Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Formulation of lipid nanoparticles containing ginsenoside Rg2 and protopanaxadiol for highly efficient delivery of mRNA†
Sin A
Park‡
a,
Dajeong
Hwang‡
 a,
Jae Hoon
Kim
a,
Seung-Yeul
Lee
a,
Jaebeom
Lee
a,
Jae Hoon
Kim
a,
Seung-Yeul
Lee
a,
Jaebeom
Lee
 bc,
Han Sang
Kim
de,
Kyung-A
Kim
bc,
Han Sang
Kim
de,
Kyung-A
Kim
 e,
Bumhee
Lim
f,
Jae-Eon
Lee
g,
Yong Hyun
Jeon
g,
Tae Jeong
Oh
e,
Bumhee
Lim
f,
Jae-Eon
Lee
g,
Yong Hyun
Jeon
g,
Tae Jeong
Oh
 a,
Jaewook
Lee
a,
Jaewook
Lee
 *a and
Sungwhan
An
*a and
Sungwhan
An
 *a
*a
aGenomictree Inc., Yuseong-gu, Daejeon, 34027, Republic of Korea. E-mail: jwlee@genomictree.com; sungwhan@genomictree.com
bDepartment of Chemistry, Chungnam National University, Yuseong-gu, Daejeon, 34134, Republic of Korea
cDepartment of Chemical Engineering and Applied Chemistry, Chungnam National University, Yuseong-gu, Daejeon, 34134, Republic of Korea
dYonsei Cancer Center, Division of Medical Oncology, Department of Internal Medicine, Yonsei University College of Medicine, Seoul, Republic of Korea
eDepartment of Internal Medicine, Graduate School of Medical Science, Brain Korea 21 Project, Yonsei University College of Medicine, Seoul, Republic of Korea
fNew Drug Development Center, Daegu-Gyeongbuk Medical Innovation Foundation (K-MEDIhub), Dong-gu, Daegu 41061, Republic of Korea
gPreclinical Research Center, Daegu-Gyeongbuk Medical Innovation Foundation (K-MEDIhub), Dong-gu, Daegu 41061, Republic of Korea
First published on 29th October 2024
Abstract
Lipid nanoparticles (LNPs) are widely recognized as crucial carriers of mRNA in therapeutic and vaccine development. The typical lipid composition of mRNA-LNP systems includes an ionizable lipid, a helper lipid, a polyethylene glycol (PEG)-lipid, and cholesterol. Concerns arise regarding cholesterol's susceptibility to oxidation, potentially leading to undesired immunological responses and toxicity. In this study, we formulated novel LNPs by replacing cholesterol with phytochemical-derived compounds, specifically ginsenoside Rg2 and its derivative phytosterol protopanaxadiol (PPD), and validated their efficacy as mRNA delivery systems. The mRNA–LNP complexes were manually prepared through a simple mixing process. The biocompatibility of these Rg2-based LNPs (Rg2-LNP) and PPD-based LNPs (PPD-LNP) was assessed through cell viability assays, while the protective function of LNPs for mRNA was demonstrated by RNase treatment. Enhanced green fluorescent protein (EGFP) mRNA delivery and expression in A549 and HeLa cells were analyzed using optical microscopy and flow cytometry. The expression efficiency of Rg2-LNP and PPD-LNP was compared with that of commercially available LNPs, with both novel formulations demonstrating superior transfection and EGFP expression. Furthermore, in vivo tests following intramuscular (I.M.) injection in hairless mice demonstrated efficient luciferase (Luc) mRNA delivery and effective Luc expression using Rg2-LNP and PPD-LNP compared to commercial LNPs. Results indicated that the efficiency of EGFP and Luc expression in Rg2-LNP and PPD-LNP surpassed that of the cholesterol-based LNP formulation. These findings suggest that Rg2-LNP and PPD-LNP are promising candidates for future drug and gene delivery systems.
Introduction
Recently, mRNA-based gene therapeutics and vaccines have garnered significant attention due to their numerous advantages, including the facile development of medicines for diseases induced by specific genes and the ability to tune target protein production in the body for therapeutic purposes.1–5 This has led to a rapid expansion of approved mRNA-based drugs.6 However, for therapeutic efficacy, mRNA must penetrate target cells and accurately translate proteins. Thus, the development of effective delivery systems is crucial.7–9Lipid nanoparticles (LNPs) stand out as notable vehicles that have significantly advanced mRNA applications in humans, with several mRNA-based therapies either approved or in clinical trials.10 Various mRNA delivery systems have been developed, including viral vectors like adeno-associated virus (AAV), lentivirus (LV) and non-viral carriers such as lipid nanoparticles (LNPs), liposomes, exosomes, polyol-based NPs, and inorganic NPs like gold or magnetic NPs.11–16 In most cases, LNP-based mRNA delivery systems have been widely used, and the FDA has approved those complex structure-based COVID-19 vaccines from Moderna and Pfizer/BioNTech.17–20 Typically, LNPs are composed of four essential components: an ionic lipid, helper lipid, PEG lipid, and sterol, and they have a specific role in structural formation. Among these, sterol is important in maintaining the lipid bilayer structure, enhancing the nanostructure stability and mRNA protection.21–24 Although cholesterol is commonly used as the sterol component, its application poses certain risks.25–28 Because cholesterol is easily oxidated and converted to oxidative derivatives, including oxysterols like 25-hydroxycholesterol and 7β-hydroxycholesterol, it exhibits potential toxicity.29–31 Moreover, it is possible that there is a risk of unexpected virus contamination because of animal-derived cholesterol.32
In this study, we have developed an alternative LNP formulation that replaces cholesterol with ginsenoside, Rg2, or its derivative phytosterol, protopanaxadiol (PPD), to address such issues. These new formulations, abundant in ginseng, mitigate the risks associated with cholesterol and leverage the significant bioactivity and biocompatibility of these phytochemicals, including anti-cancer and antioxidant effects.33–36 These beneficial properties make them suitable components for LNP formulation, with potentially synergistic effects alongside mRNA therapeutics.37–39 For therapeutic efficacy, mRNA needs to penetrate target cells and translate proteins adequately. In the present study, we have evaluated new LNPs that were Rg2-based LNPs (Rg2-LNP) and PPD-based LNPs (PPD-LNP) regarding their low toxicity, high biocompatibility, and mRNA protection ability against RNase. Also, we have demonstrated their effectiveness as a carrier for mRNA delivery and expression in vitro using enhanced green fluorescent protein (EGFP) mRNA as well as in vivo using luciferase (Luc) mRNA. Overall, the results described here show that ginsenoside and phytosterol-based LNPs could be alternatives to cholesterol-based LNPs, and consequently, Rg2-LNP and PPD-LNP exhibited considerable potential as delivery systems for mRNA therapeutics and vaccines.
Materials and methods
Materials and instruments
Ginsenoside Rg2, phytosterol protopanaxadiol (PPD), heptadecan-9-yl 8-[2-hydroxyethyl-(6-oxo-6-(undecyloxy)hexyl)amino]octanoate (ionizable lipid), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE, phospholipid), and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (PEG lipid) were purchased from Cayman Chemical (Ann Arbor, MI, USA). Also, as the positive control of LNPs, commercially available LNP preparation kits (the SM-102-LNP kit and ALC-0315-LNP kit) were purchased from Cayman Chemical. The SM-102 LNP kit contains SM-102 (ionizable lipid), distearoylphosphatidylcholine (DSPC, helper lipid), PEG lipid and cholesterol. SM-102-LNP was used for COVID-19 mRNA vaccine formulation in Moderna. We also used the ALC-0315 LNP kit to produce the other positive control LNP and it is composed of ALC-0315 (ionizable lipid), DSPC, PEG lipid and cholesterol. This LNP was also formulated for COVID-19 mRNA vaccine in Pfizer/BioNTech. RPMI, DMEM, FBS, and penicillin–streptomycin were purchased from Well Gene (Gyeongsan, Gyeongsangbuk-do, South Korea) and plates were bought from Thermo Fisher Scientific (Waltham, Massachusetts, USA) for cell culture. A549 and HeLa cells were purchased from the Korean Cell Line Bank (Seoul, South Korea). A Quant-iT Ribogreen RNA assay kit was bought from Invitrogen/Thermo Fisher Scientific (Seoul, South Korea). The in vitro luciferase assay kit was purchased from Promega (Madison, WI, USA).The size and morphology of Rg2-LNP and PPD-LNP were characterized using a nanoparticle tracking analyzer (NTA, NanoSight NS300, Malvern Panalytical, Malvern, UK), a zetasizer (Nano ZS, Malvern Panalytical, Malvern, UK) for measurement of dynamic light scattering (DLS) and a transmission electron microscope (TEM, H-7600, Hitachi, Tokyo, Japan). The cell morphology and expression of EGFP after EGFP mRNA delivery into the cell were monitored using a fluorescence optical microscope (Olympus, CKX553, Tokyo, Japan). The cell viability was measured using cell counting kit-8 (CCK-8) from Dojindo (Kumamoto, Kyushu, Japan), and the absorbance was measured using a microplate spectrophotometer (Mobi, MicroDigital, Seongnam, South Korea). The fluorescence (FL) intensity was monitored using a microplate multi-mode reader (Bio Tek Synergy HTX, Agilent, Santa Clara, CA, USA). The EGFP positive cells and the mean of FL intensities were characterized by flow cytometry (BD FACSLyric™, BD Biosciences, Franklin Lakes, NJ, USA). Male Balb/c nude mice were purchased (Orient Bio Inc, Seongnam, South Korea), and live animal imaging was performed using an IVIS Lumina III imaging system (PerkinElmer, Waltham, MA, USA).
Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 78 or 50
78 or 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 (molar ratio = ionizable lipid
39 (molar ratio = ionizable lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phospholipid
phospholipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEG lipid
PEG lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Rg2). Subsequently, Rg2-LNP was softly mixed with mRNA to construct the mRNA-Rg2-LNP complex for the delivery and expression test under various weight ratios (wmRNA
Rg2). Subsequently, Rg2-LNP was softly mixed with mRNA to construct the mRNA-Rg2-LNP complex for the delivery and expression test under various weight ratios (wmRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) wLNP), 1
wLNP), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, 1
5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and 1
10 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, and that the mixture was incubated for 15 min at room temperature (R.T.). In this case, the solvent volume ratio (VmRNA
20, and that the mixture was incubated for 15 min at room temperature (R.T.). In this case, the solvent volume ratio (VmRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) VLNP) between aqueous solution (mRNA) and ethanol (LNP) was 3
VLNP) between aqueous solution (mRNA) and ethanol (LNP) was 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After incubation, the mRNA-Rg2-LNP was sonicated for 1 min and then treated with the cell to evaluate the delivery and expression efficiency.
1. After incubation, the mRNA-Rg2-LNP was sonicated for 1 min and then treated with the cell to evaluate the delivery and expression efficiency.
To synthesize the PPD-LNP, components for the LNP formulation were mixed under varying concentrations: 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45.8 and 50
45.8 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22.9 (molar ratio = ionizable lipid
22.9 (molar ratio = ionizable lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phospholipid
phospholipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEG lipid
PEG lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PPD). The mRNA
PPD). The mRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PPD-LNP mixing ratio for further study was the same as for Rg2-LNP.
PPD-LNP mixing ratio for further study was the same as for Rg2-LNP.
To evaluate the encapsulation efficiency of Rg2-LNP and PPD-LNP using the Ribogreen RNA assay kit, the FL intensity was measured with a microplate multi-mode reader at an excitation wavelength of 480 nm and an emission wavelength of 530 nm.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phospholipid
phospholipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEG lipid
PEG lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Rg2 or PPD) were further optimized to enhance the expression efficiency, resulting in a ratio of 50
Rg2 or PPD) were further optimized to enhance the expression efficiency, resulting in a ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75
0.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 for Rg2-LNP and 50
39 for Rg2-LNP and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75
0.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22.9 for PPD-LNP.
22.9 for PPD-LNP.
In addition, after 24 h of transfection, both cell lines were harvested and resuspended in PBS with 1% FBS and then the expression efficiency was evaluated by flow cytometry.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C. The supernatant was then collected to measure relative luciferase activity using the luciferase assay system.
000g for 10 min at 4 °C. The supernatant was then collected to measure relative luciferase activity using the luciferase assay system.
Male Balb/c nude mice were sacrificed to assess the efficiency of delivery and expression in vivo. Each mouse was dosed with 10 μg of Luc mRNA encapsulated in Rg2-LNP, PPD-LNP, SM-102-LNP, or modified SM-102-LNP through an intramuscular (I.M.) injection. The molar ratios of Rg2-LNP and PPD-LNP formulation used for the Luc mRNA in vivo delivery and expression tests were 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75
0.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 and 50
39 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75
0.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22.9, respectively. The molar ratios of SM-102-LNP and modified SM-102-LNP formulations followed the manufacturer's protocol. For bioluminescence imaging (BLI), mice were administered D-luciferin via intraperitoneal injection. During imaging, anesthesia using 1–2% isoflurane gas was applied to all mice. BLI was conducted 5 min after substrate injection using the IVIS Lumina III imaging system, from 1 h to 54 h post-injection of Luc mRNA-encapsulated Rg2-LNP and PPD-LNP as well as SM-102-LNP and modified SM-102-LNP as the positive control. LIVING-IMAGE software (version 3.0, PerkinElmer) was utilized to overlay grayscale photographic images and bioluminescent color images. BLI signals were quantified in units of photons per cm2 per second per steradian (P cm−2 s−1 sr−1).
22.9, respectively. The molar ratios of SM-102-LNP and modified SM-102-LNP formulations followed the manufacturer's protocol. For bioluminescence imaging (BLI), mice were administered D-luciferin via intraperitoneal injection. During imaging, anesthesia using 1–2% isoflurane gas was applied to all mice. BLI was conducted 5 min after substrate injection using the IVIS Lumina III imaging system, from 1 h to 54 h post-injection of Luc mRNA-encapsulated Rg2-LNP and PPD-LNP as well as SM-102-LNP and modified SM-102-LNP as the positive control. LIVING-IMAGE software (version 3.0, PerkinElmer) was utilized to overlay grayscale photographic images and bioluminescent color images. BLI signals were quantified in units of photons per cm2 per second per steradian (P cm−2 s−1 sr−1).
Results and discussion
Characterization of Rg2-LNP and PPD-LNP
Following the preparation of Rg2-LNP and PPD-LNP, their physicochemical properties were characterized using various methods. The structural morphologies of Rg2-LNP, mRNA-Rg2-LNP, PPD-LNP and mRNA-PPD-LNP were observed by TEM after negative staining treatment with 2% uranyl acetate solution and they are depicted in Fig. 1 and Fig. S2.† Without mRNA, Rg2-LNP and PPD-LNP exhibited an empty vehicle structure (Fig. 1A and C). However, when mRNA was present, the structure of both LNPs changed clearly. Firstly, Rg2-LNP changed from a unilamellar to a multilamellar structure (Fig. 1A, B, and Fig. S2A†). This morphological transformation can be attributed to the amphiphilic properties of Rg2 and phospholipids, which facilitate self-assembly and enable the steroid part of Rg2 to adsorb into lipid bilayers within the LNPs.41 Additionally, it was introduced that glucose could interact with guanin in nucleic acids via hydrogen bonding.42 Another literature study reported that hydrogen bonds can form between the OH groups in glucose and the phosphate groups or nitrogenous bases in nucleotides, and this binding was characterized by FT-IR differential spectroscopy.43 These studies suggest that the hydroxyl groups of glucose in Rg2 could create hydrogen bonds with the polar functional groups of mRNAs, such as the phosphate backbone or nitrogenous bases. Therefore, the relatively higher encapsulation rate of Rg2-LNP for mRNA could be due to these hydrogen bonds. On the other hand, PPD-LNP transformed from an empty vehicle to a unilamellar structure with a nanostructured core in the presence of mRNA. This dark core indicates the self-assembly structure of mRNA, ionizable lipid, and helper lipid, which was negatively stained in the mRNA-PPD-LNP (Fig. 1D, E and Fig. S2B†). The structure of mRNA-PPD-LNP differs from that of mRNA-Rg2-LNP due to the hydrophobic properties of PPD, which interacts strongly with lipid structures. The average sizes of mRNA-Rg2-LNP and mRNA-PPD-LNP were measured by NTA to be around 117.3 ± 1.6 nm and 167.9 ± 3.9 nm, respectively (Fig. 1C and F). The sizes of the two LNPs were also measured using DLS. The average size of only Rg2-LNP was around 121 nm and mRNA-Rg2-LNP was approximately 132.9 nm, with a PDI of approximately 0.24, while the average size of PPD-LNP was about 144.7 nm and mRNA-PPD-LNP was approximately 168.9 nm, with a PDI of 0.26 (Fig. S2C and D†). In general, the presence of mRNA within the LNP structure leads to an increase in the overall size of the LNPs due to the additional payload. This occurs because mRNA can induce structural modifications, such as swelling or alterations in the arrangement of lipid layers. As a result, the size of both LNPs containing mRNA showed a slight increase. In addition, a PDI of 0.3 or less in the size measurement results indicates a homogeneous population of nanocarriers. Therefore, the two LNPs mentioned above are considered to have sufficient potential for use in drug delivery.44,45 There was some difference between the size analysis results from NTA and DLS due to the different measurement mechanisms, but both sizes were suitable for functioning as LNP carriers.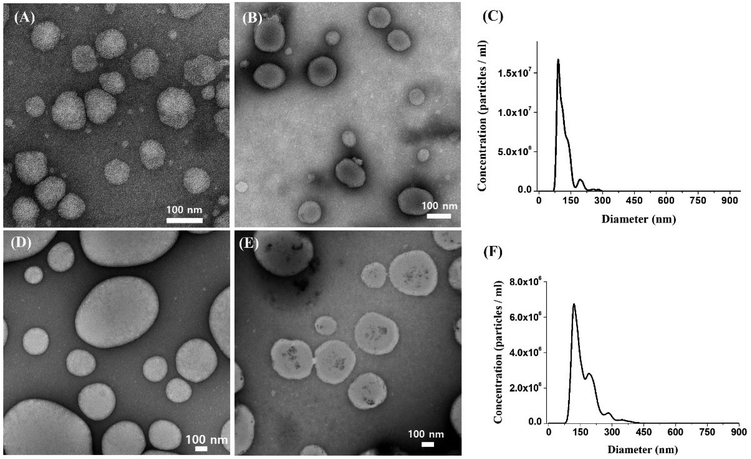 | ||
| Fig. 1 TEM images of (A) Rg2-LNP, (B) EGFP mRNA-Rg2-LNP, (D) PPD-LNP, and (E) EGFP mRNA-PPD-LNP and NTA results of (C) EGFP mRNA-Rg2-LNP and (F) EGFP mRNA-PPD-LNP. | ||
Encapsulation efficiency and mRNA stability
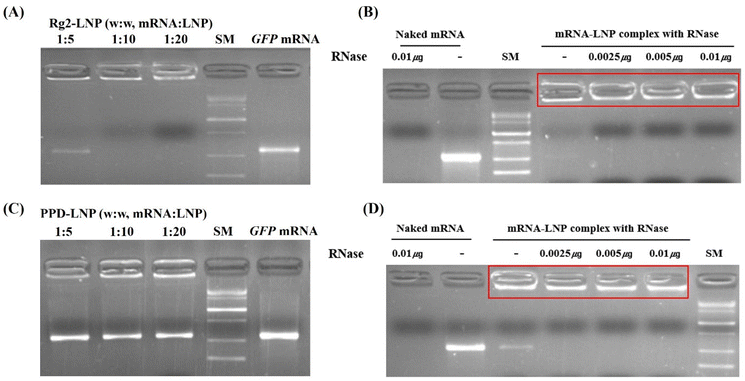
The encapsulation efficiency of Rg2-LNP and PPD-LNP was evaluated using gel electrophoresis (Fig. 2A and C). For Rg2-LNP, no residual mRNA bands were observed at weight ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and 1
10 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, indicating complete encapsulation. In contrast, PPD-LNP exhibited unencapsulated mRNA bands at all tested ratios (1
20, indicating complete encapsulation. In contrast, PPD-LNP exhibited unencapsulated mRNA bands at all tested ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, 1
5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, and 1
10, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20), though the band intensity decreased with increasing LNP mass. The encapsulation efficiency of Rg2-LNP and PPD-LNP was evaluated using the Ribogreen RNA assay, showing 81.9% and 68.7%, respectively. These mRNA-LNPs were prepared through simple hand mixing, which might result in a lower encapsulation efficiency compared to those prepared using a microfluidics system. Nevertheless, the Rg2-LNP and PPD-LNP produced by this mixing process were sufficient mRNA carriers for expressing EGFP and Luc for in vitro or in vivo experiments. However, we recognized the limitations of manual preparation, including challenges in achieving precise size uniformity and optimal mRNA encapsulation per LNP. To address these limitations, we plan to utilize microfluidic devices in future studies to enhance the development of mRNA-based therapeutics.
20), though the band intensity decreased with increasing LNP mass. The encapsulation efficiency of Rg2-LNP and PPD-LNP was evaluated using the Ribogreen RNA assay, showing 81.9% and 68.7%, respectively. These mRNA-LNPs were prepared through simple hand mixing, which might result in a lower encapsulation efficiency compared to those prepared using a microfluidics system. Nevertheless, the Rg2-LNP and PPD-LNP produced by this mixing process were sufficient mRNA carriers for expressing EGFP and Luc for in vitro or in vivo experiments. However, we recognized the limitations of manual preparation, including challenges in achieving precise size uniformity and optimal mRNA encapsulation per LNP. To address these limitations, we plan to utilize microfluidic devices in future studies to enhance the development of mRNA-based therapeutics.
On the other hand, the higher encapsulation efficiency of Rg2-LNP can be attributed to the glucose component forming hydrogen bonds with mRNA and the ionizable lipid binding through electrostatic interactions, resulting in tighter mRNA capture, as mentioned above. mRNA stability within the LNPs was assessed by analyzing mRNA integrity after RNase treatment via electrophoresis (Fig. 2B and D). Naked mRNA was completely degraded by RNase exposure, whereas mRNA within Rg2-LNP and PPD-LNP remained intact even at increased RNase concentrations (red box in Fig. 2B and D). The unencapsulated mRNA band in PPD-LNP disappeared after RNase treatment, while the encapsulated mRNA remained protected, demonstrating that both Rg2-LNP and PPD-LNP effectively shield mRNA from degradation.
Biocompatibility of Rg2-LNP and PPD-LNP
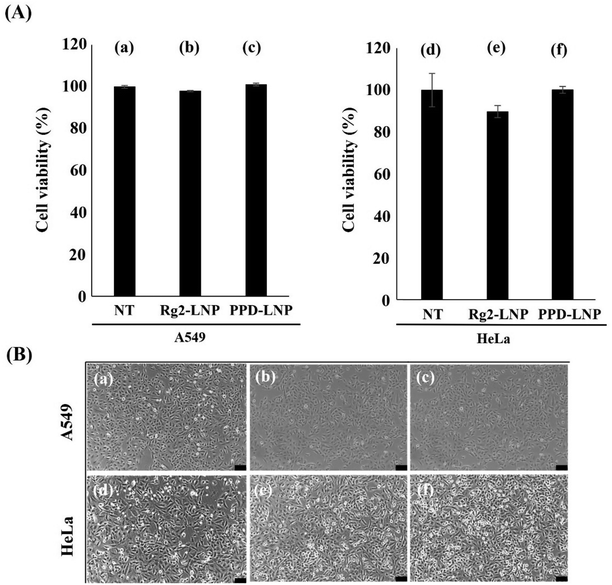
The cytotoxicity of Rg2-LNP and PPD-LNP was evaluated using a CCK-8 assay (Fig. 3 and Fig. S3†). Treatment with 0.05 μg of Rg2-LNP and PPD-LNP reduced the number of A549 and HeLa cells by a maximum of 10%, with no significant changes in cell morphology (Fig. 3A and B). Furthermore, dose-dependent cytotoxicity was assessed, revealing no significant toxicity (Fig. S3†). Additionally, it should be noted that Rg2 and PPD are phytochemicals, and those are recognized as biologically active components derived from ginseng, known for its health benefits.46–48 This suggests that the cytotoxicity of Rg2-LNP and PPD-LNP is minimal and not a critical concern for their application as gene delivery systems.In vitro mRNA delivery and expression using Rg2-LNP and PPD-LNP vehicles
The delivery and translation efficiency of EGFP mRNA encapsulated in Rg2-LNP and PPD-LNP were evaluated in A549 and HeLa cells using various molar ratios of LNP components for optimization (Fig. S4†). From these assessments, the initial molar ratios for LNP formulation were determined to be 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 for Rg2-LNP and 50
39 for Rg2-LNP and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5
1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22.9 for PPD-LNP. Firstly, in evaluating the delivery efficacy of LNPs, 250 ng of EGFP mRNA encapsulated by Rg2-LNP or PPD-LNP at a 1
22.9 for PPD-LNP. Firstly, in evaluating the delivery efficacy of LNPs, 250 ng of EGFP mRNA encapsulated by Rg2-LNP or PPD-LNP at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 weight ratio (mRNA
10 weight ratio (mRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
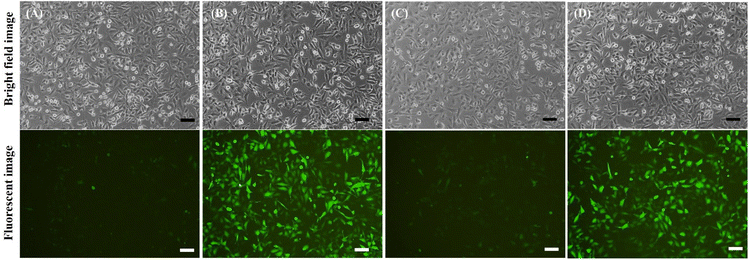
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LNP) was transfected into A549 cells. After 24 h of treatment, optical microscopy revealed a clear signal of green fluorescence in all treated cells, indicating successful delivery and translation of EGFP mRNA (Fig. 4B and C). Additionally, the delivery effectiveness and transfection efficiency of Rg2-LNP and PPD-LNP were compared with those of commercially available LNPs SM-102-LNP (Moderna) and ALC-0315-LNP (Pfizer/BioNTech), which retain the cholesterol component. After delivering EGFP mRNA with these LNPs, EGFP expression in A549 cells was carefully observed. According to the fluorescence image, the fluorescence brightness was significantly higher in cells transfected with Rg2-LNP and PPD-LNP compared to those of SM-102-LNP and ALC-0315-LNP (Fig. S5A†). In addition, the efficiency of EGFP expression was compared by flow cytometry with various LNP carriers (Fig. S5B–D†). In this case, the population of EGFP-positive A549 cells was over 95% after treatment with EGFP mRNA encapsulated Rg2-LNP, PPD-LNP, and SM-102-LNP, but it was only 25% in ALC-0315-LNP. Interestingly, the mean fluorescence intensity (MFI) was the highest in the case of PPD-LNP, followed by Rg2-LNP, which was higher than SM-102-LNP. In addition, Luc mRNA transfection yields were compared between commercial LNPs and Rg2-LNP and PPD-LNP in A549 cell lines using a bioluminescence assay (Fig. S6†). The results were similar to those of the EGFP expression trends in that relative light units (RLU) were higher for Rg2-LNP and PPD-LNP compared to SM-102-LNP and ALC-0315-LNP. Additionally, the GFP mRNA delivery and expression efficiency of modified SM-102-LNP, where the helper lipid was switched from DSPC to DOPE, was compared with Rg2-LNP and PPD-LNP, and the results are presented in Fig. S7.† In this case, brighter green fluorescence was observed in LNPs based on SM-102/DOPE/Rg2/PEG lipid (Rg2-LNP) or SM-102/DOPE/PPD/PEG lipid (PPD-LNP) than in LNPs formulated with SM-102/DOPE/cholesterol/PEG lipid (modified SM-102 LNP).
LNP) was transfected into A549 cells. After 24 h of treatment, optical microscopy revealed a clear signal of green fluorescence in all treated cells, indicating successful delivery and translation of EGFP mRNA (Fig. 4B and C). Additionally, the delivery effectiveness and transfection efficiency of Rg2-LNP and PPD-LNP were compared with those of commercially available LNPs SM-102-LNP (Moderna) and ALC-0315-LNP (Pfizer/BioNTech), which retain the cholesterol component. After delivering EGFP mRNA with these LNPs, EGFP expression in A549 cells was carefully observed. According to the fluorescence image, the fluorescence brightness was significantly higher in cells transfected with Rg2-LNP and PPD-LNP compared to those of SM-102-LNP and ALC-0315-LNP (Fig. S5A†). In addition, the efficiency of EGFP expression was compared by flow cytometry with various LNP carriers (Fig. S5B–D†). In this case, the population of EGFP-positive A549 cells was over 95% after treatment with EGFP mRNA encapsulated Rg2-LNP, PPD-LNP, and SM-102-LNP, but it was only 25% in ALC-0315-LNP. Interestingly, the mean fluorescence intensity (MFI) was the highest in the case of PPD-LNP, followed by Rg2-LNP, which was higher than SM-102-LNP. In addition, Luc mRNA transfection yields were compared between commercial LNPs and Rg2-LNP and PPD-LNP in A549 cell lines using a bioluminescence assay (Fig. S6†). The results were similar to those of the EGFP expression trends in that relative light units (RLU) were higher for Rg2-LNP and PPD-LNP compared to SM-102-LNP and ALC-0315-LNP. Additionally, the GFP mRNA delivery and expression efficiency of modified SM-102-LNP, where the helper lipid was switched from DSPC to DOPE, was compared with Rg2-LNP and PPD-LNP, and the results are presented in Fig. S7.† In this case, brighter green fluorescence was observed in LNPs based on SM-102/DOPE/Rg2/PEG lipid (Rg2-LNP) or SM-102/DOPE/PPD/PEG lipid (PPD-LNP) than in LNPs formulated with SM-102/DOPE/cholesterol/PEG lipid (modified SM-102 LNP).
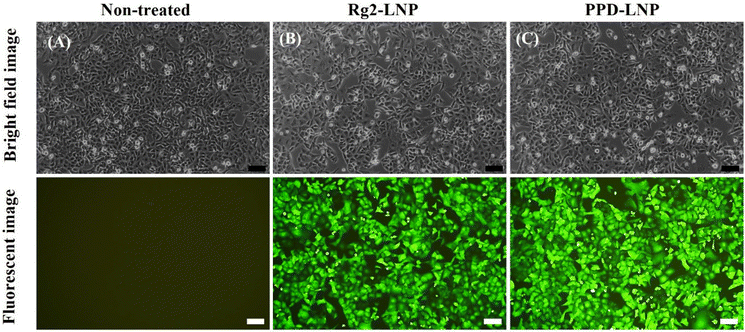 | ||
| Fig. 4 Bright field images and fluorescence images of (A) non-treated, (B) EGFP mRNA-Rg2-LNP treated and (C) EGFP mRNA-PPD-LNP treated A549 cells (scale bar: 100 μm). | ||
Optimization of mRNA delivery and transfection
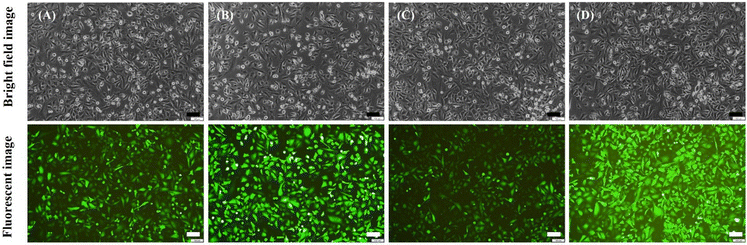
EGFP mRNA encapsulated in Rg2-LNP and PPD-LNP were also treated with HeLa cells, resulting in observable green fluorescence (Fig. 5B and C). The expression of EGFP in HeLa cells was critically influenced by the weight ratio (mRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LNP) at 1
LNP) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and 1
10 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, and a higher signal intensity of EGFP was observed at a 1
20, and a higher signal intensity of EGFP was observed at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 weight ratio (mRNA
20 weight ratio (mRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LNP) instead of 1
LNP) instead of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (Fig. 6). Interestingly, when assessing the encapsulation efficiency of LNPs by gel electrophoresis, results showed that nearly all mRNA was encapsulated in Rg2-LNP at 1
10 (Fig. 6). Interestingly, when assessing the encapsulation efficiency of LNPs by gel electrophoresis, results showed that nearly all mRNA was encapsulated in Rg2-LNP at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and 1
10 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratios, and for PPD-LNP, unencapsulated mRNA bands were similar at both ratios, so it was expected that the tendency of EGFP expression might be the same for both weight ratios in HeLa cells as in A549 cells. This indicates that the tendency of expression efficiency did not directly correspond to the encapsulation efficiency. The expression efficiency and MFI of EGFP in HeLa cells were also evaluated after transfection of EGFP mRNA using commercial LNPs, Rg2-LNP, and PPD-LNP, similar to the tests conducted in A549 cells. Flow cytometry analysis results showed that although the expression efficiency and MFI were lower in HeLa cells compared to A549 cells, PPD-LNP had the highest MFI, demonstrating a similar pattern in HeLa cells (Fig. S8†). This finding indicates that the newly developed LNPs have excellent transfection efficiency. Also, the results highlight the importance of tuning and optimizing formulation conditions for effective protein expression in target cell lines.
20 ratios, and for PPD-LNP, unencapsulated mRNA bands were similar at both ratios, so it was expected that the tendency of EGFP expression might be the same for both weight ratios in HeLa cells as in A549 cells. This indicates that the tendency of expression efficiency did not directly correspond to the encapsulation efficiency. The expression efficiency and MFI of EGFP in HeLa cells were also evaluated after transfection of EGFP mRNA using commercial LNPs, Rg2-LNP, and PPD-LNP, similar to the tests conducted in A549 cells. Flow cytometry analysis results showed that although the expression efficiency and MFI were lower in HeLa cells compared to A549 cells, PPD-LNP had the highest MFI, demonstrating a similar pattern in HeLa cells (Fig. S8†). This finding indicates that the newly developed LNPs have excellent transfection efficiency. Also, the results highlight the importance of tuning and optimizing formulation conditions for effective protein expression in target cell lines.
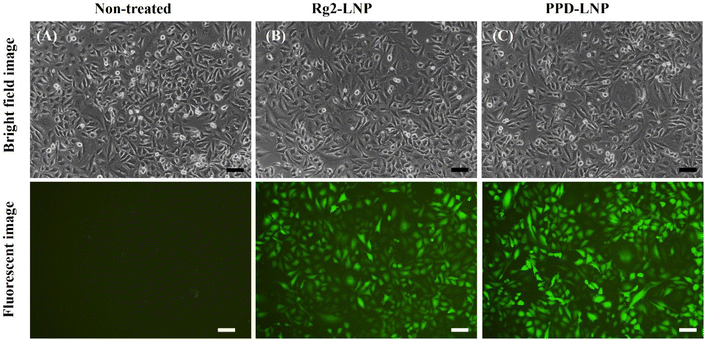 | ||
| Fig. 5 Observation of EGFP expression in HeLa cells. (A) Non-treated conditions, (B) EGFP mRNA-Rg2-LNP conditions and (C) EGFP mRNA-PPD-LNP conditions (scale bar: 100 μm). | ||
Additional optimization involved reducing the PEG lipid component in the LNP formulation, which enhanced the EGFP translation efficiency (Fig. 7B and D). It was reported that a lower surface density of PEG has been associated with increased uptake into cancer cells, including HeLa cells.49–51 In our case, also a lower PEG density improved lipid nanoparticle uptake by HeLa cells, thereby increasing EGFP translation. Furthermore, high levels of EGFP expression remained after 48 h post-treatment (Fig. S9†). These findings emphasize optimizing the mRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LNP weight ratio and the PEG lipid content based on the target cell type to achieve delivery and optimal translation efficiency.
LNP weight ratio and the PEG lipid content based on the target cell type to achieve delivery and optimal translation efficiency.
Potential of in vivo mRNA delivery and expression
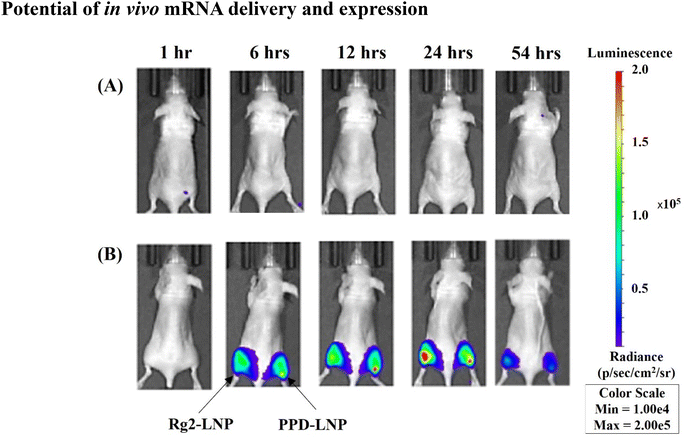
The potential for in vivo applications of Rg2-LNP and PPD-LNP was assessed using bioluminescence imaging in mice. Mice were intramuscularly injected with Luc mRNA encapsulated in Rg2-LNP and PPD-LNP. The bioluminescence imaging revealed a signal of Luc mRNA translation at 6 hours post-injection, which persisted until 54 h (Fig. 8B, Fig. S10 and S11†), while no evident signal from empty vehicles was present. To evaluate the efficiency of in vivo mRNA delivery and expression in cells of mRNA encapsulated in LNPs, Rg2-LNP, and PPD-LNP, mRNA-Luc expression was examined in hairless mice. Commercially available cholesterol-retaining SM-102-LNP (with DSPC as the helper lipid) and modified SM-102-LNP (with DOPE as the helper lipid) were used as positive controls because the population of EGFP-positive cells was comparable between SM-102-LNP, Rg2-LNP, and PPD-LNP, SM-102-LNP. According to IVIS imaging and total flux profiles after I.M. injection, the total flux of the SM-102-LNP series decreased rapidly after 6 hours (Fig. S10 and S11†).
In contrast, the expression pattern of Luc in Rg2-LNP and PPD-LNP showed a slightly delayed initial expression compared to that of SM-102-LNP, but the decrease in Luc expression was slower. Notably, Rg2-LNP had the highest flux value after 12 hours post-injection across all LNP conditions according to the profile of total flux. Additionally, the total flux of PPD-LNP was higher than that of SM-102-LNP at 12 hours post-injection (Fig. S11†). These results unequivocally demonstrate the effectiveness of Rg2-LNP and PPD-LNP in delivering mRNA and facilitating translation in vivo, instilling confidence in their potential for gene therapy applications.
Conclusion
As mRNA-based therapeutics expand, developing suitable carriers has become increasingly important. This study developed lipid nanoparticles (LNPs) incorporating ginsenoside Rg2 and its derivative PPD as sterol components, replacing cholesterol in the LNP formulation. The formulation was optimized by adjusting the molar ratio of lipid components to Rg2 or PPD and the weight ratio between mRNA and LNP. The Rg2-LNP and PPD-LNP were prepared manually through a simple mixing method, resulting in highly dispersed spherical LNPs. These carriers exhibited low toxicity, good encapsulation efficiency, and strong mRNA protection ability. Flow cytometry analysis revealed that transfection with Rg2-LNP or PPD-LNP encapsulating EGFP mRNA resulted in significantly higher transfection efficiency and MFI in A549 and HeLa cells compared to commercially available LNPs. Furthermore, evaluating mRNA delivery and expression of LNPs encapsulating Luc mRNA using hairless mice indicated that these LNPs possessed high delivery efficiency in vivo as well.In conclusion, the mRNA complexes with Rg2-LNP or PPD-LNP, synthesized through a straightforward manual mixing process, demonstrated highly effective mRNA delivery and expression, highlighting their potential as robust mRNA delivery systems in this study.
Author contributions
Sin A Park: methodology, investigation, formal analysis, and writing – original draft. Dajeong Hwang: methodology, investigation, formal analysis, and writing – original draft. Jae Hoon Kim: investigation and formal analysis. Seung-Yeul Lee: investigation and formal analysis. Jaebeom Lee: investigation, resource, and writing – review and editing. Han Sang Kim: resource, investigation, resource, and writing – review and editing. Kyung-A Kim: investigation and writing – review and editing. Bumhee Lim: resource, formal analysis, and writing – review and editing. Jae-Eon Lee: investigation, formal analysis, and writing – original draft. Yong Hyun Jeon: resource, methodology, formal analysis, and writing – review and editing. Tae Jeong Oh: project administration and writing – review and editing. Jaewook Lee: conception, methodology, investigation, formal analysis, visualization, writing – original draft, and writing – review and editing. Sungwhan An: supervision, project administration, conception, writing – original draft, and writing – review and editing.Data availability
All data are saved and located in the Network Attached Storage (NAS) system of Genomictree, and due to the confidentiality of data, the data including raw files used or analyzed during the current study are available from the corresponding authors on reasonable request.Conflicts of interest
The authors S. A Park, D. Hwang, J. H. Kim, S.-Y. Lee, T. J. Oh, J. Lee and S. An were employed by the company Genomictree, Inc, and T. J. Oh and S. An are shareholders of Genomictree, Inc. The authors S. An and J. Lee are inventors on patent applications currently pending related to the content of this manuscript. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.Acknowledgements
We thank staff members at Genomictree, Inc. for technical support, and we also thank Prof. Seung-Hoon Lee (Yong In University) for valuable advice.References
- S. Ke, A. Pandya-Jones, Y. Saito, J. J. Fak, C. B. Vågbø, S. Geula, J. H. Hanna, D. L. Black, J. E. Darnell and R. B. Darnell, Genes Dev., 2017, 31, 990–1006 CrossRef CAS PubMed
.
- A. Li, Y.-S. Chen, X.-L. Ping, X. Yang, W. Xiao, Y. Yang, H.-Y. Sun, Q. Zhu, P. Baidya, X. Wang, D. P. Bhattarai, Y.-L. Zhao, B.-F. Sun and Y.-G. Yang, Cell Res., 2017, 27, 444–447 CrossRef CAS PubMed
.
- D. M. Mauger, B. J. Cabral, V. Presnyak, S. V. Su, D. W. Reid, B. Goodman, K. Link, N. Khatwani, J. Reynders, M. J. Moore and I. J. McFadyen, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 24075–24083 CrossRef CAS PubMed
.
- G. Tavernier, O. Andries, J. Demeester, N. N. Sanders, S. C. De Smedt and J. Rejman, J. Controlled Release, 2011, 150, 238–247 CrossRef CAS PubMed
.
- Z. Meng, J. O'Keeffe-Ahern, J. Lyu, L. Pierucci, D. Zhou and W. Wang, Biomater. Sci., 2017, 5, 2381–2392 RSC
.
- C. E. Dunbar, K. A. High, J. K. Joung, D. B. Kohn, K. Ozawa and M. Sadelain, Science, 2018, 359, eaan4672 CrossRef PubMed
.
- N. Chaudhary, D. Weissman and K. A. Whitehead, Nat. Rev. Drug Discovery, 2021, 20, 817–838 CrossRef CAS PubMed
.
- J. Lutz, S. Lazzaro, M. Habbeddine, K. E. Schmidt, P. Baumhof, B. L. Mui, Y. K. Tam, T. D. Madden, M. J. Hope, R. Heidenreich and M. Fotin-Mleczek, npj Vaccines, 2017, 2, 29 CrossRef PubMed
.
- S. S. Rosa, D. M. F. Prazeres, A. M. Azevedo and M. P. C. Marques, Vaccine, 2021, 39, 2190–2200 CrossRef CAS PubMed
.
- J. Chen, J. Chen and Q. Xu, Annu. Rev. Biomed. Eng., 2022, 24, 85–109 CrossRef CAS PubMed
.
- J. Lee, J.-H. Lee, K. Chakraborty, J. Hwang and Y.-K. Lee, RSC Adv., 2022, 12, 18475–18492 RSC
.
- D. Mishra, J. R. Hubenak and A. B. Mathur, J. Biomed. Mater. Res., Part A, 2013, 101, 3646–3660 CrossRef PubMed
.
- C. Bharti, U. Nagaich, A. K. Pal and N. Gulati, Int. J. Pharm. Invest., 2015, 5, 124–133 CrossRef CAS PubMed
.
- V. Sokolova and M. Epple, Angew. Chem., Int. Ed., 2008, 47, 1382–1395 CrossRef CAS PubMed
.
- C. Heneweer, S. E. Gendy and O. Peñate-Medina, Ther. Delivery, 2012, 3, 645–656 CrossRef CAS PubMed
.
- A. Luchini and G. Vitiello, Front. Chem., 2019, 7, 343 CrossRef CAS PubMed
.
- P. R. Cullis and M. J. Hope, Mol. Ther., 2017, 25, 1467–1475 CrossRef CAS PubMed
.
- J. A. Kulkarni, D. Witzigmann, S. Chen, P. R. Cullis and R. van der Meel, Acc. Chem. Res., 2019, 52, 2435–2444 CrossRef CAS PubMed
.
-
Y. Zhao and L. Huang, in Advances in Genetics, ed. L. Huang, D. Liu and E. Wagner, Academic Press, 2014, vol. 88, pp. 13–36 Search PubMed
.
- J. E. N. Dolatabadi, H. Valizadeh and H. Hamishehkar, Adv. Pharm. Bull., 2015, 5, 151–159 CrossRef CAS PubMed
.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Nat. Rev. Mater., 2021, 6, 1078–1094 CrossRef CAS PubMed
.
- L. Schoenmaker, D. Witzigmann, J. A. Kulkarni, R. Verbeke, G. Kersten, W. Jiskoot and D. J. A. Crommelin, Int. J. Pharm., 2021, 601, 120586 CrossRef CAS PubMed
.
- J. C. Kraft, J. P. Freeling, Z. Wang and R. J. Y. Ho, J. Pharm. Sci., 2014, 103, 29–52 CrossRef CAS PubMed
.
- F. Ding, H. Zhang, J. Cui, Q. Li and C. Yang, Biomater. Sci., 2021, 9, 7534–7546 RSC
.
- A. Valenzuela, J. Sanhueza and S. Nieto, Biol. Res., 2003, 36, 291–302 CAS
.
- W. Kulig, L. Cwiklik, P. Jurkiewicz, T. Rog and I. Vattulainen, Chem. Phys. Lipids, 2016, 199, 144–160 CrossRef CAS PubMed
.
- G. Leonarduzzi, B. Sottero and G. Poli, J. Nutr. Biochem., 2002, 13, 700–710 CrossRef CAS PubMed
.
- Y. Song, J. Liu, K. Zhao, L. Gao and J. Zhao, Cell Metab., 2021, 33, 1911–1925 CrossRef CAS PubMed
.
- M. K. Pulfer and R. C. Murphy, J. Biol. Chem., 2004, 279, 26331–26338 CrossRef CAS PubMed
.
- V. M. Olkkonen, O. Béaslas and E. Nissilä, Biomolecules, 2012, 2, 76–103 CrossRef CAS PubMed
.
- J. C. Verhagen, P. ter Braake, J. Teunissen, G. van Ginkel and A. Sevanian, J. Lipid Res., 1996, 37, 1488–1502 CrossRef CAS
.
- D. W. Jayme and S. R. Smith, Cytotechnology, 2000, 33, 27–36 CrossRef CAS PubMed
.
- R. Conte, V. Marturano, G. Peluso, A. Calarco and P. Cerruti, Int. J. Mol. Sci., 2017, 18, 709 CrossRef PubMed
.
- D. Sun, S.-Y. Guo, L. Yang, Y.-R. Wang, X.-H. Wei, S. Song, Y.-W. Yang, Y. Gan and Z.-T. Wang, Acta Pharmacol. Sin., 2020, 41, 119–128 CrossRef CAS PubMed
.
- X.-R. Tan, L. Chao, K.-K. Feng, J.-Q. Le, J.-W. Shen and J.-W. Shao, J. Ind. Eng. Chem., 2022, 116, 303–309 CrossRef CAS
.
- X. Zhao, J. Wu, K. Zhang, D. Guo, L. Hong, X. Chen, B. Wang and Y. Song, Nanoscale Adv., 2022, 4, 190–199 RSC
.
- A. Vijayakumar, R. Baskaran, J.-H. Baek, P. Sundaramoorthy and B. K. Yoo, AAPS PharmSciTech, 2019, 20, 88 CrossRef CAS PubMed
.
- X. Song, L. Zang and S. Hu, Vaccine, 2009, 27, 2306–2311 CrossRef CAS PubMed
.
- C. Yue, D. Li, S. Fan, F. Tao, Y. Yu, W. Lu, Q. Chen, A. Yuan, J. Wu, G. Zhao, H. Dong and Y. Hu, Biomaterials, 2023, 301, 122291 CrossRef CAS PubMed
.
-
S. An, Republic of Korea Pat, 10-2021-0137521, 2021 Search PubMed
.
- S. L. Verstraeten, J. H. Lorent and M.-P. Mingeot-Leclercq, Front. Pharmacol., 2020, 11, 576887 CrossRef CAS PubMed
.
- E. Vengut-Climent, I. Gómez-Pinto, R. Lucas, P. Peñalver, A. Aviñó, C. Fonseca Guerra, F. M. Bickelhaupt, R. Eritja, C. González and J. C. Morales, Angew. Chem., Int. Ed., 2016, 55, 8643–8647 CrossRef CAS PubMed
.
- L. El-Mahdaoui, J. F. Neault and H. A. Tajmir-Riahi, J. Inorg. Biochem., 1997, 65, 123–131 CrossRef CAS
.
- M. Danaei, M. Dehghankhold, S. Ataei, F. Hasanzadeh Davarani, R. Javanmard, A. Dokhani, S. Khorasani and M. R. Mozafari, Pharmaceutics, 2018, 10, 57 CrossRef PubMed
.
- L. Xu, X. Wang, Y. Liu, G. Yang, R. J. Falconer and C.-X. Zhao, Adv. NanoBiomed Res., 2022, 2, 2100109 CrossRef CAS
.
- Z. A. Ratan, M. F. Haidere, Y. H. Hong, S. H. Park, J.-O. Lee, J. Lee and J. Y. Cho, J. Ginseng Res., 2021, 45, 199–210 CrossRef PubMed
.
- D. L. Huynh, N. H. Nguyen and C. T. Nguyen, Mol. Cell. Biochem., 2021, 476, 3329–3340 CrossRef PubMed
.
- S. Zhang, B. Zheng, Y. Wei, Y. Liu, L. Yang, Y. Qiu, J. Su and M. Qiu, Biomater. Sci., 2024, 12, 2672–2688 RSC
.
- S. Hak, E. Helgesen, H. H. Hektoen, E. M. Huuse, P. A. Jarzyna, W. J. M. Mulder, O. Haraldseth and C. D. L. Davies, ACS Nano, 2012, 6, 5648–5658 CrossRef CAS PubMed
.
- D. Pozzi, V. Colapicchioni, G. Caracciolo, S. Piovesana, A. L. Capriotti, S. Palchetti, S. De Grossi, A. Riccioli, H. Amenitsch and A. Laganà, Nanoscale, 2014, 6, 2782–2792 RSC
.
- C. Cruje and D. B. Chithrani, J. Nanomed. Res., 2014, 1, 27–32 Search PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01070a |
| ‡ Both contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |