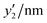Machine-learning-assisted prediction of the size of microgels prepared by aqueous precipitation polymerization†
Daisuke
Suzuki
 *ab,
Haruka
Minato
*ab,
Haruka
Minato
 ab,
Yuji
Sato
ab,
Yuji
Sato
 ab,
Ryuji
Namioka
b,
Yasuhiko
Igarashi
c,
Risako
Shibata
d and
Yuya
Oaki
ab,
Ryuji
Namioka
b,
Yasuhiko
Igarashi
c,
Risako
Shibata
d and
Yuya
Oaki
 *d
*d
aGraduate School of Environmental, Life, Natural Science and Technology, Okayama University, 3-1-1 Tsushimanaka, Kita-ku, Okayama, 700-8530, Japan. E-mail: d_suzuki@okayama-u.ac.jp
bGraduate School of Textile Science & Technology, Shinshu University, 3-15-1 Tokida, Ueda, Nagano 386-8567, Japan
cFaculty of Engineering, Information and Systems, University of Tsukuba, 1-1-1 Tennodai, Tsukuba 305-8573, Japan
dDepartment of Applied Chemistry, Faculty of Science and Technology, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan. E-mail: oakiyuya@applc.keio.ac.jp
First published on 21st October 2024
Abstract
The size of soft colloids (microgels) is essential; however, control over their size has typically been established empirically. Herein, we report a linear-regression model that can predict microgel size using a machine learning method, sparse modeling for small data, which enables the determination of the synthesis conditions for target-sized microgels.
Hydrogel nano/microparticles (nanogels/microgels) are hydrophilic or amphiphilic colloids that are highly swollen by water and are dispersed stably in aqueous solution.1 Due to their fascinating properties related to their softness and stimuli-responsiveness, their use in various applications, including controlled uptake/release of functional molecules,2 as particulate stabilizers for interfaces,3 and as soft colloidal crystals/glasses/gels,4 has been proposed.
Among the methods for producing microgels, aqueous free radical precipitation polymerization is an excellent strategy for forming microgels of uniform size under environmentally friendly and cost-effective experimental conditions.1a,5 It is widely accepted that the monomers for these polymers are water soluble, but that upon growing, the polymers become insoluble in water, which results in the formation of nuclei for the growth of microgels; these nuclei then grow until they acquire sufficient colloidal stability.2a,5c To date, tremendous efforts have been devoted to revealing the detailed mechanism of precipitation polymerization,1a,5b,c,6 which would allow the size of simple microgels (e.g., a monomer and crosslinker) obtained by precipitation polymerization to be controlled.7 However, copolymerization with various functional monomers to add further functionality to simple microgels is usually required, complicating the reaction and hence the prediction of the microgel size. In addition, various parameters including polymerization temperature and stirring conditions affect the size of the resultant microgel. Thus, in many cases, the microgel size in precipitation polymerizations has been controlled using a trial-and-error approach for each parameter based on the experience and intuition of professional researcher(s). If the size of functional microgels could be predicted, the development of applications that require precise control of the microgel size, such as targeted drug delivery and the formation of colloidal crystals composed of different microgels, would be accelerated.
Against this background, we found that machine learning (ML) is an effective way to predict the size of microgels prepared by aqueous free radical precipitation polymerization (Fig. 1). ML has been widely applied to the optimization of processes and the exploration of materials, such as controlled synthesis of nanosheet materials and discovery of new organic active materials for energy-related applications.8 However, in conventional experimental approaches, it can be difficult to collect a sufficiently large dataset to train and construct the corresponding predictors. Our group has developed a technique, which is called sparse modelling for small data (SpM-S), that combines ML and chemical insight.9 This method provides straightforward, interpretable, and generalizable models using datasets that are small compared to other ML algorithms. Here, SpM-S is applied to predict the size of microgels.
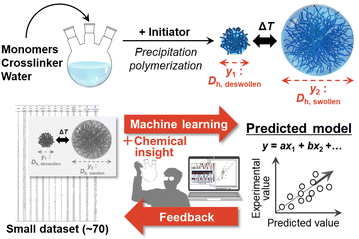 | ||
| Fig. 1 Schematic illustration of the machine-learning-assisted prediction of the microgel size developed in this study. | ||
In this study, four previously reported10 and 66 newly synthesized microgels were used to prepare the training dataset. The polymerization conditions were varied randomly (Table S1, ESI†). The hydrodynamic diameter (Dh) determined by dynamic light scattering (DLS) was employed as the size parameter for all 70 microgels (i.e., the objective variable; y1: deswollen microgels; y2: swollen microgels), since it is difficult to measure the size of microgels by means of microscopy techniques, given that the microgels are highly deformed on the solid substrates (Table S1, ESI†).11 Prior to the DLS measurements, the size uniformity was examined using scanning electron microscopy (SEM) in order to confirm the validity of calculating Dh using the Stokes–Einstein equation (Fig. 2(b)–(d) and Fig. S1–S3, ESI†). The microgels were clearly uniform in size with no secondary or individual particles, indicating that the determined Dh values are reliable. The Dh values in this study were measured at pH = 3, where the carboxyl groups in the microgels are protonated.10,12 In many cases, the obtained time-correlation functions were unreliable for highly swollen microgels with a Dh > 5 μm, and thus, these data were not used for this investigation (Table S1, ESI†). All time-correlation functions used for determining Dh are summarized in Fig. 2(c) and (d) as well as Fig. S2 and S3 (ESI†).
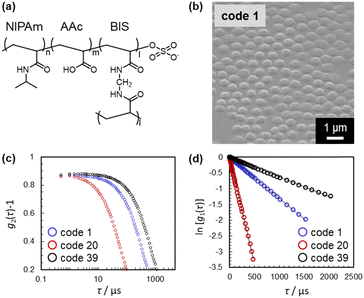 | ||
| Fig. 2 (a) Chemical structure of the microgels. (b)–(d) Representative (b) SEM image, (c) time-correlation function of the scattering intensity, g2(τ) − 1, and (d) calculated time-correlation function of the scattering electric field, ln[g1(τ)], of selected microgels. All SEM images and time-correlation functions for all microgels used in this study are summarized in Fig. S1 and S2 (ESI†). | ||
Then, ten experimental parameters and conditions, including concentration, stirring speed, and solution temperature, were set as the explanatory variables (xn) based on our chemical insight (Table 1). Here, N-isopropyl acrylamide (denoted as NIPAm, x1/mM) and crosslinker N,N′-methylenebis(acrylamide) (BIS, x2/mM) were chosen as the chemical constituents of the microgels (Fig. 2(a)). In order to add functionality to the microgels, acrylic acid (AAc, x3/mM) was selected as a model comonomer (Fig. 2(a)). Additionally, other explanatory variables that we thought would be likely to affect the microgel size, based on our empirical experience, were chosen (xn: n = 4–10) (Table 1): total monomer concentration used for the polymerization (x4/mM), concentration of the anionic surfactant sodium dodecyl sulfate (SDS) (x5/mM), concentration of sodium chloride (NaCl) (x6/mM), concentration of the water-soluble anionic initiator potassium persulfate (KPS) (x7/mM), amount of water (x8/mL), stirring speed (x9/rpm), and polymerization temperature T (x10/°C).
| No. | Explanatory variables, xn | Unit |
|---|---|---|
| 1 | N-isopropyl acrylamide (NIPAm) concentration | mM |
| 2 | N,N′-methylenebis(acrylamide) (BIS) concentration | mM |
| 3 | Acrylic acid (AAc) concentration | mM |
| 4 | Monomer concentration | mM |
| 5 | Anionic surfactant (sodium dodecyl sulfate (SDS)) concentration | mM |
| 6 | Inorganic salt (sodium chloride (NaCl)) concentration | mM |
| 7 | Aqueous anionic initiator (potassium persulfate (KPS)) concentration | mM |
| 8 | Water | mL |
| 9 | Stirring speed | rpm |
| 10 | T | °C |
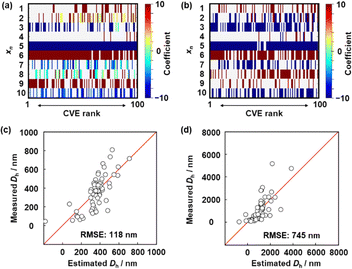
Next, the descriptors of the size of the deswollen (y1) and swollen (y2) microgels were extracted using SpM-S (Fig. 3). The detailed procedure is described in the ESI.† The potential descriptors were visualized in the weight diagram of the exhaustive search for linear regression (ES-LiR) (Fig. 3(a) and (b)).9 Here, multiple linear-regression models were prepared for all possible combinations of xn (n = 1–10) to study the contribution of each xn exhaustively, for a total of 210 − 1 (=1023) patterns. After sorting the models in ascending order of cross-validation error (CVE), the coefficients of each xn were visualized using a color scale in the weight diagram. Positive and negative coefficients are depicted using warm and cool colors, respectively (Fig. 3(a) and (b)). The more densely filled bars represent xn that are used in the models more frequently. In the present work, we selected the descriptors three steps. The potential descriptors were visually extracted from the weight diagram based on the intensity and density of the horizontal color bars (Fig. 3a and b). Then, the selection was carried out based on our chemical insight. The descriptors were finally fixed based on the prediction accuracy with adding and removing a couple of xn.
The detailed processes of the variable selection for y1 and y2 were described in the ESI† (Fig. S4 and S5, ESI†). The prediction model for y1 was constructed using these six descriptors (eqn (1)); the root-mean-squared error (RMSE) was 118 nm (Fig. 3c). In eqn (1), each xn was converted to the normalized frequency distribution (mean 0, standard deviation 1). The weight of each descriptor is represented by the coefficients.
| y1 = −91.7x5 + 57.4x6 − 2.3x7 − 7.4x10 + 33.1x1 + 38.1x9 + 356.2 | (1) |
The y2 predictor was constructed using these six descriptors (eqn (2)) with a RMSE of 745 nm (Fig. 3d).
| y2 = −17.2x2 − 341.1x5 + 435.7x6 − 252.3x7 − 96.1x10 − 189.6x3 + 1107.4 | (2) |
The descriptors in the models are partially consistent with our experience and chemical insight as experimental scientists. The anionic surfactant SDS (x5) is known to decrease the size of both the deswollen and swollen microgels (y1 and y2) during the aqueous precipitation polymerization of pNIPAm-based microgels.7a,13 In contrast, the size (y1 and y2) increases with increasing ionic strength (here, the concentration of NaCl (x6)) during polymerization.14 These findings are described by eqn (1) and (2), which provide interpretable quantitative models that are consistent with previous considerations. Although the contribution of each individual factor has been studied previously, an overall quantitative model that considers a combination of these factors has not yet been achieved.
Furthermore, although a systematic investigation of the effect of the KPS concentration (x7) has not yet been reported, based on eqn (1) and (2), it is plausible that increasing the concentration of the water-soluble initiator KPS during the polymerization decreases y1 and y2. Regarding the polymerization temperature (x10), it has been reported that temperature-programmed syntheses in which the precipitation polymerization was started at a low temperature (ca. 40 °C) and then ramped up to ca. 70 °C drastically increased the size of the resulting microgels.7b,c,15 Different from the temperature-programmed method, in the precipitation polymerization, the polymerization temperature was kept constant, and increasing the polymerization temperature (x10) decreased y1 and y2.
Finally, the preparation conditions for functional microgels with a desired size were predicted using the developed prediction model. As it is mainly microgels in the swollen state that play a crucial role, a swollen-microgel size (y2) of 500 nm was targeted. For the prediction experiments, we fixed the values of several of the polymerization conditions (xn), i.e., the concentration of AAc (x3 = 4.5 mM) and BIS (x2 = 0.75 mM, 1.50 mM, or 9.00 mM). Appropriate values for the other parameters that are included in eqn (2) under these conditions were calculated. For the parameters that are not included in eqn (2), representative values were chosen based on our experience as professional researchers: x4 = 150 mM, x8 = 100 mL, and x9 = 250 rpm. The resultant microgels all showed a size of ca. 500 nm in the swollen state regardless of the degree of crosslinking (the measured actual size of targeted microgels 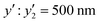 at x2 = 0.75 mM,
at x2 = 0.75 mM, 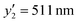 at x2 = 1.50 mM, and
at x2 = 1.50 mM, and 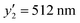 at x2 = 9.00 mM) (Table 2 and Fig. S6, ESI†). The cross validation was carried out with the addition of these new data to the original training dataset (Fig. S7, ESI†). These results demonstrate that the developed prediction method is highly reliable.
at x2 = 9.00 mM) (Table 2 and Fig. S6, ESI†). The cross validation was carried out with the addition of these new data to the original training dataset (Fig. S7, ESI†). These results demonstrate that the developed prediction method is highly reliable.
| BIS concentration | Other parameters included in eqn (2) | Target y2 = 500 nm | ||||
|---|---|---|---|---|---|---|
| x 2/mM | x 1/mM | x 5/mM | x 6/mM | x 7/mM | x 10/°C | |
| 0.75 | 144.75 | 0.50 | 0 | 3.37 | 70 | 500 |
| 1.50 | 144.00 | 0.45 | 0 | 3.38 | 70 | 511 |
| 9.00 | 163.50 | 0.10 | 0 | 2.91 | 80 | 512 |
It should be noted that eqn (1) and (2) have the potential to be further developed by adding datasets including different parameters, such as other functional comonomers, which are crucial for the development of novel microgels for new applications.12,16 Based on our previous reports,9,17 it is also likely that in addition to size, other important properties (or functions) of microgels, such as e.g., softness, can be predicted. Thus, our results represent an important first step toward a reliable method to predict microgel characteristics, which is crucial for accelerating the development of microgel science and technology.
The size of microgels prepared by aqueous free radical precipitation polymerization has been predicted using the SpM-S method to construct a prediction model using parameters that were chosen based on the authors’ experience. The size of microgels in both the deswollen and swollen state was quantitatively described by the prediction model, and the model was used successfully to determine the experimental conditions for synthesizing complicated microgels of a target size (here: 500 nm). Our findings showcase the utility of the development of machine learning with small datasets for controlling the size of colloidal particles, which is important for achieving desired functionality in applications where the size is crucial, such as drug-delivery systems and colloidal assemblies with more complex ordering. In addition, SpM-S can be applied to other small data if we can prepare a similar dataset based on the experimental results, as shown in Table S1 (ESI†).9a
D. S., H. M. and Y. O. wrote the draft of the manuscript. D. S. and Y. O. modified the manuscript. H. M., Y. S., and R. N. synthesized the microgels and contributed to microgel characterization. R. S., Y. I. and Y. O. constructed the prediction model using machine learning on small data. D. S., Y. O., H. M., and R. S. discussed the entire paper. D. S. and Y. O. designed and supervised the overall study.
D. S. and Y. O. gratefully acknowledge a Grant-in-Aid for Scientific Research (B) (24K01550) from the Japan Society for the Promotion of Science (JSPS). D. S. gratefully acknowledges a CREST grant-in-aid (JPMJCR21L2) from the Japan Science and Technology Agency (JST). The authors thank Takuma Kureha, Nahomi Matsuki, Emi Arai, and Miyuki Kuwano for assistance with the DLS measurements.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
-
(a) R. Pelton, Adv. Colloid Interface Sci., 2000, 85, 1–33 CrossRef PubMed
; (b) F. A. Plamper and W. Richtering, Acc. Chem. Res., 2017, 50, 131–140 CrossRef PubMed
; (c) D. Suzuki, K. Horigome, T. Kureha, S. Matsui and T. Watanabe, Polym. J., 2017, 49, 695–702 CrossRef
; (d) M. Karg, A. Pich, T. Hellweg, T. Hoare, L. A. Lyon, J. J. Crassous, D. Suzuki, R. A. Gumerov, S. Schneider, I. I. Potemkin and W. Richtering, Langmuir, 2019, 35, 6231–6255 CrossRef PubMed
; (e) Y. Nishizawa, K. Honda and D. Suzuki, Chem. Lett., 2021, 50, 1226–1235 CrossRef
; (f) D. Suzuki, Langmuir, 2023, 39, 7525–7529 CrossRef PubMed
; (g) Y. Gerelli, F. Camerin, S. Bochenek, M. M. Schmidt, A. Maestro, W. Richtering, E. Zaccarelli and A. Scotti, Soft Matter, 2024, 20, 3653–3665 RSC
.
-
(a) S. Nayak and L. A. Lyon, Angew. Chem., Int. Ed., 2005, 44, 7686–7708 CrossRef PubMed
; (b) Y. Hoshino, K. Imamura, M. Yue, G. Inoue and Y. Miura, J. Am. Chem. Soc., 2012, 134, 18177–18180 CrossRef PubMed
; (c) T. Kureha, Y. Nishizawa and D. Suzuki, ACS Omega, 2017, 2, 7686–7694 CrossRef PubMed
; (d) T. Kureha and D. Suzuki, Langmuir, 2018, 34, 837–846 CrossRef PubMed
; (e) S. Matsui, K. Hosho, H. Minato, T. Uchihashi and D. Suzuki, Chem. Commun., 2019, 55, 10064–10067 RSC
.
-
(a) T. Ngai, S. H. Behrens and H. Auweter, Chem. Commun., 2005, 331–333 RSC
; (b) S. Fujii, E. S. Read, B. P. Binks and S. P. Armes, Adv. Mater., 2005, 17, 1014–1018 CrossRef
; (c) D. Suzuki, S. Tsuji and H. Kawaguchi, J. Am. Chem. Soc., 2007, 129, 8088–8089 CrossRef CAS PubMed
; (d) W. Richtering, Langmuir, 2012, 28, 17218–17229 CrossRef PubMed
; (e) V. Schmitt and V. Ravaine, Curr. Opin. Colloid Interface Sci., 2013, 18, 532–541 CrossRef
; (f) T. Watanabe, M. Takizawa, H. Jiang, T. Ngai and D. Suzuki, Chem. Commun., 2019, 55, 5990–5993 RSC
; (g) Y. Nishizawa, T. Watanabe, T. Noguchi, M. Takizawa, C. Song, K. Murata, H. Minato and D. Suzuki, Chem. Commun., 2022, 58, 12927–12930 RSC
.
-
(a) T. Hellweg, C. D. Dewhurst, E. Brückner, K. Kratz and W. Eimer, Colloid Polym. Sci., 2000, 278, 972–978 CrossRef
; (b) L. A. Lyon, J. D. Debord, S. B. Debord, C. D. Jones, J. G. McGrath and M. J. Serpe, J. Phys. Chem. B, 2004, 108, 19099–19108 CrossRef
; (c) D. Suzuki, J. G. McGrath, H. Kawaguchi and L. A. Lyon, J. Phys. Chem. C, 2007, 111, 5667–5672 CrossRef
; (d) J. Mattsson, H. M. Wyss, A. Fernandez-Nieves, K. Miyazaki, Z. Hu, D. R. Reichman and D. A. Weitz, Nature, 2009, 462, 83–86 CrossRef CAS PubMed
; (e) D. Suzuki, T. Yamagata, K. Horigome, K. Shibata, A. Tsuchida and T. Okubo, Colloid Polym. Sci., 2012, 290, 107–117 CrossRef CAS
; (f) S. Minami, D. Suzuki and K. Urayama, Curr. Opin. Colloid Interface Sci., 2019, 43, 113–124 CrossRef CAS
.
-
(a) R. H. Pelton and P. Chibante, Colloids Surf., 1986, 20, 247–256 CrossRef
; (b) A. Pich and W. Richtering, Chemical Design of Responsive Microgels, Springer-Verlag, Berlin, Heidelberg, 2010, 1–37 Search PubMed
; (c) Y. Nishizawa, H. Minato, T. Inui, T. Uchihashi and D. Suzuki, Langmuir, 2021, 37, 151–159 CrossRef PubMed
.
-
(a) W. McPhee, K. C. Tam and R. Pelton, J. Colloid Int. Sci., 1993, 156, 24–30 CrossRef
; (b) F. Meunier, A. Elaïssari and C. Pichot, Polym. Adv. Technol., 1995, 6, 489–496 CrossRef
; (c) D. Duracher, A. Elaïssari and C. Pichot, J. Polym. Sci., Part A: Polym. Chem., 1999, 37, 1823–1837 CrossRef
; (d) Z. Dai and T. Ngai, J. Polym. Sci., Part A: Polym. Chem., 2013, 51, 2995–3003 CrossRef
; (e) S. Wellert, A. Radulescu, A. Carl, R. von Klitzing and K. Gawlitza, Macromolecules, 2015, 48, 4901–4909 CrossRef
; (f) O. L. J. Virtanen, M. Brugnoni, M. Kather, A. Pich and W. Richtering, Polym. Chem., 2016, 7, 5123–5131 RSC
.
-
(a) K. C. Tam, S. Ragaram and R. H. Pelton, Langmuir, 1994, 10, 418–422 CrossRef CAS
; (b) Z. Meng, M. H. Smith and L. A. Lyon, Colloid Polym. Sci., 2009, 287, 277–285 CrossRef
; (c) H. Minato, M. Murai, T. Watanabe, S. Matsui, M. Takizawa, T. Kureha and D. Suzuki, Chem. Commun., 2018, 54, 932–935 RSC
.
-
(a) S. Curtarolo, G. L. W. Hart, M. B. Nardelli, N. Mingo, S. Sanvito and O. Levy, Nat. Mater., 2013, 12, 191–201 CrossRef
; (b) K. T. Butler, J. M. Frost, J. M. Skelton, K. L. Svane and A. Walsh, Chem. Soc. Rev., 2016, 45, 6138–6146 RSC
; (c) A. Aspuru-Guzik, Digital Discovery, 2022, 1, 6–7 RSC
.
-
(a) Y. Oaki and Y. Igarashi, Bull. Chem. Soc. Jpn., 2021, 94, 2410–2422 CrossRef
; (b) H. Tobita, Y. Namiuchi, T. Komura, H. Imai, K. Obinata, M. Okada, Y. Igarashi and Y. Oaki, Energy Adv., 2023, 2, 1014–1021 RSC
; (c) Y. Haraguchi, Y. Igarashi, H. Imai and Y. Oaki, Digital Discovery, 2022, 1, 26–34 RSC
; (d) R. Mizuguchi, Y. Igarashi, H. Imai and Y. Oaki, Nanoscale, 2021, 13, 3853–3859 RSC
.
- H. Minato, M. Takizawa, S. Hiroshige and D. Suzuki, Langmuir, 2019, 35, 10412–10423 CrossRef CAS
.
-
(a) K. Horigome and D. Suzuki, Langmuir, 2012, 28, 12962–12970 CrossRef CAS
; (b) K. Horigome, T. Ueki and D. Suzuki, Polym. J., 2016, 48, 273–279 CrossRef CAS
.
- T. Kureha, D. Aoki, S. Hiroshige, K. Iijima, D. Aoki, T. Takata and D. Suzuki, Angew. Chem., Int. Ed., 2017, 56, 15393–15396 CrossRef CAS
.
-
(a) D. Suzuki, T. Yamagata, K. Horigome, K. Shibata, A. Tsuchida and T. Okubo, Colloid Polym. Sci., 2012, 290, 107–117 CrossRef CAS
; (b) Y. Nishizawa, T. Inui, R. Namioka, T. Uchihashi, T. Watanabe and D. Suzuki, Langmuir, 2022, 38, 16084–16093 CrossRef CAS
; (c) Y. Nishizawa, H. Yokoi, T. Uchihashi and D. Suzuki, Soft Matter, 2023, 19, 5068–5075 RSC
.
-
(a) D. Suzuki and R. Yoshida, Polym. J., 2010, 42, 501–508 CrossRef
; (b) D. Suzuki and K. Horigome, Langmuir, 2011, 27, 12368–12374 CrossRef
; (c) D. Suzuki and K. Horigome, J. Phys. Chem. B, 2013, 117, 9073–9082 CrossRef PubMed
; (d) T. Kawamoto, H. Minato and D. Suzuki, Soft Matter, 2024, 20, 5836–5847 RSC
.
-
(a) Z. Li, M.-H. Kwok and T. Ngai, Macromol. Rapid Commun., 2012, 33, 419–425 CrossRef PubMed
; (b) M.-H. Kwok, Z. Li and T. Ngai, Langmuir, 2013, 29, 9581–9591 CrossRef PubMed
.
-
(a) T. Hoare and R. Pelton, Macromolecules, 2004, 37, 2544–2550 CrossRef
; (b) D. Suzuki and H. Kawaguchi, Langmuir, 2005, 21, 8175–8179 CrossRef CAS PubMed
; (c) S. Nayak, D. Gan, M. J. Serpe and L. A. Lyon, Small, 2005, 1, 416–421 CrossRef CAS PubMed
; (d) D. Suzuki, H. Taniguchi and R. Yoshida, J. Am. Chem. Soc., 2009, 131, 12058–12059 CrossRef CAS
; (e) M. Keerl, J. S. Pedersen and W. Richtering, J. Am. Chem. Soc., 2009, 131, 3093–3097 CrossRef CAS PubMed
; (f) A. C. Brown, S. E. Stabenfeldt, B. Ahn, R. T. Hannan, K. S. Dhada, E. S. Herman, V. Stefanelli, N. Guzzetta, A. Alexeev, W. A. Lam, L. A. Lyon and T. H. Barker, Nat. Mater., 2014, 13, 1108–1114 CrossRef CAS
; (g) T. Kureha and D. Suzuki, Langmuir, 2018, 34, 837–846 CrossRef CAS
; (h) T. Watanabe, Y. Nishizawa, H. Minato, C. Song, K. Murata and D. Suzuki, Angew. Chem., Int. Ed., 2020, 59, 8849–8853 CrossRef CAS PubMed
; (i) F. Grabowski, V. S. Petrovskii, F. Fink, D. E. Demco, S. Herres-Pawlis, I. I. Potemkin and A. Pich, Adv. Sci., 2022, 9, 2204853 CrossRef CAS PubMed
.
-
(a) G. Nakada, Y. Igarashi, H. Imai and Y. Oaki, Adv. Theory Simul., 2019, 2, 1800180 CrossRef CAS
; (b) H. Numazawa, Y. Igarashi, K. Sato, H. Imai and Y. Oaki, Adv. Theory Simul., 2019, 2, 1900130 Search PubMed
; (c) K. Noda, Y. Igarashi, H. Imai and Y. Oaki, Adv. Theory Simul., 2020, 3, 2000084 CrossRef CAS
; (d) K. Noda, Y. Igarashi, H. Imai and Y. Oaki, Chem. Commun., 2021, 57, 5921–5924 RSC
; (e) Y. Haraguchi, Y. Igarashi, H. Imai and Y. Oaki, Adv. Theory Simul., 2021, 4, 2100158 CrossRef
; (f) K. Sakano, Y. Igarashi, H. Imai, S. Miyakawa, T. Saito, Y. Takayanagi, K. Nishiyama and Y. Oaki, ACS Appl. Energy Mater., 2022, 5, 2074–2082 Search PubMed
; (g) T. Komura, K. Sakano, Y. Igarashi, H. Numazawa, H. Imai and Y. Oaki, ACS Appl. Energy Mater., 2022, 5, 8990–8998 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc04386c |
| This journal is © The Royal Society of Chemistry 2024 |