Open Access Article
Open Access ArticleTuning foreign body response with tailor-engineered nanoscale surface modifications: fundamentals to clinical applications
Tara Chand
Yadav
 ab and
Akash
Bachhuka
ab and
Akash
Bachhuka
 *b
*b
aDepartment of Bioinformatics, Faculty of Engineering & Technology, Marwadi University, Gujarat 360003, India
bDepartment of Electronics, Electric, and Automatic Engineering, Rovira I Virgili University (URV), Tarragona, 43003, Spain. E-mail: akash.bachhuka@urv.cat; Tel: +(34) 643201339
First published on 17th July 2023
Abstract
Biomaterials are omnipresent in today's healthcare services and are employed in various applications, including implants, sensors, healthcare accessories, and drug delivery systems. Unfavorable host immunological responses frequently jeopardize the efficacy of biomaterials. As a result, surface modification has received much attention in controlling inflammatory responses since it helps camouflage the biomaterial from the host immune system, influencing the foreign body response (FBR) from protein adsorption to fibrous capsule formation. Surfaces with controlled nanotopography and chemistry, among other surface modification methodologies, have effectively altered the immune response to biomaterials. However, the field is still in its early stages, with only a few studies showing a synergistic effect of surface chemistry and nanotopography on inflammatory and wound healing pathways. Therefore, this review will concentrate on the individual and synergistic effects of surface chemistry and nanotopography on FBR modulation and the molecular processes known to modulate these responses. This review will also provide insights into crucial research gaps and advancements in various tactics for modulating FBR, opening new paths for future research. This will further aid in improving our understanding of the immune response to biomaterials, developing advanced surface modification techniques, designing immunomodulatory biomaterials, and translating discoveries into clinical applications.
Introduction
Biomaterials play a vital role in the current medical system, with applications ranging from implantable devices (orthopedics and cardiovascular) to controlled drug delivery (vaccines and genes), diagnostics, and imaging (cancer treatment).1 The implantation of a biomaterial inside the human body triggers an elaborate network of events known as the foreign body reaction (FBR), which includes plasma-material interaction, acute and chronic inflammation, matrix formation, tissue granulation, and fibrosis.2 The ability of a biomaterial to function may be jeopardized by the innate inflammatory reactions elicited by an FBR. The earliest responders to the implanted biomaterial are innate immune cells such as neutrophils, dendritic cells, macrophages, natural killer cells, T-cells, and B-cells. These cells identify the biomaterial as alien and trigger an immune response by producing cytokines and chemokines that recruit additional immune cells to the implant site. These cytokines and chemokines also activate the complement system, a protein set that works together to remove foreign invaders. When the complement system is engaged, it might create inflammation, stimulate phagocytosis, and attract extra immune cells to the implant site. The adaptive immune response begins when T-cells and B-cells recognize specific antigens on the implanted biomaterial. Antigens are molecules the immune system recognizes as foreign and causes an immunological response. T-cells help to coordinate the immune response by releasing cytokines that stimulate other immune cells, whereas B-cells create antibodies that bind to specific antigens. The adaptive immune response to the implanted biomaterial can be beneficial and deleterious. Although the immune system's reaction is necessary for tissue regeneration at the implantation site, prolonged cytokine stimulation and forming reactive oxygen species radicals and enzymes can lead to implant degradation.3 Thus, modulating the host immune system's reaction is of utmost importance when assessing outcomes and the long-term efficacy of the implanted biomaterial.Recent breakthroughs in biomaterial research have focused on customizing the shape and properties of the implant's nanoscale surface to influence the host immune system response and increase the implanted biomaterial's long-term efficacy.4 It has been established that the biomaterial's surface nanotopography modulates protein adhesion, hence influencing the binding and activation of innate immune effector cells.5 It is feasible to regulate the immune response and impact the behavior of immune cells such as macrophages, which play a critical part in the foreign body reaction, by precisely manipulating the nanoscale characteristics of the implant surface. The implant surface's nanoscale pattern can be customized to promote or inhibit various macrophage phenotypes, such as the pro-inflammatory M1 or anti-inflammatory M2 phenotypes.6 Surface features that resemble the natural extracellular matrix or give cues for immune cell interactions can be created using nanopatterning, nanogrooving, or nanotexturing techniques.4 These designed surfaces can control the release of immune-modulatory signals, modulate cytokine production, and steer macrophage polarization toward a particular phenotype. Furthermore, the nanoscale surface features of the biomaterial can be changed to improve biocompatibility and integration with the surrounding tissues. Surface changes, such as adding bioactive compounds, growth factors, or immunomodulatory drugs, can stimulate tissue regeneration while decreasing the foreign body response.7 Furthermore, antimicrobial chemicals or coatings applied to the nanoscale surface can help prevent infections and improve the long-term functioning of the implanted biomaterial.8
Understanding the interplay between macrophages and biochemical cues in a biomaterial-based foreign body response is critical in designing, fabricating, and developing immunomodulatory biomaterials. Modulating macrophages and FBR via various surface features (chemistry and nanotopography) of the implanted biomaterial have attracted a lot of attention recently.9–11 The surface nanotopography of the biomaterial influences protein adhesion, impacting the binding and activation of innate immune effector cells.12
This review paper provides a comprehensive summary of recent achievements and improvements in biomaterial research, particularly emphasizing the synergistic effects of the nanoscale surface features of implants in influencing the host immune system response. It highlights the need to modify the nanoscale shape and characteristics of implant surfaces to control immune cell behavior during the foreign body reaction. Its focus on nanoscale-level customization and its implications for biocompatibility, tissue integration, and patient outcomes make it a valuable resource for researchers, clinicians, and industry professionals interested in designing and developing immunomodulatory biomaterial implants.
1. Foreign body response (FBR) to biomaterial implants
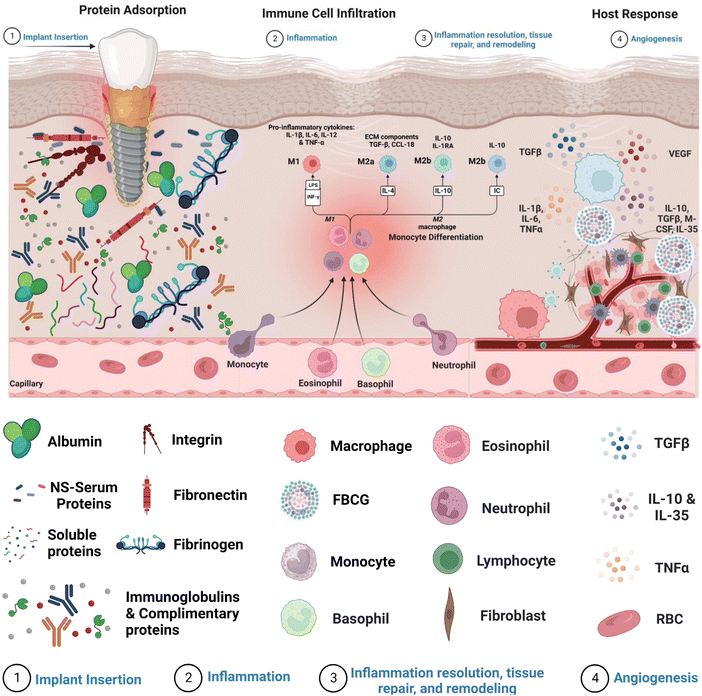
Internalization of an exogenous graft/biomaterial (medical implant devices) causes an immune response (FBR), which includes a series of actions involving several cell types and sophisticated biochemical signals involved in critical regulatory mechanisms (Fig. 1).13 FBR-mediated inflammation is initiated with the adsorption of nonspecific serum proteins onto the biomaterial's surface.14 The major serum proteins known to readily adsorb onto the biomaterial's surface are fibrinogen, fibronectin, albumin, vitronectin, immunoglobulins, and complimentary proteins.15–17 The amount and type of these adsorbed proteins influence immune cell adhesion via multiple receptors, directing their differentiation toward the inflammatory or anti-inflammatory pathway.5 Moreover, the cascade of events underlying FBR involves the activation of acute and chronic inflammation due to the interaction of blood with biomaterials and the creation of a provisional matrix.18 Early signs of acute inflammation that occur after biomaterial implantation often disappear within a week and are accompanied by a substantial inrush of neutrophils within hours of implantation. The release of cytokines soon after neutrophil extravasation attracts monocytes, which are helped further by neutrophil phagocytosis by macrophages gathered near the implant site.18 Neutrophils typically have a short lifespan, whereas monocytes can survive for extended periods by differentiating into macrophages. Furthermore, neutrophils and monocytes drawn to the implant release a variety of chemokines and pro-inflammatory cytokines that promote immune cell movement and the generation of reactive oxygen species (ROS), significantly impacting monocyte viability and macrophages.19 Chronic inflammation begins four to seven days after implant insertion, as evidenced by a proliferation of lymphocytes and macrophages surrounding the implant site. T-cells are attracted throughout the chronic inflammatory phase and have been demonstrated to significantly impact macrophage activation via a subsequent cycle. After dendritic cell stimulation, T-cells can be polarized towards TH1 or TH2 development.20 Activated T-cells can generate additional cytokines, implicitly polarizing macrophages towards a pro-inflammatory or preparative phenotype. In a pro-inflammatory state, macrophages release high levels of cytokines and ROS to break down or disintegrate the implanted biomaterial.21 The immune system's heightened response stimulates adherent macrophage fusion, producing foreign body giant cells (FBGCs). This causes the formation of a biological wall that engulfs the implant within a fibrous tissue layer of collagen, other extracellular matrix proteins, and tissue-repair cells designed to degrade the implant. However, the fibrotic capsule can impact the implanted biomaterial in good and unfavorable ways. On the one hand, it can shield host tissue from the implant, prevent infection, and lower the likelihood of implant failure. On the other hand, it can restrict the passage of nutrients and oxygen to the implant and delay its integration with the host tissue.22 Hence, minimizing the subsequent phases of the pro-inflammatory innate response cascade in the context of implant effectiveness is of the utmost importance. Table 1 depicts the various stages of foreign body reaction and their accompanied timings, allowing for an understanding of the temporal course of the host's response to biomaterial implants.| Foreign body response phases | Onset of events | Time-period |
|---|---|---|
| Implanted biomaterial | The surgical incision to implant a biomaterial or biomedical device inside the body drives tissue or organ injury, initiating an inflammatory response. | t = 0 |
| Adsorbed protein | The proteins in the blood will adhere to the biomaterial's surface, activating the blood coagulation and complement systems as well as platelets. | t > 1 s |
| Recruiting inflammatory cells | Inflammatory cells, primarily polymorphonuclear leukocytes (PMNs), are drawn to the implantation zone. Activated PMNs release chemokines as chemical attractants for monocytes, macrophages, embryonic dendritic cells, and lymphocytes. | t = 60 min |
| Cell adhesion to the biomaterial | Monocytes develop into macrophages, which cling to the outermost layer of the biomaterial and produce reactive species to break down and phagocytose it. Macrophages merge and create gigantic foreign body cells (FBGCs) in more extensive materials. | t = 1–15 days |
| Development of fibrous capsule | Transforming growth factor (TGF-b) and platelet-derived growth factor (PDGF) are factors released by macrophages and FBGCs that draw in and stimulate fibroblasts and endothelial cells, respectively. Active fibroblasts will produce collagen, resulting in the development of a fibrous capsule and leading to the encapsulation of biomaterial. | t = 3–4 weeks |
2. Acute inflammation
Acute inflammation is an important early FBR event arising from implanted biomaterials. The inflammatory reaction is triggered by the degranulation of neutrophils and mast cells, releasing histamines. Neutrophils degrade implants primarily by producing reactive oxygen species (ROS), proteolytic enzymes, and neutrophil extracellular matrix (NETs).25 These NETs are made up of a network of granular proteins, neutrophil elastase, chromatin DNA, and histones with bactericidal activity and “sticky” properties that allow pathogens to be captured and infection spread to be prevented. The role of NETs in implantable biomaterial-induced inflammation, if any, remains unknown.Concurrently, circulating monocytes are drawn to the implant site and divided into classical or M1 macrophages on the implant's surface. These activated M1 macrophages secrete a variety of pro-inflammatory cytokines such as tumor necrosis factor (TNF) α, interleukins (IL) 1β, and IL6 as well as reactive oxygen species (ROS) and degradative enzymes, like neutrophils, to destroy the implant.26,27 When macrophages cannot destroy or phagocytose these implants, they enter a state of frustrated phagocytosis, resulting in an enormous release of pro-inflammatory cytokines. Adherent macrophages gradually transform into an alternatively activated or “M2” phenotype with lesser degradative capacities, the release of anti-inflammatory cytokines, including IL-10, and tissue remodeling activity similar to wound healing processes. The transition from M1 to M2 phenotype and the mechanisms of frustrated phagocytosis induce macrophages to fuse on the biomaterial surface to form a foreign body giant cell (FBGC), which improves their phagocytic activity.28
3. Chronic inflammation
Chronic inflammation is caused by the fusing of macrophages to form FBGCs, which is aided by mast cells, basophils, and T Helper (Th) cells, through the release of IL-4 and IL-13.29 To degrade implants, these FBGCs release pro-inflammatory cytokines such as IL-6, Il-1β, TNFα, RANTES (regulated upon activation of normal T cell produced and presumably secreted), MCP-1 (monocyte chemotactic protein-1), and IL-8.30,31 Pro-inflammatory cytokine expression declines over time. In contrast, anti-inflammatory cytokines such as IL-10, transforming growth factor (TGF), and IL-1 receptor become antagonistic, marking the start of the tissue remodeling process. TGF and platelet-derived growth factors (PDGF) are produced by FBGCs, which attract fibroblasts and endothelial cells to the implant site.32 Activated fibroblasts initially secrete collagen III fibers smaller in diameter than collagen I, which are eventually replaced by more organized collagen I, providing the healing structure with additional tensile strength.33–35 Collagen production benefits wound healing and tissue remodeling, but excessive secretion causes fibrosis. In combination with FBGCs, excess collagen produces a fibrous capsule around the implant, isolating it from the rest of the body. Toxins, oxides, and nitrides secreted by giant fibrous cells in the fibrous capsule by macrophages and leukocytes can cause implant rejection or failure. This sequence of events emphasizes the relevance of macrophages, neutrophils, fibroblasts, and collagens in the foreign body cascade underlying implantation.364. Mechanisms underlying the foreign body response (FBR)
FBR is characterized by an inflammatory reaction involving macrophages at the implant site. On the other hand, excessive activation of macrophages might result in a persistent inflammatory response, eventually culminating in the production of a protective collagen-rich sheath around the implant. This procedure will continue until the implant is removed from the patient's body.37 However, the processes that drive the FBR are largely unclear, and effective techniques to prevent or minimize its deleterious impact without interfering with tissue regeneration are required. To that goal, researchers have investigated different ways, such as local delivery of anti-inflammatory or antifibrotic medicines at the interface between the device and surrounding tissue, as well as optimizing the physical features of the implant to reduce FBR. Nonetheless, these treatments have had limited effectiveness, and local administration of broad-spectrum anti-inflammatory drugs such as dexamethasone has been offered as a viable remedy.38Despite ongoing research efforts, our understanding of how surface modifications such as surface nanotopography and chemistry influence cellular responses underlying FBR needs to be completed.4 Despite this, some underlying mechanisms contributing to the FBR have been identified. The following are a few of the known mechanisms.
4.1. Cell-surface interaction via integrins
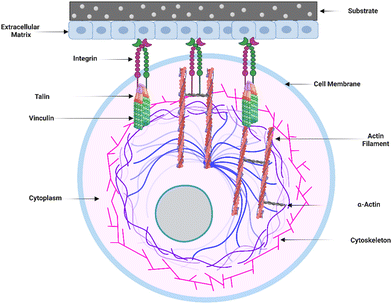
The initial adsorption of non-specific proteins onto the surface of implants sets off a chain of events that can impact postoperative inflammation and wound healing. Several proteins that bind to the surface of implanted materials have been identified, and the surface features of the implant determine the degree to which these proteins adhere. The composition and content of the adsorbed proteins can influence immune cell recognition and activation, thus regulating the foreign body response (FBR).12In vitro studies have shown that proteins are adsorbed onto biomaterial surfaces based on surface-free energy, promoting cell adhesion to the surface. Integrin surface receptors such as fibronectin, laminin, tenascin, and thrombospondin bind primarily to surface-adsorbed proteins such as fibrinogen, vitronectin, and fibronectin via RGD (Arginine–Glycine–Aspartic acid) peptide signaling pathways.39Within the extracellular matrix (ECM), integrin-mediated cell-receptor ligation occurs, resulting in integrin clustering towards the cell membrane (Fig. 2). After adhering to integrins, ECM interacts with actin filaments via adaptor proteins like talin and vinculin. This interaction between the ECM and integrins allows the cell to analyze the physical features of the ECM and change its shape as needed.23 Fibrinogen, a surface-attaching protein, was found to trigger inflammation when it binds to the integrin Mac-1 receptor.40 Macrophages bind to the tri-peptide sequence Arg-Gly-ASP (RGD) through αMβ2 integrin, the Mac-1 receptor. The integrin Mac-1 receptor and RGD-binding integrins also influenced the in vivo FBR to subcutaneously implanted polyethylene terephthalate. A fragile foreign body capsule emerged in the absence of integrin Mac-1 (about 30% thinner) or by a monitored release of an inhibiting peptide to prevent binding of the RGD-binding integrins (roughly 45% thinner). These findings show that the integrins Mac-1 and RGD-binding integrins are important in decreasing macrophage inflammatory responses to microscopic and bulk biomaterials, implying that they could be promising therapeutic targets.41
Surface modifications, such as surface chemistry, can immediately impact how macrophages interact with the implant. The precise mechanisms by which integrins regulate macrophage motility, phagocytosis, and activation remain unknown.42 However, Cha et al. demonstrated that promoting integrin-mediated connections between the collagen-derived matrix and human monocytic THP-1 cells can modulate macrophage polarity near the implant site. The implant environment appears to influence macrophage polarization by modulating the influence of effective M2 or M1 phenotypic activators such as IL-4 and lipopolysaccharide (LPS). Moreover, integrin α2β1 plays an important role, primarily because reducing its activity reduces M2 macrophage production.43 Cell adhesion to the ECM is critical for modulating cell proliferation and differentiation on biomaterial surfaces. Cell migration is enabled by the formation of thin sheets of lamellipodia and filopodia on biomaterials. Filopodia detects adsorbed protein ligands and directs cells toward the targeted ligand interaction site.44 Understanding the complicated interactions between implanted material, adsorbed proteins, and immune cells is critical for designing effective ways to prevent FBR and promote tissue regeneration in implanted devices.
4.2. Role of inflammasome in the FBR-surface cascade
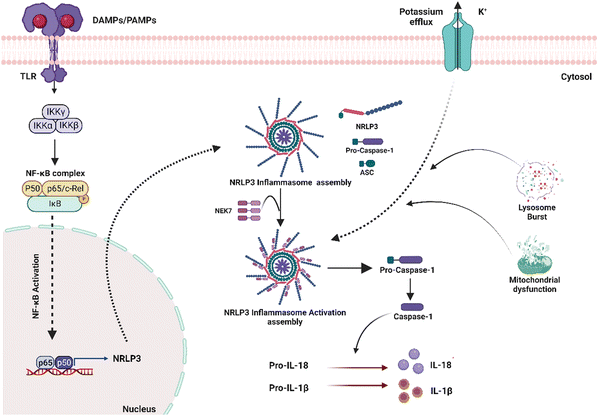
Inflammasome is an intracellular multi-complex protein in cells that secrete pro-inflammatory cytokines such as IL-1β and IL-18 (Fig. 3). Several types of inflammasomes have been identified, including nucleotide-binding oligomerization domain receptors (NOD)-like receptor (NLR-NLRP1, NLRP3, NLRP6, NLRP12), IL-1-β-converting enzyme protease-activating factor (Ipaf), and absent in melanoma (AiM2).45 Several studies have shown that surface-independent cytosolic receptors, such as the Nod-like receptor cryopyrin 3 (NLRP3), can activate and promote the FBR. The NLRP3 inflammasome is a cytosolic molecular scaffold that is especially important to biomaterials during the acute inflammatory phase of the FBR because it releases pro-inflammatory cytokines such as IL-1 and IL-18. Activation of Pannexin -1 protein via adenosine triphosphate (ATP) interaction with P2X purinoceptor 7 (P2X7) receptors, cytosolic crystalline uric acid and cholesterol, and foreign particles such as alum and asbestos can all activate the inflammasome.23 It can also be induced by the physical interaction of innate immune cell membranes with biomaterial implants. Toll-like receptor (TLR) stimulation also typically initiates NLRP3 inflammasome activation, which activates NFκB and increases NLRP3 expression, enhancing intracellular sensing. Nanoparticular carbon, polystyrene, amorphous silica, TiO2, and silver have all recently been discovered to activate the inflammasome and build an FBR. Inflammasomes recruit and activate apoptosis-associated speck-like proteins containing a CARD (ASC) and Caspase-1, which cleave pro-IL-1 and pro-IL-18 to generate IL-1 and IL-18, resulting in inflammation. Although the NLRP3 inflammasome is a powerful driver of an FBR's acute phase, it does not appear to contribute to its complete development, which depends on the ASC-mediated inflammasome.46 As a result, other ASC-dependent NLRs must also be involved in FBR progression.James Anderson's contributions to this field have been substantial, providing a pathology-based perspective on FBR in biomaterials (Table 2).23,24,47 Franz et al.48 stressed the significance of designing biomaterials that can control immune responses. In contrast, Christo et al.5 focused on the role of inflammasomes in the immune system's response to biomaterials. They created modeled substrates with precise nanotopography and surface chemistry. They discovered that the inflammasome elements ASC, NLRP3, and AIM2 were critical in modulating macrophage activation and adhesion in response to surface nanotopography and chemistry.49 Turley et al. developed different chitosan polymer mixes with varying degrees of deacetylation and discovered that a high degree of deacetylation increased NLRP3 inflammasome stimulation.50 Using biomaterials too big to be phagocytosed, researchers found that the NLRP3 inflammasome is involved in phagocytosis-independent inflammatory reactions.51 Court et al. discovered that collagen 3D scaffolds induced the development of the NLRP3 inflammasome, which resulted in increased IL-1 production in human macrophages.52 Vasconcelos et al. reported that 3D chitosan scaffolds inhibited the formation of the NLRP3 inflammasome in macrophages, contradicting a prior study that demonstrated chitosan as an inducer of the NLRP3 inflammasome in nanoscale chitosan derivatives.53 Despite its importance, there still needs to be an explanation for how different biomaterials activate the inflammasome pathway. More in-depth investigations into the involvement of different inflammasome components are required to thoroughly explain the system's stimulation in the inflammatory response to biomaterials. Nonetheless, modulating inflammasome activity is an essential method for establishing successful routes for biomaterial integration, which is a critical barrier in regenerative therapeutic therapy.
| Biomaterials | Response | Ref. |
|---|---|---|
| Carbon nanoparticles | Activation of caspase-1 increased IL-1β production | Reisetter et al.54 |
| Amino-functionalized polystyrene nanoparticles | Assembly of the NLRP3 inflammasome increased IL-1β production | Lunov et al.55 |
| Carboxyl-functionalized or non-functionalized polystyrene nanoparticles | No effect on the inflammasome pathways was observed | |
| Silica nanoparticles | Increased levels of IL-1β through inflammasome pathway activation | Gómes et al.56 |
| Titanium dioxide nanoparticles | Increased gene expression of NLRP3, caspase-1 and IL-1β | Oshaghi et al.57 |
| Chitosan-aluminum nanoparticles | Activation of the NLRP3 inflammasome increased IL-1β production | Lebre et al.58 |
| Micro-scale based biomaterials | ||
| Cobalt–Chromium–Molybdenum alloy microparticles | Irregular and larger microparticles induced higher levels of IL-1β through inflammasome pathway activation | Caicedo et al.59 |
| Hydroxyapatite microparticles | Smaller and needle-shaped microparticles lead to activation of the NLRP3 inflammasome, and increased IL-1β production | Lebre et al.60 |
| Microspheres of poly(methyl methacrylate) | Activation of caspase-1 further secretion of IL-1β | Malik et al.51 |
| Large-scale based biomaterials | ||
| Collagen 3D scaffolds | Induced assembly of the NLRP3 inflammasome increased IL-1β secretion | Court et al.52 |
| 3D chitosan scaffolds | Impaired NLRP3 inflammasome assembly | Vasconcelos et al.53 |
4.3. Protein adsorption and unfolding
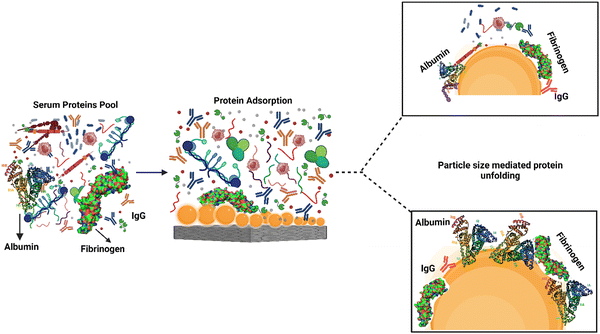
Protein adsorption and unfolding on biomaterial surfaces can influence immune cell activity significantly. Many proteins undergo structural modifications upon adsorption onto biomaterial surfaces, revealing hidden epitopes or antigenic determinants for immune cell receptors. These modifications may affect an immune cell's ability to recognize and interact with adsorbed proteins. Scavenger receptors, for example, are found on immune cells called macrophages and connect to exposed peptide sequences formed by protein unfolding on the surface. The binding of these receptors to the adsorbed proteins can activate immune cells and produce cytokines, resulting in an inflammatory reaction.61,62The roughness of biomaterial surfaces is essential in determining protein adsorption, unfolding, and the subsequent immune response. Recent research has emphasized the significance of protein unfolding in revealing hidden epitopes for immune cell receptors). For example, Visalakshan et al. studied the impact of protein unfolding in regulating fibrinogen-immune cell interactions. The researchers reported that fibrinogen adsorption on a biomaterial surface caused conformational changes that exposed hidden epitopes, resulting in greater macrophage adherence and activation.63 In another study, Dabare et al. explored the effects of surface nanotopography on albumin adsorption and unfolding behavior, as well as related immunological reactions. They observed that the percentage of immobilized albumin increased when the surface area increased due to increasing nanotopography dimensions. The protein experienced structural changes and lost alpha-helical structures. THP-1 immune cells interacted with surface-adsorbed albumin via scavenger receptors that bind to exposed peptide sequences produced by albumin unfolding on the surface. Depending on the peptide sequence revealed, the immune response might be pro-inflammatory or anti-inflammatory. This investigation's findings could lead to biomedical implants' development with tailored surface nanotopography and albumin pre-adsorption to improve biomedical implants' biocompatibility and accelerate wound healing.14 According to another study, albumin unfolds and exposes its cryptic epitopes in domains I and II to connect with the macrophage scavenger receptor-A1.64 Several studies have also found that temperature, pH, buffer concentration or composition, and nanoparticle concentration all influence albumin conformational change. With the rising temperature, native albumin transforms from reversible to irreversible in thermodynamic equilibrium. Albumin responds to pH variations by unfolding; at acidic pH, for example, domain III mostly experiences structural changes, whereas domain II becomes a molten molecule. The albumin molecule is isomerized at basic pH in domains I and II. Albumin adsorption and conformational stability have been altered by varying ionic strengths. The effect of nanotopography on albumin structural alteration and subsequent immune responses is uncommon, and more research is needed.65 A schematic representation of the protein adsorption and unfolding on nanotopography-modified surfaces has been presented in Fig. 4. The graphic depicts the nanoparticle size dependent adsorption and unfolding of proteins (albumin, fibrinogen and IgG). Understanding the relationship between protein adsorption, unfolding, and subsequent immune cell responses is necessary when designing biomaterials to promote tissue repair and regeneration.
5. Effect of a biomaterial's physicochemical properties on the FBR
A biomaterial's physicochemical properties, such as chemistry, charge, size, shape, composition, and roughness, play a vital role in modulating the events underlying the foreign body response (FBR).66–68 Understanding the link between biomaterial physicochemical attributes and the FBR is crucial for creating biomaterials that evoke a minimal immune response and enhance tissue regeneration. This review will critically review the individual and synergistic effects of these surface qualities on FBR in detail.5.1. Role of surface chemistry in regulating the FBR cascade
Surface chemistry has demonstrated increased or reduced cell adhesion based on their wettability.78 Hydrophilic surfaces have a higher attraction for cells; they can cause more inflammation and the creation of a giant fibrous capsule around implants. However, according to some research, hydrophobic surfaces promote the formation of fibrous capsules and the recruitment of inflammatory cells.79 The impact of different surface chemistry on cellular attachment and the formation of fibrous capsules around implants has been inconsistent in both in vivo and in vitro studies. This demonstrates the importance of gaining a deeper understanding of the effects of surface chemistry on biological responses to construct successful implanted device design and development.
5.2. Surface nanotopography
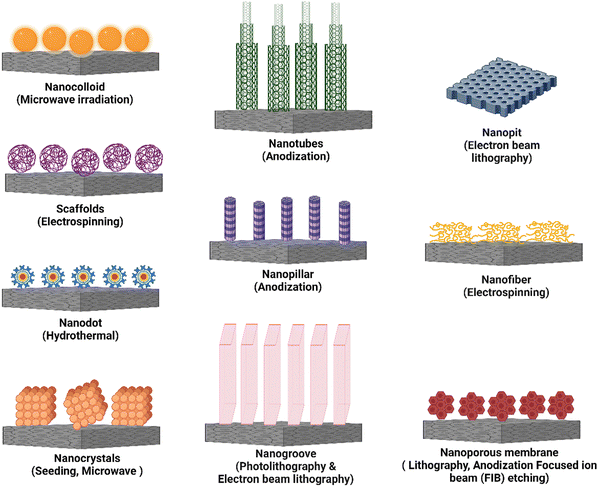
It is critical to modify surface features of the implanted biomaterial with the ability to minimize macrophage fusion, which alleviates/reduces FBR.9 Nanotopography modification is an excellent approach for developing smart multidirectional nano-interfaces that can predictably trigger and regulate the behavior of cells from many systems. Adding nanotopographical interfaces to bone biomaterials has been shown in studies to influence bone cell function and promote osteogenesis.80 To that purpose, nanotopography such as nanotubes, nanopits, nanofibers, nanodots, nanorods, nanogrooves, and nanopores have all been used. A schematic representation of these different types of nanotopography has been presented in Fig. 5. When the bio-physicochemical and mechanical parameters of these nanotopography are appropriately managed, they can increase biomaterial performance. Furthermore, nanotopography have demonstrated significant immunomodulatory effects and their direct impact on osteocytes.81 This illustrates the ability of nanotopographical alteration to influence immune cell response and lower FBR. Finally, altering the surface nanotopography (Fig. 5) of biomaterials could provide a potential option for lowering FBR and increasing the efficacy of medical implants.Nanotubes have increased bone-forming cell adhesion, diffusion, and osteogenic differentiation in orthopedic and dental applications.4,84 For example, electrochemical anodization of titanium (Ti) resulted in the self-ordering of TiO2 nanotube arrays with pore diameters ranging from 30 to 175 nm. TiO2 nanotubes with 30 nm and 80 nm pore diameters improved bone marrow-derived mesenchymal stem cell adhesion and spreading.85 Another study found that vertically architectured TiO2 nanotubes with dimensions ranging from 30 nm to 100 nm were made using pure titanium (Ti) foil and were shown to increase ER stress and osteogenic proliferation in cells.86 The use of mechanotransduction to create silicon nanotubes (VA-SiNT) resulted in a hollow structure of SiNTs that demonstrated exceptional success in Cas9 ribonucleoprotein gene delivery.87 Titania nanotubes were reported to limit immune cell growth and adhesion in blood lysate, resulting in lower levels of macrophage inflammatory protein (MIP)-1, monocyte chemotactic protein (MCP)-1, IL-6, and IL-8.88 Furthermore, TiO2 nanotube surfaces, particularly those with a diameter of 80 nm, were observed to promote macrophage adhesion and proliferation while decreasing the production of TNF-, MCP-1, and MIP-1 in macrophage cell lines, with no detectable expression of IL-1 and IL-6.89 Thin-film carbon–nanotube architectures generate nanoroughness, which can stimulate neuroendocrine PC12 cell proliferation and adhesion. This framework physically strains the 5 nm thick cell membrane, causing significant lateral displacement of subplasmalemmal secretory vesicles.90
Despite the research performed to understand the effect of nanotubes on orthopedic/dental applications, drug transport, and immune response modification. More research is required to understand the mechanisms underlying these effects and enhance nanotube-based material design. Furthermore, the potential toxicity of these substances must be thoroughly investigated to ensure their safe use in clinical settings. Developing new manufacturing techniques and integrating nanotubes with other modern technologies, such as gene editing and tissue engineering, may pave the way for the future application of nanotube-based materials in biomedicine.
These results show that the nanotopography of nanocolloids can influence protein interactions, innate immune responses, and foreign body responses. It is feasible to manage immunological reactions and inflammation by precisely tailoring the surface features of nanocolloids, such as their hydrophobicity, hydrophilicity, or ligand composition. This understanding opens up new prospects for creating nanocolloids with specific surface characteristics that can promote desired immune responses, reduce adverse reactions, and improve the biocompatibility of medical implants and devices. Further research in this area will help to build advanced nanocolloid-based therapeutic treatments and implantable biomedical devices.
The biocompatibility of implantable immunoisolation membranes is crucial for preventing fibrotic deposition, which can impair the function of encapsulated cells. Nanoscale fiber structure may aid macrophage cell regulation toward anti-inflammatory phenotypes in vitro. Wang et al. created electrospun membranes with various topographies of thermoplastic polyurethane (TPU) and micro- or nanofibers (PU-micro). Electrospun nanofibers modestly increased FBR compared to microfibrous membranes, with little activation of macrophage cells.108 To manage the macrophage phenotypic shift and minimize FBR, Song et al. inserted a short peptide termed mechano-growth factor (MGF) into an electrospun polycaprolactone (PCL) fibrous scaffold. MGF stimulates histone acetylation and increases STAT6 expression, resulting in an anti-inflammatory phenotype. A scaffold with MGF activity can inhibit FBR at the subcutaneous tissue level and prevent tissue adhesion.109 Suyeoshi et al. investigated the efficacy of nano-PGA on the kinetics of cells migrating after transplantation. The immunohistochemistry examination revealed that using nano-PGA caused an early inflammatory reaction. Nano-PGA sheets demonstrated enhanced tissue compatibility compared to regular PGA by triggering early polarization to the M2 phenotype, leading to angiogenesis and tissue repair.110 Veleirinho et al. developed PET-based electrospun non-absorbable mats for the correction of abdominal malformations in the absence of intestinal adhesion. The electrospun meshes were flexible and had excellent suture retention.111 However, mice-fed nanofibrous materials demonstrated a strong FBR, with PET and PET/chitosan groups exhibiting numerous FBGC, including nanofiber pieces. PET microfibers, on the other hand, have no discernible FBR.111 According to Cao et al., the topography of polycaprolactone (PCL) based on random and aligned electrospun nanofibers influences the FBR. Primary monocytes were collected and cultured in vitro on PCL nanofibers and PCL film. It was concluded that adhesion on the aligned scaffold was lower than on the random scaffold. The study suggests that aligned electrospun nanofibers may be a suitable scaffold for tissue engineering as they reduce host reactivity, increase tissue-scaffold integration, and induce a thinner fibrous capsule.112
Laursen et al.'s work on pelvic organ prolapse (POP) treatment is an example of a study that uses nanofibers in a medical application. To model abdominal wall healing in aged female rats, the researchers implanted electrospun polycaprolactone (PCL) meshes covered with connective tissue growth factor (CTGF)/PEG-fibrinogen (PF) and rat mesenchymal stem cells. In an abdominal repair model in aged rats, CTGF/PF-coated electrospun PCL meshes containing stem cells displayed appropriate support, biocompatibility, and no mesh-related issues.113 In another study, Mukherjee et al. created a biodegradable poly L-lactic acid-co-poly-caprolactone nanofibrous mesh (P nanomesh) for POP repair bioengineered with endometrial mesenchymal stem/stromal cells (eMSC). In vivo, these bioengineered meshes displayed excellent tissue integration and immunomodulatory effects.114 Synthetic scaffolds are necessary for regenerative medicine applications. However, foreign body reactions might block regeneration and lead to implant failure. Vacanti et al. used the anti-inflammatory medication dexamethasone to create a tissue engineering scaffold. Poly(L-lactic acid) (PLLA) and poly(–caprolactone) (PCL) electrospun fibers were generated in the presence and absence of the steroid. Dexamethasone-containing PLLA fibers generated a less severe inflammatory response, aided tissue regeneration, and allowed human mesenchymal stem cells to attach to and grow on them in vitro.115
Liu et al. created hybrid scaffolds by co-electrospinning PCL microfibers and human placental extracellular matrix (ECM) nanofibers with heparin and IL-4. In vitro and in vivo, the hybrid scaffold initially stimulated migration, NO generation, endothelial cells (ECs) tube formation, migration and maturation of vascular smooth muscle cells (VSMCs), and ECM deposition. Synthetic polymer and dECM materials' synergistic benefits revealed the new potential for producing small-diameter vascular grafts (SDVGs) and immune-regulated materials for tissue regeneration applications.116 Schoenenberger et al. investigated how external mechanical stresses and inherent topological signals from electrospun biomaterials interact to govern macrophage activation and macrophage-tendon fibroblast cross-talk. In both mechanically loaded and unloaded conditions, aligned or randomly oriented polycaprolactone nanofiber substrates were examined in vitro and in vivo. The findings revealed that macrophages might be essential mechanosensory cells in tendon regeneration. They hypothesized that biomaterial designs targeting the recruited cells biomechanical niche could influence the biological response.117 Zhu et al. developed a bi-layered vascular graft for rejuvenation using a circumferentially aligned microfiber for the internal layer and random nanofibers for the external layer, which enhances mechanical qualities and prevents bleeding after implantation. The vascular graft with a small diameter induced the regrowth of circumferentially aligned VSMCs, mimicking the configuration of native blood vessels.118
Xia et al. created a core–shell nanofibrous membrane polycaprolactone–curcumin/gelatin–tetracycline hydrochloride (PCL–Cur/GEL–TH) with antibacterial, anti-inflammatory and antioxidant properties for wound healing applications.119 In another study, the electrospinning of alginate/collagen nanofibers containing GUMS16-producing exopolysaccharide has increased cell viability and proliferation.120 Ye et al. created a prodrug by polymerizing an indomethacin-containing precursor and loading it onto electrospun poly(–caprolactone)/gelatin nanofibers. The inflammation-responsive nanofiber scaffold could inhibit cytokine secretion from the LPS-induced RAW264.7 cellular model.121
He et al. created a composite dressing replicating natural skin qualities to improve wound healing. This dressing is made up of an outside hydrophobic layer with micro–nano structures made of poly(–caprolactone)/polystyrene (PCL/PS) microspheres and an interior hydrophilic layer with curcumin-containing aligned poly(–caprolactone)/gelatin. The nanofiber alignment is essential in polarizing pro-inflammatory M1 macrophages into the healing M2 phenotype, which regulates cell proliferation, migration, and angiogenesis. Curcumin's anti-inflammatory properties encourage macrophage polarization. However, aligned nanofibers have a robust immunomodulatory capacity. Exudate buildup and dehydration are permitted by the porous matrix, lowering the risk of infection. This dressing could be helpful in the treatment of burn wounds.122
Through trilayer nanofiber scaffolds, electrospun nanofiber scaffolds show considerable potential for managing chronic inflammation in tissue regeneration.123 In contrast, multifunctional electrospun nanofiber dressings can channel surplus biofluid into improving wound healing.124 Diabetic wound tissue regeneration necessitates using a wound dressing with a porous structure and antioxidant properties (SSD-PG-PVA/KGM).125 Creating an adhesive patch of gelatin nanofibrous membranes based on hyaluronic acid and integrated with several light-emitting diode (LED) arrays has demonstrated promising results in photobiomodulation treatment for improving wound healing.126
The use of nanofiber-based scaffolds has been shown to improve bone regeneration significantly. In particular, the coating of a 3D hydrogel with nanofibers has been shown to shift the macrophage phenotype towards the M2 type, which is anti-inflammatory and pro-healing. Electrospun nanofiber polyurethane membranes produced only minor macrophage responses and few foreign body reactions when tested on RAW 264.7 macrophages.127 Immunological responses were detected on PLLA scaffolds with varying fiber sizes and alignments, with the fiber diameter determined to be a critical factor in releasing pro-inflammatory cytokines.128 Nanofibers demonstrated less inflammation than microfibers and unmodified PLLA scaffolds. As a result, the diameter of the fibers, rather than their alignment, was found to be the most critical component in macrophage activation and pro-inflammatory cytokine production.129 Incorporating an appropriate nanofiber structure could considerably increase the biocompatibility of biomaterials.
Organic nanodots of carbon and graphene have also shown potential in wound healing. These nanodots can promote adequate wound healing without scar formation by leveraging their unique features, such as creating free radicals and photothermal effects. Carbon nanomaterials and H2O2 graphene quantum dots (GQD) have been used as wound disinfection band-aids, displaying synergistic bacteria suppression and wound healing effects, and rapid wound closure.134 Xiang et al. synthesized polydopamine (PDA) and folic acid (FA) functionalized carbon nanodot-decorated ZnO, which showed sustained bactericidal activity and enhanced fibroblast growth resulting in rapid healing and closure of exposed wound surfaces.135 Furthermore, on zinc oxide surfaces, functionalized carbon nanodots decorated with polydopamine (PDA) and folic acid (FA) demonstrated prolonged bactericidal activity, increased fibroblast proliferation, and faster wound healing. These developments emphasize the potential of nanodots in improving wound treatment tactics, opening up new avenues for dealing with microbe-infested wounds, and boosting efficient healing processes. Ag nanodots have demonstrated a wide range of bactericidal capabilities, and efforts have been made to use them in wound healing.136 Although Ag nanoparticle-based wound-healing dressings such as Acticoat® have demonstrated success, more research is being conducted to reduce related toxicities and improve therapeutic efficacy. Continued efforts are being made to facilitate the translation of advanced formulations from the laboratory to clinical settings, to reduce potential toxicities and increase therapeutic efficacy in wound care. Recently, a unique technique utilizing Ag nanoparticles (21–70 nm) embedded within an organic cellulose nanocrystal matrix has been developed. These Ag nanodot-based ointments and strips have improved healing results in acute wounds, including quicker neovascularization, increased collagen accumulation, and rapid re-epithelialization. The positive results are due to the synergistic action of cellulose nanocrystals, which have a high retention capacity for wound exudate absorption, Ag nanodot's antibacterial activity absorption, and the nanodot's antibacterial activity. The nanodots stick to bacterial membranes successfully, possibly interacting with intracellular proteins.137
Gold nanodots and gold (Au) nanostructures have emerged as intriguing candidates for wound treatment owing to their inherent biocompatibility, versatility, antioxidant, and optical qualities.138 Self-assembling Au nanodots (3 nm) were coupled with antibacterial surfactin polypeptide, resulting in substantial antimicrobial action. The greater efficiency of these nanodots in engaging bacterial membranes and subsequent breakdown contributes to quicker healing and better epithelialization in MRSA-infected wounds.133 Surfactin-functionalized Au nanodots loaded with egg-white hydrolysate and 2-mercapto-1-methylimidazole have exhibited significant bactericidal capacity and improved wound healing in previous studies.139 Furthermore, photobiomodulation treatment using Au quantum dots (QDs) of 22 nm size has demonstrated encouraging outcomes in wound healing by lowering inflammation, promoting angiogenesis, and improving collagen synthesis.140
Copper (Cu) nanodots provide an alternative therapeutic method for treating chronic wounds with infections. Cu ions have therapeutic actions such as accelerating the expression of growth-promoting genes and proteins, including angiogenic vascular endothelial growth factors (VEGFs), and neutralizing the effects of hypoxia-inducible factors.141 By turning collected photons into heat, Cu-functionalized nanodots also have a regulated photothermal cidal impact on bacteria.142 Furthermore, nanodots have immunomodulatory capabilities, altering immune cell shape, adhesion, and proliferation. The effects of different nanodot array sizes on macrophage and foam cell adherence, cytoskeleton development, and pro-inflammatory gene expression have been studied. The adherence of macrophages and foam cells increased on nanodot arrays ranging from 50 nm to 100 nm, whereas it reduced on arrays ranging from 100–200 nm. Vinculin and actin filament immunofluorescence demonstrated higher cell adhesion and cytoskeleton growth on surfaces with microscopic nanotopography.101 Furthermore, the formation of titania nanodot patterns on titanium surfaces has shown modulatory effects on mesenchymal stem cell osteogenic differentiation, implying prospective orthopedic applications for regulating bone-forming cell behavior on implant surfaces.143
Nanopillar arrays have also given a new tool for researching the mechanical relationship between the cell membrane and the nucleus in living cells. Researchers could analyze subnuclear deformation in functioning cells by adding arrays of vertical nanopillars. The findings show that nuclear stiffness and the opposing impacts of actin and intermediate filaments are essential in nanopillar-induced atomic deformation.147 Furthermore, OrmoComp nanopillars of various dimensions have been created to study cortical neuron axonal development and progression. Compared to low pillars (100 nm), high OrmoComp nanopillars (400 nm) demonstrated significantly greater neurite aggregation and growth. Detailed studies of growth cone dynamics during axonal differentiation indicated that nanopillars promote elongation and early axon development, resulting in longer axons than a flat surface. The nanopillar's sidewalls provide a larger surface area for growth cone coupling, creating paxillin-rich adhesions that promote fast and sustained neurite development.148
The unusual properties of nanopillars, such as their large surface area and optical and electrical capabilities, have obtained much interest in recent years. For example, using soft lithography to create biocompatible polyurethane acrylate (PUA) nanopillars and their development with human adipose-derived stem cells (hASCs) has revealed insights into cell–nanopillar interactions such as shape alterations and cytoplasmic projections.149 Another method, which uses ultraviolet-assisted capillary force lithography (UV-CFL), has enabled the synthesis of nanopillar pattern arrays with varying densities, demonstrating that optimal nanotopographical density conditions promote bone mineralization, cell adhesion, and proliferation, potentially offering applications in bone tissue engineering.150 Furthermore, nanoengineered silk-based nanopillar arrays have been developed to regulate stem cell morphology, transforming them into ellipsoidal forms resembling chondrocytes found in natural cartilage's superficial zone. These manufactured nanopillar arrays can induce chondrogenesis and imitate the rheological properties of synovial fluids by inserting kartogenin using an ion implantation approach, making them ideal biomimetic implants for the superficial zone of natural cartilage.151 Nanopillar topography has been used successfully in stem cell-based tissue engineering to stimulate mesenchymal stem cell development and regulate stem cell fate. This method allows for the development of massive 3D cell aggregates and the upregulation of the osteogenic cellular matrix.152 Nanopillars provide a flexible and promising platform for biomedical research and applications because of their unique features and capacity to interact with cells at the nanoscale. Further research and developments in nanopillar technology can potentially transform fields such as regenerative medicine, medication delivery, and tissue engineering.
In this review, we have discussed the use of diverse nanotopography, such as nanocolloids, nanotubes, nanogrooves, nanopores, nanopits, nanofibers, and nanopillars, in controlling inflammatory responses. These nanotopography have distinct surface properties that interact with immune cells and significantly impact their behavior and functional responses. Researchers have successfully altered critical inflammatory processes by utilizing the qualities of each nanotopography, such as surface area, roughness, aspect ratio, and spatial arrangement. These include regulation of cytokine secretion, modulation of immune cell adhesion, migration and polarization, and tissue regeneration encouragement. Furthermore, the fabrication procedures used to create these different types of nanotopography differ, providing flexibility and personalization in biomedical applications. Table 3 summarizes the information from the respective sections, summarizing the impacts of these diverse nanotopography on the inflammatory response, including their specific fabrication techniques.
| Type of nanotopography | Fabrication technique | Feature size | Effect | Ref. |
|---|---|---|---|---|
| Nanotubes | Anodization | 15–100 nm, 30–100 nm | Increased angiogenesis, macrophage proliferation and cell adhesion | 82, 84 and 86 |
| Nanocolloids | Microwave irradiation, chemical method | 2–20 nm | Increased secretion of anti-inflammatory cytokines and cell adhesion | 92–94 |
| Nanopits | Electron beam lithography | 300–400 nm, 70–100 nm 10–20 nm | Increased cellular adhesion and differentiation | 96, 99 and 101 |
| Nanofiber | Electrospinning | 50–100 nm, 100–300 nm | Stimulates angiogenesis, proliferation of fibroblasts and endothelial cells | 103, 105 and 106 |
| Nanodots | Hydrothermal | 12 nm dots with 25 nm period and 20 nm lines with 40 nm period, 21–70 nm | Stimulates angiogenesis, increased fibroblast proliferation and migration | 130, 132, 133 and 137 |
| Nanopillars | Anodization, chemical etching | 15–100 nm | Increased macrophage proliferation and migration | 145 and 147 |
| Nanogrooves | Photolithography and electron beam lithography | 10–120 nm | Increased protein adsorption, increased macrophage proliferation, increased cellular adhesion and migration | 153, 156 and 158 |
6. Synergistic effect of surface features on modulating protein interaction, inflammation, and FBR
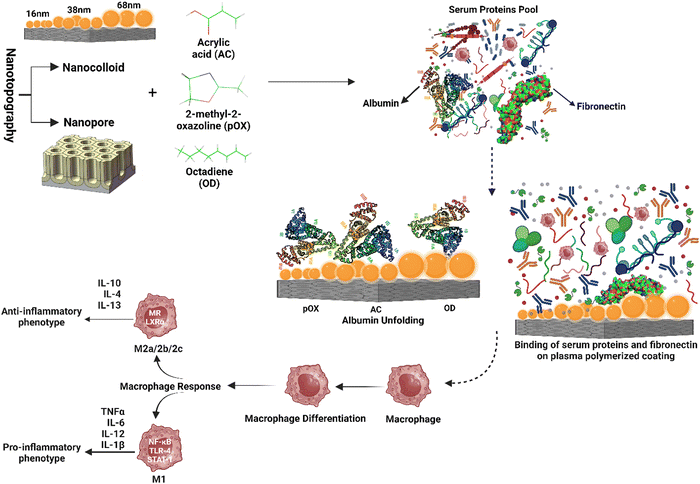
Surface features are rapidly being recognized for their synergistic impact on protein interactions and innate immune responses, which can influence the magnitude and nature of the FBR. Specific dimensions and geometries of nanostructured surfaces have been found to affect macrophage polarization, favoring the anti-inflammatory M2 phenotype and facilitating tissue remodeling. Surface chemistry modifications, such as adding cell-adhesive peptides, extracellular matrix proteins, or synthetic chemical groups, have also improved protein adsorption, immune cell adherence, and activation.33,159 However, only a handful of studies have shown the synergistic effect of nanotopography and chemistry to develop biomaterials that can actively control FBR and inflammation, resulting in improved wound healing.For instance, plasma polymerization has been used to regulate fibronectin adsorption and functionalization, revealing functional protein domains interacting with human gingival epithelial cells and promoting epithelial barrier development.160 Other research has used plasma polymerization and gold nanoparticles to create surface nanotopography and chemistry and investigate their synergistic effects on immune cell modulation via protein adsorption and functionalization. Dabare et al. investigated the synergistic effect of nanostructures and chemistry on albumin adsorption and functionalization. They discovered that as the surface area of the nanotopography increased, so did the percentage of immobilized albumin, and the protein went through structural changes and lost alpha-helical structures. THP-1 immune cells bind to exposed peptide sequences generated by albumin unfolding on the surface to engage with surface-adsorbed albumin. The immune response could be pro-inflammatory or anti-inflammatory, depending on the peptide sequence revealed.14 In a subsequent study, immune cell modulation via binding and functionalization of fibrinogen was interrogated using the same surface nanotopography and chemistry.63 In addition, another study demonstrated that protein expression, such as expression of collagens, can be controlled to a greater extent using a combination of nanotopography and chemistry, which is beneficial in regulating the FBR.35
Several other studies investigated the combined effect of nanotopography and surface chemistry on the inflammatory response; different-sized gold nanoparticles (AuNPs) were used to introduce nanotopography, while plasma polymerization was used to add surface chemistry (carboxyl, methyl, amine, and functional groups).4 Surprisingly, regardless of the surface chemistry, all surfaces showed decreased expression of pro-inflammatory cytokines. Furthermore, adding amine-coated surfaces reduced pro-inflammatory cytokine release and activated the M2 phenotype in macrophages.161 Another study found that carboxyl functional groups and nanotopography synergize in increasing osteoclast activity. Furthermore, the researchers discovered that smaller nanotopography (16 nm) with an amine coating demonstrated superior osteogenicity.162 Notably, the impact of surface nanotopography and chemistry on macrophage plasticity has also been studied. Nanotopography was discovered to be dominant in regulating the pro-inflammatory route, while surface chemistry is dominant in influencing the anti-inflammatory pathway.6 Damanik et al. developed an in vitro FBR mimicking model by employing surface microstructure to increase the biomaterial's oxygen content and surface area. The investigation provided substantial insight into using the tunable surface chemistry, roughness, wettability, and topography and their effect on cell-proliferation, cellular morphology, cellular attachment, and release of pro-inflammatory (IL-1β, IL-6) and anti-inflammatory cytokines (TGF-β1, IL-10).163 Tan et al. fabricated a vascular graft of electrospun polycaprolactone (PCL) scaffolds using PIII with surface modification encompassing cytokine interleukin-4 immobilization. Functionalized bioactive surfaces manifested overexpression of anti-inflammatory M2 genes resulting in a reduced foreign body response.164 In another study, 3D hybrid particles with modified surface nanotopography and chemistry investigated inflammatory and FBR responses in vivo. Researchers reported that collagen expression and the adhering immune cells were regulated using a combination of surface nanotopography and chemistry.165 Furthermore, the effect of surface nanotopography and silver (Ag) loading on the biomaterial-associated FBR was examined by Liu et al.166 Their results showed that the nanotopography lowered the FBR, and the Ag-loaded samples had an acceptable effect with a less severe reaction, demonstrating the promise of their technique for increasing biomaterial biocompatibility.
These examples demonstrate the ability of customized surface characteristics to influence protein interactions, innate immune responses, and the FBR. It is now possible to induce macrophage polarization towards desirable phenotypes, enhance inflammation resolution, and facilitate tissue repair by developing biomaterial surfaces with distinct nanotopographical features and personalized surface chemistry. Surface features can be precisely engineered to modify protein and immune cell binding, regulate macrophage plasticity, and alter the amount and nature of the foreign body response. These synergistic effects are depicted in a graphical representation, illustrating the intricate interplay between nanotopography and chemistry in shaping the immune response. Fig. 6 shows a graphic representation of these synergistic effects. These advancements enable precise cellular activity modulation, the controlled release of bioactive chemicals, and enhanced biomaterial integration, paving the path for sophisticated biomedical applications.
7. Clinical translation and commercialization
Closing the research-to-clinical implementation gap is critical for widely using nanotopography and surface chemistry-based biomaterials in many biological applications. Extensive preclinical and clinical research is required to demonstrate the safety, effectiveness, and long-term performance of these sophisticated biomaterials. Strong scientific proof is required for successfully commercializing and integrating these technologies into healthcare settings.Several examples of commercialized nanotopography and surface chemistry-based biomaterials demonstrate their clinical translation.
•For instance, NanoMeshTM was the first hernia repair device on the market with surface nano-modification, made of monofilament polypropylene (PP) and surface treated with accelerated neutral atom beam (ANAB) technology. The use of nanoMeshTM is approved for treating abdominal wall hernias, including inguinal (direct and indirect). (“nanoMeshTM”) is a patented hernia repair solution created and marketed by nanoMeshTM LLC. Dental implants with changed surfaces are one commercial example of nanotopography and surface chemistry-based products.167
•Hylomate's CellSense technology is another successful example. They offer two products (Hylomate Pouch and sheet) that are made of micro-engineered biosynthesized cellulose (BC) membrane and are used as a protective covering for cardiac implantable electronic devices (CIEDs). The BC membrane exhibited practicality and safety by reducing fibrotic tissue growth and enhancing access to the device pocket, potentially simplifying CIED revision operations.168
•Dental implant manufacturer Straumann offers SLACTIVE surfaces with nanotopographical features and functional coatings to improve osseointegration and long-term implant success rates.169–172
•The nanoVIS Ti surface technology utilized for spinal implants is another successful example. Compared to other surfaces commonly used in orthopedics, their implants with the nanotubular surface increased and accelerated calcified extracellular matrix production on human osteoblasts and mesenchymal stem cells at 21 days in vitro.173,174
Summary and future perspective
Surface engineering in medical implants holds enormous promise for the future. We should expect the creation of increasingly more sophisticated and personalized implant surfaces as researchers dive deeper into understanding the intricate connections between surface characteristics, proteins, immune cells, and tissues. Careful control over nanotopography, surface chemistry, and the integration of bioactive substances may be required to obtain desirable biological responses. Furthermore, advances in nanotechnology, biomaterials, and manufacturing techniques will allow for the large-scale production of implants with customized surface properties. These advancements have the potential to transform healthcare by improving implant biocompatibility, lowering problems, and improving patient outcomes. Furthermore, ongoing surface modification research will pave the way for developing innovative implants that can actively respond to their surroundings and promote tissue regeneration. We can expect significant developments that will impact the future of healthcare and enhance the lives of countless people as we continue to harness the promise of surface engineering in medical implants. Here are some possible future pathways:•Multifunctional surfaces: By combining multiple functionalities into a single surface, such as nanotopography with surface chemistry modifications and responsive materials, surfaces can respond dynamically to their surroundings, promoting precise control over cellular behavior and tissue regeneration.
•Personalized biomaterials: As 3D printing and nanofabrication techniques progress, creating patient-specific biomaterials with personalized nanotopography and surface chemistry is becoming increasingly possible. Such tailored techniques can increase biocompatibility, improve tissue integration, and promote patient-specific treatment outcomes.
•Bioactive coatings: Further study into bioactive coatings that mirror the composition and functionality of the extracellular matrix will be beneficial. These coatings can enhance biocompatibility and therapeutic results by promoting certain cellular responses, facilitating tissue regeneration, and modulating the immunological response.
In conclusion, the future of biomaterial implants lies in the synergy of additive manufacturing and nanofabrication. By integrating these technologies, researchers and manufacturers can produce increasingly sophisticated and individualized implant surfaces. This integration will allow for a better understanding of the complicated relationships between surface characteristics, proteins, immune cells, and tissues, ultimately leading to superior biological responses. Their future applications span many areas, including regenerative medicine, drug delivery systems, bioelectronic interfaces, implantable sensors and devices, customized prosthetics and implants, and smart implants. These applications can transform healthcare by providing tailored, functional, and responsive solutions that can potentially improve patient outcomes and quality of life.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
AB thanks MICINN (Spain) for Juan de la Cierva incorporation fellowship IJC-2019-042374-1 and Generalitat de Catalonia (Marie Curie Cofund) for the Beatriu de Pinos Fellowship 2021 BP 00105. All the schematics were created with http://BioRender.com.References
- D. Williams, Biomaterials, 2011, 32, 1–2 CrossRef CAS.
- A. Carnicer-Lombarte, S.-T. Chen, G. G. Malliaras and D. G. Barone, Front. Bioeng. Biotechnol., 2021, 9, 622524 CrossRef.
- N. A. Hotaling, L. Tang, D. J. Irvine and J. E. Babensee, Annu. Rev. Biomed. Eng., 2015, 17, 317–349 CrossRef CAS PubMed.
- Z. Chen, A. Bachhuka, F. Wei, X. Wang, G. Liu, K. Vasilev and Y. Xiao, Nanoscale, 2017, 9, 18129–18152 RSC.
- S. N. Christo, K. R. Diener, A. Bachhuka, K. Vasilev and J. D. Hayball, Biomed Res. Int., 2015, 2015, 342304 Search PubMed.
- A. Bachhuka, R. Madathiparambil Visalakshan, C. S. Law, A. Santos, H. Ebendorff-Heidepriem, S. Karnati and K. Vasilev, ACS Appl. Bio Mater., 2020, 3, 1496–1505 CrossRef CAS PubMed.
- H. Xue, Z. Zhang, Z. Lin, J. Su, A. C. Panayi, Y. Xiong, L. Hu, Y. Hu, L. Chen, C. Yan, X. Xie, Y. Shi, W. Zhou, B. Mi and G. Liu, Bioact. Mater., 2022, 18, 552–568 CrossRef CAS.
- T. C. Yadav, P. Gupta, S. Saini, S. Mohiyuddin, V. Pruthi and R. Prasad, ACS Omega, 2022, 7, 8350–8363 CrossRef CAS PubMed.
- K. Harawaza, B. Cousins, P. Roach and A. Fernandez, Mater. Today Bio, 2021, 12, 100152 CrossRef CAS PubMed.
- J. Li, X. Jiang, H. Li, M. Gelinsky and Z. Gu, Adv. Mater., 2021, 33, 2004172 CrossRef CAS.
- Y. He, Y. Gao, Q. Ma, X. Zhang, Y. Zhang and W. Song, J. Nanobiotechnol., 2022, 20, 510 CrossRef PubMed.
- O. Veiseh and A. J. Vegas, Adv. Drug Delivery Rev., 2019, 144, 148–161 CrossRef CAS PubMed.
- E. Marin, F. Boschetto and G. Pezzotti, J. Biomed. Mater. Res., Part A, 2020, 108, 1617–1633 CrossRef CAS PubMed.
- P. R. L. Dabare, A. Bachhuka, E. Parkinson-Lawrence and K. Vasilev, Mater. Today Adv., 2021, 12, 100187 CrossRef CAS.
- C. R. Jenney and J. M. Anderson, J. Biomed. Mater. Res., 2000, 49, 435–447 CrossRef CAS PubMed.
- T. A. Horbett, J. Biomed. Mater. Res., Part A, 2018, 106, 2777–2788 CrossRef CAS PubMed.
- D. J. Fabrizius-Homan and S. L. Cooper, J. Biomater. Sci., Polym. Ed., 1992, 3, 27–47 CrossRef.
- E. Mariani, G. Lisignoli, R. M. Borzì and L. Pulsatelli, Int. J. Mol. Sci., 2019, 20 Search PubMed.
- N. Tamassia, F. Bianchetto-Aguilera, F. Arruda-Silva, E. Gardiman, S. Gasperini, F. Calzetti and M. A. Cassatella, Eur. J. Clin. Invest., 2018, 48, e12952 CrossRef PubMed.
- R. Sridharan, A. R. Cameron, D. J. Kelly, C. J. Kearney and F. J. O’Brien, Mater. Today, 2015, 18, 313–325 CrossRef CAS.
- L. Davenport Huyer, S. Pascual-Gil, Y. Wang, S. Mandla, B. Yee and M. Radisic, Adv. Funct. Mater., 2020, 30, 1909331 CrossRef CAS.
- R. Klopfleisch and F. Jung, J. Biomed. Mater. Res., Part A, 2017, 105, 927–940 CrossRef CAS PubMed.
- J. M. Anderson, A. Rodriguez and D. T. Chang, Semin. Immunol., 2008, 20, 86–100 CrossRef CAS PubMed.
- J. M. Anderson, Annu. Rev. Mater. Res., 2001, 31, 81–110 CrossRef CAS.
- S. J. Busuttil, V. A. Ploplis, F. J. Castellino, L. Tang, J. W. Eaton and E. F. Plow, J. Thromb. Haemostasis, 2004, 2, 1798–1805 CrossRef CAS.
- C. Rosales, Front. Physiol., 2018, 9, 113 CrossRef.
- J. Kzhyshkowska, A. Gudima, V. Riabov, C. Dollinger, P. Lavalle and N. E. Vrana, J. Leukocyte Biol., 2015, 98, 953–962 CrossRef CAS.
- C. Chu, L. Liu, S. Rung, Y. Wang, Y. Ma, C. Hu, X. Zhao, Y. Man and Y. Qu, J. Biomed. Mater. Res., Part A, 2020, 108, 127–135 CrossRef CAS.
- J. R. Nakkala, Z. Li, W. Ahmad, K. Wang and C. Gao, Acta Biomater., 2021, 123, 1–30 CrossRef CAS PubMed.
- E. W. Ozpinar, A. L. Frey, G. Cruse and D. O. Freytes, Tissue Eng., Part B, 2020, 27, 590–603 CrossRef.
- Y. Chen, W. Sun, H. Tang, Y. Li, C. Li, L. Wang, J. Chen, W. Lin, S. Li, Z. Fan, Y. Cheng and C. Chen, Front. Bioeng. Biotechnol., 2022, 10 CAS.
- Z. Sheikh, P. J. Brooks, O. Barzilay, N. Fine and M. Glogauer, Materials, 2015, 8, 5671–5701 CrossRef CAS PubMed.
- A. Bachhuka, J. Hayball, L. E. Smith and K. Vasilev, ACS Appl. Mater. Interfaces, 2015, 7, 23767–23775 CrossRef CAS.
- M. Meyer, Biomed. Eng. OnLine, 2019, 18, 24 CrossRef.
- A. Bachhuka, J. D. Hayball, L. E. Smith and K. Vasilev, ACS Appl. Mater. Interfaces, 2017, 9, 5874–5884 CrossRef CAS.
- K. E. Martin and A. J. García, Acta Biomater., 2021, 133, 4–16 CrossRef CAS PubMed.
- J. B. Parker, M. F. Griffin, A. F. Spielman, D. C. Wan and M. T. Longaker, Adv. Text. Wound Care, 2022, 12, 85–96 CrossRef.
- D. G. Barone, A. Carnicer-Lombarte, P. Tourlomousis, R. S. Hamilton, M. Prater, A. L. Rutz, I. B. Dimov, G. G. Malliaras, S. P. Lacour, A. A. B. Robertson, K. Franze, J. W. Fawcett and C. E. Bryant, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2115857119 CrossRef CAS.
- J. S. Desgrosellier and D. A. Cheresh, Nat. Rev. Cancer, 2010, 10, 9–22 CrossRef CAS.
- V. K. Lishko, N. P. Podolnikova, V. P. Yakubenko, S. Yakovlev, L. Medved, S. P. Yadav and T. P. Ugarova, J. Biol. Chem., 2004, 279, 44897–44906 CrossRef CAS.
- T. D. Zaveri, J. S. Lewis, N. V. Dolgova, M. J. Clare-Salzler and B. G. Keselowsky, Biomaterials, 2014, 35, 3504–3515 CrossRef CAS PubMed.
- Y. Liu and T. Segura, Front. Bioeng. Biotechnol., 2020, 8 Search PubMed.
- B.-H. Cha, S. R. Shin, J. Leijten, Y.-C. Li, S. Singh, J. C. Liu, N. Annabi, R. Abdi, M. R. Dokmeci, N. E. Vrana, A. M. Ghaemmaghami and A. Khademhosseini, Adv. Healthcare Mater., 2017, 6, 1700289 CrossRef.
- Q. Zhao, N. Topham, J. M. Anderson, A. Hiltner, G. Lodoen and C. R. Payet, J. Biomed. Mater. Res., 1991, 25, 177–183 CrossRef CAS.
- M. Lamkanfi and V. M. Dixit, Cell, 2014, 157, 1013–1022 CrossRef CAS.
- W.-J. Zhang, S.-J. Chen, S.-C. Zhou, S.-Z. Wu and H. Wang, Front. Immunol., 2021, 12 Search PubMed.
- J. M. Anderson, ASAIO Trans, 1988, 34, 101–107 CrossRef CAS PubMed.
- S. Franz, S. Rammelt, D. Scharnweber and J. C. Simon, Biomaterials, 2011, 32, 6692–6709 CrossRef CAS PubMed.
- S. Christo, A. Bachhuka, K. R. Diener, K. Vasilev and J. D. Hayball, Sci. Rep., 2016, 6, 26207 CrossRef CAS PubMed.
- J. L. Turley, H. B. T. Moran, C. P. McEntee, K. O’Grady, N. Muñoz-Wolf, L. Jin, F. Follmann, P. Andersen, M. Andersson and E. C. Lavelle, Biomaterials, 2021, 275, 120961 CrossRef CAS.
- A. F. Malik, R. Hoque, X. Ouyang, A. Ghani, E. Hong, K. Khan, L. B. Moore, G. Ng, F. Munro, R. A. Flavell, Y. Shi, T. R. Kyriakides and W. Z. Mehal, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 20095–20100 CrossRef CAS.
- M. Court, M. Malier and A. Millet, Biomaterials, 2019, 208, 98–109 CrossRef CAS PubMed.
- D. P. Vasconcelos, C. de Torre-Minguela, A. I. Gomez, A. P. Águas, M. A. Barbosa, P. Pelegrín and J. N. Barbosa, Acta Biomater., 2019, 91, 123–134 CrossRef CAS.
- A. C. Reisetter, L. V. Stebounova, J. Baltrusaitis, L. Powers, A. Gupta, V. H. Grassian and M. M. Monick, J. Biol. Chem., 2011, 286, 21844–21852 CrossRef CAS.
- O. Lunov, T. Syrovets, C. Loos, G. U. Nienhaus, V. Mailänder, K. Landfester, M. Rouis and T. Simmet, ACS Nano, 2011, 5, 9648–9657 CrossRef CAS PubMed.
- D. M. Gómez, S. Urcuqui-Inchima and J. C. Hernandez, Innate Immun., 2017, 23, 697–708 CrossRef.
- E. Abbasi-Oshaghi, F. Mirzaei and M. Pourjafar, Int. J. Nanomed., 2019, 14, 1919–1936 CrossRef CAS PubMed.
- F. Lebre, M. C. Pedroso de Lima, E. C. Lavelle and O. Borges, Int. J. Pharm., 2018, 552, 7–15 CrossRef CAS PubMed.
- M. S. Caicedo, L. Samelko, K. McAllister, J. J. Jacobs and N. J. Hallab, J. Orthop. Res., 2013, 31, 1633–1642 CrossRef CAS PubMed.
- F. Lebre, R. Sridharan, M. J. Sawkins, D. J. Kelly, F. J. O’Brien and E. C. Lavelle, Sci. Rep., 2017, 7, 2922 CrossRef.
- M. Rahmati and M. Mozafari, Mater. Today Commun., 2018, 17, 527–540 CrossRef CAS.
- C. J. Wilson, R. E. Clegg, D. I. Leavesley and M. J. Pearcy, Tissue Eng., 2005, 11, 1–18 CrossRef CAS.
- R. M. Visalakshan, A. A. Cavallaro, M. N. MacGregor, E. P. Lawrence, K. Koynov, J. D. Hayball and K. Vasilev, Adv. Funct. Mater., 2019, 29, 1807453 CrossRef.
- J. Rozga, T. Piątek and P. Małkowski, Ann. Transplant., 2013, 18, 205–217 CrossRef CAS PubMed.
- A. H. Morris and T. R. Kyriakides, Matrix Biol., 2014, 37, 183–191 CrossRef CAS.
- H. Amani, H. Arzaghi, M. Bayandori, A. S. Dezfuli, H. Pazoki-Toroudi, A. Shafiee and L. Moradi, Adv. Mater. Interfaces, 2019, 6, 1900572 CrossRef.
- M. Rafiei, J. T. Chung and Y. Chau, Front. Drug Delivery, 2023, 3 Search PubMed.
- M. Rahmati, E. A. Silva, J. E. Reseland, C. A. Heyward and H. J. Haugen, Chem. Soc. Rev., 2020, 49, 5178–5224 RSC.
- P. Thevenot, W. Hu and L. Tang, Curr. Top. Med. Chem., 2008, 8, 270–280 CrossRef CAS.
- J. T. Yates and C. T. Campbell, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 911–916 CrossRef CAS.
- E. Ruckenstein and Z. F. Li, Adv. Colloid Interface Sci., 2005, 113, 43–63 CrossRef CAS.
- J. N. Barbosa, P. Madureira, M. A. Barbosa and A. P. Águas, J. Biomed. Mater. Res., Part A, 2006, 76A, 737–743 CrossRef CAS.
- K. M. Evans-Nguyen, L. R. Tolles, O. V. Gorkun, S. T. Lord and M. H. Schoenfisch, Biochemistry, 2005, 44, 15561–15568 CrossRef CAS PubMed.
- H. M. Rostam, S. Singh, F. Salazar, P. Magennis, A. Hook, T. Singh, N. E. Vrana, M. R. Alexander and A. M. Ghaemmaghami, Immunobiology, 2016, 221, 1237–1246 CrossRef CAS PubMed.
- A. Nair, L. Zou, D. Bhattacharyya, R. B. Timmons and L. Tang, Langmuir, 2008, 24, 2015–2024 CrossRef CAS.
- M. Shen, I. Garcia, R. V. Maier and T. A. Horbett, J. Biomed. Mater. Res., Part A, 2004, 70A, 533–541 CrossRef CAS.
- C. Y. C. Hsieh, F.-W. Hu, W.-S. Chen and W.-B. Tsai, ISRN Biomater., 2014, 2014, 718432 Search PubMed.
- K. Jung, N. Corrigan, E. H. H. Wong and C. Boyer, Adv. Mater., 2022, 34, 2105063 CrossRef CAS.
- D. J. Menzies, B. Cowie, C. Fong, J. S. Forsythe, T. R. Gengenbach, K. M. McLean, L. Puskar, M. Textor, L. Thomsen, M. Tobin and B. W. Muir, Langmuir, 2010, 26, 13987–13994 CrossRef CAS.
- J. M. Łopacińska, C. Grădinaru, R. Wierzbicki, C. Købler, M. S. Schmidt, M. T. Madsen, M. Skolimowski, M. Dufva, H. Flyvbjerg and K. Mølhave, Nanoscale, 2012, 4, 3739–3745 RSC.
- D.-H. Kim, P. P. Provenzano, C. L. Smith and A. Levchenko, J. Cell Biol., 2012, 197, 351–360 CrossRef CAS.
- N. Saito, H. Haniu, Y. Usui, K. Aoki, K. Hara, S. Takanashi, M. Shimizu, N. Narita, M. Okamoto, S. Kobayashi, H. Nomura, H. Kato, N. Nishimura, S. Taruta and M. Endo, Chem. Rev., 2014, 114, 6040–6079 CrossRef CAS.
- J. Padmanabhan and T. R. Kyriakides, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 355–370 CAS.
- K. Gulati, Surface Modification of Titanium Dental Implants, Springer International Publishing, 2023 Search PubMed.
- M. Kingsak, P. Maturavongsadit, H. Jiang and Q. Wang, Biomater. Transl. Med., 2022, 3, 221–233 Search PubMed.
- S. Mengqi, S. Wen, Z. Boxin, L. Minni, Z. Yan, W. Qun and Z. Yumei, Biomed. Mater., 2021, 16, 15024 CrossRef.
- Y. Chen, S. Aslanoglou, T. Murayama, G. Gervinskas, L. I. Fitzgerald, S. Sriram, J. Tian, A. P. R. Johnston, Y. Morikawa, K. Suu, R. Elnathan and N. H. Voelcker, Adv. Mater., 2020, 32, 2000036 CrossRef CAS.
- K. Gulati, S. Maher, D. M. Findlay and D. Losic, Nanomedicine, 2016, 11, 1847–1864 CrossRef CAS.
- P. Roy, S. Berger and P. Schmuki, Angew. Chem., Int. Ed., 2011, 50, 2904–2939 CrossRef CAS PubMed.
- J. Zhang, D. Fu, M. B. Chan-Park, L.-J. Li and P. Chen, Adv. Mater., 2009, 21, 790–793 CrossRef CAS.
- A. K. Dey, A. Gonon, E.-I. Pécheur, M. Pezet, C. Villiers and P. N. Marche, Cells, 2021, 10 Search PubMed.
- D. F. Moyano, Y. Liu, F. Ayaz, S. Hou, P. Puangploy, B. Duncan, B. A. Osborne and V. M. Rotello, Chem, 2016, 1, 320–327 CAS.
- L. Foit and C. S. Thaxton, Biomaterials, 2016, 100, 67–75 CrossRef CAS PubMed.
- H. Yang, S.-Y. Fung, S. Xu, D. P. Sutherland, T. R. Kollmann, M. Liu and S. E. Turvey, ACS Nano, 2015, 9, 6774–6784 CrossRef CAS PubMed.
- W. Gao, Y. Wang, Y. Xiong, L. Sun, L. Wang, K. Wang, H. Y. Lu, A. Bao, S. E. Turvey, Q. Li and H. Yang, Acta Biomater., 2019, 85, 203–217 CrossRef CAS PubMed.
- C. Zhao, X. Song and X. Lu, Int. J. Nanomed., 2020, 15, 3281–3290 CrossRef CAS.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS PubMed.
- T. Gong, J. Xie, J. Liao, T. Zhang, S. Lin and Y. Lin, Bone Res., 2015, 3, 15029 CrossRef CAS.
- D. Wähnert, J. Greiner, S. Brianza, C. Kaltschmidt, T. Vordemvenne and B. Kaltschmidt, Biomedicines, 2021, 9, 746 CrossRef.
- M. J. Davison, R. J. McMurray, C.-A. Smith, M. J. Dalby and R. M. D. Meek, J. Tissue Eng., 2016, 7, 2041731416652778 Search PubMed.
- M. Mohiuddin, H.-A. Pan, Y.-C. Hung and G. S. Huang, Nanoscale Res. Lett., 2012, 7, 394 CrossRef PubMed.
- M. J. Davison, R. J. McMurray, M. J. Dalby and R. M. D. Meek, Orthop. Proc., 2013, 95–B, 54 Search PubMed.
- L. Sethuram and J. Thomas, Biomed. Pharmacother., 2023, 157, 113996 CrossRef CAS.
- V. S. Waghmare, P. R. Wadke, S. Dyawanapelly, A. Deshpande, R. Jain and P. Dandekar, Bioact. Mater., 2018, 3, 255–266 CrossRef PubMed.
- T. Abdullah, K. Gauthaman, A. Mostafavi, A. Alshahrie, N. Salah, P. Morganti, A. Chianese, A. Tamayol and A. Memic, Sci. Rep., 2022, 12, 15051 CrossRef CAS PubMed.
- A. Eatemadi, H. Daraee, N. Zarghami, H. Melat Yar and A. Akbarzadeh, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 111–121 CrossRef CAS PubMed.
- M. B. Stie, M. Corezzi, A. D. Juncos Bombin, F. Ajalloueian, E. Attrill, S. Pagliara, J. Jacobsen, I. S. Chronakis, H. M. Nielsen and V. Foderà, ACS Appl. Nano Mater., 2020, 3, 1910–1921 CrossRef CAS.
- K. Wang, W.-D. Hou, X. Wang, C. Han, I. Vuletic, N. Su, W.-X. Zhang, Q.-S. Ren, L. Chen and Y. Luo, Biomaterials, 2016, 102, 249–258 CrossRef CAS.
- Y. Song, L. Li, W. Zhao, Y. Qian, L. Dong, Y. Fang, L. Yang and Y. Fan, Bioact. Mater., 2021, 6, 2983–2998 CrossRef CAS PubMed.
- Y. Sueyoshi, A. Niwa, Y. Nishikawa and N. Isogai, J. Biomed. Mater. Res., Part B, 2023, 111, 16–25 CrossRef CAS PubMed.
- B. Veleirinho, D. S. Coelho, P. F. Dias, M. Maraschin, R. Pinto, E. Cargnin-Ferreira, A. Peixoto, J. A. Souza, R. M. Ribeiro-do-Valle and J. A. Lopes-da-Silva, PLoS One, 2014, 9, e95293 CrossRef.
- H. Cao, K. Mchugh, S. Y. Chew and J. M. Anderson, J. Biomed. Mater. Res., Part A, 2010, 93A, 1151–1159 CAS.
- S. H. Laursen, S. G. Hansen, M. B. Taskin, M. Chen, L. Wogensen, J. V. Nygaard and S. M. Axelsen, J. Biomed. Mater. Res., Part B, 2023, 111, 392–401 CrossRef CAS PubMed.
- S. Mukherjee, S. Darzi, K. Paul, F. L. Cousins, J. A. Werkmeister and C. E. Gargett, Front. Pharmacol., 2020, 11 Search PubMed.
- N. M. Vacanti, H. Cheng, P. S. Hill, J. D. T. Guerreiro, T. T. Dang, M. Ma, S. Watson, N. S. Hwang, R. Langer and D. G. Anderson, Biomacromolecules, 2012, 13, 3031–3038 CrossRef CAS PubMed.
- S. Liu, L. Yao, Y. Wang, Y. Li, Y. Jia, Y. Yang, N. Li, Y. Hu, D. Kong, X. Dong, K. Wang and M. Zhu, Bioact. Mater., 2023, 21, 464–482 CrossRef CAS.
- A. D. Schoenenberger, H. Tempfer, C. Lehner, J. Egloff, M. Mauracher, A. Bird, J. Widmer, K. Maniura-Weber, S. F. Fucentese, A. Traweger, U. Silvan and J. G. Snedeker, Biomaterials, 2020, 249, 120034 CrossRef CAS PubMed.
- M. Zhu, Z. Wang, J. Zhang, L. Wang, X. Yang, J. Chen, G. Fan, S. Ji, C. Xing, K. Wang, Q. Zhao, Y. Zhu, D. Kong and L. Wang, Biomaterials, 2015, 61, 85–94 CrossRef CAS PubMed.
- D. Xia, Y. Liu, W. Cao, J. Gao, D. Wang, M. Lin, C. Liang, N. Li and R. Xu, Int. J. Mol. Sci., 2022, 23, 10983 CrossRef CAS.
- S. S. Ashraf, K. Parivar, N. Hayati Roodbari, S. Mashayekhan and N. Amini, Int. J. Biol. Macromol., 2022, 196, 194–203 CrossRef CAS.
- J. Ye, M. Gong, J. Song, S. Chen, Q. Meng, R. Shi, L. Zhang and J. Xue, Pharmaceutics, 2022, 14, 1273 CrossRef CAS PubMed.
- C. He, B. Yu, Y. Lv, Y. Huang, J. Guo, L. Li, M. Chen, Y. Zheng, M. Liu, S. Guo, X. Shi and J. Yang, ACS Appl. Mater. Interfaces, 2022, 14, 32799–32812 CrossRef CAS.
- V. Kumar, A. Kumar, N. S. Chauhan, G. Yadav, M. Goswami and G. Packirisamy, ACS Appl. Bio Mater., 2022, 5, 2726–2740 CrossRef CAS.
- X. Zhang, R. Lv, L. Chen, R. Sun, Y. Zhang, R. Sheng, T. Du, Y. Li and Y. Qi, ACS Appl. Mater. Interfaces, 2022, 14, 12984–13000 CrossRef CAS.
- M. Yu, P. Tang, Y. Tang, C. Wei, Z. Wang and H. Zhang, ACS Appl. Bio Mater., 2022, 5, 2894–2901 CrossRef CAS PubMed.
- S. Y. Lee, S. Jeon, Y. W. Kwon, M. Kwon, M. S. Kang, K.-Y. Seong, T.-E. Park, S. Y. Yang, D.-W. Han, S. W. Hong and K. S. Kim, Sci. Adv., 2023, 8, eabn1646 CrossRef PubMed.
- M. K. Haidar and H. Eroglu, Curr. Top. Med. Chem., 2017, 17, 1564–1579 CrossRef CAS.
- Y. Jia, W. Yang, K. Zhang, S. Qiu, J. Xu, C. Wang and Y. Chai, Acta Biomater., 2019, 83, 291–301 CrossRef CAS PubMed.
- J. Xie, M. R. MacEwan, A. G. Schwartz and Y. Xia, Nanoscale, 2010, 2, 35–44 RSC.
- R. Zhang, G. Jiang, Q. Gao, X. Wang, Y. Wang, X. Xu, W. Yan and H. Shen, Nanoscale, 2021, 13, 15937–15951 RSC.
- X. Tian, Y. Sun, S. Fan, M. D. Boudreau, C. Chen, C. Ge and J.-J. Yin, ACS Appl. Mater. Interfaces, 2019, 11, 4858–4866 CrossRef CAS PubMed.
- Y. Qiao, Y. Ping, H. Zhang, B. Zhou, F. Liu, Y. Yu, T. Xie, W. Li, D. Zhong, Y. Zhang, K. Yao, H. A. Santos and M. Zhou, ACS Appl. Mater. Interfaces, 2019, 11, 3809–3822 CrossRef CAS PubMed.
- W. Chen, H. Chang, J. Lu, Y. Huang, S. G. Harroun, Y. Tseng, Y. Li, C. Huang and H. Chang, Adv. Funct. Mater., 2015, 25, 7189–7199 CrossRef CAS.
- H. Sun, N. Gao, K. Dong, J. Ren and X. Qu, ACS Nano, 2014, 8, 6202–6210 CrossRef CAS.
- Y. Xiang, C. Mao, X. Liu, Z. Cui, D. Jing, X. Yang, Y. Liang, Z. Li, S. Zhu, Y. Zheng, K. W. K. Yeung, D. Zheng, X. Wang and S. Wu, Small, 2019, 15, 1900322 CrossRef PubMed.
- M. Konop, T. Damps, A. Misicka and L. Rudnicka, J. Nanomater., 2016, 2016, 47 Search PubMed.
- R. Singla, S. Soni, V. Patial, P. M. Kulurkar, A. Kumari, Y. S. Padwad and S. K. Yadav, Sci. Rep., 2017, 7, 10457 CrossRef PubMed.
- Y.-C. Yeh, B. Creran and V. M. Rotello, Nanoscale, 2012, 4, 1871–1880 RSC.
- B. Lu, F. Lu, L. Ran, K. Yu, Y. Xiao, Z. Li, F. Dai, D. Wu and G. Lan, Int. J. Biol. Macromol., 2018, 119, 505–516 CrossRef CAS PubMed.
- P. Lau, N. Bidin, S. Islam, W. N. B. W. M. Shukri, N. Zakaria, N. Musa and G. Krishnan, Lasers Surg. Med., 2017, 49, 380–386 CrossRef PubMed.
- G. Borkow, J. Gabbay, R. Dardik, A. I. Eidelman, Y. Lavie, Y. Grunfeld, S. Ikher, M. Huszar, R. C. Zatcoff and M. Marikovsky, Wound Repair Regen., 2010, 18, 266–275 CrossRef PubMed.
- X. Dai, Y. Zhao, Y. Yu, X. Chen, X. Wei, X. Zhang and C. Li, ACS Appl. Mater. Interfaces, 2017, 9, 30470–30479 CrossRef CAS PubMed.
- Y. Hong, M. Yu, J. Lin, K. Cheng, W. Weng and H. Wang, Colloids Surf., B, 2014, 123, 68–74 CrossRef CAS PubMed.
- S. Altuntas, H. K. Dhaliwal, A. E. Radwan, M. Amiji and F. Buyukserin, Biomater. Sci., 2023, 11, 181–194 RSC.
- J. Xu, H. Moon, J. Xu, J. Lim, T. Fischer, H. A. McNally, H. O. Sintim and H. Lee, ACS Appl. Mater. Interfaces, 2020, 12, 26893–26904 CrossRef CAS.
- J. Dai, Y. Lu, X.-Y. He, C. Zhong, B.-L. Lin, S. Ling, J. Gong and Y. Yao, Appl. Mater. Today, 2020, 20, 100753 CrossRef.
- L. Hanson, W. Zhao, H.-Y. Lou, Z. C. Lin, S. W. Lee, P. Chowdary, Y. Cui and B. Cui, Nat. Nanotechnol., 2015, 10, 554–562 CrossRef CAS PubMed.
- F. Milos, A. Belu, D. Mayer, V. Maybeck and A. Offenhäusser, Adv. Biol., 2021, 5, 2000248 CrossRef CAS.
- J. Kang, E.-H. Kang, Y.-S. Yun, S. Ji, I.-S. Yun and J.-S. Yeo, Appl. Microsc., 2020, 50, 26 CrossRef CAS PubMed.
- J. Kim, W.-G. Bae, K.-T. Lim, K.-J. Jang, S. Oh, K.-J. Jang, N. Li Jeon, K.-Y. Suh and J. Hoon Chung, Mater. Lett., 2014, 130, 227–231 CrossRef CAS.
- D. Dehghan-Baniani, B. Mehrjou, P. K. Chu and H. Wu, Adv. Healthcare Mater., 2021, 10, 2001018 CrossRef CAS.
- K. S. Brammer, C. Choi, C. J. Frandsen, S. Oh and S. Jin, Acta Biomater., 2011, 7, 683–690 CrossRef CAS PubMed.
- L. Zhan, L. Wang, J. Deng, Y. Zheng, Q. Ke, X. Yang, X. Zhang, W. Jia and C. Huang, Nano Res., 2023, 16, 1614–1625 CrossRef CAS.
- I. M. Bjørge, B. M. de Sousa, S. G. Patrício, A. S. Silva, L. P. Nogueira, L. F. Santos, S. I. Vieira, H. J. Haugen, C. R. Correia and J. F. Mano, ACS Appl. Mater. Interfaces, 2022, 14, 19116–19128 CrossRef.
- K.-J. Jang, S. Kim, S. Park, W. Kim, Y. Gwon, S. Park, K.-T. Lim, H. Seonwoo and J. Kim, Appl. Sci., 2020, 10 Search PubMed.
- H. N. Kim, A. Jiao, N. S. Hwang, M. S. Kim, D. H. Kang, D.-H. Kim and K.-Y. Suh, Adv. Drug Delivery Rev., 2013, 65, 536–558 CrossRef CAS.
- H. Rashidi, J. Yang and K. M. Shakesheff, Biomater. Sci., 2014, 2, 1318–1331 RSC.
- E. T. den Braber, J. E. de Ruijter, L. A. Ginsel, A. F. von Recum and J. A. Jansen, J. Biomed. Mater. Res., 1998, 40, 291–300 CrossRef CAS PubMed.
- X. Liu, Q. Feng, A. Bachhuka and K. Vasilev, ACS Appl. Mater. Interfaces, 2014, 6, 9733–9741 CrossRef CAS.
- S. Chen, Z. Huang, R. M. Visalakshan, H. Liu, A. Bachhuka, Y. Wu, P. R. L. Dabare, P. Luo, R. Liu, Z. Gong, Y. Xiao, K. Vasilev, Z. Chen and Z. Chen, Biomater. Res., 2022, 26, 88 CrossRef CAS PubMed.
- S. Chen, J. A. Jones, Y. Xu, H.-Y. Low, J. M. Anderson and K. W. Leong, Biomaterials, 2010, 31, 3479–3491 CrossRef CAS PubMed.
- K. R. Kam, L. A. Walsh, S. M. Bock, J. D. Ollerenshaw, R. F. Ross and T. A. Desai, Tissue Eng., Part A, 2014, 20, 130–138 CrossRef CAS.
- F. F. R. Damanik, T. C. Rothuizen, C. van Blitterswijk, J. I. Rotmans and L. Moroni, Sci. Rep., 2014, 4, 6325 CrossRef CAS PubMed.
- R. P. Tan, A. H. P. Chan, S. Wei, M. Santos, B. S. L. Lee, E. C. Filipe, B. Akhavan, M. M. Bilek, M. K. C. Ng, Y. Xiao and S. G. Wise, JACC, 2019, 4, 56–71 Search PubMed.
- A. Bachhuka, S. N. Christo, A. Cavallaro, K. R. Diener, A. Mierczynska, L. E. Smith, R. Marian, J. Manavis, J. D. Hayball and K. Vasilev, J. Colloid Interface Sci., 2015, 457, 9–17 CrossRef CAS PubMed.
- M. Liu, K. Wu, L. Zhao and Y. Zhang, J. Biomed. Nanotechnol., 2017, 13, 381–392 CrossRef CAS.
- K. Baylón, P. Rodríguez-Camarillo, A. Elías-Zúñiga, J. A. Díaz-Elizondo, R. Gilkerson and K. Lozano, Membranes, 2017, 7 Search PubMed.
- F. Robotti, I. Sterner, S. Bottan, J. M. Monné Rodríguez, G. Pellegrini, T. Schmidt, V. Falk, D. Poulikakos, A. Ferrari and C. Starck, Biomaterials, 2020, 229, 119583 CrossRef CAS PubMed.
- S. Heberer, S. Kilic, J. Hossamo, J.-D. Raguse and K. Nelson, Clin. Oral. Implants Res., 2011, 22, 546–551 CrossRef PubMed.
- N. Khandelwal, T. W. Oates, A. Vargas, P. P. Alexander, J. D. Schoolfield and C. Alex McMahan, Clin. Oral. Implants Res., 2013, 24, 13–19 CrossRef.
- D. Schwartz-Arad, A. Laviv and L. Levin, J. Periodontol., 2007, 78, 219–223 CrossRef PubMed.
- B. S. Kopf, S. Ruch, S. Berner, N. D. Spencer and K. Maniura-Weber, J. Biomed. Mater. Res., Part A, 2015, 103, 2661–2672 CrossRef CAS.
- R. D. Guyer, J.-J. Abitbol, D. D. Ohnmeiss and C. Yao, Spine, 2016, 41, E1146–E1150 CrossRef.
- N. Wang, H. Li, W. Lü, J. Li, J. Wang, Z. Zhang and Y. Liu, Biomaterials, 2011, 32, 6900–6911 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |