Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
SERS microscopy as a tool for comprehensive biochemical characterization in complex samples
Janina
Kneipp
 *a,
Stephan
Seifert
*a,
Stephan
Seifert
 b and
Florian
Gärber
b and
Florian
Gärber
 b
b
aDepartment of Chemistry, Humboldt-Universität zu Berlin, Brook-Taylor-Str. 2, 12489 Berlin, Germany. E-mail: janina.kneipp@chemie.hu-berlin.de
bHamburg School of Food Science, Department of Chemistry, Universität Hamburg, Grindelallee 117, 20146 Hamburg, Germany
First published on 27th June 2024
Abstract
Surface enhanced Raman scattering (SERS) spectra of biomaterials such as cells or tissues can be used to obtain biochemical information from nanoscopic volumes in these heterogeneous samples. This tutorial review discusses the factors that determine the outcome of a SERS experiment in complex bioorganic samples. They are related to the SERS process itself, the possibility to selectively probe certain regions or constituents of a sample, and the retrieval of the vibrational information in order to identify molecules and their interaction. After introducing basic aspects of SERS experiments in the context of biocompatible environments, spectroscopy in typical microscopic settings is exemplified, including the possibilities to combine SERS with other linear and non-linear microscopic tools, and to exploit approaches that improve lateral and temporal resolution. In particular the great variation of data in a SERS experiment calls for robust data analysis tools. Approaches will be introduced that have been originally developed in the field of bioinformatics for the application to omics data and that show specific potential in the analysis of SERS data. They include the use of simulated data and machine learning tools that can yield chemical information beyond achieving spectral classification.
Key learning points– SERS spectra of complex samples always reflect the very local molecular composition at the nanoscopic site of interaction of the sample with the SERS substrate rather than the general composition of a sample.– SERS-based chemical imaging can be ideally supported with other microscopic and spectroscopic imaging, including plasmon-enhanced linear and non-linear modalities, such as multiphoton-excited Raman processes. – The origin of SERS in a highly localized field opens new possibilities for high resolution mapping and imaging. – In the analysis of SERS data for biocharacterization, approaches adapted from bioinformatics that enable a reliable identification of classifiers and that acknowledge the qualitative and quantitative variations can help to identify molecule–nanostructure and molecule–molecule interactions. |
1. Introduction: explaining the task
The combination of spectrometers with microscopes has been vital to the field of Raman spectroscopy for many decades. Using a microscope to define the excitation volume in a sample and to collect the Raman scattered light has enabled the investigation and application of linear and non-linear Raman processes. Examples include the definition of sampling volumes in single living cells for spontaneous Raman scattering,1 or the widespread utilization of stimulated Raman processes, in particular coherent anti-Stokes Raman scattering (CARS)2 and stimulated Raman scattering (SRS),3 as well as the more recent development of super-resolution Raman microscopy approaches.4,5 Confining a sampling volume to the near field of a scanning probe in atomic force microscopy or scanning tunneling microscopy in tip-enhanced Raman scattering (TERS)6 has even revealed vibrations of individual molecular bonds.7In surface-enhanced Raman scattering (SERS), the excitation of localized surface plasmon resonances (LSPR) in nanostructures leads to the generation of high local optical fields in the proximity of the nanostructures, and thereby to the enhancement of the excitation field and the scattering field, by a respective frequency-dependent field enhancement factor A(ν). The enhancement in SERS is a consequence of the intensity enhancements |A(ν)|2 of each field, the so-called electromagnetic enhancement, as well as of an increased Raman cross section σRSads of the probed molecule, if it resides at the surface of the nanostructure, the so-called chemical enhancement,8–10 so that the SERS signal PSERS is
| PSERS = N·IL·|A(νL)|2·|A(νS)|2·σRSads | (1) |
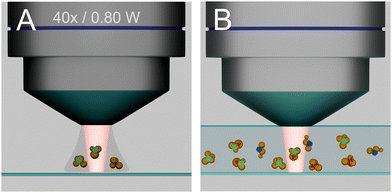
When individual nanostructures, such as particles, their aggregates, or individual nanopores,11 or a tip in TERS settings,6 are present in a probed volume in a sample, and/or when only individual or a few molecules are present,12,13 the detected SERS signal originates only from a particular nanostructure and the molecule in its proximity (Fig. 1(A)).
Differently, in complex samples that consist of different types of molecules, such as a biosample containing plasmonic nanoparticles, the inhomogeneous distribution of these SERS substrates and/or the different interaction of different molecules with them will yield selectivity in probing, and the spectra provide information from all molecules that are near an enhancing nanostructure, and not from the complete focal volume (Fig. 1(B)). In such an experiment, the size of the original excitation volume, diffraction-limited by a microscope objective, becomes unimportant with respect to the molecular information that is coming from it. The selectivity brought about by the molecule–nanostructure interaction can be unwanted or also specifically created, e.g., by adding a partitioning layer, modifying the surface potential, or functionalizing the nanostructure surface.
The outcome of different SERS experiments with the same molecule can vary with the overall sample composition, the affinity of the molecules to the particular surface of the SERS substrate, the position of the nanostructure in the sample, and/or the total surface of the SERS substrate that is available. The latter changes, e.g., when the number of plasmonic nanoparticles changes at the same molecular concentration, or when adsorbing species compete with a specific analyte for the interaction with the nanostructure.
Comparing the situation in SERS with that of non-SERS Raman experiments, strong signal fluctuations occur at low molecule concentration and when high enhancement regimes are used, due to molecular rearrangements that change both the molecule orientation and interaction and (with it) the ‘hot spots’ and the field gradients associated with them.14,15 Moreover, non-resonant Raman cross sections for different types of molecules can vary by orders of magnitude. This variation adds to the selective molecule–metal interactions with a particular SERS substrate and may render molecules with weak signals in multicomponent systems undetected.
For these reasons alone it seems easy to conceive that an application of SERS microscopy to characterize molecular composition and interaction in complex samples has remained a matter of intense debate.16 On the other hand, the fingerprint-like SERS spectra that are yielded when many different types of molecules interact with plasmonic nanostructures in the same focal volume can give quite comprehensive information on the structure and interaction of different components in a complex sample. This can work even at low molecular concentrations. The vibrational information that is obtained from SERS microspectra can provide details on molecular structure and interaction at a detail that is not provided by other microscopies or microspectroscopic tools. More importantly, the nanoscale dimensions of a SERS substrate can highlight specific, nanoscopic environments, such as small regions in cellular compartments.
In summary, several particular challenges must be addressed in order to exploit SERS microscopy for a characterization of complex biosamples. They are posed by
(i) the SERS process itself,
(ii) the control of the molecule–substrate interaction, and
(iii) the complexity of the spectral information that reflects sample composition and structure in addition to a varying enhancement.
Here, we exemplify these aspects and show that attaining a biochemical characterization of complex samples by SERS can be very useful. After a short review of typical approaches to microscopic SERS experiments where we base considerations on plasmonic substrates, that is, the nanostructures that are responsible for the SERS enhancement and their applicability in heterogeneous biological environments in Section 2, we demonstrate different possibilities to use SERS for probing the biochemical composition of such samples in Section 3. As will be briefly reviewed in Section 4, a number of ways to obtain spatially resolved SERS data, and to combine them with images generated by other spontaneous and stimulated Raman processes and other possibilities of multimodal imaging can contribute to a wide applicability of SERS in bioimaging. Some of them can also benefit from the local field enhancement that is important in linear, spontaneous SERS. Section 5 will deal with possibilities to retrieve and interpret the spectral information that is obtained in SERS experiments with complex samples.
2. Substrates for SERS experiments in biosamples
2.1. High local fields due to coupling of LSPR
Understanding, quantifying and controlling the electromagnetic enhancement has been at the heart of basic research in SERS, and has been a main initiator and driver of the broad fields of plasmonics, near field spectroscopy, and optical materials research. The electromagnetic enhancement in SERS often involves the resonance and generation of ‘hot spots’ of high field intensity that, moreover, can be highly focused, and/or that concentrate near sharp features or tips.17,18 The coupling of LSPR of individual nanostructures has been identified as being vital to strong SERS enhancements since the early reports of the effect.19–21Fig. 2(A) shows the enhanced local fields around nanostructures of gold, silver, and copper on glass surfaces for different excitation wavelengths.22 It was suggested by experiment and confirmed by theory that gaps and geometries of nanoaggregates can be optimized, resulting in structures that act as nanolenses,18 where GSERS from regions of high fields in such structures could be on the order of 1011 to 1013.12,13,23 Often, analytical, that is, experimentally determined enhancement factors for microscopic SERS substrates that contain small aggregates of individual nanostructures range from 103 to 109.24,25 This is due to the fact that only very few nanostructures can provide high local fields in experiments where SERS signals are collected from typical diffraction limited focal volumes. While those molecules residing in the hot spots are the main contributors to the spectrum, many of those that are included in the estimate of a (surface) concentration are not represented in the signal.23 A critical discussion of analytical enhancement factors of particular SERS substrates and how they can be determined is conducted in ref. 26.
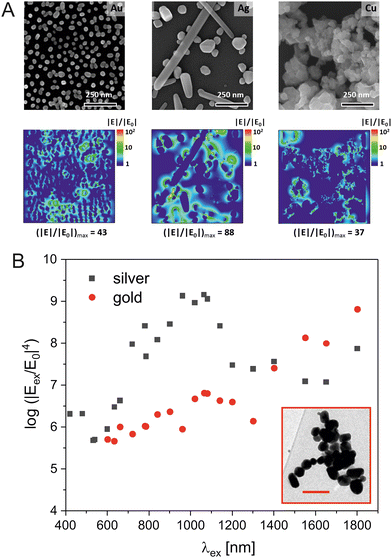 | ||
| Fig. 2 Enhancement of Raman scattering in SERS by enhanced local fields around nanostructures. (A) Scanning electron micrographs of gold, silver, and copper nanoparticles (top row) and distribution of the field enhancement around them obtained by finite-difference time-domain (FDTD) simulations in the xy-plane (bottom row). The field |E| was calculated for a wavelength of 633 nm and normalized to the incoming field |E0|. Reproduced from ref. 22 with permission from Elsevier. Copyright 2020. (B) Electromagnetic enhancement of the intensity of a SERS signal GSERS in the region of the highest field enhancement in a cluster of nanoparticles of silver (black squares) and of gold (red circles) for different wavelengths. Inset: Transmission electron micrograph of the actual silver nanoaggregate that was used for the 3D-FDTD simulations, scale bar: 200 nm. For the simulations of the enhancement by the gold nanostructure, the same nanoaggregate geometry was used as in the silver structure shown in the inset. | ||
Engineered nanosubstrates, including nanoparticles deposited on e.g., surfaces or 3D-structures,27,28 and nanostructures obtained by top-down nanofabrication have several advantages over nanoparticles in droplets, as covered in a wide range of overview works and references cited therein.29,30 They may be beneficial in the analysis of biofluids, including diagnostic applications that involve optofluidics and their incorporation into microfluidic systems.31 They can also be used as substrates for cell cultures, e.g., to enable specific sensing of the outer membrane of animal cells that are grown or deposited on them.32,33
2.2. Fitting SERS substrates to bioanalytical applications
While the SERS enhancement should be maximized through the use of different type, size, morphology and arrangement of nanostructures, the application of SERS to biological samples at the microscopic scale is mostly determined by the need to obtain interaction with the analyte within the (microscopic) probed volumes and in many cases also by the biocompatibility of the plasmonic material that is used.As examples, the surface of nanopores can be functionalized with molecules or inert coating,34 in order to warrant interaction of the molecule with the nanopore, or to achieve wetting,35 so that aqueous molecular solutions can actually be probed. Similarly, the interaction of nanoparticles in aqueous suspensions can rely on the addition of ad-ions,36 functionalization with bioorganic molecules such as alkane thiols,37 or adjusting pH, so that molecules are near the surface. Other applications rely on the fabrication of the SERS substrate within the samples, e.g. the synthesis of silver nanostructures in the presence of bacteria38 and plant samples39 or the electrochemical incorporation of the analyte into plasmonic nanostructures from their biological environment.40 The application of an electric potential in order to direct molecules towards the SERS active structure has proven as useful concept. Often, microelectrodes are used to yield the preferred interaction of molecules in complex mixtures that otherwise would escape an analysis, can provide additional enrichment41 or guide molecules through plasmonic nanopores.42
For an application of SERS in living biomaterials, such as cultured cells, gold nanostructures, being an important tool in biotechnology and diagnostics,43 appear to be the nanomaterial of choice. They are non-toxic, can be easily incorporated into biosystems, and may also provide multifunctionality and other imaging options. They give a high contrast in ultrastructural imaging by electron microscopy and X-ray nanotomography,44 and their high scattering cross section makes them ideal candidates for fast imaging by dark field microscopy.43 Moreover, gold nanoparticles have been used in photothermal therapy and drug delivery and can be functionalized in many different ways.
Even though the electromagnetic SERS enhancement obtained by individual silver nanostructures is higher than that of gold nanostructures, the latter are advantageous for analyzing biosamples. This is the case because often in a sample, nanoaggregates of gold and silver nanoparticles form that show very similar enhancement.45 The dependence of the enhancement on the LSPR for gold and silver nanoaggregates can be represented by excitation profiles across the wavelength ranges in which typical SERS experiments are conducted.46,47Fig. 2(B) shows such profiles for the example of a cluster of nanoparticles from silver and gold, obtained from the results of finite difference time domain (FDTD) simulations48,49 to determine the highest field enhancement that is found in any position of such an aggregate for excitation conditions discussed for typical SERS experiments.47 As can be seen from this plot, the wavelength range in which aggregates of gold nanoparticles provide a relatively stable electromagnetic SERS enhancement of ∼7 orders of magnitude is relatively wide, enabling even an excitation of two-photon excited effects that often use very long wavelengths around 1000 nm, such as surface enhanced hyper Raman scattering (SEHRS) (Fig. 2(B), red symbols).47,50 In an example of typical silver nanoaggregates, assuming the same size range and interparticle spacing as in the gold nanoparticles, the electromagnetic SERS enhancement is only slightly higher (Fig. 2(B), black symbols). Therefore, choosing to work with biocompatible gold nanostructures appears a good option and renders experimental conditions relatively flexible regarding the excitation wavelength. It is in agreement with numerous SERS studies on biosamples that have used excitation wavelengths in the NIR both with silver and gold nanostructures.51
The high SERS enhancement yielded by gold nanostructures is also used in combination with other materials that can have additional properties useful for bioprobing, such as improved control of the structures through magnetic properties,52 or coatings that enable specific interaction of biomolecules in ‘mixed’ samples and that can lead to partitioning of specific species, modify selectivity, and help to control aggregation, such as silica shells or ‘wrapping’ in graphene.27,53,54
2D nanomaterials and their composites with gold and also silver have been frequently applied in bioanalytical SERS.55 Apart from the use of graphene, composite structures of plasmonic particles and black phosphorous were shown to be e.g., specifically efficient intracellular SERS probes with theranostic (efficient photothermal) properties56 or sensitive sensors of exosome composition.57
The use of non-plasmonic SERS probes, such as zinc oxide semiconductor nanoprobes that rely on exciton resonances to obtain improved Raman signals of cells58 or black titanium dioxide particles59 has led to a number of applications in tumor diagnostics and other biodetection applications of SERS. Substrates consisting of titanium dioxide, such as semiconductor structures that can provide an electromagnetic field enhancement by optical bound states in the continuum,60 together with a photoinduced charge transfer, or in microspherical arrays that provide unique Mie resonances61 could be envisioned to be employed for the SERS characterization of complex samples such as cells and tissues as well.
Other non-plasmonic materials that could be further explored in bioanalytical SERS and that use the ‘chemical’ enhancement obtained by beneficial charge transfer conditions include perovskite semiconductors that improve charge transfer through vibronic coupling,62 ceric oxide, demonstrated for a sensitive detection of adenosine triphosphate (ATP),63 or multidimensional carbon-based substrates.64 The main challenge in applying the latter will be their use in the absence of any molecular resonances from probed biomolecules or without relatively high amounts of label compounds that have been added when using the chemical enhancement by carbon structures so far.65
3. Probing biological substructures of cells and tissues
Achieving an interaction of a SERS substrate and an analyte in a complex biomaterial is particularly challenging in the highly compartmentalized structure of intact cells and tissues that have membranes as efficient barriers between the environments of different organelles and cells, respectively. The different reaction spaces resulting from compartmentalization can be characterized by SERS, provided that SERS substrates can reach them. A more detailed discussion of possibilities to obtain SERS spectra from particular organelles and the type of information conveyed by such spectra are provided in ref. 66 and references cited therein. Here we give some examples on possibilities to measure SERS spectra from cellular substructures and the type of molecular information they contain.In order to probe the outer cell membrane, as well as molecules secreted from them, cells can be grown on or deposited on a plasmonic substrate.67,68 Such external substrates have been applied, e.g., to observe secretion processes upon manipulating membrane potential32,67 or to characterize the cell membrane during differentiation in different growth conditions.69 The easy access of the outer cell membrane for different types of nanoparticles has been used to study the composition of the membrane, specifically the presence of specific membrane receptors, through binding of such nanostructures.70 For specific applications, the penetration of the cell membrane by sharp structures, such as the tips of nanostars or nanopipettes has been proposed.71,72 Probing by such ‘needle-type’ SERS substrates may be particularly suitable for a microanalysis of the extracellular space, in cell cultures or even tissues, for sensing of neurotransmitters,73 or metabolites such as glucose,74 or ATP.40
Gold nanoparticles are often used in SERS experiments with living cells. Independent of their final destination in the cellular ultrastructure, they are usually taken up by the cells through the endocytic pathway, from where they must escape in order to get to other compartments, such as the nucleus or the mitochondria (Fig. 3). Insertion of SERS substrates into a cell can be verified directly by Raman microscopy, as the appearance of SERS spectra is related to the position of the plasmonic nanostructure, albeit at the diffraction-limited resolution of the Raman microscope. Nevertheless, while large amounts of nanostructures in cells and tissues usually mean that some SERS spectra can be obtained, this does not always have to be the case, specifically when the formation of nanostructure aggregates is prevented. Comparisons of the amount and signal strength of the spectra with the number of nanoparticles from spatially resolved mass spectrometry75 and their relation to morphology, size, and interparticle distance of nanostructure agglomerates in the sample are critical to the performance of an intracellular SERS substrate (Fig. 3(B)).76 Such a comparison also shows that different cell types treat the same SERS substrate differently,76 and that the surface of nanostructures and incubation conditions can be altered so that aggregate morphology is affected and interaction with the cellular environment is changed.44
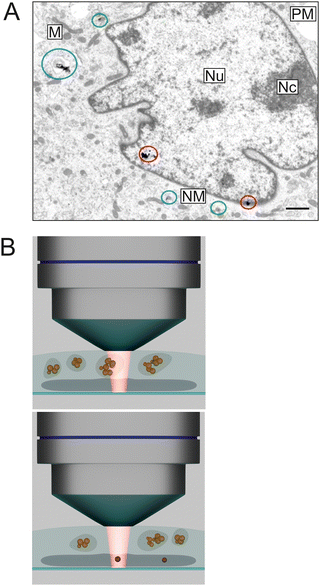 | ||
| Fig. 3 (A) Transmission electron micrograph containing different stages in the nuclear targeting of SERS probes using the endolysosomal pathway in a mouse fibroblast cell. The colored markings indicate particles in endosomes (green), escaping from the vesicles (blue), and entering the cell nucleus (red). Abbreviations: Nu, nucleus; Nc, nucleolus; NM, nuclear membrane; PM, plasma membrane; M, mitochondrion. Scale bar: 1 μm. Modified from ref. 44 with permission from ACS. Copyright 2021. (B) Schematic representing the setting in a SERS experiment after nuclear targeting. Gold nanoparticle probes contained in endolysosomes near the nucleus (top) instead of SERS probes inside the nucleus (bottom) may be sampled. | ||
While the observation of SERS signals from a cell enables subcellular localization of SERS probes and mapping of their position, the chemical information that is contained in the SERS spectra reveals their nanoscopic molecular environment. This information can be related to biomolecules that are brought into the cellular environment together with the SERS probe and that keep interacting with it, known as the nanostructure's corona. They are observed to change over time, e.g., when cellular enzymes fragment its protein component.77 More importantly, they also report molecules that are encountered by the SERS substrate, e.g., along a pathway through the cellular ultrastructure, or over time. The cellular physiology associated with processes of endolysosomal maturation are probably the most studied due to straightforward access of this compartment and their relevance to research in theranostics.78,79 As examples, tracking of autophagy events, important in cell death mechanisms, and the fusion of endolysosomal vesicles that is associated with it can be monitored in SERS experiments.80 An increased use of ATP and its stepwise conversion to adenosine monophosphate (AMP), associated with an enzyme that is crucial for endolysosomal acidification has been observed in one of the first reports on intracellular SERS,81 and cyclic AMP (cAMP) as important messenger can be detected.82 Spectra from endosomes measured with gold nanostars with long tips indicate interaction of the tips with the membrane of the endolysosomes that contain them.66 In addition to protein fragmentation that was observed in vivo in cells,77 and discrimination of spectra from endosomes of different maturation stages,79 the fusion of endolysosomes with vacuoles that contain parasites with their specific molecular environment,83 or the inhibition of enzymes in lipid breakdown (see below)84 have been characterized by SERS microspectroscopy. Approaches to retrieving such molecular information from fingerprint-like information in the spectra are discussed below.
The nuclei of cells have been a ‘popular’ target in SERS, specifically regarding their involvement in programmed cell death or structural changes in response to photo- or thermo-induced stress that could be relevant in tumor theranostics.85 Nuclear localization sequences (NLS), small peptides that usually help trafficking of proteins to the nucleus, can be used to make SERS nanoprobes escape the endolysosomal system, enter the cytoplasm, and eventually pass the nuclear pore complex (Fig. 3(A)).44 It is important to note that the conditions under which this endosomal escape and incorporation of the structures in the nucleus occurs are quite specific, and that a large fraction of the optical nanoprobes could still remain inside the endolysosomes and not escape (Fig. 3(A)).44 Consistent with results from electron microscopy and nanotomography, SERS spectra with bands related to nucleic acid vibrations could be obtained from the nuclei of cells of a sample where nuclear localization took place.44 Moreover, the attempt of nanoaggregates to escape the endosomes was accompanied by a particular biomolecular environment of the particle agglomerates.44
The possibility to probe cells and their substructures by SERS enables the application of such approaches to attain a better understanding of molecular mechanisms in tumor biology and also other pathologies such as degenerative or storage disorders. In the diagnostic context, SERS can help identify cancer biomarkers and observe the effects of anti-tumor drugs. The large number of applications, possible approaches with their advantages and drawbacks have been the subject of several comprehensive reviews and references therein, e.g., ref. 86 and 87.
4. Imaging samples with SERS
4.1. Maps from SERS microspectra of biological molecules
The signal in a SERS experiment comes from a volume limited by the nanometer-scaled local optical field generated by a nanostructure that acts as SERS substrate. The separation of several of such small volumes is often determined by the diffraction-limited resolution of the microscope. Different probed volumes in a sample, usually in the range of several tens to hundreds of femtoliters, may or may not result in a SERS spectrum, depending on the presence of a SERS substrate in them. This has consequences when SERS spectral information is used to produce images or chemical maps of a sample.In experiments where all probed volumes contain at least one ‘hot spot’ or nanostructure that gives sufficient SERS enhancement, microspectroscopic maps of the complete sample can be reconstructed. Unless ways are applied that overcome consequences of the diffraction limit (see below), further information on the number of ‘hot spots’ or their distribution within the volume cannot be generated. The definition of the excitation volume determines the separation of several of such volumes and the resolution of the resulting SERS map. In contrast, in experiments where not all sampled volumes yield a SERS spectrum, distinction can be made between probed volumes that do not contain any SERS active nanostructure and those that do. In such experiments, any SERS signal that is obtained indicates e.g., the successful delivery or targeting of a location in a cell or tissue with a SERS nanoprobe. Independent of the microscopic distribution of the SERS substrate in a sample, the spectra always reveal molecular composition, structure and interaction in the nanoscopic probed volume, near the SERS substrate. This can place specific molecular components in the ‘spotlight’ and hide others that are present as well.
The use of SERS microscopic mapping to assign specific molecular composition and structure to different microscopic regions in complex biosamples is obvious and was demonstrated by many, see, e.g., ref. 66. Since nuclear targeting usually is not exclusive and involves endosomal transport as well (Fig. 3(A)),44 applying SERS mapping within the nucleus of a cell is quite challenging, mostly since typical diffraction-limited focal volumes often include perinuclear regions of the cytoplasm with (additional) probes in endolysosomes in the same diffraction-limited spot (Fig. 3(B)).
SERS maps of the cytoplasmic region with all its organelles typically reveal the composition of endosomes, vacuoles, mitochondria, or the cytosol, depending on where the SERS active nanoparticles have been placed. As all other probed spots do not yield a spectrum, the Raman map does not give a full image of the whole sample but must be superimposed with a bright field or other type of micrograph. The small size of some of the organelles can lead to probing of more than one organelle per sampled volume (Fig. 3(B)), as is the case in early-stage endosomes that can have the size of individual small aggregates of SERS probes, and that constitute distinct biochemical reaction spaces with individual nanoenvironments and chemical composition.
When growing cells on the surface of a gold nanoisland substrate, where each micron-sized spot yields high, uniform SERS enhancement, the cell membrane that is in nm-scaled proximity of the gold structures can be probed selectively, and the full area of the cell surface can be mapped.68,69 When chemical images are generated using the intensity of bands assigned to the vibrational modes in lipids and proteins with the corresponding bright-field images of the cells, almost every spectrum, that is, every pixel, in the area of the cell contains a SERS signal.68 After analyzing such spectra collected from intact living cells, maps of SERS signals that correspond to different orders of the lipid acyl chains, different lipid polar head, such as in phosphatidylethanolamine, cholesterol, and potential anchors for proteins in the cell membrane can be reconstructed.68 They indicate micron sized areas of several sampled spots where lipids in the liquid-ordered phase and cholesterol are present in close proximity, separate from spots of more unordered lipid tails.68 The SERS maps are in agreement with suggestions of an organization of the cell membrane in micron sized domains that came from work with fixed samples or fluorescence labels but that had not been watched in unlabeled living cells. The possibility to connect the state-of-order of the lipids in the cell membrane of the live cells to the presence and structure of other molecular species can be useful to better understand the cell membrane and its biochemistry. Probing of the locally high SERS signals of cell membranes in an evanescent field, when the cells are deposited on a regular array of high local fields, can improve the optical resolution of SERS maps compared to the conventional diffraction-limited Raman microscopic maps by up to an order of magnitude further.88,89
4.2. Super-resolution in SERS microscopy
In the past three decades, super-resolution microscopy has revolutionized the microscopic imaging of biosamples. It relies on mechanisms that allow for an observation of selected emitters in volumes with an effective size that is smaller than the diffraction limit. While the super-resolution techniques that use fluorescing emitters, such as stimulated emission depletion (STED) microscopy, stochastic optical reconstruction microscopy (STORM) or photoactivated localization microscopy (PALM) need fluorophores, often introduced as labels, diminishing of the probed volumes in super-resolution Raman microscopy does not rely on external tags. Far-field super-resolution vibrational microscopy approaches that use, e.g., the saturation of local vibrations, Raman suppression, or structured illumination are discussed in excellent overviews4,90 and references therein.The SERS process relies on extremely localized high fields that provide further possibilities for selecting emitting hot spots or nanostructures in a sample to achieve super-resolution probing.91 The fluctuations or ‘blinking’ of the SERS signals due to a transient formation of hot spots can be treated in the same fashion as the blinking of fluorescence signals,15 and processed by the STORM algorithm.92 As an example, such a stochastic imaging based on the blinking behavior of SERS signals,15 can be used with dynamic illumination conditions to resolve very small biological structures, such as collagen fibrils93 and individual bacteria.94 Super-resolution SERS microscopy was also used for probing of individual nanorods and their rotation inside macrophage endolysosomes,95 and binding events of gold nanostars to receptors in the cell membrane, indicating different dynamics and uptake for nanoparticles depending on their binding mechanism to the cell surface.96
4.3. SERS in multimodal linear and non-linear imaging
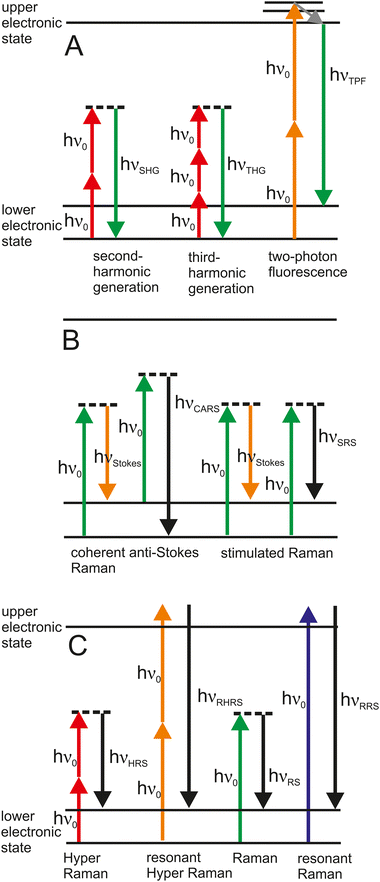
While SERS data as vibrational spectra give detailed information on molecular composition and structure, other optical signals, both linear and non-linear, are less specific but can be used to characterize the morphology of the sample or highlight some of its specifics or those of the SERS probes themselves. Therefore, combinations of SERS with e.g., elastic scattering signals, fluorescence, or second harmonic generation are particularly beneficial for microspectroscopic studies of biological objects. SERS experiments conducted in a microscope typically enable the acquisition of such other types of spectra and images from the same sample.Bioprobing that combines SERS with different modalities of fluorescence spectroscopy and imaging has developed into a broad field that bridges research in biodiagnostics with materials design and radiative decay engineering and can be exploited in different ways. Recent overviews on this topic such as97 and references therein exemplify the different ways how both effects can be used in a synergistic fashion–beyond a known quenching of fluorescence of molecules when in close proximity of a metal structure that is the prerequisite for SERS. Efficient combinations of SERS and fluorescence imaging include, among others, bimodal SERS/fluorescence probes that can benefit from a plasmonic enhancement of the fluorescence, the fluorescence-free SERS detection of a molecule in the presence of a plasmonic moiety, often exploiting molecular resonance for surface-enhanced resonant Raman scattering (SERRS) (and its fluorescence in the absence of the SERS substrate), or label-free SERS probing of certain cellular compartments while others are labeled with fluorescence probes. Apart from experiments at optimum excitation wavelengths, combined SERS and fluorescence experiments can benefit greatly from fluorescence background suppression, such as optical Kerr gating98 and shifted excitation Raman difference spectroscopy.99
Two-photon excited fluorescence imaging has been established in microscopy and yields unprecedented contrast for imaging and multiplexing of different biological fluorophores (Fig. 4(A)).100 Excitation in the near infrared, also suitable for bio-SERS experiments, can give rise to strong signals of two-photon excited luminescence of the gold nanoaggregates used as SERS substrates that can greatly exceed the two-photon fluorescence cross sections of good organic fluorophores.101,102
The scattering signals at the LSPR wavelengths of the same plasmonic nanoparticles that are used as SERS substrates enable their very sensitive localization by dark field microscopy, due to the high scattering cross-sections. Possibilities to visualize plasmonic nanoparticles by microscopy based on their scattering signal, e.g., by dark field microscopy, differential interference contrast, or interferometric scattering microscopy are discussed e.g., in ref. 43 and references therein. Hyperspectral mapping of LSPR using dark field scattering microspectra gives important information on the optical properties of the nanoaggregates and individual nanoparticles in situ in the sample.103,104 To mention only a few examples in a large body of works, imaging of plasmonic particles after their delivery into cultured cells can suggest nuclear targeting105 or indicates the binding of SERS probes to membrane receptors.106
Fast imaging of nanostructures in/and of biomaterials can be achieved with the non-linear effects of second harmonic generation (SHG) and third harmonic generation (THG).107 SHG is a parametric two-photon process, where interaction with a non-centrosymmetric structure, such as a nanoparticle or biomacromolecule produces an effective combination of two photons into a single photon of twice the energy of the incident light (Fig. 4(A)). Strongly enhanced SHG and THG have been reported in the presence of plasmonic nanoparticles.108,109 In experiments that combine SERS with SHG, nanoprobes deliver chemical information through a (linear) SERS spectrum, conveying molecular composition and structure, and also can act as harmonic probes for fast (non-linear) SHG imaging that reveals the morphological and functional structures in a sample.110 As an example, gold-coated barium titanate particles Au@BaTiO3 were used to study the endolysosomal compartment by SERS. The particles were coated by lipids, and later in the cell showed characteristic SERS signals of proteins in the endolysosomal environment. Fast image acquisition based on the strong SHG signal that is provided by the barium titanate moiety of the structures showed the positions of the probes in the cellular ultrastructure.
Non-linear processes are attractive for microscopy and spectroscopy since they can be excited with light in the near-infrared. The advantages of long excitation wavelengths include deep tissue penetration capability, reduced photodamage due to the lower photon energy, and spatially more confined probed volumes. Non-linear Raman microscopy that uses either stimulated processes such as stimulated Raman scattering (SRS)3 and coherent anti-Stokes Raman scattering (CARS),111 as well as microscopy with the spontaneous process of hyper Raman scattering (HRS)112 can make specific use of an enhancement by SERS, for reviews see ref. 50, 109 and 113 and references therein. This is the case, because their excitation relies on more than one photon, which increases a contribution by the enhancement for each excitation field. Fig. 4(B) depicts different excitations of Raman scattering that originate from a coherent interaction of photons from two lasers in SRS and CARS and the respective enhanced processes of surface-enhanced SRS (SE-SRS) and surface-enhanced CARS (SE-CARS), respectively. SE-CARS was shown to be applicable in immunohistochemical imaging of cells and tissues,114,115 and can enable temporally and spatially highly resolved probing.116,117Fig. 4(C) shows the spontaneous process of hyper Raman scattering that can be used as surface-enhanced HRS (SEHRS), in the non-resonant and the resonant case.
The spectral information in SEHRS obtained can complement that from SERS, as HRS is governed by different selection rules than spontaneous Raman scattering.50 SEHRS probing of the endolysosomal system of cells was one of the first applications of non-resonant SEHRS in biological probing,118 and the combination of SEHRS with SERS for hyperspectral imaging was demonstrated.119
5. Retrieving meaningful information from SERS microspectra
5.1. Role of molecular models
The examples in the previous sections have illustrated the wealth of molecular information that are contained in SERS microspectra of biosamples. Spectra of pure biomolecules, e.g. from lipids, proteins, and nucleic acids, collected with identical SERS substrates and excitation conditions as the experiments in the biosystem can be very helpful for the interpretation of SERS spectra of complex biomaterials where different molecular species co-occur and interact. However, the large changes that can result in SERS spectra even from small variations in the molecular environment necessitate experiments that can capture potential variance encountered in the real, complex environment as well.68,77 Different selectivity and changes in the SERS enhancement when charge, pH or concentration change, cause a variation of relative intensities of spectral bands or the absence of vibrations of particular functional groups. As a consequence, the identification of distinct molecular components by state-of-the-art multivariate chemometric tools is often not possible, in spite of the high sensitivity of SERS and the specific selectivity that is even enhanced in many experimental settings. Therefore, comprehensive data sets collected from model compounds should contain variations in e.g., concentration, secondary structure, fragmentation, varied interaction and enhancement, or specific type of molecular sequence.77,120–124 Such a collection of spectra under a large set of (relevant) experimental conditions would yield a ‘profile’ of a particular type of molecule that would allow its robust recognition in a multitude of interactions with a SERS substrate.120,125,1265.2. Pre-processing of SERS spectra
Depending on different spectral contributions and utilization of the spectra, e.g., due to plasmonic or molecular resonances or additional optical signals, SERS data may require a different amount of pre-processing.127–129 While often pre-processing is necessary to make relevant molecular information accessible at all, it may also have unintended negative impact on the analysis, such as loss of important information in the ‘background’, shifted signals, changed signal intensity, or loss in spectral resolution.130 Modern deep learning applications promise to be able to extract even more minuscule amounts of useful information from largely noisy data, albeit at the risk of overfitting to that noise.131,132Apart from experiment-specific pre-processing steps, e.g., sorting of data or background correction,127,133 smoothing, de-noising, truncating the spectra to a range of interest, or normalization can be applied,130 with variations of the outcome depending on the order in which the different procedures are applied.134–137 In order to avoid the propagation of errors and to verify whether the applied method has achieved the intended effect, inspection of spectra after individual processing steps is helpful. While there are studies on the effects of pre-processing on various kinds of spectral data for machine learning applications,138,139 detailed evaluations of pre-processing for SERS data are relatively rare.127,140 Most procedures of spectral data pre-processing also apply to SERS data, but the high variability found in spectral features of SERS data may require additional attention. As an example, enhancement factors may vary greatly within a sample and over time e.g., in suspension samples (cf.Fig. 1), where nanoparticles move in and out of a focal volume.141 Some promising approaches have been proposed for a pre-treatment of SERS data, such as wavelet-based noise reduction and correction for different artefacts or nonrelevant information.119,130,142–144 Absolute intensity values and also relative signal strengths of different vibrations in spectra become much less important and may in many cases be replaced by information of whether a signal occurs in a spectrum at all or not, converting the spectrum into a type of ‘barcode’.77,145
5.3. Analysis of complex SERS data with machine learning approaches
Since SERS experiments generate complex data with many spectra and fluctuating signals, it is useful to analyze them with methods of multivariate statistics. These methods are characterized by the fact that all spectral variables can be analyzed simultaneously. They can be divided into supervised and unsupervised approaches. The most frequently used unsupervised approach is principal component analysis (PCA).146,147 In PCA, uncorrelated principal components are obtained by linear combinations of the spectral variables to analyze the main variances in the data. However, as the main variances do not necessarily correspond to the information of interest, supervised machine learning methods can be used to train a model on specific differences. The aim of the models can be either obtaining a regression, i.e., predicting a quantitative target variable such as a concentration, or achieving classification, i.e., differentiating between groups. Many different approaches can be used to obtain corresponding models, e.g., partial least squares regression (PLS),148,149 support vector machine (SVM),150,151 random forest (RF)152 and artificial neural network (ANN).153,154 Another possibility is to combine PCA with a supervised approach, as was conducted for example for the classification of bacteria based on their SERS spectra.155 An overview of the algorithms used and current applications to different types of SERS data is given in ref. 156 and references therein.ANNs are often referred to as the approach of machine learning and artificial intelligence, because of their broad successful use in many fields.157 They have also been applied for the analysis of SERS data of several types of biological samples, e.g., for food analysis,158 forensics,159,160 palynology,161 or medical diagnostics.162 Due to the variability of SERS signals, convolutional neural networks are particularly well suited and perform very well,132 because they contain convolutional and pooling layers that summarize features that may be present in neighboring data points. However, the challenge with the application of ANNs is that they require a very large amount of training data and are often used as a ‘black box’, i.e., it is not clear how the method arrives at its results. Approaches have been developed to address these challenges, e.g., to increase the amount of training data through data augmentation,135,163 and visualization of the filters and outputs of neural networks to ‘peek into the black box’.164 For both reasons, it makes sense to also use machine learning methods other than neural networks, that may help to better reveal and analyze particular chemical differences between experiments or samples. Approaches from bioinformatics, originally developed for the analysis of other complex data, such as gene expression data, are particularly promising also for the analysis of SERS data, as will be discussed in more detail below.
5.4. Potential of approaches from bioinformatics
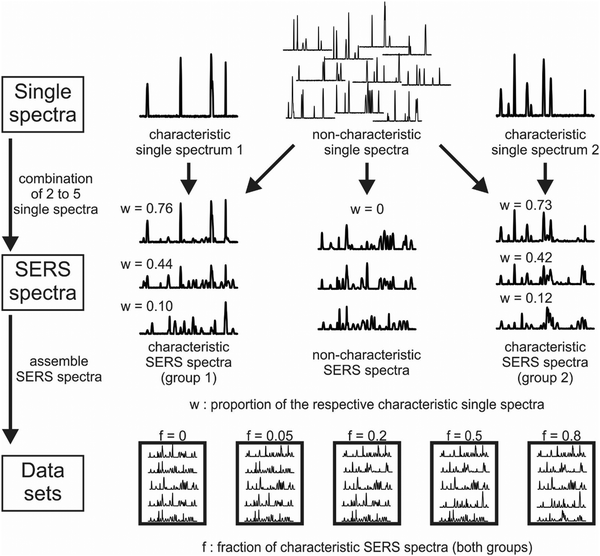 | ||
| Fig. 5 Schematic of a simulation process of SERS data. Top row: A set of single spectra were generated and used as a proxy for SERS signals occurring together in an experiment, e.g., because they originate from one molecule Middle row: They are randomly combined to simulate SERS spectra composed of different components. One respective single spectrum occurs exclusively in the spectra of one of the two data sets in order to simulate unique properties. The proportion of the characteristic single spectrum in a combined SERS spectrum is specified by a parameter w. Bottom row: In this way, numerous SERS spectra are simulated, which are then assembled in whole sets of SERS data, and the fraction of spectra that contain the characteristic spectrum is defined by a parameter f. By varying the parameters w and f, the extent of specific properties represented by the characteristic single spectra can be adjusted in the SERS spectra and data sets and the performance of machine learning methods, e.g., in spectral classification tasks, can be compared. Moreover, since the true differences between the data sets (the characteristic spectra) are known, they can be compared to the results of variable selection, relation analysis and pathway-guided approaches. Reproduced from ref. 172 with permissions from the authors (CC-BY licence). Copyright S. Seifert 2020. | ||
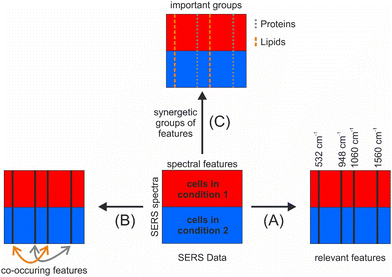
The incorporation of relationships into the selection can be achieved by utilizing so-called surrogate variables, which were originally conceived to compensate for missing values in random forests by the determination of the respective variables with the most similar performance in the nodes of the decision trees.152 For SERS data, surrogate variables may represent a spectral feature of the same molecule or specific interaction of that molecule with other molecules or the nanostructure. To analyze the mutual impact of the variables on the random forest model, surrogate variables can also be exploited by the calculation of the so-called mean adjusted agreement, a parameter that can be described as a supervised correlation coefficient.175 This parameter, which has recently been further developed to analyze qualitative variables such as those in genomic data,176 has been used for food characterization, e.g. based on nuclear magnetic resonance (NMR)177 and liquid chromatography–mass spectrometry (LC–MS) data,178,179 and to analyze relationships between mass spectrometric and spectral variables.180 Used on SERS data, the mean adjusted agreement additionally enables the analysis of co-occurring signals (Fig. 6(B)), which translates to the possibility of identifying different molecules that are located in the same nanoscopic volume in a SERS experiment. The potential of this analysis was demonstrated by the ability to study lipid-antidepressant interactions in living cells, which was previously not possible.84 In this example, antidepressants that were shown to produce very strong and distinct SERS and SEHRS spectra on different substrates, indicating a specific interaction of the molecules with the plasmonic nanostructures181 were delivered into the same compartment within living cells as the SERS probes were. In the cell, the drugs are known to inhibit acid sphingomyelinase, an important enzyme in lipid metabolism that is exclusively located in the endolysosomal environment. This enzyme inhibition leads to the accumulation of the lipid sphingomyelin, as verified by electron microscopy.84 Potential signals of the drugs inside the cells were greatly diminished or difficult to distinguish from the spectral contributions of other molecules in the endolysosomal environment.84 Moreover, sphingomyelin, as well as other lipids, are not conspicuous in SERS due to a small Raman cross-section compared to other molecules in the endolysosomal environment. Typical multivariate tools failed to identify differences between drug-treated and untreated cells. The variables selected by SMD were characteristic of the drug molecule, and the mean adjusted agreement revealed relationships with SERS signals assigned to lipid or protein molecules.84 The co-occurrence of lipid and drug signals could support the hypothesis that the tricyclic antidepressant drugs interact with the lysosomal membrane.84 Moreover, the selected and related variables identified by SMD were then used for improved chemical mapping, aiding the co-localization in the microscopic maps of the cells.
To achieve the third goal of relating signals in important groups of features (Fig. 6(C)), pathway-guided random forest methods are very promising.172 These approaches also originate from the application of bioinformatics methods in the medical field and were developed to integrate external knowledge about structure and functional relationships into the evaluation of ‘omics’ variables.182,183 They evaluate entire pathways, i.e. groups of interacting variables, regarding their importance for the analyzed outcome, instead of individual variables as done by the variable selection methods mentioned above. For SERS, these approaches enable to test for differences with respect to specific biomolecule groups such as lipids or specific compounds in mixtures that have previously been individually characterized and hence have a known SERS profile.
6. Conclusions and outlook
As discussed here, SERS spectra show the structure and composition of molecules in the interaction with nanomaterials, with unprecedented detail and sensitivity. Making use of its potential necessitates a good understanding of the experimental conditions and the possible information content of the data. In the future, specifically the combination of SERS microspectra with other linear and non-linear microspectroscopic approaches, and the possibilities to employ a plasmonic enhancement of the latter, such as studies combining SERS with CARS or SRS to SE-CARS or SE-SRS, as well as plasmon-enhanced harmonic generation microscopies appear particularly promising and will help to combine as much chemical and morphofunctional information as possible with the sensitive vibrational probing by SERS. In this regard, exploring multifunctional SERS substrates that can provide LSPR in the near infrared range and that can be combined with e.g., low dimensional materials with suitable electronic resonances, and/or high non-linear susceptibilities will be interesting.In order to understand SERS data from the complex mixtures and interactions of biomolecules in cells or tissues, it is useful to analyze SERS data of biomolecular systems at different levels of complexity, e.g., by acquiring spectra from purified compounds or their combinations. Nevertheless, the great variation that is encountered even upon small changes in a SERS experiment demands for robust data analysis tools that can take into account the representation of one particular molecule in its interactions with a SERS substrate and possibly also other molecules by a multitude, or combinations, of spectral features. As was discussed here, approaches that have been used in bioinformatics and that are becoming harnessed for the analysis of SERS data include both modeling approaches, that capture the specifics of SERS signals, as well as an adequate analysis of experimental spectra. Spectral analysis by random forest methods can -apart from a classification and identification that is common to many other machine learning tools- enable the selection of important spectral features for a direct structural interpretation, as well as the identification of co-occurring spectral features. A reliable identification of spectral features that are characteristic of molecules and their interactions can enable completely new approaches to generate image contrast in SERS microscopy.
The development of multi-purpose data analysis frameworks will rely on robust, well-controlled experiments that take into account simulated data, together with knowledge from different experiments, performed under reproducible experimental and standardized data pre-processing conditions.26,140 In future applications of SERS, the selection of spectral features by machine learning can indicate reasons for classification results, e.g., for the discrimination of different biochemical ‘states’ in healthy and diseased cells. More importantly, together with the possibilities regarding lateral (super) resolution and monitoring of temporal changes enabled by SERS, they will yield a better understanding and identification of specific interactions of molecules in biological systems.
Data availability
The Tutorial review is largely discussing published work. The original data underlying some of the figures will be made available by the authors upon reasonable request.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
J. K. would like to thank Heinrich Kittel and Arpad Dusa for help in preparation of parts of the figures, and Katrin Kneipp for discussions. The authors gratefully acknowledge funding by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - 511107129.References
- G. J. Puppels, F. F. M. de Mul, C. Otto, J. Greve, M. Robert-Nicoud, D. J. Arndt-Jovin and T. M. Jovin, Nature, 1990, 347, 301–303 CrossRef CAS PubMed.
- J.-X. Cheng, A. Volkmer and X. S. Xie, J. Opt. Soc. Am. B, 2002, 19, 1363–1375 CrossRef CAS.
- F.-K. Lu, S. Basu, V. Igras, M. P. Hoang, M. Ji, D. Fu, G. R. Holtom, V. A. Neel, C. W. Freudiger, D. E. Fisher and X. S. Xie, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11624–11629 CrossRef CAS PubMed.
- C. T. Graefe, D. Punihaole, C. M. Harris, M. J. Lynch, R. Leighton and R. R. Frontiera, Anal. Chem., 2019, 91, 8723–8731 CrossRef CAS PubMed.
- R. E. Leighton, A. M. Alperstein, D. Punihaole, W. R. Silva and R. R. Frontiera, J. Phys. Chem. B, 2023, 127, 26–36 CrossRef CAS PubMed.
- T. Deckert-Gaudig, A. Taguchi, S. Kawata and V. Deckert, Chem. Soc. Rev., 2017, 46, 4077–4110 RSC.
- R. Zhang, Y. Zhang, Z. C. Dong, S. Jiang, C. Zhang, L. G. Chen, L. Zhang, Y. Liao, J. Aizpurua, Y. Luo, J. L. Yang and J. G. Hou, Nature, 2013, 498, 82–86 CrossRef CAS PubMed.
- J. Kneipp, H. Kneipp and K. Kneipp, Chem. Soc. Rev., 2008, 37, 1052–1060 RSC.
- L. Jensen, C. M. Aikens and G. C. Schatz, Chem. Soc. Rev., 2008, 37, 1061–1073 RSC.
- S.-Y. Ding, E.-M. You, Z.-Q. Tian and M. Moskovits, Chem. Soc. Rev., 2017, 46, 4042–4076 RSC.
- C. Chen, J. A. Hutchison, P. Van Dorpe, R. Kox, I. De Vlaminck, H. Uji-i, J. Hofkens, L. Lagae, G. Maes and G. Borghs, Small, 2009, 5, 2876–2882 CrossRef CAS PubMed.
- K. Kneipp, Y. Wang, H. Kneipp, L. T. Perelman, I. Itzkan, R. R. Dasari and M. S. Feld, Phys. Rev. Lett., 1997, 78, 1667 CrossRef CAS.
- S. Nie and S. R. Emory, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- H. Xu, E. J. Bjerneld, M. Käll and L. Börjesson, Phys. Rev. Lett., 1999, 83, 4357–4360 CrossRef CAS.
- N. C. Lindquist, C. D. L. de Albuquerque, R. G. Sobral-Filho, I. Paci and A. G. Brolo, Nat. Nanotechnol., 2019, 14, 981–987 CrossRef CAS PubMed.
- J. Baumberg, S. Bell, A. Bonifacio, R. Chikkaraddy, M. Chisanga, S. Corsetti, I. Delfino, O. Eremina, C. Fasolato, K. Faulds, H. Fleming, R. Goodacre, D. Graham, M. Hardy, L. Jamieson, T. Keyes, A. Królikowska, C. Kuttner, J. Langer, C. Lightner, S. Mahajan, J. F. Masson, H. Muhamadali, M. Natan, F. Nicolson, E. Nikelshparg, K. Plakas, J. Popp, M. Porter, D. Prezgot, N. Pytlik, S. Schlücker, A. Silvestri, N. Stone, Z. Q. Tian, A. Tripathi, M. Willner and P. Wuytens, Faraday Discuss., 2017, 205, 429–456 RSC.
- P. K. Aravind, A. Nitzan and H. Metiu, Surf. Sci., 1981, 110, 189–204 CrossRef CAS.
- K. R. Li, M. I. Stockman and D. J. Bergman, Phys. Rev. Lett., 2003, 91, 227402 CrossRef PubMed.
- T. E. Furtak and J. Reyes, Surf. Sci., 1980, 93, 351–382 CrossRef CAS.
- M. I. Stockman, V. M. Shalaev, M. Moskovits, R. Botet and T. F. George, Phys. Rev. B: Condens. Matter Mater. Phys., 1992, 46, 2821–2830 CrossRef PubMed.
- F. Le, D. W. Brandl, Y. A. Urzhumov, H. Wang, J. Kundu, N. J. Halas, J. Aizpurua and P. Nordlander, ACS Nano, 2008, 2, 707–718 CrossRef CAS PubMed.
- Z. Zhang, Y. Li, J. Frisch, M. Bär, J. Rappich and J. Kneipp, J. Catal., 2020, 383, 153–159 CrossRef CAS.
- J. P. Camden, J. A. Dieringer, Y. Wang, D. J. Masiello, L. D. Marks, G. C. Schatz and R. P. Van Duyne, J. Am. Chem. Soc., 2008, 130, 12616–12617 CrossRef CAS PubMed.
- J. Kneipp, X. Li, M. Sherwood, U. Panne, H. Kneipp, M. I. Stockman and K. Kneipp, Anal. Chem., 2008, 80, 4247–4251 CrossRef CAS PubMed.
- V. Joseph, M. Gensler, S. Seifert, U. Gernert, J. P. Rabe and J. Kneipp, J. Phys. Chem. C, 2012, 116, 6859–6865 CrossRef CAS.
- S. E. J. Bell, G. Charron, E. Cortés, J. Kneipp, M. L. de la Chapelle, J. Langer, M. Procházka, V. Tran and S. Schlücker, Angew. Chem., Int. Ed., 2020, 59, 5454–5462 CrossRef CAS PubMed.
- J.-F. Li, J. R. Anema, T. Wandlowski and Z.-Q. Tian, Chem. Soc. Rev., 2015, 44, 8399–8409 RSC.
- K. R. Phillips, G. T. England, S. Sunny, E. Shirman, T. Shirman, N. Vogel and J. Aizenberg, Chem. Soc. Rev., 2016, 45, 281–322 RSC.
- M. Hardy and P. Goldberg Oppenheimer, Nanoscale, 2024, 16, 3293–3323 RSC.
- R. Pilot, R. Signorini, C. Durante, L. Orian, M. Bhamidipati and L. Fabris, Biosensors, 2019, 9, 57 CrossRef CAS PubMed.
- R. Panneerselvam, H. Sadat, E. M. Höhn, A. Das, H. Noothalapati and D. Belder, Lab Chip, 2022, 22, 665–682 RSC.
- V. Caprettini, J. A. Huang, F. Moia, A. Jacassi, C. A. Gonano, N. Maccaferri, R. Capozza, M. Dipalo and F. De Angelis, Adv. Sci., 2018, 5, 1800560 CrossRef PubMed.
- W. Wang and P. J. Vikesland, Anal. Chem., 2023, 95, 18055–18064 CrossRef CAS PubMed.
- R. Kodiyath, T. A. Papadopoulos, J. Wang, Z. A. Combs, H. Li, R. J. C. Brown, J.-L. Brédas and V. V. Tsukruk, J. Phys. Chem. C, 2012, 116, 13917–13927 CrossRef CAS.
- C. Chen, X. Xu, Y. Li, H. Jans, P. Neutens, S. Kerman, G. Vereecke, F. Holsteyns, G. Maes, L. Lagae, T. Stakenborg and P. van Dorpe, Chem. Sci., 2015, 6, 6564–6571 RSC.
- A. Stefancu, S. D. Iancu and N. Leopold, J. Phys. Chem. C, 2021, 125, 12802–12810 CrossRef CAS.
- K. E. Shafer-Peltier, C. L. Haynes, M. R. Glucksberg and R. P. Van Duyne, J. Am. Chem. Soc., 2003, 125, 588–593 CrossRef CAS PubMed.
- N. E. Dina, H. Zhou, A. Colniţa, N. Leopold, T. Szoke-Nagy, C. Coman and C. Haisch, Analyst, 2017, 142, 1782–1789 RSC.
- V. Joseph, F. Schulte, H. Rooch, I. Feldmann, I. Dörfel, W. Österle, U. Panne and J. Kneipp, Chem. Commun., 2011, 47, 3236–3238 RSC.
- H. Zhou and J. Kneipp, Vib. Spectrosc., 2024, 131, 103661 CrossRef CAS.
- H. Zhou and J. Kneipp, Anal. Chem., 2023, 95, 3363–3370 CrossRef CAS PubMed.
- Y. Zhao, M. Iarossi, A. F. De Fazio, J.-A. Huang and F. De Angelis, ACS Photonics, 2022, 9, 730–742 CrossRef CAS PubMed.
- Y. Wu, M. R. K. Ali, K. Chen, N. Fang and M. A. El-Sayed, Nano Today, 2019, 24, 120–140 CrossRef CAS.
- D. Drescher, T. Büchner, P. Schrade, H. Traub, S. Werner, P. Guttmann, S. Bachmann and J. Kneipp, ACS Nano, 2021, 15, 14838–14849 CrossRef CAS PubMed.
- K. Kneipp, H. Kneipp and J. Kneipp, Acc. Chem. Res., 2006, 39, 443–450 CrossRef CAS PubMed.
- M. D. Doherty, A. Murphy, J. McPhillips, R. J. Pollard and P. Dawson, J. Phys. Chem. C, 2010, 114, 19913–19919 CrossRef CAS.
- A. Dusa, F. Madzharova and J. Kneipp, Front. Chem., 2021, 9, 680905 CrossRef CAS PubMed.
- V. Myroshnychenko, J. Rodríguez-Fernández, I. Pastoriza-Santos, A. M. Funston, C. Novo, P. Mulvaney, L. M. Liz-Marzán and F. J. García de Abajo, Chem. Soc. Rev., 2008, 37, 1792–1805 RSC.
- Z. Zeng, Y. Liu and J. Wei, TrAC, Trends Anal. Chem., 2016, 75, 162–173 CrossRef CAS.
- F. Madzharova, Z. Heiner and J. Kneipp, Chem. Soc. Rev., 2017, 46, 3980–3999 RSC.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- T. Buchner, D. Drescher, V. Merk, H. Traub, P. Guttmann, S. Werner, N. Jakubowski, G. Schneider and J. Kneipp, Analyst, 2016, 141, 5096–5106 RSC.
- Y. Zhang, Y. Zou, F. Liu, Y. Xu, X. Wang, Y. Li, H. Liang, L. Chen, Z. Chen and W. Tan, Anal. Chem., 2016, 88, 10611–10616 CrossRef CAS PubMed.
- J.-F. Li, Y.-J. Zhang, S.-Y. Ding, R. Panneerselvam and Z.-Q. Tian, Chem. Rev., 2017, 117, 5002–5069 CrossRef CAS PubMed.
- H. Huang, W. Feng and Y. Chen, Chem. Soc. Rev., 2021, 50, 11381–11485 RSC.
- Z. Liu, H. Chen, Y. Jia, W. Zhang, H. Zhao, W. Fan, W. Zhang, H. Zhong, Y. Ni and Z. Guo, Nanoscale, 2018, 10, 18795–18804 RSC.
- C. Lin, S. Liang, Y. Peng, L. Long, Y. Li, Z. Huang, N. V. Long, X. Luo, J. Liu, Z. Li and Y. Yang, Nano-Micro Lett., 2022, 14, 75 CrossRef CAS PubMed.
- R. Haldavnekar, K. Venkatakrishnan and B. Tan, Nat. Commun., 2018, 9, 3065 CrossRef PubMed.
- J. Lin, W. Ren, A. Li, C. Yao, T. Chen, X. Ma, X. Wang and A. Wu, ACS Appl. Mater. Interfaces, 2020, 12, 4204–4211 CrossRef CAS PubMed.
- H. Hu, A. K. Pal, A. Berestennikov, T. Weber, A. Stefancu, E. Cortés, S. A. Maier and A. Tittl, Adv. Opt. Mater., 2024, 12, 2302812 CrossRef CAS.
- J. Li, Q. Xie, J. Li, L. Sun, Y. Xie, Y. Ozaki and W. Ji, Adv. Funct. Mater., 2024 DOI:10.1002/adfm.202400523.
- T. Man, W. Lai, C. Zhu, X. Shen, W. Zhang, Q. Bao, J. Chen, Y. Wan, H. Pei and L. Li, Adv. Funct. Mater., 2022, 32, 2201799 CrossRef CAS.
- C. He, L. Jiang, R. Yuan and X. Yang, Sens. Actuators, B, 2023, 374, 132777 CrossRef CAS.
- X. Tang, N. Kishimoto, Y. Kitahama, T. T. You, M. Adachi, Y. Shigeta, S. Tanaka, T. H. Xiao and K. Goda, J. Phys. Chem. Lett., 2023, 14, 10208–10218 CrossRef CAS PubMed.
- J. Lin, D. Zhang, J. Yu, T. Pan, X. Wu, T. Chen, C. Gao, C. Chen, X. Wang and A. Wu, Anal. Chem., 2023, 95, 4671–4681 CrossRef CAS PubMed.
- C. Spedalieri and J. Kneipp, Nanoscale, 2022, 14, 5314–5328 RSC.
- F. Lussier, T. Brulé, M. Vishwakarma, T. Das, J. P. Spatz and J. F. Masson, Nano Lett., 2016, 16, 3866–3871 CrossRef CAS PubMed.
- V. Zivanovic, A. Milewska, K. Leosson and J. Kneipp, Anal. Chem., 2021, 93, 10106–10113 CrossRef CAS PubMed.
- A. Milewska, O. E. Sigurjonsson and K. Leosson, ACS Appl. Bio Mater., 2021, 4, 4999–5007 CrossRef CAS PubMed.
- S. Sloan-Dennison and Z. D. Schultz, Chem. Sci., 2019, 10, 1807–1815 RSC.
- E. A. Vitol, Z. Orynbayeva, M. J. Bouchard, J. Azizkhan-Clifford, G. Friedman and Y. Gogotsi, ACS Nano, 2009, 3, 3529–3536 CrossRef CAS PubMed.
- E. I. Nikelshparg, E. S. Prikhozhdenko, R. A. Verkhovskii, V. S. Atkin, V. A. Khanadeev, B. N. Khlebtsov and D. N. Bratashov, Nanomaterials, 2021, 11, 2588 CrossRef CAS PubMed.
- P. Li, B. Zhou, X. Cao, X. Tang, L. Yang, L. Hu and J. Liu, Chem. – Eur. J., 2017, 23, 14278–14285 CrossRef CAS PubMed.
- C. Yuen and Q. Liu, J. Biophotonics, 2014, 7, 683–689 CrossRef CAS PubMed.
- D. Drescher, C. Giesen, H. Traub, U. Panne, J. Kneipp and N. Jakubowski, Anal. Chem., 2012, 84, 9684–9688 CrossRef CAS PubMed.
- D. Drescher, T. Büchner, P. Guttmann, S. Werner, G. Schneider and J. Kneipp, Nanoscale Adv., 2019, 1, 2937 RSC.
- G. P. Szekeres, M. Montes-Bayón, J. Bettmer and J. Kneipp, Anal. Chem., 2020, 92, 8553–8560 CrossRef CAS PubMed.
- K. Kneipp, A. S. Haka, H. Kneipp, K. Badizadegan, N. Yoshizawa, C. Boone, K. E. Shafer-Peltier, J. T. Motz, R. R. Dasari and M. S. Feld, Appl. Spectrosc., 2002, 56, 150–154 CrossRef CAS.
- A. Huefner, W.-L. Kuan, K. H. Müller, J. N. Skepper, R. A. Barker and S. Mahajan, ACS Nano, 2016, 10, 307–316 CrossRef CAS PubMed.
- J. Yue, Y. Shen, C. Liang, W. Shi, W. Xu and S. Xu, Anal. Chem., 2021, 93, 13038–13044 CrossRef CAS PubMed.
- J. Kneipp, H. Kneipp, M. McLaughlin, D. Brown and K. Kneipp, Nano Lett., 2006, 6, 2225–2231 CrossRef CAS PubMed.
- A. P. Silwal and H. P. Lu, ACS Omega, 2018, 3, 14849–14857 CrossRef CAS PubMed.
- V. Živanović, G. Semini, M. Laue, D. Drescher, T. Aebischer and J. Kneipp, Anal. Chem., 2018, 90, 8154–8161 CrossRef PubMed.
- V. Živanović, S. Seifert, D. Drescher, P. Schrade, S. Werner, P. Guttmann, G. P. Szekeres, S. Bachmann, G. Schneider, C. Arenz and J. Kneipp, ACS Nano, 2019, 13, 9363–9375 CrossRef PubMed.
- S. R. Panikkanvalappil, M. A. Mackey and M. A. El-Sayed, J. Am. Chem. Soc., 2013, 135, 4815–4821 CrossRef CAS PubMed.
- L. Troncoso-Afonso, G. A. Vinnacombe-Willson, C. García-Astrain and L. M. Liz-Márzan, Chem. Soc. Rev., 2024, 53, 5118–5148 RSC.
- L. Vázquez-Iglesias, G. M. Stanfoca Casagrande, D. García-Lojo, L. Ferro Leal, T. A. Ngo, J. Pérez-Juste, R. M. Reis, K. Kant and I. Pastoriza-Santos, Bioact. Mater., 2024, 34, 248–268 Search PubMed.
- Y. Oh, T. Son, S. Y. Kim, W. Lee, H. Yang, J.-R. Choi, J.-S. Shin and D. Kim, Opt. Express, 2014, 22, 27695–27706 CrossRef CAS PubMed.
- H. Lee, K. Kang, K. Mochizuki, C. Lee, K.-A. Toh, S. A. Lee, K. Fujita and D. Kim, Nano Lett., 2020, 20, 8951–8958 CrossRef CAS PubMed.
- M. Tang, Y. Han, D. Jia, Q. Yang and J.-X. Cheng, Light: Sci. Appl., 2023, 12, 137 CrossRef CAS PubMed.
- K. A. Willets, Chem. Soc. Rev., 2014, 43, 3854–3864 RSC.
- A. P. Olson, C. T. Ertsgaard, S. N. Elliott and N. C. Lindquist, ACS Photonics, 2016, 3, 329–336 CrossRef CAS.
- C. T. Ertsgaard, R. M. McKoskey, I. S. Rich and N. C. Lindquist, ACS Nano, 2014, 8, 10941–10946 CrossRef CAS PubMed.
- A. P. Olson, K. B. Spies, A. C. Browning, P. A. G. Soneral and N. C. Lindquist, Sci. Rep., 2017, 7, 9135 CrossRef PubMed.
- M. Wang, M. Chen, K. Zhanghao, X. Zhang, Z. Jing, J. Gao, M. Q. Zhang, D. Jin, Z. Dai, P. Xi and Q. Dai, Nanoscale, 2018, 10, 19757–19765 RSC.
- C. D. L. De Albuquerque and Z. D. Schultz, Anal. Chem., 2020, 92, 9389–9398 CrossRef CAS PubMed.
- Y. Hang, J. Boryczka and N. Wu, Chem. Soc. Rev., 2022, 51, 329–375 RSC.
- P. Matousek, M. Towrie, C. Ma, W. M. Kwok, D. Phillips, W. T. Toner and A. W. Parker, J. Raman Spectrosc., 2001, 32, 983–988 CrossRef CAS.
- J. Register, M. Maiwald, A. Fales, P. Strobbia, B. Sumpf and T. Vo-Dinh, J. Raman Spectrosc., 2018, 49, 1961–1967 CrossRef CAS.
- P. Mahou, M. Zimmerley, K. Loulier, K. S. Matho, G. Labroille, X. Morin, W. Supatto, J. Livet, D. Débarre and E. Beaurepaire, Nat. Methods, 2012, 9, 815–818 CrossRef CAS PubMed.
- H. Wang, T. B. Huff, D. A. Zweifel, W. He, P. S. Low, A. Wei and J.-X. Cheng, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 15752–15756 CrossRef CAS PubMed.
- J. Olesiak-Banska, M. Waszkielewicz, P. Obstarczyk and M. Samoc, Chem. Soc. Rev., 2019, 48, 4087–4117 RSC.
- P. Z. El-Khoury, A. G. Joly and W. P. Hess, J. Phys. Chem. C, 2016, 120, 7295–7298 CrossRef CAS.
- A. Al-Zubeidi, L. A. McCarthy, A. Rafiei-Miandashti, T. S. Heiderscheit and S. Link, Nanophotonics, 2021, 10, 1621–1655 CrossRef CAS.
- S. R. Panikkanvalappil, S. M. Hira, M. A. Mahmoud and M. El-Sayed, J. Am. Chem. Soc., 2014, 136, 15961–15968 CrossRef CAS PubMed.
- J. C. Fraire, M. L. Masseroni, I. Jausoro, E. M. Perassi, A. M. D. Anel and E. A. Coronado, ACS Nano, 2014, 8, 8942–8958 CrossRef CAS PubMed.
- A. Kazarine, A. A. Gopal and P. W. Wiseman, Analyst, 2019, 144, 3239–3249 RSC.
- D. Yelin, D. Oron, S. Thiberge, E. Moses and Y. Silberberg, Opt. Express, 2003, 11, 1385–1391 CrossRef PubMed.
- N. L. Gruenke, M. F. Cardinal, M. O. McAnally, R. R. Frontiera, G. C. Schatz and R. P. Van Duyne, Chem. Soc. Rev., 2016, 45, 2263–2290 RSC.
- F. Madzharova, Á. Nodar, V. Živanović, M. R. S. Huang, C. T. Koch, R. Esteban, J. Aizpurua and J. Kneipp, Adv. Funct. Mater., 2019, 29, 1904289 CrossRef CAS.
- A. Volkmer, J.-X. Cheng and X. Sunney Xie, Phys. Rev. Lett., 2001, 87, 023901 CrossRef.
- R. Shimada, H. Kano and H.-O. Hamaguchi, Opt. Lett., 2006, 31, 320–322 CrossRef CAS PubMed.
- D. V. Chulhai, Z. Hu, J. E. Moore, X. Chen and L. Jensen, Annu. Rev. Phys. Chem., 2016, 67, 541–564 CrossRef CAS PubMed.
- L. H. Machtoub, R. Pfeiffer, D. Bataveljić, P. R. Andjus and H. Jordi, e-J. Surf. Sci. Nanotechnol., 2010, 8, 362–366 CrossRef CAS.
- S. Schlücker, M. Salehi, G. Bergner, M. Schütz, P. Ströbel, A. Marx, I. Petersen, B. Dietzek and J. Popp, Anal. Chem., 2011, 83, 7081–7085 CrossRef PubMed.
- S. Yampolsky, D. A. Fishman, S. Dey, E. Hulkko, M. Banik, E. O. Potma and V. A. Apkarian, Nat. Photonics, 2014, 8, 650–656 CrossRef CAS.
- L. Ouyang, T. Meyer-Zedler, K. M. See, W. L. Chen, F. C. Lin, D. Akimov, S. Ehtesabi, M. Richter, M. Schmitt, Y. M. Chang, S. Gräfe, J. Popp and J. S. Huang, ACS Nano, 2021, 15, 809–818 CrossRef CAS PubMed.
- J. Kneipp, H. Kneipp and K. Kneipp, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 17149–17153 CrossRef CAS PubMed.
- M. Gühlke, Z. Heiner and J. Kneipp, Phys. Chem. Chem. Phys., 2016, 18, 14228–14233 RSC.
- C. Levene, E. Correa, E. W. Blanch and R. Goodacre, Anal. Chem., 2012, 84, 7899–7905 CrossRef CAS PubMed.
- P. H. L. Nguyen, B. Hong, S. Rubin and Y. Fainman, Biomed. Opt. Express, 2020, 11, 5092–5121 CrossRef CAS PubMed.
- A. Barucci, C. D'Andrea, E. Farnesi, M. Banchelli, C. Amicucci, M. De Angelis, B. Hwang and P. Matteini, Analyst, 2021, 146, 674–682 RSC.
- H. Shi, H. Wang, X. Meng, R. Chen, Y. Zhang, Y. Su and Y. He, Anal. Chem., 2018, 90, 14216–14221 CrossRef CAS PubMed.
- G. Qi, D. Wang, C. Li, K. Ma, Y. Zhang and Y. Jin, Anal. Chem., 2020, 92, 11755–11762 CrossRef CAS PubMed.
- Y. X. Leong, Y. H. Lee, C. S. L. Koh, G. C. Phan-Quang, X. Han, I. Y. Phang and X. Y. Ling, Nano Lett., 2021, 21, 2642–2649 CrossRef CAS PubMed.
- Y. Ju, O. Neumann, M. Bajomo, Y. Zhao, P. Nordlander, N. J. Halas and A. Patel, ACS Nano, 2023, 17, 21251–21261 CrossRef PubMed.
- S. Mahajan, R. M. Cole, J. D. Speed, S. H. Pelfrey, A. E. Russell, P. N. Bartlett, S. M. Barnett and J. J. Baumberg, J. Phys. Chem. C, 2010, 114, 7242–7250 CrossRef CAS.
- C. Farcau and S. Astilean, Chem. Commun., 2011, 47, 3861–3863 RSC.
- C. Wang, L. Xiao, C. Dai, A. H. Nguyen, L. E. Littlepage, Z. D. Schultz and J. Li, Sci. Rep., 2020, 10, 1460 CrossRef CAS PubMed.
- S. Guo, J. Popp and T. Bocklitz, Nat. Protoc., 2021, 16, 5426–5459 CrossRef CAS PubMed.
- J. M. Amigo, BrJAC–Braz. J. Anal. Chem., 2021, 8, 45–61 CrossRef CAS.
- F. Lussier, V. Thibault, B. Charron, G. Q. Wallace and J.-F. Masson, TrAC, Trends Anal. Chem., 2020, 124, 115796 CrossRef CAS.
- S. M. Barnett, N. Harris and J. J. Baumberg, Phys. Chem. Chem. Phys., 2014, 16, 6544–6549 RSC.
- Q. Hu, C. Sellers, J. S.-I. Kwon and H.-J. Wu, Digital Chem. Eng., 2022, 3, 100020 CrossRef PubMed.
- J. Q. Li, H. Neng-Wang, A. J. Canning, A. Gaona, B. M. Crawford, K. S. Garman and T. Vo-Dinh, Appl. Spectrosc., 2024, 78, 84–98 CrossRef CAS PubMed.
- W. Nam, H. Chen, X. Ren, M. Agah, I. Kim and W. Zhou, ACS Appl. Nano Mater., 2022, 5, 10358–10368 CrossRef CAS.
- U. Parlatan, M. O. Ozen, I. Kecoglu, B. Koyuncu, H. Torun, D. Khalafkhany, I. Loc, M. G. Ogut, F. Inci, D. Akin, I. Solaroglu, N. Ozoren, M. B. Unlu and U. Demirci, Small, 2023, 19, 2205519 CrossRef CAS PubMed.
- N. K. Afseth, V. H. Segtnan and J. P. Wold, Appl. Spectrosc., 2006, 60, 1358–1367 CrossRef CAS PubMed.
- T. Bocklitz, A. Walter, K. Hartmann, P. Rösch and J. Popp, Anal. Chim. Acta, 2011, 704, 47–56 CrossRef CAS PubMed.
- S. Fornasaro, F. Alsamad, M. Baia, L. A. E. Batista de Carvalho, C. Beleites, H. J. Byrne, A. Chiadò, M. Chis, M. Chisanga, A. Daniel, J. Dybas, G. Eppe, G. Falgayrac, K. Faulds, H. Gebavi, F. Giorgis, R. Goodacre, D. Graham, P. La Manna, S. Laing, L. Litti, F. M. Lyng, K. Malek, C. Malherbe, M. P. M. Marques, M. Meneghetti, E. Mitri, V. Mohaček-Grošev, C. Morasso, H. Muhamadali, P. Musto, C. Novara, M. Pannico, G. Penel, O. Piot, T. Rindzevicius, E. A. Rusu, M. S. Schmidt, V. Sergo, G. D. Sockalingum, V. Untereiner, R. Vanna, E. Wiercigroch and A. Bonifacio, Anal. Chem., 2020, 92, 4053–4064 CrossRef CAS PubMed.
- X. X. Han, R. S. Rodriguez, C. L. Haynes, Y. Ozaki and B. Zhao, Nat. Rev. Methods Primers, 2022, 1, 87 CrossRef.
- Z.-M. Zhang, S. Chen, Y.-Z. Liang, Z.-X. Liu, Q.-M. Zhang, L.-X. Ding, F. Ye and H. Zhou, J. Raman Spectrosc., 2010, 41, 659–669 CrossRef CAS.
- G. C. Green, A. D. C. Chan, B. S. Luo, H. Dan and M. Lin, IEEE Trans. Instrum. Meas., 2009, 58, 3713–3722 CAS.
- F. Ehrentreich, Anal. Bioanal. Chem., 2002, 372, 115–121 CrossRef CAS PubMed.
- I. S. Patel, W. R. Premasiri, D. T. Moir and L. D. Ziegler, J. Raman Spectrosc., 2008, 39, 1660–1672 CrossRef CAS PubMed.
- I. T. Jolliffe and J. Cadima, Philos. Trans. R. Soc., A, 2016, 374, 20150202 CrossRef PubMed.
- K. Pearson, London, Edinburgh Dublin Philos. Mag. J. Sci., 1901, 2, 559–572 CrossRef.
- H. Wold, J. Appl. Probab., 1975, 12, 117–142 CrossRef.
- S. Wold, M. Sjöström and L. Eriksson, Chemom. Intell. Lab. Syst., 2001, 58, 109–130 CrossRef CAS.
- B. E. Boser, I. M. Guyon and V. N. Vapnik, Proceedings of the fifth annual workshop on Computational learning theory, 1992.
- Y. Xu, S. Zomer and R. G. Brereton, Crit. Rev. Anal. Chem., 2006, 36, 177–188 CrossRef CAS.
- L. Breiman, J. Friedman, C. J. Stone and R. A. Olshen, Classification and Regression Trees, Taylor & Francis, 1984 Search PubMed.
- B. Debus, H. Parastar, P. Harrington and D. Kirsanov, TrAC, Trends Anal. Chem., 2021, 145, 116459 CrossRef CAS.
- W. S. McCulloch and W. Pitts, Bull. Math. Biophys., 1943, 5, 115–133 CrossRef.
- R. M. Jarvis and R. Goodacre, Anal. Chem., 2004, 76, 40–47 CrossRef CAS PubMed.
- D. P. dos Santos, M. M. Sena, M. R. Almeida, I. O. Mazali, A. C. Olivieri and J. E. L. Villa, Anal. Bioanal. Chem., 2023, 415, 3945–3966 CrossRef CAS PubMed.
- O. I. Abiodun, A. Jantan, A. E. Omolara, K. V. Dada, N. A. Mohamed and H. Arshad, Heliyon, 2018, 4, e00938 CrossRef PubMed.
- J. Zhu, X. Jiang, Y. Rong, W. Wei, S. Wu, T. Jiao and Q. Chen, Food Chem., 2023, 414, 135705 CrossRef CAS PubMed.
- J. Chen, P. Wang, Y. Tian, R. Zhang, J. Sun, Z. Zhang and J. Gao, J. Biophotonics, 2023, 16, e202200254 CrossRef PubMed.
- X. Sha, G. Fang, G. Cao, S. Li, W. Hasi and S. Han, Analyst, 2022, 147, 5785–5795 RSC.
- S. Seifert, V. Merk and J. Kneipp, J. Biophotonics, 2016, 9, 181–189 CrossRef CAS PubMed.
- X. Shao, H. Zhang, Y. Wang, H. Qian, Y. Zhu, B. Dong, F. Xu, N. Chen, S. Liu, J. Pan and W. Xue, Nanomedicine, 2020, 29, 102245 CrossRef CAS PubMed.
- S.-h Luo, W.-l Wang, Z.-f Zhou, Y. Xie, B. Ren, G.-k Liu and Z.-q Tian, Anal. Chem., 2022, 94, 10151–10158 CrossRef CAS PubMed.
- J. Q. Li, P. V. Dukes, W. Lee, M. Sarkis and T. Vo-Dinh, J. Raman Spectrosc., 2022, 53, 2044–2057 CrossRef CAS PubMed.
- P. S. Gromski, H. Muhamadali, D. I. Ellis, Y. Xu, E. Correa, M. L. Turner and R. Goodacre, Anal. Chim. Acta, 2015, 879, 10–23 CrossRef CAS PubMed.
- F. Degenhardt, S. Seifert and S. Szymczak, Briefings Bioinf., 2019, 20, 492–503 CrossRef PubMed.
- P. J. Castaldi, I. J. Dahabreh and J. P. A. Ioannidis, Briefings Bioinf., 2011, 12, 189–202 CrossRef PubMed.
- F. Seyednasrollah, A. Laiho and L. L. Elo, Briefings Bioinf., 2015, 16, 59–70 CrossRef CAS PubMed.
- A.-L. Boulesteix, S. Lauer and M. J. Eugster, PLoS One, 2013, 8, e61562 CrossRef CAS PubMed.
- J. Wright, A. Y. Yang, A. Ganesh, S. S. Sastry and Y. Ma, IEEE Trans. Pattern Anal. Mach. Intell., 2009, 31, 210–227 Search PubMed.
- G. K. Sandve and V. Greiff, Bioinformatics, 2022, 38, 4994–4996 CrossRef CAS PubMed.
- S. Seifert, Sci. Rep., 2020, 10, 5436 CrossRef PubMed.
- S. Janitza, E. Celik and A.-L. Boulesteix, Adv. Data Anal. Classif., 2018, 12, 885–915 CrossRef.
- M. B. Kursa and W. R. Rudnicki, J. Stat. Softw., 2010, 36, 1–13 CrossRef.
- S. Seifert, S. Gundlach and S. Szymczak, Bioinformatics, 2019, 35, 3663–3671 CrossRef CAS PubMed.
- L. F. Voges, L. C. Jarren and S. Seifert, Bioinformatics, 2023, btad471, DOI:10.1093/bioinformatics/btad471.
- S. Wenck, T. Mix, M. Fischer, T. Hackl and S. Seifert, Metabolites, 2023, 13, 1075 CrossRef CAS PubMed.
- H. Lösel, M. Arndt, S. Wenck, L. Hansen, M. Oberpottkamp, S. Seifert and M. Fischer, Talanta, 2024, 271, 125598 CrossRef PubMed.
- S. Wenck, M. Creydt, J. Hansen, F. Gärber, M. Fischer and S. Seifert, Metabolites, 2022, 12, 5 CrossRef CAS PubMed.
- H. Lösel, J. Brockelt, F. Gärber, J. Teipel, T. Kuballa, S. Seifert and M. Fischer, Metabolites, 2023, 13, 882 CrossRef PubMed.
- V. Živanović, F. Madzharova, Z. Heiner, C. Arenz and J. Kneipp, J. Phys. Chem. C, 2017, 121, 22958–22968 CrossRef.
- S. Seifert, S. Gundlach, O. Junge and S. Szymczak, Bioinformatics, 2020, 36, 4301–4308 CrossRef CAS PubMed.
- G. S. Eichler, M. Reimers, D. Kane and J. N. Weinstein, Genome Biol., 2007, 8, R187 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |