Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Agent-based learning of materials datasets from the scientific literature†
Mehrad
Ansari
 ab and
Seyed Mohamad
Moosavi
ab and
Seyed Mohamad
Moosavi
 *ab
*ab
aAcceleration Consortium, University of Toronto, Toronto, Ontario M5S 3E5, Canada. E-mail: mohamad.moosavi@utoronto.ca
bDepartment of Chemical Engineering & Applied Chemistry, University of Toronto, Toronto, Ontario M5S 3E5, Canada
First published on 28th October 2024
Abstract
Advancements in machine learning and artificial intelligence are transforming the discovery of materials. While the vast corpus of scientific literature presents a valuable and rich resource of experimental data that can be used for training machine learning models, the availability and accessibility of these data remains a bottleneck. Accessing these data by manual dataset creation is limited due to issues in maintaining quality and consistency, scalability limitations, and the risk of human error and bias. Therefore, in this work, we develop a chemist AI agent, powered by large language models (LLMs), to overcome these limitations by autonomously creating structured datasets from natural language text, ranging from sentences and paragraphs to extensive scientific research articles and extract guidelines for designing materials with desired properties. Our chemist AI agent, Eunomia, can plan and execute actions by leveraging the existing knowledge from decades of scientific research articles, scientists, the Internet and other tools altogether. We benchmark the performance of our approach in three different information extraction tasks with various levels of complexity, including solid-state impurity doping, metal–organic framework (MOF) chemical formula, and property relationships. Our results demonstrate that our zero-shot agent, with the appropriate tools, is capable of attaining performance that is either superior or comparable to the state-of-the-art fine-tuned material information extraction methods. This approach simplifies compilation of machine learning-ready datasets for the applications of discovery of various materials, and significantly eases the accessibility of advanced natural language processing tools for novice users in natural language. The methodology in this work is developed as open-source software on https://github.com/AI4ChemS/Eunomia.
1 Introduction
The past decade's extraordinary achievements in leveraging machine learning for chemical discovery highlight the power of accessible knowledge and structured data.1–3 However, a significant portion of chemical knowledge, particularly the experimental ones, is scattered across the scientific literature in an unstructured format.4 Researchers face challenges in effectively utilizing existing knowledge for design of experiments, as well as in comprehending the entirety of previous studies in a field. Thus, the development of methodologies to extract information from the literature and convert it into structured data will play a fundamental role in advancing the machine learning for molecules and materials.Natural Language Processing (NLP) is a powerful tool for extracting information from the scientific literature. Conventional NLP methods have been used in materials and chemical sciences5–10 for named entity recognition. However, these methods are limited in other NLP tasks that are needed for a general-purpose data extraction tool, including co-reference resolution, relation extraction, template filling, argument mining, and entity linking. To better understand these NLP terminologies, let us consider an example taken from an abstract of a materials paper11 in the field of metal–organic frameworks (MOFs):
“An isoreticular series of cobalt-adeninate bio-MOFs (bio-MOFs-11–14) is reported. The pores of bio-MOFs-11–14 are decorated with acetate, propionate, butyrate, and valerate, respectively. The nitrogen (N2) and carbon dioxide (CO2) adsorption properties of these materials are studied and compared. The isosteric heats of adsorption for CO2are calculated, and the CO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) N2selectivities for each material are determined. As the lengths of the aliphatic chains decorating the pores in bio-MOFs-11–14 increase, the BET surface areas decrease from 1148 m2g−1to 17 m2g−1while the CO2
N2selectivities for each material are determined. As the lengths of the aliphatic chains decorating the pores in bio-MOFs-11–14 increase, the BET surface areas decrease from 1148 m2g−1to 17 m2g−1while the CO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) N2selectivities predicted from ideal adsorbed solution theory at 1 bar and 273 K for a 10
N2selectivities predicted from ideal adsorbed solution theory at 1 bar and 273 K for a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) 90 CO2
90 CO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) N2mixture range from 73
N2mixture range from 73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) 1 for bio-MOF-11 to 123
1 for bio-MOF-11 to 123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) 1 for bio-MOF-12 and finally to 107
1 for bio-MOF-12 and finally to 107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) 1 for bio-MOF-13. At 298 K, the selectivities are 43
1 for bio-MOF-13. At 298 K, the selectivities are 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) 1 for bio-MOF-11, 52
1 for bio-MOF-11, 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) 1 for bio-MOF-12, and 40
1 for bio-MOF-12, and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) 1 for bio-MOF-13. The water stability of bio-MOFs-11–14 increases with increasing aliphatic chain length.”
1 for bio-MOF-13. The water stability of bio-MOFs-11–14 increases with increasing aliphatic chain length.”
• Named entity recognition involves identifying and classifying the specific entities within the text into predefined categories (i.e., chemical compounds: “bio-MOFs-11–14”, “acetate”, experimental conditions: “1 bar”, “273 K”, “10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 CO2
90 CO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N2 mixture”).
N2 mixture”).
• Co-reference resolution focuses on finding all expressions that refer to the same entity in the text. As an example, phrases like “these materials”, “each material” are references that relate back to the bio-MOFs-11–14 mentioned in the first sentence.
• Relation extraction involves extracting semantic relationships from the text, which usually occur between two or more entities (i.e., the impact of “aliphatic chain lengths” on “BET surface areas” and “CO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N2 selectivities”).
N2 selectivities”).
• Template filing is an efficient approach to extract and structure complex information from text. As an example: material name: bio-MOFs-11–14.
• Argument mining focuses on the automatic identification and extracts the reasoning presented within the text. As an example, the “increase in the water stability” of the mentioned MOFs is connected to the “increasing length of the aliphatic chains”.
• Entity linking takes one step further than named entity recognition and distinguishes between similarly named entities (i.e., the term “bio-MOFs” would be linked to databases or literature that describes these materials in detail).
The emergence of Large Language Models (LLMs) or foundation models shows a great promise in tackling these complex NLP tasks.7,12–15 Huang et al.16 fine-tuned a language model (BERT) on battery publications to extract device-level information from a paragraph that contains one device only. Dunn et al.7 showed that fine-tuned LLMs using 100–1000 data points can perform relation extraction as well as template filling, enabling conversion of the extracted information into user-defined output formats. Despite these promising results, these methods require training data, limiting their ease of use and broad applicability. Moreover, LLM based approaches have not been explored for more intricate challenges, such as argument mining and co-reference resolution. These tasks are critical for practically using NLP for automated database development. For example, in one article, multiple materials might be discussed and authors use abbreviations like “compound 1” or simply “1” in the entire research manuscript for referencing after initially defining the chemical compound in the introduction section. Additionally, description of material properties often comes with various interpretations, limiting using rigid name entity matching. As implementations of standalone LLMs fall short in addressing these intricate tasks, new methods are needed to enable reliable information extraction. An effective approach is to augment LLMs with domain-specific toolkits. These specialized tools offer precise answers, thus addressing the inherent limitations of LLMs in specific domains, and enhancing their overall performance and applicability.17–20
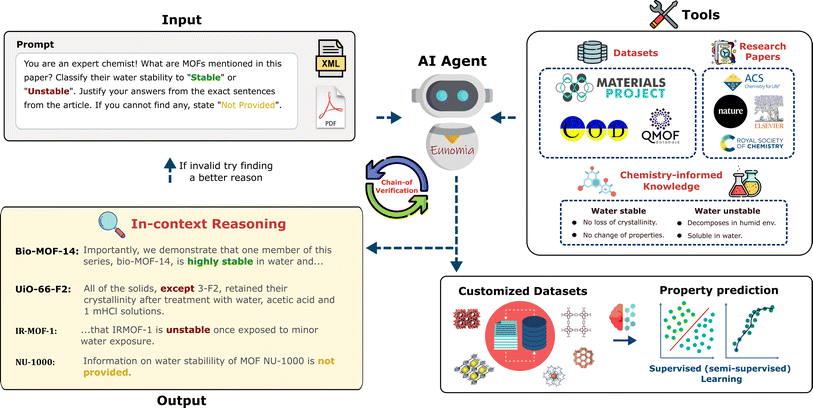
In this work, we introduce an autonomous AI agent, Eunomia, augmented with chemistry-informed tools, designed to extract materials science-relevant information from unstructured text and convert it into structured databases. With an LLM at its core, our AI agent is capable of strategizing and executing actions by tapping into a wealth of knowledge from academic publications, domain-specific experts, the Internet, and other user-defined resources (see Fig. 1). We show that this method streamlines data extraction, achieving remarkable accuracy and performance solely with a pre-trained LLM (GPT-4 (ref. 21)), eliminating the need for fine-tuning. It offers adaptability by accommodating a variety of information extraction tasks through natural language text prompts for new output schemas and reducing the risk of hallucinations through a chain-of-verification processes. This capability extends beyond what a standalone LLM can offer. Eunomia simplifies the development of tailored datasets from the literature for domain experts, eliminating the need for extensive programming, NLP, or machine learning expertise.
This manuscript is organized as follows: benchmarking and evaluating the model performance on three different materials' NLP tasks with varying level of complexity are represented in Section 3. This is followed by Section 4, with a discussion on the implications of our findings, the advantages and limitations of our approach, as well as suggested directions for future work. Finally, in Section 5, we describe our methodology on the agent's toolkits and evaluation metrics.
2 AI agent
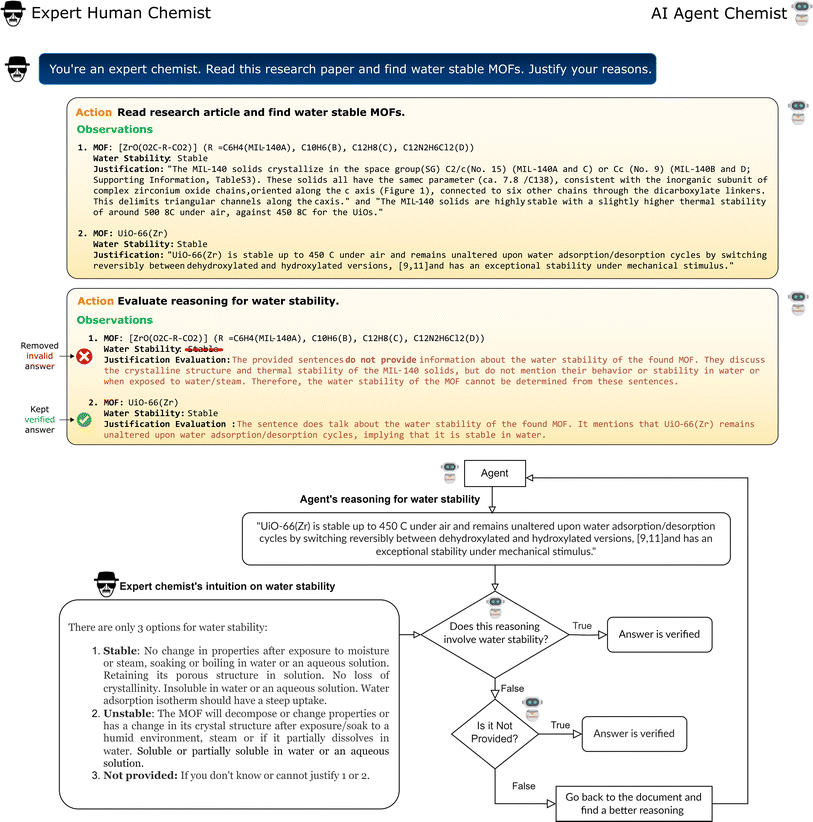
In the realm of artificial intelligence, an “agent” is an autonomous entity capable of taking action based on its environment. In this work, we developed a chemist AI agent, Eunomia, to autonomously extract information from the scientific literature (Fig. 1). We use an LLM to serve as the brain of our agent.22 The LLM is equipped with advanced capabilities like planning and tool use to act beyond just a text generator, and act as a comprehensive problem solver, enabling effective interactions with the environment. We use ReAct architecture23 for planning, enabling both reasoning and action. Our agent can interact with external sources like knowledge bases or environments to obtain more information. These knowledge bases are developed as toolkits (see Methods section for details) allowing the agent to extract relevant information from research articles, publicly available datasets, and built-in domain-specific chemical knowledge, ensuring its proficiency in playing the role of an expert chemist. We use OpenAI's GPT-4 (ref. 21) with a temperature of zero as our LLM and LangChain24 for the application framework development (note the choice of LLM is only a hyperparameter and other LLMs can also be used with our agent). The application of LLMs in developing autonomous agents is a growing area of research,18,23,25–28 with a detailed survey available in ref. 29 for further insights.In addition to the standard search and text manipulation tools, we have implemented a Chain-of-Verification (CoV) tool to enhance the robustness of our AI agent against hallucination. Hallucination in an LLM refers to the generation of content that strays from factual reality or includes fabricated information.30 In the CoV approach, the agent iteratively assesses its responses to ensure they remain logically consistent and coherent (see Methods section 5.1.2 for details). This addition helps particularly with eliminating mistakenly extracted data related to semantically similar properties. An illustrative example is the case of stability of materials, where thermal, mechanical, and chemical stabilities might be confused by the agent. Fig. 2 illustrates how CoV process works in action: the agent is tasked to identify MOFs and the corresponding water stability data in a paper. The agent initially misclassifies a thermally stable MOF as water-stable, but then it corrects this mistake by a comprehensive review using the CoV tool. This tool improves the performance of the agent and ensures robust data extraction.
3 Case studies
We evaluate the performance of our AI agent by benchmarking it across three different materials' NLP tasks, with increasing task complexity (Table 1). In our assessment, we evaluate a broad range of text lengths, including sentences, paragraphs, and entire manuscripts, along with various NLP tasks outlined in the Introduction, which form the basis for defining complexity. The first case study focuses on assessing the agent's performance on NLP tasks of lower complexity, specifically named entity recognition and relation extraction. For this, we use our agent to extract the relationships of host-to-many dopants from a single sentence. The second case study, with medium NLP complexity, involves obtaining MOFs' chemical formula and their corresponding guest species from a paragraph with multiple sentences. Finally, the third case study centers on predicting a given property of MOFs based on the context coming from a materials research paper. The property of interest in our work is water stability. This case study aims to, in addition to named entity recognition and relation extraction, evaluate the co-reference resolution and argument mining proficiency of our AI agent, tailored for chemists, which involves a high level of NLP complexity. In all case studies, our chemist AI agent, Eunomia, is a zero-shot learner that is equipped with the Doc Search tool (see Section 5.1.1). We have also conducted additional experiments by equipping Eunomia with the chain-of-verification (CoV) tool, as described in Section 5.1.2. This is referred to as Eunomia + CoV from here on.| Case study | Context | NLP tasks | Task complexity |
|---|---|---|---|
| (1) Relationship of host-to-many dopants | Sentence | Named entity recognition, relation extraction | Easy |
| (2) MOF formula and guest species relationship | Paragraph | Named entity recognition, relation extraction | Medium |
| (3) MOF property relationship | Research paper | Named entity recognition, relation extraction, template filing, argument mining, entity linking | Hard |
To fairly compare the performance of our agent with the state-of-the-art fine-tuned LLM methods, the evaluation methodology for the first two case studies mirrors precisely that of ref. 7 (see Section 5.2 for details), serving as a benchmark reference. In the following section, ref. 7 is referred to as LLM-NERRE, which involves fine-tuning a pre-trained LLM (GPT-3 (ref. 31)) on 100–1000 sets of manually annotated examples, and then using the model to extract information from unstructured text.
3.1 Case study 1: relationship of host-to-many dopants (Easy)
This case study aims to extract structured information about solid-state impurity doping from a single sentence. The objective is to identify the two entities “host” and “dopant”. “Host” refers to the foundational crystal, sample, or category of material characterized by essential descriptors in its proximate context, such as “ZnO2 nanoparticles”, “LiNbO3”, or “half-Heuslers”. “Dopant” means the elements or ions that represent a minority component, deliberately introduced impurities, or specific atomic-scale defects or carriers of electric charge like “hole-doped” or “S vacancies”. A single host can be combined with multiple dopants, through individual doping or simultaneous doping with different species, and one dopant can associate with various host materials. The text may contain numerous dopant–host pairings within the same sentence, and also instances of dopants and hosts that do not interact.Eunomia shows an excellent performance in this task, demonstrating the effectiveness of our approach in named entity recognition and relation extraction. Performance comparison between our chemist AI agent (Eunomia), and LLM-NERRE can be found in Table 2. In this setting, the same definition of hosts and dopants given above is passed to Eunomia via the input prompt, while LLM-NERRE is fine-tuned on 413 sentences. The testing set contains 77 sentences. Notably, in both tasks, Eunomia + CoV exceeds the performance of LLM-NERRE in terms of the F1 score. This clearly demonstrates the effectiveness of our approach compared to fine-tuning, which can be labor-intensive and error-prone. We instruct Eunomia not to make up answers, which lead to a more cautious outcome, wherein uncertain or unclear inputs yield no output. As an example, in the sentence “An anomalous behavior of the emission properties of alkali halides doped with heavy impurities, stimulated new efforts for its interpretation, invoking delicate and sophisticated mechanisms whose interest transcends the considered specific case.”, the ground-truth host materials is “alkali halides”. However, due to the nature of exact-word matching metric implemented in ref. 7 a cautious agent with no predictions for the host entity will be penalized with two false negatives, one for each word in the ground-truth, leading to lower recall score.
3.2 Case study 2: MOF formula and guest species relationship (Medium)
The goal of this case study is to identify MOF formula and guest species from unstructured text, as a paragraph with multiple sentences. The MOF formula refers to the chemical formula of a MOF, which is an important piece of information for characterizing and identifying MOFs. The guest species, on the other hand, are chemical species that have been incorporated, stored, or adsorbed in the MOF. These species are of interest because MOFs are often used for ion and gas separation, and the presence of specific guest species can affect the performance of the MOF. We limit our method to stand-alone Eunomia without CoV due to the complexity of defining a chemistry-informed CoV verification tool for this specific task. It should be noted that ref. 7 also included results on the identification of synthesis descriptions and applications pertaining to MOFs. However, as the metric of exact-word matching reported in ref. 7 does not fairly and adequately reflect the model performance for the multi-word (>2 words) nature of these outputs, we have limited our benchmarking to the MOF formula and guest species identification only.Table 3 shows the performance comparison between Eunomia and LLM-NERRE on the MOF formula and guest species relationship extraction task. While Eunomia shows a superior performance on the MOF formula compared to LLM-NERRE, the relatively low performance of both approaches is related to the nature of the exact word matching. Using semantic similarity would be a more appropriate indicator in this context. On the guest species entity, while Eunomia shows a high recall (0.923), precision is relatively poor (0.429). This can be attributed to how the exact-word matching metrics have been defined in ref. 7, where precision is majorly lowered by the presence of the extra unmatched predicted words (false positives), while recall remains high because all ground truth items were found in the predicted words.
3.3 Case study 3: MOF property relationship (Hard)
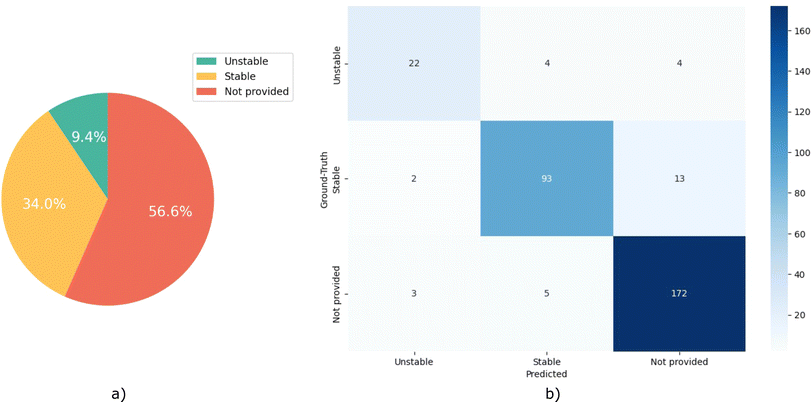
This case study aims to mimic a practical scenario of developing datasets from the scientific literature, where we evaluate the agent's performance on extracting MOF's water stability. To excel in this goal, the agent must identify all MOFs mentioned within the research paper, evaluate their water stability, and support these evaluations using exact sentences derived from the document. Such tasks are inherently linked to the NLP functions of named entity recognition, co-reference resolution, relation extraction, and argument mining. This is particularly a challenging task as researchers report the water stability in various ways, using phrases ranging from “the material remains crystalline in humid conditions” to “the MOF is stable in wide range of pH”, or “the material is not soluble in water”.For this case study, we created a hand-labeled dataset based on a selection of 101 materials research papers, which contain a selection of 371 MOFs. Three expert chemists manually read through and review each paper, pinpointing the MOFs referenced within. In the 3-way redundancy for data labeling, we carefully considered all three perspectives and insights, and finalized the dataset by averaging the outcomes. A portion of these articles are selected considering the original work by Burtch et al.,32 where they developed a dataset of MOF names and their water stability by manually reading 111 research articles. To mimic the practical data extraction scenario, in which the agent is passed many articles, many of which do not contain the desired information, we included articles with no information about water stability. Each MOF in our set is assigned to one of the three classes of “Stable”, “Unstable”, and “Not provided”. Fig. 3a presents the distribution of the classes within this dataset.
For this case study, we have established criteria to characterize water-stable MOFs, drawing from the study by Burtch et al.32 and our own chemical intuition. A water-stable MOF should meet the following criteria:
• No alteration in properties after being subjected to moisture or steam, or when soaked or boiled in water or an aqueous solution.
• Preservation of its porous architecture in liquid (water) environments.
• Sustained crystallinity without loss of structural integrity in aqueous environments.
• Insoluble in water or aqueous solutions.
• Exhibiting a pronounced rise in its water adsorption isotherm.
These water stability guidelines are defined as rules to Eunomia within the input prompt, as well as in its equipped CoV tool.
Eunomia with CoV tool retrieves most (yield of 86.20%) of the reported MOFs and shows an excellent performance (accuracy of 0.91) in inferring their water stability. This high yield and accuracy demonstrates the capability of our approach in extracting desired knowledge from the natural text. As expected, in the absence of CoV, there is a marginal decrease in accuracy to 0.86, along with a yield reduction to 82.70%. Note that with the CoV in place, and upon further review of the paper for better reasoning sentences, the agent tends to discover additional MOFs that were initially missed during the first retrieval attempt. Thus, this leads to an increased yield compared to running the model without CoV. Taking into account the confusion matrix in Fig. 3b, it is evident that our agent adopts a cautious approach in its predictions. This is reflected in the substantial number of “Not provided” predictions which, upon comparison with the actual ground-truth class, indicates a propensity of the agent to acknowledge the insufficiency of information for making a definitive prediction, rather than mistakenly categorizing samples into the incorrect “Stable” or “Unstable” classes, and contaminating the agent's resulting dataset with unwanted noise.
It is interesting to leverage our AI agent to extract information that goes beyond mere categorical and numerical labels. One practical case is to retrieve design guidelines for enhancing the properties of materials from research papers. To demonstrate this, we tasked Eunomia to extract material design rules for developing more water-stable MOFs using information from a research paper.33 By inputting domain-specific knowledge in natural language, the AI agent planned and executed actions, and provided a summary of the information contained in the scientific paper (see Fig. 4). This example showcases Eunomia's capability in various information retrieval tasks, which are inaccessible using other NLP methods. We envision that this approach can ease data retrieval from lengthy texts, enabling researchers to access and process information more efficiently.
 | ||
| Fig. 4 Extracting guidelines for designing materials with desired properties. The AI agent is tasked to play the role of an expert chemist and identify design guidelines for water-stable MOFs by obtaining context from a research paper.33 Additional chemistry-informed knowledge on water-stability is also provided to the agent as natural language input within the query. Utilizing this knowledge along with insights from the research paper, Eunomia suggests incorporating ethyl ester groups and phosphonate ester linkers, and adding organic tethers for enhanced protection against water-induced degradation. | ||
4 Discussion
We presented a high performing and robust method for extracting domain specific information from complex, unstructured text – ranging from sentences and paragraphs to extensive research articles – using AI agents. Scientists and researchers can use our open-source application to effortlessly develop tailored datasets for their specific areas and use them for downstream predictive tasks. It is important to note that given the zero-shot learning setting of our approach, the model's performance heavily relies on the quality of the input query. For complex problems involving iterative reasoning tasks, it is often crucial to generate explicit chain-of-thought (CoT) steps to achieve high accuracy in the final outputs. Therefore, the input query should include detailed, comprehensive step-by-step instructions.Our method shows close to perfect performance on the benchmarks we developed. Given that LLMs are continually improving, we believe that the reported performance represents the minimum expected from this workflow. As the performance of the model is already quite high (almost saturates the benchmarks), experimenting with newer models could potentially yield slightly better results. However, we note that at the current stage of open-source LLMs, the limited context window size restricts the use of detailed queries. Furthermore, agents powered by open-source LLMs often struggle with task planning and following a given set of instructions accurately. With the recent efforts on training LLMs to internalize these CoT steps,34,35 we expect that our approach will be even more feasible with open-source LLMs. While currently the cost of querying large datasets may become expensive, we expect the rapid advancements in LLMs will also diminish this cost.
Unlike other methods that follow a pipeline-based or end-to-end approach, our agent-based method could appeal to domain experts due to its minimal demand for programming skills, NLP and machine learning knowledge. Users are not required to rigidly define an output schema or engage in the meticulous task of creating manual annotations for the purpose of fine-tuning. Rather, they can simply prompt the agent with more context and describe how their desired output should be formatted in natural language. Moreover, the agent can easily be extended and equipped with other tools (e.g., Dataset Search, CSV Generator, etc.) to be adapted to other problems. For example, we showed that, by equipping the agent with the chain-of-verification tool (CoV), we can minimize hallucinations and improve the agent's performance. Similarly, by including reasoning tools, we can ask the agent to explain its reasoning based on the provided context to develop more transparent workflows for the LLM-based methods, and reduce their known “black-box” nature. This, simultaneously, offers a great opportunity for human-in-the-loop oversight, especially for tasks of critical importance.
Our results reveal an important observation: while large language models are few-shot learners,31 AI agents with appropriate tools and instructions are capable of being zero-shot learners. This brings an excellent opportunity to boost the performance of standalone LLMs across various domain-specific tasks without having to go through labor-intensive fine-tuning processes. A future thorough and systematic analysis of prompt sensitivity can provide valuable insights into this observation.
5 Methods
5.1 Agent toolkits
The semantic similarity search is ranked using Maximum Marginal Relevance (MMR)38 based on cosine similarity, defined as
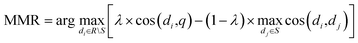 | (1) |
It is important to note that in some unsuccessful experiments, we observed that the AI agent repeatedly referred back to the document, even after pinpointing the correct answer. Although this minor issue remained unresolved, we introduced an iteration limit for the agent to avoid unnecessary model running costs.
5.2 Evaluation metrics
Multiple metrics have been defined to assess the agent's performance across different case studies. Precision, recall and F1 score are defined as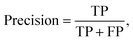 | (2) |
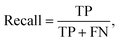 | (3) |
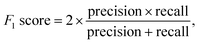 | (4) |
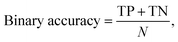 | (5) |
In case studies 1 and 2 (Sections 3.1 and 3.2), the evaluation metrics used are precisely those defined in the work of ref. 7. In particular, they assessed named entity relationships on a word-to-word matching basis by initially decomposing an entity E into a collection of k words separated by whitespace, denoted as E = {w1, w2, w3, …, wk}. For evaluating entities in named entity recognition exclusively, they enumerated the words that are identical in both the true entity set Etrue and the test entity set Etest as true positives (Etrue ∩ Etest), and the distinct elements in each set as false positives (Etest\Etrue) or false negatives (Etrue\Etest). For instance, if the true entity is “Bi2Te3 thin film” and the predicted entity is “Bi2Te3 film sample”, they noted two true positive matches (“Bi2Te3”, “film”), one false negative (“thin”), and one false positive (“sample”). An exceptional case arises for formula-type entities critical to material identification, whereby Etest must encompass all wi interpreted as stoichiometries to consider any wi ∈ Etest as correct. For example, with “Bi2Te3 thin film” as Etrue and “thin film” as Etest, three false negatives would be registered. For more details on the scoring metrics and the case studies, readers are encouraged to refer to ref. 7.
For our last case study in Section 3.3 (predicting the water stability of MOFs), the ternary accuracy is defined as
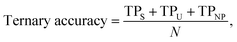 | (6) |
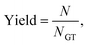 | (7) |
Data availability
All data (including the dataset used for case study 3) and code used to produce results in this study, and examples on how to use our approach, are publicly available in the following GitHub repository: https://github.com/AI4ChemS/Eunomia. The methodology in this work is also developed as an open-source application on https://eunomia.streamlit.app.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This research was undertaken thanks in part to funding provided to the University of Toronto's Acceleration Consortium from the Canada First Research Excellence Fund - Grant number CFREF-2022-00042. SMM acknowledges the support by the Natural Sciences and Engineering Research Council of Canada (NSERC) under grant number RGPIN-2023-04232. The authors thank Alexander Dunn and Andrew S. Rosen for their assistance on the implementation of LLM-NERRE. The authors also thank Haoning Yuan for assisting in manual curation of the water stability dataset used in Section 3.3.References
- S. M. Moosavi, K. M. Jablonka and B. Smit, The role of machine learning in the understanding and design of materials, J. Am. Chem. Soc., 2020, 142(48), 20273–20287 CrossRef CAS PubMed.
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev and A. Walsh, Machine learning for molecular and materials science, Nature, 2018, 559(7715), 547–555 CrossRef CAS PubMed.
- B. Sanchez-Lengeling and A. Aspuru-Guzik, Inverse molecular design using machine learning: generative models for matter engineering, Science, 2018, 361(6400), 360–365 CrossRef CAS PubMed.
- H. M. Sayeed, W. Smallwood, S. G. Baird and T. D. Sparks, NLP meets materials science: Quantifying the presentation of materials data in literature, Matter, 2024, 7(3), 723–727 CrossRef.
- L. Weston, V. Tshitoyan, J. Dagdelen, O. Kononova, A. Trewartha and K. A. Persson, et al., Named entity recognition and normalization applied to large-scale information extraction from the materials science literature, J. Chem. Inf. Model., 2019, 59(9), 3692–3702 CrossRef CAS PubMed.
- M. C. Swain and J. M. Cole, ChemDataExtractor: a toolkit for automated extraction of chemical information from the scientific literature, J. Chem. Inf. Model., 2016, 56(10), 1894–1904 CrossRef CAS PubMed.
- J. Dagdelen, A. Dunn, S. Lee, N. Walker, A. S. Rosen, G. Ceder, K. A. Persson and A. Jain, Structured information extraction from scientific text with large language models, Nat. Commun., 2024, 15(1), 1418 CrossRef CAS PubMed.
- A. Nandy, C. Duan and H. J. Kulik, Using machine learning and data mining to leverage community knowledge for the engineering of stable metal–organic frameworks, J. Am. Chem. Soc., 2021, 143(42), 17535–17547 CrossRef CAS PubMed.
- E. Kim, K. Huang, A. Saunders, A. McCallum, G. Ceder and E. Olivetti, Materials synthesis insights from scientific literature via text extraction and machine learning, Chem. Mater., 2017, 29(21), 9436–9444 CrossRef CAS.
- L. T. Glasby, K. Gubsch, R. Bence, R. Oktavian, K. Isoko and S. M. Moosavi, et al., DigiMOF: A Database of Metal–Organic Framework Synthesis Information Generated via Text Mining, Chem. Mater., 2023, 4510–4524 CrossRef CAS PubMed.
- T. Li, D. L. Chen, J. E. Sullivan, M. T. Kozlowski, J. K. Johnson and N. L. Rosi, Systematic modulation and enhancement of CO2:N2 selectivity and water stability in an isoreticular series of bio-MOF-11 analogues, Chem. Sci., 2013, 4(4), 1746–1755 RSC.
- M. P. Polak and D. Morgan, Extracting accurate materials data from research papers with conversational language models and prompt engineering, Nat. Commun., 2024, 15(1), 1569 CrossRef CAS PubMed.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, ChatGPT chemistry assistant for text mining and the prediction of MOF synthesis, J. Am. Chem. Soc., 2023, 145(32), 18048–18062 CrossRef CAS PubMed.
- Z. Zheng, Z. He, O. Khattab, N. Rampal, M. A. Zaharia, C. Borgs, J. T. Chayes and O. M. Yaghi, Image and data mining in reticular chemistry powered by GPT-4V, Digital Discovery, 2024, 3(3), 491–501 RSC.
- K. M. Jablonka, Q. Ai, A. Al-Feghali, S. Badhwar, J. D. Bocarsly and A. M. Bran, et al., 14 Examples of How LLMs Can Transform Materials Science and Chemistry: A Reflection on a Large Language Model Hackathon, Digital Discovery, 2023, 1233–1250 RSC.
- S. Huang and J. M. Cole, BatteryBERT: a pretrained language model for battery database enhancement, J. Chem. Inf. Model., 2022, 62(24), 6365–6377 CrossRef CAS PubMed.
- A. D. White, G. M. Hocky, H. A. Gandhi, M. Ansari, S. Cox and G. P. Wellawatte, et al., Assessment of chemistry knowledge in large language models that generate code, Digital Discovery, 2023, 2(2), 368–376 RSC.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller, Augmenting large language models with chemistry tools, Nature Machine Intelligence, 2024, 6, 1–11 CrossRef PubMed.
- D. A. Boiko, R. MacKnight, B. Kline and G. Gomes, Autonomous chemical research with large language models, Nature, 2023, 624(7992), 570–578 CrossRef CAS PubMed.
- J. Lála, O. O'Donoghue, A. Shtedritski, S. Cox, S. G. Rodriques and A. D. White, PaperQA: Retrieval-Augmented Generative Agent for Scientific Research, arXiv, 2023, preprint, arXiv:231207559, DOI:10.48550/arXiv.2312.07559.
- OpenAI, GPT-4 Technical Report, 2023 Search PubMed.
- T. Kojima, S. S. Gu, M. Reid, Y. Matsuo and Y. Iwasawa, Large language models are zero-shot reasoners, Adv. Neural Inf. Process. Syst., 2022, 35, 22199–22213 Search PubMed.
- S. Yao, J. Zhao, D. Yu, N. Du, I. Shafran, K. Narasimhan, et al., ReAct: Synergizing Reasoning and Acting in Language Models, arXiv, 2022, preprint, arXiv:221003629, DOI:10.48550/arXiv.2210.03629.
- H. Chase, LangChain, 2022, available from: https://github.com/langchain-ai/langchain Search PubMed.
- R. Nakano, J. Hilton, S. Balaji, J. Wu, L. Ouyang, C. Kim, et al., WebGPT: browser-assisted question-answering with human feedback, arXiv, 2021, preprint, arXiv:211209332, DOI:10.48550/arXiv.2112.09332.
- T. Schick, J. Dwivedi-Yu, R. Dessì, R. Raileanu, M. Lomeli, L. Zettlemoyer, et al., Toolformer: language models can teach themselves to use tools, arXiv, 2023, preprint, arXiv:230204761, DOI:10.48550/arXiv.2302.04761.
- P. Lu, B. Peng, H. Cheng, M. Galley, K. W. Chang, Y. N. Wu, et al., Chameleon: plug-and-play compositional reasoning with large language models, arXiv, 2023, preprint, arXiv:230409842, DOI:10.48550/arXiv.2304.09842.
- Y. Qin, S. Hu, Y. Lin, W. Chen, N. Ding, G. Cui, et al., Tool learning with foundation models, arXiv, 2023, preprint, arXiv:230408354, DOI:10.48550/arXiv.2304.08354.
- Z. Xi, W. Chen, X. Guo, W. He, Y. Ding, B. Hong, et al., The Rise and Potential of Large Language Model Based Agents: A Survey, arXiv, 2023, preprint, arXiv:230907864, DOI:10.48550/arXiv.2309.07864.
- V. Rawte, A. Sheth and A. Das, A survey of hallucination in large foundation models, arXiv, 2023, preprint, arXiv:230905922, DOI:10.48550/arXiv.2309.05922.
- T. Brown, B. Mann, N. Ryder, M. Subbiah, J. D. Kaplan and P. Dhariwal, et al., Language models are few-shot learners, Adv. Neural Inf. Process. Syst., 2020, 33, 1877–1901 Search PubMed.
- N. C. Burtch, H. Jasuja and K. S. Walton, Water stability and adsorption in metal–organic frameworks, Chem. Rev., 2014, 114(20), 10575–10612 CrossRef CAS PubMed.
- J. M. Taylor, R. Vaidhyanathan, S. S. Iremonger and G. K. Shimizu, Enhancing water stability of metal–organic frameworks via phosphonate monoester linkers, J. Am. Chem. Soc., 2012, 134(35), 14338–14340 CrossRef CAS PubMed.
- Y. Deng, Y. Choi and S. Shieber, From Explicit CoT to Implicit CoT: Learning to Internalize CoT Step by Step, arXiv, 2024, preprint, arXiv:240514838, DOI:10.48550/arXiv.2405.14838.
- G. Feng, B. Zhang, Y. Gu, H. Ye, D. He and L. Wang, Towards revealing the mystery behind chain of thought: a theoretical perspective, Adv. Neural Inf. Process. Syst., 2024, 36, 70757–70798 Search PubMed.
- R. Greene, T. Sanders, L. Weng and A. Neelakantan, New and improved embedding model, OpenAI Blog, 2022, https://openai.com/blog/new-and-improved-embedding-model Search PubMed.
- J. Johnson, M. Douze and H. Jégou, Billion-scale similarity search with GPUs, IEEE Transactions on Big Data, 2019, 7(3), 535–547 Search PubMed.
- J. Carbonell and J. Goldstein, The use of MMR, diversity-based reranking for reordering documents and producing summaries, in Proceedings of the 21st Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, 1998, pp. 335–336 Search PubMed.
- S. Dhuliawala, M. Komeili, J. Xu, R. Raileanu, X. Li, A. Celikyilmaz, et al., Chain-of-verification reduces hallucination in large language models, arXiv, 2023, preprint, arXiv:230911495, DOI:10.48550/arXiv.2309.11495.
- B. Zhang, Y. Tan, Y. Shen, A. Salem, M. Backes, S. Zannettou, et al., Breaking Agents: Compromising Autonomous LLM Agents Through Malfunction Amplification, arXiv, 2024, preprint, arXiv:240720859, DOI:10.48550/arXiv.2407.20859.
- A. Jain, S. P. Ong, G. Hautier, W. Chen, W. D. Richards and S. Dacek, et al., Commentary: The Materials Project: a materials genome approach to accelerating materials innovation, APL Mater., 2013, 1(1), 011002 CrossRef.
- A. Merkys, A. Vaitkus, A. Grybauskas, A. Konovalovas, M. Quirós and S. Gražulis, Graph isomorphism-based algorithm for cross-checking chemical and crystallographic descriptions, J. Cheminf., 2023, 15(1), 25 Search PubMed.
- A. Vaitkus, A. Merkys and S. Gražulis, Validation of the crystallography open database using the crystallographic information framework, J. Appl. Crystallogr., 2021, 54(2), 661–672 CrossRef CAS PubMed.
- M. Quirós, S. Gražulis, S. Girdzijauskaitė, A. Merkys and A. Vaitkus, Using SMILES strings for the description of chemical connectivity in the Crystallography Open Database, J. Cheminf., 2018, 10(1), 1–17 Search PubMed.
- A. Merkys, A. Vaitkus, J. Butkus, M. Okulič-Kazarinas, V. Kairys and S. Gražulis, COD::CIF::Parser: an error-correcting CIF parser for the Perl language, J. Appl. Crystallogr., 2016, 49(1), 292–301 CrossRef CAS PubMed.
- S. Gražulis, A. Merkys, A. Vaitkus and M. Okulič-Kazarinas, Computing stoichiometric molecular composition from crystal structures, J. Appl. Crystallogr., 2015, 48(1), 85–91 CrossRef PubMed.
- S. Gražulis, A. Daškevič, A. Merkys, D. Chateigner, L. Lutterotti and M. Quiros, et al., Crystallography Open Database (COD): an open-access collection of crystal structures and platform for world-wide collaboration, Nucleic Acids Res., 2012, 40(D1), D420–D427 CrossRef PubMed.
- S. Gražulis, D. Chateigner, R. T. Downs, A. Yokochi, M. Quirós and L. Lutterotti, et al., Crystallography Open Database–an open-access collection of crystal structures, J. Appl. Crystallogr., 2009, 42(4), 726–729 CrossRef PubMed.
- R. T. Downs and M. Hall-Wallace, The American Mineralogist crystal structure database, Am. Mineral., 2003, 88(1), 247–250 CAS.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, The Cambridge structural database, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72(2), 171–179 CrossRef CAS PubMed.
- A. S. Rosen, S. M. Iyer, D. Ray, Z. Yao, A. Aspuru-Guzik and L. Gagliardi, et al., Machine learning the quantum-chemical properties of metal–organic frameworks for accelerated materials discovery, Matter, 2021, 4(5), 1578–1597 CrossRef CAS.
- A. S. Rosen, V. Fung, P. Huck, C. T. O'Donnell, M. K. Horton and D. G. Truhlar, et al., High-throughput predictions of metal–organic framework electronic properties: theoretical challenges, graph neural networks, and data exploration, npj Comput. Mater., 2022, 8(1), 112 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00252k |
| This journal is © The Royal Society of Chemistry 2024 |