Open Access Article
Open Access ArticleFast detection of penicillium rot and the conservation status of packaged citrus fruit using an optical array sensor†
Alessia
Cavallaro
 a,
Rossella
Santonocito
a,
Rossella
Santonocito
 a,
Roberta
Puglisi
a,
Andrea
Pappalardo
a,
Roberta
Puglisi
a,
Andrea
Pappalardo
 ab,
Federico
La Spada
c,
Rossana
Parlascino
c,
Mario
Riolo
c,
Santa Olga
Cacciola
c,
Nunzio
Tuccitto
ab,
Federico
La Spada
c,
Rossana
Parlascino
c,
Mario
Riolo
c,
Santa Olga
Cacciola
c,
Nunzio
Tuccitto
 *a and
Giuseppe
Trusso Sfrazzetto
*a and
Giuseppe
Trusso Sfrazzetto
 *ab
*ab
aDepartment of Chemical Sciences, University of Catania, Viale A. Doria 6, 95123 Catania, Italy. E-mail: giuseppe.trusso@unict.it
bINSTM Udr of Catania, Viale Andrea Doria 6, 95125, Catania, Italy
cDepartment of Chemical Sciences, University of Catania, Viale A. Doria 6, 95123 Catania, Italy
First published on 30th October 2024
Abstract
A novel optical array sensor designed to detect the conservation status of citrus fruit as well as contamination of ripened fruits by green mold incited by the fungus Penicillium digitatum is reported here. The device demonstrates high sensitivity, specificity, and cost-effectiveness, making it suitable for integration into the citrus fruit supply chain, including production and packaging systems.
In the last few years, the production and packaging of agricultural products for human consumption, such as fruits and vegetables, have become more stringent as regards quality and security. In this respect, the global market demands assurance of the product quality inside the packaging, particularly its freshness, natural scents, and long shelf-life. A major constraint to the successful trade of citrus fruit is the occurrence of severe postharvest diseases affecting fruits throughout the supply chain.1 In this respect, the citrus green mold caused by Penicillium digitatum is a wound-mediated disease considered the main cause of post-harvest citrus fruit spoilage resulting in 90% of the total post-harvest losses.2–5 Recent studies focussed on volatile organic compounds (VOCs) and other secondary metabolites produced in healthy and P. digitatum-infected citrus fruits.6,7 Specifically, VOCs emitted by healthy citrus fruit are responsible for their characteristic fresh and zesty smell, including limonene, myrcene, α-pinene and β-pinene, sabinene, γ-terpinene, linalool, neral, and geranial.6 These VOCs are synthesized and emitted by the fruit as part of its natural metabolic processes. The profile of VOCs changes significantly when citrus fruits are infected by P. digitatum. The infection leads to the production of specific VOCs that are different from those in healthy fruits, including limonene and γ-terpinene, which are in general correlated with the presence of wound of fruit peel, ethanol, which increases during infection due to fermentation processes, and ethyl acetate, 2-methyl-1-propanol, 1-butanol, 2 and 3-methyl-1-butanol, hexanal, octanal and nonanal.6 Additionally, during the infective process, specific VOCs related to the fungal metabolism are emitted: (E)-2-hexenal, a volatile compound associated with green and leafy aromas, often indicative of tissue damage and stress response; 1-methoxy-3-methylbenzene, which has a strong, medicinal odor; diethyl carbonate provides a sweet and fruity scent, while ethyl 2-phenylacetate contributes a floral and honey-like aroma; and propyl octanoate and ethyl decanoate, both esters, add fruity and sweet notes.6 Therefore, changes in the VOC profile can be recorded and can be used as a biomarker to differentiate between healthy and infected fruits.
Traditional analytical techniques used to detect VOCs are gas-chromatography based techniques and electrochemical analyses, which show extremely high selectivity and sensitivity. However, they suffer from some practical limitations, such as high costs, complex sampling pre-treatments, and the requirement of specialized operators. Likewise, the food industries require faster and easier detection methods that are cost-effective and capable of being integrated seamlessly into existing production and packaging lines without requiring extensive modifications.
Remarkably, a recent work reported by Zhao et al.8 presented a chemiresistive gas sensor for the detection of VOCs based on bimetallic nanosphere-functionalized electrospun nanofibers. They achieved a selective and sensitive detection (at the ppb level), which is of huge relevance both for environmental and diagnostic aims, in the gas sensing field. However, the need for user-friendly and portable devices that provide real-time monitoring capabilities is critical to ensure timely intervention and minimize waste. In this context, smart methodologies can be used to monitor the presence of VOCs associated with the state of ripening. In particular, biomolecules, such as nucleotides,9–11 proteins and oligopeptides, can be used as “olfactory receptors”.12–15
These sensors are extremely specific and sensitive, but show some limitations related to the possibility to detect a limited number of VOCs, low stability and high costs of these biomolecules.16 In fact, the stability of these biomolecules is critical, due to wide variability of environmental conditions, extreme in some cases, to which the packages are exposed. In contrast, synthetic molecular optical sensors are stable under a wide range of conditions, and practical to use, due to the possibility to monitor the change of colour or emission in the presence of an analyte. In addition, low costs, high sensitivity, and a fast response make optical devices more convenient for practical use. Colorimetric sensors are employed to monitor VOCs, with barcodes on a paper17 using chemically sensitive dyes, or aromatic compounds able to interact with π-electrons.18–21
Selectivity is one of the main targets in the sensing field, in particular when complex natural sources must be analysed. This target has been successfully achieved employing array technology, a sensing device containing different probes able to interact with analytes with different affinities.22 This technology is inspired by nature, particularly the human nose and tongue, in which thousand receptors interact with molecules that give a characteristic odour or taste, respectively. In the array, the total response of all receptors elaborated by multivariate statistical analysis leads to a fingerprint of the desired analyte, also in a complex mixture.23–25
Here, a new optical array sensor based on fluorescent organic molecules, able to monitor the storage status of citrus fruit inside a closed packaging, is reported. In addition, this device is able to detect the green mold of the fruit induced by P. digitatum,26 representing the first optical device, and to the best of our knowledge, to monitor simultaneously the conservation status and potential contamination of the citrus packaging content.
The optical array was designed to contain 20 fluorescent probes, able to exploit multiple non-covalent interactions with analytes. The chosen families of probes are BODIPYs, naphthalimides and rhodamines: they have a wide absorption and emission range and can be excited at a wavelength of 365 nm. The synthesised probes, represented in Fig. 1, are: RhB (1), RhBP (2), RhBM (3), OBP (4), MBP (5), PBP (6), OBEP (7), MBEP (8), PBEP (9), phenanthrene (10), RhB-di-Pico (11), RhB-di-EA (12), Naph-di-EA (13), Naph-di-iBu (14), BDPy-di-NH2 (15), BDPy-Pico (16), BDPy-EA (17), BDPy-Ar (18), BDPy-OH (19) and BDPy-di-EA (20). Synthesis and characterization are detailed in the ESI.† Each probe of the array consists of a fluorophore and a recognition site (represented in black and red, respectively, in Fig. 1), designed to establish non-specific and non-covalent interactions, such as the hydrogen bond, CH–π, π–π, ion–dipole and dipole–dipole, with the analytes.
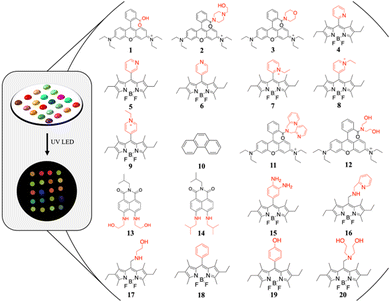 | ||
| Fig. 1 Schematic representation of the sensor array, with recognition sites highlighted in red. The real device under the UV LED is shown on the left. | ||
The emission of the probes was collected by an optical fibre, under excitation using an UV LED (λex 365 nm) and a CCD (a silicon-based multi-channel array detector of the visible range) as a detector.
We selected such an optical fibre-based device due to its versatility for real-time monitoring, by virtue of its advantages of cost-effectiveness, flexibility, chemical inertness, and remote and multiplexed detection capability.
The employed assembled device consists of (i) a light source (λex = 365 nm), (ii) an optical fibre which transmits light to the probes and then collects their emission, (iii) a hyperspectral detection unit (CCD) and (iv) a computer to visualize the fluorescence spectra (ESI,† Fig. S1).
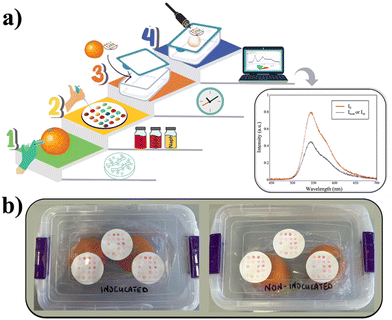
The synthesised fluorescent probes were dropped onto a polyamide circular support (1.5 μL of a 1 mM solution in CHCl3), and the solvent was evaporated in the air. Three supports were attached to the cover's internal part of a plastic transparent box with two oranges inoculated with P. digitatum, and the same was done in another box with two oranges inoculated with sterile distilled water (s.d.w.), as reported in Fig. 2b. Thus, array-based labels were exposed to volatile analytes naturally produced by oranges inside the two boxes: citrus rotting and ripening related VOCs in the first and only ripening in the second.6 Monitoring of the label was performed for four consecutive days every 24 h placing the optical fibre-based analysis probe above each spot. This allowed us to obtain the visible spectrum of each sensitive molecular probe (a detailed description of the device fabrication and measurement protocol is provided in the ESI,† pages S10–S13). Temperature inside the box was maintained at 25 °C, while humidity increased with the development of the fungus in the box with inoculated oranges, making the measurements impossible beyond the fourth day. Fig. 2a shows a schematic representation of whole steps involved in sensing using the fluorescent array, further detailed in the ESI† (Fig. S3).
Five emission spectra of each spot were collected: the first is the emission before exposure to any analyte (I0) and the following four are emissions after 1, 2, 3 and 4 days, respectively, indicated as Inon (“non-inoculated”) and Iin (“inoculated”).
The entire experiment was performed three times, using three different samples of inoculated and non-inoculated oranges, thus obtaining data mediated from nine independent measurements (Tables S1 and S2, ESI†).
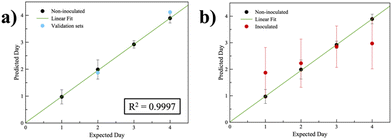
Partial least squares regression (PLS) was applied to obtain a multivariate response of the array for the quantification of analytes produced from healthy oranges (meaning not inoculated). Due to the different VOC composition variations over time, each probe detects these variations leading to a characteristic fingerprint for each day. The plot in Fig. 3a shows the correlation between the actually passed days (expected day) and the predicted day by the model, where each point is represented as a mean of nine values and each error bar is the standard deviation calculated on the replicates. A linear fit was achieved with R2 = 0.9997, representing a good linearity between the expected and predicted day, making us understand that the sensor array is able to identify and quantify ripening of oranges.
The PLS model was applied to predict, as a test set, the ripening day by means of the fluorescence data acquired from the spots of the array placed in the healthy orange box used as the false unknown validation dataset. Data are reported in blue dots in Fig. 3a. Indisputably, it is also possible to determine ripeness in the case of samples obtained from independent measurement with respect to those with which the PLS algorithm was trained. We applied the PLS model to a dataset acquired from the label placed inside boxes containing “inoculated” oranges. Calculated ripening days are reported in red dots in Fig. 3b. Otherwise, if the fruit is contaminated, the emitted VOCs make the model unusable. The results are extremely scattered within each day of exposure.
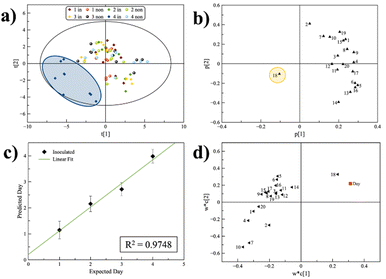
Note the large standard deviation obtained from 9 replicates. We do not detect a significant difference among days. This result shows a difference in the behaviour of the fruit in the case of incipient rot and suggests, indirectly, that the molecular sensor array is somehow able to detect if the fruit is unhealthy. To this end, a principal component analysis (PCA) was performed. Fig. 4a reports the scores plot of the first two PCs. We found that the “inoculated” sample at the fourth day is different at a statistically relevant level both compared to the other days and to the “non-inoculated” sample. This confirms that the array is sensitive to rot even though the PCA method is unsupervised, and the model is not forced to discriminate between samples on individual days of analysis. Loadings plot, reported in Fig. 4b, revealed that the variable that best statistically represents the indicated cluster is the fluorescence intensity variation related to the probe named 18, corresponding to BDPy-Ar, which is a Bodipy with an aryl as the recognition site. As previously described, several VOCs contain unsaturated carbon atoms that can interact with the aryl moiety of probe 18 by the π–π interaction, leading to a change of the HOMO–LUMO gap and a variation of the emission in the solid state.
We applied the PLS, as a supervised approach, to see if the degree of disease progression could also be assessed in the case of rot. PLS regression of “inoculated” samples shows a good linear correlation (R2 = 0.9748, Fig. 4c) between the expected and predicted days, thus making possible the distinction of samples presenting the fungus. Fig. 4d reports the loadings plot of the first two PCs of PLS. The plot shows that, again, variable 18 is strongly correlated with variable Y of the PLS shown in the red labelled ‘day’ in the plot. Although this is valid for the first two PCs, a good linear regression depends on many more variables. The model uses 7 PCs. In fact, we tried to test whether the univariate intensity of the single probe 18 was sufficient to quantify the temporal evolution of the rot with poor results.
To this end, the VIP (variable importance of projection) histogram from the PLS of the “inoculated” sample was obtained (ESI,† Fig. S6), whose values reflect the importance of each term in the model. Terms in the model with a VIP > 1 are important.
Particularly, probes 10, 7, 4, 1, 15, 20, 18 and 9 have a VIP value > 1, and they are united by the presence of aromatic substituents in the recognition site (except for 20). This result emphasizes the contribution of more than one probe to the quantification of rot metabolites in citrus. In detail, it seems that the selected probes are capable of recognizing double bonds in rotting metabolites, exploiting π–π and cation-π interactions assisted by hydrogen bonding. Indeed, recently Rovetto et al. reported the presence of mycotoxins, such as patulin, during the advanced stages of fruit spoilage.7 This study further confirms that the fluorescence-based sensor array can effectively monitor the progression of disease in citrus fruits. The correlation between expected and predicted days of ripening and spoilage described in this study agrees with the temporal evolution of metabolite production reported by Rovetto et al.
The development of the optical array sensor based on fluorescent organic molecules represents a significant advancement in the real-time monitoring of the citrus fruit conservation status and the detection of green mold caused by P. digitatum. This sensor effectively differentiates between healthy and infected fruits by analyzing VOCs, thus ensuring the quality and safety of citrus during storage and transportation. Importantly, this non-destructive method can be used for monitoring the health status of the fruit without the need to open or visually inspect the packaging. The high sensitivity, specificity, and cost-effectiveness of this device make it a practical solution for integration into existing production and packaging lines, addressing the critical need for user-friendly and rapid detection methods in the food industry. Interestingly, the application of this array could be extended to the rapid selection of fruits based on their ripeness degree, provided that appropriate standards are defined experimentally. It has an advantage over other methods (see ESI,† Table S4) used at present to determine the quality standards, because these methods are destructive, and the results of colorimetric methods are relative, as the color of citrus fruit peel is influenced by the temperature. The successful application of the technology developed in this study highlights its potential for broader use in monitoring various agricultural products, ultimately contributing to reduced waste and improved food safety.
This work has been partially funded by European Union (NextGeneration EU), through the MUR-PNRR project SAMOTHRACE (ECS00000022). N. T. and S. O. C. acknowledge the following project for partial funding: BiOrangePack (smart and innovative packaging, post-harvest rot management and shipping of organic citrus fruit funded under the PRIMA Section 2 Multitopics 2019 Call).
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- Y. Chavan, K. Paul and N. Kolekar, Food Production, Diversity, and Safety Under Climate Change, in Advances in Science, Technology & Innovation, ed. R. Chakraborty, P. Mathur and S. Roy, Springer, Switzerland, 2024, pp. 319–331 Search PubMed.
- M. Ismail and J. Zhang, Outlooks Pest Manage., 2004, 15, 29–35 CrossRef.
- J. H. Costa, J. M. Bazioli, J. G. de Moraes Pontes and T. P. Fill, Fungal Biol., 2019, 123, 584–593 CrossRef CAS PubMed.
- D. Macarisin, L. Cohen, A. Eick, G. Rafael, E. Belausov, M. Wisniewski and S. Droby, Phytopathology, 2007, 97, 1491–1500 CrossRef CAS PubMed.
- M. F. Perez, J. Perez Ibarreche, A. S. Isas, M. Sepulveda, J. Ramallo and J. R. Diba, Biol. Control, 2017, 115, 135–140 CrossRef.
- J. Wu, J. Cao, J. Chen, L. Huang, Y. Wang, C. Sun and C. Sun, Food Chem., 2023, 412, 135524 CrossRef CAS PubMed.
- E. I. Rovetto, C. Luz, F. La Spada, G. Meca, M. Riolo and S. O. Cacciola, Toxins, 2023, 15, 407 CrossRef CAS PubMed.
- H. Zhao, J. Li, X. She, Y. Chen, M. Wang, Y. Wang, A. Du, C. Tang, C. Zou and Y. Zhou, ACS Sens., 2024, 9, 2183–2193 CrossRef CAS PubMed.
- Z. Kai, Q. Chang, C. Xing, B. Zhang and J. Liu, Mater. Sci. Eng., C, 2009, 29, 1191–1195 CrossRef.
- S. M. Khamis, R. A. Jones, A. Johnson, G. Preti, J. Kwak and A. Gelperin, AIP Adv., 2012, 2, 171103–171135 Search PubMed.
- S. Gaggiotti, S. Palmieri, F. Della Pelle, M. Sergi, A. Cichelli, M. Mascini and D. Compagnone, Sens. Actuators, B, 2020, 308, 127697 CrossRef CAS.
- R. Khadka, N. Aydemir, C. Carraher, C. Hamiaux, J. Cheema, J. Malmström, A. Kralice and J. Travas-Sejdic, Biosens. Bioelectron., 2019, 126, 207–213 CrossRef CAS PubMed.
- M. Nardiello, C. Scieuzo, R. Salvia, D. Farina, A. Franco, J. A. Cammack, J. K. Tomberlin, P. Falabella and K. C. Persaud, Nanotechnology, 2022, 33, 205501 CrossRef PubMed.
- K. C. Park, S. A. Ochieng, J. Zhu and T. C. Baker, Chem. Senses, 2002, 27, 343–352 CrossRef PubMed.
- S. Sankaran, S. Panigrahi and S. Mallik, Biosens. Bioelectron., 2011, 26, 3103–3109 CrossRef CAS.
- X. Gong, J. Huang, Y. Xu, Z. Li, L. Li, D. Li, T. Belwal, P. Jeandet, Z. Luo and Y. Xu, Trends Food Sci., 2023, 131, 61–76 CrossRef CAS.
- E. G. Bakhoum, M. H. M. Cheng and R. A. Kyle, IEEE Trans. Instrum. Meas., 2016, 65, 1707–1715 CAS.
- R. P. Reji, G. Marappan, Y. Sivalingam and V. J. Surya, Mater. Lett., 2022, 306, 130945 CrossRef CAS.
- H. Lin, J.-J. Lin, Z.-X. Man, H.-J. Jin, F. Y. H. Kutsanedzie and Q.-S. Chen, Food Anal. Methods, 2020, 13, 2192–2203 CrossRef.
- N. Tuccitto, G. Catania, A. Pappalardo and G. Trusso Sfrazzetto, Chem. – Eur. J., 2021, 27, 13715–13718 CrossRef CAS PubMed.
- R. Santonocito, A. Cavallaro, R. Puglisi, M. Intravaia, R. M. Toscano, A. Pappalardo and G. Trusso Sfrazzetto, Curr. Org. Chem., 2023, 27, 876–882 CrossRef CAS.
- Z. Li and K. S. Suslick, Acc. Chem. Res., 2021, 54, 950–960 CrossRef CAS PubMed.
- R. Santonocito, R. Parlascino, A. Cavallaro, R. Puglisi, A. Pappalardo, F. Aloi, A. Licciardello, N. Tuccitto, S. O. Cacciola and G. Trusso Sfrazzetto, Sens. Actuators, B, 2023, 393, 134305 CrossRef CAS.
- R. Santonocito, A. Cavallaro, R. Puglisi, A. Pappalardo, N. Tuccitto, M. Petroselli and G. Trusso Sfrazzetto, Chem. – Eur. J., 2024, 30, e202401201 CrossRef CAS PubMed.
- E. Butera, A. Zammataro, A. Pappalardo and G. Trusso Sfrazzetto, ChemPlusChem, 2021, 86, 681–695 CrossRef PubMed.
- E. I. Rovetto, F. La Spada, F. Aloi, M. Riolo, A. Pane, M. Garbelotto and S. O. Cacciola, J. Plant Pathol., 2024, 106, 411–437 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Method of detection, syntheses of probes, preparation of fungal inoculum, fruit inoculation and packaging, and sensing by the fluorescent array. See DOI: https://doi.org/10.1039/d4cc04700a |
| This journal is © The Royal Society of Chemistry 2024 |