Open Access Article
Open Access ArticleWeak effects of prebiotically plausible peptides on self-triphosphorylation ribozyme function†
Joshua T.
Arriola
a,
Shayan
Poordian
a,
Estefanía Martínez
Valdivia
b,
Tommy
Le
a,
Luke J.
Leman
 c,
Joan G.
Schellinger
b and
Ulrich F.
Müller
c,
Joan G.
Schellinger
b and
Ulrich F.
Müller
 *a
*a
aDepartment of Chemistry & Biochemistry, University of California, San Diego, La Jolla, CA 92093, USA. E-mail: ufmuller@ucsd.edu
bDepartment of Chemistry & Biochemistry, University of San Diego, San Diego, CA 92110, USA
cDepartment of Chemistry, The Scripps Research Institute, La Jolla, CA 92037, USA
First published on 12th September 2024
Abstract
Catalytic RNAs (ribozymes) were central to early stages of life on earth. The first ribozymes probably emerged in the presence of prebiotically generated peptides because amino acids can be generated under abiotic conditions, and amino acids can oligomerize into peptides under prebiotically plausible conditions. Here we tested whether the presence of prebiotically plausible peptides could have aided the emergence of ribozymes, by an in vitro selection of self-triphosphorylation ribozymes from random sequence in the presence of ten different octapeptides. These peptides were composed of ten different, prebiotically plausible amino acids, each as mixture of D- and L-stereoisomers. After five rounds of selection and high throughput sequencing analysis, ten ribozymes that appeared most promising for peptide benefits were tested biochemically for possible benefits from each of the ten peptides. The strongest peptide benefit enhanced ribozyme activity by 2.6-fold, similar to the effect from an increase in the pH by one-half unit. Four arbitrarily chosen ribozymes from a previous selection without peptides showed no significant change in their activity in the presence of the ten peptides. Therefore, the used prebiotically plausible peptides – peptides without evolutionarily optimized sequence, without cationic or aromatic side chains – did not provide a strong benefit for the emergence of ribozyme activity. This finding stands in contrast to previously identified polycationic peptides, conjugates between peptides and polyaromatic hydrocarbons, and modern mRNA encoded proteins, all of which can strongly increase ribozyme function. The results are discussed in the context of origins of life.
Introduction
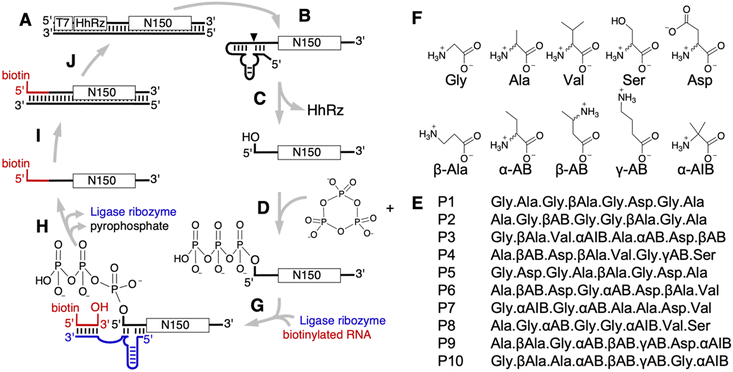
Early stages of life on earth likely relied on catalytic RNAs to promote self-replication and evolution.1–4 This idea is supported by the finding that many of today's enzyme cofactors are nucleotides or variants thereof,5,6 and by the idea that RNA's ability to store information as well as catalyze reactions could have been sufficient for an early RNA-dominated life form that was later overtaken by the contemporary, interdependent DNA–RNA–protein systems.1–4 The strongest support for this hypothesis is the ribosome, which is a highly evolved ribozyme and synthesizes all genetically encoded proteins in today's life forms.7,8 This means that the last universal common ancestor (LUCA) – which contained a complex ribosome9 – was preceded by an evolving molecular system with sophisticated catalytic RNAs. However, it is unclear how a system based on sophisticated RNAs emerged in a prebiotic environment:10 based on current prebiotic model reactions for nucleotide synthesis and non-enzymatic RNA polymerization11–13 and highly controlled experiments that select ribozymes from random sequence,14,15 it appears unlikely that sophisticated ribozymes emerged de novo in a prebiotic environment. One approach to address this challenge is to investigate the possible help of molecules that likely existed on prebiotic earth but don't have that function in today's biochemistry.16 An example for this approach lies in α-hydroxy acids, which can catalyze peptide bond formation in wet-dry cycling conditions.17 Analogously, we explore here whether prebiotically plausible peptides containing five biological and five non-biological amino acids, as mixtures of D- and L-isomers, could have aided the function of catalytic RNAs. Prebiotically plausible peptides are fundamentally different from ribosome-synthesized peptides and proteins because they contain a set of prebiotically plausible amino acids that only partially overlap with biological amino acids, they contain both (biological) L and (non-biological) D stereoisomers, and their amino acid sequence was not encoded and therefore not optimized by Darwinian evolution.Amino acids and peptides can be generated under prebiotically plausible conditions, much easier than the building blocks of RNA (compare ref. 17–20 to ref. 12 and 21–23). Since amino acids appear as side products in reactions that may have given rise to nucleotides,24 it is reasonable to assume that the first catalytic RNAs emerged in the presence of amino acids and peptides. Such peptides might have benefitted early ribozymes: peptides can increase the thermodynamic stability of duplex RNA25 and improve ribozyme interactions with their RNA substrate either by direct interactions26 or by co-localizing the ribozyme and its RNA substrate within RNA/peptide coacervates.27 Most of these previous studies used L-amino acids with a high content of arginine and lysine. The exception is a study that also explored the effects of an ornithine decapeptide on the function of a polymerase ribozyme, and found the same beneficial effect as a lysine decapeptide.26 For both decapeptides, the DL-peptides resulted in the same benefits as the all-L peptides, showing that at least in this case the amino acid chirality is not critical. While ornithine is prebiotically plausible, it is not one of the more frequent amino acids.19 Therefore, a prebiotic function of ornithine decapeptides would likely have required enrichment steps such as coacervate formation with polycationic RNAs. Most previous studies on peptide benefits for ribozymes tested the effect of peptides on individual, existing ribozymes that were selected in the absence of peptides. The exception is a recent in vitro selection study of new ribozymes in the presence of octapeptides composed of the 20 biological L-amino acids, which found that conjugates of octapeptides with polyaromatic hydrocarbons can lead to ∼900-fold enhancement of ribozyme activity.28 However, these peptides consisted exclusively of L-amino acids, and included amino acids that would probably not have existed before a macromolecule-catalyzed metabolism. To directly address the question whether prebiotically plausible peptides could have aided the emergence of the first ribozymes, it is necessary to probe RNA sequence space for the existence of ribozymes – for example by in vitro selection from random sequence – in the presence of prebiotically plausible peptides that consist of prebiotically plausible amino acids, and include the chiral amino acids as D- and L-stereoisomers.
The model system used in this study to probe RNA sequence space for the existence of ribozymes is an in vitro selection for self-triphosphorylation ribozymes.29 This in vitro selection system previously yielded more than 300 different ribozyme clusters from 1.7 × 1014 different sequences,30 and has performed robustly in five different selection contexts.28,29,31–33 Self-triphosphorylation ribozymes are relevant for prebiotic scenarios because they catalyze the reaction between their 5′-hydroxyl group and the prebiotically plausible energy source ‘cyclic trimetaphosphate’ (cTmp)34,35 to generate 5′-triphosphates. The analogous nucleoside 5′-triphosphates (NTPs) are used today as energy currency in all biological organisms, and were thus likely at the core of life forms even before LUCA. Together, the robust performance and prebiotic relevance make this in vitro selection setup an ideal model system to study what molecules can increase the frequency of ribozymes in RNA sequence space, in different chemical environments.
Here we describe the in vitro selection of self-triphosphorylation ribozymes in the presence of ten octapeptides, each composed of D/L mixtures of ten different, prebiotically plausible amino acids, and with the N-terminal amino group as their only positive charge. After high throughput sequencing analysis of the selected RNA sequences, ten promising ribozymes were tested biochemically with each of the ten peptides individually. The strongest peptide benefit to a ribozyme was a 2.6-fold increase in activity, less than the benefit from increasing the pH by one unit. Similarly, four ribozymes that were previously selected in the absence of peptides were not strongly affected by the same ten peptides. The results are discussed in the context of RNA-centered early life forms.
Results
To test whether prebiotically plausible peptides could help the emergence of ribozymes, we performed an in vitro selection of self-triphosphorylating ribozymes in the presence of peptide octamers containing ten different prebiotically plausible amino acids, covering five biological and five non-biological amino acids (Fig. 1). The set of prebiotically available amino acids has been studied in prebiotic model reactions,18–20,36–38 and several approaches comparing the amino acid's chemical complexity and the comparisons of their codons.39 Because the current study is focused on the earliest self-replicating systems long before LUCA, the results from prebiotic model reactions were prioritized. The frequency of the ten amino acids in the ten peptides used in this study was based on their abundance in a representative prebiotic model reaction that was analyzed by modern LC-MS, and should be seen as one of the many, similar plausible distributions for prebiotic scenarios:18,19 22 × glycine, 14 × alanine, 9 × aspartate, 8 × b-alanine, 6 × a-amino butyric acid, 6 × b-amino butyric acid, 5 × valine, 5 × a-amino isobutyric acid, 3 × g-amino butyric acid, 2 × serine. All amino acids with a chiral center were represented as mixtures of D- and L-isomers. The presence of α-, β-, and γ-amino acids generated backbones with different spacing between peptide bonds. The sequences of the peptides were designed manually, with an effort to evenly distribute the ten amino acids over the ten peptides, and the eight positions. Each of the peptides contained between three and six chiral amino acids both as D- and L-isomers, with a total of 232 stereoisomers in the ten peptides. The length of eight amino acids per peptide was chosen because this length is accessible via wet-dry cycling reactions in the presence of α-hydroxy acids.16,17 All ten octapeptides were well soluble at a concentration of at least 20 mM in water.The in vitro selection was conducted as described earlier29,33 (Fig. 1), with an effective complexity of 4.2 × 1014 sequences in the first selection round. The RNAs were challenged to catalyze the nucleophilic attack of their 5′-hydroxyl group to the phosphorus of cyclic trimetaphosphate (cTmp), thereby generating a 5′-triphosphate. The incubation with cTmp was done in the presence of the described ten peptides, each at 1 mM concentration. Depending on the number of stereoisomers for a given peptide, the concentration for each stereoisomer was lower. For example, peptides with 3 chiral positions and therefore 8 stereoisomers contained only 1/8 of that concentration – 125 μM – for each steroisomer. Importantly, there was no selection pressure to utilize these peptides: the procedure only selected for self-triphosphorylation catalysis. Therefore, if a large fraction of the selected ribozymes would depend on peptides, it would show that evolving RNAs adapt to the presence of peptides, and suggest that prebiotic peptides may increase the frequency of ribozymes in sequence space. RNAs that successfully catalyzed the reaction during the incubation with cTmp and the ten peptides, and therefore carried a 5′-triphosphate, were isolated via ligation to a biotinylated oligonucleotide and streptavidin-coated magnetic beads, reverse transcribed, and PCR amplified to complete one cycle of selection. After four rounds of selection, the RNA library was dominated by catalytically active sequences as judged by the decrease of PCR cycles required after reverse transcription (Fig. S1, ESI†).
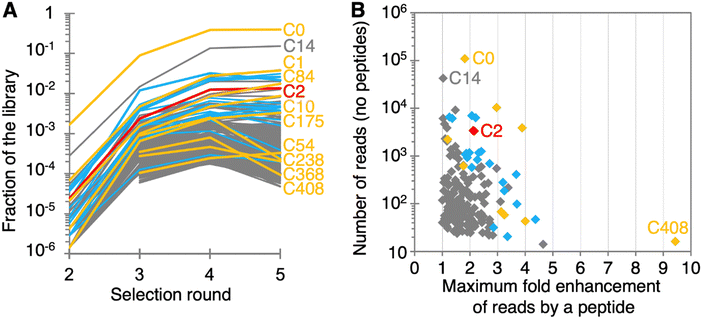
High throughput sequencing (HTS) analysis showed that more than 200 sequence clusters were enriched in the selection protocol (Fig. 2A). To identify whether individual RNAs were benefitting from the presence of peptides, a fifth selection round was appended, in 22 different variants that differed only in the presence of peptides. The first variant was in the presence of all ten peptides, each at 1 mM concentration. The second variant was done in the absence of peptides. The remaining 20 variants were done in the presence of each one of the ten peptides, at either 1 mM or 10 mM concentration. After HTS analysis of these 22 sub-libraries and the four preceding selection rounds, the enrichment of sequence clusters in the presence of individual peptides was determined (Fig. S2, ESI†). To identify ribozymes that may benefit from peptides in their activity, the increase of reads in the presence of individual peptides was determined. No cluster with at least 20 reads showed a stronger peptide-mediated read enhancement than 4-fold, suggesting that none of the ribozyme clusters benefitted more than 4-fold from peptides (the 9.4-fold peptide benefit for cluster 408 could be a result of statistical error because this cluster contained only 16 reads). To test the outcome with the ribozymes that appeared most likely to have a peptide benefit, 30 sequences with a high enrichment in the presence of peptides, and a number of reads without peptides that was high enough for statistical purposes were chosen for further analysis (colored symbols in Fig. 2B; sequences in Fig. S3, ESI†).
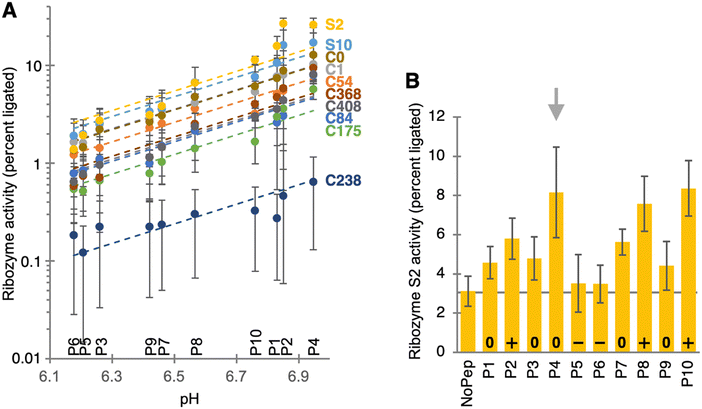
To identify peptide/ribozyme pairs where the peptide may increase ribozyme activity, the 30 chosen ribozyme sequences were tested for catalytic activity under selection conditions, in the absence and presence of the specific peptides that were suggested as beneficial by HTS analysis (Fig. S4A, ESI†). The strongest peptide benefit for ribozyme activity was 1.65-fold, consistent with the idea that under selection conditions, these peptides mediated little benefit to the ribozymes. To test whether peptides could benefit selected ribozymes under suboptimal reaction conditions, the pH in the assay was decreased to pH 7.0, which facilitated about 2-fold peptide benefit for ribozyme S2 (Fig. S4B, ESI†). For additional insight into ribozyme S2, it was truncated at its 3′-terminus, reducing its length from 201 nucleotides in the original isolate to 180 nucleotides while increasing ribozyme activity (Fig. S5, ESI†), and its secondary structure was probed using SHAPE (Fig. S6, ESI†).
Because the HTS data appeared unreliable to identify small peptide benefits, the biochemical assays were expanded to a larger number of ribozyme/peptide combinations. The activity of ten different ribozymes was measured in the presence of the ten individual peptides, in 100 ribozyme/peptide combinations (Fig. 3A). The effect of the ten peptide preparations on the activity of the ten ribozymes was largely described by their influence on the reaction pH. This pH influence was mostly due to an unintended carryover of ammonium hydrogen carbonate from the purification procedure: NH4HCO3 leads to a pH increase in solution because CO2 has a higher vapor pressure than NH3 in aqueous solution. For nine of the ten ribozymes, the logarithm of ribozyme activity was well-correlated with the reaction pH with a slope of 1.0. This slope is indicative of a single deprotonation event as the rate limiting step of catalysis, and was observed previously for a self-triphosphorylation ribozyme.29 This deprotonation is likely the deprotonation of the 5′-hydroxyl group, which is rate limiting for its nucleophilic attack on a phosphorus of cyclic trimetaphosphate. However, ribozyme S2 showed significant deviations from this correlation (yellow symbols in Fig. 3A), with increases of activity in the presence of peptides P4 and P2. Therefore, ribozyme S2 was studied in more detail.
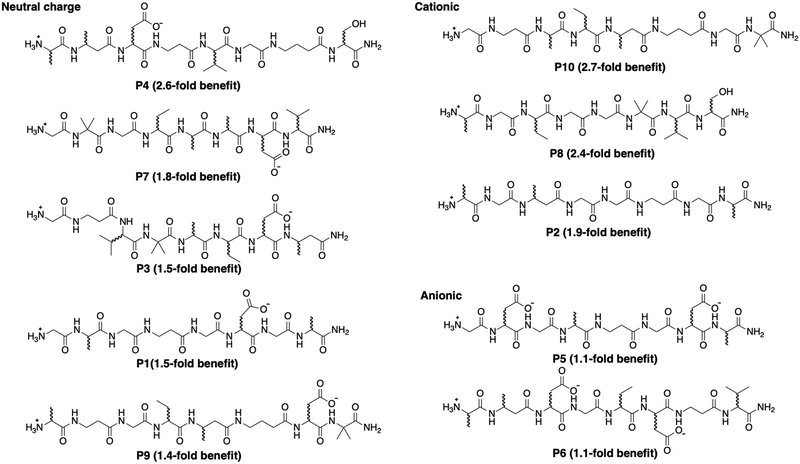
To measure the peptide influence on the activity of ribozyme S2 without the confounding influence of ammonium hydrogen carbonate (Fig. 3B), all peptide preparations were subjected to an additional, 12-hour long lyophilization procedure. The remaining influence of peptide preparations on the reaction pH was therefore caused by the influence of the peptide's amino and carboxy groups, reflected in the peptide's overall charge (the carboxy terminus was used as amide and therefore uncharged). When ribozyme S2 was then incubated with cTmp in the presence or absence of the re-purified peptides, the three peptides with an overall positive charge (P2, P8, P10) resulted in the expected 2- to 3-fold increase in ribozyme activity. Peptides 5 and 6 carried an overall negative charge and did not increase ribozyme activity. In contrast, peptide 4 mediated a 2.6-fold increase in ribozyme activity. Peptides P1, P3, P7 and P9 also did not carry an overall charge and also appeared to increase ribozyme activity by an average of 1.6 ± 0.2-fold. Together, this means that these uncharged peptides mediate a less than 2-fold, nonspecific benefit for ribozyme activity, and that peptide 4 increased the activity of ribozyme S2 slightly more, due to specific interactions of peptide 4 with the ribozyme.
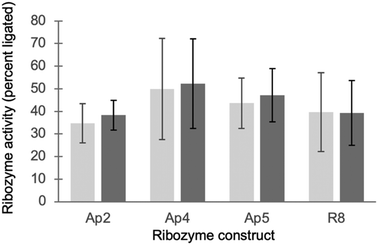
All ribozymes tested above had been selected in the presence of the ten studied peptides, and might therefore have adapted to the presence of these peptides. To test whether ribozymes that were selected in the absence of peptides would be influenced by the peptides, four ribozymes that were selected in the presence of only 50 mM MgCl2, 100 mM Na3cTmp, and 50 mM Tris/HCl pH 8.329,30 were tested in the same buffer but in the presence of the peptides used in this study (Fig. 4). None of the four ribozymes showed a peptide-mediated increase or decrease in activity. Together, these data show that ten octapeptides consisting of D/L mixtures from prebiotically plausible amino acids did not strongly increase, or decrease the activity of self-triphosphorylation ribozymes.
Discussion
To test whether prebiotically plausible peptides can increase the frequency and/or activity of ribozymes in RNA sequence space, an in vitro selection for self-triphosphorylation ribozymes was conducted in the presence of ten prebiotically plausible peptides. This selection identified hundreds of ribozyme clusters, and 10 sequences were analyzed biochemically with each individual peptide for possible benefits. Most peptide effects on ribozyme activity were explained by their effect on reaction pH. A less than 2-fold, non-specific benefit for ribozyme activity was found for four uncharged peptides, while the uncharged peptide P4 mediated a 2.6-fold benefit for ribozyme S2. The data suggest for the specific system used (self-triphosphorylation ribozymes in the presence of ten specific octamers at 1 mM concentration, at 50 mM free Mg2+ concentration and pH 8.3) the peptides provided only weak benefits for the frequency of ribozymes in sequence space, and the activity of the selected ribozymes.The chemical structure of the ten peptides (Fig. 5) allows to speculate what features of the peptides may be responsible for the beneficial effect especially of peptide 4 on ribozyme S2. However, the small differences between the benefits of uncharged peptides could be caused by interactions weaker than a single hydrogen bond, therefore the current data set is insufficient to pin-point the feature(s) of peptide P4 mediating the 2.6-fold benefit for ribozyme S2.
This study explored how prebiotically plausible peptides could have influenced the emergence and function of catalytic RNAs. The interpretation of this mostly negative result with respect to the origin of life – no RNA/peptide couples were identified where peptides increased RNA activity more than 3-fold – needs to be done with care, given that only ten specific octapeptides were studied, that ribozymes were selected only for self-triphosphorylation activity, that all experiments were done with relatively high Mg2+ concentration (50 mM free Mg2+), and that only peptide concentrations up to 10 mM were explored. In addition, it needs to be considered that peptides with encoded sequences were likely not present at the emergence of an RNA world and only generated at the end of an RNA world, when a precursor to the ribosome was able to polymerize amino acids with some sequence fidelity. Given these constraints, it is useful to consider the two extreme hypotheses: the hypothesis that prebiotically plausible peptides were essential for catalytic RNAs is likely incorrect because many ribozymes have been selected, and are active in the absence of peptides. The hypothesis that peptides had no influence on the rise of an RNA world organism is likely also incorrect because peptides likely existed in the same environment in which RNA emerged, and some peptides can support ribozyme function (see introduction and the two paragraphs below). The question is whether peptides in a prebiotic environment could have supported the emergence and function of catalytic RNAs. The narrow interpretation of the results in the present study is that the type of ribozymes studied here (self-triphosphorylation ribozymes) does not benefit more than 3-fold from the ten used octapeptides, under the specific conditions. While only ten octapeptides were studied, it needs to be considered that prebiotically plausible peptides were not synthesized by a ribosome, and did not have an encoded sequence. In the absence of such sequence encoding, the representation of the ten used prebiotically amino acids by the ten used peptidescan serve as an 'average view' that suggests that most peptides based on these ten amino acids are unlikely to strongly affect the emergence of self-triphosphorylation ribozymes, and perhaps many catalytic RNAs. However, it is possible that other sequences of the same amino acids would benefit self-triphosphorylation and other ribozymes. For this to be relevant in a prebiotic scenario, local enrichment by a process of chemical evolution would have to be postulated. For example, polycationic peptides could become enriched in coacervates with RNA40 and aromatic amino acids may form amyloids or other aggregates, especially if they are homochiral.41 This situation changes after the appearance of the first proto-ribosome and amino acid metabolism, when amino acid sequences can be encoded, when homochiral as well as aromatic amino acids become available, and when the benefits seen in today's proteins can complement the function of catalytic RNAs.42–44
If cationic amino acids such as ornithine and 2,4-diamino butyric acid could have been abundant in prebiotic peptides then the corresponding peptides may have changed the outcome of this study. These amino acids were not included in the current study because their abundance in prebiotic model reactions is far below that of the ten used amino acids.19 However, it is well documented that peptides that are rich in the cationic amino acids arginine or lysine can improve ribozyme activity by increasing the thermodynamic stability of RNA/RNA duplexes,25 stabilizing the sequence non-specific binding of an RNA substrate by a ribozyme,26 or by forming coacervates with the RNA that lead to better RNA/RNA interactions.27 Peptides containing multiple cationic and multiple hydrophobic amino acids can enhance the activity of polymerase ribozymes through the formation of aggregates.45 All of these studies required multiple cationic amino acids to benefit ribozyme function, and the requirement of multiple rare amino acids further reduces the peptide's expected abundance in a prebiotic setting. However, very rare peptides could have become enriched and co-localized with RNA by partitioning of polycationic peptides in coacervates with polyanionic RNA. It remains to be seen whether polycationic peptides can be generated, and enriched in prebiotic model reactions in sufficient abundance to provide their benefit to catalytic nucleic acids.
While aromatic amino acids are rare or absent in prebiotic model reactions,19 peptides containing multiple aromatic amino acids are able to aggregate in amyloids and then interact sequence specifically with short RNA fragments.41 Moreover, aromatic moieties have been shown to benefit ribozyme activity: conjugates between polyaromatic hydrocarbons and peptides are able to increase the activity of some ribozymes by 100-fold or more.28 Since insoluble organic matter is the dominant form of carbon in meteorites, and rich in polyaromatic hydrocarbons,46,47 it is possible that the prebiotic environment was rich in polyaromatic hydrocarbons, and present in prebiotic peptides. This may have acted as ‘prebiotic equivalent’ to the aromatic amino acids present in modern proteins. However, it is unclear whether this usefulness of PAHs for ribozymes is a rare phenomenon or whether it may have been wide-spread and important during an RNA world. Future work will show what kind of prebiotically plausible molecules could have aided the emergence and function of an RNA world organism.
Materials and methods
Synthesis and purification of peptides
Peptides were synthesized on rink amide PS resin (0.77 mmol g−1, Novabiochem) and Rink Amide ChemMatrix (0.45 mmol g−1, Biotage) following the Fmoc/tBu strategy using a microwave-assisted peptide synthesizer (Alstra, Biotage). Side chain protecting groups for amino acids were as follows: Ser(tBu), His(Trt), Lys(Boc), Asp(OtBu), Glu(OtBu). Unnatural amino acids used for the syntheses of prebiotically plausible peptides were Fmoc-D-Ala-OH, Fmoc-D-Asp-OH (OtBu), Fmoc-D-Val-OH, Fmoc-D-Ser-OH(tBu), Fmoc-β-alanine, Fmoc-α-L-aminobutyric acid, Fmoc-α-D-aminobutyric acid, Fmoc-L-β-aminobutyric acid, Fmoc-D-β-aminobutyric acid, γ-aminobutyric acid, Fmoc-α-amino butyric acid. For the peptides containing prebiotically plausible residues, a racemic mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used for each amino acid reagents. Typical coupling reactions were performed at 0.100–0.200 mmol scale with 3.0 equiv. of Fmoc-protected amino acids, 2.94 equiv. of HBTU and 6.0 equiv. of NMM for 5 min at 75 °C. All reagents were pre-dissolved in DMF at 0.5 M. Fmoc deprotections were performed with 20% piperidine in DMF for 10 min at rt. Washing was performed after every deprotection and coupling step using DMF. For the syntheses of some sequences, the deprotection and coupling steps described above were either performed twice or with increased reagent equivalence to ensure reaction completion. Cleavage of the peptide sequences from the solid support with concomitant side chain deprotection was accomplished by placing the resin in fritted SPE tube and treating with TFA cleavage cocktail (∼20 mL g−1) containing 90
1) was used for each amino acid reagents. Typical coupling reactions were performed at 0.100–0.200 mmol scale with 3.0 equiv. of Fmoc-protected amino acids, 2.94 equiv. of HBTU and 6.0 equiv. of NMM for 5 min at 75 °C. All reagents were pre-dissolved in DMF at 0.5 M. Fmoc deprotections were performed with 20% piperidine in DMF for 10 min at rt. Washing was performed after every deprotection and coupling step using DMF. For the syntheses of some sequences, the deprotection and coupling steps described above were either performed twice or with increased reagent equivalence to ensure reaction completion. Cleavage of the peptide sequences from the solid support with concomitant side chain deprotection was accomplished by placing the resin in fritted SPE tube and treating with TFA cleavage cocktail (∼20 mL g−1) containing 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 TFA
2.5 TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dimethoxybenzene
dimethoxybenzene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O
H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS for 2 hour. Cleaved peptides were then precipitated in cold ether, centrifuged, dissolved in methanol and reprecipitated in cold ether (3×).
TIS for 2 hour. Cleaved peptides were then precipitated in cold ether, centrifuged, dissolved in methanol and reprecipitated in cold ether (3×).
HPLC characterization and purification of peptides were carried out at rt on analytical (Jupiter C18 5 μm, 300 Å, 150 × 4.6 mm by Phenomenex, Torrance, CA) and semi-preparative columns (Aquasil C18 5 μm, 100 Å, 150 × 10 mm by Keystone Scientific Inc., Waltham, MA) with Prostar 325 Dual Wavelength UV-Vis Detector from Agilent Technologies with Varian pumps (Santa Clara, CA) with detection set at 225 and 406 nm. Peptides were eluted from column following a gradient using mobile phases A: 0.1% TFA in H20 and B: 0.1% TFA in CH3CN. MS analyses were obtained on a LTQ ESI-MS spectrometer (San Jose, CA). Solutions were prepared in either methanol or methanol/water (formic acid 1%) with flow rate of 10 μL min−1, spray voltage at 4.50 kV, capillary temperature at 300 °C, capillarity voltage at 7.00 V, tube lens at 135.00 V. Purified peptides were characterized by analytical HPLC (with purity typically greater than 90%) and MS (either by direct injection or LC-MS).
To remove a possible carryover of TFA, peptides were dissolved in a total volume of 2 to 5 mL 100 mM (NH4)HCO3, and frozen as a thin film on the inside of a glass bulb cooled in a dry ice/isopropanol bath. After lyophilizing the frozen solution to dryness in oil vacuum (∼2 mbar), the process was repeated once with 100 mM (NH4)HCO3, and once or twice with water. The desiccated peptide was weighed, and dissolved in water to a stock concentration of 10 mM.
In vitro selection
A 183 nucleotide (nt) long DNA oligomer that included 150 positions with randomized sequence and flanked by 5′- and 3′-constant regions was purchased from integrated DNA technologies (IDT). 5′-GAGACCGCAAGAGAC(N150)CATGGTTCAGACTACAAC-3′ A 82 nt long oligomer was used to add the promoter sequence of T7 RNA polymerase and the sequence of the hammerhead ribozyme (underlined) to the 5′ end of the 183-mer. 5′-AATTTAATACGACTCACTATA![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GAGACCGCAAGAGAC-3′. Amplification with the 67-mer and a short 3′ primer generated the double stranded DNA pool. qPCR was used to determine the amount of amplifiable DNA and this number was used to estimate that the starting DNA pool consisted of 4.2 × 1014 sequences. The DNA pool was transcribed into RNA using T7 RNA polymerase under standard conditions. 100 nM of purified RNA sub-pool was incubated with 50 mM Tris/HCl, 3.3 mM NaOH, 100 mM MgCl2, 50 mM of freshly dissolved trimetaphosphate and 1 mM each of ten prebiotically plausible peptides (10 mM total). This mixture was allowed to incubate for 3 hours at room temperature. The reaction was quenched by ethanol precipitation. The RNA pellet was washed with chilled water and desalted using P30 Tris/HCl spin columns (Bio-Rad). Active ribozymes were ligated to a biotinylated oligomer (biotin-d(GTAACATAATGA)rU) using the R3C ligase ribozyme whose arms were designed to anneal to the 5′ constant region of the pool RNA and the biotinylated oligomer. A mixture containing 1.3 μM of desalted pool RNA, 1 μM ligase ribozyme, 1.2 μM biotinylated oligomer, 100 mM KCl, 100 mM Tris/HCl, and 60 pM triphosphorylated RNA was heated to 65 °C for 2 minutes then cooled to 30 °C at a rate of 0.1 °C per second. The triphosphorylated RNA was used to generate a small amount of ligation product and thereby reduce the number of PCR cycles, and prevent artifacts during PCR amplification. After heat renaturation, an equal volume of a mix consisting of 40% (w/v) PEG8000, 4 mM Spermidine, and 50 mM MgCl2 was added to the ligase reaction mixture. This mixture was incubated at 30 °C for 3 hours. After the incubation step, the reaction was quenched by adding final concentrations of 13.9 mM sodium/EDTA, 50 mM Tris/HCl, 50 mM KCl, 0.012% (w/v) Triton X-100, and 1.19 μM of a 61 nt oligomer that was complementary to the ligase ribozyme (5′-AGTAACATAATGACTTCAACCCATTCAAACTGTTCTTACGAACAATCGAGCACAAGAGAC-3′). This mixture was heated at 50 °C for 10 minutes. Streptavadin magnetic beads (Promega) were washed thrice with 20 mM HEPES/KOH pH7.2, 0.01% (w/v) Triton X-100 and 50 mM KCl. Biotinylated RNA was then captured by mixing the RNA with the magnetic beads and rotating end-over-end at room temperature for at least 30 minutes. A magnetic rack was used to focus the beads and the beads were washed twice with a solution containing 0.01% (w/v) Triton X-100 and 20 mM NaOH. Captured RNA was eluted from the beads by incubating the beads in a solution of 25 mM Tris/HCl pH 8.5, 1.56 mM EDTA and 96% formamide at 65 °C for 3 minutes. The beads were removed by immediate centrifugation, and the supernatant was concentrated by ethanol precipitation. 1 μg of tRNA was used as precipitation carrier. Captured RNA was resuspended in 10 mM Tris/HCl, pH 8.3. The RNA was reverse transcribed using superscript III (Invitrogen) and a reverse transcription primer (5′-GTTGTAGTCTGAACCATG-3′) complementary to the 3′ constant region. The products were then PCR amplified with 5′ and 3′ primers that could bind to the sequence of the biotinylated oligomer and the 3′ constant region of the pool respectively. A second PCR amplification was performed to add the T7 promoter sequence and the hammerhead ribozyme sequence to the DNA.
GAGACCGCAAGAGAC-3′. Amplification with the 67-mer and a short 3′ primer generated the double stranded DNA pool. qPCR was used to determine the amount of amplifiable DNA and this number was used to estimate that the starting DNA pool consisted of 4.2 × 1014 sequences. The DNA pool was transcribed into RNA using T7 RNA polymerase under standard conditions. 100 nM of purified RNA sub-pool was incubated with 50 mM Tris/HCl, 3.3 mM NaOH, 100 mM MgCl2, 50 mM of freshly dissolved trimetaphosphate and 1 mM each of ten prebiotically plausible peptides (10 mM total). This mixture was allowed to incubate for 3 hours at room temperature. The reaction was quenched by ethanol precipitation. The RNA pellet was washed with chilled water and desalted using P30 Tris/HCl spin columns (Bio-Rad). Active ribozymes were ligated to a biotinylated oligomer (biotin-d(GTAACATAATGA)rU) using the R3C ligase ribozyme whose arms were designed to anneal to the 5′ constant region of the pool RNA and the biotinylated oligomer. A mixture containing 1.3 μM of desalted pool RNA, 1 μM ligase ribozyme, 1.2 μM biotinylated oligomer, 100 mM KCl, 100 mM Tris/HCl, and 60 pM triphosphorylated RNA was heated to 65 °C for 2 minutes then cooled to 30 °C at a rate of 0.1 °C per second. The triphosphorylated RNA was used to generate a small amount of ligation product and thereby reduce the number of PCR cycles, and prevent artifacts during PCR amplification. After heat renaturation, an equal volume of a mix consisting of 40% (w/v) PEG8000, 4 mM Spermidine, and 50 mM MgCl2 was added to the ligase reaction mixture. This mixture was incubated at 30 °C for 3 hours. After the incubation step, the reaction was quenched by adding final concentrations of 13.9 mM sodium/EDTA, 50 mM Tris/HCl, 50 mM KCl, 0.012% (w/v) Triton X-100, and 1.19 μM of a 61 nt oligomer that was complementary to the ligase ribozyme (5′-AGTAACATAATGACTTCAACCCATTCAAACTGTTCTTACGAACAATCGAGCACAAGAGAC-3′). This mixture was heated at 50 °C for 10 minutes. Streptavadin magnetic beads (Promega) were washed thrice with 20 mM HEPES/KOH pH7.2, 0.01% (w/v) Triton X-100 and 50 mM KCl. Biotinylated RNA was then captured by mixing the RNA with the magnetic beads and rotating end-over-end at room temperature for at least 30 minutes. A magnetic rack was used to focus the beads and the beads were washed twice with a solution containing 0.01% (w/v) Triton X-100 and 20 mM NaOH. Captured RNA was eluted from the beads by incubating the beads in a solution of 25 mM Tris/HCl pH 8.5, 1.56 mM EDTA and 96% formamide at 65 °C for 3 minutes. The beads were removed by immediate centrifugation, and the supernatant was concentrated by ethanol precipitation. 1 μg of tRNA was used as precipitation carrier. Captured RNA was resuspended in 10 mM Tris/HCl, pH 8.3. The RNA was reverse transcribed using superscript III (Invitrogen) and a reverse transcription primer (5′-GTTGTAGTCTGAACCATG-3′) complementary to the 3′ constant region. The products were then PCR amplified with 5′ and 3′ primers that could bind to the sequence of the biotinylated oligomer and the 3′ constant region of the pool respectively. A second PCR amplification was performed to add the T7 promoter sequence and the hammerhead ribozyme sequence to the DNA.
HTS analysis
Illumina sequencing primers were added to the ends of rounds 2–5 pools. Pools were sequenced on MiSeq with 10% PhiX. FastQ files were processed using custom Python scripts for rounds 2–5 (including all sub-rounds in round 5). The usearch merge function was used to merge forward and revers reads. Sequences not containing both constant regions or short randomized regions (<135 nt in length or 10% of 150) were first filtered out. The file was reformatted to Fasta for easier downstream processing. The total counts for each sequence were calculated. The usearch clustering algorithm was used to group similar sequences into clusters. Clusters were defined as sequences that shared at least 75% sequence identity and had at least 100 reads in a given round. Clusters and sequences were tracked through each round. 189 sequences were evaluated further. For each sequence, the fold enrichment in the round 5 sub-rounds were determined by normalizing to the no peptide sub-round.Generation of 3′ truncations
3′ truncations were generated by PCR using Taq DNA polymerase and a series of 3′ primers. The 3′ primers annealed to different portions of the ribozyme sequence of interest to generate a series of truncations with desired deletions from the 3′ end. DNA templates of each truncation were transcribed using T7 RNA polymerase, and purified by denaturing PAGE.Self-triphosphorylation activity assays
Purified ribozyme (5 μM) was incubated with 50 mM Tris/HCl, 100 mM MgCl2, 50 mM of freshly dissolved trimetaphosphate, and either a mixture of peptides, individual peptides or water as a control. This mixture was incubated for 3 hours at room temperature. The products were ligated to a radiolabeled oligomer (γ32P-d(GTAACATAATGA)rU) using an R3C ligase ribozyme whose arms were designed to anneal to the 5′ constant region of the pool RNA and the oligomer. Following the incubation with buffer, Tmp and peptides, the mixture was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in a solution that contained a final concentration of 0.5 μM ligase ribozyme, 0.5 μM biotinylated oligomer, 100 mM KCl, 100 mM Tris/HCl, and 15 mM sodium EDTA. This mixture was heat renatured at 65 °C for 2 minutes then cooled to 30 °C at a rate of 0.1 °C per second. After heat renaturation, an equal volume of a mix consisting of 40% PEG8000 (w/v), 4 mM Spermidine, and 50 mM MgCl2 was added to the ligase reaction mixture. This mixture was incubated at 30 °C for 2 hours. After the incubation step, the reaction was quenched by ethanol precipitation. The reaction products were resolved on a denaturing 15% polyacrylamide denaturing gel.
10 in a solution that contained a final concentration of 0.5 μM ligase ribozyme, 0.5 μM biotinylated oligomer, 100 mM KCl, 100 mM Tris/HCl, and 15 mM sodium EDTA. This mixture was heat renatured at 65 °C for 2 minutes then cooled to 30 °C at a rate of 0.1 °C per second. After heat renaturation, an equal volume of a mix consisting of 40% PEG8000 (w/v), 4 mM Spermidine, and 50 mM MgCl2 was added to the ligase reaction mixture. This mixture was incubated at 30 °C for 2 hours. After the incubation step, the reaction was quenched by ethanol precipitation. The reaction products were resolved on a denaturing 15% polyacrylamide denaturing gel.
SHAPE probing
Selective 2′hydroxyl acylation analyzed by primer extension (SHAPE) was performed on the 180-nucleotide long ribozyme isolated in this study, with 1-methyl-7-nitro-2H-3,1-benzoxazine-2,4(1H)-dione (1M7) as the chemical probe. 50 pmol of ribozyme were incubated in buffer containing 50 mM MES/NaOH pH 6.0, 100 mM MgCl2, 3.3 mM NaOH, 50 mM Tmp, and 10 mM peptide 4 at room temperature for 2 minutes. This mixture was incubated at room temperature for 2 minutes or 2 hours. 1M7 was added to this solution such that its final concentration was 2 mM and the concentration of DMSO was 1% of the solution. A negative control was prepared with no 1M7 and 1% DMSO. Both samples were incubated at room temperature for 3 minutes. The reaction was quenched by ethanol precipitation and resuspended in 10 μL of 5 mM Tris/HCl pH 8.0. The products were reverse transcribed using superscript III reverse transcriptase (Invitrogen) according to manufacturer's instructions and trace amounts of a radiolabeled primers were added. Three reverse transcription primers were used to probe different portions of the ribozyme. The sequences of these primers are (5′-TTAGTCTGAACCATGA-3′, annealed to pos. 165–180); (5′-CGATGTTGACATTTTC-3′, annealed to pos. 112–127); and (5′-AAACTCAAGTGGGTTT-3′, annealed to pos. 53–68). The RNA template in each sample was degraded by alkaline hydrolysis, by incubating for 5 minutes at 80 °C in a solution containing 750 mM NaOH. The reaction was quenched by adding a two-fold stoichiometric excess of acetic acid to generate a NaOAc/HOAc buffer with a final concentration of 300 mM. The products were ethanol precipitated, resuspended in formamide loading buffer and resolved on a denaturing 10% polyacrylamide gel. The RNA secondary structure was predicted with help by the secondary structure prediction tool RNAstructure (ReuterMathews10). To do this, the SHAPE data was mapped onto an initial secondary structure prediction and manually adjusted to better represent the SHAPE data.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Aeronautics and Space Association (NASA) with the award NNX16AJ27G to UFM. We would like to acknowledge Kevin J. Sweeney for advice on performing the in vitro selection in the presence of peptides, and on the HTS analysis.References
- A. Rich, On the problems of evolution and biochemical information transfer. Horizons in biochemistry, Academic Press Inc., New York, NY, 1962, pp. 103–126 Search PubMed.
- F. H. C. Crick, The origin of the genetic code, J. Mol. Biol., 1968, 38, 367–379 CrossRef CAS PubMed.
- C. R. Woese, The genetic code. The molecular basis for genetic expression, Harper & Row, New York, 1967 Search PubMed.
- L. E. Orgel, Evolution of the genetic apparatus, J. Mol. Biol., 1968, 38, 381–393 CrossRef CAS PubMed.
- H. B. White 3rd, Coenzymes as fossils of an earlier metabolic state, J. Mol. Evol., 1976, 7(2), 101–104 CrossRef CAS PubMed.
- A. D. Goldman and B. Kacar, Cofactors are Remnants of Life's Origin and Early Evolution, J. Mol. Evol., 2021, 89(3), 127–133 CrossRef CAS PubMed.
- H. F. Noller, V. Hoffarth and L. Zimniak, Unusual resistance of peptidyl transferase to protein extraction procedures, Science, 1992, 256(5062), 1416–1419 CrossRef CAS PubMed.
- P. Nissen, J. Hansen, N. Ban, P. B. Moore and T. A. Steitz, The structural basis of ribosome activity in peptide bond synthesis, Science, 2000, 289(5481), 920–930 CrossRef CAS PubMed.
- A. J. Crapitto, A. Campbell, A. Harris and A. D. Goldman, A consensus view of the proteome of the last universal common ancestor, Ecol. Evol., 2022, 12(6), e8930 CrossRef PubMed.
- P. Pavlinova, C. N. Lambert, C. Malaterre and P. Nghe, Abiogenesis through gradual evolution of autocatalysis into template-based replication, FEBS Lett., 2022, 1873–3468, 14507 Search PubMed.
- S. Stairs, A. Nikmal, D. K. Bucar, S. L. Zheng, J. W. Szostak and M. W. Powner, Divergent prebiotic synthesis of pyrimidine and 8-oxo-purine ribonucleotides, Nat. Commun., 2017, 8, 15270 CrossRef CAS PubMed.
- S. Becker, J. Feldmann, S. Wiedemann, H. Okamura, C. Schneider and K. Iwan, et al., Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides, Science, 2019, 366(6461), 76–82 CrossRef CAS PubMed.
- F. R. Bowler, C. K. Chan, C. D. Duffy, B. Gerland, S. Islam and M. W. Powner, et al., Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation, Nat. Chem., 2013, 5(5), 383–389 CrossRef CAS PubMed.
- D. P. Bartel and J. W. Szostak, Isolation of new ribozymes from a large pool of random sequences, Science, 1993, 261(5127), 1411–1418 CrossRef CAS PubMed.
- A. Akoopie, J. T. Arriola, D. Magde and U. F. Müller, A GTP-synthesizing ribozyme selected by metabolic coupling to an RNA polymerase ribozyme, Sci. Adv., 2021, 7(41), eabj7487 CrossRef CAS PubMed.
- R. Krishnamurthy, Giving Rise to Life: Transition from Prebiotic Chemistry to Protobiology, Acc. Chem. Res., 2017, 50(3), 455–459 CrossRef CAS PubMed.
- J. G. Forsythe, S. S. Yu, I. Mamajanov, M. A. Grover, M. Krishnamurthy and F. M. Fernandez, et al., Ester-mediated amide bond formation driven by wet-dry cycles: a possible path to polypeptides on the prebiotic Earth, Angew. Chem., Int. Ed., 2015, 54(34), 9871–9875 CrossRef CAS PubMed.
- S. L. Miller, A production of amino acids under possible primitive earth conditions, Science, 1953, 117(3046), 528–529 CrossRef CAS PubMed.
- E. T. Parker, H. J. Cleaves, J. P. Dworkin, D. P. Glavin, M. Callahan and A. Aubrey, et al., Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(14), 5526–5531 CrossRef CAS PubMed.
- C. S. Foden, S. Islam, C. Fernández-García, L. Maugeri, T. D. Sheppard and M. W. Powner, Prebiotic synthesis of cysteine peptides that catalyze peptide ligation in neutral water, Science, 2020, 370(6518), 865–869 CrossRef CAS PubMed.
- M. W. Powner, B. Gerland and J. D. Sutherland, Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions, Nature, 2009, 459(7244), 239–242 CrossRef CAS PubMed.
- W. Huang and J. P. Ferris, Synthesis of 35–40 mers of RNA oligomers from unblocked monomers. A simple approach to the RNA world, Chem. Commun., 2003,(12), 1458–1459 RSC.
- L. Li, N. Prywes, C. P. Tam, D. K. O’Flaherty, V. S. Lelyveld and E. C. Izgu, et al., Enhanced Nonenzymatic RNA Copying with 2-Aminoimidazole Activated Nucleotides, J. Am. Chem. Soc., 2017, 139(5), 1810–1813 CrossRef CAS PubMed.
- S. Islam and M. W. Powner, Prebiotic systems chemistry: Complexity overcoming clutter, Chem., 2017, 2(4), 470–501 CAS.
- M. Frenkel-Pinter, J. W. Haynes, A. M. Mohyeldin, M. C, A. B. Sargon and A. S. Petrov, et al., Mutually stabilizing interactions between proto-peptides and RNA, Nat. Commun., 2020, 11(1), 3137 CrossRef CAS PubMed.
- S. Tagami, J. Attwater and P. Holliger, Simple peptides derived from the ribosomal core potentiate RNA polymerase ribozyme function, Nat. Chem., 2017, 9(4), 325–332 CrossRef CAS PubMed.
- R. R. Poudyal, C. D. Keating and P. C. Bevilacqua, Polyanion-Assisted Ribozyme Catalysis Inside Complex Coacervates, ACS Chem. Biol., 2019, 14(6), 1243–1248 CrossRef CAS PubMed.
- K. J. Sweeney, T. Le, M. Z. Jorge, J. G. Schellinger, L. J. Leman and U. F. Müller, Peptide conjugates with polyaromatic hydrocarbons can benefit the activity of catalytic RNAs, Chem. Sci., 2023, 14, 10318–10328 RSC.
- J. E. Moretti and U. F. Muller, A ribozyme that triphosphorylates RNA 5′-hydroxyl groups, Nucleic Acids Res., 2014, 42(7), 4767–4778 CrossRef CAS PubMed.
- A. Pressman, J. E. Moretti, G. W. Campbell, U. F. Muller and I. A. Chen, Analysis of in vitro evolution reveals the underlying distribution of catalytic activity among random sequences, Nucleic Acids Res., 2017, 45(14), 8167–8179 CrossRef CAS PubMed.
- G. F. Dolan, A. Akoopie and U. F. Muller, A Faster Triphosphorylation Ribozyme, PLoS One, 2015, 10(11), e0142559 CrossRef PubMed.
- J. T. Arriola and U. F. Müller, A combinatorial method to isolate short ribozymes from complex ribozyme libraries, Nucleic Acids Res., 2020, 48(20), e116–e116 CrossRef CAS PubMed.
- K. J. Sweeney, X. Han and U. F. Müller, A ribozyme that uses lanthanides as cofactor, Nucleic Acids Res., 2023, gkad513 Search PubMed.
- M. A. Pasek, T. P. Kee, D. E. Bryant, A. A. Pavlov and J. I. Lunine, Production of potentially prebiotic condensed phosphates by phosphorus redox chemistry, Angew. Chem., Int. Ed., 2008, 47(41), 7918–7920 CrossRef CAS PubMed.
- M. A. Pasek, J. P. Harnmeijer, R. Buick, M. Gull and Z. Atlas, Evidence for reactive reduced phosphorus species in the early Archean ocean, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(25), 10089–10094 CrossRef CAS PubMed.
- W. Löb, Über das Verhalten des Formamids unter der Wirkung der stillen Entladung Ein Beitrag zur Frage der Stickstoff-Assimilation, Ber. Dtsch. Chem. Ges., 1913, 46(1), 684–697 CrossRef.
- D. Ring, Y. Wolman, N. Friedmann and S. L. Miller, Prebiotic Synthesis of Hydrophobic and Protein Amino Acids, Proc. Natl. Acad. Sci. U. S. A., 1972, 69(3), 765–768 CrossRef CAS PubMed.
- Y. Wolman, W. J. Haverland and S. L. Miller, Nonprotein Amino Acids from Spark Discharges and Their Comparison with the Murchison Meteorite Amino Acids, Proc. Natl. Acad. Sci. U. S. A., 1972, 69(4), 809–811 CrossRef CAS PubMed.
- E. N. Trifonov, The triplet code from first principles, J. Biomol. Struct. Dyn., 2004, 22(1), 1–11 CrossRef CAS PubMed.
- F. P. Cakmak, S. Choi, M. O. Meyer, P. C. Bevilacqua and C. D. Keating, Prebiotically-relevant low polyion multivalency can improve functionality of membraneless compartments, Nat. Commun., 2020, 11(1), 5949–5959 CrossRef CAS PubMed.
- S. K. Rout, R. Cadalbert, N. Schröder, J. Wang, J. Zehnder and O. Gampp, et al., An Analysis of Nucleotide–Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA, J. Am. Chem. Soc., 2023, 145(40), 21915–21924 CrossRef CAS PubMed.
- H. Hartman and T. Smith, The Evolution of the Ribosome and the Genetic Code, Life, 2014, 4(2), 227–249 CrossRef PubMed.
- N. J. Reiter, A. Osterman, A. Torres-Larios, K. K. Swinger, T. Pan and A. Mondragon, Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA, Nature, 2010, 468(7325), 784–789 CrossRef CAS PubMed.
- C. L. Will and R. Luhrmann, Spliceosome structure and function, Cold Spring Harbor Perspect. Biol., 2011, 3(7), a003707 CAS.
- P. Li, P. Holliger and S. Tagami, Hydrophobic-cationic peptides modulate RNA polymerase ribozyme activity by accretion, Nat. Commun., 2022, 13(1), 3050 CrossRef CAS PubMed.
- G. D. Cody, E. Heying, C. M. O. Alexander, L. R. Nittler, A. L. D. Kilcoyne and S. A. Sandford, et al., Establishing a molecular relationship between chondritic and cometary organic solids, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(48), 19171–19176 CrossRef CAS PubMed.
- C. M. O. Alexander, M. Fogel, H. Yabuta and G. D. Cody, The origin and evolution of chondrites recorded in the elemental and isotopic compositions of their macromolecular organic matter, Geochim. Cosmochim. Acta, 2007, 71(17), 4380–4403 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00129j |
| This journal is © The Royal Society of Chemistry 2024 |