Open Access Article
Open Access ArticleHigh throughput electronic detection of biomarkers using enzymatically amplified metallization on nanostructured surfaces†
Hanhao
Zhang‡
,
Neda
Rafat‡
,
Josiah
Rudge
,
Sai Preetham
Peddireddy
,
Yoo Na
Kim
,
Taaseen
Khan
and
Aniruddh
Sarkar
 *
*
Georgia Institute of Technology, 315 Ferst Dr NW, Atlanta, GA 30332, USA. E-mail: aniruddh.sarkar@bme.gatech.edu
First published on 31st October 2024
Abstract
Enzyme-linked immunosorbent assays are commonly used for clinical biomarker detection. However, they remain resource-intensive and difficult to scale globally. Here we present a miniaturized direct electronic biosensing modality which generates a simple and sensitive, quantitative, resistive readout of analyte binding in immunoassays. It utilizes the enhanced metallization generated by synergistic catalytic activity of nanostructured surfaces, created using gold nanoparticles, with enzymatic metallization, catalyzed by analyte-bound enzyme-labeled antibodies, to create a connected metal layer between microelectrodes. Based on this scheme, we develop a portable, high-throughput electronic biomarker detection device and platform which allows testing 96 different low volume (3 μL) clinical samples in a handheld device. We find an analyte concentration-dependent tunable digital switch-like behavior in the measured resistance of this device. We use this system to further explore the mechanism of enhanced metallization and find optimal parameters. Finally, we use this platform to perform quantitative measurement of viral antigen-specific antibody titers from convalescent COVID-19 patient serum.
1. Introduction
The severe diagnostics shortage during the early stages of the COVID-19 pandemic underscored the inadequacy of current centralized laboratory-based diagnostics infrastructure in scaling rapidly to meet global needs during emerging infectious disease outbreaks.1 Arguably, lack of timely and easy-to-use diagnostics contributed to the failure of early public health efforts to contain the global spread of COVID-19. Sensitive and specific yet inexpensive and globally, rapidly scalable diagnostics modalities, especially those that are field-deployable and can be used at the point-of-care (POC), can help in early control of infectious disease outbreaks such as COVID-19, Ebola, MERS, H1N1 etc.2 Even now, as the global deployment of COVID-19 vaccines has occurred, scalable methods for sero-surveillance of biomarkers related to prior infection and vaccine efficacy are needed, especially due to the risk of emergence of vaccine-evading viral variants.3 Multiplexed and high-throughput antibody-based biomarker measurements can enable this and these can be used as prognostic biomarkers as well.4,5Currently, enzyme-linked immunosorbent assays (ELISA) are the gold standard in quantitative detection of multiple categories of biomarkers.6 ELISAs provide high sensitivity (i.e. low limit of detection, LOD), specificity, repeatability and quantitative ability.7 However, they are based on sensitive optical measurement of enzymatic reaction products and require specialized instruments such as plate readers, which use optics, optical detectors (e.g. photomultiplier tubes) and automatic moving stages, all of which add to their high cost (usually >$25,000) and size that restricts their usage to centralized laboratories by trained personnel. Efforts have been made to develop portable and/or inexpensive ELISAs,8 but a cost-versus-performance trade-off often exists due to the cost of optical components, especially when miniaturized. This is exacerbated by the lower absorbance path lengths as sample volumes are scaled down, requiring further higher optical detection sensitivity in miniaturized versions.9 An inexpensive alternative is the lateral flow assay (LFA) which often uses nano-conjugated antibodies (e.g. with gold nanoparticles) to return a output, based on nanoparticle aggregation, that can be directly observed with the naked eye.10 LFAs are easy to use as well and are widely disseminated for POC diagnostics. However, LFAs are often less sensitive and their result is binary (positive/negative)2 which restricts their diagnostic value.
Electrical and electrochemical detection principles can be more suitable for POC diagnostics. Instead of the use of intermediate optics, they convert immunobinding directly to an electronic signal.11–18 Their size and cost can both be scaled down via miniaturization and integration using microfabrication-based mass manufacturing.11 These properties help them overcome disadvantages of traditional ELISA including sample volume requirement and larger size/cost of the required hardware.19,20 Current electrochemical biosensors however often still remain too complex to fabricate21,22 or use especially in a high-throughput format, and are still rarely used in the clinic.23
Here, we set out to develop a miniaturize and broadly applicable direct electronic biosensing modality that can exploit the sensitivity afforded by enzymatic amplification and yet generate a simple, yet sensitive and quantitative electronic readout in an inexpensive portable platform. Gold nanoparticle (AuNP) labeled probes are widely used in LFAs and silver reduction catalyzed by them has been used for higher sensitivity in optical24 and electronic25,26 detection. We have recently made a counter-intuitive observation that instead of using AuNP-labeled probes, immobilizing the AuNPs on the chip surface creating a nanostructured catalytic surface provides >100× higher sensitivity when coupled with enzymatic metallization.27 The mechanism underlying this enhanced enzymatic metallization on AuNP-labeled nanostructured surfaces, however, remains unexplored. Unlike the various works where AuNPs serve as the independent substrate for silver deposition to occur on their surfaces,28,29 here a combined effect from AuNPs, microscale interdigitated electrodes (μIDEs) and glass that form the nanostructure alters the amount of silver deposition. Understanding it could hold the key to its optimization using various properties of AuNPs or other nanomaterials and lead to higher sensitivity, ease-of-use and eventual clinical translation. Since it is the combined effect of different parameters that determines the performance of the above system, a high-throughput version of the above chip and corresponding hardware are necessary for efficient screening. Additionally, such an easy-to-use portable chip and system would enable the use of this method by others as well as help progress this detection modality towards clinical applications.
In this work, we first developed a high-throughput micro-electrode array system for ELISA (or EASyELISA) microchip to explore, optimize and clinically apply the above AuNP-driven electronic immunoassay. This high-throughput EASyELISA microchip enables simultaneous rapid testing of 96 different small volume (3 μL) clinical samples, matching the throughput of conventional 96-well ELISA plates. We then used it to explore the combined effect of electrode size, AuNP size and concentration on the performance of the microchip by running an immunoassay for SARS-CoV-2 viral antigen-specific antibodies. We observed a tunable digital switch-like behavior of the resistive readout versus antibody concentration where the switching threshold was found to be tunable using electrode gap or AuNP size and concentration. Finally, we developed a handheld cellphone-interfaced electronic reader for the EASyELISA microchip which could automatically analyze 96 individual electrodes, thus building a full handheld plate reader equivalent device and system. We then used the chip to measure quantitative electronic readouts of SARS-CoV-2 Spike (S)-specific antibodies from convalescent COVID-19 patient serum (n = 5) and pre-pandemic healthy serum (n = 3), performing serial dilution curves for titer measurements, showing clear distinction between the two patient classes.
2. Materials and methods
Soda lime glass wafers were purchased from UniversityWafer (Boston, MA). Phosphate buffer saline (PBS) and deionized water (DIW) were purchased from Fisher Scientific (Hampton, NH). Sodium hydroxide, poly-L-lysine solution, Bovine Serum Albumin (BSA), and gold nanoparticles (AuNPs: 5 nm/10 nm/100 nm) were purchased from Sigma Aldrich (St. Louis, MO). Tween 20 and reagent alcohol were purchased from VWR (Radnor, PA). Polydimethylsiloxane (PDMS) sheet was purchased from Greene Rubber Company (Woburn, MA). Pierce Bovine Serum Albumin, Biotinylated, and HRP-Conjugated Streptavidin were purchased from ThermoFisher (Waltham, MA). Enzymatic metallization substrate solution (EnzMet) was purchased from CedarLane (Burlington, Canada). SARS-CoV-2 (2019-nCoV) spike recombinant protein was purchased from Immune Technology (New York, NY). Recombinant Anti-SARS-CoV-2 Spike Glycoprotein S1 antibody was purchased from Abcam (Cambridge, United Kingdom). Mouse Anti-Human IgG Fc-HRP was purchased from Southern Biotech (Birmingham, AL). Clinical serum samples (healthy/SARS-Cov-2 patient) were purchased from RayBiotech (Peachtree Corners, GA). Vendor obtained serum from donors with informed consent after IRB approval (PROTOCOL NO: SOP-TF-PH-002 STERLING IRB ID: 8291-BZhang). Details are on file and available with the vendor.2.1 Microfabrication of EASyELISA microchips
Soda lime glass wafers were sonicated in acetone then isopropyl alcohol for 5 minutes each. Cleaned wafers were soaked in piranha solution for 20 minutes. After rinsing with DIW and drying, NR-9 1500PY Photoresist (Futurrex, Franklin, NJ) was spin coated and baked at 150 °C for 2.5 minutes. Coated wafers then went through UV exposure (Karl Suss MA-6, Suss MicroTec, Germany), 1 minute baking at 100 °C and development with RD-6 (Futurrex, Franklin, NJ). After 30 seconds of plasma descum (Vision 320 RIE, Plasma-therm, St. Petersburg, FL), 10 nm of titanium plus 100 nm of gold was deposited via e-beam evaporation (Denton Explorer, Denton Vacuum, Moorestown, NJ). Gold-coated wafers were then sonicated in acetone for 5 minutes to perform liftoff. After rinsing with isopropyl alcohol and drying, the wafers were diced (DAD3360, DISCO, Japan) to obtain individual chips.2.1.1.1 SARS-CoV-2 antibody assay. Solution containing AuNPs of different sizes and concentrations were incubated in the wells for 45 minutes. 25 μg ml−1 of SARS-Cov-2 Spike protein solution were added and incubated in the wells overnight at 4 °C. The chips were blocked as above. Different dilutions of anti-SARS-Cov-2 spike human antibody were added to the wells and incubated for 1 hour. 1 μg ml−1 of horseradish peroxidase-conjugated anti-human IgG (anti-IgG-HRP) was then added and incubated for 30 minutes. Enzmet addition, drying and storage were performed as above. When running the assay with clinical samples, the concentration of SARS-Cov-2 Spike protein was raised to 50 μg ml−1 and dilutions of patient/healthy samples in PBS were used instead of commercial antibody. All other procedures remained the same.
2.2 Assembly of the portable reader
PmodIA impedance analyzer (Digilent), Analog multiplexer (ADG731, Analog Devices), Arduino Nano, Bluetooth module (HC05, DSD Tech) and consumables (connector pins etc.) were purchased from Digikey. Casings were printed with Form 2 (Formlabs) and F170 (Stratasys).2.3 Data acquisition and analysis
Optical images were taken via stereomicroscope (AmScope, CA), Scanning Electron Microscopy (SEM) images were taken via SU8010 SEM (Hitachi, Japan), and resistances were measured with a digital multimeter (Fig. 1, 3 and 5) or with the custom handheld reader. Data was analyzed with ImageJ and GraphPad Prism. All data presented are the arithmetic mean of at least 2 technical repeats and error bars represent standard deviation.3. Results and discussion
3.1 Demonstration of electronic detection using nanostructure surfaces
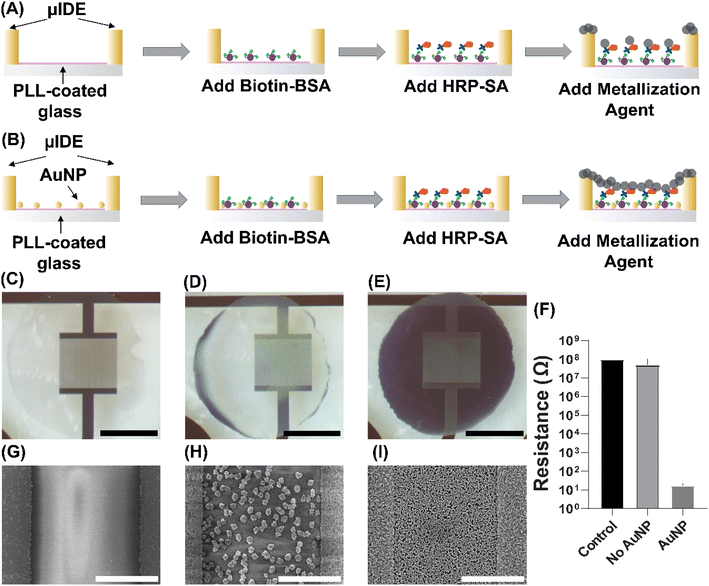
We first performed a model binding assay using biotinylated bovine serum albumin (biotin-BSA) as a capture agent and Horseradish Peroxidase labeled streptavidin (HRP-SA) as analyte. Microscale interdigitated electrodes (width and inter-electrode gap: 10 μm) were made on glass wafers using standard microfabrication methods. The μIDEs were coated with a poly-L-lysine (PLL) adhesive layer and stored in a desiccator. Before usage, a layer of laser-cut PDMS sheet was added on the top of the chip, forming microwells around the μIDEs. For the group without AuNPs (Fig. 1A), solution containing Biotin-BSA was directly added into the wells. HRP-SA and enzymatic metallization substrate were separately added following protocol in Methods section, resulting in silver deposition across the surface of μIDEs. For the second group (Fig. 1B), AuNPs (10 nm, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50× diluted in DI water from an OD = 1 stock, see ESI Table 1†) were first immobilized on the surface of μIDEs. The stable immobilization via charge-based AuNP – lysine binding has been confirmed by various works.30,31 Biotin-BSA, HRP-SA and metallization substrate were subsequently added following the same protocol as the previous group. Significant differences in deposited silver darkness were observed from the post-assay optical images (Fig. 1C–E) of the μIDEs. There was no silver deposited for the control group (no biotin-BSA or HRP-SA) (Fig. 1C). Small amount of silver was found for