DOI:
10.1039/D4AN01082E
(Critical Review)
Analyst, 2024,
149, 5739-5761
Phenoxy-1,2-dioxetane-based activatable chemiluminescent probes: tuning of photophysical properties for tracing enzymatic activities in living cells
Received
10th August 2024
, Accepted 1st November 2024
First published on 1st November 2024
Abstract
The use of chemiluminophores for tracing enzymatic activities in live-cell imaging has gained significant attention, making them valuable tools for diagnostic applications. Among various chemiluminophores, the phenoxy-1,2-dioxetane scaffold exhibits significant structural versatility and its activation is governed by the chemically initiated electron exchange luminescence (CIEEL) mechanism. This mechanism can be initiated by enzymatic activity, changes in pH, or other chemical stimuli. The photophysical properties of phenoxy-1,2-dioxetanes can be fine-tuned through the incorporation of different substituents on the phenolic ring and by anchoring them with specific triggers. This review discusses the variations in physicochemical properties, including emission maxima, quantum yield, aqueous solubility, and pKa, as influenced by structural modifications, thereby establishing a comprehensive structure–activity relationship. Furthermore, it categorises the probes based on different enzyme classes, such as hydrolase-sensitive probes, oxidoreductase-responsive probes, and transferase-activatable phenoxy-1,2-dioxetanes, offering a promising platform technology for the early diagnosis of diseases and disorders. The summary section highlights key opportunities and limitations associated with applying phenoxy-1,2-dioxetanes in achieving precise and effective enzyme assays.
 Jagpreet Singh Sidhu | Dr Jagpreet Sidhu is an Assistant Professor at Birla Institute of Technology and Science (BITS) Pilani, Pilani Campus in the Pharmacy Department. He received his PhD from IIT Ropar, India in 2020. During his doctoral program, he specialized in the development of activatable fluorogenic probes tailored for disease diagnostic applications. During his postdoctoral program at IISc Bangalore, he worked on the synthesis of optical reporters for tracing enzymatic activity in living cells. He also worked on the synthesis of deuterated synthetic fatty acids to establish the catalytic mechanism of membrane-bound enzymes. He developed interdisciplinary approaches that integrate organic synthesis, chemical biology, protein engineering, and biochemical analysis. |
 Mandeep K. Chahal | Dr Mandeep K. Chahal completed her PhD at the IIT Roorkee, India, in 2017, working on the synthetic chemistry of substituted porphyrins and their applications in sensing. Further on, she worked as a post-doctoral research fellow with Dr Jonathan P. Hill, Prof. Katsuhiko Ariga and Dr Shinsuke Ishihara at the National Institute for Materials Science (NIMS), Japan. She moved to the University of Southampton, in the UK, where she completed a Royal Society Newton International Fellowship working with Prof. Steve Goldup on mechanically interlocked molecules (MIMs). From 2023, she has been working as a Lecturer at the University of Kent; here, she is exploring diverse applications in the functionalisation of tetrapyrrole macrocycles. |
1 Introduction
Activatable sensing systems have become increasingly prevalent in medical science to trace diverse bioinformatics in the context of location, time, and environment in living subjects.1 Such systems are sensitive to external stimuli such as enzymes, biomolecules, or other cellular events and give the readout signal.2 The “always on” sensing probes display unchanging optical signals in the presence or absence of target species and suffer from high background signals. In contrast, an activatable probe changes its output signal once it comes into contact with the target of interest and thus provides a better signal-to-background ratio.3,4 Optically detected chemical probes emerge as promising candidates for sensing and diagnosis due to their advantages, including high specificity, low detection thresholds, rapid response times, and straightforward technical implementation.5,6 The last decade has witnessed significant progress in cellular imaging techniques to reveal living-cell secrets and disclose undiscovered disease information.7,8 However, high energy from excitation sources causes permanent damage to living tissues, while low signal-to-noise ratios and autofluorescence backgrounds limit the applications of fluorescence-based sensors in clinical samples.9,10 Moreover, the inadequate penetration of light into deep biological tissues primarily caused by scattering and absorption of high energy short waves is also a serious concern.11,12 Unlike fluorescence, chemiluminescence is an optical imaging technique in which a luminophore generates light without requiring excitation energy from an external source.13,14 It is a chemical reaction-based light emission phenomenon that can occur in a wide range of chemical events even inside living systems, where excited-state intermediates subsequently break down to release the energy in the form of visible light.15,16 The unique feature of chemiluminescence results in exceptional sensitivity by eliminating the need for excitation energy/source.17 As a result, it can mitigate the photobleaching, light scattering, background disturbance, and autofluorescence of biomolecules.18,19 Thus, chemiluminescent probes have been actively explored for the non-invasive imaging of cellular events in living cells. Leveraging the potential of chemiluminescence, innovative imaging, and therapeutic approaches continues to emerge, necessitating a comprehensive and current assessment of advancements within this dynamic research domain.
The majority of these chemiluminescent probes are categorized into four groups:20 1. luminol and its derivatives, 2. cypridina luciferin derivatives, 3. peroxyoxalate esters, and 4. Phenoxy-1,2-dioxetanes (Fig. 1). The luminescence of luminol and peroxyoxalate derivatives is generally triggered by oxidizing agents.21,22 In such cases, the probe undergoes oxidation, forming a highly energetic and unstable intermediate. Subsequently, the intermediate decomposes to generate luminogenic species that emit photons as it returns to its ground state.23,24 These probes are commonly used for imaging hydrogen peroxide (H2O2) because it serves both as an oxidizing agent and as a target analyte in numerous research studies.25,26 Furthermore, cypridina-based chemiluminescent probes have been reported to be activated in the presence of singlet oxygen species.27 As a result, most cypridina-based and luminol-based probes have been developed to trace cellular events linked to oxidizing species.28 However, it is crucial to note that in addition to these analytes, enzymes play an extremely significant role in nearly every cellular pathway. Enzymes are ubiquitous polypeptide biomolecules that catalyse selective and specific biochemical reactions.29,30 A diverse array of enzymes, including esterases, oxidases, reductases, proteases, and transferases are key biocatalysts and central figures in the chemistry of living organisms. Their involvement ranges from facilitating abnormal cellular growth to triggering inflammatory responses to a diseased state.31,32 Therefore, understanding the physiology of enzymes in disease pathogenesis offers invaluable insights for developing targeted therapeutic interventions and diagnostics. The diagnosis of disease using enzyme-triggering motifs provides a promising approach to designing activatable probes. Presently, a plethora of enzyme-activatable optical probes has been documented.33 In this regard, the luminescence properties of phenoxy-1,2-dioxetanes have been widely explored in the literature to facilitate the tracing of enzymatic activities.34,35 Its luminescence properties can be tailored as per the specific enzyme's catalytic nature. Unlike other chemiluminophores, phenoxy-1,2-dioxetanes can be modified so that they become directly activated by the biochemical reactions of the enzymes without any assistance from oxidizing species.36–38 Interestingly, their emission has been controlled by shielding of the phenolate with a protecting group, which can be only activated in an enzymatic event, therefore spontaneously initiating chemiluminescence.39 For several decades, triggerable dioxetanes of this kind have been employed in commercially available in vitro assays. Recently, there has been a huge surge of research interest in the utilisation of chemiluminescent phenoxy-1,2-dioxetanes for in vivo imaging, mainly due to their biocompatibility and potential to emit sufficiently bright light essential for live animal imaging.40–43
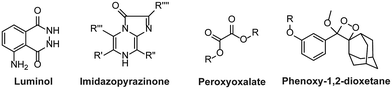 |
| | Fig. 1 Chemical scaffolds of chemiluminescence probes. | |
Over the last decade, the structural properties of phenoxy-1,2-dioxetanes have been extensively explored through derivatisation with various functional groups. These structural changes result in fast chemo-excitation, pKa adjustment, improved quantum yields, and emission enhancement in physiological buffers.44,45 As such, this review summarises the recent advancements in phenoxy-1,2-dioxetane derivatives tailored for monitoring enzymatic activities. It also explores the impact of different functional groups on their photophysical properties including emission maxima, quantum yields, and further outlines the designing principles of corresponding systems. Additionally, the review discusses potential challenges and outlines future directions for developing chemiluminescent technologies.
2 Photophysical and structural properties of phenoxy-1,2-dioxetanes
Over the years, phenoxy-1,2-dioxetanes have emerged as powerful light-emitting probes for bio-imaging applications, especially for activatable sensing.46 The key advancement in the chemistry of these compounds occurred approximately three decades ago, marked by the Schaap group's discovery of triggerable dioxetanes.36,37 Initial investigations revealed the involvement of phenoxy-1,2-dioxetane as an inherently unstable intermediate in numerous chemiluminescent systems.47 Over time, certain phenoxy-1,2-dioxetanes were developed that can be selectively activated by a specific analyte of interest, which results in chemiluminescence.41,48,49 The activation of the 1,2-dioxetane probe occurs via the chemically initiated electron exchange luminescence (CIEEL) mechanism, where chemiexcitation is initiated through phenolate-dioxetane biradical intermediates (Fig. 2). These biradical intermediates undergo further decomposition through the intramolecular back electron transfer (BET) mechanism and release energy to excite a benzoate ester. Ultimately, it returns to the ground state and emits light of a specific wavelength.18,50–54 Unlike conventional chemiluminophores, the chemiexcitation processes of phenoxy-1,2-dioxetanes rely not only on oxidative species, but can also have chemiluminescence triggered by enzymes.19,55
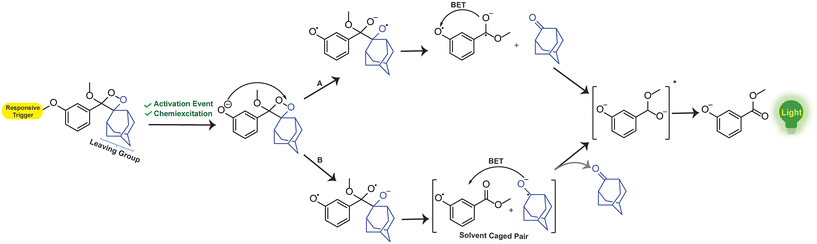 |
| | Fig. 2 Chemiexcitation activation mechanism of phenoxy-1,2-dioxetane-based probes through CIEEL. | |
However, to activate first-generation phenoxy-1,2-dioxetanes for chemiluminescence, a higher pH value (approximately 10) was required, which hampered their applications for live animal imaging.56 Hence, to decrease the pKa of phenol-dioxetane, Lippert and coworkers synthesised both halogenated and unsubstituted derivatives of phenoxy-1,2-dioxetane (CL-1a, CL-1b, and CL-1c).41 This work revealed that the acidity of phenol is increased due to the presence of ortho halogens and resulted in the highest chemiluminescence at pH 7.4, therefore confirming that lowering the pKa of phenol is accountable for enhancing chemiluminescence at physiological pH (Fig. 3). Furthermore, to improve the chemiluminescence in an aqueous medium at neutral pH, Higuchi and coworkers introduced an acetamide group ortho to phenol (CL-2).57CL-2 is expected to have a lower pKa value due to intramolecular NH⋯O hydrogen bonding interactions in both neutral and anionic states. Recently, Pu and coworkers reported benzoxazole-substituted phenoxy-1,2-dioxetane probes (CL-3a and CL-3b), to study the role of intramolecular hydrogen bonding in their chemiluminescence response.58 They have found that the presence of the benzoxazole moiety resulted in both increased chemiluminescence half-lives (up to ∼33-fold) and 8.2-fold greater brightness in comparison to the classical methylacrylate-phenoxyl-1,2-dioxetane derivative (CL-4) in aqueous solution. Although halogens and H-bonding groups containing probes were activatable at physiological pH in aqueous solution, their chemiluminescence quantum yields were not improved significantly due to the quenching effect of water molecules. Shabat and coworkers have adopted multiple strategies to improve aqueous emission and quantum yield. They hypothesised that enhancement in quantum yield of phenoxy-1,2-dioxetnae could be achieved by incorporating an electron acceptor, namely an acrylic ester/acrylonitrile group ortho to the phenolate donor.59 To investigate the role of electron-withdrawing groups (EWGs), they synthesised acryl-substituted phenoxy-benzoate derivatives, CL-5 and CL-6.59 The presence of EWGs significantly improved their emission in water, with ΦCL (chemiluminescence quantum yield) values reaching up to 40% (Table 1). These findings suggested that the introduction of an acryl group into the phenoxy-1,2-dioxetane could elevate its ΦCL under physiological conditions. Furthermore, replacing acryl ester and acrylonitrile with acrylic acid (CL-5d) substantially amplified the chemiexcitation rate in aqueous solution. Additionally, the presence of chlorine at the ortho position reduced the pKa of the phenolic OH, resulting in an enhanced extinction coefficient. The increase in extinction coefficient was attributed to the higher concentration of phenoxide ions in an aqueous medium induced by the chlorine substituent. However, this alteration did not affect the emission wavelength and had a minor impact on the quantum yield. The improvement in the kinetic profile of luminophores is generally accompanied by a decrease in t1/2 of luminophores (Table 1). In summary, the luminophores substituted with an acryl group (with or without chlorine) demonstrate notably intense chemiluminescent emission upon deprotonation at pH 7.4, with their ΦCL reaching up to 3000 times the value seen for the unmodified structures.
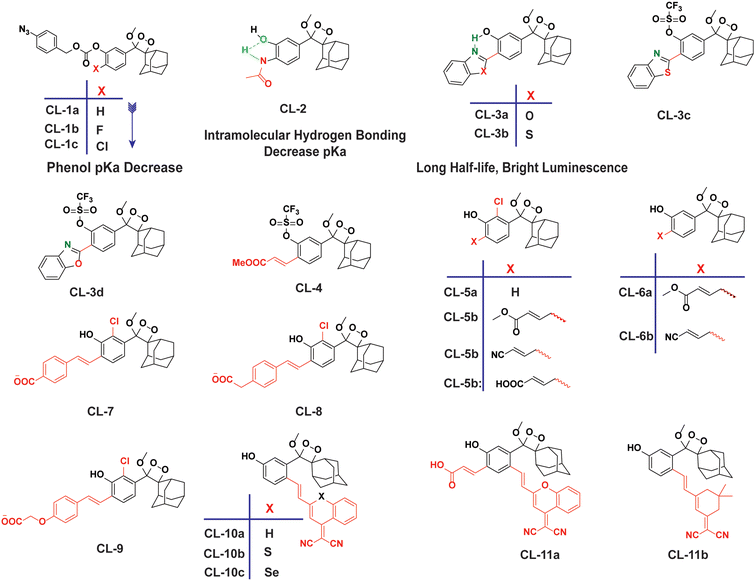 |
| | Fig. 3 Chemical structural frameworks of phenoxy-1,2-dioxetane derivatives (CL-1 to CL-11). | |
Table 1 Photo-physical properties of CL-2 to CL-11
| Probe |
λ
max CL (nm) |
T
1/2
|
Φ
CL (×10−2 Einstein per mol) |
Test condition |
Ref. |
|
CL-2
|
450 |
— |
2.7 × 10−4 |
pH 11.0 |
57
|
|
CL-3b
|
515 |
23.2 h |
— |
PBS (10% DMSO) |
58
|
|
CL-3c
|
515 |
121 min |
0.189 |
PBS (pH 7.4, 10% DMSO) |
58
|
|
CL-3d
|
515 |
129 min |
0.137 |
PBS (pH 7.4, 10% DMSO) |
58
|
|
CL-4
|
515 |
3.6 min |
0.023 |
PBS (pH 7.4, 10% DMSO) |
58
|
|
CL-5a
|
470 |
17 min |
0.003 |
PBS (pH 7.4, 5% DMSO) |
11
|
|
CL-5b
|
540 |
∼7 min |
2.5 |
PBS (pH 7.4, 5% DMSO) |
11
|
|
CL-5c
|
525 |
10 min |
9.8 |
PBS (pH 7.4, 5% DMSO) |
|
|
CL-5d
|
510 |
85 s |
0.098 |
In PBS (10% DMSO) |
|
|
CL-6a
|
540 |
23 min |
2.3 |
PBS (pH 7.4, 5% DMSO) |
|
|
CL-6b
|
525 |
22 min |
7.4 |
PBS (pH 7.4, 5% DMSO) |
|
|
CL-7
|
535 |
8.4 s |
4.2 |
PBS (pH 7.4, 10% DMSO) |
60
|
|
CL-8
|
500 |
5.1 s |
1.4 |
PBS (pH 7.4, 10% DMSO) |
60
|
|
CL-9
|
490 |
3.2 s |
0.6 |
PBS (pH 7.4, 10% DMSO) |
60
|
|
CL-10a
|
660 |
178 min |
0.82 |
PBS (pH 7.4, 10% FBS) |
61
|
|
CL-10b
|
760 |
— |
0.23 |
PBS (pH 7.4, 1% DMSO) |
62
|
|
CL-10c
|
780 |
— |
0.12 |
PBS (pH 7.4, 1% DMSO) |
62
|
|
CL-11a
|
690 |
53 min |
1.125 |
PBS (pH 7.4, 10% FBS) |
61
|
|
CL-11b
|
650 |
14 min |
2.90 |
PBS (10% DMSO) |
63
|
Furthermore, in specific chemiluminescence bioassays, the absolute quantum yield of a luminophore is not the primary factor determining the efficacy of a chemiluminescent probe. Instead, what matters significantly is the rate at which chemiexcitation occurs within the free phenolate-dioxetane. Luminophores exhibiting rapid chemiexcitation kinetics are highly sought after because they have the potential to enhance the sensitivity of chemiluminescent analytical bioassays. A faster release of photons leads to a higher signal-to-noise ratio within a shorter timeframe, ultimately resulting in improved sensitivity. To improve the chemiexcitation rate, Shabat and coworkers stabilised the phenoxy radical with styryl substituents (CL-7, CL-8, and CL-9).60 The chemiexcitation of the styryl derivatives happened approximately two orders of magnitude higher than the chemiexcitation of CL-5 and CL-6, as well as the unsubstituted Schaap's dioxetane (CL-5a). Furthermore, the change in the pH did not affect the half-lives and thus proved that chemiexcitation is an intrinsic factor without any impact on pKa. With these structural modifications, the physicochemical properties of phenoxy-1,2-dioxetane derivatives were improved to visualise enzymatic activities in living cells. So, by considering all these parameters, the probes have been tailored for the sensing of enzymes, as discussed in the next section.
To employ Schaap's dioxetanes for live-cell imaging, it is crucial to enhance aqueous solubility and adjust their light emission towards the near-infrared (NIR) region. NIR wavelengths are highly advantageous for live-cell imaging as they can penetrate deeply into tissues and encounter less scattering than light with shorter wavelengths.64,65 In pursuit of live-cell imaging, Shabat and coworkers conjugated dicyanomethylene-4H-chromene (DCMC) at the para position of the phenol donor in a phenoxy-dioxetane probe (CL-10a) that resulted in NIR emission at 660 nm with comparative quantum yield.61 A particularly remarkable luminophore possessing a DCMC acceptor along with an acrylic acid substituent at the ortho position of the phenol (CL-11a) exhibited a faster kinetics (t1/2 53 minutes) profile than CL-10a (t1/2 178 minutes). Faster kinetics is attributed to a lower pKa value for phenol, which ultimately affects the generation of phenolate species. Additionally, substituting an oxygen atom with sulfur in the dicyanomethylene-4H-chromene ring (CL-10b) leads to improvement both in NIR emission (760 nm) and also chemiluminescence quantum yield (Table 1). Interestingly, the substitution of sulfur with selenium (CL-10c) further increased the emission wavelength (780 nm); however, it compromised the ΦCL value (0.12 × 10−2 Einstein per mol). The increment in emission energy was explained based on a decrease in the energy gap between the ground state and the excited state of the molecule. Among the three heteroatoms, the inclusion of sulfur resulted in the brightest chemiluminescence and the highest signal-to-background ratio (SBR), of approximately 415.1, being exhibited. Further, the conjugation of 2-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile to phenoxy-1,2-dioxetane (CL-11b) compromised the emission wavelength (650 nm) but improved the ΦCL (4.6 × 10−2 Einstein per mol) beyond that of CL-11a.63 Thereby, it is worth playing around with extended π-conjugation in the molecular structure of phenoxy-1,2-dioxetanes for tuning the spectroscopic parameters. The chemiluminescence spectroscopic data of phenoxy-1,2-dioxetanes are summarised in Table 1. The molecular structures and chemiluminescence parameters of phenoxy-1,2-dioxetanes with different ortho substituents are summarised in Table 2. Based on the photophysical properties and the chemical nature of the chemiluminophores, the structure–activity relationship has been illustrated in Fig. 4.
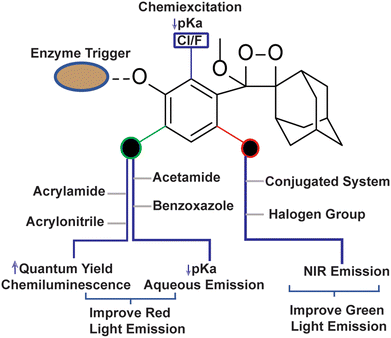 |
| | Fig. 4 Structure-activity relationship of phenoxy-1,2-dioxetane to tune physico-chemical Parameters. | |
Table 2 Spectroscopic parameters of phenoxy-1,2-dioxetane derivatives in PBS (100 mM), pH 7.4, 10% DMSO43
| Phenoxy-1,2-dioxetane |
λ
max CL [nm] |
t
1/2
|
Relative brightness |
Φ
CL % |
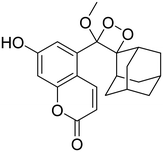
|
460 nm |
16 h |
1 |
55 |
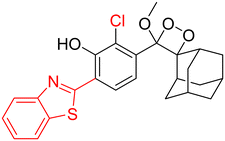
|
490 nm |
10.3 h |
0.70 |
20 |
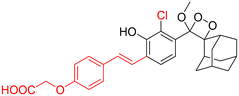
|
495 nm |
3.4 s |
480 |
0.58 |
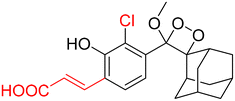
|
510 nm |
1.6 min |
245 |
9.8 |
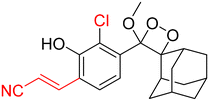
|
520 nm |
20 min |
41 |
20 |
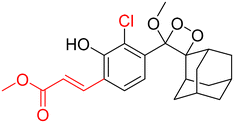
|
550 nm |
10.5 min |
16 |
4.8 |
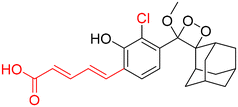
|
540 nm |
15.6 s |
206 |
1.9 |
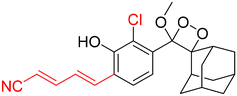
|
590 nm |
2 min |
39 |
1.1 |
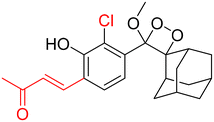
|
600 nm |
13.5 min |
0.85 |
0.42 |
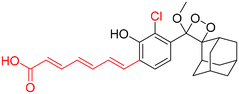
|
600 nm |
3.3 s |
65 |
0.14 |
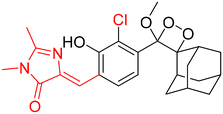
|
610 nm |
11.9 min |
0.25 |
0.073 |
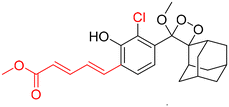
|
610 nm |
31.2 s |
8.3 |
0.5 |
|
660 nm |
5 min |
0.24 |
0.13 |
|
670 nm |
18 s |
1.2 |
0.012 |
|
710 nm |
17 min |
0.61 |
0.38 |
3 Enzyme-triggered phenoxy-1,2-dioxetane
3.1 Hydrolase-selective chemiluminescent probe
Fibroblast activation protein-alpha (FAPα).
FAPα (EC3.4.21.B28) is a transmembrane type II serine proteolytic enzyme that participates in the hydrolysis of peptide bonds and is involved in multiple pathogeneses, including cancer.66,67 It possesses the capability to break down various components of the extracellular matrix (ECM) and has dipeptidyl peptidase (DPP) activity. Degradation of the ECM is a key component of the process influencing the growth, invasion, and metastasis of tumors. It is also implicated in the excessive growth observed at the boundaries of keloid wounds and contributes to the invasive nature of keloids.68,69 Therefore, tracing of FAPα activity holds significant importance in disease diagnosis. Recently, Cao and coworkers developed the first phenoxy-1,2-dioxetane-based chemiluminescent probes (CL-12a, CL-12b, and CL-12c) bearing acrylic ester and chlorine groups ortho to the phenol for the detection of FAPα.70 The phenolic OH was masked with glycine–proline dipeptide to restrain the chemiexcitation of the luminophore. FAPα hydrolysed the peptide bond after the proline, forming a phenoxy-dioxetane (through a rearrangement involving 1,6 elimination) that rapidly decomposes via the CIEEL process to an excited benzoate ester, which decayed to the ground state with the release of green light energy. The results suggested that probes exhibited highly selective and sensitive detection of FAPα with a limit of detection of 0.785, 0.965, and 0.587 ng mL−1, using CL-12a, CL-12b, and CL-12c, respectively. Among all the probes, CL-12a showed the highest selectivity with 33-fold and 121-fold emission enhancement toward FAPα over structurally similar enzymes, i.e. PREP (prolyl oligopeptidase) and DPIV (dipeptidase IV), respectively, while CL-12b and CL-12c remained moderately to weakly selective, which might be due to the inappropriate interactions of the probe at the active pocket of the enzyme. Subsequently, the stronger chemiluminescence signal in the cytoplasm of CL-12a-treated HepG2 cells compared to LO2 cells indicated higher expression of FAPα in cancerous cells than in healthy ones (Fig. 5A). The selectivity of probes for endogenous enzymes was analysed by treating the cell lines with different concentrations of FAPα inhibitor (SP-13786). Furthermore, CL-12a was directly injected into the tumor and also injected into the tumor pre-treated with FAPα inhibitor (Fig. 5B). The higher chemiluminescence reported in the tumor site also proved the selectivity for endogenous FAPα and its applications for visualization of cancer cells.
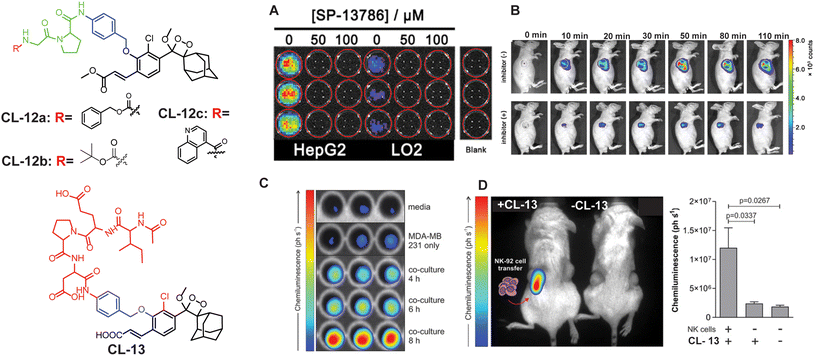 |
| | Fig. 5 Molecular structure of CL-12 and CL-13. (A) Chemiluminescence images of HepG2 and LO2 upon treatment with CL-12a. (B) Chemiluminescence response of CL-12 in animal models at different time intervals with and without inhibitor. (C) Chemiluminescence images of CL-13 incubated with media only, MDA-MB-231 cells alone, and co-cultures of MDA-MB-231 cells with NK-92 cells for the indicated times. (D) Chemiluminescence quantification in animal models. (A) and (B) are reproduced with permission.70 Copyright © 2021 American Chemical Society. (C) and (D) reprinted with permission.71 Copyright © 2021 Wiley-VCH. | |
Granzyme B.
Granzyme B is a type of serine protease that is released into cancer cells by natural killer cells to trigger apoptotic cell death.71–73 Through its proteolytic activity, granzyme B targets various intracellular proteins involved in apoptotic signalling pathways, ultimately dismantling the target cells. Recent studies provided insights into the multifaceted roles of granzyme B in immune regulation, inflammation, and tissue homeostasis.74 To assay this enzyme, a probe (CL-13) was designed and synthesised by Vendrell and coworkers.71CL-13 was meticulously crafted to target granzyme B with utmost precision and superior signal clarity amidst background noise. The probe comprises a phenolic hydroxyl group linked to an Ile-Glu-Pro-Asp peptide through a self-immolating linker. The authors found that within 10 minutes the incubation of CL-13 with granzyme B led to cleavage of the peptide linkage accompanied by 139-fold chemiluminescence enhancement at 520 nm. The varying concentrations of granzyme B showed linear enhancement in chemiluminescence with a detection limit of 0.7 nM. Furthermore, as shown in Fig. 5C, bright emission in MDA-MB cells co-cultured with NK cells as opposed to MDA-MB-231 signified higher expression of granzyme B in NK cells. Subsequently, CL-13 was administered into the tumor-contained NK cells, and live animal images were recorded immediately (Fig. 5D). Bright chemiluminescence signals were reported exclusively in the tumor area administered with NK cells via injection.
3.2 Aminopeptidases
Aminopeptidase N (APN; EC 3.4.11.2).
Aminopeptidase N (APN) is an extracellular protease enzyme responsible for the cleavage of amino acid residues from the N-terminal of a substrate.75 It is a widely distributed transmembrane ectoenzyme, participating in various physiological events, including cell migration, cell survival, angiogenesis, and viral uptake.76 It is a well-known biomarker of hematopoietic cells, and its elevated expression facilitates tumor cell invasion.77 The correlation between its expression levels and invasive capacity underscores APN's potential as a therapeutic target for cancer pathogenesis and its diagnosis.78 Recently, numerous activatable chemiluminescence strategies have been adopted for monitoring APN activities. Lian and coworkers constructed an APN-triggered phenoxy-1,2-dioxetane derivative probe, CL-14.79 The native chemiluminescence of the phenoxy-1,2-dioxetane nucleus was quenched by masking phenolic OH with an L-alanine motif through para-amino benzyl alcohol as a self-immolative linker. The probe was also decorated with an acrylonitrile group ortho to the phenol to improve quantum yield and luminosity time. Once hydrolysed by APN, the probe was rapidly activated and showed 26-fold chemiluminescence enhancement with a detection limit of 0.53 ng mL−1. Chemiluminescence signals linearly correlated to cell densities of HepG-2 in the range of 0.2 × 104 to 2.0 × 104. The primary approach to treat malignancies is through surgical procedures, which often provide the most effective chance for cure in many cases. The success of the surgery depends on the thoroughness of identifying and removing all aspects of the tumor, including its borders, microscopic elements, and any metastatic lesions. If the surgical resection is incomplete, there is a risk of the malignant tumors recurring. In this particular report, probe solution was sprayed around the tumor for in situ tumor imaging. The chemiluminescence emission intensity within the tumor tissue in animal models quickly increased, with a tumor-to-normal tissue ratio of 1.2 × 106 being recorded (Fig. 6A) followed by image-guided surgical removal of tumor tissue that ensured the complete excision of cancerous cells (Fig. 6B). Instead of acrylonitrile, Gao and coworkers reported an acrylic ester-based chemiluminescent probe (CL-15) for image-guided tumor surgery.80 It showed almost 190-fold chemiluminescence enhancement at 540 nm within 20 minutes of co-existence with APN. Moreover, its LOD (0.056 ng mL−1) showed almost ten times higher sensitivity than that of CL-14. The chemiluminescence signal was detectable even at 20 mm thickness of chicken breast tissue with an extremely low background signal (174.2 ± 30.2). Therefore, CL-15 was employed to distinguish tumor tissues from normal tissues by directly spraying its aqueous solution on HepG2 xenograft tumors ex vivo, with normal liver tissue as control. It exhibited a high tumor-to-normal tissue ratio (T/N 160), which may be a promising approach for precise tumor resection. In an orthotopic HepG2-tumor mouse model, a luminescence signal reached a maximum value within 10 minutes post-injection at the tumor site and, lasting for over 40 minutes (Fig. 6C and D), it may facilitate the removal of identified tumors under chemiluminescence guidance. Furthermore, Cao and coworkers replaced alanine with leucine to switch off the chemiexcitation and thus developed the CL-16 probe.81 It showed 53-fold emission enhancement upon incubation with APN, and the detection limit was reported to be 0.068 U mL−1. Cellular studies revealed that the probe is permeable through the cell membrane and its chemiluminescence was switched on in response to endogenous APN. The probe solution when injected into the tumor area showed a drastic increase in chemiluminescence intensity within 5 minutes of incubation and attained saturation in 20 minutes, while the tumor treated with an APN inhibitor did not show any noticeable emission. Based on the structure of these probes, we conclude that the presence of an acrylic ester group instead of an acrylonitrile group improved the detection limit with a better signal-to-noise ratio. This might be due to the more hydrophobic nature of the acrylic ester than the acrylic acid facilitating cell-membrane permeability.
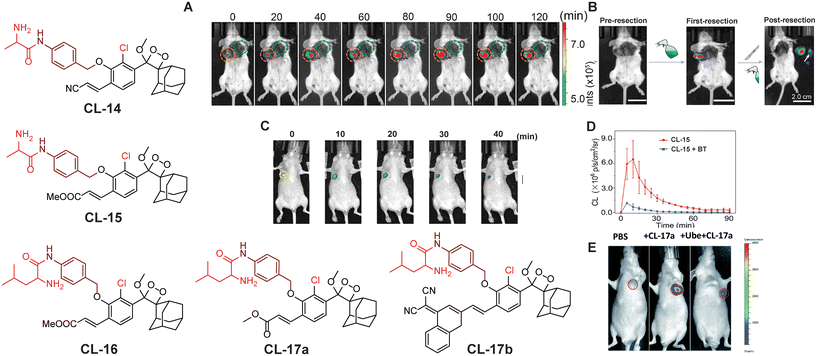 |
| | Fig. 6 Chemiluminescence probes (CL-14 to CL-17b) for aminopeptidase N and leucine aminopeptidase. (A) Chemiluminescence images of 4T1-tumor-mice after spraying of CL-14. (B) Image-guided tumor surgery using CL-14. Reprinted with permission.79 Copyright © 2022 Elsevier B.V. (C) Chemiluminescence images of a HepG2 xenograft after intra-tumoral injection of CL-15. (D) Quantification of the chemiluminescence signal of CL-15. (E) Live animal images of HepG2 tumor-containing mice. (C) and (D) reprinted with permission.80 Copyright © 2022 Wiley-VCH GmbH. (E) is reproduced from ref. 83 with permission from the Royal Society of Chemistry. Copyright © 2022 Royal Society of Chemistry. | |
Leucine amino peptidase (LAP).
LAP (EC 3.4.11.1) is a proteolytic enzyme that belongs to M1 and M17 peptidases, which catalyse the cleavage of the peptide bond between the N-termini leucine residues in proteins.82 Understanding the enzymatic activity of LAP is crucial for elucidating its role in physiological processes and exploring its potential as a target for therapeutic intervention in diseases where dysregulation of peptide metabolism is implicated. Cheng and coworkers explored chemiluminescence probes (CL-17a and CL-17b) that incorporated L-leucine into phenoxy-1,2-dioxetane to target LAP.83In vivo imaging is preferably achieved using NIR chemiluminescence emission because of its superior depth penetration. CL-17b behaves as an NIR chemiluminescent probe because the π-electron system of the phenolic luminophore has been extended by inserting a dicyano methyl chromone group ortho to the phenol. The luminescence of CL-17a (a conventional probe) attained emission saturation at 550 nm within 20 minutes of LAP catalytic action. However, the NIR-emitting CL-17b remained inactive under similar conditions. Computational studies demonstrated that CL-17b faced steric hindrance when reaching the enzyme's active site due to its larger size, while CL-17a interacts through hydrogen bonding and thus is catalysed by LAP. It showed a signal-to-noise value of approximately 1260 with a detection limit of 0.008 U mL−1. The greater chemiluminescence signal of HePG-2 cells compared to LO2 cells upon treatment with CL-17a justified the application for the differentiation of cancer cells from normal ones. Saturation of the chemiluminescence signal 10 minutes after injection into the tumor has also been reported by these authors. The selectivity of the probe was predicted by inhibiting LAP activity with Ube, followed by CL-17a treatment (Fig. 5E). Furthermore, cancerous liver tissue showed higher chemiluminescence intensity than normal tissues establishing the application potential of CL-17a for detection of LAP in clinical samples.
Prostate-specific antigen (PSA, EC 3.4. 21.77).
Prostate-specific antigen is a serine protease enzyme that hydrolyses the amide linkage of peptide substrates after glutamine. It is a key biomarker in the realm of prostate cancer diagnosis, prognosis, and therapeutic monitoring.84,85 Initially, it was identified for its role in liquefying seminal fluid but later garnered widespread attention due to its remarkable specificity to the prostate gland. Over the years, PSA has transitioned from a mere physiological marker to a crucial tool in the early detection of prostate cancer and semen analysis in criminal cases.86 To target PSA, Portnoy and coworkers employed the Mu-HSSKLQ polypeptide, as a PSA trigger.87 The chemiluminescence of ortho acrylic acid-substituted phenoxy-1,2-dioxetane is quenched with this peptide (CL-18). The cleavage of peptide bonds in the presence of PSA has led to a 157-fold change in emission intensity and 63-fold higher signal-to-noise ratio has been achieved as compared to a commercially available fluorescent probe. It distinctly underscores the superior PSA-detection capability of CL-18 over fluorescent probes. Real-time semen samples showed 14-fold chemiluminescence compared to the control. Furthermore, the probe maintained its effectiveness even after three days (Fig. 7A and B) following sample preparation of human semen residues on fabric.
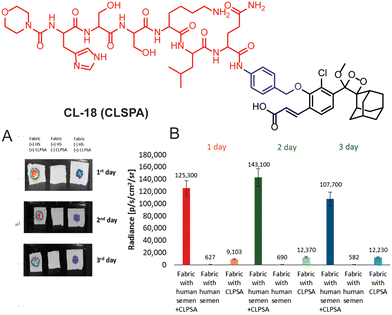 |
| | Fig. 7 (A) Chemiluminescence images of human semen incubated with CL-18 at different time intervals. (B) Chemiluminescence emission intensity quantification of (A). Reprinted with permission.87 Copyright © 2020 American Chemical Society. | |
Cathepsin B.
Cathepsin B is a lysosomal cysteine protease (EC 3.4.22.1) responsible for protein degradation, antigen processing, and tissue remodeling.88,89 It is a dipeptidyl carboxypeptidase that removes two amino acid residues from the C-terminus of a peptide substrate. The occluding loop of the active pocket contains two histidine residues located at positions 110 and 111. These histidine residues act on the C-terminal carboxylate of the substrate, thereby aiding in the facilitation of the enzyme's exopeptidase activity.90 Within lysosomes, cathepsins are involved in the digestion of proteins and peptides, breaking them down into amino acids that can be reused for building new proteins or generating energy. It is linked to various health issues such as neck, cervical, colon, breast, and ovarian cancers, as well as neurodegenerative diseases and inflammatory disorders. This makes it a promising target for therapeutics.91–93 To trace its activity, Shabat and coworkers chose valine-Cit dipeptide as a substrate for cathepsin B.94CL-19 is comprised of a conventional phenoxy-1,2-dioxetane scaffold bearing a cathepsin B-sensitive moiety, while CL-20 is additionally decorated with an acrylic ester to enhance its chemiluminescence efficiency. The CL-20 was further modified with 17-mer polyethylene glycol (PEG) and KRKGC peptide to improve its aqueous solubility and cell permeability (CL-21 and CL-22). PEG is well known for its hydrophilic nature and has been extensively explored for drug delivery applications.95 The peptide (KRKGC) comprises three positively charged residues, which enhance its solubility in an aqueous system and improve the cell membrane permeability.96 With such modifications, CL-21 showed better chemiluminescence response than CL-19 and CL-20, while CL-22 had even better chemiluminescence than CL-21. The superior chemiluminescence efficiency is attributed to the hydrophilic nature of these probes. Remarkably, CL-22 showed a limit of detection 76.29 U mL−1, which was 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold superior to its fluorescent analog. Due to the large expression of cathepsin B, RAW 264.7 and CT26 cells, except for 3T3 (the control group), showed bright chemiluminescence upon treatment with CL-22.97 Furthermore, to achieve NIR emission, 12-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile conjugated at the para position of phenoxyl-1,2-dioxetane (CL-23) and the dipeptide “phenylalanine-lysine” were employed as a trigger for cathepsin B.63 The conjugation of an extended π system at the para position increased the chemiluminescence quantum yield along with emission wavelength. The catalytic action of cathepsin B led to 75.5-fold chemiluminescence enhancement at 650 nm within 30 minutes of incubation (Fig. 8A). For in vivo applications, the probe solution was injected intraperitoneally into 4T1-bearing tumor mouse models. The chemiluminescence emission attained maxima within 10 minutes of injection and it was 4-fold higher than the mouse pre-treated with cathepsin B inhibitor. Laparotomy was conducted 30 minutes after probe injection to illustrate the potential for precise tumor resection guided by chemiluminescence diagnosing deeply buried tumors (Fig. 8B). The fluorescence signal remained undetectable in the intestinal region compared to the chemiluminescence signal. The higher signal-to-background ratio (S/B 7.6) was reported for image-guided surgery in cathepsin B expressive tissues.
000-fold superior to its fluorescent analog. Due to the large expression of cathepsin B, RAW 264.7 and CT26 cells, except for 3T3 (the control group), showed bright chemiluminescence upon treatment with CL-22.97 Furthermore, to achieve NIR emission, 12-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile conjugated at the para position of phenoxyl-1,2-dioxetane (CL-23) and the dipeptide “phenylalanine-lysine” were employed as a trigger for cathepsin B.63 The conjugation of an extended π system at the para position increased the chemiluminescence quantum yield along with emission wavelength. The catalytic action of cathepsin B led to 75.5-fold chemiluminescence enhancement at 650 nm within 30 minutes of incubation (Fig. 8A). For in vivo applications, the probe solution was injected intraperitoneally into 4T1-bearing tumor mouse models. The chemiluminescence emission attained maxima within 10 minutes of injection and it was 4-fold higher than the mouse pre-treated with cathepsin B inhibitor. Laparotomy was conducted 30 minutes after probe injection to illustrate the potential for precise tumor resection guided by chemiluminescence diagnosing deeply buried tumors (Fig. 8B). The fluorescence signal remained undetectable in the intestinal region compared to the chemiluminescence signal. The higher signal-to-background ratio (S/B 7.6) was reported for image-guided surgery in cathepsin B expressive tissues.
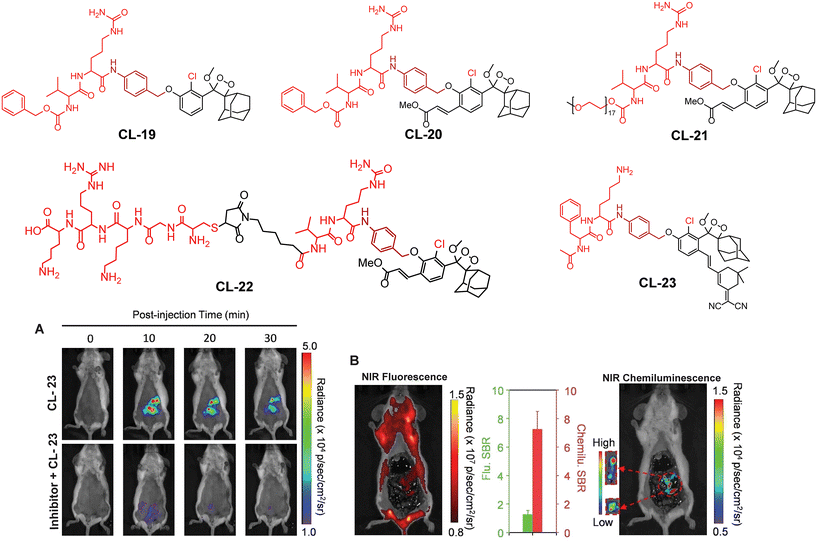 |
| | Fig. 8 Chemical structure of probes CL-19 to CL-23. (A) Images of mice acquired at different time intervals after intraperitoneal injection of CL-23. (B) Fluorescence and chemiluminescence images of the intestine after laparotomy. (A) and (B) are reprinted with permission.63 Copyright © 2022 Wiley-VCH GmbH. | |
Galactosidase.
Beta-galactosidase (β-Gal, EC.3.2.1.23) is a hydrolytic enzyme found in cell lysosomes that catalyses the breakdown of glycosidic linkages of oligosaccharides.98 It is generally synthesised as an 88 kDa precursor protein followed by its transfer to the lysosome where it is further processed to a 64 kDa active protein.99,100 The detection of β-Gal levels in the human body is critical in biomarking for diagnosing primary ovarian cancer and renal disease.101,102 Elevated β-Gal activity can lead to the cleavage of the glycosidic bond within the amino polysaccharide side chain and ultimately causes the breakdown of macromolecular proteoglycans and thus facilitates cancer metastasis.103 Consequently, the development of β-Gal activatable probes holds significant importance, not only for assessing its activity but also for disease diagnosis. Numerous initiatives have been undertaken to monitor the increased activity of β-Gal in various preclinical cancer models. Due to the distinctive specificity for β-galactosyl bonds, it is possible to tailor the chemiluminescence of phenoxy-1,2-dioxetane for β-Gal detection. A common approach involves the functionalisation of phenolic OH with β-galactose as a protective group. β-Gal cleaves the β-galactose on addition of the substrate and thus triggers chemiluminescence.
This approach has been used by various research groups to develop numerous chemiluminescence probes. Initially, the chemiluminescence signals from Schaap's dioxetanes were enhanced through a process where energy from the excited state of the chemiluminophore was transferred to a nearby fluorophore.104,105 This energy transfer led to increased light emission, serving as an indirect method to amplify chemiluminescence signals. Following a similar approach, Shabat and coworkers designed and synthesised Schaap's dioxetane-fluorophore conjugates (CL-24, CL-25, and CL-26) as NIR probes for detecting β-galactosidase (β-Gal) through covalent attachment of fluorophores.106 Surprisingly, these probes suffered from light-induced photo-decomposition except for CL-24, which is a basic Schaap's-dioxetane derivative without tethering dye. CL-25, linked to fluorescein, was particularly unstable under light exposure (t1/2 = 45 minutes), compared to the more stable CL-26 (t1/2 = 6 hours). Upon β-Gal activation, the emission spectrum of CL-26 closely matched its fluorescence spectrum, confirming energy transfer from phenoxy-dioxetane to the fluorophore. CL-26 produced a 25-fold chemiluminescence increase at 714 nm after β-Gal activation, while CL-25 exhibited a 100-fold increase at 535 nm compared to CL-24. Furthermore, CL-25 and CL-26 were administered intraperitoneally into mice, with and without ex vivo exposure to β-Gal. Strikingly, CL-26 generated a robust in vivo chemiluminescence image, whereas CL-25 remained inactive without any chemiluminescence signal (Fig. 9). These results highlight the in vivo imaging superiority of the NIR-emitting CL-26 compared to the short-wavelength emitting CL-25 probe.
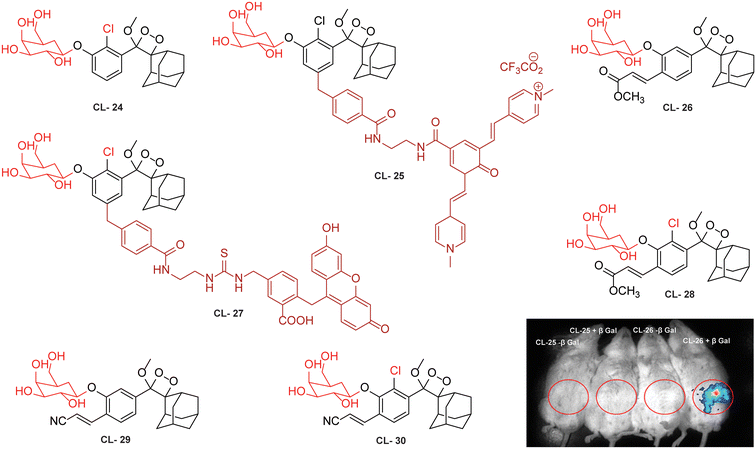 |
| | Fig. 9 Molecular structure of probes CL-24 to CL-30. Whole body images after 15 minutes of intraperitoneal injection of CL-25 and CL-26. (Probe solution incubated with β-Gal for 30 minutes prior to injection.) Reprinted with permission.106 Copyright © 2016 American Chemical Society. | |
Probes using energy transfer mechanisms are structurally composed of two components: a 1,2-dioxetane moiety and a fluorescent dye. This combination increases the overall dimensions of the probe, which in turn limits its ability to access the active pockets of the enzyme. In contrast, a probe that consists of a smaller molecular structural framework enhances the binding interactions at the enzyme's catalytic site. Additionally, direct emitting probes exhibit superior photostability, and their chemical synthesis is comparatively straightforward. By exploring the structure–activity relationship, Shabat and coworkers modified benzoate species with different electron-withdrawing groups (EWGs) to modulate the chemiluminescence efficiency and aqueous solubility of phenoxy-dioxetane probes.59 They addressed the steric hindrance from ortho substituents by incorporating a self-immolative linker between the phenolic oxygen and the β-Gal substrate (probes CL-27, CL-28, CL-29, and CL-30). In the presence of β-Gal, CL-27 showed a 500-fold stronger emission signal than CL-24 in aqueous media. Interestingly, CL-28 had similar emission amplification but with an improved kinetics profile, highlighting the role of the chlorine group in chemiexcitation. When the acrylic ester group was replaced with acrylonitrile (CL-30), 1800-fold higher chemiluminescence was observed compared to CL-24. The detection limits and kinetics parameters for these probes were not reported.
For further improvements in photophysical properties, the same group made significant advancements in developing NIR chemiluminescence turn-on probes by integrating an acceptor substituent with extended π-electron systems.61 This integration allows increased emission efficiency, resulting in more sensitive and effective probes for biological imaging and detection applications. Probe CL-31 was designed explicitly for β-Gal and exhibited a characteristic chemiluminescence kinetics profile with a 17-fold signal-to-noise ratio at 660 nm. The chemiluminescence intensity of transfected cells treated with CL-31 was approximately 14 times higher than that of wild-type cells (Fig. 10A). Furthermore, Pu and coworkers developed an NIR-emitting luminescent probe (CL-32) by conjugating dicyanomethylene-4H-benzothiopyran with a phenoxy-1,2-dioxetane moiety.62 It showed 85.6-fold chemiluminescence enhancement when treated with β-Gal, with a detection limit of 73 mU L−1. SKOV3cells exhibited a 13-fold increase in emission compared to HeLa cells within 30 minutes of treatment. In SKOV3 tumor-bearing mice, the chemiluminescence signal increased progressively after intratumoral injection, and at 20 minutes, SKOV3 tumors displayed a 6.5-fold higher signal compared to HeLa tumors (Fig. 10B). Despite these promising results, accurate delivery of such probes into cancer cells is a very challenging issue. Numerous strategies have been adopted to improve the cell specificity including anchoring a cell surface receptor-specific moiety to a cellular tracker.107,108 In this regard, Shabat and coworkers anchored the tumor cell-specific peptide “CGKRK” to probe via a maleimide-based linker.109 This approach not only improved the aqueous solubility but also directed the probe toward tumor cells.109 The effectiveness of two probes, CL-33a and CL-33b, in imaging β-Gal activity was tested in mice with CT26 tumors. Direct injection of these probes into tumor sites showed that CT26-LacZ tumors injected with CL-33b exhibited a chemiluminescence signal approximately 20 times stronger than that of CT26-WT tumors, while CL-33a remained inactive (Fig. 10C). This significant enhancement in chemiluminescence was attributed to the lower pKa of the phenoxy-1,2-dioxetane bearing a chlorine substituent, which promoted chemiexcitation in the mildly acidic tumor microenvironment.
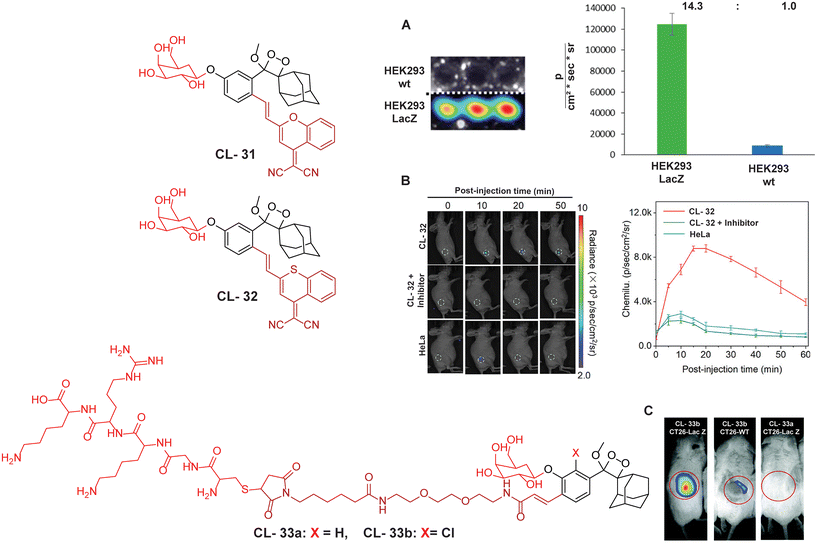 |
| | Fig. 10 Molecular structure of probes CL-31 to CL-33. (A) Imaging of cell line upon treatment with CL-31 and quantification of emission intensity. Figure reproduced with permission.61 Copyright © 2017, American Chemical Society (B) In vivo imaging of tumor-bearing mice upon treatment with CL-32. Reproduced with permission.62 Copyright© 2020 Wiley-VCH GmbH. (C) In vivo imaging of tumor-bearing mice upon treatment with CL-33b. Reproduced with permission.109 Copyright © 2018 Royal Society of Chemistry. | |
Monitoring low drug concentrations in individual patients is a challenge, impeding clinicians’ ability to personalise treatments.110–112 As a result, there is an apparent necessity for novel cancer therapies that not only minimise side effects but also enable real-time imaging of drug bio-distribution. One promising approach is to analyse drug release through chemiluminescence, which offers a superior signal-to-noise ratio in living tissues. Shabat and coworkers devised a theranostic prodrug, CL-34, to deliver monomethyl auristatin E in response to β-Gal activity.113 The activation of this prodrug by β-Gal was accompanied by direct emission of green light, with a linear correlation between light emission at 555 nm and the release of the free drug. The activation of the prodrug with chemiluminescence enhancement is shown in Fig. 11A. In HEK293-LacZ cells, treatment with the prodrug led to a 25-fold increase in chemiluminescence compared to wild-type HEK293 cells (Fig. 11B), and the IC50 was reduced by a factor of 20 in HEK293-LacZ cells. Increased chemiluminescence in HEK293-LacZ versus wild-type HEK293 revealed higher β-Gal expression. Additionally, mice bearing CT26-LacZ tumors treated with CL-34 showed approximately five times higher chemiluminescence compared to mice treated with the control prodrug (Fig. 11C).
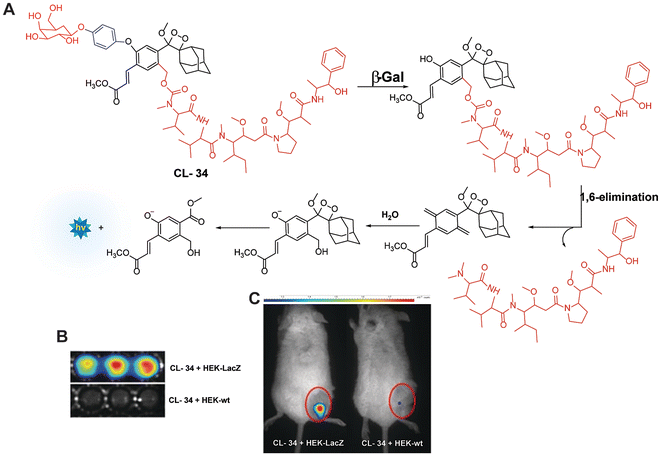 |
| | Fig. 11 (A) Molecular mechanism of drug release from CL-34 in the presence of β-Gal. (B) Chemiluminescence images of mammalian cells upon treatment with prodrug. (C) Whole-body imaging of mice upon treatment with prodrug CL-34.113 Copyright © 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | |
Esterases (EC 3.1).
Esterases are classified under the hydrolase class, capable of hydrolysis of esters into alcohols and acids. A wide range of enzymes, including acetylcholinesterase, carboxylesterase, lipase, etc. are classified under the hydrolase enzyme class.114,115 They perform critical operations in biological systems such as ester metabolism, substance transportation, and gene expression. Their absence or abnormal expression leads to the pathogenesis of Wolman disease, cancer, and hyperlipidemia.116 As a result, the development of novel and efficient esterase assays has considerable importance for the diagnosis and staging of numerous ailments. Recently, to monitor its activity, Chen and coworkers masked the phenolic OH of phenoxy-1,2-dioxetane with acetoxymethyl ether (CL-35). Hydrolysis of the ester bond in the presence of esterase was followed by release of phenoxy1,2-dioxetane through a 1,6-elimination rearrangement reaction, as shown in Fig. 12A. The probe's emission reached saturation at 540 nm within 20 minutes, with a detection limit calculated to be 1.90 × 10−3 U mL−1.117 Chemiluminescence intensity showed a linear relationship with varying esterase concentrations. Additionally, a time-dependent change in emission was observed at the injection site when the probe solution, with or without esterase, was injected subcutaneously into animal models (Fig. 12B).
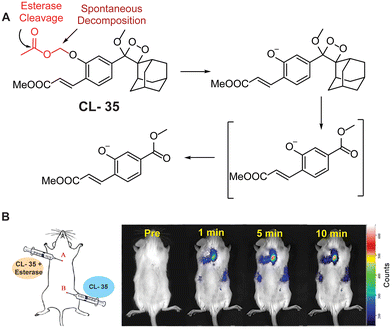 |
| | Fig. 12 (A) Activation mechanism of CL-35 in the presence of esterase. (B) Chemiluminescence monitoring in animal models. Reprinted with permission.117 Copyright © 2022 Elsevier B.V. | |
3.3 Oxidoreductase
Nitroreductase (NTR).
NTR is a flavin-containing redox enzyme that participates in the reduction of nitro-containing compounds in organisms.118 Under hypoxic conditions, it facilitates the electron transfer reduction of nitro groups to hydroxylamines, subsequently converting them into amines in the presence of nicotinamide adenine dinucleotide (NADH).119 Based on their catalytic activity, NTRs are categorised into two types: type I, which are oxygen-insensitive, and type II, which are oxygen-sensitive.120 Type II NTRs facilitate the one-electron transfer reduction of nitro groups, producing superoxide anions. Type I NTRs are further subdivided into major and minor groups based on their cofactor preferences. This class of enzymes functions as homodimers with flavin mononucleotide (FMN)-binding sites necessary for their activity. They operate through a ping-pong bi–bi redox mechanism, utilising a nicotinamide cofactor to transfer electrons and reduce nitroaromatic compounds.121,122 Due to their close association with hypoxic environments in living cells, NTRs have been extensively studied for imaging of cancer cells and drug delivery applications. In tumor microenvironments, hypoxia often leads to increased levels of NTR; however, it is scarcely expressed in normal tissues.123,124 Numerous NTR-responsive chemiluminescent probes and drug delivery systems have been developed, and extensively utilised for cellular imaging.125,126
Lippert and coworkers utilised the para-nitrobenzyl moiety as a triggering substrate (CL-36 and CL-37) for the imaging of NTR.42 The para-nitrobenzyl group underwent self-immolative cleavage when exposed to an enzyme in the presence of NADH. This process led to liberation of phenolate-1,2-dioxetane, which subsequently underwent the CIEEL mechanism and emits light. Furthermore, the chemiluminescence intensity increased with the addition of 10% Emerald II. Emerald II is an aqueous solution that contains a polymer that imparts a hydrophobic environment for the chemiluminophore and thus diminishes the quenching effect of water molecules. Probe CL-37 showed significantly improved signal-to-noise ratio (172-fold) and selectivity over biothiols compared to CL-36. The limit of detection calculated for CL-37 was 1.9 ng mL−1. In experiments with tumor xenograft SCID/BALB-C mice, hypoxic conditions were maintained in environments with 21% and 100% oxygen. Notably, the animals exposed to oxygen-deficient conditions exhibited brighter chemiluminescence than those in oxygen-rich environments (Fig. 13A). However, the authors found that use of cationic polymers caused toxicity to living tissues and thus limited their clinical applicability.127 To address this issue, Zhang and coworkers synthesised a water-soluble probe (CL-38) by introducing an electron-withdrawing carboxylate substituent ortho to the NTR trigger.128 This probe showed 6000-fold chemiluminescence enhancement upon incubation with NTR/NADH even without an additional enhancer and had a detection limit of 0.947 ng mL−1, significantly surpassing that of CL-36. Furthermore, the probe solution was injected intratumorally to visualise the degree of hypoxia in A459 tumor-bearing mouse models. The chemiluminescence of CL-38 correlated directly with the tumor's hypoxic state (Fig. 13B), revealing a mean photon flux intensity in hypoxic mice 4.5 times greater than in mice with sufficient oxygen. While probes CL-37 and CL-38 responded to tissue oxygenation in animal models, studies of their use on cell lines were not explored. To fill this gap, Lippert and coworkers reported the effectiveness of CL-39 and CL-40 chemiluminescent reporters for hypoxia across various settings: in vitro, within cellular environments, and in vivo.129CL-40 showed 37-fold chemiluminescence enhancement in 80 minutes in rat liver microsomes, while CL-39 exhibited an extraordinary 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold enhancement in just 20 minutes. The drastic increase in chemiluminescence response and ultra-sensitivity of CL-39 was attributed to acetoxymethyl ester, which improved its cellular uptake. Additionally, CL-40 was successfully utilised to discriminate between hypoxic and normoxic conditions in A549 cells, with images taken 60 minutes post-injection revealing distinct contrasts between hypoxic and well-oxygenated tissues (Fig. 13C). The photon flux intensity (1 × 107 photons per s) of CL-39 was 100 times greater than that of CL-37 and CL-NTR. These findings suggest that positioning of EWG substituents at the para position of phenolic OH is a critical component of the process to improve chemiluminescence emission and sensitivity. Moreover, integrating hydrophobic groups, such as esters, alongside EWGs could further facilitate cellular uptake. Future structural modifications may enhance the photophysical properties and cellular uptake of phenoxy-1,2-dioxetane derivatives.
000-fold enhancement in just 20 minutes. The drastic increase in chemiluminescence response and ultra-sensitivity of CL-39 was attributed to acetoxymethyl ester, which improved its cellular uptake. Additionally, CL-40 was successfully utilised to discriminate between hypoxic and normoxic conditions in A549 cells, with images taken 60 minutes post-injection revealing distinct contrasts between hypoxic and well-oxygenated tissues (Fig. 13C). The photon flux intensity (1 × 107 photons per s) of CL-39 was 100 times greater than that of CL-37 and CL-NTR. These findings suggest that positioning of EWG substituents at the para position of phenolic OH is a critical component of the process to improve chemiluminescence emission and sensitivity. Moreover, integrating hydrophobic groups, such as esters, alongside EWGs could further facilitate cellular uptake. Future structural modifications may enhance the photophysical properties and cellular uptake of phenoxy-1,2-dioxetane derivatives.
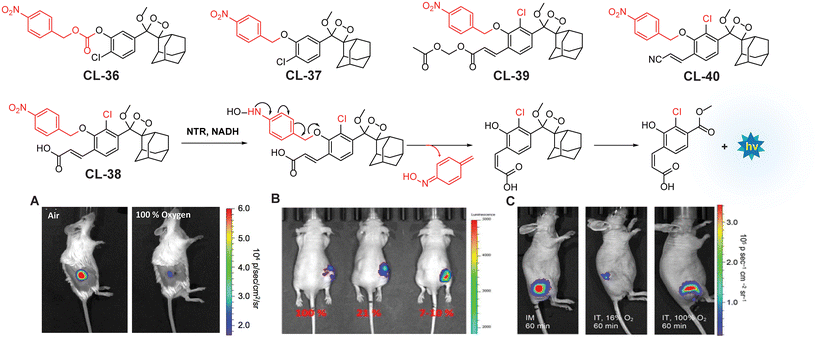 |
| | Fig. 13 Molecular structure of probes CL-36 to CL-40. (A) Images of living H1299 lung-tumor-bearing mice 1.5 min after administering intratumoral injections of CL-37 and 10% Emerald II enhancer. Figure reproduced with permission.42 Copyright © 2016 American Chemical Society. (B) Images of tumor (A549) within 10 minutes of intratumoral injection of CL-38. Reproduced with permission.128 Copyright © 2018 American Chemical Society. (C) Images of animal models bearing tumor after injection with CL-40. Figure reproduced with permission.129 Copyright © 2019 American Chemical Society. | |
NAD(P)H quinone oxidoreductase 1 (DT-diaphorase; EC 1.6.5.2).
NAD(P)H quinone oxidoreductase 1 is an intracellular flavoenzyme that regulates the redox state by facilitating the two-electron reduction reaction of quinones toward their respective hydroquinones.130 It works almost with similar efficiency in the presence of cofactors NADPH or NADH. It is a homodimer having two active pockets where the FAD cofactor is bound tightly.131 Its significant overexpression (5–200-fold) in numerous types of tumors makes it responsible for multiple carcinogenic processes. This heightened expression in tumors highlights the necessity for developing sensitive probes that can detect cancer subtypes associated with high NQO1 levels.132 To track NQO1 activity in both in vitro and in vivo models, chemiluminophores are generally conjugated with quinone-based substrates that undergo bio-reduction. Kim and coworkers investigated chemiluminescence of phenoxy-1,2-dioxetane by covalently tethering it with trimethyl-locked quinone through a self-immolative linker (CL-41).133 This probe, when bio-catalysed by NQO1, showed 130-fold chemiluminescence enhancement at 515 nm, with a detection limit of 51 ng mL−1. The quinone reduction mechanism followed by light emission is shown in Fig. 14A. Additionally, A549 cells showed 34 times more chemiluminescence than H596 cells when treated with CL-41. NQO1 activity was also visualised in A549 and H596-derived xenografts, where A549 tumor-bearing mice showed strongly bright chemiluminescence while no apparent signal was obtained in H596 ones (Fig. 14B). Shabat and coworkers extended this work by synthesising acrylonitrile (CL-42) and styryl-substituted probes (CL-43) to detect varying concentrations of NADH in the presence of NQO1. CL-42 showed more rapid chemiexcitation than CL-41, with a limit of detection of 8 nM, while CL-43 exhibited 4-fold higher sensitivity, achieving a detection limit of 2 nM. This improvement is attributed to the styryl group, which enhances the reactivity of phenolate anions. Later, the same group designed the probe CL-44 to improve the emission wavelength and aqueous solubility.43 It showed remarkable 310-fold emission enhancement at 610 nm in response to NQO1. However, mammalian cell line studies were not conducted with this probe.
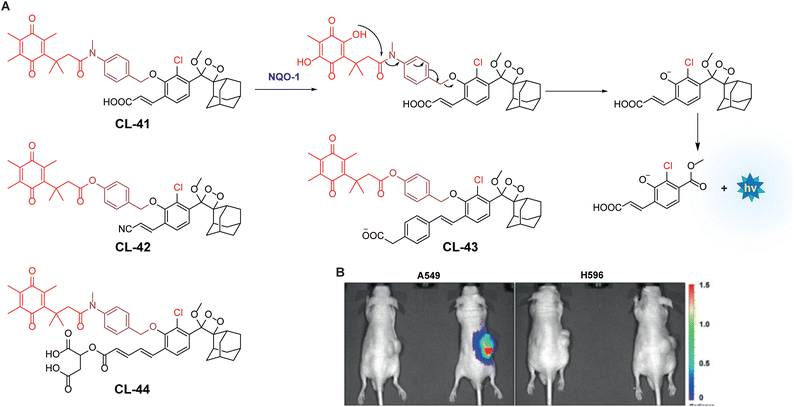 |
| | Fig. 14 (A) Activation mechanism of CL-41 in the presence of NQO1. (B) In vivo imaging of NQO1 upon treatment with CL-41. Reprinted with permission.133 Copyright © 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | |
Lysyl oxidase (LOX).
Lysyl oxidase is a protein-lysine 6-oxidase (EC 1.4.3.13), which catalyses the deamination reaction of amino acids to promote cross-linking in collagen and elastin tissues, thereby enhancing the structural integrity of the ECM proteins.134,135 The enzyme's catalytic site contains copper-binding domains, which are essential for its activity. LOX is an ECM protein secreted by tumor cells to foster tumor growth and metastasis.136,137 Consequently, LOX has garnered significant attention as a potential therapeutic target in cancer treatment, and efforts are underway to develop a synthetic chemiluminescent reporter for monitoring LOX activity. Pu and coworkers developed activatable chemiluminescent probes for targeted photodynamic cancer therapy (PDT).138 They modified the malononitrile group of dicyanomethylene-4H-benzopyran-phenoxyl-dioxetane by replacing it with benzoylacetonitrile, 1,3-dimethylbarbituric acid, and 1,3-diethyl-2-thiobarbituric acid to narrow the energy gap. Three different probes (CL-45a, CL-45b, and CL-45c) have been developed and further modified to track LOX activity. These probes exhibited chemiluminescence emissions redshifted to 700, 738, and 742 nm with half-lives of 178, 62, and 114 minutes, respectively. Based on the highest single oxygen-generating capacity of CL-45b for PDT, its chemiluminescence was masked with propylamine to target LOX (CL-46). Upon activation by LOX within 30 minutes, CL-46 showed a remarkable 19.1-fold enhancement in chemiluminescence, with a limit of detection of 0.013 U mL−1. Mice bearing 4T1 tumors were injected with a probe intratumorally followed by immediate chemiluminescence imaging for 80 minutes, achieving peak brightness in just 3 minutes (Fig. 15). The tumor-to-background ratio was reported to be 14.6, which was 3.04 times higher than that observed in the β-aminopropionitrile (LOX inhibitor) group. Subsequently, light irradiation for PDT resulted in complete inhibition of tumor growth.
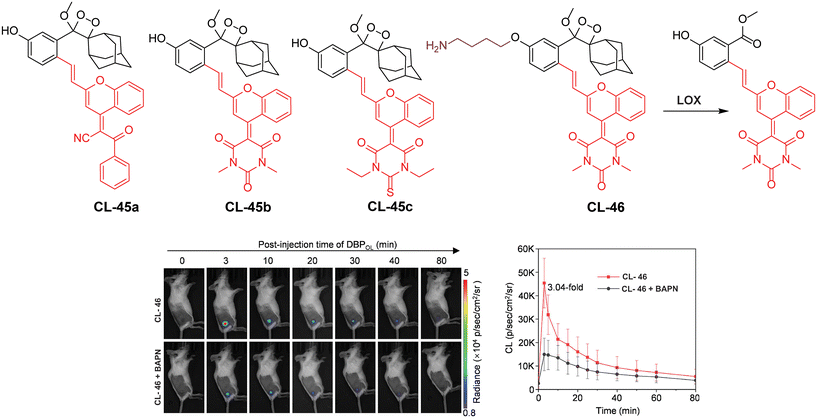 |
| | Fig. 15 Molecular structure of probes CL-45 and CL-46. Chemiluminescence images of animal models upon treatment with CL-46 and quantification of emission intensity.138 Copyright © 2023 Wiley-VCH GmbH. | |
Tyrosinase.
Tyrosinase is a copper-containing metalloenzyme (EC 1.14.18.1) involved in the synthesis of melanin pigment in the skin.139 It catalyses the ortho-hydroxylation of monophenol to diphenol, followed by formation of the corresponding quinone, and leads to an unusual accumulation of melanin in skin cells.140 Due to its ability to oxidise small phenolic molecules, the chemiluminophores are modified with triggering units containing para- or meta-phenol groups. Based on the enzyme–substrate reaction mechanism, Sessler and coworkers developed tyrosine-based chemiluminescent probes (CL-47, CL-48, and CL-49).141 In this system, tyrosinase-mediated catalysis leads to the formation of stable ortho-benzoquinone intermediates, resulting in a poor chemiluminescence response. However, when thiols are introduced, they fully reduce the intermediate through 1,6-elimination, releasing benzoate intermediates, resulting in robust chemiluminescence (Fig. 16). The addition of thiols to the reaction solution improved the chemiexcitation by 430-fold compared to the response with tyrosinase alone. Thiol conjugation to benzoquinone promoted the 1,6-elimination reaction and resulted in enhanced chemiexcitation. For CL-47, the detection limit was determined to be 0.1 U mL−1 with varying enzyme concentrations in the presence of glutathione. The inferior response of CL-48 compared to CL-47 is attributed to its sterically hindered structure. Additionally, the presence of a carboxylic group limited cell permeability, leading to further modification of CL-47 by adding a 2-dimethylaminoethyl group to create CL-49. Notably, CL-49 exhibited a 2.6-fold higher chemiluminescent response in B16 cells compared to EMT6 cells, indicating its ability to differentiate between cell lines. However, the inherent limitation of the ‘AND’ logic operation lies in the inability to monitor tyrosinase activity independently without adding thiols. This highlights significant demands for a system capable of monitoring two species through independent emission channels.
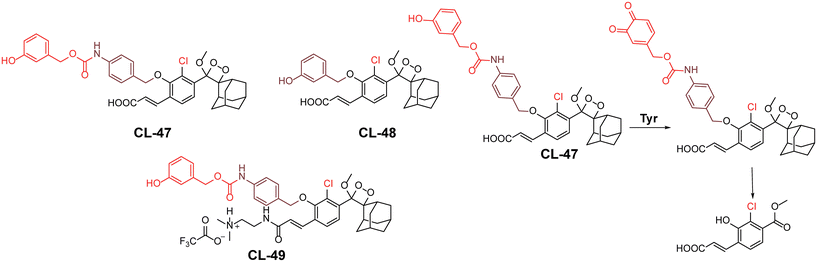 |
| | Fig. 16 Molecular structure of probes CL-47 to CL-49 and the reaction mechanism outline for tyrosinase detection. | |
3.4 Transferase
Gamma-glutamyl transpeptidase (EC 2.3.2.2) is a cell membrane-bound transferase enzyme crucial for maintaining glutathione and cysteine homeostasis by breaking down γ-glutamate in glutathione.142,143 Abnormal activity of this enzyme is associated with liver dysfunction, asthma, diabetes, and the pathogenesis of cancer.144 Therefore, researchers have been tracing its activity noninvasively in living cells and animal models to identify the diseased cells. Ye and coworkers employed modified Schaap's dioxetane substituted with an acrylic ester to track the activity of glutamyl transpeptidase (CL-50).145 Similar to previous probes, this probe incorporates a phenoxy-1,2-dioxetane structure anchored with the recognition unit of γ-glutamic acid, facilitating interaction with the enzyme's active site (Fig. 17A). It showed a significant 876-fold increase in chemiluminescence within 30 minutes of incubation with glutamyl transpeptidase. The chemiluminescence peaked in intensity within 15 minutes and remained detectable for over 45 minutes in OVCAR5 and U87MG cells. In these cancer cells, the average chemiluminescence intensity was approximately 29-fold and 23-fold higher, respectively, compared to HUVEC cells. In tumor-bearing mice, intravenous administration showed a gradual increase in chemiluminescence up until 12 minutes before returning close to background levels after 1 hour (Fig. 17B). Inhibition of glutamyl transpeptidase activity with GGsTop in U87MG tumors suppressed the chemiluminescence during imaging. Quantitative analysis revealed that the intensity of chemiluminescence in cancerous cells treated with CL-50 was 2.1 times greater than in tumors treated with GGsTop. A summary of the phenoxy-1,2-dioxetane derivatives is provided in Table 3.
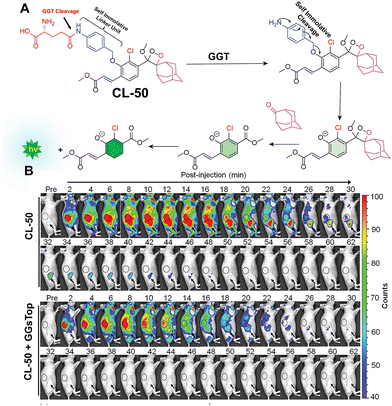 |
| | Fig. 17 (A) Activation mechanism of CL-50 upon incubation with glutamyl transpeptidase. (B) Real-time chemiluminescence images of U87MG-tumor-bearing mice after injection of CL-50. Reprinted with permission.145 Copyright © 2019 American Chemical Society. | |
Table 3 Summary of phenoxy-1,2-dioxetane derivatives discussed in this review
| Probe |
Enzyme |
LOD |
Mice/injection |
Fold enhancement |
λ
em (nm) |
Cell type |
Φ
CL (Einstein per mol) |
T
1/2
|
Ref. |
|
CL-12a
|
FAPα |
0.785 ng mL−1 |
6-Week-old male BALB/c nude mice, IT |
121 |
— |
HepG2 |
— |
— |
70
|
|
CL-12c
|
FAPα |
0.965 ng mL−1 |
— |
— |
— |
HepG2 |
— |
— |
|
CL-12c
|
FAPα |
0.587 ng mL−1 |
— |
— |
— |
HepG2 |
— |
— |
|
CL-13
|
Granzyme B |
0.7 nM |
NSG mice, IT |
139 |
520 |
NK cells |
— |
— |
71
|
|
CL-14
|
Aminopeptidase N |
0.53 ng mL−1 |
Nude BABL/c mice |
26 |
510 |
HepG2 |
— |
— |
79
|
|
CL-15
|
Aminopeptidase N |
0.056 ng mL−1 |
3–5-Week-old BALB/c nude mice |
— |
540 |
HepG2 |
— |
— |
80
|
|
CL-16
|
Aminopeptidase N |
0.068 U mL−1 |
6-Week-old BALB/c nude mice, IT |
53 |
560 |
HepG2 and LO2 |
— |
— |
81
|
|
CL-17a
|
LAP |
0.008 U mL−1 |
4–5-Week-old BALB/c nude mice, IT |
1260 |
550 |
HepG2 and LO2 |
— |
— |
83
|
|
CL-17b
|
LAP |
Inactive |
— |
— |
— |
— |
— |
— |
|
CL-18
|
PSA |
— |
— |
157 |
|
|
— |
— |
87
|
|
CL-22
|
Cathepsin B |
76.29 mU mL−1 |
— |
— |
540 |
RAW 246.7, CT26, and 3T3 |
— |
— |
97
|
|
CL-23
|
Cathepsin B |
— |
— |
75.5 |
650 |
4T1 |
— |
— |
63
|
|
CL-24
|
β-Gal |
— |
— |
— |
— |
— |
— |
— |
|
CL-25
|
β-Gal |
4.0 × 10−3 units per mL |
BALB/c female mice, IP |
100 |
535 |
HEK293-LacZ |
3.8 × 10−3 |
170 min |
106
|
|
CL-26
|
β-Gal |
— |
BALB/c female mice, IP |
25 |
714 |
HEK293-LacZ |
9.0 × 10−4 |
100 minutes |
|
CL-31
|
β-Gal |
— |
— |
17 |
660 |
HEK293-LacZ |
0.82 × 10−2 |
178 minutes |
61
|
|
CL-32
|
β-Gal |
73 mU mL−1 |
Adult female Ncr nude mice, IT |
85.6 |
760 |
SKOV3 |
0.23 × 10−2 |
62 seconds |
62
|
|
CL-33a
|
β-Gal |
— |
7-Week-old BALB/c female mice, IT |
— |
|
CT26 |
— |
— |
109
|
|
CL-33b
|
β-Gal |
— |
7-Week old BALB/c female mice, IT |
30 |
|
CT26 |
— |
— |
|
CL-34
|
β-Gal |
— |
BALB/c, IT |
25 |
555 |
HEK293-LacZ |
— |
— |
113
|
|
CL-35
|
Esterase |
1.90 × 10−3 U mL−1 |
— |
— |
540 |
HeLa |
— |
— |
117
|
|
CL-37
|
NTR |
1.9 ng mL−1 |
6-Week old BALB/c female mice, IT |
172 |
545 |
— |
— |
— |
42
|
|
CL-38
|
NTR |
0.947 ng mL−1 |
SCID/BALB-C, IT |
6000 |
— |
— |
— |
— |
128
|
|
CL-39
|
NTR |
— |
— |
60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
516 |
— |
— |
— |
129
|
|
CL-40
|
NTR |
— |
Athymic nude mice |
37 |
525 |
A549 |
— |
— |
|
CL-41
|
NQO1 |
51 ng mL−1 |
|
130 |
515 |
A549 |
— |
— |
132
|
|
CL-42
|
NQO1 |
8 nM for NADH |
— |
— |
— |
— |
— |
— |
60
|
|
CL-43
|
NQO1 |
2 nM for NADH |
— |
— |
— |
— |
— |
— |
|
CL-44
|
NQO1 |
— |
— |
310 |
610 |
— |
— |
— |
43
|
|
CL-46
|
LOX |
0.013 U mL−1 |
BALB/c mice (female, 5 weeks old), IT |
19.1 |
720 |
4T1 |
— |
— |
138
|
|
CL-47
|
Tyrosinase |
0.1 U mL−1. |
— |
430 |
540 |
B16 |
— |
— |
141
|
|
CL-50
|
γ-Glutamate |
16 mL U−1 |
Female, athymic, 6–8 weeks old, BALB/c mice |
876 |
— |
U87MG |
— |
— |
145
|
4 Conclusion and perspective
This review highlights the advancements in phenoxy-1,2-dioxetanes as chemiluminophores for diagnostic applications, focusing on a multidisciplinary approach that combines expertise in organic chemistry, biochemistry, cell biology, and photophysics. Tables 1 and 2 present various design strategies for chemiluminophores aimed at enhancing the photophysical properties of phenoxy-1,2-dioxetanes, such as emission maxima, quantum yields, and pKa values. Meanwhile, Table 3 provides an overview of different 1,2-dioxetane chemiluminophores, categorised based on their selectivity for specific enzyme classes relevant to various diseases and their use in bio-imaging applications.
Despite the advances in activatable probes for bio-imaging, research on chemiluminescence-based approaches remains in its early stages. The reported chemiluminescent probes for imaging endogenous enzymes meet several obstacles including low emission intensity, emission at shorter wavelengths, and shorter luminescence lifetimes. Therefore, extensive investigation into structural modifications of phenoxy-1,2-dioxetanes is needed to enhance chemiexcitation, emission wavelength, and luminescence half-life. Previously, various methods have been employed to achieve near-infrared (NIR) emission for deep-tissue imaging, including the conjugation of fluorescent dyes with phenoxy-1,2-dioxetane and the addition of electron-withdrawing groups (EWGs) at the ortho and para positions of the phenol group. However, these approaches increase the overall size of the probe, limiting its ability to enter the enzyme's active site and often resulting in compromised quantum yields. Therefore, more structural modifications are necessary to improve the brightness. Additionally, tuning the pKa of the phenolic OH group is critically important for effective bio-imaging at physiological pH. The kinetics parameters, including the Michaelis–Menten constant and Vmax, are crucial in determining the affinity of these probes for particular enzymes. However, this study was not conducted for the chemiluminescent probes discussed in this review. Evaluating parameters such as Kcat and the substrate's association constant with the enzyme could significantly enhance the design strategies for enzyme-specific probes.
Among the wide range of enzymes involved in cellular metabolism and disease progression, only a few have garnered attention for the development of chemiluminescent probes. The continuous advancements in chemical biology and medicinal chemistry are expected to enhance our understanding of the precise reaction sites within enzymes and biomarkers, paving the way for the development of new chemiluminescent enzyme probes. We anticipate that these probes will emerge as indispensable tools in both diagnostics and theranostics. This review provides valuable insights for aspiring researchers entering this field.
Data availability
No primary research results, software, or code have been included and no new data were generated or analysed as part of this review.
Conflicts of interest
All the authors declare no conflict of interest.
Acknowledgements
J. S. Sidhu acknowledges the BITS-Pilani, Pilani campus for providing infrastructure and research facilities. MKC would like to thank the Royal Society of Chemistry (R23-0850952021) and University of Kent for funding.
References
- M. Zhao, B. Li, H. Zhang and F. Zhang, Chem. Sci., 2021, 12, 3448–3459 RSC.
- V. Naresh and N. Lee, Sensors, 2021, 21, 1109 CrossRef CAS PubMed.
- S. He, J. Song, J. Qu and Z. Cheng, Chem. Soc. Rev., 2018, 47, 4258–4278 RSC.
- E. A. Owens, M. Henary, G. El Fakhri and H. S. Choi, Acc. Chem. Res., 2016, 49, 1731–1740 CrossRef CAS PubMed.
- J. A. Thomas, Chem. Soc. Rev., 2015, 44, 4494–4500 RSC.
- A. Fernández and M. Vendrell, Chem. Soc. Rev., 2016, 45, 1182–1196 RSC.
- S. Yoon, S. Y. Cheon, S. Park, D. Lee, Y. Lee, S. Han, M. Kim and H. Koo, Biomater. Res., 2022, 26, 57 CrossRef PubMed.
- A. Ji, H. Lou, J. Li, Y. Hao, X. Wei, Y. Wu, W. Zhao, H. Chen and Z. Cheng, Chem. Sci., 2024, 15, 3339–3348 RSC.
- Y. Shi, Y. Hu, N. Jiang and A. K. Yetisen, ACS Sens., 2022, 7, 1615–1633 CrossRef CAS PubMed.
- W. L. Rice, D. M. Shcherbakova, V. V. Verkhusha and A. T. N. Kumar, Cancer Res., 2015, 75, 1236–1243 CrossRef CAS PubMed.
- O. Green, T. Eilon, N. Hananya, S. Gutkin, C. R. Bauer and D. Shabat, ACS Cent. Sci., 2017, 3, 349–358 CrossRef CAS PubMed.
- Y. Yang, S. Wang, L. Lu, Q. Zhang, P. Yu, Y. Fan and F. Zhang, Angew. Chem., Int. Ed., 2020, 59, 18380–18385 CrossRef CAS PubMed.
- Y. Liu, J. Tan, M. Wan, L. Zhang and X. Yao, ACS Omega, 2020, 5, 15922–15930 CrossRef CAS PubMed.
- A. Lyu, Y. Wang and H. Cui, Anal. Chem., 2023, 95, 7914–7923 CrossRef CAS PubMed.
- A. Roda, M. Mirasoli, E. Michelini, M. Di Fusco, M. Zangheri, L. Cevenini, B. Roda and P. Simoni, Biosens. Bioelectron., 2016, 76, 164–179 CrossRef CAS PubMed.
- S. Emdadi, M. H. Sorouraddin and L. Denanny, Analyst, 2021, 146, 1326–1333 RSC.
- X. Lu, X. Song, Q. Wang, W. Hu, W. Shi, Y. Tang, Z. Wu, Q. Fan and W. Huang, RSC Adv., 2020, 10, 11861–11864 RSC.
- U. Haris and A. R. Lippert, ACS Sens., 2023, 8, 3–11 CrossRef CAS PubMed.
- H. Gunduz, T. Almammadov, M. Dirak, A. Acari, B. Bozkurt and S. Kolemen, RSC Chem. Biol., 2023, 4, 675–684 RSC.
- Y. Yang and F. Zhang, Analysis Sensing, 2021, 1, 75–89 CrossRef CAS.
- A. V. Romanyuk, I. D. Grozdova, A. A. Ezhov and N. S. Melik-Nubarov, Sci. Rep., 2017, 7, 3410 CrossRef PubMed.
- M. A. Tzani, D. K. Gioftsidou, M. G. Kallitsakis, N. V. Pliatsios, N. P. Kalogiouri, P. A. Angaridis, I. N. Lykakis and M. A. Terzidis, Molecules, 2021, 26, 7664 CrossRef CAS PubMed.
- L. Yue and Y.-T. Liu, J. Phys. Chem. B, 2020, 124, 7682–7693 CrossRef CAS PubMed.
- H. An, C. Guo, D. Li, R. Liu, X. Xu, J. Guo, J. Ding, J. Li, W. Chen and J. Zhang, ACS Appl. Mater. Interfaces, 2020, 12, 17230–17243 CrossRef CAS PubMed.
- R. C. Allen, Antioxidants, 2022, 11, 518 CrossRef CAS PubMed.
- H. Karatani, Anal. Sci., 2022, 38, 613–621 CrossRef CAS PubMed.
- J. Zhang, C. Wickizer, W. Ding, R. Van, L. Yang, B. Zhu, J. Yang, Y. Wang, Y. Wang, Y. Xu, C. Zhang, S. Shen, C. Wang, Y. Shao and C. Ran, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2310131120 CrossRef CAS PubMed.
- D. Calabria, M. Guardigli, M. Mirasoli, A. Punzo, E. Porru, M. Zangheri, P. Simoni, E. Pagnotta, L. Ugolini, L. Lazzeri, C. Caliceti and A. Roda, Anal. Biochem., 2020, 600, 113760 CrossRef CAS PubMed.
- C. Bourlieu, T. Astruc, S. Barbe, J.-G. Berrin, E. Bonnin, R. Boutrou, V. Hugouvieux, S. Le Feunteun and G. Paës, Biotechnol. Adv., 2020, 41, 107546 CrossRef CAS PubMed.
- J. S. Sidhu, N. Kaur and N. Singh, Biosens. Bioelectron., 2021, 191, 113441 CrossRef CAS PubMed.
- M. R. Bond and J. A. Hanover, Annu. Rev. Nutr., 2013, 33, 205–229 CrossRef CAS PubMed.
- R. Gupta, R. K. Ambasta and P. Kumar, Life Sci., 2020, 243, 117278 CrossRef CAS PubMed.
- J. Zhang, X. Chai, X.-P. He, H.-J. Kim, J. Yoon and H. Tian, Chem. Soc. Rev., 2019, 48, 683–722 RSC.
- M. David, Q. Jaber, M. Fridman and D. Shabat, Chem. – Eur. J., 2023, 29, e202300422 CrossRef CAS PubMed.
- H. N. Kagalwala, R. T. Reeves and A. R. Lippert, Curr. Opin. Chem. Biol., 2022, 68, 102134 CrossRef CAS PubMed.
- A. P. Schaap, R. S. Handley and B. P. Giri, Tetrahedron Lett., 1987, 28, 935–938 CrossRef CAS.
- A. P. Schaap, T.-S. Chen, R. S. Handley, R. DeSilva and B. P. Giri, Tetrahedron Lett., 1987, 28, 1155–1158 CrossRef CAS.
- A. P. Schaap, M. D. Sandison and R. S. Handley, Tetrahedron Lett., 1987, 28, 1159–1162 CrossRef CAS.
- S. Gutkin, S. Gandhesiri, A. Brik and D. Shabat, Bioconjugate Chem., 2021, 32, 2141–2147 CrossRef CAS PubMed.
- L. Liu and R. P. Mason, PLoS One, 2010, 5, e12024 CrossRef PubMed.
- J. Cao, R. Lopez, J. M. Thacker, J. Y. Moon, C. Jiang, S. N. S. Morris, J. H. Bauer, P. Tao, R. P. Mason and A. R. Lippert, Chem. Sci., 2015, 6, 1979–1985 RSC.
- J. Cao, J. Campbell, L. Liu, R. P. Mason and A. R. Lippert, Anal. Chem., 2016, 88, 4995–5002 CrossRef CAS PubMed.
- S. Gutkin, R. Tannous, Q. Jaber, M. Fridman and D. Shabat, Chem. Sci., 2023, 14, 6953–6962 RSC.
- S. Gutkin, R. Tannous, Q. Jaber, M. Fridman and D. Shabat, Chem. Sci., 2023, 14, 6953–6962 RSC.
- A. Fu, Y. Mao, H. Wang and Z. Cao, J. Pharm. Biomed. Anal., 2021, 204, 114266 CrossRef CAS PubMed.
-
J. C. Hummelen, T. M. Luider and H. Wynberg, in Methods Enzymol., Academic Press, 1986, vol. 133, pp. 531–557 Search PubMed.
- L. F. M. L. Ciscato, F. H. Bartoloni, A. S. Colavite, D. Weiss, R. Beckert and S. Schramm, Photochem. Photobiol. Sci., 2014, 13, 32–37 CrossRef CAS PubMed.
- M. Yang, J. Zhang, D. Shabat, J. Fan and X. Peng, ACS Sens., 2020, 5, 3158–3164 CrossRef CAS PubMed.
- K. J. Bruemmer, O. Green, T. A. Su, D. Shabat and C. J. Chang, Angew. Chem., 2018, 130, 7630–7634 CrossRef.
- M. Vacher, I. F. Galván, B.-W. Ding, S. Schramm, R. Berraud-Pache, P. Naumov, N. Ferré, Y.-J. Liu, I. Navizet, D. Roca-Sanjuán, W. J. Baader and R. Lindh, Chem. Rev., 2018, 118, 6927–6974 CrossRef CAS PubMed.
- W. Adam, I. Bronstein, A. V. Trofimov and R. F. Vasil'ev, J. Am. Chem. Soc., 1999, 121, 958–961 CrossRef CAS.
- L. F. M. L. Ciscato, F. H. Bartoloni, D. Weiss, R. Beckert and W. J. Baader, J. Org. Chem., 2010, 75, 6574–6580 CrossRef CAS PubMed.
- H. Takakura, Molecules, 2021, 26, 1618 CrossRef CAS PubMed.
- P. Farahani, M. A. Oliveira, I. F. Galván and W. J. Baader, RSC Adv., 2017, 7, 17462–17472 RSC.
- C. Dodeigne, L. Thunus and R. Lejeune, Talanta, 2000, 51, 415–439 CrossRef CAS PubMed.
- I. Bronstein, B. Edwards and J. C. Voyta, J. Biolumin. Chemilumin., 1989, 4, 99–111 CrossRef CAS PubMed.
- Y. Hisamatsu, T. Fukiage, K. Honma, A. G. Balia, N. Umezawa, N. Kato and T. Higuchi, Org. Lett., 2019, 21, 1258–1262 CrossRef CAS PubMed.
- J. Huang, P. Cheng, C. Xu, S. S. Liew, S. He, Y. Zhang and K. Pu, Angew. Chem., Int. Ed., 2022, 61, e202203235 CrossRef CAS PubMed.
- O. Green, T. Eilon, N. Hananya, S. Gutkin, C. R. Bauer and D. Shabat, ACS Cent. Sci., 2017, 3, 349–358 CrossRef CAS PubMed.
- N. Hananya, J. P. Reid, O. Green, M. S. Sigman and D. Shabat, Chem. Sci., 2019, 10, 1380–1385 RSC.
- O. Green, S. Gnaim, R. Blau, A. Eldar-Boock, R. Satchi-Fainaro and D. Shabat, J. Am. Chem. Soc., 2017, 139, 13243–13248 CrossRef CAS PubMed.
- J. Huang, Y. Jiang, J. Li, J. Huang and K. Pu, Angew. Chem., Int. Ed., 2021, 60, 3999–4003 CrossRef CAS PubMed.
- X. Wei, J. Huang, C. Zhang, C. Xu, K. Pu and Y. Zhang, Angew. Chem., Int. Ed., 2023, 62, e202213791 CrossRef CAS PubMed.
- G. Blum, G. von Degenfeld, M. J. Merchant, H. M. Blau and M. Bogyo, Nat. Chem. Biol., 2007, 3, 668–677 CrossRef CAS PubMed.
- E. A. Mason, R. Lopez and R. P. Mason, Opt. Mater. Express, 2016, 6, 1384–1392 CrossRef CAS.
- A. A. Fitzgerald and L. M. Weiner, Cancer Metastasis Rev., 2020, 39, 783–803 CrossRef CAS PubMed.
- Z. Yu, Y. Huang, H. Chen, Z. Jiang, C. Li, Y. Xie, Z. Li, X. Cheng, Y. Liu, S. Li, Y. Liang and Z. Wu, ACS Pharmacol. Transl. Sci., 2023, 6, 1745–1757 CrossRef CAS PubMed.
- L. Zhang, W. Ying, Z. Sheng, L. Lv, J. Gao, Y. Xue and L. Liu, Anal. Biochem., 2022, 655, 114859 CrossRef CAS PubMed.
- K. Dienus, A. Bayat, B. F. Gilmore and O. Seifert, Arch. Dermatol. Res., 2010, 302, 725–731 CrossRef CAS PubMed.
- A. Fu, H. Wang, T. Huo, X. Li, W. Fu, R. Huang and Z. Cao, Anal. Chem., 2021, 93, 6501–6507 CrossRef CAS PubMed.
- J. I. Scott, S. Gutkin, O. Green, E. J. Thompson, T. Kitamura, D. Shabat and M. Vendrell, Angew. Chem., Int. Ed., 2021, 60, 5699–5703 CrossRef CAS PubMed.
- I. Voskoboinik, J. C. Whisstock and J. A. Trapani, Nat. Rev. Immunol., 2015, 15, 388–400 CrossRef CAS PubMed.
- J. A. Trapani, Genome Biol., 2001, 2 DOI:10.1186/gb-2001-2-12-reviews3014.
- J. E. Davis, V. R. Sutton, M. J. Smyth and J. A. Trapani, Cell Death Differ., 2000, 7, 973–983 CrossRef CAS PubMed.
- S. A. Amin, N. Adhikari and T. Jha, J. Med. Chem., 2018, 61, 6468–6490 CrossRef PubMed.
-
A. J. Turner, Membrane alanyl aminopeptidase, in Handbook of Proteolytic Enzymes, 2004, pp. 289–294. DOI:10.1016/B978-0-12-079611-3.50077-X. Epub 2012 Dec 2.
- M. Alfalah, M. P. Krahn, G. Wetzel, S. von Hörsten, C. Wolke, N. Hooper, T. Kalinski, S. Krueger, H. Y. Naim and U. Lendeckel, J. Biol. Chem., 2006, 281, 11894–11900 CrossRef CAS PubMed.
- S.-X. Cui, X.-J. Qu, Z.-H. Gao, Y.-S. Zhang, X.-F. Zhang, C.-R. Zhao, W.-F. Xu, Q.-B. Li and J.-X. Han, Cancer Lett., 2010, 292, 153–162 CrossRef CAS PubMed.
- X. Shi, Y. Deng, X. Liu, G. Gao, R. Wang and G. Liang, Biosens. Bioelectron., 2022, 208, 114212 CrossRef CAS PubMed.
- Y. Liu, J. Zeng, Q. Li, M. Miao, Z. Song, M. Zhao, Q. Miao and M. Gao, Adv. Opt. Mater., 2022, 10, 2102709 CrossRef CAS.
- R. Sun, X. Wu, Y. Mao, H. Wang, C. Bian, P. Lv, Z. Zhao, X. Li, W. Fu, J. Lu and Z. Cao, Luminescence, 2022, 37, 1335–1342 CrossRef CAS PubMed.
- M. B. Harbut, G. Velmourougane, S. Dalal, G. Reiss, J. C. Whisstock, O. Onder, D. Brisson, S. McGowan, M. Klemba and D. C. Greenbaum, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, E526–E534 CrossRef CAS PubMed.
- B. Wang, Z. Chen, X. Cen, Y. Liang, L. Tan, E. Liang, L. Zheng, Y. Zheng, Z. Zhan and K. Cheng, Chem. Sci., 2022, 13, 2324–2330 RSC.
- M. J. Ahrens, P. A. Bertin, E. F. Vonesh, T. J. Meade, W. J. Catalona and D. Georganopoulou, Prostate, 2013, 73, 1731–1737 CrossRef CAS PubMed.
- A. P. Drabovich, P. Saraon, K. Jarvi and E. P. Diamandis, Nat. Rev. Urol., 2014, 11, 278–288 CrossRef CAS PubMed.
- P. Suttipasit and S. Wongwittayapanich, J. Forensic Leg. Med., 2018, 54, 102–108 CrossRef PubMed.
- S. Gutkin, O. Green, G. Raviv, D. Shabat and O. Portnoy, Bioconjugate Chem., 2020, 31, 2488–2493 CrossRef CAS PubMed.
- V. Turk, V. Stoka, O. Vasiljeva, M. Renko, T. Sun, B. Turk and D. Turk, Biochim. Biophys. Acta, Proteins Proteomics, 2012, 1824, 68–88 CrossRef CAS PubMed.
- M. C. Yoon, V. Hook and A. J. O'Donoghue, Biochemistry, 2022, 61, 1904–1914 CrossRef CAS PubMed.
- T. R. Lambeth, Z. Dai, Y. Zhang and R. R. Julian, RSC Chem. Biol., 2021, 2, 606–611 RSC.
- A. Amritraj, K. Peake, A. Kodam, C. Salio, A. Merighi, J. E. Vance and S. Kar, Am. J. Pathol., 2009, 175, 2540–2556 CrossRef CAS PubMed.
- H. Dong, Y. Qin, Y. Huang, D. Ji and F. Wu, Neurochem. Int., 2019, 126, 178–186 CrossRef CAS PubMed.
- K. Ditaranto, T. L. Tekirian and A. J. Yang, Neurobiol. Dis., 2001, 8, 19–31 CrossRef CAS PubMed.
- M. E. Roth-Konforti, C. R. Bauer and D. Shabat, Angew. Chem., Int. Ed., 2017, 56, 15633–15638 CrossRef CAS PubMed.
- J. M. Harris and R. B. Chess, Nat. Rev. Drug Discovery, 2003, 2, 214–221 CrossRef CAS PubMed.
- J. A. Hoffman, E. Giraudo, M. Singh, L. Zhang, M. Inoue, K. Porkka, D. Hanahan and E. Ruoslahti, Cancer Cell, 2003, 4, 383–391 CrossRef CAS PubMed.
- M. E. Roth-Konforti, C. R. Bauer and D. Shabat, Angew. Chem., Int. Ed., 2017, 56, 15633–15638 CrossRef CAS PubMed.
- S. Chen, X. Ma, L. Wang, Y. Wu, Y. Wang, W. Fan and S. Hou, Sens. Actuators, B, 2023, 379, 133272 CrossRef CAS.
- A. T. Hoogeveen, F. W. Verheijen and H. Galjaard, J. Biol. Chem., 1983, 258, 12143–12146 CrossRef CAS PubMed.
- S. Zhang, J. McCarter, Y. Okamura-Oho, F. Yaghi, A. Hinek, S. Withers and J. Callahan, Biochem. J., 1994, 304, 281–288 CrossRef CAS PubMed.
- M. Li, M. Yang and W.-H. Zhu, Mater. Chem. Front., 2021, 5, 763–774 RSC.
- D. Asanuma, M. Sakabe, M. Kamiya, K. Yamamoto, J. Hiratake, M. Ogawa, N. Kosaka, P. L. Choyke, T. Nagano, H. Kobayashi and Y. Urano, Nat. Commun., 2015, 6, 6463 CrossRef CAS PubMed.
- J. Zhang, P. Cheng and K. Pu, Bioconjugate Chem., 2019, 30, 2089–2101 CrossRef CAS PubMed.
- J. Y. Park, J. Gunpat, L. Liu, B. Edwards, A. Christie, X.-J. Xie, L. J. Kricka and R. P. Mason, Luminescence, 2014, 29, 553–558 CrossRef CAS PubMed.
- J.-C. Tseng and A. L. Kung, J. Biomed. Sci., 2015, 22, 45 CrossRef PubMed.
- N. Hananya, A. Eldar Boock, C. R. Bauer, R. Satchi-Fainaro and D. Shabat, J. Am. Chem. Soc., 2016, 138, 13438–13446 CrossRef CAS PubMed.
- J. Zalejski, J. Sun and A. Sharma, J. Imaging, 2023, 9, 192 CrossRef PubMed.
- F. Zheng, W. Xiong, S. Sun, P. Zhang and J. J. Zhu, Nanophotonics, 2019, 8, 391–413 CrossRef CAS.
- T. Eilon-Shaffer, M. Roth-Konforti, A. Eldar-Boock, R. Satchi-Fainaro and D. Shabat, Org. Biomol. Chem., 2018, 16, 1708–1712 RSC.
- L. J. van ‘t Veer and R. Bernards, Nature, 2008, 452, 564–570 CrossRef PubMed.
- R. N. Woodring, E. G. Gurysh, E. M. Bachelder and K. M. Ainslie, ACS Appl. Bio Mater., 2023, 6, 934–950 CrossRef CAS PubMed.
- M. I. Khan, M. I. Hossain, M. K. Hossain, M. H. K. Rubel, K. M. Hossain, A. M. U. B. Mahfuz and M. I. Anik, ACS Appl. Bio Mater., 2022, 5, 971–1012 CrossRef CAS PubMed.
- S. Gnaim, A. Scomparin, S. Das, R. Blau, R. Satchi-Fainaro and D. Shabat, Angew. Chem., Int. Ed., 2018, 57, 9033–9037 CrossRef CAS PubMed.
- L. Feng, Z. M. Liu, J. Hou, X. Lv, J. Ning, G. B. Ge, J. N. Cui and L. Yang, Biosens. Bioelectron., 2015, 65, 9–15 CrossRef CAS PubMed.
- S. F. Sousa, M. J. Ramos, C. Lim and P. A. Fernandes, ACS Catal., 2015, 5, 5877–5887 CrossRef CAS.
- C. Morisseau, Int. J. Mol. Sci., 2022, 23, 4870 CrossRef PubMed.
- F. Wang, Y. Wang, J. Zhang, S. Zheng, B. Xie, S. Lu, J. Zhou, C. Wang, F. Wang, M. Jiang and X. Chen, Sens. Actuators, B, 2023, 375, 132880 CrossRef CAS.
- X. Liu, S. Zeng, M. Zhang, M. Jiang, Y. S. Kafuti, P. Shangguan, Y. Yu, Q. Chen, J. Wang, X. Peng, J. Yoon and H. Li, Chem. Commun., 2022, 58, 11438–11441 RSC.
- C. Weng, H. Yang, B. S. Loh, M. W. Wong and W. H. Ang, J. Am. Chem. Soc., 2023, 145, 6453–6461 CrossRef CAS PubMed.
- E. M. Williams, R. F. Little, A. M. Mowday, M. H. Rich, J. V. E. Chan-Hyams, J. N. Copp, J. B. Smaill, A. V. Patterson and D. F. Ackerley, Biochem.
J., 2015, 471, 131–153 CrossRef CAS PubMed.
- D. Liu, T. N. Wanniarachchi, G. Jiang, G. Seabra, S. Cao, S. D. Bruner and Y. Ding, RSC Chem. Biol., 2022, 3, 436–446 RSC.
- R. S. Boddu, O. Perumal and D. K, Biotechnol. Appl. Biochem., 2021, 68, 1518–1530 CAS.
- W. Wang, J. Cai, N.-K. Wong, M. Hong, J. Deng, L. Jin, Y. Ran, Y. Zhang, Y. Zhou and B.-O. Guan, Analyst, 2022, 147, 1449–1456 RSC.
- A. Chevalier, Y. Zhang, O. M. Khdour, J. B. Kaye and S. M. Hecht, J. Am. Chem. Soc., 2016, 138, 12009–12012 CrossRef CAS PubMed.
- S. A. Yoon, J. Chun, C. Kang and M. H. Lee, ACS Appl. Bio Mater., 2021, 4, 2052–2057 CrossRef CAS PubMed.
- W. Sun, M. Tong, G. Liu, X. Wang, N. Fan, X. Song, D. Yang and D. Zhang, Results Chem., 2021, 3, 100177 CrossRef CAS.
- R. Tannous, O. Shelef, S. Gutkin, M. David, T. Leirikh, L. Ge, Q. Jaber, Q. Zhou, P. Ma, M. Fridman, U. Spitz, K. N. Houk and D. Shabat, ACS Cent. Sci., 2024, 10, 28–42 CrossRef CAS PubMed.
- J. Sun, Z. Hu, R. Wang, S. Zhang and X. Zhang, Anal. Chem., 2019, 91, 1384–1390 CrossRef CAS PubMed.
- L. S. Ryan, J. Gerberich, J. Cao, W. An, B. A. Jenkins, R. P. Mason and A. R. Lippert, ACS Sens., 2019, 4, 1391–1398 CrossRef CAS PubMed.
- A. L. Pey, C. F. Megarity and D. J. Timson, Biosci. Rep., 2019, 39 DOI:10.1042/BSR20191874.
- M. Faig, M. A. Bianchet, P. Talalay, S. Chen, S. Winski, D. Ross and L. M. Amzel, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 3177–3182 CrossRef CAS PubMed.
- S. R. Punganuru, H. R. Madala, V. Arutla, R. Zhang and K. S. Srivenugopal, Sci. Rep., 2019, 9, 8577 CrossRef PubMed.
- S. Son, M. Won, O. Green, N. Hananya, A. Sharma, Y. Jeon, J. H. Kwak, J. L. Sessler, D. Shabat and J. S. Kim, Angew. Chem., Int. Ed., 2019, 58, 1739–1743 CrossRef CAS PubMed.
- S. R. Pinnell and G. R. Martin, Proc. Natl. Acad. Sci. U. S. A., 1968, 61, 708–716 CrossRef CAS PubMed.
- S. D. Vallet, M. Guéroult, N. Belloy, M. Dauchez and S. Ricard-Blum, ACS Omega, 2019, 4, 8495–8505 CrossRef CAS PubMed.
- T. Liburkin-Dan, S. Toledano and G. Neufeld, Int. J. Mol. Sci., 2022, 23, 6249 CrossRef CAS PubMed.
- L. Leung, D. Niculescu-Duvaz, D. Smithen, F. Lopes, C. Callens, R. McLeary, G. Saturno, L. Davies, M. Aljarah, M. Brown, L. Johnson, A. Zambon, T. Chambers, D. Ménard, N. Bayliss, R. Knight, L. Fish, R. Lawrence, M. Challinor, H. Tang, R. Marais and C. Springer, J. Med. Chem., 2019, 62, 5863–5884 CrossRef CAS PubMed.
- J. Huang, C. Zhang, X. Wang, X. Wei and K. Pu, Angew. Chem., Int. Ed., 2023, 62, e202303982 CrossRef CAS PubMed.
- R. J. Obaid, E. U. Mughal, N. Naeem, A. Sadiq, R. I. Alsantali, R. S. Jassas, Z. Moussa and S. A. Ahmed, RSC Adv., 2021, 11, 22159–22198 RSC.
- T.-S. Chang, Materials, 2012, 5, 1661–1685 CrossRef CAS.
- O. Shelef, A. C. Sedgwick, S. Pozzi, O. Green, R. Satchi-Fainaro, D. Shabat and J. L. Sessler, Chem. Commun., 2021, 57, 11386–11389 RSC.
-
M. H. Hanigan, in Advances in Cancer Research, ed. D. M. Townsend and K. D. Tew, Academic Press, 2014, vol. 122, pp. 103–141 Search PubMed.
- P. N. Brennan, J. F. Dillon and E. B. Tapper, Liver Int., 2022, 42, 9–15 CrossRef CAS PubMed.
- M. B. West, Y. Chen, S. Wickham, A. Heroux, K. Cahill, M. H. Hanigan and B. H. M. Mooers, J. Biol. Chem., 2013, 288, 31902–31913 CrossRef CAS PubMed.
- R. An, S. Wei, Z. Huang, F. Liu and D. Ye, Anal. Chem., 2019, 91, 13639–13646 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a,
Gurjot
Kaur
b,
Atharva Rajesh
Chavan
*a,
Gurjot
Kaur
b,
Atharva Rajesh
Chavan
 a,
Mandeep K.
Chahal
c and
Rajeev
Taliyan
a
a,
Mandeep K.
Chahal
c and
Rajeev
Taliyan
a




![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold superior to its fluorescent analog. Due to the large expression of cathepsin B, RAW 264.7 and CT26 cells, except for 3T3 (the control group), showed bright chemiluminescence upon treatment with CL-22.97 Furthermore, to achieve NIR emission, 12-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile conjugated at the para position of phenoxyl-1,2-dioxetane (CL-23) and the dipeptide “phenylalanine-lysine” were employed as a trigger for cathepsin B.63 The conjugation of an extended π system at the para position increased the chemiluminescence quantum yield along with emission wavelength. The catalytic action of cathepsin B led to 75.5-fold chemiluminescence enhancement at 650 nm within 30 minutes of incubation (Fig. 8A). For in vivo applications, the probe solution was injected intraperitoneally into 4T1-bearing tumor mouse models. The chemiluminescence emission attained maxima within 10 minutes of injection and it was 4-fold higher than the mouse pre-treated with cathepsin B inhibitor. Laparotomy was conducted 30 minutes after probe injection to illustrate the potential for precise tumor resection guided by chemiluminescence diagnosing deeply buried tumors (Fig. 8B). The fluorescence signal remained undetectable in the intestinal region compared to the chemiluminescence signal. The higher signal-to-background ratio (S/B 7.6) was reported for image-guided surgery in cathepsin B expressive tissues.
000-fold superior to its fluorescent analog. Due to the large expression of cathepsin B, RAW 264.7 and CT26 cells, except for 3T3 (the control group), showed bright chemiluminescence upon treatment with CL-22.97 Furthermore, to achieve NIR emission, 12-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile conjugated at the para position of phenoxyl-1,2-dioxetane (CL-23) and the dipeptide “phenylalanine-lysine” were employed as a trigger for cathepsin B.63 The conjugation of an extended π system at the para position increased the chemiluminescence quantum yield along with emission wavelength. The catalytic action of cathepsin B led to 75.5-fold chemiluminescence enhancement at 650 nm within 30 minutes of incubation (Fig. 8A). For in vivo applications, the probe solution was injected intraperitoneally into 4T1-bearing tumor mouse models. The chemiluminescence emission attained maxima within 10 minutes of injection and it was 4-fold higher than the mouse pre-treated with cathepsin B inhibitor. Laparotomy was conducted 30 minutes after probe injection to illustrate the potential for precise tumor resection guided by chemiluminescence diagnosing deeply buried tumors (Fig. 8B). The fluorescence signal remained undetectable in the intestinal region compared to the chemiluminescence signal. The higher signal-to-background ratio (S/B 7.6) was reported for image-guided surgery in cathepsin B expressive tissues.





![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold enhancement in just 20 minutes. The drastic increase in chemiluminescence response and ultra-sensitivity of CL-39 was attributed to acetoxymethyl ester, which improved its cellular uptake. Additionally, CL-40 was successfully utilised to discriminate between hypoxic and normoxic conditions in A549 cells, with images taken 60 minutes post-injection revealing distinct contrasts between hypoxic and well-oxygenated tissues (Fig. 13C). The photon flux intensity (1 × 107 photons per s) of CL-39 was 100 times greater than that of CL-37 and CL-NTR. These findings suggest that positioning of EWG substituents at the para position of phenolic OH is a critical component of the process to improve chemiluminescence emission and sensitivity. Moreover, integrating hydrophobic groups, such as esters, alongside EWGs could further facilitate cellular uptake. Future structural modifications may enhance the photophysical properties and cellular uptake of phenoxy-1,2-dioxetane derivatives.
000-fold enhancement in just 20 minutes. The drastic increase in chemiluminescence response and ultra-sensitivity of CL-39 was attributed to acetoxymethyl ester, which improved its cellular uptake. Additionally, CL-40 was successfully utilised to discriminate between hypoxic and normoxic conditions in A549 cells, with images taken 60 minutes post-injection revealing distinct contrasts between hypoxic and well-oxygenated tissues (Fig. 13C). The photon flux intensity (1 × 107 photons per s) of CL-39 was 100 times greater than that of CL-37 and CL-NTR. These findings suggest that positioning of EWG substituents at the para position of phenolic OH is a critical component of the process to improve chemiluminescence emission and sensitivity. Moreover, integrating hydrophobic groups, such as esters, alongside EWGs could further facilitate cellular uptake. Future structural modifications may enhance the photophysical properties and cellular uptake of phenoxy-1,2-dioxetane derivatives.



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000




















