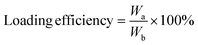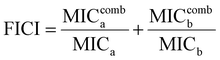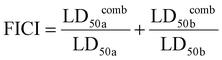Quaternary ammonium-tethered hyperbranched polyurea nanoassembly synergized with antibiotics for enhanced antimicrobial efficacy†
Yanwen
Feng
a,
Jiang
Bian
a,
Guoyi
Yu
a,
Pei
Zhao
*b and
Jun
Yue
 *a
*a
aSchool of Biomedical Engineering, Shenzhen Campus of Sun Yat-sen University, Shenzhen, Guangdong 518107, P. R. China. E-mail: yuejun3@mail.sysu.edu.cn
bLaboratory Animal Center, Shenzhen Campus of Sun Yat-sen University, Shenzhen, Guangdong 518107, P. R. China. E-mail: zhaopei55@mail.sysu.edu.cn
First published on 29th December 2023
Abstract
The effective transportation of antibiotics to bacteria embedded within a biofilm consisting of a dense matrix of extracellular polymeric substances is still a challenge in the treatment of bacterial biofilm associated infections. Here, we developed an antibiotic nanocarrier constructed from quaternary ammonium-tethered hyperbranched polyureas (HPUs-QA), which showed high loading capacity for a model antibiotic, rifampicin, and high efficacy in the transportation of rifampicin to biofilms. The rifampicin-loaded HPUs-QA nanoassembly (HPUs-Rif/QA) demonstrated a synergistic antimicrobial effect in killing planktonic bacteria and eradicating the corresponding biofilms. Compared to the treatment of bacteria-infected chronic wounds by either HPUs-QA or rifampicin alone, HPUs-Rif/QA showed superior efficacy in promoting wound healing by more effectively inhibiting bacteria colonization. This study highlights the potential of the HPUs-QA nanoassembly in synergistic action with antibiotics for the treatment of biofilm associated infections.
Introduction
Bacterial biofilms are stable and dense membranes of bacterial aggregates formed by bacteria adhering to each other or to the surface of a material, and embedding in their self-produced extracellular polymeric substances (EPS).1 Infections caused by bacterial biofilms have caused serious global health problems.2 Conventional antibiotic therapy remains one of the most important treatments for bacterial infections,3–5 however, conventional clinical antibiotics have limited effect on infections caused by biofilms, and the key reason is that they are unable to break through the barrier of biofilm EPS.6–8 Biofilm formation can prevent the penetration of antibiotics to reach pathogenic bacteria, as well as facilitating inter-colony signaling, helping bacteria to evade host immune attack, and enzymes in biofilms have a trapping and destructive effect on antibiotics.9–12 Moreover, the complicated microenvironment of biofilms, such as acidic conditions and high expression of enzymes, can cause antibiotic degradation or inactivation,13,14 leading to the use of a several times greater amount of antibiotic than required. However, treating biofilm-associated infections with excessive antibiotics can not only bring serious toxicity problems, but also induce the development of antibiotic resistance.15 In general, bacteria within a biofilm are 10–1000 times more resistant to antibiotics than planktonic bacteria,16–18 and horizontal gene transfer within the biofilm will allow the spread of clones of evolved drug-resistant genes to other strains.19–21To overcome the biofilm EPS barrier, various strategies have been developed, including the use of biofilm dispersants or EPS degradation agents for enhanced antibiotic penetration.22–29 Biofilm dispersants can be classified into two types based on their sites of action. The first type includes enzymes or surfactants that target the EPS, and consists mainly of polysaccharides, extracellular DNA, and proteins. Polysaccharides enzymes, DNases or proteases are used to degrade the EPS, while surfactants disrupt the interaction between the EPS components, leading to the dissolution of the biofilm.30 The second type comprises small molecular antibacterial agents that target bacteria within the biofilm. Examples of such agents include antimicrobial peptides, quaternary ammonium salts, antibacterial lipids, phenazines, quinolones, nitric oxide, and nitrogen oxide free radicals. These dispersants can both break down mature biofilms and exhibit a certain degree of bactericidal activity.31,32
Although using specific biofilm dispersants or EPS degradation agents has been proved as an effective way to improve antibiotic penetration, their roles are still limited on the surface of the biofilm and a high risk of side effects might occur if used without protection. In recent years, nanomaterials have been shown to have a great effect on antibiotic delivery for biofilm eradication and the killing of dormant bacteria.33–37 Additionally, these nanomaterials protect antibiotics from degradation and inactivation by enzymes in the EPS, thereby improving their bioavailability.38–40 Numerous studies have demonstrated that nanomaterials with unique physical and chemical properties offer new strategies for the treatment of bacterial diseases, enhancing the effectiveness of antibacterial drugs and overcoming bacterial resistance caused by biofilms. Currently, there is an increased focus on the biosafety of materials, leading to the need for the development of safer and more efficient advanced biosafety materials to prevent and treat pathogenic microbial infections.41,42 In this regard, polymers have garnered significant attention for their exceptional drug-loading capacity and biocompatibility.43–49 Among them, hyperbranched polymers, characterized by their numerous end groups and internal cavities, have emerged as highly promising nanocarriers.50,51
For certain antibiotics that target the interior of bacteria (e.g. rifampicin, vancomycin, and streptomycin), the thick bacterial cell wall serves as a secondary barrier against antibiotics,52–54 which is an often overlooked problem in conventional antibiotic delivery systems. The cell wall is crucial in maintaining bacterial cell integrity, and previous research has shown that cationic antibacterial polymers can adsorb, penetrate, and destroy bacterial cell walls, resulting in bacterial death.55 Recently, we developed a synthetic antimicrobial polymer based on quaternary ammonium (QA)-tethered hyperbranched polyureas (HPUs-QA), which demonstrated a much higher biofilm penetration capacity and an enhanced cell permeability via punching holes on bacterial cell walls, compared to linear QA-containing polymers.56 Aside from deep penetration and bactericidal abilities, HPUs-QA also offer advantages in terms of ease of synthesis and terminal modifications, and high drug loading efficiency due to their porous interior structure. By tuning the molecular weight of hyperbranched polyureas, HPUs-QA themselves may form a stable nanoassembly in water due to their amphiphilic character. Therefore, we hypothesize that HPUs-QA may not only act as a potential antibiotic carrier to achieve deeper biofilm penetration, but also be able to assist the transport of antibiotics through the bacterial cell wall, leading to a synergistic antimicrobial effect.
To prove our hypothesis and illustrate the potential of HPUs-QA in synergizing with antibiotics for enhanced antimicrobial efficacy, here we prepared an HPUs-QA nanoassembly encapsulated with rifampicin (Rif), an intra-bacterial targeting antibiotic model. The synergistic effects of HPUs-QA with rifampicin in killing planktonic bacteria and eradicating biofilms were systematically evaluated. Furthermore, the efficacy of the Rif-loaded HPUs-QA nanoassembly (HPUs-Rif/QA) in the treatment of bacteria-infected chronic wounds was studied to illustrate its potential applications in promoting the healing of chronic wounds.
Results and discussion
Preparation of the HPUs-Rif/QA nanoassembly
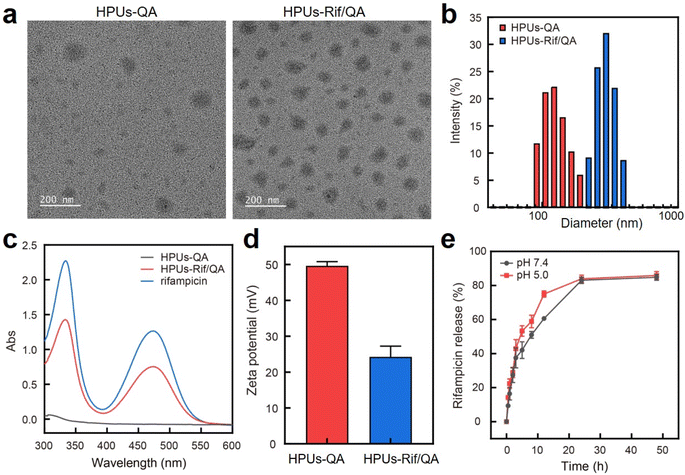
In this study, QA-tethered hyperbranched polyureas (HPUs-QA) were prepared via the one-pot synthesis of a polyurea framework and post-polymerization modifications (Scheme 1a) according to our recent publication.56 The molecular weight of the hyperbranched polyureas and the grafting ratio of QA on the basis of 1H NMR spectra (Fig. S1 and S2, in ESI†) were calculated as ∼8.7 kDa and 40%, respectively. The amphiphilic HPUs-QA was able to form a nanoassembly in water with an irregular spherical shape (Fig. 1a). Then, the sequential nanoprecipitation method57 (Scheme 1b) was utilized to load the hydrophobic antibiotic rifampicin (Rif) within the HPUs-QA nanoassembly (HPUs-Rif/QA). Briefly, HPUs-QA and rifampicin were dissolved in a mixture of organic solvents. During the addition of the antisolvent, the hydrophobic rifampicin was first precipitated to form particles, and then the addition of a large number of branched structures of HPUs-QA were able to encapsulate the rifampicin in the cavity to form a shell–core drug-carrying micelle structure. The rifampicin nanoprecipitate acts as a hydrophobic core, and quaternary ammonium compounds with short carbon chains form the shell layer of the micelles.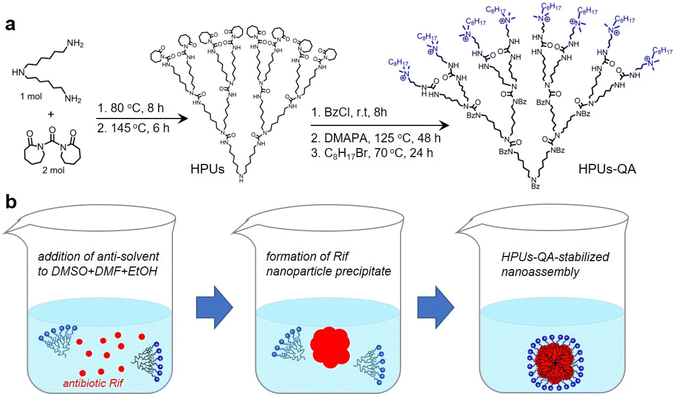 | ||
| Scheme 1 Schematic illustrations of the synthesis of HPUs-QA (a) and the preparation of the Rif-loaded HPUs-QA nanoassembly via a sequential nanoprecipitation method (b). | ||
After rifampicin loading, the morphology of the nanoassembly remained the same (Fig. 1a), but the overall diameter increased from ∼150 nm to ∼295 nm as assessed by dynamic light scattering (Fig. 1b). The size measured using DLS was larger than that observed in the TEM images, primarily because the TEM images provided the actual size of the sample in its dried state, while the size measured by DLS represented the hydrodynamic diameter in a hydrated state. Consequently, the nanoparticles exhibited a larger hydrodynamic volume due to the solvent effect in the hydrated state. This loading method increased the drug loading capacity and improved the encapsulation rate of the carrier. The characteristic absorption of rifampicin in the UV-vis spectra of the HPUs-QA nanoassembly clearly indicated the successful encapsulation of rifampicin (Fig. 1c). Through a calibration curve of rifampicin absorbance as a function of standard rifampicin concentration (Fig. S3, ESI†), the drug loading efficiency of the HPUs-Rif/QA was calculated as ∼31%, and the encapsulation efficiency reached as high as 93%. In addition, both the HPUs-QA and HPUs-Rif/QA showed a positive surface charge (Fig. 1d), facilitating their penetration into the biofilm.
Next, we examined the drug release profiles of HPUs-Rif/QA in PBS at pH 7.4 or pH 5.0 (Fig. 1e). The results showed that ∼80% of the rifampicin was released over 24 h incubation and there was a slightly increased release rate with an acidic buffer than under neutral conditions, while the overall amounts of drug released after 24 h were almost the same.
Synergistic effect of bacteria-growth inhibition between HPUs-QA and rifampicin
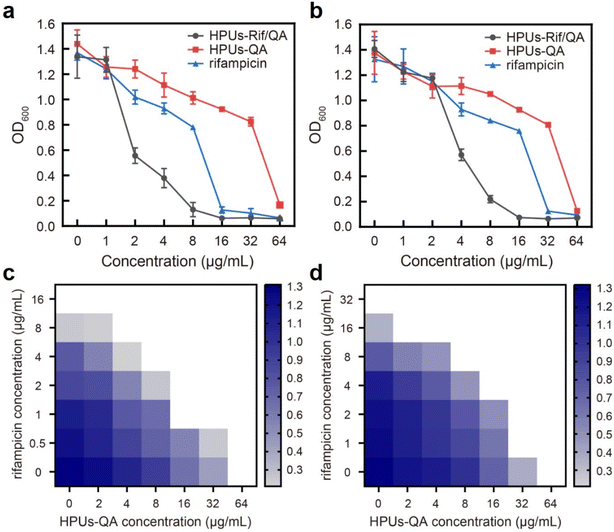
To investigate whether the HPUs-Rif/QA could exhibit an enhanced bacteria-growth inhibition effect, we determined the minimum inhibitory concentrations (MIC) of HPUs-Rif/QA, free rifampicin and HPUs-QA at a series of concentrations by measuring the optical density at 600 nm (OD600) of the bacterial suspension after overnight incubation. Two typical rifampicin-targeting bacterial strains, E. coli and P. aeruginosa, were chosen for the investigation. As shown in Fig. 2a and b, HPUs-Rif/QA exhibited a much higher bacteria-growth inhibition efficacy than either free rifampicin or HPUs-QA alone. Quantitatively, the MICs of free rifampicin, HPUs-QA and HPUs-Rif/QA were 16, 64 and 8 μg mL−1 for E. coli, while those for P. aeruginosa were 32, 64 and 16 μg mL−1, respectively.To determine whether there is a synergistic effect between HPUs-QA and rifampicin on bacteria-growth inhibition, we determined their fractional inhibitory concentration index (FICI) via the ‘chequerboard’ experiment,58 in which a two-dimensional array of serial concentrations of HPUs-QA and rifampicin was used as the basis for FICI calculation (Fig. 2c and d). Generally, synergy was defined by FICI ≤ 0.5 and antagonism by FICI > 4, while FICI between 0.5 and 1 was interpreted as addition, and FICI between 1 and 4 as indifference.59 The results showed that the FICI values between rifampicin and HPUs-QA were calculated as 0.25 and 0.375 for E. coli and P. aeruginosa, respectively, indicating that they had a synergistic growth-inhibition effect against both E. coli and P. aeruginosa. The synergistically increased bacteria-inhibition effect could be ascribed to the enhanced permeability of the bacterial cell wall by HPUs-QA that facilitated the entry of rifampicin, as indicated in our recent publication.56
HPUs-Rif/QA displayed a synergistic antibiofilm effect
A deep penetration of antibiotics into biofilms is a prerequisite for their high-efficiency intra-biofilm bacterial eradication. To investigate whether rifampicin is able to penetrate deeply within a biofilm with the assistance of HPUs-QA, we probed the distribution of rifampicin, either in its free form, or with it encapsulated in the HPUs-QA nanoassembly, by confocal fluorescence imaging. For visualization, rifampicin was pre-labeled with a fluorophore, fluorescein isothiocyanate (FITC, Fig. S4, ESI†). In our previous study, we found that hyperbranched polyureas had a tailorable nontraditional intrinsic fluorescence emitting from the blue to green gap spectral range,60 which is convenient for probing the localizations of HPUs-QA within the biofilms. As shown in Fig. 3, the locations of free RifFITC or the RifFITC-encapsulated HPUs-QA nanoassembly (HPUs-RifFITC/QA) within the biofilm were analyzed via Z-stack confocal fluorescence images, where green and red fluorescence indicate SYTO 59-stained biofilm and FITC channels, respectively. Compared to the free RifFITC, which was distributed only on the surface of the biofilm, HPUs-RifFITC/QA displayed a much deeper penetration within the E. coli biofilm after just a short time of incubation (0.5 h). In addition, most of the fluorescence signals of RifFITC and HPUs-QA were overlapping (Fig. S5, ESI†), indicating that HPUs-QA and RifFITC formed a nanocomplex and penetrated together into the biofilm.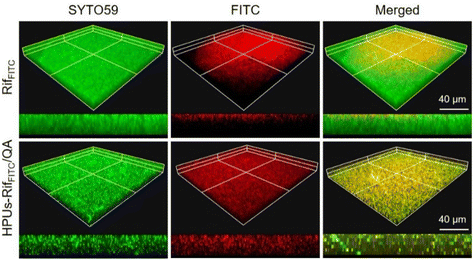 | ||
| Fig. 3 Confocal fluorescence microscopy image of the E. coli biofilm post-treatment with FITC-labeled rifampicin (RifFITC) or the RifFITC-loaded HPUs-QA nanoassembly. | ||
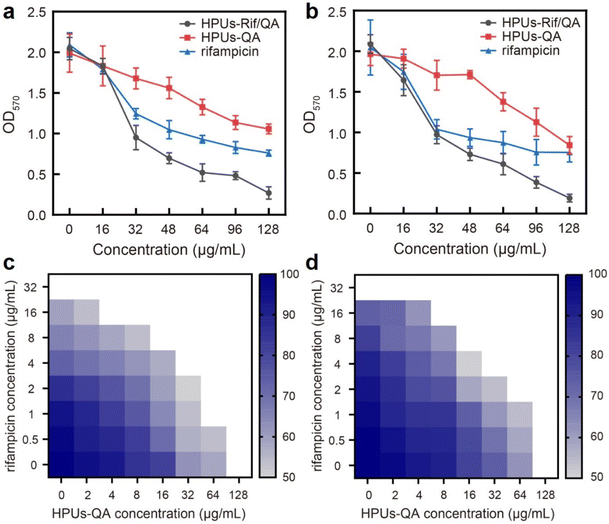
Next, to investigate whether the enhanced penetration of HPUs-Rif/QA could lead to an enhanced antibiofilm efficacy, we measured the residual biomass of E. coli or P. aeruginosa biofilms post-treatment with a series of concentrations of HPUs-Rif/QA, HPUs-QA or free rifampicin. As shown in Fig. 4a and b, the biomasses all decreased with the increase of sample concentration, and when the concentration was higher than 32 μg mL−1, HPUs-Rif/QA showed a much higher rate of biofilm clearance than the other two formulations. For quantitative analysis, the half lethal dose (LD50) was defined as the material concentration required to remove 50% of the biofilm. Results showed that the LD50 values of free rifampicin, HPUs-QA and HPUs-Rif/QA for E. coli biofilms were 64 μg mL−1, 128 μg mL−1 and 32 μg mL−1, respectively. For P. aeruginosa, HPUs-Rif/QA showed a similar trend of biofilm clearance efficacy.
To evaluate whether there is a synergistic antibiofilm effect between HPUs-QA and rifampicin, we determined the FICI via the ‘chequerboard’ experiment (Fig. 4c and d), where the LD50 values of combined treatments of biofilms with HPUs-QA and rifampicin under a two-dimensional array of serial concentrations were obtained for calculation. The results showed that for both E. coli and P. aeruginosa biofilms, the FICI was calculated as 0.375, an indication of a synergistic antibiofilm effect.
HPUs-Rif/QA exhibited higher efficacy in the treatment of bacteria-infected chronic wounds
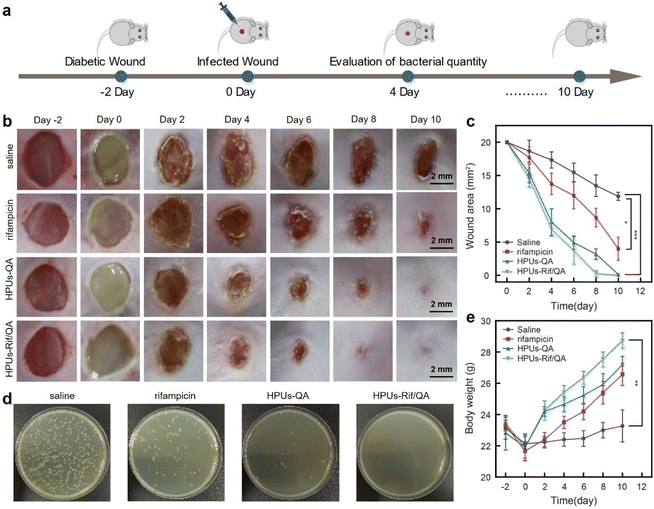
To investigate whether the synergistic antibiofilm effect of HPUs-QA and rifampicin could achieve better therapeutic effects against bacterial infections in vivo, we constructed a skin wound infection model on the backs of diabetic mice, followed by administering and recording the wound area and body weight changes of each mouse every other day (Fig. 5a). Two days after infecting the wounds with methicillin-resistant Staphylococcus aureus, a slimy gel-like shiny pimple on the surface of the wound bed, indicative of significant wound biofilm and inflammation, appeared for all mice (Fig. 5b). Subsequently, HPUs-Rif/QA solution at a biosafe concentration (128 μg mL−1, Fig. S6, in ESI†), rifampicin or HPUs-QA at an equivalent concentration to HPUs-Rif/QA was added dropwise to cover the whole wound area for evaluation. On day 4, the epidermis of the affected area of the mice in the HPUs-QA and HPUs-Rif/QA groups tended to dry out, and the tissues around the edge of wounds became less reddish, suggesting a relieving of the inflammatory response. On day 6, a significant difference between the unhealed wound areas of the different groups appeared, with the order rifampicin > HPUs-QA > HPUs-Rif/QA (Fig. 5c). On day 10, a complete closure of the wounds from the HPUs-Rif/QA group was observed, while there were still 8.7 mm2 and 3.2 mm2 wounds left unhealed for the rifampicin and HPUs-QA groups, which was probably due to their lower efficiency in biofilm clearance.To quantitatively analyze the residual bacteria at the wound site, one mouse was randomly selected from each group on day 4 and a swab of the wound from each group was collected, followed by quantification of viable bacteria using the agar plating method. As shown in Fig. 5d, no visible bacterial colonies could be found on the surface of the culturing plates from the HPUs-Rif/QA group. However, there were ∼5.4% and ∼26.7% of colonies from the HPUs-QA and rifampicin groups respectively, in proportion to those from the saline group. These results indicated that HPUs-Rif/QA had a higher efficiency in promoting the healing of bacteria-infected wounds than either free rifampicin or HPUs-QA via inhibiting the bacteria colonization.
Next, the therapeutic effect of HPUs-Rif/QA on the bacteria-infected wounds was further verified by analyzing the body weight changes of the mice (Fig. 5e). In the first two days following the bacteria challenge, all the mice experienced a rapid decrease in body weight, probably due to the post-surgical constitutional weakness and inappetence. Subsequently, the mice gradually began to gain weight as the treatment progressed, indicating restoration of their physical condition and return to a normal diet. After 2 days of treatment, the body weights of mice from the HPUs-Rif/QA and HPUs-QA groups had recovered to their preoperational level, and the trend of weight gain was then maintained, while the weight gain rate for mice from the HPUs-Rif/QA group was always faster than that for mice from the HPUs-QA group. In contrast, the body weights in the rifampicin and saline groups increased very slowly, which could be due to the infection-induced anorexia. The changes in body weight reflected that HPUs-Rif/QA could accelerate the health recovery of the mice from the infection state.
Finally, wound skin tissues were collected and stained with hematoxylin–eosin (H&E, Fig. 6a) as well as Masson's trichrome stain (Fig. 6b) to evaluate wound healing efficacy and the quality of regenerated skin from a histological perspective. For better comparison, healthy skin tissues were also collected and stained under the same conditions, where there were no inflammatory cells, but with intact hair follicles, plenty of collagen fibers and clear dermal boundaries. Compared to those infected wounds treated either with saline or rifampicin, where there was still a large quantity of inflammatory cells but only a small number of collagen fibers, the wound sites in the HPUs-QA group displayed a significant reduction in inflammatory cells, more collagen fibers (blue color in Fig. 6b), and a small number of follicular structures. More importantly, for HPUs-Rif/QA treatment, nearly no inflammatory cells could be found around the wounds and the highest density of collagen fibers and healthy hair follicles was clearly seen, almost approaching the dermal structure of normal skin, indicating the substantial completion of the proliferative phase. Taken all together, the combination of HPUs-QA with rifampicin was superior to their individual treatments in promoting wound healing.
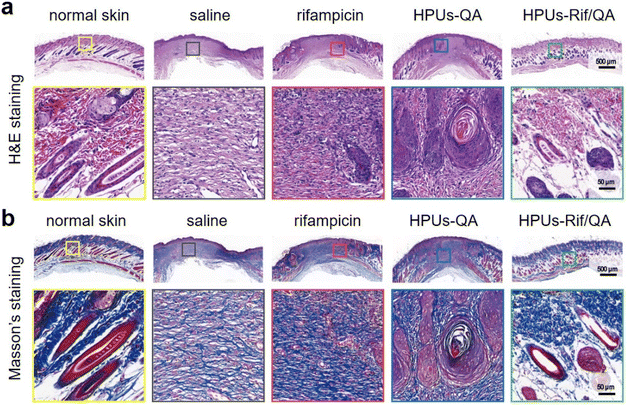 | ||
| Fig. 6 Photomicrographs of hematoxylin and eosin (H&E) (a), and Masson's (b) stained skin lesions, where red and blue indicate muscle fibers and collagen fibers, respectively. | ||
Conclusion
An HPUs-QA nanoassembly based on quaternary ammonium-tethered hyperbranched polyureas was prepared, and showed high efficacy in transporting antibiotic rifampicin to the biofilm. The rifampicin-loaded HPUs-QA nanoassembly demonstrated promising synergistic effects in killing planktonic bacteria and eradicating biofilms, and showed superior efficacy in promoting the healing of bacteria-infected wounds compared to the individual treatments, by inhibiting bacteria colonization.Experimental
Materials and bacterial strains
All organic solvents used in the experiments were employed without any prior treatment. Bis(hexamethylenetriamine) (BHTA), 3-dimethylaminopropylamine (DMAPA), benzoyl chloride, triethylamine, dimethylformamide (DMF), ethyl ether, trichloromethane (CHCl3), dichloromethane (DCM), dimethyl sulfoxide (DMSO), methanol, and ethanol were purchased from Sigma-Aldrich. Carbonyl biscaprolactam (CBC) was obtained from Actu-All Chemicals, and 1-bromooctane was sourced from Sahn Chemistry. Anhydrous sodium sulfate, rifampicin and crystal violet were procured from McLean, while sodium chloride was acquired from Tianjin Zhiyuan Chemical Reagent Co., Ltd. Deuterated dimethyl sulfoxide (DMSO-d6) and deuterochloroform (CDCl3) were purchased from Qingdao Tenglong Microwave Technology Co., Ltd. Tryptone soybean broth (TSB) powder was sourced from Solarbio Life Science, and agar power was obtained from Guangzhou Huankai Microbial Technology Co., Ltd. SYTO59 and a bacterial Live/Dead BacLight Bacterial Viability Kit were purchased from Thermo Scientific. Escherichia coli (E. coli, ATCC25922), Pseudomonas aeruginosa (P. aeruginosa, ATCC27853), and methicillin-resistant Staphylococcus aureus (MRSA, ATCC43300) were purchased from Guangzhou Scissorhands Gene Technology Co., Ltd and cultured in the TSB medium at 37 °C. Finally, all experimental mice (BALB/c) were purchased from Sun Yat-Sen University Laboratory Animal Center, Guangzhou, China. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Sun Yat-sen University and approved by the Animal Ethics Committee of Sun Yat-sen University.Preparation and characterization of HPUs-QA and HPUs-Rif/QA
First, 3.24 g of BHTA (15 mmol) and 7.584 g of CBC (30 mmol) were dissolved in 100 mL of DMF. After purging the mixture with argon to remove oxygen, it was stirred at 85 °C for 8 h under an argon atmosphere. Subsequently, the temperature was raised to 145 °C and the reaction was allowed to continue for an additional 6 h. Most of the DMF was then removed under reduced pressure. The remaining mixture was then dissolved in CHCl3 and subjected to washing with saturated aqueous sodium chloride five times to eliminate impurities and residual DMF. The organic layer was collected, dried over anhydrous sodium sulfate, and filtered. The solvent was then removed under reduced pressure. Finally, the obtained product was further purified by precipitating in a large amount of ethyl ether and dried under vacuum, resulting in viscous liquid HPUs. Next, 1.5 g of HPUs (0.15 mmol), 50 mg of benzoyl chloride (0.35 mmol), and 75 mg of triethylamine (0.7 mmol) were dissolved in 20 mL of DCM and stirred at room temperature for 8 h. After removing the solvent under reduced pressure, the product was further purified by precipitating in a large amount of ethyl ether and dried under vacuum, leading to the formation of hyperbranched polyurea with the first end of the secondary amine substituted. Finally, 1.5 g of product obtained from the previous reaction was reacted with an excess amount of DMAPA in 20 mL of DMF at 125 °C for 48 h. Then, the reaction temperature was lowered to 70 °C, and 1-bromooctane was added to the reaction system, allowing the reaction to continue for 24 h. The solvent was removed under reduced pressure, and the resulting product was further purified by precipitating in a large amount of ethyl ether and dried under vacuum to obtain HPUs-QA. 1H NMR characterizations of the above products at each step were performed using an Agilent VNMRS600 spectrometer with DMSO-d6 or CDCl3 as the solvent.To prepare HPUs-Rif/QA, a mixture of three solvents (DMSO, DMF, and ethanol) was prepared in a ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. Then, 30 mg of rifampicin and 15 mg of HPUs-QA were weighed and dissolved in 10 mL of the solvent mixture to form a solution. Next, 190 mL of ultrapure water was added to the solution as an antisolvent and vigorously stirred. The resulting mixture was then transferred to a dialysis bag (Mw 3500) and dialyzed with ultrapure water for 48 h before freeze-drying. HPUs-QA were prepared by the same method.
4. Then, 30 mg of rifampicin and 15 mg of HPUs-QA were weighed and dissolved in 10 mL of the solvent mixture to form a solution. Next, 190 mL of ultrapure water was added to the solution as an antisolvent and vigorously stirred. The resulting mixture was then transferred to a dialysis bag (Mw 3500) and dialyzed with ultrapure water for 48 h before freeze-drying. HPUs-QA were prepared by the same method.
The particle size and zeta potential of the HPUs-QA and HPUs-Rif/QA were determined using a Malvern Laser Particle Sizer (Malvern, UK). To confirm the successful loading of rifampicin by the HPUs-QA, the UV-visible spectra of rifampicin, HPUs-QA and HPUs-Rif/QA were measured using a UV-visible spectrophotometer (Shimadzu, Japan). The morphologies of the HPUs-QA and HPUs-Rif/QA were observed using transmission electron microscopy (JEM F200, Japan) after negative-staining with phosphotungstic acid.
Determination of drug loading and encapsulation efficiency
First, 10 mg of rifampicin were dissolved in dimethyl sulfoxide (DMSO) to prepare a 1 mg mL−1 solution, which was then diluted into a series of standard solutions with concentrations of 30 μg mL−1, 25 μg mL−1, 20 μg mL−1, 15 μg mL−1, 10 μg mL−1, and 5 μg mL−1. The absorbance of these solutions was measured at 339 nm using a UV-visible spectrophotometer (Beckman Coulter, USA). A standard curve of rifampicin was generated by linear fitting the absorbance values to the corresponding drug concentrations (Fig. S1, ESI†):| Abs = 0.0338 × C + 0.0103 (R2 = 0.9992, n = 6) |
To assess the drug loading content and encapsulation efficiency of HPUs-Rif/QA, 9 mL of DMSO was added to 1 mL of aqueous polymer micelles to disrupt the nanomicelles loaded with the drug, thus releasing the rifampicin. Subsequently, the absorbance at 339 nm was measured and used to determine the loading of HPUs-QA by substituting it into the standard curve.
where Wa is the weight of rifampicin content in the micelles, Wb is the weight of the nanocomplexes, and Wc is the total weight of rifampicin administered.
Drug release assay
To assess the in vitro drug release of HPUs-Rif/QA, 5 mL of the sample was loaded into a dialysis bag with a molecular weight cutoff of 3500 and immersed in 150 mL of release media at two different pH values (pH 7.4 PBS and pH 5.0 PBS). Subsequently, the rifampicin release assay was conducted in a thermostatic shaking incubator at 37 °C. At designated time intervals (0.5 h, 1 h, 2 h, 3 h, 5 h, 8 h, 12 h, 24 h, and 48 h), 3 mL of release medium was withdrawn and replaced with an equal volume of fresh release medium. The withdrawn release medium at each time point was then analyzed for absorbance at 339 nm. The concentration of rifampicin was determined from the standard curve to determine the total amount of rifampicin released.Assessment of synergistic antibacterial activity
To evaluate the antibacterial activity of different agents (rifampicin, HPUs-QA, HPUs-Rif/QA), their minimum inhibitory concentrations (MIC) against E. coli and P. aeruginosa were determined. The bacteria were inoculated into fresh TSB medium and incubated for 12 hours at 37 °C in a shaking incubator. Afterwards, they were diluted with fresh TSB medium to obtained a bacterial solution with a concentration of 1 × 106 colony forming units per milliliter (CFU per mL). The antibacterial agents were prepared in varying concentrations and mixed with the bacterial solution. The mixture was transferred into 96-well plates and incubated at 37 °C in a shaking incubator. Following a 12-hour incubation period, the optical density at 600 nm (OD600) of the samples was measured using a microplate reader (BioTek Synergy 4, USA) to determine the MICs.To assess the synergistic antibacterial activity between rifampicin and HPUs-QA, two-fold serial dilutions of HPUs-QA and rifampicin were prepared in advance. Subsequently, 100 μL of HPUs-QA (x-axis direction) and 100 μL rifampicin (y-axis direction) were mixed to create a 7 × 7 matrix in 96-well plates. Each well contained a final volume of 200 μL and 1 × 106 CFU per mL bacteria. After a 12-hour incubation at 37 °C in a shaker incubator, the absorbance at 600 nm was measured using a microplate reader (BioTek Synergy 4, USA). The antibacterial fractional inhibitory concentration index (FICI) between HPUs-QA and rifampicin was calculated using the following equation:
Biofilm permeability assessments
For biofilm permeability assessments, 100 μL of the bacterial suspension at a concentration of 1 × 107 CFU per mL was inoculated into each well of a 96-well plate. The plate was then incubated at 37 °C for 1 hour under stationary conditions to allow initial adhesion of the bacteria to the bottom of the wells. Then, the suspension was carefully removed. Next, 200 μL of TSB was added to each well and incubated statically at 37 °C for 2 days, with fresh TSB replacement at the 24-hour mark. Once the biofilm had formed, the medium was discarded, and the plates were washed with PBS to remove any planktonic bacteria. The resulting biofilm can be used for subsequent testing experiments.To visualize the colocalization of HPUs-QA with rifampicin in the biofilms, we prepared FITC-labeled rifampicin (RifFITC). A mixture containing 30 mg of rifampicin and 10 mg of FITC was dissolved in 10 mL of DMSO and stirred in darkness for 8 hours. Subsequently, the mixture was dialyzed with ultrapure water for 24 hours before undergoing freeze-drying. The RifFITC compound was then subjected to 1H NMR characterization using an Agilent VNMRS600 spectrometer, with DMSO-d6 serving as the solvent. The RifFITC compound was encapsulated in HPUs-QA, and the resulting complex was co-incubated with mature biofilms. Fluorescence images were captured using a confocal laser scanning microscope (Olympus, Japan).
To compare the biofilm penetration capability of free rifampicin with HPUs-Rif/QA, the biofilms were separately incubated with RifFITC and HPUs-RifFITC/QA. Subsequently, the biofilms were stained with SYTO59, and after washing away any unbound dye with PBS, the biofilms were observed and photographed along the z-axis direction using confocal laser scanning microscopy.
Synergistic antibiofilm assessments
Similarly, to evaluate whether there is a synergistic antibiofilm effect between HPUs-QA and rifampicin, 100 μL of HPUs-QA and 100 μL of rifampicin with different concentrations were mixed to create an 8 × 8 matrix in 96-well plates containing pre-formed biofilms. These solutions were then incubated with pre-formed biofilm for 24 hours. Residual biomass in the biofilms was measured using crystal violet staining. After carefully discarding the medium, the biofilm was washed twice with PBS and fixed with 100 μL of methanol. After 20 minutes, the methanol was discarded and the biofilm was left to dry at room temperature. Subsequently, 100 μL of a 1% crystal violet solution was added to each well and the biofilm was stained for 20 minutes. The crystal violet solution was then removed, the biofilm was washed with PBS, and 100 μL of 95% ethanol was added to dissolve the biofilm. After 40 minutes, the supernatant was transferred to a new 96-well plate. Finally, the optical density at 570 nm was measured using a microplate reader (BioTek Synergy 4, USA). The antibiofilm FICI between HPUs-QA and rifampicin was calculated based on the following equation:where LD50
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a and LD50
a and LD50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b represent the concentrations of the corresponding components that inhibit 50% biofilm formation when used alone, while LD50
b represent the concentrations of the corresponding components that inhibit 50% biofilm formation when used alone, while LD50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) comba and LD50
comba and LD50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) combb represent the concentrations of the corresponding components that inhibit 50% biofilm formation when the two components are used together.
combb represent the concentrations of the corresponding components that inhibit 50% biofilm formation when the two components are used together.
In vivo antibiofilm infection assay
The purpose of this section is to describe the methodology and procedures for the in vivo antibiofilm infection assay using bacterial biofilm-infected diabetic chronic wound models.Male BALB/c mice (5–6 weeks old, obtained from Sun Yat-Sen University Laboratory Animal Center, Guangzhou, China) were used for the experiments. Streptozocin dissolved in PBS at a concentration of 10 mg mL−1 was injected intraperitoneally at a dose of 50 mg kg−1 after a 12-hour fast. The mice were then fed normal food, and the injection was continued for 5 days. Blood glucose levels of the mice were monitored, and mice with blood glucose higher than 11.1 mM were considered as the type I diabetic mice model. Once the mice had been anaesthetised, their backs were shaved and sterilised with alcohol. A 5 mm wound was created on each mouse's back, and then 10 μL of MRSA bacterial suspension (1 × 108 CFU per mL) was dropped onto the wound. The bacteria were allowed to colonize and grow for 48 hours, during which time the wound was protected with sterile gauze.
Next, the mice with biofilm-infected wounds were randomly divided into four groups, with 5 mice in each group. The control group received saline treatment, the positive control group received rifampicin and HPUs-QA treatment, and the experimental group received HPUs-Rif/QA treatment. Every 24 hours, HPUs-Rif/QA solution at a biosafe concentration (128 μg mL−1, 20 μL), rifampicin or HPUs-QA at an equivalent concentration to HPUs-Rif/QA and saline were applied to the infected wounds of each respective group. Measurements and assessments were conducted every 48 hours. The wound areas and the weights of the mice were measured, and photographs were taken to record any changes in the wounds. If there was a significant difference in wound infection and size between the experimental and control groups, one mouse was sacrificed in each group for further analysis.
Wound tissue was dissected and placed in sterile PBS. Bacteria on each mouse wound were evaluated using the TSB-agar plate dilution method. After wounds in the experimental group were almost healed, all the mice were sacrificed, and all wounds were collected for histological analysis using H&E and Masson's staining. Overall, these procedures allowed for the in vivo assessment of the efficacy of various treatments in reducing biofilm infection in diabetic chronic wounds.
Statistical analysis
The data was presented as mean ± S.E.M., with a sample size of n = 3, for the particle size and zeta potential test, drug release test, anti-suspension bacteria test, antibiofilm test, and treatment of chronic wound test. The unpaired two-tailed t-test was employed to determine the significant difference (p-value). A p-value of less than 0.05 was considered to indicate statistically significant differences. GraphPad Prism 8 software was utilized for the statistical analysis.Conflicts of interest
There are no financial conflicts of interest to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (52273163, 52103103), Science and Technology Planning Project of Guangdong Province (2023A0505050119), and Shenzhen Science and Technology Program (JCYJ20230807111200002).References
- H.-C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Biofilms: an emergent form of bacterial life, Nat. Rev. Microbiol., 2016, 14(9), 563–575 CrossRef CAS PubMed.
- J. L. Del Pozo, Biofilm-related disease, Expert Rev. Anti-Infect. Ther., 2018, 16(1), 51–65 CrossRef CAS PubMed.
- H. Wu, C. Moser, H.-Z. Wang, N. Høiby and Z.-J. Song, Strategies for combating bacterial biofilm infections, Int. J. Oral Sci., 2015, 7(1), 1–7 CrossRef CAS PubMed.
- L. Radlinski and B. P. Conlon, Antibiotic efficacy in the complex infection environment, Curr. Opin. Microbiol., 2018, 42, 19–24 CrossRef CAS PubMed.
- Q. Xu, X. Hu and Y. Wang, Alternatives to Conventional Antibiotic Therapy: Potential Therapeutic Strategies of Combating Antimicrobial-Resistance and Biofilm-Related Infections, Mol. Biotechnol., 2021, 63(12), 1103–1124 CrossRef CAS PubMed.
- Y. Liu, L. Q. Shi, L. Z. Su, H. C. van der Mei, P. C. Jutte, Y. J. Ren and H. J. Busscher, Nanotechnology-based antimicrobials and delivery systems for biofilm-infection control, Chem. Soc. Rev., 2019, 48(2), 428–446 RSC.
- S. E. Birk, A. Boisen and L. H. Nielsen, Polymeric nano- and microparticulate drug delivery systems for treatment of biofilms, Adv. Drug Delivery Rev., 2021, 174, 30–52 CrossRef CAS PubMed.
- W. Yin, Y. Wang, L. Liu and J. He, Biofilms: The Microbial “Protective Clothing” in Extreme Environments, Int. J. Mol. Sci., 2019, 20(14), 3423 CrossRef CAS PubMed.
- O. Ciofu, E. Rojo-Molinero, M. D. Macià and A. Oliver, Antibiotic treatment of biofilm infections, APMIS, 2017, 125(4), 304–319 CrossRef PubMed.
- J. L. del Pozo and R. Patel, The Challenge of Treating Biofilm-associated Bacterial Infections, Clin. Pharmacol. Ther., 2007, 82(2), 204–209 CrossRef CAS PubMed.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Bacterial biofilms: from the Natural environment to infectious diseases, Nat. Rev. Microbiol., 2004, 2(2), 95–108 CrossRef CAS PubMed.
- K. Lewis, Persister cells, dormancy and infectious disease, Nat. Rev. Microbiol., 2007, 5(1), 48–56 CrossRef CAS PubMed.
- T. Wang, E. J. Cornel, C. Li and J. Du, Drug delivery approaches for enhanced antibiofilm therapy, J. Controlled Release, 2023, 353, 350–365 CrossRef CAS PubMed.
- C. W. Hall and T.-F. Mah, Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria, FEMS Microbiol. Rev., 2017, 41(3), 276–301 CrossRef CAS PubMed.
- B. Malaekeh-Nikouei, B. S. Fazly Bazzaz, E. Mirhadi, A. S. Tajani and B. Khameneh, The role of nanotechnology in combating biofilm-based antibiotic resistance, J. Drug Delivery Sci. Technol., 2020, 60, 101880 CrossRef CAS.
- A. Vishwakarma, F. Dang, A. Ferrell, H. A. Barton and A. Joy, Peptidomimetic Polyurethanes Inhibit Bacterial Biofilm Formation and Disrupt Surface Established Biofilms, J. Am. Chem. Soc., 2021, 143(25), 9440–9449 CrossRef CAS PubMed.
- Y. Gao, J. Wang, M. Chai, X. Li, Y. Deng, Q. Jin and J. Ji, Size and Charge Adaptive Clustered Nanoparticles Targeting the Biofilm Microenvironment for Chronic Lung Infection Management, ACS Nano, 2020, 14(5), 5686–5699 CrossRef CAS PubMed.
- F. A. Al-Wrafy, A. A. Al-Gheethi, S. K. Ponnusamy, E. A. Noman and S. A. Fattah, Nanoparticles approach to eradicate bacterial biofilm-related infections: A critical review, Chemosphere, 2022, 288, 132603 CrossRef CAS PubMed.
- Z. Wang, X. Liu, Y. Duan and Y. Huang, Infection microenvironment-related antibacterial nanotherapeutic strategies, Biomaterials, 2022, 280, 121249 CrossRef CAS PubMed.
- D. Sharma, L. Misba and A. U. Khan, Antibiotics versus biofilm: an emerging battleground in microbial communities, Antimicrob Resist Infect Control, 2019, 8(1), 76 CrossRef PubMed.
- Z. Pang, R. Raudonis, B. R. Glick, T.-J. Lin and Z. Cheng, Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies, Biotechnol. Adv., 2019, 37(1), 177–192 CrossRef CAS PubMed.
- A. G. Gomez and Z. Hosseinidoust, Liposomes for Antibiotic Encapsulation and Delivery, ACS Infect. Dis., 2020, 6(5), 896–908 CrossRef PubMed.
- H. Le, C. Arnoult, E. De, D. Schapman, L. Galas, D. L. Cerf and C. Karakasyan, Antibody-Conjugated Nanocarriers for Targeted Antibiotic Delivery: Application in the Treatment of Bacterial Biofilms, Biomacromolecules, 2021, 22(4), 1639–1653 CrossRef CAS.
- C. A. Omolo, D. Hassan, N. Devnarain, Y. Jaglal, C. Mocktar, R. S. Kalhapure, M. Jadhav and T. Govender, Formulation of pH responsive multilamellar vesicles for targeted delivery of hydrophilic antibiotics, Colloids Surf., B, 2021, 207, 112043 CrossRef CAS PubMed.
- Y. Wang, Q. Yuan, W. Feng, W. D. Pu, J. Ding, H. J. Zhang, X. Y. Li, B. Yang, Q. Dai, L. Cheng, J. Y. Wang, F. J. Sun and D. L. Zhang, Targeted delivery of antibiotics to the infected pulmonary tissues using ROS-responsive nanoparticles, J. Nanobiotechnol., 2019, 17(1), 103 CrossRef.
- Y. Q. Rao, Y. Y. Sun, P. Y. Li, M. Xu, X. A. Chen, Y. L. Wang, Y. Chen, X. Deng, S. H. Yu and H. Y. Hu, Hypoxia-sensitive adjuvant loaded liposomes enhance the antimicrobial activity of azithromycin via phospholipase-triggered releasing for Pseudomonas aeruginosa biofilms eradication, Int. J. Pharm., 2022, 623, 121910 CrossRef CAS PubMed.
- S. Tian, H. C. van der Mei, Y. Ren, H. J. Busscher and L. Shi, Co-Delivery of an Amyloid-Disassembling Polyphenol Cross-Linked in a Micellar Shell with Core-Loaded Antibiotics for Balanced Biofilm Dispersal and Killing, Adv. Funct. Mater., 2022, 32(51), 2209185 CrossRef CAS.
- H.-M. Ren, L. Han, L. Zhang, Y.-Q. Zhao, C. Lei, Z. Xiu, N. Zhao, B. Yu, F. Zhou, S. Duan and F.-J. Xu, Inhalable responsive polysaccharide-based antibiotic delivery nanoparticles to overcome mucus barrier for lung infection treatment, Nano Today, 2022, 44, 101489 CrossRef CAS.
- Y. N. Albayaty, N. Thomas, M. Jambhrunkar, M. Al-Hawwas, A. Kral, C. R. Thorn and C. A. Prestidge, Enzyme responsive copolymer micelles enhance the anti-biofilm efficacy of the antiseptic chlorhexidine, Int. J. Pharm., 2019, 566, 329–341 CrossRef CAS PubMed.
- K. P. Rumbaugh and K. Sauer, Biofilm dispersion, Nat. Rev. Microbiol., 2020, 18(10), 571–586 CrossRef CAS PubMed.
- J. L. Del Pozo, Novel treatment dynamics for biofilm-related infections, Expert Rev. Anti-Infect. Ther., 2021, 19(11), 1443–1456 CrossRef CAS PubMed.
- A. D. Verderosa, M. Totsika and K. E. Fairfull-Smith, Bacterial Biofilm Eradication Agents: A Current Review, Front. Chem., 2019, 7, 824 CrossRef CAS PubMed.
- Z. Geng, Z. Cao and J. Liu, Recent advances in targeted antibacterial therapy basing on nanomaterials, Exploration, 2023, 3(1), 20210117 CrossRef PubMed.
- D. He, T. Yang, W. Qian, C. Qi, L. Mao, X. Yu, H. Zhu, G. Luo and J. Deng, Combined photothermal and antibiotic therapy for bacterial infection via acidity-sensitive nanocarriers with enhanced antimicrobial performance, Appl. Mater. Today, 2018, 12, 415–429 CrossRef.
- M. Liu, D. He, T. Yang, W. Liu, L. Mao, Y. Zhu, J. Wu, G. Luo and J. Deng, An efficient antimicrobial depot for infectious site-targeted chemo-photothermal therapy, J. Nanobiotechnol., 2018, 16(1), 23 CrossRef PubMed.
- X. Huang, T. Li, X. C. Zhang, J. Deng and X. T. Yin, Bimetallic palladium@copper nanoparticles: Lethal effect on the Gram-negative bacterium Pseudomonas aeruginosa, Mater. Sci. Eng., C, 2021, 129, 112392 CrossRef CAS.
- X. Jin, Z. Ou, G. Zhang, R. Shi, J. Yang, W. Liu, G. Luo, J. Deng and W. Wang, A CO-mediated photothermal therapy to kill drug-resistant bacteria and minimize thermal injury for infected diabetic wound healing, Biomater. Sci., 2023, 11(18), 6236–6251 RSC.
- S. Fulaz, S. Vitale, L. Quinn and E. Casey, Nanoparticle–Biofilm Interactions: The Role of the EPS Matrix, Trends Microbiol., 2019, 27(11), 915–926 CrossRef CAS PubMed.
- K. Forier, K. Raemdonck, S. C. De Smedt, J. Demeester, T. Coenye and K. Braeckmans, Lipid and polymer nanoparticles for drug delivery to bacterial biofilms, J. Controlled Release, 2014, 190, 607–623 CrossRef CAS PubMed.
- M. Imran, S. K. Jha, N. Hasan, A. Insaf, J. Shrestha, J. Shrestha, H. P. Devkota, S. Khan, N. Panth, M. E. Warkiani, K. Dua, P. M. Hansbro, K. R. Paudel and Y. Mohammed, Overcoming Multidrug Resistance of Antibiotics via Nanodelivery Systems, Pharmaceutics, 2022, 14(3), 586 CrossRef CAS PubMed.
- J. Ding, H. Xiao and X. Chen, Advanced biosafety materials for prevention and theranostics of biosafety issues, Biosaf. Health, 2022, 4(2), 59–60 CrossRef PubMed.
- Y. Yu, J. Ding, Y. Zhou, H. Xiao and G. Wu, Biosafety chemistry and biosafety materials: A new perspective to solve biosafety problems, Biosaf. Health, 2022, 4(1), 15–22 CrossRef PubMed.
- F. Huang, X. Cai, X. Hou, Y. Zhang, J. Liu, L. Yang, Y. Liu and J. Liu, A dynamic covalent polymeric antimicrobial for conquering drug-resistant bacterial infection, Exploration, 2022, 2(5), 20210145 CrossRef PubMed.
- K. Deng, Y. Li, X. Liang, C. Shen, Z. Zeng and X. Xu, Virus-inspired nanoparticles as versatile antibacterial carriers for antibiotic delivery against Gram-negative and Gram-positive bacteria, Chin. Chem. Lett., 2022, 33(3), 1619–1622 CrossRef CAS.
- Y.-H. Lao, M. Li, M. A. Gao, D. Shao, C.-W. Chi, D. Huang, S. Chakraborty, T.-C. Ho, W. Jiang, H.-X. Wang, S. Wang and K. W. Leong, HPV Oncogene Manipulation Using Nonvirally Delivered CRISPR/Cas9 or Natronobacterium gregoryi Argonaute, Adv. Sci., 2018, 5(7), 1700540 CrossRef PubMed.
- Y. Fang, Z. Liu, H. Wang, X. Luo, Y. Xu, H. F. Chan, S. Lv, Y. Tao and M. Li, Implantable Sandwich-like Scaffold/Fiber Composite Spatiotemporally Releasing Combretastatin A4 and Doxorubicin for Efficient Inhibition of Postoperative Tumor Recurrence, ACS Appl. Mater. Interfaces, 2022, 14(24), 27525–27537 CrossRef CAS PubMed.
- Y. Huang, L. Mu, X. Zhao, Y. Han and B. Guo, Bacterial Growth-Induced Tobramycin Smart Release Self-Healing Hydrogel for Pseudomonas aeruginosa-Infected Burn Wound Healing, ACS Nano, 2022, 16(8), 13022–13036 CrossRef CAS PubMed.
- L. Qiao, Y. Liang, J. Chen, Y. Huang, S. A. Alsareii, A. M. Alamri, F. A. Harraz and B. Guo, Antibacterial conductive self-healing hydrogel wound dressing with dual dynamic bonds promotes infected wound healing, Bioact. Mater., 2023, 30, 129–141 CAS.
- Y. Yang, M. Li, G. Pan, J. Chen and B. Guo, Multiple Stimuli-Responsive Nanozyme-Based Cryogels with Controlled NO Release as Self-Adaptive Wound Dressing for Infected Wound Healing, Adv. Funct. Mater., 2023, 33(31), 2214089 CrossRef CAS.
- A. Kavand, N. Anton, T. Vandamme, C. A. Serra and D. Chan-Seng, Synthesis and functionalization of hyperbranched polymers for targeted drug delivery, J. Controlled Release, 2020, 321, 285–311 CrossRef CAS PubMed.
- Y. Wang and S. M. Grayson, Approaches for the preparation of non-linear amphiphilic polymers and their applications to drug delivery, Adv. Drug Delivery Rev., 2012, 64(9), 852–865 CrossRef CAS PubMed.
- P. A. Lambert, Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria, J. Appl. Microbiol., 2002, 92(s1), 46S–54S CrossRef.
- R. E. Impey, D. A. Hawkins, J. M. Sutton and T. P. Soares da Costa, Overcoming Intrinsic and Acquired Resistance Mechanisms Associated with the Cell Wall of Gram-Negative Bacteria, Antibiotics, 2020, 9(9), 623 CrossRef CAS PubMed.
- S. Wang, Y. Gao, Q. Jin and J. Ji, Emerging antibacterial nanomedicine for enhanced antibiotic therapy, Biomater. Sci., 2020, 8(24), 6825–6839 RSC.
- X. Wang, M. Zhang, T. Zhu, Q. Wei, G. Liu and J. Ding, Flourishing Antibacterial Strategies for Osteomyelitis Therapy, Adv. Sci., 2023, 10(11), e2206154 CrossRef PubMed.
- Y. Luo, G. Yu, X. Liu, Y. Feng, P. Zhao and J. Yue, Engineering Cationic Hyperbranched Polyureas for Combating Bacterial Biofilms, Chem. Mater., 2023, 35(12), 4705–4716 CrossRef CAS.
- Y. Liu, G. Yang, T. Baby, Tengjisi, D. Chen, D. A. Weitz and C.-X. Zhao, Stable Polymer Nanoparticles with Exceptionally High Drug Loading by Sequential Nanoprecipitation, Angew. Chem., Int. Ed., 2020, 59(12), 4720–4728 CrossRef CAS.
- F. C. Odds, Synergy, antagonism, and what the chequerboard puts between them, J. Antimicrob. Chemother., 2003, 52(1), 1 CrossRef CAS PubMed.
- M. Balouiri, M. Sadiki and S. K. Ibnsouda, Methods for in vitro evaluating antimicrobial activity: A review, J. Pharm. Anal., 2016, 6(2), 71–79 CrossRef PubMed.
- Y. Luo, G. Yu, F. Liu, Y. Feng, P. Zhao and J. Yue, Structure-Dependent Nontraditional Intrinsic Fluorescence of Aliphatic Hyperbranched Polyureas, Bioconjugate Chem., 2022, 33(7), 1319–1327 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H NMR spectra of HPUs and HPUs-QA; calibration curve of rifampicin absorbance as a function of standard rifampicin concentrations; characterization of FITC-labeled rifampicin; confocal fluorescence images of HPUs-RifFITC/QA-treated biofilm; cytotoxicity and hemocompatibility of HPUs-QA. See DOI: https://doi.org/10.1039/d3bm01519j |
| This journal is © The Royal Society of Chemistry 2024 |