Open Access Article
Open Access ArticleCompact lens-free imager using a thin-film transistor for long-term quantitative monitoring of stem cell culture and cardiomyocyte production†
Taishi
Kakizuka
 ab,
Tohru
Natsume
c and
Takeharu
Nagai
ab,
Tohru
Natsume
c and
Takeharu
Nagai
 *ab
*ab
aSANKEN, The University of Osaka, Mihogaoka 8-1, Ibaraki, Osaka 567-0047, Japan. E-mail: ng1@sanken.osaka-u.ac.jp
bTransdimensional Life Imaging Division, Institute for Open and Transdisciplinary Research Initiatives, The University of Osaka, Yamadaoka 2-1, Suita, Osaka 565-0871, Japan
cCellular and Molecular Biotechnology Research Institute, National Institute of Advanced Industrial Science and Technology, 2-3-26 Aoumi, Koto-ku, Tokyo 135-0064, Japan
First published on 14th October 2024
Abstract
With advancements in human induced pluripotent stem cell (hiPSC) technology, there is an increasing demand for quality control techniques to manage the long-term process of target cell production effectively. While monitoring systems designed for use within incubators are promising for assessing culture quality, existing systems still face challenges in terms of compactness, throughput, and available metrics. To address these limitations, we have developed a compact and high-throughput lens-free imaging device named INSPCTOR. The device is as small as a standard culture plate, which allows for the installation of multiple units within an incubator. INSPCTOR utilises a large thin-film transistor image sensor, enabling simultaneous observation of six independent culture environments, each approximately 1 cm2. With this device, we successfully monitored the confluency of hiPSC cultures and identified the onset timing of epithelial-to-mesenchymal transition during mesodermal induction. Additionally, we quantified the beating frequency and conduction of hiPSC-derived cardiomyocytes by using high-speed imaging modes. This enabled us to identify the onset of spontaneous beating during differentiation and assess chronotropic responses in drug evaluations. Moreover, by tracking beating frequency over 10 days of cardiomyocyte maturation, we identified week-scale and daily-scale fluctuations, the latter of which correlated with cellular metabolic activity. The metrics derived from this device would enhance the reproducibility and quality of target cell production.
Introduction
The recent advances in human induced pluripotent stem cell (hiPSC) technology have enabled the production of various cell types constituting our human body, accelerating progress in regenerative medicine and drug discovery.1–3 The process of inducing target cells from hiPSCs is a prolonged procedure, lasting from days to weeks and sometimes even months. Furthermore, the optimised culture conditions and induction efficiency are known to vary from lab to lab, from one cell line to another, and sometimes depending on subtle differences in sample handling.4,5 Therefore, optimisation of the induction conditions is often required for each experimental site, and strict control of the entire process is essential for achieving high reproducibility. Consequently, lab automation has attracted attention as a technology for efficiently handling a large number of cell culture samples with high reproducibility.6–8 On the other hand, it is crucial not only to handle cell culture efficiently and reproducibly but also to evaluate their quality effectively. If the induction process is evaluated on the end products alone, the entire process, which may have taken several weeks, might be rendered useless. The ability to monitor and evaluate quality during the induction process enables early assessment of culture conditions, rapid detection of abnormalities, and rigorous reproducibility, including at intermediate points.9–12 Thus, systems capable of long-term monitoring and quality evaluation of cell culture are of significant value.Imaging devices installed inside incubators serve as the primary technology for long-term cell culture monitoring.13 At the same time, several requirements must be met to effectively monitor hiPSCs culture and their differentiation induction. The first requirement is compactness since incubators offer limited space. In particular, miniaturised microscopes and lens-free imaging systems meet this demand. Miniaturised microscopes are designed to maintain as high resolution as conventional microscopes in a smaller system, making them useful for analysing detailed structures.14–17 Lens-free imaging systems offer a wide field of view, further miniaturisation, and cost-effectiveness since they do not require magnification and objective lenses.18–24
High throughput is also essential, given the need for parallel culture under multiple conditions, such as optimisation of differentiation induction. High-throughput devices capable of observing multiple regions using scanning mechanisms have been reported and commercialised.25,26 For instance, products such as the provi-CM20 and CM30 by Evident are representative and are already in use.27 Additionally, technologies have been developed that allow simultaneous observation of multiple wells in a culture plate by aligning compact imaging units (illumination, lenses, and sensors) to match the positions of the wells.28–30 However, most of these devices are large and expensive, posing a challenge to miniaturisation. While these devices can be effectively utilised in labs equipped with large incubators and ample funding, there is still a significant demand for compact and high-throughput devices.
Finally, providing effective metrics for quality control is a crucial requirement. Previously, many in-incubator monitoring systems have attempted to achieve high resolution and implement fluorescence observation.15,16,19,20,28,29 For example, Son et al. developed an upright-type miniaturised microscope array mountable on a 12-well plate, achieving a single-cell resolution fluorescence imaging system that is relatively compact and high-throughput while the field of view (FOV) is small.28 Such technology is expected to be utilised for detailed dynamic analysis at the cellular level. However, from the perspective of quality control in cell production processes for regenerative medicine and drug discovery, these devices may not be suitable. For instance, confluency is the most common parameter in cell culture monitoring, where a wide FOV that can capture many hiPSC colonies is more important than resolution. In addition, label-free quantifications are more crucial than fluorescence observation. Therefore, it is challenging to find a monitoring system that simultaneously meets all these requirements above.
Here, we propose an in-incubator cell imaging device that is compact, high-throughput, and capable of providing quantitative metrics for quality control of hiPSC culture and the induction processes of target cells. This device utilises lens-free imaging and notably uses a thin-film transistor (TFT) image sensor. TFT technology, primarily developed to meet the market demands of liquid crystal displays,31,32 is also widely used in biological research.33–35 Applications include electrophysiological studies,36 X-ray imaging,37,38 bacterial colony classification,39 and biochemical detection such as glucose sensors,40,41 immunosensors,42 and DNA sensors.43 TFTs have the advantage of enabling the cost-effective production of large image sensors.32,39 In this study, we developed an image sensor with a large FOV by combining TFT and photodiode arrays. By placing cell culture chambers on the sensor, six independent culture environments can be simultaneously monitored with an FOV of approximately 1 cm2. Additionally, by implementing lens-free imaging with dark-field illumination from the rear and scattered light detection, we reduced the illumination system's size, achieving a device volume comparable to standard cell culture plates such as 6-well plates. We named this device INSPCTOR (in-incubator specialised compact lens-free TFT cell monitor).
A potential disadvantage of the TFT image sensor is its large pixel pitch, which reduces spatial resolution. At the same time, during the hiPSCs culture and target cell inductions, colony-level morphological changes and coordinated dynamic behaviours at the population level occur.44 Therefore, even if it cannot analyse morphology at the single-cell level, achieving a wide FOV can potentially allow for the acquisition of effective metrics for quality control of the culture. To demonstrate the effectiveness of INSPCTOR, we quantified the confluency of hiPSC culture and analysed morphological changes at the colony level caused by epithelial-to-mesenchymal transition (EMT) during mesoderm induction. In addition, by implementing high-speed imaging modes, we quantified the beating dynamics of hiPSC-derived cardiomyocytes and tracked their fluctuations throughout their long-term production processes. These results indicate that INSPCTOR is a practical, high-throughput, and compact device for long-term monitoring and quality evaluation of hiPSC culture and its differentiation induction processes, highlighting its potential contribution to regenerative medicine and drug discovery.
Materials and methods
Fabrication of thin-film transistor image sensor panel
INSPCTOR's image sensor consists of a low-temperature poly-silicon thin-film transistor (LTPS-TFT) circuit and an amorphous silicon (a-Si) pin photodiode formed on a glass substrate (Fig. S1†). Both the LTPS-TFT and the a-Si pin photodiode were fabricated using standard photolithography processes for liquid crystal display production. The LTPS-TFT channel layer (poly-Si) was deposited using chemical vapour deposition (CVD) and subsequently crystallised through an excimer laser annealing process. The a-Si pin photodiode was stacked on top of the TFT using CVD. The upper electrode of the a-Si pin photodiode is a transparent indium tin oxide electrode, allowing the photodiode to absorb light from above. The top layer is covered with a transparent acrylic resin for surface protection. The surface of the acrylic resin has been plasma-treated to improve the adherence of the cultured cells. All the above manufacturing processes took place at Kyocera Corporation.Construction of the base module of INSPCTOR
The base module of INSPCTOR consists of an LED light source, a control board, a power board, a Li-ion battery, and a connection interface for the sensor module (Fig. S2†). The LED light source utilises a planar light source commonly used in LCD displays. The control board is equipped with an integrated circuit comprising a field programmable gate array (FPGA), a microcontroller unit (MCU), and memory, and it can be connected to the sensor module via pogo pins. The control board performs various functions, such as receiving control signals from a PC, driving the sensor, reading light signals from the sensor, converting the light signals from analogue to digital, and transmitting the light signals to the PC. Communication with the PC is conducted via Bluetooth. The power board is connected to the control board through an FFC connector, manages the lithium-ion battery's voltage control, and supplies power to the control board and the light source. The outer frame of the base module is fabricated by machining polycarbonate. All the above manufacturing processes took place at Kyocera Corporation.HiPSC culture
The hiPSC line 585A1 (ref. 45) was maintained according to the methods described in a previous report.46 Specifically, 6-well culture plates were coated with 3 μg ml−1 iMatrix-511 (982021, MATRIXOME) in D-PBS for 1 hour at 37 °C, and then hiPSCs were cultured in StemFit medium (AK02N, Ajinomoto). The medium was changed daily, and cells were passaged using TrypLE Select (A1285901, Thermo Fisher Scientific) every 4 days. Cell counting was performed using Countess (Thermo Fisher Scientific). At each passage, 2 × 105 cells were seeded. For the first 24 hours post-passage, 10 μM Y-27632 (259-00613, FUJIFILM Wako) was added to the StemFit medium. When culturing on INSPCTOR, the sensor was coated with 5 μg ml−1 iMatrix-511 in D-PBS for 1 hour at 37 °C, and the desired number of cells was passaged. After the passage, cells were incubated in StemFit containing 10 μM Y-27632 for 24 hours, followed by daily medium changes without Y-27632.Fluorescent staining and observation of hiPSCs
Two days post-seeding on the sensor module, hiPSC images were captured using INSPCTOR. Subsequently, cytoplasmic staining or nuclear staining was performed by adding 2 μM CMFDA (223-02151, FUJIFILM Wako) or 1 μM Kakshine47 to the StemFit medium, respectively. Cells were incubated at 37 °C for 30 minutes, washed with PBS, and fixed with 4% PFA at room temperature for 15 minutes. The cytoplasm-stained cells were examined under a conventional upright microscope. For nuclear-stained cells, after removing the culture chamber, cells were mounted with Fluomount-G (0100-01, Southern Biotech), covered with a glass slide, and then high-resolution fluorescence imaging was conducted using AMATERAS.48Mesoderm induction
We used a base medium consisting of RPMI-1640 with HEPES (189-02145, FUJIFILM Wako) supplemented with B27 supplement without insulin (A1895601, Thermo Fisher Scientific) and penicillin–streptomycin (168-23191, FUJIFILM Wako), designated as RPMI/B27-ins. On the second day after seeding hiPSCs on INSPCTOR, the culture medium was switched to RPMI/B27-ins with 6 μM CHIR99021 (038-23101, FUJIFILM Wako) to initiate mesoderm differentiation. Control samples were switched to RPMI/B27-ins without CHIR. The medium was not changed during the subsequent 48 hour observation on INSPCTOR.Cardiomyocyte differentiation induction
Cardiomyocyte differentiation was induced based on temporal modulation of Wnt signalling in monolayer culture.49,50 RPMI/B27-ins containing 50 μg ml−1 ascorbate is denoted as RPMI/B27-ins/asc. On day 0, the third day after hiPSC seeding, the medium was switched to RPMI/B27-ins/asc supplemented with 6 μM CHIR99021 to initiate differentiation. On day 1, the medium was changed to RPMI/B27-ins/asc. On day 3, for ventricular-like cardiomyocyte induction, the medium was switched to RPMI/B27-ins/asc containing 5 μM IWP-2 (034-24301, FUJIFILM Wako), 5 μM XAV-939 (247-00951, FUJIFILM Wako), and 1 μM BMS-453 (19076, Cayman Chemical).50,51 For atrial-like cardiomyocyte induction, the medium was switched to RPMI/B27-ins/asc containing 5 μM IWP-2, 5 μM XAV-939, and 1 μM retinoic acid (186-01114, FUJIFILM Wako).50–52 From day 5 onwards, both ventricular-like and atrial-like cardiomyocytes were cultured under the same conditions. On day 5, the medium was changed to RPMI/B27-ins/asc. Subsequently, the medium was changed every other day to RPMI-1640 with HEPES containing B27 supplement (17504044, Thermo Fisher Scientific) and penicillin–streptomycin, which is denoted as RPMI/B27. On day 11 or 12, cells were detached using TrypLE Select and reseeded onto dishes or sensors coated with Matrigel (354277, Corning). After seeding, cells were incubated in RPMI/B27 containing 10% KnockOut Serum Replacement (10828010, Thermo Fisher Scientific) for 24 hours, followed by medium changes every other day with new RPMI/B27. For glucose-free medium, no glucose RPMI-1640 (185-02865, FUJIFILM Wako) containing B27 supplement, 5 mM sodium DL-lactate (L4263, Merck), and penicillin–streptomycin was used.Drug treatment on cardiomyocytes
On day 23 of ventricular-like cardiomyocyte induction, cells were passaged using TrypLE Select and seeded at 2 × 105 cells per well on the sensor. After seeding, cells were cultured in RPMI/B27 containing 10% KnockOut Serum Replacement for 24 hours, followed by medium changes every other day with RPMI/B27. Drug response assays were conducted on day 30 of the induction. The medium was changed just before the experiment. Drug concentrations were adjusted by replacing half of the medium with a medium containing adequate concentrations of isoproterenol (I0260, Tokyo Chemical Industry) and propranolol (168-28071, Fujifilm). Measurements were taken 15 minutes after the medium change. All conditions were sequentially measured using the same sample, starting from the lowest to the highest concentration as follows: control, 10 nM Iso., 30 nM Iso., 100 nM Iso., 300 nM Iso., 300 nM Iso. and 300 nM Pro., 300 nM Iso. and 1 μM Pro., and 300 nM Iso. and 3 μM Pro.Image processing and quantification
To evaluate INSPCTOR's detection accuracy, we aligned the fluorescence image with the corresponding INSPCTOR image, manually identifying the same locations in both. Given the fluorescence image's 1.1 μm pixel pitch, it was segmented into a 59 pixel grid, matching each grid area with an INSPCTOR pixel. Colony areas in the fluorescence image were delineated using a Gaussian filter for smoothing followed by baseline subtraction with the rolling ball algorithm, binarising the image by setting an intensity threshold, applying the watershed algorithm to segment, and extracting spatially connected areas as individual colonies. For confluency quantification, the rolling ball algorithm was used to obtain and subtract a baseline from the image, and the same threshold was manually set across all conditions. Regions of interest (ROI) were set within the well, excluding the chamber edges. The proportion of pixels with signals exceeding the threshold was calculated. For quantification of EMT during mesoderm induction, the Sobel filter was used to compute the first derivative and visualise the spatial gradient. ROIs were set within the well, excluding the chamber edges. The average value of the first derivative image in each ROI was divided by the mean brightness of the original image and then normalised with the value of the first time frame. To quantify cardiomyocyte beating dynamics, low pass filtering was applied to the temporal intensity fluctuations in each pixel to remove high-frequency noise and identify the baseline. The change ratio from the baseline was calculated pixel by pixel to normalise the intensity fluctuations. A fast Fourier transform analysis was performed on the pixel-level intensity fluctuations, and the dominant frequency in the movie was analysed by averaging the results from all pixels. Peak detection was performed on the pixel-level intensity fluctuation to visualise the contraction conduction based on the differences in peak timing. All the processes are carried out using Fiji53 and Python 3.7.Results
Design of INSPCTOR
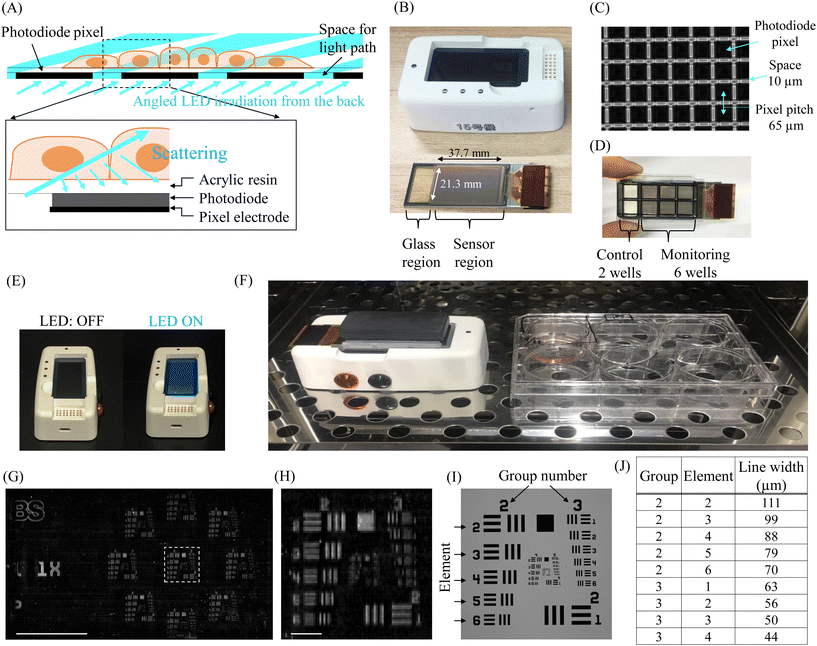
INSPCTOR is a lens-free imaging device designed for the long-term monitoring of hiPSC culture and the production processes of target cells within an incubator. It is designed to employ a novel illumination strategy, which illuminates the cells from the rear and detects the scattered light using a TFT image sensor, optimising light capture (Fig. 1A). The device comprises a sensor module for cell culture and light detection, as well as a base module for light irradiation, data storage, and transfer (Fig. 1B, S1 and S2†).The sensor module includes an image sensor deposited on a glass substrate and a connection to the base module. The image sensor pixels are composed of low-temperature poly-silicon thin-film transistors (LTPS-TFT) and thin-film amorphous Si photodiodes (Fig. S1†). 580 × 328 pixels are arrayed with a pitch of 65 μm, yielding a sensor area of approximately 37.7 × 21.3 mm (Fig. 1B and C). A 10 μm gap between the pixels allows light irradiation from the rear (Fig. 1A and C).
Due to advancements in TFT manufacturing technology, the sensor module can be mass-produced cost-effectively and is designed as a disposable consumable.32,39 Consequently, cells can be cultured directly on the sensor, resulting in proximity to the sensor corresponding to the thickness of the surface acrylic resin layer of 4 μm (Fig. 1A). The size of the sensor module is approximately 76 × 26 mm, equivalent to a standard slide glass. As a result, common cell culture chambers can be attached. When a typical 8-well type chamber (#30118, SPL LIFE SCIENCES) is placed, the sensor can cover an area equivalent to 6 wells (Fig. 1D). Therefore, the six independent culture environments of approximately 1cm2 can be simultaneously monitored. The remaining two wells are covered only by glass, allowing observation of control samples with a conventional inverted microscope (Fig. 1D).
The base module features an LED irradiation surface, an internal battery, data storage, and a connection to the sensor module (Fig. 1B and E, S2†). The proximity of cells to the sensor in the sensor module reduces image blurring, which might otherwise be caused by distance. This proximity eliminates the need for coherent light illumination or computational image reconstruction, often required in lens-free imaging.24 Therefore, to minimize the device and simplify the imaging processes, we employed a novel illumination strategy. Specifically, we employed dark-field illumination from the rear and scattering detection, thereby miniaturising the base module's illumination system. To suppress background signals during cell observation and enhance the detection of scattered light, the LEDs are designed to irradiate obliquely through a louvre film (Fig. 1A). Data obtained from the sensor module is saved in the base module. Light irradiation, data acquisition, and data transfer are remotely controlled by an external PC connected via Bluetooth low energy (BLE). If a direct BLE connection with the PC outside the incubator proves challenging, a BLE antenna wired to the PC can be installed inside the incubator (Fig. S3†). These features enable real-time monitoring of the culture.
Fig. 1F illustrates the appearance of INSPCTOR installed inside an incubator. The XY plane is approximately 48 cm2 (9.5 × 5.0 cm), with a maximum height of about 4.5 cm to the top of the lid, occupying a volume of less than 220 cm3. Compared to a standard 6-well plate (e.g., 12.8 × 8.5 × 2.2 cm), the XY plane of INSPCTOR is less than half the size, yet the height is approximately double, achieving a compact design with a comparable volume. Therefore, despite limited space inside an incubator, multiple devices can be easily installed, facilitating effective high-throughput monitoring.
To assess the spatial resolution of INSPCTOR, we analysed a USAF 1951 target pattern (R3L1S4P, Thorlabs) on transparent glass comprising three lines of specific widths that reflect light at intervals matching their widths (Fig. 1G–J). We evaluated whether these lines could be distinctly recognised (Fig. 1I and J). By removing the sensor module's cell culture chamber and substituting a glass slide with the pattern, Fig. 1G illustrates the full FOV where INSPCTOR detected light from the pattern. An enlarged view (Fig. 1H) reveals that vertical lines of 56 μm width (group 3, element 2) and horizontal lines of 79 μm width (group 2, element 5) were distinguishable, supporting that INSPCTOR spatial resolution approaches its pixel pitch, aligning with the optical detection principle of the device.
In the following sections, we will present examples of applications of INSPCTOR for long-term monitoring of hiPSC culture and its differentiation induction.
Long-term confluency monitoring of hiPSC culture
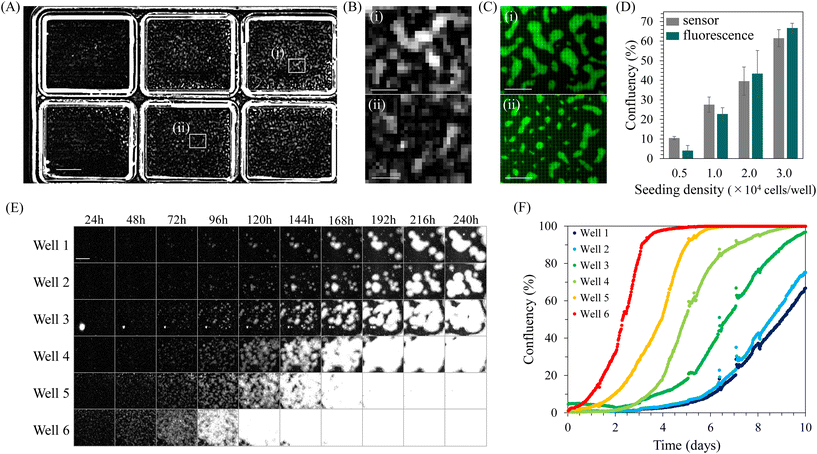
Confluency is a widely used quantitative indicator for monitoring cell cultures. Particularly in hiPSC cultures, quantifying confluency is valuable for determining the appropriate passage timing for maintaining undifferentiated cells45,46 and adjusting the culture density for optimal differentiation induction.49–52,54,55 Therefore, we attempted to quantify confluency during hiPSC culture using INSPCTOR.INSPCTOR detects scattered light, producing an image with a dark background and bright regions where scattering objects such as cells and chamber wells exist. Fig. 2A shows a full FOV image of undifferentiated hiPSCs cultured on and observed by INSPCTOR. Six culture areas can be easily identified from the scattered light at the edges of the wells. Since hiPSCs proliferate, forming dense colonies, they are expected to scatter light strongly and can be sufficiently detected by the resolution of INSPCTOR (Fig. S4†).
Indeed, distributed bright signals were detected within the wells (Fig. 2A). Upon enlarging specific regions within wells, signals presumably originating from hiPSC colonies were identified with a sufficient signal-to-noise ratio (Fig. 2B). To confirm that these signals originate from hiPSC colonies, cytoplasm of the same sample was fluorescently stained and observed under a conventional upright microscope (Fig. 2C). Comparing the two, it was found that regions of hiPSC colonies confirmed by fluorescence are also detected by INSPCTOR despite the coarser images due to the pixel pitch (Fig. 2B and C).
To evaluate the cell detection accuracy of INSPCTOR, we performed a detailed comparison with a nuclear-stained fluorescence image offering subcellular resolution (Fig. S5†). Initially, we quantified the fluorescence intensity for areas corresponding to each INSPCTOR pixel and found a positive correlation with INSPCTOR's pixel intensity (Fig. S5G†). We established thresholds for signal intensities to assess cell presence, using fluorescence-based judgment as a benchmark. INSPCTOR achieved approximately 90% detection accuracy (Fig. S5G†). An investigation into detection errors revealed that false positives often arose from background noise (cyan arrowheads in Fig. S5B and E†) or from INSPCTOR's overestimation of large colony contours (cyan arrowheads in Fig. S5C and F†). False negatives primarily occurred when INSPCTOR missed small colony regions (white arrowheads in Fig. S5C and F†). Further analysis demonstrated INSPCTOR's signal intensity with colony size, detecting all colonies over 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μm2, though efficiency diminished with smaller colonies (Fig. S5H and I†). These findings confirm INSPCTOR's high detection accuracy for hiPSCs, which typically form large colonies, despite some inaccuracies.
000 μm2, though efficiency diminished with smaller colonies (Fig. S5H and I†). These findings confirm INSPCTOR's high detection accuracy for hiPSCs, which typically form large colonies, despite some inaccuracies.
Next, we quantified confluency from the images obtained by INSPCTOR and compared the results with those quantified based on fluorescence images. As a result, INSPCTOR showed confluency values comparable to those quantified from fluorescence images (Fig. 2D). This result demonstrates the sufficient capability of INSPCTOR to quantify the confluency of hiPSC culture.
Given the above results, we next conducted long-term monitoring of confluency during the undifferentiated culture of hiPSCs. Here, hiPSCs were cultured at six different seeding densities, and images were captured every 30 minutes for 10 days (Fig. 2E and F). Fig. 2E displays the extracted regions of each well, showing the colonies grow larger over time. Plotting quantified confluencies successfully visualised the difference in timing of increase depending on the seeding density (Fig. 2F). These results indicate that INSPCTOR can be effectively utilised for long-term quantitative monitoring of confluency during hiPSC culture.
Monitoring of EMT during mesoderm induction
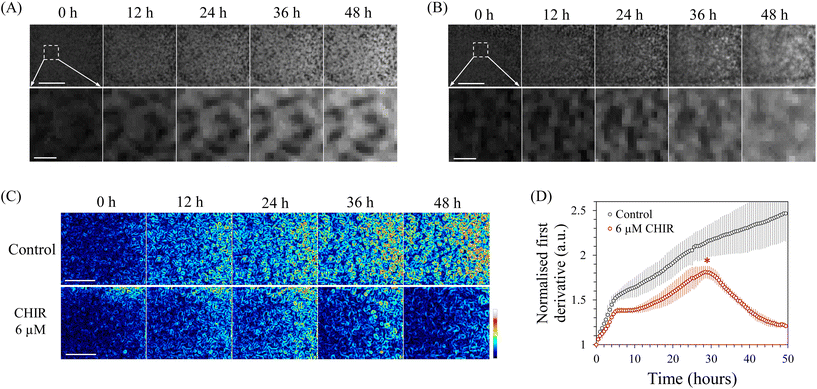
Next, we attempted to quantify changes in the cellular state during differentiation induction using INSPCTOR. As an example, we specifically observed mesoderm induction from hiPSCs. The mesoderm is a germ layer that gives rise to various cells and organs, such as the heart, blood cells, kidney, muscle, osteocytes, and adipocytes. It can be induced from pluripotent stem cells in vitro.54–56 During the induction, EMT occurs as an initial cell state change. Due to EMT, cells tightly bound in colonies acquire migratory properties, spreading outside the colonies, causing the colony morphology to collapse and cells to distribute in a flattened manner.55Here, we attempted to quantify this EMT using INSPCTOR. HiPSCs were induced into mesoderm through Wnt signal stimulation using 6 μM CHIR99021 (CHIR), and the process was monitored every 30 minutes over a period of 48 hours. In a control sample without CHIR, the morphology of colonies remained largely unchanged over 48 hours, accompanied by an increase in average intensity (Fig. 3A). In contrast, in the CHIR-treated samples, colonies collapsed in their shape during the latter stage, and the original morphology was lost after 48 hours (Fig. 3B). We used a first derivative filter to calculate the local spatial gradient and quantified this morphological change (Fig. 3C and S6†). Strong gradients around the contour of the maintained colonies were detected in the control sample (Fig. S6A†), while a smaller gradient was quantified in the CHIR-treated group (Fig. S6B†), indicating that morphological differences were successfully quantified. Fig. 3C and D showed that the local gradient in the control increased monotonically over time, suggesting continued cell proliferation and the formation of denser colonies. In contrast, the spatial gradient in the CHIR-treated samples decreased from the midway point around 30 hours after mesoderm induction (Fig. 3D asterisk). This indicates that the acquisition of migratory properties and changes in colony shape due to EMT begin at this time.
These results demonstrate that INSPCTOR can accurately identify the initiation timing of EMT, a typical cellular state change occurring during mesoderm induction from hiPSCs. By using this timing as an indicator, researchers can effectively conduct intermediate evaluation and assess the reproducibility of the differentiation induction process.
Quantification of beating dynamics of hiPSC-derived cardiomyocytes
Next, to illustrate how we can quantify functional indicators of differentiation products, we attempted to quantify the beating dynamics of cardiomyocytes. Cardiomyocytes are a type of cell that can be effectively induced from hiPSCs in vitro and thus are expected to play significant roles in drug discovery and regenerative medicine.57–59 HiPSC-derived cardiomyocytes exhibit spontaneous beating at the cellular level.60 Their beating dynamics are closely related to electrophysiology and are useful for evaluating cellular states and drug responsiveness.61First, to accurately capture the rapid beating dynamics of cardiomyocytes, we implemented high-speed imaging modes. The maximum imaging speed is determined by the image readout speed, which can be improved by reducing the FOV. Here, we implemented two FOV restriction modes: single-area and six-area modes. The single-area mode limited the FOV to a small area, 2.08 × 2.08, 3.12 × 3.12, or 4.16 × 4.16 mm square, which enabled observations at 10.8, 10.1, or 8.2 fps respectively (Table 1). In the six-area mode, six different areas corresponding to the six wells on the sensor can be captured at high speeds simultaneously, allowing for the comparison of multiple culture conditions. Each area was 0.78 × 0.78, 1.04 × 1.04, or 1.56 × 1.56 mm square in size, achieving 25.4, 12.9, or 8.8 fps respectively (Table 2).
| Area (pixel) | Area (mm) | Speed | |
|---|---|---|---|
| Mode 1 | 32 × 32 | 2.08 × 2.08 | 10.8 fps |
| Mode 2 | 48 × 48 | 3.12 × 3.12 | 10.1 fps |
| Mode 3 | 64 × 64 | 4.16 × 4.16 | 8.2 fps |
| Area (pixel) | Area (mm) | Speed | |
|---|---|---|---|
| Mode 1 | 12 × 12 | 0.78 × 0.78 | 25.4 fps |
| Mode 2 | 16 × 16 | 1.04 × 1.04 | 12.9 fps |
| Mode 3 | 24 × 24 | 1.56 × 1.56 | 8.8 fps |
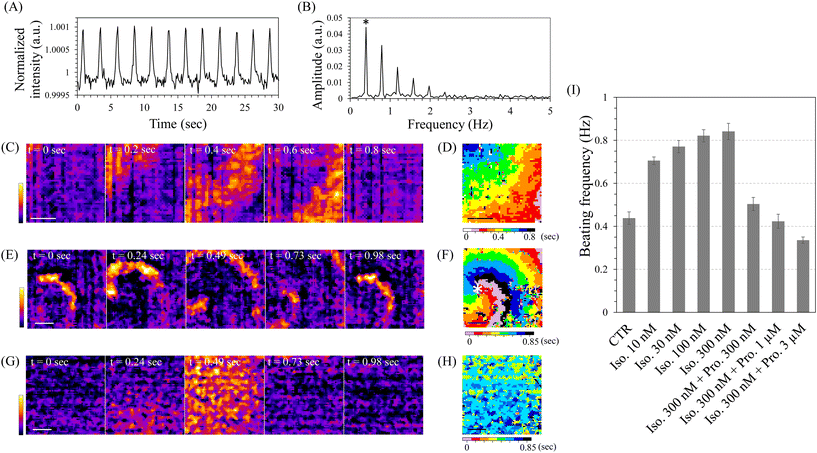
Using these high-speed imaging modes, we observed cardiomyocytes induced from hiPSCs directly on the sensor. At days 10–11 of cardiomyocyte induction, spontaneous contraction-relaxation cycles were observed in most of the cells within the control well, as observed under a conventional inverted microscope, indicating highly efficient cardiomyocyte induction (Movie S1†). High-speed imaging with the single-area mode of INSPCTOR revealed periodic increases in image intensity (Fig. 4A). This increase likely reflects enhanced scattered light intensity when the cardiomyocytes contract and become denser. Analysing this intensity change with the fast Fourier transform (FFT) enabled quantification of the beating frequency (Fig. 4B). As a result, multiple peaks appeared at frequencies that were integer multiples of the lowest frequency observed. This occurs because the original intensity change displayed sharp spikes, unlike a sine curve (Fig. S7†). The peak showing the lowest frequency corresponded to the beating frequency (Fig. 4B asterisk). In addition, the Single-area mode with a FOV exceeding 4 mm2 allowed for spatiotemporal analysis, which enabled the visualisation of the beating conduction patterns (Fig. 4C and D). Interestingly, around days 10–11 of the differentiation induction, reentry patterns characterised by circulating beating propagation were observed in approximately half of the samples (Fig. 4E and F, Movie S2†). The reentry pattern is thought to arise from fragmented propagation waves in heterogeneous environments.62 The observed reentry was likely due to temporary spatial heterogeneity occurring during the cardiomyocyte differentiation process. These results demonstrate that INSPCTOR can effectively quantify the beating frequency and conduction as functional indicators of cardiomyocytes derived from hiPSCs.
Next, we assessed the drug response in cardiomyocytes using INSPCTOR by quantifying their beating frequency. We reseeded hiPSC-derived cardiomyocytes as monolayer cultures with spatially uniform distribution (Movie S3†). These cells were then used on day 30 of cardiomyocyte induction. No reentry patterns were observed, and the propagation speed was significantly faster than that on days 10–11, showing almost simultaneous contractions (Fig. 4G and H). Here, we analysed the chronotropic response to β-adrenergic receptor perturbation using isoproterenol as the agonist and propranolol as the antagonist (Fig. 4I). As a result, when the isoproterenol was applied, a concentration-dependent increase in beating frequency was confirmed. Moreover, maintaining a high concentration of isoproterenol and subsequently adding the propranolol led to a concentration-dependent decrease in beating frequency. These results are consistent with the reported chronotropic response to isoproterenol and propranolol.58 Therefore, it can be concluded that INSPCTOR can evaluate drug response based on the beating frequency of hiPSC-derived cardiomyocytes.
Detection of beating onset during cardiomyocyte differentiation from hiPSCs
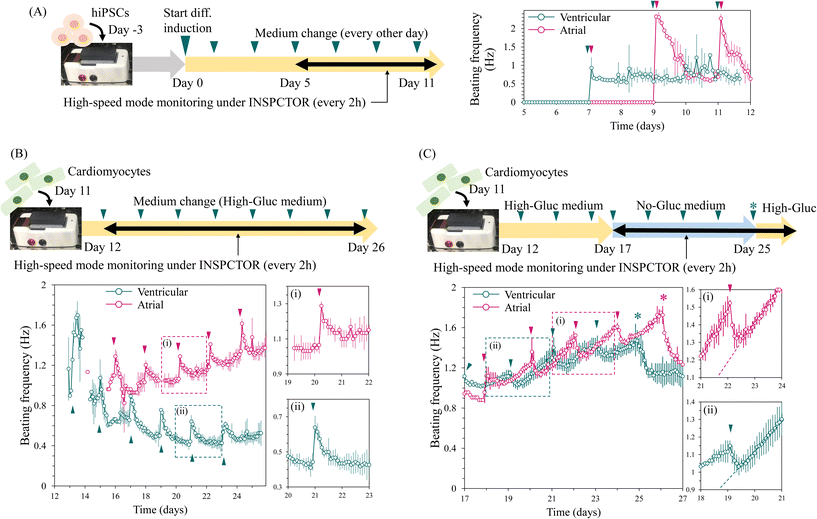
In the context of cardiomyocyte production for regenerative medicine and drug discovery, efficient and rapid induction reduces production costs and is thus valuable. When optimising the induction conditions, the timing of beating initiation indicates the speed of cardiomyocyte induction. Therefore, we attempted to detect the onset of spontaneous beating during cardiomyocyte induction using INSPCTOR. Here, we observed two differentiation conditions that induced cardiomyocytes with a bias toward atrial- and ventricular-like phenotype. We monitored these conditions from day 5 to 12 of differentiation using the six-area mode with high-speed imaging at 8.8 fps, capturing images every two hours.In the atrial and ventricular induction conditions, initiation of spontaneous beating was successfully detected on days 9 and 7, respectively (Fig. 5A). Generally, hiPSC-derived cardiomyocytes initiate beating between days 7 and 12, which is consistent with our results.49–52 The observed beating frequencies in the ventricular condition fluctuated between 0.6 and 1 Hz (Fig. 5A green plots). In contrast, the atrial condition initially exhibited a slightly higher frequency of approximately 2.3 Hz, which rapidly decreased to below 1.0 Hz. Similar fluctuations repeated after the medium change (Fig. 5A magenta plots). These results demonstrate that INSPCTOR can detect the onset of spontaneous beating and the differences in beating frequency fluctuations. Thus, these timing and patterns can be utilised to assess the speed, reproducibility, and quality of cardiomyocyte induction.
Long-term monitoring of beating frequency during cardiomyocyte maturation
Next, we conducted a long-term time-lapse observation of the maturation process of cardiomyocytes. A prolonged culture is essential for producing mature cardiomyocytes.63 Here, cardiomyocytes induced toward atrial- and ventricular-like phenotypes were reseeded on the sensor on day 11 of the induction and observed for approximately two weeks starting from day 12. Images were captured every 2 hours at 8.8 fps using the six-area mode (Fig. 5B). As a result, stable beatings were consistently detected from day 14 under the ventricular condition and from day 15 under the atrial condition. The cell state recovery and the time until reaching the confluence monolayer upon proliferation (Fig. S8†) may contribute to this delayed detection of beating after passage. Subsequently, cardiomyocytes exhibited two major patterns in the fluctuation of beating frequency: long-term and short-term changes.As for the long-term change, in the ventricular condition, the beating frequency gradually decreased to stabilise around 0.5 Hz (Fig. 5B green plots). In contrast, under the atrial condition, the frequency gradually increased and reached 1.4 Hz by day 26 but had not yet stabilised (Fig. 5B magenta plots). HiPSC-derived atrial cardiomyocytes typically exhibit a higher beating frequency than ventricular cardiomyocytes, which is consistent with our results.52 However, whether the frequency has stabilised can only be determined through long-term monitoring, as demonstrated by our observation.
The short-term change was characterised by rapid frequency increases observed every other day, followed by a gradual return to baseline within approximately one day (Fig. 5B-i and ii). This frequency increase coincided with the timing of medium changes, as indicated by arrowheads in Fig. 5B. We speculate that this change could be attributed to fluctuations in pH resulting from glucose metabolism. It is known that the beating frequency of cardiomyocytes is pH-dependent; it increases under alkaline conditions and decreases under acidic conditions.64,65 Additionally, lactate accumulates in the medium due to glucose metabolism of cardiomyocytes, leading to a decrease in medium pH.66–68 The culture medium contained 5 mM glucose (high-Gluc medium). Therefore, the observed phenomenon is consistent with the assumption that the medium, initially neutral after a medium change, gradually becomes acidic due to lactate accumulation from glucose metabolism.
To test this hypothesis, we used a medium without glucose (Fig. 5C). Cardiomyocytes are known to metabolise lactate to produce ATP via the TCA cycle and survive for extended periods without glucose.66 Thus, we switched to a glucose-free, 5 mM lactate (no-Gluc) medium from day 17 in the ventricular condition and from day 18 in the atrial condition. Interestingly, in the no-Gluc medium condition, the frequency rapidly decreased upon each medium change (Fig. 5C). After reaching the baseline, the frequency increased almost linearly (Fig. 5C-i and ii). These changes may reflect a decrease in lactate concentration upon metabolic consumption and a shift towards alkaline pH over the two days before the medium change. These observations do not contradict our hypothesis that metabolic activity leads to changes in pH and beating frequency. In addition, under both atrial and ventricular conditions in the no-Gluc medium, the frequency continued to increase without stabilising but began to decrease after the medium was changed to the high-Gluc medium (Fig. 5C asterisk). This suggests that metabolic differences that occurred in the no-Gluc medium induced the long-term and non-stabilizing increase in beating frequency.
These results demonstrate that INSPCTOR effectively tracks beating frequency during the maturation process of cardiomyocytes. The long-term changes allow us to determine if the beating frequency has stabilised, which indicates the appropriate timing of downstream analysis, such as drug evaluation. Additionally, the daily responses to medium changes suggest the capability to quantify metabolic-dependent fluctuations. Thus, further research is anticipated to provide more concrete interpretations and practical indicators of cellular metabolic state in the future.
Discussion
We have developed a lens-free imaging device named INSPCTOR. Despite being as compact as a standard 6-well plate, it can simultaneously observe six independent culture environments. This throughput is attributable to the strength of TFT technology, which enables the cost-effective production of large image sensors.32,39 Additionally, INSPCTOR does not require an objective lens and utilises LEDs for illumination, making it an inexpensive device. Multiple devices can be installed at a low cost, enabling easy expansion of monitoring samples. Particularly in long-term differentiation induction processes, it is common for multiple batches to be cultured in parallel, as the next batch often begins before the completion of a previous batch. Therefore, rather than increasing the throughput by enlarging the device, utilising multiple compact devices with adequate throughput is more practical for operational efficiency. Based on these factors, INSPCTOR can be considered an effective and affordable device for monitoring differentiation induction within the limited space of an incubator.An ideal incubator imaging system for quality control in long-term cell culture combines compact size, high throughput, and a larger FOV. Current systems fall into three categories: single-FOV, multi-FOV, and scanning mechanism systems. Single-FOV systems, while compact and capable of a wide FOV, often fall short of throughput capabilities.14–24 On the other hand, systems with scanning mechanisms excel in throughput and FOV but typically struggle with bulkiness.25–27 This study introduces a multi-FOV system designed to address these limitations, offering a balanced solution that maintains compactness while enhancing throughput and maximising FOV.
Several multi-FOV in-incubator imaging systems have been developed to improve observation throughput by incorporating dedicated imaging units for each culture well. For instance, miniaturised modular-array microscopy (MAM) utilises 12 small imaging units to serve a 12-well plate,28 while EmSight employs imaging units designed for Fourier ptychographic microscopy to simultaneously observe six samples in a 6-well plate.29 Each system, however, faces distinct challenges. MAM, targeted primarily at single-cell analysis, offers a limited FOV of 0.75 × 0.45 mm and requires individual focusing for each well, struggling with focal drift during medium exchanges due to temperature fluctuations. Conversely, EmSight, despite providing a wide FOV and high spatial resolution through variable illumination angles and digital refocusing, involves complex hardware with six sets of objective lenses, tube lenses, and CMOS cameras, complicating miniaturisation and simplification efforts.
Unlike traditional array-type multi-FOV systems that require a dedicated imaging unit for each well, INSPCTOR employs a simpler, more efficient design, utilising just one image sensor and a single planar light source. This system features innovative backlight illumination through pixel gaps, which avoids the shadows cast by chamber walls typical in top-down illumination setups. This enables uniform lighting across all wells without the need for individual light sources. The lens-free design of INSPCTOR also contributes to its compact size, as it eliminates the bulk of typically added lenses. Additionally, the use of a large TFT image sensor allows for multi-FOV imaging with a single sensor, simplifying the overall design. A significant advantage of INSPCTOR is its ability to culture cells directly on the sensor, removing the necessity for focusing mechanisms and thus preventing focal drift during prolonged observations. These advancements render INSPCTOR not only simpler and smaller but also more practical compared to other multi-FOV systems.
As highlighted previously, the INSPCTOR system achieves the simultaneous observation of six wells with a large FOV while retaining the simplicity and compactness typical of a single-FOV system. This design effectively fulfils the three key requirements for optimal in-incubator long-term cell culture monitoring: compact size, high throughput, and a larger FOV. Hence, INSPCTOR exemplifies a high-level integration of these critical attributes, making it an exemplary tool for advanced quality control in long-term cell culture.
INSPCTOR is designed for the disposable use of the sensor module. Unlike applications involving long-term differentiation induction with low subculture frequency, handling cells with high subculture frequency could significantly increase the costs associated with disposable components. The sensor module can withstand autoclave sterilisation and can be reused if properly cleaned. Furthermore, without intensive cleaning, reusing the same well by reseeding cells after detachment with trypsinisation, as a common practice to economise on culture plates, is feasible. However, in either case, remaining dead cells or debris can scatter light and become a noise source, so caution is required for reuse. Theoretically, it is also possible to reuse the sensor module by applying a thin, easily peelable film to the surface. However, reflections due to differences in refractive index between the film and the image sensor surface would need to be managed, possibly using optical adhesives. Moreover, a thick film layer can blur images, necessitating either an extremely thin film or computational methods to correct blurring. Overall, reusing the sensor module seems feasible, depending on the approach and purpose.
One of the main challenges with our INSPCTOR is the low image resolution caused by the 65 μm pitch of the TFT image sensor. Due to the large pixel size, some cells may not be directly illuminated. In fact, a positive correlation was observed between colony size and INSPCTOR signal intensity; however, detection efficiency significantly decreased for small colonies that were the same size or smaller than the pixel size (Fig. S5†). On the other hand, light scattered by the illuminated cells can indirectly irradiate surrounding cells, potentially enhancing detection efficiency. Indeed, the signal intensity in large colonies increased, enabling reliable colony detection, although it often extended beyond the actual borders of the colonies (Fig. S5†).
The primary concern is how many valuable metrics can be obtained given this resolution. Moreover, unlike in studies of molecular mechanisms with fluorescence imaging, non-label quantification is required for quality control in differentiation induction. Despite the above concerns, we successfully demonstrated the acquisition of multiple useful metrics in addition to confluency. For example, INSPCTOR has demonstrated its effectiveness during mesoderm induction. Mesoderm induction is a shared initial process of producing various mesodermal-derived cell types. This induction period strongly influences subsequent fate determination, making the optimisation of this process crucial.10,12,49–52,54–56 By quantifying changes in colony morphology, INSPCTOR successfully identified the onset timing of EMT during mesoderm induction. This can serve as a versatile metric in various mesodermal-derived cell differentiation induction processes. In the future, elucidating the relationship between EMT quantification and target cell induction efficiency could provide additional metrics for evaluating induction quality beyond just reproducibility. Additionally, by implementing high-speed imaging modes in INSPCTOR, we effectively monitored the onset timing of spontaneous beating, conduction patterns, and beating frequency over extended periods during cardiomyocyte production. The onset timing of spontaneous beating serves as an indicator of cardiomyocyte induction speed. The difference in conduction patterns is known to cause variations in maturation speed,69 potentially making them valuable indicators for assessing maturation. Furthermore, reentry, where conduction circulates in space, is considered a mechanism for various arrhythmias,70,71 suggesting potential applications for evaluating drug efficacy against arrhythmias. Additionally, we demonstrated that beating frequency fluctuation reflects metabolic activity. Changes in metabolic state are known to be essential for the maturation of cardiomyocytes.63 Although a detailed analysis was not conducted in this study, quantifying the dynamics of beating frequency fluctuation could indicate cellular metabolic state and be used to determine maturity in the future.
Our TFT image sensor has a 65 μm pitch (390 ppi), but recent developments are advancing toward higher resolution TFTs in response to the demand for head-mounted displays. State-of-the-art high-resolution TFTs include Samsung's 2250 ppi (ref. 72) and BOE's 2117 ppi.73 Although these are designed for colour displays, they would correspond to a pixel pitch of approximately 4 μm if used for monochrome applications like INSPCTOR. The pixel size and photodiode spacing are determined by the precision of photolithography processes, and the aforementioned ultra-high-resolution TFTs achieve a pixel spacing of approximately 1 μm. The development of high-resolution imaging devices using such high-resolution TFT image sensors is anticipated in the future.
Looking ahead, INSPCTOR is anticipated to be versatile in monitoring diverse cell populations. Although not ideal for analysing small or intricate structures, its design excels in observing larger objects due to its wide FOV. Consequently, its potential applications include monitoring diverse types such as organoids,2 wound healing assays,74 bacterial colonies,39 and so on.
Conclusion
In this study, we developed INSPCTOR, a compact and high-throughput imaging device, for quality control in long-term stem cell culture and differentiation. By quantifying images and movies captured by INSPCTOR without the need for staining, we successfully obtained effective metrics, even from low-resolution images derived from its TFT image sensors. We demonstrated these capabilities by monitoring hiPSC culture, mesodermal differentiation, and cardiomyocyte production. In the future, we expect that incorporating cutting-edge image and movie analysis, such as deep learning, will enable the provision of effective metrics for various other types of differentiation induction processes.9,10,39,75 We believe this research will significantly contribute to advancements in regenerative medicine and drug discovery.Data availability
A set of imaging data for this article is available at SSBD:repository at https://doi.org/10.24631/ssbd.repos.2024.06.364.Author contributions
Conceptualisation: T. Nagai, T. Natsume, and T. Kakizuka. Device design: T. Natsume. Sample preparation: T. Kakizuka. Data acquisition: T. Kakizuka. Data analysis and visualisation: T. Kakizuka. Manuscript writing – original draft: T. Kakizuka. Manuscript writing – review & editing: T. Kakizuka, T. Nagai, and T. Natsume.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to Mr Sho Nakamitsu, Mr Kotaro Kobayashi, Mr Eiji Kanda, and Mr Seiji Matsuda of Kyocera Corporation, Japan, for their vigorous cooperation in manufacturing INSPCTOR and its control software. This work was supported in part by The Uehara Memorial Foundation (T. Nagai), Takeda Science Foundation (T. Nagai), Global Center for Medical Engineering and Informatics (MEI) Grant 2021 (T. Kakizuka), Artificial Intelligence Research Center (AIRC-SANKEN) Grant 2021 (T. Kakizuka), and Osaka University Institute for Datability Science (IDS) interdisciplinary co-creation projects 2023 (T. Kakizuka).References
- K. Takahashi, et al., Induction of pluripotent stem cells from adult human fibroblasts by defined factors, Cell, 2007, 131(5), 861–872 CrossRef CAS PubMed.
- J. Kim, B.-K. Koo and J. A. Knoblich, Human organoids: model systems for human biology and medicine, Nat. Rev. Mol. Cell Biol., 2020, 21(10), 571–584 CrossRef CAS PubMed.
- G. Liu, et al., Advances in pluripotent stem cells: history, mechanisms, technologies, and applications, Stem Cell Rev. Rep., 2020, 16(1), 3–32 CrossRef PubMed.
- L. P. Freedman, et al., Reproducibility: changing the policies and culture of cell line authentication, Nat. Methods, 2015, 12(6), 493–497 CrossRef CAS PubMed.
- V. Volpato and C. Webber, Addressing variability in iPSC-derived models of human disease: guidelines to promote reproducibility, Dis. Models Mech., 2020, 13(1), dmm042317 CrossRef CAS PubMed.
- D. Paull, et al., Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells, Nat. Methods, 2015, 12(9), 885–892 CrossRef CAS PubMed.
- M. Daniszewski, et al., Automated cell culture systems and their applications to human pluripotent stem cell studies, SLAS Technol., 2018, 23(4), 315–325 CrossRef CAS PubMed.
- G. N. Kanda, et al., Robotic search for optimal cell culture in regenerative medicine, eLife, 2022, 11, e77007 CrossRef CAS PubMed.
- D. E. Desa, T. Qian and M. C. Skala, Label-free optical imaging and sensing for quality control of stem cell manufacturing, Curr. Opin. Biomed. Eng., 2023, 25, 100435 CrossRef CAS PubMed.
- X. Yang, et al., A live-cell image-based machine learning strategy for reducing variability in PSC differentiation systems, Cell Discovery, 2023, 9(1), 53 CrossRef CAS PubMed.
- J. Lee, et al., Large-scale smart bioreactor with fully integrated wireless multivariate sensors and electronics for long-term in situ monitoring of stem cell culture, Sci. Adv., 2024, 10(7), eadk6714 CrossRef CAS PubMed.
- T. Qian, et al., Label-free imaging for quality control of cardiomyocyte differentiation, Nat. Commun., 2021, 12(1), 4580 CrossRef CAS PubMed.
- K. Yang, et al., Recent development of portable imaging platforms for cell-based assays, Biosens. Bioelectron., 2019, 124, 150–160 CrossRef PubMed.
- S. B. Kim, et al., A mini-microscope for in situ monitoring of cells, Lab Chip, 2012, 12(20), 3976–3982 RSC.
- Y. S. Zhang, et al., A cost-effective fluorescence mini-microscope for biomedical applications, Lab Chip, 2015, 15(18), 3661–3669 RSC.
- D. Jin, et al., Compact wireless microscope for in-situ time course study of large scale cell dynamics within an incubator, Sci. Rep., 2015, 5(1), 18483 CrossRef CAS PubMed.
- M. P. Walzik, et al., A portable low-cost long-term live-cell imaging platform for biomedical research and education, Biosens. Bioelectron., 2015, 64, 639–649 CrossRef CAS PubMed.
- S. B. Kim, et al., A cell-based biosensor for real-time detection of cardiotoxicity using lensfree imaging, Lab Chip, 2011, 11(10), 1801–1807 RSC.
- G. Zheng, et al., The ePetri dish, an on-chip cell imaging platform based on subpixel perspective sweeping microscopy (SPSM), Proc. Natl. Acad. Sci. U. S. A., 2011, 108(41), 16889–16894 CrossRef CAS PubMed.
- C. Han, et al., Wide field-of-view on-chip Talbot fluorescence microscopy for longitudinal cell culture monitoring from within the incubator, Anal. Chem., 2013, 85(4), 2356–2360 CrossRef CAS PubMed.
- I. Pushkarsky, et al., Automated single-cell motility analysis on a chip using lensfree microscopy, Sci. Rep., 2014, 4(1), 4717 CrossRef PubMed.
- S. V. Kesavan, et al., High-throughput monitoring of major cell functions by means of lensfree video microscopy, Sci. Rep., 2014, 4(1), 5942 CrossRef CAS PubMed.
- O. Mudanyali, et al., Compact, light-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications, Lab Chip, 2010, 10(11), 1417–1428 RSC.
- A. Ozcan and E. McLeod, Lensless imaging and sensing, Annu. Rev. Biomed. Eng., 2016, 18, 77–102 CrossRef CAS PubMed.
- G. Gürkan and K. Gürkan, Incu-stream 1.0: an open-hardware live-cell imaging system based on inverted bright-field microscopy and automated mechanical scanning for real-time and long-term imaging of microplates in incubator, IEEE Access, 2019, 7, 58764–58779 Search PubMed.
- T. Osaki, et al., Flatbed epi relief-contrast cellular monitoring system for stable cell culture, Sci. Rep., 2017, 7(1), 1897 CrossRef PubMed.
- A. J. Stout, et al., Simple and effective serum-free medium for sustained expansion of bovine satellite cells for cell cultured meat, Commun. Biol., 2022, 5(1), 466 CrossRef CAS PubMed.
- J. Son, B. Mandracchia and S. Jia, Miniaturized modular-array fluorescence microscopy, Biomed. Opt. Express, 2020, 11(12), 7221–7235 CrossRef CAS PubMed.
- J. Kim, et al., Incubator embedded cell culture imaging system (EmSight) based on Fourier ptychographic microscopy, Biomed. Opt. Express, 2016, 7(8), 3097–3110 CrossRef CAS PubMed.
- A. C. S. Chan, et al., Parallel Fourier ptychographic microscopy for high-throughput screening with 96 cameras (96 Eyes), Sci. Rep., 2019, 9(1), 11114 CrossRef PubMed.
- P. K. Weimer, The TFT a new thin-film transistor, Proc. IRE, 1962, 50(6), 1462–1469 Search PubMed.
- A. Yan, et al., Thin-Film Transistors for Integrated Circuits: Fundamentals and Recent Progress, Adv. Funct. Mater., 2024, 34(3), 2304409 CrossRef CAS.
- A. Tixier-Mita, et al., Review on thin-film transistor technology, its applications, and possible new applications to biological cells, Jpn. J. Appl. Phys., 2016, 55(4S), 04EA08 CrossRef.
- D. Wang, et al., Thin-film transistor arrays for biological sensing systems, Flexible Printed Electron., 2022, 7(2), 023004 CrossRef CAS.
- A. Kumar, A. K. Goyal and N. Gupta, thin-film transistors (TFTs) for highly sensitive biosensing applications: a review, ECS J. Solid State Sci. Technol., 2020, 9(11), 115022 CrossRef CAS.
- F. A. Shaik, et al., TFT sensor array for real-time cellular characterization, stimulation, impedance measurement and optical imaging of in-vitro neural cells, Biosens. Bioelectron., 2020, 169, 112546 CrossRef CAS PubMed.
- X. Ou, et al., Recent development in x-ray imaging technology: Future and challenges, Research, 2021, 2021, 9892152 CrossRef CAS PubMed.
- A. Nathan, et al., Amorphous silicon detector and thin film transistor technology for large-area imaging of X-rays, Microelectron. J., 2000, 31(11–12), 883–891 CrossRef.
- Y. Li, et al., Deep learning-enabled detection and classification of bacterial colonies using a thin-film transistor (TFT) image sensor, ACS Photonics, 2022, 9(7), 2455–2466 CrossRef CAS.
- Z.-T. Zhu, et al., A simple poly (3, 4-ethylene dioxythiophene)/poly (styrene sulfonic acid) transistor for glucose sensing at neutral pH, Chem. Commun., 2004, 1556–1557 RSC.
- J. Liu, M. Agarwal and K. Varahramyan, Glucose sensor based on organic thin film transistor using glucose oxidase and conducting polymer, Sens. Actuators, B, 2008, 135(1), 195–199 CrossRef CAS.
- P. I. Reyes, et al., ZnO thin film transistor immunosensor with high sensitivity and selectivity, Appl. Phys. Lett., 2011, 98, 17 Search PubMed.
- P. Estrela, et al., Field effect detection of biomolecular interactions, Electrochim. Acta, 2005, 50(25–26), 4995–5000 CrossRef CAS.
- Y. Sasai, Cytosystems dynamics in self-organization of tissue architecture, Nature, 2013, 493(7432), 318–326 CrossRef CAS PubMed.
- K. Okita, et al., An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells, Stem Cells, 2013, 31(3), 458–466 CrossRef CAS PubMed.
- M. Nakagawa, et al., A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells, Sci. Rep., 2014, 4(1), 3594 CrossRef PubMed.
- K. Uno, N. Sugimoto and Y. Sato, N-aryl pyrido cyanine derivatives are nuclear and organelle DNA markers for two-photon and super-resolution imaging, Nat. Commun., 2021, 12(1), 2650 CrossRef CAS PubMed.
- T. Ichimura, et al., Exploring rare cellular activity in more than one million cells by a transscale scope, Sci. Rep., 2021, 11(1), 16539 CrossRef CAS PubMed.
- X. Lian, et al., Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(27), E1848–E1857 CrossRef CAS PubMed.
- S. Hamad, et al., Generation of human induced pluripotent stem cell-derived cardiomyocytes in 2D monolayer and scalable 3D suspension bioreactor cultures with reduced batch-to-batch variations, Theranostics, 2019, 9(24), 7222 CrossRef CAS PubMed.
- Q. Zhang, et al., Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals, Cell Res., 2011, 21(4), 579–587 CrossRef CAS PubMed.
- L. Cyganek, et al., Deep phenotyping of human induced pluripotent stem cell–derived atrial and ventricular cardiomyocytes, JCI Insight, 2018, 3, 12 CrossRef PubMed.
- J. Schindelin, et al., Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9(7), 676–682 CrossRef CAS PubMed.
- D. Evseenko, et al., Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(31), 13742–13747 CrossRef CAS PubMed.
- A. Q. Lam, et al., Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers, J. Am. Soc. Nephrol., 2014, 25(6), 1211–1225 CrossRef CAS PubMed.
- K. M. Loh, et al., Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types, Cell, 2016, 166(2), 451–467 CrossRef CAS PubMed.
- P. Liang, et al., Drug screening using a library of human induced pluripotent stem cell–derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity, Circulation, 2013, 127(16), 1677–1691 CrossRef CAS PubMed.
- N. Yokoo, et al., The effects of cardioactive drugs on cardiomyocytes derived from human induced pluripotent stem cells, Biochem. Biophys. Res. Commun., 2009, 387(3), 482–488 CrossRef CAS PubMed.
- M. Kawamura, et al., Feasibility, safety, and therapeutic efficacy of human induced pluripotent stem cell-derived cardiomyocyte sheets in a porcine ischemic cardiomyopathy model, Circulation, 2012, 126(11 suppl 1), S29–S37 CAS.
- J. J. Kim, et al., Mechanism of automaticity in cardiomyocytes derived from human induced pluripotent stem cells, J. Mol. Cell. Cardiol., 2015, 81, 81–93 CrossRef CAS PubMed.
- T. Hayakawa, et al., Image-based evaluation of contraction–relaxation kinetics of human-induced pluripotent stem cell-derived cardiomyocytes: Correlation and complementarity with extracellular electrophysiology, J. Mol. Cell. Cardiol., 2014, 77, 178–191 CrossRef CAS PubMed.
- S. V. Pandit and J. Jalife, Rotors and the dynamics of cardiac fibrillation, Circ. Res., 2013, 112(5), 849–862 CrossRef CAS PubMed.
- E. Karbassi, et al., Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine, Nat. Rev. Cardiol., 2020, 17(6), 341–359 CrossRef PubMed.
- P.-P. van Bogaert, J. S. Vereecke and E. E. Carmeliet, The effect of raised pH on pacemaker activity and ionic currents in cardiac Purkinje fibers, Pfluegers Arch., 1978, 375, 45–52 CrossRef CAS PubMed.
- H. Satoh and I. Seyama, On the mechanism by which changes in extracellular pH affect the electrical activity of the rabbit sino-atrial node, J. Physiol., 1986, 381(1), 181–191 CrossRef CAS PubMed.
- S. Tohyama, et al., Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes, Cell Stem Cell, 2013, 12(1), 127–137 CrossRef CAS PubMed.
- G. D. Lopaschuk and J. S. Jaswal, Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation, J. Cardiovasc. Pharmacol., 2010, 56(2), 130–140 CrossRef CAS PubMed.
- K. Jr, C. Stephen, S. Purohit and R. Tian, Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes, Circ. Res., 2013, 113(5), 603–616 CrossRef PubMed.
- J. Li, et al., Circulating re-entrant waves promote maturation of hiPSC-derived cardiomyocytes in self-organized tissue ring, Commun. Biol., 2020, 3(1), 122 CrossRef PubMed.
- G. Tse, Mechanisms of cardiac arrhythmias, J. Arrhythm., 2016, 32(2), 75–81 CrossRef PubMed.
- A. G. Kléber and Y. Rudy, Basic mechanisms of cardiac impulse propagation and associated arrhythmias, Physiol. Rev., 2004, 84(2), 431–488 CrossRef PubMed.
- H. S. Lee, et al., 46-1: Invited Paper: Large-area Ultra-high Density 5.36 10Kx6K 2250 ppi Display, SID Symposium Digest of Technical Papers, 2018, vol. 491 Search PubMed.
- J. Xie, et al., 90-3: Invited Paper: Ultra High PPI VR Display Devices, SID Symposium Digest of Technical Papers, 2024, vol. 551 Search PubMed.
- J. E. N. Jonkman, et al., An introduction to the wound healing assay using live-cell microscopy, Cell Adhes. Migr., 2014, 8(5), 440–451 CrossRef PubMed.
- E. Moen, et al., Deep learning for cellular image analysis, Nat. Methods, 2019, 16(12), 1233–1246 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00528g |
| This journal is © The Royal Society of Chemistry 2024 |