Deciphering the spectroscopic and thermodynamic aspects of binding of biologically important antioxidants with the alkali induced state of human serum albumin†
Received
20th September 2024
, Accepted 1st November 2024
First published on 1st November 2024
Abstract
Protein–ligand interactions are crucial for developing and identifying novel therapeutic targets. In this study, we investigate the interaction of the alkali induced state of human serum albumin (pH 11.2) with three hydroxycinnamic acid derivatives (HCDs), ferulic acid (FA), sinapic acid (SA) and trans-o-coumaric acid, which are biologically important antioxidants, and compare the outcomes with the results obtained at physiological pH (7.4). This study aims to explore the interaction of altered protein conformation with small molecules. Spectroscopic characterization methods show that the conformation of HSA and the ionic properties of HCDs are pH-dependent. Fluorescence, FRET and lifetime measurements reveal that the binding of HCDs with HSA is different at both pH 7.4 and 11.2. Despite the moderate binding of HCDs to HSA, circular dichroism and thermal denaturation studies report no conformational changes in HSA in the presence of HCDs. Isothermal titration calorimetry is employed to assess their binding based on structure and energetics using thermodynamic parameters. Standard molar enthalpy change (ΔH0m) and standard molar entropy change (ΔS0m) values vary with the change of pH from 7.4 to 11.2 with the contributions from the exothermicity and hydrophobicity of functional and aromatic groups of HCDs. Ferulic acid (FA) and sinapic acid (SA) binding to HSA is entropically driven, whereas trans-o-coumaric acid (CA) acid binding is enthalpically favourable. Our ITC studies also reveal that the involvement of –OH functional groups present in CA in binding with HSA is greater than that present in FA and SA at pH 11.2. Overall, this experimental study shows the comparable binding strength of HCDs to both the alkali-induced state of HSA and native HSA (pH 7.4). However, the mechanism of their binding is different.
Introduction
The interaction of biomacromolecules with biologically important small molecules is essential for understanding their binding, function and stability.1,2 The progress of fundamental research and drug development also depends on their interactions. Numerous investigations have been done on various molecules, such as drugs, metal ions, bioinorganic complexes, natural dyes, polymers, and ionic liquids, and their interactions with proteins. Mostly, these studies cover therapeutics, disorders, biosensing, and their applications. To improve the biological and pharmacological properties of ligands, the molecular understanding of protein–ligand interactions is necessary. In recent years, hydroxycinnamic acid derivatives (HCDs), which are plant bioactive compounds, have shown to possess antioxidant, anticarcinogenic, antimutagenic, and antimicrobial properties.3 They are phenolic acid compounds and include ferulic acid, sinapic acid, caffeic acid, p-coumaric acid, and chlorogenic acid found in cereals, coffee, tea, wine, fruits, and vegetables. Their molecular structures are depicted in Fig. 1 with cinnamic acid as the major group with –OH and –OCH3 groups positioned at different locations. These compounds have applications mostly in the food sector.4,5 The phenolic groups in HCDs act as antioxidants, contributing to the longer shelf life of dietary proteins. Apart from their applications in the food industry, they have potential therapeutic properties. Several studies have been done to investigate the anticancer properties of ferulic acid (FA) and sinapic acid (SA) bound to serum albumins and other proteins based on their binding affinity, binding site and cell studies.6,7 Human serum albumin (HSA), a major transport protein, is involved in the delivery of many endogenous and exogenous molecules such as hormones, vitamins, and drugs in the body.8 Although it is not a therapeutic protein, its transport abilities make it a suitable model to explore drug–protein interactions and potential pharmaceutical applications.4 Many studies have been conducted on how pH affects the stability and conformation of proteins.9–11 It is reported that the normal (N) form of human serum albumin (HSA) exists in the pH range of 4.0 to 8.0.12 The expanded (E) form exists at acidic pH below 2.0 and the aged (A) form of HSA is found in the pH range over 10.0.13 These pH-induced perturbations in protein conformations also allow HSA to interact with the bound ligands with different binding sites. These findings can offer experimental evidence for stronger binding affinities and the role of pH in the protein structure and function. Likewise, pH has a critical role in modulating the properties of hydroxycinnamic acid derivatives (HCDs), and therefore it can also influence the protein–ligand interactions.14,15 Despite their many properties, the applications of HCDs have been restricted by their poor solubility and stability.16 In addition, these HCDs, as a platform for drug delivery, require better binding structural features to develop new therapeutic approaches. Various research groups have reported on how pH affects the stability, solubility, structure and pKa values of HCDs. F. Liu et al. have investigated the impact of polyphenols (quercetagetin, chlorogenic acid and EGCG) on the food protein at pH 7.4 and 9.0.17 They have observed improved function and solubility of this protein by conjugating it with polyphenols under alkaline treatment. Another research group has studied the effects of heat treatment and high pH on various plant phenolic compounds.18 They have reported that ferulic acid and trans-cinnamic acid among them are stable and resist high pH-induced degradation. The existence of the mono-anionic form of ferulic acid at physiological pH has also been reported.19 On the other hand, the effect of pH on another phenolic acid, trans-p-coumaric acid (p-CAH2), has been studied, which demonstrates the presence of double anionic form (p-CA2−) when pH is over 9.0, whereas single anionic form (p-CH−) dominates at pH 6.0 and 7.0.5 B. Smyk et al. have reported three ionic species of sinapic acid (SAH2) spectroscopically over a pH range of 1.9–11.5.20 They have observed that mostly two ionic forms SAH2 and SAH− are in equilibrium at pH 2.2 to 6.6. When the pH is beyond 6.6, SA2− ionic form exists in equilibrium with SH− ionic species. These HCDs have two pKa values, which are around 4.3 and 8.8 as reported in the literature21 and also determine the charge on the HCDs with respect to pH. Fig. S1 (ESI†) displays the ionic structures of these HCDs with the pH variation. The isoelectric point (pI) of HSA lies between 4.6 and 4.9, and therefore overall surface charge on the protein is negative at both physiological and alkaline pH.22 Thus the change in pH alters the function and structure of both HSA and HCDs. The aim of this work is to investigate how alkaline pH can affect the interaction of HCDs with HSA since most of the experimental studies available in the literature have investigated physiological pH. To understand the protein's structure and stability at the molecular level, it is important to provide perturbation to the system in the form of pH, temperature or addition of cosolvents.23,24 Even extreme pH levels are capable of forming partially folded states involved in neurodegenerative disorders.9,11,25 Various biophysical methods and techniques such as NMR, chromatographic methods, isothermal titration calorimetry, X-ray crystallography, electrophoresis, UV-vis and fluorescence methods, and differential scanning calorimetry have been utilized to understand protein–ligand interactions. In this work, spectroscopic and calorimetric techniques have been used to determine the effect of HCDs on HSA at pH 7.4 and 11.2 with a focus on their binding results. For investigating conformational changes of the protein, CD and fluorescence methods have been employed. Time-correlated single photon counting (TCSPC) experiments have been conducted to examine the nature of fluorescence quenching. Moreover, the interactions of HCDs with HSA have been explored using ultrasensitive isothermal calorimetry (ITC). This experimental study highlights the nature and strength of their binding and examines the role of the functionality of HCDs and energetics based on thermodynamic data.
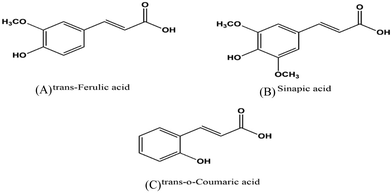 |
| | Fig. 1 Chemical structures of hydroxycinnamic acid derivatives (A), (B) and (C). | |
Materials and methods
Human serum albumin (96–98%) and monobasic potassium phosphate (>99%) were procured from the Sigma-Aldrich Chemical Company, USA. trans-Ferulic acid (trans-4-hydroxy-3-methoxycinnamic acid), sinapic acid (3,5-dimethoxy-4-hydroxycinnamic acid), and trans-o-coumaric acid (trans-2-hydroxycinnamic acid) were purchased from Tokyo Chemical Industry Co., Ltd. These hydroxycinnamic acid derivatives (HCDs) have a purity percentage of >99%. Mass measurements were carried out on a Sartorius BP 211D digital balance with a readability of 0.01 mg. Double-distilled water was used for the preparation of the 20 × 10−3 mol dm−3 phosphate buffer at pH 7.4 and 11.2, respectively. The reported pH of the buffer solutions was maintained by the addition of aqueous HCl or NaOH solution using an Equiptronics EQ614A pH meter at room temperature. Every buffer solution was completely degassed before the preparation of the samples for the experiments. Prior to every experiment, the protein solution at the respective pH was dialyzed overnight with a three-time change of buffer at 4 °C. The degassed buffer solution was used for the preparation of every solution to be used in the experiments. Protein concentration using the molar extinction coefficient, E1%280 = 5.3,26 was determined using a Jasco V-550 double-beam spectrophotometer.
Fluorescence spectroscopy
Fluorescence measurements were conducted on a Cary Eclipse spectrofluorimeter (Varian, USA) using a 1 cm path-length quartz cell. HSA was maintained at 5 × 10−6 M at pH 7.4 and 11.2. The protein was excited at 295 nm in the absence and presence of HCDs by fixing excitation and emission slit widths at 5 nm. Background emissions coming from all the components other than the protein were subtracted to obtain the final protein emission spectra. To check for reproducibility, these fluorescence experiments were performed twice. Proteins in solution upon excitation will have less fluorescence intensity if the additives show UV absorption at both the excitation and emission wavelengths.27 This phenomenon is known as the inner filter effect (IFE). Using the following equation (eqn (1)), the fluorescence intensity of protein with the IFE was rectified.| | | Fcor = Fobs × 10(A295+Aem)/2 | (1) |
where Fcor and Fobs are the corrected and observed fluorescence intensities in experiments. A295 and Aem represent the absorbance at 295 nm and the emission wavelength (λmax) of all components, respectively.
Time-resolved fluorescence spectroscopy
Time-resolved fluorescence measurements were performed using a DeltaFlex Time-Correlated Single Photon Counting (TCSPC) spectrometer, with λex = 295 nm, λem = 347 (pH 7.4) and λem = 343 nm (pH 11.2). The full width at half maximum of the instrument response function is 70 ps, and the resolution is 13 ps per channel. The data were fitted to the tri-exponential function (eqn (2)) after deconvolution of the instrument response function by an iterative re-convolution technique. This was achieved using the EzTime software, using reduced chi-square and weighted residuals as parameters for goodness of the fit.| | 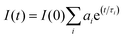 | (2) |
Here, τi and ai are the lifetime and amplitude or preexponential of the ith component, respectively.
Circular dichroism (CD) spectroscopy
The Jasco J1500 spectropolarimeter was used to perform the CD experiments. A 0.2 cm path length cuvette containing 5 × 10−6 M HSA was used to record the far UV-CD (190–260 nm) spectra. The N2 gas in the CD instrument was continuously purged throughout the whole experiment. The reaction time of 4 seconds and a scan rate of 100 nm min−1 were selected, respectively. All CD spectra were baseline corrected. Three accumulations on average were run for each CD spectrum. The thermal denaturation experiments were done using CD spectroscopy to measure the ellipticity in millidegrees at 222 nm in a temperature interval of 25–95 °C. The following equation (eqn (3)) was used to determine the fraction of the denatured protein as a function of temperature:| | 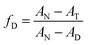 | (3) |
Here, AN and AD are the ellipticities of native and denatured proteins at 222 cm and AT is the ellipticity at temperature T, respectively. The analysis of the plots of fraction denatured (fD) versus temperature (T) was performed by using the EXAM program developed by Kirchoff.28 The analysis provides the values of transition temperature (Tm) and enthalpy of unfolding (ΔuH) accompanying the thermal unfolding.
Isothermal titration calorimetry (ITC)
Thermodynamic studies of the interaction between HSA and hydroxycinnamic acid derivatives (HCDs) were performed by using a Nano ITC purchased from TA Instruments, USA. To avoid the formation of air bubbles during titration, thoroughly degassed sample solutions were used before filling the samples into the ITC cell and syringe. The sample cell and ITC syringe have the volume capacity of 300 and 50 μL, respectively. The syringe was filled with HCD solution and titrated against 0.13 × 10−3 M HSA in the sample cell. The experiments consisted of 24 consecutive injections of 2 μL each and the time interval between consecutive injections is 300 s. The ITC studies were performed twice for the reproducibility at pH 7.4 and 11.2 at 298.15 K. The main ITC profiles were corrected by conducting the dilution ITC experiment of protein and HCD solution and analysed to obtain the thermodynamic parameters using the NanoAnalyze software.
Results and discussion
Spectroscopic characterization of HSA at pH 7.4 and 11.2
The CD data at pH 11.2 reveal a small decrease in secondary and tertiary structural contents of HSA, which suggests that the protein has undergone conformational changes. Furthermore, the CD thermal data recorded at 222 nm provide transition temperature (Tm) values for HSA. The Tm of HSA at pH 7.4 is (69.9 ± 0.3) °C, which reduces to (57.1 ± 0.1) °C at pH 11.2. Spectroscopically, the values of enthalpies of unfolding (ΔuH) for HSA are obtained as (302.9 ± 2.1) and (261.7 ± 1.8) kJ mol−1 at pH 7.4 and 11.2, respectively. The calculated values of Tm and enthalpy of unfolding are in accordance with the literature.26 The reason for the reduced thermostability might be due to the existence of a less stable aged (A) form of HSA beyond the pH of 9.0.29 In fluorescence data, the reduced intensity with a slight blue shift in the emission spectrum indicates that Trp 214 in HSA at pH 11.2 is in a more non-polar environment with respect to HSA at pH 7.4. This indicates internalization of the tryptophan residue around pH 11.2.11 A change in pH can perturb the conformation of the protein. This can result in exposing those regions of the protein to the solvent and forming the partially folded state of the protein. 8-Anilino-1-naphthalenesulfonic acid (ANS) is a hydrophobic dye that can bind to solvent accessible nonpolar parts of the protein.30 It is used for the identification of partially folded states of the protein.31 The HSA–ANS complex displays increased ANS fluorescence intensity upon excitation at 365 nm at pH 7.4. Moreover, HSA at pH 11.2 upon interaction with ANS shows decreased fluorescence intensity with a slight blue shift. The altered conformation and developed net negative charges (pI of HSA, 4.7) reduce the ANS binding sites on the protein at pH 11.2.11 The spectroscopic data indicate that different conformational forms of HSA exist at pH 7.4 and 11.2. The spectroscopic characterization of HSA at pH 7.4 and 11.2 is shown in Fig. 2.
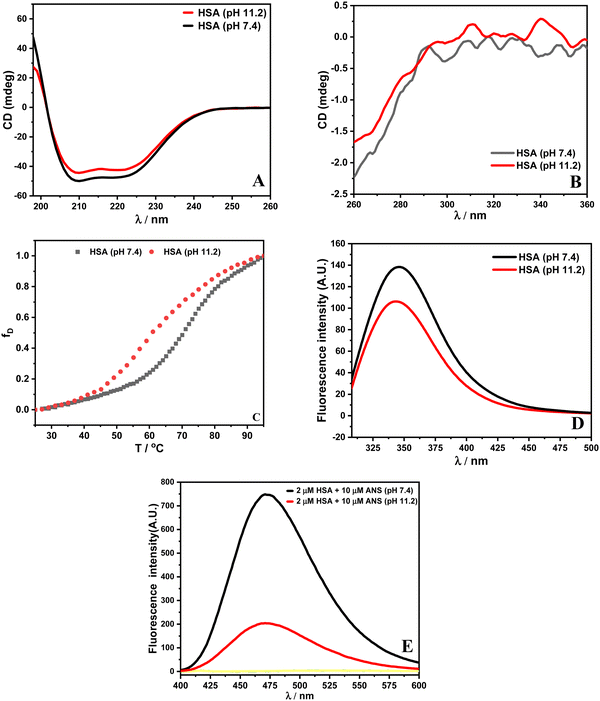 |
| | Fig. 2 Plots of (A) far UV-CD spectra, (B) near UV-CD spectra, (C) thermal denaturation and (D) fluorescence spectrum upon excitation of HSA at 295 nm and (E) ANS spectral studies with λex = 365 nm. | |
Spectroscopic characterization of hydroxycinnamic acid derivatives at pH 7.4 and 11.2
Fig. 3 and Fig. S2 (ESI†) show the UV-vis and fluorescence emission spectra of hydroxycinnamic acid derivatives (HCDs) with increasing molarities at 11.2 and 7.4, respectively. There is a shift observed in the absorbance peaks to the longer wavelength when pH is raised from 7.4 to 11.2. In the case of ferulic acid, the absorption peak is at 225 nm and broad absorption ranges from 250 to 350 nm at pH 7.4, which is shifted to 250–400 nm at pH 11.2. Similarly, absorption from 250 to 350 nm shifts to 300–400 nm with the change in pH from 7.4 to 11.2 for sinapic acid. No shift in the absorption peaks has been observed for trans-coumaric acid. Shifting of the absorption peak to longer wavelength regions can be due to the existence of double anionic forms of HCDs, which are pH-dependent.5,19,20 Different ionic forms of HCDs at different pH values are given in Fig. S1 (ESI†). In the literature, it is reported that the electronic transition from HOMO−1 to the LUMO is associated with the absorption band around 290 nm, and the HOMO to LUMO transition is associated with the band near 325 nm.32 The energy gap between the HOMO and LUMO is reduced at pH 11.2, which causes the absorption maxima of HCDs to shift to longer wavelengths, and this effect is also amplified by the presence of –OH and –OCH3 groups as electron donors. In addition, fluorescence experiments have been performed. The hydroxycinnamic acid derivatives are excited at 295 nm to investigate whether their fluorescence spectra overlap with that of protein HSA. It is observed that the λmax of ferulic acid is 423 and 463 nm at pH 7.4 and 11.2, respectively. For sinapic acid, it shifts from 425 nm with a shoulder peak around 460 nm (pH 7.4) to 480 nm (pH 11.2). No shift in λmax ≈ 500 nm for trans-o-coumaric acid has been observed upon excitation when pH is changed from 7.4 to 11.2. However, the double anionic form of trans-p-coumaric acid, when the pH is beyond 9.0, has been reported in the literature.5 The resulting fluorescence emission of HCDs is due to the conjugation of the styrene double bond (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), methoxy group (–OCH3) and the phenol ring.33 Both the UV and fluorescence studies indicate that the properties of the HCDs also change with changes in pH depending on the functional groups present in HCDs.
CH–), methoxy group (–OCH3) and the phenol ring.33 Both the UV and fluorescence studies indicate that the properties of the HCDs also change with changes in pH depending on the functional groups present in HCDs.
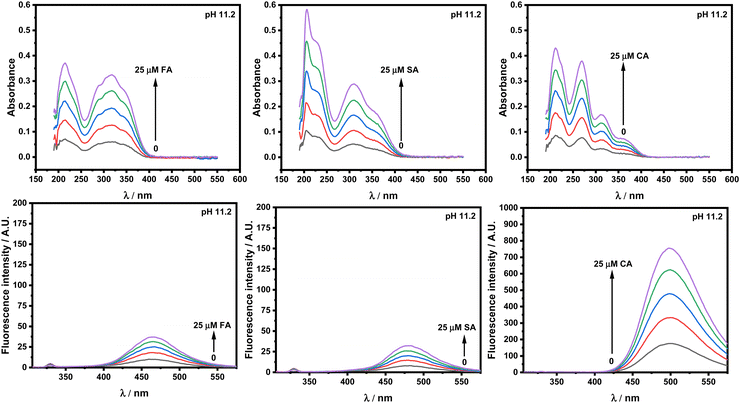 |
| | Fig. 3 Absorbance and fluorescence intensity plots of ferulic acid (FA), sinapic acid (SA) and trans-o-coumaric acid (CA) at pH 11.2 at 298.15 K. | |
Fluorescence spectroscopic studies
Protein excitation at 295 nm provides unique information on the tryptophan (Trp) residue(s) microenvironment. The emission maximum (λmax) of HSA in the fluorescence spectra at pH 7.4 upon excitation has been recorded at 347 nm.26 Any shift in the λmax implies change in the environment of the Trp residue(s). Fig. 4 presents the emission spectra of HSA at pH 7.4 and 11.2 respectively. The addition of FA, SA and CA shows quenching in the fluorescence intensity at both the pH values.
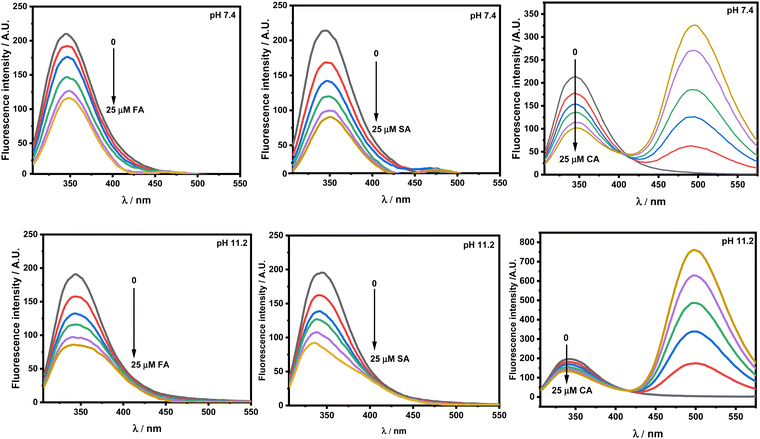 |
| | Fig. 4 Emission spectra of 5 × 10−6 M HSA upon excitation at 295 nm in the presence of ferulic acid (FA), sinapic acid (SA) and trans-o-coumaric acid (CA) at pH 7.4 and 11.2 at 298.15 K. | |
There is red- and blue-shift observed in the emission spectra of HSA with increasing concentration of HCDs at pH 7.4 and 11.2, respectively. Generally, red- and blue-shifts give an idea about Trp residues moving into the polar and non-polar environments, as their emission fluorescence is solvent-sensitive.10,34 These shifts in the presence of HCDs indicate the conformational changes in the protein. The λmax of HSA is slightly red-shifted by HCDs from 347 nm to 353 nm, suggesting the exposure of Trp-214 to a polar environment at pH 7.4. It is blue-shifted towards 330 nm (broad fluorescence peak) at 11.2, which means there is internalization of the Trp 214 residue in HSA by HCDs. Furthermore, we have cross-checked these shifts in λmax by obtaining the fluorescence spectra of tryptophan amino acid in the presence of increasing concentration of HCDs at pH 7.4 and 11.2 (see Fig. S6, ESI†). The red shift in the spectra has been observed at both pH values. This suggests that the shifting of λmax of HSA at pH 11.2 towards a lower wavelength might be due to the emission peaks of HCDs shifting towards the region of the emission peak of HSA. The broadness in the emission peak of HSA at pH 11.2 has been observed with the addition of HCDs at higher concentration. These peaks have been deconvoluted using the Peak Analyser method of Origin 2021. After deconvolution, two fluorescence peaks at λmax ≈ 330 nm for HSA and other at λmax more than 350 nm for HCDs have been observed. The deconvulated peaks in the emission spectra at every concentration of HCDs at pH 11.2 are shown in Fig. 5 and Fig. S3–S5 (ESI†).
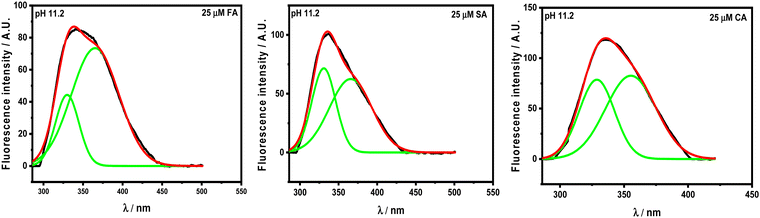 |
| | Fig. 5 Deconvoluted fluorescence peaks in the emission spectra of 5 × 10−6 M HSA in the presence of 25 × 10−6 M HCDs. | |
Significant non-radiative energy transfer has been seen from the donor (Trp in HSA) to the acceptor (CA) at pH 7.4 and 11.2 in Fig. 4. This happens when the absorption spectra of the acceptor and the emission spectrum of the donor coincide. Based on the Förster non-radiative energy transfer hypothesis, the distance between the donor and acceptor can be calculated.27Fig. 6 shows the overlap region of the emission spectra of the protein and the absorbance spectra of CA at pH 7.4 and 11.2.
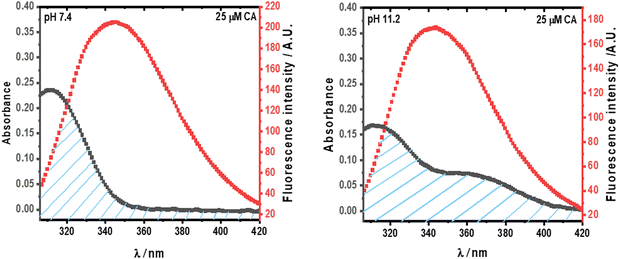 |
| | Fig. 6 The overlap between the emission spectra of HSA (5 × 10−6 M) and the absorption spectra of CA (25 × 10−6 M). | |
The efficiency of this energy transfer from the donor to the acceptor can be determined using the following equation (eqn (4)):
| | 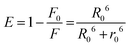 | (4) |
where
F0 and
F represent the fluorescence intensities of HSA in the absence and presence of the acceptor, respectively. The average distance between the acceptor and donor is denoted by
r0. When the energy transfer efficiency is 50%, the critical distance is represented by
R0. The following equation (
eqn (5)) can be used to estimate the value of
R0:
| | | R06 = 8.79 × 10−25·k2·n−4·Φ·J (λ) | (5) |
where
k2 = 2/3 represents the dipole's spatial orientation for random orientation in fluid solution,
Φ denotes the fluorescence quantum yield of the donor, which is 0.118 for HSA in the absence of the acceptor. In the wavelength range where spectral overlap is important,
n represents the average refractive index of the medium; in this example,
n = 1.4. The following equation (
eqn (6)) measures the spectral overlap integral, or
J(
λ), between the absorption spectrum of the acceptor and the emission spectrum of the donor:
| | 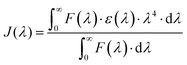 | (6) |
where
F(
λ) represents the normalised fluorescence intensity at a wavelength (
λ) of the donor. The extinction coefficient of the acceptor at the wavelength is denoted by
ε(
λ). As seen in
Fig. 4, the overlapping region of the HSA emission spectrum and the CA absorption spectrum were integrated to determine the value of
J(
λ). The calculated values of
J(
λ) and other parameters related to non-radiative energy transfer are tabulated in
Table 1. Based on the rule 0.5
R0 <
r0 < 1.5
R0, the results have shown that the distance between HSA (donor) and CA (acceptor) at both pH values was less than 8 nm, indicating a high chance of energy transfer from HSA to CA.
Table 1 Parameters related to non-radiative transfer for the HSA–CA complex at pH 7.4 and 11.2 at 298.15 K
|
|
J cm−3 L−1 mol−1 |
E/% |
R
0/nm |
r
0/nm |
| pH 7.4 |
|
|
(1.10 ± 0.02) × 10−15 |
52.09 ± 0.31 |
1.64 ± 0.01 |
1.62 ± 0.01 |
|
|
| pH 11.2 |
|
|
(2.18 ± 0.05) × 10−15 |
33.58 ± 0.12 |
1.84 ± 0.03 |
2.06 ± 0.02 |
The strength of fluorescence quenching and binding of HCDs with HSA at pH 7.4 and 11.2 has been further explored by classic Stern–Volmer, modified Stern–Volmer and double logarithmic plots using eqn (7)–(9). respectively.
| | 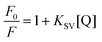 | (7) |
| | 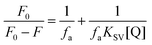 | (8) |
| | 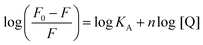 | (9) |
Here,
F0 is the fluorescence intensity of HSA,
F is the fluorescence intensity of HSA in the presence of HCDs,
KSV is the Stern–Volmer quenching constant,
fa is the fraction of the fluorophore that is accessible to the quencher, [Q] is the concentration of quencher,
KA is the association constant and
n is the number of set of binding sites. The values of
KSV indicate the accessibility of the fluorophore to the quencher.
The Stern–Volmer and modified Stern–Volmer plots of 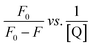 and double logarithmic plots of
and double logarithmic plots of 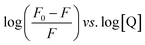 are shown in Fig. 7. For every HCD that interacts with the fluorophore (Trp214), the value of KSV is of the order of 104. The parameters associated with the fluorescence quenching studies for the binding of HCDs with HSA are shown in Table 2 at both pH values. The fluorescence data show a single set of binding sites (n ≈ 1) with the fractional accessibility (fa) of the tryptophan residue, which is almost one at both pH values.26 The magnitude of the association constant (KA) of HCDs with HSA is almost identical at pH 7.4 and 11.2. The interaction studies of bovine serum albumin with hydroxycinnamic acid derivatives at physiological pH have been conducted, showing the values of association constants (KA) in the order as follows: p-CA > FA > SA.4 This indicates that the strength of binding differs from protein to protein based on its conformation and affinity for the ligands and the pH conditions. Here, the change in the values of KA with the pH might be due to the differently exposed interacting amino acid groups in HSA. HCDs binding with the BSA protein at neutral pH have also been characterised by NMR methodology. This indicates the involvement of hydrogen bonding and hydrophobic interactions with the protein in their binding process.4
are shown in Fig. 7. For every HCD that interacts with the fluorophore (Trp214), the value of KSV is of the order of 104. The parameters associated with the fluorescence quenching studies for the binding of HCDs with HSA are shown in Table 2 at both pH values. The fluorescence data show a single set of binding sites (n ≈ 1) with the fractional accessibility (fa) of the tryptophan residue, which is almost one at both pH values.26 The magnitude of the association constant (KA) of HCDs with HSA is almost identical at pH 7.4 and 11.2. The interaction studies of bovine serum albumin with hydroxycinnamic acid derivatives at physiological pH have been conducted, showing the values of association constants (KA) in the order as follows: p-CA > FA > SA.4 This indicates that the strength of binding differs from protein to protein based on its conformation and affinity for the ligands and the pH conditions. Here, the change in the values of KA with the pH might be due to the differently exposed interacting amino acid groups in HSA. HCDs binding with the BSA protein at neutral pH have also been characterised by NMR methodology. This indicates the involvement of hydrogen bonding and hydrophobic interactions with the protein in their binding process.4
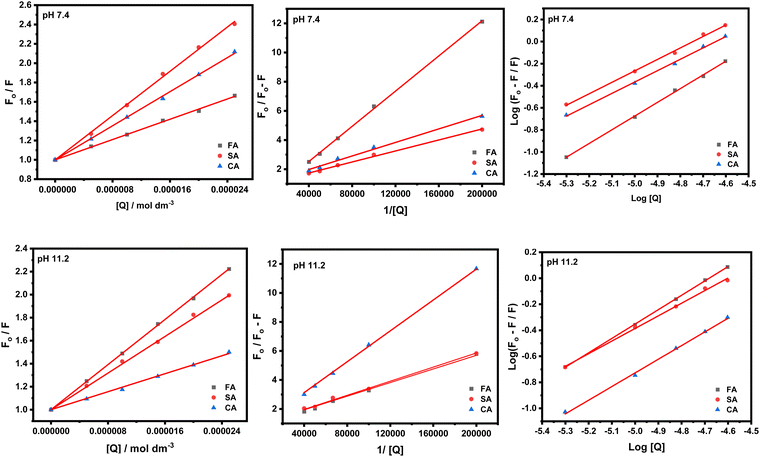 |
| | Fig. 7 Stern–Volmer, modified Stern–Volmer and double logarithmic plots associated with the quenching of 5 × 10−6 M HSA at pH 7.4 and 11.2 in the presence of HCDs at 298.15 K. | |
Table 2 Quenching and binding parameters associated with interaction of HCDs with HSA obtained from Stern–Volmer, modified Stern–Volmer and double logarithmic plots at pH 7.4 and 11.2 at 298.15 K
| HCDs |
Stern–Volmer equation |
Modified Stern–Volmer equation |
Double logarithmic equation |
|
K
SV/M−1 |
K
SV/M−1 |
f
a
|
K
A/M−1 |
n
|
| pH 7.4 |
|
|
|
|
|
| FA |
(2.94 ± 0.12) × 104 |
(3.10 ± 0.11) × 104 |
1.03 ± 0.02 |
(4.42 ± 0.11) ×105 |
1.26 ± 0.08 |
| SA |
(5.10 ± 0.13) × 104 |
(4.98 ± 0.11) × 104 |
1.01 ± 0.02 |
(1.25 ± 0.12) × 105 |
1.07 ± 0.11 |
| CA |
(4.23 ± 0.13) × 104 |
(4.16 ± 0.10) × 104 |
1.00 ± 0.01 |
(7.08 ± 0.17) × 104 |
1.04 ± 0.01 |
|
|
| pH 11.2 |
| FA |
(5.09 ± 0.10) × 104 |
(4.85 ± 0.13) × 104 |
1.02 ± 0.02 |
(2.51 ± 0.10) × 105 |
1.15 ± 0.12 |
| SA |
(4.10 ± 0.12) × 104 |
(4.09 ± 0.12) × 104 |
1.00 ± 0.01 |
(4.07 ± 0.15) × 105 |
0.99 ± 0.10 |
| CA |
(2.05 ± 0.13) × 104 |
(1.98 ± 0.12) × 104 |
1.01 ± 0.02 |
(4.71 ± 0.15) × 104 |
1.08 ± 0.12 |
Lifetime measurement study: nature of quenching
HSA fluorescence decay curves at pH 7.4 and 11.2 have been obtained, which have their excitation wavelengths at 295 nm. These decay curves have been better fitted to the sum of three exponentials with chi square values (χ2) ≃ 1. Both the lifetimes and pre-exponential parameters at pH 7.4 are tabulated in Table 3. The Trp 214 residue of HSA upon excitation forms three substructures.29 Emission from these three substructures gives rise to three lifetime (τ) values. It is reported that two of the three lifetimes result from substructures formed by free tryptophan in solution. The third lifetime belongs to the substructure resulting from tryptophan interaction with the surrounding microenvironment in the protein. These substructure populations are defined by the pre-exponential parameters that are dependent on the fluorophore environment and protein structure. Lifetimes and pre-exponential values are dependent on the pH and thus on the protein form.13,35 Lifetime measurements at different pH values along the emission wavelengths allow us to differentiate the various forms of HSA and the nature of quenching.
Table 3 Lifetime (τ), amplitude (α) and average lifetime (〈τ〉) values and fluorescence decay profiles of 5 × 10−6 M HSA in the presence of HCDs at pH 7.4 at 298.15 K
| Additives/μM |
Lifetime/ns |
Amplitude (%) |
Avg. lifetime/ns |
|
τ
1
|
τ
2
|
τ
3
|
α
1
|
α
2
|
α
3
|
〈τ〉 |
χ
2
|
| Ferulic acid |
| 0 |
2.61 |
6.92 |
0.46 |
38 |
35 |
27 |
5.55 |
1.07 |
| 5 |
1.94 |
6.15 |
0.26 |
34 |
36 |
30 |
4.97 |
1.06 |
| 15 |
1.73 |
5.84 |
0.17 |
29 |
28 |
43 |
4.21 |
1.09 |
| 25 |
1.49 |
5.39 |
0.10 |
29 |
16 |
55 |
3.20 |
1.10 |
|
|
| Sinapic acid |
| 0 |
2.61 |
6.92 |
0.46 |
38 |
35 |
27 |
5.55 |
1.07 |
| 5 |
1.92 |
6.16 |
0.24 |
35 |
38 |
27 |
5.09 |
1.02 |
| 15 |
1.79 |
5.90 |
0.17 |
32 |
33 |
35 |
4.58 |
1.04 |
| 25 |
1.56 |
5.51 |
0.10 |
13 |
11 |
76 |
2.91 |
1.10 |
|
|
|
p-Coumaric acid |
| 0 |
2.61 |
6.92 |
0.46 |
38 |
35 |
27 |
5.55 |
1.07 |
| 5 |
1.76 |
5.25 |
0.27 |
32 |
32 |
36 |
4.33 |
1.02 |
| 15 |
1.55 |
4.90 |
0.17 |
32 |
30 |
38 |
4.01 |
0.97 |
| 25 |
1.21 |
4.12 |
0.13 |
13 |
13 |
74 |
2.82 |
1.04 |
The lifetime values for native HSA observed at pH 7.4 are τ1 = 2.61 (38%), τ2 = 6.92 ns (35%) and τ3 = 0.46 ns (27%) with the average lifetime (〈τ〉) of 5.55 ns. Vos et al. have analysed their HSA lifetime results using three exponentials: 3.7 ns (29%), 7.3 ns (30%), and 0.11 ns (41%), resulting in an average lifetime of 6 ns, utilising a 300 nm excitation.36 Fluorescence lifetimes that have been reported in the literature for HSA (pH 6.8) upon excitation at 295 nm are 7.03 ns (30%), 3.17 ns (38%), and 0.68 ns (32%), with an average lifetime of 5.32 ns.37 The average lifetime value of HSA at pH 11.2 is reduced to 3.79 ns. The lower lifetime values indicate the existence of a less stable conformation of HSA at pH 11.2 than at pH 7.4. The trend of lifetime values decreases with increasing concentration of HCDs at pH 7.4 while being constant at pH 11.2 shown in Fig. 8. This shows that the nature of quenching is dynamic at pH 7.4 and static at pH 11.2. Here, the values of average lifetime of HSA depend upon the pH as well as the excitation wavelength of the system. Lifetimes and pre-exponential parameters at pH 11.2 are given in Table 4. Overall, this suggests that there are collisions between the emitting moiety (HSA) and the quencher (HCDs) at pH 7.4. There is the ground state-complex formation between HSA and HCDs (quencher) at pH 11.2. Different types of quenching suggest the distinct mechanism of their binding, which is due to differences in pH, conformation and surface charges of proteins and HCDs.
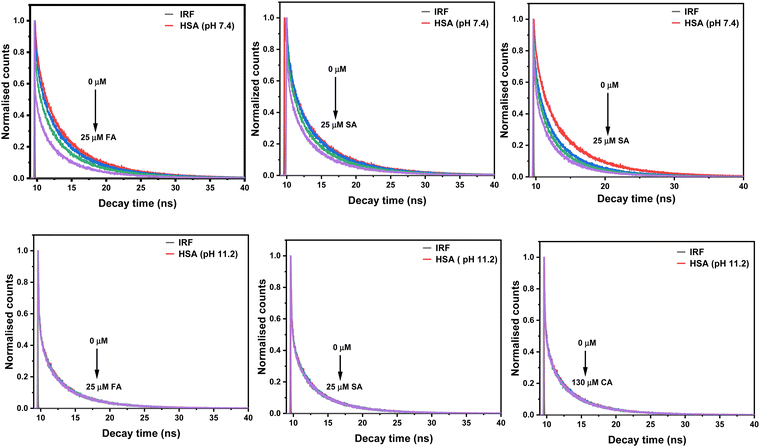 |
| | Fig. 8 Fluorescence decay profiles of 5 × 10−6 M HSA in the presence of different concentrations of HCDs at 298.15 K. | |
Table 4 Lifetimes (τ), amplitudes (α) and average lifetimes (〈τ〉)and fluorescence decay profiles of 8 × 10−6 mol dm−3 HSA in the presence of HCDs at pH 7.4 at 298.15 K
| Additives/μM |
Lifetime/ns |
Amplitude (%) |
Avg. lifetime/ns |
|
τ
1
|
τ
2
|
τ
3
|
α
1
|
α
2
|
α
3
|
〈τ〉 |
χ
2
|
| Ferulic acid |
| 0 |
1.57 |
5.15 |
0.15 |
24 |
26 |
50 |
3.79 |
1.05 |
| 5 |
1.71 |
5.23 |
0.17 |
26 |
26 |
48 |
3.87 |
1.10 |
| 15 |
1.55 |
5.17 |
0.14 |
26 |
26 |
48 |
3.79 |
1.08 |
| 25 |
1.49 |
5.09 |
0.15 |
25 |
28 |
47 |
3.88 |
1.06 |
|
|
| Sinapic acid |
| 0 |
1.57 |
5.15 |
0.15 |
24 |
26 |
50 |
3.79 |
1.05 |
| 5 |
1.56 |
5.21 |
0.14 |
23 |
24 |
53 |
3.68 |
1.13 |
| 15 |
1.55 |
5.15 |
0.15 |
24 |
26 |
50 |
3.79 |
1.05 |
| 25 |
1.46 |
5.08 |
0.13 |
22 |
25 |
53 |
3.66 |
1.07 |
|
|
|
p-Coumaric acid |
| 0 |
1.57 |
5.15 |
0.15 |
24 |
26 |
50 |
3.79 |
1.05 |
| 5 |
1.55 |
5.15 |
0.13 |
24 |
26 |
50 |
3.81 |
1.06 |
| 15 |
1.56 |
5.21 |
0.13 |
23 |
24 |
53 |
3.68 |
1.04 |
| 25 |
1.58 |
5.15 |
0.14 |
24 |
26 |
50 |
3.78 |
1.08 |
Analysis of conformational changes in HSA
UV-vis spectroscopic studies.
The UV-vis spectra of the protein show two absorption peaks, one at 208 nm and the other at 280 nm. The peak located at 208 nm with strong absorption belongs to n–π* transition. This is linked to changes in the conformation of the peptide bond that occur during α-helix transformation. The weak absorption peak at 280 nm corresponds to π–π* transition, which includes aromatic amino acid residues such as Trp, Tyr, and Phe residues. Fig. 9 displays the absorbance spectra of HSA in the presence of HCDs at both pH values. With the gradual addition of HCDs to HSA, a drop in the intensity of the absorption peak at 208 nm with a slight red shift has been observed. However, the absorption band at 280 nm has not been nearly changed. This suggests that the change in the secondary structure of HSA has no significant effect on the environment of Trp, Tyr, and Phe residues. This spectroscopic study in correlation with fluorescence confirms that the environments of aromatic residues in HSA at both pH values are less altered.
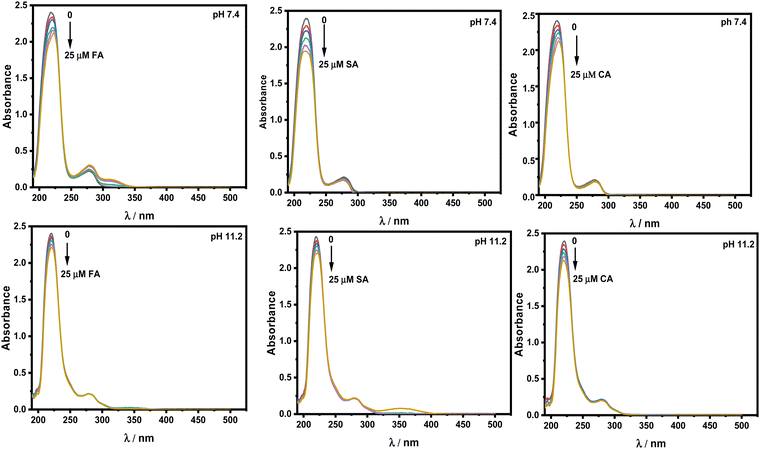 |
| | Fig. 9 Absorbance spectra of 5 × 10−6 M HSA in the presence of increasing concentration of HCDs at pH 7.4 and 11.2 at 298.15 K. | |
Circular dichroism spectroscopic studies.
In order to determine how these HCDs affect the secondary structure of HSA at pH 7.4 and 11.2, CD spectroscopy has been employed. HSA is a globular protein mainly composed of 67% α-helical conformation with a very low β-sheet content.38 The α-helical conformation of the protein is characterised by the CD peaks at 208 and 222 nm.39 Any alteration in these peak regions results in a change in the α-helicity, which is what causes the protein's overall conformation to be disturbed. A drop in the negative values of ellipticity (mdeg) often denotes a decrease in the α-helical conformation and its stability. The CD spectra at pH 7.2 and 11.2 are shown in Fig. 10.
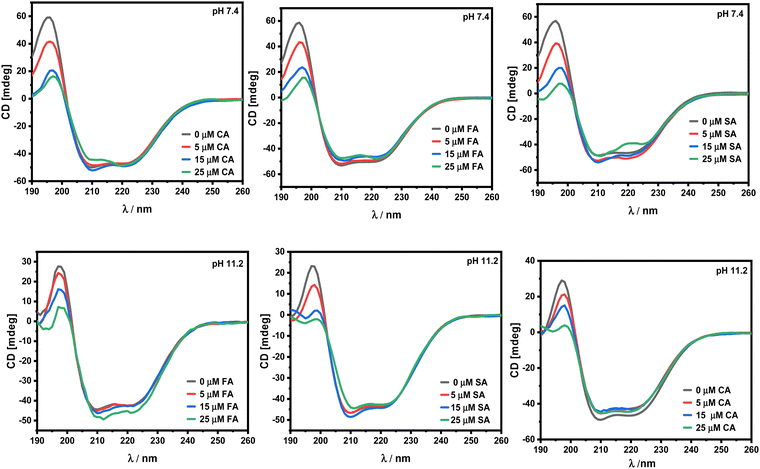 |
| | Fig. 10 CD spectra of 5 × 10−6 M HSA in the presence of increasing concentration of HCDs from 0 to 25 μM at pH 7.4 and 11.2 at 298.15 K. | |
Both HCDs and HSA vary in their structural properties with the change in pH and here it is found that the secondary structure of HSA does not get affected by its interaction with HCDs. In the literature, FA is reported to have less impact on changing the secondary structure of BSA upon binding.40 The involvement of non-covalent interactions of these phenolic compounds has no effect on the secondary structure of the proteins.41 A phenolic OH group on ferulic acid, p-coumaric acid, and sinapic acid can reduce the α-helix structure of HSA by 5–6%. Furthermore, chlorogenic acid and caffeic acid with two phenolic OH groups in their structure have decreased 7–9% of the α-helix structure of HSA. These HCDs depending on the number of availability of –OH groups can cause conformational changes in proteins by forming hydrogen bonds with the C, O and N groups of the major polypeptide chain in HSA when they enter hydrophobic binding cavities (subdomain IIA or IIIA).42 In this study, only slight alterations in the secondary structure of HSA have been reported even at pH 11.2, emphasising the significance of hydrogen bonding of phenolic hydroxyl groups of these HCDs with the protein.
Thermal denaturation studies of HSA.
Furthermore, thermal denaturation studies of HSA in the presence of HCDs have been performed by measuring mean residual ellipticity at 222 nm over a temperature range of 25 °C to 95 °C at pH 7.4 and 11.2. Fig. 11 shows the plots of fraction denatured (fD) of HSA against the temperature. CD data imply that the addition of HCDs in increasing concentration does not affect the secondary structure of HSA. A change in pH from 7.4 to 11.2 drops the transition temperature of HSA by 12.8 °C conferring the alkaline pH-induced conformational instability in HSA. The conformation of protein is altered and its binding affinity to HCDs must be different. The addition of increasing concentration of HCDs to HSA at both pH values does not vary Tm and ΔuH that much. The values of transition temperature (Tm) and enthalpy of unfolding (ΔuH) are tabulated in Table 5. Their binding has been studied by various research groups at physiological pH.7,43 Our ITC and fluorescence binding studies have shown the moderate binding of HCDs to HSA. The CD thermal and spectroscopic data reveal that the HCDs upon binding with HSA keep the conformation of the protein almost intact (mostly secondary structure) even though the same concentration ratio of HCDs to HSA is maintained in every experiment.
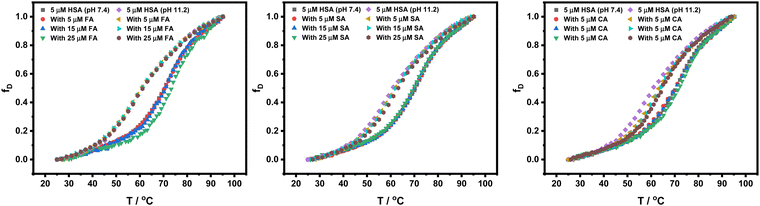 |
| | Fig. 11 Fraction denaturation (fD) plots of 5 × 10−6 M HSA against the temperature in the presence of HCDs at pH 7.4 and 11.2. | |
Table 5 Values of transition temperature (Tm) and enthalpy of unfolding (ΔuH) of 5 × 10−6 M HSA in the presence of HCDs
| Additive/(μM) |
T
m/°C |
ΔuH/kJ mol−1 |
| pH 7.4 |
pH 11.2 |
pH 7.4 |
pH 11.2 |
| Ferulic acid |
| 0 |
69.9 ± 0.3 |
57.1 ± 0.1 |
302.9 ± 2.1 |
261.7 ± 1.8 |
| 5 |
68.4 ± 0.2 |
57.7 ± 0.3 |
307.5 ± 1.7 |
238.8 ± 1.5 |
| 15 |
67.9 ± 0.1 |
58.3 ± 0.2 |
306.3 ± 2.3 |
238.9 ± 2.1 |
| 25 |
68.4 ± 0.8 |
59.2 ± 0.4 |
294.1 ± 2.1 |
242.8 ± 2.3 |
|
|
| Sinapic acid |
| 0 |
69.9 ± 0.1 |
57.1 ± 0.1 |
302.9 ± 2.1 |
261.7 ± 1.8 |
| 5 |
69.7 ± 0.1 |
58.2 ± 0.1 |
302.7 ± 1.5 |
254.8 ± 1.4 |
| 15 |
68.3 ± 0.6 |
59.7 ± 0.3 |
311.1 ± 1.2 |
252.9 ± 2.1 |
| 25 |
68.9 ± 0.1 |
60.2 ± 0.2 |
313.2 ± 1.1 |
259.1 ± 2.5 |
|
|
|
p-Coumaric acid |
| 0 |
69.9 ± 0.3 |
57.1 ± 0.1 |
302.9 ± 2.1 |
261.7 ± 1.8 |
| 5 |
69.3 ± 0.1 |
61.2 ± 0.3 |
303.1 ± 1.7 |
272.9 ± 1.6 |
| 15 |
68.5 ± 0.1 |
61.9 ± 0.1 |
312.5 ± 2.3 |
273.6 ± 2.3 |
| 25 |
70.1 ± 0.2 |
63.1 ± 0.1 |
314.6 ± 2.9 |
279.1 ± 2.5 |
Isothermal titration calorimetric studies: thermodynamic aspects.
Isothermal titration calorimetry is a useful technique for studying the interactions of small molecules with proteins.44–46 The experiments have been performed to explore the nature and energetics of HCD binding to HSA at both pH values. Various research groups have performed the interaction studies of these HCDs with proteins using spectroscopic techniques with a focus on their binding properties based on mostly spectroscopic research.4,7,42,47 The mechanistic information based on thermodynamics using ITC at alkaline pH has not been explored yet. For binding studies (even weak interactions), the ITC technique is more reliable than the fluorescence spectroscopic analysis. Spectroscopic approaches detect the change in enthalpy indirectly based on the mechanism describing the ligand binding process; calorimetry allows one to directly determine the change in enthalpy accompanying a process at a certain temperature. The upper panel in Fig. 12 and 13 shows the raw ITC data and the lower one is fitted and corrected ITC profiles. The ITC data are analysed by fitting a single set of binding site model to the experimental data points using the NanoAnalyze software. The ITC dilution profiles of HSA and HCDs, which are subtracted from the main ITC profiles, are shown in Fig. S8 and S7 (ESI†), respectively. The thermodynamic signatures such as the standard molar enthalpy change (ΔH0m), standard molar entropy change (ΔS0m), standard molar Gibbs free energy change (ΔG0m), n (stoichiometry) and association constant (KA), have been obtained from analysed ITC data. These parameters are tabulated in Tables 5 and 6.
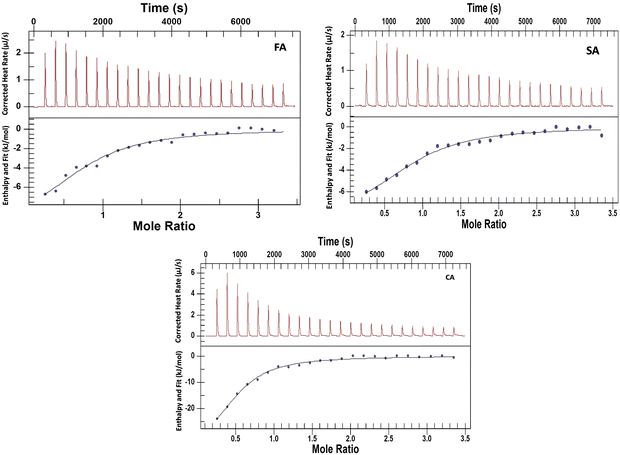 |
| | Fig. 12 ITC profiles associated with the titration of 2.5 mM HCDs (syringe) against 0.13 mM HSA taken inside the ITC cell at pH 7.4 at 298.15 K. | |
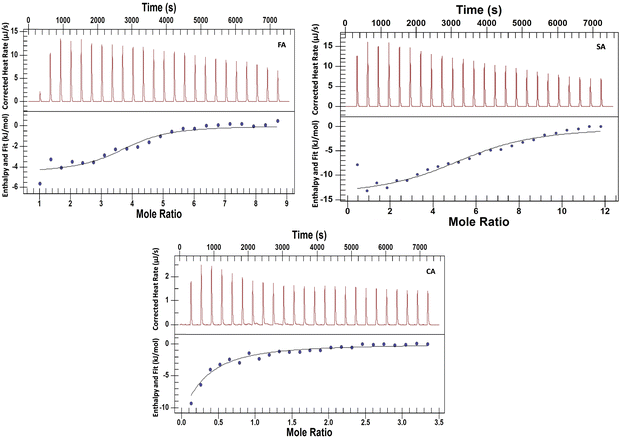 |
| | Fig. 13 ITC profiles of the titration of 6.5 mM FA, 7.8 mM SA and 2.5 mM CA against 0.13 mM HSA taken inside the ITC cell at pH 11.2 at 298.15 K. All the HCD solutions are taken in the ITC syringe. | |
Table 6 Thermodynamic parameters associated with the interaction of HCDs with HSA at pH 7.4 at 298.15 K
| Additive (pH 7.4) |
ΔH0m (kJ mol−1) |
ΔS0m (J mol−1 K−1) |
n
|
K (M−1) |
ΔG0m (kJ mol−1) |
| Ferulic acid |
−10.90 ± 1.23 |
47.41 ± 0.30 |
0.90 ± 0.02 |
(2.43 ± 0.12) × 104 |
−25.03 ± 0.50 |
| Sinapic acid |
−9.07 ± 1.50 |
54.58 ± 0.11 |
0.85 ± 0.02 |
(2.74 ± 0.11) × 104 |
−25.34 ± 0.30 |
| Coumaric acid |
−39.41 ± 1.73 |
−42.64 ± 0.71 |
0.51 ± 0.01 |
(4.75 ± 0.10) × 104 |
−26.70 + 0.30 |
Ross and Subramanian have well addressed the thermodynamics of protein association with ligands.48 They have reported ΔH0m > 0 and ΔS0m > 0 for interactions that are mostly hydrophobic, ΔH0m ∼ 0 and ΔS0m > 0 for electrostatic interactions and ΔH0m < 0 and ΔS0m < 0 when the van der Waals and H-bond interactions are involved. Here, the values of ΔH0m change with the change in pH from 7.4 to 11.2. This indicates that the surface properties of both proteins and HCDs change with the pH and hence their interactions. These HCDs can interact electrostatically with proteins at neutral pH because of their low pKa carboxylic acid groups, which deprotonate first, rather than their high pKa –OH groups.21,49 Negative values of ΔH0m suggest that their interactions with HSA at pH 7.4 and 11.2 are predominantly polar in nature. Lower values of ΔH0m for the interactions of FA and SA with HSA have been observed as compared to CA at both pH values. Exothermic contributions to ΔH0m values have been reduced by FA, whereas they are increased by SA and CA with the increase in pH from 7.4 to 11.2.
Low exothermicity in the values of ΔH0m clearly indicates the contribution of hydrophobicity. This can be attributed to the interaction of aromatic moieties of HCDs with the non-polar residues of the protein such as aromatic amino acids. Overall, the exothermicity in the values of ΔH0m is high for CA at pH 11.2 among these HCDs, suggesting the major contribution of polar interactions. Binding of CA with HSA involves the negative entropic change (ΔS0m) values, which suggest the role of hydrogen bonding in their interactions. Hydroxyl groups of phenolic compounds play a significant role in the binding of HCDs with the proteins.50 Previously, the hydrogen bonds have been reported by the interaction of the phenolic hydroxyl group of these HCDs with C, O, N and H of the main polypeptide chain of the protein.51 Association of FA and SA with HSA involves the desolvation process, which is indicated by the positive entropic changes (ΔS0m). Binding of three HCDs is spontaneous in nature as observed by the negative standard molar Gibbs free energy change (ΔG0m). Overall, the complexation of HCDs with HSA not only involves non-covalent interactions, such as electrostatic, hydrogen bonding, hydrophobic and π–π stacking, but also involves the desolvation and solvation processes. Although the magnitude of their binding constant (KA) is the same (see Tables 6 and 7), their mechanism of binding to HSA is different at both pH values. Previously, utilising FTIR and fluorescence methods, Kang et al. have investigated FA interaction with HSA at physiological pH and reported a binding constant of 2.23 × 104 M−1.47 Likewise, Shuang Li et al. have also reported the value of KA to be 1.793 × 104 M−1 with a stoichiometric ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by spectroscopic techniques.43 A range of values for the binding constant from 1.92 × 103 M−1 to 6.87 × 108 M−1 at pH 7.0 have been observed when the molar ratio of SA to HSA was varied between 0.1 and 4.26 and 4.26 and 30, indicating a non-specific interaction between them.42 Jiang min et al. have reported the binding of CA with human serum albumin (pH 7.0) by a spectroscopic method, KA = 1.1 × 105 M−1.7 Mostly, these research groups have focussed on the interactions based on the spectroscopic studies at physiological pH. In our study, the analysis of the ITC profile reveals that the interaction of FA and SA with HSA is entropically favourable, whereas the interaction of CA with HSA is enthalpically favourable. For FA and SA, hydrophobic contribution is more than that for CA, which involves the desolvation process. Predominance of H-bond formation is seen from the negative value of ΔS0m, which shows conformation restriction of HSA upon binding. This indicates that the involvement of –OH groups in CA with polar amino acid residues is more as compared to FA and SA. With the increase of pH, the mode of binding of these HCDs changes, indicating the change in the conformation of HSA, which is different at both pH values. Overall, the mechanistic insights into binding of HCDs with HSA obtained calorimetrically along with the thermodynamic parameters and the type of quenching based on TCSPC data at pH 7.4 and 11.2 at 298.15 K are presented in Fig. 14. This shows the predominance of the functional and aromatic groups of hydroxycinnamic acid derivatives in their molecular structure in the binding process with HSA, which are responsible for their enthalpically or entropically driven binding and dynamic or static type of quenching on the basis of protein–HCD complex formation.
1 by spectroscopic techniques.43 A range of values for the binding constant from 1.92 × 103 M−1 to 6.87 × 108 M−1 at pH 7.0 have been observed when the molar ratio of SA to HSA was varied between 0.1 and 4.26 and 4.26 and 30, indicating a non-specific interaction between them.42 Jiang min et al. have reported the binding of CA with human serum albumin (pH 7.0) by a spectroscopic method, KA = 1.1 × 105 M−1.7 Mostly, these research groups have focussed on the interactions based on the spectroscopic studies at physiological pH. In our study, the analysis of the ITC profile reveals that the interaction of FA and SA with HSA is entropically favourable, whereas the interaction of CA with HSA is enthalpically favourable. For FA and SA, hydrophobic contribution is more than that for CA, which involves the desolvation process. Predominance of H-bond formation is seen from the negative value of ΔS0m, which shows conformation restriction of HSA upon binding. This indicates that the involvement of –OH groups in CA with polar amino acid residues is more as compared to FA and SA. With the increase of pH, the mode of binding of these HCDs changes, indicating the change in the conformation of HSA, which is different at both pH values. Overall, the mechanistic insights into binding of HCDs with HSA obtained calorimetrically along with the thermodynamic parameters and the type of quenching based on TCSPC data at pH 7.4 and 11.2 at 298.15 K are presented in Fig. 14. This shows the predominance of the functional and aromatic groups of hydroxycinnamic acid derivatives in their molecular structure in the binding process with HSA, which are responsible for their enthalpically or entropically driven binding and dynamic or static type of quenching on the basis of protein–HCD complex formation.
Table 7 Thermodynamic parameters associated with the interaction of HCDs with HSA at pH 11.2 at 298.15 K
| Additive (pH 11.2) |
ΔH0m (kJ mol−1) |
ΔS0m (J mol−1 K−1) |
n
|
K (M−1) |
ΔG0m (kJ mol−1) |
| Ferulic acid |
−4.58 ± 0.61 |
74.09 ± 0.34 |
3.71 ± 0.10 |
(4.67 ± 0.16) × 104 |
−26.66 ± 0.31 |
| Sinapic acid |
−14.29 ± 1.70 |
30.90 ± 0.21 |
5.67 ± 0.12 |
(1.31 ± 0.10) × 104 |
−23.51 ± 0.42 |
| Coumaric acid |
−65.23 ± 2.13 |
−140.1 ± 0.30 |
1.34 ± 0.08 |
(1.29 ± 0.12) × 104 |
−23.47 ± 0.30 |
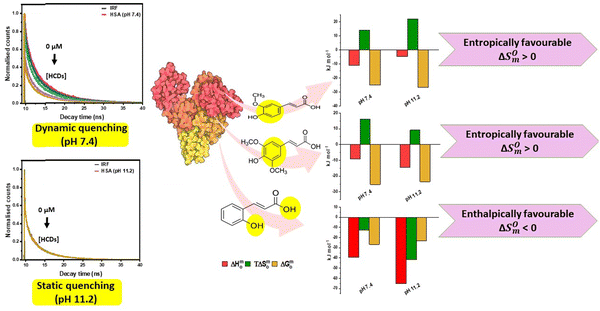 |
| | Fig. 14 Mechanistic insights into binding of HCDs with HSA obtained from ITC and TCSPC studies at pH 7.4 and 11.2 at 298.15 K. | |
The stoichiometry (n) of binding of HCDs with HSA increases with the change in pH from 7.4 to 11.2. For FA and SA at pH 7.4, binding stoichiometry is almost 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while for CA, it is 1
1, while for CA, it is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. This indicates that 1 molecule of CA is shared by 2 molecules of HSA. This is one of the reasons for the predominance of hydrogen bonding and more restriction of the conformation of HSA upon binding with CA indicated by negative entropic changes. The stoichiometry becomes approx. 3.7, 5.6 and 1.3 for FA, SA and CA binding with HSA at pH 11.2, respectively. The increased value of stoichiometry again indicates altered conformation, surface properties and the availability of more sites for HSA and HCDs. Our ITC experiments reveal that the hydrogen bonding and polar forces predominate in HCD–protein interactions at both physiological and alkaline pH values; nevertheless, their modes of binding are distinct.
2. This indicates that 1 molecule of CA is shared by 2 molecules of HSA. This is one of the reasons for the predominance of hydrogen bonding and more restriction of the conformation of HSA upon binding with CA indicated by negative entropic changes. The stoichiometry becomes approx. 3.7, 5.6 and 1.3 for FA, SA and CA binding with HSA at pH 11.2, respectively. The increased value of stoichiometry again indicates altered conformation, surface properties and the availability of more sites for HSA and HCDs. Our ITC experiments reveal that the hydrogen bonding and polar forces predominate in HCD–protein interactions at both physiological and alkaline pH values; nevertheless, their modes of binding are distinct.
Conclusions
The present study aims to investigate how antioxidants interact with the alkali-induced state of human serum albumin. UV and fluorescence results indicate the change in the conformation of the protein and different ionic structures of HCDs with the change in pH. Fluorescence data reveal the moderate binding of HCDs to HSA, which is further explained based on FRET analysis and deconvolution methods. In addition, lifetime measurements show the dynamic and static quenching at pH 7.4 and 11.2, respectively. UV-vis and circular dichroism studies do not show significant conformational changes in HSA upon binding with HCDs at both pH values. No conformational change is further verified by the thermal denaturation experiments. The thermodynamics of their binding and energetics are explored using ultrasensitive isothermal titration calorimetry (ITC). The calorimetric data reveal that the binding of HCDs to HSA is predominantly electrostatic and polar in nature. The values of the standard enthalpy change (ΔH0m) and the standard entropy change (ΔS0m) vary with the change of pH from 7.4 to 11.2. The exothermicity in ΔH0m values increases at pH 11.2 for CA binding to HSA compared to FA and SA. The contribution of hydrophobicity of the aromatic moiety of FA and SA bound to HSA is greater than that of CA. This is why ferulic acid (FA) and sinapic acid (SA) binding to HSA is entropically favourable, which causes desolvation upon their binding. For trans-o-coumaric acid (CA), binding to HSA is enthalpically favourable, which is due to the predominance of H-bonding formation and restriction of conformation of HSA upon binding. Overall, the strength of binding (KA) of HCDs to HSA is almost similar at both pH values. However, the spectroscopic and calorimetric investigations demonstrate that their binding mechanisms are different at both pH values. This experimental study presents the optimised HSA–HCD binding data at alkaline pH, which can be useful for deriving the guidelines for rational drug design. No conformational changes in HSA upon moderate binding with HCDs can be detected. These HCDs, which have not yet received adequate attention, can be used as a platform for applications in the drug delivery and pharmaceutical sectors.
Author contributions
Anjali Maheshwari: formal analysis; investigation; methodology; validation; writing – original draft; writing – reviewing and editing and Nand Kishore: formal analysis, funding acquisition, investigation, methodology, project administration, validation, writing – reviewing and editing.
Data availability
The data supporting this article have been included as part of the ESI.†
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgements
The authors are thankful to the Indian Institute of Technology, Bombay, and Council of Scientific and Industrial Research (CSIR) for providing financial support. The authors also acknowledge the Central Facility of Department of Chemistry and IRCC at the Indian Institute of Technology Bombay, Mumbai for providing instrument facilities.
References
- N. Keswani, S. Choudhary and N. Kishore, Interaction of weakly bound antibiotics neomycin and lincomycin with bovine and human serum albumin: biophysical approach, J. Biochem., 2010, 148, 71–84 CrossRef.
- T. Topală, A. Bodoki, L. Oprean and R. Oprean, Bovine serum albumin interactions with metal complexes, Clujul Med., 2014, 87, 215–219 Search PubMed.
- L. Yang, J. Nao and X. Dong, The Therapeutic Potential of Hydroxycinnamic Acid Derivatives in Parkinson's Disease: Focus on In Vivo Research Advancements, J. Agric. Food Chem., 2023, 71, 10931–10951 Search PubMed.
- X. L. Jin, X. Wei, F. M. Qi, S. S. Yu, B. Zhou and S. Bai, Characterization of hydroxycinnamic acid derivatives binding to bovine serum albumin, Org. Biomol. Chem., 2012, 10, 3424–3431 RSC.
- M. Putschögl, P. Zirak and A. Penzkofer, Absorption and emission behaviour of trans-p-coumaric acid in aqueous solutions and some organic solvents, Chem. Phys., 2008, 343, 107–120 CrossRef.
- W. F. Zhiwei Huang, H. Chen, P. Tan, M. Huang, H. Shi, B. Sun, Y. Cheng, T. Li, Z. Mou and Q. Li, Sinapic acid inhibits pancreatic cancer proliferation, migration, and invasion via downregulation of the AKT/Gsk-3β signal pathway, Drug Dev. Res., 2022, 83, 721–734 Search PubMed.
- M. Jiang, M. X. Xie, D. Zheng, Y. Liu, X. Y. Li and X. Chen, Spectroscopic studies on the interaction of cinnamic acid and its hydroxyl derivatives with human serum albumin, J. Mol. Struct., 2004, 692, 71–80 CrossRef.
- D. C. Carter and J. X. Ho, Structure of serum albumin, Adv. Protein Chem., 1994, 45, 153–176 CrossRef.
- A. Maheshwari and N. Kishore, pH-dependent interactions of biologically important metal ions with hen egg white lysozyme based on its hydration properties: thermodynamic and mechanistic insights, Int. J. Biol. Macromol., 2024, 259, 129297 CrossRef CAS PubMed.
- Anjali and N. Kishore, Influence of amino acids on alkaline pH induced partially folded molten globule like intermediate of bovine serum albumin: conformational and thermodynamic insights, J. Mol. Liq., 2022, 368, 120599 CrossRef CAS.
- P. Prasanthan and N. Kishore, Alkali induced unique partially folded state of bovine serum albumin: qualitative and quantitative insights, Int. J. Biol. Macromol., 2019, 138, 252–261 CrossRef CAS.
- M. M. Gomari, N. Rostami, D. R. Faradonbeh, H. R. Asemaneh, G. Esmailnia, S. Arab, M. Farsimadan, A. Hosseini and N. V. Dokholyan, Evaluation of pH change effects on the HSA folding and its drug binding characteristics, a computational biology investigation, Proteins: Struct., Funct., Bioinf., 2022, 90, 1908–1925 CrossRef CAS PubMed.
- M. Amiri, K. Jankeje and J. R. Albani, Characterization of human serum albumin forms with pH. Fluorescence lifetime studies, J. Pharm. Biomed. Anal., 2010, 51, 1097–1102 CrossRef CAS PubMed.
- Q. Wang, Y. Tang, Y. Yang, L. Lei, X. Lei, J. Zhao, Y. Zhang, L. Li, Q. Wang and J. Ming, The interaction mechanisms, and structural changes of the interaction between zein and ferulic acid under different pH conditions, Food Hydrocolloids, 2022, 124, 107251 CrossRef CAS.
- G. Xing, X. Rui, D. Wang, M. Liu, X. Chen and M. Dong, Effect of Fermentation pH on Protein Bioaccessibility of Soymilk Curd with Added Tea Polyphenols As Assessed by in Vitro Gastrointestinal Digestion, J. Agric. Food Chem., 2017, 65, 11125–11132 CrossRef CAS.
- S. Das and A. B. H. Wong, Stabilization of ferulic acid in topical gel formulation via nanoencapsulation and pH optimization, Sci. Rep., 2020, 10, 1–18 CrossRef.
- F. Liu, C. Ma, D. J. McClements and Y. Gao, A comparative study of covalent and non-covalent interactions between zein and polyphenols in ethanol-water solution, Food Hydrocolloids, 2017, 63, 625–634 CrossRef.
- M. Friedman and H. Jurgens, Effect of pH on the Stability of Plant Phenolic Compounds, J. Agric. Food Chem., 2000, 48, 2101–2110 CrossRef PubMed.
- D. H. Truong, N. T. A. Nhung and D. Q. Dao, Iron ions chelation-based antioxidant potential vs. pro-oxidant risk of ferulic acid: a DFT study in aqueous phase, Comput. Theor. Chem., 2020, 1185, 112905 CrossRef.
- B. Smyk and R. Drabent, Spectroscopic investigation of the equilibria of the ionic forms of sinapic acid, Analyst, 1989, 114, 723–726 RSC.
- I. Aguilar-Hernández, N. K. Afseth, T. López-Luke, F. F. Contreras-Torres, J. P. Wold and N. Ornelas-Soto, Surface enhanced Raman spectroscopy of phenolic antioxidants: a systematic evaluation of ferulic acid, p-coumaric acid, caffeic acid and sinapic acid, Vib. Spectrosc., 2017, 89, 113–122 CrossRef.
- T. P. H. Hutapea, K. A. Madurani, M. Y. Syahputra, M. N. Hudha, A. N. Asriana, Suprapto and F. Kurniawan, Albumin: source, preparation, determination, applications, and prospects, J. Sci.: Adv. Mater. Devices, 2023, 8, 100549 Search PubMed.
- M. Hossain, N. Huda and A. K. Bhuyan, A surprisingly simple three-state generic process for reversible protein denaturation by trifluoroethanol, Biophys. Chem., 2022, 291, 106895 CrossRef.
- N. S. Jha, E. Judy and N. Kishore, 1,1,1,3,3,3-Hexafluoroisopropanol and 2,2,2-trifluoroethanol act more effectively on protein in combination than individually: thermodynamic aspects, J. Chem. Thermodyn., 2018, 121, 39–48 CrossRef.
- G. Rabbani, E. Ahmad, N. Zaidi, S. Fatima and R. H. Khan, PH-Induced Molten Globule State of Rhizopus niveus Lipase is More Resistant Against Thermal and Chemical Denaturation Than Its Native State, Cell Biochem. Biophys., 2012, 62, 487–499 CrossRef.
- P. Prasanthan and N. Kishore, Unusual human serum albumin fibrillation inhibition by ketoprofen and fenoprofen: mechanistic insights, J. Mol. Recognit., 2012, 34, e2937 CrossRef.
- J. H. Shi, Q. Wang, D. Q. Pan, T. T. Liu and M. Jiang, Characterization of interactions of simvastatin, pravastatin, fluvastatin, and pitavastatin with bovine serum albumin: multiple spectroscopic and molecular docking, J. Biomol. Struct. Dyn., 2017, 35, 1529–1546 CrossRef.
-
W. H. Kirchoff, EXAM, US Dep. Energy, Thermodyn. Div. Natl. Inst. Stand. Technol., Gaithersburg, MD, USA, 1988.
- M. Amiri, K. Jankeje and J. R. Albani, Origin of fluorescence lifetimes in human serum albumin. Studies on native and denatured protein, J. Fluoresc., 2010, 20, 651–656 CrossRef.
- R. Ghosh and N. Kishore, Physicochemical Insights into the Role of Drug Functionality in Fibrillation Inhibition of Bovine Serum Albumin, J. Phys. Chem. B, 2020, 124, 8989–9008 CrossRef PubMed.
- M. Engelhard and P. A. Evans, Kinetics of interaction of partially folded proteins with a hydrophobic dye: evidence that molten globule character is maximal in early folding intermediates, Protein Sci., 1995, 4, 1553–1562 CrossRef PubMed.
- C. Martin, J. L. Bruneel, F. Castet, A. Fritsch, P. L. Teissedre, M. Jourdes and F. Guillaume, Spectroscopic and theoretical investigations of phenolic acids in white wines, Food Chem., 2017, 221, 568–575 CrossRef.
- A. Belay, E. Libnedengel, H. K. Kim and Y. H. Hwang, Effects of solvent polarity on the absorption and fluorescence spectra of chlorogenic acid and caffeic acid compounds: determination of the dipole moments, Luminescence, 2016, 31, 118–126 CrossRef PubMed.
- T. Imoto, L. S. Forster, J. A. Rupley and F. Tanaka, Fluorescence of lysozyme: emissions from tryptophan residues 62 and 108 and energy migration, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 1151–1155 CrossRef.
- K. Jankeje, M. Amiri and J. R. Albani, Relation between Human Serum Albumin Structure and Fluorescence Decay Parameters of Tryptophan Residue 214, Encycl. Anal. Chem., 2017, 1–19 Search PubMed.
- K. Vos, A. van Hoek and A. J. W. G. Visser, Application of a reference convolution method to tryptophan fluorescence in proteins: a refined description of rotational dynamics, Eur. J. Biochem., 1987, 165, 55–63 CrossRef.
- J. K. A. Kamal and D. V. Behere, Spectroscopic studies on human serum albumin and methemalbumin: optical, steady-state, and picosecond time-resolved fluorescence studies, and kinetics of substrate oxidation by methemalbumin, J. Biol. Inorg. Chem., 2002, 7, 273–283 CrossRef.
- K. Murayama and M. Tomida, Heat-induced secondary structure and conformation change of bovine serum albumin investigated by Fourier transform infrared spectroscopy, Biochemistry, 2004, 43, 11526–11532 CrossRef.
- P. Prasanthan and N. Kishore, Alkali induced unique partially folded state of bovine serum albumin: qualitative and quantitative insights, Int. J. Biol. Macromol., 2019, 138, 252–261 CrossRef PubMed.
- Y. Yang, S. Wang, X. Liu, W. Zhang, W. Tong, H. Luo and L. Zhao, Interactions of ferulic acid and ferulic acid methyl ester with endogenous proteins: determination using the multi-methods, Heliyon, 2024, 10, e24605 CrossRef.
- H. Ojha, K. Mishra, M. I. Hassan and N. K. Chaudhury, Spectroscopic and isothermal titration calorimetry studies of binding interaction of ferulic acid with bovine serum albumin, Thermochim. Acta, 2012, 548, 56–64 CrossRef.
- Y. Liu, M. X. Xie, M. Jiang and Y. D. Wang, Spectroscopic investigation of the interaction between human serum albumin and three organic acids, Spectrochim. Acta, Part A, 2005, 61, 2245–2251 CrossRef PubMed.
- S. Li, K. Huang, M. Zhong, J. Guo, W. Z. Wang and R. Zhu, Comparative studies on the interaction of caffeic acid, chlorogenic acid and ferulic acid with bovine serum albumin, Spectrochim. Acta, Part A, 2010, 77, 680–686 CrossRef PubMed.
- K. Rajarathnam and J. Rösgen, Isothermal titration calorimetry of membrane proteins – Progress and challenges, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 69–77 CrossRef.
- R. Ghosh and N. Kishore, Physicochemical Insights into the Stabilization of Stressed Lysozyme and Glycine Homopeptides by Sorbitol, J. Phys. Chem. B, 2018, 122, 7839–7854 CrossRef PubMed.
- H. Su and Y. Xu, Application of ITC-based characterization of thermodynamic and kinetic association of ligands with proteins in drug design, Front. Pharmacol., 2018, 9, 1–7 CrossRef PubMed.
- J. Kang, Y. Liu, M. X. Xie, S. Li, M. Jiang and Y. D. Wang, Interactions of human serum albumin with chlorogenic acid and ferulic acid, Biochim. Biophys. Acta, Gen. Subj., 2004, 1674, 205–214 CrossRef PubMed.
- P. D. Ross and S. Subramanian, Thermodynamics of Protein Association Reactions: Forces Contributing to Stability, Biochemistry, 1981, 20, 3096–3102 CrossRef PubMed.
- D. Malyshev, R. Öberg, L. Landström, P. O. Andersson, T. Dahlberg and M. Andersson, pH-induced changes in Raman, UV-vis absorbance, and fluorescence spectra of dipicolinic acid (DPA), Spectrochim. Acta, Part A, 2022, 271, 120869 CrossRef PubMed.
- H. Yilmaz, B. Gultekin Subasi, H. U. Celebioglu, T. Ozdal and E. Capanoglu, Chemistry of Protein-Phenolic Interactions Toward the Microbiota and Microbial Infections, Front. Nutr., 2022, 9, 1–16 Search PubMed.
- S. Schefer, M. Oest and S. Rohn, Interactions between phenolic acids, proteins, and carbohydrates—influence on dough and bread properties, Foods, 2021, 10, 1–29 CrossRef PubMed.
|
| This journal is © the Owner Societies 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  *
*
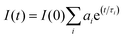
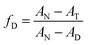

![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), methoxy group (–OCH3) and the phenol ring.33 Both the UV and fluorescence studies indicate that the properties of the HCDs also change with changes in pH depending on the functional groups present in HCDs.
CH–), methoxy group (–OCH3) and the phenol ring.33 Both the UV and fluorescence studies indicate that the properties of the HCDs also change with changes in pH depending on the functional groups present in HCDs.




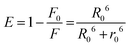
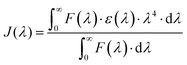
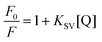
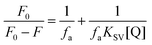
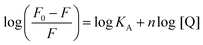
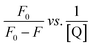 and double logarithmic plots of
and double logarithmic plots of 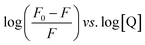 are shown in Fig. 7. For every HCD that interacts with the fluorophore (Trp214), the value of KSV is of the order of 104. The parameters associated with the fluorescence quenching studies for the binding of HCDs with HSA are shown in Table 2 at both pH values. The fluorescence data show a single set of binding sites (n ≈ 1) with the fractional accessibility (fa) of the tryptophan residue, which is almost one at both pH values.26 The magnitude of the association constant (KA) of HCDs with HSA is almost identical at pH 7.4 and 11.2. The interaction studies of bovine serum albumin with hydroxycinnamic acid derivatives at physiological pH have been conducted, showing the values of association constants (KA) in the order as follows: p-CA > FA > SA.4 This indicates that the strength of binding differs from protein to protein based on its conformation and affinity for the ligands and the pH conditions. Here, the change in the values of KA with the pH might be due to the differently exposed interacting amino acid groups in HSA. HCDs binding with the BSA protein at neutral pH have also been characterised by NMR methodology. This indicates the involvement of hydrogen bonding and hydrophobic interactions with the protein in their binding process.4
are shown in Fig. 7. For every HCD that interacts with the fluorophore (Trp214), the value of KSV is of the order of 104. The parameters associated with the fluorescence quenching studies for the binding of HCDs with HSA are shown in Table 2 at both pH values. The fluorescence data show a single set of binding sites (n ≈ 1) with the fractional accessibility (fa) of the tryptophan residue, which is almost one at both pH values.26 The magnitude of the association constant (KA) of HCDs with HSA is almost identical at pH 7.4 and 11.2. The interaction studies of bovine serum albumin with hydroxycinnamic acid derivatives at physiological pH have been conducted, showing the values of association constants (KA) in the order as follows: p-CA > FA > SA.4 This indicates that the strength of binding differs from protein to protein based on its conformation and affinity for the ligands and the pH conditions. Here, the change in the values of KA with the pH might be due to the differently exposed interacting amino acid groups in HSA. HCDs binding with the BSA protein at neutral pH have also been characterised by NMR methodology. This indicates the involvement of hydrogen bonding and hydrophobic interactions with the protein in their binding process.4





![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by spectroscopic techniques.43 A range of values for the binding constant from 1.92 × 103 M−1 to 6.87 × 108 M−1 at pH 7.0 have been observed when the molar ratio of SA to HSA was varied between 0.1 and 4.26 and 4.26 and 30, indicating a non-specific interaction between them.42 Jiang min et al. have reported the binding of CA with human serum albumin (pH 7.0) by a spectroscopic method, KA = 1.1 × 105 M−1.7 Mostly, these research groups have focussed on the interactions based on the spectroscopic studies at physiological pH. In our study, the analysis of the ITC profile reveals that the interaction of FA and SA with HSA is entropically favourable, whereas the interaction of CA with HSA is enthalpically favourable. For FA and SA, hydrophobic contribution is more than that for CA, which involves the desolvation process. Predominance of H-bond formation is seen from the negative value of ΔS0m, which shows conformation restriction of HSA upon binding. This indicates that the involvement of –OH groups in CA with polar amino acid residues is more as compared to FA and SA. With the increase of pH, the mode of binding of these HCDs changes, indicating the change in the conformation of HSA, which is different at both pH values. Overall, the mechanistic insights into binding of HCDs with HSA obtained calorimetrically along with the thermodynamic parameters and the type of quenching based on TCSPC data at pH 7.4 and 11.2 at 298.15 K are presented in Fig. 14. This shows the predominance of the functional and aromatic groups of hydroxycinnamic acid derivatives in their molecular structure in the binding process with HSA, which are responsible for their enthalpically or entropically driven binding and dynamic or static type of quenching on the basis of protein–HCD complex formation.
1 by spectroscopic techniques.43 A range of values for the binding constant from 1.92 × 103 M−1 to 6.87 × 108 M−1 at pH 7.0 have been observed when the molar ratio of SA to HSA was varied between 0.1 and 4.26 and 4.26 and 30, indicating a non-specific interaction between them.42 Jiang min et al. have reported the binding of CA with human serum albumin (pH 7.0) by a spectroscopic method, KA = 1.1 × 105 M−1.7 Mostly, these research groups have focussed on the interactions based on the spectroscopic studies at physiological pH. In our study, the analysis of the ITC profile reveals that the interaction of FA and SA with HSA is entropically favourable, whereas the interaction of CA with HSA is enthalpically favourable. For FA and SA, hydrophobic contribution is more than that for CA, which involves the desolvation process. Predominance of H-bond formation is seen from the negative value of ΔS0m, which shows conformation restriction of HSA upon binding. This indicates that the involvement of –OH groups in CA with polar amino acid residues is more as compared to FA and SA. With the increase of pH, the mode of binding of these HCDs changes, indicating the change in the conformation of HSA, which is different at both pH values. Overall, the mechanistic insights into binding of HCDs with HSA obtained calorimetrically along with the thermodynamic parameters and the type of quenching based on TCSPC data at pH 7.4 and 11.2 at 298.15 K are presented in Fig. 14. This shows the predominance of the functional and aromatic groups of hydroxycinnamic acid derivatives in their molecular structure in the binding process with HSA, which are responsible for their enthalpically or entropically driven binding and dynamic or static type of quenching on the basis of protein–HCD complex formation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while for CA, it is 1
1, while for CA, it is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. This indicates that 1 molecule of CA is shared by 2 molecules of HSA. This is one of the reasons for the predominance of hydrogen bonding and more restriction of the conformation of HSA upon binding with CA indicated by negative entropic changes. The stoichiometry becomes approx. 3.7, 5.6 and 1.3 for FA, SA and CA binding with HSA at pH 11.2, respectively. The increased value of stoichiometry again indicates altered conformation, surface properties and the availability of more sites for HSA and HCDs. Our ITC experiments reveal that the hydrogen bonding and polar forces predominate in HCD–protein interactions at both physiological and alkaline pH values; nevertheless, their modes of binding are distinct.
2. This indicates that 1 molecule of CA is shared by 2 molecules of HSA. This is one of the reasons for the predominance of hydrogen bonding and more restriction of the conformation of HSA upon binding with CA indicated by negative entropic changes. The stoichiometry becomes approx. 3.7, 5.6 and 1.3 for FA, SA and CA binding with HSA at pH 11.2, respectively. The increased value of stoichiometry again indicates altered conformation, surface properties and the availability of more sites for HSA and HCDs. Our ITC experiments reveal that the hydrogen bonding and polar forces predominate in HCD–protein interactions at both physiological and alkaline pH values; nevertheless, their modes of binding are distinct.

