Open Access Article
Open Access ArticleMonitoring lipid alterations in Drosophila heads in an amyotrophic lateral sclerosis model with time-of-flight secondary ion mass spectrometry†
Minh Uyen Thi
Le‡
ab,
Jeong Hyang
Park‡§
c,
Jin Gyeong
Son‡
 a,
Hyun Kyung
Shon
a,
Sunho
Joh
a,
Chang Geon
Chung¶
c,
Jae Ho
Cho
c,
Alexander
Pirkl
d,
Sung Bae
Lee
*c and
Tae Geol
Lee
*ab
a,
Hyun Kyung
Shon
a,
Sunho
Joh
a,
Chang Geon
Chung¶
c,
Jae Ho
Cho
c,
Alexander
Pirkl
d,
Sung Bae
Lee
*c and
Tae Geol
Lee
*ab
aBio-imaging Team, Safety Measurement Institute, Korea Research Institute of Standard and Science (KRISS), Daejeon 34113, Republic of Korea. E-mail: tglee@kriss.re.kr; Fax: +82-42-868-5033; Tel: +82-42-868-5003
bDepartment of Nano Science, University of Science and Technology (UST), Daejeon 34113, Republic of Korea
cDepartment of Brain Sciences, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu 42988, Republic of Korea. E-mail: sblee@dgist.ac.kr
dIonTOF Technologies GmbH, Helsenbergstrasse 15, 48149 Münster, Germany
First published on 19th December 2023
Abstract
Lipid alterations in the brain are well-documented in disease and aging, but our understanding of their pathogenic implications remains incomplete. Recent technological advances in assessing lipid profiles have enabled us to intricately examine the spatiotemporal variations in lipid compositions within the complex brain characterized by diverse cell types and intricate neural networks. In this study, we coupled time-of-flight secondary ion mass spectrometry (ToF-SIMS) to an amyotrophic lateral sclerosis (ALS) Drosophila model, for the first time, to elucidate changes in the lipid landscape and investigate their potential role in the disease process, serving as a methodological and analytical complement to our prior approach that utilized matrix-assisted laser desorption/ionization mass spectrometry. The expansion of G4C2 repeats in the C9orf72 gene is the most prevalent genetic factor in ALS. Our findings indicate that expressing these repeats in fly brains elevates the levels of fatty acids, diacylglycerols, and ceramides during the early stages (day 5) of disease progression, preceding motor dysfunction. Using RNAi-based genetic screening targeting lipid regulators, we found that reducing fatty acid transport protein 1 (FATP1) and Acyl-CoA-binding protein (ACBP) alleviates the retinal degeneration caused by G4C2 repeat expression and also markedly restores the G4C2-dependent alterations in lipid profiles. Significantly, the expression of FATP1 and ACBP is upregulated in G4C2-expressing flies, suggesting their contribution to lipid dysregulation. Collectively, our novel use of ToF-SIMS with the ALS Drosophila model, alongside methodological and analytical improvements, successfully identifies crucial lipids and related genetic factors in ALS pathogenesis.
1. Introduction
Amyotrophic lateral sclerosis (ALS) is a late-onset neurodegenerative disease characterized by a gradual loss of motor neurons in the spinal cord, brainstem, and motor cortex.1 The GGGGCC (G4C2) hexanucleotide repeat expansion mutation in the intron of the C9orf72 gene is the leading genetic cause of familial ALS (C9-ALS).2–4 ALS patients suffer from gradual muscle weakness and atrophy throughout the body, which ultimately leads to death within 2–5 years from the onset of symptoms. Once ectopic neuronal cell death and muscle atrophy become prevalent in the late stages of the disease, the disease progression cannot be reversed. However, the pathogenic mechanisms and associated neuropathic features in the early stages of the disease remain unclear.To understand the pathogenic mechanisms and associated neuropathic features of ALS, researchers have profiled altered levels of proteins,5 mRNAs, and lipids in afflicted neurons of both human patients and animal models.6,7 Although lipids are a major component of cell membranes, energy sources, and a functional element of living organisms,8,9 only a handful of studies have investigated lipid alteration in ALS10–15 due in part to technical limitations. For example, Cutler et al. reported that the levels of sphingomyelin, ceramides, and cholesterol esters were increased in the spinal cord of the SOD1 ALS mouse model and in ALS patients compared to controls.15 Such changes in lipids have been conventionally assessed through extracted samples using liquid chromatography tandem mass spectrometry, inevitably limiting our understanding of site-specific alterations in lipid profiles associated with ALS. Analytical approaches involving mass spectrometry imaging (MSI) have emerged as powerful tools for linking spatial information with molecular changes in biomedical research. MSI techniques also facilitate label-free mapping by analyzing intrinsic information within the sample itself. Since changes in site-specific lipids may reflect pathological and metabolic alterations during disease progression, monitoring these changes presents a promising approach for disease studies. Therefore, identifying spatiotemporal alteration of lipids in ALS may provide valuable insights into understanding the pathogenic mechanisms of the disease.
In this regard, it is noteworthy that we have recently introduced matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) as an MSI technique in conjunction with the Drosophila ALS model and successfully identified a number of phospholipids associated with ALS pathogenesis in the Drosophila ALS model expressing PR80 dipeptide repeat proteins.16 However, the selective ionization process of molecules specific to the matrix used, a characteristic unique to the MALDI-TOF technique,17 necessitates additional MSI techniques that enable a broader spectrum of molecular detection. This expansion is crucial for a comprehensive understanding of the overall lipid alterations pathogenically associated with ALS. In contrast to aforementioned MALDI-TOF, time-of-flight secondary ion mass spectrometry (ToF-SIMS) allows observation in the low mass region around m/z 100–500 without matrix interference. This is achieved by utilizing a matrix-free liquid metal ion beam with high lateral resolution. Due to these analytical features, ToF-SIMS has been applied to a variety of biological samples, ranging from cell to tissue analysis. Ostrowski et al. applied SIMS to the analysis of single-cell imaging, demonstrating lipid changes with cell surface curvature during mating in Tetrahymena thermophile.18 Tian et al. conducted a comparative study of mouse brain tissue, comparing normal and injured samples using GCIB-SIMS, and discovered an enhancement in signals such as cardiolipin and gangliosides after injury.19 In addition, studies have been conducted with various types of tissues, including lipidomics in kidney20 tissue and the distribution of inorganic nanoparticles in lung tissue.21 Notably, ToF-SIMS makes it possible to study images of small samples, such as Drosophila heads, wherein local lipid alterations can be detected, revealing their site-specific roles in disease pathogenesis or symptoms.22–24 Considering that the detectable mass range and image resolution in SIMS analysis are influenced by the type of ion beam, improvements in detection and analysis are expected to be achieved through the appropriate selection of the ion beam, such as liquid metal ion beam or gas cluster ion beam.
In this study, we aimed to explore the potential of ToF-SIMS for monitoring disease-related lipid changes in a C9-ALS Drosophila model, focusing on the early stages of disease progression. Our research was designed to build upon and enhance previous studies, including our own,16 by implementing several key methodological and analytical improvements. These enhancements included the adoption of ToF-SIMS as a complementary technique to MALDI-TOF for mapping lipid changes in ALS, the use of a Bi3+ ion beam to achieve high-resolution imaging at the micron level, and the application of matrix-free ionization to detect significant signals in the small molecule region within the m/z 200–600 range. From a biological perspective, we sought to investigate variations in specific lipid levels during the course of ALS progression using a genetic model. For this purpose, we employed the well-established (G4C2)36C9orf72 gene model to analyze the spatial distribution of ALS-related lipid changes with ToF-SIMS. The expanded G4C2 model is widely used for pathogenic mechanism studies related to both ALS and frontotemporal dementia (FTD).3 As mentioned earlier, despite C9-ALS being the primary genetic cause of ALS, the mechanisms behind lipid alterations associated with C9-ALS remain unclear and have only recently begun to be reported.25,26 Our results indicate a significant increase in lipid levels during the initial 5 days of (G4C2)36 expression, even preceding the onset of motor dysfunction. Notably, we observed the accumulation of fatty acids (FAs), triacylglycerols (TAGs), and ceramides (Cer) in the fat body region outside the brain following (G4C2)36 expression, indicative of the isolation and storage of lipids. Our data suggest that lipid accumulation just outside the brain is an early manifestation of disease progression, likely originating from neurons, and contributes to C9-ALS pathogenesis in flies. The successful identification of novel lipids associated with ALS pathogenesis, achieved through the methodological and analytical enhancements in this study, holds promise for future research, extending beyond ALS and applicable to various diseases.
2. Material and methods
2.1. Animal and sample preparation
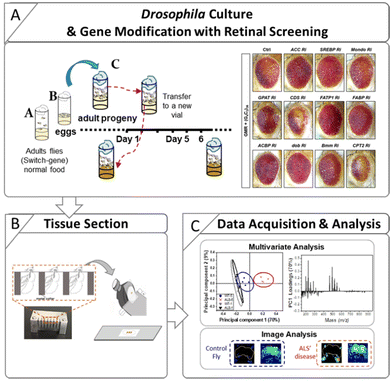
Control flies (w1118) and an ALS Drosophila model carrying an expanded (G4C2)36 repeat in the C9orf72 gene were used in this study. The inducible GeneSwitch Gal4 system (elav-GS) was also used so that the (G4C2)36 gene could be expressed upon RU486 treatment (mifepristone, Sigma-Aldrich, USA). The control and ALS flies were the progeny of elav-GS flies crossed with w1118 flies and (G4C2)36 flies, respectively. The preparation steps for the Drosophila used in this study are described in Scheme 1. The preparation procedure for ALS flies with RU486 treatment is as follows. Parents for the cross [w1118 with elav-GS flies and (G4C2)36 with elav-GS flies] were put into vials with normal media (Bloomington Formulation, Genesee Scientific, Cat #. 66-112) (Scheme 1A) and then removed after 3 days. After the eggs hatched, the larvae were fed with normal media. After eclosion, the adult progenies were transferred into new normal media vials at 25 °C, after which male flies were collected and fed with RU486-treated media. The flies were collected for analysis after 1, 3, 5, and 9 days from the start point of the RU486 media. The flies were attached to a fly collar and embedded in 10% gelatin (G1890, Sigma-Aldrich, USA). Subsequently, the gelatin block was frozen in liquid nitrogen and stored at −80 °C until the cryosection procedure.Similarly, gelatin blocks of RNAi fly stocks were prepared to study genetic interactions. The following lines were used in this study: w1118, (BL5905), UAS-(G4C2)36 (BL58688), elavGS-gal4 (BL43642), UAS-Luciferase (BL35788), GMR-gal4 (BL84247), UAS-ACC RNAi (BL34885), UAS-SREBP RNAi (BL25975), UAS-Mondo RNAi (BL27059), UAS-GPAT RNAi (BL61335), UAS-CDS RNAi (BL58118), UAS-FATP1 RNAi (BL50709), UAS-FABP RNAi (BL34685), UAS-ACBP RNAi (BL67020), UAS-dob RNAi (BL65925), UAS-Bmm RNAi (BL25926), and UAS-CPT2 RNAi (BL62455) from Bloomington Drosophila Stock Center (BDSC).
The sample sections were prepared by the tape-supported method as reported in our previous work.27 A cryostat CM 3050 S (Leica, Nussloch, Germany) was used to section the Drosophila samples with a thickness of 14 μm. The sections were then attached to SUS substrates using conductive carbon double-sided tape (NEM tape, Nisshin Co., Ltd, Japan), as shown in Scheme 1B. The samples were freeze-dried on a pre-chilled aluminum block in a vacuum under 1 × 10−2 torr for 2 h. The dried samples were then stored at −80 °C in vacuum-sealed bags to keep them fresh until ToF-SIMS analysis.
2.2. Instrumentation and analysis
ToF-SIMS experiments were performed using a TOF-SIMS V instrument (IONTOF GmbH, Muenster, Germany) equipped with a pulsed 25 keV Bi3+ primary ion beam. The Bi3+ clusters were employed to analyze the sample in positive and negative ion mode, while the beam was operated in bunched mode to obtain the highest possible mass resolution with an approximate primary ion current of 0.1 pA. Images were acquired on an area of 500 × 500 μm2 by scanning with 256 × 256 pixels in random mode.For lipid identification, a hybrid SIMS instrument was employed (Hybrid SIMS, IONTOF GmbH Muenster, Germany) using a 20 keV Ar3000+ cluster primary ion beam with an ion current of 5.5 pA. A ToF analyzer and an Orbitrap (Q Exactive HF, Thermo Fisher Scientific GmbH, Bremen, Germany) were used in combination for better resolving power. An area of 500 × 500 μm2 was scanned with 50 × 50 pixels in sawtooth mode at a pixel resolution of 10 μm. By using the Orbitrap, a mass resolution of 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z 200 was achieved. Cluster signal patterns of a silver foil were used to calibrate the mass scale of the Orbitrap analyzer.
000 at m/z 200 was achieved. Cluster signal patterns of a silver foil were used to calibrate the mass scale of the Orbitrap analyzer.
Images of 500 × 500 μm2 were obtained with an ion dose of 8 × 1011 ions cm−2 (TOF-SIMS V) and 1.7 × 1013 ions cm−2 (Hybrid SIMS). During the experiments, an electron flood gun was used for charge compensation. All SIMS data were analyzed with the Surface Lab 7.0 software suite (IONTOF GmbH, Muenster, Germany). In the current study, we aimed to achieve more accurate external calibration by employing the Drosophila sample as calibration material and conducting additional measurements using the Hybrid instrument's Orbi SIMS mode. In order to increase the mass accuracy of the Bi3+ measurement spectra from TOF-SIMS V, Ar3000+ measurement was performed using the Hybrid SIMS on the same Drosophila sample, after which the Bi3+ measurement spectra were recalibrated based on the Ar3000+ value (Fig. S1†). Hybrid SIMS is more accurate in identifying specific molecules because it is equipped with an Orbitrap analyzer and has about 30 times higher mass resolution than ToF-SIMS. Utilizing accurate mass values from the Orbitrap analyzer, we were able to significantly reduce the average mass error from 57.57 to 12.18 for positive ions and from 210.74 to 67.35 for negative ions. The results of the mass deviation calculations are summarized for each ion and shown in Table S1.† Mass calibration was performed using C2H5+, C3H7+, C5H12N+, C5H13NPO3+, and C5H15NPO4+ peaks in positive mode and C4H−, C6H−, C16H31O2−, and C18H35O2− peaks in negative ion mode. Identification of all molecules was based on precisely verified m/z values. Lipid identification was then carried out by the best fit of experimental m/z values with theoretical m/z values from the LipidMaps database (LIPID MAPS: https://www.lipidmaps.org).
2.3. Negative geotaxis assay
In the negative geotaxis assay, 15 flies of each genotype at 1 day after eclosion were collected and transferred to an acrylic cylinder (3 cm diameter, 18 cm height). To prevent escape, the top of the cylinder was sealed with sponges. After acclimation (30 min), the flies were placed on the inner bottom surface of the cylinder by lightly tapping (5 times) against a foam pad. Climbing ability was analyzed by measuring the number of flies that arrived at a target line (10 cm from the bottom of the cylinder) within 5 s.2.4. Retinal screening
The fly eyes (left eyes only) were imaged on day 1 after eclosion using a Leica microscope (Leica M205). Images were taken at 160× magnification immediately upon dissection.2.5. qRT-PCR
Total RNA was extracted from the heads of Drosophila (n = 20) using easy blue solution (iNtRON Biotechnology, Korea) with the following genotypes: +/elavGS-gal4, (G4C2)36/+; elavGS-gal4/+. The concentration of the total RNA of each sample was measured with a NanoDrop spectrophotometer. The mRNA level of the elav gene was used as a control. qRT-PCR was performed using SYBR reagent (qPCRBIO SyGreen Mix Separate-ROX, PCR Biosystems Ltd, UK) and a Real Time PCR Detection System (CFX96, Bio-Rad, USA). The primer sequences for each gene were as follows: FATP1 Ri, 5′ FATP1: ATGGGCTGGATTTTTGCTGTG and 3′ FATP1: GTCGCACCCGCTATGTAGAAC; ACBP Ri, 5′ ACBP: CAGGCGGCAGTCAATGTGATA and 3′ ACBP: GAACAGCCCGTAGAACTTGAG; SREBP Ri, 5′ SREBP: AGTCGCCGCTTCTCGTCTA, and 3′ SREBP: TGTATGGTGGCTGTTGGTTGG, dob Ri, 5′ dob: GTCCAGGAGATACAACGGCAT and 3′ dob: TCCTCGGGCAACCAACTTC; Bmm Ri, 5′ Bmm: TCCCTGGGTCCCTTCAGTC and 3′ Bmm: TCGTCTGGTAGATGCTTCTGTA.2.6. Statistical analysis
A principal component analysis (PCA) is often used to facilitate the discrimination of the most characteristic spectral data and the significant signals; detailed descriptions of PCA are given in previous publications.30,31 The PCA in the current work was performed using the NESAC/BIO MVA Toolbox running in MATLAB (MathWorks, Inc., Natick, USA). The scores are plotted within the 95% confidence limit. Peak selection was done manually from the peaks in the range m/z 100–900. To reduce the influence of background signals, the selected peaks were normalized by the sum of their corresponding peak intensities to account for the fluctuations between the spectra.3. Results and discussion
3.1. Monitoring of lipid changes in ALS Drosophila by ToF-SIMS analysis
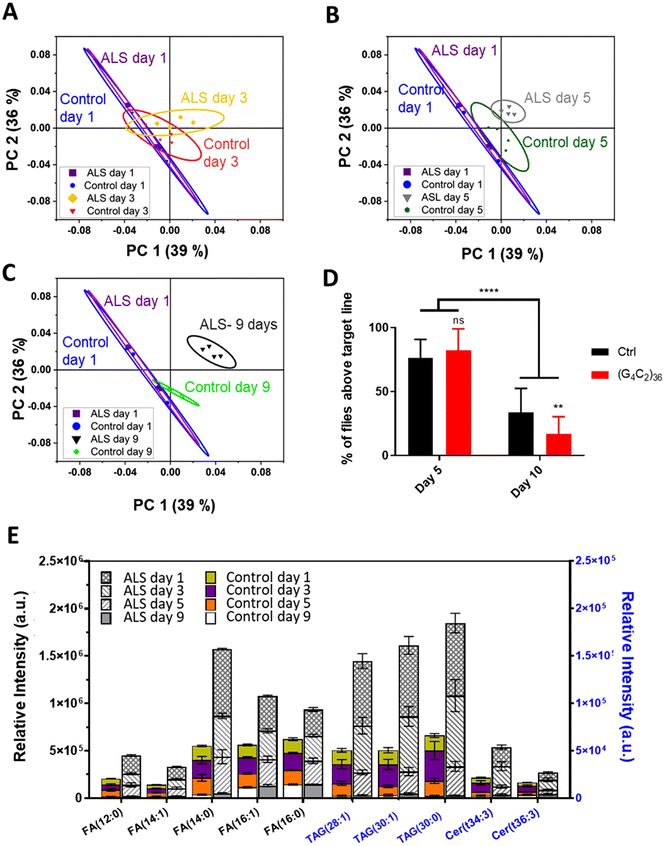
To identify a molecular basis for lipid alteration in ALS, we expressed (G4C2)36 repeats in Drosophila, using the well-characterized C9-ALS fly model.28 With the C9-ALS model, ToF-SIMS analysis was conducted and PCA was used to gauge the extent of lipid changes during disease progression. PCA, a type of multivariate analysis, is a powerful regression method used to extract meaningful information through dimensionality reduction of complex mass spectral data. For the PCA, we selected peaks within the m/z range of 100–900 at positive mode and negative mode, and combined peak lists used to study the changes in the lipid levels in entire Drosophila head samples. Each spectra was normalized to the total ion counts of the corresponding ion mode. Using the gene switch, we identified temporal changes in the Drosophila lipids that occurred with the onset of ALS. To check the level of lipids in the early stages of C9-ALS, we selected flies at 1, 3, 5, and 9 days after eclosion, which correspond to the timepoint of gene expression. To synchronize the expression of the ALS gene with the growth stages of the adult flies, we used RU486-treated medium to activate the gene switch. This allowed us to align the timing of gene expression with the date the flies started growing in the RU486-treated medium after being transferred from the normal medium as adults. Fig. 1 shows representative scores of the PCA results used for differentiating the extracted mass spectra from the different groups. We only plot the first principal component (PC1) and the second principal component (PC2) because they accounted for 75% of the total variance in the experiment.As the scores plotted in Fig. 1 demonstrate, PC1 showed increasing separation between the control and ALS samples with age, suggesting an imbalance of lipid content in ALS flies. More specifically, we analyzed the difference between the control and ALS flies at days (A) 3 (B) 5, and (C) 9 relative to day 1 control and ALS flies. The results indicated that a clear difference in lipids between control and ALS flies began on day 5 and became more evident by day 9. On day 1 and day 3, the ALS flies did not show significant differences in lipid levels compared to the control flies. Although a segregation of the control and ALS by PCA was observed from day 3 onward, Fig. 1A indicates a substantial overlap of the 95% confidence level ellipses.
Next, we checked whether the ALS flies showed locomotive defects on day 5. Surprisingly, we found no significant difference in their climbing ability compared to the control flies (Fig. 1D); however, by day 10, the difference became apparent (Fig. 1D). Together, these data demonstrate that the changes in lipid content preceded discernible motor symptoms in the ALS flies, suggesting that altered lipid content might be an important early pathological feature of ALS.
We next identified the types of lipids that were upregulated in the “pre-symptomatic” day 5 flies. According to the highest loading values, the molecular candidates that contributed the most to these results were the fatty acid (FA), triacylglycerol (TAG), and ceramide (Cer) groups (Table S2†). The lipids of these three groups increased rapidly in the ALS flies compared to control flies starting at day 3 and onward until at least day 9. This is remarkable, considering that motor dysfunction was not observed until day 10. Characteristically, the three groups of lipids, namely FAs, TAGs, and Cer, began to change on day 3 and showed significant differences on day 5. This pathological change that occurred quite early compared with behavioral changes in Drosophila suggests that lipid imbalance is closely linked to ALS.
3.2. Alterations in lipid levels and their distribution in Drosophila in the early stage of ALS
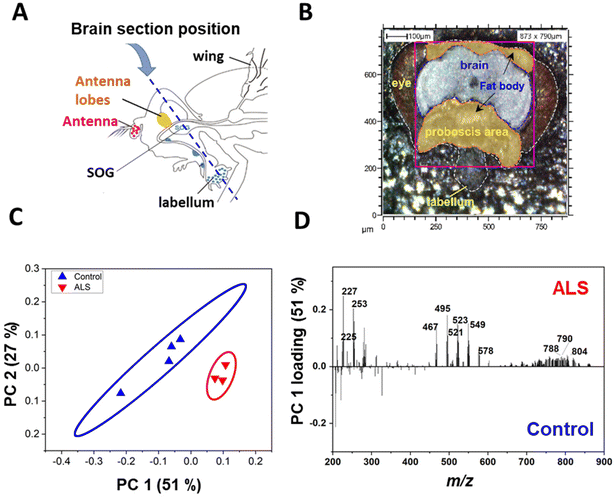
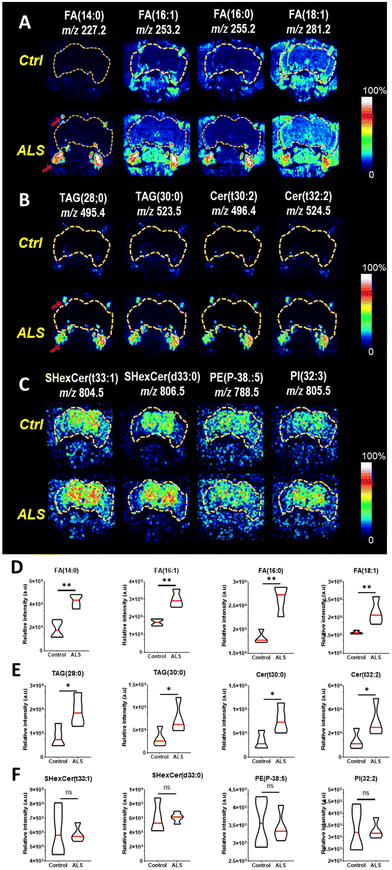
The C9-ALS C9orf72 gene model used in this study targets the gene that most commonly causes ALS/FTD. ALS involves the loss of motor neurons in the upper brain and spinal cord, and FTD is a brain disease in which nerve cells in the frontal and temporal lobes are lost.29 Because motor neurons and nerve cells are clustered in the head region, the onset of ALS/FTD should be associated with pathological changes in lipids of the head. We examined where in the head the three aforementioned lipid classes accumulated in C9-ALS flies. To obtain spatiotemporal information about the lipids in the heads of fruit flies, Drosophila heads were sectioned coronally, which included the labellum, head fat body, and the brain (Fig. 2A). Among the sampled Drosophila heads, mass imaging measurements were carried out on the fat body and brain regions, as indicated by the pink dotted line in the optical image (Fig. 2B). Mass imaging was carried out with a highly focused Bi3+ ion beam, which enabled an improved image resolution compared to conventional gas cluster ion beams in ToF-SIMS19 or laser beams in MALDI-TOF.30To confirm the difference in lipid levels between control and ALS flies on day 5, PCA was performed again after excluding all other days. In the day 5 samples, PC1 clearly separated the control and ALS samples, confirming a lipid imbalance in the ALS flies (Fig. 2C). Molecular candidates that contributed the most to the distinction between the two groups can be identified from the PC1 loading values (Fig. 2D). The detailed discriminant ions as significant contributors to the classification are summarized in Table S1.† Molecular candidates contributing to this distinction were identified to belong to the FA and TAG groups, including myristic acid at m/z 227.2 FA (14:0), palmitoleic acid at m/z 253.2FA (16:1), palmitic acid at m/z 255.2FA (16:0), oleic acid at m/z 281.2FA (18:1), TAG (28:0) at m/z 495.4, and TAG (30:0) at m/z 523.5. Also, Cer (t30:2) at m/z 496.4, Cer (t32:3) at m/z 524.4, Cer-derivatives, and phospholipids showed variation between control and ALS flies.
Next, the distribution of upregulated lipid species in the ALS flies was determined by ToF-SIMS image analysis. We employed tape-supported sampling method for image analysis, effectively preserving the spatial information of molecules within the sample.27 The ToF-SIMS images indicated that FAs (Fig. 3A), TAGs, and Cer (Fig. 3B), which were highly abundant in the ALS flies were accumulated near the proboscis area, specifically in the fat body regions. The fat body regions surrounding the brain contain many glial cells, and an imbalance in lipid metabolism causes the accumulation of lipid droplets (LDs) in the glial cells.31 The LD-rich glial cells are less sensitive to harmful reactive oxygen species (ROS) activity than neurons, so they are considered the main site of FA storage and lipid metabolism in the brain.32 Neuronal FAs that are not been used during the electron transport chain (ETC)-mediated synthesis of adenosine triphosphate (ATP) are transported to glial cells by apolipoproteins. As the accumulation of FAs is toxic to neuronal cells, FAs must be stored as TAGs in LDs. Interestingly, we found a highly localized distribution of FAs and TAGs in the fat body regions (Fig. 3A and B). Also, the presence of Cer, a key component of biosynthesis and catabolism, was also observed in the fat body regions in the ALS flies. It has been shown that increased production and accumulation of Cer in mouse models induced apoptosis in cortical and motor neurons,33,34 and accordingly, their accumulation may contribute to ALS pathogenesis.35 We also performed two-tailed tests on each group of four fruit flies to test for statistical significance, and we found that all FA, TAG, and Cer subunits showed a significant increase in ALS compared to controls (Fig. 3D and E). Based on the reported results, the localization of FAs, TAGs, and Cer in the fly model we identified may be considered a strategy to avoid the potential effects of toxic lipid accumulation in neurons that may lead to neurodegeneration. In Fig. 3C, we also found increased sulfatides hexosyl ceramide (SHexCer), a type of sphingolipid, and phosphatidylethanolamine (PE) and phosphoinositol (PI), types of phospholipids in the brain area of ALS flies. It is well known that dysfunction of sphingolipid and phospholipid metabolism causes the pathogenesis of neurodegenerative diseases.36–38 Notably, the distribution of these molecules was site-specific, and its intensity in the ToF-SIMS images was higher than that of the control. However, statistical comparisons of the total intensity of images of the four flies showed that they did not have a significant difference in ALS compared to controls (Fig. 3F).
In this study, we observed increases in fatty acids (FAs), tiacylglycerols (TAGs), and ceramides (Cer) in the fat body regions of ALS flies. These lipids were concentrated in the fat body regions, potentially as a mechanism to mitigate neurotoxicity caused by lipid accumulation. Furthermore, we detected elevated levels of sphingolipids and phospholipids in the brain regions, known to accompany changes in neurodegenerative diseases. Our results, analyzed using a Bi3+ ion beam, demonstrated that differences in the relative low molecular weight region (m/z 200–600) were region-specific and statistically significant. However, in the m/z 600–900 range, although region-specific differences were observed, statistical distinctions were less clear, suggesting that for lipid metabolism above m/z 600, methods such as our previous use of MALDI-TOF16 may offer advantages in studying fruit flies. In this regard, these findings align well with our intention of this study design to optimize methodological and analytical settings for understanding overall lipid alterations implicated in ALS by complementing our prior approach.16 Taken together, while a variety of lipid changes are associated with ALS, we were able to confirm that they are site-specific. This information can enhance our understanding of the relationship between lipid metabolism and ALS.
3.3. Monitoring lipid changes through the regulation of lipid-related genes involved in ALS
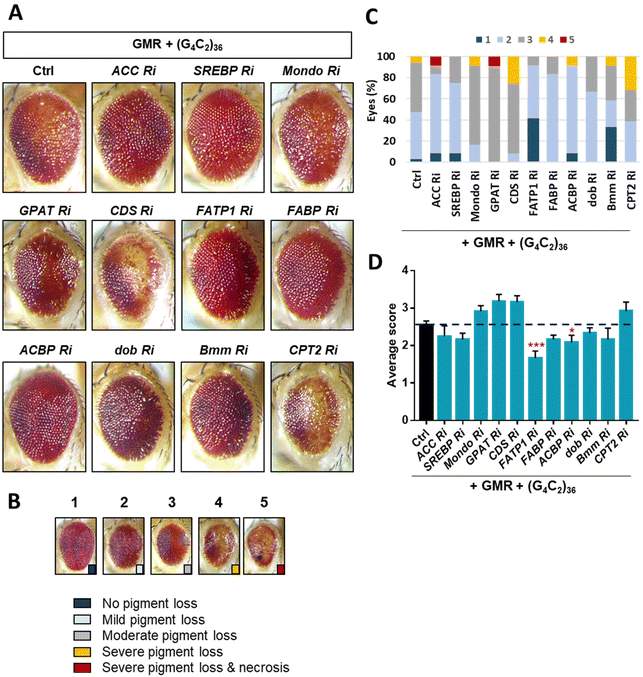
ToF-SIMS analysis allowed us to monitor the changes in the lipid levels in a fly model of ALS. The major lipids that were increased by day 5 were FAs, TAGs, and Cer. To identify how these lipids were increased, we examined lipid regulators involved in lipid metabolism such as lipid biosynthesis, trafficking, and lipolysis (Fig. S2†). By knocking down regulatory genes, we sought to identify genes that could modify the levels of FAs, TAGs, and Cer in the ALS flies. To do this, we performed an RNAi (Ri) screen of lipid regulators that modify (G4C2)36-induced retinal degeneration, given that expression of expanded G4C2 repeat in Drosophila eyes induces obvious degenerative changes in the overall appearance of the eyes characterized by pigment loss and ectopic necrosis of ommatidium (Fig. 4A). Among the regulatory genes, FATP1 Ri and ACBP Ri considerably suppressed the loss of pigment in the eyes expressing (G4C2)36 (Fig. 4C and D). These data suggest a possibility that (G4C2)36 causes toxicity in the ALS flies partly through upregulating FATP1 and ACBP mRNAs. To test this hypothesis, we conducted qRT-PCR experiments to measure the mRNA levels of FATP1 and ACBP as well as other regulatory genes that showed a slight rescue of eye phenotypes, namely SREBP, dob, and Bmm. Interestingly, FATP1 and ACBP mRNA levels were significantly increased compared to the control (Fig. 5A and B). However, SREBP, dob, and Bmm mRNA levels did not show any noticeable change (Fig. 5C and Fig. S3†).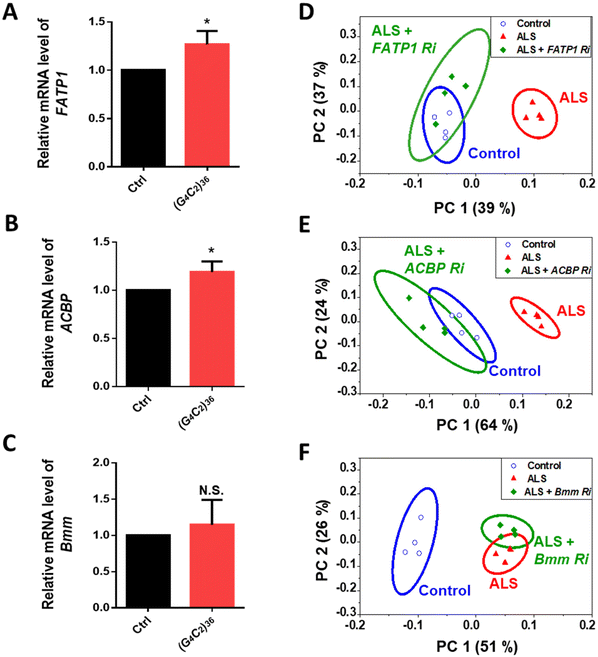 | ||
| Fig. 5 (A–C) Quantification of mRNA levels of denoted genes in Drosophila heads expressing ALS-associated (G4C2)36 on day 5 after the induction of (G4C2)36 expression by RU486 treatment. *p < 0.05, N.S., not significant, p > 0.05 by two-tailed t-test; error bars ± SEM; n ≥ 3 replicates. (D–F) PCA of the whole head region of the genetic models corresponding to A–C on day 5. In the scores plot of PC1 and PC2, the ellipses represent the 95% confidence interval. The clusters associated with control flies are presented in blue, while the clusters associated with ALS flies are presented in red. Loading plots for PC1 of each PCA results are shown in Fig. S5.† PC1 separates control samples from ALS samples [genotype: Ctrl, +/elavGS-gal4, ALS, (G4C2)36/+; elavGS-gal4/+]. | ||
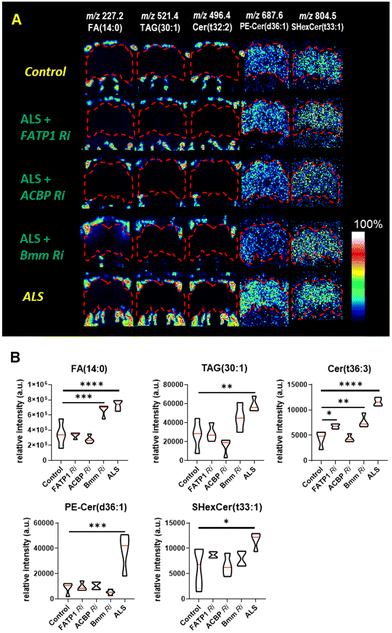
Through the Ri screening for lipid regulators that could modify the retinal degeneration caused by (G4C2)36, we found that FATP1, ACBP, and Bmm showed the least pigment loss. These results suggest a possibility that the Ri of the three genes may also alter lipid levels in ALS flies as well. To confirm the lipid changes upon knockdown of these genes, we performed ToF-SIMS analysis and conducted PCA on three groups: control, ALS, and ALS + Ri of lipid regulators (RNAi of FATP1, ACBP, and Bmm). The PCA score plots revealed that ALS + FATP1 Ri and ALS + ACBP Ri were similar to the control (Fig. 5D and E), while ALS + Bmm Ri was similar to ALS (Fig. 5F). The FAs, TAGs, and Cer were identified as the main factors that influenced the similar grouping between FATP1 Ri and ACBP Ri and the control. Details of the PC1 loading values are shown in Table S3.†
We discovered that FATP1 Ri and ACBP Ri primarily affected the fatty acid pathways of the ALS flies, particularly in the fat body regions rather than in the brain. It is worth noting that in the ToF-SIMS images showing the relative lipid abundance (Fig. 6A), the levels of FA (14:0), TAG (28:1), and Cer (t36:3) were decreased in FATP1 Ri and ACBP Ri compared to the ALS flies, while in bmm Ri, the levels of these lipids were only slightly decreased compared to the ALS flies. The differences are also evident in the violin plots showing the differences in FA (14:0), TAG (28:1), and Cer (t36:3) were decreased in FATP1 Ri and ACBP Ri compared to the ALS flies, while in bmm Ri, the levels of these lipids were not significantly decreased compared to the ALS flies (Fig. 6B). In ALS + bmm Ri, we observed that the PE-Cer level was significantly reduced in the brain region compared to the ALS model and other ALS + Ri models (Fig. 6A and B). We suspected that the decreased PE-Cer level in the brain also contributed to recovering the eye degeneration of the ALS model (Fig. 4D). However, based on the effects of the FATP1 Ri and ACBP Ri models on the eye degeneration (Fig. 4D) and the PCA results (Fig. 5D and E), it can be inferred that FAs, TAGs, and Cer are the main contributors to the C9-ALS model.
By knockdown of the lipid regulators, we discovered two candidate genes that can regulate the lipids that are increased in ALS. Also, by performing mass spectrometry analysis on the FATP1 Ri and ACBP Ri models, we found that we could significantly lower the three lipid groups of FAs, TAGs, and Cer, which showed the largest increase in ALS. As demonstrated here, the molecular imaging approach using Bi3+ in ToF-SIMS has a significant advantage in revealing spatial information about molecules undergoing changes in the ALS disease model with high-resolution MSI. But on the other hand, ToF-SIMS has a weak ion beam current and it operates in a rastering mode, taking a longer time to obtain sufficient signals from secondary ions compared to MALDI-TOF, which ionizes with a laser source. The extended measurement time makes it challenging to apply to screening methods that require rapid analysis of large-area samples or to intergroup analysis of biological samples with substantial sample sizes. In addition, ToF-SIMS is an ionization method specialized in the low-mass range, from m/z 200–600, making the measurement mass range a critical consideration. Although the cluster ion beam in SIMS allows for the ionization of larger molecules, the difficulty of focusing the beam makes it challenging to image samples as small as, for example, a fruit fly. However, it is possible to image samples about the size of a mouse. In contrast, MALDI-TOF can detect a wide range of masses while maintaining an image resolution of about 10 microns, but with this approach there is a tendency for matrix-dependent ionization, making it crucial to select the appropriate matrix for the type of molecule that should be ionized. Collectively, we anticipate that combining MSI analysis of small samples or localized regions and low molecular ranges with gene knockdown studies will be a promising approach to uncover pathological pathways associated with different disease information, which may not be obtainable by other analytical approaches.
4. Conclusions
In the study of ALS, one essential aspect is the understanding of the pathological processes. In this work, we showed that ToF-SIMS measurement and PCA result can reveal significant details regarding the content and spatial information of specific lipid groups in ALS flies. Alteration of the lipid profiles was observed by day 5 in the fly model, notably before locomotive defects were seen on day 10. Specifically, the MSI approach using ToF-SIMS helped us identify differences in the lipid levels throughout the brain and its nearby regions between control and ALS flies. The visual distributions of ToF-SIMS imaging revealed a close relationship between the increase in fat body lipid droplets and the accumulation of FAs, TAGs, and Cer in the fat body regions of ALS fly heads.With the support of ToF-SIMS and PCA results, our findings point out that the pathways of fatty acid binding proteins and Acyl-CoA-binding proteins strongly affect changes in the lipid metabolism of ALS flies. With ToF-SIMS imaging, decreases in the high levels of FAs, TAGs, and Cer were observed in lipid droplets localized in the fat body regions. In contrast, in the bmm Ri model, we found that the suppression of the lipolysis process of storage TAGs had a relatively small influence on reducing the toxicity of metabolic disorders in our ALS-linked Drosophila models. Therefore, the current approach combining ToF-SIMS analysis with genetic models can be a useful tool for further studies on the causal relationship between the identified lipids upregulated during disease progression, leading to a better understanding of the pathogenic mechanisms of ALS/FTD and thus helping to identify potential targets for therapeutic intervention, particularly at the early stages.
Author contributions
Conceptualization: J. G. Son, T. G. Lee; formal analysis: M. U. T. Le, J. H. Park; project administration: S. B. Lee; funding acquisition: T. G. Lee; methodology: H. K. Shon, Alexander Pirkl; verification: C. G. Chung, S. Joh, J. H. Cho; writing – original draft: M. U. T. Le, J. H. Park; writing – review & editing: J. G. Son, S. B. Lee. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Development of Measurement Standards and Technology for Biomaterials and Medical Convergence funded by the Korea Research Institute of Standards and Science (No. KRISS-2023-GP2023-0007), the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korean government (MSIT) (No. 2021RC1C1011598), and the Nano Material Technology Development Program (No. 2016M3A7B6908929 and 2018M3D1A1058814) of the National Research Foundation (NRF) funded by the Ministry of Science and ICT, Republic of Korea.References
- J. P. Taylor, R. H. Brown Jr. and D. W. Cleveland, Decoding ALS: from genes to mechanism, Nature, 2016, 539(7628), 197–206, DOI:10.1038/nature20413.
- E. Majounie, A. E. Renton, K. Mok, E. G. Dopper, A. Waite, S. Rollinson, A. Chio, G. Restagno, N. Nicolaou and J. Simon-Sanchez, et al., Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study, Lancet Neurol., 2012, 11(4), 323–330, DOI:10.1016/S1474-4422(12)70043-1.
- A. E. Renton, E. Majounie, A. Waite, J. Simon-Sanchez, S. Rollinson, J. R. Gibbs, J. C. Schymick, H. Laaksovirta, J. C. van Swieten and L. Myllykangas, et al., A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD, Neuron, 2011, 72(2), 257–268, DOI:10.1016/j.neuron.2011.09.010.
- M. DeJesus-Hernandez, I. R. Mackenzie, B. F. Boeve, A. L. Boxer, M. Baker, N. J. Rutherford, A. M. Nicholson, N. A. Finch, H. Flynn and J. Adamson, et al., Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS, Neuron, 2011, 72(2), 245–256, DOI:10.1016/j.neuron.2011.09.011.
- M. R. Jain, W. W. Ge, S. Elkabes and H. Li, Amyotrophic lateral sclerosis: Protein chaperone dysfunction revealed by proteomic studies of animal models, Proteomics: Clin. Appl., 2008, 2(5), 670–684, DOI:10.1002/prca.200780023.
- R. R. Nair, S. Corrochano, S. Gasco, C. Tibbit, D. Thompson, C. Maduro, Z. Ali, P. Fratta, A. A. Arozena and T. J. Cunningham, et al., Uses for humanised mouse models in precision medicine for neurodegenerative disease, Mamm. Genome, 2019, 30(7–8), 173–191, DOI:10.1007/s00335-019-09807-2.
- R. Mejzini, L. L. Flynn, I. L. Pitout, S. Fletcher, S. D. Wilton and P. A. Akkari, ALS Genetics, Mechanisms, and Therapeutics: Where Are We Now?, Front. Neurosci., 2019, 13, 1310, DOI:10.3389/fnins.2019.01310.
- K. S. Cho, S. M. Bang and A. Toh, Lipids and Lipid Signaling in Drosophila Models of Neurodegenerative Diseases, in Omega-3 Fatty Acids in Brain and Neurological Health, 2014, pp. 327–336 Search PubMed.
- M. Jafurulla, G. A. Kumar, B. D. Rao and A. Chattopadhyay, A Critical Analysis of Molecular Mechanisms Underlying Membrane Cholesterol Sensitivity of GPCRs, Adv. Exp. Med. Biol., 2019, 1115, 21–52, DOI:10.1007/978-3-030-04278-3_2.
- A. B. Chaves-Filho, I. F. D. Pinto, L. S. Dantas, A. M. Xavier, A. Inague, R. L. Faria, M. H. G. Medeiros, I. Glezer, M. Y. Yoshinaga and S. Miyamoto, Alterations in lipid metabolism of spinal cord linked to amyotrophic lateral sclerosis, Sci. Rep., 2019, 9(1), 11642, DOI:10.1038/s41598-019-48059-7.
- H. Blasco, C. Veyrat-Durebex, C. Bocca, F. Patin, P. Vourc’h, J. K. Nzoughet, G. Lenaers, C. R. Andres, G. Simard and P. Corcia, et al., Lipidomics Reveals Cerebrospinal-Fluid Signatures of ALS, Sci. Rep., 2017, 7(1), 17652, DOI:10.1038/s41598-017-17389-9.
- F. Schmitt, G. Hussain, L. Dupuis, J. P. Loeffler and A. Henriques, A plural role for lipids in motor neuron diseases: energy, signaling and structure, Front. Cell. Neurosci., 2014, 8, 25, DOI:10.3389/fncel.2014.00025.
- G. Hussain, H. Anwar, A. Rasul, A. Imran, M. Qasim, S. Zafar, M. Imran, S. K. S. Kamran, N. Aziz and A. Razzaq, et al., Lipids as biomarkers of brain disorders, Crit. Rev. Food Sci. Nutr., 2020, 60(3), 351–374, DOI:10.1080/10408398.2018.1529653.
- C. Veyrat-Durebex, C. Bris, P. Codron, C. Bocca, S. Chupin, P. Corcia, P. Vourc'h, R. Hergesheimer, J. Cassereau and B. Funalot, et al., Metabo-lipidomics of Fibroblasts and Mitochondrial-Endoplasmic Reticulum Extracts from ALS Patients Shows Alterations in Purine, Pyrimidine, Energetic, and Phospholipid Metabolisms, Mol. Neurobiol., 2019, 56(8), 5780–5791, DOI:10.1007/s12035-019-1484-7.
- R. G. Cutler, W. A. Pedersen, S. Camandola, J. D. Rothstein and M. P. Mattson, Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis, Ann. Neurol., 2002, 52(4), 448–457, DOI:10.1002/ana.10312.
- H. J. Jang, M. U. T. Le, J. H. Park, C. G. Chung, J. G. Shon, G. S. Lee, J. H. Moon, S. B. Lee, J. S. Choi, T. G. Lee and S. Yoon, Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging of Phospholipid Changes in a Drosophila Model of Early Amyotrophic Lateral Sclerosis, J. Am. Soc. Mass Spectrom., 2021, 32(10), 2534–2545, DOI:10.1021/jasms.1c00167.
- N. T. N. Phan, A. S. Mohammadi, M. D. Pour and A. G. Ewing, Laser Desorption Ionization Mass Spectrometry Imaging of Drosophila Brain Using Matrix Sublimation versus Modification with Nanoparticles, Anal. Chem., 2016, 88(3), 1734–1741, DOI:10.1021/acs.analchem.5b03942.
- S. G. Ostrowski, C. T. V. Bell, N. Winograd and A. G. Ewing, Mass Spectrometric Imaging of Highly Curved Membranes During Tetrahymena Mating, Science, 2004, 305(5680), 71–73, DOI:10.1123/science.1099791.
- H. Tian, L. J. Sparvero, A. A. Amoscato, A. Bloom, H. Bayir, V. E. Kagan and N. Winograd, Gas Cluster Ion Beam Time-of-Flight Secondary Ion Mass Spectrometry High-Resolution Imaging of Cardiolipin Speciation in the Brain: Identification of Molecular Losses after Traumatic Injury, Anal. Chem., 2017, 89(8), 4611–4619, DOI:10.1021/acs.analchem.7b00164.
- I. Abbas, M. Noun, D. Touboul, D. Sahali, A. Brunelle and M. Ollero, Kidney Lipidomics by Mass Spectrometry Imaging: A Focus on the Glomerulus, Int. J. Mol. Sci., 2019, 20(7) DOI:10.3390/ijms20071623.
- L. Veith, J. Bottner, A. Vennemann, D. Breitenstein, C. Engelhard, J. Meijer, I. Estrela-Lopis, M. Wiemann and B. Hagenhoff, Detection of ZrO(2) Nanoparticles in Lung Tissue Sections by Time-of-Flight Secondary Ion Mass Spectrometry and Ion Beam Microscopy, Nanomaterials, 2018, 8(1), 44, DOI:10.3390/nano8010044.
- N. T. Phan, J. S. Fletcher and A. G. Ewing, Lipid structural effects of oral administration of methylphenidate in Drosophila brain by secondary ion mass spectrometry imaging, Anal. Chem., 2015, 87(8), 4063–4071, DOI:10.1021/acs.analchem.5b00555.
- N. T. N. Phan, M. Munem, A. G. Ewing and J. S. Fletcher, MS/MS analysis and imaging of lipids across Drosophila brain using secondary ion mass spectrometry, Anal. Bioanal. Chem., 2017, 409(16), 3923–3932, DOI:10.1007/s00216-017-0336-4.
- E. A. Bonnin and S. O. Rizzoli, Novel Secondary Ion Mass Spectrometry Methods for the Examination of Metabolic Effects at the Cellular and Subcellular Levels, Front. Behav. Neurosci., 2020, 14, 124, DOI:10.3389/fnbeh.2020.00124.
- P. Andres-Benito, E. Gelpi, M. Jove, N. Mota-Martorell, E. Obis, M. Portero-Otin, M. Povedano, A. Pujol, R. Pamplona and I. Ferrer, Lipid alterations in human frontal cortex in ALS-FTLD-TDP43 proteinopathy spectrum are partly related to peroxisome impairment, Neuropathol. Appl. Neurobiol., 2021, 47(4), 544–563, DOI:10.1111/nan.12681.
- L. Lucy, R. M. Kevin, P. Nagireddy, M. Mirjana and J. B. Hugo, The Glia-Neuron Lactate Shuttle and Elevated ROS Promote Lipid Synthesis in Neurons and Lipid Droplet Accumulation in Glia via APOE/D, Cell Metab., 2017, 26(5), 719–737, DOI:10.1016/j.cmet.2017.08.024.
- M. U. T. Le, J. G. Son, H. K. Shon, J. H. Park, S. B. Lee and T. G. Lee, Comparison between thaw-mounting and use of conductive tape for sample preparation in ToF-SIMS imaging of lipids in Drosophila microRNA-14 model, Biointerphases, 2018, 13(3), 03B414, DOI:10.1116/1.5019597.
- S. Mizielinska, S. Gronke, T. Niccoli, C. E. Ridler, E. L. Clayton, A. Devoy, T. Moens, F. E. Norona, I. O. C. Woollacott and J. Pietrzyk, et al., C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins, Science, 2014, 345(6201), 1192–1194, DOI:10.1126/science.1256800.
- R. Ferrari, D. Kapogiannis, E. D. Huey and P. Momeni, FTD and ALS: a tale of two diseases, Curr. Alzheimer Res., 2011, 8(3), 273–294, DOI:10.2174/156720511795563700.
- M. K. Andersen, T. S. Hoiem, B. S. R. Claes, B. Balluff, M. Martin-Lorenzo, E. Richardsen, S. Krossa, H. Bertilsson, R. M. A. Heeren and M. B. Rye, et al., Spatial differentiation of metabolism in prostate cancer tissue by MALDI-TOF MSI, Cancer Metab., 2021, 9(1), 9, DOI:10.1186/s40170-021-00242-z.
- V. Kis, B. Barti, M. Lippai and M. Sass, Specialized Cortex Glial Cells Accumulate Lipid Droplets in Drosophila melanogaster, PLoS One, 2015, 10(7), e0131250, DOI:10.1371/journal.pone.0131250.
- D. Yang, X. Wang, L. Zhang, Y. Fang, Q. Zheng, X. Liu, W. Yu, S. Chen, J. Ying and F. Hua, Lipid metabolism and storage in neuroglia: role in brain development and neurodegenerative diseases, Cell Biosci., 2022, 12(1), 106, DOI:10.1186/s13578-022-00828-0.
- S. Willaime, P. Vanhoutte, J. Caboche, Y. Lemaigre-Dubreuil, J. Mariani and B. Brugg, Ceramide-induced apoptosis in cortical neurons is mediated by an increase in p38 phosphorylation and not by the decrease in ERK phosphorylation, Eur. J. Neurosci., 2001, 13(11), 2037–2046, DOI:10.1046/j.0953-816x.2001.01581.x.
- F. Irie and Y. Hirabayashi, Application of exogenous ceramide to cultured rat spinal motoneurons promotes survival or death by regulation of apoptosis depending on its concentrations, J. Neurosci. Res., 1998, 54(4), 475–485, DOI:10.1002/(sici)1097-4547(19981115)54:4<475::Aid-jnr5>3.0.Co;2-p.
- G. McCluskey, C. Donaghy, K. E. Morrison, J. McConville, W. Duddy and S. Duguez, The Role of Sphingomyelin and Ceramide in Motor Neuron Diseases, J. Pers. Med., 2022, 12(9), 1418, DOI:10.3390/jpm12091418.
- Y. Dong and V. W. Yong, Oxidized phospholipids as novel mediators of neurodegeneration, Trends Neurosci., 2022, 45(6), 419–429, DOI:10.1016/j.tins.2022.03.002.
- A. Shamim, T. Mahmood, F. Ahsan, A. Kumar and P. Bagga, Lipids: An insight into the neurodegenerative disorders, Clin. Nutr. Exp., 2018, 20, 1–19, DOI:10.1016/j.yclnex.2018.05.001.
- S. Hernandez-Diaz and S. F. Soukup, The role of lipids in autophagy and its implication in neurodegeneration, Cell Stress, 2020, 4(7), 167–186, DOI:10.15698/cst2020.07.225.
Footnotes |
| † Electronic supplementary information (ESI) available: Two types of ion beam comparison spectra on fly heads, retina screening results of the denoted RNAi lipid regulators, qRT-PCR of SREBP and dob genes, PCA loading plot of gene modified model, mass deviation after external calibration, PCA loading value for ALS and gene modified model (PDF). See DOI: https://doi.org/10.1039/d3an01670f |
| ‡ These authors contributed equally. |
| § Present address: MitoImmune Therapeutics Inc., Gangnam-gu, Seoul, 06123, Republic of Korea. |
| ¶ Present address: Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA. |
| This journal is © The Royal Society of Chemistry 2024 |