the μIDE without AuNPs (Fig. 1D). However, a uniform dark layer of silver covered the entire μIDE that had AuNPs pre-immobilized (Fig. 1E). This layer of silver acted as a conductor, connecting the gap between μIDEs as a bridge. With connected μIDEs, a closed circuit would be formed when performing resistance measurement. As detailed in previous work,27 such a decrease in resistance after the assay suggested the presence of target molecules in the sample. A huge difference in resistance was observed only for the group with HRP-SA and AuNP immobilization (Fig. 1F). Scanning electron microscopy (SEM) revealed further striking nanoscale differences. In the control μIDE: there were small silver deposits on the gold electrodes but none on the glass surface between electrodes (Fig. 1G). In the μIDE with HRP-SA but without AuNPs: electrode surfaces were found to have denser silver deposits but the glass surface between electrodes was found to have sparse nanoparticulate silver deposits (∼400 nm) which had a characteristic ‘desert-rose’ shape, but they were unconnected from each other (Fig. 1H). This shape has been reported by others previously32,33 as characteristic for enzyme-catalyzed silver nanoparticle formation. Finally, in the μIDEs with HRP-SA and AuNP immobilization: while the electrode surfaces were found to have denser silver deposits here too, a starkly different silver morphology was observed on the glass surface between electrodes, with a denser layer with much larger number of smaller nano-particulates which merged to form a connected but perforated mesh-like layer (Fig. 1I). This change in silver morphology and density indicates a change in deposition mechanism to a synergistic activity between enzymatic and AuNP-catalyzed silver deposition.
50× diluted in DI water from an OD = 1 stock, see ESI Table 1†) were first immobilized on the surface of μIDEs. The stable immobilization via charge-based AuNP – lysine binding has been confirmed by various works.30,31 Biotin-BSA, HRP-SA and metallization substrate were subsequently added following the same protocol as the previous group. Significant differences in deposited silver darkness were observed from the post-assay optical images (Fig. 1C–E) of the μIDEs. There was no silver deposited for the control group (no biotin-BSA or HRP-SA) (Fig. 1C). Small amount of silver was found for the μIDE without AuNPs (Fig. 1D). However, a uniform dark layer of silver covered the entire μIDE that had AuNPs pre-immobilized (Fig. 1E). This layer of silver acted as a conductor, connecting the gap between μIDEs as a bridge. With connected μIDEs, a closed circuit would be formed when performing resistance measurement. As detailed in previous work,27 such a decrease in resistance after the assay suggested the presence of target molecules in the sample. A huge difference in resistance was observed only for the group with HRP-SA and AuNP immobilization (Fig. 1F). Scanning electron microscopy (SEM) revealed further striking nanoscale differences. In the control μIDE: there were small silver deposits on the gold electrodes but none on the glass surface between electrodes (Fig. 1G). In the μIDE with HRP-SA but without AuNPs: electrode surfaces were found to have denser silver deposits but the glass surface between electrodes was found to have sparse nanoparticulate silver deposits (∼400 nm) which had a characteristic ‘desert-rose’ shape, but they were unconnected from each other (Fig. 1H). This shape has been reported by others previously32,33 as characteristic for enzyme-catalyzed silver nanoparticle formation. Finally, in the μIDEs with HRP-SA and AuNP immobilization: while the electrode surfaces were found to have denser silver deposits here too, a starkly different silver morphology was observed on the glass surface between electrodes, with a denser layer with much larger number of smaller nano-particulates which merged to form a connected but perforated mesh-like layer (Fig. 1I). This change in silver morphology and density indicates a change in deposition mechanism to a synergistic activity between enzymatic and AuNP-catalyzed silver deposition.
We hypothesize that in absence of the AuNPs, the HRP molecules themselves nucleate silver deposition sites which grow into unconnected larger nano-particulates (∼400 nm), while the rest of the surface remains uncovered. This deposition is likely quenched by the eventual inactivation of the enzymatic activity by silver deposition itself. However, with the nanostructured catalytic surface resulting from AuNP immobilization, we hypothesize that the AuNPs act preferentially as nucleation sites for the HRP-catalyzed metallization. Thus, a denser and connected silver layer with smaller length-scale results which covers most of the surface. The density and uniformity of silver are reflected by the resistance across μIDEs. After enough silver is deposited on the glass area between two electrodes, the silver acts as a conducting bridge, forming a gold electrode–silver–gold electrode closed circuit. Measured resistance of the μIDEs showed an open circuit (10 MΩ) for both the control μIDE and the μIDE with HRP-SA but without AuNPs, whereas the resistance dropped to 100 Ω post-assay for the μIDE with HRP-SA and AuNPs. This establishes the dry-stable, simple resistive readout of this assay with six orders of magnitude of change in resistance which enables its easy and inexpensive measurement in a fully portable system.
3.2 Development of high-throughput electronic detection chip and portable reader system
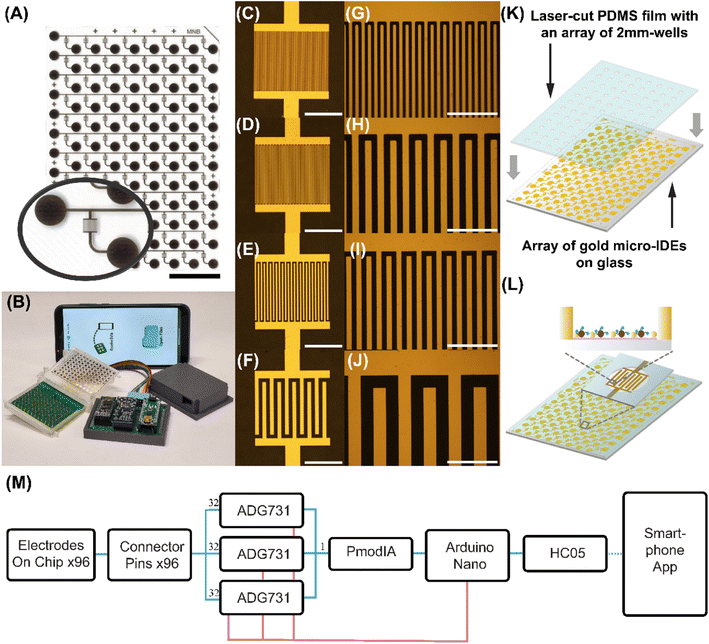
We then fabricated the high-throughput EASyELISA chip with 96 μIDE pairs (Fig. 2A). This credit-card sized glass chip (76 mm × 55 mm) had 12 rows × 8 columns, with every pair of adjacent columns having μIDE pairs with the same electrode gaps and widths (w = g = 5 μm, 10 μm, 20 μm and 50 μm, Fig. 2C–J). Additionally, the inter-column gap made the chip compatible with standard multi-channel pipettes for high-throughput assays. This chip was then visually aligned and assembled, via reversible bonding, with a thin (0.01′′) laser-cut PDMS microwell array (Fig. 2K) which enables loading of small volumes (∼1.5–3 μL) of samples and reagents reliably in a high-throughput yet leak-free manner. This enables performing 96 different assays on the individual μIDEs (Fig. 2L) on a single chip. Compared to conventional ELISA, here a 10–80-fold reduction of the sample volume to only 3 μL each is achieved. Also, washing and drying of the chip can be done by placing the entire chip in wash buffer and micro-centrifuging or blow drying respectively. This saves the effort of additional pipetting and draining steps or the need for any pumps or tubing needed for traditional microfluidic chips and is more akin to standard slide-based assays.34,35Additionally, to enable ease of rapid electronic measurement in a POC-compatible manner, an inexpensive handheld measurement system was built (Fig. 2B). A custom printed circuit board was designed with 96 spring-mounted pogo-pins and assembled inside a 3D-printed enclosed connector which enabled easy connection to all 96 μIDEs. As shown in the block diagram (Fig. 2M), signal from the connectors was routed via three 32-plex analog multiplexers (ADG731) which allow the digital selection of a single μIDE for measurement by an Arduino Nano. The selected output is then connected to a PmodIA Impedance analyzer module. This module has an AD5933 12 bit impedance converter chip with a frequency generator which excited the device under test and an analog-to-digital converter to capture the response. A discrete Fourier transform is performed automatically on chip and the real and imaginary parts of the response can be relayed via serial communication, to an Arduino Nano and sent to a smartphone via a Bluetooth module. A custom Android application was developed which not only controlled the above operations but also visualized the received data and enabled its cloud-based storage. Details of the design of the chip, system and application can be found via links in ESI.† By measuring resistance value for eight different resistors ranging from 100 ohms to 210![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ohms with both the reader and the multimeter, we found a high correlation of r = 0.999958 between the two methods (Fig S1†), proving equivalence of the two in our application. We note that this reader hardware itself is effectively a portable 96-channel impedance analyzer or LCR meter which can be useful beyond its application with the EASyELISA chip here as well.
000 ohms with both the reader and the multimeter, we found a high correlation of r = 0.999958 between the two methods (Fig S1†), proving equivalence of the two in our application. We note that this reader hardware itself is effectively a portable 96-channel impedance analyzer or LCR meter which can be useful beyond its application with the EASyELISA chip here as well.
3.3 Optimization of electronic immunoassay on high-throughput electronic detection chip
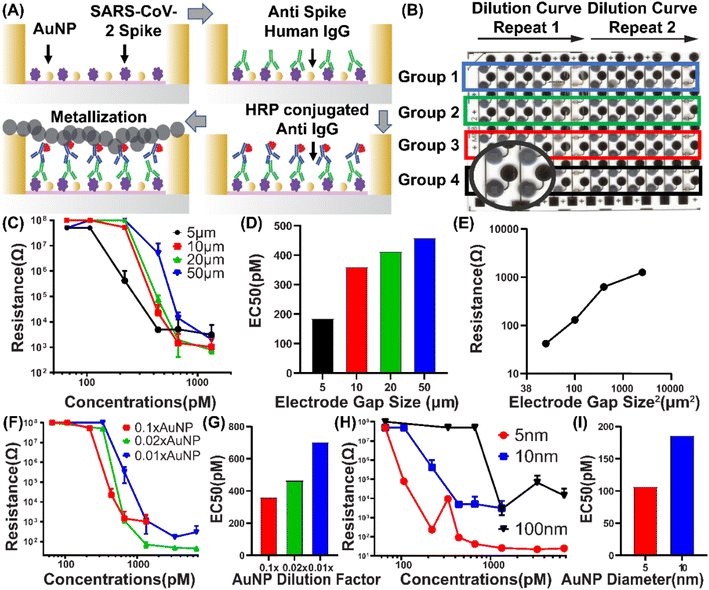
We then applied the EASyELISA chip to a SARS-CoV-2 viral antigen-specific antibody detection assay, with an anti-Spike monoclonal antibody (anti-S mAb) as the target and performed a parametric study and optimization (Fig. 3A). To study the interaction between the AuNPs, μIDEs and the enzymatic metallization (quantified by measured resistance readout), we selected inter-electrode gap, g (while maintaining w = g), concentration and size of AuNPs as the three target parameters. First, the effect of varying g was explored with AuNPs of a particular size and concentration (10 nm, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10× diluted). AuNPs and S antigen were immobilized on the chip as above, followed by incubation with serial dilutions of anti-S mAbs and then with HRP-labeled anti-human IgG probe followed by the metallization. As shown in Fig. 3B, four groups of 6-point serial sample dilution curves with two replicates each were generated on a single chip. Within the first six electrodes in each group, the concentration of sample decreased from left to right. A systematic variation in the darkness of the deposited silver in the microwells, correlated with the sample dilutions, was observed. Resistance measurement results from this experiment are shown in Fig. 3C. μIDE resistances are found to reduce systematically with increasing anti-S mAb concentration. Resistance versus concentration curves are observed to form the typical sigmoidal shape seen in traditional ELISAs. The top of the curve (>50 MΩ) corresponds to low anti-S mAb concentrations and the bottom (10 kΩ) corresponds to high concentrations of anti-S mAb. This establishes the operation of the high-throughput EASyELISA chip and assay with a direct resistive electronic readout.
10× diluted). AuNPs and S antigen were immobilized on the chip as above, followed by incubation with serial dilutions of anti-S mAbs and then with HRP-labeled anti-human IgG probe followed by the metallization. As shown in Fig. 3B, four groups of 6-point serial sample dilution curves with two replicates each were generated on a single chip. Within the first six electrodes in each group, the concentration of sample decreased from left to right. A systematic variation in the darkness of the deposited silver in the microwells, correlated with the sample dilutions, was observed. Resistance measurement results from this experiment are shown in Fig. 3C. μIDE resistances are found to reduce systematically with increasing anti-S mAb concentration. Resistance versus concentration curves are observed to form the typical sigmoidal shape seen in traditional ELISAs. The top of the curve (>50 MΩ) corresponds to low anti-S mAb concentrations and the bottom (10 kΩ) corresponds to high concentrations of anti-S mAb. This establishes the operation of the high-throughput EASyELISA chip and assay with a direct resistive electronic readout.
Notably, here, the large (>5000-fold) resistance change observed occurs over ∼5–10-fold change in anti-S mAb concentration. This can be contrasted with the 25–100-fold change in analyte concentration over which typical ELISA optical signal vs. concentration sigmoidal curves run from top to bottom.6 Thus, EASyELISA is found to have a sharper transition and switch-like characteristic compared to traditional ELISAs. The switching point is defined as the point where the slope of resistance increases dramatically. Physically, it is the concentration of sample whose corresponding silver changes from a uniform dense layer to a sparse layer. The silver particles are no longer closely attached to each other, making electrons harder to travel between gold electrodes when a voltage is applied. This loose connection of silver particle results in resistance increasing dramatically. This could indicate a potential cooperativity-like mechanism underlying the silver deposition. Additionally, it also relates back to the nanoscale morphology of the deposited silver which grows as separate particles and then merges rather than just growing thicker as a continuous layer.
Five parameter asymmetric logistic curves were fitted to the data and the extracted EC50 values, which represent a metric for the switching points, were found to systematically increase with the electrode gap (Fig. 3C). We infer from this that the connectivity of the nanoscale mesh-like silver layer (Fig. 1I), may have a characteristic microscale length-scale as well which grows with analyte concentration and once it increases above the electrode gap size, closed electrode is observed. Thus, μIDEs with lower gap switch at lower analyte concentrations. Overall, smaller inter-electrode gap sizes thus provide better sensitivity or lower LODs. Beyond the switching point, at a given anti-S mAb concentration, resistances for μIDEs were found to vary with the electrode gap 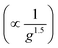 . Theoretically, a
. Theoretically, a 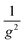 dependence is expected due to the increase in number of electrodes in a given area. This indicates that there may be other secondary effects involved such as the effect of electrode gap on silver deposition.
dependence is expected due to the increase in number of electrodes in a given area. This indicates that there may be other secondary effects involved such as the effect of electrode gap on silver deposition.
Finally, we compared the effect of AuNP size by running the serial anti-S mAb dilutions with 5 nm, 10 nm and 100 nm AuNPs (at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10× dilution) on g = 5 μm electrodes. All the assays showed switch-like characteristics (Fig. 3H). The switching point was found to shift to lower sample concentrations as the AuNP size was reduced (Fig. 3I). However, unlike with the earlier parameters, here, we see AuNP sizes also drive much larger differences in resistance e.g. entire impedance curve of 5 nm AuNPs (red) is below that of the 10 nm AuNPs (blue). On the other hand, the 100 nm AuNPs do not drive the μIDE resistance to a fully closed low impedance state, whose resistance reached hundred-million ohms range when the concentration was still around nano molar. Similar trends were seen with μIDEs with g = 10 μm (Fig. S3†). Overall, this is indicative of lower AuNP sizes driving significantly higher silver deposition in a sample and thus enzyme concentration-dependent manner. This could be due to higher AuNP counts or higher effective areas or area-to-volume ratios of the smaller AuNPs contributing to the higher synergistic catalytic activity of the nanostructured surface generated by them. Earlier work on catalytic properties of AuNPs has also observed similar trends with size.36 Here, this indicates that 5 nm AuNPs will provide the highest sensitivity. The EC50 of the S mAb serial dilution curve with 5 nm AuNPs was ∼100pM indicating a 10–100 pM scale LOD.
10× dilution) on g = 5 μm electrodes. All the assays showed switch-like characteristics (Fig. 3H). The switching point was found to shift to lower sample concentrations as the AuNP size was reduced (Fig. 3I). However, unlike with the earlier parameters, here, we see AuNP sizes also drive much larger differences in resistance e.g. entire impedance curve of 5 nm AuNPs (red) is below that of the 10 nm AuNPs (blue). On the other hand, the 100 nm AuNPs do not drive the μIDE resistance to a fully closed low impedance state, whose resistance reached hundred-million ohms range when the concentration was still around nano molar. Similar trends were seen with μIDEs with g = 10 μm (Fig. S3†). Overall, this is indicative of lower AuNP sizes driving significantly higher silver deposition in a sample and thus enzyme concentration-dependent manner. This could be due to higher AuNP counts or higher effective areas or area-to-volume ratios of the smaller AuNPs contributing to the higher synergistic catalytic activity of the nanostructured surface generated by them. Earlier work on catalytic properties of AuNPs has also observed similar trends with size.36 Here, this indicates that 5 nm AuNPs will provide the highest sensitivity. The EC50 of the S mAb serial dilution curve with 5 nm AuNPs was ∼100pM indicating a 10–100 pM scale LOD.
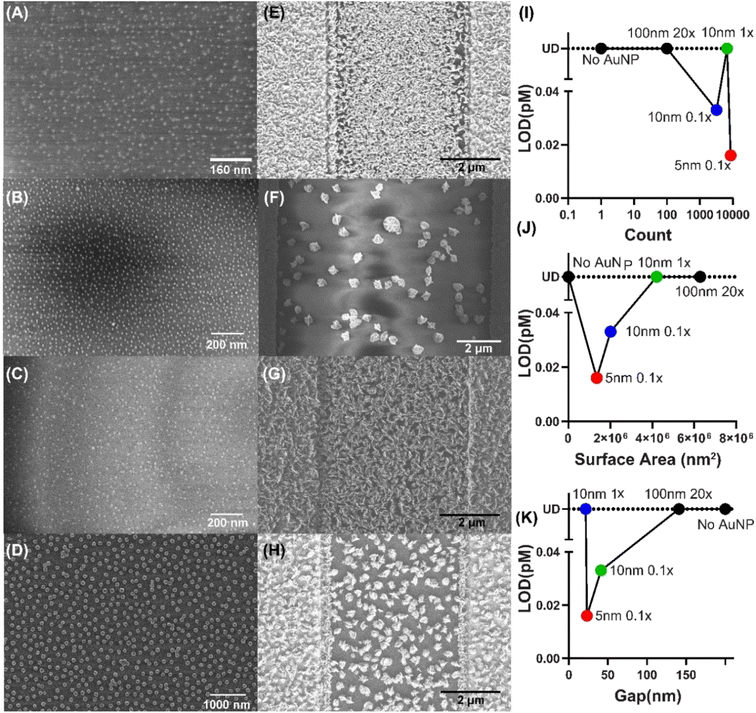
Next, given the complex variation of LOD with AuNP parameters observed above, we investigated how AuNPs affect silver deposition by performing SEM imaging on them (Fig. 4a–d) and their corresponding silver metallization (Fig. 4E–H). We analyzed the immobilized AuNP count, surface area and average inter-particle gap of each of the AuNP conditions from SEM images and studied the variation of the assay LOD with these parameters. Results showed that the AuNP count or surface area occupied by AuNPs alone does not determine the amount of silver deposition and LOD (Fig. 4I and J).
Finally, we studied the effect of inter-AuNP gap size (Fig. 4K) and observed that there may be an optimal inter-particle gap which results in the lowest (i.e. best) LOD.
We hypothesize that enzymatic silver metallization at the AuNPs as nucleation sites, then grows around these seeds which then merge and form the connected mesh eventually. Thus, AuNPs further apart result in higher analyte LODs as higher enzyme concentrations are needed to fill the higher inter-particle gaps. On the other hand, the S protein, used as capture antigen here, is reported to be ∼21 nm in length.37–39 Thus, it is plausible that too small an inter-particle gap blocks the capture antigen binding. Overall, we conclude that a dense layer of small AuNPs with gap size slightly larger than 20 nm provides the highest enzymatic silver metallization and best LOD.
3.4 Clinical immunoassay for electronic detection of viral antigen-specific antibodies
Given the above results, we selected 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
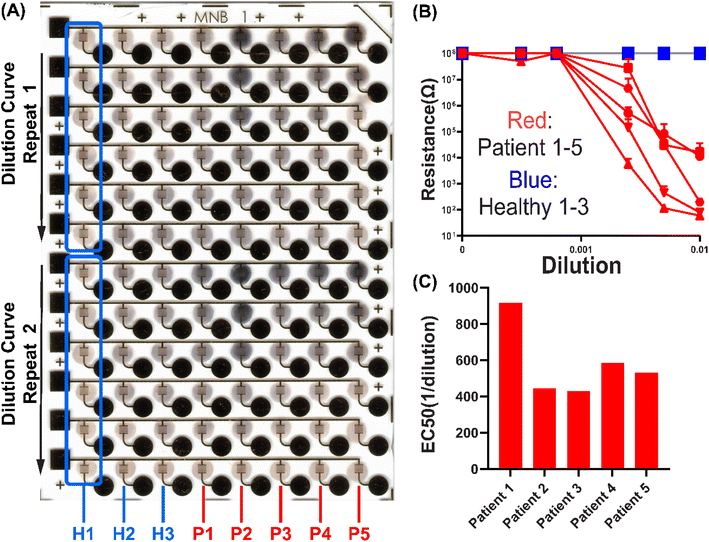
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 × 5 nm AuNPs on μIDEs with 5 μm gaps to perform a clinical immunoassay. Dilution curves of 5 convalescent COVID-19 patient serum samples and 3 pre-pandemic healthy serum samples, with two technical repeats, were run on a single chip (Fig. 5). Clear differences in silver metallization were observed between patient and healthy samples (Fig. 5A). Resistance readings for all healthy samples were all around 100 MΩ at all dilutions while lower dilution-dependent resistances were obtained from all patient samples (Fig. 5B). Fitted EC50s are obtained as anti-S antibody titers for the patient serum samples (Fig. 5C). This shows that the EASyELISA microchip developed here can be used for clinical immunoassays with quantitative readout of titers of antibody-based biomarkers.
10 × 5 nm AuNPs on μIDEs with 5 μm gaps to perform a clinical immunoassay. Dilution curves of 5 convalescent COVID-19 patient serum samples and 3 pre-pandemic healthy serum samples, with two technical repeats, were run on a single chip (Fig. 5). Clear differences in silver metallization were observed between patient and healthy samples (Fig. 5A). Resistance readings for all healthy samples were all around 100 MΩ at all dilutions while lower dilution-dependent resistances were obtained from all patient samples (Fig. 5B). Fitted EC50s are obtained as anti-S antibody titers for the patient serum samples (Fig. 5C). This shows that the EASyELISA microchip developed here can be used for clinical immunoassays with quantitative readout of titers of antibody-based biomarkers.
4. Conclusion
Here we presented a miniaturized electronic biosensing modality which generates a simple yet sensitive resistive readout. It utilizes the enhanced metallization generated by the synergistic catalytic activity of a nanostructured surface created using AuNPs with enzymatic metallization. We developed a high-throughput microchip which allows testing 96 different low volume (3 μL) clinical samples with the ease-of-use afforded by multichannel pipette compatibility and a handheld cellphone-interfaced reader, which enabled the below advantages: compared to conventional 96-well ELISA, it requires less sample volume and smaller/cheaper portable hardware; compared to lateral flow assay, it enables quantitative instead of binary result. Compared to field-effect transistor-based electrochemical biosensors21,22,40 (Table S1†), it is easier to fabricate and enables dry measurement/storage and re-measurement. Due to its unique metallization-based sensing mechanism, the major disadvantages include repeatability and signal degradation due to oxidation if the chip is not handled properly.Using this chip, we found a tunable digital switch-like response characteristic in antibody detection, where the switching threshold is tunable using various AuNP and device parameters. We also explored the mechanism of synergistic enhancement of metallization and optimized it via high-throughput parametric variation. We found that small electrode gaps and AuNP sizes enhance the assay sensitivity. Higher AuNP density, or lower inter-AuNP gaps, also enhanced sensitivity up to a certain optimum. Overall, we found evidence supporting a proposed mechanism that enzyme-catalyzed metallization is nucleated on AuNPs on the surface and grows to merge as an electrically connected mesh-like layer. Further work is needed to establish this mechanism via generalizing to other nanomaterials (e.g. nanowires) and capture molecules. Finally, using this platform we also demonstrated the detection and quantitation of antiviral antigen-specific antibodies from convalescent COVID-19 patient serum. The silver deposited on the chip is dry-stable, which provides a way to preserve the result of a clinical immunoassay directly on the chip as well. However, one current limitation of this is, due to oxidation of the deposited metal on the electrode, the resistance readings from the chip can drift in the long term (∼days) unless stored in air-tight containers with inert gas, limiting the stability of the result in case repeated measurement is needed. However, ELISA suffers from the same drawback as the liquid inside the well could evaporate, modifying the amount of absorbance unless properly sealed and stored. Further future work would include applying this platform to other binding-based assays of clinical utility (including DNA, RNA, cells, etc.) and test the specificity if required by application, thus developing a versatile POC-compatible electronic biosensing system.
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding contact: Aniruddh Sarkar (E-mail: aniruddh.sarkar@bme.gatech.edu). The data supporting this article have been included as part of the ESI.† All the code and AutoCAD designs of the chips can be accessed using the Github links in ESI file.† All data reported used in this work will be shared by the corresponding authors on request.Author contributions
H. Zhang: conceptualization, methodology, investigation, validation, data curation, formal analysis, visualization, software, writing – original draft, writing – review & editing. N. Rafat: conceptualization, methodology, investigation, validation, formal analysis, writing – review & editing. J. Rudge: investigation, validation, data curation, formal analysis, software, writing – review & editing. S. Peddireddy: investigation, formal analysis, writing – review & editing. Y. Kim: software, formal analysis, writing – review & editing. T. Khan: investigation, writing – review & editing. A. Sarkar: conceptualization, methodology, formal analysis, writing – review & editing resources, supervision, project administration, funding acquisition. All authors reviewed the manuscript.Conflicts of interest
H. Z., N. R. and A. S. are co-inventors of a patent application related to the nanoparticle-enhancement method for electronic biomarker detection.Acknowledgements
This work was funded by the National Institute of Health (NIH, R01AI182322). Microfabrication of chips was done with support from staff and facilities of the Institute for Electronics and Nanotechnology (IEN) at Georgia Tech.References
- O. Vandenberg, D. Martiny, O. Rochas, A. van Belkum and Z. Kozlakidis, Nat. Rev. Microbiol., 2021, 19, 171–183 CrossRef PubMed.
- Y. Zhou, Y. Wu, L. Ding, X. Huang and Y. Xiong, TrAC, Trends Anal. Chem., 2021, 145, 116452 CrossRef PubMed.
- C. D. C., Overview of Testing for SARS-CoV-2, https://www.cdc.gov/covid/hcp/clinical-care/overview-testing-sars-cov-2.html, (accessed Nov 08 2024) Search PubMed.
- S. P. Peddireddy, S. A. Rahman, A. R. Cillo, G. M. Vijay, A. Somasundaram, C. J. Workman, W. Bain, B. J. McVerry, B. Methe, J. S. Lee, P. Ray, A. Ray, T. C. Bruno, D. A. A. Vignali, G. D. Kitsios, A. Morris, H. Singh, A. Sarkar and J. Das, Cell Rep., 2022, 39(13), 111020 CrossRef PubMed.
- A. Saha, T. Chakraborty, J. Rahimikollu, H. Xiao, L. B. Pereira De Oliveira, T. W. Hand, S. Handali, W. E. Secor, L. A. O. Fraga, J. K. Fairley, J. Das and A. Sarkar, Sci. Transl. Med., 2024, 16(765) DOI:10.1126/scitranslmed.adk7832.
- J. R. Crowther, Methods Mol. Biol., 2000, 149, 1–413 Search PubMed.
- S. Hosseini, P. Vázquez-Villegas, M. Rito-Palomares and S. O. Martinez-Chapa, in Enzyme-linked Immunosorbent Assay (ELISA): from A to Z, ed. S. Hosseini, P. Vázquez-Villegas, M. Rito-Palomares and S. O. Martinez-Chapa, Springer Singapore, Singapore, 2018, pp. 67–115, DOI:10.1007/978-981-10-6766-2_5.
- A. Zhdanov, J. Keefe, L. Franco-Waite, K. R. Konnaiyan and A. Pyayt, Biosens. Bioelectron., 2018, 103, 138–142 CrossRef.
- K. Grabowska, X. Wang, A. Jacobsson and J. Dillner, J. Immunol. Methods, 2002, 271, 1–15 CrossRef.
- B. G. Andryukov, AIMS Microbiol., 2020, 6, 280–304 Search PubMed.
- K. Kikkeri, D. Wu and J. Voldman, Lab Chip, 2021, 22, 100–107 RSC.
- S. S. Mahshid, S. E. Flynn and S. Mahshid, Biosens. Bioelectron., 2021, 176, 112905 CrossRef PubMed.
- J. Mok, M. N. Mindrinos, R. W. Davis and M. Javanmard, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 2110–2115 CrossRef.
- J. Rudge, M. Hoyle, N. Rafat, A. Spitale, M. Honan and A. Sarkar, ACS Omega, 2023, 8(25), 22934–22944 CrossRef PubMed.
- M. Seifert, E. Vargas, V. Ruiz-Valdepenas Montiel, J. Wang, T. C. Rodwell and A. Catanzaro, Sci. Rep., 2021, 11, 19193 CrossRef PubMed.
- R. Zeng, M. Qiu, Q. Wan, Z. Huang, X. Liu, D. Tang and D. Knopp, Anal. Chem., 2022, 94, 15155–15161 CrossRef.
- Z. Yu, H. Gong, J. Xu, Y. Li, Y. Zeng, X. Liu and D. Tang, Anal. Chem., 2022, 94, 3418–3426 CrossRef PubMed.
- X. Wang, H. Wang, X. Wan, Q. Wei, Y. Zeng and D. Tang, Biosens. Bioelectron., 2025, 267, 116749 CrossRef.
- S. Lv, K. Zhang, L. Zhu, D. Tang, R. Niessner and D. Knopp, Anal. Chem., 2019, 91, 12055–12062 CrossRef PubMed.
- Y. Wang, R. Zeng, S. Tian, S. Chen, Z. Bi, D. Tang and D. Knopp, Anal. Chem., 2024, 96, 13663–13671 CrossRef PubMed.
- D. S. Juang, C.-H. Lin, Y.-R. Huo, C.-Y. Tang, C.-R. Cheng, H.-S. Wu, S.-F. Huang, A. Kalnitsky and C.-C. Lin, Biosens. Bioelectron., 2018, 117, 175–182 CrossRef PubMed.
- C. K. Tang, A. Vaze, M. Shen and J. F. Rusling, ACS Sens., 2016, 1, 1036–1043 CrossRef.
- C. D. Chin, V. Linder and S. K. Sia, Lab Chip, 2012, 12, 2118–2134 RSC.
- T. Laksanasopin, T. W. Guo, S. Nayak, A. A. Sridhara, S. Xie, O. O. Olowookere, P. Cadinu, F. Meng, N. H. Chee, J. Kim, C. D. Chin, E. Munyazesa, P. Mugwaneza, A. J. Rai, V. Mugisha, A. R. Castro, D. Steinmiller, V. Linder, J. E. Justman, S. Nsanzimana and S. K. Sia, Sci. Transl. Med., 2015, 7, 273re271 Search PubMed.
- Y. Liu, D. Zhang, E. C. Alocilja and S. Chakrabartty, Nanoscale Res. Lett., 2010, 5, 533–538 CrossRef PubMed.
- S. J. Park, T. A. Taton and C. A. Mirkin, Science, 2002, 295, 1503–1506 CrossRef.
- N. Rafat, H. Zhang, J. Rudge, Y. N. Kim, S. P. Peddireddy, N. Das and A. Sarkar, Small, 2022, 18, 2203309 CrossRef.
- H. Liu, B. Liu, P. Huang, Y. Wu, F. Y. Wu and L. Ma, Mikrochim. Acta, 2020, 187, 551 CrossRef.
- J. Chen, A. A. Jackson, V. M. Rotello and S. R. Nugen, Small, 2016, 12, 2469–2475 CrossRef.
- N. P. Goode, M. Shires and A. M. Davison, Histochemistry, 1992, 98, 67–72 Search PubMed.
- K. Rahme, G. Minassian, E. Ghanem, M. Nakhl, R. El Hage, E. Souaid and J. Holmes, A Simple Synthesis of Polymer Coated Gold and Silver Nanoparticles in Water for Potential Use in Biomedical Applications, TechConnect Briefs, 2018, vol. 1, pp. 87–90, ISBN: 978-0-9975117-8-92018 Search PubMed.
- T. Schüler, A. Steinbrück, G. Festag, R. Möller and W. Fritzsche, J. Nanopart. Res., 2009, 11, 939–946 CrossRef.
- H. Schneidewind, T. Schüler, K. K. Strelau, K. Weber, D. Cialla, M. Diegel, R. Mattheis, A. Berger, R. Möller and J. Popp, Beilstein J. Nanotechnol., 2012, 3, 404–414 CrossRef.
- N. Rafat, L. Brewer, N. Das, D. J. Trivedi, B. K. Kaszala and A. Sarkar, ACS Sens., 2023, 8(2), 534–542 CrossRef.
- G. Zhou, S. Bergeron and D. Juncker, J. Proteome Res., 2015, 14, 1872–1879 CrossRef.
- B. Hvolbæk, T. V. W. Janssens, B. S. Clausen, H. Falsig, C. H. Christensen and J. K. Nørskov, Nano Today, 2007, 2, 14–18 CrossRef.
- L. Tai, G. Zhu, M. Yang, L. Cao, X. Xing, G. Yin, C. Chan, C. Qin, Z. Rao, X. Wang, F. Sun and Y. Zhu, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2112703118 CrossRef PubMed.
- Y. Cai, J. Zhang, T. Xiao, H. Peng, S. M. Sterling, R. M. Walsh, S. Rawson, S. Rits-Volloch and B. Chen, Science, 2020, 369, 1586–1592 CrossRef.
- B. Turoňová, M. Sikora, C. Schürmann, W. J. H. Hagen, S. Welsch, F. E. C. Blanc, S. von Bülow, M. Gecht, K. Bagola, C. Hörner, G. van Zandbergen, J. Landry, N. T. D. de Azevedo, S. Mosalaganti, A. Schwarz, R. Covino, M. D. Mühlebach, G. Hummer, J. Krijnse Locker and M. Beck, Science, 2020, 370, 203–208 CrossRef PubMed.
- A. C. Glavan, D. C. Christodouleas, B. Mosadegh, H. D. Yu, B. S. Smith, J. Lessing, M. T. Fernández-Abedul and G. M. Whitesides, Anal. Chem., 2014, 86, 11999–12007 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Links to Github for hardware design files/code. Comparison between current work and existing works on detection assays utilizing enzyme, metal deposition and gold (Table S1). Properties of AuNP stock solutions (Table S2); correlation between resistance reading obtained from the portable reader and that of a digital multimeter (Fig. S1); resistance of μIDE using 1 × 10 nm AuNP on 5 μm electrodes (Fig. S2); resistance of μIDE using different AuNP sizes on 10 μm electrode gap (Fig. S3); AuNP image analysis (Fig. S4). Procedure of microfabricating the EASyELISA chip (Fig. S5); close-up views of the portable reader (Fig. S6). See DOI: https://doi.org/10.1039/d4ay01657b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |