Open Access Article
Open Access ArticleDynamic selection of high-affinity aptamers using a magnetically activated continuous deflection microfluidic chip†
Ke-Zhu
Yang
,
Meng
Wang
,
Ming-Yue
Gao
,
Yong-Tao
Wang
and
Zhi-Ling
Zhang
 *
*
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, 430072, People's Republic of China. E-mail: zlzhang@whu.edu.cn
First published on 8th February 2024
Abstract
To accelerate the discovery of high-affinity aptamers, a magnetically activated continuous deflection (MACD) chip was designed. The MACD chip could achieve dynamic selection in a continuous flow, which meant that the binding and separation were carried out consecutively. Dynamic selection could make selection efficient. Low-affinity sequences could be eluted in time and high-affinity sequences could be enriched via dynamic selection. The stringency of the conditions could be further increased by lowering the target concentration in the dynamic selection. Finally, a C.al3 aptamer with high-affinity and high-specificity for Candida albicans (C. albicans) was obtained through six rounds of selection. Its dissociation constant (Kd) was 7.9 nM. This demonstrated that dynamic selection using a MACD chip was an effective method for high-affinity aptamer selection.
Since aptamers were first screened from oligonucleotide libraries,1 researchers have screened a large number of aptamers including small molecules,2 proteins,3 viruses,4 and bacteria.5 Aptamers have the advantages of easy chemical synthesis, easy modification, strong affinity, and high specificity.6 Based on these advantages, aptamers have been widely used in targeted drug delivery,7 cell imaging,8 biological molecular recognition, detection,9etc. Therefore, there is an urgent need to develop a method to accelerate the discovery of high-affinity and high-specificity aptamers.
In general, aptamers are obtained through SELEX in vitro. Due to the iterative nature of aptamer selection, aptamer generation requires a large amount of time, labor, and resources, which makes it inefficient and often leads to the failure of screening.10 For effective isolation of high-quality aptamers from high-complexity libraries, the key is to improve the binding efficiency and separation efficiency. Microfluidic SELEX (M-SELEX) could achieve the key due to its advantages of accurate flow control, high automation, and real-time monitoring.11,12 Recently, many M-SELEX methods have been developed to increase the efficiency of aptamer selection, such as bead-based microfluidic SELEX,13 sol–gel microfluidic SELEX,14 and microarray chip SELEX.15 Moreover, theoretical models and experimental studies have suggested that the use of a small number of targets during library incubation can form highly stringent competitive binding and promote the enrichment of high-affinity aptamers.16,17 Recent studies suggest that M-SELEX could obtain high-affinity aptamers within fewer rounds by using a small amount of target molecules.18,19 Therefore, the efficiency of selection could be improved by M-SELEX combined with the use of low target concentrations.
In our previous work, a magnetism-controlled microfluidic chip was developed for the efficient discovery of aptamers and the efficiency of selection was improved via trapping MNs on a microscale.20 Besides, a corresponding V-target lattice structure was developed to enhance the efficiency of the binding process.21 However, this method is static selection, that is, the target is fixed, which could not separate low-affinity sequences in time and affect the enrichment of high-affinity sequences, so its efficiency of selection still needs to be improved. On this previous base, a dynamic selection in a continuous flow was developed to accelerate the selection efficiency using a MACD chip. Dynamic selection made the screening process much easier, which relied on the movement of the targets labeled with MNs through different solution phases in a continuous flow. Low-affinity sequences could be eluted in time when the targets flowed through the washing phase and high-affinity sequences could be enriched when they flowed through the library phase. The stringency of the conditions was increased by lowering the target concentration in dynamic selection. C. albicans is the most widespread fungal pathogen in the world.22 The overgrowing of C. albicans can lead to diseases ranging from oral or vaginal candidiasis to life-threatening systemic candidiasis.23 Unfortunately, few aptamers have been discovered for C. albicans. Therefore, C. albicans was chosen to be a target for dynamic selection using the MACD chip. A high-affinity aptamer for C. albicans was obtained after six rounds of selection and its Kd could reach the nanomolar level. The selected aptamer had potential application in inhibiting C. albicans biofilm formation. The MACD chip provided a novel approach for aptamer discovery via dynamic selection.
The dynamic selection process using a MACD chip is displayed in Fig. 1. When the targets labeled with MNs, pretreated DNA library, and washing buffer were pumped into the MACD chip to form a sample phase, library phase, and washing phase, respectively, the three fluids would flow in a laminar manner due to the low Reynolds number. The laminar effect was simulated with red, blue, and black ink (Fig. S4, ESI†). Under the combined action of magnetic force (Fm) and hydrodynamic drag force (Fd), the targets labeled with MNs would move along the nickel strip and sequentially flow through the library phase and washing phase. Therefore, dynamic selection in a continuous flow where the binding and separation were carried out consecutively was achieved. First, low-affinity and high-affinity sequences would randomly bind to the targets when they first flowed through the library phase. Then, the low-affinity sequences would be washed off under the flow force when targets with low-affinity and high-affinity sequences flowed through the washing phase. Next, the high-affinity sequences would bind to the targets more easily than the low-affinity sequences under competition when the targets secondly flowed through the library phase. Finally, low-affinity sequences would be further removed when targets flowed through the washing phase for the last time. Via dynamic selection, high-affinity sequences would be sufficiently enriched compared to static selection.
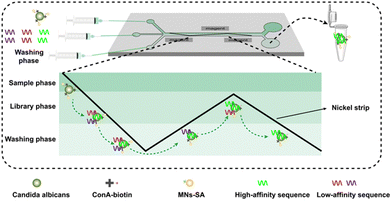 | ||
| Fig. 1 Overview of the selection process using a magnetically activated continuous deflection chip for high-affinity aptamer selection. | ||
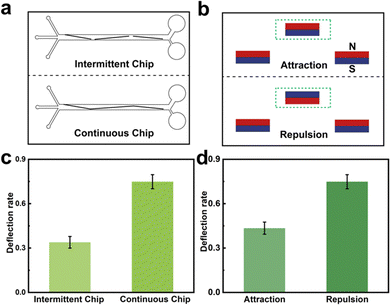
To achieve the movement of MNs in different solutions, two kinds of chips were designed: the intermittent chip and the continuous chip (Fig. 2a). The difference between the two chips was the connection form of the nickel strip. The deflection effect of the chip was characterized by the deflection rate, which was calculated by the difference in UV absorption of MNs at 600 nm before and after the chip. MNs were pumped into the intermittent chip and the continuous chip at 2 μL min−1 and MNs collected from the outlet were measured by using a UV spectrophotometer. The deflection rate of the intermittent chip was 33.9% (Fig. 2c). The reason for the low deflection rate was that some MNs would agglomerate at the end of the intermittent strip (Fig. S5, ESI†). The deflection rate of the continuous chip was 74.9%. In addition, the magnetic orientation on each side of the chip was optimized. Fig. 2b shows the schematic of the two forms of magnet placement. The deflection rates of the two placements were 43.5% and 74.9%, respectively (Fig. 2d). Therefore, the continuous chip and the magnets which were placed in the same pole on both sides were chosen for subsequent experiments.
MNs could move along the nickel strip mainly depending on the precise manipulation of the magnetic field on a micrometer scale. Therefore, it was essential to simulate the magnetic field and force distribution in the micrometer region. COMSOL Multiphysics software was used to simulate the change of magnetic flux density and magnetic field gradient (|(B·∇)B|) in the chip. From the result of the simulated color map, |(B·∇)B| had obvious strong and weak changes along the direction of the nickel strip (Fig. S6a, ESI†). To further observe the influence of the introduction of the nickel strip on the|(B·∇)B|, numerical simulations were carried out for the different microregions along the green lines, which crossed over the nickel strips. It could be found that the value of |(B·∇)B| could reach 6000 T/(T/m) (Fig. S6b–d, ESI†). When the targets labeled with MNs were pumped into the MACD chip, they would experience Fm and Fd. MNs could move along the nickel strip in different directions under the action of Fm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ and Fd (Fig. S7, ESI†). The force analysis on the MNs is presented in S.7, ESI.† Red fluorescent MNs (RFMNs) were used to verify the deflection feasibility. Images were taken in five different parts of the MACD chip. There was distinct red fluorescence at the edge of the nickel strip in each image (Fig. S8, ESI†). This indicated that MNs could move along the nickel. The experimental results corresponded to the results of numerical simulation.
θ and Fd (Fig. S7, ESI†). The force analysis on the MNs is presented in S.7, ESI.† Red fluorescent MNs (RFMNs) were used to verify the deflection feasibility. Images were taken in five different parts of the MACD chip. There was distinct red fluorescence at the edge of the nickel strip in each image (Fig. S8, ESI†). This indicated that MNs could move along the nickel. The experimental results corresponded to the results of numerical simulation.
The reaction of streptavidin (SA) and biotin was used as a model to explore the binding reaction in the MACD chip (Fig. S9a, ESI†). The SA protein was chemically conjugated to MNs successfully. The hydrodynamic size of the MNs changed from 518.2 nm to 587.7 nm (Fig. S9b, ESI†) and the zeta potential (ζ) of the MNs changed from −27.4 mV to −10.7 mV (Fig. S9c, ESI†). The MNs-SA solution, biotinylated DNA (5′-Alexa Fluor 488-AGCGTCGAATACACTACAG-biotin-3′), and 1 × PBS (pH 7.2) were pumped into the chip at 2 μL min−1 for reacting during 1.5 h. MNs were collected and observed under an inverted fluorescence microscope. The surface of the MNs had a distinct green fluorescence (Fig. S9d, ESI†), which demonstrated that MNs-SA could successfully react with biotin in the MACD chip. Therefore, the MACD chip provided the possibility of successful binding in a continuous flow using the MACD chip.
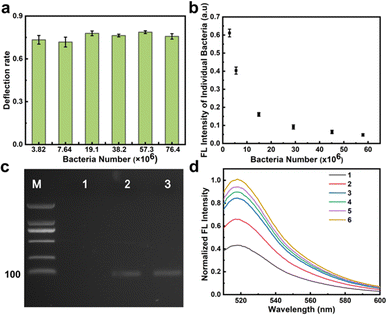
C. albicans was successfully conjugated to MNs-SA (Fig. S10, ESI†). The amounts of ConA-biotin and MNs-SA were optimized during the coupling process (Fig. S11, ESI†). MNs-C. albicans complexes were incubated with syto9 dye and then were pumped into the deflection chip at 2 μL min−1. There was distinct green fluorescence at the edge of the nickel strip in different parts of the deflection zone (Fig. S12, ESI†). This indicated that the MNs-C. albicans complex could move along the nickel strip to achieve movement in different solutions. The low-affinity sequences could be eluted in the washing buffer (S.13, ESI†). Experimental and theoretical models24,25 have shown that low target concentrations could form highly stringent competition conditions and promote the enrichment of high-affinity aptamers to facilitate the selection of aptamers. Therefore, it was important to optimize the number of targets. The target concentration could be controlled during the screening process by pumping different numbers of targets into the MACD chip. The different absorption of C. albicans at 260 nm before and after the MACD chip was used to calculate the deflection rate. All deflection rates were between 70 and 80%, indicating that the deflection was relatively stable (Fig. 3a). As the number of bacteria decreased, the average fluorescence intensity of individual bacteria increased (Fig. 3b). This showed that sequences bound by individual bacteria were increased as the number of bacteria decreased via dynamic selection. The binding efficiency of the bacteria could be improved by decreasing the number of bacteria for aptamer selection. When the number of bacteria was as low as 3.82 × 106, the bound sequences were difficult to be amplified using traditional PCR (Fig. 3c). Taking into account the time cost, 7.64 × 106C. albicans was chosen for dynamic selection. Under these conditions, about 1400 C. albicans flowed through the library phase per second. The amount was low enough in aptamer selection and was beneficial to facilitate the selection of the high-affinity aptamer.
To remove the interference factor and improve the specificity of the selection, Saccharomyces cerevisiae (S. cerevisiae) is chosen as the negative selection and the result is shown in Fig. S14a, ESI.† Obvious green fluorescence on the surface of the bacteria collected in the reservoir pool of the positive unit was observed under the fluorescence microscope, indicating that the aptamer was successfully bound to the bacteria (Fig. S14b, ESI†). The secondary library was prepared by the following steps: first, the bound sequences were amplified and the amplified product was captured by MNs-SA (Fig. S15a, ESI†), the amount of MNs-SA was optimized to capture the full amplification product (Fig. S15c, ESI†), then the amplified product was denatured under the action of NaOH (Fig. S15b, ESI†), and the released ssDNA library in the supernatant was purified by 10 K ultrafiltration (Fig. S15d, ESI†). With the increase of the selection rounds, the enriched fluorescence intensity gradually increased, and after the sixth round, the fluorescence signal did not increase significantly and the screening was stopped (Fig. 3d).
After six rounds of dynamic selection, 20 clones were randomly selected for cloning and sequencing. The homology of the obtained sequences was analyzed by using Clustalx software (Fig. S16, ESI†). Sequences with relatively high homology and frequency of occurrence, namely C.al2, C.al3, C.al9, and C.al10, were selected for affinity testing. The Kd values of aptamers C.al2, C.al3, C.al9, and C.al10 were measured to be 21.5 nM, 7.9 nM, 50.2 nM, and 9.2 nM, respectively (Fig. 4a–d). The affinity of C.al3 screened by this method was higher than that of most other bacterial aptamers (Table S1, ESI†). Notably, the affinity of C.al3 was 10-fold higher compared to that of the C. albicans selected so far.26
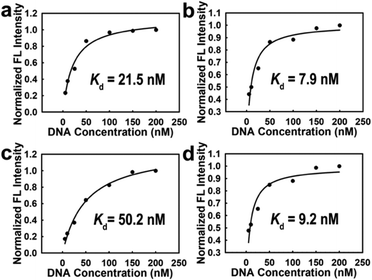 | ||
| Fig. 4 Determination of the dissociation constants for C. albicans with (a) C.al2, (b) C.al3, (c) C.al9, and (d) C.al10. | ||
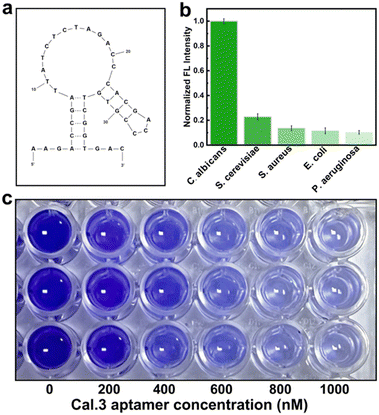
The C.al3 aptamer with the highest affinity was selected for the specificity and membrane inhibition experiment. The secondary structure of C.al3 was simulated by using mfold web (Fig. 5a). As shown in Fig. 5b, C.al3 could specifically recognize C. albicans compared to other bacteria. The formation of a biofilm increased pathogenic microorganisms such as C. albicans due to adverse environments, and the host's immune and antimicrobial drug resistance, and it was a major cause of chronic infection, and recurrent infections.27 So the inhibition effect of the aptamer (C.al3) on the growth of C. albicans biofilms was investigated, and three independent crystal violet staining biofilm assays were performed. With the increase of the C.al3 concentration, the crystal violet color in a 96-well plate gradually became lighter, indicating that the bacterial biofilm content gradually decreased (Fig. 5c). These results suggested that C.al3 could specifically recognize C. albicans and it had a certain application potential to inhibit the formation of C. albicans biofilms.
In summary, a MACD chip was developed to screen aptamers for C. albicans. The targets labeled with MNs would move along a nickel strip and sequentially flow through a library phase and washing phase in the MACD chip. Based on this, dynamic selection in continuous flows could be implemented. The efficiency of selection was improved by the elution of low-affinity sequences to facilitate the enrichment of high-affinity sequences. The stringency of the selection was increased by lowering the target concentration via dynamic selection. A C.al3 aptamer with high-affinity and high-specificity for C. albicans was obtained. The MACD chip could have the potential to provide an efficient method for bacterial aptamer selection.
This work was supported by the National Natural Science Foundation of China (22074107 and 22274118).
Conflicts of interest
There are no conflicts to declare.Notes and references
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- H. Qu, A. T. Csordas, J. Wang, S. S. Oh, M. S. Eisenstein and H. T. Soh, ACS Nano, 2016, 10, 7558–7565 CrossRef CAS PubMed.
- H. I. Lin, C. C. Wu, C. H. Yang, K. W. Chang, G. B. Lee and S. C. Shiesh, Lab Chip, 2015, 15, 486–494 RSC.
- H. C. Lai, C. H. Wang, T. M. Liou and G. B. Lee, Lab Chip, 2014, 14, 2002–2013 RSC.
- E. E. F. Brown, A. Cooper, C. Carrillo and B. Blais, Front. Microbiol., 2019, 10, 456 CrossRef PubMed.
- R. Nutiu and Y. Li, Angew. Chem., Int. Ed., 2005, 44, 1061–1065 CrossRef CAS PubMed.
- C. Wang, B. Liu, J. Lu, G. Zhang and A. Lu, J. Nanosci. Nanotechnol., 2014, 14, 501–512 CrossRef CAS PubMed.
- F. Xia, A. He, H. Zhao, Y. Sun, Q. Duan, S. J. Abbas, J. Liu, Z. Xiao and W. Tan, ACS Nano, 2022, 16, 169–179 CrossRef CAS PubMed.
- M. Ilgu and M. Nilsen-Hamilton, Analyst, 2016, 141, 1551–1568 RSC.
- S. Qian, D. Chang, S. He and Y. Li, Anal. Chim. Acta, 2022, 1196, 339511 CrossRef CAS PubMed.
- H. Y. Dong, Q. H. Xie, D. W. Pang, G. Chen and Z. L. Zhang, Chem. Commun., 2021, 57, 3555–3558 RSC.
- J. Qian, X. Lou, Y. Zhang, Y. Xiao and H. T. Soh, Anal. Chem., 2009, 81, 5490–5495 CrossRef PubMed.
- X. Lou, J. Qian, Y. Xiao, L. Viel, A. E. Gerdon, E. T. Lagally, P. Atzberger, T. M. Tarasow, A. J. Heeger and H. T. Soh, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2989–2994 CrossRef CAS PubMed.
- J. Y. Ahn, S. Lee, M. Jo, J. Kang, E. Kim, O. C. Jeong, T. Laurell and S. Kim, Anal. Chem., 2012, 84, 2647–2653 CrossRef CAS PubMed.
- X. Liu, H. Li, W. Jia, Z. Chen and D. Xu, Lab Chip, 2017, 17, 178–185 RSC.
- Y. J. Seo, S. Chen, M. Nilsen-Hamilton and H. A. Levine, Bull. Math. Biol., 2010, 72, 1623–1665 CrossRef PubMed.
- O. Alkhamis and Y. Xiao, J. Am. Chem. Soc., 2023, 145, 194–206 CrossRef CAS PubMed.
- D. Chang, Z. Wang, C. D. Flynn, A. Mahmud, M. Labib, H. Wang, A. Geraili, X. Li, J. Zhang, E. H. Sargent and S. O. Kelley, Nat. Chem., 2023, 15, 773–780 CrossRef CAS PubMed.
- S. S. Oh, K. M. Ahmad, M. Cho, S. Kim, Y. Xiao and H. T. Soh, Anal. Chem., 2011, 83, 6883–6889 CrossRef CAS PubMed.
- S. L. Hong, Y. T. Wan, M. Tang, D. W. Pang and Z. L. Zhang, Anal. Chem., 2017, 89, 6535–6542 CrossRef CAS PubMed.
- Y. T. Wang, M. Wang, K. Z. Yang and Z. L. Zhang, Sens. Diagn., 2023, 2, 418–426 RSC.
- S. M. Noble, S. French, L. A. Kohn, V. Chen and A. D. Johnson, Nat. Genet., 2010, 42, 590–598 CrossRef CAS PubMed.
- M. Swidergall and S. LeibundGut-Landmann, Mucosal Immunol., 2022, 15, 829–836 CrossRef CAS PubMed.
- H. A. Levine and M. Nilsen-Hamilton, Comput. Biol. Chem., 2007, 31, 11–35 CrossRef CAS PubMed.
- Y. Ding and J. Liu, J. Am. Chem. Soc., 2023, 145, 7540–7547 CrossRef CAS PubMed.
- X. L. Tang, Y. Hua, Q. Guan and C.-H. Yuan, Eur. J. Clin. Microbiol. Infect. Dis., 2016, 35, 587–595 CrossRef CAS PubMed.
- C. J. Nobile and A. D. Johnson, Annu. Rev. Microbiol., 2015, 69, 71–92 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc00229f |
| This journal is © The Royal Society of Chemistry 2024 |