Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Pseudouridine and N1-methylpseudouridine as potent nucleotide analogues for RNA therapy and vaccine development
Lyana L. Y.
Ho
ab,
Gabriel H. A.
Schiess
c,
Pâmella
Miranda
cd,
Gerald
Weber
 c and
Kira
Astakhova
c and
Kira
Astakhova
 *a
*a
aTechnical University of Denmark, 2800 Kongens Lyngby, Denmark. E-mail: kiraas@kemi.dtu.dk
bThe Hong Kong Polytechnic University, 11 Yuk Choi Rd, Hung Hom, Hong Kong
cDepartamento de Física, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brazil
dPrograma Interunidades de Pós-Graduação em Bioinformática, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brazil
First published on 19th March 2024
Abstract
Modified nucleosides are integral to modern drug development, serving as crucial building blocks for creating safer, more potent, and more precisely targeted therapeutic interventions. Nucleobase modifications often confer antiviral and anti-cancer activity as monomers. When incorporated into nucleic acid oligomers, they increase stability against degradation by enzymes, enhancing the drugs’ lifespan within the body. Moreover, modification strategies can mitigate potential toxic effects and reduce immunogenicity, making drugs safer and better tolerated. Particularly, N1-methylpseudouridine modification improved the efficacy of the mRNA coding for spike protein of COVID-19. This became a crucial step for developing COVID-19 vaccine applied during the 2020 pandemic. This makes N1-methylpseudouridine, and its “parent” analogue pseudouridine, potent nucleotide analogues for future RNA therapy and vaccine development. This review focuses on the structure and properties of pseudouridine and N1-methylpseudouridine. RNA has a greater structural versatility, different conformation, and chemical reactivity than DNA. Watson–Crick pairing is not strictly followed by RNA that has more unusual base pairs and base-triplets. This requires detailed structural studies and structure–activity relationship analyses for RNA, also when modifications are incorporated. Recent successes in this direction are revised in this review. We describe recent successes with using pseudouridine and N1-methylpseudouridine in mRNA drug candidates. We also highlight remaining challenges that need to be solved to develop new mRNA vaccines and therapies.
Introduction
Deoxyribo- and ribonucleic acids (DNA and RNA) represent a class of natural compounds central to the life on Earth. Their main functions are to store, transmit genetic information and to allow species adopt, survive, and evolve.Nucleosides are monomers of DNA and RNA (the latter illustrated in Fig. 1A). They have two distinct structural elements: nitrogen heterocycle, called nucleobase, and a ribose carbohydrate. Each element serves as a scaffold for multiple non-covalent interactions, vital to biological performance of nucleic acids. Nucleobase provides with π-stacking and hydrogen bonding interactions, forming nucleic acid helices and universal genetic code.
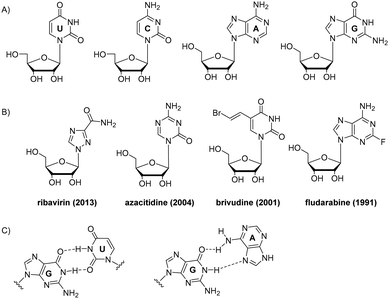 | ||
| Fig. 1 Chemical structures of natural RNA nucleotides (A), examples of nucleoside drugs (year of FDA approval) (B), and wobble RNA base pairs: GU and GA (C). | ||
Given crucial biological importance of nucleic acids, interference with their synthesis and function creates powerful therapeutic interventions. This can be achieved via chemical modification of nucleosides.
Multiple nucleoside drugs with nucleobase modifications have been developed. Several examples are shown in Fig. 1B. Modified nucleoside monomers include drugs for treating viral infections (ribavirin, idoxuridine, trifluridine, brivudine), cancer (azacitidine, fludarabine, 6-mercaptopurine, decitabine), and immunosuppressive drug azathioprine, used in organ transplantation and autoimmune diseases.1 Due to their altered nucleobase structure, these drugs interfere with DNA synthesis, transcription and/or translation leading to cell death.1
As to oligonucleotide drugs, it mostly ribose and phosphate backbone modifications that have been incorporated. Chemical modifications in these drugs result in enhanced stability, improved pharmacokinetics, reduced toxicity, increased target specificity, or resistance to degradation, thereby improving their therapeutic potential.2
Over the last two decades, there has been a growing interest in RNA drug development. RNA differs from DNA by having a 2′-hydroxyl group in ribose sugar, which gives rise to unique conformational features, specific hydration, and electrostatic properties of RNA (Fig. 1A).1 Another structural feature of RNA is that uracil nucleobase does not contain the major groove 5-methyl group as that in DNA nucleotide thymidine. As a result of this, RNA duplexes are more compact than B-DNA helices. Ribose nucleotides tend to adopt the C3′-endo sugar pucker resulting in compact A-form RNA duplexes with eleven base pairs per turn.1
RNA has various alternative base pairings, such as G:U wobble pair and A:C pair, which differ from DNA. G:U pair has an exocyclic amino group that acts as a prominent hydrogen bond donor in the minor grooves, important for recognising proteins and other ligands (Fig. 1C).3 G:U pair frequently occurs, such as in codon–anticodon interaction which forms the genetic code between tRNA and mRNA. The hydrogen bonding of G:U are strongly dependent on their next neighbour pairs, ranging from very weak bonding when in tandem configuration to strong if located at the end of a double strand.4 G:A pairs are ubiquitous at RNA helices termini, in loops and folds of tertiary structure. Among G:A, G:G and A:A pairs, sheared G:A pairs that include a Hoogsteen interaction are most common (Fig. 1C).3
RNA nucleotides form multiple interactions with nucleobases and sugars. G-tetrad and G-quartet are similar for DNA and RNA but more diversity is found in RNA. Quadruple interaction can be formed in highly folded RNA molecules as well.3
RNA folding is complex and sensitive to temperature and buffer composition. Metal ions like Mg2+ and K+ stabilize RNA tertiary structure.3 As a result of stable structure, misfolding of RNA could become a severe problem with e.g., therapeutics and vaccine candidates.3
Pseudouridine (Ψ) is an abundant post-transcriptional modification of mRNA which together with N1-methylpseudouridine (N1-Me-Ψ), became a milestone in developing mRNA drug candidates. This review describes Ψ and N1-Me-Ψ nucleobase analogues, their impact on RNA structure and appealing properties in RNA vaccines and therapeutic candidates. We also give highlights on most potent applications of modified mRNA achieved so far, discuss future directions and remaining challenges.
Pseudouridine
The first known and one of the most abundant RNA modifications is the pseudouridine (Ψ), or 5-ribosyluracil, which is a post-transcriptional modification and an isomer of uridine (U) (Fig. 2A). Pseudouridine contains a C–C base-sugar bond due to the uracil base is attached to the sugar by a C1′–C5 bond unlike a C1′–N1 glycosidic linkage (Fig. 2), enhancing the base rotation. Moreover, it has an additional ring nitrogen atom (N1 imino atom), which behaves as an additional hydrogen bond donor.5,6 The replacement of U by Ψ promotes a C3′-endo sugar conformation and increases the local base stacking, thermodynamically stabilizing RNA duplexes.5,7,8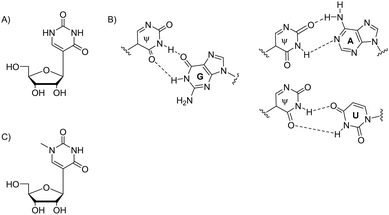 | ||
| Fig. 2 Chemical structure of Ψ (A), its base pairing with G, A and U nucleobases (B); chemical structure of N1-methyl-Ψ (C). | ||
Pseudouridine (Ψ) sites have been identified in different types of natural mRNA, such as tRNA, rRNA, snRNA and mRNA.9–13 Intracellular Ψ formation is catalyzed by a class of enzymes known as pseudouridine synthases.14 Pseudouridine synthases can be classified into two main families: stand-alone pseudouridine synthases and pseudouridine synthase domains within larger proteins. Stand-alone pseudouridine synthases include TruA found in bacteria and TruB found in bacteria and yeast. In eukaryotes, several pseudouridine synthase domains have been found. Eucaryotic H/ACA box small nucleolar ribonucleoproteins (snoRNPs) have a dyskerin (Cbf5) component which catalyzes pseudouridylation in rRNA, snRNA, and telomerase RNA. Nop10 is another component of H/ACA snoRNPs which is involved in the pseudouridylation activity.14
In humans, there are multiple enzymes catalyzing pseudouridylation. RPUSD (RNA pseudouridylate synthase domain-containing) family consists of several enzymes involved in Ψ formation. RPUSD1 and RPUSD2 are involved in mitochondrial RNA pseudouridylation. The PUS family consists of enzymes with pseudouridine synthase activity towards multiple RNA species, represented by PUS1 and PUS7.
Quantitative Ψ detection remains being challenging. Therefore its biological role is still not completely understood. Incorporated into mRNA, Ψ increases its stability and modifies various cellular and biological processes, such as transcription, pre-mRNA splicing and mRNA translation.14 Ψ was also found in stop codons.15
Pseudouridylation is a crucial post-transcriptional modification that influences the structural stability and function of transfer RNA (tRNA) as well. With nanopore sequencing it has been shown that Ψ modifications are present in diverse positions of tRNA.16 As a result of a modification in the hisT gene, the tRNA E. coli and Salmonella typhimurium lacks Ψ38, Ψ39 and Ψ40 positions and may have a reduced polypeptide chain elongation rate (20–25%) and longer cell division time (30%). In addition, inhibition of Ψ38 and Ψ39, as a result of DEG1 gene disruption, in S. cerevisiae leads to reduced growth rate.9
Notably, pseudouridylation of tRNA affects its methylation at other positions. In tRNAPhe of S. cerevisiae, Ψ55 positively influences the introduction of methylated nucleotides, m5U54 and m1A58.16
Aberrant pseudouridine modifications in tRNA can impact translation accuracy and efficiency, potentially contributing to disease pathogenesis.10,11,17–20 For example, mutations in the PUS1 gene, which encodes a pseudouridine synthase, are associated with mitochondrial myopathy and sideroblastic anaemia.19 Abnormal pseudouridylation has been implicated in neurological disorders.20 Altered tRNA modifications, including pseudouridylation, have been observed in various cancer types.9,10,17,18
Nucleic acid function is guided by non-covalent interactions between polynucleotides and with cellular protein machinery.3 Therefore 2D and 3D structure of RNA is central to its interactions and function. Regarding impact on RNA structure, two hydrogen bond donors by N1 and N3 imine protons, and two hydrogen bond acceptors are present in Ψ (Fig. 2B). Therefore, Ψ acts as a universal nucleobase by enhancing stability when it pairs with A, G, U or C in a double helix. The relationship between Ψ-modified mRNA structure and stability is supported by multiple examples of in vitro-synthesised, more stable, Ψ-containing mRNAs, and the natural Ψ-containing mRNAs’ increased half-life time, as in eukaryotic parasite Toxoplasma gondii.21,22
NMR structural studies found that Ψ stacks better than U due to the C1′–C5 glycoside bond in Ψ that has higher rotational lability than the C1′–N1 bond in U.5,23 Ψ enhances local RNA base stacking in both single- and double-stranded conformations and promotes an increase in neighbouring stacking stabilization at nucleotide level.7,9 In addition, Ψ induces a more rigid phosphodiester backbone in its vicinity, increasing the base stacking neighbouring, perhaps due to C–C isomerization in Ψ.9,14 Moreover, the base stacking enhancement is considered the most important contribution of Ψ to RNA stability.7,24–27
To investigate more the impact of Ψ on RNA structure, thermodynamic studies were conducted.5 The data revealed that Ψ stabilizes the RNA duplexes when U is replaced and Ψ–A, Ψ–G, Ψ–U and Ψ–C pairs are formed. The effect of pseudouridylation is more significant when it occurs at the internal positions of the RNA structure, usually releasing more negative free energy. Enhanced thermodynamic stability by Ψ is around 0.5 kcal mol−1, except in 5′-CΨG/3′GAC and 5′-GΨC/3′CGG, nearly 2.5 and 1.5 kcal mol−1 is recorded respectively. Ψ contribution is, in most cases, weaker than a typical hydrogen bond (2–10 kcal mol−1), which indicates that the enhanced stability is a result of a different mechanism.28 This endorses the hypothesis that the contribution of Ψ to the stability arises from the C1′–C5 bond properties. Current NMR data5 suggest that the N1H in the 5′-CΨG/3′-GAC trimer is in a weak hydrogen bond while there is no evidence for hydrogen bonding in the 5′-GΨC/3′-CGG trimer.5 Ψ biological roles, such as nonsense suppression of stop codons and non-synonymous translation, might be better understood by identifying contexts where N1H is in an active hydrogen bond.29 The 3D structures and computational studies of these outlier duplexes could be interesting to investigate due to little knowledge of the interactions that are favourable to stacking.
Ψ–A, Ψ–G, Ψ–U and Ψ–C base pairs have shown enhancement of base stacking and also sequence context dependence.5,6,30 High level quantum mechanical (QM) methods have shown a clear dependence of the change in the base stacking energies concerning the sequence context with a range from −1.59 to 0.23 kcal mol−1. However, the change from U–A to Ψ–A base pair not always stabilizes the stacking interactions of the duplex.6,30 Yet experimental data of internal and terminal Ψ–A pair has shown greater stability in comparison to predicted data of U–A pair in the same duplexes, on average 1.7 and 1.0 kcal mol−1, respectively.30
Terminal Ψ–A, –C, –G and –U pairs show stability enhancement in both 5′ and 3′ terminals in a similar way to U–A, –C, –G and –U pairs, respectively. Curiously, those Ψ pairs at 3′ terminal reach slightly higher energies than the U pairs, except Ψ–A pair. Ψ–C pair contributes with more stability than U–C pair only when added at 3′ terminal. On the other hand, only Ψ–U pair shows a more stable behaviour than U pair in 5′ terminal. Similarly, Ψ–G pairs have higher stabilization when at 3′ terminal (3.22 kcal mol−1) in comparison to U–G pairs (2.44 kcal mol−1). Finally, any internal Ψ pair enhances the thermodynamic stability of RNA duplexes. Mainly Ψ–A and Ψ–G pairs have large energies, within a 0.4–0.8 kcal mol−1, when compared to A–U and G–U pairs, perhaps due to the favourable base stacking and hydrogen bonds of Ψ.5,31
RNA N1-methylpseudouridine structure
N1-Methylpseudouridine (N1-methyl-Ψ) is another naturally occurring RNA nucleotide analogue32 which has an extra hydrogen bond donor in the nucleobase (Fig. 2C). This N1-modified structure can be found in natural 18S rRNA and tRNA, not only in humans but also in archaea and eukaryotes. Research shows that RNAs of Thermococcales and Nanoarchaea33 include N1-methyl-Ψ.34,35N1-Methyl-Ψ biosynthetic pathway begins with converting U to Ψ, being catalyzed by the aforementioned pseudouridine synthase Pus10.36,37S-Adenosylmethionine (SAM)-dependent pseudouridine N3-methyltransferase YbeA in eubacteria38,39 and N1-specific pseudouridine methyltransferase Nep1 are two enzymes that can further methylate Ψ. Nep1 catalyse the N1-specific pseudouridine methylation of position 1191 (Saccharomyces cerevisiae numbering, nucleotide 913 in M. jannaschii) in RNA.40,41Ψ- and N1-methyl-Ψ modified RNA duplexes have different stability which has been confirmed with multiple thermal denaturation studies.5N1-Methyl-Ψ differs from Ψ by having a methyl group to replace the extra hydrogen at N1-position, so it no longer has the universal base character like Ψ (Fig. 2). N1-Methyl-Ψ only forms the traditional Watson–Crick pair but both Ψ and N1-methyl-Ψ have the C5–C1′ bond that allows rotation between the nucleobase and sugar, achieving better base-pairing, base-stacking and duplex stability.42
A recent molecular dynamics study shows that N1-methyl-Ψ induces a higher stabilization effect of the dsRNA, due to stronger stacking and base pair interactions, than Ψ. Moreover, N1-methyl-Ψ:A pair have a stronger binding interaction than both U:A and Ψ:A pairs in the majority of neighbours context.43
Overall, N1-methyl-Ψ is more like uridine in translation coding but at the same time, behaves like Ψ that let the mRNA not trigger the immune response which is of critical importance to RNA vaccines and therapies, described below.
mRNA drug development
mRNA holds a potential to treat a vast spectrum of diseases and to act as a vaccine, by producing the desired protein in vivo and acting as adjuvant.44 Key steps in mRNA drug development include: sequence design, chemical modification, formulation, testing in vitro and in vivo, and finally, trials.Currently there are two approved mRNA vaccines, by Moderna and Pfizer.45,46 According to FDA, January 2024, there are 56 mRNA drugs in the clinical pipeline worldwide, with R&D mainly focused on vaccines, accounting for about 84%, while therapeutic drugs account for about 16%. Except for the mRNA COVID-19 vaccine, which is urgently marketed, most others are in the early stages.47
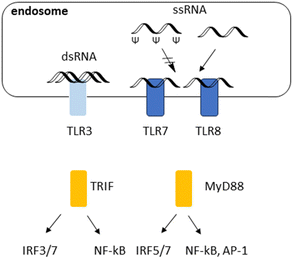
Over the last decade, therapeutic mRNAs and mRNA vaccines have been encountering several major obstacles. First, long single (ss) or double stranded (ds) RNAs in the cytosol are commonly derived from the genome of RNA viruses or intermediate products that are generated during viral replication, leading to immune response.48 Introducing Ψ and N1-Me-Ψ has been a breakthrough to overcome this challenge. Immune responses to RNA and their inhibition with Ψ are illustrated in Fig. 3. TLR7 and TLR8 recognize single stranded (ss) RNA. These TLRs preferentially recognize polyuridine (polyU) and guanosine/uridine-rich (GU-rich) sequences. TLR7 and TLR8 also recognize RNA degradation products and require free guanosines and uridines, respectively, for maximum activation. Ψ and N1-methyl-Ψ could optimise mRNA performance by reduced immunogenicity and effective protein translation. Reduced binding to TLRs due to Ψ.
Delivery has been another main challenge to mRNA therapeutics and vaccines, that could not be solved solely with chemical modification of mRNA. mRNA is rapidly degraded in vivo and poorly uptaken by cells. Delivery systems like lipid nanoparticles (LNPs) have high efficiency, low toxicity and are applicable to various cell types. LNP-mRNA nanoparticle formulations have been successful and reached clinical applications.44–46 LNPs have encapsulation efficiency reaching 90%, and can be targeted through surface modification.49,50
On-going research on LNP mRNA covers multiple potential applications. Main direction in current trials is cancer vaccines. Very recently Moderna and Merck published successful results of phase IIb study on melanoma cancer vaccine.51 However it is a limiting factor that MC3 dLin-MC3-DMA (also known as C12-200) cationic formulation used in COVID-19 vaccine is patented and cannot be broadly used for other mRNA drug candidates. To overcome this, there are attempts to develop alternative formulations. Cationic lipid formulations using DOTAP (1,2-dioleoyl-3-trimethylammonium-propane), PEGylated lipids and DOPE (dioleoylphosphatidylethanolamine) are being tested.52,53
The delivery tools for mRNA are not limited to LNP. Naturally occurring vesicles, such as exosomes, can be used to carry mRNA.52 These vesicles are derived from cells and besides mRNA encapsulation, can facilitate cell-specific targeting. Encapsulation efficiency of exosomes is lower than for LNP, however exosomes are less toxic and have higher target specificity.52
Polymeric nanoparticles and dendrimers have been actively explored for the mRNA delivery.53 Polymeric nanoparticles, such as those made from polyethyleneimine (PEI), poly(lactic-co-glycolic acid) (PLGA), or chitosan, can form complexes with siRNA through electrostatic interactions. These nanoparticles protect mRNA from degradation and can enhance cellular uptake.53,54 Other examples of reported nanoparticles for mRNA include inorganic nanoparticles, such as gold or silica nanoparticles, hybrid nanoparticles, virus-like nanoparticles and peptide-based nanoparticles.55–58 They are extensively reviewed elsewhere.
Overall, there is no universal solution to all nucleic acid delivery tasks. Choosing the most suitable delivery system depends on the specific requirements of the therapeutic application, including the targeted tissue, desired release kinetics, and safety considerations.
N1-Methylpseudouridine impact on COVID-19 mRNA vaccine
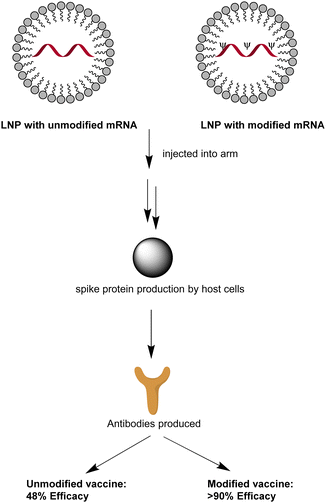
A key principle of the mRNA-based vaccination is that under low dosage, non-modified mRNA encodes the antigen while acting as an adjuvant (Fig. 4). Restricted to a maximum of 12 μg dosage by patients’ tolerance in late-stage clinical trials, unmodified mRNA vaccine CVnCoV maintained only 48% efficacy, regardless of the disease severity.45 In contrast, 30 μg Pfizer-BioNTech or 100 μg Moderna's mRNA vaccines could demonstrate around 95% high protection rate against COVID-19 after modification with N1-methyl-Ψ.45Vaccines composed of unmodified and modified mRNA compared after being injected into the muscle of the upper arm.45 An immune response would be created and N1-methylpseudouridylated mRNA demonstrates over 90% efficacy against COVID-19 symptoms, much higher than the unmodified one which is beneath 50%.
Clearly, N1-methyl-Ψ has different impact on product protein translation compared to Ψ. According to Kim et al. (2022), N1-methyl-Ψ does not necessarily change the decoding accuracy in a reconstituted system.59 Neither would it increase the probability of miscoded peptides, nor stabilise the mismatched RNA-duplex formed. It only has a slight tendency in increasing errors when reverse transcription occurs.
Remaining challenges in therapeutic mRNA development
mRNA development needs to be supported by research on mRNA structure since it is closely related to the function of mRNA. Nonetheless, mRNA structure prediction is still unreliable despite combining the usage of thermodynamic stability and evolutionary covariation information.60 The conserved mRNA structure could be predicted by the combination of three features: using significant covariation, negative evolutionary information and a plethora of probabilistic folding algorithms which incorporate those positive covariations into a single structure.39Another challenge is how to produce exogenous long mRNA while incorporating N1-methyl-Ψ flexibly on a large scale. Getting inspiration from nature and utilizing enzymes is an effective method. In this approach, fragments can become the building blocks of plasmids for COVID-19 vaccine synthesis. According to past studies, T7 polymerase could effectively build RNAs longer than 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nucleotides perfectly without error61,62 while tolerating non-natural NTPs. m1Ψ triphosphate can be incorporated into RNA by polymerases providing long modified RNA molecules (Fig. 5).63 This allows producing large quantities of ling modified RNA, of tremendous benefit to RNA drug development and commercialization.
000 nucleotides perfectly without error61,62 while tolerating non-natural NTPs. m1Ψ triphosphate can be incorporated into RNA by polymerases providing long modified RNA molecules (Fig. 5).63 This allows producing large quantities of ling modified RNA, of tremendous benefit to RNA drug development and commercialization.
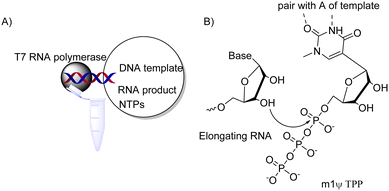 | ||
| Fig. 5 Producing N1-methyl-Ψ-modified mRNA by in vitro transcription. (A) Key components of enzymatic reaction and RNA product; NTP = nucleotide triphosphate;63 (B) enzymatic 3′ → 5′ elongation of RNA during in vitro transcription. | ||
Conclusions
Overall, mRNA therapy is a cutting-edge medical approach that uses synthetic mRNA molecules to prevent and treat various conditions. This technology has gained significant attention and recognition, particularly due to its role in the development of COVID-19 vaccines.45 mRNA therapy has the potential to treat a wide range of diseases, including infectious diseases, cancer, genetic disorders, and more.64–66 By introducing synthetic mRNA into the body, it's possible to instruct cells to produce specific proteins that can correct or combat disease. As to vaccine development, mRNA technology gained widespread attention during the COVID-19 pandemic when companies like Pfizer-BioNTech and Moderna developed highly effective COVID-19 vaccines using this approach.45 The vaccines contain mRNA that encodes the spike protein of the SARS-CoV-2 virus, which triggers an immune response, providing protection against the virus.45One of the advantages of mRNA therapy is its rapid development and adaptability. Creating synthetic mRNA for a new target can be faster and more scalable than traditional vaccine or drug development processes. With LNP technology, efficient delivery can be achieved as well.64–66
Despite its promise, mRNA therapy faces challenges, including ensuring the stability and delivery of mRNA molecules to target cells, managing immune responses, and addressing potential side effects. These pitfalls can be overcome by using chemical analogues of RNA nucleotides. Pseudouridine (Ψ) is an important modification that is conserved naturally in the RNA structure. Studies showed that Ψ-modification allows mRNA to resist intrinsic immune responses67 and Ψ-derivatives could further improve mRNA properties, such as stability and efficacy of translation.
To apply this technology to vaccine and therapy development, deeper knowledge on the impact of Ψ and its analogues on RNA structure is required. Computational approaches, thermodynamics and structural investigations with e.g., NMR would be significant next steps in this direction.
Author contributions
The review is conceptualized by GHA Schiess and K. Astakhova. LLY Ho and K. Astakhova made the initial draft. All authors contributed to finalising the review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Danish Foundation for Independent Research (award 3105-00141B) to KAS; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) to GHAS and PM; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (Fapemig) to GW.Notes and references
- E. De Clercq and G. Li, Approved Antiviral Drugs over the Past 50 Years, Clin. Microbiol. Rev., 2016, 29(3), 695–747 CrossRef PubMed.
- T. C. Roberts, R. Langer and M. J. A. Wood, Advances in oligonucleotide drug delivery, Nat. Rev. Drug Discovery, 2020, 19(10), 673–694 CrossRef CAS PubMed.
- Nucleic Acids in Chemistry and Biology, ed. G. M. Blackburn, M. J. Gait, D. Loakes and D. Williams, Royal Society of Chemistry, 2006 Search PubMed.
- T. D. Amarante and G. Weber, Evaluating hydrogen bonds and base stacking of single, tandem and terminal GU mismatches in RNA with a mesoscopic model, J. Chem. Inf. Model., 2016, 56(1), 101–109 CrossRef CAS PubMed.
- E. Kierzek, M. Malgowska, J. Lisowiec, D. H. Turner, Z. Gdaniec and R. Kierzek, The contribution of pseudouridine to stabilities and structure of RNAs, Nucleic Acid Res., 2014, 42(5), 3492–3501 CrossRef CAS PubMed.
- I. Deb, Ł. Popenda and J. Sarzyńska, et al., Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair, Sci. Rep., 2019, 9, 16278 CrossRef PubMed.
- D. R. Davis, Stabilization of RNA stacking by pseudouridine, Nucleic Acid Res., 1995, 23(24), 5020–5026 CrossRef CAS PubMed.
- K. B. Hall and L. W. McLaughlin, Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods, Biochemistry, 1991, 30(7), 1795–1801 CrossRef CAS PubMed.
- M. Charette and M. W. Gray, Pseudouridine in RNA: what, where, how, and why, IUBMB Life, 2000, 49(5), 341–352 CrossRef CAS PubMed.
- X. Xue, Z. Wang, Y. Wang and X. Zhou, Disease Diagnosis Based on Nucleic Acid Modifications, ACS Chem. Biol., 2023, 18(10), 2114–2127 CrossRef CAS PubMed.
- P. Nombela, B. Miguel-López and S. Blanco, The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities, Mol. Cancer, 2021, 20(1), 1–30 CrossRef PubMed.
- S. Tavakoli, M. Nabizadeh, A. Makhamreh, H. Gamper, C. A. McCormick, N. K. Rezapour, Y. M. Hou, M. Wanunu and S. H. Rouhanifard, Semi-quantitative detection of pseudouridine modifications and type I/II hypermodifications in human mRNAs using direct long-read sequencing, Nat. Commun., 2023, 14(1), 334 CrossRef CAS PubMed.
- Q. Dai, L. S. Zhang, H. L. Sun, K. Pajdzik, L. Yang, C. Ye, C. W. Ju, S. Liu, Y. Wang, Z. Zheng, L. Zhang, B. T. Harada, X. Dou, I. Irkliyenko, X. Feng, W. Zhang, T. Pan and C. He, Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution, Nat. Biotechnol., 2023, 41(3), 344–354 CrossRef CAS PubMed.
- S. H. Boo and Y. K. Kim, The emerging role of RNA modifications in the regulation of mRNA stability, Exp. Mol. Med., 2020, 52(3), 400–408 CrossRef CAS PubMed.
- J. Song, L. Dong, H. Sun, N. Luo, Q. Huang, K. Li, X. Shen, Z. Jiang, Z. Lv, L. Peng, M. Zhang, K. Wang, K. Liu, J. Hong and C. Yi, CRISPR-free, programmable RNA pseudouridylation to suppress premature termination codons, Mol. Cell, 2023, 83(1), 139–155 CrossRef CAS PubMed e9.
- M. C. Lucas, L. P. Pryszcz, R. Medina, I. Milenkovic, N. Camacho, V. Marchand, Y. Motorin, L. Ribas de Pouplana and E. M. Novoa, Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing, Nat. Biotechnol., 2024, 42(1), 72–86 CrossRef CAS PubMed.
- N. M. Martinez, A. Su, M. C. Burns, J. K. Nussbacher, C. Schaening, S. Sathe, G. W. Yeo and W. V. Gilbert, Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing, Mol. Cell, 2022, 82(3), 645–659 CrossRef CAS PubMed e9.
- J. A. Stockert, A. Gupta, B. Herzog, S. S. Yadav, A. K. Tewari and K. Yadav, Predictive value of pseudouridine in prostate cancer, Am. J. Clin. Exp. Urol., 2019, 7(4), 262–272 Search PubMed.
- E. Fernandez-Vizarra, A. Berardinelli, L. Valente, V. Tiranti and M. Zeviani, Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA), BMJ Case Rep., 2009, 2009, bcr05.2009.1889 Search PubMed.
- T. Y. Lin, R. Smigiel, B. Kuzniewska, J. J. Chmielewska, J. Kosińska, M. Biela, A. Biela, A. Kościelniak, D. Dobosz, I. Laczmanska, A. Chramiec-Głąbik, J. Jeżowski, J. Nowak, M. Gos, S. Rzonca-Niewczas, M. Dziembowska, R. Ploski and S. Glatt, Destabilization of mutated human PUS3 protein causes intellectual disability, Hum. Mutat., 2022, 43(12), 2063–2078 CrossRef CAS PubMed.
- M. J. Holmes, L. R. Padgett, M. S. Bastos and W. J. Sullivan Jr, m6A RNA methylation facilitates pre-mRNA 3'-end formation and is essential for viability of Toxoplasma gondii, PLoS Pathog., 2021, 17(7), e1009335 CrossRef CAS PubMed.
- M. A. Nakamoto, A. F. Lovejoy, A. M. Cygan and J. C. Boothroyd, mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii, RNA, 2017, 23(12), 1834–1849 CrossRef CAS PubMed.
- N. B. Leontis and E. Westhofm, Analysis of RNA motifs, Curr. Opin. Struct. Biol., 2003, 13(3), 300–308 CrossRef CAS PubMed.
- D. R. Davis, Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies), in Modification and Editing of RNA, ed. H. Grosjean and R. Benne, 1998, pp. 85–102 Search PubMed.
- D. R. Davis, C. A. Veltri and L. Nielsen, An RNA model system for investigation of pseudouridine stabilization of the codon-anticodon n interaction in tRNALys, tRNAHis and tRNATyr, J. Biomol. Struct. Dyn., 1998, 15, 1121–1132 CrossRef CAS PubMed.
- C. S. Yarian, M. M. Basti, R. J. Cain, G. Ansaria, R. H. Guenther, E. Sochacka, G. Czerwinska, A. Malkiewics and P. F. Agris, Structural and functional roles of the N1- and N3-protons of Ψ at tRNA's position 39, Nucleic Acids Res., 1999, 27, 3543–3549 CrossRef CAS PubMed.
- P. C. Durant and D. R. Davis, Stabilization of the anticodon n stemloop of tRNALys,3 by and A+–C base pair and by pseudouridine, J. Mol. Biol., 1999, 285, 115–131 CrossRef CAS PubMed.
- T. W. Martin and Z. S. Derewenda, The name is bond—H bond, Nat. Struct. Biol., 1999, 6(5), 403–406 CrossRef CAS PubMed.
- H. Monajemi and S. M. Zain, How stop codon pseudouridylation induces nonsense suppression, Comput. Theor. Chem., 2021, 1204, 113414 CrossRef CAS.
- G. A. Hudson, R. J. Bloomingdale and B. M. Znosko, Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides, RNA, 2013, 19(11), 1474–1482 CrossRef CAS PubMed.
- N. Sugimoto, R. Kierzek, S. M. Freier and D. H. Turner, Energetics of internal GU mismatches in ribooligonucleotide helixes, Biochemistry, 1986, 25(19), 5755–5759 CrossRef CAS PubMed.
- R. C. Brand, et al., Biosynthesis of a Hypermodified Nucleotide in Saccharomyces Carlsbergensis 17S and HeLa-Cell 18S Ribosomal Ribonucleic Acid, Biochem. J., 1978, 169(1), 71–77 CrossRef CAS PubMed.
- J. Urbonavicius, et al., Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer, Mol. Microbiol., 2007, 67(2), 323–335 CrossRef PubMed.
- H. Pang, M. Ihara, Y. Kuchino, S. Nishimura, R. Gupta, C. R. Woese and J. A. McCloskey, Structure of a modified nucleoside in archaebacterial trna which replaces ribosylthymine. 1-Methylpseudouridine, J. Biol. Chem., 1982, 257(7), 3589–3592 CrossRef CAS PubMed.
- R. Gupta, Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs, J. Biol. Chem., 1984, 259(15), 9461–9471 CrossRef CAS PubMed.
- P. Gurha and R. Gupta, Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA, RNA, 2008, 14(12), 2521–2527 CrossRef CAS PubMed.
- J. P. Wurm, et al., Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs, RNA, 2012, 18(3), 412–420 CrossRef CAS PubMed.
- R. Rya, et al., Identification of pseudouridine methyltransferase in Escherichia coli, RNA, 2008, 14(10), 2223–2233 CrossRef PubMed.
- E. Purta, et al., YbeA is the m3Ψ methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA, RNA, 2008, 14(10), 2234–2344 CrossRef CAS PubMed.
- J. P. Wurm, et al., The ribosome assembly factor NEP1 responsible for Bowen–Conradi syndrome is a pseudouridine-N1-specific methyltransferase, Nucleic Acid Res., 2010, 37(7), 2387–2398 CrossRef PubMed.
- B. Meyer, et al., The Bowen–Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Ψ1191 in yeast 18S rRNA, Nucleic Acids Res., 2011, 39(4), 1526–1537 CrossRef CAS PubMed.
- E. Westhof, Pseudouridines or how to draw on weak energy differences, Biochem. Biophys. Res. Commun., 2019, 520(4), 702–704 CrossRef CAS PubMed.
- N. Dutta, I. Deb, J. Sarzynska and A. Lahiri, Structural and thermodynamic consequences of base pairs containing pseudouridine and N1-methylpseudouridine in RNA duplexes, bioRxiv, 2023.
- C. Wang, Y. Zhang and Y. Dong, Lipid nanoparticle-mRNA formulations for therapeutic applications, Acc. Chem. Res., 2021, 54(23), 4283–4293 CrossRef CAS PubMed.
- P. Morais, H. Adachi and Y. T. Yu, The critical contribution of pseudouridine to mRNA COVID-19 vaccines, Front. Cell. Dev. Biol., 2021, 9, 789427 CrossRef PubMed.
- Y. N. Lamb, BNT162b2 mRNA COVID-19 Vaccine: First Approval, Drugs, 2021, 81(4), 495–501 CrossRef CAS PubMed.
- Web: https://www.fda.gov; accessed January 21st 2024.
- T. Uehata and O. Takeuchi, RNA Recognition and Immunity-Innate Immune Sensing and Its Posttranscriptional Regulation Mechanisms, Cells, 2020, 9(7), 1701 CrossRef CAS PubMed.
- M. Herrera-Barrera, R. C. Ryals, M. Gautam, A. Jozic, M. Landry, T. Korzun, M. Gupta, C. Acosta, J. Stoddard, R. Reynaga, W. Tschetter, N. Jacomino, O. Taratula, C. Sun, A. K. Lauer, M. Neuringer and G. Sahay, Peptide-guided lipid nanoparticles deliver mRNA to the neural retina of rodents and nonhuman primates, Sci Adv., 2023, 9(2), eadd4623 CrossRef PubMed.
- D. Shi, S. Toyonaga and D. G. Anderson, In Vivo RNA Delivery to Hematopoietic Stem and Progenitor Cells via Targeted Lipid Nanoparticles, Nano Lett., 2023, 23(7), 2938–2944 CrossRef CAS PubMed.
- Web: https://www.merck.com/news/moderna-and-merck-announce-mrna-4157-v940-an-investigational-personalized-mrna-cancer-vaccine-in-combination-with-keytruda-pembrolizumab-met-primary-efficacy-endpoint-in-phase-2b-keynote-94/; accessed January 21st 2024.
- S. J. Tsai, N. A. Atai, M. Cacciottolo, J. Nice, A. Salehi, C. Guo, A. Sedgwick, S. Kanagavelu and S. J. Gould, Exosome-mediated mRNA delivery in vivo is safe and can be used to induce SARS-CoV-2 immunity, J. Biol. Chem., 2021, 297(5), 101266 CrossRef CAS PubMed.
- N. Gretskaya, M. Akimov, D. Andreev, A. Zalygin, E. Belitskaya, G. Zinchenko, E. Fomina-Ageeva, I. Mikhalyov, E. Vodovozova and V. Bezuglov, Multicomponent Lipid Nanoparticles for RNA Transfection, Pharmaceutics, 2023, 15(4), 1289 CrossRef CAS PubMed.
- L. S. Mbatha, F. Maiyo, A. Daniels and M. Singh, Dendrimer-Coated Gold Nanoparticles for Efficient Folate-Targeted mRNA Delivery In Vitro, Pharmaceutics, 2021, 13(6), 900 CrossRef CAS PubMed.
- A. Suberi, M. K. Grun, T. Mao, B. Israelow, M. Reschke, J. Grundler, L. Akhtar, T. Lee, K. Shin, A. S. Piotrowski-Daspit, R. J. Homer, A. Iwasaki, H. W. Suh and W. M. Saltzman, Polymer nanoparticles deliver mRNA to the lung for mucosal vaccination, Sci. Transl. Med., 2023, 15(709), eabq0603 CrossRef CAS PubMed.
- H. Shin, S. Kang, C. Won and D. H. Min, Enhanced Local Delivery of Engineered IL-2 mRNA by Porous Silica Nanoparticles to Promote Effective Antitumor Immunity, ACS Nano, 2023, 17(17), 17554–17567 CrossRef CAS PubMed.
- M. Segel, B. Lash, J. Song, A. Ladha, C. C. Liu, X. Jin, S. L. Mekhedov, R. K. Macrae, E. V. Koonin and F. Zhang, Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery, Science, 2021, 373(6557), 882–889 CrossRef CAS PubMed.
- D. Grant-Serroukh, M. R. Hunter, R. Maeshima, A. D. Tagalakis, A. M. Aldossary, N. Allahham, G. R. Williams, M. Edbrooke, A. Desai and S. L. Hart, Lipid-peptide nanocomplexes for mRNA delivery in vitro and in vivo, J. Controlled Release, 2022, 348, 786–797 CrossRef CAS PubMed.
- K. Q. Kim, et al., N1-methylpseudouridine found within COVID-19 mRNA vaccines produces faithful protein products, Cell Rep., 2022, 40(9), 111300 CrossRef CAS PubMed.
- E. Rivas, RNA structure prediction using positive and negative evolutionary information, PLoS Comput. Biol., 2020, 16(10), e1008387 CrossRef CAS PubMed.
- M. Chamberlin, J. McGrath and L. Waskell, New RNA polymerase from Escherichia coli infected with bacteriophage T7, Nature, 1970, 228, 227–231 CrossRef CAS PubMed.
- M. Chamberlin, et al., Isolation of bacterial and bacteriophage RNA polymerases and their use in synthesis of RNA in vitro, Methods Enzymol., 1983, 101, 540–568 CAS.
- I. H. Goldberg and M. Rabinowitz, The incorporation of 5-ribosyluracil triphosphate into RNA in nuclear extracts of mammalian cells, Biochem. Biophys. Res. Commun., 1961, 6, 394–398 CrossRef CAS PubMed.
- C. J. Knudson, P. Alves-Peixoto, H. Muramatsu, C. Stotesbury, L. Tang, P. J. C. Lin, Y. K. Tam, D. Weissman, N. Pardi and L. J. Sigal, Lipid-nanoparticle-encapsulated mRNA vaccines induce protective memory CD8 T cells against a lethal viral infection, Mol. Ther., 2021, 29(9), 2769–2781 CrossRef CAS PubMed.
- J. Ramos da Silva, K. Bitencourt Rodrigues and G. Formoso, et al., Single immunizations of self-amplifying or non-replicating mRNA-LNP vaccines control HPV-associated tumors in mice, Sci. Transl. Med., 2023, 15(686), eabn3464 CrossRef CAS PubMed.
- E. Kenjo, H. Hozumi, Y. Makita, K. A. Iwabuchi, N. Fujimoto, S. Matsumoto, M. Kimura, Y. Amano, M. Ifuku, Y. Naoe, N. Inukai and A. Hotta, Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice, Nat. Commun., 2021, 12(1), 7101 CrossRef CAS PubMed.
- K. Karikó, et al., Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA, Immunity, 2005, 23, 165–175 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |