Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Machine learning-guided high throughput nanoparticle design†
Ana
Ortiz-Perez‡
 a,
Derek
van Tilborg‡
a,
Derek
van Tilborg‡
 ab,
Roy
van der Meel
ab,
Roy
van der Meel
 a,
Francesca
Grisoni
a,
Francesca
Grisoni
 *ab and
Lorenzo
Albertazzi
*ab and
Lorenzo
Albertazzi
 *a
*a
aInstitute for Complex Molecular Systems (ICMS), Department of Biomedical Engineering, Eindhoven University of Technology, PO Box 513, 5600 MB Eindhoven, The Netherlands. E-mail: l.albertazzi@tue.nl
bCentre for Living Technologies, Alliance TU/e, WUR, UU, UMC Utrecht, Princetonlaan 6, 3584 CB, Utrecht, The Netherlands. E-mail: f.grisoni@tue.nl
First published on 3rd June 2024
Abstract
Designing nanoparticles with desired properties is a challenging endeavor, due to the large combinatorial space and complex structure–function relationships. High throughput methodologies and machine learning approaches are attractive and emergent strategies to accelerate nanoparticle composition design. To date, how to combine nanoparticle formulation, screening, and computational decision-making into a single effective workflow is underexplored. In this study, we showcase the integration of three key technologies, namely microfluidic-based formulation, high content imaging, and active machine learning. As a case study, we apply our approach for designing PLGA-PEG nanoparticles with high uptake in human breast cancer cells. Starting from a small set of nanoparticles for model training, our approach led to an increase in uptake from ∼5-fold to ∼15-fold in only two machine learning guided iterations, taking one week each. To the best of our knowledge, this is the first time that these three technologies have been successfully integrated to optimize a biological response through nanoparticle composition. Our results underscore the potential of the proposed platform for rapid and unbiased nanoparticle optimization.
Introduction
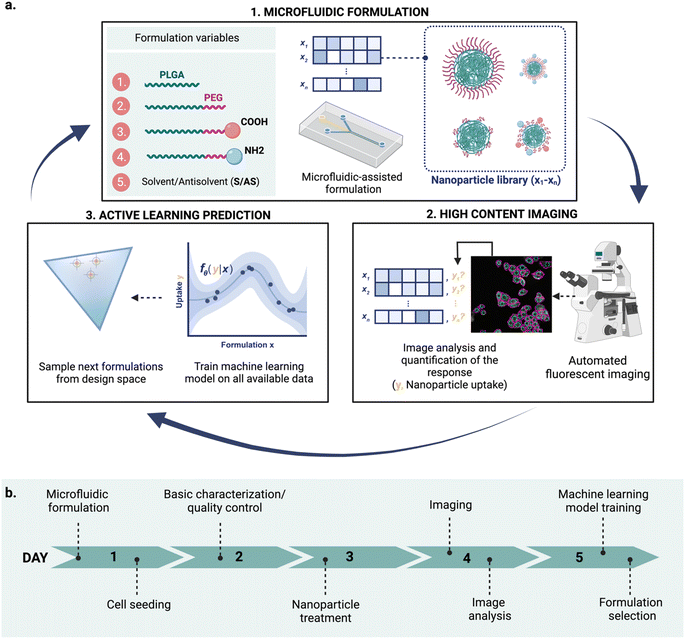
Nanomedicines are relevant for a variety of biomedical applications,1 from diagnosis2 and disease prevention3 to novel therapeutic approaches.4 Nanomedicine platforms with a wide range of physicochemical properties can be engineered using a variety of materials,5,6 by tuning nanoparticle composition and formulation variables.7,8 These properties, in turn, influence nanoparticle fate and their ability to cross biological barriers.5,9,10 This versatility opens opportunities to build tailored carriers for a specific application and patient populations5 but also poses a great challenge towards the design of optimal materials. The resulting enormous combinatorial design space – realistically consisting of thousands of formulations for a single nanoparticle type – makes formulation exploration a daunting task. Thus, we need efficient ways to navigate this vast space, in a time- and cost-effective manner. Novel tools for high-throughput formulation and screening, as well as data-driven computational methods for nanoparticle design hold a great promise to revolutionize the current landscape of material discovery.However, integrating these tools into a single robust, rapid, and effective workflow is still an open question. In this study, we combined three key technologies: microfluidic formulation, high content imaging, and active machine learning into an iterative workflow to accelerate nanoparticle design (Fig. 1).
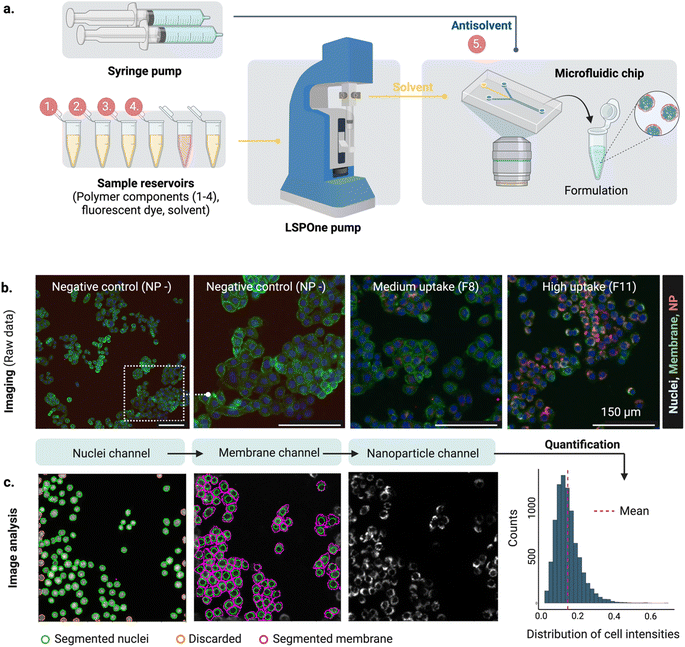
Microfluidics offers a versatile platform for rapid and reproducible production of highly monodispersed nanoparticles11,12 compared to standard bulk formulation. Control over formulation parameters, such as the solvent mixing rate, is achieved by handling small volumes of liquids in highly controlled environments. The solvent mixing rate drives the formulation of several self-assembling nanoparticles including amphiphilic lipids and polymers13 and controls physical properties like size.
In parallel, the spread of fluorescence-based microscopy together with the rapid development of bio-image analysis tools14 and automation has enabled the high throughput screening of nanocarriers using high content imaging (HCI). HCI combines automated fluorescence imaging and analysis, providing quantitative multiparametric data from images.15,16
HCI-based assays can then be used to understand the impact of the nanoparticle on the cell, including uptake,17,18 endosomal escape,19 or cytotoxicity,20 assisting the rational design of nanoparticles.
Finally, machine learning can be used to guide nanoparticle development21,22 with the aim of reducing the number of nanoparticle formulations needed to optimize a response. Since the number of available data is often highly limited, specific machine learning strategies like active machine learning are particularly suited for this task.23–27 By operating in an iterative fashion, active machine learning uses model predictions to decide which samples should be screened and added to the training data to update the model in the next cycle.28,29 This allows models to reach a desired response faster by screening fewer samples. Furthermore, the iterative nature of active learning makes it fitting for integration with automated design platforms where nanoparticles designs are optimized sequentially.
Although these techniques have been widely explored on their own, combining their advantages can potentially accelerate nanoparticle design. Here, we demonstrate an integrated and semi-automated iterative workflow for rapid nanoparticle design (Fig. 1a), combining the strengths of (1) microfluidic-assisted nanoparticle formulation, (2) HCI, and (3) active machine learning. We apply this iterative approach to find poly(lactic-co-glycolic acid)-polyethylene glycol (PLGA-PEG) compositions that yield a high uptake in MDA-MB-468 human breast cancer cells. Owing to its modular character, the approach can be adapted to explore other nanoparticle formulations and responses of interest beyond uptake.
Results & discussion
As a case study, we focused on PLGA-PEG nanoparticles. PLGA-PEG is an amphiphilic block copolymer that self-assembles into nanospheres via nanoprecipitation. This type of formulation can be easily adapted to the microfluidic format, which offers several advantages over traditional formulation, such as size tunability by controlling the fluidic parameters.8,30,31 In addition, PLGA-PEG has excellent biocompatibility and high tunability. The base polymer can be manufactured with different properties, such as molecular weight or functional end-groups. For creating a library of non-targeted PLGA-PEG based nanoparticles, we chose to vary four different building blocks (PLGA, PLGA-PEG, PLGA-PEG-COOH, PLGA-PEG-NH2) and one process variable (the flow rate ratio between the solvent and antisolvent). By varying building components that directly influence physicochemical properties (size, PEGylation, and charge), we aim to maximize their uptake in a model of human breast cancer.Platform for nanoparticle design
Our proposed workflow is constituted of three components (microfluidics formulation, HCI and machine learning), each of which contributes to the ‘experimental cycle’ represented in Fig. 1a. Each cycle can be performed in a week (Fig. 1b), allowing for rapid design iterations. The three components of our platform are the following.At each cycle, the three technologies work complementary as follows: (a) microfluidics technology is used for nanoparticle production, (b) the obtained nanoparticles are analyzed using HCI for property determination, and (c) the experimental results are used to train the machine learning model, which is then used to suggest what to formulate next. The optimal learning strategy (exploration vs. exploitation) over cycles is not predetermined and can be adjusted upon learned insights. Choosing between exploration and exploitation is case-dependent and it is ultimately decided by the scientist.
Designing PLGA-PEG nanoparticles for high uptake
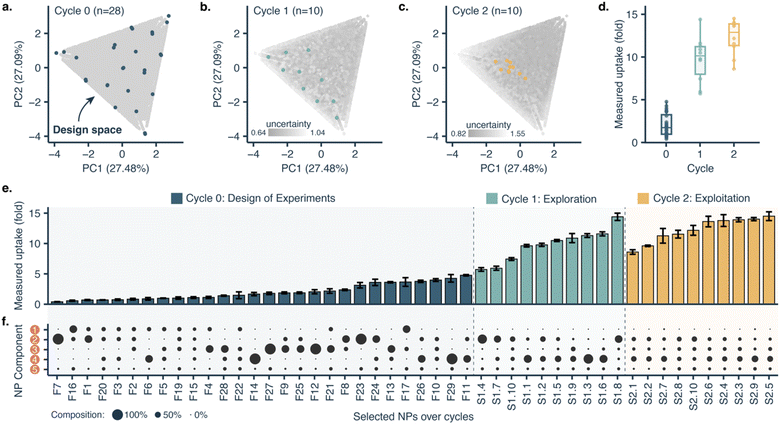
The proposed design platform was used to perform three cycles. Per each cycle, nanoparticles were produced in the microfluidics platform with the chosen formulation. Hydrophobic fluorescent dyes (DiD) were incorporated into the polymer mixture to allow for estimation of cell accumulation. Cell uptake was determined via HCI and expressed as fold-increase accumulation (compared to the uptake control, a 100% PLGA-PEG nanoparticle formulated in bulk).The measured response per nanoparticle was used to train the Bayesian neural network for uptake prediction. The trained model was then used to select the next cycle formulations from a virtual library of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nanoparticles spanning the entire design space homogenously. Formulations were considered only if their predicted polydispersity index (PDI) was lower than a predetermined threshold (PDI < 0.2, predicted with a different machine learning model, see Materials and methods). Filtering via PDI is a form of quality control ensuring the produced nanoparticles are colloidally stable and suitable for biomedical applications such as drug delivery. As a learning strategy, we started by exploring the uncertain areas of the design space (exploration), after which we aimed to find high response nanoparticles (exploitation). As a result, the study was executed in three cycles, as described below (Fig. 3).
000 nanoparticles spanning the entire design space homogenously. Formulations were considered only if their predicted polydispersity index (PDI) was lower than a predetermined threshold (PDI < 0.2, predicted with a different machine learning model, see Materials and methods). Filtering via PDI is a form of quality control ensuring the produced nanoparticles are colloidally stable and suitable for biomedical applications such as drug delivery. As a learning strategy, we started by exploring the uncertain areas of the design space (exploration), after which we aimed to find high response nanoparticles (exploitation). As a result, the study was executed in three cycles, as described below (Fig. 3).
With only three full cycles we were able to move from a mean uptake of 2.03 ± 1.28-fold in the initial set to 12.30 ± 2.02-fold in the last cycle. The maximal uptake improved from 4.77-fold in the first cycle to 14.50-fold in the last cycle.
Model interpretation
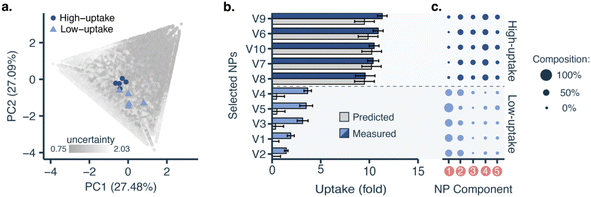
To fully leverage what the machine learning model learned from the data, we applied it to interrogate nanoparticle composition–function relationships. We retrained the model with all the data generated from all cycles and used it to select five nanoparticles with low predicted uptake and five with high predicted uptake from the virtual library for further formulation and screening (Fig. 4a). Although the model was better attuned to high-uptake formulations, it was able to identify both high-uptake (10.54 ± 0.66-fold) and low-uptake nanoparticles (2.74 ± 0.99-fold), with statistically significant differences (p < 0.001, two-tailed t-test, Fig. 4b). This shows that the model has learnt relevant formulation–uptake relationships.Low- and high-uptake nanoparticles showed statistically significant differences (p < 0.001, two-tailed t-test) in the content of three polymers (Fig. 4c): (1) PLGA (low-uptake: 53 ± 9%, high-uptake: 5 ± 5%), (2) PLGA-PEG-COOH (low-uptake: 3 ± 5%, high-uptake: 24.4 ± 4.8%), and (3) PLGA-PEG-NH2 (low-uptake: 10 ± 6%, high-uptake: 25 ± 5%). This is also reflected in the predictions over the whole design space (see ESI†). Furthermore, low-uptake nanoparticles were found to be more monodisperse (PDI = 0.059 ± 0.013) and bigger (size = 154.5 ± 21.4 nm) than high-uptake nanoparticles (PDI = 0.122 ± 0.015, size = 114.0 ± 5.2 nm). Polydispersity or nanoparticle heterogeneity was traditionally seen as an undesired property. However, this intrinsic heterogeneity can be considered a structural parameter contributing to nanoparticle fate and biological function.40
Conclusions and outlook
In this work, we demonstrate a nanoparticle design platform combining three complementary technologies, namely microfluidics-assisted formulation, high content imaging, and machine learning. These three technologies have been tuned to work synergistically within an active learning framework, where the results of each experimental cycle are used to inform the next.As a proof-of-concept, we applied our approach for designing PLGA-PEG nanoparticles with high uptake in MDA-MB-468 human breast cancer cells. With only two experimental cycles of 5 days each, we were able to triple the measured uptake from ∼5-fold to ∼15-fold. The resulting model was able to generate low uptake and high uptake nanoparticles based on their composition. Such a model could be used for exploring the relationships between the nanoparticle components and their function as an ‘hypothesis generator’. These results demonstrate the potential of this approach to efficiently navigate complex design spaces of multicomponent nanoparticles.
Owing to its modularity, this approach can be further expanded to tackle virtually any nanoparticle formulation. In the future, we will apply this approach for designing nanoparticles with relevant translational properties, such as selective cytotoxicity in cancer cells, or the capability to deliver functional cargo to target cells. Moreover, the approach is generalizable to a range of nanomaterials and can be expanded to models with different biological complexities, e.g., cell lines, patient-derived organoids, or organs-on-a-chip. Our approach demonstrates the potential of closed-loop platforms for rapid and iterative nanoparticle optimization driven by machine learning.
Materials and methods
Nanoparticle formulation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 wt
1 wt![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) wt ratio. The mixture was degassed in a desiccator, poured over the master mold, degassed once more and baked overnight at 60 °C. After elastomer curation, the PMDS chips were peeled off from the master mold, the inlets and outlets were punched with a 1.2 mm biopsy puncher and stored in a dust-free environment. On the same day of formulation, to keep surface hydrophilicity, the PDMS replica was freshly bonded to a clean 25 × 75 mm glass slide using oxygen plasma (at 20 W for 30 s), achieved with an Emitech K1050X Plasma Asher from Quorum (East Sussex, UK).
wt ratio. The mixture was degassed in a desiccator, poured over the master mold, degassed once more and baked overnight at 60 °C. After elastomer curation, the PMDS chips were peeled off from the master mold, the inlets and outlets were punched with a 1.2 mm biopsy puncher and stored in a dust-free environment. On the same day of formulation, to keep surface hydrophilicity, the PDMS replica was freshly bonded to a clean 25 × 75 mm glass slide using oxygen plasma (at 20 W for 30 s), achieved with an Emitech K1050X Plasma Asher from Quorum (East Sussex, UK).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, at room temperature under stirring (700 rpm). The resulting nanoparticle solution (1 mg ml−1) was left under stirring (400 rpm) on a shaker overnight at room temperature, protected from light, to let the acetonitrile evaporate.
10, at room temperature under stirring (700 rpm). The resulting nanoparticle solution (1 mg ml−1) was left under stirring (400 rpm) on a shaker overnight at room temperature, protected from light, to let the acetonitrile evaporate.
Nanoparticle characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) before measurement. Nanoparticle uptake standard was always included on the plate. Fluorescence coefficients to correct for differences in fluorescence intensity were calculated for each batch in comparison to the uptake standard.
10) before measurement. Nanoparticle uptake standard was always included on the plate. Fluorescence coefficients to correct for differences in fluorescence intensity were calculated for each batch in comparison to the uptake standard.
Cells were seeded at a density of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 in an “ibiTreat” μ-Plate 96 well back (Cat no. 89626) from IBIDI (Gräfelfing, Germany), cultured for 38–48 h before nanoparticle treatment. Half an hour before starting the treatment, nanoparticle stock solutions were pre-incubated with human serum (1
000 cells per cm2 in an “ibiTreat” μ-Plate 96 well back (Cat no. 89626) from IBIDI (Gräfelfing, Germany), cultured for 38–48 h before nanoparticle treatment. Half an hour before starting the treatment, nanoparticle stock solutions were pre-incubated with human serum (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v
1, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) for 30 min at 37 °C. Cells were washed three times with serum-free phenol-free DMEM media, and each nanoparticle condition (pre-incubated with serum) was added to each well at a working concentration of 50 μg ml−1. The resulting “incubation media” contained 5% human serum. After 23.5 h, the cells were counterstained with Hoechst and Alexa Fluor™ 488 WGA at 37 °C. After 24 h, cells were washed with serum-free media 3 times, fixed with PFA 2% (diluted in DPBS 1x), for 10 min, at room temperature. After fixation, cells were washed three times with DPBS 1x and stored at 4 °C protected from light until imaging.
v) for 30 min at 37 °C. Cells were washed three times with serum-free phenol-free DMEM media, and each nanoparticle condition (pre-incubated with serum) was added to each well at a working concentration of 50 μg ml−1. The resulting “incubation media” contained 5% human serum. After 23.5 h, the cells were counterstained with Hoechst and Alexa Fluor™ 488 WGA at 37 °C. After 24 h, cells were washed with serum-free media 3 times, fixed with PFA 2% (diluted in DPBS 1x), for 10 min, at room temperature. After fixation, cells were washed three times with DPBS 1x and stored at 4 °C protected from light until imaging.
Machine learning and computation
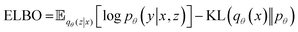 | (1) |
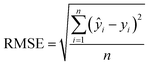 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 virtual formulations were sampled from of the 1.85 × 108 theoretically possible formulations (see ESI†).
000 virtual formulations were sampled from of the 1.85 × 108 theoretically possible formulations (see ESI†).
Data availability
All code, trained models, and results are available on GitHub: https://github.com/molML/Nano_Particles_Active_Learning. All raw screening data are available on: https://zenodo.org/records/8289605.Author contributions
Conceptualization: AOP, DvT, LA, FG; methodology: AOP, DvT, LA, FG; experiments (wet-lab): AOP; experiments (computational): DvT; formal analysis and investigation: AOP, DvT, with contributions from all authors; writing – original draft: AOP, DvT; writing – review & editing: all authors.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We would like to thank Emiel Visser for providing the MATLAB script to automate the LSPOne pump and for an Eppendorf holder CAD design. We would also like to thank Stijn Haenen for 3D printing the Eppendorf holders. AOP and LA are supported by NWO through a Vidi Grant (192.028). RvdM is supported by a Vidi grant (19861) from the Dutch Research Council (NWO). FG acknowledges the support of the Centre for Living Technologies.References
- B. Pelaz, C. Alexiou, R. A. Alvarez-Puebla, F. Alves, A. M. Andrews, S. Ashraf, L. P. Balogh, L. Ballerini, A. Bestetti, C. Brendel, S. Bosi, M. Carril, W. C. W. Chan, C. Chen, X. Chen, X. Chen, Z. Cheng, D. Cui, J. Du, C. Dullin, A. Escudero, N. Feliu, M. Gao, M. George, Y. Gogotsi, A. Grünweller, Z. Gu, N. J. Halas, N. Hampp, R. K. Hartmann, M. C. Hersam, P. Hunziker, J. Jian, X. Jiang, P. Jungebluth, P. Kadhiresan, K. Kataoka, A. Khademhosseini, J. Kopeček, N. A. Kotov, H. F. Krug, D. S. Lee, C.-M. Lehr, K. W. Leong, X.-J. Liang, M. Ling Lim, L. M. Liz-Marzán, X. Ma, P. Macchiarini, H. Meng, H. Möhwald, P. Mulvaney, A. E. Nel, S. Nie, P. Nordlander, T. Okano, J. Oliveira, T. H. Park, R. M. Penner, M. Prato, V. Puntes, V. M. Rotello, A. Samarakoon, R. E. Schaak, Y. Shen, S. Sjöqvist, A. G. Skirtach, M. G. Soliman, M. M. Stevens, H.-W. Sung, B. Z. Tang, R. Tietze, B. N. Udugama, J. S. VanEpps, T. Weil, P. S. Weiss, I. Willner, Y. Wu, L. Yang, Z. Yue, Q. Zhang, Q. Zhang, X.-E. Zhang, Y. Zhao, X. Zhou and W. J. Parak, Diverse Applications of Nanomedicine, ACS Nano, 2017, 11(3), 2313–2381, DOI:10.1021/acsnano.6b06040.
- M. Murar, L. Albertazzi and S. Pujals, Advanced Optical Imaging-Guided Nanotheranostics towards Personalized Cancer Drug Delivery, Nanomaterials, 2022, 12(3), 399, DOI:10.3390/nano12030399.
- C. Feng, Y. Li, B. E. Ferdows, D. N. Patel, J. Ouyang, Z. Tang, N. Kong, E. Chen and W. Tao, Emerging Vaccine Nanotechnology: From Defense against Infection to Sniping Cancer, Acta Pharm. Sin. B, 2022, 12(5), 2206–2223, DOI:10.1016/j.apsb.2021.12.021.
- J. Nam, S. Son, K. S. Park, W. Zou, L. D. Shea and J. J. Moon, Cancer Nanomedicine for Combination Cancer Immunotherapy, Nat. Rev. Mater., 2019, 4(6), 398–414, DOI:10.1038/s41578-019-0108-1.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Engineering Precision Nanoparticles for Drug Delivery, Nat. Rev. Drug Discovery, 2021, 20(2), 101–124, DOI:10.1038/s41573-020-0090-8.
- R. T. Stiepel, E. Duggan, C. J. Batty and K. M. Ainslie, Micro and Nanotechnologies: The Little Formulations That Could, Bioeng. Transl. Med., 2023, 8(2), e10421, DOI:10.1002/btm2.10421.
- G. Yamankurt, E. J. Berns, A. Xue, A. Lee, N. Bagheri, M. Mrksich and C. A. Mirkin, Exploration of the Nanomedicine-Design Space with High-Throughput Screening and Machine Learning, Nat. Biomed. Eng., 2019, 3(4), 318–327, DOI:10.1038/s41551-019-0351-1.
- P. M. Valencia, E. M. Pridgen, M. Rhee, R. Langer, O. C. Farokhzad and R. Karnik, Microfluidic Platform for Combinatorial Synthesis and Optimization of Targeted Nanoparticles for Cancer Therapy, ACS Nano, 2013, 7(12), 10671–10680, DOI:10.1021/nn403370e.
- E. Blanco, H. Shen and M. Ferrari, Principles of Nanoparticle Design for Overcoming Biological Barriers to Drug Delivery, Nat. Biotechnol., 2015, 33(9), 941–951, DOI:10.1038/nbt.3330.
- W. Poon, B. R. Kingston, B. Ouyang, W. Ngo and W. C. W. Chan, A Framework for Designing Delivery Systems, Nat. Nanotechnol., 2020, 15(10), 819–829, DOI:10.1038/s41565-020-0759-5.
- P. M. Valencia, O. C. Farokhzad, R. Karnik and R. Langer, Microfluidic Technologies for Accelerating the Clinical Translation of Nanoparticles, Nat. Nanotechnol., 2012, 7(10), 623–629, DOI:10.1038/nnano.2012.168.
- S. J. Shepherd, D. Issadore and M. J. Mitchell, Microfluidic Formulation of Nanoparticles for Biomedical Applications, Biomaterials, 2021, 274, 120826, DOI:10.1016/j.biomaterials.2021.120826.
- Y. Liu, G. Yang, D. Zou, Y. Hui, K. Nigam, A. P. J. Middelberg and C.-X. Zhao, Formulation of Nanoparticles Using Mixing-Induced Nanoprecipitation for Drug Delivery, Ind. Eng. Chem. Res., 2020, 59(9), 4134–4149, DOI:10.1021/acs.iecr.9b04747.
- R. Haase, E. Fazeli, D. Legland, M. Doube, S. Culley, I. Belevich, E. Jokitalo, M. Schorb, A. Klemm and C. Tischer, A Hitchhiker's Guide through the Bio-Image Analysis Software Universe, FEBS Lett., 2022, 596(19), 2472–2485, DOI:10.1002/1873-3468.14451.
- M. Mattiazzi Usaj, E. B. Styles, A. J. Verster, H. Friesen, C. Boone and B. J. Andrews, High-Content Screening for Quantitative Cell Biology, Trends Cell Biol., 2016, 26(8), 598–611, DOI:10.1016/j.tcb.2016.03.008.
- D. J. Brayden, S.-A. Cryan, K. A. Dawson, P. J. O'Brien and J. C. Simpson, High-Content Analysis for Drug Delivery and Nanoparticle Applications, Drug Discovery Today, 2015, 20(8), 942–957, DOI:10.1016/j.drudis.2015.04.001.
- B. Yang, C. J. Richards, T. B. Gandek, I. de Boer, I. Aguirre-Zuazo, E. Niemeijer and C. Åberg, Following Nanoparticle Uptake by Cells Using High-Throughput Microscopy and the Deep-Learning Based Cell Identification Algorithm Cellpose, Front. nanotechnol., 2023, 5 DOI:10.3389/fnano.2023.1181362.
- M. B. Cutrona and J. C. Simpson, A High-Throughput Automated Confocal Microscopy Platform for Quantitative Phenotyping of Nanoparticle Uptake and Transport in Spheroids, Small, 2019, 15(37), 1902033, DOI:10.1002/smll.201902033.
- Y. Rui, D. R. Wilson, S. Y. Tzeng, H. M. Yamagata, D. Sudhakar, M. Conge, C. A. Berlinicke, D. J. Zack, A. Tuesca and J. J. Green, High-Throughput and High-Content Bioassay Enables Tuning of Polyester Nanoparticles for Cellular Uptake, Endosomal Escape, and Systemic in Vivo Delivery of mRNA, Sci. Adv., 2022, 8(1) DOI:10.1126/sciadv.abk2855.
- S. Kelly, M. H. Byrne, S. J. Quinn and J. C. Simpson, Multiparametric Nanoparticle-Induced Toxicity Readouts with Single Cell Resolution in HepG2 Multicellular Tumour Spheroids, Nanoscale, 2021, 13(41), 17615–17628, 10.1039/D1NR04460E.
- X. Chen and H. Lv, Intelligent Control of Nanoparticle Synthesis on Microfluidic Chips with Machine Learning, NPG Asia Mater., 2022, 14(1), 69, DOI:10.1038/s41427-022-00416-1.
- H. Tao, T. Wu, M. Aldeghi, T. C. Wu, A. Aspuru-Guzik and E. Kumacheva, Nanoparticle Synthesis Assisted by Machine Learning, Nat. Rev. Mater., 2021, 6(8), 701–716, DOI:10.1038/s41578-021-00337-5.
- F. Mekki-Berrada, Z. Ren, T. Huang, W. K. Wong, F. Zheng, J. Xie, I. P. S. Tian, S. Jayavelu, Z. Mahfoud, D. Bash, K. Hippalgaonkar, S. Khan, T. Buonassisi, Q. Li and X. Wang, Two-Step Machine Learning Enables Optimized Nanoparticle Synthesis, npj Comput. Mater., 2021, 7(1), 55, DOI:10.1038/s41524-021-00520-w.
- K. Abdel-Latif, R. W. Epps, F. Bateni, S. Han, K. G. Reyes and M. Abolhasani, Self-Driven Multistep Quantum Dot Synthesis Enabled by Autonomous Robotic Experimentation in Flow, Adv. Intell. Syst., 2021, 3(2), 2000245, DOI:10.1002/aisy.202000245.
- O. Voznyy, L. Levina, J. Z. Fan, M. Askerka, A. Jain, M.-J. Choi, O. Ouellette, P. Todorović, L. K. Sagar and E. H. Sargent, Machine Learning Accelerates Discovery of Optimal Colloidal Quantum Dot Synthesis, ACS Nano, 2019, 13(10), 11122–11128, DOI:10.1021/acsnano.9b03864.
- D. Van Tilborg, H. Brinkmann, E. Criscuolo, L. Rossen, R. Özçelik and F. Grisoni, Deep Learning for Low-Data Drug Discovery: Hurdles and Opportunities, preprint, Chemrxiv, 2024, DOI:10.26434/chemrxiv-2024-w0wvl.
- Z. Bao, J. Bufton, R. J. Hickman, A. Aspuru-Guzik, P. Bannigan and C. Allen, Revolutionizing Drug Formulation Development: The Increasing Impact of Machine Learning, Adv. Drug Delivery Rev., 2023, 202, 115108, DOI:10.1016/j.addr.2023.115108.
- A. Krause, A. Singh and C. Guestrin, Near-Optimal Sensor Placements in Gaussian Processes: Theory, Efficient Algorithms and Empirical Studies, J. Mach. Learn. Res., 2008, 9(2), 235–284 Search PubMed.
- D. Reker and G. Schneider, Active-Learning Strategies in Computer-Assisted Drug Discovery, Drug Discovery Today, 2015, 20(4), 458–465, DOI:10.1016/j.drudis.2014.12.004.
- R. Karnik, F. Gu, P. Basto, C. Cannizzaro, L. Dean, W. Kyei-Manu, R. Langer and O. C. Farokhzad, Microfluidic Platform for Controlled Synthesis of Polymeric Nanoparticles, Nano Lett., 2008, 8(9), 2906–2912, DOI:10.1021/nl801736q.
- A. G. Mares, G. Pacassoni, J. S. Marti, S. Pujals and L. Albertazzi, Formulation of Tunable Size PLGA-PEG Nanoparticles for Drug Delivery Using Microfluidic Technology, PLoS One, 2021, 16(6), e0251821, DOI:10.1371/journal.pone.0251821.
- C. McQuin, A. Goodman, V. Chernyshev, L. Kamentsky, B. A. Cimini, K. W. Karhohs, M. Doan, L. Ding, S. M. Rafelski, D. Thirstrup, W. Wiegraebe, S. Singh, T. Becker, J. C. Caicedo and A. E. Carpenter, CellProfiler 3.0: Next-Generation Image Processing for Biology, PLoS Biol., 2018, 16(7), e2005970, DOI:10.1371/journal.pbio.2005970.
- A. Alijagic, N. Scherbak, O. Kotlyar, P. Karlsson, X. Wang, I. Odnevall, O. Benada, A. Amiryousefi, L. Andersson, A. Persson, J. Felth, H. Andersson, M. Larsson, A. Hedbrant, S. Salihovic, T. Hyötyläinen, D. Repsilber, E. Särndahl and M. Engwall, A Novel Nanosafety Approach Using Cell Painting, Metabolomics, and Lipidomics Captures the Cellular and Molecular Phenotypes Induced by the Unintentionally Formed Metal-Based (Nano)Particles, Cells, 2023, 12(2), 281, DOI:10.3390/cells12020281.
- B. Settles, Active Learning Literature Survey; Technical Report, University of Wisconsin-Madison Department of Computer Sciences, 2009, https://minds.wisconsin.edu/handle/1793/60660 (accessed 2023-08-04) Search PubMed.
- A. Graves, Practical Variational Inference for Neural Networks, In Advances in Neural Information Processing Systems, ed. J. Shawe-Taylor, R. Zemel, P. Bartlett, F. Pereira and K. Q. Weinberger, Curran Associates, Inc., 2011, vol. 24 Search PubMed.
- Y. Ovadia, E. Fertig, J. Ren, Z. Nado, D. Sculley, S. Nowozin, J. Dillon, B. Lakshminarayanan and J. Snoek, Can You Trust Your Model’ s Uncertainty? Evaluating Predictive Uncertainty under Dataset Shift, In Advances in Neural Information Processing Systems, Curran Associates, Inc., 2019, vol. 32 Search PubMed.
- A. Wu, S. Nowozin, E. Meeds, R. E. Turner, J. M. Hernández-Lobato and A. L. Gaunt, Deterministic Variational Inference for Robust Bayesian Neural Networks, 2019 Search PubMed.
- T. A. Meyer, C. Ramirez, M. J. Tamasi and A. J. Gormley, A User's Guide to Machine Learning for Polymeric Biomaterials, ACS Polym. Au, 2023, 3(2), 141–157, DOI:10.1021/acspolymersau.2c00037.
- Z. Bao, F. Yung, R. J. Hickman, A. Aspuru-Guzik, P. Bannigan and C. Allen, Data-Driven Development of an Oral Lipid-Based Nanoparticle Formulation of a Hydrophobic Drug, Drug Delivery Transl. Res., 2023 DOI:10.1007/s13346-023-01491-9.
- J.-M. Rabanel, V. Adibnia, S. F. Tehrani, S. Sanche, P. Hildgen, X. Banquy and C. Ramassamy, Nanoparticle Heterogeneity: An Emerging Structural Parameter Influencing Particle Fate in Biological Media?, Nanoscale, 2019, 11(2), 383–406, 10.1039/C8NR04916E.
- L. V. Jospin, H. Laga, F. Boussaid, W. Buntine and M. Bennamoun, Hands-On Bayesian Neural Networks—A Tutorial for Deep Learning Users, IEEE Comput. Intell. Mag., 2022, 17(2), 29–48, DOI:10.1109/MCI.2022.3155327.
- D. P. Kingma and J. Ba, A Method for Stochastic Optimization, arXiv, Preprint, 2014, arXiv:1412.6980, DOI:10.48550/ARXIV.1412.6980.
- C. Bishop, Pattern Recognition and Machine Learning, Springer, 2006 Search PubMed.
- T. Chen and C. Guestrin, XGBoost: A Scalable Tree Boosting System, In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, ACM, San Francisco California USA, 2016, pp. 785–794, doi: DOI:10.1145/2939672.2939785.
- F. Zhdanov, Diverse Mini-Batch Active Learning, arXiv, Preprint, 2019, arXiv:1901.05954v1, DOI:10.48550/ARXIV.1901.05954.
- A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Kopf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai and S. Chintala, PyTorch: An Imperative Style, High-Performance Deep Learning Library, In Advances in Neural Information Processing Systems, ed. H. Wallach, H. Larochelle, A. Beygelzimer, F. d. Alché-Buc, E. Fox and R. Garnett, Curran Associates, Inc., 2019, vol. 32 Search PubMed.
- E. Bingham, J. P. Chen, M. Jankowiak, F. Obermeyer, N. Pradhan, T. Karaletsos, R. Singh, P. Szerlip, P. Horsfall and N. D. P. Goodman, Deep Universal Probabilistic Programming, J. Mach. Learn. Res., 2019, 1–6 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, A. Müller, J. Nothman, G. Louppe, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and É. Duchesnay, Scikit-Learn: Machine Learning in Python, arXiv, Preprint, 2012, arXiv:1201.0490v4, DOI:10.48550/ARXIV.1201.0490.
- Team, R. D. C. R.: A Language and Environment for Statistical Computing, 2010 Search PubMed.
- H. Wickham, Data Analysis, In ggplot2: Elegant Graphics for Data Analysis, ed. H. Wickham, Use R.!, Springer International Publishing, Cham, 2016, pp. 189–201, DOI:10.1007/978-3-319-24277-4_9.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00104d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |