Open Access Article
Open Access ArticleCapture of RNA G-quadruplex structures using an L-RNA aptamer†
Sin Yu
Lam‡
 a,
Mubarak Ishaq
Umar‡
ab,
Haizhou
Zhao
a,
Jieyu
Zhao
a and
Chun Kit
Kwok
a,
Mubarak Ishaq
Umar‡
ab,
Haizhou
Zhao
a,
Jieyu
Zhao
a and
Chun Kit
Kwok
 *ac
*ac
aDepartment of Chemistry and State Key Laboratory of Marine Pollution, City University of Hong Kong, Kowloon Tong, Hong Kong SAR 999077, China
bRNA Molecular Biology Group, National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), National Institutes of Health, Bethesda, MD, USA
cShenzhen Research Institute of City University of Hong Kong, Shenzhen, China. E-mail: ckkwok42@cityu.edu.hk
First published on 29th August 2024
Abstract
G-quadruplexes (dG4 and rG4) are nucleic acid secondary structures formed by the self-assembly of certain G-rich sequences, and they have distinctive chemical properties and play crucial roles in fundamental biological processes. Small molecule G4 ligands were shown to be crucial in characterizing G4s and understanding their functions. Nevertheless, concerns regarding the specificity of these synthetic ligands for further investigation of G4s, especially for rG4 isolation purposes, have been raised. In comparison to G4 ligands, we propose a novel magnetic bead-based pulldown assay that enables the selective capture of general rG4s using functionalized L-Apt.4-1c from both simple buffer and complex media, including total RNA and the cell lysate. We found that our L-RNA aptamer can pulldown general rG4s with a higher efficiency and specificity than the G4 small molecule ligand BioTASQ v.1 in the presence of non-target competitors, including dG4 and non-G4 structures. Our findings reveal that biotinylated L-aptamers can serve as effective molecular tools for the affinity-based enrichment of rG4 of interest using this new assay, which was also verified by quantitative reverse transcription-polymerase chain reaction (RT-qPCR) on endogenous transcripts. This work provides new and important insights into rG4 isolation using a functionalized L-aptamer, which can potentially be applied in a transcript-specific or transcriptome-wide manner in the future.
Introduction
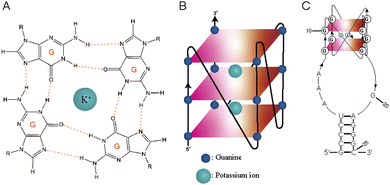
RNA G-quadruplexes (rG4s) are non-canonical secondary structures that form through association of guanine (G)-rich RNA sequences. Four guanine bases (Gs) interact with each other to form square planar G-quartet structures (Fig. 1A), which stack to form G-quadruplex (G4) structures (Fig. 1B). These structures are stabilized in the presence of central monovalent cations (K+ > Na+ > Li+).1,2 G-quadruplexes (G4s) are thermostable and play vital roles in various cellular activities, including replication, transcription, and translation, in diverse organisms, encompassing humans, mice, plants, viruses, yeasts, and bacteria.3–9 Due to their biological significance, G4s have been recognized as promising targets in cancer research and antimicrobial studies.10,11 Targeting G4s is crucial for studying their cellular roles,12 and over the years, several classes of G4 targeting tools have been developed, which include G4-specific antibodies, peptides, and chemicals.13Among the G4 targeting tools mentioned above, several G4-specific tools have shown high affinity for G4s (dG4s and rG4) and are useful for stabilizing and characterizing them.14–18 These tools include antibodies like BG4 and 1H6 as well as peptides like cyclic RHAU23, b-G4pep3, and pep11.19–22 Small molecule G4 ligands, such as N-methyl mesoporphyrin IX (NMM), QUMA-1, and Thioflavin T (ThT), have also demonstrated preferential binding to both DNA and RNA G4s.23–25 They are widely used for G4 detection due to their ability to increase fluorescence intensity upon binding. Pyridostatin (PDS) is an example that has been reported to stabilize telomeric G4 and subsequently disrupt the telomerase activity of human cells.26 Template-assembled synthetic G-quartets (TASQs) are biomimetic ligands that target G4s and have been extensively studied and evolved into different variants like CyTASQ, BioTASQ, and BioCyTASQ,27 having applications in G4 isolation, visualization and imaging systems.28,29 Müller et al.12 and Sperti et al.30 performed in vitro G4 pull-down using the biotinylated PDS and TASQs, respectively, for further investigation of G4s. While these studies have been significant in advancing G4 ligands as molecular tools for G4s, it is important to note that these G4 ligands cannot readily distinguish dG4 and rG4 targets. Additionally, the synthesis and preparation of some of the G4 ligands can be challenging, presenting a critical bottleneck for further development and application of G4 tools, as well as general characterization of G4s. As a potential solution and application, we propose an alternative strategy for targeting specific rG4s, which involves the use of L-RNA aptamers.
Aptamers are short, single-stranded DNA or RNA oligonucleotides that bind to their target of interest with high affinity and specificity.31,32 Natural aptamers (D-oligonucleotides) are prone to nuclease-mediated degradation in biological media.33,34 To overcome this issue, modified aptamers have been developed using a mirror-image SELEX selection process that eventually generates unnatural L-DNA/L-RNA aptamers that have great biostability.35–37 Our group previously selected and reported a series of novel L-RNA aptamers, including L-Ap3-7,38L-Apt.4-1c,39 and L-Apt.8f40 through rG4-SELEX for targeting TERRA rG4, hTERC rG4, and APP rG4, respectively. These L-aptamers exhibited great binding affinity to their targets and high specificity over dG4s and non-G4s as shown in previous studies. To address the specificity limitation of traditional G4 ligands, L-aptamers offer advantages such as high rG4 specificity, ease of synthesis, low cytotoxicity and low immunogenicity.31 These advantages make L-aptamers a promising alternative in our proposed approach for further rG4 research. Among these L-aptamers, L-Apt.4-1c is the shortest L-RNA aptamer developed so far with 25 nucleotides, exhibiting high affinity binding to D-hTERC rG4 and preferential binding to rG4s, but not to dG4s and non-G4s. Our structural analysis also suggested that the G-rich regions of the aptamer folds into a unique two G-quartet rG4 structure (Fig. 1C).39,41 Herein, we introduce a novel pulldown approach for isolating a specific rG4 in vitro using L-Apt.4-1c from both simple buffer and complex biological media, including total RNA and the cell lysate. This aptamer has great potential to serve as a substitute for G4 ligands for certain applications related to rG4s. We report that the pulldown efficiency of L-Apt.4-1c is high for a specific rG4 in both the absence and the presence of different non-targets. Notably, our results demonstrate that L-Apt.4-1c is highly effective in isolating rG4 of interest, and its performance is better compared to the G4 small molecule ligand BioTASQ v.1.
Results and discussion
Design of pulldown assay
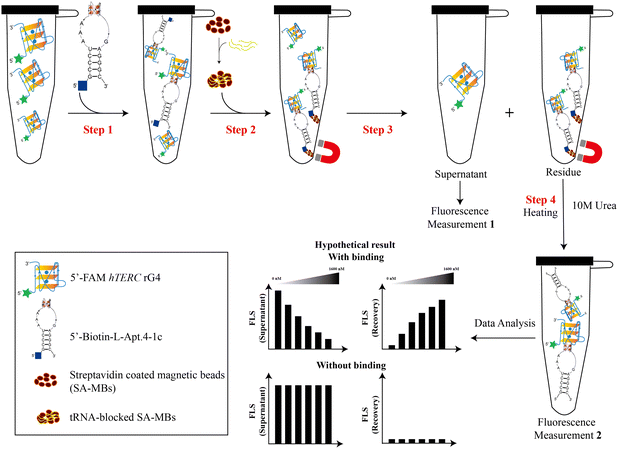
Our group recently reported that L-Apt.4-1c exhibits high affinity for general D-rG4s, such as D-NRAS rG4, D-APP rG4, and D-hTERC rG4. Among these rG4s, D-hTERC rG4 was a positive target of L-Apt.4-1c in rG4-SELEX, with a dissociation constant (Kd) of 59.1 ± 11.9 nM.39 To investigate whether L-RNA aptamers can serve as molecular tools that have equal or even superior performance to G4 ligands, we designed a novel pulldown assay (both non-competitive and competitive) to specifically pulldown FAM-D-rG4 targets using streptavidin-coated magnetic beads (SA-MBs) coupled with Biotin-l-Apt.4-1c (Fig. 2). We first appended the biotin label at the 5′ position of L-Apt.4-1c as the biotin functional group is needed for interacting with the streptavidin-coated magnetic beads for pulldown purposes. To evaluate the impact of biotin labeling of L-Apt.4-1c on the binding affinity towards 5′-FAM-D-hTERC rG4, we conducted a binding experiment using MST, and determined the Kd value to be 72.2 ± 14.6 nM (Fig. S1, ESI†). This Kd value is comparable to the value obtained from the unlabelled L-Apt.4-1c and FAM-D-hTERC rG4 interaction, indicating that the functionalization of L-Apt.4-1c with 5′-biotin does not affect its interaction with D-hTERC rG4, and thus we can use it for the development of the pulldown assay. To investigate whether other FAM-D-rG4 targets are also feasible in our pulldown assay, we selected FAM-D-NRAS rG4 and FAM-D-APP rG4 as two additional rG4s, with the Kd values of 75.0 ± 10.2 nM (Fig. S2, ESI†) and 55.8 ± 4.52 nM (Fig. S3, ESI†) towards Biotin-L-Apt.4-1c, respectively.For this new pulldown assay, we have devised the workflow as follows: first, FAM-D-rG4 targets (including hTERC rG4, NRAS rG4, and APP rG4) (100 nM) or other non-targets were incubated in 1× annealing buffer with different concentrations of biotinylated substrates (including Biotin-L-Apt.4-1c and BioTASQ v.1): 0 nM, 2.5 nM, 120 nM, 300 nM, 800 nM, and 1600 nM as non-competitive assay (Fig. 2, step 1). Similar experiments were performed using BioTASQ v.1 to compare the pulldown efficiency and specificity for rG4 shown by the biotin-L-RNA aptamer versus the G4 small molecule ligand. Next, to ensure that the pulldown of rG4 targets by the Biotin-L-aptamer is not affected by competitors, we performed competitive assay. For competitive assay, FAM-D-hTERC rG4 and Biotin-L-Apt.4-1c were incubated with either 1× or 10× unlabelled competitors in step 1 (Fig. S4, ESI†). A few non-targets with different structures were chosen as competitors (including RNA hairpin, hTELO dG4 with hybrid topology, and a mixture of poly rA, rU, and rC) due to their lack of binding with Biotin-L-Apt.4-1c. To validate the specificity of the pulldown by the biotin-L-aptamer for rG4, FAM-hTERC dG4 was chosen as one of the competitors. FAM-hTERC dG4 is the DNA version of FAM-hTERC rG4 with parallel topology, which weakly binds to Biotin-L-Apt.4-1c, with a Kd value of 221 ± 57.6 nM (Fig. S5, ESI†). The pulldown assay relies on the biotinylated complex (like Biotin-L-Apt.4-1c–FAM-D-hTERC rG4) attachment to yeast tRNA blocked SA-MBs (Fig. 2, step 2). The supernatant (unbound FAM-labelled target) was eluted from the biotinylated bound complex (residue) using a magnetic rack upon binding (Fig. 2, step 3). The unbound FAM-labelled supernatant was measured and denoted as Measurement 1 (Fig. 2). Last, the residues (bound) in the magnetic beads were washed with 1× annealing buffer and incubated at 95 °C for 15 minutes with 10 M urea to release biotinylated complexes (bound) (Fig. 2, step 4). Measurement 1 (supernatant) was conducted to ascertain the amount of unbound target or non-target in the presence of 5′-biotin-L-Apt.4-1c, while measurement 2 (recovery) quantified the amount of target or non-target bound to 5′-Biotin-L-Apt.4-1c. The fluorescence intensity was normalized to 100% and more details of the data analysis can be found in the Method section in the ESI.†
Evaluation of the binding capacity of L-Apt.4-1c in a simple buffer
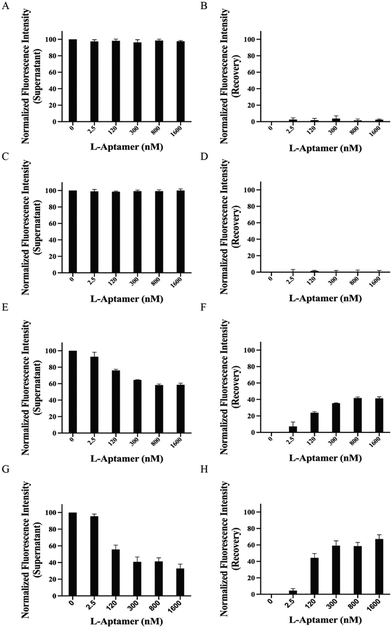
To examine the capacity and specificity of L-Apt.4-1c to pulldown rG4s specifically, we first performed a non-competitive pulldown assay using FAM-non-targets, including RNA hairpin, hTELO dG4, and hTERC dG4.39 Based on the data, we observed that both the RNA hairpin (Fig. 3A and B) and hTELO dG4 (Fig. 3C and D) cannot be pulled down by the biotin-L-aptamer due to the absence of binding. This was indicated by the steady fluorescence intensity of the supernatant (approximately 100%) and the absence of recovery (approximately 0%). Theoretically, the fluorescence intensity of the supernatant decreased with increasing concentration of the biotin-L-aptamer (Fig. 2) upon binding with its corresponding target. Since FAM-hTERC dG4 showed weak binding with Biotin-l-Apt.4-1c (Fig. S5, ESI†), the fluorescence intensity showed an increasing trend, but only recovered to 41.4 ± 1.95% at 1600 nM Biotin-L-Apt.4-1c (Fig. 3E and F). To investigate the effectiveness of the biotin-L-aptamer in isolating the rG4 of interest, we performed a non-competitive assay using FAM-hTERC rG4, FAM-NRAS rG4, and FAM-APP rG4.39hTERC rG4 exhibited an approximately 3 times stronger binding affinity than hTERC dG4 with Biotin-L-Apt.4-1c. The pulldown product of FAM-hTERC rG4-Biotin-L-Apt.4-1c showed a recovery of 4.47 ± 2.60% when the concentration of Biotin-L-Apt.4-1c ranged from 0 to 2.5 nM (Fig. 3G and H). Remarkably, a significant increase in the fluorescence intensity recovery was observed from 4.47 ± 2.60% to 44.3 ± 5.23% within the range of 2.5 to 120 nM, which aligns with the Kd value from MST. As the concentration of Biotin-l-Apt.4-1c exceeded the Kd value from 300 to 1600 nM, there appeared to be a gradual saturation at 67.2 ± 5.38% of the fluorescence intensity with an increasing trend. Similar increasing trends were observed in the pulldown of FAM-NRAS rG4 and FAM-APP rG4. The fluorescence intensity recovery increased from 24.5 ± 3.10% to 66.1 ± 3.84% (Fig. S6, ESI†) and from 14.6 ± 2.78% to 48.8 ± 3.59% (Fig. S7, ESI†) within the range of 2.5 to 1600 nM, respectively. To avoid false positive results in isolating FAM-rG4s using the biotin-L-aptamer, we conducted a similar set of experiments as a negative control, except that we used Biotin-l-Apt.4-1c M9, which is a mutated version of Biotin-L-Apt.4-1c. Our group conducted the mutagenesis experiment by EMSA and found that some point mutations of this L-aptamer showed no binding, especially the Gs potentially involved in the formation of the G-quartet structure.39 Among these mutants, we chose the point mutation from G9 to A9 (named Biotin-L-Apt.4-1c M9) for our pulldown assay, which resulted in the absence of an increasing trend as the concentration of Biotin-l-Apt.4-1c M9 was increased (Fig. S8, ESI†). In short, our non-competitive pulldown assay reflected that Biotin-l-Apt.4-1c isolates general rG4s with the highest fluorescence recovery, highlighting the validity of this novel assay and illustrating the capturing ability of this L-aptamer for general rG4s against other non-targets.To identify whether Biotin-L-Apt.4-1c specifically recognizes general rG4 structures against rG4 mutants, we performed competitive pulldown assays using the biotin-L-aptamer for the isolation of rG4s in the presence of their corresponding rG4 mutants. In each rG4, we mutated some Gs to As in the respective sequences to disrupt the formation of the rG4s (Table S1, ESI†). The fluorescence intensities showed an increasing trend in the recovery and gradually saturated at 1600 nM for the isolation of hTERC rG4, NRAS rG4, and APP rG4 in the presence of their corresponding rG4 mutants, with values of 53.7 ± 5.94% (Fig. S9, ESI†), 61.5 ± 1.57% (Fig. S10, ESI†), and 42.5 ± 2.26% (Fig. S11, ESI†), respectively. The fluorescence recoveries in the presence of rG4 mutants were similar to the non-competitive results, which showed that Biotin-L-Apt.4-1c was able to distinguish between rG4 structures or rG4 mutants to avoid false positive results. To further display the pulldown efficiency of Biotin-L-Apt.4-1c for FAM-hTERC rG4 in the presence of non-targets, we employed unlabelled structural and non-structural competitors, including hTERC dG4, hTELO dG4, and a mixture of poly rA, rC, and rU. The unlabelled non-targets were utilized to compete with FAM-hTERC rG4 when incubating with Biotin-L-Apt.4-1c (Fig. S4, ESI†). Meanwhile, to investigate the influence of the structured competitor amount, we used two different ratios of hTERC rG4 to non-target dG4s: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (1×) and 1
1 (1×) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
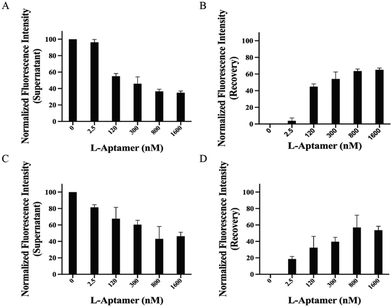
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (10×). We incubated Biotin-L-Apt.4-1c with FAM-hTERC rG4 in the presence of either 1× or 10× unlabelled hTELO dG4 or hTERC dG4 competitor. Upon binding with the non-targets, the recovery of fluorescence intensity will be approximately 0% due to the absence of the FAM label in the 5′ position. In the presence of the 1× structural competitor, we observed that there is a significant increase in the recovery of fluorescence intensity (3.78 ± 3.53% to 44.9 ± 3.20% and 11.9 ± 2.55% to 50.3 ± 1.68%) within the range of 2.5 to 120 nM Biotin-L-Apt.4-1c upon binding towards FAM-hTERC rG4 in the presence of 1× hTELO dG4 and 1× hTERC dG4, respectively, which gradually reached saturation at 65.1 ± 2.08% and 67.1 ± 2.45% at 1600 nM (Fig. 4 and Fig. S12, ESI†). Similarly, an increasing trend of fluorescence intensity was observed, with 66.6 ± 3.23% and 56.7 ± 16.1% recovered at 1600 nM in the presence of 10× hTERC dG4 and 10× hTELO dG4, respectively (Fig. S12 and S13, ESI†). Although both hTELO dG4 and hTERC dG4 serve as dG4 non-targets, FAM-hTELO dG4 cannot be pulled down by the biotin-L-aptamer (Fig. 3D), while FAM-hTERC dG4 can be pulled down by the biotin-L-aptamer (Fig. 3F). However, the results here showed that the inclusion of the 1× or 10× unlabelled hTELO dG4 or hTERC dG4 resulted in a comparable fluorescence recovery (Fig. 4 and Fig. S12, ESI†) when compared to the recovery at different concentrations of L-aptamer in non-competitive assay (Fig. 3H), suggesting that the presence and the quantity of non-targets, whether they bind or do not bind with L-Apt.4-1c, do not significantly affect the pulldown efficiency for FAM-hTERC rG4 shown by Biotin-L-Apt.4-1c. Therefore, the pulldown of Biotin-L-Apt.4-1c is specific for rG4 and likely dependent on the binding affinity.
10 (10×). We incubated Biotin-L-Apt.4-1c with FAM-hTERC rG4 in the presence of either 1× or 10× unlabelled hTELO dG4 or hTERC dG4 competitor. Upon binding with the non-targets, the recovery of fluorescence intensity will be approximately 0% due to the absence of the FAM label in the 5′ position. In the presence of the 1× structural competitor, we observed that there is a significant increase in the recovery of fluorescence intensity (3.78 ± 3.53% to 44.9 ± 3.20% and 11.9 ± 2.55% to 50.3 ± 1.68%) within the range of 2.5 to 120 nM Biotin-L-Apt.4-1c upon binding towards FAM-hTERC rG4 in the presence of 1× hTELO dG4 and 1× hTERC dG4, respectively, which gradually reached saturation at 65.1 ± 2.08% and 67.1 ± 2.45% at 1600 nM (Fig. 4 and Fig. S12, ESI†). Similarly, an increasing trend of fluorescence intensity was observed, with 66.6 ± 3.23% and 56.7 ± 16.1% recovered at 1600 nM in the presence of 10× hTERC dG4 and 10× hTELO dG4, respectively (Fig. S12 and S13, ESI†). Although both hTELO dG4 and hTERC dG4 serve as dG4 non-targets, FAM-hTELO dG4 cannot be pulled down by the biotin-L-aptamer (Fig. 3D), while FAM-hTERC dG4 can be pulled down by the biotin-L-aptamer (Fig. 3F). However, the results here showed that the inclusion of the 1× or 10× unlabelled hTELO dG4 or hTERC dG4 resulted in a comparable fluorescence recovery (Fig. 4 and Fig. S12, ESI†) when compared to the recovery at different concentrations of L-aptamer in non-competitive assay (Fig. 3H), suggesting that the presence and the quantity of non-targets, whether they bind or do not bind with L-Apt.4-1c, do not significantly affect the pulldown efficiency for FAM-hTERC rG4 shown by Biotin-L-Apt.4-1c. Therefore, the pulldown of Biotin-L-Apt.4-1c is specific for rG4 and likely dependent on the binding affinity.
To comprehensively examine the effect of non-structured competitors, we incubated Biotin-L-Apt.4-1c with FAM-hTERC rG4 in the presence of a mixture of 1× unlabelled poly rA, rC, and rU. The final concentration of each non-structured competitor in the mixture was 100 nM. The results showed a similar decreasing trend in the fluorescence intensity for the supernatant (Fig. S14A, ESI†) and an increasing trend for recovery (2.68 ± 1.06% to 38.5 ± 1.66%) in the range of 2.5 to 120 nM Biotin-L-Apt.4-1c upon binding to FAM-hTERC rG4 in the presence of non-structured competitors (Fig. S14B, ESI†). In the range of 300 to 1600 nM Biotin-L-Apt.4-1c, the recovery of fluorescence intensity gradually reached saturation at 62.2 ± 2.13%, which is also similar to the presence of either 1× or 10× unlabelled hTELO dG4 (Fig. 4 and Fig. S13, ESI†). Our results demonstrated that, despite the presence of 1× or 10× non-target competitors, Biotin-L-Apt.4-1c exhibits excellent efficacy in pulling down FAM-hTERC rG4. These findings further highlight the utility of the L-Apt.4-1c pulldown assay by providing additional evidence of the selectivity and sensitivity of Biotin-Apt.4-1c towards FAM-hTERC rG4.
Comparison of L-Apt.4-1c and BioTASQ v.1 in terms of pulldown efficacy
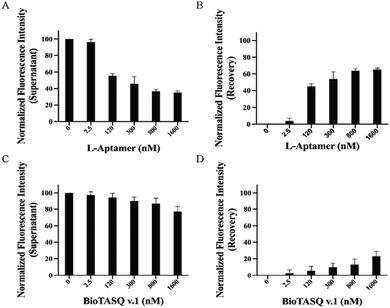
To compare the isolating ability of Biotin-L-Apt.4-1c and the biotinylated G4 small molecule ligands for FAM-hTERC rG4, we performed both non-competitive and competitive assays using a biotinylated G4 small molecule ligand to pulldown FAM-hTERC rG4. BioTASQ v.1, a biotinylated ligand known to isolate G4s,12 was employed as the representative of a G4 small molecule ligand in our assay. First, we performed an MST assay to determine the binding affinity for both BioTASQ v.1-hTERC rG4 and BioTASQ v.1-hTELO dG4 interactions, and the Kd values were found to be 225 ± 61.5 nM (Fig. S15A, ESI†) and 226 ± 47.7 nM (Fig. S15B, ESI†). Different from L-Apt.4-1c, BioTASQ v.1 showed a similar binding affinity towards both hTERC rG4 and hTELO dG4, but about 3 times weaker than that of Biotin-L-Apt.4-1c. To verify the ability of G4 ligands to isolate their binding targets, we conducted a non-competitive assay. However, the results showed a weak increasing trend to isolate either hTERC rG4 or hTELO dG4 with increasing concentration of BioTASQ v.1, which is at 30.6 ± 7.93% (Fig. S16, ESI†) and 13.8 ± 1.68% (Fig. S17, ESI†) at 1600 nM, respectively. Our results revealed that the isolating ability of the L-aptamer was approximately twice that of BioTASQ v.1 (Fig. 3H); therefore, we concluded that the pulldown efficacy for either dG4 or rG4 using BioTASQ v.1 was poor upon binding. As a control, we performed the same experiment on a RNA hairpin, and no binding was found with approximately 0% of the recovery of fluorescence intensity (Fig. S18, ESI†), which verified that BioTASQ v.1 can only distinguish rG4 and dG4. As a second control, we also conducted a competitive pulldown assay to assess whether BioTASQ v.1 specifically isolates FAM-hTERC rG4 in the presence of unlabelled hTELO dG4 (1× and 10×) and non-G4 competitors (a mixture of poly rA, rC, and rU). Compared to the absence of unlabelled hTELO dG4 (Fig. S17, ESI†), the presence of 1× and 10× unlabelled hTELO dG4 competitors recovered 22.9 ± 6.04% (Fig. 4) and 17.3 ± 6.84% (Fig. S13, ESI†) of fluorescence intensity, suggesting that the pulldown efficiency is slightly affected by the amount of dG4 competitor. However, the pulldown efficiency for FAM-hTERC rG4 shown by BioTASQ v.1 in the presence of a mixture of non-G4s (45.5 ± 4.90%) (Fig. S19, ESI†) is comparable to that of the absence of unlabelled competitors (30.6 ± 7.93%) (Fig. S16, ESI†). It is reasoned that hTERC rG4 (target) and hTELO dG4 (competitor) have a similar Kd value to BioTASQ v.1, and hence a larger amount of competitor (10×) with a similar affinity would greatly interfere with the pulldown efficiency. Our approach suggested that L-Apt.4-1c competes with state-of-the-art G4 small molecule ligands while offering its specificity to general rG4s even in the presence of dG4, providing great potential to substitute small molecule G4 ligands in further investigations into rG4s based on our new approach.Evaluation of the binding capacity of L-Apt.4-1c in complex media
To mimic the isolation of specific rG4 using L-aptamers under physiological conditions, we carried out an extensive investigation in two distinctive media as competitors, including total RNA and the cell lysate. First, total RNA represents the collection of all RNA molecules, including ribosomal RNAs (rRNAs), messenger RNAs (mRNAs), and other non-coding RNAs (ncRNAs), which provides valuable information about the potential impact of cellular RNA molecules on the isolation efficiency for hTERC rG4 using L-Apt.4-1c. We found that 49.7 ± 16.3% fluorescence recovery was observed at 1600 nM 5′-Biotin-L-Apt.4-1c in the presence of total RNA (Fig. 5A and B). This result showed that L-Apt.4-1c is highly selective for hTERC rG4 in a complex mixture of different RNA species. To assess whether L-Apt.4-1c isolates hTERC rG4 in a biologically relevant solution, we employed the cell lysate as a competitor, a mixture of cellular components, including metabolites, nucleic acids, proteins, etc. Our result showed that 53.6 ± 4.84% of fluorescence recovery was observed at 1600 nM 5′-Biotin-L-Apt.4-1c in the presence of cell lysate (Fig. 5C and D). Interestingly, the fluorescence recovery was comparable to the presence of 10× hTELO dG4 (56.7 ± 16.1%) (Fig. S13, ESI†), indicating that the interaction between 5′-Biotin-L-Apt.4-1c and FAM hTERC rG4 is not significantly inhibited in various biological media. However, total RNA and the cell lysate are two complex media that contain RNA of different sizes, sequences and structures, along with other biomolecules in the case of cell lysate. To further verify the effectiveness of pulldown by Biotin-L-Apt.4-1c, hTERC rG4 and APP rG4 enrichment was monitored among several rG4s in the endogenous pulldown (in either total RNA or the cell lysate) using Biotin-L-Apt.4-1c, followed by RT-qPCR. In total RNA pulldown, hTERC rG4 and APP rG4 were determined at threshold cycles (Ct) of 25.3 cycles and 23.4 cycles (Fig. S20A, ESI†), respectively. To compare the rG4 enrichment, endogenous pulldown was also performed using the biotin-L-mutant, which yielded much larger Ct values of >40 and 38.7 cycles, respectively. The large difference in Ct values indicated that rG4s can be enriched using Biotin-L-Apt.4-1c. Similarly, the hTERC rG4 and APP rG4 in cell lysate pulldown also showed enrichment, albeit weaker, with Ct values of 26.3 and 25.7 cycles using Biotin-L-Apt.4-1c and 28.6 and 28.0 cycles using the biotin-L-mutant (Fig. S20B, ESI†). It is likely that cell lysate samples are more complex in nature, and therefore the enrichment observed is less obvious than that observed using total RNA samples. To sum up, our approach can specifically pulldown rG4 using the L-aptamer from both simple buffer and complex media with the evidence of both competitive pulldown analysis and RT-qPCR results, and this result has significant implications for further investigation of rG4 structures in diverse systems.Conclusion
In conclusion, this work developed a magnetic-bead based pulldown platform by leveraging L-Apt.4-1c as a general rG4 binder with high affinity and specificity. Our platform allows both non-competitive and competitive analyses for the interaction between rG4 targets (or non-rG4 targets) and the L-aptamer (or G4 small molecule ligands). Furthermore, we also investigated the endogenous pulldown efficiency for hTERC rG4 using Biotin-L-Apt.4-1c, demonstrating its ability to enrich the target rG4 from total RNA or the cell lysate. In the future, L-aptamers can also be generated using efficient SELEX-based approaches, such as rG4-SELEX38 and G4-SELEX-seq,42 which typically require a few rounds of selection, and the resultant L-aptamer can be functionalized for affinity capture or isolation, as well as for other applications such as imaging. To sum up, our findings indicate that L-Apt.4-1c exhibits superior pulldown efficiency and specificity for general rG4s compared to BioTASQ v.1. This work provides a valuable platform for the identification and characterization of rG4s and shows the capability for further enhancing the potential of L-aptamers as highly promising molecular tools for exploring and understanding the rG4 structure in a transcript-specific and transcriptome-wide manner.Data availability
The data underlying this article are available in the article and in the ESI.† Further data will be shared upon reasonable request to the corresponding author.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
This work was supported by the NSFC Excellent Young Scientists Fund (Hong Kong and Macau) Project [32222089]; Research Grants Council of the Hong Kong SAR, China Projects [CityU 11100123, CityU 11100222, CityU 11100421]; Croucher Foundation Project [9509003]; State Key Laboratory of Marine Pollution Seed Collaborative Research Fund [SCRF/0037, SCRF/0040, SCRF/0070]; and City University of Hong Kong projects [7030001, 6000827, 9678302] to C. K. K. We thank Prof. David Monchaud for sharing the BioTASQ v.1 ligand.Notes and references
- J. T. Davis, Angew. Chem., Int. Ed., 2004, 43, 668–698 CrossRef CAS PubMed.
- C. K. Kwok and C. J. Merrick, Trends Biotechnol., 2017, 35, 997–1013 CrossRef CAS PubMed.
- F. Dumetz, E. Y. Chow, L. M. Harris, S. W. Liew, A. Jensen, M. I. Umar, B. Chung, T. F. Chan, C. J. Merrick and C. K. Kwok, Nucleic Acids Res., 2021, 49, 12486–12501 CrossRef CAS PubMed.
- X. Yang, J. Cheema, Y. Zhang, H. Deng, S. Duncan, M. I. Umar, J. Zhao, Q. Liu, X. Cao, C. K. Kwok and Y. Ding, Genome Biol., 2020, 21, 226 CrossRef CAS PubMed.
- X. Shao, W. Zhang, M. I. Umar, H. Y. Wong, Z. Seng, Y. Xie, Y. Zhang, L. Yang, C. K. Kwok and X. Deng, mBio, 2020, 11(1), e02926-19 CrossRef PubMed.
- X. Chen, J. Yuan, G. Xue, S. Campanario, D. Wang, W. Wang, X. Mou, S. W. Liew, M. I. Umar, J. Isern, Y. Zhao, L. He, Y. Li, C. J. Mann, X. Yu, L. Wang, E. Perdiguero, W. Chen, Y. Xue, Y. Nagamine, C. K. Kwok, H. Sun, P. Munoz-Canoves and H. Wang, Nat. Commun., 2021, 12, 5043 CrossRef CAS PubMed.
- N. Bohalova, A. Cantara, M. Bartas, P. Kaura, J. Stastny, P. Pecinka, M. Fojta and V. Brazda, Int. J. Mol. Sci., 2021, 22, 3433 CrossRef CAS PubMed.
- G. Qin, C. Zhao, Y. Liu, C. Zhang, G. Yang, J. Yang, Z. Wang, C. Wang, C. Tu, Z. Guo, J. Ren and X. Qu, Cell Discovery, 2022, 8, 86 CrossRef CAS PubMed.
- R. Hansel-Hertsch, D. Beraldi, S. V. Lensing, G. Marsico, K. Zyner, A. Parry, M. Di Antonio, J. Pike, H. Kimura, M. Narita, D. Tannahill and S. Balasubramanian, Nat. Genet., 2016, 48, 1267–1272 CrossRef CAS PubMed.
- L. M. Harris and C. J. Merrick, PLoS Pathog., 2015, 11, e1004562 CrossRef PubMed.
- R. Hänsel-Hertsch, M. Di Antonio and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2017, 18, 279–284 CrossRef PubMed.
- S. Muller, S. Kumari, R. Rodriguez and S. Balasubramanian, Nat. Chem., 2010, 2, 1095–1098 CrossRef PubMed.
- T. Santos, G. F. Salgado, E. J. Cabrita and C. Cruz, Pharmaceuticals, 2021, 14 Search PubMed.
- K. H. Ngo, R. Yang, P. Das, G. K. T. Nguyen, K. W. Lim, J. P. Tam, B. Wu and A. T. Phan, Chem. Commun., 2020, 56, 1082–1084 RSC.
- H. Fernando, S. Sewitz, J. Darot, S. Tavare, J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2009, 37, 6716–6722 CrossRef CAS PubMed.
- H. Fernando, R. Rodriguez and S. Balasubramanian, Biochemistry, 2008, 47, 9365–9371 CrossRef CAS PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- S. M. Javadekar, N. M. Nilavar, A. Paranjape, K. Das and S. C. Raghavan, DNA Res., 2020, 27, 1–17 CrossRef CAS PubMed.
- X. Mou and C. K. Kwok, J. Am. Chem. Soc., 2023, 145, 18693–18697 CrossRef CAS PubMed.
- P. M. Yangyuoru, M. Di Antonio, C. Ghimire, G. Biffi, S. Balasubramanian and H. Mao, Angew. Chem., 2015, 127, 924–927 CrossRef PubMed.
- A. Henderson, Y. Wu, Y. C. Huang, E. A. Chavez, J. Platt, F. B. Johnson, R. M. Brosh, Jr., D. Sen and P. M. Lansdorp, Nucleic Acids Res., 2017, 45, 6252 CrossRef CAS PubMed.
- B. Heddi, V. V. Cheong, H. Martadinata and A. T. Phan, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 9608–9613 CrossRef CAS PubMed.
- S. Xu, Q. Li, J. Xiang, Q. Yang, H. Sun, A. Guan, L. Wang, Y. Liu, L. Yu, Y. Shi, H. Chen and Y. Tang, Sci. Rep., 2016, 6, 24793 CrossRef CAS PubMed.
- J. Ren, H. Qin, J. Wang, N. W. Luedtke, E. Wang and J. Wang, Anal. Bioanal. Chem., 2011, 399, 2763–2770 CrossRef CAS PubMed.
- H. Arthanari, S. Basu, T. L. Kawano and P. H. Bolton, Nucleic Acids Res., 1998, 26, 3724–3728 CrossRef CAS PubMed.
- R. Rodriguez, S. Müller, J. A. Yeoman, C. Trentesaux, J.-F. Riou and S. Balasubramanian, J. Am. Chem. Soc., 2008, 130, 15758–15759 CrossRef CAS PubMed.
- M. Nikan and J. C. Sherman, Angew. Chem., Int. Ed., 2008, 47, 4900–4902 CrossRef CAS PubMed.
- F. R. Sperti, T. Charbonnier, P. Lejault, J. Zell, C. Bernhard, I. E. Valverde and D. Monchaud, ACS Chem. Biol., 2021, 16, 905–914 CrossRef CAS PubMed.
- I. Renard, M. Grandmougin, A. Roux, S. Y. Yang, P. Lejault, M. Pirrotta, J. M. Y. Wong and D. Monchaud, Nucleic Acids Res., 2019, 47, 5502–5510 CrossRef CAS PubMed.
- F. Sperti Rota, T. Charbonnier, P. Lejault, J. Zell, C. Bernhard, I. E. Valverde and D. Monchaud, ACS Chem. Biol., 2021, 16, 905–914 CrossRef PubMed.
- A. Ozer, J. M. Pagano and J. T. Lis, Mol. Ther.--Nucleic Acids, 2014, 3, e183 CrossRef CAS PubMed.
- A. B. Iliuk, L. Hu and W. A. Tao, Anal. Chem., 2011, 83, 4440–4452 CrossRef CAS PubMed.
- A. V. Lakhin, V. Z. Tarantul and L. V. Gening, Acta Naturae, 2013, 5, 34–43 CrossRef CAS PubMed.
- P. Rothlisberger and M. Hollenstein, Adv. Drug Delivery Rev., 2018, 134, 3–21 CrossRef CAS PubMed.
- D. Ji, H. Feng, S. W. Liew and C. K. Kwok, Trends Biotechnol., 2023, 1360–1384 CrossRef CAS PubMed.
- J. P. Elskens, J. M. Elskens and A. Madder, Int. J. Mol. Sci., 2020, 21 Search PubMed.
- B. E. Young, N. Kundu and J. T. Sczepanski, Chemistry, 2019, 25, 7981–7990 CrossRef CAS PubMed.
- C. Y. Chan and C. K. Kwok, Angew. Chem., Int. Ed., 2020, 59, 5293–5297 CrossRef CAS PubMed.
- M. I. Umar and C. K. Kwok, Nucleic Acids Res., 2020, 48, 10125–10141 CrossRef CAS PubMed.
- H. Zhao, H. Y. Wong, D. Ji, K. Lyu and C. K. Kwok, ACS Appl. Mater. Interfaces, 2022, 14, 30582–30594 CrossRef CAS PubMed.
- C.-Y. Chan and C. K. Kwok, Angew. Chem., Int. Ed., 2020, 59, 5293–5297 CrossRef CAS PubMed.
- D. Ji, J. H. Yuan, S. B. Chen, J. H. Tan and C. K. Kwok, Nucleic Acids Res., 2023, 51, 11439–11452 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00161c |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |