Open Access Article
Open Access ArticleLeveraging natural language processing to curate the tmCAT, tmPHOTO, tmBIO, and tmSCO datasets of functional transition metal complexes†
Ilia Kevlishvili a,
Roland G. St. Michel
a,
Roland G. St. Michel ab,
Aaron G. Garrison
ab,
Aaron G. Garrison a,
Jacob W. Toney
a,
Jacob W. Toney a,
Husain Adamji
a,
Husain Adamji a,
Haojun Jia
a,
Haojun Jia ac,
Yuriy Román-Leshkov
ac,
Yuriy Román-Leshkov ac and
Heather J. Kulik
ac and
Heather J. Kulik *ac
*ac
aDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. E-mail: hjkulik@mit.edu
bDepartment of Materials Science and Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
cDepartment of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
First published on 26th June 2024
Abstract
The breadth of transition metal chemical space covered by databases such as the Cambridge Structural Database and the derived computational database tmQM is not conducive to application-specific modeling and the development of structure–property relationships. Here, we employ both supervised and unsupervised natural language processing (NLP) techniques to link experimentally synthesized compounds in the tmQM database to their respective applications. Leveraging NLP models, we curate four distinct datasets: tmCAT for catalysis, tmPHOTO for photophysical activity, tmBIO for biological relevance, and tmSCO for magnetism. Analyzing the chemical substructures within each dataset reveals common chemical motifs in each of the designated applications. We then use these common chemical structures to augment our initial datasets for each application, yielding a total of 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 631 compounds in tmCAT, 4599 in tmPHOTO, 2782 in tmBIO, and 983 in tmSCO. These datasets are expected to accelerate the more targeted computational screening and development of refined structure–property relationships with machine learning.
631 compounds in tmCAT, 4599 in tmPHOTO, 2782 in tmBIO, and 983 in tmSCO. These datasets are expected to accelerate the more targeted computational screening and development of refined structure–property relationships with machine learning.
1. Introduction
Transition metal complexes (TMCs) are used in a wide variety of applications, including as homogeneous catalysts1–3 for fine chemical synthesis3–6 and in advanced sensor, display, and data storage devices.3,7 The electronic and steric properties of a TMC are influenced by those of the constituent metal and ligands.7,8 However, the variety of metal identities and oxidation states that may be used in conjunction with ligands of different connectivity and charge results in a combinatorial design space too large to sample exhaustively.3,7,9,10 Density functional theory (DFT) calculations are often leveraged in high-throughput virtual screening (HTVS) campaigns to explore chemical space in search of new molecules with desired properties, although the cost of such calculations limits the number of complexes that may be investigated.1,3,7,9,11,12 Such campaigns can be further accelerated through machine learning, which relies on large datasets of experimental and computational results for training.7,9,11Prior efforts have been made to curate datasets of TMCs,13–16 their constituent ligands,17–23 and relevant reactions.4,24–27 Many of these datasets are based on entries from the Cambridge Structural Database (CSD),28 a digital repository of experimental crystal structures, including molecular crystals of thousands of TMCs. However, challenges exist with current datasets, which primarily fall into one of two classes. The first class of datasets are exceptionally large but contain properties of limited relevance to applications-oriented molecular discovery.10,13,24,29–36 The second class of datasets are highly focused, containing relevant information very specific to local regions of chemical space, and as such are not easily generalized to new chemical applications.14,19–23,27,37,38 The transition metal quantum mechanics (tmQM) dataset is an example of a large, nonspecific dataset of interest to transition metal chemistry, containing 86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 665 mononuclear TMCs.16 These structures were extracted from the CSD and subjected to additional filtering by retaining only structures with at least one C and H atom, and only those which contain allowed non-metal elements (i.e., B, Si, N, P, As, O, S, Se, F, Cl, Br, and I). Furthermore, oxidation states were assumed for metals to ensure closed-shell character where possible and only structures that had a net charge of no more than +1 or less than −1 were retained. Their geometries were optimized at the semiempirical extended tight binding (xTB) level of theory, and DFT single-point energy calculations were performed on the resulting structures. While the tmQM is a valuable dataset in computational chemistry workflows for investigating TMCs,17,39,40 a key limitation is the absence of a mapping between molecular structures and the relevant areas of chemistry. This hinders further investigation into structures that are particularly promising for applications in catalysis, photochemistry, or other fields of interest. In contrast, datasets such as the ligand knowledge base (LKB) curated in pioneering work by Fey et al.19–22 and kraken later developed by Gensch et al.23 are examples of detailed, applications-focused datasets with limited transferability. Both datasets primarily consist of organophosphorus ligands, include relevant physicochemical descriptors useful in building quantitative structure–property relationships, and are based on commercial and virtual libraries.19–23,41 However, these do not generalize well to other areas of chemistry beyond organophosphorus ligands, exemplifying a limitation of such datasets and a need for large datasets linked to targeted chemical applications.
665 mononuclear TMCs.16 These structures were extracted from the CSD and subjected to additional filtering by retaining only structures with at least one C and H atom, and only those which contain allowed non-metal elements (i.e., B, Si, N, P, As, O, S, Se, F, Cl, Br, and I). Furthermore, oxidation states were assumed for metals to ensure closed-shell character where possible and only structures that had a net charge of no more than +1 or less than −1 were retained. Their geometries were optimized at the semiempirical extended tight binding (xTB) level of theory, and DFT single-point energy calculations were performed on the resulting structures. While the tmQM is a valuable dataset in computational chemistry workflows for investigating TMCs,17,39,40 a key limitation is the absence of a mapping between molecular structures and the relevant areas of chemistry. This hinders further investigation into structures that are particularly promising for applications in catalysis, photochemistry, or other fields of interest. In contrast, datasets such as the ligand knowledge base (LKB) curated in pioneering work by Fey et al.19–22 and kraken later developed by Gensch et al.23 are examples of detailed, applications-focused datasets with limited transferability. Both datasets primarily consist of organophosphorus ligands, include relevant physicochemical descriptors useful in building quantitative structure–property relationships, and are based on commercial and virtual libraries.19–23,41 However, these do not generalize well to other areas of chemistry beyond organophosphorus ligands, exemplifying a limitation of such datasets and a need for large datasets linked to targeted chemical applications.
The curation of a chemically targeted and synthetically accessible TMC dataset relies on systematically reviewing literature on the TMCs that are contained within existing databases. The broad scope of TMC literature, however, would make manual processing arduous, prompting the use of natural language processing (NLP) techniques42,43 for efficient analysis. NLP has been utilized extensively in the extraction of material properties and material synthesis parameters from the literature.37,44–51 More recently, large language models (LLMs) coupled with prompt engineering have gained increasing popularity in automating scientific text mining for chemical information due to their more user-friendly nature.52–56 A crucial aspect of text mining for classifying text based on chemical domain involves topic modeling,57 which is the identification of underlying themes in large sets of scientific text. For tasks of this nature, prompt engineering typically requires a priori definition of the possible latent topics. Nevertheless, LLMs can still be leveraged to obtain contextualized embeddings of the text that capture semantic information58,59 and subsequently cluster text based on semantic similarity.60,61 Here, each cluster corresponds to a latent topic, as facilitated by algorithms like bidirectional encoder representations from transformers for topic modeling (BERTopic).62 Simpler topic modeling approaches, such as latent Dirichlet allocation (LDA),63 which utilizes bag-of-words and statistical patterns of co-occurring words to infer latent topics, can also be employed to cluster manuscripts in a corpus. While these unsupervised NLP methods have been leveraged for summarizing research trends in chemistry,64 with an emphasis on biochemical and medicinal research65–68 as well as in the classification of large biomolecular datasets,69 they have yet to be extended to the space of transition metal chemistry and in the development of application-specific TMC datasets.
To construct chemically targeted TMC datasets, we conduct text mining on manuscripts associated with synthesizable TMCs from the tmQM database, focusing only on their titles and abstracts, and leverage both simple NLP tools as well as transformer models to process the text. Using topic modeling, we segment the structures in the tmQM database based on distinct chemistry applications. Through this process, we introduce four new TMC datasets – tmCAT containing catalytically-relevant TMCs, tmPHOTO with photoactive TMCs, tmSCO comprising TMCs with magnetic properties, and tmBIO containing biologically-relevant TMCs. Additionally, we performed substructure analysis to compare trends in metal-local structures among tmQM TMCs and the four curated datasets to subsequently enrich each chemistry-specific dataset by adding additional tmQM TMCs that could potentially be suitable for the given application.
2. Computational details
2.1. Corpus curation and text pre-processing
Manuscript titles and abstracts used for supervised learning as well as unsupervised clustering were obtained from a corpus that was curated in November 2020.70,71 Most manuscripts in this corpus were retrieved directly using the ArticleDownloader package.72 Manuscripts from the Royal Society of Chemistry (RSC), Wiley-VCH, the American Association for the Advancement of Science (AAAS), Springer, and Nature were obtained directly. Articles from the American Chemical Society (ACS) were obtained via a direct download agreement between ACS and the Massachusetts Institute of Technology. We created a secondary corpus of abstracts for manuscripts that could not be obtained with the ArticleDownloader package by scraping article URLs using the BeautifulSoup package v.4.12.2.73 We used the HTML title to retrieve manuscript titles. To obtain the manuscript abstract, we parsed the HTML paragraphs and retrieved the first paragraph that contained more than 400 characters. From a set of 100 randomly selected DOIs, we manually validated the abstract and title retrieval procedure, which shows that this approach can be used to retrieve titles and abstracts at a high rate (ESI Table S1†). However, this procedure can in rare cases (i.e., 1 case out 100 tested) lead to retrieval of an introduction paragraph instead of the abstract. Because the secondary corpus contains some degree of impurity, it was only used to identify more catalysis-relevant complexes based on the abstract with a trained classifier model (see Section 3.1).Abstracts were preprocessed using the NLTK v.3.8.1 package.74 The text was cleaned by lowercasing and removing punctuation, URLs, and numbers. The cleaned text was then tokenized using a regular-expression tokenizer, RegexpTokenizer, implemented in the NLTK package. Tokenized text was filtered using stop words with standard English stop words. For unsupervised clustering, an additional set of stop words was introduced to avoid clustering based on chemical languages (see Section 3.3). The filtered text was stemmed with the Snowball Stemmer and lemmatized with the WordNet Lemmatizer, both implemented in the NLTK package.
2.2. Featurization
For the classification model, we featurized the corpus using the term-frequency inverse-document-frequency (TF-IDF) vectorizer implemented in the scikit-learn v.1.4.0 package.75 The TF-IDF vectorizer accounts for the frequency of a given token within a document and its frequency in a collection of documents, assigning lower weight to common tokens across the entire corpus. Only tokens that appeared in at least 10 documents were retained, and the vector included mono-, bi-, and trigrams. The feature vector was fit using only the training set to avoid data leakage from inverse document frequency weighting. To reduce the feature vector length, we evaluated the χ2 score of each feature using the training set and retained only the 300 most important features as computed by the χ2 test. For unsupervised learning using BERTopic, we used the Sentence Transformers v2.2.2 (ref. 76) package for transforming abstracts into a feature vector. The semantic embedding was done using the sentence transformer sentence-BERT (SBERT) model,76 which converted the title and abstract into a 768-dimensional vector. Embedding was done using a pre-trained Siamese BERT network, all-mpnet-base-v2 transformer. Embeddings can be compared using cosine similarity. We then reduced the dimensionality of this vector to a five-dimensional mapping using uniform manifold approximation and projection (UMAP),77 which was selected because dimensionality can be reduced in UMAP using the same cosine metric as in the SBERT embedding. We generated a CountVectorizer feature vector of the corpus for unsupervised learning with latent Dirichlet allocation (LDA).78 LDA utilizes a bag of words vector, which consists of the overall token count for each document. Furthermore, LDA requires a predefined number of clusters. We decomposed the corpus into 20 clusters to maintain relative consistency with the 23–25-cluster size identified by BERTopic. The count vector was generated using scikit-learn and consists of a vector of term count length per document.While BERT-based models can, in principle, be applied without text preprocessing, this in practice leads to clustering by transition metal or material (ESI Table S2†). To avoid this, we introduced stop words before text processing to avoid dependence on specific materials or metals, with a full list of stop words provided on Zenodo.79
2.3. NLP models
The catalysis classifier is a random forest classifier implemented in the scikit-learn v.1.4.0 package. A random 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 train/test split was used to evaluate the model performance on a set-aside test set. Hyperparameter optimization does not significantly affect the model performance. Full grid search cross-validation summary can be accessed through the Zenodo repository.79 Default random forest hyperparameters were used for training without hyperparameter optimization, with the exception of the minimum samples required to split a node that was set to 10, and the number of trees that was set to 1000 (ESI Table S3†). The minimum sample split was increased from the default to avoid overfitting, and the number of models in the ensemble was increased to improve model performance. Dimensionality reduction of high-dimensional feature vectors for unsupervised clustering (5-dimensional vector) and visualization (2-dimensional vector) were carried out using uniform manifold approximation and projection for dimension reduction (UMAP) with the UMAP package v0.5.5.77 We use the 5-dimensional vector to identify dense clusters, and we interpret topic assignments through cluster-level TF-IDF vectors. Clustering on the reduced 5-dimensional vector was carried out using HDBSCAN,80 a hierarchical density-based clustering algorithm, using the HDBSCAN v0.8.33 package. The topic assignment of dense clusters was achieved using the BERTopic package v0.16.0 with a modified class-based TF-IDF vector (c-TF-IDF).62 Clustering using LDA was carried out with the implementation in the scikit-learn package.
20 train/test split was used to evaluate the model performance on a set-aside test set. Hyperparameter optimization does not significantly affect the model performance. Full grid search cross-validation summary can be accessed through the Zenodo repository.79 Default random forest hyperparameters were used for training without hyperparameter optimization, with the exception of the minimum samples required to split a node that was set to 10, and the number of trees that was set to 1000 (ESI Table S3†). The minimum sample split was increased from the default to avoid overfitting, and the number of models in the ensemble was increased to improve model performance. Dimensionality reduction of high-dimensional feature vectors for unsupervised clustering (5-dimensional vector) and visualization (2-dimensional vector) were carried out using uniform manifold approximation and projection for dimension reduction (UMAP) with the UMAP package v0.5.5.77 We use the 5-dimensional vector to identify dense clusters, and we interpret topic assignments through cluster-level TF-IDF vectors. Clustering on the reduced 5-dimensional vector was carried out using HDBSCAN,80 a hierarchical density-based clustering algorithm, using the HDBSCAN v0.8.33 package. The topic assignment of dense clusters was achieved using the BERTopic package v0.16.0 with a modified class-based TF-IDF vector (c-TF-IDF).62 Clustering using LDA was carried out with the implementation in the scikit-learn package.
All machine learning models, scripts, Jupyter notebooks, datasets, and their associated structures are provided on Zenodo.79 Geometry assignment was carried out using functionality implemented in the molSimplify geometry_changes branch, and the script is included in the Zenodo repository.79
3. Results and discussion
3.1. Catalytic transition metal dataset
To curate a dataset of catalytically relevant transition metal complexes, we utilized the tmQM dataset81 consisting of 86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 665 unique CSD refcodes as a starting point, and we obtained the manuscripts associated with each CSD refcode using the ArticleDownloader package.72 Through this procedure, we curated a corpus consisting of 28
665 unique CSD refcodes as a starting point, and we obtained the manuscripts associated with each CSD refcode using the ArticleDownloader package.72 Through this procedure, we curated a corpus consisting of 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 394 unique manuscripts, accounting for 50
394 unique manuscripts, accounting for 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 968 crystals in the dataset. To utilize natural language processing models for identifying catalysis manuscripts, we focused on manuscript abstracts because they are information-dense texts that tend to avoid the discussion of broader topics and are therefore well suited for identifying whether a manuscript will discuss catalysis (i.e., versus introductions). Furthermore, abstracts are publicly available, typically on the article’s DOI-accessible HTML webpage, which enables retrieval of abstracts that could not be obtained using the ArticleDownloader package.72 If abstracts could not be extracted automatically from the manuscript (here, this occurred for 4682 of 28
968 crystals in the dataset. To utilize natural language processing models for identifying catalysis manuscripts, we focused on manuscript abstracts because they are information-dense texts that tend to avoid the discussion of broader topics and are therefore well suited for identifying whether a manuscript will discuss catalysis (i.e., versus introductions). Furthermore, abstracts are publicly available, typically on the article’s DOI-accessible HTML webpage, which enables retrieval of abstracts that could not be obtained using the ArticleDownloader package.72 If abstracts could not be extracted automatically from the manuscript (here, this occurred for 4682 of 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 394 manuscripts), we used titles instead.
394 manuscripts), we used titles instead.
To train a sentiment analysis model that could identify whether an abstract is associated with catalysis, we first identified the subset of manuscripts related to catalysis based solely on whether the manuscript titles contained the keyword “catal” but did not contain false-positive keywords (e.g., “uncatal”, “acid catal” or “base catal”, ESI Table S3†). These steps produced a 4585-manuscript subset where titles were confidently labeled as addressing catalysis (ESI Table S3†). We then confidently identified non-catalytic manuscripts by excluding manuscripts with catalytic and associated keywords (i.e., “catal”, “turnover”, and “polymer”) that occurred at least once in either the title or abstract (ESI Table S4†). This non-catalytic set consists of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 557 manuscripts, the majority of manuscripts not identified as catalytic in our initial step, leaving only 3252 unlabeled manuscripts. To analyze the label assignment, we randomly sampled 50 catalysis and 50 non-catalysis manuscripts and checked them manually. We find that 48 of the positive and 49 of the negative labels were correctly assigned using our approach.
557 manuscripts, the majority of manuscripts not identified as catalytic in our initial step, leaving only 3252 unlabeled manuscripts. To analyze the label assignment, we randomly sampled 50 catalysis and 50 non-catalysis manuscripts and checked them manually. We find that 48 of the positive and 49 of the negative labels were correctly assigned using our approach.
Despite our efforts to carefully label manuscripts confidently, simple pattern matching leads to a small number of incorrect label assignments. Our approach to avoid false positive labels by only looking at patterns in the title is expected to miss true positive hits, motivating a more robust approach for identifying catalyst-focused manuscripts. We next pursued a more systematic approach by developing a classifier model using natural language processing. Prior to training the classifier, we first created a balanced dataset of catalysis and non-catalysis manuscripts by subsampling the non-catalytic manuscripts to achieve a balanced set of 4585 non-catalysis manuscripts to match the 4585 catalysis manuscripts. Among the set of 9170 manuscripts, 1312 manuscripts (364 catalysis, 948 non-catalysis) did not have a defined abstract, and the title was instead used for training the classifier model. We then separated this set of 9170 manuscripts into training and test sets using a stratified random split of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
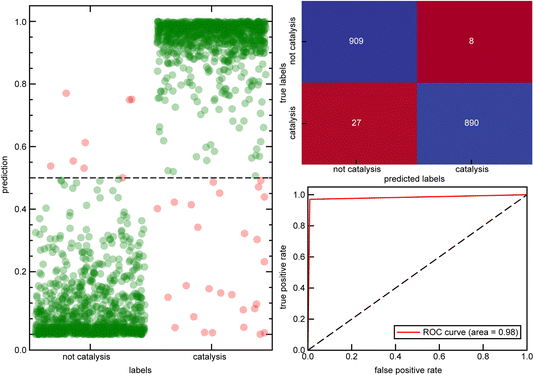
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. Using the abstract text of each of these papers, we preprocessed the text by elimination of uppercase letters, removal of punctuation, lemmatization, stemming, and removal of stop words to make the text suitable for natural language processing tasks (see Computational details). We first featurized the papers based on the term frequency-inverse document frequency (TF-IDF) vectorizer82 from the training set (see Computational details). Subsequently, a random forest classifier model was trained on the reduced TF-IDF feature vectors to predict whether an abstract is related to catalysis, achieving a high accuracy of 0.97 and strong separation between the two classes and ROC-AUC of 0.98 (Fig. 1).
20. Using the abstract text of each of these papers, we preprocessed the text by elimination of uppercase letters, removal of punctuation, lemmatization, stemming, and removal of stop words to make the text suitable for natural language processing tasks (see Computational details). We first featurized the papers based on the term frequency-inverse document frequency (TF-IDF) vectorizer82 from the training set (see Computational details). Subsequently, a random forest classifier model was trained on the reduced TF-IDF feature vectors to predict whether an abstract is related to catalysis, achieving a high accuracy of 0.97 and strong separation between the two classes and ROC-AUC of 0.98 (Fig. 1).
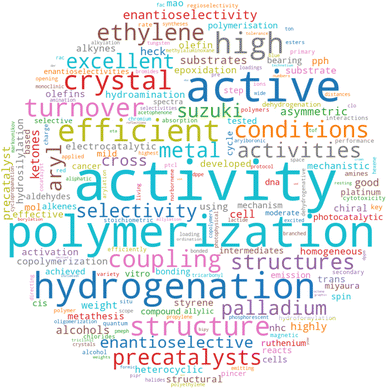
Despite the strong overall performance, we next analyzed the specific cases where the model failed to ensure that it was a suitable tool for confidently assigning a catalysis focus to manuscripts. Out of 1834 unique abstracts in the test set, 35 were incorrectly labeled. Only eight of these abstracts were given a false positive label and were manually inspected to prevent dataset contamination. Among these eight abstracts, five had labels that were inaccurately assigned by the rule-based method (i.e., missing the “catal” and additional keywords in the abstract/title) and thus should have been labeled as catalysis manuscripts. Additionally, one entry lacked an abstract, and its title had been used for training instead, likely contributing to the failure of the model to correctly predict the non-catalytic label for the manuscript. Furthermore, 27 complexes were given false negative labels. Among these, 14 had labels that were inaccurately assigned by the rule-based method. This analysis reveals that only a negligible fraction of manuscripts were wrongly labeled as catalysis-focused by the classifier model, and our method can detect complexes that the rule-based approach missed. Similarly, the model can detect true positive labels that were missed by the rule-based method. However a negligible but non-zero number of catalysis manuscripts might be missed by the model. Detailed analysis of all false labels are provided in the Zenodo repository.79 Unsurprisingly, the feature importance analysis of TF-IDF vectors showed that the most crucial features are keywords directly related to catalysis. However, several other significant word features were also identified, including activity, polymerization, hydrogenation, coupling, selectivity, Suzuki, and enantioselective, among others (Fig. 2). To test the effectiveness of these additional tokens related to catalysis, we developed a separate random forest model that was trained on a TF-IDF feature vector that excluded the direct catalysis keywords (ESI Table S5†). This second random forest model still achieved 89% accuracy, demonstrating that other relevant keywords effectively identify catalysis-related manuscripts (ESI Table S6 and Fig. S1†).
Given the promising performance of the classifier, we utilized the random forest model trained on the full TF-IDF feature vector to identify additional catalysis papers from the superset of all unlabeled manuscripts. This unlabeled set comprised any manuscript from the clean corpus not included in the original training/test set, which includes 3252 manuscripts that were not labeled as either catalysis or not-catalysis, the excluded non-catalytic manuscripts absent from the subsampled set, and manuscripts titles/abstracts mined from HTML source if they could not be obtained through the ArticleDownloader package. In total, this set comprised 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 449 manuscripts associated with 30
449 manuscripts associated with 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 345 unique CSD refcodes. By applying the random forest classifier to this unlabeled set, we identified 6208 additional manuscripts in the corpus as associated with catalysis (ESI Fig. S2 and S3†). With this added set, we identify the final tmCAT dataset, which consists of 10
345 unique CSD refcodes. By applying the random forest classifier to this unlabeled set, we identified 6208 additional manuscripts in the corpus as associated with catalysis (ESI Fig. S2 and S3†). With this added set, we identify the final tmCAT dataset, which consists of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 793 unique manuscripts and the structures of 19
793 unique manuscripts and the structures of 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 associated TMCs. While NLP will identify abstracts that are associated with catalysis, it is not necessarily the case that all crystal structures associated with these papers are relevant for catalysis (i.e., they could correspond to non-catalytic cations or catalyst precursors), and further analysis of the chemical composition of the dataset is merited (see Section 3.4).
250 associated TMCs. While NLP will identify abstracts that are associated with catalysis, it is not necessarily the case that all crystal structures associated with these papers are relevant for catalysis (i.e., they could correspond to non-catalytic cations or catalyst precursors), and further analysis of the chemical composition of the dataset is merited (see Section 3.4).
3.2. Comparing catalysis and non-catalysis complexes
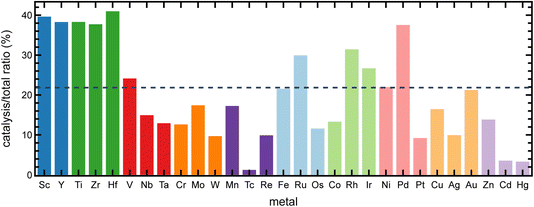
We next quantified differences in the composition between catalytically-relevant tmCAT complexes and the broader tmQM dataset. To make this comparison, we first examined the distribution of transition metals in the tmQM and the tmCAT subset. Based on their frequency in the tmCAT dataset relative to the whole tmQM set, group 4 metals (i.e., Ti, Zr, and Hf) are most disproportionately used for catalysis, which can likely be attributed to the large number of papers and structures related to olefin polymerization with homogeneous Ziegler–Natta catalysts in our set (Fig. 3). Group 3 metals (i.e., Sc and Y) are also extensively studied for catalysis (Fig. 3). These metals are present in compounds that are often used as post-metallocene olefin polymerization catalysts83 and lactone ring-opening polymerization catalysts.84 These findings indicate the prevalence of polymerization catalysts in the tmCAT subset, which we also explore further with additional text analysis in Section 3.3. Furthermore, Pd, Ru, Ir, Rh, and Au metals are abundant in our set, as would be expected because of their relatively frequent use in catalysis applications.85–88 Conversely, more earth-abundant transition metals are less commonly used in catalysis despite recent efforts towards sustainable catalysis using these metals.89 While iron- and nickel-based catalysts have been studied relatively frequently, cobalt-based catalysts are notably underrepresented, especially in comparison to isovalent Ir and Rh species that are more represented in the catalysis dataset. This analysis underscores the continued need to advance catalysis toward using more earth-abundant metals.Next, we analyzed the differences and similarities between the tmCAT and the rest of the tmQM dataset (i.e., the non-catalytic portion) in terms of descriptors that had been computed during the curation of the tmQM dataset.81 We first focus on electronic descriptors, such as molecular orbital energetics90 and metal charges that are commonly employed in the screening of transition metal catalysts.90,91 The relevant descriptors available in the original tmQM set81 include the HOMO and LUMO energies, the HOMO–LUMO gap, and the transition metal center partial charge. These properties were computed using the TPSSh meta-GGA hybrid functional with empirical D3(BJ) dispersion correction and a def2-SVP basis set based on GFN2-xTB optimized geometries. Interestingly, the distribution of these descriptors shows no major differences between the catalytic (tmCAT) subset and the non-catalytic subset of the tmQM dataset (ESI Fig. S4–S7†). These observations suggest that when considering a diverse set of complexes, with wide-ranging transition metals, oxidation states, coordination environments, and ligands, descriptors based on frontier orbitals or metal partial charges may be insufficient for inferring reactivity. These observations are consistent with past work showing that frontier orbital energies alone struggle to generalize to catalyst activities across multiple metals and oxidation states.92
We next considered geometric descriptors that evaluate the steric environment defined by ancillary ligands93 as another commonly employed class of descriptors for catalyst screening. We would expect an active catalyst (i.e., not a precatalyst) to feature an open site that can lead to the association of a reactant to the active site. However, active catalysts with open metal sites are usually not energetically stable intermediates and tend to have a sacrificial ligand or a solvent coordinated to the open site. To compare tmCAT structures to the non-catalytic subset of tmQM, we computed the percent buried volume of the metal for all complexes in the tmQM dataset. Analysis of the distribution of this parameter shows no significant difference in buried volume between the tmCAT and the rest of the tmQM dataset (ESI Fig. S8†). We anticipate this lack of distinction is attributable to the fact that deposited crystal structures are likely precatalysts that need to undergo an activation process to form an active catalyst, meaning that geometric descriptors on CSD structures are unlikely to be useful for identifying catalysis-capable complexes.
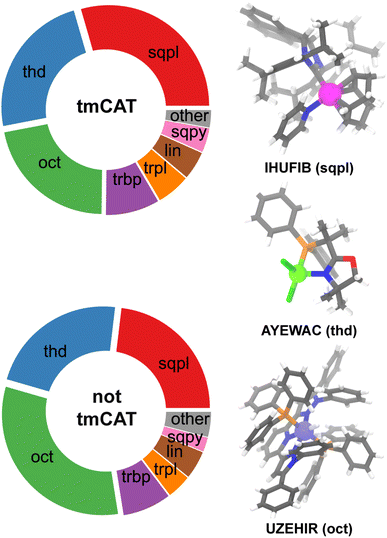
Beyond steric metrics, one might anticipate other differences in the metal-local coordination that might distinguish the tmCAT and non-catalysis tmQM subsets. We hypothesized that even though pre-catalyst complexes should be heavily featured in tmCAT, some noticeable differences could still be observed between catalysis and non-catalysis datasets when comparing coordination geometries because some geometries are less probable for precatalysts (e.g., those with six monodentate ligands or three bidentate ligands). We assigned metal coordination geometries by examining the geometric deviations from possible ideal transition metal geometries and assigning a geometric class with the lowest deviation (see Computational details). When a haptic ligand is encountered (e.g., an alkene bound via its π bond to a metal center), a single occupancy was assigned at the geometric centroid of the haptic ligand. The most noticeable difference between tmCAT and the non-catalysis tmQM subset is due to the significant reduction in the number of octahedral complexes in tmCAT accompanied by a significant enhancement in the frequency of square planar complexes (Fig. 4). Despite a lack of difference between tmCAT and the non-catalytic tmQM in terms of steric descriptors, this enhancement of square planar over octahedral structures is consistent with our expectation of enhancing coordinatively unsaturated complexes in the tmCAT dataset as well as the fact that more octahedral structures are likely to be less compatible with catalysis due to higher-denticity ligands. Furthermore, the relative frequencies of other coordinatively unsaturated geometries, such as square pyramidal and trigonal planar complexes are also enhanced in the tmCAT dataset.
3.3. Unsupervised learning with natural language processing maps CSD structures to other applications
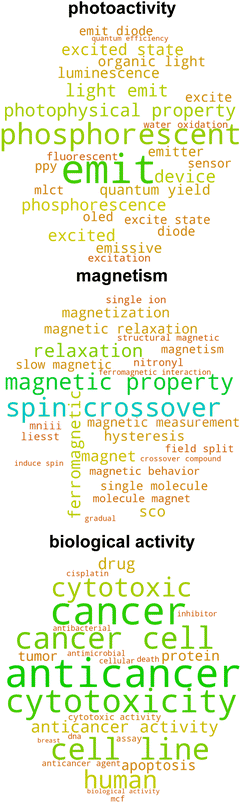
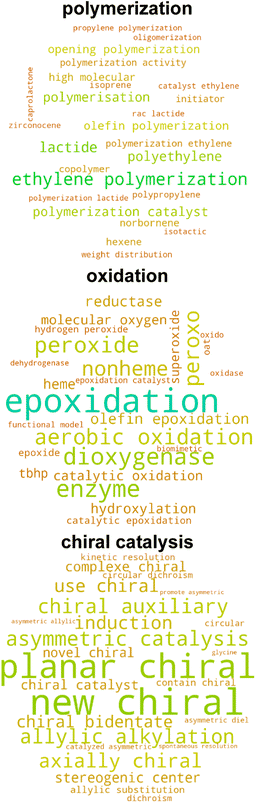
Given that the catalysis classifier only assigns approximately 25% of tmQM complexes to the tmCAT dataset, we next aimed to identify if the remaining 75% could be at least partly assigned to other application areas for TMCs by carrying out a more expansive review of the full corpus of papers in the tmQM set. Defining a topical subset within a corpus using supervised learning necessitates curation of a smaller subset, either manually or by defining heuristic rules for identifying positive and negative examples. On the other hand, by using unsupervised learning on the text, it is possible to identify clusters within a corpus that are semantically similar. Analyzing these clusters can lead to an improved understanding of different possible applications and topics covered by the corpus. A common approach for this purpose in natural language processing is topic modeling,94 an unsupervised learning approach that can both cluster a corpus and identify topics associated with each cluster. We applied topic modeling to our full corpus of manuscript titles and abstracts to identify latent application topics, offering a more comprehensive view of the underlying themes and connections within the corpus.We utilized the BERTopic model,62 a method that clusters bodies of text based on their semantic embedding and then assigns topics using a modified, class-based TF-IDF (c-TF-IDF) vector or other interpreter (see Computational details). Importantly, we identified several subtopics that could be associated with non-catalytic applications, including biological activity, photoactivity, magnetism, self-assembly, and X-ray crystal structure characterization (Fig. 5 and S9 ESI†). Additionally, many of the topics uncovered by this analysis are complementary to our earlier labeling of catalyst papers, as other uncovered subtopics include several catalysis-related areas with more specific applications including polymerization, hydrogenation, chiral catalysis, and cross-coupling (Fig. 6 and S10 ESI†). Even though the number of subtopic clusters and cluster composition is distinct across different models (i.e., because there are multiple topic models that differ based on a random seed), all major subtopic clusters we identified are conserved. Using this additional information, we now introduce three additional datasets consisting of complexes associated with manuscripts consistently categorized across all five models as follows: (i) tmPHOTO, which consists of photoactivity-associated complexes, (ii) tmSCO, which consists of compounds exhibiting properties relevant to studies of magnetism, and (iii) tmBIO, which consists of complexes with biologically relevant activities. We identified each of these datasets by inspecting the most significant tokens associated with each of the clusters. For example, the tmPHOTO set is derived from the cluster that consists of manuscript abstracts that discuss phosphorescence, emission, and quantum yield, indicating that photophysical activity is discussed throughout these manuscripts. Similarly, we identified that tmSCO manuscript abstracts are associated with tokens that are related to spin-crossover, magnetic properties and hysteresis, all keywords that are related to changes in the spin state of a TMC. Likewise, we determined that manuscript abstracts associated with the tmBIO cluster discuss properties such as cytotoxicity, cancer, cell, and apoptosis, which are all related to biological activity relevant to pharmaceutical applications.
Based on how semantic embedding works, we expect it to place/arrange subtopics with greater similarity closer to each other in the reduced dimensional space. To analyze the performance of the unsupervised learning approach and visualize how different topics relate to each other, we employed UMAP for further dimensionality reduction on the SBERT embedding, reducing the embeddings to two dimensions better suited for visualization while retaining the global structure of the data for distance comparison. We find that catalysis sub-topics are predominantly clustered closely to each other, suggesting that the wording used throughout catalysis-associated abstracts is highly similar. Furthermore, catalysis topics that are more closely related to each other, such as polymerization and metathesis, or hydrogenation and hydroboration/boration, each of which is related to olefin functionalization, are more closely clustered (Fig. 7). As expected, the other, non-catalysis topics are more distant in the UMAP-reduced space (Fig. 7). The biologically active cluster, arguably the most different from other applications due to its relevance in biological and pharmaceutical applications, stands out as the most distinct cluster. Manuscripts related to photoactivity and magnetism are comparatively clustered close to each other, which can be expected because both topics are associated with the transition to an excited state via external stimulus (Fig. 7). Alternative dimensionality reduction techniques, such as t-distributed stochastic neighbor embedding (t-SNE),95 lead to similar conclusions, although we avoided using t-SNE because it is less effective at preserving the global data structure (ESI Fig. S11†).
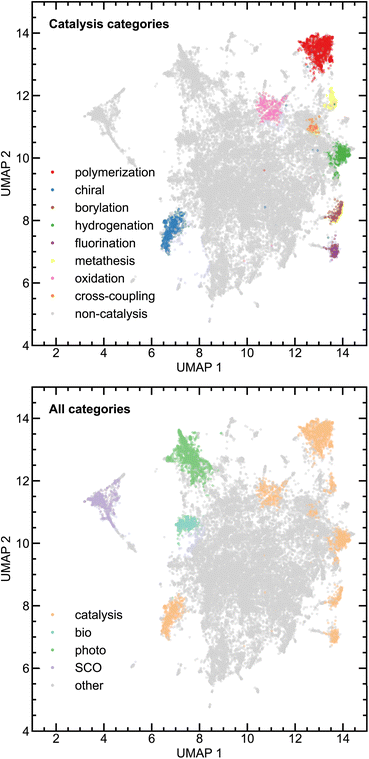 | ||
| Fig. 7 UMAP dimensionality reduction of SBERT embedding vectors colored by different cluster topics for different catalysis applications (top) and general applications (bottom). | ||
To support the findings from the BERTopic model, we employed an additional topic modeling approach, latent Dirichlet allocation (LDA).78 LDA is a Bayesian method that iteratively assigns two probabilities: one indicating that a given token belongs to a topic and the other indicating that a topic belongs to a document. This LDA analysis produces semantically similar clusters to BERTopic (ESI Fig. S12†). Using the LDA approach, several catalysis clusters are identified, including polymerization, chiral catalysis, cross-coupling catalysis, olefin functionalization catalysis, and mechanism-focused catalysis. Some non-catalysis topics are also conserved, such as photoactivity, biological activity, magnetism, and X-ray crystal structure characterization. Feature reduction of the token count vector, obtained using UMAP reduction with Hellinger distance, leads to a similar mapping, demonstrating the close relationship between the identified clusters and the shorter distances between catalysis-related clusters (ESI Fig. S13†).
Even with the expanded subsets identified by unsupervised clustering using BERTopic or using the LDA analysis, a significant portion of the tmQM complexes are either unlabeled or assigned to difficult-to-interpret clusters. Furthermore, these datasets curated using simple natural language processing methods are not fully context-aware and don’t account for more detailed information present in the manuscript, such as discussion of failed experimental attempts, meaning that they may contain “negative” examples. That is, these subsets could contain complexes that were either used as counterexamples, found to be ineffective for a given activity, or represent a precursor structure that was crystallized before the in situ assembly of chemically relevant species. Accordingly, we carried out analysis of the structures present in each dataset to both support the composition of these datasets and enhance them with chemically relevant species (see Section 3.4). Such species might not have been originally used for a given application and therefore be missed by NLP approaches, but due to chemical similarity, they could have a complementary application.
3.4. Analysis of common metal-centered motifs in curated datasets
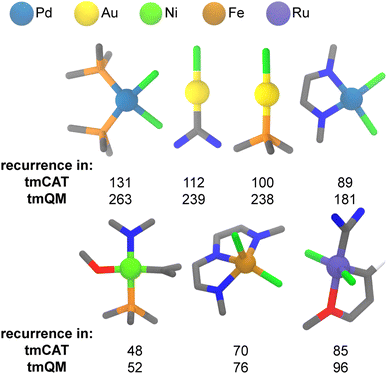
It is intuitive that strong relationships should exist between the metal, its coordination environment in a TMC and the associated property or application. As a representative example, linear gold chlorides coordinated to phosphine or N-heterocyclic carbene (NHC)-type ligands are commonly utilized in catalysis96,97 but would not be expected to be relevant for spin-crossover because the Au metal is closed-shell. We thus carried out an analysis of the metal-centered subgraphs to contrast trends in metal-local structure in the overall tmQM set to those in the tmCAT, tmPHOTO, tmBIO, and tmSCO subsets. We carried out this analysis after excluding lanthanide complexes because they are relatively poorly represented across all datasets. To capture the metal-local environment, we computed metal-centered subgraphs with a truncation of 2 bond paths away (i.e., d = 2) from the metal on the molecular graph. The radius of 2 was chosen because it captures metal-local electronic character while also reducing the diversity of possible combinations. We obtained these subgraphs on the CSD-reported connectivity using the molSimplify package,98 and we then determined the uniqueness of these subgraphs by computing and comparing the edge-attributed molecular graph hashes of each metal-centered substructure. The Python script, resulting substructures, and graph hashes are all provided on Zenodo.79We first analyzed common structural motifs in the tmCAT dataset. Breaking down the dataset into structural motifs at d = 2 shows that while most complexes are unique when analyzing metal-local character, several structural motifs appear relatively frequently in the tmCAT dataset (ESI Fig. S14†). Overall, 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 complexes in the tmCAT dataset are represented by 10
250 complexes in the tmCAT dataset are represented by 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 094 unique d = 2 (i.e., metal-local) substructures. In particular, palladium dichlorides bound to either two phosphine ligands or to bidentate nitrogen-coordinating ligands are very common in the tmCAT dataset, appearing 131 and 89 times, respectively (Fig. 8). These metal-local motifs are likely derived from complexes that have been widely studied for their ability to catalyze cross-coupling reactions.99 Similarly, linear gold chlorides bound to NHC or phosphine ligands are common in the tmCAT dataset, appearing 112 and 100 times, respectively (Fig. 8). Linear gold complexes are known to catalyze various π-functionalization and annulation reactions, among others.100 The high abundance of these gold and palladium motifs is in line with their occurrence in the tmQM superset, where the popularity of these complexes has led to their widespread examination in many contexts. On the other hand, several frequently occurring motifs were identified that are almost exclusively studied for catalysis (Fig. 8). These include nickel catalysts with four mixed N, C, O, and P coordinating ligands, i.e., where each ligand type coordinates the metal once; iron dichloride catalysts with tridentate nitrogen coordinating ligands; and ruthenium dichloride catalysts with an NHC ligand and carbene ligand with a chelating oxygen group101 (i.e., likely derived from the second-generation Hoveyda–Grubbs catalyst102). Surprisingly, all these catalysts have been primarily studied for polymerization: Ni complexes are utilized for ethylene copolymerization with carbon monoxide,103 Fe complexes are used as catalysts for linear homo-polymerization of ethylene for the synthesis of high-density polyethylene,104 and Hoveyda–Grubbs catalysts are utilized for ring-opening metathesis polymerization (ROMP).105 This highlights how some motifs, despite their limited range of application, have been the focus of a great deal of study as a result of their industrial relevance.
094 unique d = 2 (i.e., metal-local) substructures. In particular, palladium dichlorides bound to either two phosphine ligands or to bidentate nitrogen-coordinating ligands are very common in the tmCAT dataset, appearing 131 and 89 times, respectively (Fig. 8). These metal-local motifs are likely derived from complexes that have been widely studied for their ability to catalyze cross-coupling reactions.99 Similarly, linear gold chlorides bound to NHC or phosphine ligands are common in the tmCAT dataset, appearing 112 and 100 times, respectively (Fig. 8). Linear gold complexes are known to catalyze various π-functionalization and annulation reactions, among others.100 The high abundance of these gold and palladium motifs is in line with their occurrence in the tmQM superset, where the popularity of these complexes has led to their widespread examination in many contexts. On the other hand, several frequently occurring motifs were identified that are almost exclusively studied for catalysis (Fig. 8). These include nickel catalysts with four mixed N, C, O, and P coordinating ligands, i.e., where each ligand type coordinates the metal once; iron dichloride catalysts with tridentate nitrogen coordinating ligands; and ruthenium dichloride catalysts with an NHC ligand and carbene ligand with a chelating oxygen group101 (i.e., likely derived from the second-generation Hoveyda–Grubbs catalyst102). Surprisingly, all these catalysts have been primarily studied for polymerization: Ni complexes are utilized for ethylene copolymerization with carbon monoxide,103 Fe complexes are used as catalysts for linear homo-polymerization of ethylene for the synthesis of high-density polyethylene,104 and Hoveyda–Grubbs catalysts are utilized for ring-opening metathesis polymerization (ROMP).105 This highlights how some motifs, despite their limited range of application, have been the focus of a great deal of study as a result of their industrial relevance.
Next, we expanded our substructure analysis to the photochemistry-relevant tmPHOTO subset. Analysis of metal identity in tmPHOTO reveals that iridium, platinum, and copper complexes are significantly amplified in this dataset (ESI Fig. S15†). Iridium106 and platinum107 complexes have been common targets for photophysical applications due to spin–orbit coupling that allows intersystem-crossing, which leads to high quantum yields. On the other hand, copper complexes108 have been explored as an earth-abundant alternative to more rare and expensive iridium and platinum complexes. Substructure mapping shows that 3043 complexes in the tmPHOTO dataset are represented by 1150 unique d = 2 structural motifs. Several commonly recurring substructures can be observed in the tmPHOTO set (ESI Fig. S16†). These include iridium complexes with a coordination number of six with two bidentate C⁁N coordinating ligands and two oxygen-coordinating ligands (i.e., structural analogs of Ir(ppy)2(acac)), which is a substructure that appears in tmPHOTO 101 times (ESI Fig. S17†). Platinum complexes with a coordination number of four, a bidentate C⁁N type ligand, and two oxygen-coordinating ligands are other commonly recurring structural motifs (i.e., structural analogs of Pt(ppy)(acac)), with this substructure appearing 89 times in tmPHOTO. Another similarly commonly recurring motif is a 4-coordinate copper complex with mixed nitrogen and phosphorus coordinating atoms, including a bidentate nitrogen ligand, which appears 89 times in the tmPHOTO dataset (ESI Fig. S17†). Examples of structural motifs that are almost exclusively studied for photophysical properties include Ir complexes with two bidentate C⁁N type ligands and a bidentate N⁁N type ligand, as well as platinum complexes with one bidentate C NHC-type ligand and two oxygen-coordinating ligands (ESI Fig. S17†).
Moving on to the spin-crossover relevant subset, we note that there are necessary differences for TMCs that exhibit switchable magnetic behavior. Here, iron, manganese, nickel, and cobalt complexes occur with higher relative frequency in the tmSCO set than in the tmQM superset (ESI Fig. S15†). These metals are all third-row transition metals that tend to have relatively low d-orbital splitting energy (Δ) and, depending on the oxidation state, they are expected to have multiple accessible spin states. Substructure mapping shows that 834 complexes in the tmSCO dataset are represented by 534 unique d = 2 structural motifs. A few d = 2 structural motifs are representative of this dataset through multiple recurrences (ESI Fig. S18†). These recurring motifs include manganese complexes with four nitrogen-coordinating ligands, including a bidentate sp3 hybridized ligand and two oxygen-coordinating ligands, which appear 39 times in tmSCO and only six additional times (i.e., 45 in total) across the entire tmQM dataset. This highlights how these complexes are nearly exclusively targeted for magnetic properties. Another common structural motif includes an iron center with four sp2 hybridized nitrogen coordinating ligands and two nitrogen coordinating ligands that could be either isocyanides or cyanates (ESI Fig. S19†). These complexes appear in the tmSCO set 27 times, and in the tmQM superset a total of 44 times. Interestingly, a relatively common structural motif includes iron bound to a tetradentate nitrogen-coordinating ligand with two additional isocyanides/cyanate ligands, which appears in the tmQM dataset 9 times, all of which are in the tmSCO subset, suggesting that these motifs have only been studied for applications related to magnetism (ESI Fig. S19†).
Finally, we analyzed the substructures in the biological activity subset, which we expect to be the most diverse due to the broad nature of this set. Ruthenium and platinum are the most heavily represented metals in the tmBIO dataset (ESI Fig. S15†). Overall, substructure mapping shows that 1808 complexes in the tmBIO dataset are represented by 974 unique d = 2 structural motifs. A high interest in platinum for biological applications can be attributed to cisplatin,109 i.e., cis-diamminedichloroplatinum(II), the first inorganic small molecule approved as a pharmaceutical anti-cancer drug. Cisplatin-resistant cancers110 have led to the search for alternate platinum complexes as anti-cancer drugs111 and are represented in the tmBIO dataset. Furthermore, the high toxicity of cisplatin has led to the search for alternate inorganic and organometallic complexes that could be used as anti-cancer medications. In particular, the high promise of ruthenium arene 1,3,5-triaza-7-phosphaadamantane (RAPTA)112 compounds has led to a significant effort in screening ruthenium-based piano stool complexes as potential anti-cancer drugs.113 These efforts are consistent with the makeup of the tmBIO dataset. In fact, the three most commonly recurring structural motifs feature ruthenium arene complexes with one chloride and two additional ancillary ligands (ESI Fig. S20 and S21†). These complexes, cumulatively, appear in the tmBIO dataset 82 times and 208 in the tmQM dataset. Furthermore, a motif that is closely related to cisplatin, with platinum, two chloride ligands, a single ammonia, and an organic N-coordinating ligand, appears in the tmBIO set 17 times and has been exclusively studied for biological applications (ESI Fig. S21†).
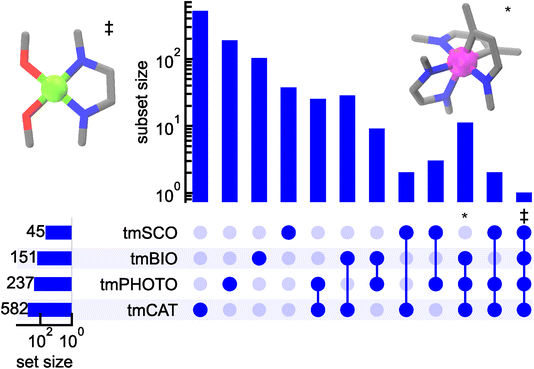
Finally, we analyzed if any structural motifs appear frequently across different datasets to identify if they are frequently studied for multiple applications. To achieve this, we first created a subset of each dataset, consisting of commonly recurring motifs in each of the dataset (i.e. five or more recurrences for tmCAT and 3 or more recurrences for other datasets). We then analyzed overlaps among the datasets. These motifs are mostly exclusive to a given set, with more than 82% of metal-centered motifs only appearing in one of the four sets exclusively (Fig. 9). However, a single metal-centered substructure appears at least five times across all datasets. This motif consists of a nickel metal center with mixed N⁁N,O,O coordinating atoms and is reminiscent of salen complexes, despite the inability of radius 2 subgraphs to capture the entirety of tetradentate salen ligands (Fig. 9, left inset). Salen complexes are a common enzyme mimic often used in asymmetric catalysis114 but could be used as targets for photoactive complexes due to the rigid nature of the ligand, as magnetic complexes with the nickel as the metal center, and the square planar coordination environment of salen ligands with Ni complex can be targets for DNA intercalation. Furthermore, several structural motifs were identified to reside among tmCAT, tmPHOTO, and tmBIO sets but not tmSCO. A noteworthy example includes an iridium motif with two bidentate C⁁N-type ligands and single bidentate N⁁N type ligand (i.e., Ir(ppy)2(bpy) analogs). These complexes are commonly used as triplet sensitizers, which can be applied for photocatalysis115 to access excited states and for biological applications to target reactive singlet oxygen formation116 (Fig. 9, right inset).
Analysis of common application-specific structural motifs reveals that, for a given motif, there are complexes within the tmQM dataset that contain the motif, but the associated manuscripts do not indicate the complex has been assessed for that specific application. Therefore, we supplemented each dataset using structural mapping to identify chemically similar structures to add to our data subsets. To complete this augmentation, we note that for most of these application-specific datasets, multidentate ligands can play an important role, such as in defining the steric environment and introducing added stability for catalysis or inhibiting thermal relaxation pathways for photoactive compounds. This is consistent with several common multidentate motifs (i.e., five-membered rings) when considering d = 2 substructures. However, d = 2 substructures cannot capture bidentate ligands that form six-membered metallacycles. Therefore, to avoid introduction of less relevant complexes in each dataset, we identify matching metal-centered substructures in the tmQM dataset with d = 3. Unsurprisingly, increasing the radius of metal-centered substructures leads to an increase in the number of recurring substructures, with, e.g., 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 696 unique d = 3 motifs (vs. 10
696 unique d = 3 motifs (vs. 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 094 for d = 2) in the tmCAT dataset (ESI Fig. S22†). To only introduce additional structures with high relevance to a given application, only motifs with high recurrence (i.e., 5 or more for tmCAT, 3 or more for the other subsets) were supplemented. Using structural mapping, we augmented the tmCAT, tmPHOTO, tmSCO, and tmBIO datasets with 2381, 1556, 149, and 974 additional chemically relevant complexes, respectively. The final, application-specific datasets we curated can be accessed on Zenodo.79
094 for d = 2) in the tmCAT dataset (ESI Fig. S22†). To only introduce additional structures with high relevance to a given application, only motifs with high recurrence (i.e., 5 or more for tmCAT, 3 or more for the other subsets) were supplemented. Using structural mapping, we augmented the tmCAT, tmPHOTO, tmSCO, and tmBIO datasets with 2381, 1556, 149, and 974 additional chemically relevant complexes, respectively. The final, application-specific datasets we curated can be accessed on Zenodo.79
4. Conclusions
In summary, we employed natural language processing techniques, both supervised and unsupervised, to link experimentally synthesized compounds in the large and diverse tmQM database, which consists of 86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 665 TMCs, to their respective applications. Using the manuscript abstracts, we first trained a classifier model to identify manuscripts that are related to catalysis with an accuracy of 0.97. Using this model, we curated a dataset of catalysis-related transition metal complexes, called tmCAT, which initially consisted of 19
665 TMCs, to their respective applications. Using the manuscript abstracts, we first trained a classifier model to identify manuscripts that are related to catalysis with an accuracy of 0.97. Using this model, we curated a dataset of catalysis-related transition metal complexes, called tmCAT, which initially consisted of 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 unique complexes. Analysis of common electronic and geometric descriptors revealed that commonly used descriptors fail to distinguish between catalytic and non-catalytic sets. However, the analysis of coordination geometry of catalytically relevant complexes showed that geometries with open metal sites were significantly enhanced in the tmCAT set.
250 unique complexes. Analysis of common electronic and geometric descriptors revealed that commonly used descriptors fail to distinguish between catalytic and non-catalytic sets. However, the analysis of coordination geometry of catalytically relevant complexes showed that geometries with open metal sites were significantly enhanced in the tmCAT set.
Using topic modeling, an unsupervised clustering method often used in natural language processing, we further curated three additional initial datasets: tmPHOTO, a dataset consisting of 3043 unique complexes with photophysical relevance, tmBIO, a dataset consisting of 1808 unique complexes with biological relevance, and tmSCO, a dataset consisting of 834 unique complexes with relevance to magnetism. Analyzing the chemical substructures within each dataset identified frequently targeted complexes for their designated applications, such as bidentate N⁁N palladium dichlorides for catalysis, iridium complexes with two C⁁N ligands or platinum complexes with one C⁁N ligand for photophysics, and platinum dichlorides or ruthenium piano-stool complexes for biologically relevant complexes. By mapping these substructures to their applications, we identified previously synthesized complexes that had strong chemical similarity to those already identified for each application. We used these additional complexes to supplement the textually curated datasets, leading to 2381 additional tmCAT complexes, 1556 additional tmPHOTO complexes, 974 additional tmBIO complexes, and 149 additional tmSCO complexes in the final data sets.
The curated tmCAT, tmPHOTO, tmBIO, and tmSCO datasets are expected to enable more focused high-throughput computational screening and development of predictive machine learning models while still allowing for exploration across diverse chemical spaces. The language models employed in this study also have the potential for broader application, such as to curate subsets of other classes of materials, including metal–organic frameworks.
Data availability
Data for this article, including the analysis of coordination geometries and substructure analysis, the corpus curated for identifying catalysts, the resulting datasets of all complexes, and the machine learning models for the curation are available in a Zenodo repository at https://zenodo.org/records/11404217. The data supporting this article, including relevant figures and tables, have also been included as part of the supplementary information.†. The supplementary information comprises: validation of abstracts and titles obtained from HTML; all keywords used to screen for catalysis manuscripts in the title; all keywords used to screen for catalysis manuscripts in the abstract and title; additional stop words included in the secondary TF-IDF vectorizer; receiver operating curve of the secondary catalysis classifier; the performance metrics of the secondary catalysis classifier; distribution of the predicted probabilities of the manuscript related to catalysis in the unlabeled set in the corpus; distribution of the predicted probabilities of the manuscript related to catalysis in the HTML-mined corpus; density distribution of HOMO orbital energies; density distribution of LUMO orbital energies; density distribution of HOMO–LUMO gap energies; density distribution of the metal charge; density distribution of buried volume of metal; the five largest clusters of the BERTopic model using text without the introduction of stop words; wordclouds of two additional non-catalysis topics; wordclouds of five additional catalysis-related topics; t-SNE embedding of SBERT embedding vectors; topic breakdown using latent Dirichlet allocation; UMAP embedding of count vectorizer feature vector; frequency of molecular subgraph recurrence in the tmCAT dataset; frequency of the five most common transition metals in tmPHOTO, tmBIO and tmSCO datasets; frequency of molecular subgraph recurrence in the tmPHOTO dataset; representative substructures of the tmPHOTO dataset; frequency of molecular subgraph recurrence in the tmSCO dataset; representative substructures of the tmSCO dataset; frequency of molecular subgraph recurrence in the tmBIO dataset; representative substructures of tmBIO dataset; frequency of d = 3 molecular subgraph recurrence in the tmCAT dataset.Author contributions
Conceptualization – I. K. and H. J. K.; software – I. K., A. G. G., and R. G. S. M.; formal analysis – I. K., A. G. G., R. G. S. M., H. A., H. J., J. W. T.; data curation – I. K. and H. A.; writing – original draft – I. K., H. A., and J. W. T.; writing – review and editing – I. K., A. G. G., R. G. S. M., H. A., H. J., J. W. T., and H. J. K.; supervision – H. J. K.; funding acquisition – H. J. K. and Y. R.-L.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was primarily supported by the National Science Foundation under grant number CBET-1846426. The authors also acknowledge funding from the Office of Naval Research (N00014-20-1-2150). Y. R.-L. acknowledges support from the US Department of Energy, Office of Basic Energy Sciences, under award no. DE-SC0016214. H. J. K. holds an Alfred P. Sloan Fellowship in Chemistry and a Simon Family Faculty Research Innovation Fund, which supported this work. The authors thank Adam H. Steeves for providing a critical reading of the manuscript.References
- N. Fey, Lost in Chemical Space? Maps to Support Organometallic Catalysis, Chem. Cent. J., 2015, 9, 38 CrossRef PubMed.
- J. Jover and N. Fey, The Computational Road to Better Catalysts, Chem.–Asian J., 2014, 9, 1714–1723 CrossRef CAS PubMed.
- M. Swart and M. Gruden, Spinning around in Transition-Metal Chemistry, Acc. Chem. Res., 2016, 49, 2690–2697 CrossRef CAS PubMed.
- S. W. Krska, D. A. DiRocco, S. D. Dreher and M. Shevlin, The Evolution of Chemical High-Throughput Experimentation to Address Challenging Problems in Pharmaceutical Synthesis, Acc. Chem. Res., 2017, 50, 2976–2985 CrossRef CAS PubMed.
- B. S. Takale, F.-Y. Kong and R. R. Thakore, Recent Applications of Pd-Catalyzed Suzuki–Miyaura and Buchwald–Hartwig Couplings in Pharmaceutical Process Chemistry, Organics, 2022, 3, 1–21 CrossRef CAS.
- C. S. Allardyce, A. Dorcier, C. Scolaro and P. J. Dyson, Development of Organometallic (Organo-Transition Metal) Pharmaceuticals, Appl. Organomet. Chem., 2005, 19, 1–10 CrossRef CAS.
- A. Nandy, C. Duan, M. G. Taylor, F. Liu, A. H. Steeves and H. J. Kulik, Computational Discovery of Transition-Metal Complexes: From High-Throughput Screening to Machine Learning, Chem. Rev., 2021, 121, 9927–10000 CrossRef CAS PubMed.
- C. A. Tolman, Steric Effects of Phosphorus Ligands in Organometallic Chemistry and Homogeneous Catalysis, Chem. Rev., 1977, 77, 313–348 CrossRef CAS.
- N. Fey and J. M. Lynam, Computational Mechanistic Study in Organometallic Catalysis: Why Prediction Is Still a Challenge, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2022, 12, e1590 CAS.
- J.-L. Reymond, L. Ruddigkeit, L. Blum and R. van Deursen, The Enumeration of Chemical Space, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2012, 2, 717–733 CAS.
- J. P. Janet, S. Ramesh, C. Duan and H. J. Kulik, Accurate Multiobjective Design in a Space of Millions of Transition Metal Complexes with Neural-Network-Driven Efficient Global Optimization, ACS Cent. Sci., 2020, 6, 513–524 CrossRef CAS PubMed.
- J. A. Hageman, J. A. Westerhuis, H. W. Frühauf and G. Rothenberg, Design and Assembly of Virtual Homogeneous Catalyst Libraries – Towards in Silico Catalyst Optimisation, Adv. Synth. Catal., 2006, 348, 361–369 CrossRef CAS.
- A. Nandy, M. G. Taylor and H. J. Kulik, Identifying Underexplored and Untapped Regions in the Chemical Space of Transition Metal Complexes, J. Phys. Chem. Lett., 2023, 14, 5798–5804 CrossRef CAS PubMed.
- S. DiLuzio, V. Mdluli, T. U. Connell, J. Lewis, V. VanBenschoten and S. Bernhard, High-Throughput Screening and Automated Data-Driven Analysis of the Triplet Photophysical Properties of Structurally Diverse, Heteroleptic Iridium(III) Complexes, J. Am. Chem. Soc., 2021, 143, 1179–1194 CrossRef CAS PubMed.
- R. N. Motz, E. M. Lopato, T. U. Connell and S. Bernhard, High-Throughput Screening of Earth-Abundant Water Reduction Catalysts toward Photocatalytic Hydrogen Evolution, Inorg. Chem., 2021, 60, 774–781 CrossRef CAS PubMed.
- D. Balcells and B. B. Skjelstad, tmQM Dataset-Quantum Geometries and Properties of 86k Transition Metal Complexes, J. Chem. Inf. Model., 2020, 60, 6135–6146 CrossRef CAS PubMed.
- S. S. Chen, Z. Meyer, B. Jensen, A. Kraus, A. Lambert and D. H. Ess, Realigands: A Ligand Library Cultivated from Experiment and Intended for Molecular Computational Catalyst Design, J. Chem. Inf. Model., 2023, 63, 7412–7422 CrossRef CAS PubMed.
- I. Kevlishvili, C. Duan and H. J. Kulik, Classification of Hemilabile Ligands Using Machine Learning, J. Phys. Chem. Lett., 2023, 14, 11100–11109 CrossRef CAS PubMed.
- N. Fey, A. C. Tsipis, S. E. Harris, J. N. Harvey, A. G. Orpen and R. A. Mansson, Development of a Ligand Knowledge Base, Part 1: Computational Descriptors for Phosphorus Donor Ligands, Chem.–Eur. J., 2006, 12, 291–302 CrossRef CAS PubMed.
- R. A. Mansson, A. H. Welsh, N. Fey and A. G. Orpen, Statistical Modeling of a Ligand Knowledge Base, J. Chem. Inf. Model., 2006, 46, 2591–2600 CrossRef CAS PubMed.
- J. Jover, N. Fey, J. N. Harvey, G. C. Lloyd-Jones, A. G. Orpen, G. J. J. Owen-Smith, P. Murray, D. R. J. Hose, R. Osborne and M. Purdie, Expansion of the Ligand Knowledge Base for Monodentate P-Donor Ligands (LKB-P), Organometallics, 2010, 29, 6245–6258 CrossRef CAS.
- J. Jover, N. Fey, J. N. Harvey, G. C. Lloyd-Jones, A. G. Orpen, G. J. Owen-Smith, P. Murray, D. R. Hose, R. Osborne and M. Purdie, Expansion of the Ligand Knowledge Base for Chelating P,P-Donor Ligands (LKB-PP), Organometallics, 2012, 31, 5302–5306 CrossRef CAS PubMed.
- T. Gensch, G. Dos Passos Gomes, P. Friederich, E. Peters, T. Gaudin, R. Pollice, K. Jorner, A. Nigam, M. Lindner-D’Addario, M. S. Sigman and A. Aspuru-Guzik, A Comprehensive Discovery Platform for Organophosphorus Ligands for Catalysis, J. Am. Chem. Soc., 2022, 144, 1205–1217 CrossRef CAS PubMed.
- S. M. Kearnes, M. R. Maser, M. Wleklinski, A. Kast, A. G. Doyle, S. D. Dreher, J. M. Hawkins, K. F. Jensen and C. W. Coley, The Open Reaction Database, J. Am. Chem. Soc., 2021, 143, 18820–18826 CrossRef CAS PubMed.
- D. T. Ahneman, J. G. Estrada, S. Lin, S. D. Dreher and A. G. Doyle, Predicting Reaction Performance in C-N Cross-Coupling Using Machine Learning, Science, 2018, 360, 186–190 CrossRef CAS PubMed.
- M. Fitzner, G. Wuitschik, R. J. Koller, J. M. Adam, T. Schindler and J. L. Reymond, What Can Reaction Databases Teach Us About Buchwald-Hartwig Cross-Couplings?, Chem. Sci., 2020, 11, 13085–13093 RSC.
- D. Perera, J. W. Tucker, S. Brahmbhatt, C. J. Helal, A. Chong, W. Farrell, P. Richardson and N. W. Sach, A Platform for Automated Nanomole-Scale Reaction Screening and Micromole-Scale Synthesis in Flow, Science, 2018, 359, 429–434 CrossRef CAS PubMed.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, The Cambridge Structural Database, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 171–179 CrossRef CAS PubMed.
- G. Montavon, M. Rupp, V. Gobre, A. Vazquez-Mayagoita, K. Hansen, A. Tkatchenko, K.-R. Müller and O. A. von Lilienfeld, Machine Learning of Molecular Electronic Properties in Chemical Compound Space, New J. Phys., 2013, 15, 095003 CrossRef CAS.
- A. M. Virshup, J. Contreras-Garcia, P. Wipf, W. Yang and D. N. Beratan, Stochastic Voyages into Uncharted Chemical Space Produce a Representative Library of All Possible Drug-Like Compounds, J. Am. Chem. Soc., 2013, 135, 7296–7303 CrossRef CAS PubMed.
- R. Ramakrishnan, P. O. Dral, M. Rupp and O. A. von Lilienfeld, Quantum Chemistry Structures and Properties of 134 Kilo Molecules, Sci. Data, 2014, 1, 140022 CrossRef CAS PubMed.
- M. Rupp, A. Tkatchenko, K. R. Muller and O. A. von Lilienfeld, Fast and Accurate Modeling of Molecular Atomization Energies with Machine Learning, Phys. Rev. Lett., 2012, 108, 058301 CrossRef PubMed.
- J.-L. Reymond, R. van Deursen, L. C. Blum and L. Ruddigkeit, Chemical Space as a Source for New Drugs, Med. Chem. Commun., 2010, 1, 30–38 RSC.
- R. Ramakrishnan, M. Hartmann, E. Tapavicza and O. A. von Lilienfeld, Electronic Spectra from TDDFT and Machine Learning in Chemical Space, J. Chem. Phys., 2015, 143, 084111 CrossRef PubMed.
- L. Ruddigkeit, R. van Deursen, L. C. Blum and J. L. Reymond, Enumeration of 166 Billion Organic Small Molecules in the Chemical Universe Database GDB-17, J. Chem. Inf. Model., 2012, 52, 2864–2875 CrossRef CAS PubMed.
- H. Kneiding, A. Nova and D. Balcells, Directional Multiobjective Optimization of Metal Complexes at the Billion-Scale with the tmQMg-L Dataset and PL-MOGA Algorithm, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-k3tf2-v2.
- P. Schwaller, A. C. Vaucher, T. Laino and J.-L. Reymond, Prediction of Chemical Reaction Yields Using Deep Learning, Mach. Learn.: Sci. Technol., 2021, 2, 015016 Search PubMed.
- P. Schwaller, A. C. Vaucher, T. Laino and J.-L. Reymond, Data Augmentation Strategies to Improve Reaction Yield Predictions and Estimate Uncertainty, NeurIPS, 2020, DOI:10.26434/chemrxiv.13286741.v1.
- H. Jin and K. M. Merz Jr, Modeling Zinc Complexes Using Neural Networks, J. Chem. Inf. Model., 2024, 64, 3140–3148 CrossRef CAS PubMed.
- A. G. Garrison, J. Heras-Domingo, J. R. Kitchin, G. Dos Passos Gomes, Z. W. Ulissi and S. M. Blau, Applying Large Graph Neural Networks to Predict Transition Metal Complex Energies Using the tmQM_wB97MV Data Set, J. Chem. Inf. Model., 2023, 63, 7642–7654 CrossRef CAS PubMed.
- T. Gensch, S. R. Smith, T. J. Colacot, Y. N. Timsina, G. Xu, B. W. Glasspoole and M. S. Sigman, Design and Application of a Screening Set for Monophosphine Ligands in Cross-Coupling, ACS Catal., 2022, 12, 7773–7780 CrossRef CAS.
- K. R. Chowdhary, in Fundamentals of Artificial Intelligence, ed. K. R. Chowdhary, Springer India, New Delhi, 2020, pp. 603–649, DOI:10.1007/978-81-322-3972-7_19.
- M. Krallinger, O. Rabal, A. Lourenço, J. Oyarzabal and A. Valencia, Information Retrieval and Text Mining Technologies for Chemistry, Chem. Rev., 2017, 117, 7673–7761 CrossRef CAS PubMed.
- E. Kim, K. Huang, A. Saunders, A. McCallum, G. Ceder and E. Olivetti, Materials Synthesis Insights from Scientific Literature via Text Extraction and Machine Learning, Chem. Mater., 2017, 29, 9436–9444 CrossRef CAS.
- E. Kim, K. Huang, S. Jegelka and E. Olivetti, Virtual Screening of Inorganic Materials Synthesis Parameters with Deep Learning, npj Comput. Mater., 2017, 3, 53 CrossRef.
- M. C. Swain and J. M. Cole, ChemDataExtractor: A Toolkit for Automated Extraction of Chemical Information from the Scientific Literature, J. Chem. Inf. Model., 2016, 56, 1894–1904 CrossRef CAS PubMed.
- J. Mavracic, C. J. Court, T. Isazawa, S. R. Elliott and J. M. Cole, ChemDataExtractor 2.0: Autopopulated Ontologies for Materials Science, J. Chem. Inf. Model., 2021, 61, 4280–4289 CrossRef CAS PubMed.
- S. Park, B. Kim, S. Choi, P. G. Boyd, B. Smit and J. Kim, Text Mining Metal–Organic Framework Papers, J. Chem. Inf. Model., 2018, 58, 244–251 CrossRef CAS PubMed.
- Z. Jensen, E. Kim, S. Kwon, T. Z. H. Gani, Y. Román-Leshkov, M. Moliner, A. Corma and E. Olivetti, A Machine Learning Approach to Zeolite Synthesis Enabled by Automatic Literature Data Extraction, ACS Cent. Sci., 2019, 5, 892–899 CrossRef CAS PubMed.
- E. Kim, Z. Jensen, A. van Grootel, K. Huang, M. Staib, S. Mysore, H.-S. Chang, E. Strubell, A. McCallum, S. Jegelka and E. Olivetti, Inorganic Materials Synthesis Planning with Literature-Trained Neural Networks, J. Chem. Inf. Model., 2020, 60, 1194–1201 CrossRef CAS PubMed.
- A. Nandy, C. Duan and H. J. Kulik, Using Machine Learning and Data Mining to Leverage Community Knowledge for the Engineering of Stable Metal–Organic Frameworks, J. Am. Chem. Soc., 2021, 143, 17535–17547 CrossRef CAS PubMed.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, ChatGPT Chemistry Assistant for Text Mining and the Prediction of MOF Synthesis, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef CAS PubMed.
- K. M. Jablonka, P. Schwaller, A. Ortega-Guerrero and B. Smit, Leveraging Large Language Models for Predictive Chemistry, Nat. Mach. Intell., 2024, 6, 161–169 CrossRef.
- M. P. Polak and D. Morgan, Extracting Accurate Materials Data from Research Papers with Conversational Language Models and Prompt Engineering, Nat. Commun., 2024, 15, 1569 CrossRef CAS PubMed.
- S. Liu, T. Wen, A. Pattamatta and D. J. Srolovitz, A Prompt-Engineered Large Language Model, Deep Learning Workflow for Materials Classification, arXiv, 2024, preprint, arXiv:2401.17788, DOI:10.48550/arXiv.2401.17788.
- K. Hatakeyama-Sato, N. Yamane, Y. Igarashi, Y. Nabae and T. Hayakawa, Prompt Engineering of GPT-4 for Chemical Research: What Can/Cannot Be Done?, Sci. Technol. Adv. Mater.: Methods, 2023, 3, 2260300 Search PubMed.
- I. Vayansky and S. A. P. Kumar, A Review of Topic Modeling Methods, Inf. Syst., 2020, 94, 101582 CrossRef.
- E. H. Huang, R. Socher, C. D. Manning and A. Y. Ng, Proceedings of the 50th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Jeju, Republic of Korea, 2012, pp. 873–882 Search PubMed.
- T. Mikolov, K. Chen, G. Corrado and J. Dean, Efficient Estimation of Word Representations in Vector Space, arXiv, 2013, preprint, arXiv:1301.3781, DOI:10.48550/arXiv.1301.3781.
- T. Mikolov, I. Sutskever, K. Chen, G. S. Corrado and J. Dean, Distributed Representations of Words and Phrases and Their Compositionality, NeurIPS, 2013, vol. 26 Search PubMed.
- M. Kusner, Y. Sun, N. Kolkin and K. Weinberger, in Proceedings of the 32nd International Conference on Machine Learning, ed. F. Bach, and D. Blei, PMLR: Proceedings of Machine Learning Research, 2015, vol. 37 Search PubMed.
- M. Grootendorst, BERTopic: Neural Topic Modeling with a Class-Based TF-IDF Procedure, arXiv, 2022, preprint, arXiv:2203.05794, DOI:10.48550/arXiv.2203.05794.
- D. M. Blei, A. Y. Ng and M. I. Jordan, Latent Dirichlet Allocation, J. Mach. Learn. Res., 2003, 3, 993–1022 Search PubMed.
- S. Huang and J. M. Cole, ChemDataWriter: A Transformer-Based Toolkit for Auto-Generating Books That Summarise Research, Digital Discovery, 2023, 2, 1710–1720 RSC.
- H. Jelodar, Y. Wang, C. Yuan, X. Feng, X. Jiang, Y. Li and L. Zhao, Latent Dirichlet Allocation (LDA) and Topic Modeling: Models, Applications, a Survey, Multimed. Tools Appl., 2019, 78, 15169–15211 CrossRef.
- H. J. Kang, C. Kim and K. Kang, Analysis of the Trends in Biochemical Research Using Latent Dirichlet Allocation (LDA), Processes, 2019, 7, 379 CrossRef CAS.
- B. X. Tran, C. A. Latkin, N. Sharafeldin, K. Nguyen, G. T. Vu, W. W. S. Tam, N.-M. Cheung, H. L. T. Nguyen, C. S. H. Ho and R. C. M. Ho, Characterizing Artificial Intelligence Applications in Cancer Research: A Latent Dirichlet Allocation Analysis, JMIR Med. Inform., 2019, 7, e14401 CrossRef PubMed.
- M. Karabacak, P. Jagtiani, A. Jain, F. Panov and K. Margetis, Tracing Topics and Trends in Drug-Resistant Epilepsy Research Using a Natural Language Processing-Based Topic Modeling Approach, Epilepsia, 2024, 65, 861–872 CrossRef PubMed.
- N. Schneider, N. Fechner, G. A. Landrum and N. Stiefl, Chemical Topic Modeling: Exploring Molecular Data Sets Using a Common Text-Mining Approach, J. Chem. Inf. Model., 2017, 57, 1816–1831 CrossRef CAS PubMed.
- A. Nandy, M. G. Taylor and H. J. Kulik, Identifying Underexplored and Untapped Regions in the Chemical Space of Transition Metal Complexes, J. Phys. Chem. Lett., 2023, 14, 5798–5804 CrossRef CAS PubMed.
- M. G. Taylor, T. Yang, S. Lin, A. Nandy, J. P. Janet, C. Duan and H. J. Kulik, Seeing Is Believing: Experimental Spin States from Machine Learning Model Structure Predictions, J. Phys. Chem. A, 2020, 124, 3286–3299 CrossRef CAS PubMed.
- E. Kim, K. Huang, S. Jegelka and E. Olivetti, Virtual Screening of Inorganic Materials Synthesis Parameters with Deep Learning, npj Comput. Mater., 2017, 3, 53 CrossRef.
- L. Richardson, Beautiful Soup Documentation, 2007 Search PubMed.
- S. Bird, E. Klein and E. Loper, Natural Language Processing with Python: Analyzing Text with the Natural Language Toolkit, O’Reilly Media, Inc., 2009 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss and V. Dubourg, Scikit-Learn: Machine Learning in Python, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- N. Reimers and I. Gurevych, Sentence-BERT: Sentence Embeddings Using Siamese BERT-Networks, arXiv, 2019, preprint, arXiv:1908.10084, DOI:10.48550/arXiv.1908.10084.
- L. McInnes, J. Healy and J. Melville, UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction, arXiv, 2018, preprint, arXiv:1802.03426, DOI:10.48550/arXiv.1802.03426.
- D. M. Blei, A. Y. Ng and M. I. Jordan, Latent Dirichlet Allocation, J. Mach. Learn. Res., 2003, 3, 993–1022 Search PubMed.
- Zenodo Repository for Leveraging Natural Language Processing to Curate the tmCAT, tmPHOTO, tmBIO, and tmSCO Datasets of Functional Transition Metal Complexes, https://zenodo.org/records/11404217, accessed May 31 2024 Search PubMed.
- R. J. G. B. Campello, D. Moulavi and J. Sander, Advances in Knowledge Discovery and Data Mining, Berlin, Heidelberg, 2013, pp. 160–172 Search PubMed.
- D. Balcells and B. B. Skjelstad, tmQM Dataset—Quantum Geometries and Properties of 86k Transition Metal Complexes, J. Chem. Inf. Model., 2020, 60, 6135–6146 CrossRef CAS PubMed.
- M. Lan, C.-L. Tan, H.-B. Low and S.-Y. Sung, in Special Interest Tracks and Posters of the 14th International Conference on World Wide Web, Association for Computing Machinery, Chiba, Japan, 2005, DOI:10.1145/1062745.1062854.
- P. M. Zeimentz, S. Arndt, B. R. Elvidge and J. Okuda, Cationic Organometallic Complexes of Scandium, Yttrium, and the Lanthanoids, Chem. Rev., 2006, 106, 2404–2433 CrossRef CAS PubMed.
- X. Wang, J. L. Brosmer, A. Thevenon and P. L. Diaconescu, Highly Active Yttrium Catalysts for the Ring-Opening Polymerization of ε-Caprolactone and δ-Valerolactone, Organometallics, 2015, 34, 4700–4706 CrossRef CAS.
- A. Biffis, P. Centomo, A. Del Zotto and M. Zecca, Pd Metal Catalysts for Cross-Couplings and Related Reactions in the 21st Century: A Critical Review, Chem. Rev., 2018, 118, 2249–2295 CrossRef CAS PubMed.
- S. P. Nolan and H. Clavier, Chemoselective Olefin Metathesis Transformations Mediated by Ruthenium Complexes, Chem. Soc. Rev., 2010, 39, 3305–3316 RSC.
- M. Iglesias and L. A. Oro, A Leap Forward in Iridium–NHC Catalysis: New Horizons and Mechanistic Insights, Chem. Soc. Rev., 2018, 47, 2772–2808 RSC.
- W. Zi and T. F. Dean, Recent Advances in Enantioselective Gold Catalysis, Chem. Soc. Rev., 2016, 45, 4567–4589 RSC.
- J. Chen and Z. Lu, Asymmetric Hydrofunctionalization of Minimally Functionalized Alkenes via Earth Abundant Transition Metal Catalysis, Org. Chem. Front., 2018, 5, 260–272 RSC.
- P. Liao, R. B. Getman and R. Q. Snurr, Optimizing Open Iron Sites in Metal–Organic Frameworks for Ethane Oxidation: A First-Principles Study, ACS Appl. Mater. Interfaces, 2017, 9, 33484–33492 CrossRef CAS PubMed.
- P. C. Andrikopoulos, C. Michel, S. Chouzier and P. Sautet, In Silico Screening of Iron-Oxo Catalysts for CH Bond Cleavage, ACS Catal., 2015, 5, 2490–2499 CrossRef CAS.
- A. Nandy, J. Zhu, J. P. Janet, C. Duan, R. B. Getman and H. J. Kulik, Machine Learning Accelerates the Discovery of Design Rules and Exceptions in Stable Metal–Oxo Intermediate Formation, ACS Catal., 2019, 9, 8243–8255 CrossRef CAS.
- K. Wu and A. G. Doyle, Parameterization of Phosphine Ligands Demonstrates Enhancement of Nickel Catalysis via Remote Steric Effects, Nat. Chem., 2017, 9, 779–784 CrossRef CAS PubMed.
- I. Vayansky and S. A. P. Kumar, A Review of Topic Modeling Methods, Inf. Syst., 2020, 94, 101582 CrossRef.
- L. Van der Maaten and G. Hinton, Visualizing Data Using t-SNE, J. Mach. Learn. Res., 2008, 9, 2579–2605 Search PubMed.
- N. Marion and S. P. Nolan, N-Heterocyclic Carbenes in Gold Catalysis, Chem. Soc. Rev., 2008, 37, 1776–1782 RSC.
- S. P. Nolan, The Development and Catalytic Uses of N-Heterocyclic Carbene Gold Complexes, Acc. Chem. Res., 2011, 44, 91–100 CrossRef CAS PubMed.
- E. I. Ioannidis, T. Z. H. Gani and H. J. Kulik, molSimplify: A Toolkit for Automating Discovery in Inorganic Chemistry, J. Comput. Chem., 2016, 37, 2106–2117 CrossRef CAS PubMed.
- N. Hazari, P. R. Melvin and M. M. Beromi, Well-Defined Nickel and Palladium Precatalysts for Cross-Coupling, Nat. Rev. Chem., 2017, 1, 0025 CrossRef CAS PubMed.
- D. Campeau, D. F. León Rayo, A. Mansour, K. Muratov and F. Gagosz, Gold-Catalyzed Reactions of Specially Activated Alkynes, Allenes, and Alkenes, Chem. Rev., 2021, 121, 8756–8867 CrossRef CAS PubMed.
- S. Gessler, S. Randl and S. Blechert, Synthesis and Metathesis Reactions of a Phosphine-Free Dihydroimidazole Carbene Ruthenium Complex, Tetrahedron Lett., 2000, 41, 9973–9976 CrossRef CAS.
- G. C. Vougioukalakis and R. H. Grubbs, Ruthenium-Based Heterocyclic Carbene-Coordinated Olefin Metathesis Catalysts, Chem. Rev., 2010, 110, 1746–1787 CrossRef CAS PubMed.
- Z. Bie, B.-G. Li, J. Hu and Z. Yao, Studies on Alternating Copolymerization of Ethylene and Carbon Monoxide Using Nickel-Based Catalyst: Cocatalyst and the Polarity of Solvent, Macromol. React. Eng., 2022, 16, 2100047 CrossRef CAS.
- V. C. Gibson, C. Redshaw and G. A. Solan, Bis(Imino)Pyridines: Surprisingly Reactive Ligands and a Gateway to New Families of Catalysts, Chem. Rev., 2007, 107, 1745–1776 CrossRef CAS PubMed.
- C. W. Bielawski and R. H. Grubbs, Living Ring-Opening Metathesis Polymerization, Prog. Polym. Sci., 2007, 32, 1–29 CrossRef CAS.
- C. Yang, F. Mehmood, T. L. Lam, S. L.-F. Chan, Y. Wu, C.-S. Yeung, X. Guan, K. Li, C. Y.-S. Chung, C.-Y. Zhou, T. Zou and C.-M. Che, Stable Luminescent Iridium(III) Complexes with Bis(N-Heterocyclic Carbene) Ligands: Photo-Stability, Excited State Properties, Visible-Light-Driven Radical Cyclization and CO2 Reduction, and Cellular Imaging, Chem. Sci., 2016, 7, 3123–3136 RSC.
- J. Kalinowski, V. Fattori, M. Cocchi and J. A. G. Williams, Light-Emitting Devices Based on Organometallic Platinum Complexes as Emitters, Coord. Chem. Rev., 2011, 255, 2401–2425 CrossRef CAS.
- M. S. Lazorski and F. N. Castellano, Advances in the Light Conversion Properties of Cu(I)-Based Photosensitizers, Polyhedron, 2014, 82, 57–70 CrossRef CAS.
- A. W. Prestayko, J. C. D’Aoust, B. F. Issell and S. T. Crooke, Cisplatin (cis-Diamminedichloroplatinum II), Cancer Treat. Rev., 1979, 6, 17–39 CrossRef CAS PubMed.
- M. Kartalou and J. M. Essigmann, Mechanisms of Resistance to Cisplatin, Mutat. Res., 2001, 478, 23–43 CrossRef CAS PubMed.
- R. B. Weiss and M. C. Christian, New Cisplatin Analogues in Development, Drugs, 1993, 46, 360–377 CrossRef CAS PubMed.
- B. S. Murray, M. V. Babak, C. G. Hartinger and P. J. Dyson, The Development of RAPTA Compounds for the Treatment of Tumors, Coord. Chem. Rev., 2016, 306, 86–114 CrossRef CAS.
- S. Swaminathan, J. Haribabu, N. Balakrishnan, P. Vasanthakumar and R. Karvembu, Piano Stool Ru(II)-Arene Complexes Having Three Monodentate Legs: A Comprehensive Review on Their Development as Anticancer Therapeutics over the Past Decade, Coord. Chem. Rev., 2022, 459, 214403 CrossRef CAS.
- B. de Castro, R. Ferreira, C. Freire, H. García, E. J. Palomares and M. J. Sabater, Photochemistry of Nickel Salen Based Complexes and Relevance to Catalysis, New J. Chem., 2002, 26, 405–410 RSC.
- K. Teegardin, J. I. Day, J. Chan and J. Weaver, Advances in Photocatalysis: A Microreview of Visible Light Mediated Ruthenium and Iridium Catalyzed Organic Transformations, Org. Process Res. Dev., 2016, 20, 1156–1163 CrossRef CAS PubMed.
- R. Bevernaegie, B. Doix, E. Bastien, A. Diman, A. Decottignies, O. Feron and B. Elias, Exploring the Phototoxicity of Hypoxic Active Iridium(III)-Based Sensitizers in 3D Tumor Spheroids, J. Am. Chem. Soc., 2019, 141, 18486–18491 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fd00087k |
| This journal is © The Royal Society of Chemistry 2024 |