Development of a molecular glue-based Lin28 degrader to regulate cellular proliferation and stemness†
Minha
Lee‡
,
Wan Gi
Byun‡
,
Sumin
Son
and
Seung Bum
Park
 *
*
CRI Center for Chemical Proteomics, Department of Chemistry, Seoul National University, Seoul, 08826, Korea. E-mail: sbpark@snu.ac.kr
First published on 20th September 2024
Abstract
Let-7 microRNAs (miRNAs) regulate cellular processes including stemness and proliferation. Lin28, an RNA-binding protein, controls let-7 miRNA biogenesis and is a key factor in maintaining stem cell properties. We developed SB1349, a novel molecular glue-based degrader targeting Lin28. SB1349 induces Lin28 degradation through a proteasome-dependent pathway, enhances let-7 miRNA levels, and downregulates oncogenes c-Myc and IMP1. SB1349 also promotes the differentiation in neuroblastoma cells, highlighting its potential as a therapeutic agent for various diseases.
Let-7 microRNAs (miRNAs) are critical regulators of various cellular processes, including stemness and cellular proliferation. Specifically, let-7 miRNAs inhibit stem cell self-renewal and promote differentiation by targeting genes that maintain pluripotency.1,2 In cancer cells, let-7 miRNAs exhibit significant anticancer activity by repressing oncogenes and impeding uncontrolled proliferation.3 These 22-nucleotide miRNAs bind to the 3′ untranslated regions (3′ UTRs) of their target mRNAs, blocking the translation of diverse oncogenic target mRNAs such as c-Myc and IMP1.4,5
The RNA-binding protein Lin28 regulates the biogenesis of let-7 miRNAs. Primary let-7 (pri-let-7) transcripts are processed by the enzymes DROSHA and DICER to generate precursor (pre-let-7) and mature let-7 miRNAs, respectively. Lin28 binds to the loop regions of pri-let-7 and pre-let-7, blocking their maturation and promoting their degradation. Lin28 suppresses the level of let-7 miRNAs, which function as tumor suppressors through these processes.6 Additionally, Lin28 is recognized as a stemness factor essential for maintaining the self-renewal capacity of stem cells by regulating genes involved in pluripotency and proliferation.7
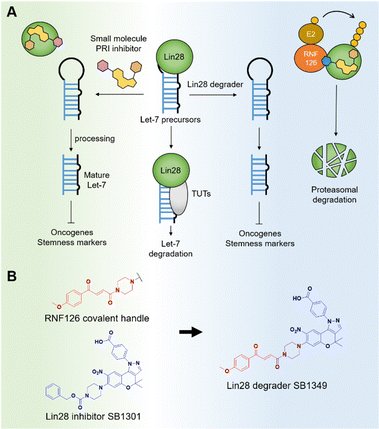
Given its profound biological and pharmacological importance, the Lin28–let-7 axis has emerged as a significant potential anticancer target across various disciplines, including chemical biology and medicinal chemistry.8 The impact of let-7 miRNAs on multiple oncogenic pathways has spurred significant research efforts toward developing small-molecule Lin28 inhibitors.9 These inhibitors have shown promise in treating various diseases, including cancer and non-alcoholic fatty liver disease (NAFLD).10,11 In the context of cancer, these inhibitors downregulate oncogenes and target cancer stem cells (Fig. 1A). Previously, our group discovered a benzopyranylpyrazole-based Lin28–let-7 interaction inhibitor, named SB1301, through a FRET-based binding assay utilizing a site-specific protein labeling strategy.9a This protein–RNA interaction (PRI) inhibitor, SB1301, exhibited Lin28-inhibiting activity in multiple cell lines and has been utilized as a basis for developing new modality Lin28 inhibitors, such as peptide-SB1301 conjugates.12 Despite these significant advancements, there have been no reports of a small-molecule degrader targeting Lin28.
For this reason, we decided to develop a novel molecular glue-based small-molecule degrader of Lin28 in this study. Our approach leverages the unique properties of a but-2-ene-1,4-dione (fumarate) as a molecular glue degrader handle. A recent study revealed that introducing the fumarate handle to the exit vector on the nitrogen atom of piperazine can convert small-molecule ligands into molecular glue degraders, primarily by recruiting the RING-family E3 ubiquitin ligase RNF126.13 Inspired by the study and the chemical structure of SB1301, we substituted the benzyloxycarbonyl group on the piperazine of SB1301 with the fumarate handle, obtaining SB1349, a molecular glue-based Lin28 degrader (Fig. 1B).
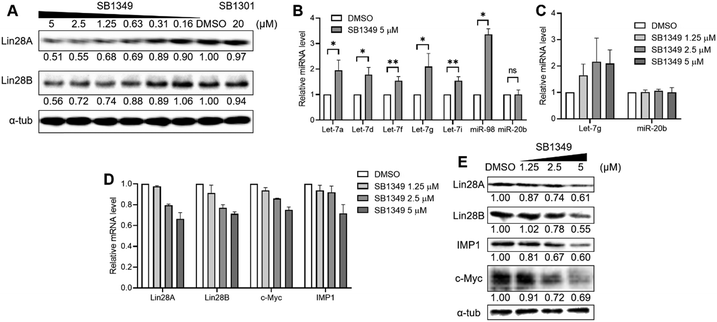
We first evaluated whether SB1349 could induce the degradation of Lin28 proteins. Given that the Lin28 PRI inhibitor SB1301 disrupts the interactions of the two Lin28 paralogs (Lin28A and Lin28B) with let-7 members,14 we quantified the levels of both paralogs with western blot analysis. Treatment of human choriocarcinoma JAR cells, expressing both Lin28A and Lin28B, with SB1349 demonstrated a dose-dependent degradation of both paralogs of Lin28 (Fig. 2A and Fig. S1, ESI†). Interestingly, SB1301 showed a negligible effect on the expression of Lin28 proteins at a 20 μM concentration under the tested conditions (Fig. 2A). We also confirmed that SB1349 could reduce the cellular levels of Lin28 proteins in other cell lines, such as human ovarian teratocarcinoma PA-1 cells (expressing Lin28A exclusively) and human embryonic kidney 293T cells (expressing Lin28B exclusively) (Fig. S2, ESI†). A water-soluble tetrazolium (WST)-based cell viability assay confirmed that SB1349 has no significant cytotoxicity at tested concentrations (Fig. S3, ESI†). To further validate these findings, we performed TaqMan-based quantitative RT-PCR (qRT-PCR), which demonstrated that SB1349 increased the cellular levels of various mature let-7 members, including let-7a, d, f, g, i, and miR-98. In contrast, SB1349 had minimal impact on miR-20b, a miRNA not associated with Lin28 (Fig. 2b). The increase in let-7 miRNA members was dose-dependent, while miR-20b levels remained unaffected (Fig. 2C and Fig. S4, ESI†).
Next, we investigated the impact of SB1349 on the cellular expressions of let-7 target genes to explore the downstream effects of Lin28 degradation. First, we conducted qRT-PCR to measure the mRNA levels of the let-7 target oncogenes. Dose-dependent treatment with SB1349 reduced the levels of both c-Myc and IMP1 mRNAs (Fig. 2D). We also examined the mRNA levels of Lin28A and Lin28B. Since Lin28 and let-7 are engaged in a negative feedback loop, inhibiting Lin28 function and thereby increasing mature let-7 levels should result in the downregulation of Lin28 itself. As expected, the treatment of JAR cells with SB1349 led to a dose-dependent reduction in the mRNA levels of both Lin28A and Lin28B (Fig. 2D). However, the treatment with SB1301 at 5 μM did not effectively reduce the mRNA levels of these let-7 targets compared to the same concentration of SB1349 (Fig. S5, ESI†). Correspondingly, western blot analysis revealed a dose-dependent decrease in the expression of both c-Myc and IMP1 proteins upon treatment with SB1349 (Fig. 2E). These data indicated that SB1349 effectively degrades Lin28 proteins and demonstrates better activity than SB1301. By promoting Lin28 degradation, SB1349 upregulates mature let-7 levels and downregulates target oncogenes, c-Myc and IMP1.
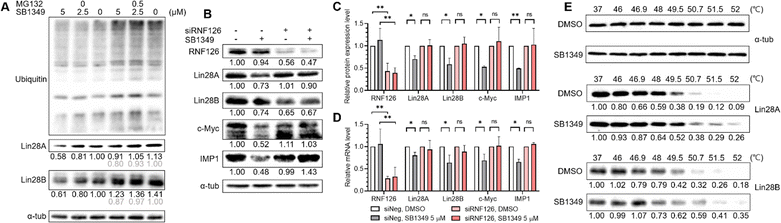
To elucidate the molecular mechanism of SB1349-mediated degradation of Lin28, we performed western blot analysis in the presence of MG132, a proteasome inhibitor. MG132 treatment led to an accumulation of ubiquitinated proteins, confirming the effective inhibition of proteasomal activity. MG132 reduced the SB1349-mediated degradation of Lin28A and Lin28B (Fig. 3A and Fig. S6, ESI†), suggesting that SB1349 promotes Lin28 degradation through a proteasome-dependent pathway.
We then investigated whether SB1349-mediated Lin28 degradation depends on the E3 ligase RNF126. Knockdown of RNF126 using siRNA diminished the degradation effect of SB1349 on both Lin28A and Lin28B, while its degradation activity was maintained in cells treated with negative control siRNA (Fig. 3B and C). In RNF126-knocked down JAR cells, the enhanced cellular levels of mature let-7g were significantly decreased, corroborating the involvement of RNF126 in this process (Fig. S7, ESI†). Furthermore, the downregulation of Lin28A, Lin28B, c-Myc, and IMP1 mRNAs by SB1349 was abolished upon RNF126 knockdown, and the reduction in c-Myc and IMP1 protein expression were also diminished (Fig. 3B–D). These results indicated that RNF126 is essential for the SB1349-mediated degradation of Lin28.
Next, we conducted cellular thermal shift assay (CETSA) experiments to confirm SB1349's direct binding to Lin28A and Lin28B within a cellular context. The treatment with SB1349 enhanced the thermal stability of both Lin28A and Lin28B (Fig. 3E and Fig. S8a, ESI†). Consistently, SB1301 also exhibited thermal stability stabilization for both Lin28 paralogs (Fig. S8b, ESI†). These results demonstrate that SB1349 induces RNF126-dependent proteasomal degradation of Lin28 by directly binding to Lin28.
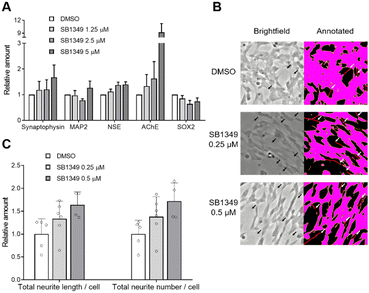
Considering that Lin28 is a key regulator of stem cell characteristics through both let-7-dependent and let-7-independent pathways, a small-molecule degrader of Lin28 could be a valuable tool for inducing differentiation. Therefore, we investigated the potential application of SB1349 in inducing the differentiation of SH-SY5Y, a human-derived neuroblastoma cell line expressing Lin28B. Since primary mammalian neurons are difficult to propagate, transforming immortalized undifferentiated cells into neuron-like cells is beneficial for neuroscience research.15 We first assessed whether SB1349 can degrade Lin28 protein in SH-SY5Y cells. As expected, SB1349 dose-dependently reduced the levels of Lin28B (Fig. S9, ESI†). Next, qRT-PCR experiments confirmed that SB1349 treatment increased the expression of various neuronal markers, including synaptophysin, microtubule-associated protein-2 (MAP2), and neuron-specific enolase (NSE),16 by 1.68-, 1.27-, and 1.40-fold respectively, at 5 μM (Fig. 4A). Additionally, we checked the mRNA level of acetylcholinesterase (AChE), which acts as a primary cholinesterase to regulate neurotransmission. AChE expression also increased, aligning with known differentiation patterns in SH-SY5Y cells.17 Correspondingly, the stem cell marker SOX2 expression was downregulated,16 indicating a successful transition from a stem cell state to a more differentiated neuronal state. Intriguingly, the SB1349 treatment exhibited similar or better differentiation patterns compared to the SB1301 treatment (Fig. S10, ESI†).
Having established that SB1349 altered the expression of multiple mRNAs toward a differentiated state, we investigated the morphological changes induced by SB1349. It is well-known that undifferentiated SH-SY5Y cells, which tend to grow in clusters, become more dispersed and morphologically similar to primary neurons, with extended neurite length and an increased number of neurites after differentiation.15,16,18 After treating the cells with compounds for 3 days, we imaged and analyzed them using NeuroQuantify. This deep learning-based image analysis software detects and quantifies neurons and neurites in phase-contrast microscopic images.19 We first validated its use by demonstrating that treatment with 10 μM retinoic acid (RA), the most commonly used agent for SH-SY5Y differentiation, significantly increased both the total neurite length per cell and the total number of neurites per cell (Fig. S11 and S12, ESI†). Following this validation, we applied the same analysis to SB1349. We observed that it similarly enhanced both the total neurite length and the total neurite number per cell in a dose-dependent manner (Fig. 4B, C and Fig. S11 and S12, ESI†), indicating that SB1349 could induce the differentiation of SH-SY5Y cells by degrading the Lin28 stem cell factor.
In summary, we have developed SB1349, a novel molecular glue-based small-molecule degrader targeting Lin28. Our study demonstrated that SB1349 induces a dose-dependent degradation of Lin28A and Lin28B, enhances the levels of mature let-7 miRNAs, and downregulates the let-7 target oncogenes c-Myc and IMP1. Mechanistic investigations revealed that SB1349 induces Lin28 degradation through a proteasome-dependent pathway and requires the E3 ligase RNF126 for its activity. Biophysical studies confirmed the direct interaction of SB1349 with Lin28 proteins in the cellular environment. Additionally, application studies highlighted its potential to promote cellular differentiation of neurites. Overall, these findings provide a foundation for further exploration of SB1349 as a therapeutic agent, with the potential to target Lin28 in various disease contexts beyond cancer. Furthermore, this study suggests that with further improvements, such as reducing cytotoxicity, SB1349 could be utilized as a chemical tool or therapeutic strategy to promote differentiation in stem and progenitor cell models.
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean Government (Ministry of Science & ICT) (RS-2024-00438764 to S.B.P.) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (RS-2023-00273284 to W.G.B.). JAR and SH-SY5Y cells were obtained from the Korean Cell Line Bank (KCLB).
Data availability
All data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- I. Büssing, F. J. Slack and H. Grobhans, Trends Mol. Med., 2008, 14, 400–409 CrossRef PubMed.
- I. Ibarra, Y. Erlich, S. K. Muthuswamy, R. Sachidanandam and G. J. Hannon, Genes Dev., 2007, 21, 3238–3243 CrossRef CAS PubMed.
- B. Boyerinas, S. M. Park, A. Hau, A. E. Murmann and M. E. Peter, Endocr. – Relat. Cancer, 2010, 17, F19–36 CAS.
- V. B. Sampson, N. H. Rong, J. Han, Q. Yang, V. Aris, P. Soteropoulos, N. J. Petrelli, S. P. Dunn and L. J. Krueger, Cancer Res., 2007, 67, 9762–9770 CrossRef CAS PubMed.
- N. Degrauwe, M. L. Suvà, M. Janiszewska, N. Riggi and I. Stamenkovic, Genes Dev., 2016, 30, 2459–2474 CrossRef CAS PubMed.
- J. E. Thornton and R. I. Gregory, Trends Cell Biol., 2012, 22, 474–482 CrossRef CAS PubMed.
- J. Balzeau, M. R. Menezes, S. Cao and J. P. Hagan, Front. Genet., 2017, 8, 31 Search PubMed.
- W. G. Byun, D. Lim and S. B. Park, Curr. Opin. Chem. Biol., 2022, 68, 102149 CrossRef CAS PubMed.
- (a) D. Lim, W. G. Byun, J. Y. Koo, H. Park and S. B. Park, J. Am. Chem. Soc., 2016, 138, 13630–13638 CrossRef CAS PubMed; (b) D. Lim, W. G. Byun and S. B. Park, ACS Med. Chem. Lett., 2018, 9, 1181–1185 CrossRef CAS PubMed; (c) M. Roos, U. Pradere, R. P. Ngondo, A. Behera, S. Allegrini, G. Civenni, J. A. Zagalak, J. R. Marchand, M. Menzi, H. Towbin, J. Scheuermann, D. Neri, A. Caflisch, C. V. Catapano, C. Ciaudo and J. Hall, ACS Chem. Biol., 2016, 11, 2773–2781 CrossRef CAS PubMed; (d) L. Borgelt, F. Li, P. Hommen, P. Lampe, J. Hwang, G. L. Goebel, S. Sievers and P. Wu, ACS Med. Chem. Lett., 2021, 12, 893–898 CrossRef CAS PubMed; (e) M. Radaeva, C. H. Ho, N. Xie, S. Zhang, J. Lee, L. Liu, N. Lallous, A. Cherkasov and X. Dong, Cancers, 2022, 14, 5687 CrossRef CAS PubMed; (f) L. Wang, R. G. Rowe, A. Jaimes, C. Yu, Y. Nam, D. S. Pearson, J. Zhang, X. Xie, W. Marion, G. J. Heffron, G. Q. Daley and P. Sliz, Cell Rep., 2018, 23, 3091–3101 CrossRef CAS PubMed; (g) D. A. Lorenz, T. Kaur, S. A. Kerk, E. E. Gallagher, J. Sandoval and A. L. Garner, ACS Med. Chem. Lett., 2018, 9, 517–521 CrossRef CAS PubMed.
- Q. Zhang, M. Shi, R. Zheng, H. Han, X. Zhang and F. Lin, Eur. J. Pharmacol., 2023, 956, 175935 CrossRef CAS PubMed.
- E. Lekka, A. Kokanovic, S. Mosole, G. Civenni, S. Schmidli, A. Laski, A. Ghidini, P. Iyer, C. Berk, A. Behera, C. V. Catapano and J. Hall, Nat. Commun., 2022, 13, 7940 CrossRef CAS PubMed.
- P. Hommen, J. Hwang, F. Huang, L. Borgelt, L. Hohnen and P. Wu, ChemBioChem, 2023, 24, e202300376 CrossRef CAS PubMed.
- E. S. Toriki, J. W. Papatzimas, K. Nishikawa, D. Dovala, A. O. Frank, M. J. Hesse, D. Dankova, J. G. Song, M. Bruce-Smythe, H. Struble, F. J. Garcia, S. M. Brittain, A. C. Kile, L. M. McGregor, J. M. McKenna, J. A. Tallarico, M. Schirle and D. K. Nomura, ACS Cent. Sci., 2023, 9, 915–926 CrossRef CAS PubMed.
- E. Piskounova, C. Polytarchou, J. E. Thornton, R. J. LaPierre, C. Pothoulakis, J. P. Hagan, D. Iliopoulos and R. I. Gregory, Cell, 2011, 147, 1066–1079 CrossRef CAS PubMed.
- M. M. Shipley, C. A. Mangold and M. L. Szpara, J. Visualized Exp., 2016, 108, e53193 Search PubMed.
- S. Gurunathan and J. H. Kim, Int. J. Mol. Sci., 2017, 18, 2549 CrossRef PubMed.
- L. Pulkrabkova, L. Muckova, M. Hrabinova, A. Sorf, T. Kobrlova, P. Jost, D. Bezdekova, J. Korabecny, D. Jun and O. Soukup, Arch. Toxicol., 2023, 97, 2209–2217 CrossRef CAS PubMed.
- J. Kovalevich and D. Langford, Methods Mol. Biol., 2013, 1078, 9–21 CrossRef CAS PubMed.
- K. M. Dang, Y. J. Zhang, T. Zhang, C. Wang, A. Sinner, P. Coronica and J. K. S. Poon, arXiv, 2023, preprint, arXiv:2310.10978 DOI:10.48550/arXiv.2310.10978.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures, bioassay protocols, additional figures, and the characterization of all new compounds. See DOI: https://doi.org/10.1039/d4cc03614j |
| ‡ M. Lee and W. G. Byun contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |