Open Access Article
Open Access ArticleEffects of steric hindrance from single-stranded overhangs on target-strand loading into the Cas12a active site†
Heyjin
Son‡
a,
Youngjae
Kang‡
bc,
Yo Han
Song
d,
Jaeil
Park
e and
Sanghwa
Lee
 *bc
*bc
aGenome Editing Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon 34141, Republic of Korea
bDepartment of Medical Life Sciences, College of Medicine, The Catholic University of Korea, Seoul 06591, Republic of Korea. E-mail: sanghwalee@catholic.ac.kr
cDepartment of Medical Sciences, Graduate School of The Catholic University of Korea, Seoul 06591, Republic of Korea
dDepartment of Chemistry, Gwangju Institute of Science and Technology (GIST), Gwangju 61005, Republic of Korea
eAdvanced Photonics Research Institute, Gwangju Institute of Science and Technology, Gwangju 61005, Republic of Korea
First published on 17th October 2024
Abstract
CRISPR-Cas12a, an RNA-guided DNA endonuclease, induces double-strand breaks by cleaving the non-target strand (NTS) first, followed by the target strand (TS). Using single-molecule FRET with alternating-laser excitation, we found that steric hindrance from the 3′ overhangs of both the cleaved NTS and crRNA impedes TS loading into the catalytic core. Our study highlights the direct involvement of both 3′ NTS and crRNA overhangs in TS cleavage, offering insights into regulatory strategies for Cas12a cleavage reactions.
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) protein system, originally identified as a bacterial immune defence mechanism, has opened up a new horizon in genome engineering.1–3 Cas12a, a member of the class 2 type V CRISPR system, has gained significant attention due to its unique properties, positioning it as an alternative to Cas9. Cas12a differs markedly from Cas9 in several ways: it has the ability to self-process pre-crRNA, targets T-rich protospacer adjacent motifs (PAMs), generates sticky ends upon cleavage, and exhibits indiscriminate single-stranded DNA nuclease activity upon subsequent target DNA cleavage.4–6 The most notable distinction, however, is Cas12a's ability to generate double-stranded breaks using a single catalytic site. Unlike Cas9, which employs separate nuclease domains for the non-target strand (NTS) and the target strand (TS), Cas12a sequentially cleaves the NTS and then the TS using its single RuvC catalytic site. This sequential cleavage mechanism has been elucidated using single-molecule FRET (smFRET) and extensive biochemical assays.7–10
Despite its promising potential, the CRISPR-Cas12a system faces challenges that limit its broader application. One significant issue is the undesired consequences caused by double-stranded DNA breaks, which necessitates the development of a Cas12a nickase that can precisely cleave only one strand.11 The development of a Cas12a nickase would facilitate Cas12a-based prime editing, a cutting-edge genome editing technique that enables more precise editing with fewer side effects.12–14 In this respect, a detailed understanding of the sequential DNA cleavage by Cas12a is essential. However, the current understanding of the DNA cleavage mechanism mediated by CRISPR-Cas12a is insufficient to effectively control the sequential cleavage process. Particularly, the molecular details of TS cleavage remain unclear due to the dynamic nature of the intermediate states.
In this study, we utilized smFRET with alternating-laser excitation (ALEX) to elucidate the molecular intricacies governing the steric inhibition by 3′ overhangs of cleaved NTS and crRNA in TS cleavage by Acidaminococcus sp. Cas12a (AsCas12a). Using the smFRET-ALEX assay, we systematically investigated the individual reaction steps of the Cas12a ternary complex in real time and assessed changes in the kinetics of TS cleavage by varying the length and sequence of either the cleaved NTS or crRNA overhangs. Specifically, we observed that pre-removal of 3′ NTS and crRNA overhangs facilitated the loading of TS into the catalytic site, resulting in an acceleration of TS cleavage. These findings provide a deeper understanding of the mechanism underlying Cas12a-mediated double-stranded DNA cleavage using a single catalytic site, paving the way for precise and efficient genome editing applications.
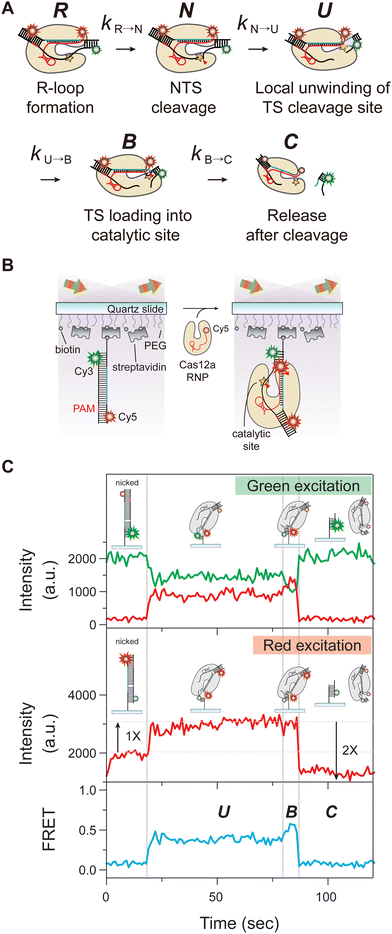
We previously reported that after NTS cleavage, the TS cleavage reaction proceeds via two consecutive steps: local unwinding of the PAM-distal region containing the putative cleavage site, followed by loading of the exposed TS into the catalytic core (Fig. 1A).15 To further investigate the involvement of the cleaved NTS and crRNA overhangs in TS cleavage using the smFRET-ALEX, we prepared the Cas12a ternary complex mimicking post-NTS cleavage states and labelled with one Cy3 and two Cy5 fluorophores as illustrated in Fig. 1B. This assay allows us to directly monitor the conformational dynamics of the Cas12a ternary complex during the DNA cleavage reaction and the simultaneous release of Cy5-labelled Cas12a ribonucleoprotein (RNP) and cleaved DNA fragment after double-stranded DNA cleavage. In this experiment, we successfully observed the conformational dynamics and the binding/release of the Cas12a complex via changes in Cy3-Cy5 FRET and Cy5 stoichiometry, respectively (Fig. 1C). Under Cy3 excitation with a green laser, the two consecutive steps, the local unwinding of PAM-distal region containing the putative TS cleavage site (U state) and the subsequent binding of the exposed TS at the catalytic site (B state), were clearly identified as changes in FRET efficiency. Meanwhile, we observed a single Cy5 signal on the immobilized DNA substrates under Cy5 excitation with a red laser. However, upon binding of the Cas12a with Cy5-labeled crRNA, the Cy5 intensity was doubled because an additional Cy5 fluorophore of crRNA was added to the Cas12a complex. Finally, the double Cy5 signal on the Cas12a ternary complex disappeared under Cy5 excitation, indicating that the Cy5 double-labelled complex with Cas12a RNP and cleaved DNA fragment was released simultaneously after the DNA cleavage.
Structural and single-molecule studies on Cas12a revealed that a crRNA-TS heteroduplex of up to 20 base pairs is initially formed and further exposure of the putative cleavage site of the TS is required for TS loading into the catalytic site of Cas12a.15–18 Based on this finding, it is speculated that base pairing between the 3-nucleotide (nt) 3′-end of the crRNA and its complementary region of the TS may impede the cleavage-competent TS loading, thereby reducing TS cleavage activity. To test this hypothesis, we performed smFRET-ALEX assays using crRNA variants of different length and sequence: 18 nt, 20 nt, 23 nt, and 23 nt with 5 mismatches at the 3′-end of the crRNA. In all variants, the minimal crRNA-TS heteroduplex required for TS cleavage was successfully formed. On the other hand, the rate of transition from U to B states, indicating TS loading into the catalytic site, increased as the number of 3′-end nucleotides of crRNAs base-paired to the putative TS cleavage site in the PAM-distal region decreased (Fig. 2B). Consistent with this, a previous study using single-molecule magnetic tweezers and a gel-based assay for Lachnospiraceae bacterium Cas12a (LbCas12a)-mediated target strand cleavage showed that TS cleavage was significantly increased when using 20 nt crRNA and 24 nt crRNA with 4 mismatches at the 3′-end.17 However, in contrast to the result for the mismatched crRNA, our assay interestingly revealed that the U to B transition rate for the 23 nt crRNA with 5 mismatches at the 3′-end was similar to that of the fully matched 23 nt crRNA. This discrepancy may be due to different experimental approaches using a different Cas12a orthologue, but should be re-examined in an independent experiment. Meanwhile, the rate of transition from B to C states showed no significant correlation with the length and sequence of the 3′ crRNA overhang, indicating that TS catalysis is largely unaffected by the crRNA overhang once the TS is already loaded into the catalytic site (Fig. 2C). Taken together, our findings unexpectedly suggest that base pairing of the putative TS cleavage site is not the origin of the inhibition of TS loading and the resulting reduction in TS cleavage. In conclusion, we suggest that the 3′ crRNA overhang flanking the crRNA-TS heteroduplex sterically inhibits TS loading into the catalytic site and thus reduces TS cleavage efficiency.
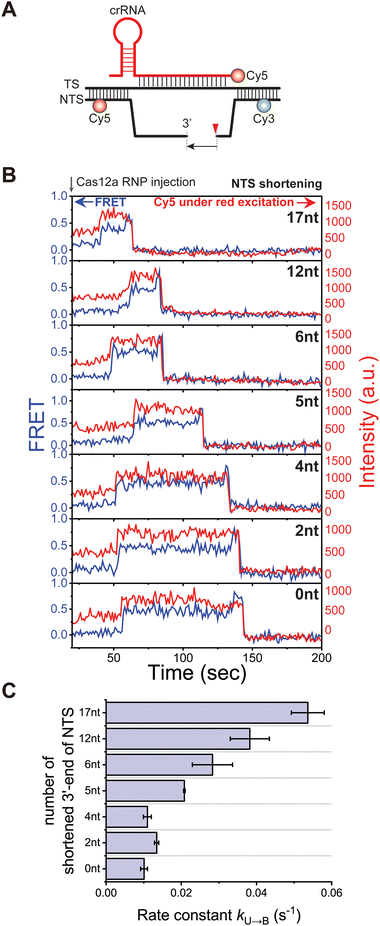
Based on this result, we next asked whether the cleaved NTS overhang also acts as the other inhibitory strand for TS loading into the catalytic site. To address this question, we observed the conformational dynamics during the TS cleavage reaction using target DNA substrates with the shortened 3′-end of the cleaved NTS (Fig. 3A). Representative time traces of FRET (blue) under Cy3 excitation and Cy5 intensity (red) under Cy5 excitation are shown in Fig. 3B, clearly identifying the individual steps of the TS cleavage reaction, the U to B and the B to C transitions. Kinetic analysis of the individual reaction steps at varying the number of truncated nucleotides at the 3′-end of the cleaved NTS revealed that the rate of U to B transition changed dramatically when five or more nucleotides were truncated (Fig. 3C). As the number of truncated nucleotides increased, the rate of U to B transition gradually increased. These results suggest that the 3′ overhang of cleaved NTS also sterically inhibits the loading of TS into the catalytic site, thereby reducing the efficiency of TS cleavage. In particular, it is noteworthy that removal of up to 4 nucleotides from the 3′-end of the cleaved NTS barely changed the rate of TS loading. This result can be understood in the same context with the fact that the NTS is further shortened by 4 nucleotides due to the trimming activity of Cas12a, allowing the TS to load into the catalytic site for catalysis.19
Our study elucidates the molecular mechanism underlying the sequential DNA cleavage activity of CRISPR-Cas12a, focusing on the steric hindrance imposed by the 3′ overhangs of both the cleaved NTS and crRNA during TS cleavage. Using single-molecule FRET with alternating laser excitation (smFRET-ALEX), we found that these overhangs directly impede TS loading into the catalytic site, thereby reducing TS cleavage efficiency. These findings provide important insights into the CRISPR-Cas12a mechanism and pave the way for the development of improved Cas12a-based tools, such as more precise genome editing and more sensitive molecular diagnostics.
This work was supported by the National Research Foundation of Korea (NRF) grants (RS-2023-00218318, and RS-2024-00349323) and Genome editing research program (RS-2023-00263285) funded from the Korea government (MSIT). This research was also supported in part by the Basic Medical Science Facilitation Program through the Catholic Medical Center of the Catholic University of Korea funded by the Catholic Education Foundation, in part by the Catholic Medical Center Research Foundation made in the program year of 2023, and in part by the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (KGM5382423).
Data availability
The data supporting this study have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, Science, 2007, 315, 1709–1712 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- J. D. Sander and J. K. Joung, Nat. Biotechnol., 2014, 32, 347–355 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed.
- I. Fonfara, H. Richter, M. Bratovič, A. Le Rhun and E. Charpentier, Nature, 2016, 532, 517–521 CrossRef CAS PubMed.
- B. Paul and G. Montoya, Biomed. J., 2020, 43, 8–17 CrossRef PubMed.
- J. C. Cofsky, D. Karandur, C. J. Huang, I. P. Witte, J. Kuriyan and J. A. Doudna, eLife, 2020, 9, e55143 CrossRef CAS PubMed.
- Y. Jeon, Y. H. Choi, Y. Jang, J. Yu, J. Goo, G. Lee, Y. K. Jeong, S. H. Lee, I.-S. Kim, J.-S. Kim, C. Jeong, S. Lee and S. Bae, Nat. Commun., 2018, 9, 2777 CrossRef PubMed.
- S. Stella, P. Mesa, J. Thomsen, B. Paul, P. Alcón, S. B. Jensen, B. Saligram, M. E. Moses, N. S. Hatzakis and G. Montoya, Cell, 2018, 175, 1856–1871.e1821 CrossRef CAS PubMed.
- D. C. Swarts and M. Jinek, Mol. Cell, 2019, 73, 589–600.e584 CrossRef CAS PubMed.
- M. Kosicki, K. Tomberg and A. Bradley, Nat. Biotechnol., 2018, 36, 765–771 CrossRef CAS PubMed.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby, A. Raguram and D. R. Liu, Nature, 2019, 576, 149–157 CrossRef CAS PubMed.
- P. J. Chen and D. R. Liu, Nat. Rev. Genet., 2023, 24, 161–177 CrossRef CAS PubMed.
- R. Liang, Z. He, K. T. Zhao, H. Zhu, J. Hu, G. Liu, Q. Gao, M. Liu, R. Zhang, J.-L. Qiu and C. Gao, Nat. Biotechnol., 2024 DOI:10.1038/s41587-023-02095-x.
- H. Son, J. Park, I. Hwang, Y. Jung, S. Bae and S. Lee, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2113747118 CrossRef CAS PubMed.
- D. Dong, K. Ren, X. Qiu, J. Zheng, M. Guo, X. Guan, H. Liu, N. Li, B. Zhang, D. Yang, C. Ma, S. Wang, D. Wu, Y. Ma, S. Fan, J. Wang, N. Gao and Z. Huang, Nature, 2016, 532, 522–526 CrossRef CAS PubMed.
- M. M. Naqvi, L. Lee, O. E. T. Montaguth, F. M. Diffin and M. D. Szczelkun, Nat. Chem. Biol., 2022, 18, 1014–1022 CrossRef CAS PubMed.
- D. C. Swarts, J. van der Oost and M. Jinek, Mol. Cell, 2017, 66, 221–233.e224 CrossRef CAS PubMed.
- I. Strohkendl, F. A. Saifuddin, J. R. Rybarski, I. J. Finkelstein and R. Russell, Mol. Cell, 2018, 71, 816–824.e813 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc04716h |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |