Table 2). Good microbial growth was observed for most of the Gram-negative microorganisms at a substrate concentration of 100 mg L−1 except for E. coli and R. planticola which were both inhibited by this substrate. Brown-coloured microbial colonies were observed where growth had occurred and the colour intensity was generally strong, except for S. typhimurium which produced a relatively weak colouration. Of the Gram-positive microorganisms, only L. monocytogenes, E. faecium and E. faecalis grew in the presence of this substrate resulting in the formation of moderately intense, brown-coloured colonies.
We have also investigated the effect of attaching an additional electron-withdrawing ester group onto the alkenyl component of substrate 15c and hence compound 19 was prepared. After reduction of substrate 19 by a nitroreductase, the resulting amine's lone pair of electrons would be mesomerically associated with both the ester and pyridinium groups giving additional conjugation relative to compound 14c. Unfortunately, this substrate inhibited the growth of all the microorganisms and thus no coloured colonies were observed (data not shown). The alkenyl moiety in structure 19 now possesses three pendant electron-deficient groups and hence may be susceptible to Michael addition by bio-nucleophiles which may account for this substrate's disappointing results. It was envisaged that the presence of the ester group and an un-quaternised pyridine ring (as opposed to a quaternised ring) might overcome this problem and compound 17 was therefore evaluated as a potential nitroreductase substrate (Table 3 and Fig. 8). This substrate enabled the formation of dark green/blue-coloured microbial colonies with most Gram-negative bacteria when nitroreductase activity was present (Fig. 8). Noteworthy was the observation that the growth of three Gram-positive bacteria (L. monocytogenes, E. faecium and E. faecalis) was augmented compared to other substrates examined in this study and strong, blue-coloured microbial colonies were formed. In view of the interesting results obtained from heterocycle 17, its furyl analogue 18 was also prepared and evaluated (Table 3 and Fig. 8). This compound was associated with a similar microbial growth profile to its thienyl analogue; however, the depth of the resulting microbial colonies’ pink colour was not as visually intense as its thienyl-counterpart.
| Substrate | 17 | 18 | |||
|---|---|---|---|---|---|
| Colony colour | Dark green | Pink | |||
| Substrate conc. (mg L−1) | 100 | 100 | |||
| Colony growth (G)a and colour intensity (CI)b | G | CI | G | CI | |
| a + good growth, +/− weak growth, Tr. trace of growth, − no growth. Growth on control plate was + (Gram-negative species) and + (Gram-positive species and yeasts). b + strong colour, +/− weak colour, Tr. trace of colour, − no colour. c NCTC: National Collection of Type Cultures; ATCC: American Type Culture Collection; NCPF: National Collection of Pathogenic Fungi. d Formerly named Klebsiella pneumoniae. e Formerly named Bacillus subtilis. f Blue. | |||||
| Microorganism/referencec | |||||
| Gram-negative microorganisms | |||||
| 1 | Escherichia coli | − | − | − | − |
| NCTC 10418 | |||||
| 2 | Raoultella planticola | − | − | − | − |
| NCTC 9528 | |||||
| 3 | Providencia rettgeri | + | + | + | +/− |
| NCTC 7475 | |||||
| 4 | Enterobacter cloacae | + | + | Tr. | Tr. |
| NCTC 11936 | |||||
| 5 | Serratia marcescens | + | + | + | + |
| NCTC 10211 | |||||
| 6 | Salmonella typhimurium | Tr. | − | − | − |
| NCTC 74 | |||||
| 7 | Pseudomonas aeruginosa | + | + | + | + |
| NCTC 10662 | |||||
| 8 | Yersinia enterocolitica | + | + | + | + |
| NCTC 11176 | |||||
| 9 | Burkholderia cepacia | + | + | + | + |
| NCTC 10743 | |||||
| 10 | Acinetobacter baumannii | + | +f | + | + |
| NCTC 12156 | |||||
| Gram-positive microorganisms | |||||
| 11 | Streptococcus pyogenes | − | − | − | − |
| NCTC 8306 | |||||
| 12 | Staphylococcus aureus (MRSA) | Tr. | − | − | − |
| NCTC 11939 | |||||
| 13 | Staphylococcus aureus (MSSA) | Tr. | − | − | − |
| NCTC 6571 | |||||
| 14 | Staphylococcus epidermidis | Tr. | − | − | − |
| NCTC 11047 | |||||
| 15 | Listeria monocytogenes | + | +f | + | + |
| NCTC 11994 | |||||
| 16 | Enterococcus faecium | + | +f | + | + |
| NCTC 7171 | |||||
| 17 | Enterococcus faecalis | + | +f | + | + |
| NCTC 775 | |||||
| 18 | Bacillus atrophaeus | − | − | − | − |
| ATCC 9372 | |||||
| Yeasts | |||||
| 19 | Candida albicans | − | − | − | − |
| ATCC 90028 | |||||
| 20 | Candida glabrata | − | − | − | − |
| NCPF 3943 | |||||
Conclusions
In this study, we have synthesised a range of novel nitroreductase substrates which have been studied as potential microbial diagnostic reagents. These substrates were evaluated against 20 clinically important microorganisms in Columbia agar. Most Gram-negative microorganisms reduced most substrates thereby producing coloured colonies. The growth of Gram-positive microorganisms and yeasts was generally poorer (or completely inhibited) compared to their Gram-negative counterparts and hence coloured colonies were not observed in many cases. A range of different coloured colonies could be visualised depending upon the structure of the substrate and good colour contrast with the agar background could be obtained. The chromophores produced as a result of nitroreductase activity were localised within the microbial colonies and did not diffuse into the medium. We envisage that such nitroreductase substrates could be used to highlight the presence of Gram-negative pathogens that are lacking in glycosidase activity (or other hydrolases of use for specific identification) while differentiating competing flora using complementary substrates for known hydrolases.Author contributions
Conceptualisation; ALJ, JDP, SO, SPS, synthetic work and data analysis; ALJ, AJ, VLS, HES, SPS, VTT, GT, microbiological work and data analysis, MC-R, VC, SO, JDP, CR-D; project management JDP, SO, SPS, GT, writing manuscript JDP, SPS, GT.Conflicts of interest
The Freeman Hospital and Northumbria University receive ongoing funding from bioMérieux for the development and evaluation of enzyme substrates and culture media.Acknowledgements
We thank bioMérieux SA for generous financial support and the EPSRC UK National Mass Spectrometry Facility at Swansea University for high resolution mass spectra.References
- M. Manafi, W. Kneifel and S. Bascomb, Microbiol. Mol. Biol. Rev., 1991, 55, 335–348 CrossRef CAS PubMed.
- J. D. Perry and A. M. Freydière, J. Appl. Microbiol., 2007, 103, 2046–2055 CrossRef CAS PubMed.
- S. Orenga, A. L. James, M. Manafi, J. D. Perry and D. H. Pincus, J. Microbiol. Methods, 2009, 79, 139–155 CrossRef CAS PubMed.
- L. Váradi, J. L. Luo, D. E. Hibbs, J. D. Perry, R. J. Anderson, S. Orenga and P. W. Groundwater, Chem. Soc. Rev., 2017, 46, 4818–4832 RSC.
- A. L. James, J. D. Perry, M. Ford, L. Armstrong and F. K. Gould, Appl. Environ. Microbiol., 1996, 62, 3868–3870 CrossRef CAS PubMed.
- A. L. James, J. D. Perry, M. Ford, L. Armstrong and F. K. Gould, J. Appl. Microbiol., 1997, 82, 532–536 CrossRef CAS PubMed.
- J. D. Perry, M. Ford, J. Taylor, A. L. Jones, R. Freeman and F. K. Gould, J. Clin. Microbiol., 1999, 37, 766–768 CrossRef CAS PubMed.
- P. A. Smith, D. Mellors, A. Holroyd and C. Gray, Lett. Appl. Microbiol., 2001, 32, 78–82 CrossRef CAS PubMed.
- M. Cellier, A. L. James, S. Orenga, J. D. Perry, A. K. Rasul, S. N. Robinson and S. P. Stanforth, Bioorg. Med. Chem., 2014, 22, 5249–5269 CrossRef CAS PubMed.
- M. D. Roldán, E. Pérez-Reinado, F. Castillo and C. Moreno-Vivián, FEMS Microbiol. Rev., 2008, 32, 474–500 CrossRef PubMed.
- Y.-L. Qi, L. Guo, L. L. Chen, H. Li, Y.-S. Yang, A.-Q. Jiang and H.-L. Zhu, Coord. Chem. Rev., 2020, 421, 213460 CrossRef CAS.
- D. C. Stein, E. Carrizosa and S. Dunham, BMC Microbiol., 2009, 9, 239 CrossRef PubMed.
- Y. J. Debets-Ossenkopp, R. G. Pot, D. J. van Westerloo, A. Goodwin, C. M. Vandenbroucke-Grauls, D. E. Berg, P. S. Hoffman and J. G. Kusters, Antimicrob. Agents Chemother., 1999, 43, 2657–2662 CrossRef CAS PubMed.
- A. F. Bedernjak, P. W. Groundwater, M. Gray, A. L. James, S. Orenga, J. D. Perry and R. J. Anderson, Tetrahedron, 2013, 69, 8456–8462 CrossRef CAS.
- L. Tian, H. Feng, Z. Dai and R. Zhang, J. Mater. Chem. B, 2021, 9, 53–79 RSC.
- Z. Li, X. Gao, W. Shi, X. Li and H. Ma, Chem. Commun., 2013, 49, 5859–5861 RSC.
- J. W. Yoon, S. Kim, Y. Yoon and M. H. Lee, Dyes Pigm., 2009, 171, 107779 CrossRef.
- S. Xu, Q. Wang, Q. Zhang, L. Zhang, L. Zuo, J.-D. Jiang and H.-Y. Hu, Chem. Commun., 2017, 53, 11177–11180 RSC.
- A. L. James, S. Orenga, J. Perry, V. L. Salwatura and S. Stanforth, US Pat., 0129205, 2012 Search PubMed.
- O. Fabrega, A. L. James, S. Orenga, J. Perry, V. L. Salwatura and S. Stanforth, US Pat., 0129204, 2012 Search PubMed.
- A. L. James, J. D. Perry, C. Jay, D. Monget, J. W. Rasburn and F. K. Gould, Lett. Appl. Microbiol., 2001, 33, 403–408 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb00715d |
| ‡ Deceased. |
| This journal is © The Royal Society of Chemistry 2023 |