 Open Access Article
Open Access ArticleMicrofluidics for macrofluidics: addressing marine-ecosystem challenges in an era of climate change
Fangchen
Liu
 a,
Cyril
Deroy
a,
Cyril
Deroy
 a and
Amy E.
Herr
a and
Amy E.
Herr
 *ab
*ab
aDepartment of Bioengineering, University of California, Berkeley, California 94158, USA. E-mail: aeh@berkeley.edu
bChan Zuckerberg Biohub, San Francisco, California 94158, USA
First published on 2nd August 2024
Abstract
Climate change presents a mounting challenge with profound impacts on ocean and marine ecosystems, leading to significant environmental, health, and economic consequences. Microfluidic technologies, with their unique capabilities, play a crucial role in understanding and addressing the marine aspects of the climate crisis. These technologies leverage quantitative, precise, and miniaturized formats that enhance the capabilities of sensing, imaging, and molecular tools. Such advancements are critical for monitoring marine systems under the stress of climate change and elucidating their response mechanisms. This review explores microfluidic technologies employed both in laboratory settings for testing and in the field for monitoring purposes. We delve into the application of miniaturized tools in evaluating ocean-based solutions to climate change, thus offering fresh perspectives from the solution-oriented end of the spectrum. We further aim to synthesize recent developments in technology around critical questions concerning the ocean environment and marine ecosystems, while discussing the potential for future innovations in microfluidic technology. The purpose of this review is to enhance understanding of current capabilities and assist researchers interested in mitigating the effects of climate change to identify new avenues for tackling the pressing issues posed by climate change in marine ecosystems.
1. Introduction
Climate change and the emission of greenhouse gasses are profoundly impacting ocean properties and marine ecosystems, as detailed in the recent report by the Fifth National Climate Assessment.1 The associated physical and biogeochemical effects include ocean warming and marine heatwaves, sea-level rise, ocean acidification, and ocean deoxygenation. Since 1993, ocean warming has accelerated, with a significant increase in heat uptake, doubling in that period.2 While sea levels are projected to rise between 0.43 m and 0.84 m by 2100,3 the pH of surface ocean waters has been decreasing at varying rates across different regions since the 1980s, indicating widespread ocean acidification.4 Additionally, the upper ocean layer has experienced a 2% loss in oxygen from 1970 to 2010, with predictions of continued decline.4These physical and biochemical changes are leading to numerous threats to marine life and ecosystems, such as shifting species distributions and changing interactions, habitat loss, species loss, and increases in marine diseases and harmful algal blooms (HABs). Species across the marine food web are redistributing geographically in response to changing ocean conditions, impacting ecosystem functions and biodiversity.5 As climate-change stressors exceed the adaptive capacities of marine organisms, some experience habitat loss and species extinction. Skeleton- and shell-forming organisms, for example, are particularly vulnerable due to decreased saturation levels of calcium carbonate resulting from lower pH levels in seawater.4 Coral reefs, which are vital habitats supporting a diverse range of marine species, are rapidly degrading under the strain of ocean warming, leaving little time for recovery. This degradation not only destroys ecosystems but also affects approximately half a billion people globally who rely on coral reefs for food, income, and protection from waves.6 Furthermore, the frequency and severity of HABs and diseases caused by marine pathogens are on the rise, leading to significant environmental and economic costs.7,8 This complex interplay of climatic factors underscores the urgent need for comprehensive understanding and action to mitigate the impacts of climate change on marine environments.
Amidst the significant perturbations caused by climate change, the ocean presents potential solutions for mitigating its effects through means such as renewable energy generation and CO2 storage and sequestration.9,10 Given that oceans cover 71% of the Earth's surface, these marine environments hold substantial opportunities for harnessing renewable energy from waves, tides, ocean currents, thermal gradients, and salinity gradients.11 The theoretical potential of ocean energy is estimated to range between 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 terawatt hours (TW h) to 130
000 terawatt hours (TW h) to 130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 TW h of electricity per year, which could support more than double the current global-energy demand. However, currently less than 0.01% of this estimated potential is being utilized.12 For this potential to be realized, marine energy technologies need to advance in terms of technological readiness and cost-effectiveness, and the environmental impacts of such power-generation systems need careful consideration.9,11,13
000 TW h of electricity per year, which could support more than double the current global-energy demand. However, currently less than 0.01% of this estimated potential is being utilized.12 For this potential to be realized, marine energy technologies need to advance in terms of technological readiness and cost-effectiveness, and the environmental impacts of such power-generation systems need careful consideration.9,11,13
The ocean also plays a crucial role in carbon sequestration. Together with the terrestrial biosphere, they absorb about half of the CO2 emissions from human activities, with more than 50% of CO2 absorbed by terrestrial plants subsequently cycled into the ocean, making it a significant carbon sink.14,15 Enhancing oceanic CO2 uptake through chemical or biological approaches—marine carbon dioxide removal (CDR)—is viewed as a viable strategy to reduce atmospheric CO2 and meet near-future global climate goals.16,17 However, the efficacy, durability, and environmental impacts of these ocean-based CDR techniques must be thoroughly assessed before widespread deployment.
Increasingly, microfluidic tools and technologies are contributing to study of the impacts of climate change on marine ecosystems and underpinning ocean-based solutions. Recent review articles have highlighted the application of microfluidics in the environmental field, including marine environments. Fukuba and Fujii provided a comprehensive review of lab-on-a-chip platforms tailored to biochemical measurements in the ocean, emphasizing in situ applications.18 Datta et al. discussed perspectives on using microfluidic approaches to reduce carbon emissions in energy generation.19 Aryal et al. explored microfluidic analytical devices applied in monitoring various environments, including air, water, and soil matrices.20
This review synthesizes these insights, anchored in the critical challenges facing ocean and marine ecosystems due to climate change. In section 2, we cover technologies for both laboratory manipulation and field monitoring, as well as system integration, to elucidate the effects of climate change on the ocean and the adaptive responses of marine organisms (Table 1). Moreover, in section 3, we discuss the use of miniaturized tools to evaluate ocean-based solutions to climate change, offering fresh perspectives from the solution-oriented end of the spectrum (Table 2). These insights reveal the problems caused by climate change and highlight several opportunities for further positive impacts through microfluidic tools.
| Device name | Application | Highlights | Lab or field study | Ref. |
|---|---|---|---|---|
| Marine pH quantification lab-on-a-chip | Measurement of seawater pH using colorimetry with meta-cresol purple indicator dye | - High sensitivity (0.002 pH units) and reproducibility | Field | 22 |
| - Suitable for long-term, submerged operation | ||||
| - Compact size (227.5 cm3) and low power consumption | ||||
| Automated alkalinity analyzer | In situ, autonomous monitoring of seawater total alkalinity for ocean carbon cycle studies and carbon dioxide removal verification | - High accuracy (−0.17 ± 24 μmol kg−1) and precision (16 μmol kg−1) | Field | 23 |
| - Low reagent consumption and energy-efficient | ||||
| - Suitable for long-term deployments with minimal maintenance | ||||
| Microfluidic continuous flow DIC sensor | Continuous and autonomous measurement of total dissolved inorganic carbon (TCO2) in ocean water | - High precision and accuracy in measuring dissolved inorganic carbon | Field | 24 |
| - Suitable for long-term deployment in ocean floats for continuous monitoring | ||||
| - Minimal reagent payload, efficient for extended use | ||||
| Microfluidic DIC conductivity sensor | Measurement of dissolved inorganic carbon (DIC) in seawater using a microfluidic device with membrane separation of CO2 | - Requires low reagent and sample volumes | Field | 25 |
| - Integrates with autonomous ocean profiling systems | ||||
| - Effective conductimetric determination, potential for miniaturization | ||||
| Microfluidic impedance cytometer for algal calcification | Measurement of the calcification state of single coccolithophore cells by assessing the ratio of particulate inorganic carbon (PIC) to particulate organic carbon (POC) | - Real-time data, integratable with other lab-on-a-chip systems | Lab | 27 |
| - Small sample volumes, efficient and cost-effective for large-scale studies | ||||
| - Insights into calcification variation within populations | ||||
| Microfluidic behavioral analyzer | Quantitative analysis of marine zooplankton preferences and responsiveness to varying environmental conditions such as pH, salinity, and chemical gradients | - Real-time, high-throughput analysis of individual and population-specific behavioral responses | Lab | 28 |
| - Creates stable, controlled environmental gradients | ||||
| Microfluidic rheotactic analyzer | Evaluation of rheotactic behaviors of dinoflagellates (Karlodinium veneficum) in different flow conditions | - Creates stable, controlled environmental gradients | Lab | 29 |
| - Compact and portable design | ||||
| Microfluidic hatching platform | Real-time observation of oxygen changes during the hatching process of Artemia in response to temperature and salinity variations | - Real-time monitoring of metabolic and morphological changes | Lab | 31 |
| - High control over microenvironmental conditions | ||||
| - Suitable for analyzing the impact of climate change on aquatic species | ||||
| Microfluidic coral culture platform | Long-term culture and physiological study of single coral polyps | - Controlled environment for long-term culture | Lab | 32 |
| - Real-time monitoring of coral polyp growth and metabolic processes | ||||
| - Reduces environmental stress during culture | ||||
| Miniaturized coral polyp culture platform | Long-term culture and real-time monitoring of individual coral polyps, focusing on their response to temperature changes and environmental stressors | - Controlled microenvironment for dynamic culture | Lab | 33 |
| - Continuous perfusion and temperature control | ||||
| - Real-time monitoring of metabolic activity and stress responses | ||||
| Multi-gradient coral polyp culture chip | Culturing and analyzing coral polyps under varying concentration and temperature gradients to study their growth and physiological responses | - Creates multiple environmental gradients (temperature and concentration) simultaneously | Lab | 34 |
| - Real-time monitoring of coral polyp responses under stress | ||||
| - Stable, controlled microenvironment for extended culture periods | ||||
| Coral-on-a-chip platform | Live-imaging microscopy and study of coral physiological processes, including calcification, coral-pathogen interactions, and coral bleaching under controlled environmental conditions | - Controlled microenvironment for long-term coral polyp observation | Lab | 35 |
| - Real-time, high-resolution imaging of coral physiological processes | ||||
| - Facilitates coral-pathogen interactions and bleaching studies at single-cell resolution | ||||
| Microfluidic coral infection (MCI) platform | Visualizing and studying the early stages of coral infection by Vibrio coralliilyticus using NanoSIMS and microfluidics to track bacterial penetration and dispersal in coral tissues | - Real-time, high-resolution imaging of infection dynamics at the microscale | Lab | 36 |
| - Studies pathogen interactions with coral at tissue and single-cell levels | ||||
| - Insights into early infection stages and coral immune responses | ||||
| Traptasia microfluidic device | Live imaging and analysis of Aiptasia larvae to study coral and anemone bleaching mechanisms, specifically algal symbiont expulsion under environmental stress | - Real-time, high-throughput imaging of algal expulsion | Lab | 37 |
| - Study highly motile organisms in a stable trapping array | ||||
| - Simple to use, minimal equipment, suitable for non-specialists | ||||
| Vibrio chemotaxis and chemokinesis microfluidic platform | Studying the effects of temperature on the motility behaviors (chemotaxis and chemokinesis) of the coral pathogen Vibrio coralliilyticus, particularly its response to coral mucus | - Controlled environment for pathogen motility studies | Lab | 50 |
| - Real-time, high-resolution imaging of bacterial responses to temperature and coral mucus | ||||
| - Evaluates how elevated temperatures affect pathogen motility and infection | ||||
| Microfluidic T-maze | Studying phenotypic heterogeneity in bacterial chemotaxis by exposing bacteria to a sequence of decisions in a T-maze to quantify their chemotactic sensitivity coefficient | - Real-time, high-resolution imaging of bacterial movement and decision-making | Lab | 51 |
| - Quantifies phenotypic heterogeneity in chemotactic sensitivity | ||||
| - Compares different sources of heterogeneity affecting chemotactic performance | ||||
| High-throughput chemotaxis screening device | Investigating the chemotactic responses of heterotrophic bacteria to metabolites released from virus-infected cyanobacteria, using high-throughput screening to quantify bacterial attraction to specific metabolites during various stages of viral infection | - Real-time, high-resolution imaging and quantification of bacterial responses | Lab | 52 |
| - Insights into chemical interactions between virus-infected cyanobacteria and heterotrophic bacteria | ||||
| - Identifies compounds driving chemotactic responses, aiding nutrient and carbon cycling understanding | ||||
| Single-molecule tracking microfluidic device | Single-molecule fluorescence imaging and tracking of bacterial regulatory proteins (TcpP and ToxR) in Vibrio cholerae during real-time environmental changes, such as pH shifts | - Real-time observation of single-molecule dynamics in live bacterial cells | Lab | 53 |
| - High-resolution imaging and detailed analysis of molecular interactions | ||||
| - First use of single-molecule tracking in live bacterial cells within a microfluidic device | ||||
| Aquaculture disease detection system | Rapid detection of RNA-based aquaculture diseases using reverse transcription polymerase chain reaction (RT-PCR) | - High sensitivity and specificity for detecting multiple aquaculture pathogens | Lab | 54 |
| - Integrates micro temperature control and microfluidic modules for precise, automated sample handling | ||||
| - Portable with low power consumption | ||||
| Dual-sample microfluidic LAMP Chip | Rapid and simultaneous detection of multiple waterborne pathogenic bacteria using loop-mediated isothermal amplification (LAMP) | - Integrates automated sample handling and LAMP reaction in a microfluidic chip | Lab | 55 |
| - Reduces reagent and sample consumption | ||||
| - Provides rapid results within 35 minutes | ||||
| Double-layer microfluidic biochip for harmful algae detection | Ultrasensitive, rapid, and portable detection of harmful algal blooms (HABs) using a double-layer microfluidic biochip integrated with graphene oxide nanosheets | - High sensitivity and specificity for detecting multiple harmful algae species | Lab | 56 |
| - Rapid detection within 45 minutes | ||||
| - Integrates photoluminescence detection system for accurate quantification | ||||
| Microfluidic concentration gradient generator for ballast water treatment | Real-time detection and treatment of microalgae in ballast water using laser-induced chlorophyll fluorescence and concentration gradient generator | - Integrates chemical treatment for effective reduction of microalgae population | Lab | 57 |
| - Provides rapid and automated detection and treatment | ||||
| - Reduces reagent and sample consumption | ||||
| Automated intelligent microfluidic platform (AIMP) | Real-time, on-site sampling, detection, and monitoring of microalgae using a combination of automated system control, intelligent data analysis, and user interaction | - Portable and economical for field applications | Field | 58 |
| - Capable of long-term monitoring of microalgae production (e.g., astaxanthin from Haematococcus Pluvialis) | ||||
| - Integrates machine learning for high precision | ||||
| Automated flow cytometer | High-resolution analysis of North Sea phytoplankton community | - Automated high-frequency sampling with high spatial resolution (2.2 ± 1.8 km) | Field | 60 |
| - Analyzes a wide range of cell sizes (1 to 800 μm) | ||||
| - Significant correlation with traditional water sampling methods | ||||
| CytoSub flow cytometer | High-frequency monitoring of ciliates and dinoflagellates in coastal aquatic systems | - High-frequency, real-time plankton analysis | Field | 61 |
| - Accurate biomass estimation correlated with microscopy | ||||
| - Effective for early warning and environmental monitoring | ||||
| Microfluidic immunoassay chip for algal toxins | Rapid detection of algal toxins (microcystin, saxitoxin, and cylindrospermopsin) using immuno-enzyme assays | - High sensitivity and specificity for detecting multiple algal toxins | Lab | 62 |
| - Automated sample handling and immuno-enzyme reactions in a microfluidic chip | ||||
| - Provides rapid results within 25 minutes | ||||
| Lab-on-a-disc (LOAD) toxin sensor | In situ detection of algal toxins (microcystin-LR, saxitoxin, domoic acid) using a centrifugal microfluidic platform with immunofluorescence detection | - Automated sample handling and immunofluorescence detection in a centrifugal microfluidic platform | Field | 63 |
| - Provides rapid results within 30 minutes with minimal user interaction | ||||
| - Portable and easy to operate, suitable for field applications | ||||
| Portable phosphate sensor | Accurate detection of phosphate in water samples | - High accuracy with detection errors of <2% for standard solutions | Field | 71 |
| - Effective in filtering out interference from bubbles, light intensity, and salinity | ||||
| - Shortened detection time to 80 seconds with LOD of 0.4 μM | ||||
| Lab-on-chip nutrient sensor | In situ analysis of nitrate and phosphate in the deep sea | - High precision with LOD of 0.03 μM for nitrate and 0.016 μM for phosphate | Field | 72 |
| - Capable of operating at depths >4800 m | ||||
| - Automated and compact design for integration with profiling floats and autonomous underwater vehicles | ||||
| Inlaid microfluidic nitrite analyzer | Continuous flow nitrite determination in marine environments | - High sensitivity with LOD of 94 nM | Field | 73 |
| - Minimal reagent consumption | ||||
| - Automated, capable of high-frequency sampling (10 samples per hour) | ||||
| Portable lab-on-chip nitrate sensor | Rapid nitrate determination in water samples | - High reduction ratio of 94.8% using 3D double microstructured assisted reactors DMARs | Field | 74 |
| - Fast detection time of 115 seconds per sample | ||||
| - Low reagent consumption (26.8 μL per sample) and minimal use of toxic reagents | ||||
| Multi-nutrient seawater analyzer | Simultaneous determination of phosphate, silicic acid, and nitrate plus nitrite in seawater | - High precision with detection limits of 0.18 μM for phosphate, 0.15 μM for silicic acid, 0.45 μM for nitrate, and 0.35 μM for nitrite | Field | 75 |
| - Effective in long-term deployment with a 46 day field test | ||||
| - Robust against salinity variations and fouling effects | ||||
| Wireless microfluidic sensor for metal ion detection | Real-time detection of metal ions in water using low-temperature cofired ceramic (LTCC) technology | - High sensitivity with detection limits as low as 5 μM | Field | 76 |
| - Wireless detection and differentiation of various metal ions | ||||
| - Portable and suitable for monitoring industrial wastewater | ||||
| Portable lead ion detector | Rapid detection of hazardous Pb2+ in various environmental samples | - High sensitivity with LOD of 0.00464 μg L−1 | Field | 77 |
| - Reduces assay time by leveraging thermocapillary convection | ||||
| - Integrates a smartphone-based electrochemical workstation for field use | ||||
| Microfluidic microplastic identifier | Label-free identification of small-sized microplastics in seawater | - Achieves 93% accuracy using a convolutional neural network (CNN) | Field | 78 |
| - Effective for microplastics smaller than 50 μm | ||||
| - Integrates machine learning with Raman spectroscopy for high precision | ||||
| Integrated microfluidic microplastics analyzer | Automated analysis of microplastics from environmental samples | - Conducts sample digestion, filtration, and counting within the microfluidic chip | Field | 79 |
| - Low-cost and low demand for laboratory equipment | ||||
| - Suitable for continuous on-site inspection of microplastics |
| Device name | Application | Highlights | Lab or field study | Ref. |
|---|---|---|---|---|
| Microfluidic benthic microbial fuel cell (MBMFC) | In situ power generation from marine sediments using electrogenic bacteria | - High power density (30–120 mW m−2) with 8-fold improvement in steady-state production | Field | 88 |
| - Layered microfluidic design confines bacteria within 90 μm, enhancing electron capture | ||||
| - Scalable proof-of-concept for renewable underwater power sources | ||||
| Photo fermentative biohydrogen microbioreactor | Photo fermentative biohydrogen generation using a microbioreactor | - High biohydrogen production with an optimal illumination intensity of 6000 1× | Lab | 96 |
| - Three-dimensional numerical modeling for performance evaluation | ||||
| - Enhanced understanding of biofilm growth and light transfer effects on hydrogen production | ||||
| Integrated digital microfluidic bioreactor | Fully automatic screening of microalgal growth and stress-induced lipid accumulation | - High-throughput digital microfluidic system for encapsulating microalgal cells | Lab | 108 |
| - Automatic nutrient gradient generation and lipid accumulation analysis | ||||
| - Significant reduction in labor and time compared to traditional methods | ||||
| Droplet-based microfluidic screening system | Screening and sorting of microalgal populations for strain engineering | - High-throughput analysis and sorting of microalgal cells | Lab | 109 |
| - Real-time fluorescence detection for phenotype assessment | ||||
| - Efficient identification of genetically modified strains | ||||
| VIV-EEL energy capture device | Harnessing ocean current energy through vortex induced vibration | - Efficient energy capture at low flow velocities (starting at 0.3 m s−1) | Field | 110 |
| - High energy capture efficiency due to optimal cylinder size and flow field disturbance | ||||
| - Provides a stable power supply for underwater systems | ||||
| Underwater flag-like triboelectric nanogenerator (UF-TENG) | Harvesting ocean current energy at extremely low velocity conditions | - Low startup velocity (0.133 m s−1) for energy harvesting | Field | 112 |
| - High power density and enhanced performance with vortex street effect | ||||
| - Simple structure and cost-effective compared to traditional generators | ||||
| Bioinspired poly (ionic liquid) membrane | Efficient salinity gradient energy harvesting through bioinspired hierarchical nanoporous membranes | - High chemical stability and rapid ion transport | Lab | 115 |
| - Effective osmotic energy conversion in both aqueous and organic solutions | ||||
| - Potential for large-scale production and practical applications | ||||
| Nanofluidic membrane energy harvester | Capturing energy from salinity gradients using optimized nanochannels and a bipolar soft layer | - High energy conversion efficiency with optimal nanochannel geometry | Lab | 116 |
| - Enhanced ion transport with bipolar soft layer | ||||
| - Significant improvement in power output with increased concentration ratio | ||||
| Nanofluidic salinity gradient energy converter | Investigating impacts of membrane fouling on energy conversion | - Enhanced energy conversion efficiency with fouling near the high concentration side | Lab | 117 |
| - Fouling at low concentration end significantly reduces performance | ||||
| - 26.47% increase in electric power with optimal fouling conditions | ||||
| Microfluidic salinity gradient-induced electricity generator | Synergistic freshwater and electricity generation using solar steam | - High evaporation efficiency (88%) and simultaneous electricity generation | Lab | 118 |
| - Stable voltage output (150–250 mV) with varying salinity gradients | ||||
| - Salt harvesting capability (0.33 kg m−2 h−1) | ||||
| Hybrid microfluidic-differential carbonator (μ-DC) | Optimization of CO2 biofixation by microalgae | - High biofixation rate of 0.2416 g L−1 d−1 with optimal conditions | Lab | 119 |
| - Significant impact of CO2 concentration and microalgae to media ratio | ||||
| - High predictive accuracy of models with R2 > 0.8 and low error rates |
2. Evaluating the impact of climate change on marine properties and ecosystems
2.1. Changing ocean properties – acidification and carbon chemistry
The ocean, as the largest reservoir absorbing carbon dioxide from the atmosphere, is significantly impacted by increasing CO2 concentrations due to climate change, leading to ocean acidification. Microfluidic systems and miniaturized (bio)sensors are crucial for analyzing in situ and sampled seawater for pH, carbonate chemistry, and other measurements relevant to the ocean carbon cycle such as particulate carbon.21 These systems require minimal sample and reagent volumes, making microfluidic tools suitable for remote and long-term ocean monitoring. The versatility of microfluidic systems opens possibilities for developing rugged, stable, field-deployable systems designed for harsh ocean environments.Seawater pH stands as a critical metric for monitoring ocean acidification, a phenomenon that profoundly influences marine organisms. Pinto et al. developed a lab-on-a-chip device tailored for in situ and autonomous pH measurement of seawater (Fig. 1A).22 Leveraging a poly(dimethylsiloxane) (PDMS)-based microfluidic chip and an integrated optical-electronic system for colorimetric analysis, the device reported precise and reproducible pH measurements. By utilizing meta-cresol purple (mCP) as an indicator dye and measuring pH-sensitive optical absorption, the authors achieved remarkable resolution of 0.002 pH units for the 7.5–8.2 seawater pH range, with a total detection time of about 8 min. Optimization of microfluidic channel geometry, indicator dye concentration, and seawater volumes further enhanced performance. This innovation holds promise for submerged deployment, owing to its pressure-independent operability and potential for integration into autonomous underwater vehicles (AUVs).
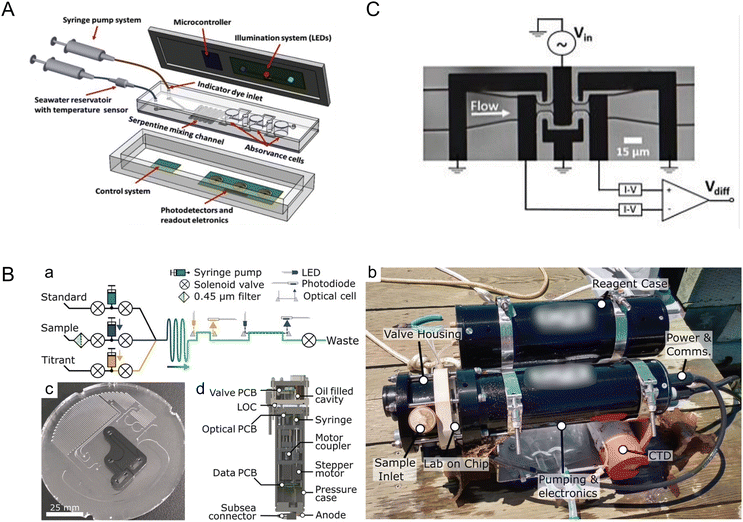 | ||
| Fig. 1 Monitoring ocean acidification and carbon chemistry with microfluidics. (A) Conceptual design of a lab-on-chip (LOC) device for marine pH quantification by colorimetry by Pinto et al., illustrating the microfluidic and optical electronic system (reproduced from ref. 22 with permission from Elsevier, copyright 2019). (B) Design of a microfluidic LOC total alkalinity analyzer by Sonnichsen et al. (a) Fluid schematic showing pumps, valves, optical cells, and optical components. While titrating, each point is built from fresh sample and titrant. Fluid paths during sample pH determination are shown with solid arrows and optical paths with dashed arrows. (b) Photograph of the alkalinity analyzer with a reagent case immediately after a jetty experiment, with a co-deployed CTD sensor. (c) Image of the LOC platform in production, showing the channels and inlaid optical cells. This layer is subsequently bonded with a capping layer to form the closed system. (d) Analyzer cross section showing location of major mechanical and electrical components (reproduced from ref. 23, CC BY 4.0, 2023). (C) Microfluidic impedance cytometry by de Bruijn et al. Overview of the differential impedance measurement setup showing the coplanar electrode pairs, which are shielded by the ground electrodes. The channel height is 10 μm (reproduced from ref. 27, CC BY 4.0, 2022). | ||
While measuring the pH of a fluid assesses proton concentration, alkalinity assesses the capacity for seawater to buffer acidity, making alkalinity pivotal in understanding the ocean's capacity to counteract rising atmospheric CO2 levels. Sonnichsen et al. developed an automated microfluidic analyzer capable of in situ monitoring of seawater total alkalinity (Fig. 1B).23 By employing stepper-motor driven syringe pumps for precise mixing and thorough characterization of temperature effects, the system demonstrated high accuracy and reliability even under field conditions, with 25 days of cumulative operation. This analyzer, validated against laboratory measurements, showcased appropriate precision and accuracy, making the technology suitable for continuous ocean monitoring missions.
Continuously measuring dissolved inorganic carbon (DIC) concentration is paramount for monitoring ocean acidification and modeling climate change. Tweedie et al. introduced a microfluidic sensor for continuous in situ DIC measurement in seawater.24,25 To measure DIC in seawater samples, excess acid is usually added to convert DIC ions to CO2 gas that is then measured using various methods including coulometry, mass spectrometry, infrared methods, and conductimetry.26 The microfluidic sensor developed by Tweedie et al. utilized the conductimetric method, which is more suitable for miniaturization as compared to others.25 In the system, extracted CO2 gas was mixed with an alkaline receiving solution which led to reduction of conductivity proportional to the added CO2 concentration. This sensor, leveraging a microfluidic conductivity cell with thin-film electrodes and a membrane-based gas exchange cell, achieved remarkable precision and accuracy. By incorporating asymmetric Y-meters fabricated with channel dimensions down to ∼75 μm, the system precisely metered reagents for sample acidification and CO2 liberation, thus reporting DIC concentration measurements of 1000–3000 μmol kg−1 with a precision of ∼0.2% standard deviation for peak height measurements at a concentration of 2000 μmol kg−1. Furthermore, with low reagent volumes (∼500 μL) facilitated by incorporating a planar membrane and sputter-deposited Ti/Au electrodes onto a thermoplastic manifold, this system could seamlessly integrate into ocean-float platforms for long-term DIC monitoring.
Moreover, understanding particulate carbon dynamics, especially the role of calcifying algae, is crucial in comprehending the ocean carbon cycle. De Bruijn et al. presented a microfluidic impedance cytometer for rapid, non-invasive analysis of calcifying algae, allowing for insights into the particulate inorganic carbon to particulate organic carbon (PIC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POC) ratio (Fig. 1C).27 This method involved mixing cells with polystyrene reference beads and processing the mixture through a microfluidic chip, where impedance measurements were taken at high frequencies and analyzed using MATLAB. Achieving a throughput of 2.5–20.0 cells per second, this method differentiated cells, beads, debris, and dead cells, with a linear correlation observed between the measured electrical phase and the PIC
POC) ratio (Fig. 1C).27 This method involved mixing cells with polystyrene reference beads and processing the mixture through a microfluidic chip, where impedance measurements were taken at high frequencies and analyzed using MATLAB. Achieving a throughput of 2.5–20.0 cells per second, this method differentiated cells, beads, debris, and dead cells, with a linear correlation observed between the measured electrical phase and the PIC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POC ratio, enabling cellular-level calcification analysis in response to environmental changes. These advancements demonstrate the significant potential of microfluidic technologies in enhancing our understanding and monitoring of oceanic carbon dynamics.
POC ratio, enabling cellular-level calcification analysis in response to environmental changes. These advancements demonstrate the significant potential of microfluidic technologies in enhancing our understanding and monitoring of oceanic carbon dynamics.
2.2. Changing distribution and behavior of marine species – plankton
Shifting ocean properties induce changes in the distribution and behavior of marine species within the intricate web of marine life. Microfluidic devices offer a unique capability to create controlled microenvironments that mimic these changing ocean conditions, making them particularly useful for studying the responses of marine plankton to various environmental stressors. Given plankton's fundamental role at the base of the marine food web and their size compatibility with microfluidic compartments, plankton are an ideal focus for such studies.In the investigation of marine plankton responses to chemical alterations in their environment, Ramanathan et al. effectively employed two microfluidic devices—a laminar flow device and a chemical gradient device—to observe and quantify the behavior of zooplankton under varying pH and salinity conditions (Fig. 2).28 These devices provided controlled and stable chemical environments that closely mimicked natural ecological conditions. The study was able to quantify the zooplankton's pH preferendum and determine the minimum [H+] and [NaCl] required to trigger behavioral changes, such as alterations in distribution. Furthermore, the research revealed population heterogeneity, identifying subpopulations within the zooplankton species Platynereis dumerilii that exhibited different pH preferences, highlighting the nuanced ecological dynamics driven by chemical changes in the ocean.
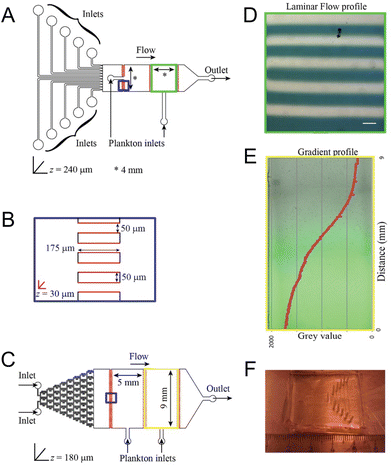 | ||
| Fig. 2 Microfluidic tools for investigating the behavior of marine plankton in response to climate change as reported by Ramanathan et al. (A) Laminar flow device with ten individual inlets for different chemicals and two 4 × 4 mm chambers for loading plankton. (B) A magnified scheme of the shallow channels connecting adjacent chambers (blue squares in the geometries). Regions in red are manufactured to a depth of 30 μm to prevent plankton from being flushed out. (C) Gradient generator device with a “Christmas tree” gradient generator and two 9 × 5 mm chambers for loading plankton. (D) Laminar flow as observed inside the chamber with a flow rate of 400 μl h−1. A blue dye was used in every alternate stream for visualization. Scale bar represents 400 μm. (E) Gradient established within the Christmas tree gradient generator using a flow rate of 100 μl h−1. Fluorescein was used to visualize and quantify the gradient. (F) A photograph of the laminar flow microfluidic device (reproduced from ref. 28, CC BY 4.0, 2015). | ||
In the context of changing oceanic dynamics influenced by climate change, Li et al. conducted a study to investigate the rheotactic behavior of the dinoflagellate species Karlodinium veneficum using a microfluidic device.29 Employing controlled flow conditions, both steady and oscillatory, the researchers aimed to elucidate how these hydrodynamic environments influence the swimming behaviors of dinoflagellates. The steady flow conditions were regulated by a syringe pump, while oscillatory flows were generated using a cam follower.30 By tracking the trajectories and orientations of dinoflagellates within these environments, the study aimed to quantify the interaction between planktonic motility and hydrodynamic conditions. This approach provided valuable insights into how environmental factors, particularly flow dynamics, may influence the distribution and abundance of planktonic organisms, contributing to a deeper understanding of ecosystem responses to changing oceanic conditions.
In addition to studying plankton motility, microfluidics also facilitates the investigation of plankton reproductive responses to environmental factors. Dey et al. employed a microfluidic platform equipped with an integrated oxygen sensor and a digital stereo microscope to monitor the hatching process of Artemia, or brine shrimp – a common live feed used in aquaculture – in real time.31 Operating under precisely controlled temperature and salinity conditions, the researchers observed morphological changes and measured oxygen consumption rates during the hatching stages. The findings indicated that higher temperatures enhanced the metabolic resumption of dormant cysts, while lower temperatures and salinities prolonged the differentiation stage of hatching. This innovative platform offers a method to study the impacts of climate change on the hatching processes of marine plankton, highlighting how microfluidic technologies can significantly contribute to our understanding of marine life under changing climatic conditions.
2.3. Loss of algal habitats and loss of species – coral reefs
Coral reefs are crucial ecosystems that support a diverse range of marine organisms, including fish, shellfish, turtles, and marine mammals. However, these vital habitats face accelerated degradation due to the twin threats of ocean warming and acidification, phenomena intensified by ongoing climate change. In response, microfluidic platforms have become essential tools, offering capabilities for in situ observation of coral symbiotic interactions and the monitoring of dynamic responses within these critical microenvironments. Additionally, the integration of high-throughput analysis facilitates detailed studies on the diversity and functions of symbiotic partners, further enriching our understanding of coral ecosystems.Microfluidics presents a well-suited approach to manipulating the microenvironments of coral by precisely controlling nutrient availability and replicating the natural flow conditions found in aquatic HABs. Key to the success of these platforms are features such as high substance transfer rates, low shear rates, and exact temperature control, all essential for the successful culturing of coral polyps on chips. Pang et al. developed a microfluidic platform designed for culturing single coral polyps and monitoring their growth over extended periods (Fig. 3A–C).32 This platform maintains a constant laminar flow, which facilitates the controlled high transfer rates of oxygen and bicarbonate essential for normal coral polyp processes over more than fifteen days of cultivation.
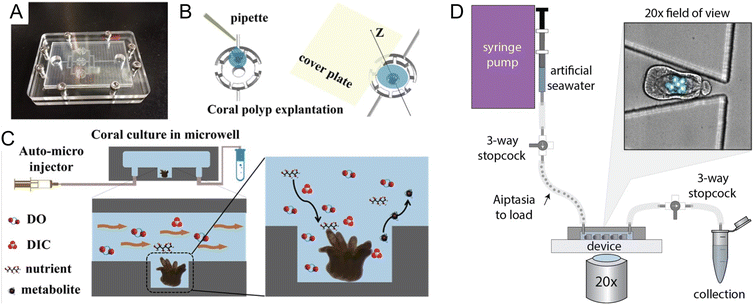 | ||
| Fig. 3 Microfluidic tools for cultivation and manipulation of coral and sea anemone species. (A–C) Culture of single coral polyps in a microchip by Pang et al. (A) Photo of the microfluidic chip with three microwells. The microchip was sealed with the cover glass using eight screws at the peripheral area. (B) Schematic presentation of the explantation of the coral polyp in the microwell. (C) A micro-pump was used to provide a fluid environment for the coral polyp culture. Design of the microwells in the chip can allow for a rapid exchange of soluble substrates (DO (Dissolved Oxygen), DIC, nutrients, and waste) and to reduce the effect of shear stress on the single coral polyps (reproduced from ref. 32, CC BY 4.0, 2020). (D) Schematic of the ‘Traptasia’ device and associated hardware for loading, trapping, and imaging individual Aiptasia larva, developed by Treuren et al. Organisms are loaded downstream of a stopcock assembly and connected to the device inlet. An upstream syringe pump provides constant fluid flow for trapping as well as nutrient and/or treatment delivery to the larva. Inset shows sample field of view with stably trapped Aiptasia larva (brightfield) and algae symbionts (blue-green, chlorophyll autofluorescence) (reproduced from ref. 37, CC BY 4.0, 2019). | ||
Similarly, Luo et al. introduced a temperature-adjustable coral-polyp-on-chip device that allows researchers to study over a 3-day culture period the effects of elevated temperatures on coral bleaching, providing valuable insights into how corals respond to thermal stress.33 Zhou et al. reports a simulation-based study of a coral-polyp culture chip design that includes chemical concentration and temperature gradients.34 Numerical simulation informs a design-based understanding of the conditions that can be generated on chip, so as to allow eventual study of the growth and physiological responses of coral polyps under various environmental pressures. These and similar efforts promise to offer a deeper understanding of coral adaptation and resilience under climate change-induced stresses.
The integration of microfluidic technologies with advanced imaging and analysis techniques has made possible the real-time observation of symbiotic interactions and dynamic responses within coral microenvironments. Shapiro et al. utilized a microfluidic platform to visualize the calcification and skeletogenesis of coral polyps by incorporating the fluorescent dye calcein into the growing calcium carbonate crystals.35 This method allows for precise monitoring of coral growth and skeletal development under various conditions. In a similar application, Gibbin et al. designed a microfluidic platform specifically for coral infection studies.36 This setup facilitated the inoculation of coral fragments with bacterial pathogens for subsequent visualization of pathogen penetration and dispersal within coral tissues. Moreover, Treuren et al. developed a device called “Traptasia”, designed for live imaging of Aiptasia larvae, a model system for coral symbiosis (Fig. 3D).37 This device allows for the isolation and time-lapse imaging of individual larvae and their algal symbionts, providing insights into the processes of coral and sea anemone bleaching under various environmental stressors.
Beyond imaging, the combination of microfluidics with cutting-edge analytical tools offers a promising pathway for exploring the complex interactions between corals and their symbiotic partners. While this area remains relatively nascent, the use of -omic methodologies such as single-cell RNA sequencing (RNA-seq) has started to reveal the diversity of cell types within corals and sea anemones. This approach is instrumental in generating comprehensive cell type atlases, as demonstrated by studies from Sebé-Pedrós et al. and Levy et al.38,39 Additionally, a range of molecular techniques and experimental strategies, as reviewed by Engelberts et al., have been employed or show potential for further exploration within coral research.40 These include metatranscriptomics,41 metaproteomics,42 metabolomics,43 stable isotope probing,44 fluorescence in situ hybridization (FISH),45 Raman microspectroscopy,46 nanoscale secondary ion mass spectrometry (NanoSIMS),47 MALDI-MS imaging,48 and synchrotron radiation Fourier-transform infrared (SR-FTIR) spectral microscopy.49 Each of these techniques contributes uniquely to the understanding of microbial interactions and symbiotic functions within coral reef holobionts, providing critical insights into their resilience or susceptibility to environmental changes.
2.4. Increasing marine diseases and harmful algal blooms (HABs)
Climate change has intensified challenges in marine ecosystems, particularly with the rise in marine pathogens and HABs. Advanced microfluidic systems, integrated with biosensors and molecular probes, are crucial for detecting and analyzing these threats, providing detailed insights into the behaviors of harmful microorganisms.Microfluidic platforms can precisely control chemical conditions making these platforms instrumental in studies of chemotaxis. Chemotaxis is a crucial behavior for bacteria navigating the ocean's fluidic environment. Garren et al. designed a microfluidic channel to create a transient gradient of coral mucus via molecular diffusion for observation of a coral pathogen's swimming behavior at various temperatures.50 The research demonstrated that elevated seawater temperatures (≥23 °C) significantly improved the pathogen's chemotactic ability, while their chemokinetic ability was enhanced when temperature further increased to 30 °C, suggesting more efficient host-seeking behaviors in warmer oceanic conditions. Salek et al. transformed the classical T-maze, conventionally used in animal ecology, into a microfluidic device that tracks the movement of individual Escherichia coli cells through chemical gradients.51 Utilizing phase contrast microscopy and MATLAB-based image processing, the authors observed these movements with single-cell resolution. Mathematical modeling and 2D numerical simulations helped analyze bacterial movement and calculate concentration profiles across various junctions of the maze, uncovering considerable variability in chemotactic sensitivity within a genetically identical clonal population. Additionally, Henshaw et al. implemented a transient chemical gradient microfluidic device to investigate the chemotactic responses of bacteria to exudates of viral-infected cyanobacteria.52 The authors' findings indicated that bacteria are drawn to intact, viral-infected cyanobacteria, suggesting that exudates from single phage-infected cyanobacteria can attract nearby bacterial communities. This behavior suggests that chemotaxis may play a crucial role in facilitating interactions between cyanobacteria and heterotrophic bacteria, potentially aiding in the breakdown of cyanobacterial blooms during the natural succession of HABs.
Beyond observing cellular behaviors like chemotaxis, microfluidic systems provide platforms for real-time examination of bacterial molecular responses to environmental changes. In 2023, Calkins et al. utilized a modified “mother machine” microfluidic device paired with chitosan-coated coverslips to conduct single-molecule tracking of crucial virulence transcription factors in a water-borne pathogen.53 The authors' findings revealed that a decrease in pH led to an increase in the subcellular diffusivity of the transcription factor, which in turn triggered virulence. With access to appropriate genetic constructs of specific strains, this microfluidic methodology can be applied to study bacterial regulatory mechanisms at the single-molecule level under varying environmental conditions.
The rapid detection of marine pathogens and species that form HABs is enhanced by the integration of microfluidic systems with molecular tools. Lien et al. engineered an integrated microfluidic system that automates the reverse transcription polymerase chain reaction (RT-PCR) to swiftly identify diseases in aquaculture.54 This system incorporates micro temperature sensors and array-type micro heaters to ensure precise and consistent temperature conditions for running four parallel RT-PCR processes. Pneumatic micropumps, microvalves, and microchannels facilitate the automatic transport of samples and reagents within the system. Utilizing random primers in the RT process simplifies chip design and decreases the consumption of RT products, boosting the system efficiency. The system achieved a detection limit of 10 copies per μL in 2.5 h, as validated with four types of purified RNA samples. This technique is well-suited for detecting RNA-based disease markers in water samples. Further developments include the work of Jin et al., who created a microfluidic chip integrated with loop-mediated isothermal amplification (LAMP) that can quickly and simultaneously detect 10 types of waterborne pathogenic bacteria.55 The chip is capable of independently analyzing two samples, each with 11 reaction wells, with a detection limit for genomic DNA ranging from 0.07 to 8.4 pg μL−1 and a reaction time of 35 min. This setup is ideal for on-site detection and routine monitoring of multiple marine pathogens. Additionally, Li et al. developed a microfluidic chip using graphene oxide (GO) nanosheets to detect six species of harmful algal bloom-forming algae.56 The chip uses single-strand DNA (ssDNA) probes, tailored to the 18S rDNA gene fragments of the algae species. The fluorescence signal from these probes is initially quenched by the GO nanosheets and is restored when complementary ssDNA from a sample binds and detaches the fluorophore from the GO. This detection process, which completes in 45 min, achieves a sensitivity limit of 108 aM, demonstrating its effectiveness and speed in monitoring harmful algal bloom species.
While molecular-based detection methods typically require DNA or RNA extraction from seawater samples, microfluidic systems also offer direct detection based on various cellular properties, enhancing compatibility with in situ analysis. Wang et al. designed a microfluidic chip that integrates impedance pulse sensing and LED light-induced chlorophyll fluorescence (LED-LICF) to identify marine nonindigenous microorganisms in a ship's ballast waters.57 Impedance pulse sensing, or resistance pulse sensing, measures the size, shape, and volume of microalgae and bacteria, while LED-LICF quantifies the chlorophyll fluorescence in microalgae. This label-free detection system can be tailored for in situ assessment of seawater samples, providing a well-characterized signal response from targeted species. In another study, Zheng et al. developed the automated intelligent microalgae phenotyping (AIMP) platform to detect microalgae based on size and morphology across four species (Fig. 4).58 AIMP employs laser-cut microfluidic chips for sample handling, coupled with a low-cost USB portable microscope and an OpenCV-based image stitching function for panoramic imaging. The platform uses machine learning, specifically the YOLOv5 architecture. Additionally, the AIMP platform can monitor the accumulation of astaxanthin, a red pigment, in an algal species over a 30 day period.
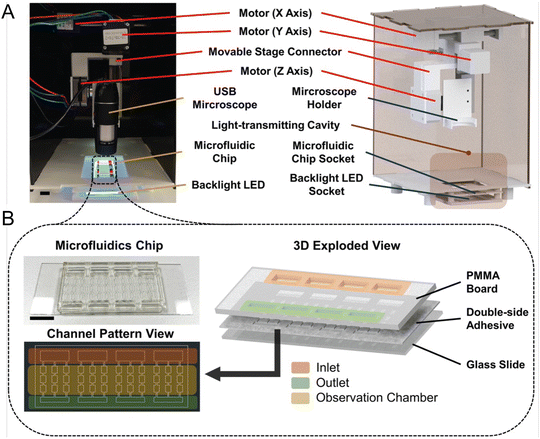 | ||
| Fig. 4 Automated and intelligent microfluidic platform (AIMP) for detecting HABs, as reported by Zheng et al. (A) Actual image and design schematic of the imaging part of the AIMP. (B) Schematic and image of the microfluidic chip used to load and observe microalgae (reproduced from ref. 58, CC BY 4.0, 2023). | ||
Parallel to this area of microfluidic cellular-detection innovation, in situ flow cytometry has been successfully applied for monitoring aquatic phytoplankton.59 Thyssen et al. utilized an automated scanning flow cytometer (SFC) connected to a PocketFerryBox to investigate the phytoplankton community structure in the North Sea with high spatial resolution, identifying ten phytoplankton groups based on optical pulse shapes and estimating their abundance and size.60 Similarly, Pereira et al. employed a submersed CytoSense flow cytometry instrument (CytoSub) capable of reaching depths up to 200 m to monitor plankton communities in real time, capturing detailed variability data on ciliates and dinoflagellates over a 12 h period.61 These methods illustrate the potential for developing and integrating image recognition models to automatically classify plankton communities, further enhancing the utility and scope of in situ marine biological monitoring.
In addition to the direct detection of organisms in seawater samples, the monitoring of marine system health can also be enhanced through the detection of biochemical markers. Antibody-based microfluidic immunoassays have been developed specifically for this purpose. These assays are capable of detecting toxins associated with HABs, including microcystin, saxitoxin, cylindrospermopsin, and domoic acid. Notable studies by Zhang et al.62 and Maguire et al.63 have successfully applied these immunoassays in identifying these critical toxins. Immunoassays provide a valuable tool for assessing the ecological impact of HABs and aid in the timely management of associated risks to marine health.
2.5. Damaging ocean pollutants – nutrients, heavy metals, and microplastics
Beyond altering marine environments and organisms, climate change is amplifying the effects of pollutants including nutrients, heavy metals, and microplastics. For instance, changing precipitation patterns leads to increased nutrient runoffs from land, which contributes to eutrophication and alters the marine nutrient cycles;64,65 increasing ocean acidification due to climate change may interact with heavy metals to make them more toxic for marine organisms;66,67 and changing ocean physical and chemical properties can impact the degradation and dispersion of microplastics, as well as their accumulation in the food chain.68–70 Microfluidics, excellent in efficient chemical and particle analysis, serves as a powerful tool for detecting these pollutants.Microfluidics provide tools for more portable detection of nutrients in water samples, as well as in situ nutrient detection in the ocean.71–75 Zhu et al. developed a portable phosphate sensor combining a microfluidic gradient Fabry-Pérot array (FPA) and smart phone control (Fig. 5A).71 Fabry-Pérot cavities were deployed to increase sensitivity of absorbance measurements by optical interferences between two reflecting surfaces. The microfluidic bidirectional gradient of water sample and chromogenic agent removes the need for standard curve characterization, reducing detection time to 80 s, while achieving a limit of detection (LOD) of 0.4 μM, comparable to traditional instruments. As for continuous in situ nutrient analysis, Beaton et al. reported a family of lab-on-chip colorimetric analyzers for nitrate and phosphate measurements from surface ocean to as deep as 4800 m.72 A microfluidic mixer based on secondary inertial flows was designed to efficiently mix reagents and samples on the chip, contributing to the detection accuracy. The nitrate sensors had a LOD of 0.030 ± 0.005 μM (mean ± 1σ), and the phosphate sensors had a LOD of 0.016 ± 0.003 μM (mean ± 1σ). To operate at elevated hydrostatic pressures and low temperatures experienced in the deep sea, an oil-filled hydraulically pressure-compensated housing was designed. Another critical feature of microfluidic in situ analysis is the high-frequency monitoring of nutrient fluctuations. Motahari et al. proposed an automated microfluidic nitrite analyzer with a sampling frequency of at least 10 samples per hour through continuous flow and reagent injection.73 An inlaid optical unit was designed to facilitate the inspection of the chip fabrication quality, which enabled colorimetric absorbance determination of nitrite. Reagent injection frequency, injection volume, and plug length were optimized, and resulted in nanomolar LOD (94 nM) with minimal reagent usage (20 μL).
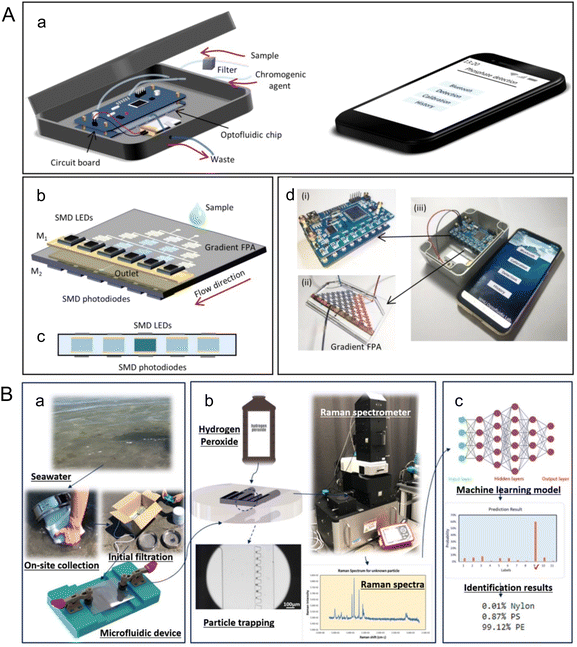 | ||
| Fig. 5 Analyzing marine pollutants using microfluidics. (A) A portable microfluidic phosphate sensor by Zhu et al. (a) Schematic of the smart phosphate sensor. It mainly consists of a gradient FPA, a circuit board, and a smartphone. (b) Gradient FPA. The width of the channel is 400 μm, and the height is 250 μm. (c) Cross-sectional view of the absorbance cells, which are coated by gold films (M1 and M2) to form a FP microcavity. The SMD LEDs (880 nm) and the SMD phototransistors are welded to the circuit board. The optical length is 250 μm. (d) Image of the circuit board (i), the gradient FPA (ii), and the smart phosphate sensor (iii) (reproduced from ref. 71 with permission from the American Chemical Society, copyright 2020). (B) A label-free microfluidic approach to identify small-sized marine microplastics by Gong et al. (a) An assembled microfluidic device is utilized for on-site collection of samples from seawater, which undergoes initial filtration using a 45 μm filter. (b) After safely transporting the device back to the laboratory, a solution of 30% (v/v) hydrogen peroxide is injected into the device, where it is left for 16 h to facilitate sample processing. Subsequently, Raman spectra are acquired from the processed samples. (c) Machine learning identification process incorporated in the microfluidic-based method for marine microplastic analysis (reproduced from ref. 78, CC BY 4.0, 2023). | ||
Heavy metals can be measured using microfluidic tools in a portable and time efficient way with reasonable sensitivity as compared to conventional methods such as inductively coupled plasma mass spectrometry (ICP-MS). Liang et al. developed a wireless microfluidic sensor capable of detecting metal ions including Pb2+, Cd2+, Mg2+, Ca2+, Na+, and K+, where Pb2+ and Cd2+ are commonly considered water pollutants.76 The liquid analyzing device was designed based on the capacitive-inductive (LC) resonant circuit, which was integrated with the microfluidic channel. The integration was enabled by the low-temperature cofired ceramic (LTCC) technology, an alternative to common microfluidic materials such as silicon, glass, and PDMS, with improved properties. The metal ions and their concentrations in pure water solutions were measured by the response behavior of the wireless microfluidic sensor with a limit of detection (LOD) of 5 μM, suitable for industrial wastewater monitoring. Further lowering the detection limit, Ma et al. developed a microfluidic electrochemical sensing platform with the LOD of Pb2+ at 0.00498 μg L−1 in river water samples.77 Thermocapillary convection promoted electrolyte flow and electron transfer in the device, which reduced the detection time, while working electrodes made of 3D Ag-rGO-f-Ni(OH)2/NF composites could absorb more targets, which amplified the signal. To monitor metal pollutants in seawater, where background ion concentrations may be high and pollutant ion concentrations may be relatively low, further development of microfluidic tools will be needed.
Microplastics are small pieces of plastic less than 5 mm in length. Detection of microplastic pollutants can take advantage of microfluidic technologies' unique ability to manipulate and detect particles at micro- and nano- scales.78,79 Gong et al. introduced a microfluidic approach for detecting microplastics in seawater through trapping and identification processes (Fig. 5B).78 Sieve-like traps of different sizes were designed to trap microplastics of corresponding size ranges down to 6 μm. Raman spectroscopy and machine learning were used to measure and analyze readouts from the microfluidic device for label free microplastic identification. The application of a convolutional neural network (CNN) was found to give an identification accuracy of 93%.
3. Ocean-based solutions to climate change
3.1. Ocean-based renewable energy
Transitioning from fossil fuels to renewable energy sources is essential for reducing greenhouse gas emissions and mitigating climate change. Among the innovative approaches in this transition, microfluidic devices are emerging as a promising technology to harness renewable energy from the ocean. These devices can utilize biological sources such as marine microbes, as well as non-biological systems like waves, currents, and salinity gradients. Microfluidics and micro-, nano-scale systems can manipulate, control, and interact with chemicals, cells, and other substances of interest at a small scale, which provides unprecedented views of physical, chemical, and biological events.80 As for energy applications, microfluidic technologies can be leveraged through their characteristics including high surface-to-volume ratios, capacity for high temperature and high pressure experiments, and length scales characteristic of microbes and fluids.81While many microfluidic technologies for marine energy are at an early stage where cost may not be available or relevant, cost remains a critical concern for energy technologies. In this regard, microfluidics may offer a few advantages which could help reduce costs compared to traditional approaches, including (1) low reagent consumption and minimal human input (time, automation) and (2) the possibility for mass production (with the optimization of technology).
Marine sediments offer a naturally controlled environment that supports the native benthic microorganisms used in microbial fuel cells. Microscale devices can be particularly useful in optimizing these microorganisms as they can be easily implemented in the natural environment and can control the electrode layout with cell-scale precision. For instance, Nguyen et al. developed a benthic microbial fuel cell that confines bacteria within ∼90 μm of the anode, significantly enhancing electron capture efficiency and power production (Fig. 6A).88 The device incorporates electrodes of the fractal H-architecture (H-fractal) that consists of a pattern where fractal binary splitting is applied to make the distance from a centroid to any node in a planar rectangle equal. H-fractal electrodes were designed to help remove the bias in the electric resistivity due to variability in the spatial distribution of the bacteria. This device was tested under laboratory-created sediment conditions and achieved steady-state power production levels ranging from 20–80 mW m−2, as compared to 8–10 mM m−2 from typical membrane-less microbial fuel cells. Future developments could focus on adapting this technology for use in natural marine sediments and further optimizing the system to increase power output, thereby making a more substantial contribution to renewable energy portfolios.
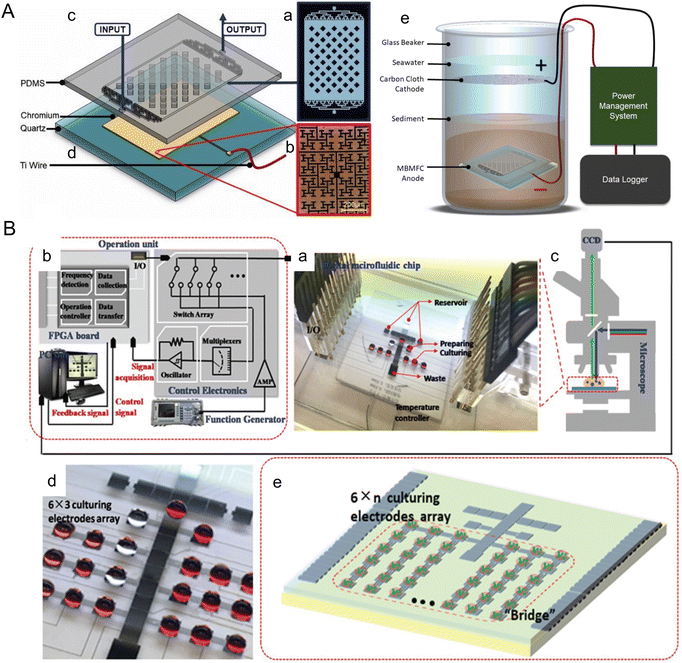 | ||
| Fig. 6 Microfluidic tools harnessing marine energy from microorganisms. (A) A microfluidic benthic microbial fuel cell (MBMFC) developed by Nguyen et al. (a–d) MBMFC components anode chip exploded view, assembled view, and photograph as follows: (a) schematic of the mold pattern for the PDMS layer showing microchannels/cavities (white) and solid silicone (black), (b) microscope image of the H-fractal chrome anode patterned on a quartz substrate, (c) 3D model of PDMS top layer molded with microchannels and dome cavity, (d) 3D model of quartz substrate with H-fractal, patterned chrome electrode. H-fractal electrodes consist of a pattern with fractal binary splitting that helps remove the bias in the electric resistivity due to variability of the spatial distribution of the bacteria. (e) In situ experimental set-up of the full MBMFC (reproduced from ref. 88 with permission from Elsevier, copyright 2021). (B) A digital microfluidic (DMF) bioreactor for automatic screening of microalgal growth and lipid production by Wang et al. (a) Image of the DMF chip, a patterned 24-electrode array which is connected to (b) the operation unit with position feedback for actuating and mounted onto (c) a microscope for visual monitoring. (d) Image of the DMF chip with a 6 × 3 culturing electrode array. (e) Schematic of the DMF chip with a 6 × n culturing electrode array (reproduced from ref. 108 with permission from John Wiley and Sons, copyright 2020). | ||
To optimize conditions for biohydrogen production effectively, microbioreactors can be employed. Microbioreactors are miniaturized cultivation systems widely utilized in biotechnological processes suitable for high-throughput optimization and precise environmental control. Their application in biohydrogen production, particularly from a modeling perspective, has gained attention fairly recently.95 Notably, Aghajani Delavar and Wang performed 3D modeling using the lattice Boltzmann method to assess the effects of illumination and fluid velocity on biohydrogen production through photofermentation in a microbioreactor.96 This modeling approach can provide critical insights that guide the experimental design and development of microbioreactors, potentially enhancing the efficiency and scalability of biohydrogen production from marine sources.
Microfluidic approaches offer the unique ability to manipulate and analyze cells, which have been widely applied to enhance microalgae biofuel production. There are rich review papers with comprehensive descriptions of these tools.103–107 Here, we highlight a couple of platforms published within the past five years with a focus on marine algae. Wang et al. introduced a digital microfluidic (DMF) system for automatic screening of microalgal growth and lipid accumulation under stress conditions (Fig. 6B).108 Microalgae cells were successfully cultivated in the discrete droplets that functioned as micro-bioreactors. Capacitance-based position estimator, electrode-saving-based compensation, and deterministic splitting approach were incorporated to optimize droplet manipulation. The system was able to screen 4 marine microalgae species for lipid production after a 5 day cultivation. Yu et al. developed a droplet-based microfluidic method for analyzing and screening microalgal populations, which can be integrated into genetic transformation workflows for strain engineering.109 A laser sheet illumination system was used to measure chlorophyll fluorescence intensity, which uniformly covered the sorting channel and enhanced the sorting performance. Wild type and genetically engineered cells of two marine algae species were sorted in a high throughput manner using this device at a rate of 1.2 × 105 cells per h, which could reduce transformation workflow from ∼7 weeks to 4 weeks.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 J within 90 s.
000 J within 90 s.
In contrast to piezoelectric generators, Wang et al. introduced an Underwater Flag-like Triboelectric Nanogenerator (UF-TENG) to capture energy from low-velocity ocean currents through flow-induced vibrations (Fig. 7A).112 The UF-TENG consists of a strip of PTFE membrane, 50 μm thick, sandwiched between two conductive ink-coated PET membranes, each 25 μm thick. The differing bending modulus of these materials causes the PTFE to alternately contact two electrodes during vibration. This interaction generates a transient current by transferring electrostatically induced charges upon contact. The UF-TENG had a critical start up flow velocity of 0.133 m s−1, and a peak output power of 52.3 μW at 0.461 m s−1. Impressively, a series of six UF-TENG units successfully powered an underwater thermometer, demonstrating the potential of such technologies for use in self-powered marine wireless sensing systems, enhancing monitoring and data collection capabilities in marine environments.
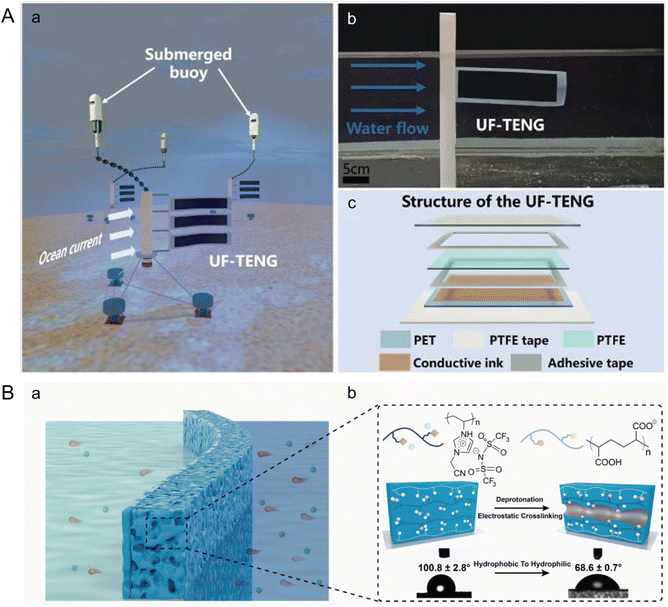 | ||
| Fig. 7 Microfluidics leveraging ocean waves and salinity gradients to harness marine energy. (A) Structure of an underwater flag-like triboelectric nanogenerator (UF-TENG) for harvesting ocean current energy by Wang et al. (a) Prototypes of the UF-TENG and self-powered submerged buoys. (b) UF-TENG in flowing water. (c) Detailed structure of the UF-TENG (reproduced from ref. 112 with permission from Elsevier, copyright 2021). (B) Bioinspired poly (ionic liquid) (PIL) membrane for efficient salinity gradient energy harvesting by Hu et al. (a) Schematic of the salinity gradient energy harvesting by employing the PIL membrane. (b) Molecular formulas of the two polyelectrolytes that make up the PIL membrane, schematic diagram of the formation mechanism of nanochannels, and its change in contact angle. The PIL membrane becomes hydrophilic after forming its channel (reproduced from ref. 115 with permission from Elsevier, copyright 2022). | ||
To improve these nanofluidic membranes further, enhancing their efficiency and facilitating their deployment, Dartoomi et al. proposed modeling the geometry of the nanofluidic channels and the bipolar layer to better evaluate ion transfer and power production.116 Additionally, Mao et al. emphasized the need to investigate the effects of membrane fouling, which can significantly impact the efficiency and lifespan of the energy conversion systems.117
Moreover, the concept of using salinity gradients for energy generation has been combined with freshwater production. Wani et al. developed a dual-purpose paper-based microfluidic system capable of generating both electricity and freshwater simultaneously throughout the day.118 This system maintains a salinity gradient in paper-based two-legged channels between saltwater and tap water, while also performing interface solar steam generation using a connected evaporator. A maximum output power density of 9.9 mW m−2 was achieved by connecting four channels in series. While at the prototype stage, this integrated approach not only provides a sustainable energy source but also addresses the critical need for freshwater, demonstrating the potential of microfluidic technologies to tackle multiple environmental challenges concurrently.
3.2. Carbon storage and sequestration in ocean ecosystems/sediments
As ocean ecosystems and sediments naturally function as carbon sinks, researchers are exploring solutions to leverage these systems for removing excess CO2 from the atmosphere. Solutions span a range of technologies, including chemical methods like ocean alkalinity enhancement and electrochemical approaches, as well as biological strategies such as nutrient fertilization, artificial upwelling and downwelling, seaweed cultivation, and the restoration of ocean and coastal ecosystems.9,16 Microfluidics, in particular, offers promising applications in enhancing carbon storage and sequestration in ocean-related systems. Microfluidic technology can improve the efficiency of CO2 biofixation by microalgae, a process critical for converting CO2 into biomass, thus effectively removing CO2 from the atmosphere. Additionally, microfluidic systems have the potential to shed light on the role of the microbial carbon pump in the ocean carbon cycle. This pump involves the transfer of carbon from surface waters to the deep ocean through microbial processes, contributing significantly to long-term carbon storage. By enabling precise control and monitoring of microenvironments, microfluidic devices can facilitate detailed studies of these complex biochemical pathways and interactions, enhancing our understanding and optimization of natural and engineered carbon sequestration methods.CO2 fixation through photosynthesis offers a dual benefit of reducing atmospheric CO2 levels while simultaneously accumulating biomass, which can be optimized using advanced technologies. Microfluidic devices are proving invaluable for enhancing the performance of CO2 fixation by microalgae in a high-throughput and controlled manner. Abdulla Yusuf et al. utilized a hybrid microfluidic-differential carbonator (μ-DC) to fine-tune the culture conditions for green microalgae, aiming to maximize CO2 biofixation rates.119 Key variables such as light intensity, CO2 concentration, and the ratio of microalgae to media were initially explored using a full factorial design. Subsequently, response surface methodology and face-centered central composite design were applied to optimize these critical factors.
Furthermore, microbial communities are essential in marine carbon sequestration through the microbial carbon pump mechanism, which involves the production of refractory dissolved organic carbon that resists remineralization and reversion to CO2, thereby forming a significant carbon reservoir.120–123 Microscale systems provide a platform to study the turnover of organic carbon at scales relevant to marine microbial ecology. For instance, Enke et al. utilized polysaccharide microbeads to investigate the degradation of particulate organic matter by marine bacterial communities on particle surfaces.124 Their findings highlighted that community composition significantly influences the half-life of these particles, with multispecies communities generally exhibiting longer half-lives. These insights not only enhance our understanding of microbial interactions but also contribute to strategies aimed at augmenting marine carbon sequestration.
4. Discussion
The use of microfluidics in studying the diverse impacts of climate change on ocean and marine systems is increasingly centered around in situ monitoring of environmental properties and ecosystems, with applications in energy generation. Microfluidics, coupled with miniaturized sensor systems, offers precise flow control and requires small sample volumes while maintaining detection limits comparable to traditional bulk methods. The compactness and flexibility of these systems make them ideal for long-term monitoring in remote oceanic locations. For effective field deployment, optimizing device performance is crucial, including enhancing device lifespan and improving the spatiotemporal resolution of analysis under natural ocean conditions such as pressure, temperature, salinity, and pH.18 Furthermore, the integration of energy harvesting devices directly on the chip could significantly enhance the versatility of in situ sensing applications. Looking forward, the development of automatic and intelligent data analysis capabilities for real-time monitoring data could facilitate smarter management of ocean resources.Microfluidic tools are also exceptionally well-suited to simulate microscale marine organisms, providing valuable insights into their behavior under climate-related perturbations. There is significant potential for these tools to advance the cultivation and study of non-model organisms, which are abundant in marine ecosystems. Marine plankton such as bacteria, phytoplankton, and zooplankton are particularly amenable to investigation in micro- and milli-fluidic systems due to their size scale. One commonly studied aspect is the motility of an organism, typically observed through real-time imaging. While microfluidic studies often focus on well-characterized, lab-raised strains, there is a growing interest in the biological community to explore “non-model” organisms that are challenging to manage in lab settings but could provide answers to questions unapproachable by existing model systems.125 For instance, many environmental microbes are non-culturable yet play critical ecological roles. To explore these organisms, a controlled perturbation module could be integrated into an in situ detection workflow, allowing for behavioral tests alongside identification.
Coral, another example of a non-model marine organism, has seen development in microfluidic platforms designed to provide a means for intermediate- and longer-term husbandry of coral polyps by precisely controlling the physical and chemical environment. However, both the long-term culture and reproduction of corals remain challenging. Microfluidic systems, through methods such as fractional-factorial experiment design, could help determine optimal conditions for prolonged culture and potentially reproduction. Even without the capability to induce reproduction of new generations, prolonged culture on a chip would significantly contribute to our understanding of coral responses to environmental stressors, offering a new avenue for coral conservation and study under the pressures of climate change.
Microfluidics, with its capabilities at small scales and controlled (extreme) conditions, is poised to significantly advance energy research,81 particularly in harnessing renewable energy from ocean systems. This technology is instrumental in developing methods to produce bioenergy through microfluidic benthic microbial fuel cells, fermentative biohydrogen generation, and marine microalgae biomass, as well as harvesting physical energy from ocean waves, currents, and salinity gradients. In marine systems, microfluidic energy harvesting can power in situ sensing and monitoring systems, enabling long-term measurements in remote ocean environments. Additionally, these technologies support the development of large-scale energy production. For instance, optimizing biohydrogen production could provide a clean biofuel source, and nanofluidic channels with high efficiency could be integrated into power plants that utilize salinity gradients. Moreover, another area of marine energy with potential for further exploration is energy derived from ocean thermal gradients. Microfluidic applications frequently require precise thermal control,126,127 and thermal gradients can induce flow within microfluidic chips.128 These systems could potentially harness thermal gradients from natural sources such as deep-sea hydrothermal vents or artificial sources like heat generated by ocean-operating equipment and platforms.
While the ocean offers significant capacity for carbon sequestration, microfluidic technologies have predominantly been applied to study CO2 fixation by photosynthesis. The turnover of fixed organic matter by marine bacterial communities could be further explored using microscale systems. Besides investigating ocean carbon flux through biological systems, another crucial aspect of marine carbon dioxide removal (CDR) involves chemical approaches that utilize carbon chemistry and alkalinity to enhance CO2 storage in seawater. The first deployment of an electrochemical marine CDR system was recently reported at PNNL in Washington.129 Microfluidics has also been used to study CO2 sequestration in saline aquifers by simulating the geo-surface environment and testing the effects of pressure and salinity.130–132 Similar experiments could be designed for chemical CO2 sequestration in oceans to optimize sequestration performance. Moreover, as marine carbon cycling is a complex process that involves photosynthetic fixation and microbial conversion through biological processes, as well as chemical processes of CO2 absorption and dissolution, understanding their synergistic effects is crucial to maximizing ocean carbon storage.122 Microfluidic devices, with their ability to manipulate microorganisms and integrate electrochemical systems, can be engineered to investigate these various processes and their interactions, thereby enhancing our understanding and effectiveness of marine-based climate mitigation strategies.
Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.Conflicts of interest
There are no conflicts to declare.References
- K. E. Mills, E. B. Osborne, R. J. Bell, C. S. Colgan, S. R. Cooley, M. C. Goldstein, R. B. Griffis, K. Holsman, M. Jacox and F. Micheli, in Fifth National Climate Assessment, ed. A. R. Crimmins, C. W. Avery, D. R. Easterling, K. E. Kunkel, B. C. Stewart and T. K. Maycock, U.S. Global Change Research Program, Washington, DC, USA, 2023, ch. 10 Search PubMed.
- N. L. Bindoff, W. W. L. Cheung, J. G. Kairo, J. Arístegui, V. A. Guinder, R. Hallberg, N. Hilmi, N. Jiao, M. S. Karim, L. Levin, S. O'Donoghue, S. R. P. Cuicapusa, B. Rinkevich, T. Suga, A. Tagliabue and P. Williamson, in The Ocean and Cryosphere in a Changing Climate: Special Report of the Intergovernmental Panel on Climate Change, ed. H.-O. Pörtner, D. C. Roberts, V. Masson-Delmotte, P. Zhai, M. Tignor, E. Poloczanska, K. Mintenbeck, A. Alegría, M. Nicolai, A. Okem, J. Petzold, B. Rama and N. M. Weyer, Cambridge University Press, Cambridge, 2019, ch. 5, pp. 447–588 Search PubMed.
- M. Oppenheimer, B. C. Glavovic, J. Hinkel, R. V. D. Wal, A. K. Magnan, A. Abd-Elgawad, R. Cai, M. Cifuentes-Jara, R. M. DeConto, T. Ghosh, J. Hay, F. Isla, B. Marzeion, B. Meyssignac and Z. Sebesvari, in The Ocean and Cryosphere in a Changing Climate: Special Report of the Intergovernmental Panel on Climate Change, ed. H.-O. Pörtner, D. C. Roberts, V. Masson-Delmotte, P. Zhai, M. Tignor, E. Poloczanska, K. Mintenbeck, A. Alegría, M. Nicolai, A. Okem, J. Petzold, B. Rama and N. M. Weyer, Cambridge University Press, Cambridge, 2019, ch. 4, pp. 321–446 Search PubMed.
- J. G. Canadell, P. M. S. Monteiro, M. H. Costa, L. C. D. Cunha, P. M. Cox, A. V. Eliseev, S. Henson, M. Ishii, S. Jaccard, C. Koven, A. Lohila, P. K. Patra, S. Piao, J. Rogelj, S. Syampungani, S. Zaehle and K. Zickfeld, in Climate Change 2021 – The Physical Science Basis: Working Group I Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, ed. V. Masson-Delmotte, P. Zhai, A. Pirani, S. L. Connors, C. Péan, S. Berger, N. Caud, Y. Chen, L. Goldfarb, M. I. Gomis, M. Huang, K. Leitzell, E. Lonnoy, J. B. R. Matthews, T. K. Maycock, T. Waterfield, O. Yelekçi, R. Yu and B. Zhou, Cambridge University Press, Cambridge, 2021, ch. 5, pp. 673–816 Search PubMed.
- G. T. Pecl, M. B. Araújo, J. D. Bell, J. Blanchard, T. C. Bonebrake, I. C. Chen, T. D. Clark, R. K. Colwell, F. Danielsen, B. Evengård, L. Falconi, S. Ferrier, S. Frusher, R. A. Garcia, R. B. Griffis, A. J. Hobday, C. Janion-Scheepers, M. A. Jarzyna, S. Jennings, J. Lenoir, H. I. Linnetved, V. Y. Martin, P. C. McCormack, J. McDonald, N. J. Mitchell, T. Mustonen, J. M. Pandolfi, N. Pettorelli, E. Popova, S. A. Robinson, B. R. Scheffers, J. D. Shaw, C. J. B. Sorte, J. M. Strugnell, J. M. Sunday, M.-N. Tuanmu, A. Vergés, C. Villanueva, T. Wernberg, E. Wapstra and S. E. Williams, Science, 2017, 355, eaai9214 CrossRef PubMed.
- T. P. Hughes, M. L. Barnes, D. R. Bellwood, J. E. Cinner, G. S. Cumming, J. B. C. Jackson, J. Kleypas, I. A. van de Leemput, J. M. Lough, T. H. Morrison, S. R. Palumbi, E. H. van Nes and M. Scheffer, Nature, 2017, 546, 82–90 CrossRef CAS PubMed.
- C. A. Burge, C. Mark Eakin, C. S. Friedman, B. Froelich, P. K. Hershberger, E. E. Hofmann, L. E. Petes, K. C. Prager, E. Weil, B. L. Willis, S. E. Ford and C. D. Harvell, Annu. Rev. Mar. Sci., 2014, 6, 249–277 CrossRef PubMed.
- C. A. Heil and A. L. Muni-Morgan, Front. Ecol. Evol., 2021, 9, 646080 CrossRef.
- OPC, Ocean Climate Action Plan, U.S. government, United States of America, 2023 Search PubMed.
- C. Voigt, in The Environmental Rule of Law for Oceans: Designing Legal Solutions, ed. F. M. Platjouw and A. Pozdnakova, Cambridge University Press, Cambridge, 2023, pp. 17–30 Search PubMed.
- Y. Sang, H. B. Karayaka, Y. Yan, N. Yilmaz and D. Souders, in Comprehensive Energy Systems, ed. I. Dincer, Elsevier, Oxford, 2018, pp. 733–769 Search PubMed.
- IRENA, Innovation outlook: Ocean energy technologies, Abu Dhabi, 2020 Search PubMed.
- Tethys_PNNL, Marine Energy, https://tethys.pnnl.gov/technology/marine-energy, (accessed May 22, 2024) Search PubMed.
- P. Friedlingstein, M. O'Sullivan, M. W. Jones, R. M. Andrew, J. Hauck, A. Olsen, G. P. Peters, W. Peters, J. Pongratz, S. Sitch, C. Le Quéré, J. G. Canadell, P. Ciais, R. B. Jackson, S. Alin, L. E. O. C. Aragão, A. Arneth, V. Arora, N. R. Bates, M. Becker, A. Benoit-Cattin, H. C. Bittig, L. Bopp, S. Bultan, N. Chandra, F. Chevallier, L. P. Chini, W. Evans, L. Florentie, P. M. Forster, T. Gasser, M. Gehlen, D. Gilfillan, T. Gkritzalis, L. Gregor, N. Gruber, I. Harris, K. Hartung, V. Haverd, R. A. Houghton, T. Ilyina, A. K. Jain, E. Joetzjer, K. Kadono, E. Kato, V. Kitidis, J. I. Korsbakken, P. Landschützer, N. Lefèvre, A. Lenton, S. Lienert, Z. Liu, D. Lombardozzi, G. Marland, N. Metzl, D. R. Munro, J. E. M. S. Nabel, S. I. Nakaoka, Y. Niwa, K. O'Brien, T. Ono, P. I. Palmer, D. Pierrot, B. Poulter, L. Resplandy, E. Robertson, C. Rödenbeck, J. Schwinger, R. Séférian, I. Skjelvan, A. J. P. Smith, A. J. Sutton, T. Tanhua, P. P. Tans, H. Tian, B. Tilbrook, G. van der Werf, N. Vuichard, A. P. Walker, R. Wanninkhof, A. J. Watson, D. Willis, A. J. Wiltshire, W. Yuan, X. Yue and S. Zaehle, Earth Syst. Sci. Data, 2020, 12, 3269–3340 CrossRef.
- M. Hori, C. J. Bayne and T. Kuwae, in Blue Carbon in Shallow Coastal Ecosystems: Carbon Dynamics, Policy, and Implementation, ed. T. Kuwae and M. Hori, Springer Singapore, Singapore, 2019, pp. 1–31 Search PubMed.
- NASEM, A Research Strategy for Ocean-based Carbon Dioxide Removal and Sequestration, The National Academies Press, Washington, DC, 2022 Search PubMed.
- NASEM, Negative Emissions Technologies and Reliable Sequestration: A Research Agenda, The National Academies Press, Washington, DC, 2019 Search PubMed.
- T. Fukuba and T. Fujii, Lab Chip, 2021, 21, 55–74 RSC.
- S. S. Datta, I. Battiato, M. A. Fernø, R. Juanes, S. Parsa, V. Prigiobbe, E. Santanach-Carreras, W. Song, S. L. Biswal and D. Sinton, Lab Chip, 2023, 23, 1358–1375 RSC.
- P. Aryal, C. Hefner, B. Martinez and C. S. Henry, Lab Chip, 2024, 24, 1175–1206 RSC.
- S. M. Bushinsky, Y. Takeshita and N. L. Williams, Curr. Clim. Change Rep., 2019, 5, 207–220 CrossRef PubMed.
- V. C. Pinto, C. F. Araújo, P. J. Sousa, L. M. Gonçalves and G. Minas, Sens. Actuators, B, 2019, 290, 285–292 CrossRef CAS.
- C. Sonnichsen, D. Atamanchuk, A. Hendricks, S. Morgan, J. Smith, I. Grundke, E. Luy and V. J. Sieben, ACS Sens., 2023, 8, 344–352 CrossRef CAS PubMed.
- M. Tweedie, A. Macquart, J. Almeida, B. Ward and P. Maguire, Meas. Sci. Technol., 2020, 31, 065104 CrossRef CAS.
- M. Tweedie, D. Sun, D. R. Gajula, B. Ward and P. D. Maguire, Microfluid. Nanofluid., 2020, 24, 37 CrossRef CAS PubMed.
- R. H. Byrne, Environ. Sci. Technol., 2014, 48, 5352–5360 CrossRef CAS PubMed.
- D. S. de Bruijn, D. B. Van de Waal, N. R. Helmsing, W. Olthuis and A. van den Berg, Glob. Chall., 2023, 7, 2200151 CrossRef PubMed.
- N. Ramanathan, O. Simakov, C. A. Merten and D. Arendt, PLoS One, 2015, 10, e0140553 CrossRef PubMed.
- S.-W. Li, P.-H. Lin, T.-Y. Ho, C.-H. Hsieh and C.-L. Sun, Sci. Rep., 2021, 11, 11105 CrossRef CAS PubMed.
- C.-L. Sun, H.-C. Lee and R.-X. Kao, Exp. Fluids, 2012, 52, 23–35 CrossRef CAS.
- P. Dey, T. M. Bradley and A. Boymelgreen, Sci. Rep., 2023, 13, 6370 CrossRef CAS PubMed.
- A.-P. Pang, Y. Luo, C. He, Z. Lu and X. Lu, Sci. Rep., 2020, 10, 6964 CrossRef CAS PubMed.
- Y. Luo, J. Zhao, C. He, Z. Lu and X. Lu, Micromachines, 2020, 11, 127 CrossRef PubMed.
- S. Zhou, E. S. Fu, B. Chen and H. Yan, Micromachines, 2022, 13, 832 CrossRef PubMed.
- O. H. Shapiro, E. Kramarsky-Winter, A. R. Gavish, R. Stocker and A. Vardi, Nat. Commun., 2016, 7, 10860 CrossRef CAS PubMed.
- E. Gibbin, A. Gavish, I. Domart-Coulon, E. Kramarsky-Winter, O. Shapiro, A. Meibom and A. Vardi, BMC Microbiol., 2018, 18, 39 CrossRef CAS PubMed.
- W. Van Treuren, K. K. Brower, L. Labanieh, D. Hunt, S. Lensch, B. Cruz, H. N. Cartwright, C. Tran and P. M. Fordyce, Sci. Rep., 2019, 9, 9275 CrossRef PubMed.
- A. Sebé-Pedrós, B. Saudemont, E. Chomsky, F. Plessier, M.-P. Mailhé, J. Renno, Y. Loe-Mie, A. Lifshitz, Z. Mukamel, S. Schmutz, S. Novault, P. R. H. Steinmetz, F. Spitz, A. Tanay and H. Marlow, Cell, 2018, 173, 1520–1534.e1520 CrossRef PubMed.
- S. Levy, A. Elek, X. Grau-Bové, S. Menéndez-Bravo, M. Iglesias, A. Tanay, T. Mass and A. Sebé-Pedrós, Cell, 2021, 184, 2973–2987.e2918 CrossRef CAS PubMed.
- J. P. Engelberts, S. J. Robbins, K. Damjanovic and N. S. Webster, Mar. Biol., 2021, 168, 175 CrossRef.
- C. Daniels, S. Baumgarten, L. Yum, C. Michell, T. Bayer, C. Arif, C. Roder, E. Weil and C. Voolstra, Front. Mar. Sci., 2015, 2, 62 Search PubMed.
- M. Liu, L. Fan, L. Zhong, S. Kjelleberg and T. Thomas, ISME J., 2012, 6, 1515–1525 CrossRef CAS PubMed.
- A. Williams, E. N. Chiles, D. Conetta, J. S. Pathmanathan, P. A. Cleves, H. M. Putnam, X. Su and D. Bhattacharya, Sci. Adv., 2021, 7, eabd4210 CrossRef CAS PubMed.
- P. Tremblay, J. F. Maguer, R. Grover and C. Ferrier-Pagès, J. Exp. Biol., 2015, 218, 1223–1234 Search PubMed.
- P. Schimak Mario, M. Kleiner, S. Wetzel, M. Liebeke, N. Dubilier and M. Fuchs Bernhard, Appl. Environ. Microbiol., 2016, 82, 62–70 CrossRef CAS PubMed.
- P. Kubryk, J. S. Kölschbach, S. Marozava, T. Lueders, R. U. Meckenstock, R. Niessner and N. P. Ivleva, Anal. Chem., 2015, 87, 6622–6630 CrossRef CAS PubMed.
- T. Krueger, J. Bodin, N. Horwitz, C. Loussert-Fonta, A. Sakr, S. Escrig, M. Fine and A. Meibom, Sci. Rep., 2018, 8, 12710 CrossRef PubMed.
- C. Kopp, M. Wisztorski, J. Revel, M. Mehiri, V. Dani, L. Capron, D. Carette, I. Fournier, L. Massi, D. Mouajjah, S. Pagnotta, F. Priouzeau, M. Salzet, A. Meibom and C. Sabourault, Zoology, 2015, 118, 125–131 CrossRef CAS PubMed.
- K. Petrou, D. A. Nielsen and P. Heraud, Front. Mar. Sci., 2018, 5, 110 CrossRef.
- M. Garren, K. Son, J. Tout, J. R. Seymour and R. Stocker, ISME J., 2016, 10, 1363–1372 CrossRef CAS PubMed.
- M. M. Salek, F. Carrara, V. Fernandez, J. S. Guasto and R. Stocker, Nat. Commun., 2019, 10, 1877 CrossRef PubMed.
- R. J. Henshaw, J. Moon, M. R. Stehnach, B. P. Bowen, S. M. Kosina, T. R. Northen, J. S. Guasto and S. A. Floge, bioRxiv, 2023, preprint, DOI:10.1101/2023.10.24.563588.
- A. L. Calkins, L. M. Demey, B. M. Rosenthal, V. J. DiRita and J. S. Biteen, Anal. Chem., 2023, 95, 774–783 CAS.
- K.-Y. Lien, S.-H. Lee, T.-J. Tsai, T.-Y. Chen and G.-B. Lee, Microfluid. Nanofluid., 2009, 7, 795 CrossRef CAS.
- J. Jin, L. Duan, J. Fu, F. Chai, Q. Zhou, Y. Wang, X. Shao, L. Wang, M. Yan, X. Su, Y. Zhang, J. Pan and J. Chen, Anal. Methods, 2021, 13, 2710–2721 RSC.
- P. Li, L. Qiang, Y. Han, Y. Chu, J. Qiu, F. Song, M. Wang, Q. He, Y. Zhang, M. Sun, C. Li, S. Song, Y. Liu, L. Han and Y. Zhang, Micromachines, 2022, 13, 1759 CrossRef PubMed.
- J. Wang, G. Wang, M. Chen, Y. Wang, G. Ding, Y. Zhang, Y. Kang and X. Pan, Algal Res., 2019, 42, 101593 CrossRef.
- J. Zheng, T. Cole, Y. Zhang, D. Yuan Bayinqiaoge and S.-Y. Tang, Lab Chip, 2024, 24, 244–253 RSC.
- J. S. Erickson, N. Hashemi, J. M. Sullivan, A. D. Weidemann and F. S. Ligler, Anal. Chem., 2012, 84, 839–850 CrossRef CAS PubMed.
- M. Thyssen, S. Alvain, A. Lefèbvre, D. Dessailly, M. Rijkeboer, N. Guiselin, V. Creach and L. F. Artigas, Biogeosciences, 2015, 12, 4051–4066 CrossRef.
- G. C. Pereira, A. R. Figueiredoa and N. F. F. Ebeckena, Braz. J. Microbiol., 2018, 78, 240–247 CAS.
- J. Zhang, S. Liu, P. Yang and G. Sui, Lab Chip, 2011, 11, 3516–3522 RSC.
- I. Maguire, J. Fitzgerald, B. Heery, C. Nwankire, R. O'Kennedy, J. Ducrée and F. Regan, ACS Omega, 2018, 3, 6624–6634 CrossRef CAS PubMed.
- T. DeVries, Nat. Geosci., 2018, 11, 15–16 CrossRef CAS.
- D. A. Hutchins and A. Tagliabue, Nat. Geosci., 2024, 17, 495–502 CrossRef CAS.
- P. Jin, J. Zhang, J. Wan, S. Overmans, G. Gao, M. Ye, X. Dai, J. Zhao, M. Xiao and J. Xia, Front. Mar. Sci., 2021, 8, 801889 CrossRef.
- A. Botté, C. Seguin, J. Nahrgang, M. Zaidi, J. Guery and V. Leignel, Ecotoxicology, 2022, 31, 194–207 CrossRef PubMed.
- F. Haque and C. Fan, iScience, 2023, 26, 107649 CrossRef CAS PubMed.
- H. V. Ford, N. H. Jones, A. J. Davies, B. J. Godley, J. R. Jambeck, I. E. Napper, C. C. Suckling, G. J. Williams, L. C. Woodall and H. J. Koldewey, Sci. Total Environ., 2022, 806, 150392 CrossRef CAS PubMed.
- X.-F. Wei, W. Yang and M. S. Hedenqvist, Nat. Commun., 2024, 15, 2052 CrossRef CAS PubMed.
- J. Zhu, G. Han, X. Hu, Y. Zuo, L. Chen, F. Wang, Y. Yang, F. Jiang, C. Sun, W. Zhao and X. Han, ACS Sens., 2020, 5, 1381–1388 CrossRef CAS PubMed.
- A. D. Beaton, A. M. Schaap, R. Pascal, R. Hanz, U. Martincic, C. L. Cardwell, A. Morris, G. Clinton-Bailey, K. Saw, S. E. Hartman and M. C. Mowlem, ACS Sens., 2022, 7, 89–98 CrossRef CAS PubMed.
- S. Motahari, S. Morgan, A. Hendricks, C. Sonnichsen and V. Sieben, Micromachines, 2024, 15, 519 CrossRef PubMed.
- F. Wang, J. Zhu, X. Hu, L. Chen, Y. Zuo, Y. Yang, F. Jiang, C. Sun, W. Zhao and X. Han, Lab Chip, 2021, 21, 1109–1117 RSC.
- M. F. Altahan, M. Esposito and E. P. Achterberg, Sensors, 2022, 22, 3479 CrossRef CAS PubMed.
- Y. Liang, M. Ma, F. Zhang, F. Liu, T. Lu, Z. Liu and Y. Li, ACS Omega, 2021, 6, 9302–9309 CrossRef CAS PubMed.
- S. Ma, W. Zhao, Q. Zhang, K. Zhang, C. Liang, D. Wang, X. Liu and X. Zhan, J. Hazard. Mater., 2023, 448, 130923 CrossRef CAS PubMed.
- L. Gong, O. Martinez, P. Mesquita, K. Kurtz, Y. Xu and Y. Lin, Sci. Rep., 2023, 13, 11011 CrossRef CAS PubMed.
- M. Zhang, X. Wang, Y. Zhang and Y. Fan, Anal. Chim. Acta, 2023, 1261, 341237 CrossRef CAS PubMed.
- J. Dou, Z. Yang, B. Singh, B. Ma, Z. Lu, J. Xu and Y. He, Sci. Total Environ., 2024, 937, 173597 CrossRef CAS PubMed.
- D. Sinton, Lab Chip, 2014, 14, 3127–3134 RSC.
- D. R. Lovley, Nat. Rev. Microbiol., 2006, 4, 497–508 CrossRef CAS PubMed.
- B. E. Logan and J. M. Regan, Trends Microbiol., 2006, 14, 512–518 CrossRef CAS PubMed.
- B. E. Logan, ChemSusChem, 2012, 5, 988–994 CrossRef CAS PubMed.
- P. Pandey, V. N. Shinde, R. L. Deopurkar, S. P. Kale, S. A. Patil and D. Pant, Appl. Energy, 2016, 168, 706–723 CrossRef CAS.
- J. R. Trapero, L. Horcajada, J. J. Linares and J. Lobato, Appl. Energy, 2017, 185, 698–707 CrossRef CAS.
- Y. Gao, M. Mohammadifar and S. Choi, Adv. Mater. Technol., 2019, 4, 1900079 CrossRef CAS.
- T. Nguyen, Y. M. Arias-Thode, A. Obraztsova, A. Sarmiento, A. Stevens-Bracy, D. Grbovic and E. P. Kartalov, J. Environ. Chem. Eng., 2021, 9, 105659 CrossRef CAS.
- J. D. Holladay, J. Hu, D. L. King and Y. Wang, Catal. Today, 2009, 139, 244–260 CrossRef CAS.
- A. Reungsang, N. Zhong, Y. Yang, S. Sittijunda, A. Xia and Q. Liao, in Bioreactors for Microbial Biomass and Energy Conversion, ed. Q. Liao, J.-S. Chang, C. Herrmann and A. Xia, Springer Singapore, Singapore, 2018, pp. 221–317 Search PubMed.
- A. Adessi, E. Corneli and R. De Philippis, in Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects, ed. P. C. Hallenbeck, Springer International Publishing, Cham, 2017, pp. 321–350 Search PubMed.
- J. Cai and G. Wang, Int. J. Hydrogen Energy, 2012, 37, 15070–15080 CrossRef CAS.
- J. Cai, G. Wang and G. Pan, Int. J. Hydrogen Energy, 2012, 37, 4057–4067 CrossRef CAS.
- Y. J. Kim, H. S. Lee, E. S. Kim, S. S. Bae, J. K. Lim, R. Matsumi, A. V. Lebedinsky, T. G. Sokolova, D. A. Kozhevnikova, S.-S. Cha, S.-J. Kim, K. K. Kwon, T. Imanaka, H. Atomi, E. A. Bonch-Osmolovskaya, J.-H. Lee and S. G. Kang, Nature, 2010, 467, 352–355 CrossRef CAS PubMed.
- L. J. Frey and R. Krull, in Microfluidics in Biotechnology, ed. J. Bahnemann and A. Grünberger, Springer International Publishing, Cham, 2022, pp. 67–100 Search PubMed.
- M. Aghajani Delavar and J. Wang, Renewable Energy, 2022, 181, 1034–1045 CrossRef CAS.
- M. Wang, X. Ye, H. Bi and Z. Shen, Biotechnol. Biofuels Bioprod., 2024, 17, 10 CrossRef PubMed.
- M. Roy and K. Mohanty, Algal Res., 2019, 44, 101683 CrossRef.
- J. R. Ziolkowska, in Biofuels for a More Sustainable Future: Life Cycle Sustainability Assessment and Multi-Criteria Decision Making, ed. J. Ren, A. Scipioni, A. Manzardo and H. Liang, 2020, ch. 1, pp. 1–19 Search PubMed.
- M. A. Borowitzka, J. Appl. Phycol., 2013, 25, 743–756 CrossRef CAS.
- Y. Maeda, T. Yoshino, T. Matsunaga, M. Matsumoto and T. Tanaka, Curr. Opin. Biotechnol., 2018, 50, 111–120 CrossRef CAS PubMed.
- Z. H.-Y. Tay, F.-L. Ng, T.-C. Ling, M. Iwamoto and S.-M. Phang, 3 Biotech, 2022, 12, 148 CrossRef PubMed.
- Y.-J. Juang and J.-S. Chang, Biotechnol. J., 2016, 11, 327–335 CrossRef CAS PubMed.
- H. S. Kim, T. P. Devarenne and A. Han, Algal Res., 2018, 30, 149–161 CrossRef.
- P. Bodénès, H.-Y. Wang, T.-H. Lee, H.-Y. Chen and C.-Y. Wang, Biotechnol. Biofuels, 2019, 12, 33 CrossRef PubMed.
- F. Liu, A. Giometto and M. Wu, Anal. Bioanal. Chem., 2021, 413, 2331–2344 CrossRef CAS PubMed.
- A. B. Alias, S. Mishra, G. Pendharkar, C.-S. Chen, C.-H. Liu, Y.-J. Liu and D.-J. Yao, Molecules, 2022, 27, 1910 CrossRef CAS PubMed.
- Y. Wang, H. Zhao, X. Liu, W. Lin, Y. Jiang, J. Li, Q. Zhang and G. Zheng, Biotechnol. Bioeng., 2021, 118, 294–304 CrossRef CAS PubMed.
- Z. Yu, K. Geisler, T. Leontidou, R. E. B. Young, S. E. Vonlanthen, S. Purton, C. Abell and A. G. Smith, Algal Res., 2021, 56, 102293 CrossRef PubMed.
- Y. Zhou, F. Kong, H. Liu, Y. Jin, H. Chen and C. Sun, Renewable Energy, 2024, 225, 120325 CrossRef.
- G. W. Taylor, J. R. Burns, S. A. Kammann, W. B. Powers and T. R. Welsh, IEEE J. Oceanic Eng., 2001, 26, 539–547 CrossRef.
- Y. Wang, X. Liu, T. Chen, H. Wang, C. Zhu, H. Yu, L. Song, X. Pan, J. Mi, C. Lee and M. Xu, Nano Energy, 2021, 90, 106503 CrossRef CAS.
- Z. Zhang, L. Wen and L. Jiang, Nat. Rev. Mater., 2021, 6, 622–639 CrossRef CAS.
- W. Xin, L. Jiang and L. Wen, Acc. Chem. Res., 2021, 54, 4154–4165 CrossRef CAS PubMed.
- Y. Hu, Y. Teng, Y. Sun, P. Liu, L. Fu, L. Yang, X.-Y. Kong, Q. Zhao, L. Jiang and L. Wen, Nano Energy, 2022, 97, 107170 CrossRef CAS.
- H. Dartoomi, M. Khatibi and S. N. Ashrafizadeh, Langmuir, 2022, 38, 10313–10330 CrossRef CAS PubMed.
- R. Mao, X. Chen, R. Long, X. Liu, Z. Liu and W. Liu, Chem. Eng. Sci., 2023, 281, 119156 CrossRef CAS.
- T. A. Wani, P. Kaith, P. Garg and A. Bera, ACS Appl. Mater. Interfaces, 2022, 14, 35802–35808 CrossRef CAS PubMed.
- H. Abdulla Yusuf, S. M. Z. Hossain, A. A. Khamis, H. T. Radhi and A. S. Jaafar, J. CO2 Util., 2020, 42, 101291 CrossRef.
- N. Jiao, G. J. Herndl, D. A. Hansell, R. Benner, G. Kattner, S. W. Wilhelm, D. L. Kirchman, M. G. Weinbauer, T. Luo, F. Chen and F. Azam, Nat. Rev. Microbiol., 2010, 8, 593–599 CrossRef CAS PubMed.
- N. Jiao and Q. Zheng, Appl. Environ. Microbiol., 2011, 77, 7439–7444 CrossRef CAS PubMed.
- N. Jiao, T. Luo, Q. Chen, Z. Zhao, X. Xiao, J. Liu, Z. Jian, S. Xie, H. Thomas, G. J. Herndl, R. Benner, M. Gonsior, F. Chen, W.-J. Cai and C. Robinson, Nat. Rev. Microbiol., 2024, 22, 408–419 CrossRef CAS PubMed.
- S. N. Ahmad, T. A. Mir, T. Shareef, S. Pattnaik and S. A. Lone, in Microbiomes and the Global Climate Change, ed. S. A. Lone and A. Malik, Springer Singapore, Singapore, 2021, pp. 17–29 Search PubMed.
- T. N. Enke, G. E. Leventhal, M. Metzger, J. T. Saavedra and O. X. Cordero, Nat. Commun., 2018, 9, 2743 CrossRef PubMed.
- J. J. Russell, J. A. Theriot, P. Sood, W. F. Marshall, L. F. Landweber, L. Fritz-Laylin, J. K. Polka, S. Oliferenko, T. Gerbich, A. Gladfelter, J. Umen, M. Bezanilla, M. A. Lancaster, S. He, M. C. Gibson, B. Goldstein, E. M. Tanaka, C.-K. Hu and A. Brunet, BMC Biol., 2017, 15, 55 CrossRef PubMed.
- M. B. Kulkarni and S. Goel, Sens. Actuators, A, 2022, 341, 113590 CrossRef CAS.
- A. Khater, O. Abdelrehim, M. Mohammadi, A. Mohamad and A. Sanati-Nezhad, TrAC, Trends Anal. Chem., 2021, 138, 116234 CrossRef CAS.
- W. Dai, X. Xia, X. Ding, X. Wei, X. Zhu, W. Xue, Z. Hou and Y. Cao, Adv. Mater. Technol., 2024, 9, 2301986 CrossRef CAS.
- EERE, First-of-its-Kind Marine Carbon Dioxide Removal Device Deployed in Washington State, U.S. Department of Energy, Washington, 2024 Search PubMed.
- Y. Q. Zhang, A. Sanati-Nezhad and S. H. Hejazi, Lab Chip, 2018, 18, 285–295 RSC.
- T.-H. M. Ho, J. Yang and P. A. Tsai, Lab Chip, 2021, 21, 3942–3951 RSC.
- A. Sell, H. Fadaei, M. Kim and D. Sinton, Environ. Sci. Technol., 2013, 47, 71–78 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |
