Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Generating knowledge graphs through text mining of catalysis research related literature†
Alexander S.
Behr
 *a,
Diana
Chernenko
a,
Dominik
Koßmann
b,
Arjun
Neyyathala
c,
Schirin
Hanf
*a,
Diana
Chernenko
a,
Dominik
Koßmann
b,
Arjun
Neyyathala
c,
Schirin
Hanf
 c,
Stephan A.
Schunk
c,
Stephan A.
Schunk
 d and
Norbert
Kockmann
d and
Norbert
Kockmann
 a
a
aDepartment of Biochemical and Chemical Engineering, Laboratory of Equipment Design, TU Dortmund University, Dortmund, Germany. E-mail: alexander.behr@tu-dortmund.de
bDepartment of Computer Science, Pattern Recognition Group, TU Dortmund University, Dortmund, Germany
cInstitute for Inorganic Chemistry, Karlsruhe Institute of Technology, Karlsruhe, Germany
dhte GmbH, Heidelberg, Germany
First published on 5th August 2024
Abstract
Structured research data management in catalysis is crucial, especially for large amounts of data, and should be guided by FAIR principles for easy access and compatibility of data. Ontologies help to organize knowledge in a structured and FAIR way. The increasing numbers of scientific publications call for automated methods to preselect and access the desired knowledge while minimizing the effort to search for relevant publications. While ontology learning can be used to create structured knowledge graphs, named entity recognition allows detection and categorization of important information in text. This work combines ontology learning and named entity recognition for automated extraction of key data from publications and organization of the implicit knowledge in a machine- and user-readable knowledge graph and data. CatalysisIE is a pre-trained model for such information extraction for catalysis research. This model is used and extended in this work based on a new data set, increasing the precision and recall of the model with regard to the data set. Validation of the presented workflow is presented on two datasets regarding catalysis research. Preformulated SPARQL-queries are provided to show the usability and applicability of the resulting knowledge graph for researchers.
Introduction
Effective data management is crucial for innovation and knowledge development by integrating and repurposing data and empowering the community to utilize them. High quality data management enables continuous data reuse, guided by the “FAIR” principles – findability, accessibility, interoperability, and reusability – ensuring easy access and compatibility. These principles streamline data management, enhancing research impact and fostering a cycle of knowledge dissemination across various domains.1–3The increasing annual volume of publications and data related to catalysis research poses challenges for knowledge extraction from it, particularly within constrained time frames. In particular, Scopus lists 186![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 744 articles for the keyword “catalysis” in the time between 2013 and 2023.4 Challenges in catalysis research are intertwined with the complexities of information retrieval, data systematization, and structuring. As the quantity of information grows, manual execution of these processes becomes increasingly time-consuming. Here, strategies of digital transitions for catalysis can help to increase the value of obtained data.5 Furthermore, application of language model search for synthesis planning in heterogeneous catalysis allowed for systematic literature search and accelerated catalytic synthesis planning.6
744 articles for the keyword “catalysis” in the time between 2013 and 2023.4 Challenges in catalysis research are intertwined with the complexities of information retrieval, data systematization, and structuring. As the quantity of information grows, manual execution of these processes becomes increasingly time-consuming. Here, strategies of digital transitions for catalysis can help to increase the value of obtained data.5 Furthermore, application of language model search for synthesis planning in heterogeneous catalysis allowed for systematic literature search and accelerated catalytic synthesis planning.6
Information structuring and systematization can be achieved through the utilization of ontologies. An ontology serves as a data model, depicting a collection of concepts and the relationships among these concepts within a specific domain.7 In this framework, terms are organized hierarchically as classes and subclasses, with each class linked to other classes through properties. This allows for automated classification of research papers with regard to those classes, increasing FAIRness of the classified texts.
Ontology learning (OL) from text is the (semi-)automated process of ontology creation or reuse for enrichment or population purposes. In recent years, several OL approaches have been developed to automate the construction of ontologies. Heuristic and conceptual clustering is one of the statistical-based approaches used for grouping the concepts based on the semantic distance between them to build towards hierarchies. This method was employed in previous work8 for knowledge extraction from catalysis-related texts for the automatic creation of a taxonomy with important terms extracted from texts. However, the resulting hierarchy is still missing specific interrelations between the terms, and concepts lack proper characterization through axioms. This proves that it is important to integrate the relation extraction in the process of OL. One other tool OntoCmaps9 is an OL system, with which non-taxonomic relations can be recognized with dependency structure analysis and ontologies are constructed in the form of concept maps, which are not domain-specific and can contain not necessary information.
To extract valuable information from publications in the field of catalysis research, which can be considered as a named entity recognition (NER) task, a pre-trained model, CatalysisIE,10 was used. This allows for identification of key information from a text based on pre-trained classes, such as names of people. The authors of this model constructed the first benchmark data set for knowledge extraction from the scientific literature in catalysis using active learning to generate a candidate sentence pool for annotation purposes. With this, extracted entities can be categorized into six categories: catalyst, reaction, reactant, product, characterization, and treatment. For the text span representation, pre-trained SciBERT11 models were used. The parameters of SciBERT were optimized for catalysis-related information extraction (IE) by undergoing the domain adaptation using a corpus consisting of 10.4 million words.10
The objective of this work is to facilitate the acquisition of information for catalysis research. This is obtained through the design of a tool for the automatic systematization of data extracted from scholarly publications into knowledge graphs. The construction of the knowledge graphs is based on an ontology, which allows for higher data FAIRness by structured relations and conceptual classification of knowledge. Additionally, the content of these publications is preserved in the form of terms deemed relevant to catalysis research. Utilizing CatalysisIE, the extracted entities can be categorized into the six concepts. After preprocessing, the abstracts from scholarly publications can be extracted with natural language processing (NLP) techniques. NLP techniques enable computers to interpret and generate human language, such as scientific texts. For IE, the pretrained model by the authors of CatalysisIE10 can be used. Furthermore, the CatalysisIE model is trained on the complemented dataset presented in this work.
Ontologies can be queried using SPARQL12 queries, e.g. formulated in Python functions. SPARQL is a structured query language used to retrieve data stored within databases, especially for triplet-based data, such as ontologies. Automatically generated knowledge graphs containing information for the retrieval of the publications can also later be queried for retrieval of publications.
Methods
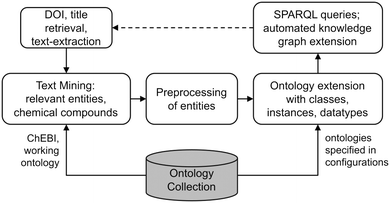
The overall framework proposed in this work starts with scholarly text, which is retrieved from publisher repositories, like Scopus.4 On this data, text mining is applied to extract relevant entities and extracted entities are compared to substance classes from the ChEBI13 ontology to mitigate synonyms. The extracted entities are then searched for in already existing ontologies stemming from a collection of ontologies of the domain of knowledge. After preprocessing of the found entities, an ontology gets selected and extended by the necessary classes, instances and datatypes. Finally, the resulting knowledge graph contains the semantic information on the analyzed abstracts, allowing for deeper insights and knowledge extraction by SPARQL queries. Additionally, the outcome of the queries can be used to find new publications and reiterate the framework with the newly added publications, yielding a growing knowledge graph. The overview on the proposed framework as discussed in this work is depicted in Fig. 1.Data retrieval
To start with the data retrieval, PDF files containing scholarly publications are processed one by one to extend a working ontology with entities of fundamental character extracted from the abstracts.In this work, abstracts were processed assuming that this part of the initial text contains important information about the content of the article and the output is less affected by noisy and repetitive information. The noisy information could be, for example, the previous studies usually mentioned in the introduction section. Abstracts and publication titles were retrieved using text extraction techniques directly from PDF-files. Furthermore, the publicly available CrossRef REST API and publishers' API for metadata retrieval were used.14 The CrossRef REST API was integrated with the habanero Python package,15 which fulfils the role of a low-level client that provides functionalities for querying and response handling. The pybliometrics package,16 an API-wrapper to access Scopus, is used for abstract scraping, which implies the process of automatically mining data or collecting information from the internet, along with pdfdataextractor17 for abstract extraction directly from PDF files. When an abstract is not able to be retrieved from a PDF, the HTML-version of the publication and thus its abstract are retrieved based on the DOI.
After text preprocessing, entities relevant to the field of catalysis research are extracted from the text using a pre-trained model and labelled with one of the six categories: catalyst, reaction, reactant, product, characterization, and treatment.
Text mining and preprocessing
In the next step, chemical entities are detected in the extracted entities using chemdataextractor (CDE).18 After detection, chemical compounds are split into components using regular expression (regex) functions.19 Support materials such as “ZSM-5” and “MCM-41” introduce abbreviations and are considered as an exception because they are abbreviations that cannot directly be resolved to their chemical compounds and thus not processed further. Hence, their pattern of capital letters followed by a hyphen and integers is matched using regex functions. Synonymy is one challenge of NER in chemistry: the same concept can be expressed using different terms. This challenge can be observed when chemical entities are extracted from texts. To address this issue, a method is developed to identify synonyms and match them to the corresponding compounds in the ontology. The goal of the process is to obtain compound names that can be used to identify the chemical entities from the input list of possible chemical compounds extracted from the text. Chemical compounds and their components are identified using extracted synonyms of existing ChEBI terms, InChiKeys, and by querying the PubChem20 database using the pubchempy Python package.21 If only one compound is found in PubChem, its IUPAC name is used for further identification of the chemical entity. However, if multiple compounds are retrieved, both the SMILES notation and the corresponding IUPAC names are presented to the user. This allows the user to select the compound that best matches the chemical entity being searched for, or to use the searched name for identification if there is no match.An example of compound identification with user feedback for “Rh2O3” is shown in Fig. 2(a). In cases where no compounds could be retrieved from PubChem and no synonyms were found in ChEBI, the user is prompted to confirm whether the compound exists. If the user answers “no”, the presumed chemical entity is skipped. By responding “yes”, the user confirms the existence of the entity and it will be identified with the name provided to the user (see Fig. 2(b)).
Ontology learning
The process of OL is divided into three sub-processes. The first sub-process is the extension of an ontology with terms from other ontologies. In the second phase of the process, terms that do not exist in the input ontologies are generated using categories of extracted entities and the context information. Finally, newly created classes are populated with instances, and relations between instances will be created. However, further preprocessing is required before these steps can be performed.As the main working ontology, the Allotrope Foundation Ontology (AFO)22 is selected. In this ontology, object properties important for the ontology extension process are created manually (Table 4 in the appendix), while classes and data properties are created automatically. After preprocessing of the extracted entities based on their labelled roles, the working ontology is extended with classes and instances using existing terms in other ontologies. Furthermore, rule-based approaches in combination with syntactic dependency parsing are pursued. The initial preprocessing of the extracted entities includes the POS taggers, lemmatization, tokenization, and the use of regular expressions. In specific steps, such as lemmatization, modules from spaCy23 are used. Thus, the preprocessing allows prevention of the creation of synonyms as independent concepts in the ontology and creation of relations based on the context in the text. For example, given an entity “RhCo/SiO2” labelled “catalyst”, “RhCo” is recognised as a catalyst, which is supported on “SiO2”. This relation will be added later in the ontology to the corresponding classes of the compounds. To recreate the hierarchical structure within the ontology, syntactic dependency parsing was utilized as a preprocessing step to define the “head” – word of the entity and use it for search in ontologies. For example, the “head” of “packed bed methanation” is “methanation”.
In this work, some existing ontologies were used in the workflow for ontology extension. The primary selection criterion for these ontologies was their relevance to the domain of catalysis research. The selection of ontologies for this work, which are listed along with a short description in Table 1, is based on the overview from the collection of ontologies for catalysis research presented in ref. 24.
| Ontology acronym | Full name | Description |
|---|---|---|
| BFO25 | Basic Formal Ontology | A small ontology is designed for use in analysis and integration in scientific or other domains. It does not contain physical, chemical, or other domain-specific terms and is well used as a top-level ontology |
| AFO22 | Allotrope Foundation Ontology | AFO is a domain-specific ontology, offering a standardized vocabulary and semantic framework for the representation of laboratory analytical processes. AFO is aligned with BFO as a top-level ontology. Reasoning can be only provided by the HermiT reasoner |
| ChEBI13 | Chemical Entities of Biological Interest | ChEBI represents a vocabulary with a focus on small molecular entities and contains such information as InChiKeys, CAS numbers, and exact and related synonyms of chemical compounds |
| MOP26 | Molecular Process Ontology | Domain ontology contains good conceptual descriptions of molecular processes, such as crystallization and methylation |
| RXNO27 | Name Reaction Ontology | Domain ontology is strongly connected to MOP. It contains more than 500 classes representing organic reactions with good conceptual description |
Ontology extension is based on the reuse of the classes existing in other ontologies to properly characterize concepts and reuse existing frameworks and axioms. The reuse of ontology terms creates links between data, making the ontology more valuable.28
For example, ChEBI contains more than 3 million axioms. Thus, only relevant subsets of ontologies were reused. After searching for classes within the working ontology using the owlready2 Python package,29 missing chemical compounds and possible reaction types are searched for in other ontologies as listed in Table 1. To accomplish this, a nested dictionary is used, containing all IRIs of terms alongside their corresponding labels, prefLabels, altLabels, and the names used within the ontology. Applying a class information extraction process that incorporates functions from the owlready2 package, the dictionary was generated for 22 ontologies relevant to the domain of catalysis research.24 Once the dictionary is loaded into the Python environment, it is searched for classes missing in the working ontology. If one of the labels, prefLabels, altLabels, or names matches the searched entity, the corresponding IRI is added to the dictionary along with the matching value. The IRIs of found terms that are still missing in the working ontology are stored in automatically created text files. The names of the text files include the acronym of the source ontology for further reference.
Ontology extension
The first part of the ontology extension is implemented using ROBOT,28 an open-source library and command-line tool for automating ontology development tasks. ROBOT provides diverse ontology processing commands, including class extraction and merging of ontologies. Using the Syntactic Locality Module Extractor (SMLE) method, a subset of an ontology is created by starting with the “seed” term, the IRI of which is defined in the created text-file and adding related terms necessary to maintain logical relationships.This ensures that the module retains all the same logical entailments in the full ontology, providing consistency in the ontology subset. The chosen SMLE approach is the BOTTOM module, which contains the terms in the seed, their corresponding superclasses, and the interrelations between them. As the name implies, the class hierarchy is built from the bottom up, gathering the superclasses of the selected class. Thus, for each ontology, a separate subset of relevant classes is created in rdf/xml (owl) format.
The second task for which ROBOT is used in this work is to merge the created subsets of classes and the main ontology into a single ontology with a single .owl-file. Thus, the merging process is used to update the working ontology within existing terms of other ontologies.
Because some of the merged ontologies are aligned with different top-level ontologies, terms that theoretically share the same definition are located at different positions within the class hierarchy. For example, the OBO is the top-level ontology of ChEBI, while the AFO is aligned with the BFO. Both ontologies have, for example, the term “atom”, but at different positions in the class hierarchy. Another factor why the same terms are represented differently is related to the granularity problem of ontologies. This issue arises because ontologies often adopt different levels of details when representing identical knowledge to support different applications.30
Since all of the utilized ontologies are connected to the domain of catalysis24 and chemistry,31 terms with identical designation are assumed equivalent. The equivalence of classes indicates that respective classes share all their instances, and the descriptions of both classes are interlinked. However, the use of the equivalence relation does not imply class equality. Both relations are defined differently in OWL. Equality is denoted by “owl:sameAs”, while the equivalence is represented by owl:equivalentClass. Class equality can only be defined by the description language OWL-Full, and owlready2 supports only equivalence.29
To identify terms with the same designation that originate from different ontologies and consequently have different IRIs, the mappings created in previous work24 are used in the processing. These mappings represent all terms shared by two ontologies according to the same IRIs or the same set of labels, prefLabels, names, or altLabels. After merging ontologies to reuse existing terms, the process of creating new classes and subsequently populating the working ontology with new instances is initialized. First, a new instance of a publication is generated as an instance of the “publication” superclass. The DOI and title of the publication retrieved at the beginning of the process are added to the publication instance as datatypes using the “has doi” and “has title” datatype properties, respectively. Extracted chemical compounds that do not exist in the working ontology after merging are then created as new classes within the working ontology, utilizing the context information of the new compounds. Chemical compounds that can be further broken down into compounds and atoms, such as “Al2O3” or “titanium dioxide”, or those that are recognized as compounds using pubchempy are created as subclasses of the “molecule” class.
Support material entities, which represent a combination of two or more carrier compounds, like “TiO2–SiO2”, or materials such as “MCM-41” are created as subclasses of the “support material” class. Each newly created class and instance are automatically assigned a generated name linked to the number of the processed publications in the working ontology.
Entities from the “Reactant”, “Product” and “Catalyst” categories that represent specific types of chemical entities, such as “light olefin” and “vapour phase propene”, are created as instances of the corresponding chemical compound. Extracted and preprocessed catalyst entities are created as instances of the “chemical substance” class.
Chemical entities, which represent catalysts in the form of
“<catalytic compound>/<support compound>” or
“<catalytic compound>@<support compound>”
are labelled in the ontology
“<catalytic compound> supported on <support compound>”
and linked with their chemical compounds based on their roles in the entity using the “catalytic component of” and “support component of” object properties. A schematic example of interconnections within the ontology is shown in Fig. 3.
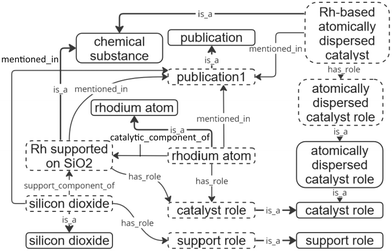 | ||
| Fig. 3 Examples of created entities and assigned relations. Entities within dashed boxes represent instances, while continuous bordered boxes represent classes. | ||
Table 4 in the appendix lists the object properties and their inverse properties that need to be defined within the working ontology in order to assign the relations between the newly created entities.
The creation of the classes corresponding to the catalyst types is based on the creation of subclasses of the term “catalyst role”, which already exists in the AFO ontology. Roles in ontologies are used to reduce the amount of object properties and thus to speed up reasoning. The corresponding roles of terms are provided as classes in the ontology and terms are linked to them via the “has role” object property. The hierarchical structuring of the catalyst roles is based on the content of the classes extracted from the entities. For instance, within the text corpus, the extracted class of an entity after preprocessing might be a “dispersed catalyst role”, while the catalyst type corresponding to another entity is an “atomically dispersed catalyst role”. Since the second class is identical to the first but with an additional word, it is considered a subclass of the first class. In case no entity from the text corpus has a “dispersed catalyst role” as an extracted class, then an “atomically dispersed catalyst role” is created as a subclass of the “catalyst role”.
Chemical reactions are created as subclasses of the previously extracted reaction “heads”. If there is no corresponding reaction found in other ontologies, a new class is created as a subclass of “chemical reaction (molecular)”, which is also a class within the AFO. For each class created after the merging that corresponds to an extracted entity or a chemical compound, an instance is created with an automatically generated name.
The label of the instance is the same as the label of its corresponding class. The same procedure is applied to the newly created classes. All classes and instances, once created, can be reused for ontology extension with the respective publications. The newly created classes of chemical compounds are linked to their corresponding components via “has part” relations at the instance level.
Created instances are linked to their roles according to the categories and the context using the “has role” relation. The used roles include the “support role”, “reactant role”, “product role”, and “catalyst role”, and all are created as subclasses of the “catalyst role”.
Finally, all created and used instances that are mentioned in the processed publication are linked to the instance of the publication through the “mentioned in” object property. Entities labelled “Characterization” and “Treatment” are added to annotations of publications as comment.
SPARQL queries
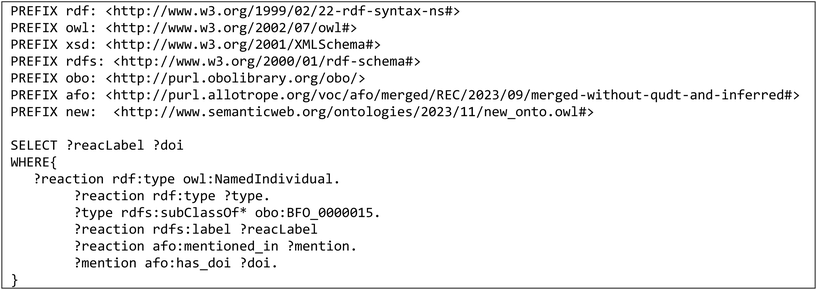
Once the ontology is extended with publications of interest, it can be queried for relevant information extracted from the publications using SPARQL queries.32 For ease of use, some SPARQL queries are pre-formulated in Python functions, the output of which depends on the information required as input by the user.The following competency questions were implemented as SPARQL queries and can thus be easily retrieved from the knowledge graph resulting from the extension of the ontology. The corresponding SPARQL queries are numbered and exemplary input and output of the queries are listed in Table 5 in the appendix and an exemplary SPARQL query is listed in Table 6 in the appendix:
• Give me a list of reactions (1), reactants, support materials, catalysts, and products mentioned in one specific publication, which is a part of the knowledge graph, in one list (2) or separately,
• Retrieve the abstracts from publications in the ontology (3),
• Give me a list of DOIs of publications from the working ontology, which mention the same reactions (4) or the specific reaction (5) or catalyst (6),
• Give me a list of reactions, reactants, support materials, catalysts, and products mentioned in all publications of the knowledge graph (7),
• I need a list of all possible synonyms for the extracted reactants (8), support materials (9), catalysts (10), and products (11) in the form of chemical entities,
• I need possible catalysts where the support material from this paper can be used (12).
The retrieved entities can be used to query Scopus for new publications with similar context. Using the pybliometrics Python package, the search is performed, leading to a query, which has the same structure as a query that works in the Scopus advanced search. With the chosen query type ‘TITLE-ABS-KEY()’ (as depicted exemplarily in Fig. 4), the search is performed within the titles, abstracts, and keywords of the publication.
 | ||
| Fig. 4 Two types of query formulations for the advanced search for further publications in Scopus, executed by the Python API. | ||
Since there are multiple ways to name a specific chemical compound, to avoid a large number of possible queries and at the same time allow diversity in the naming of chemical compounds, a trade name or common name and a formula listed in the class annotations of a chemical compound are used for queries' formulation. Moreover to exclude mismatches, the publication will be skipped if during text mining no reaction was found in the text. After a query is executed, its results are downloaded and cached to speed up the subsequent analysis.
After the results are concatenated into one table, duplicates are removed from it. As Scopus contains records of articles published since 1970, an option to filter the results by publication date is integrated into the process, to allow for the inclusion of primarily newer publications into the knowledge graph. Utilization of the pandas Python module33 allows the resulting DataFrames to be stored as sheets in an Excel file.
Results and discussion
The initial dataset (dataset 1) used in this work consists of 14 articles on the topic of catalysis in liquid phase hydroformylation and 9 articles on catalysis in gas phase hydroformylation. The hydroformylation of olefins using syngas was selected as a use case as it is one of the most important chemical transformations in the industry.34 However, to narrow down the literature pool, the focus was set on heterogeneous catalysts based on RhxAy systems, since such systems have shown to be very promising alternatives to homogeneous and purely Rh-based heterogeneous hydroformylation catalysts.35 These articles were processed together for the extension of the AFO as the working ontology. The articles were provided in PDF format from different publishers, being Elsevier, ACS Publications, Brookhaven National Laboratory, Nature, and Royal Society of Chemistry. After the finalization of the initial ontology extension process, the ontology is used for the retrieval of new publications from Scopus. As a manual benchmark, a sample of 50 randomly selected publications from the list with unique query results was analysed.Moreover, a set of 28 publications on methanation processes (dataset 2) was used to evaluate how well the created tool works on the different types of catalysed reactions. Hereby the focus was laid on the heterogeneously catalysed conversion of carbon monoxide and carbon dioxide to methane via hydrogenation, which is important for the production of synthetic natural gas. In particular, the valorization of CO2 together with renewable hydrogen might be considered an integral sustainable path towards the production of renewable gaseous fuels.36 For that, an extension of an alternate ontology setup similar to the first dataset was performed.
The dataset for training of the model was complemented with 151 sentences manually labelled in label-studio37 from 18 abstracts of papers to the topic of hydroformylation in the liquid and gas phase. Checkpoints from the model trained by the authors of CatalysisIE and the model trained on the complemented dataset were compared with each other.
To evaluate the difference in the prediction of the checkpoints, ten manually labelled abstracts from papers to the same topic were compared to predictions of both models. Since it is important to gain as many correct distinct predictions from the text as possible to be able to describe the content of the publication using extracted entities, the recall R of the model was evaluated with the number of true positives TP and false negatives FN using eqn (1). To obtain the true positives and false negatives, the amount of distinct entities was counted and compared with the number of distinct entities from the prediction after qualitative manual labelling of the texts. This comparison for each extracted abstract from dataset 1 is shown in Table 9 in the appendix.
Besides recall, the precision Pr was selected for evaluation of multi-label classification. Because class imbalances are present in the dataset, the precision was calculated using eqn (2) with the number of positives Pi instead of true positives TP and the number of used labels N. Furthermore, the standard deviation σ of the precision was selected as a metric and calculated using eqn (3). The sum of true positives corresponds to the number of correctly predicted instances. Precision and its standard deviation were calculated for the six categories for each of the abstracts.
Since the information about the quantity of the extracted distinct entities is important for the knowledge graph extension, it was evaluated using the aforementioned recall metric.
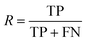 | (1) |
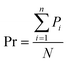 | (2) |
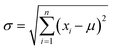 | (3) |
Extraction of sequences was treated as a binary classification problem, where the sum of TP is correctly extracted from distinct entities and is independent from the assigned label. The sum of true positives and false negatives is the total number of distinct manually labelled entities in the text. The metrics were calculated for the ten manually labelled abstracts from papers to the same topic and compared to predictions of both checkpoints I and II. Here, checkpoint I addresses the complemented model, while checkpoint II addresses the pre-trained checkpoint, provided by the developers of CatalysisIE. The resulting metrics are listed in Table 2. Deviations in the metrics of the fourth publication may be due to formatting errors in the retrieved abstract, causing extracted tokens to end with citation numbers (e.g., “catalysts9”), thus not being counted as found entities.
| TP + FN | CP | TP | R | Pr | σ | |
|---|---|---|---|---|---|---|
| 1 | 12 | I | 11 | 91.7 | 83.3 | 40.8 |
| II | 10 | 83.3 | 66.7 | 51.6 | ||
| 2 | 15 | I | 14 | 93.3 | 93.3 | 14.9 |
| II | 12 | 80.0 | 80.0 | 44.7 | ||
| 3 | 15 | I | 13 | 86.7 | 75.6 | 43.3 |
| II | 12 | 70.0 | 73.3 | 43.5 | ||
| 4 | 16 | I | 6 | 37.5 | 35.0 | 23.8 |
| II | 6 | 37.5 | 38.7 | 30.7 | ||
| 5 | 15 | I | 10 | 66.7 | 64.6 | 9.9 |
| II | 7 | 66.0 | 61.8 | 26.7 | ||
| 6 | 15 | I | 13 | 86.7 | 58.3 | 49.2 |
| II | 14 | 93.3 | 66.7 | 51.6 | ||
| 7 | 37 | I | 33 | 89.2 | 88.2 | 21.7 |
| II | 24 | 64.9 | 62.7 | 16.3 | ||
| 8 | 29 | I | 25 | 86.2 | 68.2 | 10.5 |
| II | 24 | 82.8 | 64.7 | 11.4 | ||
| 9 | 25 | I | 12 | 48.0 | 63.6 | 26.0 |
| II | 13 | 52.0 | 68.6 | 25.1 | ||
| 10 | 6 | I | 6 | 100.0 | 100.0 | 0.0 |
| II | 6 | 100.0 | 100.0 | 0.0 |
The entities labelled “Characterization” were predicted least accurately. Additionally, there were no “Treatment” labels in the evaluation dataset. Overall, the model trained on the expanded dataset (CP I) was better at predicting entities labelled “Catalyst”. The average recall of the newly trained model for ten abstracts is equal to 86.67% with a standard deviation of 20.85% and shows a high average precision of 71.90%. In comparison, the recall of the old model (CP II) is 80.00% with a standard deviation of 19.37% and an average precision of 66.67%. In both cases only in one text, precision and recall fall under 50%. Furthermore, for the ten publications shown in Table 2, in the cases where CP I achieved higher precision, σ was lower. This indicates that the dispersion across the different classes in relation to Pr has decreased and therefore the model makes more stable predictions across the classes.
To investigate the performance of the extended model further, ten abstracts from dataset 2 are labelled manually and classified with CP I and CP II to evaluate the metrics as in Table 2. The resulting metrics are presented in more detail in Table 7. An average recall of 82.81% with a standard deviation of 22.39% and an average precision of 71.46% was achieved for CP I. Furthermore, an average recall of 79.47% with a standard deviation of 19.92% and an average precision of 73.20% was achieved for CP II. Thus, the extended model can also be applied on dataset 2.
Title recognition by 19 out of 23 processed PDFs from dataset 1 was successful and 26 from 28 publications from dataset 2 could be recognized correctly. Publications of “Royal Society of Chemistry” could not be correctly recognized because the layout of the publications is not integrated in the workflow of the used pdfdataextractor package.
The AFO was chosen as the initial ontology, because of its linkage to the chemical domain and well-defined structure in the class hierarchy. Table 8 lists the terms and textual definitions assigned as equivalent in ontologies for both datasets, which exist in the AFO and are merged into the working ontology from ChEBI.
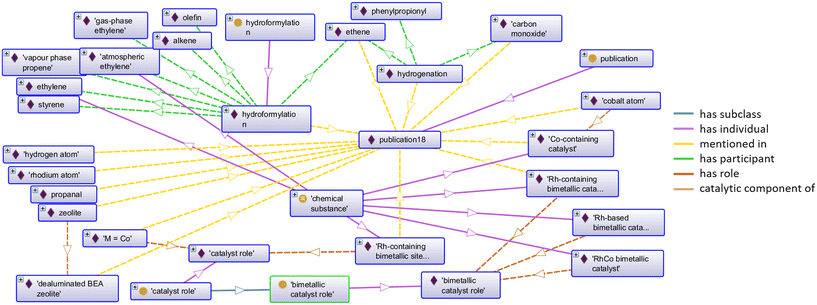
Chemicals which could not be found in PubChem or in ChEBI are created as instances of the class “chemical substance”. For dataset 1, the ontology is extended with 53 instances of “chemical substance”. Dataset 2 results in 55 instances of “chemical substance” that were also created automatically. Each of the generated instances representing extracted entities and their chemical components is provided with a connection to the publication in which it is mentioned and linked to the corresponding roles as shown in an excerpt of the resulting ontology in Fig. 5. The reactions that are mentioned within the publication are listed, including the respective participants of the reactions within the knowledge base (upper area of the figure). The individual “cobalt atom”, for example, is connected with the individual “Co-containing catalyst” via the object property “catalytic component of” (right area of the figure), thus indicating the suitable catalytic component of the concept extracted from text. Furthermore, the role of a “bimetallic catalyst role” is asserted to the three individuals on the bottom right of Fig. 5. The class “bimetallic catalyst role” is created as a subclass of the “catalyst role”, which in turn also has an individual that is connected to other substances via the “has role” object property (bottom left of the figure).
The knowledge graph with publications from dataset 1 was extended by 48 classes from the other ontologies, including their superclasses and interrelations. In total, 331 new classes, 9 new object properties, 2 new data properties, and 155 new individuals were added to the working ontology. From the new classes, 288 were merged from other ontologies, while none of the new individuals were merged from other ontologies, as expected. The new object and data properties were merged from other ontologies.
In the knowledge graph with dataset 2, 39 classes from other ontologies were imported from other ontologies together with their respective superclasses and interrelations. With this, 222 new classes, 4 new object properties, 2 new data properties, and 130 new individuals were added to the working ontology. Here, 198 from the 222 new classes were merged from other ontologies, while also none of the new individuals were merged from other ontologies. The new object and data properties were merged from the other ontologies listed in Table 1 and counted without the ones already presented in Table 4 in the appendix. The explained ontology metrics are listed in Table 3.
| Metric | Initial ontology | Extended ontology dataset 1 | Extended ontology dataset 2 |
|---|---|---|---|
| Classes | 3116 | 3447 | 3338 |
| Instances | 47 | 203 | 178 |
| Logical axioms | 5755 | 6936 | 6596 |
| SubClassOf | 4823 | 5372 | 5174 |
| Equivalent classes | 178 | 188 | 185 |
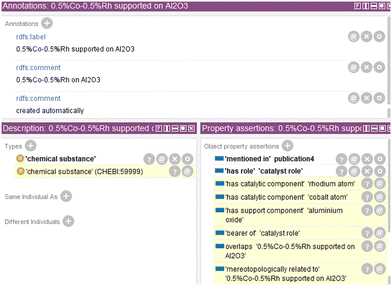
Fig. 6 shows the individual “0.5% Co–0.5% Rh supported on Al2O3” in an excerpt of Protégé after reasoning with HermiT.38 The implicit knowledge is highlighted in yellow, showing an increased semantic expressiveness for the individual describing the catalyst complex. Thus, the individual now also can be found when searching the knowledge graph, e.g., for catalysts that contain cobalt.
Most of the terms in both knowledge graphs originate from the ChEBI ontology and identify chemical compounds and atoms. But also, the classes for such terms as “hydrogenation”, “hydroformylation”, and “acylation” are reused from the RXNO ontology. In the current process, entities representing some chemical groups, such as “phenolic substances” or “phenolic species”, can be recognized with the text mining module, but the extension of the ontology with them is not implemented. This includes, for example, entities such as “phenolic substances”, “phenolic species”, and “alkyl species” which are usually classified as products or reactants in the text. Such entities cannot be queried in PubChem, and in ChEBI, the presumed superclasses are placed in different positions. All queries are formulated within the functions in the module “queries” provided in the GitHub repository of this work39 and can be executed by the Jupiter notebook “user_queries.ipynb” contained in the repository. It contains descriptions of code cells, which execute specific queries, which can answer competency questions formulated in Methods. In addition, some examples of executed functions are also provided in the notebook.
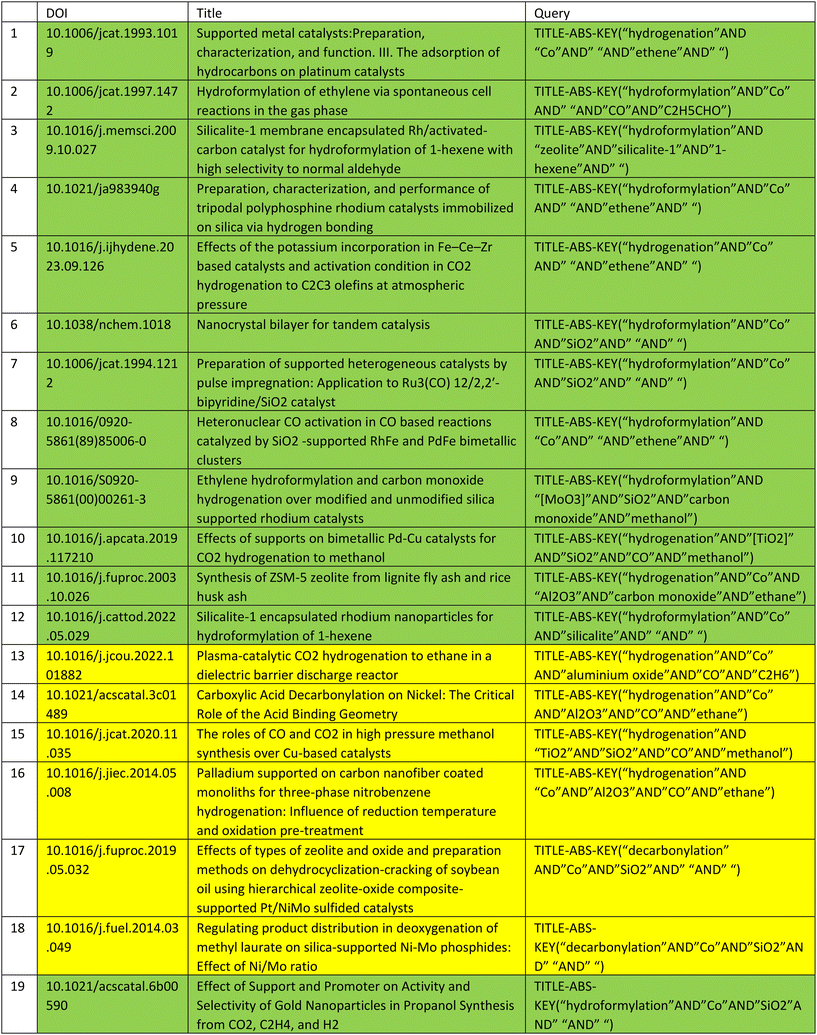
The querying process was evaluated on the generated graph from dataset 1. All of the 19 publications of dataset 1 could be queried without any issues. From 1603 queries retrieved from the ontology with dataset 1, 1092 publications were retrieved, from which 603 publications were distinct publications. This means that 68.1% of queries were formulated correctly and resulted in retrieval and digestion of publications for further integration into the knowledge graph and 44.8% of retrieved publications were duplicates, and thus did not need to be integrated again to the knowledge graph. To constrain the resulting number of publications a bit and focus on currency, the result is filtered for publications that were published in the period between 01.01.2019 and 31.12.2023. This results in 71 unique publications with relevance to the content contained in dataset 1. The time frame can be customized according to user preferences within the querying process. After analysing the queries with the corresponding results, it is noticeable that queries are case insensitive. For example, the query to the Scopus API “TITLE-ABS-KEY (“hydroformylation” AND “Co” AND” “AND “alkene” AND “Aldehyde”)” retrieved among others the publication “Interfacial Tandem Catalysis for Ethylene Carbonylation and C–C Coupling to 3-pentanone on Rh/Ceria”, which according to the query should contain “Co” within the title, abstract or keywords. After analysing the abstract manually, it was determined that it does not contain “Co” and “cobalt” but “CO” instead. This can be attributed to the lack of case sensitivity in the Scopus API. For dataset 2, the Scopus API was queried with 1272 queries which resulted in 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 892 publications from which 9092 were distinct. This corresponds to 25.33% of all retrieved publications, thus leading to 9092 new publications deemed as semantic related to the content of dataset 2.
892 publications from which 9092 were distinct. This corresponds to 25.33% of all retrieved publications, thus leading to 9092 new publications deemed as semantic related to the content of dataset 2.
Listings of the resulting publications are found in the provided GitHub repository of this work39 in the “output” directory.
To further rate the quality of the query result, a random sample in size of 50 publications from the resulting filtered list of 731 publications similar to those of dataset 1 was selected for the evaluation of the queried content. The list of chosen publications for evaluation is provided in the appendix in Table 9.
These publications are rated as similar to the publications in the knowledge graph (of dataset 1) if the following requirements are fulfilled, based on the evaluation of their abstracts and titles:
• Heterogeneous or homogeneous catalysis or catalysts are mentioned.
• Hydroformylation or hydrogenation is mentioned.
• Rh-, Co-, and Ni-based catalysts with silica, zeolite or aluminium oxide as a support material are mentioned.
According to these restrictions, 34 out of 50 publications of the sample provided are rated as similar to the content of the publications within the knowledge graphs, which is equal to 68% accuracy.
Conclusions
This work shows a very elegant way to interconnect information extraction from natural language as provided in scientific contributions with ontologies. A pre-trained model (CatalysisIE) is used for the information extraction from text in combination with regular expressions. To further optimize the classification from the text, CatalysisIE was refined by further training with a self-labeled dataset demonstrating that minimal input from domain experts is required for this fine-tuning. The resulting two models were compared against each other, showing that the further trained model yielded better results. Thus, if information extraction is used to extend ontologies, an extended dataset on the respective domain is needed to enhance precision and recall of the model. With this, six different categories could be extracted from the scientific literature, allowing for the respective classification of the publications that were used as the knowledge-base. Future work could extend this information extraction model to more categories, such as conversion. Two knowledge graphs were created in an automated fashion based on two data sets. These knowledge graphs allowed finding further publications in the same domain of research by querying the Scopus API. Furthermore, SPARQL-queries were preformulated for the created knowledge graphs, giving researchers fast access to the asserted knowledge.The quality of matched articles to the content of the knowledge graphs was evaluated with dataset 1, dealing with hydroformylation reactions. A random sample of 50 publications was investigated in more detail and abstracts, keywords, and titles were screened for the mention of catalysts, hydrogenation or hydroformylation, and whether Rh-, Co-, and Ni-based catalysts with silica, zeolite or aluminium oxide as a support material were mentioned. With these quite strict criteria and with the lack of case sensitivity in the Scopus API, 68% accuracy was achieved for the random sample. This allows for more structured searches of relevant scientific literature in the domain of catalysis research, which is highly important, especially in this domain, as research is quite heterogeneous and the number of relevant publications in the field is quite high. However, a more thorough post-processing of these found publications needs to be conducted, e.g. by a post-processing that conducts a case-sensitive automated search of the respective keywords in the extracted abstracts from Scopus, to improve the accuracy of the output related publications.
Appendix
| Object property | Reverse object property | ||
|---|---|---|---|
| Name | Rdfs:Label | Name | Rdfs:Label |
| supported_on | Supported on | support_material_of | Support material of |
| support_component_of | Support component of | has_support_component | Has support component |
| catalytic_component_of | Catalytic component of | has_catalytic_component | Has catalytic component |
| mentioned_in | Mentioned in | Mentions | Mentions |
| Query no. | Input parameters | Output |
|---|---|---|
| 1 | Doi = r‘10.1021/acsami.0c21749.s001’ | [‘hydroformylation’] |
| 2 | list_type = ‘all’ | [‘hydroformylation’, ‘olefin’, ‘rhodium atom’, ‘Rh-based atomically dispersed catalyst’, ‘Rh supported on ZnO modified with Pi’, ‘zinc oxide’, ‘phosphate ion’, ‘aldehyde’, ‘linear aldehyde’] |
| Doi = r‘10.1021/acsami.0c21749.s001’ | ||
| 3 | Doi = r‘10.1021/acsami.0c21749.s001’ | Abstract: in the study of heterogeneity of homogeneous processes, effective control of the microenvironment of active sites… |
| 4 | Doi = r‘10.1021/acsami.0c21749.s001’ | [[‘10.1021/acscatal.1c02014.s001’], [‘10.1021/acscatal.0c04684.s001’], [‘10.1021/acscatal.1c00705.s002’], [‘10.1021/acscatal.1c04359’],…] |
| 5 | Reac = “hydrogenation”, Doi = none | [[‘10.1021/acscatal.0c04684.s001’], [‘10.1021/acscatal.1c00705.s002’],…] |
| 6 | Cat = “RhCo”, Doi = none | [[‘10.1021/acscatal.0c04684.s001’], [‘10.1021/acscatal.1c00705.s002’], |
| 7 | list_type = ‘all’ | [‘styrene’, ‘cobalt atom’, ‘0.5% Co–0.5% Rh supported on Al2O3’, ‘Co-containing catalyst’, ‘aluminum oxide’, ‘hydroformylation’,…] |
| Doi = none | ||
| 8 | list_type = ‘reactant’ | [‘olefin’] |
| Doi = r‘10.1021/acsami.0c21749.s001’ | ||
| 9 | list_type = ‘product’ | [‘Aldehyd’, ‘RCHO’, ‘aldehidos’, ‘aldehydes’, ‘Aldehyde’, ‘aldehydum’, ‘an aldehyde’, ‘RC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)H’, ‘aldehyde’, ‘aldehido’, ‘linear aldehyde’] O)H’, ‘aldehyde’, ‘aldehido’, ‘linear aldehyde’] |
| Doi = r‘10.1021/acsami.0c21749.s001’ | ||
| 10 | Doi = r‘10.1021/acsami.0c21749.s001’ | [[‘45Rh’, ‘rhodium’, ‘Rh’, ‘rodio’, ‘Rh(111)’, ‘rhodium atom’]], [[‘Rh on ZnO modified with Pi’, ‘Rh supported on ZnO modified with Pi’]] |
| 11 | Doi = r‘10.1021/acsami.0c21749.s001’ | [[‘zinc oxide’, ‘oxyde de zinc’, ‘Zinkoxid’, ‘oxido de cinc’, ‘ZnO’], [‘phosphate ions’, ‘Pi’, ‘phosphate’, ‘phosphate ion’]] |
| 12 | Doi = r‘10.1021/acscatal.1c02014.s001’ | [[‘silicon dioxide’, ‘Rh supported on SiO2 catalyst’], [‘silicon dioxide’, ‘Rh2P nanoparticle supported on SiO2 support material’], [‘silicon dioxide’, ‘Rh7Co1P4 supported on SiO2’],…] |
| only_doi = false |
| TP + FN | CP | TP | R | Pr | σ | |
|---|---|---|---|---|---|---|
| 1 | 15 | I | 13 | 86.7 | 68.3 | 32.5 |
| II | 12 | 80.0 | 61.7 | 36.1 | ||
| 2 | 19 | I | 15 | 78.9 | 82.7 | 28.9 |
| II | 15 | 78.9 | 82.7 | 28.9 | ||
| 3 | 9 | I | 9 | 100.0 | 93.3 | 14.9 |
| II | 9 | 100.0 | 93.3 | 14.9 | ||
| 4 | 11 | I | 6 | 54.5 | 66.7 | 28.9 |
| II | 7 | 63.6 | 70.8 | 26.0 | ||
| 5 | 6 | I | 6 | 100.0 | 100.0 | 0.0 |
| II | 6 | 100.0 | 100.0 | 0.0 | ||
| 6 | 14 | I | 8 | 57.1 | 43.8 | 51.5 |
| II | 8 | 57.1 | 43.8 | 51.5 | ||
| 7 | 12 | I | 12 | 100.0 | 73.6 | 29.7 |
| II | 11 | 91.7 | 72.6 | 42.6 | ||
| 8 | 12 | I | 7 | 58.3 | 69.3 | 41.3 |
| II | 6 | 50.0 | 49.3 | 46.6 | ||
| 9 | 9 | I | 4 | 44.4 | 69.0 | 27.0 |
| II | 5 | 55.6 | 73.8 | 25.1 | ||
| 10 | 15 | I | 15 | 100.0 | 92.6 | 13.5 |
| II | 15 | 100.0 | 92.6 | 13.5 |
| Term label | IRIs + Definitions | |
|---|---|---|
| In the AFO | In ChEBI | |
| ‘Chemical substance’ | http://purl.allotrope.org/ontologies/material#AFM_0001097 | http://purl.obolibrary.org/obo/CHEBI_33250 |
| A chemical substance is a portion of material that is matter of constant composition best characterized by the entities (molecules, formula units, atoms) it is composed of [IUPAC] | A chemical entity constituting the smallest component of an element having the chemical properties of the element | |
| ‘Anion’ | http://purl.allotrope.org/ontologies/material#AFM_0000161 | http://purl.obolibrary.org/obo/CHEBI_22563 |
| An anion (−) is an ion with more electrons than protons, giving it a net negative charge (since electrons are negatively charged and protons are positively charged) | A monoatomic or polyatomic species having one or more elementary charges of the electron | |
| ‘Ion’ | http://purl.allotrope.org/ontologies/material#AFM_0000077 | http://purl.obolibrary.org/obo/CHEBI_24870 |
| An ion is an atom or molecule in which the total number of electrons is not equal to the total number of protons, giving the atom or molecule a net positive or negative electrical charge | A molecular entity having a net electric charge | |
| ‘Role’ | http://purl.obolibrary.org/obo/BFO_0000023 | http://purl.obolibrary.org/obo/CHEBI_50906 |
| B is a role means: b is a realizable entity and b exists because there is some single bearer that is in some special physical, social, or institutional set of circumstances in which this bearer does not have to be and b is not such that, if it ceases to exist, then the physical make-up of the bearer is thereby changed [BFO] | A role is particular behavior which a material entity may exhibit | |
| ‘Cation’ | http://purl.allotrope.org/ontologies/material#AFM_0000189 | http://purl.obolibrary.org/obo/CHEBI_36916 |
| A cation (+) is an ion with fewer electrons than protons, giving it a positive charge | A monoatomic or polyatomic species having one or more elementary charges of the proton | |
| ‘Group’ | http://purl.obolibrary.org/obo/BFO_0000023 | http://purl.obolibrary.org/obo/CHEBI_24433 |
| A group is an aggregate of people | A defined linked collection of atoms or a single atom within a molecular entity | |
| ‘Atom’ | http://purl.allotrope.org/ontologies/material#AFM_0001028 | http://purl.obolibrary.org/obo/CHEBI_33250 |
| An atom is a smallest particle still characterizing a chemical element. It consists of a nucleus of a positive charge carrying almost all its mass (more than 99.9%) and Z electrons determining its size | A chemical entity constituting the smallest component of an element having the chemical properties of the element | |
| ‘Chemical substance’ | http://purl.allotrope.org/ontologies/material#AFM_0001097 | http://purl.obolibrary.org/obo/CHEBI_59999 |
| A chemical substance is a portion of material that is matter of constant composition best characterized by the entities (molecules, formula units, atoms) it is composed of | A chemical substance is a portion of matter of constant composition, composed of molecular entities of the same type or of different types | |
Data availability
Data for this article and the associated codes are available on GitHub at https://github.com/AleSteB/CatalysisIE_Knowledge_Graph_Generator. The checkpoint of the extended CatalysisIE model is available on Zenodo at https://zenodo.org/records/12634956.Author contributions
A. S. B.: conceptualization, data curation, methodology, validation, supervision, writing – original draft, writing – review & editing, visualization. D. C.: conceptualization, data curation, methodology, software, validation, investigation, writing – original draft, writing – review & editing. D. K.: methodology, writing – review & editing. A. N.: data curation. S. H.: data curation, writing – review & editing. S. A. S.: writing – review & editing. N. K.: conceptualization, funding acquisition, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the Deutsche Forschungsgemeinschaft (DFG) for funding this research as part of the Nationale Forschungsdateninfrastruktur (NFDI) initiative (grant no.: NFDI/2-1-2021). A.S.B. thanks the networking program ‘Sustainable Chemical Synthesis 2.0’ (SusChemSys 2.0) for the support and fruitful discussions across disciplines.References
- D. W. Hook, S. J. Porter and C. Herzog, Dimensions: Building Context for Search and Evaluation, Front. Res. Metr. Anal., 2018, 3 DOI:10.3389/frma.2018.00023.
- M. D. Wilkinson, et al., The FAIR Guiding Principles for scientific data management and stewardship, Sci. Data, 2016, 3, 160018, DOI:10.1038/sdata.2016.18.
- A. Salazar, B. Wentzel, S. Schimmler, R. Gläser, S. Hanf and S. A. Schunk, How Research Data Management Plans Can Help in Harmonizing Open Science and Approaches in the Digital Economy, Chemistry, 2023, 29(9), e202202720, DOI:10.1002/chem.202202720.
- B. V. Elsevier, Scopus, 2024. Accessed: February 2024. [Online]. Available: https://www.scopus.com/.
- C. P. Marshall, J. Schumann and A. Trunschke, Achieving Digital Catalysis: Strategies for Data Acquisition, Storage and Use, Angew. Chem., Int. Ed., 2023, 62(30), e202302971, DOI:10.1002/anie.202302971.
- M. Suvarna, A. C. Vaucher, S. Mitchell, T. Laino and J. Pérez-Ramírez, Language models and protocol standardization guidelines for accelerating synthesis planning in heterogeneous catalysis, Nat. Commun., 2023, 14(1), 7964, DOI:10.1038/s41467-023-43836-5.
- S. Mishra and S. Jain, A Study of Various Approaches and Tools on Ontology, in 2015 IEEE International Conference on Computational Intelligence & Communication Technology, Ghaziabad, India, 2015, pp. 57–61 Search PubMed.
- A. S. Behr, M. Völkenrath and N. Kockmann, Ontology extension with NLP-based concept extraction for domain experts in catalytic sciences, Knowl. Inf. Syst., 2023, 65(12), 5503–5522, DOI:10.1007/s10115-023-01919-1.
- A. Zouaq, D. Gasevic and M. Hatala, Towards open ontology learning and filtering, Information Systems, 2011, 36(7), 1064–1081, DOI:10.1016/j.is.2011.03.005.
- Y. Zhang, C. Wang, M. Soukaseum, D. G. Vlachos and H. Fang, Unleashing the Power of Knowledge Extraction from Scientific Literature in Catalysis, J. Chem. Inf. Model., 2022, 62(14), 3316–3330, DOI:10.1021/acs.jcim.2c00359.
- I. Beltagy, K. Lo and A. Cohan, SciBERT: A Pretrained Language Model for Scientific Text, EMNLP. [Online]. Available: https://arxiv.org/pdf/1903.10676.pdf.
- W3C Sparql 1.1. [Online]. Available: https://www.w3.org/TR/sparql11-update/.
- J. Hastings, et al., ChEBI in 2016: Improved services and an expanding collection of metabolites, Nucleic Acids Res., 2016, 44(D1), D1214–D1219, DOI:10.1093/nar/gkv1031.
- CrossRef, CrossRef API Documentation. Accessed: 2024.
- S. Chamberlain, J. Maupetit, S. Peak, C. Talbert, D. Himmelstein and K. Niemeyer, Habanero: Python client for the Crossref API, 2024, Accessed: 2024. [Online]. Available: https://github.com/sckott/habanero.
- M. E. Rose and J. R. Kitchin, pybliometrics: Scriptable bibliometrics using a Python interface to Scopus, SoftwareX, 2019, 10, 100263, DOI:10.1016/j.softx.2019.100263.
- M. Zhu and J. M. Cole, PDFDataExtractor: A Tool for Reading Scientific Text and Interpreting Metadata from the Typeset Literature in the Portable Document Format, J. Chem. Inf. Model., 2022, 62(7), 1633–1643, DOI:10.1021/acs.jcim.1c01198.
- M. C. Swain and J. M. Cole, ChemDataExtractor: A Toolkit for Automated Extraction of Chemical Information from the Scientific Literature, J. Chem. Inf. Model., 2016, 56(10), 1894–1904, DOI:10.1021/acs.jcim.6b00207.
- Python Software Foundation, re - Regular expression operations, 2024.
- S. Kim, et al., PubChem 2023 update, Nucleic Acids Res., 2023, 51(D1), D1373–D1380, DOI:10.1093/nar/gkac956.
- M. Swain, PubChemPy: A way to interact with PubChem in Python, 2014, [Online]. Available: https://github.com/mcs07/PubChemPy.
- Allotrope Foundation, Allotrope Foundation Ontologies. Accessed: 2022.
- I. Montani et al., spaCy: Industrial-strength Natural Language Processing in Python, 2022.
- A. S. Behr, H. Borgelt and N. Kockmann, Ontologies4Cat: investigating the landscape of ontologies for catalysis research data management, J. Cheminf., 2024, 16(1), 16, DOI:10.1186/s13321-024-00807-2.
- R. Arp, B. Smith and A. D. Spear, Building ontologies with Basic Formal Ontology, Massachusetts Institute of Technology, Cambridge, Massachusetts, 2015 Search PubMed.
- C. Batchelor, Molecular Process Ontology (MOP). [Online]. Available: https://github.com/rsc-ontologies/rxno.
- C. Batchelor, Chemical Reactions Ontology (RXNO). [Online]. Available: https://github.com/rsc-ontologies/rxno.
- R. C. Jackson, J. P. Balhoff, E. Douglass, N. L. Harris, C. J. Mungall and J. A. Overton, ROBOT: A Tool for Automating Ontology Workflows, BMC Bioinf., 2019, 20(1), 407, DOI:10.1186/s12859-019-3002-3.
- J.-B. Lamy, Owlready: Ontology-oriented programming in Python with automatic classification and high level constructs for biomedical ontologies, Artif. Intell. Med., 2017, 80, 11–28, DOI:10.1016/j.artmed.2017.07.002.
- P. Sun and S. Zhang, Identifying Granularity Differences between Large Biomedical Ontologies through Rules, AMIA Annu. Symp. Proc., 2010, 2010, 927–931 Search PubMed.
- P. Strömert, J. Hunold, A. Castro, S. Neumann and O. Koepler, Ontologies4Chem: the landscape of ontologies in chemistry, Pure Appl. Chem., 2022, 94(6), 605–622, DOI:10.1515/pac-2021-2007.
- SPARQL 1.1 Query Language, ed. E. Prud'hommeaux, S. Harris and A. Seaborne, W3C, 2013, [Online] Available: https://www.w3.org/TR/sparql11-query Search PubMed.
- W. McKinney, Data Structures for Statistical Computing in Python, in Proceedings of the 9th Python in Science Conference, Austin, Texas, 2010, pp. 56–61 Search PubMed.
- Y. Liu, et al., Rhodium nanoparticles supported on silanol-rich zeolites beyond the homogeneous Wilkinson's catalyst for hydroformylation of olefins, Nat. Commun., 2023, 14(1), 2531, DOI:10.1038/s41467-023-38181-6.
- S. Hanf, L. Alvarado Rupflin, R. Gläser and S. Schunk, Current State of the Art of the Solid Rh-Based Catalyzed Hydroformylation of Short-Chain Olefins, Catalysts, 2020, 10(5), 510, DOI:10.3390/catal10050510.
- K. Ghaib, K. Nitz and F.-Z. Ben-Fares, Chemical Methanation of CO 2 : A Review, ChemBioEng Rev., 2016, 3(6), 266–275, DOI:10.1002/cben.201600022.
- M. Tkachenko, M. Malyuk, A. Holmanyuk and N. Liubimov, Label Studio: Data labeling software.
- B. Motik, R. Shearer, G. Stoils and I. Horrocks, HermiT OWL Reasoner: The New Kid on the OWL Block, University of Oxford, Accessed: May 14 2022. [Online]. Available: https://www.hermit-reasoner.com/.
- A. S. Behr and D. Chernenko, CatalysisIE Knowledge Graph Generator. [Online]. Available: https://github.com/AleSteB/CatalysisIE_Knowledge_Graph_Generator.
Footnote |
| † Electronic supplementary information (ESI) available: https://github.com/AleSteB/CatalysisIE_Knowledge_Graph_Generator. See DOI: https://doi.org/10.1039/d4cy00369a |
| This journal is © The Royal Society of Chemistry 2024 |