Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Reversible bonding in thermoplastic elastomer microfluidic platforms for harvestable 3D microvessel networks†
Byeong-Ui
Moon
 *ab,
Kebin
Li
ab,
Lidija
Malic
*ab,
Kebin
Li
ab,
Lidija
Malic
 abc,
Keith
Morton
ab,
Han
Shao
d,
Lauren
Banh
efg,
Sowmya
Viswanathan
efg,
Edmond W. K.
Young
abc,
Keith
Morton
ab,
Han
Shao
d,
Lauren
Banh
efg,
Sowmya
Viswanathan
efg,
Edmond W. K.
Young
 de and
Teodor
Veres
*abd
de and
Teodor
Veres
*abd
aMedical Devices, Life Sciences Division, National Research Council of Canada, Boucherville, QC J4B 6Y4, Canada. E-mail: Ben.Moon@nrc-cnrc.gc.ca; Teodor.Veres@cnrc-nrc.gc.ca
bCenter for Research and Applications in Fluidic Technologies (CRAFT), Toronto, ON M5S 3G8, Canada
cDepartment of Biomedical Engineering, McGill University, Montreal, QC H3A 2B4, Canada
dDepartment of Mechanical & Industrial Engineering, University of Toronto, Toronto, ON M5S 3G8, Canada
eInstitute of Biomedical Engineering, University of Toronto, Toronto, ON M5S 3G9, Canada
fOsteoarthritis Research Program, Division of Orthopedic Surgery, Schroeder Arthritis Institute, University Health Network, ON M5T 0S8, Canada
gKrembil Research Institute, University Health Network, ON M5T 0S8, Canada
First published on 18th September 2024
Abstract
Transplantable ready-made microvessels have therapeutic potential for tissue regeneration and cell replacement therapy. Inspired by the natural rapid angiogenic sprouting of microvessels in vivo, engineered injectable 3D microvessel networks are created using thermoplastic elastomer (TPE) microfluidic devices. The TPE material used here is flexible, optically transparent, and can be robustly yet reversibly bonded to a variety of plastic substrates, making it a versatile choice for microfluidic device fabrication because it overcomes the weak self-adhesion properties and limited manufacturing options of poly(dimethylsiloxane) (PDMS). By leveraging the reversible bonding characteristics of TPE material templates, we present their utility as an organ-on-a-chip platform for forming and handling microvessel networks, and demonstrate their potential for animal-free tissue generation and transplantation in clinical applications. We first show that TPE-based devices have nearly 6-fold higher bonding strength during the cell culture step compared to PDMS-based devices while simultaneously maintaining a full reversible bond to (PS) culture plates, which are widely used for biological cell studies. We also demonstrate the successful generation of perfusable and interconnected 3D microvessel networks using TPE–PS microfluidic devices on both single and multi-vessel loading platforms. Importantly, after removing the TPE slab, microvessel networks remain intact on the PS substrate without any structural damage and can be effectively harvested following gel digestion. The TPE-based organ-on-a-chip platform offers substantial advantages by facilitating the harvesting procedure and maintaining the integrity of microfluidic-engineered microvessels for transplant. To the best of our knowledge, our TPE-based reversible bonding approach marks the first confirmation of successful retrieval of organ-specific vessel segments from the reversibly-bonded TPE microfluidic platform. We anticipate that the method will find applications in organ-on-a-chip and microphysiological system research, particularly in tissue analysis and vessel engraftment, where flexible and reversible bonding can be utilized.
Introduction
Microfluidic cell culture systems offer a myriad of benefits over conventional Petri dish or well-plate-based cell culture platforms by providing precise spatiotemporal control of complex 2D and 3D cell and tissue microenvironments using microscale geometries and fluidic operations.1–5 Unlike traditional static cell culture methods, microfluidic devices allow complex, dynamic, and perfusable microenvironments, enabling them to more accurately mimic in vivo conditions. Microfluidic cell culture systems have now advanced to more physiologically relevant microfluidic organ-on-a-chip (OOC) devices that can recapitulate organ-level functions.6 Numerous OOC devices have been developed over the last few years, including lung-on-a-chip,7–9 beating heart-on-a-chip,10–12 peristalsis motion in a gut-on-a-chip,13,14 cancer tumor-on-a-chip, vessels-on-a-chip,15 and many others. In general, microfluidic cell culture systems can be useful for a variety of applications including high-throughput cell-based assays, pre-clinical drug testing, and biofabrication of tissue constructs for regenerative medicine.16Microfluidic vascular networks have been used for a variety of biomedical applications including to model vascularized organs and study vascular diseases. Through self-assembly of endothelial cells (ECs) in 3D environments, perfusable vessel networks can be created and applied to study the effects of hemodynamic flow on vessel network formation and regression, as well as to examine signaling pathways involved in vessel remodeling.17,18 Using organ specific ECs and local vessel supporting cell types (i.e. perivascular cells, stromal cells, parenchymal cells and stem cells), vascularized tissue models also have been established for the brain plexus,19,20 heart,21 lung,22 and liver23 to study cell–cell interactions, immune responses, and responses to drugs and inflammatory stimuli. In addition to single-organ modeling, vessel networks can be used to join multiple on-chip organ compartments to study complex multi-organs processes, such as intestine-vessel-liver interaction models, to study toxicity effects and organ metabolism.24 The emergences of microfluidic vessel networks have thus enabled the generation of various vascularized models that can be scaled up to achieve high throughput formats for drug screening applications.25,26
In terms of regenerative medicine, microvessels are amenable to in vivo delivery and implantation in a clinical setting. Recently, Sun et al. also showed that “ready-made” or pre-fabricated microvessels can be used for in vivo vascularization for cardiomyocyte engraftment.27,28 Their findings revealed that transplanted microvessels enhanced tissue engraftment and led to cardiac functional recovery, highlighting their potential use for cell replacement therapies. Inspired by this potential of transplantable microvessels, we demonstrate here a strategy to use microfluidic-based microvessel networks as a convenient way to construct ready-made (herein termed “microfluidic-engineered”) microvessels, eliminating the need for animal sacrifice. This approach would be able to produce off-the-shelf, injectable microfluidic-engineered microvessels while preserving both the mechanical properties and biological functions of the microvessels.29 Previous studies have also demonstrated the use of injectable microgel and microfluidic tube-based approaches to enhance tissue vascularization and angiogenesis processes,30,31 which show therapeutic potential in vascular regeneration, wound healing, vessel engraftment and small organ transplantation (such as pancreatic islets).
Currently, poly(dimethylsiloxane) (PDMS) is the most popular device material for microfluidic devices due to its simple fabrication process. However, inherent physical and chemical properties of PDMS can cause absorption of small hydrophobic drug molecules or metabolites that can lead to adverse experimental effects and potential artifacts in biological assays.32,33 Furthermore, while manufacturability and scale-up of PDMS is possible, commercialization with PDMS is considered a major challenge due to barriers in mass production processes. Alternative materials that can combine advantages of manufacturability with those of common polymers thermoplastics would greatly improve the commercialization potential of OOC devices and their adoption for pre-clinical applications.34 Thermoplastic elastomers (TPE) are a class of thermoplastic materials that show great potential as an alternative for microfluidic devices and cell culture applications due to their broad spectrum of material properties.35–37 TPEs are optically transparent, soft, flexible, and stretchable, all of which are desirable properties for OOC devices. TPEs are also compatible with other plastic materials and manufacturing processes, thus facilitating a viable path to commercialization efforts. In particular, the ability of TPE materials to be reversibly bonded makes them highly versatile, allowing for conformal attachment to other thermoplastic materials. Previously, our group has demonstrated TPE-based, reversibly bonded microfluidic platforms for applications in protein patterning,38 modular droplet formation,39 and cell patterning.40 Herein, we sought to capitalize on reversible bonding mechanism of TPE-based devices to make tissue-engineered microvessels on-chip that can be made available as off-the-shelf injectable tissue-laden entities.
Regarding the reversible bonding methods, several examples of fabrication techniques have been shown for cell culture applications on different materials. Chu et al., demonstrated a non-plasma treatment fabrication process to create a reversibly sealed microfluidic device using a new PDMS-based adhesive polymer mixture, which was then applied to cardiomyocyte patterning.41 Pitingolo et al., showed a PDMS-coated poly (methyl methacrylate) (PMMA) microfluidic spheroid culture device using permanent magnets to seal the two stacks of PMMA.42 The temporarily bonded device was detached by removing the magnets from the inserts, allowing collection of the spheroid sample for off-chip analysis. Funano et al., showed reversible glass–glass bonding microfluidic devices using a wet water drop method and binding clip pressure for cell recovery applications.43 Recent advances in highlighting reversible bonding in microfluidic technology have been reviewed by Zhang and colleagues.44
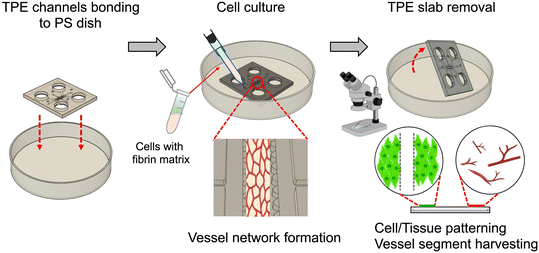
In this paper, we demonstrated the use of TPE microfluidic devices to facilitate microfluidic cell and tissue culture and employed a reversible bonding technique to retrieve cells and tissues. We specifically focused on the formation and recovery 3D microvessel networks in a controlled microfluidic environment with potential to achieve animal-free tissue generation and transplantation for clinical applications (Fig. 1). First, we conducted a study to assess the bonding strength of TPE with a polystyrene (PS)/glass substrate and characterized the properties of the TPE material. Subsequently, we demonstrated microfluidic cell culture and cell handling on TPE–PS devices, studying the effectiveness of cell recovery. Finally, we showcased an OOC application of TPE–PS microfluidic devices, specifically focusing on 3D microfluidic-engineered vessel network formation by investigating the integrity of single and multi-vessel loading on a chip, which can be applied for organ-to-organ interactions and tissue patterning applications. To our knowledge, this is the first demonstration of 3D OOC and tissue handling using an openable TPE-based microfluidic platform with reversible bonding to achieve tissue generation, recovery and extraction.
Results and discussion
Bonding strength of the TPE–PS device
Prior to using the TPE slab for cell culture, we first examined the bonding strength of TPEs on the PS and glass substrates. TPE bonding strength was assessed by measuring delamination pressures in microchannel devices (see ESI† Fig. S1). A single layer of microfluidic devices was prepared in both PDMS and TPE-based materials and bonded against PS/glass substrates with and without post-assembly heat treatment at both 37 °C and 65 °C. For both materials, the most effective bonding results were achieved by meticulously eliminating any air bubbles trapped between the two surfaces.As shown in Table 1, the bonding strength of the TPE–PS device is nearly 6-fold higher than the PDMS–PS device while TPE–glass device is over 3-fold higher than the PDMS–glass device with identical microfluidic design. In addition, the bonding strength of the TPE–PS/glass devices is significantly enhanced further using a post-assembly heat treatment. In contrast, post-assembly heat treatment has no significant effect on PDMS–PS bonding strength. We believe that the superior bonding strength of TPE materials on PS/glass substrates derives from its intrinsic physical properties. The rubber-like TPE film is composed of styrene–ethylene–butylene–styrene (SEBS) and exhibits strong van der Waals interactions at the interface between SEBS and PS/glass.45 This stronger bond between TPE and PS/glass substrates is attributed to the intrinsic adhesive and cohesive properties resulting from the reorganization and interpenetration of mobile branched polymer chains at the interface. Since the glass transition temperature of TPE is below −50 °C, this macromolecular motion and the reorganization of the soft polymer segments can occur at the interface even at room temperature as well as at post-heat treatment temperatures.46 However, the hydrophobic nature of PDMS, due to the CH3 groups on its surface, requires O2 plasma treatment to form bridging Si–O–Si bonds at the interface, resulting in irreversible bonding. For subsequent cell experiments, the bonded TPE–PS devices were heat-treated overnight at either 37 °C in a CO2 incubator or at 65 °C in an oven.
| Device | No post assembly-heat treatment | Post assembly-heat treatment at 37 °C | Post assembly-heat treatment at 65 °C |
|---|---|---|---|
| PDMS–PS | 31 ± 0.5 kPa | 33 ± 0.9 kPa | 35 ± 0.3 kPa |
| TPE–PS | 181 ± 1.2 kPa | 211 ± 4.3 kPa | 282 ± 11.6 kPa |
| PDMS–glass | 31.7 ± 3.0 kPa | 50.3 ± 0.8 kPa | 63.6 ± 2.5 kPa |
| TPE–glass | 95.6 ± 2.2 kPa | 138.2 ± 2.7 kPa | 157.4 ± 4.2 kPa |
Comparison study of protein absorption on PDMS and TPE channels
We evaluated protein absorption on the TPE material using TPE–glass bonded devices and compared them with the PDMS–glass bonded devices, as TPE is expected to have fewer protein absorption issues. To confirm this, we exposed fluorescein isothiocyanate (FITC)-dextran solution to the devices and compared their fluorescence intensities. The resulting measurements show that the TPE–glass bonded substrate exhibits significantly lower fluorescence intensity compared to the PDMS–glass substrate (see ESI† Fig. S2). These results are consistent with the other studies on Rhodamine B adsorption in the microchannels,45 and open polymer disks,47 confirming that TPE materials are a better choice than PDMS for applications with fewer protein-related issues, such as drug-screening microfluidic devices.33,48Cell culture on TPE–PS platform and cell recovery study
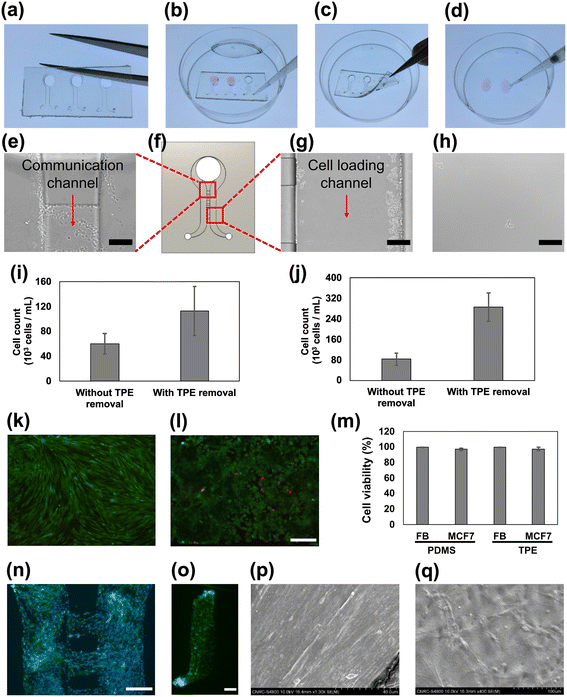
Microfluidic cell culture has been previously demonstrated on TPE-based constructs, using the described simple bonding process.49,50 Here, we advance the study of TPE-based microfluidic cell culture and handling by incorporating a reversible bonding method to harvest the cultured cells after removing the TPE slab. We began by replicating the TPE slabs from the epoxy mold using a hot embossing process (Fig. 2a). To prepare the device for cell culture, the slab was attached to the PS dish without any post-processing. After two days of cell culture, we manually removed the TPE slab using forceps (Fig. 2b and c). A drop of trypsin–EDTA was introduced to the cultured regions and the trypsinized cells were subsequently collected (Fig. 2d). For comparison study, we also performed on-chip trypsinization without removing the TPE slab. Fig. 2e, g and h shows the resulting images for fibroblast cells (FBs) and human breast cancer cells (MCF-7) after trypsinization and with and without TPE removal. We observed that unharvested cells remain within the channel and between communicating channels when trypsinization is performed through the microfluidic channels (see also ESI† Fig. S3). However, when the TPE microfluidic devices is removed prior to trypsinization, more cells were removed and recovered from the PS surface. As such, by employing trypsinization after TPE removal resulted in significantly higher cell recovery for both FBs and MCF-7 cells (Fig. 2i and j) compared to those in removed and recovered from within the channels. We believe this method to easily remove the channel constructs could also be beneficial to microfluidic in vitro culture or cell sorting systems where rare cells need to be efficiently handled and harvested.51 Although other reversible bonding methods have been demonstrated in microfluidic devices using PDMS materials with mechanical clamping,52 chemical treatment,53 optimized mixing ratio of PDMS base and curing agent, and fine-tuning O2 plasma treatments,54 they still require tuning to find the optimal conditions for reversible PDMS bonding. Our approach, employing TPE material-based microfluidic devices, is notable for its direct compatibility with a range of thermoplastic substrates without the need for complicated optimization processes.We also evaluated the cytotoxicity of cells cultured in TPE channels and compared with PDMS devices by examining the viability of cells. Fig. 2k and l shows representative images of the resulting cell cytotoxicity experiments in TPE bonded devices with the FBs and MCF-7 cells. Both TPE- and PDMS-based channels exhibited over 95% cell viability, showing that the TPE material is highly biocompatible and suitable for cell culture platforms (Fig. 2m). In addition, we showcase a practical implementation of our reversible bonding methodology by employing for 2D cell patterning of FBs. After achieving confluence of FBs in both the main and communication channels, we removed the TPE slab using the reversible bonding mechanism and proceeded to stain the cells as illustrated in Fig. 2a–d. Fig. 2n shows the stained image of Hoechst and FITC-phalloidin after TPE device removal, demonstrating that reversible bonding could be also used for cell patterning and migration applications.55,56
3D tissue culture
Microfluidic approaches to on-chip 3D tissue formation offers a highly relevant 3D physiological microenvironment that enables various studies from fundamental cellular crosstalk to OOC applications. To assess the effectiveness of our TPE materials for 3D cell culture, FBs were cultured for two days in a fibrin gel matrix composed of collagen, fibrinogen and thrombin. Subsequently, the TPE slab was gently removed using forceps. Notably, upon removing TPE slab, we observed that the FBs-embedded fibrin gel remained intact and attached to the hard, unstructured PS dish substrate. Fig. 2o shows the image of FITC-phalloidin stained cells confirming that FBs-embedded in fibrin gel retain the shape of the entire microfluidic channels and remain attached to the PS surface.While fluorescence microscopy enables labeling of specific structures or markers within the tissue, scanning electron microscopy (SEM) offers high-resolution view of surface morphology to examine cell and tissue ultra-structure. Combining fluorescence microscopy and SEM may provide a more comprehensive view of a tissue sample by revealing both its internal structure and surface features. Fig. 2p and q show examples of FB tissue SEM images taken after TPE slab removal. The compacted tissue sample appeared dense with elongated shapes as a result of surface treatment. In contrast, the non-compacted tissue sample displayed spreading and smooth surface characteristics. Our reversible TPE bonding approach thus shows full access to the tissue surface features and topography by enabling SEM images outside the device and may be applicable to other in vitro microtissue and non-biological sample analyses.57
Generation of 3D microvessel networks
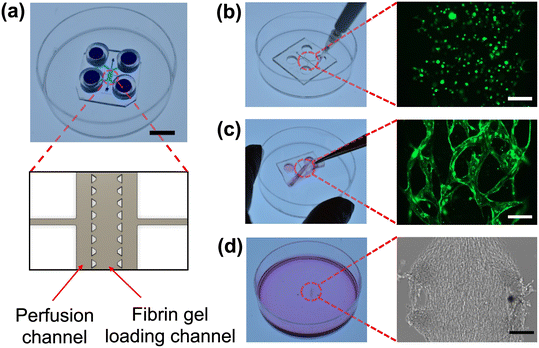
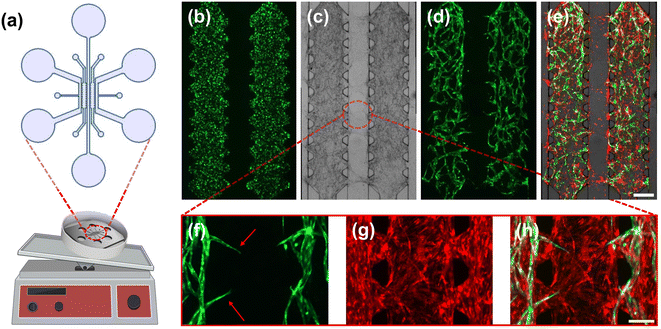
Vasculogenesis and the vessel network growth were examined by co-culturing GFP-expressing human umbilical vein endothelial cells (GFP-HUVECs) and RFP-expressing human lung fibroblasts (RFP-FBs) in a TPE-PS microfluidic device. For vasculogenesis, the TPE–PS microfluidic device consists of one central channel for fibrin gel loading and two side channels for media perfusion (Fig. 3a).58,59 There are additional connecting input ports designed for removing air bubbles during the flow of culture media within the perfusion channels. After the cells have acclimatized to in the fibrin matrix on day 0, we attached TPE tubes on all the inlet and outlet reservoir to increase the reservoir volume to ∼150 μL. These TPE tubes are also reversibly bonded to the TPE slab and both are simultaneously removed for the tissue harvesting process.For vasculogenesis experiments, we seeded a mixture of GFP-HUVECs and RFP-FBs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 ratio) in a fibrin gel and examined the microvessel network formation over 8 days. Cells were loaded into the micropillar-guided central channel using a pipette tip (Fig. 3b). On day 4, we observed that GFP-expressed HUVECs began to cluster and self-assemble into vessel-like structure and were developed further until the last day of culture. For the subsequent procedure, we removed the TPE slab using forceps to allow harvesting (Fig. 3c). To harvest the microfluidic-engineered microvessels from the device, ensuring the accessibility of the tissue for the digestion steps is crucial. Interestingly, upon removal of the TPE, the tissue as a whole remained intact on the PS substrate, preserving the patterns of the device design (Fig. 3d).
0.75 ratio) in a fibrin gel and examined the microvessel network formation over 8 days. Cells were loaded into the micropillar-guided central channel using a pipette tip (Fig. 3b). On day 4, we observed that GFP-expressed HUVECs began to cluster and self-assemble into vessel-like structure and were developed further until the last day of culture. For the subsequent procedure, we removed the TPE slab using forceps to allow harvesting (Fig. 3c). To harvest the microfluidic-engineered microvessels from the device, ensuring the accessibility of the tissue for the digestion steps is crucial. Interestingly, upon removal of the TPE, the tissue as a whole remained intact on the PS substrate, preserving the patterns of the device design (Fig. 3d).
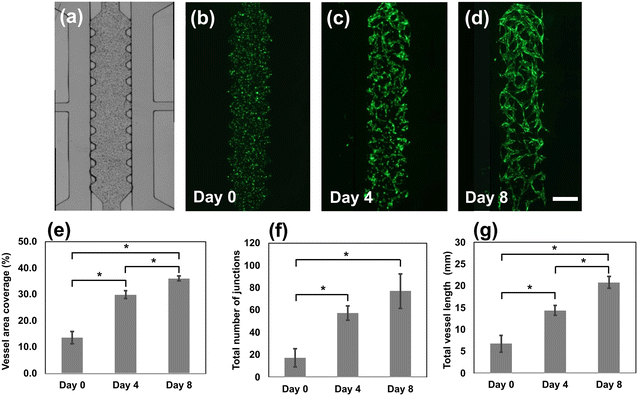
Fig. 4 depicts the progression of vessel network formation in the microfluidic channels and provides quantitative analysis of cell culture over an 8 day period. From the images, we measured the vessel area coverage, the total number of junctions and the total vessel length. The vessel area coverage increased significantly from 14% (day 0) to 30%, and 36% by days 4 and 8, respectively. Interconnected vessel networks began to form on day 3 and continued to develop until the last day of culture. The total number of junctions increased from 17 (day 0), to 54 (day 4) and then to 77 (day 8). The total length of the vessels also increased dramatically, from 14 mm on day 4 to 22 mm on day 8. In comparison to previous work performed using PDMS-based devices where the formation of the vascular networks spanned 4–7 days,58,60 the TPE-based vessel network formation appeared to entail a slightly longer period. We hypothesize that the slower development of the vessel networks may stem from diminished oxygen exchange within the device, attributed to the lower gas permeability of TPE materials compared to PDMS materials (see ESI† Table S1 for oxygen permeability study). The oxygen permeability of TPE is two orders of magnitude lower than PDMS but significantly higher than PS. Improvements in direct gas exchange with the culture media are possible using an open-top microfluidic chip design, or by incorporating of a TPE membrane sandwiched device to enhance oxygen exchange in the templated cell culture microenvironments.61,62 The main takeaway is that we were nevertheless able to successfully create full, functional microvessel networks using TPE microfluidic devices.
3D perfusable microvessel networks
We investigated 3D luminal and perfusable microvessel networks to verify that the developed microvessel structure displays behavior similar to that observed in vivo. For these experiments, we loaded cells at a mixing ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
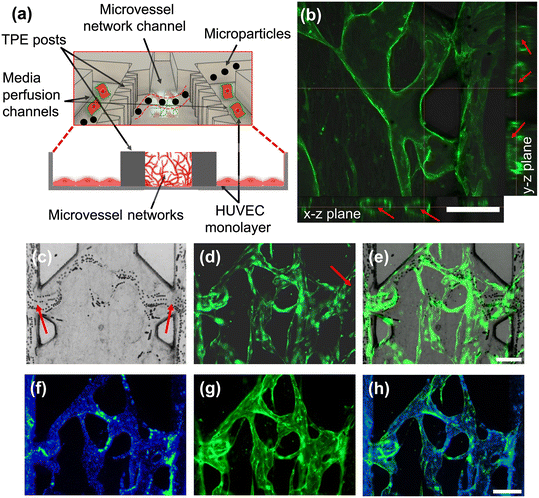
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, consisting of GFP-HUVECs and RFP-FBs, respectively, during the cell seeding step to support opening of the vessels between the TPE posts.58,63 On day 6, we cultured a HUVEC monolayer in the side channels to create opening connections to the luminal microvessels (Fig. 5a). Before assessing perfusion capacity using microparticles, the 3D luminal microvessel structures were examined using confocal microscopy. Fig. 5b shows images of confocal z-stacks revealing distinct 3D tubular architecture and organization of the microvessel network in both the x–z and y–z planes.
1, consisting of GFP-HUVECs and RFP-FBs, respectively, during the cell seeding step to support opening of the vessels between the TPE posts.58,63 On day 6, we cultured a HUVEC monolayer in the side channels to create opening connections to the luminal microvessels (Fig. 5a). Before assessing perfusion capacity using microparticles, the 3D luminal microvessel structures were examined using confocal microscopy. Fig. 5b shows images of confocal z-stacks revealing distinct 3D tubular architecture and organization of the microvessel network in both the x–z and y–z planes.
To assess the perfusability of the networks, perfusion experiments were conducted by introducing different diameter microparticles into the side channels. Fig. 5a captures the flow of 10 μm diameter microparticles through the luminal microvessel network in the middle channel carried by hydrostatic flow from one side channels to the other. Specifically, we observed that the microparticles flowed into the luminal microvessel network as they approached the luminal opening ports, where hydrodynamic resistance is lower (Fig. 5c–e and ESI† Movie S1). Furthermore, we confirmed the perfusability and integrity of the microvessel networks using 0.62 μm blue fluorescence beads. The sub-micron particles remained confined within the network, indicating no leakage inside of the vessel lumens (Fig. 5f–h). Thus, our results demonstrated that the TPE-based microfluidic devices can support the growth of 3D microvessel networks and the formation of microfluidic vessel-on-a-chip models. Additional investigation is needed to evaluate the permeability and barrier functions relevant to drug screening applications.60
Harvesting pre-fabricated, microfluidic-engineered microvessels
To demonstrate the reversible bonding capacity of the extraction and utilization of engineered vessel networks for downstream analysis and off-the-shelf microvessel harvesting and engraftment applications, on day 9 of the tissue culture, we gently removed the TPE slab as previously mentioned (see “Cell culture on TPE–PS platform and cell recovery study” section). Upon removal of the TPE, the vessel networks remained intact on the PS substrate without damage or defects in the structures (see ESI† Fig. S4). The cells were patterned around the TPE posts in the bright-field images and the vessel networks revealed distinctively in GFP fluorescence signals on PS substrate. Analysis of the vessel area coverage, the total number of vessel network junctions and the total vessel length also showed no significant difference before and after TPE removal. We also observed sustained cell culture after the removal of the controlling microstructures: in the absence of microstructures and after two additional culture days, areas originally patterned with TPE post structures were quickly covered by migrating FBs. The cells continued to grow in the outward direction under static flow conditions (see ESI† Fig. S5). Endothelial sprouting and tips also appeared outside of the patterned area, indicating that the main network of vessels within the central channel is stable. Therefore, microfluidic-based cell culture environments allow the intricate spatial patterns and enable the control of interstitial flow within the channels.64The reversible bonding has the potential to enhance microfluidic-based in vitro tissue sample preparation for immunological and histological staining by allowing easily access to the microchannels and their contents. In contrast, traditional irreversibly bonded PDMS devices enclose the microchannels completely, necessitating the use of a scalpel to separate the PDMS material from the substrate, which can induce tissue damage or disrupt their structure.65 We compared Hoechst tissue staining between an irreversibly bonded PDMS–glass device and a reversibly bonded TPE–PS device. Results showed that staining of the entire tissue was more efficient after the TPE slab removal, because the reagents are more freely accessible to the whole patterned area (see ESI† Fig. S6). This approach to immunostaining may be particularly useful for in vitro microfluidic tissue culture systems where end-point analyses and downstream off-chip assessments are required.66
To demonstrate the feasibility of harvesting microvessel from the produced microfluidic-engineered microvessels, we extracted vessel network tissue from the reversibly-bonded microfluidic devices. Microvessels from six devices were pooled together, followed by tissue digestion through dissociation in a collagenase and DNase solution. The microvessel segments obtained from this procedure were then recovered and observed under the microscope (see ESI† Fig. S7). The retrieved vessel segments ranged in size from ∼50 to 200 μm. The significance of retrieving engineered microvessel using TPE-based microfluidic devices lies in its potential implications, providing a platform for generating and successfully handling microvessel networks without the laborious and intricate processes required for animal handling, rodent dissection, and sacrifice, further raising ethical concerns thereof. Given that transplanted microvessels isolated from rat adipose tissue have shown promising outcomes in supporting in vivo vascularization of cardiomyocyte engraftment,27 and functionality of human islets,67 the microfluidic-engineered microvessel approach may offer a more convenient method for achieving animal-free tissue generation and transplantation. Although other reversible bonding technologies could be potentially employed for in vitro microvessel formation, harvesting, and tissue transplantation applications, our work, to the best of our knowledge, marks the first confirmation of successful retrieval of organ-specific vessel segments from a reversibly-bonded TPE microfluidic platform (see Table S2† for other approaches).
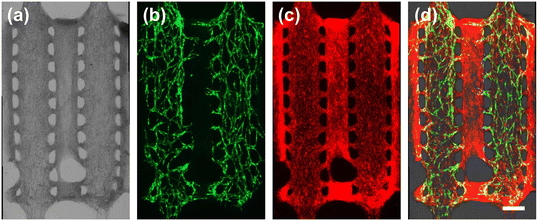
Multi-vessel platform for parallel co-culture of vascularized tissues
To showcase the ability to load and culture multiple vessel networks simultaneously on a TPE–PS microfluidic platform, we designed and built devices with two identical cell loading chambers arranged in parallel, so that culture media could flow through both middle and side channels (Fig. 6a). The single vessel chamber design includes a vessel-forming chamber, with the gap between the two cell-loading channels set to 500 μm to assess angiogenic sprouting. Fig. 6 shows bright-field and fluorescence micrographs after cell seeding (Fig. 6b) and the resulting vessel network formation on day 8 (Fig. 6c–e). A dense layer of FBs covered the entirety of the two chambers (Fig. 6e). Simultaneously, the GFP-HUVECs contribute to the creation of a 3D lumens by forming vessel connections (Fig. 6d). In particular, we observed that FBs demonstrate rapid cell migration behavior and migrate outside of the main chamber to cover the media flow channels. This FBs action may serve the process of HUVEC sprouting (Fig. 6c and e) for the following experiments.68,69Subsequently, angiogenic sprouting experiments were conducted by placing the TPE–PS device on a platform rocker. This was done to induce hydrostatic flow through the channels because angiogenic sprouting is promoted by flow-mediated shear stress (Fig. 6a).70Fig. 6f–h shows sprouting angiogenesis directed toward the center of the channel, with GFP-HUVECs initiating sprouting from the pre-existing vessel network. Using our approach, future investigations are envisaged to study the impact of hydrostatic flow direction on angiogenic sprouting71 and to perform vessel-to-vessel communication studies.
Next, we explored the reversible bonding characteristics of TPE using the multi-vessel chamber platform. Interestingly, Fig. 7 shows the multi-vessel chambers after the removal of the TPE slab, revealing intact microvessels and FBs adhered to the flat PS substrate. This patterned matrix could serve as an open tissue construct and may find applications in scenarios such as blood vessel-to-spheroid communication studies within open microfluidic systems.72–74
Conclusions
In this paper, we presented a novel approach to fabricating all-polymer microfluidic devices for cell handling and OOC systems applications. This method allows for the recovery of cell cultures and OOC constructs using the reversible bonding between microstructured TPE films and planar PS substrates. The fabricated TPE–PS devices surpass the bonding strength of similar PDMS devices, while maintaining intact and high viability cell and tissue cultures. The TPE–PS platform was shown to support cell culture over an 8 day period and enable the development of interconnected microvessel networks. This platform was also leveraged to generate perfusable 3D microvessel networks that remained intact, allowing microfluidic-engineered microvessel recovery and extraction. By removing the TPE-based microfluidic slab, we further demonstrated facile harvesting and manipulation of patterned cells and tissues formed within the TPE–PS platform. Future investigation will involve cultivating and studying specialized vascularized micro-organs within on-chip tissue chambers through co-culturing different cell types. We envision that our method will facilitate implementation of assays and analyses for organ-on-chip research and find applications in pharmacology and regenerative medicine.75,76Experimental section
TPE–PS bonded microfluidic devices fabrication
To facilitate monolayer cell culture on TPE slab fabrication, we adopted a three-layered device design (Fig. 2f).77 The depth of the communication channel, cell seeding channel and inlet/outlet ports correspond to 25 μm, 200 μm and 400 μm, respectively. For the vasculogenesis devices (Fig. 3), we designed one cell loading channel and two media perfusion channels with four reservoirs. Unlike other vasculogenesis designs, additional side input ports were added to facilitate cell seeding and culture. The detailed device designs with corresponding dimensions are shown in Fig. S8.† The preparation of silicon master molds for all devices presented was performed using SU-8 photoresists and standard photolithography. Briefly, SU-8 photoresist (Gersteltec, Pully, Switzerland) was spin-coated onto a 6 inch silicon wafer (Silicon Quest International, Santa Clara, CA), pre-baked and exposed using UV light at 365 nm (EVG 620 Mask Aligner, Austria) through a photomask. The photomask was generated using computer-aided design (CAD) software (AutoCAD 2022, Autodesk, Inc., Dan Rafael, CA) and printed onto a high-resolution transparency sheet (Fineline Imaging, Colorado Springs, CO). Following the UV exposure, the SU-8 photoresist was post-baked and subsequently developed in propylene glycol monomethyl ether acetate (PGMEA; Sigma-Aldrich, Oakville, ON). Finally, the patterned resist microstructures were rinsed with PGMEA and isopropanol, and the wafer was dried with a stream of nitrogen gas. An epoxy mold was fabricated from the SU-8/silicon master using an intermediate PDMS replica (Sylgard 184; Dow Corning, Midland, MI). The PDMS liquid prepolymers (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w elastomer base/curing agent) were poured onto the master mold and cured at 85 °C for 2 h. The PDMS replica was separated from the master mold and then used as a template to create an epoxy mold for the TPE hot embossing process. The epoxy mold was replicated from an intermediate PDMS mold by thermal curing a mixture of Conapoxy FR 1080 part A (resin) and part B (hardener) (ELANTAS PDG Inc., St. Louis, MO) in a ratio 75
1 w/w elastomer base/curing agent) were poured onto the master mold and cured at 85 °C for 2 h. The PDMS replica was separated from the master mold and then used as a template to create an epoxy mold for the TPE hot embossing process. The epoxy mold was replicated from an intermediate PDMS mold by thermal curing a mixture of Conapoxy FR 1080 part A (resin) and part B (hardener) (ELANTAS PDG Inc., St. Louis, MO) in a ratio 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 by weight for 8 h at 80 °C in an oven after degassing the mixture in a vacuum chamber. After peeling off the PDMS template from the cured epoxy, the epoxy mold was hard-baked for 3 h at 180 °C.
60 by weight for 8 h at 80 °C in an oven after degassing the mixture in a vacuum chamber. After peeling off the PDMS template from the cured epoxy, the epoxy mold was hard-baked for 3 h at 180 °C.
TPE received in the form of pellets (Mediprene OF 400 M, Åmål, Sweden) was extruded at 165 °C to form TPE sheets about 2 mm thick. The devices microstructures were hot-embossed (EVG® 520HE, EV Group, Schärding, Austria) into these TPE sheets using the epoxy mold at an applied force of 6000 N at a temperature of 170 °C for 5 min.
Finally, the hot-embossed TPE substrates were cut to size using titanium scissors (Fiskars Group, Espoo, Finland) in order to fit into PS Petri dishes (60 × 15 mm, Ultident Scientific Inc., Saint-Laurent, Canada). Access holes for the inlets/outlets and reservoirs were punched in the TPE using 1 mm diameter biopsy punches (Integra Miltex, Inc., Rietheim-Weilheim, Germany) and a ¼ inch Mayhew punch (Mayhew Steel Products, Inc., Turners Falls, MA), respectively. The TPE slabs were assembled by simply attaching and pressing the TPE slab onto the PS Petri dish being careful to remove any air bubbles, followed by heat treatment at 37 °C or 65 °C, depending on the experimental requirements.
The short TPE tubes shown in Fig. 3a were made using 4 mm thick TPE sheets. The TPE was flattened using hot embossing and punched using ¼ inch and ⅜ inch punches (Mayhew Steel Products, Inc., Turners Falls, MA) to make inner and outer holes, respectively. The prepared TPE tubes were then directly attached to the TPE slab reservoirs after cell seeding and removed prior to the tissue harvesting step.
SEM sample preparation and imaging
The surface morphology of 3D FB samples were obtained after the TPE slab removal by scanning electron microscopy (SEM, Hitachi, S-4800). The 3D FB samples were subjected to the dehydration process by sequentially transferring them to different ethanol concentrations (30%, 50%, 70%, 90%). Upon drying, the samples were carefully mounted onto an aluminum stub and coated with a 10 nm platinum thin film (Leica EM ACE600, Leica) to avoid charging effects during the SEM observation. The SEM images were acquired using accelerating voltages set to 10 keV.TPE–PS bonding strength tests
TPE bonding strength was assessed by measuring delamination pressures in a simple device comprised of a small chamber having a diameter of 0.8 mm with a 100 μm wide microchannel. The experimental setup consisted of an in-house-built pneumatic pressure testing box equipped with 16 independent channels (see ESI† Fig. S1). The delamination pressure was defined as the pressure under which the edge of the chamber started detaching from the substrate. The delamination process was monitored under an inverted microscope (Eclipse TE-2000-U, Nikon, Japan). Delamination test devices were prepared in both PDMS and TPE-based materials and bonded against PS/glass substrates both with and without heat treatments at 37 °C in a cell culture incubator (HERAcell viso 160i; Thermo Fisher, Waltham, MA) or at 65 °C in an oven (Binder, Thermo Fisher Scientific, Waltham, MA).Cell culture
Human lung FBs (NHLF, Lonza, Basel, Switzerland) and human breast carcinoma cell line (MCF-7, ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle medium (DMEM) with 10% FBS, penicillin (100 units per mL) and streptomycin (100 μg mL−1) and used at passage numbers of P4–P7 and P7–P10, respectively. RFP-FBs (RFP-HLF, Angio-Proteomie, Boston, MA) and GFP-HUVECs (HUVEC, Angio-Proteomie) were cultured in DMEM with 10% FBS, penicillin (100 units per mL) and streptomycin (100 μg mL−1) and EC growth media kits (EGM-2 Bulletkit, Lonza), respectively. The passage numbers of RFP-FBs and GFP-HUVECs were P4–P6. All cells were maintained at 37 °C and 5% CO2 in an incubator.Microfluidic cell seeding and culture
For cell recovery application experiments, we used lung FBs (NHLFs) and breast cancer cells (MCF-7). The TPE–PS devices were first cleaned with 70% ethanol and deionized (DI) water (rinsed three times) and then coated with 0.05 mg mL−1 fibronectin (Sigma-Aldrich) in PBS for 30 min at room temperature. For monolayer cell culture experiments, the FBs and MCF-7 were seeded on each side of the TPE–PS channels at a concentration of 2 × 106 cells per mL. Prior to cell seeding, the channels were filled with culture media and then a 4 μL volume of suspended cells were seeded on each side of the microchannels. The seeded cells were cultured for two days with culture media replaced after 24 h. To remove the TPE slab from the PS substrate, we used forceps to lift the slab from the PS substrate. The trypsinization process was performed after two days of cell culture using a 0.05% trypsin–EDTA solution. A drop of 30 μL of trypsin–EDTA solution was added to both cell-cultured regions of the TPE-removed substrate and the TPE–PS channels and incubated for 4 min in the CO2 incubator. Cells were counted to compare the recovery achieved with and without removal of the TPE slab. For 3D FB gel formation, we used a fibrin gel matrix composed of 2.5 mg mL−1 fibrinogen, 0.2 mg mL−1 collagen type I, 1 U mL−1 thrombin 2 μg mL−1 aprotinin and culture media at a cell concentration of 7 × 106 cells per mL. The FB embedded gel was loaded only one side of the channel. Followed by 2 days of cell culture, the TPE slab was gently removed using forceps.For microvessel network formation experiments, we used RFP-FBs and GFP-HUVECs. The TPE–PS devices were obtained by first cleaning the TPE slab. This was accomplished by soaking the substrate in 70% ethanol for 30 min, followed by DI water rinsing under the hood. The slab was then attached to the PS dish and covered with the lid. Subsequently, the device was placed in the 65 °C oven to increase the bonding strength of the device. Three days prior to on-chip culture, RFP-FBs and GFP-HUVECs were thawed and prepared for cell seeding. We used the RFP-FBs and GFP-HUVECs cell at concentrations of 7.5 × 106 cells per mL and 10 × 106 cells per mL, respectively. On the day of seeding, the cells were suspended in a freshly prepared 2.5 mg mL−1 fibrinogen (Sigma-Aldrich) in PBS solution with supplemented 0.2 mg mL−1 collagen type I (VWR, Mississauga, ON), 10 μg mL−1 Aprotinin (Sigma-Aldrich) and EGM-2 solution. Before loading the cells, the prepared solution was mixed with 1 U mL−1 thrombin (Sigma-Aldrich). Immediately after, the mixture of fibrin gel matrix was gently loaded into the loading channel. The gel was polymerized for 10 min in the CO2 incubator. After gelation, EGM-2 culture media was added to each side of the wells. Using the bubble removal input ports, we aspirated any observed air bubbles so that the culture media could flow through the perfusion channels. Once the cells were attached on the plate, we added additional TPE reservoirs in each well. We supplied 150 μL and 50 μL upper and lower reservoirs, respectively. The medium was changed daily.
To demonstrate microvasculature network perfusability, we first coated the device surface with Quick coating solution (Angio-Proteomie) and seeded GFP-HUVECs on the side channels via the bubble removal port on day 6. We ensured there was no flow during this cell seeding process and waited until the cells were settled on the surface. The monolayer of cells continued to culture, and by day 9, we observed that microparticles entered the vessel, leading to the opening of the lumen and subsequent flow through the vessel.
For multi-vessel chamber experiments, we followed a similar experimental protocol as the one described for the microvessel network formation studies. In addition, we placed the multi-vessel chamber device on the rocking plate (OrganoFlow®, MIMETAS, Oegstgeest, The Netherlands) on day 8 to introduce hydrostatic flow. The rocker was set to move every 10 min time interval with a 20-degree angle. The cells were cultured for additional 4 days with media changed daily.
Cell viability assay, imaging and analysis
We performed cell cytotoxicity experiments using LIVE/DEAD Cell Imaging Kit (cat. # R37601 Invitrogen, Waltham, MA). To assess cell viability, a mixture of live and dead reagents was directly introduced either in the TPE–PS channels or onto the cell patterned area after TPE slab removal. Following 20 min incubation at room temperature, fluorescence images were acquired with an inverted EVOS FL Auto Cell Imaging System (Life Technologies, Carlsbad, CA) using 4× and 10× objectives. The captured images were analyzed using ImageJ (NIH, Bethesda, MD) after post-processing. Cell viability was determined by calculating the ratio of living cells to the total cell count.GFP-HUVECs were cultured and used to quantify microvessel growth by assessing vessel area coverage, total number of junctions, and total vessel length. Images were captured on days 0, 4, and 8 using the EVOS FL microscope with a 4× objective. The microscope GFP settings were at 90% intensity with a 250 ms exposure time. The acquired images were analyzed using Angiotool software (version 0.6a, National Cancer Institute, Bethesda, MD) to assess the characteristics of microvessel growth.78
Vessel perfusion experiments and 3D confocal imaging
To assess the perfusable microvessel network, we performed microparticle tracking to confirm the opening of the vessels and perfusion through the lumen. After 6 days of vasculogenesis vessel formation, we seeded a HUVEC monolayer onto the side channels and cultured them for another 3 days to verify microvessel opening next to the TPE posts. For perfusion experiments, we suspended 10 μm PS microparticles (Phosphorex, Hopkinton, MA) in the culture media at a concentration of approximately 3 × 105 microparticles per mL and introduced them into the side channel of the device, allowing gravity-driven flow in the vessel networks. Additionally, we tested microvessel network perfusion experiments with 0.62 μm blue, fluorescent microbeads (Fluoro-Max, ThermoFisher) to confirm that there was no perfusion flow outside of the luminal flow.Immunostaining
For monolayer immunostaining experiments, the TPE slab was first removed from the PS substrate. All staining reagents were then directly deposited on top of the FBs. The cells were fixed with 4% formaldehyde for 20 min and washed with PBS. Then, the cells were permeabilized with 0.2% Triton X-100 treatment. After washing with PBS, the cells were exposed to FITC-conjugated phalloidin (Sigma-Aldrich) and Hoechst 33342 (ThermoFisher) for 1 h. Phalloidin and Hoechst were diluted in a PBS blocking buffer (ThermoFisher) at dilution ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 1
100 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, respectively. The stained cells were examined and imaged under the inverted EVOS FL Auto Cell Imaging System.
500, respectively. The stained cells were examined and imaged under the inverted EVOS FL Auto Cell Imaging System.
To examine the 3D cross-sectional view of the microvessel networks, the microvessels were stained with CD31 (mouse anti-human CD-31, Cedarlane, Burlington, Canada) by introducing the reagents into the side channels of the microfluidic device. Briefly, the formed tissue was fixed with 4% formaldehyde and treated with 0.2% Triton X-100, as mentioned above. After fixation and Triton X-100 treatment, the tissue was washed 3 times. CD-31 primary antibody solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution with PBS blocking buffer) was then loaded into the channels and left overnight in a 4 °C refrigerator. After washing with PBS, goat anti-mouse secondary antibody (Alexa Fluor™ 488, ThermoFisher) diluted at a ratio of 1
50 dilution with PBS blocking buffer) was then loaded into the channels and left overnight in a 4 °C refrigerator. After washing with PBS, goat anti-mouse secondary antibody (Alexa Fluor™ 488, ThermoFisher) diluted at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 with the PBS blocking buffer was introduced into the channels. The sample was shielded from light and the incubation was performed for 1 h at room temperature, the stained tissue was examined using a Ti Eclipse inverted confocal microscope (Nikon, Melville, NY).
100 with the PBS blocking buffer was introduced into the channels. The sample was shielded from light and the incubation was performed for 1 h at room temperature, the stained tissue was examined using a Ti Eclipse inverted confocal microscope (Nikon, Melville, NY).
Statistical analysis
The analyzed data are presented as mean ± standard deviation (SD) unless otherwise stated. For statistical analysis characterizing microvessel networks (Fig. 4), we performed a one-way ANOVA test followed by post hoc Bonferroni correction using Microsoft Excel. Statistical significance was set at p < 0.05 for comparisons among three groups. The unpaired two-tailed t-test was used to assess the statistical significance between two independent groups.Data availability
The authors confirm that the data supporting the findings of this work are available within the article and its ESI† materials. Raw data and additional data that support the findings of this study are available from the corresponding author (B.-U. Moon), upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support from the Centre for Research and Applications in Fluidic Technologies (CRAFT) Project Award. Some figures were created with https://www.biorender.com.References
- E. W. K. Young and D. J. Beebe, Chem. Soc. Rev., 2010, 39, 1036–1048 RSC.
- X. Li, A. V. Valadez, P. Zuo and Z. Nie, Bioanalysis, 2012, 4, 1509–1525 CrossRef CAS PubMed.
- B. D. Cardoso, E. M. S. Castanheira, S. Lanceros-Méndez and V. F. Cardoso, Adv. Healthcare Mater., 2023, e2202936 CrossRef PubMed.
- Q. Smith and S. Gerecht, Curr. Opin. Chem. Eng., 2014, 3, 42–50 CrossRef PubMed.
- C. O'Connor, E. Brady, Y. Zheng, E. Moore and K. R. Stevens, Nat. Rev. Mater., 2022, 7, 702–716 CrossRef PubMed.
- G. Vunjak-Novakovic, K. Ronaldson-Bouchard and M. Radisic, Cell, 2021, 184, 4597–4611 CrossRef CAS PubMed.
- D. Huh, Ann. Am. Thorac. Soc., 2015, 12, S42–S44 CrossRef PubMed.
- S. Park and E. W. K. Young, Adv. Mater. Technol., 2021, 6, 2100828 CrossRef CAS.
- S. Park, J. Newton, T. Hidjir and E. W. K. Young, Lab Chip, 2023, 23, 3671–3682 RSC.
- A. Marsano, C. Conficconi, M. Lemme, P. Occhetta, E. Gaudiello, E. Votta, G. Cerino, A. Redaelli and M. Rasponi, Lab Chip, 2016, 16, 599–610 RSC.
- O. Mastikhina, B.-U. Moon, K. Williams, R. Hatkar, D. Gustafson, O. Mourad, X. Sun, M. Koo, A. Y. L. Lam, Y. Sun, J. E. Fish, E. W. K. Young and S. S. Nunes, Biomaterials, 2020, 233, 119741 CrossRef CAS PubMed.
- Q. Wu, P. Zhang, G. O'Leary, Y. Zhao, Y. Xu, N. Rafatian, S. Okhovatian, S. Landau, T. A. Valiante, J. Travas-Sejdic and M. Radisic, Biofabrication, 2023, 15, 035023 CrossRef PubMed.
- H. J. Kim, D. Huh, G. Hamilton and D. E. Ingber, Lab Chip, 2012, 12, 2165–2174 RSC.
- K. K. Lee, H. A. McCauley, T. R. Broda, M. J. Kofron, J. M. Wells and C. I. Hong, Lab Chip, 2018, 18, 3079–3085 RSC.
- N. Walji, S. Kheiri and E. W. K. Young, Adv. Biol., 2021, 5, 2101080 CrossRef CAS PubMed.
- N. Hakimi, R. Cheng, L. Leng, M. Sotoudehfar, P. Q. Ba, N. Bakhtyar, S. Amini-Nik, M. G. Jeschke and A. Günther, Lab Chip, 2018, 18, 1440–1451 RSC.
- H. Sano, M. Watanabe, T. Yamashita, K. Tanishita and R. Sudo, Biofabrication, 2020, 12, 045008 CrossRef CAS PubMed.
- K. Haase, F. Piatti, M. Marcano, Y. Shin, R. Visone, A. Redaelli, M. Rasponi and R. D. Kamm, Biomaterials, 2022, 280, 121248 CrossRef CAS PubMed.
- J. Lim, S. Rhee, H. Choi, J. Lee, S. Kuttappan, T. T. Yves Nguyen, S. Choi, Y. T. Kim and N. L. Jeon, Mater. Today Bio, 2023, 22, 100773 CrossRef CAS PubMed.
- T. Russell, Q. Dirar, Y. Li, C. Chiang, D. T. Laskowitz and Y. Yun, PLoS One, 2023, 18, 0288025 Search PubMed.
- U. Arslan, M. Brescia, V. Meraviglia, D. M. Nahon, R. W. J. van Helden, J. M. Stein, F. E. van den Hil, B. J. van Meer, M. Vila Cuenca, C. L. Mummery and V. V. Orlova, Stem Cell Rep., 2023, 18, 1394–1404 CrossRef CAS PubMed.
- O. Jung, Y. T. Tung, E. Sim, Y. C. Chen, E. Lee, M. Ferrer and M. J. Song, Biofabrication, 2022, 14, 025012 CrossRef PubMed.
- S. Ya, W. Ding, S. Li, K. Du, Y. Zhang, C. Li, J. Liu, F. Li, P. Li, T. Luo, L. He, A. Xu, D. Gao and B. Qiu, ACS Appl. Mater. Interfaces, 2021, 13, 32640–32652 CrossRef CAS PubMed.
- N. Xu, H. Lin, J. M. Lin, J. Cheng, P. Wang and L. Lin, Anal. Chem., 2023, 95, 17064–17072 CrossRef CAS PubMed.
- D. T. T. Phan, X. Wang, B. M. Craver, A. Sobrino, D. Zhao, J. C. Chen, L. Y. N. Lee, S. C. George, A. P. Lee and C. C. W. Hughes, Lab Chip, 2017, 17, 511–520 RSC.
- S. R. Lee, Y. Kim, S. Kim, J. Kim, S. Park, S. Rhee, D. Park, B. Lee, K. Baek, H. Y. Kim and N. L. Jeon, Microsyst. Nanoeng., 2022, 8, 126 CrossRef PubMed.
- X. Sun, J. Wu, B. Qiang, R. Romagnuolo, M. Gagliardi, G. Keller, M. A. Laflamme, R.-K. Li and S. S. Nunes, Sci. Transl. Med., 2020, 12, eaax2992 CrossRef CAS PubMed.
- X. Sun, Y. Aghazadeh and S. S. Nunes, Nat. Protoc., 2022, 17, 2721–2738 CrossRef CAS PubMed.
- H. H. G. Song, R. T. Rumma, C. K. Ozaki, E. R. Edelman and C. S. Chen, Cell Stem Cell, 2018, 22, 340–354 CrossRef CAS PubMed.
- C. Li, X. Han, Z. Ma, T. Jie, J. Wang, L. Deng and W. Cui, Adv. Healthcare Mater., 2022, 11, 2101836 CrossRef CAS PubMed.
- S. Chen, B. Han, Y. Zhao, Y. Ren, S. Ai, M. Jin, Y. Song, X. Qu and X. Wang, Polym. Test., 2023, 125, 108109 CrossRef CAS.
- K. J. Regehr, M. Domenech, J. T. Koepsel, K. C. Carver, S. J. Ellison-Zelski, W. L. Murphy, L. A. Schuler, E. T. Alarid and D. J. Beebe, Lab Chip, 2009, 9, 2132–2139 RSC.
- B. J. van Meer, H. de Vries, K. S. A. Firth, J. van Weerd, L. G. J. Tertoolen, H. B. J. Karperien, P. Jonkheijm, C. Denning, A. P. IJzerman and C. L. Mummery, Biochem. Biophys. Res. Commun., 2017, 482, 323–328 CrossRef CAS PubMed.
- D. E. Ingber, Nat. Rev. Genet., 2022, 23, 467–491 CrossRef CAS PubMed.
- M. D. Borysiak, K. S. Bielawski, N. J. Sniadecki, C. F. Jenkel, B. D. Vogt and J. D. Posner, Lab Chip, 2013, 13, 2773–2784 RSC.
- J. Lachaux, C. Alcaine, B. Gómez-Escoda, C. M. Perrault, D. O. Duplan, P. Y. J. Wu, I. Ochoa, L. Fernandez, O. Mercier, D. Coudreuse and E. Roy, Lab Chip, 2017, 17, 2581–2594 RSC.
- J. Lachaux, H. Salmon, F. Loisel, N. Arouche, I. Ochoa, L. L. Fernandez, G. Uzan, O. Mercier, T. Veres and E. Roy, Adv. Mater. Technol., 2019, 4, 1800308 CrossRef.
- D. Brassard, L. Clime, K. Li, M. Geissler, C. Miville-Godin, E. Roy and T. Veres, Lab Chip, 2011, 11, 4099–4107 RSC.
- A. H. McMillan, J. Mora-Macías, J. Teyssandier, R. Thür, E. Roy, I. Ochoa, S. De Feyter, I. F. J. Vankelecom, M. B. J. Roeffaers and S. C. Lesher-Pérez, Nano Sel., 2021, 2, 1385–1402 CrossRef CAS.
- B.-U. Moon, K. Morton, K. Li, C. Miville-Godin and T. Veres, Processes, 2021, 9, 1–11 CrossRef.
- M. Chu, T. T. Nguyen, E. K. Lee, J. L. Morival and M. Khine, Lab Chip, 2017, 17, 267–273 RSC.
- G. Pitingolo, P. Nizard, A. Riaud and V. Taly, Sens. Actuators, B, 2018, 274, 393–401 CrossRef CAS.
- S. I. Funano, N. Ota and Y. Tanaka, Lab Chip, 2021, 21, 2244–2254 RSC.
- Y. Zhang, K. Sun, Y. Xie, K. Liang, J. Zhang and Y. Fan, Rev. Sci. Instrum., 2023, 94, 061501 CrossRef CAS PubMed.
- M. D. Borysiak, K. S. Bielawski, N. J. Sniadecki, C. F. Jenkel, B. D. Vogt and J. D. Posner, Lab Chip, 2013, 13, 2773–2784 RSC.
- J. Lachaux, C. Alcaine, B. Gómez-Escoda, C. M. Perrault, D. O. Duplan, P. Y. J. Wu, I. Ochoa, L. Fernandez, O. Mercier, D. Coudreuse and E. Roy, Lab Chip, 2017, 17, 2581–2594 RSC.
- K. Domansky, J. D. Sliz, N. Wen, C. Hinojosa, G. Thompson, J. P. Fraser, T. Hamkins-Indik, G. A. Hamilton, D. Levner and D. E. Ingber, Microfluid. Nanofluid., 2017, 21, 107 CrossRef.
- Y. Wang, Y. Gao, Y. Pan, D. Zhou, Y. Liu, Y. Yin, J. Yang, Y. Wang and Y. Song, Acta Pharm. Sin. B, 2023, 13, 2483–2509 CrossRef PubMed.
- A. H. McMillan, E. K. Thomée, A. Dellaquila, H. Nassman, T. Segura and S. C. Lesher-Pérez, Micromachines, 2020, 11, 1–19 CrossRef PubMed.
- S. Schneider, E. J. S. Brás, O. Schneider, K. Schlünder and P. Loskill, Micromachines, 2021, 12, 575 CrossRef.
- M. Zborowski and J. J. Chalmers, Anal. Chem., 2011, 83, 8050–8056 CrossRef CAS PubMed.
- Q. Chen, G. Li, Y. Nie, S. Yao and J. Zhao, Microfluid. Nanofluid., 2014, 16, 83–90 CrossRef CAS.
- S. Sonney, N. Shek and J. M. Moran-Mirabal, Biomicrofluidics, 2015, 9, 1–11 CrossRef PubMed.
- K. Y. Song, H. Zhang, W. J. Zhang and A. Teixeira, Microfluid. Nanofluid., 2018, 22, 135 CrossRef.
- H. Zhong, L. Xuan, D. Wang, J. Zhou, Y. Li and Q. Jiang, RSC Adv., 2017, 7, 21837–21847 RSC.
- L. Zhao, T. Guo, L. Wang, Y. Liu, G. Chen, H. Zhou and M. Zhang, Anal. Chem., 2018, 90, 777–784 CrossRef CAS PubMed.
- K. O. Rojek, M. Ćwiklińska, J. Kuczak and J. Guzowski, Chem. Rev., 2022, 122, 16839–16909 CrossRef CAS PubMed.
- S. Kim, H. Lee, M. Chung and N. L. Jeon, Lab Chip, 2013, 13, 1489–1500 RSC.
- J. S. Jeon, S. Bersini, M. Gilardi, G. Dubini, J. L. Charest, M. Moretti and R. D. Kamm, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 214–219 CrossRef CAS PubMed.
- D. T. T. Phan, X. Wang, B. M. Craver, A. Sobrino, D. Zhao, J. C. Chen, L. Y. N. Lee, S. C. George, A. P. Lee and C. C. W. Hughes, Lab Chip, 2017, 17, 511–520 RSC.
- D. Brassard, L. Clime, K. Li, M. Geissler, C. Miville-Godin, E. Roy and T. Veres, Lab Chip, 2011, 11, 4099–4107 RSC.
- Q. Li, K. Niu, D. Wang, L. Xuan and X. Wang, Lab Chip, 2021, 22, 2682–2694 RSC.
- K. Soon, M. Li, R. Wu, A. Zhou, N. Khosraviani, W. D. Turner, J. D. Wythe, J. E. Fish and S. S. Nunes, Biomaterials, 2022, 288, 121729 CrossRef CAS PubMed.
- V. van Duinen, S. J. Trietsch, J. Joore, P. Vulto and T. Hankemeier, Curr. Opin. Biotechnol., 2015, 35, 118–126 CrossRef CAS PubMed.
- C. F. E. Jones, S. Di Cio, J. T. Connelly and J. E. Gautrot, Front. Bioeng. Biotechnol., 2022, 10, 915702 CrossRef PubMed.
- S. Yasotharan, S. Pinto, J. G. Sled, S. S. Bolz and A. Günther, Lab Chip, 2015, 15, 2660–2669 RSC.
- Y. Aghazadeh, F. Poon, F. Sarangi, F. T. M. Wong, S. T. Khan, X. Sun, R. Hatkar, B. J. Cox, S. S. Nunes and M. C. Nostro, Cell Stem Cell, 2021, 28, 1936–1949 CrossRef CAS PubMed.
- J. A. Whisler, M. B. Chen and R. D. Kamm, Tissue Eng., Part C, 2014, 20, 543–552 CrossRef CAS PubMed.
- J. B. Tefft, C. S. Chen and J. Eyckmans, APL Bioeng., 2021, 5, 016102 CrossRef CAS PubMed.
- S. Kim, M. Chung, J. Ahn, S. Lee and N. L. Jeon, Lab Chip, 2016, 16, 4189–4199 RSC.
- V. Vickerman and R. D. Kamm, Integr. Biol., 2012, 4, 863–874 CrossRef CAS.
- S. Oh, H. Ryu, D. Tahk, J. Ko, Y. Chung, H. K. Lee, T. R. Lee and N. L. Jeon, Lab Chip, 2017, 17, 3405–3414 RSC.
- Q. Zhang, S. Feng, L. Lin, S. Mao and J. M. Lin, Chem. Soc. Rev., 2021, 50, 5333–5348 RSC.
- D. S. Y. Lin, S. Rajasekar, M. K. Marway and B. Zhang, ACS Biomater. Sci. Eng., 2021, 7, 2964–2972 CrossRef CAS PubMed.
- A. S. Mao and D. J. Mooney, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 14452–14459 CrossRef CAS PubMed.
- G. Simitian, M. Virumbrales-Muñoz, C. Sánchez-De-Diego, D. J. Beebe and D. Kosoff, Lab Chip, 2022, 22, 3618–3636 RSC.
- A. B. Theberge, J. Yu, E. W. K. Young, W. A. Ricke, W. Bushman and D. J. Beebe, Anal. Chem., 2015, 87, 3239–3246 CrossRef CAS PubMed.
- E. Zudaire, L. Gambardella, C. Kurcz and S. Vermeren, PLoS One, 2011, 6, 1–12 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00530a |
| This journal is © The Royal Society of Chemistry 2024 |