Open Access Article
Open Access ArticleDevelopment of an in vitro platform for the analysis of contractile and calcium dynamics in single human myotubes†
Camila
Vesga-Castro
 ab,
Laura
Mosqueira-Martín
ab,
Laura
Mosqueira-Martín
 cde,
Paul
Ubiria-Urkola
ab,
Pablo
Marco-Moreno
cde,
Paul
Ubiria-Urkola
ab,
Pablo
Marco-Moreno
 cde,
Klaudia
González-Imaz
cde,
Jorge
Rendon-Hinestroza
cd,
Ainara
Vallejo-Illarramendi
cde,
Klaudia
González-Imaz
cde,
Jorge
Rendon-Hinestroza
cd,
Ainara
Vallejo-Illarramendi
 *cde and
Jacobo
Paredes
*ab
*cde and
Jacobo
Paredes
*ab
aTecnun School of Engineering, University of Navarra, Manuel de Lardizábal 13, 20018 San Sebastián, Spain. E-mail: jparedes@unav.es
bBiomedical Engineering Center, University of Navarra, Campus Universitario, 31080, Pamplona, Spain
cGroup of Neuroscience, Department of Pediatrics, Faculty of Medicine and Nursing, Donostia Hospital, UPV/EHU, 20014 San Sebastian, Spain. E-mail: ainara.vallejo@ehu.eus
dGroup of Neuromuscular Diseases, IIS Biogipuzkoa, 20014 San Sebastian, Spain
eCIBERNED, Ministry of Science and Innovation, Instituto de Salud Carlos III, 28031 Madrid, Spain
First published on 4th September 2024
Abstract
In vitro myotube cultures are widely used as models for studying muscle pathophysiology, but their limited maturation and heterogeneity pose significant challenges for functional analyses. While they remain the gold standard for studying muscle function in vitro, myotube cultures do not fully recapitulate the complexity and native features of muscle fibers, which may compromise their ability to predict in vivo outcomes. To promote maturation and decrease heterogeneity, we have incorporated engineered structures into myotube cultures, based on a PDMS thin layer with micrometer-sized grooves (μGrooves) placed over a glass substrate. Different sizes and shapes of μGrooves were tested for their ability to promote alignment and fusion of myoblasts and enhance their differentiation into myotubes. A 24 hour electrical field stimulation protocol (4 V, 6 ms, 0.1 Hz) was used to further promote myotube maturation, after which several myotube features were assessed, including myotube alignment, width, fusion index, contractile function, and calcium handling. Our results indicate superior calcium and contractile performance in μGrooved myotubes, particularly with the 100 μm-width 700 μm-long geometry (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). This platform generated homogeneous and isolated myotubes that reproduced native muscle features, such as excitation–contraction coupling and force-frequency responses. Overall, our 2D muscle platform enables robust high-content assays of calcium dynamics and contractile readouts with increased sensitivity and reproducibility compared to traditional myotube cultures, making it particularly suitable for screening therapeutic candidates for different muscle pathologies.
1). This platform generated homogeneous and isolated myotubes that reproduced native muscle features, such as excitation–contraction coupling and force-frequency responses. Overall, our 2D muscle platform enables robust high-content assays of calcium dynamics and contractile readouts with increased sensitivity and reproducibility compared to traditional myotube cultures, making it particularly suitable for screening therapeutic candidates for different muscle pathologies.
Introduction
Skeletal muscle is a highly complex and hierarchical structure composed of aligned and multinucleated muscle fibers.1 This particular organization enables the muscle to fulfill its main function, which is the generation of contractile force. Muscle performance may be compromised in some conditions, for instance, after major injuries, aging, or genetic diseases, such as muscular dystrophies.2–4 To develop new therapeutic approaches for these disorders, representative and reproducible in vitro models are required to perform drug screening and mechanistic studies.5–9 Yet, the generation of in vitro muscle models is particularly challenging since they must reproduce physiological muscle development and boost the maturation of the engineered tissues. These processes require an adequate substrate and fine control of the microenvironment, including culture media and physicochemical signals, in order to enable the generation and organization of contractile units.The evaluation of these in vitro models typically relies on molecular and cellular analyses, such as the evaluation of fusion index, striation pattern, acetylcholine receptor aggregation, and myogenic marker expression.10–13 Myotube cultures have provided valuable insights into key molecular processes, such as glucose uptake14 and calcium handling,15 and have been extensively used for muscle disease modeling.3,16,17 However, standard 2D myotube cultures exhibit lower maturation levels compared to native muscle.1,18 In addition, these cultures are heterogeneous in terms of morphology and maturation, leading to the high variability and low reproducibility observed in these models.19
To address these challenges, different in vitro platforms have been proposed to promote myoblast organization, fusion, and differentiation into aligned myotubes. These platforms usually incorporate geometrical cues such as micropatterning of proteins,10,20–22 electrospun fibers,23,24 modified surfaces,25–32 or substrates with tissue-like stiffness,14,18,33,34 which aim to decrease morphological variability and improve maturation and cell alignment. In addition, chronic stimulation has demonstrated a potentiating effect on maturation and contractile capacity.35–37 Despite all the efforts made so far, important challenges remain unresolved with respect to 2D muscle platforms. Among them, the high heterogeneity of myotube cultures is one of the main factors affecting data dispersion, reliability and reproducibility. Overcoming this challenge is necessary to advance in the development of new treatments for muscle diseases.
Calcium plays a fundamental role in muscle fiber function and is essential for muscle contraction. The excitation–contraction (E–C) coupling mechanism underlies muscle contraction and involves the activation of voltage-gated calcium channels (DHPR) upon depolarization of the muscle membrane. This triggers the release of calcium from the sarcoplasmic reticulum via RyR channels, leading to the generation of force through the interaction of contractile proteins.4 Measuring calcium dynamics in myotubes can provide valuable functional insight into pathogenic mechanisms of muscle diseases. Therefore, calcium handling is used as a main functional readout in 2D muscle models,8,25 typically reporting basal calcium levels and calcium transients induced by chemical or physical stimulation. Contractility is another relevant functional readout, which can be evaluated, for instance, by image motion analysis of myotubes during electrical stimulation38 or by evaluating substrate deformation.3,7,39 Both outcomes provide highly relevant physiological information about the functional performance of muscle in vitro systems that is complementary to gene and protein expression analyses. However, to the best of our knowledge, no studies have compared calcium handling and contractile performance of myotube cultures, so far.19 Moreover, there is very limited information regarding contractile readouts in current myotube models, which is likely due to low myotube maturation, and their tendency for detachment, especially under tetanic conditions.19
In this study, we aimed to develop a 2D human muscle system with reduced data variability and high-throughput potential for the assessment of functional outcomes. To this end, we designed a platform comprising isolated, aligned, homogeneous, and contractile human myotubes. Our platform consists of an 80 μm-thick PDMS layer with micrometer-sized grooves (μGrooves) placed onto a glass substrate. Confinement of myoblasts within the μGrooves homogenizes myoblast alignment, fusion, and differentiation. We tested different μGroove geometries for their ability to generate low scattered data sets. Our study provides a step-by-step description for the generation of a 2D in vitro human muscle platform enabling the assessment of relevant functional outcomes such as calcium dynamics and contractility. This platform has been extensively characterized and optimized and would be useful for high-throughput drug screening and disease modeling.
Experimental
Fabrication of μGrooved substrates
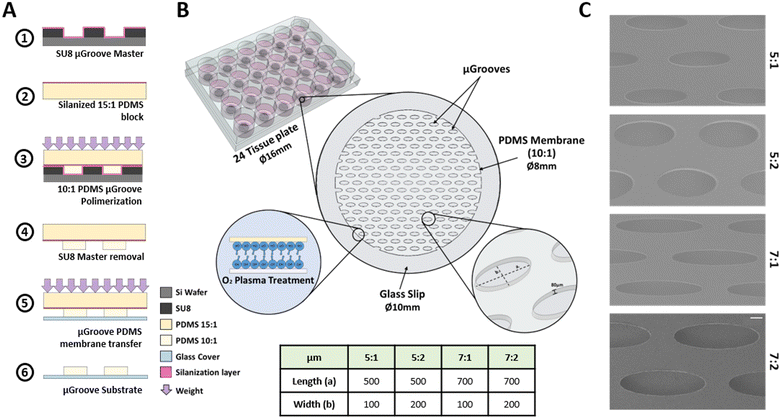
The culture platform consists of a glass slip substrate covered with a PDMS membrane containing the μGrooves features. This membrane was fabricated following an adapted method.40 The fabrication protocol, depicted in Fig. 1, consists of the following processes. Step 1: 80 μm height SU8 μGrooves masters were fabricated by photolithography (2025 MicroChem). Step 2: a 3 mm-thick 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio PDMS block was prepared to enable the membrane generation (poly(dimethylsiloxane) Sylgard 184 silicone, Dow Corning, 0002-04-000002). Both master and PDMS blocks were silanized overnight with trichloro(1H,1H,2H,2H-perfluorooctyl)silane (448931-10G, Sigma). Step 3: then, PDMS (10
1 ratio PDMS block was prepared to enable the membrane generation (poly(dimethylsiloxane) Sylgard 184 silicone, Dow Corning, 0002-04-000002). Both master and PDMS blocks were silanized overnight with trichloro(1H,1H,2H,2H-perfluorooctyl)silane (448931-10G, Sigma). Step 3: then, PDMS (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) was poured onto the silanized PDMS and immediately put in contact with the SU-8 master. 600 g weights were used to ensure complete and homogeneous contact between the master and the PDMS block, and left overnight at 60 °C until polymerization was completed. Step 4: masters were then removed and the PDMS membrane together with glass coverslips were treated with oxygen plasma. Step 5: both treated surfaces were put together facilitating the transfer of the PDMS membrane onto the glass coverslips and removing the PDMS block. The coverslips with the PDMS membrane were kept overnight at 60 °C to enhance the bonding. Step 6: last, the PDMS block was carefully separated leaving the final culturing platform as illustrated in Fig. 1B. μGrooves substrates were stored at room temperature (RT) in sealed 24 well-plates.
1 ratio) was poured onto the silanized PDMS and immediately put in contact with the SU-8 master. 600 g weights were used to ensure complete and homogeneous contact between the master and the PDMS block, and left overnight at 60 °C until polymerization was completed. Step 4: masters were then removed and the PDMS membrane together with glass coverslips were treated with oxygen plasma. Step 5: both treated surfaces were put together facilitating the transfer of the PDMS membrane onto the glass coverslips and removing the PDMS block. The coverslips with the PDMS membrane were kept overnight at 60 °C to enhance the bonding. Step 6: last, the PDMS block was carefully separated leaving the final culturing platform as illustrated in Fig. 1B. μGrooves substrates were stored at room temperature (RT) in sealed 24 well-plates.
Four different elliptical geometries of μGrooves with different length-to-width ratios were fabricated, namely: 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (500 μm/100 μm), 5
1 (500 μm/100 μm), 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (500 μm/200 μm), 7
2 (500 μm/200 μm), 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (700 μm/100 μm), and 7
1 (700 μm/100 μm), and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (700 μm/200 μm). Characterization of SU8 masters (height and dimensions) were performed with a profilometer (KLA-Tencor, P6 Model, and Smart WLI, gbs) using the Profiler 7.31 software. μGrooved PDMS replicas were characterized by image analysis: the acquisition was performed with a Leica microscope (Leica Microsystems, DMIL LED with a DFC345 FX camera), and the analysis with ImageJ software (NIH) (Fig. S1†). Scanning electron microscopy images of PDMS μGrooves were also taken to complete the dimensional characterization (SEM, at 5 kV, Fig. 1C). The number of μGrooves/well was calculated in AutoCAD. The expected cells were calculated by dividing μGroove area by the area of a myoblast.
2 (700 μm/200 μm). Characterization of SU8 masters (height and dimensions) were performed with a profilometer (KLA-Tencor, P6 Model, and Smart WLI, gbs) using the Profiler 7.31 software. μGrooved PDMS replicas were characterized by image analysis: the acquisition was performed with a Leica microscope (Leica Microsystems, DMIL LED with a DFC345 FX camera), and the analysis with ImageJ software (NIH) (Fig. S1†). Scanning electron microscopy images of PDMS μGrooves were also taken to complete the dimensional characterization (SEM, at 5 kV, Fig. 1C). The number of μGrooves/well was calculated in AutoCAD. The expected cells were calculated by dividing μGroove area by the area of a myoblast.
Before cell seeding, μGrooves were sterilized with 70% ethanol and UV-light. Subsequently, μGroove passivation was performed by incubation for 48 hours with growth media at 37 °C, allowing conditioning of the PDMS to ensure its stability during the assays.
Cell cultures
For our in vitro muscle platform, we used an immortalized human myoblast cell line (8220),41 kindly provided by Dr. Vincent Mouly (Myology Institute Paris, France). Cells were grown and differentiated as previously described.12 Briefly, cells were seeded at 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 myoblasts/well onto 24-well plates carrying μGrooved coverslips, previously coated for one hour at RT with either 25 μg ml−1 fibronectin (F1141, Sigma) or 0.5% gelatin (G1890, Sigma). Myoblasts were grown in skeletal muscle cell growth medium (SGM, Pelobiotech), supplemented with 10% fetal bovine serum (Gibco-Invitrogen). This high-density cell culture (200
000 myoblasts/well onto 24-well plates carrying μGrooved coverslips, previously coated for one hour at RT with either 25 μg ml−1 fibronectin (F1141, Sigma) or 0.5% gelatin (G1890, Sigma). Myoblasts were grown in skeletal muscle cell growth medium (SGM, Pelobiotech), supplemented with 10% fetal bovine serum (Gibco-Invitrogen). This high-density cell culture (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2) allowed for a homogeneous cell distribution within the μGrooves and also contributed to faster myotube formation due to fast and synchronous monolayer formation in the first 24 hours after seeding. High-density cell cultures benefit myotube formation, as increased cell-to-cell contacts enhance cell alignment, fusion and subsequent formation of mature myotubes.42
000 cells per cm2) allowed for a homogeneous cell distribution within the μGrooves and also contributed to faster myotube formation due to fast and synchronous monolayer formation in the first 24 hours after seeding. High-density cell cultures benefit myotube formation, as increased cell-to-cell contacts enhance cell alignment, fusion and subsequent formation of mature myotubes.42
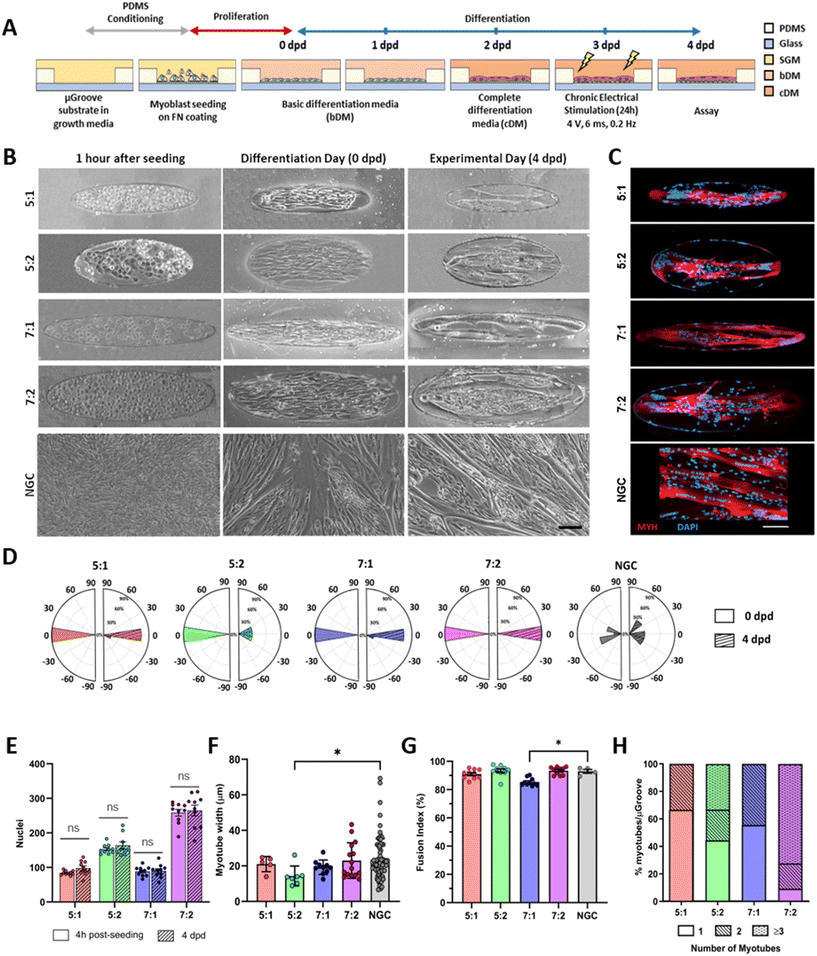
Upon confluence, the growth medium was replaced with basic differentiation medium (bDM) to induce myotube differentiation. After 2 days in differentiation (dpd), when the first multinucleated myotubes appeared, the medium was replaced with complete differentiation media (cDM) carrying different growth factors. The components of the skeletal muscle media, (reference and catalog number) are described in Fig. S2.† At 3 dpd coverslips were placed into 6 well-plates and exposed to chronic electrical stimulation (4 V, 6 ms pulse width, and 0.2 Hz) for 24 hours using the C-PACE EP System (IonOptix Westwood, MA 02090 USA). Protocol scheme is shown in Fig. 2A.
Immunofluorescence
Samples were fixed in 4% paraformaldehyde for one hour at RT and washed three times with PBS for 5 minutes. Samples were blocked with PBS containing 0.02% NaN3, 0.5% Triton X100, 2% goat serum (G9023, Sigma) and 2% bovine serum albumin (BSA) for one hour at RT. Next, anti-myosine heavy chain (MYH) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, IC4470F, R&D), anti-ryanodine receptor 1 (1
25, IC4470F, R&D), anti-ryanodine receptor 1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, #ARR-001, Alomone Labs), and CaV1.1 monoclonal 1A (1
50, #ARR-001, Alomone Labs), and CaV1.1 monoclonal 1A (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, MA3-920, Thermo Fisher Scientific) primary antibodies in blocking buffer were incubated overnight at 4 °C. After three PBS washes of 5 minutes, the samples were incubated with Alexa Fluor 647 or 555-conjugated secondary antibodies (1
50, MA3-920, Thermo Fisher Scientific) primary antibodies in blocking buffer were incubated overnight at 4 °C. After three PBS washes of 5 minutes, the samples were incubated with Alexa Fluor 647 or 555-conjugated secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400; A-31571, A-21428 or A-21235, Invitrogen) for one hour at RT. Subsequently, nuclei were stained with DAPI (0.1 μg mL−1, #D9542, Sigma-Aldrich) for 10 minutes at RT, and coverslips were washed and mounted onto microscope slides using ProLong™ Diamond Antifade Mountant (#P36930, Thermo Fisher Scientific). Image acquisition was performed with a LSM 980 confocal microscope with Airyscan2 with a 10× and 25× objectives (Carl Zeiss, Germany).
400; A-31571, A-21428 or A-21235, Invitrogen) for one hour at RT. Subsequently, nuclei were stained with DAPI (0.1 μg mL−1, #D9542, Sigma-Aldrich) for 10 minutes at RT, and coverslips were washed and mounted onto microscope slides using ProLong™ Diamond Antifade Mountant (#P36930, Thermo Fisher Scientific). Image acquisition was performed with a LSM 980 confocal microscope with Airyscan2 with a 10× and 25× objectives (Carl Zeiss, Germany).
Evaluation of contractility
μGrooves, and NGC were transferred to a six-well plate (glass bottom, P06-1.5H-N, Cellvis) filled with 37 °C imaging buffer (IB, 125 mM NaCl, 1.2 mM MgSO4, 5 mM KCl, 25 mM HEPES, 6 mM Glucose, 2 mM CaCl2, pH 7.4). Myotube contractility was analyzed at different voltages (8–40 V, at 2 ms, 0.1 Hz) using an electrical stimulator (C-PACE EP Unit, IonOptix, Westwood, MA). The contractility–frequency relationship was determined by inducing 1 s train of pulses at 20 V and different frequencies (0.1, 1 to 5, 10, 30, and 60 Hz). Myotube contractility was recorded with an inverted microscope (ECLIPSE Ti-S/L100, Nikon) equipped with a 10× objective at 100 frames per second (fps). Videos of single myotubes were cropped into three regions of 50 × 20 μm, which were subsequently subjected to pixel movement analysis. Each area was directly processed with the ImageJ Macro MUSCLEMOTION43 and the results were averaged, providing the contractility profile for each myotube. This approach resulted in better signal-to-noise ratios compared to full-frame video analysis. Peak amplitude (PA) for both twitch (0.1 Hz) and tetanic contractions (10 Hz), and twitch kinetics, time to peak (TTP), and half relaxation time (RT50), were calculated as depicted in ESI† Fig. S4.Calcium imaging
Basal cytosolic calcium levels were measured following a previously described protocol with minor modifications.15,41 Briefly, myotubes grown onto μGrooves substrates were loaded with 4 μM Fura-2 AM (F1221, Thermo Fisher) and 0.02% pluronic acid (F1225, Thermo Fisher) in culture medium for 30 minutes at 37 °C. Cells were washed twice in Ringer's buffer and incubated for 30 minutes to complete de-esterification. Myotubes were subjected to electrical stimulation to elicit twitch (2 ms, 20 V, 0.1 Hz) and tetanic responses (2 ms, 20 V, 10 Hz), using the same protocol as the one described for contractility experiments. Basal intracellular calcium concentration of myotube responders was calculated by using the 380/340 Fura-2 AM fluorescent ratios. In situ calibration was performed with 10 μM ionomycin (I3909-1ML, Sigma-Aldrich) in 2 mM Ca2+, followed by 100 mM EGTA (E3889, Sigma) in Ca2+ free buffer, using a Fura-2 AM dissociation constant of 224 nM. For the evaluation of Ca2+ transients, myotubes were loaded with 5 μM Fluo-8 AM (Ref. 21082, AAT Bioquest) and 2.5 mM probenecid (Ref. 20062, AAT Bioquest) for 30 minutes at 37 °C. Videos were acquired with an ECLIPSE Ti-S/L100 microscope (Nikon) equipped with a 10× S-Fluor objective and attached to a lambda-DG4 illumination system and an Orca-Flash2.8 camera (Hamamatsu). The ΔF/F ratio was used to plot Ca2+ fluxes and all subsequent analyses were performed with the NisElements-AR software, as previously described. Average intensity of three circular regions of interest (ROIs, 10 μm diameter) per myotube was used.Statistical analysis
Statistical analysis was performed using Graph Prism 10 (GraphPad Software, San Diego, CA, USA). Data are presented as mean ± standard deviation (SD). Data distribution was evaluated using the Shapiro–Wilk and D'Agostino & Pearson omnibus normality tests. Equal SDs were not assumed unless with paired test. For data following a normal distribution, t-test with Welch's correction or paired t-test were used. When comparing more than two groups, Welch ANOVA tests were used followed by Dunnett's T3 post hoc test. For data that did not follow a normal distribution Mann Whitney test and and Wilcoxon matched-pairs signed rank test were used for two group comparisons, and when comparing more than two groups, Kruskal–Wallis test followed by Dunn's post hoc test was used. For all statistical analyses, the adjusted p-value of less than 0.05 was considered statistically significant. Data marked with an † was calculated by the authors from published papers.Results and discussion
Fabrication and characterization of μGrooves
Characterization of SU8 masters, with a targeted height of 80 μm, resulted in experimental height values of 76.63 ± 6.13 μm, which correspond to the PDMS membrane thickness. The average dimensions of each geometry were 506.8 ± 9.5 or 701.8 ± 15.9 μm for length, and 117.1 ± 12.3 or 211.9 ± 9.5 μm for width. The width/length ratio and area for each geometry are detailed in Fig. S1A and B.† During the fabrication process we observed suboptimal geometry definition in some membranes, such as partially open structures or torn μGroove walls (Fig. S1C†). These issues are likely caused by non-uniform weight distribution, harsh delamination processes, or scratches (Fig. S1C†). Therefore, it is crucial to control these factors in order to promote uniform coating of glass substrates and myoblast growth within the μGrooves. In particular, the direction of peeling-off during delamination is of utmost importance and should be performed in the direction of the main axis of the geometries to preserve the structure of the μGrooves. Additionally, increasing the weight up to 600 g improves the contact between PDMS layers during polymerization. Overall, our fabrication yield was around 60%, which resulted in more than ninety functional μGrooves per glass slide.Morphological analysis
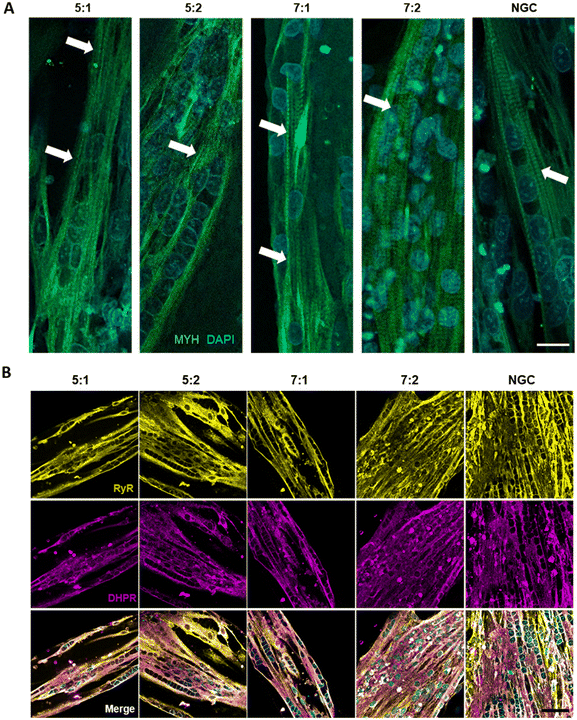
Cells were seeded at high density onto the substrate to confine them within the μGrooves in confluency. Low-cell density cultures were also performed (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2; data not shown). However, 24 hours after seeding, cultures were heterogeneous within the same condition. High cell-density cultures, on the other hand, promoted cellular alignment and differentiation. We found that μGrooved structures facilitated cellular alignment both at early stages of differentiation (within the first 24 hours, Fig. 2B and S3A†) and at later stages, when myotubes with striation pattern were observed at 4 dpd in all conditions, (Fig. 3A). The presence of well-organized cross-striations indicate the assembly of MYH into sarcomeres and the formation of mature myotubes.44–46Fig. 2B shows representative contrast-phase images of cultures at 0 dpd and 4 dpd, for four different geometries and non-μGrooved controls (NGC), while Fig. S3A† depicts a representative day-by-day sequence of these cultures. We found that μGrooved cells aligned along the longitudinal axes of the geometries, while cells grown on non-grooved substrates did not exhibit preferential alignment, as evidenced by the higher dispersion observed in the polar histograms representing the relative orientation of cells in each culture condition at 0 dpd and 4 dpd (Fig. 2D). The majority of myoblasts and myotubes in the μGrooves aligned with a value of less than 10°, which conforms to the established criterion for cellular alignment.20,32 Specifically, the alignment for myoblasts at 0 dpd was 1.7 ± 1.5°, 3.6 ± 4.2°, 1.6 ± 1.2°, and 3.8 ± 2.6° for 5
000 cells per cm2; data not shown). However, 24 hours after seeding, cultures were heterogeneous within the same condition. High cell-density cultures, on the other hand, promoted cellular alignment and differentiation. We found that μGrooved structures facilitated cellular alignment both at early stages of differentiation (within the first 24 hours, Fig. 2B and S3A†) and at later stages, when myotubes with striation pattern were observed at 4 dpd in all conditions, (Fig. 3A). The presence of well-organized cross-striations indicate the assembly of MYH into sarcomeres and the formation of mature myotubes.44–46Fig. 2B shows representative contrast-phase images of cultures at 0 dpd and 4 dpd, for four different geometries and non-μGrooved controls (NGC), while Fig. S3A† depicts a representative day-by-day sequence of these cultures. We found that μGrooved cells aligned along the longitudinal axes of the geometries, while cells grown on non-grooved substrates did not exhibit preferential alignment, as evidenced by the higher dispersion observed in the polar histograms representing the relative orientation of cells in each culture condition at 0 dpd and 4 dpd (Fig. 2D). The majority of myoblasts and myotubes in the μGrooves aligned with a value of less than 10°, which conforms to the established criterion for cellular alignment.20,32 Specifically, the alignment for myoblasts at 0 dpd was 1.7 ± 1.5°, 3.6 ± 4.2°, 1.6 ± 1.2°, and 3.8 ± 2.6° for 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 7
2, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 7
1, and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 geometries, respectively. As for myotubes at 4 dpd, the alignment values were 6.5 ± 4.5°, 10.8 ± 9.1°, 2.5 ± 1.6°, and 2.5 ± 1.8° for 5
2 geometries, respectively. As for myotubes at 4 dpd, the alignment values were 6.5 ± 4.5°, 10.8 ± 9.1°, 2.5 ± 1.6°, and 2.5 ± 1.8° for 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 7
2, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 7
1, and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 geometries, respectively. The alignment angle for NGC could not be calculated due to the lack of a reference axis. However, the data represented in the polar histograms clearly show a higher dispersion of alignment compared to the μGrooved myotubes. Our findings are similar to a previous study with human primary myotubes, where higher myotube alignment was observed in myotubes differentiated on micropatterned and nanopatterned surfaces compared to standard surfaces.22,25,32 Overall, our data demonstrate that the alignment of myoblasts and myotubes is higher as the width-to-length ratio decreases, with 7
2 geometries, respectively. The alignment angle for NGC could not be calculated due to the lack of a reference axis. However, the data represented in the polar histograms clearly show a higher dispersion of alignment compared to the μGrooved myotubes. Our findings are similar to a previous study with human primary myotubes, where higher myotube alignment was observed in myotubes differentiated on micropatterned and nanopatterned surfaces compared to standard surfaces.22,25,32 Overall, our data demonstrate that the alignment of myoblasts and myotubes is higher as the width-to-length ratio decreases, with 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometry showing the highest myotube alignment. These results are consistent with previous studies reporting that myotube and myoblast alignment is highly dependent on the width of the substrates21,26,29,31,32,34,47,48 and substrate geometry.10,23,25,31,32
1 geometry showing the highest myotube alignment. These results are consistent with previous studies reporting that myotube and myoblast alignment is highly dependent on the width of the substrates21,26,29,31,32,34,47,48 and substrate geometry.10,23,25,31,32
We next proceeded to evaluate the number of nuclei, myotube width, and fusion index, which are commonly used indicators of myotube differentiation, maturation and cell death.12,41 First, we determined cell survival in the μGrooves and found no significant differences in the total number of nuclei at 4 hours post-seeding and at 4 dpd within the same μGroove geometries (Fig. 2E). This is indicative of cell survival throughout the culture lifespan in the different geometries. The highest number of nuclei was found in the 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 geometry, followed by the 5
2 geometry, followed by the 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 geometry, while the 5
2 geometry, while the 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 7
1 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometries showed similar numbers of nuclei (Fig. 2C, E and S3B†). These data are in line with the calculated μGroove areas of the geometries (Fig. S1A†).
1 geometries showed similar numbers of nuclei (Fig. 2C, E and S3B†). These data are in line with the calculated μGroove areas of the geometries (Fig. S1A†).
Regarding myotube width, we found similar values in μGrooved and NGC myotubes (20.1 ± 7.7 μm and 23.5 ± 11.8 μm, respectively) (Fig. 2F and S7A†), with myotubes in the 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 geometry being slightly thinner compared to NGC myotubes (Kruskal–Wallis test, *p < 0.05). Our results are consistent with other studies in C2C12 and primary human myotubes grown on nanopatterned surfaces reporting myotube widths of around 20 μm.13,23,25 Conversely, other studies in human myotubes have reported higher myotube widths in micropatterned compared to non-patterned substrates in association with increased fusion index,22 supporting that myotube width is highly dependent on the cell type and closely related to its fusion index. Interestingly, μGrooved myotubes presented higher homogeneity as reflected by a lower coefficient of variation (CV). In particular, in 5
2 geometry being slightly thinner compared to NGC myotubes (Kruskal–Wallis test, *p < 0.05). Our results are consistent with other studies in C2C12 and primary human myotubes grown on nanopatterned surfaces reporting myotube widths of around 20 μm.13,23,25 Conversely, other studies in human myotubes have reported higher myotube widths in micropatterned compared to non-patterned substrates in association with increased fusion index,22 supporting that myotube width is highly dependent on the cell type and closely related to its fusion index. Interestingly, μGrooved myotubes presented higher homogeneity as reflected by a lower coefficient of variation (CV). In particular, in 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 7
1 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometries with 100 μm-widths, the CV was reduced to 20.3% and 21%, respectively compared to 50.2% in NGCs, representing a decrease in variability of around 60% (Fig. S7B†). These results are consistent with previous studies on claiming higher myotube variability in diameter and shape on flat surfaces compared to patterned substrates.22,23,31 Indeed, primary human myotubes differentiated on micropatterned22 and nanopatterned23 substrates present a 24%† and 30%† CV, respectively compared to a 50%† CV in non-patterned myotubes.22
1 geometries with 100 μm-widths, the CV was reduced to 20.3% and 21%, respectively compared to 50.2% in NGCs, representing a decrease in variability of around 60% (Fig. S7B†). These results are consistent with previous studies on claiming higher myotube variability in diameter and shape on flat surfaces compared to patterned substrates.22,23,31 Indeed, primary human myotubes differentiated on micropatterned22 and nanopatterned23 substrates present a 24%† and 30%† CV, respectively compared to a 50%† CV in non-patterned myotubes.22
Regarding myoblast fusion, calculated as the percentage of nuclei within the myotube, our cultures showed an overall high fusion index ranging from 85% to 93%. These values are much higher than those reported in previous studies (<65%),25,30,34,36 likely due to the optimized differentiation media12 and the intrinsic nature of the immortalized myoblast line 8220, which we selected for its high differentiation and maturation potential.41,49 Microenvironment and cellular interactions are essential for myoblast fusion. Previous studies have shown that micropatterning may increase or decrease the myoblast fusion index, depending on the specific geometry.10,22,34 Our analysis revealed a slightly reduced fusion index in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGrooves compared to NGC at 4 dpd (9%, Fig. 2G, *p < 0.05). 7
1 μGrooves compared to NGC at 4 dpd (9%, Fig. 2G, *p < 0.05). 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGrooves present the lowest wide-to-length ratio compared to other geometries (0.143, **p < 0.01, ****p < 0.0001, Fig. S1A†). Thus, the mild reduction of myoblast fusion in 7
1 μGrooves present the lowest wide-to-length ratio compared to other geometries (0.143, **p < 0.01, ****p < 0.0001, Fig. S1A†). Thus, the mild reduction of myoblast fusion in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGrooves may be explained by the specific distribution of forces in this geometry.10 All in all, this effect is minor and does not impact overall myotube differentiation and maturation, as shown by the preserved myotube width in 7
1 μGrooves may be explained by the specific distribution of forces in this geometry.10 All in all, this effect is minor and does not impact overall myotube differentiation and maturation, as shown by the preserved myotube width in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGrooves (Fig. 2F). As for the other geometries, we did not observe any significant differences in the fusion index compared to NGC (Fig. 2G). In contrast, other studies have shown enhanced myoblast fusion index in micropatterned substrates.22,30 However, these studies were performed in human myoblasts with low fusion index. Interestingly, a recent study in human myoblasts with high fusion index reports no changes in myoblast fusion between control and microgrooved substrates, which validates our results for most geometries.25
1 μGrooves (Fig. 2F). As for the other geometries, we did not observe any significant differences in the fusion index compared to NGC (Fig. 2G). In contrast, other studies have shown enhanced myoblast fusion index in micropatterned substrates.22,30 However, these studies were performed in human myoblasts with low fusion index. Interestingly, a recent study in human myoblasts with high fusion index reports no changes in myoblast fusion between control and microgrooved substrates, which validates our results for most geometries.25
Subsequently, we evaluated the number of myotubes generated in each geometry, since our goal was to generate single isolated myotubes per μGroove. Our analysis revealed that the width of the μGroove has a significant impact on myotube formation, as previously reported.29 Indeed, 100 μm widths resulted in the formation of single myotubes in over 50% of the μGrooves, whereas 200 μm widths generally presented more than one myotube per μGroove such as the 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 geometry with multiple myotubes in over 70% of the μGrooves (Fig. 2H). The average number of myotubes per μGroove was 1.33 ± 0.48, 1.88 ± 0.90, 1.44 ± 0.52, 2.63 ± 0.67 for 5
2 geometry with multiple myotubes in over 70% of the μGrooves (Fig. 2H). The average number of myotubes per μGroove was 1.33 ± 0.48, 1.88 ± 0.90, 1.44 ± 0.52, 2.63 ± 0.67 for 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 7
2, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 7
1 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 respectively. The effect of the substrate size on myotube development has been investigated in previous works, with similar results to the ones reported in this study. For instance, Shimizu et al. found that the number of myotubes generated in micropatterns increased with the micropattern width, with average myotube numbers of 1.1, 1.6, and 3.6 for widths of 50 μm, 100 μm, and 200 μm, respectively.29 Similarly, S. Zatti et al. suggested an optimal width range of 30–100 μm for micropatterned structures, as values below this range may hinder myoblast differentiation due to pattern size and reduced cell quantity.34 In a different culture setup based on thin films, Sun et al. reached similar conclusions and proposed a width of 100 μm to maximize myotube formation.48
2 respectively. The effect of the substrate size on myotube development has been investigated in previous works, with similar results to the ones reported in this study. For instance, Shimizu et al. found that the number of myotubes generated in micropatterns increased with the micropattern width, with average myotube numbers of 1.1, 1.6, and 3.6 for widths of 50 μm, 100 μm, and 200 μm, respectively.29 Similarly, S. Zatti et al. suggested an optimal width range of 30–100 μm for micropatterned structures, as values below this range may hinder myoblast differentiation due to pattern size and reduced cell quantity.34 In a different culture setup based on thin films, Sun et al. reached similar conclusions and proposed a width of 100 μm to maximize myotube formation.48
In summary, our findings indicate that μGrooved substrates promote the development of single aligned myotubes with higher homogeneity compared to standard myotube cultures (NGC). Consistent with prior research,10 our results highlight the μGroove width as a crucial factor influencing myotube morphology. Other studies have explored the use of mechanical cues to enhance the homogeneity of myotube cultures, both in terms of morphology and function demonstrating higher homogeneity of myotube size and maturation on a micropatterned platform termed MyoScreen (Cytoo, France).22 Their findings in relation to cell alignment (<10°), fusion index (69%), and myotube width (median of 28 μm) are comparable to our results. However, the MyoScreen platform often generates myotube clusters (usually 2 or 3 are observed), similar to our results obtained with 200 μm-width geometries. Remarkably, the Myoscreen platform has a high percentage of myotube detachment after acetylcholine stimulation (40–60%), whereas myotubes in our μGroove platform myotubes were able to withstand repeated tetanic stimulations with minimal detachment (<15%). This indicates that the micropatterning technique may not be optimal for assays requiring repeated contractions or tetanic stimulations.
Contractility
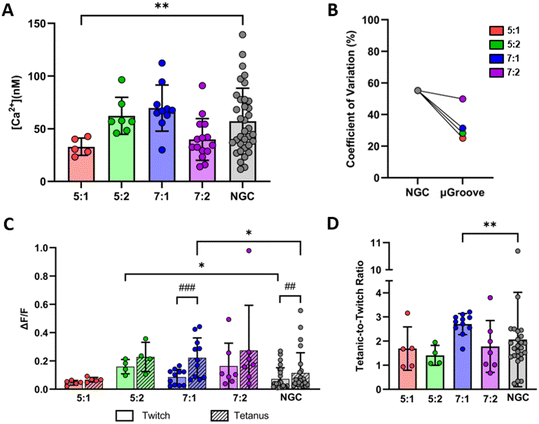
First, we characterized the contractile capacity of our cultures as an indicator of myotube maturation. As illustrated in ESI† Fig. S6 and Video S1, our cultures exhibited spontaneous contractions with a smaller peak amplitude compared to those induced by electrical stimulation. Similarly, previous studies have reported spontaneous contractility in myotubes,25,44–46,50 which is generally considered to indicate a relatively high maturation level and has been shown to correlate with mature cross-striated patterning. Spontaneous contractions in 2D cultures are not usually observed in human myotubes until later stages of differentiation, often after 10 dpd or later.25,45 However, in our hands, we observed spontaneous contractions starting at 4 dpd after 24 hours chronic stimulation (4 V, 6 ms, 0.2 Hz), both in micropatterned and NGC myotubes. This is consistent with previous studies showing that chronic electrical stimulation improves myotube maturation.19,44,51Next, we evaluated contractility as a main functional outcome, by using single pulses or a train of electric pulses to elicit twitch and tetanic contractions, respectively. We first analyzed the percentage of myotubes that exhibited twitch contractions in μGrooves and NGC. Fig. S5A† illustrates the effect of μGrooves on myotube contractility. Our results indicate that the percentage of responder myotubes is higher in μGrooves (74.9 ± 10.7%) compared to NGC (47.5 ± 38.9%) at maximal voltage (40 V) and it is consistent with previous studies.25,52 This reflects a higher efficiency of E–C coupling, which is associated with a higher maturation of μGrooved myotubes. Moreover, CV at 40 V was much higher in the NGC substrates (76%) compared to μGrooved substrates (33.35%), which indicates a higher homogeneity of myotubes in the μGrooves (Fig. S5A†). Similar findings were reported for electrically stimulated human primary myotubes grown on nanopatterned surfaces, where myotubes responders were calculated based on calcium transients. In this study, more than 95% of nanopatterned-myotubes responded to electrical stimulation compared to less that 45% of myotubes grown on flat surfaces.25
Next, we studied the excitability threshold of μGrooved and NGC myotubes by measuring their contractile response to voltage increments ranging from 8 to 40 V, as shown in Fig. S5A.† We observed that within this range, the proportion of myotube responders in μGrooves increased proportionally with the applied voltage, while NGC myotubes did not display such behaviour. This fact may be because 8 V is a sufficiently high value for all functional myotubes of the NGC to respond to stimulation. Therefore, it is reasonable to expect that there is no variation in this ratio. In the case of grooves, these values of electric potential are conditioned by the presence of the membrane. To verify if this effect was caused by the PDMS membrane acting as an electrical insulator, we performed electric field simulations at 8, 10, and 20 V (Fig. S5B and C†). Our results show that the μGrooved PDMS membrane indeed acted as an electrical insulator, reducing the potential that reaches the μGrooves by up to 60% for 20 V, while the electric field in the NGC remained homogeneous on the substrate surface. This indicates that only 12 V out of the 20 V pulse generated by the C-Pace actually reached the myotubes within the μGrooves. To address this limitation, some researchers have incorporated conductive particles such as carbon nanotubes or graphene into PDMS, resulting in a significant increase in electrical permittivity that is proportional to the concentration of carbon nanocomposites.53,54 This approach has been shown to achieve a dielectric constant (∈) approximately 700 times higher than that of pure PDMS, without compromising the mechanical properties of the polymer.55 This technique could be implemented in our platform to enhance the conductive properties of μGrooves, particularly in experiments requiring chronic stimulation.
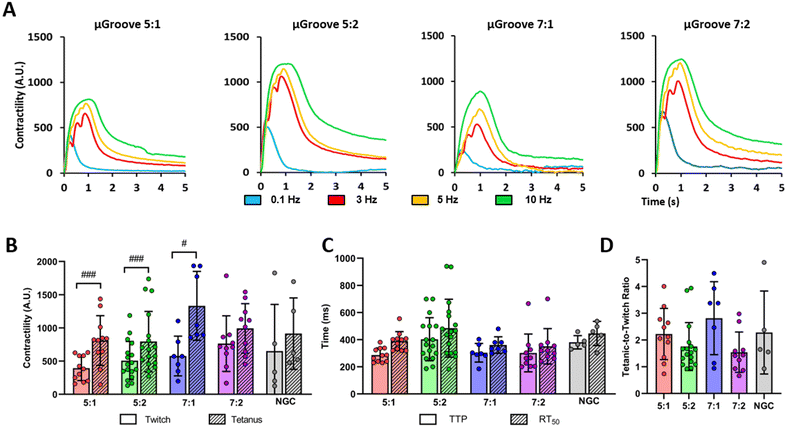
For subsequent contractile experiments, we chose to use 20 V pulses as this voltage elicited a response in more than 50% of myotubes, while minimizing adverse effects such as overstimulation, electroporation, or electrochemical damage compared to using 40 V.8,9 The electrical stimulation protocol was selected based on previous studies8,9,50 and optimized for maximal myotube responses with minimal signs of fatigue or detachment after tetanic stimulation. Fig. 4A shows representative contractile traces of μGrooved myotubes in response to electrical stimulation. We observed an increase in contractility in all geometries in response to increasing frequencies (0.1, 3, 5, 10 Hz). We calculated the peak amplitude of these contractions for both twitch and tetanic stimuli (Fig. 4B) and found significant increases between tetanic and twitch contractions for 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 7
2 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometries (#p < 0.05, ##p < 0.01, ###p < 0.001). We also calculated the time to peak (TTP) and half-relaxation time (RT50) for twitch contractions (Fig. 4C). However, no significant differences were observed between the different μGroove geometries and NGC for any of the values analyzed. Notably, the CV of the peak amplitude for twitch and tetanic contractions was almost 50% lower in the 5
1 geometries (#p < 0.05, ##p < 0.01, ###p < 0.001). We also calculated the time to peak (TTP) and half-relaxation time (RT50) for twitch contractions (Fig. 4C). However, no significant differences were observed between the different μGroove geometries and NGC for any of the values analyzed. Notably, the CV of the peak amplitude for twitch and tetanic contractions was almost 50% lower in the 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 μGroove geometry compared to its NGC, as illustrated in Fig. S7C.† Measuring tetanic contractions in 2D muscle models is usually very challenging due to the tendency of contractile myotubes to detach prematurely. In fact, in the literature, only 27% of 2D in vitro skeletal muscle studies report tetanic contractile responses.19 However, in this study, we were able to evaluate both twitch and tetanic contractions in our μGroove platform, enabling a more comprehensive analysis of the contractile function.
2 μGroove geometry compared to its NGC, as illustrated in Fig. S7C.† Measuring tetanic contractions in 2D muscle models is usually very challenging due to the tendency of contractile myotubes to detach prematurely. In fact, in the literature, only 27% of 2D in vitro skeletal muscle studies report tetanic contractile responses.19 However, in this study, we were able to evaluate both twitch and tetanic contractions in our μGroove platform, enabling a more comprehensive analysis of the contractile function.
Finally, the tetanic-to-twitch contractile ratio was calculated as an indicator of muscle maturity and performance.19 The 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometry exhibited the highest tetanic-to-twitch ratio (2.82 ± 1.36), with approximately 57% of the myotubes showing a ratio over 3 (Fig. 4D). The rest of the groups presented ratios ranging from 1.5 to 2.3. The tetanic-to-twitch ratio of 7
1 geometry exhibited the highest tetanic-to-twitch ratio (2.82 ± 1.36), with approximately 57% of the myotubes showing a ratio over 3 (Fig. 4D). The rest of the groups presented ratios ranging from 1.5 to 2.3. The tetanic-to-twitch ratio of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGrooved myotubes is notably higher compared to the ones reported in other 2D in vitro studies that use human cell lines (e.g. 2.01; 0.91),8,9 but still below the range typically observed in native tissue (between 4 and 10).56 These findings underscore the necessity for exploring alternative approaches to create muscle models that more closely resemble physiological conditions. Co-culturing in μGrooved platforms is a potential method that could enhance maturity and adhesion of myotubes, while preserving their homogeneity.
1 μGrooved myotubes is notably higher compared to the ones reported in other 2D in vitro studies that use human cell lines (e.g. 2.01; 0.91),8,9 but still below the range typically observed in native tissue (between 4 and 10).56 These findings underscore the necessity for exploring alternative approaches to create muscle models that more closely resemble physiological conditions. Co-culturing in μGrooved platforms is a potential method that could enhance maturity and adhesion of myotubes, while preserving their homogeneity.
One limitation of the current study is that the electrical stimulation used for functional evaluation was applied to the entire plate, even though only one myotube was imaged at a time. This can lead to a gradual loss of contractile performance over time, particularly during tetanic contractility. To prevent any fatigue effects, we restricted the number of stimulations on each μGroove substrate to a maximum of 10. However, to exploit the full high-throughput capability of the platform, it would be desirable to integrate electrodes that enable the stimulation of individual myotubes.
Calcium handling
Calcium handling analysis was used to characterize the fitness and functional status of myotube cultures. First, we evaluated the basal cytosolic calcium concentration in the different μGroove geometries and their respective controls, as illustrated in Fig. 5A. The basal calcium concentration in μGrooved myotubes ranged between 30 and 70 nM, with the highest levels found in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGroove geometry (69.6 ± 21.9 nM), followed by 5
1 μGroove geometry (69.6 ± 21.9 nM), followed by 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (62.2 ± 17.5 nM), 7
2 (62.2 ± 17.5 nM), 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (39.8 ± 19.9 nM), and 5
2 (39.8 ± 19.9 nM), and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (32.9 ± 8.21 nM). Notably, μGrooved myotubes on 5
1 (32.9 ± 8.21 nM). Notably, μGrooved myotubes on 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometry showed a significantly reduced basal calcium levels compared to NGCs (*p < 0.01). Most interestingly, different variability was observed between the μGroove geometries and the NGC datasets. Overall, we found a higher CV in NGC myotubes (55.2%) than in μGrooved myotubes (34%, Fig. 5B and S7D†). In particular, the most relevant reductions in CV were observed for 5
1 geometry showed a significantly reduced basal calcium levels compared to NGCs (*p < 0.01). Most interestingly, different variability was observed between the μGroove geometries and the NGC datasets. Overall, we found a higher CV in NGC myotubes (55.2%) than in μGrooved myotubes (34%, Fig. 5B and S7D†). In particular, the most relevant reductions in CV were observed for 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 7
2 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometries, with decreases of 54.7%, 48.9% and 43.1%, respectively compared to NGCs. In contrast, similar CV for the 7
1 geometries, with decreases of 54.7%, 48.9% and 43.1%, respectively compared to NGCs. In contrast, similar CV for the 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 μGroove geometry dataset was observed compared to NGC. While these results are expected due to the greater morphological homogeneity observed in μGrooved myotubes, the meaning behind the differences in basal calcium observed in the 5
2 μGroove geometry dataset was observed compared to NGC. While these results are expected due to the greater morphological homogeneity observed in μGrooved myotubes, the meaning behind the differences in basal calcium observed in the 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGroove geometry remains uncertain. One possibility is that these differences may be attributed to variations in myotube maturation levels among the different geometries. In this line, it has been previously reported that resting cytosolic calcium presents a profile of evolution with a progressive increase up to a maximum level followed by a gradual decrease in human myotubes.57 Another possibility is that basal intracellular calcium levels may be affected by the myotube area or by the number of nuclei within each myotube. Further investigation is necessary to confirm these hypotheses.
1 μGroove geometry remains uncertain. One possibility is that these differences may be attributed to variations in myotube maturation levels among the different geometries. In this line, it has been previously reported that resting cytosolic calcium presents a profile of evolution with a progressive increase up to a maximum level followed by a gradual decrease in human myotubes.57 Another possibility is that basal intracellular calcium levels may be affected by the myotube area or by the number of nuclei within each myotube. Further investigation is necessary to confirm these hypotheses.
Subsequently, we evaluated calcium transients in response to electrical stimuli, following a similar protocol as the one used for contractility assessment. As shown in Fig. 5C, both NGC and μGrooved myotubes exhibited twitch and tetanic calcium transients at 0.1 Hz and 10 Hz, respectively. Analysis of peak amplitude of calcium transients revealed that myotubes grown in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGroove geometry presented the highest increase between tetanic and twitch contractions (paired Student's t-test; ###p < 0.001). Additionally, tetanic response in 7
1 μGroove geometry presented the highest increase between tetanic and twitch contractions (paired Student's t-test; ###p < 0.001). Additionally, tetanic response in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGrooved myotubes were significantly increased compared to NGC myotubes by 2-fold (*p < 0.05). We next calculated the tetanic-to-twitch ratio for calcium responses (Fig. 5D). As expected, the 7
1 μGrooved myotubes were significantly increased compared to NGC myotubes by 2-fold (*p < 0.05). We next calculated the tetanic-to-twitch ratio for calcium responses (Fig. 5D). As expected, the 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGroove geometry demonstrated the best performance with a ratio of 2.70 ± 0.43, compared to 2.07 ± 5.20 in NGC (*p < 0.05). Furthermore, this dataset presented a CV of 16%, which is the lowest among all the groups. These data are in line with the contractility results, where myotubes in 7
1 μGroove geometry demonstrated the best performance with a ratio of 2.70 ± 0.43, compared to 2.07 ± 5.20 in NGC (*p < 0.05). Furthermore, this dataset presented a CV of 16%, which is the lowest among all the groups. These data are in line with the contractility results, where myotubes in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometry also exhibited the best performance regarding the tetanic-to-twitch ratio, which further supports our conclusions. It should be noted that in this study all the values were included, although it could be argued that tetanic-to-twitch ratio values <1 could be considered as non-valid.
1 geometry also exhibited the best performance regarding the tetanic-to-twitch ratio, which further supports our conclusions. It should be noted that in this study all the values were included, although it could be argued that tetanic-to-twitch ratio values <1 could be considered as non-valid.
Overall, these results demonstrate that 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μGrooves enhance myotube maturation and homogeneity compared to standard cultures in several morphological and functional parameters. However, the 7
1 μGrooves enhance myotube maturation and homogeneity compared to standard cultures in several morphological and functional parameters. However, the 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dataset variability is still high in some parameters, such as twitch and tetanic-to-twitch ratio. The number of replicates for each condition may differ due to the chosen inclusion criteria for a minimum of 5 samples, and that myotubes should be able to complete the entire functional assessment up to at least 10 Hz. Further optimization of the platform, such as incorporating microelectrodes into the system or increasing culture lifespan with chronic stimulation and matrix overlayers,12,25 may allow for lower data variability.
1 dataset variability is still high in some parameters, such as twitch and tetanic-to-twitch ratio. The number of replicates for each condition may differ due to the chosen inclusion criteria for a minimum of 5 samples, and that myotubes should be able to complete the entire functional assessment up to at least 10 Hz. Further optimization of the platform, such as incorporating microelectrodes into the system or increasing culture lifespan with chronic stimulation and matrix overlayers,12,25 may allow for lower data variability.
Conclusions
In this study, we propose a 2D platform that utilizes μGrooved structures to promote the alignment and differentiation of myoblasts into single, aligned, and mature myotubes. Our results demonstrate that this platform generates myotubes with higher homogeneity in morphology, contractile performance, and calcium handling compared to traditional cultures.Overall, the best performance was observed in myotubes confined to the 100 μm-wide μGrooves, specifically in the 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometry, where myotubes exhibited superior contractile and calcium responses compared to other geometries and to non-grooved controls. Indeed, myotubes in the 7
1 geometry, where myotubes exhibited superior contractile and calcium responses compared to other geometries and to non-grooved controls. Indeed, myotubes in the 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 geometry showed higher tetanic-to-twitch ratio values both for contractility and calcium transients. The consistency of the calcium handling values with the contractility analysis provides further evidence for the reliability of our μGroove platform.
1 geometry showed higher tetanic-to-twitch ratio values both for contractility and calcium transients. The consistency of the calcium handling values with the contractility analysis provides further evidence for the reliability of our μGroove platform.
Additionally, the design of μGrooved substrates is compatible with chronic stimulation and enables functional evaluation of myotubes via contractility and calcium handling analysis, offering a comprehensive characterization of 2D muscle models. Moreover, our platform is very flexible and can be used in combination with other designs. For example, a neuromuscular system could be easily achieved by adding a matrigel overlay on top of the μGrooves, and motoneurons or explants could be added to this overlay. This system would allow long term coculture to study the formation of neuromuscular junction development. In addition, this platform could also be compatible with microelectrode recordings.
Our platform represents a promising approach for generating physiologically relevant in vitro muscle models that can be used to better understand muscle development and disease, and to identify new therapeutic targets for muscle weakness. The high-content, high-throughput potential and reduced heterogeneity makes this platform a powerful tool for drug screening and muscle engineering research. Future studies could explore the use of different substrate materials, cell types, electrical stimulation optimization, and other improvements to further optimize the platform and elucidate underlying mechanisms of muscle physiopathology.
Data availability
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.Author contributions
CVC, JRH and JP designed and fabricated the μGrooves, CVC, LMM, KGI and PMM performed experiments, PUU performed the electrical simulations, CVC, JP, PMM, and AVI analyzed the data, CVC wrote the draft, JP and AVI wrote and edited the manuscript, and all authors reviewed the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Gobierno Vasco (AV-I, 2022111045 and IT1732-22), Diputación Foral de Gipuzkoa (JP, 2018-CIEN-000086-01), and Ministerio de Ciencia e Innovación (AVI, PID2020-11978RB-I00). CVC held a PhD fellowship from Tecnun and is currently holding a Postdoctoral fellowship from the Basque Government, LMM and KGI hold a PhD fellowship from UPV/EHU, and PMM holds a PhD fellowship from Jesús Gangoiti Barrera Foundation. The authors thank Maria Bikuna-Izaguirre for technical help with SEM images, and Unai Lertxundi for profilometer image characterization of μGrooved substrates. The authors would also like to thank Dr. Vincent Mouly and the Platform for Immortalization of Human Cells (Myology Institute) for sharing human immortalized myoblasts.References
- E. P. Widmaier, H. Raff and K. T. Strang, Vander's Human Phisiology – The Mechanisms of Body Function, McGraw-Hill, New York, NY, 11th edn, 2008 Search PubMed.
- G. Chandra, A. Defour, K. Mamchoui, K. Pandey, S. Mishra, V. Mouly, S. C. Sreetama, M. Mahad Ahmad, I. Mahjneh, H. Morizono, N. Pattabiraman, A. K. Menon and J. K. Jaiswal, Dysregulated calcium homeostasis prevents plasma membrane repair in Anoctamin 5/TMEM16E-deficient patient muscle cells, Cell Death Discovery, 2019, 5(118), 1–15, DOI:10.1038/s41420-019-0197-z.
- A. P. Nesmith, M. A. Wagner, F. S. Pasqualini, B. B. O'Connor, M. J. Pincus, P. R. August and K. K. Parker, A human in vitro model of Duchenne muscular dystrophy muscle formation and contractility, J. Cell Biol., 2016, 215(1), 47–56, DOI:10.1083/jcb.201603111.
- A. Vallejo-Illarramendi, I. Toral-Ojeda, G. Aldanondo and A. López de Munain, Dysregulation of calcium homeostasis in muscular dystrophies, Expert Rev. Mol. Med., 2014, 16(October 2014), e16, DOI:10.1017/erm.2014.17.
- H. Vandenburgh, J. Shansky, F. Benesch-Lee, V. Barbata, J. Reid, L. Thorrez, R. Valentini and G. Crawford, Drug-screening platform based on the contractility of tissue-engineered muscle, Muscle Nerve, 2008, 37(4), 438–447, DOI:10.1002/mus.20931.
- A. Alave Reyes-Furrer, S. De Andrade, D. Bachmann, H. Jeker, M. Steinmann, N. Accart, A. Dunbar, M. Rausch, E. Bono, M. Rimann and H. Keller, Matrigel 3D bioprinting of contractile human skeletal muscle models recapitulating exercise and pharmacological responses, Commun. Biol., 2021, 4, 1183, DOI:10.1038/s42003-021-02691-0.
- K. Wilson, M. Das, K. J. Wahl, R. J. Colton, J. Hickman and W.-L. Zhou, Measurement of Contractile Stress Generated by Cultured Rat Muscle on Silicon Cantilevers for Toxin Detection and Muscle Performance Enhancement, PLoS One, 2010, 5(6), 1–11, DOI:10.1371/journal.pone.0011042.
- Z. Al Tanoury, J. F. Zimmerman, J. Rao, D. Sieiro, H. M. McNamara, T. Cherrier, A. Rodríguez-Delarosa, A. Hick, F. Bousson, C. Fugier, F. Marchiano, B. Habermann, J. Chal, A. P. Nesmith, S. Gapon, E. Wagner, V. A. Gupta, R. Bassel-Duby, E. N. Olson, A. E. Cohen, K. K. Parker and O. Pourquié, Prednisolone rescues Duchenne muscular dystrophy phenotypes in human pluripotent stem cell-derived skeletal muscle in vitro, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(28), 1–12, DOI:10.1073/pnas.2022960118/-/DCSupplemental.
- J. W. Santoso, X. Li, D. Gupta, G. C. Suh, E. Hendricks, S. Lin, S. Perry, J. K. Ichida, D. Dickman and M. L. McCain, Engineering skeletal muscle tissues with advanced maturity improves synapse formation with human induced pluripotent stem cell-derived motor neurons, APL Bioeng., 2021, 5(3), 036101, DOI:10.1063/5.0054984.
- P. Bajaj, B. Reddy, L. Millet, C. Wei, P. Zorlutuna, G. Bao and R. Bashir, Patterning the differentiation of C2C12 skeletal myoblasts, Integr. Biol., 2011, 3(9), 897, 10.1039/c1ib00058f.
- K. Shimizu, H. Kassai, Y. Kamei, K. Yamamoto, T. Nagashima, T. Maekawa, H. Akiyama and H. Honda, Alignment of Skeletal Muscle Cells Facilitates Acetylcholine Receptor Clustering and Neuromuscular Junction Formation with Co-Cultured Human iPSC-Derived Motor Neurons, Cells, 2022, 11(23), 3760, DOI:10.3390/cells11233760.
- I. Toral-Ojeda, G. Aldanondo, J. Lasa-Elgarresta, H. Lasa-Fernandez, C. Vesga-Castro, V. Mouly, A. L. de Munain and A. Vallejo-Illarramendi, A Novel Functional In Vitro Model That Recapitulates Human Muscle Disorders, in Muscle Cell and Tissue – Current Status of Research Field InTech, 2018, DOI:10.5772/intechopen.75903.
- H. S. Yang, B. Lee, J. H. Tsui, J. Macadangdang, S.-Y. Jang, S. G. Im and S.-H. Kim, Electroconductive Nanopatterned Substrates for Enhanced Myogenic Differentiation and Maturation, Adv. Healthcare Mater., 2015, 5(1), 137–145, DOI:10.1002/adhm.201500003.
- H. Kaji, T. Ishibashi, K. Nagamine, M. Kanzaki and M. Nishizawa, Electrically induced contraction of C2C12 myotubes cultured on a porous membrane-based substrate with muscle tissue-like stiffness, Biomaterials, 2010, 31(27), 6981–6986, DOI:10.1016/j.biomaterials.2010.05.071.
- I. Toral-Ojeda, G. Aldanondo, J. Lasa-Elgarresta, H. Lasa-Fernández, R. Fernández-Torrón, A. López de Munain and A. Vallejo-Illarramendi, Calpain 3 deficiency affects SERCA expression and function in the skeletal muscle, Expert Rev. Mol. Med., 2016, 18, e7, DOI:10.1017/erm.2016.9.
- V. Cerrada, I. García-Consuegra, J. Arenas and M. E. Gallardo, Creation of an iPSC-Based Skeletal Muscle Model of McArdle Disease Harbouring the Mutation c.2392T>C (p.Trp798Arg) in the PYGM Gene, Biomedicines, 2023, 11(9), 2434, DOI:10.3390/biomedicines11092434.
- J. Tarum, H. Degens, M. D. Turner, C. Stewart, C. Sale and L. Santos, Modelling Skeletal Muscle Ageing and Repair In Vitro, J. Tissue Eng. Regener. Med., 2023, 2023, 9802235, DOI:10.1155/2023/9802235.
- A. Bettadapur, G. C. Suh, N. A. Geisse, E. R. Wang, C. Hua, H. A. Huber, A. A. Viscio, J. Y. Kim, J. B. Strickland and M. L. McCain, Prolonged Culture of Aligned Skeletal Myotubes on Micromolded Gelatin Hydrogels, Sci. Rep., 2016, 6, 1–14, DOI:10.1038/srep28855.
- C. Vesga-Castro, J. Aldazabal, A. Vallejo-Illarramendi and J. Paredes, Contractile force assessment methods for in vitro skeletal muscle tissues, eLife, 2022, 11, e77204, DOI:10.7554/eLife.77204.
- L. Altomare, M. Riehle, N. Gadegaard, M. Tanzi and S. Farè, Microcontact Printing of Fibronectin on a Biodegradable Polymeric Surface for Skeletal Muscle Cell Orientation, Int. J. Artif. Organs, 2010, 33(8), 535–543, DOI:10.1177/039139881003300804.
- R. M. Duffy, Y. Sun and A. W. Feinberg, Understanding the Role of ECM Protein Composition and Geometric Micropatterning for Engineering Human Skeletal Muscle, Ann. Biomed. Eng., 2016, 44(6), 2076–2089, DOI:10.1007/s10439-016-1592-8.
- J. Young, Y. Margaron, M. Fernandes, E. Duchemin-Pelletier, J. Michaud, M. Flaender, O. Lorintiu, S. Degot and P. Poydenot, MyoScreen, a High-Throughput Phenotypic Screening Platform Enabling Muscle Drug Discovery, SLAS Discovery, 2018, 23(8), 1–17, DOI:10.1177/2472555218761102.
- J. S. Choi, S. J. Lee, G. J. Christ, A. Atala and J. J. Yoo, The influence of electrospun aligned poly(ε-caprolactone)/collagen nanofiber meshes on the formation of self-aligned skeletal muscle myotubes, Biomaterials, 2008, 29(19), 2899–2906, DOI:10.1016/j.biomaterials.2008.03.031.
- I. Kim, S. S. Lee, A. Ghosh, S. J. Ferguson and O. Bar-Nur, A Self-Renewing Biomimetic Skeletal Muscle Construct Engineered using Induced Myogenic Progenitor Cells, Adv. Funct. Mater., 2024, 34(1), 2300571, DOI:10.1002/adfm.202300571.
- J. Brunetti, S. Koenig, A. Monnier and M. Frieden, Nanopattern surface improves cultured human myotube maturation, Skeletal Muscle, 2021, 11(12), 1–14, DOI:10.1186/s13395-021-00268-3.
- V. Hosseini, S. Ahadian, S. Ostrovidov, G. Camci-Unal, S. Chen, H. Kaji, M. Ramalingam and A. Khademhosseini, Engineered Contractile Skeletal Muscle Tissue on a Microgrooved Methacrylated Gelatin Substrate, Tissue Eng., Part A, 2012, 18(23–24), 2453–2465, DOI:10.1089/ten.tea.2012.0181.
- M. T. Lam, S. Sim, X. Zhu and S. Takayama, The effect of continuous wavy micropatterns on silicone substrates on the alignment of skeletal muscle myoblasts and myotubes, Biomaterials, 2006, 27(24), 4340–4347, DOI:10.1016/j.biomaterials.2006.04.012.
- P. Molnar, W. Wang, A. Natarajan, J. W. Rumsey and J. J. Hickman, Photolithographic Patterning of C2C12 Myotubes using Vitronectin as Growth Substrate in Serum-Free Medium, Biotechnol. Prog., 2007, 23(1), 265–268, DOI:10.1021/bp060302q.
- K. Shimizu, H. Sasaki, H. Hida, H. Fujita, K. Obinata, M. Shikida and E. Nagamori, Assembly of skeletal muscle cells on a Si-MEMS device and their generative force measurement, Biomed. Microdevices, 2010, 12(2), 247–252, DOI:10.1007/s10544-009-9379-4.
- P.-Y. Wang, H.-T. Yu and W.-B. Tsai, Modulation of alignment and differentiation of skeletal myoblasts by submicron ridges/grooves surface structure, Biotechnol. Bioeng., 2010, 106(2), 285–294, DOI:10.1002/bit.22697.
- J. Gingras, R. M. Rioux, D. Cuvelier, N. A. Geisse, J. W. Lichtman, G. M. Whitesides, L. Mahadevan and J. R. Sanes, Controlling the Orientation and Synaptic Differentiation of Myotubes with Micropatterned Substrates, Biophys. J., 2009, 97(10), 2771–2779, DOI:10.1016/j.bpj.2009.08.038.
- J. L. Charest, A. J. García and W. P. King, Myoblast alignment and differentiation on cell culture substrates with microscale topography and model chemistries, Biomaterials, 2007, 28(13), 2202–2210, DOI:10.1016/j.biomaterials.2007.01.020.
- L. T. Denes, L. A. Riley, J. R. Mijares, J. D. Arboleda, K. McKee, K. A. Esser and E. T. Wang, Culturing C2C12 myotubes on micromolded gelatin hydrogels accelerates myotube maturation, Skeletal Muscle, 2019, 9(1), 17, DOI:10.1186/s13395-019-0203-4.
- S. Zatti, A. Zoso, E. Serena, C. Luni, E. Cimetta and N. Elvassore, Micropatterning Topology on Soft Substrates Affects Myoblast Proliferation and Differentiation, Langmuir, 2012, 28(5), 2718–2726, DOI:10.1021/la204776e.
- M. Juhas and N. Bursac, Roles of adherent myogenic cells and dynamic culture in engineered muscle function and maintenance of satellite cells, Biomaterials, 2014, 35(35), 9438–9446, DOI:10.1016/j.biomaterials.2014.07.035.
- T. Uchimura, T. Asano, T. Nakata, A. Hotta and H. Sakurai, A muscle fatigue-like contractile decline was recapitulated using skeletal myotubes from Duchenne muscular dystrophy patient-derived iPSCs, Cell Rep. Med., 2021, 2(6), 1–16, DOI:10.1016/j.xcrm.2021.100298.
- K. Yoshioka, A. Ito, M. Arifuzzaman, T. Yoshigai, F. Fan, K. I. Sato, K. Shimizu, Y. Kawabe and M. Kamihira, Miniaturized skeletal muscle tissue fabrication for measuring contractile activity, J. Biosci. Bioeng., 2021, 131(4), 434–441, DOI:10.1016/j.jbiosc.2020.11.014.
- A. Badu-Mensah, X. Guo, C. W. McAleer, J. W. Rumsey and J. J. Hickman, Functional skeletal muscle model derived from SOD1-mutant ALS patient iPSCs recapitulates hallmarks of disease progression, Sci. Rep., 2020, 10(1), 14302, DOI:10.1038/s41598-020-70510-3.
- H. Hamaguchi, T. S. Matsui, S. Deguchi, Y. Furuichi, N. L. Fujii and Y. Manabe, Establishment of a system evaluating the contractile force of electrically stimulated myotubes from wrinkles formed on elastic substrate, Sci. Rep., 2022, 12, 13818, DOI:10.1038/s41598-022-17548-7.
- D. Huh, H. J. Kim, J. P. Fraser, D. E. Shea, M. Khan, A. Bahinski, G. A. Hamilton and D. E. Ingber, Microfabrication of human organs-on-chips, Nat. Protoc., 2013, 8(11), 2135–2157, DOI:10.1038/nprot.2013.137.
- J. Lasa-Elgarresta, L. Mosqueira-Martín, K. González-Imaz, P. Marco-Moreno, G. Gerenu, K. Mamchaoui, V. Mouly, A. López de Munain and A. Vallejo-Illarramendi, Targeting the Ubiquitin-Proteasome System in Limb-Girdle Muscular Dystrophy With CAPN3 Mutations, Front. Cell Dev. Biol., 2022, 10, 822563, DOI:10.3389/fcell.2022.822563.
- V. Mudera, A. S. T. Smith, M. A. Brady and M. P. Lewis, The effect of cell density on the maturation and contractile ability of muscle derived cells in a 3D tissue-engineered skeletal muscle model and determination of the cellular and mechanical stimuli required for the synthesis of a postural phenotype, J. Cell. Physiol., 2010, 225(3), 646–653, DOI:10.1002/jcp.22271.
- L. Sala, B. J. van Meer, L. G. J. Tertoolen, J. Bakkers, M. Bellin, R. P. Davis, C. Denning, M. A. E. Dieben, T. Eschenhagen, E. Giacomelli, C. Grandela, A. Hansen, E. R. Holman, M. R. M. Jongbloed, S. M. Kamel, C. D. Koopman, Q. Lachaud, I. Mannhardt, M. P. H. Mol, D. Mosqueira, V. V. Orlova, R. Passier, M. C. Ribeiro, U. Saleem, G. L. Smith, F. L. Burton and C. L. Mummery, Musclemotion, Circ. Res., 2018, 122(3), 5–16, DOI:10.1161/CIRCRESAHA.117.312067.
- C. E. Torgan and M. P. Daniels, Regulation of Myosin Heavy Chain Expression during Rat Skeletal Muscle Development In Vitro, Mol. Biol. Cell, 2001, 12, 1499–1508, DOI:10.1091/mbc.12.5.1499.
- K. Fujiwara, R. Yamamoto, T. Kubota, A. Tazumi, T. Sabuta, M. P. Takahashi and H. Sakurai, Mature Myotubes Generated From Human-Induced Pluripotent Stem Cells Without Forced Gene Expression, Front. Cell Dev. Biol., 2022, 10, 886879, DOI:10.3389/fcell.2022.886879.
- K. J. M. Boonen, K. Y. Rosaria-Chak, F. P. T. Baaijens, D. W. J. Van Der Schaft and M. J. Post, Essential environmental cues from the satellite cell niche: Optimizing proliferation and differentiation, Am. J. Physiol., 2009, 296(6), 1338–1345, DOI:10.1152/ajpcell.00015.2009.
- K. Nagamine, T. Kawashima, T. Ishibashi, H. Kaji, M. Kanzaki and M. Nishizawa, Micropatterning contractile C2C12 myotubes embedded in a fibrin gel, Biotechnol. Bioeng., 2010, 105(6), 1161–1167, DOI:10.1002/bit.22636.
- Y. Sun, R. Duffy, A. Lee and A. W. Feinberg, Optimizing the structure and contractility of engineered skeletal muscle thin films, Acta Biomater., 2013, 9(8), 7885–7894, DOI:10.1016/j.actbio.2013.04.036.
- J. M. Aizpurua, J. I. Miranda, A. Irastorza, E. Torres, M. Eceiza, M. Sagartzazu-Aizpurua, P. Ferrón, G. Aldanondo, H. Lasa-Fernández, P. Marco-Moreno, N. Dadie, A. López de Munain and A. Vallejo-Illarramendi, Discovery of a novel family of FKBP12 “reshapers” and their use as calcium modulators in skeletal muscle under nitro-oxidative stress, Eur. J. Med. Chem., 2021, 213, 113160, DOI:10.1016/j.ejmech.2021.113160.
- R. G. Dennis and P. E. Kosnik, Excitability and isometric contractile properties of mammalian skeletal muscle constructs engineered in vitro, In Vitro Cell. Dev. Biol.: Anim., 2000, 36(5), 327–335, DOI:10.1290/1071-2690(2000)036<0327:EAICPO>2.0.CO;2.
- C. A. Chapotte-Baldacci, C. Cognard, P. Bois, A. Chatelier and S. Sebille, Handling a mature calcium signature through optogenetics improves the differentiation of primary murine myotubes, Cell Calcium, 2022, 103, 102546, DOI:10.1016/j.ceca.2022.102546.
- Y. Takayama, A. Wagatsuma, T. Hoshino and K. Mabuchi, Simple micropatterning method for enhancing fusion efficiency and responsiveness to electrical stimulation of C2C12 myotubes, Biotechnol. Prog., 2015, 31(1), 220–225, DOI:10.1002/btpr.2003.
- T. Nair, J. T. Symanowski and H. M. Gach, Comparison of complex permittivities of isotonic colloids containing single-wall carbon nanotubes of varying chirality, Bioelectromagnetics, 2012, 33(2), 134–146, DOI:10.1002/bem.20689.
- H. Shivashankar, A. M. Kevin, S. B. S. Manohar and S. M. Kulkarni, Investigation on dielectric properties of PDMS based nanocomposites, Phys. B, 2021, 602, 412357, DOI:10.1016/j.physb.2020.412357.
- G. Liu, Y. Chen, M. Gong, X. Liu, Z.-K. Cui, Q. Pei, J. Gu, C. Huang and Q. Zhuang, Enhanced dielectric performance of PDMS-based three-phase percolative nanocomposite films incorporating a high dielectric constant ceramic and conductive multi-walled carbon nanotubes, J. Mater. Chem. C, 2018, 6(40), 10829–10837, 10.1039/C8TC03868F.
- C. S. Cheng, Y. El-Abd, K. Bui, Y.-E. Hyun, R. H. Hughes, W. E. Kraus and G. A. Truskey, Conditions that promote primary human skeletal myoblast culture and muscle differentiation in vitro, Am. J. Physiol., 2014, 306(4), C385–C395, DOI:10.1152/ajpcell.00179.2013.
- N. Imbert, C. Cognard, G. Duport, C. Gulllou and G. Raymond, Abnormal calcium homeostasis in Duchenne muscular dystrophy myotubes contracting in vitro, Cell Calcium, 1995, 18(3), 177–186, DOI:10.1016/0143-4160(95)90062-4.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00442b |
| This journal is © The Royal Society of Chemistry 2024 |