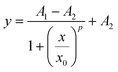Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Single-molecule microfluidic assay for prostate-specific antigen based on magnetic beads and upconversion nanoparticles†
Dorota
Sklenárová‡
a,
Antonín
Hlaváček‡
*b,
Jana
Křivánková
b,
Julian C.
Brandmeier
 ac,
Julie
Weisová
b,
Michal
Řiháček
d,
Hans H.
Gorris
ac,
Julie
Weisová
b,
Michal
Řiháček
d,
Hans H.
Gorris
 a,
Petr
Skládal
a and
Zdeněk
Farka
a,
Petr
Skládal
a and
Zdeněk
Farka
 *a
*a
aDepartment of Biochemistry, Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic. E-mail: farka@mail.muni.cz
bInstitute of Analytical Chemistry of the Czech Academy of Sciences, Veveří 97, 602 00 Brno, Czech Republic. E-mail: hlavacek@iach.cz
cInstitute of Analytical Chemistry, Chemo- and Biosensors, University of Regensburg, Universitätsstraße 31, 930 53 Regensburg, Germany
dDepartment of Laboratory Methods, Faculty of Medicine, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic
First published on 24th June 2024
Abstract
Early-stage diagnosis of prostatic carcinoma is essential for successful treatment and, thus, significant prognosis improvement. In laboratory practice, the standard non-invasive diagnostic approach is the immunochemical detection of the associated biomarker, prostate-specific antigen (PSA). Ultrasensitive detection of PSA is essential for both diagnostic and recurrence monitoring purposes. To achieve exceptional sensitivity, we have developed a microfluidic device with a flow-through cell for single-molecule analysis using photon-upconversion nanoparticles (UCNPs) as a detection label. For this purpose, magnetic microparticles (MBs) were first optimized for the capture and preconcentration of PSA and then used to implement a bead-based upconversion-linked immunoassay (ULISA) in the microfluidic device. The digital readout based on counting single nanoparticle-labeled PSA molecules on MBs enabled a detection limit of 1.04 pg mL−1 (36 fM) in 50% fetal bovine serum, which is an 11-fold improvement over the respective analog MB-based ULISA. The microfluidic technique conferred several other advantages, such as easy implementation and the potential for achieving high-throughput analysis. Finally, it was proven that the microfluidic setup is suitable for clinical sample analysis, showing a good correlation with a reference electrochemiluminescence assay (recovery rates between 97% and 105%).
Introduction
The sensitive and reliable detection of clinically relevant biomarkers is essential for disease diagnosis. Prostate-specific antigen (PSA) is the most important biomarker of prostatic carcinoma, which is the leading cause of death within the male population worldwide caused by oncologic diseases.1 Due to their robustness and widespread use, immunoassays are considered the gold standard of diagnostic methods.2,3 The most common immunoassay technique is the enzyme-linked immunosorbent assay (ELISA), which uses a microtiter plate (MTP) as the solid phase for biomolecule immobilization, together with an enzyme-based signal generation element.4,5 However, as biologically active compounds and various disease biomarkers may be present in exceptionally low levels,6,7 the sensitivity of conventional ELISA is often insufficient for early-stage disease diagnosis.8,9 Thus, it is necessary to search for novel solid phases, labels, and detection techniques with the potential to improve the limit of detection (LOD) within the assays.10Magnetic microparticles (MBs) represent a promising alternative solid phase in immunoassays. They typically consist of one or more magnetite (Fe3O4) nanocores surrounded by a polymer layer. The resulting core–shell structure is usually larger than 1 μm in diameter and has superparamagnetic properties.11 MBs additionally provide a large surface-to-volume ratio, low toxicity, and biocompatibility.12 Surface-modified MBs have found a plethora of applications, including drug delivery, cancer therapy,13,14 and immunoassays.15–17 For immunoassays, the MBs are commonly conjugated with antibodies, allowing for the magnetic preconcentration and separation of the MB-bound immunocomplexes from sample matrix and unbound reagents.18
Different kinds of nanomaterials can be used as advanced immunoassay labels. In particular, photon-upconversion nanoparticles (UCNPs) exhibiting anti-Stokes luminescence (i.e., the ability to convert low-energy NIR excitation to higher energy emission)19,20 are gaining popularity, as they enable sensitive detection without optical background interferences.21 As UCNPs are typically synthesized in organic solvents, a surface modification and conjugation with biorecognition molecules, such as antibodies or streptavidin, are required to render them dispersible in aqueous media and make them available as immunoassays labels.22–24 Highly sensitive UCNP-based assays for the detection of various analytes were previously reported.25 Digital immunoassays based on detecting and counting individual immunocomplexes are a promising way to achieve exceptional sensitivity and further improve assay performance.26 For example, we used epiluminescence microscopy to count individual UCNP-labeled immunocomplexes on the MTP surface.19
While MTP-based immunoassays typically require relatively large sample volumes and offer only limited throughput, modern miniaturization techniques reduce the sample consumption and may also improve other assay parameters (e.g., sensitivity, signal-to-background (S/B) ratio, and working range).27,28 In particular, microfluidics only requires minimal amounts of sample and enables rapid sample processing with a high throughput. The fundamental principle of microfluidics lies in the manipulation of fluids in channels of sub-millimeter dimensions, ranging from hundreds to tens of micrometers,29 allowing for the manipulation of exceptionally small volumes (10−9 to 10−18 L).30 There are several reports of microfluidic devices for immunoanalysis based on conventional organic fluorophore labels in the literature.31–33 Microfluidics-based methods for magnetic bead-based immunoassays have also been previously reported.34–36 In all cases, however, the immunoreaction took place on a chip, which required applying an external magnetic force during the experiments and prolonged the readout. Thus, in order to detect single analyte molecules as labeled complexes while maintaining the advantages of microfluidics (e.g., high throughput, effective manipulation, low sample consumption), it is necessary to develop simple yet effective techniques such as droplet microfluidics for digital detection.37,38
We have previously reported the use of MBs in immunoassays for the detection of PSA based on various labels to fully optimize the MB-based upconversion-linked immunosorbent assay (ULISA) and to explore the possibilities for sensitivity enhancement.10 Moreover, we have employed massively parallel spectroscopy of UCNPs for detecting single analyte molecules free in dispersion.39 To the best of our knowledge, however, no studies have been conducted on the digital detection combining UCNP labels with MBs.
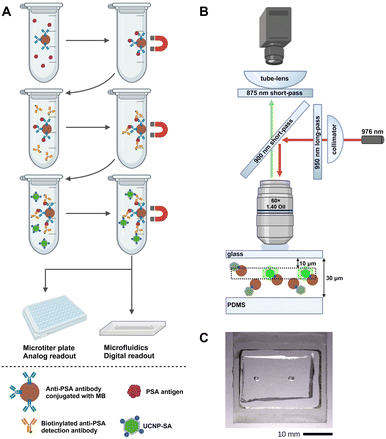
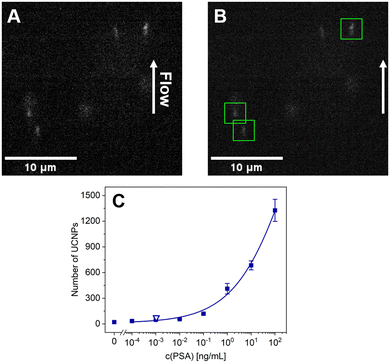
In this work, we have developed an MB-based ULISA for the single-molecule detection of PSA utilizing a flow-through microfluidic device. The immunoassay was performed in microtubes to enable sample introduction into the device (Fig. 1A). The samples were introduced to a microfluidic chip with a flow-through cell and individual immunocomplexes were counted (Fig. 1B), utilizing a microfluidic chip with a flow-through cell (Fig. 1C).
Following our previous report on detecting single UCNPs in a free dispersion,39 the method developed here allows for counting single labels on the freely dispersed MBs. Moreover, it enables the single-molecule readout in liquid, the native environment of proteins, eliminating the risk of possible structural changes or aggregation, which may cause an increase in non-specific binding and complicate the evaluation of results. Thus, while maintaining simplicity in terms of operation, the microfluidic setup allowed for enhancing the assay sensitivity further.
Materials and methods
Chemicals and reagents
Fetal bovine serum, sodium borate decahydrate, and tris(hydroxymethyl)aminomethane were purchased from Merck (USA). SuperBlock TBS, streptavidin (SA), Tween 20, and tosyl-activated Dynabeads MyOne (diameter of 1 μm) were obtained from Thermo Fischer Scientific (USA). PSA antigen (ab78528) and mouse anti-PSA monoclonal antibody (ab403) were purchased from Abcam (UK). Goat anti-PSA polyclonal antibody (AF1344) and biotinylated goat anti-PSA polyclonal antibody (BAF1344) were obtained from R&D Systems (USA). Human serum albumin (HSA) and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (USA). Carcinoembryonic antigen (CEA) was obtained from HyTest (Finland). All other chemicals were purchased in the highest available quality from either Penta (Czech Republic) or Carl Roth (Germany). Clinical samples of human serum were provided with written consent from all participants; the study was approved by the Ethics Committee of the University Hospital Brno (project number 24/22, document number 04-160322/EK). The reference data on PSA concentrations were obtained by the Elecsys electrochemiluminescence immunoassay analyzer (Roche Diagnostics, Germany).The solutions used included coating buffer (50 mM NaHCO3/Na2CO3, 0.05% NaN3; pH 9.6), phosphate buffer (50 mM NaH2PO4/Na2HPO4; pH 7.4), phosphate-buffered saline (PBS; phosphate buffer with 150 mM NaCl; pH 7.4), washing buffer (50 mM Tris, 0.05% NaN3, 0.05% Tween 20, 1 mM KF; pH 7.5), and assay buffer (50 mM NaH2PO4/Na2HPO4, 10% SuperBlock, 0.05% NaN3, 0.01% Tween 20, 150 mM NaCl, 1 mM KF; pH 7.5). All buffers were prepared using deionized water and filtered using a 0.22 μm membrane (Millipore Express Plus, Merck, Germany).
The protocols for the synthesis and characterization of UCNPs (NaYF4:Yb3+,Er3+) and their bioconjugates with streptavidin and the anti-PSA polyclonal antibody, conjugation of MBs with the anti-PSA monoclonal antibody, and the procedure of MTP-based ULISA for the analog readout are provided in the ESI.†
Preparation of samples for MB-based ULISA
During the sample preparation for both the analog and the digital MB-based experiments, the entire immunocomplex formation was carried out in microtubes. After each step, the samples were incubated for 1 h at room temperature under shaking on a MultiVortex V-32 (Biosan, Latvia). Between the individual steps, the microtubes were placed on magnetic holders to separate the formed MB-bound immunocomplexes from unbound reagents and the sample matrix. Three washing steps consisting of 3 min of magnetic separation, supernatant removal and addition of washing buffer were followed by the final supernatant removal and the addition of the next reagent solution. To compare the data from the analog readout to those obtained with the microfluidic device, two identical sets of samples were prepared to carry out both assays.First, serial dilutions of the PSA antigen (10−4 to 102 ng mL−1) in the volume of 450 μL were prepared in 50% bovine serum in assay buffer. The total sample volume was enough for approximately 10 to 20 microfluidic measurements, depending on the setup parameters (utilizing 20 to 40 μL of sample per measurement); however, a larger volume was used in the sample preparation steps to enable easier manipulation with the MBs. The procedure for the clinical sample analysis was identical to the one carried out to obtain the calibration curve. The spiked serum was mixed with 50 μL of the MB-ab403 bioconjugate (50 μg mL−1) to capture the PSA from the sample. In the next step, the BAF1344 biotinylated anti-PSA polyclonal antibody (500 μL, 0.25 μg mL−1) was added to each microtube. Then, the UCNP-SA bioconjugates (500 μL, 2 μg mL−1) were added, followed by the resuspension of the MB-bound immunocomplexes in 500 μL of assay buffer. The samples for the microfluidic measurements were stored at 4 °C until the readout was performed. The second set was immediately transferred to a non-binding MTP (Greiner Bio-One, Austria) and, after short shaking at 1000 rpm, left to dry at room temperature.
It was not necessary to remove the MBs not carrying PSA and UCNPs from the solution, as the measured signal was provided only by the specifically bound UCNP-SA label. The MBs not carrying the labeled immunocomplex did not display any luminescence and, therefore, did not disrupt the obtained data. Furthermore, as the microfluidic setup allowed for the specific counting of the immunocomplex-bound UCNPs, no further purification of the MBs during the sample preparation was necessary.
Upconversion luminescence scanning in MTPs
After the drying, both the MTP- and MB-based ULISA plates were placed in an upconversion scanner Upcon S-Pro (Labrox, Finland) and the upconversion luminescence was measured in each well using the following optical setup: excitation filter at 976 ± 60 nm, emission filter at 540 ± 30 nm, D800 dichroic mirror, excitation spot size of 4 mm, excitation time of 500 ms, and 100% laser power. For the MTP-based ULISA, the wells were raster-scanned each by 64 points (8 × 8 grid), with the distance between the first and the last row of 2.1 mm. For the MB-based ULISA, the MTP wells were raster-scanned each by 36 points (6 × 6 grid), with the distance between the first and the last row of 4.5 mm. The truncated average was calculated by excluding 25% of the lowest and highest values. In the analysis, the average upconversion luminescence signals and the standard deviations (error bars in the graphs) were calculated from 3 MTP wells.Preparation of the microfluidic device
The microfluidic channel was molded of polydimethylsiloxane (PDMS) with a rectangular cross-section of 200 μm in width and 30 μm in depth. The PDMS mold was attached to a glass substrate (thickness of 170 μm). Both the capillary and the microfluidic chip were thoroughly washed with deionized water between the application of individual samples. To maintain the flow of the sample through the capillary and the chip, a pressure of 1 kPa was applied directly to the cuvette via a custom-designed pressure controller.Single-molecule readout utilizing the microfluidic device
For the microfluidic measurements, the microtubes were stored at 4 °C and transferred to room temperature directly before use. The MB mixtures were transferred into a custom-made cuvette attached to a shaker. The samples were continuously mixed to ensure uniform distribution of the MBs throughout the measurement. The samples were then transferred from the cuvette into a capillary (diameter of 50 μm) attached to the output of the cuvette on one side and the input of the microfluidic chip on the other side.For each measurement, the microscope objective CFI Plan Apo Lambda (60× magnification, NA 1.40; Nikon, Japan) was focused 10 μm below the substrate glass slide of the chip. A 976 nm laser (10 W; Roithner Lasertechnik, Austria) with a 950 nm long-pass filter served as the excitation source; the beam was reflected to the microscope objective via a 900 nm short-pass dichroic mirror. The power output measured from the microscope objective was set to 1.3 W, resulting in the average excitation intensity of 9.4 kW cm−2 in the detection point, which was characterized as reported previously.39 An 875 nm short-pass filter was inserted before the microscope tube lens, enabling the detection of both the green and the red emission from the UCNPs as a single spot. The images of UCNPs from the microfluidic chip were taken by a Zyla 5.5 sCMOS camera (Andor Technology – Oxford Instruments, UK) using the sensor area of 1024 px × 1024 px (imaging area of 111 μm × 111 μm in the sample plane). As the imaging area did not fully cover the flow region, the resulting number of UCNPs represents an estimated number.
The MicroManager software (Laboratory for Optical and Computational Instrumentation, University of Wisconsin, USA) was used to control the camera. With an exposure time of 5 ms, 10 images were recorded per second, and three sets of 500 frames were recorded for each sample. For counting individual immunocomplexes, the images of single UCNPs were localized using a U-net convolutional neural network,40 which was trained and tested as reported previously with minor modifications (Fig. S1†).39 In the analysis, the average numbers of UCNPs and standard deviations (error bars in the graphs) were calculated from 3 independent measurements.
Data evaluation
OriginPro 2022 (OriginLab, USA) was utilized for the data evaluation. The calibration curves were plotted into scatter graphs and fitted with a 4-parameter logistic function:
The LODs were estimated from the regression curves as the concentrations corresponding to the sum of the background value obtained by the logistic fit (A1) and three times the standard deviation of the blank (s0):
| y(LOD) = A1 + 3s0 |
Results and discussion
Characterization of the UCNPs and the MBs
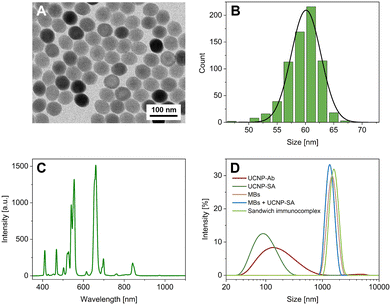
UCNPs (NaYF4:Yb3+,Er3+) modified with streptavidin via an alkyne-PEG-neridronate linker served as labels for both the MTP- and MB-based ULISA assays. The UCNPs were visualized using transmission electron microscopy (Fig. 2A and B), and their emission spectrum was measured (Fig. 2C). Moreover, the conjugates were characterized via dynamic light scattering (DLS; Fig. 2D and S2†) and nanoparticle tracking analysis (NTA; Fig. S3†). In the DLS, the size distribution based on the intensity of scattered light showed that the most represented nanoparticle size was 141.8 ± 11.2 nm for the UCNP-Ab bioconjugate and 91.3 ± 0.2 nm for the UCNP-SA bioconjugate. The polydispersity index (PdI) has reached the values of 0.26 ± 0.02 and 0.17 ± 0.01 for the UCNP-Ab and UCNP-SA, respectively. The PdI values for monodisperse systems lie within the range of 0.05–0.1, while polydisperse systems reach values of 0.4 and higher.41 Thus, both UCNP bioconjugates were on the verge of mono- and polydispersity. The results of the UCNP-Ab analysis revealed a second peak at higher values (∼4 μm), showing the presence of some aggregates in the sample. The UCNP-SA showed only a single size fraction and a lower PdI, confirming its suitability as an immunoassay label.Utilizing the DLS software, the number-based distribution was also evaluated (Fig. S2†). The maximum in the number distribution was found at 50.7 ± 5.9 nm and 43.8 ± 0.2 nm for UCNP-Ab and UCNP-SA, respectively. The data showed only one size fraction for both bioconjugates, proving that the second peak in the UCNP-Ab intensity distribution represented only a small number of strongly scattering particles.
Furthermore, MBs in various environments were characterized via DLS. The sizes determined from the intensity-based distribution (Fig. 2D) were 1357 ± 206 nm for unbound MBs, 1481 ± 156 nm for the mixture of MBs and UCNP-SA, and 1505 ± 143 nm for the full sandwich immunocomplex of MBs with PSA and UNCPs. Number-based distribution (Fig. S2†) revealed sizes of 1329 ± 193 nm, 1446 ± 142 nm, and 1478 ± 140 nm, respectively. It must be noted that relatively large standard deviations were obtained, caused by the sedimentation of the relatively large MBs during the measurement. Nevertheless, the comparable size values confirm that the presence of PSA and UCNP-SA did not cause any aggregation of the MB dispersion.
Analog readout of the MTP- and MB-based ULISA
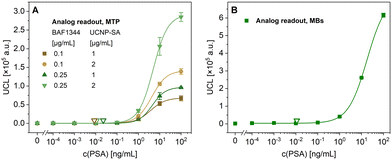
To confirm the functionality of the bioconjugates, PSA in 50% bovine serum was first detected in an MTP-based assay, testing different concentrations of biotinylated anti-PSA polyclonal antibody (0.1 and 0.25 μg mL−1) and UCNP-SA conjugate (1 and 2 μg mL−1) (Fig. 3A). The optimal performance was observed when using 0.25 μg mL−1 of the biotinylated anti-PSA antibody and 2 μg mL−1 of UCNP-SA, resulting in an LOD of 11 pg mL−1 and a working range from 1.7 to 14.8 ng mL−1. The S/B ratio was 625, confirming minimal levels of non-specific binding and, thus, exceptionally low background.The MB-based ULISA experiments were optimized regarding the MB concentration (Fig. S4A†), the ratio of MBs and UCNPs (Fig. S4B†), and the evaluation of assay performance in the whole concentration range of fetal bovine serum (Fig. S4C†). The data showed that the MB concentration of 50 μg mL−1 combined with 2 μg mL−1 of UCNP-SA was optimal for the sensitive detection of PSA and that serum did not significantly affect the assay performance. The assay specificity was further verified by studying the effect of three other biomolecules, including human serum albumin (HSA), bovine serum albumin (BSA), and carcinoembryonic antigen (CEA), on the MB-based ULISA (Fig. S5†). The results showed no cross-reactivity with the other biomolecules, demonstrating the exceptional specificity of the assay.
Afterward, the MB-based ULISA was optimized for immunocomplex formation in microtubes, and the data were analyzed utilizing the analog readout mode (Fig. 3B). The assay reached an LOD of 11 pg mL−1 and a working range from 4.5 to 71.1 ng mL−1. The results showed a minimal background signal in 50% serum, with an S/B ratio of 353. The assay parameters were comparable to the MTP-based ULISA, though the S/B ratio was 1.8 times lower. The possible explanation lies within the use of the MBs, which yielded a higher maximum signal, albeit also a higher background. In our previously published study,10 the LOD achieved by an MB-based ULISA of 3.87 pg mL−1 was comparable, albeit slightly lower. In the cited paper, the assay was performed within the MTP wells with automatic washing, whereas here, the assay format revolved around the use of microtubes with manual washing in order to introduce the samples into the microfluidic device effectively. The difference in the assay procedures, as well as the lower MB concentration utilized here to prevent the microfluidic channel from clogging, may also have resulted in the variations.
Optimizations of the microfluidic setup
To obtain reliable data utilizing the microfluidic device, several optimization steps were required, which were carried out as follows. The first microfluidic measurements were conducted using a capillary with a diameter of 30 μm and 80 μg mL−1 of MBs. This capillary size, however, was too small such that the MB-bound immunocomplexes started to clog the capillary and did not reach the flow cell (data not shown). After increasing the capillary diameter to 50 μm and lowering the MB concentration to 50 μg mL−1 to improve capillary flow, it was possible to continue with the optimization steps.The optimization measurements (Fig. S6†) were performed at the UCNP-SA concentration of 2 μg mL−1. However, in this experiment, the MBs carrying higher PSA concentrations (>1 ng mL−1) tended to adsorb to the PDMS surface of the flow cell. Therefore, the laser beam was not able to penetrate through the otherwise transparent PDMS, and the accumulated energy absorbed by the MB layer damaged parts of the microfluidic chip due to the excess heat. Consequently, the obtained data were inconsistent, with large standard deviations because the flow cell did not function appropriately, resulting in an LOD of 40 pg mL−1 and a narrow working range from 0.07 to 0.9 ng mL−1. Moreover, the average number of UCNPs was below 120, even for the highest PSA concentration, resulting in a low S/B ratio of 12. Albeit higher UCNP-SA concentrations could further improve the LOD, they were not tested, as it was previously reported that individual particle counting in an MTP-based single-molecule ULISA was rather tricky at high label concentrations.9 Moreover, a high label concentration could lead to aggregate formation within the microfluidic channel and thus decrease the reliability of the method.
In order to confirm that the chip damage was caused mainly by the MBs and not by the UCNP-SA label, a direct bioconjugate of the UCNPs with the AF1344 anti-PSA polyclonal antibody (UCNP-Ab) was prepared. After optimizing the UCNP-Ab concentration in an MTP-based assay (Fig. S7†), the concentration of 12 μg mL−1 was used for the microfluidic setup. The direct conjugate allowed for a one-step detection without using a biotinylated antibody, though its most effective concentration was 6 times higher than the effective concentration of the SA-UCNP label. The lower required label concentration in the case of UCNP-SA compared to UCNP-Ab is connected with the higher affinity of the streptavidin-biotin interaction compared to the weaker antibody–antigen interaction.9
To further confirm that the chip damage was induced by the MBs and not the UCNPs, absorption spectra of both dispersions in concentrations identical to those utilized in the microfluidic measurements were analyzed using the Specord 210+ spectrometer (Analytik Jena, Germany). The results (Fig. S10†) showed that the absorbance of UCNPs under NIR wavelength of 980 nm was negligible (value close to zero) in comparison to that of the MBs (0.35 a.u.), confirming that MBs were responsible for absorbing the excitation light leading to the thermal damage of the chip.
More intense mixing was introduced to prevent the MBs from sedimenting within the sample mixture. Even though the first measurements (Fig. S8A†) showed improved standard deviations, the resulting data were inconsistent. The relatively high background led to the S/B of 3.3, a very narrow working range of 13 to 21 ng mL−1, and a relatively high LOD of 13.0 ng mL−1. As the unfavorable results may have been caused by the high UCNP label concentration, it was lowered to 4 μg mL−1 for the subsequent optimizations. Fig. S8B† shows an improvement in both the curve and the parameters, reaching the S/B ratio of 21, a working range from 2.6 to 22.8 ng mL−1, and an LOD of 0.39 ng mL−1. However, due to the relatively high LOD as well as the concentration of the UCNP-Ab compared to the UCNP-SA required for a successful assay outcome, it was concluded that the UCNP-Ab conjugate was not suitable for this setup.
Therefore, identical measurements were performed utilizing the UCNP-SA conjugate at the previously optimized concentration of 2 μg mL−1. During these measurements, the MBs did not adsorb to the chip surface; however, the obtained curves (Fig. S9A†) again showed a similar outcome concerning the large standard deviations. Upon further analysis of the camera record, it was observed that due to the relatively high pressure applied to the cuvette and the downward slope of the capillary, the motion of the MB-bound UCNPs through the flow cell was too fast, causing them to appear as elongated lines instead of spots (Fig. S9B†). This has rendered the AI calculator incapable of reliably counting the UCNP labels attached to the MBs. Therefore, for the subsequent experiments, the tilt of the capillary was adjusted, and an otherwise identical measurement utilizing the UCNP-SA conjugate was performed.
The viscosity of the samples in various concentrations did not affect the measurements, as the viscosity is given almost exclusively by the buffer composition. All experiments were carried out in the same buffer, and the sample matrix – possibly causing viscosity changes – was washed out in the previous assay steps. Therefore, the viscosity changes can be neglected in our experimental setting. Moreover, the PSA concentrations in the samples were generally low.
The optimal flow rate was approximately 1 μm per 5 ms of exposure time, corresponding to 200 μm s−1. This flow rate was set by applying a pressure of 1 kPa to the cuvette. With a cross-section of the microfluidic channel of 30 μm × 200 μm, this is approximately 0.0012 μL s−1 (neglecting the slower liquid flow around the microfluidic channel walls in this approximation).
Optimized digital assay based on microfluidic device
The optimized assay conditions and measurement parameters were implemented in microfluidic PSA detection to validate the reliability and functionality of the setup. The obtained calibration curve (Fig. 4) has shown a trend similar to the one achieved with the analog readout of the MB-ULISA. With the digital microfluidics-based measurement, the data were not dependent on integrated signal but rather on counting single UCNPs, corresponding to individual sandwich immunocomplexes. Thus, the aggregate influence was effectively eliminated, consequently yielding a lower background. The background signal reduction is beneficial for low analyte concentrations, albeit analog readout may be more suitable for higher concentrations.19The 4-parameter logistic fit of the digital readout utilizing the microfluidic device (Fig. 4C) resulted in a low LOD of 1.04 pg mL−1 (36 fM). The obtained value marks an 11-fold improvement compared to the analog readout, thus significantly enhancing the assay sensitivity. The recurrence of prostate cancer may be indicated by an increase of PSA levels within an exceptionally narrow range of low concentrations (0.1–0.2 ng mL−1), often challenging to detect via conventional methods.42 The LOD achieved by the microfluidics-based assay verifies its practical potential for disease recurrence monitoring, which was later demonstrated via clinical sample analysis. The working range of the assay is not discussed here due to the absence of saturation in the curve, rendering the S/B ratio incomputable within the context of relevance.
As an alternative means of data evaluation, the results were plotted in a double logarithmic scale and fitted utilizing a linear function (Fig. S11†). However, as the plot did not yield any improvement (LOD of 12 pg mL−1), it was concluded that all the data would be fitted via the 4-parameter logistic function.
Due to the dependency of the digital readout precision on sampling error, the limited number of observed binding events (n) may not be representative of all binding events.9 This uncertainty can be calculated from the n and is known as Poisson noise. The Poisson noise values for all the concentrations are shown in Table S1.† The obtained data show that Poisson noise decreased with growing concentration, thus confirming the expected outcome. The calculated values were low and comparable to the experimental standard deviations within the curve. As reported previously, the Poisson noise may be further reduced by increasing the imaging area or analysis time, leading to a larger amount of data to be analyzed.9
It must be noted that at the concentration corresponding to the LOD, there is a relatively high surplus of PSA molecules over MBs. Considering the binding capacity, size, and weight of a single MB,43,44 each MB can bind approximately 6.0 × 104 to 7.8 × 104 antibody molecules. Therefore, with 50 μg mL−1 of MBs and a PSA concentration of 1.04 pg mL−1, there are approximately 400 PSA molecules per MB, which corresponds to a fractional antibody occupancy of 0.5–0.7%. Even though agreeing with the literature,45 the calculation, together with the experimental results, suggests relatively low efficiency of PSA binding and labeling. Therefore, the LOD might be further improved by increasing the immunochemical reaction efficiency, e.g., by optimizing the assay procedure or using different immunoreagents.
Analysis of clinical samples
Clinical samples of human serum were analyzed in order to demonstrate the practical potential of the microfluidics-based setup (Table 1). For the analysis, the samples were mixed with 50% bovine serum in assay buffer to improve the blocking and decrease the potential differences between the individual clinical samples. Furthermore, this allowed approaching the PSA levels near the EC50 value of the assay, as this concentration renders the analysis most reliable in terms of precision (Table S2†). An MB-based ULISA was performed, and the prepared samples were introduced to a non-binding MTP for analog readout and to the microfluidic device for digital readout. The data obtained via the analog readout show that the assay was marginally outside the 10% range of the reference electrochemiluminescence assay. The digital microfluidics assay was within 10% of the reference method, showing recovery rates between 97 and 105%, which was better than the analog readout. The results proved the reliability of the developed setup and confirmed its suitability for clinical applications.| Reference [ng mL−1] | Analog readout | Digital readout | ||
|---|---|---|---|---|
| Found [ng mL−1] | Recovery rate [%] | Found [ng mL−1] | Recovery rate [%] | |
| 2.08 | 1.82 ± 0.03 | 87.8 ± 1.6 | 2.0 ± 0.7 | 97 ± 33 |
| 9.23 | 10.0 ± 0.3 | 108.5 ± 2.8 | 8.9 ± 0.6 | 97 ± 6 |
| 30.64 | 34.1 ± 1.6 | 111 ± 5 | 32 ± 6 | 105 ± 20 |
Comparison of MB-based microfluidic ULISA with other assays for PSA
Table 2 summarizes the parameters of PSA assays reported in the literature. Several studies were conducted on the use of UCNPs for the single-molecule immunochemical detection of PSA. For instance, our previous work reported on an MTP-based ULISA for single-molecule detection utilizing widefield epiluminescence microscopy.19 The UCNP-Ab conjugates were used as a label in a sandwich assay format, reaching the LOD of 1.2 pg mL−1 in 25% serum. In the follow-up study, the binding kinetics were improved by using biotinylated antibody and UCNP-SA conjugates, obtaining an exceptionally low LOD of 23 fg mL−1 in 25% serum.9 However, the data acquisition was labor-intensive, with a low throughput and time-consuming evaluation. Additionally, MTP-based immunoassays and the corresponding readout techniques require rather large sample volumes in hundreds of microliters.| Technique | LOD | Working range | Ref. |
|---|---|---|---|
| Analog and digital MTP-based ULISA (UCNP-Ab) | 20.3 pg mL−1 | 0.1–10 ng mL−1 | 19 |
| 1.2 pg mL−1 | 0.01–1 ng mL−1 | ||
| Analog and digital MTP-based ULISA (UCNP-SA) | 380 fg mL−1 | 1–1000 pg mL−1 | 9 |
| 23 fg mL−1 | 0.1–100 pg mL−1 | ||
| Upconversion- and infrared absorption-based assay utilizing UCNP nanoprobes | 0.05 pg mL−1 | 0.00002–50 ng mL−1 | 46 |
| 15.5 pg mL−1 | 0.02–200 ng mL−1 | ||
| SERS-based droplet microfluidics | Below 0.1 ng mL−1 | 0.05–200 ng mL−1 | 47 |
| MB-based droplet microfluidics | 7 pg mL−1 | Not provided | 38 |
| Magnetic microfluidic chip with a magnetoimpedance sensor | 0.1 ng mL−1 | 0.1–20 ng mL−1 | 48 |
| Analog MTP-based ULISA | 10.8 pg mL−1 | 1.7–14.8 ng mL−1 | This work |
| Analog MB-based ULISA | 11.1 pg mL−1 | 4.5–71.1 ng mL−1 | This work |
| Digital MB-based ULISA | 1.04 pg mL−1 | 13–100 ng mL−1 | This work |
Another study focused on the development of nanoprobes based on silica-coated UCNPs bioconjugated with an anti-PSA antibody, utilizing upconversion luminescence and infrared absorption readout.46 The obtained LODs were 0.05 pg mL−1 for upconversion luminescence and 15.5 pg mL−1 for infrared absorption, with a sample consumption of 2 mL. Even though the LOD obtained via the upconversion luminescence measurement was lower than the one reported here, the sample consumption and reagent concentrations were significantly higher. Here, although the volume of the prepared samples was 500 μL to enable easier manipulation with the MBs, the sample volume required for one microfluidic measurement was only 20–40 μL, depending on the measurement setup. Moreover, the sample preparation procedure offers potential for future optimizations to minimize sample consumption. Additionally, unlike the previously published studies on the UCNP-based single-molecule detection,19,49 the setup developed here renders the detection simple in terms of operation while enabling detection in free dispersion of MB-bound immunocomplexes.
There are also reports on various microfluidic formats for PSA detection. Gao et al.47 developed a surface-enhanced Raman scattering (SERS)-based droplet microfluidics device, achieving an LOD below 0.1 ng mL−1 (no further specification available). Others used of carboxylated MBs in a quartz capillary-based droplet microfluidic chip.38 The MB-bound sandwich immunocomplexes were introduced to a droplet-generating microfluidic chip, where the enzyme label reacted with a fluorescent substrate, achieving an LOD of 7 pg mL−1. In a study utilizing an integrated magnetic microfluidic chip with a magnetoimpedance sensor,48 the authors described a chip containing a reaction platform for immunocomplex formation. As immunomagnetic beads were used as a label, the presence of magnets was required throughout the experiments. The beads modified with streptavidin and biotinylated anti-PSA antibody were trapped in a self-assembled film within the chip. The generated stray magnetic field was measured by a magnetoimpedance sensor, reaching the LOD of 0.1 ng mL−1.
The LOD achieved via the flow-through microfluidic device developed here (1.04 pg mL−1, 36 fM) represents a significant improvement in comparison to each of the discussed microfluidic studies. In addition to the simple experiment design, no need for magnetic separation during the measurements, and easy implementation of the device itself, the method demonstrates sensitivity enhancement utilizing the UCNP-based labels. The comparative literature analysis further reinforces the conclusion that the method developed in our study exhibits significant sensitivity and represents a foundation for further development of microfluidics-based immunoassays.
Conclusions
This study reports the development of a microfluidics-based device with a flow-through cell for single-molecule detection of PSA. By advancing our previous study for detecting single UCNPs,39 it was possible for the first time to perform single-molecule detection in a free dispersion of MB-bound immunocomplexes without the need for enzyme-based signal amplification. The microfluidic setup consisted of a PDMS chip with a flow-through cell, allowing for single-molecule microfluidic measurements of the immunocomplexes in an MB dispersion for the first time. An MB-based ULISA was developed and optimized for immunocomplex formation in microtubes. Then, two identical sets of samples were prepared to compare the analog readout utilizing an upconversion luminescence scanner and digital readout based on microfluidics. The analog readout of the MTP- and MB-based ULISA reached comparable LODs of 10.8 pg mL−1 and 11.1 pg mL−1, respectively. The LOD obtained in the microfluidic measurement reached the value of 1.04 pg mL−1 (36 fM). This result represents an 11-fold improvement compared to the analog readout, demonstrating the exceptional sensitivity of the microfluidics-based approach. Moreover, the setup only required small amounts of sample (20–40 μL per measurement) and allowed for rapid sample processing. With future optimizations, e.g., based on the fabrication of a multi-channel flow-through chip, the method holds significant potential for high-throughput analysis. Finally, clinical samples of human serum were analyzed, and the results were compared to those obtained by a standard electrochemiluminescence method. The data showed a good correlation in both the analog and the digital readout, proving the reliability of the microfluidic setup. The performance of the microfluidics-based assay allowed for increased sensitivity as well as suitability for clinical applications (e.g., disease recurrence monitoring), which are qualities essential for diagnosis and post-operation treatment.Data availability
The data supporting this article have been included as part of the ESI.† Additional data are available upon request.Author contributions
D. S.: methodology, investigation, manuscript writing – original draft, manuscript writing – review & editing, A. H.: development of instrumentation and software for microchip assay, nanoparticle synthesis, proposing the detection of single immunochemical complexes on MBs with UCNP labels, manuscript writing – review & editing; J. K.: microfluidic chip fabrication; J. C. B.: nanoparticle surface functionalization and bioconjugate preparation; J. W.: nanoparticle synthesis; M. R.: validation, formal analysis; H. H. G.: manuscript writing – review & editing; P. S.: manuscript writing – review & editing; Z. F.: supervision, conceptualization, methodology, manuscript writing – original draft, manuscript writing – review & editing.Conflicts of interest
The authors declare they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.Acknowledgements
We thank Ekaterina Makhneva for the help with the preparation of UCNP bioconjugates and optimizations of the MB-based ULISA. Table of contents graphic was created with https://BioRender.com. The work was funded by grant 21-03156S from the Czech Science Foundation. Dorota Sklenárová thanks Brno Ph.D. Talent Scholarship funded by the Brno City Municipality. The publication was produced with support for the long-term conceptual development of the research organizations from the Institute of Analytical Chemistry of the Czech Academy of Sciences (RVO: 68081715).Notes and references
- Y.-Q. Yang, Y.-C. Yang, M.-H. Liu and Y.-H. Chan, Anal. Chem., 2020, 92, 1493–1501 CrossRef CAS.
- G. Ward, A. Simpson, L. Boscato and P. E. Hickman, Clin. Biochem., 2017, 50, 1306–1311 CrossRef CAS PubMed.
- Z. Farka, T. Juřík, D. Kovář, L. Trnková and P. Skládal, Chem. Rev., 2017, 117, 9973–10042 CrossRef CAS PubMed.
- S. Gan and K. Patel, J. Invest. Dermatol., 2013, 133, e12 CrossRef CAS PubMed.
- S. Hosseini, P. Vázquez-Villegas, M. Rito-Palomares and S. O. Martinez-Chapa, in Enzyme-linked Immunosorbent Assay (ELISA): From A to Z, ed. S. Hosseini, P. Vázquez-Villegas, M. Rito-Palomares and S. O. Martinez-Chapa, Springer, Singapore, 2018, pp. 67–115 Search PubMed.
- X. Pei, B. Zhang, J. Tang, B. Liu, W. Lai and D. Tang, Anal. Chim. Acta, 2013, 758, 1–18 CrossRef CAS PubMed.
- J.-J. Xu, W.-W. Zhao, S. Song, C. Fan and H.-Y. Chen, Chem. Soc. Rev., 2014, 43, 1601–1611 RSC.
- X. Zhang, Y. Yu, J. Shen, W. Qi and H. Wang, Talanta, 2020, 212, 120794 CrossRef CAS PubMed.
- M. J. Mickert, Z. Farka, U. Kostiv, A. Hlaváček, D. Horák, P. Skládal and H. H. Gorris, Anal. Chem., 2019, 91, 9435–9441 CrossRef CAS PubMed.
- E. Makhneva, D. Sklenárová, J. C. Brandmeier, A. Hlaváček, H. H. Gorris, P. Skládal and Z. Farka, Anal. Chem., 2022, 94, 16376–16383 CrossRef CAS PubMed.
- T. Seki, Y. Seki, N. Iwata and S. Furumi, Materials, 2022, 15, 4943 CrossRef CAS PubMed.
- G. Tataru, M. Popa and J. Desbrieres, Int. J. Pharm., 2011, 404, 83–93 CrossRef CAS PubMed.
- L. Tonthat, Y. Yamamoto, F. Aki, H. Saito and K. Mitobe, IEEE Trans. Magn., 2018, 54, 1–6 Search PubMed.
- M.-C. Zhong, L. Gong, D. Li, J.-H. Zhou, Z.-Q. Wang and Y.-M. Li, Appl. Phys. Lett., 2014, 105, 181112 CrossRef.
- J. Xu, W. Yin, Y. Zhang, J. Yi, M. Meng, Y. Wang, H. Xue, T. Zhang and R. Xi, Food Chem., 2012, 134, 2526–2531 CrossRef CAS PubMed.
- L. F. Huergo, K. A. Selim, M. S. Conzentino, E. C. M. Gerhardt, A. R. S. Santos, B. Wagner, J. T. Alford, N. Deobald, F. O. Pedrosa, E. M. de Souza, M. B. Nogueira, S. M. Raboni, D. Souto, F. G. M. Rego, D. L. Zanette, M. N. Aoki, J. M. Nardin, B. Fornazari, H. M. P. Morales, V. A. Borges, A. Nelde, J. S. Walz, M. Becker, N. Schneiderhan-Marra, U. Rothbauer, R. A. Reis and K. Forchhammer, ACS Sens., 2021, 6, 703–708 CrossRef CAS.
- Z. Gao, M. Xu, L. Hou, G. Chen and D. Tang, Anal. Chem., 2013, 85, 6945–6952 CrossRef CAS PubMed.
- Z. Wang, R. Cai, Z. Gao, Y. Yuan and T. Yue, Compr. Rev. Food Sci. Food Saf., 2020, 19, 3802–3824 CrossRef CAS.
- Z. Farka, M. J. Mickert, A. Hlaváček, P. Skládal and H. H. Gorris, Anal. Chem., 2017, 89, 11825–11830 CrossRef CAS.
- S. Mohanty and A. M. Kaczmarek, Chem. Soc. Rev., 2022, 51, 6893–6908 RSC.
- Z. Farka, M. J. Mickert, Z. Mikušová, A. Hlaváček, P. Bouchalová, W. Xu, P. Bouchal, P. Skládal and H. H. Gorris, Nanoscale, 2020, 12, 8303–8313 RSC.
- M. Pastucha, E. Odstrčilíková, A. Hlaváček, J. C. Brandmeier, V. Vykoukal, J. Weisová, H. H. Gorris, P. Skládal and Z. Farka, IEEE J. Sel. Top. Quantum Electron., 2021, 27, 1–11 Search PubMed.
- S. Wilhelm, M. Kaiser, C. Würth, J. Heiland, C. Carrillo-Carrion, V. Muhr, O. S. Wolfbeis, W. J. Parak, U. Resch-Genger and T. Hirsch, Nanoscale, 2015, 7, 1403–1410 RSC.
- X. Zhu, J. Zhang, J. Liu and Y. Zhang, Adv. Sci., 2019, 6, 1901358 CrossRef CAS PubMed.
- M. V. DaCosta, S. Doughan, Y. Han and U. J. Krull, Anal. Chim. Acta, 2014, 832, 1–33 CrossRef CAS PubMed.
- Z. Farka, M. J. Mickert, M. Pastucha, Z. Mikušová, P. Skládal and H. H. Gorris, Angew. Chem., Int. Ed., 2020, 59, 10746–10773 CrossRef CAS PubMed.
- A. H. C. Ng, U. Uddayasankar and A. R. Wheeler, Anal. Bioanal. Chem., 2010, 397, 991–1007 CrossRef CAS.
- C.-C. Lin, J.-H. Wang, H.-W. Wu and G.-B. Lee, J. Assoc. Lab. Autom., 2010, 15, 253–274 CrossRef CAS.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS PubMed.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed.
- M. Zhao, X. Li, Y. Zhang, Y. Wang, B. Wang, L. Zheng, D. Zhang and S. Zhuang, Food Chem., 2021, 339, 127857 CrossRef CAS PubMed.
- X. Yu, Y. Xia, Y. Tang, W.-L. Zhang, Y.-T. Yeh, H. Lu and S.-Y. Zheng, Small, 2017, 13, 1700425 CrossRef PubMed.
- J. Kong, L. Jiang, X. Su, J. Qin, Y. Du and B. Lin, Lab Chip, 2009, 9, 1541 RSC.
- C. T. Lim and Y. Zhang, Biosens. Bioelectron., 2007, 22, 1197–1204 CrossRef CAS PubMed.
- H. Aghamohammadi, K. E. Thomas, S. Srikant, J. Deglint, A. Wong and M. Poudineh, Lab Chip, 2023, 23, 3245–3257 RSC.
- M. Y. H. Tang and H. Cheung Shum, Lab Chip, 2016, 16, 4359–4365 RSC.
- E. Huang, D. Huang, Y. Wang, D. Cai, Y. Luo, Z. Zhong and D. Liu, Biosens. Bioelectron., 2022, 195, 113684 CrossRef CAS PubMed.
- Z. Zeng, J. Tian, Z. Ren, Y. Yang, Q. Gong, R. Sun, X. Zhang, W. Liu and C. Chen, J. Sep. Sci., 2023, 46, 2300471 CrossRef CAS.
- A. Hlaváček, K. Uhrová, J. Weisová and J. Křivánková, Anal. Chem., 2023, 95, 12256–12263 CrossRef.
- O. Ronneberger, P. Fischer and T. Brox, in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, ed. N. Navab, J. Hornegger, W. M. Wells and A. F. Frangi, Springer International Publishing, Cham, 2015, pp. 234–241 Search PubMed.
- S. Bhattacharjee, J. Controlled Release, 2016, 235, 337–351 CrossRef CAS PubMed.
- G. Ploussard, N. Fossati, T. Wiegel, A. D'Amico, M. S. Hofman, S. Gillessen, N. Mottet, S. Joniau and D. E. Spratt, Eur. Urol. Oncol., 2021, 4, 150–169 CrossRef PubMed.
- J. P. Fouquet, D. Sikpa, R. Lebel, R. Sibgatulin, M. Krämer, K.-H. Herrmann, A. Deistung, L. Tremblay, J. R. Reichenbach and M. Lepage, Magn. Reson. Imaging, 2022, 92, 67–81 CrossRef CAS PubMed.
- T. Liao, F. Yuan, H. Yu and Z. Li, Anal. Methods, 2016, 8, 1577–1585 RSC.
- H. H. Gorris and T. Soukka, Anal. Chem., 2022, 94, 6073–6083 CrossRef CAS PubMed.
- K. Wang, R. Ni, X. Xing, X. Wen, J. Liu, Y. Ding and X. Hong, Microchim. Acta, 2020, 187, 516 CrossRef CAS.
- R. Gao, Z. Cheng, A. J. deMello and J. Choo, Lab Chip, 2016, 16, 1022–1029 RSC.
- Z. Feng, S. Zhi, L. Guo, Y. Zhou and C. Lei, Microchim. Acta, 2019, 186, 252 CrossRef PubMed.
- Y. Chen, O. Shimoni, G. Huang, S. Wen, J. Liao, H. T. T. Duong, M. Maddahfar, Q. P. Su, D. G. Ortega, Y. Lu, D. H. Campbell, B. J. Walsh and D. Jin, Cytometry, Part A, 2022, 101, 400–410 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Protocols for the preparation and characterization of UCNP and MB conjugates and processing of micrographs, results of the UCNP characterization and immunoassay optimization. See DOI: https://doi.org/10.1039/d4lc00346b |
| ‡ The authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |