Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Application of molecularly imprinted polymers (MIPs) as environmental separation tools
Despina A.
Gkika
a,
Athanasia K.
Tolkou
a,
Dimitra A.
Lambropoulou
 b,
Dimitrios N.
Bikiaris
b,
Dimitrios N.
Bikiaris
 b,
Petros
Kokkinos
c,
Ioannis K.
Kalavrouziotis
c and
George Z.
Kyzas
b,
Petros
Kokkinos
c,
Ioannis K.
Kalavrouziotis
c and
George Z.
Kyzas
 *a
*a
aHephaestus Laboratory, Department of Chemistry, International Hellenic University, Kavala, Greece. E-mail: dgkika@chem.ihu.gr; tolkatha@chem.ihu.gr; kyzas@chem.ihu.gr
bDepartment of Chemistry, Aristotle University of Thessaloniki, Thessaloniki, Greece. E-mail: dlambro@chem.auth.gr; dbic@chem.auth.gr
cSchool of Science and Technology, Hellenic Open University, Patras, Greece. E-mail: pkokkin@eap.gr; ikalabro@eap.gr
First published on 21st November 2023
Abstract
Suitable sorbents are required for the effective enhancement of sample extraction. Molecular imprinted polymers (MIPs) and related techniques can be utilized to create sorbents that possess specialized binding capabilities for target analytes, exhibiting high selectivity, and other unique attributes such as thermochemical stability, reusability, and sensitivity, thus aligning with the principles of green chemistry. These attributes can be customized; hence sample preparation can be carried out using a variety of methods and can be applied to a broad spectrum of samples, including environmental, biological, and food samples. Numerous techniques have emerged for the production of MIPs, that have their individual advantages and disadvantages. This review places particular emphasis on the interactions between primary functional groups and monomers and how these functional groups impact MIP performance. Additionally, we offer insights into how functional groups can significantly enhance the imprinting effect, resulting in a markedly increased imprinting factor and specific rebinding capacity. This work initially discusses the headway made in synthesis approaches and the applications of MIPs over the past five years. Then, provide a comprehensive overview of the common challenges encountered and the environmental applications of MIPs. The significance of the availability of various polymerization mechanisms and use of diverse functional molecules and cross-linkers is emphasized.
1. Introduction
Investigating different analytes using innovative polymers has become a central focus of research endeavors.1 This direction has yielded positive outcomes in numerous areas, such as detecting environmental contaminants and enhancing the functionality of electrochemical devices.2,3 Molecular recognition is of crucial importance in biological processes and currently garners significant attention from researchers, due to its relevance in catalytic, separation and sensing applications.1 Molecular recognition requires the establishment of non-covalent interactions between antibodies and antigens (referred to as “guest” and “host” respectively). These interactions encompass various forces such as hydrogen bonds and van der Waals forces.4 This interaction holds immense significance in the biomedical and biotechnological fields.The emergence of Molecular Imprinting Technology (MIT) can be traced back to its introduction by Wulff and Sarhan in the early 1970s.5 Subsequently, a multitude of researchers have played a pivotal role in furthering its progress. MIT entails the polymerization of a functional monomer in the company of a crosslinker encircling a template molecule that necessitates imprinting. In the initial stages, a pre-complex develops between the template molecule and the functional monomer, followed by polymerization occurring around this pre-complex. This process generates three-dimensional voids, facilitating precise molecular recognition.6 This molecular imprinting technique imparts selectivity to recognition sites within synthetic polymers, using template molecules that can be atoms, ions, molecules, complexes, or micro-organisms. To enable the molecules to be recognized, the template must be removed, creating empty spaces.7
The distinguishing feature of molecular recognition lies in its ability to specifically target particular molecules.8,9 Biomedical applications heavily rely on this principle 10. However, there are certain limitations10 such as limited lifespan, instability under extreme conditions, increased costs, and challenging adaptation to a large-scale level. To address these challenges, molecular imprinting was introduced as a solution, ultimately giving rise to the emergence of MIPs.11
Biomarkers play a pivotal role in providing crucial information about various diseases, including cancer. However, detecting these biomarkers at low concentrations in complex matrices poses significant technical challenges.12 Molecularly imprinted polymer-based sensors have emerged as an attractive solution for clinical applications due to their cost-effectiveness, reusability, high stability, and resilience to physical and chemical factors. These sensors exhibit exceptional sensitivity to even minor structural changes in biomolecules 6. MIP-based biosensors offer numerous diagnostic possibilities for the detection of various cancer biomarkers, including proteins such as PSA, Myo, CA15-3, HER-2, and CA-125, as well as small molecules like 5-HIAA and neopterin. This versatility is attributed to their robustness, sensitivity, and cost-effective analysis of biomarkers.12 Additionally, Surface Plasmon Resonance (SPR) sensors, which can serve as an alternative to traditional albumin monitoring approaches, can also be adapted for real-time detection and monitoring of various other proteins.7
The research community's interest in the field of MIPs is not a recent development. K. Mosbach et al.13 were reportedly the first to mention an “imprinted polymer” in 1984. G. Wulff also utilized the term “imprinted polymer” in a paper published in 1985,14 although he had previously published works as part of a series titled “enzyme-analog built polymers” since 1973.14 Since then, MIPs have captured conceptual attention15 due to their widespread use, resulting from desirable attributes such as high physical stability, robustness, excellent reusability, and cost-effective synthesis.16 Over the past 20 years, a significant number of research works, review articles and other publications have been released on the subject,17,18 demonstrating their applicability in multiple domains, including catalysis,19 sensors,20 solid phase extraction,21 drug delivery,22 water and wastewater treatment, environmental and biomedical sensing,23 and chromatography.24 Furthermore, computational modeling studies have emerged, advancing the rational design and outcomes of MIPs.25
MIPs are synthetic polymers that are carefully constructed to possess specific pores matching the structure of the target material, displaying a remarkable selectivity toward it.26 These pores are created through polymerization using a monomer a crosslinker and initiator.27 Porogens also have a significant part in MIPs as they influence the volume of the pores and how selective the resulting material is. They serve as a stable medium for the solubility of other components and contribute to the porous structure of the polymer film. The selection of various porogens can be made according to the interactions observed between the template material and the monomer.28 Research has demonstrated that MIPs produced using imprinting technology possess the ability to readily recognize molecules, mimicking natural units such as biological receptors and antibodies.
In recent times, MIPs have emerged as valuable tools for the recovery of environmental pollutants due to their adsorption capabilities and adaptability to specific target pollutants, setting them apart from carbonaceous materials. Recently, MIPs have gained attention in the recovery of environmentally persistent pollutants (EPs). Unlike carbonaceous materials, MIPs serve as adsorbents and possess the capability to adapt to the specific pollutant of interest. This adaptability means that MIPs can be custom-designed for a particular molecule of interest. The MIP preparation process involves the mixing of monomers with the chosen pollutant during polymerization, followed by the removal of this molecule using an appropriate solvent. As a result, an imprinted polymer is created with cavities that mimic the shape and volume of the pollutant. This affinity enables the MIP to selectively attract the target molecule, making it more efficient in adsorbing EPs with enhanced selectivity, specificity, and the ability to concentrate large volumes of pollutants using a minimal amount of MIP.29 Sorbents based on MIPs provide several benefits, including the ability to be reused, cost-efficiency, and effective isolation of the desired analyte from the sample.29 Moreover, they allow for the use of natural solvents, promoting a green approach when utilizing MIPs.30,31
However, the concept of MIPs is characterized by its inherent complexity. Firstly, MIPs technology represents a multidimensional research area. As previously mentioned, it was initially utilized for the recognition of chemical and biological molecules. However, the scope of MIPs extends far beyond molecular recognition and has found applications in diverse fields such as drug delivery, purification, chemo-biosensing and catalysis.32,33 Secondly, the intricacy of MIPs is further enhanced by the involvement of a wide range of academic disciplines interested in this research domain, including polymer and material chemistry, biochemistry, economics, and other multidisciplinary approaches.34 This interdisciplinary perspective promotes varied exploration and evaluation methodologies, resulting in diverse approaches to studying and assessing MIPs technology.
The exceptional selectivity and recognition capabilities of MIPs make them highly suitable for utilization as adsorbents during sample preparation,35 due to offering relatively inexpensive and reusable solutions, facilitating the efficient removal of the target molecule. These adsorbents are particularly favored in extraction reliant methods. Moreover, MIPs can be combined with naturally sourced solvents to promote environmentally friendly practices.31,36 MIP-based sample preparation finds significant application in environmental analysis, especially in the analysis of water samples preparation methods. Numerous literature reviews have delved on the applications of MIPs in environmental water sample analysis.37,38
Considering the aforementioned factors, the complexity of MIPs and the continually increasing publication rate make it an intriguing research area. There is a need to identify the underlying sources of influence and conduct a methodological review of the attributes and advancements within this field.
Over the past four years, several notable reviews have concentrated on the synthesis and environmental uses of Molecularly Imprinted Polymers (MIPs). Kamyab et al. provided an overview of the current advancements in carbon-based MIPs for detecting hazardous pollutants with high sensitivity and selectivity in aqueous settings.39 The following year, Metwally et al. conducted a review of various MIP applications over the previous five years, focusing on the detection of various types of water and wastewater contaminants and proposing future application approaches.16 Kadhem et al. delved into the progress of imprinted molecular technologies, particularly their application in environmental and biomedical sensors.40 Han et al. delved into the fundamental principles, recent advancements, and diverse applications of MIPs in various fields, including environmental remediation, wastewater purification, food analysis, chemical and biological sensing, precise drug delivery, protein identification and purification, as well as hormone removal.41 Villarreal-Lucio et al. aimed to gather information on MIPs’ use as adsorbents for environmental pollutants and their subsequent utilization in preconcentration and pollutant analysis.42 In a recent study, Kaya et al. presented a comprehensive review of the most current applications and emerging patterns in the use of MIP-based sample preparation techniques for analyzing environmental water samples.43
To date, MIPs have found widespread use in environmental applications. However, their environmental application has typically centered on various aspects, including the synthesis of MIPs, preparation methods (such as solid-phase extraction and adsorption), analysis methods (e.g., liquid chromatography-mass spectrometry and high-performance liquid chromatography), matrices (including wastewater, river water, and lake water), and MIP components (crosslinkers, templates, solvents, and monomers). One notable challenge in this regard is optimizing the interaction between monomers and functional groups. MIPs comprise several well-known components, with the monomer's primary role being to provide functional groups that facilitate complex formation. Given the dearth of comprehensive reviews on the application of MIPs as environmental separation tools, especially regarding the critical role of functional groups, this review is structured to outline the applications of MIPs as environmental separation tools. It places particular emphasis on the interactions between primary functional groups and monomers and how these functional groups impact MIP performance. Additionally, we offer insights into how functional groups can significantly enhance the imprinting effect, resulting in a markedly increased imprinting factor and specific rebinding capacity. This review serves a dual purpose: firstly, to provide current insights into the utilization of MIPs as tools for environmental separation, and secondly, to assess the role of functional groups as a parameter influencing MIP performance.
In essence, this review encapsulates the noteworthy advancements achieved in the application of MIPs as environmental separation tools over the past five years. This review highlights its novelty that influence the molecular imprinting process, notably evaluating the parameter of functional groups and its effect on MIP performance. Additionally, we delve into the mechanisms and synthesis strategies of MIPs, presenting their applications as tools for environmental separation. Lastly, we extend our discussion to explore the future prospects of this captivating and rewarding research field.
This review takes a more contemporary approach by incorporating the latest available studies. It is crucial for future research to assess recent advancements in technology and novel approaches. As a result, this review conducts a thorough assessment of the latest environmental uses of MIPs in sample preparation methodologies, offering a comprehensive insight into both MIPs and the primarily utilized sample preparation approaches.
2. Components of MIPs
In molecular imprinting, the template molecule refers to the material that interacts with the monomer. When it is subsequently extracted from the polymer matrix, newly formed cavities that match in shape and functional groups appear. The template should to exhibit significant physicochemical interactions with the monomer. However, it should be noted that not all types of molecules can be used as templates. Molecules affected by high temperature levels or the presence of groups that impact the polymerization process may be considered unsuitable for this role.44Functional monomers significantly impact the production of all MIPs, by forming compounds with the templates. The monomers provide functional hydrogen bonds that impact the structural stability and porosity of the resulting material.45 The cross-linking degree in polymers is essential for the structural stability of the end result, as well as its morphology and the stability of the binding location. Therefore, the crosslinker is often used in abundance, as opposed to the monomer.
Initiators play an important role in MIPs, particularly in the polymerization of free radicals. They can be activated through a thermal or photochemical process, generating free radicals. It is important to consider the stability of the template molecule when selecting the appropriate initiator. If the template molecule is unstable, it would not be suitable for polymerization using thermally or photochemically activated initiators, respectively. In cases where low-temperature polymerization is necessary, photochemically active initiators are employed.44
3. Mechanism approaches and synthesis strategies of MIPs
3.1 Mechanisms
Molecular imprinting can be broadly categorised as covalent, semi- or non-covalent, based on the binding modes among the template and the monomers. During the preparation of MIPs, careful consideration must be given to the selection and proportion of templates, monomers, cross-linkers and other parameters that might affect the process.46The covalent approach to molecular imprinting was pioneered by Wulff and his colleagues.47 In this method, the template and the monomer form reversible covalent bonds, resulting in a highly stable compound. This approach ensures a uniform distribution of binding locations within the produced polymer, leading to improved sensitivity and high yields. However, the major drawback is the limited amount of template molecules capable of forming reversible covalent bonds with functional monomers, which restricts the widespread use of this method.44
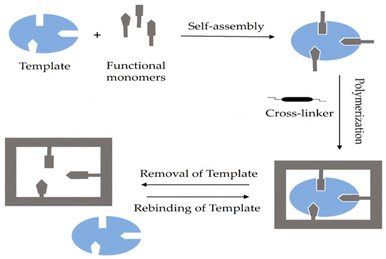
In contrast, the non-covalent imprinting approach, introduced by Mosbach et al.,48 involves the formulation of a pre-polymerization compound of the template and the functional group through weak electrostatic interactions and hydrogen bonds. This approach closely resembles the biorecognition process.44Fig. 1 illustrates the schematic diagram of the non-covalent imprinting mechanism.49
 | ||
| Fig. 1 Diagram illustrating the non-covalent method. Reproduced from ref. 49 with permission from MDPI, copyright 2023. | ||
In the non-covalent imprinting strategy, the creation of a complex between the template and functional monomer depends on weak non-covalent forces, such as van der Waals interactions, hydrogen bonding, π–π stacking, dipole–dipole interactions, and ion–dipole interactions, among others. These techniques utilize readily accessible components and, therefore, demand minimal synthetic complexity.50
The principal methodologies employed for the development of Molecularly Imprinted Polymers (MIPs) can be classified into two main categories, depending on the interactions that occur between the template molecule and the monomer's functional groups during the pre-polymerization phase. Additionally, this discussion encompasses variations of these methods, such as semi-covalent imprinting, metal-mediated imprinting, and interactions based on host–guest inclusion. In aqueous environments, the recognition of template molecules in non-covalently imprinted polymers is significantly influenced by hydrophobic interactions. These hydrophobic interactions are particularly valuable for detecting nonpolar compounds in polar solvents, as demonstrated by MIPs utilizing cyclodextrins (CD-MIPs). Cyclodextrins are cyclic oligosaccharides characterized by their hydrophilic outer surface and hydrophobic central cavity, which consists of α-1,4-linked α-D-glucopyranose units. They readily form non-covalent complexes with a wide variety of host–guest molecules. The interaction between cyclodextrins and the target guest is easily reversible and functions effectively in polar solvents like water. This approach has yielded successful results in imprinting various compounds, including oligopeptides, amino acids, peptides, steroids, and more.50
Takeuchi and colleagues engineered a photosensitive functional monomer, comprising diaminopyridine and azobenzene components. A custom-designed monomer was created to selectively recognize porphyrin derivatives containing carboxylic acids as their template molecules. The scientists noted that several hydrogen bonds could form between the template and the functional monomer, facilitating the exact arrangement of the functional monomer next to the template during the polymerization procedure. This precise alignment led to the creation of specialized imprinted pockets that perfectly matched the target molecule's shape.51
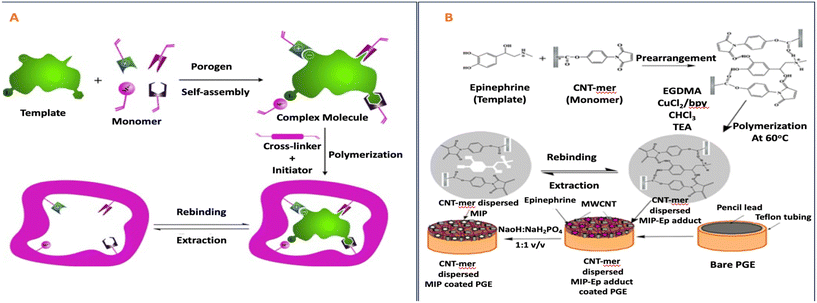
The research team led by Varenne undertook a study involving the design and characterization of molecularly imprinted polymers. These polymers were developed for the purpose of selectively extracting oxazepam at low concentrations from environmental water and human urine samples using solid-phase extraction techniques. In this study, methacrylic acid (MAA) was employed as the monomer due to its capacity to engage in polar interactions with NOR and OXA. Additionally, the research explored the influence of the crosslinker's composition by synthesizing MIPs with two variants: one using ethylene glycol dimethacrylate (EGDMA), a commonly utilized crosslinker with MAA for MIP synthesis (referred to as MIP/NIP 1), and the other employing divinylbenzene (DVB), selected for its ability to establish π–π interactions with compounds possessing aromatic groups (referred to as MIP/NIP 2).52
The term “self-assembly” was introduced based on the observed adaptability in the template-monomer interaction. This inherent flexibility, combined with the ease of removing the template, has generated significant interest among scientists, making the non-covalent option the predominant choice in molecular imprinting.
Scientists have acknowledged the advantages of both methods and have devised a hybrid approach for molecular imprinting. In this hybrid protocol, the rebinding process primarily relies on the non-covalent interaction, while the covalent method is employed during the initial imprinting step.44
3.2 Synthesis strategies
To generate molecularly imprinted polymers (MIPs), a process is undertaken to create a polymer matrix with embedded cavities that possess functional sites. This is accomplished by combining the template with the monomer while a porogenic solvent that does not participate in the chemical reactions is present (Fig. 2).53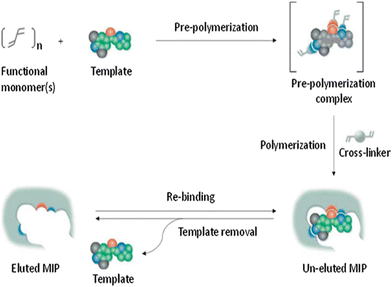 | ||
| Fig. 2 Diagram illustrating the creation of MIPs. Reproduced from ref. 53 with permission from Elsevier, copyright 2023. | ||
In molecular imprinting, the process of polymerization can be categorized depending on the mechanisms utilized for polymerizing the template-monomer compound and cross-linked monomers (Table 1). Example categories are bulk, precipitation, emulsion and suspension polymerization, among various other methods. Among these methods, bulk (traditional) polymerization,54 is the most commonly employed due to being simple.
| Preparation methods | Polymerization method | Process | Pros | Cons |
|---|---|---|---|---|
| Free radical polymerization | Bulk polymerization | Polymerization is conducted in an organic solvent, followed by subsequent sieving and grinding.72 | The process is rather simple.73 | Sieving and grinding are labor-heavy and have high duration times.74Mechanical manipulation often associated with limited selectivity and reproducibility.74 |
| Precipitation polymerization | Polymerization takes place in a solution; precipitation occurs once the polymer forms, rendering it insoluble in the solution.72 | Production of MIPs at a scale of 50–100 nm in a large scale.75 | Several factors influence the particle dimensions, such as the speed of stirring, and the temperature levels of the process.76 | |
| Suspension polymerization | Polymerization is carried out in an aqueous medium.72 | Production of monodispersed MIPs in a large scale.77 | The presence of surfactants can potentially lead to toxicity concerns.79 | |
| Single-step polymerization process, formation of spherical particles.78 | Large particle size (hundreds of micrometers), diminished recognition capability.78 | |||
| Emulsion polymerization | Polymerization necessitates surfactants to emulsify both the organic and aqueous phases.55 | Production of MIPs in a large scale80. | Large size particles in the micro range80 | |
| High yield, water-soluble.78 | Residual surfactants, limited imprinting efficiency.78 | |||
| Multistep swelling | The initiator's oil-in-water emulsion is utilized to generate spherical particles, which subsequently undergo swelling.72 | Produces spherical, uniform MIP.71 | Long process. Requires extensive optimization of each step.71 | |
| Electropolymerization | MIPs are deposited onto the surface of electrode materials in the presence of a template.81 | A simple, speedy, and cost-effective method for creating MIPs (ref. 50). | This polymerization technique is characterized by high costs. Achieving optimization in the MIP coating process is intricate and demands a substantial amount of time.81 | |
| Sol gel polymerization | Sol gel polymerization | This process involves dissolving a metal oxide precursor in a low molecular weight solvent medium, catalyzed by a hydrolysis (water) and polycondensation step.82 | Easy to operate at ambient temperature levels.83 | The absence of a suitable polymerization method and functional monomer selection.50 |
| Surface imprinting technique | Surface imprinting technique | Thin layers of molecularly imprinted polymers are grafted onto surfaces during polymerization.72 | Targeted molecule can more easily access the binding sites.84 | Time-consuming process.85 |
| Controlled living polymerization | Atom transfer radical polymerization | A modified iteration of Kharasch addition, enabling the creation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 adducts between alkyl halides and alkenes.86 1 adducts between alkyl halides and alkenes.86 |
Broad spectrum of initiators.78 | Toxicity of the halide initiator.78 |
| Adaptability to diverse monomers.78 | Vulnerability of the reduced metal catalyst to oxygen.78 | |||
| Mild reaction conditions.78 | ||||
| Reversible addition–fragmentation chain-transfer | A specialized transfer agent is used, enabling the swift degenerative transfer of propagating radicals.86 | Can be applied to various categories of free radical polymerization. | Low stability of the chain transfer agent compound in the reaction medium. When reactions are performed in aqueous media, the reactants easily suffer transformations by hydrolysis or by nucleophilic attacks, even when macro-CTAs are formed.87 | |
| Its efficacy is not reliant on a specific catalyst or environmental settings.78 |
In this method, the template, the monomer, and other components are combined and sealed together. To prevent the loss of imprinting complex, external inert gases like nitrogen are used to remove oxygen. The resulting polymers are in block form and are subsequently crushed and sieved. However, this process has drawbacks such as non-regular shape and dimensions, potential damage to the imprinted cavities during crushing and grinding, and a large input requirement. These factors contribute to low production of MIPs and potential template leakage.55
In precipitation polymerization, the compounds are formulated in a significant quantity of solvent, and then the end result is transported to a precipitation medium where they are not soluble and precipitate at the bottom. This approach allows control over the particle size but still results in irregular shapes. It requires a larger amount of solvents compared to bulk polymerization.56 MIPs created using the bulk approach have a monolithic structure. If these monoliths are not ready to use, they must undergo a tedious and time-consuming process of being crushed, grinded, and sieved to procure microparticles for the sorption process.57 In contrast, precipitation polymerization yields MIPs that manifest as spherical beads with uniform sizes ranging from nano to sub-micrometers, demonstrating excellent suitability for solid-phase extraction (SPE) in conventional applications. When polymerization occurs, the template becomes enveloped by the interconnected chains of the polymer network. By subsequently extracting the template molecule using a solvent, well-defined cavities with precise dimensions, shapes, and active surface sites are created.57
Suspension polymerization, pioneered by Mayes and Mosbach,58 entails the dissolution of all necessary polymerization components in an appropriate organic solvent, which is subsequently blended with another solvent. Through vigorous stirring, small droplets are formed during the polymerization process.59 The resultant particle size produced through this method ranges from 10 to 100 μm.60
Emulsion polymerization occurs in an oil/water system consisting of two phases.56 In this method, the monomer, crosslinking agent and template are initially emulsified in an aqueous solution, followed by the disperse phase where stabilizers are added. This approach has a high polymerization rate; however, it is challenging, costly, and often results in low polymer purity. The use of water can also negatively impact its effectiveness.55
Cui et al. produced hydrophilic MIPs by cross-linking copper cations and styrene-methacrylic acid copolymer. Hydrogen bonding and metal chelation enabled the formation of a cross-linked network where tetracycline served as the template molecule61 (Fig. 3).
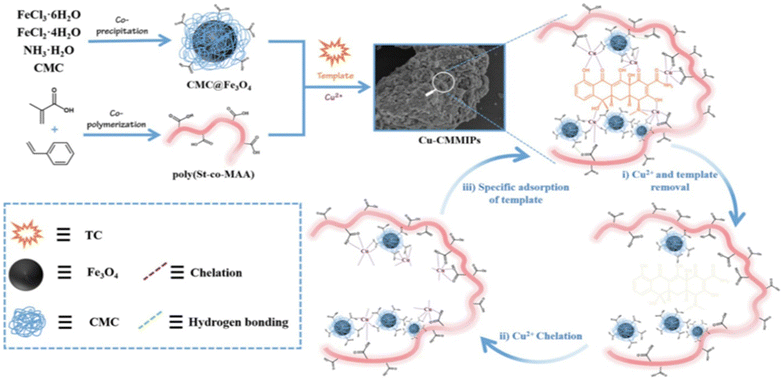 | ||
| Fig. 3 Illustration demonstrating the fabrication of Cu-MMIPs. Reproduced from ref. 61 with permission from Elsevier copyright 2023. | ||
The surface imprinting technique involves the application of imprinted layers onto porous silica or a solid medium using various mechanisms. This method offers simplicity, versatility, a large imprinted surface area, controlled morphology and dimensions, and a high number of binding sites.62 It surpasses traditional polymerization methods in terms of enhanced reproducibility, selectivity, and sensitivity. However, large-scale production of surface-imprinted polymers still faces limitations.33Compared to bulk imprinting, it offers better access to the binding locations, achieving fast recognition and minimizing the diffusion distance between the target molecule and the binding site, while steric hindrance is limited. This provides notable benefits when imprinting bio-macromolecules.63 Surface-imprinted nanomaterials facilitate a quick template transfer, thereby enhancing the binding capacity and kinetics. Additionally, they can integrate other functions, providing additional features.64 Stimuli-responsive MIPs (SR-MIPs) are intelligent compounds that can respond to a particular stimulus such as pH, temperature, magnetic fields by undergoing significant changes in their morphology or other attributes, resembling natural receptors. Two main methods are employed for synthesizing SR-MIPs: grafting/incorporating responsive materials into MIPs and including responsive substances in the MIP framework. The former is primarily used for magnetic SR-MIPs.32 Photo-irradiation is a frequently utilized stimulus due to its convenience and controllability. Photo-responsive materials have gained significant attention for their potential applications in areas such as optics and memory storage and processing.50Fig. 4 depicts the various new imprinting technologies and strategies for the preparation of TCM-related MIPs.65
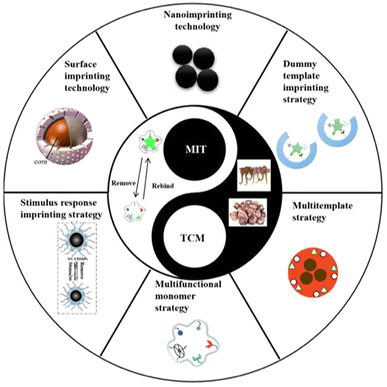 | ||
| Fig. 4 Recent advancements in imprinting technologies and strategies applied in the development of MIPs specifically designed for traditional Chinese medicine. Reproduced from ref. 65 with permission from MDPI, copyright 2023. | ||
Nanoimprinting technology involves the preparation of MIPs with nano-sized dimensions.66 These nano-MIPs eliminate the need for further processing and exhibit high mechanical strength, ensuring the durability of their recognition locations. The nanoscale dimensions of MIPs result in a large specific surface area, allowing for almost full extraction of the template and maximizing the available binding locations, that can be found typically on or close to the surface, providing notable binding abilities. Additionally, the accessibility of template molecules to the molecularly imprinted sites is enhanced, leading to fast binding dynamics. Therefore, nanoimprinting technology holds great potential for a variety of applications and can contribute to the stability of molecularly imprinted sensors by synthesizing more robust nanomaterials.67
To address the issue of template molecule leakage, the dummy template imprinting strategy is commonly employed. This strategy involves using a structurally similar molecule to the target molecule as the template for imprinting. By doing so, specific recognition by MIPs is maintained while avoiding interference caused by template molecule leakage.68 This approach is particularly useful when working with expensive, chemically unstable, or potentially hazardous template molecules. The successful utilization of dummy template MIPS has been demonstrated for the extraction of pesticides in cinchona.69
The objective of the multi-functional monomer imprinting approach is to enhance the bonding between the target and the monomer, by employing multiple monomers that have interactions with the template.70 This method is an effective means to enhance the selectivity and adsorption ability of MIPs. It offers a valuable approach to imprinting various analytes and enhancing selectivity. However, continuous exploration is needed to determine the appropriate selection and combination of existing functional monomers, as well as the design and synthesis of new monomers, in order to fully take advantage of their synergistic nature.67
Multi-step swelling polymerization is a meticulous and laborious approach employed to produce spherical, uniform MIPs. The polymerization proceeds gradually in sequential steps, starting with small polymeric seeds like polystyrene. With each step, particle swelling increases. In spite of the long duration of the process and the requirement for extensive optimization of each polymerization step, multi-step polymerization demonstrates versatility with various templates.71
Electro-polymerization facilitates the direct creation of thin MIP films with the desirable thickness on the electrode by regulating the amount of electro-polymerization cycles applied. This approach presents a straightforward, rapid, and cost-efficient technique for preparing MIPs.83
Controlled or living polymerization denotes a polymerization procedure that advances in a regulated fashion. In this approach, adverse reactions occur at remarkably low frequencies, and the degree of polymerization of the resulting polymers increases proportionally with monomer conversion. The concept of ‘living’ polymerization made its debut in 1956, thanks to the groundbreaking research by M. Szwarc,88 who introduced the idea of living anionic polymerization to the scientific community. This pivotal discovery laid the foundation for modern polymer science, enabling the elongation of polymer chains with consecutive blocks and resulting in polymers with exceptionally low polydispersities. For a significant period, living anionic polymerization remained the primary method for producing block copolymers. It was only many years later that this ‘living’ approach was extended to include cationic (ring-opening) polymerization of heterocyclic compounds and the polymerization of vinyl monomers.86
However, it wasn't until the emergence of contemporary controlled radical polymerization (CRP) techniques that these ionic processes were surpassed in providing precise control over molecular weight and molecular weight distribution in synthetic polymers. Nevertheless, they faced limitations due to their susceptibility to impurities and moisture, as well as their restricted applicability to specific monomers (i.e., aprotic and apolar), which limited their utility.86
Otsu in 198289,90 coined the term “Controlled Radical Polymerization” when discussing the use of dithiocarbamates as initiators. These initiators undergo reversible photo-dissociation, resulting in the formation of a propagating alkyl radical and a dormant dithiocarbamyl species. The introduction of substances capable of deactivating propagating radicals in a reversible and selective manner, rather than initiating new polymer chains, marked a significant milestone in achieving control over radical polymerization. Otsu coined the term “iniferters” to describe these compounds, combining “initiator”, “transfer agent”, and “terminator” to emphasize their sequential roles in the polymerization process.86
As an evolving polymerization technique, controlled/living radical precipitation polymerization (CRPP) incorporates the CRP mechanism into established precipitation polymerization systems. This fusion combines the benefits of CRP with the framework of conventional precipitation polymerization. We will explore various CRPP approaches that have been devised.91Both atom transfer radical polymerization (ATRP), and reversible addition–fragmentation chain transfer (RAFT) are well-suited techniques for achieving the goal of creating uniform polymeric materials.92
In recent times, there has been substantial interest in CRPs owing to their capacity to precisely synthesize well-defined polymers with predetermined molecular weights and low molar mass dispersions. CRPs combine the advantages of traditional free radical polymerization with the characteristics of living polymerization, making them a highly explored subject in polymer science. While CRPs provide control over the synthesis process akin to living anionic polymerization, they do so with a slightly reduced level of precision. Nevertheless, CRPs hold a distinctive advantage in operating under gentler and less restrictive reaction conditions, making them applicable to a broader spectrum of monomers. The attainment of control in CRPs is contingent upon establishing a dynamic equilibrium between active and dormant species, resulting in exceedingly low radical concentrations within the polymerization systems and consequently minimal radical termination, ultimately leading to regulated polymerizations.93
The core principle underlying controlled radical polymerization (CRP) revolves around maintaining an equilibrium between actively progressing free radicals or active species and dormant ones. This balance serves to minimize chain-breaking reactions and the sudden initiation of multiple polymer chains. Among the various CRP techniques, the most effective ones include ATRP, stable free radical polymerization (SFRP), nitroxide-mediated polymerization (NMP), and RAFT. The presence of CRP “living” groups on the polymer's surface is of paramount importance because these “living” groups facilitate advanced surface modifications, such as the incorporation of hydrophilic comonomers.91
ATRP stemmed from atom transfer radical addition (ATRA), a modified iteration of Kharasch addition, enabling the creation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 adducts between alkyl halides and alkenes.86
1 adducts between alkyl halides and alkenes.86
ATRP has undergone thorough examination and offers distinct benefits, including a broad spectrum of initiators, adaptability to diverse monomers, and the use of mild reaction conditions. Nevertheless, ATRP faces obstacles in terms of industrial implementation, chiefly stemming from the toxicity of the halide initiator and the vulnerability of the reduced metal catalyst to oxygen. Consequently, its large-scale application necessitates careful consideration of environmental contamination risks and the potential expense associated with equipment needed to maintain controlled oxygen levels within the polymerization vessel.78 It functions by establishing an equilibrium between addition and fragmentation through the use of thiocarbonylthio compounds to control the progression of radical polymerization.78
RAFT has evolved into a remarkably successful CRP technique recognized for its broad utility and straightforward implementation. In RAFT polymerization, a specialized transfer agent is used, enabling the swift degenerative transfer of propagating radicals. This pivotal chain-transfer function competes effectively with propagation, resulting in a notable reduction in termination occurrences. Within RAFT polymerization, propagating radicals swiftly engage with the RAFT agent, rendering it more reactive than typical vinyl monomers. This proficient degenerative transfer process becomes the predominant mechanism, leading to a decreased frequency of propagation events and a decelerated polymerization rate.86
Within RAFT polymerization, propagating radicals swiftly engage with the RAFT agent, rendering it more reactive than typical vinyl monomers. This proficient degenerative transfer process becomes the predominant mechanism, leading to a decreased frequency of propagation events and a decelerated polymerization rate.86 A significant advantage for industrial applications is its flexibility, which can be applied to various categories of free radical polymerization. Its efficacy is not reliant on a specific catalyst or environmental settings.78
3.3. Factors that impact the molecular imprinting process
There are various factors affecting the molecular imprinting process, influencing the properties and performance of MIPs. Careful selection and optimization of each factor is essential to achieve the highest efficiency and favorable characteristics in MIPs. Parameters such as functional groups, the type and quantity of templates, monomers, cross-linkers, and porogens are thoroughly examined in this section.The preference for a functional monomer over a conventional one stems from its incorporation of Y functional groups, enabling interactions with the template molecule through H-bonds, dipole–dipole interactions, and ionic bonding, ultimately culminating in the formation of a template-monomer complex.94 The selection of the appropriate functional monomer is crucial, and three types are commonly employed: (i) acidic compounds such as methacrylic acid, (ii) alkaline compounds like 4-vinyl pyridine, and (iii) pH-neutral compounds such as styrene.95
The choice of an appropriate functional monomer that can engage favorably with the template molecule is crucial. In cases where the template molecule is acidic, it is often advantageous to select a functional monomer with basic functional groups, and vice versa. For instance, methacrylic acid is preferred for a basic template, while vinylpyridine is suitable for an acidic template. However, this selection becomes more complex when the template molecule lacks functional groups that enable well-defined interactions with other molecules.96 The template's functional group, whether it be amino, carboxyl, hydroxyl, amide, or ester, plays a vital role in MIP performance. Through the creation of custom-designed binding sites, MIPs not only recognize the size and shape of the template but also respond to the functional groups present on the molecule.97
The molecular imprinting process encompasses various interactions, including hydrogen bonding, dipole–dipole interactions, and ionic interactions. These interactions between the template molecule and the functional groups within the polymer matrix drive molecular recognition, allowing the resulting polymer to selectively recognize and bind to the template molecules. Therefore, comprehending the chemical interactions that occur during MIP development is of paramount importance.94Fourier-transform infrared spectroscopy (FTIR) serves as a suitable technique for monitoring the functional groups within MIPs. FTIR spectra enable the assessment of non-covalent interactions like hydrogen bonding that take place within MIPs.98
Numerous examples underscore the significance of functional groups in influencing MIP performance (Table 2).
| Analytes (matrix) | Template | Functional group | Functional monomer | Crosslinker | Porogen (solvent) | Ref. |
|---|---|---|---|---|---|---|
| Key: EGDMA: ethylene glycol dimethacrylate, NSAIDs: non-steroidal anti-inflammatory drugs, MAA: methacrylic acid, ACN: acetonitrile, MNZ: miconazole, DCM: dichloromethane, MBAA: N,N-methylenebis(acrylamide), DCF: diclofenac: Benzodiazepines, E2: 17β-estradiol, NFX: norfloxacin, TC: tetracycline, CR: Congo red, 2-VP: 2-vinylpyridin. | ||||||
| Spiked blood serum and river water | 2-Phenylphenol (agricultural fungicide) | Alkane functional group | Styrene | Divinyl benzene | ACN | 99 |
| Environmental water samples | MNZ | Azole group | MAA | EGDMA | ACN | 100 |
| Environmental and biological samples | BZDs | Aromatic groups | MAA | EGDMA & Divinylbenzene | DCM/toluene | 52 |
| Different real water samples | DCF | Carboxyl group | MAA | MBAA | Water/MeCN, MeCN, chloroform, toluene/MeCN | 101 |
| Environmental water samples | E2 | Amino, hydroxyl, and phenyl groups | Dopamine | Dopamine | ACN | 102 |
| Wastewater | Fenoprofen | Pyridine group | 2-VP | EGDMA | ACN | 103 |
| Environmental water samples | TC | Carboxyl and hydroxyl groups | Styrene-methacrylic acid | Cu2+ | — | 61 |
| Wastewater | CR | Acidic functional groups | MAA | Methyl acrylic acid ethylene glycol | ACN | 104 |
| Wastewater | Diclofenac, naproxen, and ibuprofen | Carboxylic group | MAA and 2-VP | EGDMA | Toluene | 105 |
| Seawater samples | NFX | Hydrogen bonding and ionic bonding | MAA | EGDMA | ACN | 106 |
For instance, Bakhtiar et al. prepared a MIP for 2-phenylphenol using styrene as the functional monomer and divinyl benzene as the cross-linker via precipitation polymerization. FT-IR spectroscopy of the MIP particles revealed peaks corresponding to C–H stretching from alkane functional groups. Batch binding assays with template molecules and analogs possessing similar functional groups indicated that the MIP's high selectivity depended largely on the binding sites created by functional groups.99
In another study, Sun et al. developed molecularly imprinted polymers for climbazole (CBZ) using either micomazole (MNZ) or CBZ as templates. They employed methacrylic acid (MAA) as the functional monomer to establish strong hydrogen interactions with the azole group.100 Varenne's research team conducted an investigation into the development and characterization of MIPs designed for the selective solid-phase extraction of oxazepam from trace concentrations found in environmental water and human urine samples. Methacrylic acid (MAA) was selected as the monomer due to its capacity to engage in polar interactions with NOR and OXA. Furthermore, the study delved into the influence of the crosslinker's nature by producing MIPs with either ethylene glycol dimethacrylate (EGDMA), commonly used in conjunction with MAA for MIP synthesis (referred to as MIP/NIP 1), or divinylbenzene (DVB), chosen for its ability to establish π–π interactions with compounds containing aromatic groups (referred to as MIP/NIP 2).52
Amaly and co-workers designed uniformly sized molecularly imprinted polymer nanospheres through precipitation polymerization, employing methacrylic acid (MAA) as the functional monomer and N,N-methylenebis(acrylamide) (MBAA) as a crosslinker for the removal of diclofenac (DFC), serving as a model for pharmaceutical pollutants. MAA, a commonly used functional monomer in noncovalent imprinting, exhibits an excellent ability to interact with DFC through hydrogen bonding and acid–base interactions between acidic carboxylate groups and the basic (–N–) lone pair of DFC. Rebinding experiments confirmed that increasing the introduction of carboxyl groups from MAA significantly enhanced the imprinting effect, leading to a substantially increased imprinting factor and a specific rebinding capacity of up to 450 mg g−1 after 15 minutes.101
Recently, Tian's research team introduced an innovative approach for fabricating magnetic molecularly imprinted nanobeads (E2-MMINs) designed to target 17β-estradiol (E2). These E2-MMINs were synthesized by applying molecularly imprinted polymers onto the surface of magnetic nanobeads in aqueous solvents. In this technique, carboxyl group-functionalized Fe3O4 nanoparticles were employed as carriers, while E2 served as the template molecule, and dopamine was used as the functional monomer. The selection of dopamine was motivated by E2's hydrophobic characteristics, as dopamine, with its amino, hydroxyl, and phenyl groups, could engage in interactions with E2 through hydrogen bonding and π–π stacking. Furthermore, dopamine readily underwent oxidation to produce thin polydopamine layers on solid carriers under mildly alkaline conditions at room temperature.102
Mbhele et al. synthesized a molecularly imprinted polymer used as a solid-phase extraction (SPE) sorbent for selectively isolating fenoprofen in aqueous samples. The choice of 2-vinylpyridine as the functional monomer was driven by its ability to establish weak hydrogen bonds between its basic nitrogen atom and the carboxyl hydrogen atoms of acidic pharmaceuticals. Moreover, the pyridine group formed stacking electrostatic interactions with the aromatic rings of nonsteroidal anti-inflammatory drugs (NSAIDs), including fenoprofen. These compounds, including fenoprofen, ketoprofen, gemfibrozil, and triclosan, contain hydroxyl groups capable of binding with the nitrogen atom of the 2-vinylpyridine functional monomer. Given the close physicochemical resemblance of these compounds to fenoprofen, they had the potential to compete for binding sites within MIP cavities imprinted with fenoprofen.103
Cui and colleagues devised a straightforward method to synthesize hydrophilic molecularly imprinted polymers (MIPs) with full physical cross-linking. Copper ions (Cu2+) and a styrene-methacrylic acid copolymer (poly(St-co-MAA)) were employed as cross-linkers in this process. Initially, magnetic nanoparticles coated with CMC (CMC@Fe3O4) interacted with the template tetracycline (TC), forming the initial cross-linking network through hydrogen bonds and coordination bonds, with Cu2+ serving as the cross-linker. After TC was removed, the resulting magnetic hydrophilic imprinted polymers (Cu-CMMIPs) retained imprinted cavities that matched TC in size, dimensions, and functional groups. In Cu-CMMIPs, the absorption peak of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O shifted to 1600 cm−1, indicating the involvement of carboxyl groups from CMC in coordination bonds formation. Additionally, a weak absorption band at 630 cm−1 was attributed to Cu–O stretching vibrations, demonstrating successful coordination between Cu2+ and the polymer network. This strategy offered several distinct features, including the hydrophilicity imparted by the abundant carboxyl and hydroxyl groups in the imprinted network.61
O shifted to 1600 cm−1, indicating the involvement of carboxyl groups from CMC in coordination bonds formation. Additionally, a weak absorption band at 630 cm−1 was attributed to Cu–O stretching vibrations, demonstrating successful coordination between Cu2+ and the polymer network. This strategy offered several distinct features, including the hydrophilicity imparted by the abundant carboxyl and hydroxyl groups in the imprinted network.61
Yuan et al. prepared MIPs via a one-step swelling polymerization method. Polystyrene microspheres served as seeds, Congo red (CR) as template molecules, methacrylic acid as the functional monomer, methyl acrylic acid ethylene glycol ester as the crosslinker, acetonitrile as the solvent, and azobisisobutyronitrile as the initiator. The interactions between the functional groups (–COOH and –NH2) in the pores, created by the functional monomers, played a crucial role in providing adsorption selectivity for CR on CR-MIP.104
J. Meléndez-Marmolejo et al. synthesized a series of molecularly imprinted polymers (MIPs) for diclofenac, naproxen, and ibuprofen using different polymerization approaches. Two functional monomers, methacrylic acid (MAA) and 2-vinylpyridine (2-VP), were tested, and ethylene glycol dimethacrylate (EGDMA) was used as the crosslinker. These MIPs were designed to adsorb the target molecules through chemisorption bonds, involving chemical reactions or chemical-like forces between the functional groups of the adsorbent and the template. Diclofenac, which features a benzene ring and a carboxylic group similar to 2-vinylpyridine, was found to have high extraction capacity but low selectivity when using 2-vinylpyridine as the monomer. Although ibuprofen shares structural similarities with diclofenac and naproxen, its extraction capacity was lower, indicating that the presence of specific functional groups played a significant role in conformational memory. Therefore, MIP recognition depended not only on molecule size but also on the distribution of their functional groups.105
Li and collogues devised a novel approach for the purification and preconcentration of norfloxacin (NFX) in seawater samples using molecularly imprinted solid-phase extraction (MISPE). The functional monomer, methacrylic acid (MAA), and crosslinker, ethylene glycol dimethacrylate (EGDMA), were carefully chosen for this purpose. MAA was specifically chosen as the functional monomer due to its ability to promote hydrogen bonding and ionic interactions in the porogen prior to polymerization.106
In a study by Simon et al.,109 it was demonstrated that templates with maximum two functional groups exhibited remarkable selectivity, implying that the predominant mechanism for molecular recognition is shape selectivity. This observation may be ascribed to a potential clash between shape selectivity and pre-organization of functional groups during the procedure, or it could suggest an incomplete realization of the re-binding behavior. Noteworthy is the notable enhancement in efficiency for templates containing three or more functional groups, as the spacing between these groups is increased.
Two significant insights were put forth by Spivak et al.110 concerning the selectivity of MIPs, both of which are affected by the cavity's shape. Firstly, steric constraints emerge when a molecule exceeds the size limit of an imprinted site designed using a smaller template. Secondly, for optimal interactions within the binding locations, the chosen morphologies must not exceed the size of the template. Additionally, branching topologies demonstrate superior selectivity compared to linear hydrocarbons, whereby simple chain groups with more than 8 carbons lose their recognition capability entirely.
 | ||
| Fig. 6 A illustrates a schematic representation of the preparation and recognition process for a MIP. Reproduced from ref. 113 with permission from Elsevier, copyright 2023. Fig. 4.B depicts a schematic representation of the synthesis of CNT-mer dispersed MIP and the fabrication of CNT-mer dispersed MIP-modified PGE (electrode1). In this study, the use of CNT-mers in the MIP matrix required a solvent that facilitated their homogeneous dispersion. The synthetic protocol for preparing template-free MIPs using the activator produced by electron transfer for atom-transfer radical polymerization method is presented. Reproduced from ref. 114 with permission from Elsevier, copyright 2023. | ||
Ideally, monomers with the ability to demonstrate cage effects or strongly interact in the specific solvent are preferred as they enhance the capacity and homogeneity of the binding cavities. Moreover, the monomers should be suitable for the chosen polymerization method.115 Furthermore, the monomers used in MIPs should possess thermal and chemical stability, resisting environmental influences.116,117 Zhang et al.74 evaluated the use of UV-vis spectra for the selection of MIPs. Monomers should have the capacity to bond with the template. Non-covalent interactions are more commonly employed in MIP synthesis due to their relatively weaker intensity.118 These interactions are typically based on weak forces such, resulting in easy template extraction. Non-covalent interactions are rather simple and efficient, although they can be easily disrupted during the separation of the template-monomer complex. In contrast, covalent interactions involve slower binding and more difficult template extraction. The most frequently used monomers include but are not limited to MAA, 2-VP and 4-VP.119
In 2010, Nunez et al.120 utilized 4-VP and MAA in their study to develop the molecularly imprinted solid-phase extraction (MISPE) process. The selection was made considering the capacity of non-steroidal anti-inflammatory drugs (NSAIDs) to establish hydrogen bonds. This characteristic contributes to achieving a high extraction efficiency. Therefore, 2-VP and 4-VP are frequently employed as monomers for NSAIDs.121 On the other hand, MAA and other polar monomers demonstrate limited effectiveness in establishing hydrogen bonds with templates that are polar or have at least weak polarity. In such scenarios, hydrophobic monomers can be utilized, although it is more advantageous to design and synthesize monomers that possess morphologies complementing the template.122
Holland et al.126 and Rosengren et al.127 observed similar results, indicating that lower concentrations of cross-linkers result in better binding ability. Gavrilovic et al.128 suggest that adding functional cross-linkers to traditional polymerization reagents can improve “shape selectivity”. By adopting this approach, the quantity of binding sites is augmented, and their uniformity is enhanced, thus underscoring the remarkable potential of these functional cross-linkers. The monomer to crosslinker ratio is particularly significant as it affects the physical attributes of the produced MIPs, according to Mueller.129 Porogens are frequently used to enhance the surface area and capacity of MIPs. The available surface area is greatly influenced by the template, monomer and cross-linker solubility. If the complex or polymer precipitates, phase separation can diminish the surface area, whereas the use of non-solvents that cause emulsion formation can generate pores or cracks. Incorporating solid porogens, such as dissolvable salt particles, proves to be an effective method for augmenting surface area. The surface area, pore dimensions, binding ability, and morphology of the pre-polymerization complex are influenced by the properties of the cross-linker. Research has shown that intermolecular forces among the template, monomer, cross-linker, and solvent drive the formation of the pre-polymerization complex.129 Similar to porogens, this can lead to phase separation, leading to denser morphologies and more limited surface area. Typically, the presence of distinct pre-polymerization complexes with varying structures is observed, as molecules with similar characteristics demonstrate comparable binding capacities.
According to Guyot and Bartholin,130 solvents with higher solubility are advantageous for liquid-based applications like solid phase extraction (SPE) because they increase cavity accessibility owing to larger surface areas. However, employing a minimal quantity of solvent may yield a polymer with reduced surface area, resulting in a thicker and less permeable material.131 Sellergren et al.132 noted that the use of dichloromethane resulted in smaller pores and surface area.
In a study by Spivak et al.,133 it was found that the best outcomes were often achieved when using a polymer based on chloroform with limited porosity. This suggests that the material's identification properties are not directly linked to the polymer's topology. Furthermore, the potential solvent polarity plays a significant role, with a solvent exhibiting stable polarity being essential for facilitating the assembly of monomer-template interactions. In cases where the template and monomers interact primarily through hydrophobic interactions, water is often preferred as the solvent,134 whereas polar solvents can assist in the imprinting of hydrophilic or weakly polar templates.135
Surya et al.139 devised an electrochemical biomimetic sensor for ciprofloxacin detection. They utilized a chitosan gold nanoparticle-modified Molecularly Imprinted Polymer (Ch-AuMIP) to modify a glassy carbon electrode (GCE) for constructing the sensor. To characterize Ch-AuMIP, various properties were examined using Fourier transform infrared spectroscopy (FT-IR), scanning electron microscopy (SEM), atomic force microscopy (AFM), and cyclic voltammetry (CV). FT-IR analysis revealed the presence of intramolecular hydrogen bonding in chitosan between the –NH2 and –OH groups, evidenced by a broadband between 2400 cm−1 and 3400 cm−1, along with an absorbance band at 1650 cm−1, confirming the presence of an amide group in chitosan. AFM showed a high roughness in the MIP sample, indicating the presence of active sites, which facilitated the conversion of analyte adsorption into electrical changes during CV and DPV measurements. SEM images displayed a surface area with numerous cavities on Ch-AuMIP.
Wei et al.140 introduced a molecularly imprinted fluorescence sensor using carbon quantum dots (CQDs) with surface modification by acrylic acid for tetracycline detection. The sensor underwent characterization through transmission electron microscopy (TEM) and FT-IR. Tetracycline, acrylamide (AM), ethylene glycol dimethacrylate (EGDMA), and azobisisobutyronitrile (AIBN) were employed as the template molecule, functional monomer, cross-linker, and initiator, respectively. TEM analysis revealed CQDs coated with polymers, resulting in an irregular structure with agglomeration due to strong adhesion between the polymer layers and CQDs. FTIR spectra confirmed the successful elution of the template molecule from MIPs-AA/CQDs. Characteristic peaks at 3440 and 1610 cm−1 indicated imine group vibrations of the functional monomer (acrylamide), while the peak at 1675 cm−1 indicated carbonyl group vibrations of acrylamides, confirming the successful preparation of MIPs-acrylic acid/CQDs with good selectivity and sensitivity for tetracyclines.
Shaheen et al.141 synthesized two-dimensional graphitic carbon nitride nanosheets (g-C3N4) through microwave-assisted methods and incorporated them into MIPs using chloramphenicol as a template to create a novel mass-sensitive sensor for chloramphenicol detection. Structural characterizations and phase identification of g-C3N4 were conducted using X-ray diffraction (XRD), while FTIR characterized the chemical bonds within g-C3N4. Scanning electron microscopy revealed 2D lamellar structures of g-C3N4 with wrinkles and irregular folds, and atomic force microscopy confirmed sheet layers approximately 30 nm thick. The sensor demonstrated excellent sensitivity and selectivity, offering a practical method for detecting chloramphenicol residues in complex samples.
Zhang et al.142 developed surface plasmon resonance (SPR) sensors based on Molecularly Imprinted Polymers (MIPs) using a surface-immobilized initiator approach with kanamycin (KAN) as the template, 4-vinylbenzeneboronic acid as the functional monomer, and EGDMA as the cross-linking agent. Characterization of the sensor under optimal conditions involved SEM and FT-IR. AFM was used to examine the surface morphologies of the bare chip, MUA-modified film, and MIP film. The MIP film exhibited smoother and more regular topography, indicating the formation of evenly distributed holes. FTIR spectrometry revealed successful binding of 4-vinylbenzeneboronic acid to the MIP or non-imprinted polymer (NIP) film, with slight shifts in absorption peak positions in the MIP film after template molecule removal.
4. Applications
4.1 MIPs-solid-phase extraction
MIPs offer practicality in processing and analyzing a wide range of soluble samples. The solid-phase extraction (SPE) process plays a crucial role in MIPs applications, as it serves as the fundamental method for extracting, pre-concentrating, and analyzing emerging pollutants. MIPs are primarily utilized in the pre-concentration and extraction of target pollutants from various water samples, including wastewater.42 This approach employs materials like C18, HLB, and ion-exchange stationary phases to extract compounds with a wide range of physicochemical characteristics, including solubility, pKa, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, and functional groups. To enhance selectivity in solid-phase extraction (SPE) and improve analytical reproducibility and sensitivity, molecularly imprinted polymers (MIPs) can be utilized as sorbents. The most commonly employed application of MIPs in sample preparation is MIP-SPE, which entails filling cartridges with the synthesized sorbent using either dry or slurry packing methods. The selective extraction of analytes is achieved through a series of standard SPE steps, including conditioning, sample loading, washing, and elution.143
P, and functional groups. To enhance selectivity in solid-phase extraction (SPE) and improve analytical reproducibility and sensitivity, molecularly imprinted polymers (MIPs) can be utilized as sorbents. The most commonly employed application of MIPs in sample preparation is MIP-SPE, which entails filling cartridges with the synthesized sorbent using either dry or slurry packing methods. The selective extraction of analytes is achieved through a series of standard SPE steps, including conditioning, sample loading, washing, and elution.143
SPE is a well-established sample preparation technique,143 which conventionally involves four steps: (a) SPE column activation, (b) transportation of the sample solution to the column, (c) rinsing with a suitable solvent, and (d) utilizing a solvent to elute the target analyte. The SPE process is straightforward, time-efficient, and cost-effective in terms of solvent usage.46Table 3 provides a short presentation of the recent applications of sample preparation techniques based on MIPs in terms of environmental sample analysis.
| Polymerization Method | MIP | Target analytes/compounds | Analytical technique | Recovery | Environmental medium | Ref. |
|---|---|---|---|---|---|---|
| Emulsion polymerization | Magnetic hydrophilic imprinted polymers (Cu-CMMIPs) | Tetracycline/antibiotic | High-performance liquid chromatography combined with diode-array detection | 91.2% to 101.3% | Lake water | 61 |
| Bulk polymerization | Methacrylic acid-MIPs | Oxazepam/anxiolytic drug | Liquid chromatography with mass spectrometry | 88.0%–92.0% | Tap, mineral and river water | 52 |
| Precipitation polymerization method | MIP-nanospheres | Diclofenac/NSAIDs | — | — | Lake and river water | 101 |
| Bulk polymerization | E2-MMINs-Hydrophilic magnetic molecularly imprinted nanobeads | 17β-Estradiol (E2)/Estrogen | High-performance liquid chromatography with ultraviolet detection | 99.7%–102.5% | Lake water | 102 |
| Precipitation polymerization | MAA-EGDMA MIP | Norfloxacin/antibiotic | High-performance liquid chromatography | 77.6%–86.8% | Seawater | 106 |
| In situ polymerization | MIPs-E1 | Estrone (E1), estradiol (E2), estriol (E3), testosterone (TTR), androstenedione (ADD) and boldenone (BLD)/estrogens | High-performance liquid chromatography | 88.5%–105.1% | Sewage plant water | 146 |
| Bulk polymerization | Bulk polymerization MIP | Oxytetracycline/antibiotic | High-performance liquid chromatography | 80.78%–93.75% | Soil | 147 |
| Bulk polymerization | Magnetic molecularly imprinted polymer (MMIP) | Norfloxacin/antibiotic | High-performance liquid chromatography | 94.5%–95.0% | Wastewater | 148 |
| Bulk polymerization | Bulk polymerization MIP | Sertraline/antidepressant | High-performance liquid chromatograph combined with a diode array detector | — | Wastewater | 149 |
| Bulk polymerization | Bulk polymerization MIP | Ketoprofen/NSAIDS | High-performance liquid chromatography | — | Wastewater | 150 |
In addition to their highly selective adsorption of target templates, imprinted polymers possess another intriguing characteristic known as re-adsorption. After an imprinted polymer has been utilized for a particular application, it needs to be cleaned and can then be reused. This sustainable approach not only prolongs the lifespan of the polymer or material but also reduces the overall analysis costs. From a practical perspective, reusing the adsorbent diminishes the expenses associated with treatment and lessens the environmental impact of spent adsorbents.144
The utilization of microtechniques in combination with more selective sorbents represents one of the most recent advancements in sample preparation based on Molecularly Imprinted Polymers (MIPs). This approach aligns with the principles of green analytical chemistry, allowing for the reuse of sorbents, the substitution of harmful solvents with eco-friendly alternatives, and the reduction of both solvent and sorbent usage. In accordance with green analytical chemistry principles, the introduction of new materials in microtechnology also leads to reduced sorbent requirements, resulting in lower precursor and solvent consumption during sorbent material manufacturing.43,145
This approach resulted in a high degree of hydrophilicity, thanks to the abundant presence of carboxyl and hydroxyl groups. Additionally, the incorporation of magnetic structures allowed for magnetic separation throughout the extraction phase. The synthesized MIPs, referred to as MIP(Cu-CMMIPs), exhibited remarkable selectivity and effectiveness in the extraction of tetracycline from real lake water samples, displaying a substantial adsorption capacity. The linear range of detection expanded between 10 to 2500 μg L−1 and 3.75 μg L−1, respectively. Recovery research studies yielded satisfactory values, ranging from approximately 91% to 101%. Overall, this approach demonstrated favorable enrichment effectiveness and is adequately hydrophilic and compatible with water.61
Varenne et al., attempted the solid-phase extraction of oxazepam (OXA) via MIPs. Oxazepam is known for its persistent structure, which makes its complete removal from surface waters challenging. Therefore, it is crucial to identify the harmful residues present in environmental waters that pose risks to human health. The researchers developed MIPs with high selectivity that could remove OXA even at very low concentrations in environmental settings. The bulk polymerization method was utilized for synthesis. The efficiency of the fabricated MIPs was juxtaposed to that of a typical sorbent using river water samples. The results revealed that the MIPs exhibited a 30% higher sensitivity than the commonly used sorbents. This approach shows promise as a removal method for environmental samples.52
N. Amaly employed a precipitation polymerization method to design uniformly MIP nanospheres for the elimination of diclofenac (DFC), a model pharmaceutical pollutant. Methacrylic acid was utilized as the monomer. Rebinding tests demonstrated that the incorporation of carboxyl groups from methacrylic acid significantly enhanced the imprinting effect. The molecularly imprinted polymers exhibited an adsorption capacity of over 85% even after seven regeneration cycles, indicating their potential for multiple reuse.101
X. Tian developed an innovative type of magnetic molecularly imprinted nanobeads, known as E2-MMINs, specifically designed for 17β-estradiol (E2). The synthesized materials were thoroughly evaluated in terms of production and adsorption conditions, as well as their physicochemical properties. Notably, they featured thin imprinting layers, a stable crystal structure, and the ability for rapid magnetic separation. The nanobeads exhibited swift kinetics, a high binding capacity of approximately 41.5 mg g−1, satisfactory specificity (imprinting factor of around 8.1), and excellent reusability (with an adsorption efficiency of over 95% even after ten cycles). Moreover, when combined with high-performance liquid chromatography, the method achieved an impressively low detection limit of 0.008 μg L−1. The effectiveness of this approach was successfully demonstrated through its application to water samples.102
Li developed a very selective and efficient approach for the purification and pre-concentration of norfloxacin (NFX), using molecularly imprinted solid-phase extraction (MISPE). The MIP was created through precipitation polymerization and exhibited excellent adsorption capacity for NFX. An innovative approach was implemented for the quantification of NFX in seawater by combining offline MISPE with high-performance liquid chromatography and diode array detection. The columns demonstrated recoveries close to 78% for spiked seawater samples, with a relative standard deviation below 5.6% and a detection limit of 0.027 μg L−1.106
X. Wang used (MIPs)-coated solid-phase microextraction (SPME) fibers for the determination of Estrone (E1). The MIPs-E1 coating was synthesized via in situ polymerization on the inner and outer surfaces of the capillary. This method offers almost twice the enrichment capacity as opposed to in-tube SPME (IT-SPME). Under optimized conditions, the low limits of detection (max 0.8 μg L−1) and quantification (max 2.6 μg L−1) were achieved. The method exhibited good reproducibility, with a fiber-to-fiber relative standard deviation ranging between approximately 4.5 and 8%. The recoveries ranged from 88.5% to 105%. Furthermore, the produced fiber demonstrated great adsorption ability and specific selectivity in comparison to commercial alternatives.146
Cui developed a new and efficient pretreatment process for the analysis of bioavailable oxytetracycline (OTC) found in soils using a MIP synthesized through bulk polymerization. For comparison purposes, the total extractable content of OTC in soils was measured using an Oasis HLB SPE method. The MIP method achieved OTC recoveries ranging from approximately 80% to 94%. In assessing the intensity of OTC pollution and evaluating potential environmental hazards, the newly introduced method provided more valuable data than the Oasis HLB SPE method.147
In a separate investigation, composites of magnetic MIPs (MMIPs) were successfully synthesized, cross-linked using Fe3O4, and later coated with oleic acid. This innovative approach enabled the selective elimination of norfloxacin from waste and drinking water using NORF-MMIP, which is based on surface molecular imprinting and magnetic separation. The primary objective was to address environmental pollution. The adsorption performance of the fabricated material at room temperature was evaluated through various tests, noting that the maximum adsorption ability was achieved at 35 °C reaching approximately 42 mg g−1.148
Gornic, examined the potential of MIPs as adsorbents for the elimination of SSRIs in water treatment. Sertraline was specifically selected as the template. The structure of the MIPs was customized towards selectivity for sertraline. Notably, the maximum capacity was found to be approximately 72 mg g−1, while the highest imprinting factor reached about 3.7. Furthermore, the MIPs exhibited cross-reactivity towards other SSRIs. The fabricated MIPs could be reused. Despite their relatively smaller surface area the MIPs generally displayed superior sorption ability than activated carbon in wastewater samples.149
A MIP specifically targeting ketoprofen was fabricated and utilized for adsorbing the desired compound from water. The synthesis process employed the bulk polymerization approach at elevated temperatures (ranging between 60 to 80 °C), incorporating ketoprofen as the template. A non-imprinted polymer (NIP) was also created following a similar procedure, but without the presence of ketoprofen. Findings suggested that the interactions between the template and the monomer were primarily driven by hydrogen bonding, a finding corroborated by experimental results. The extraction efficiency was notably high, exceeding 90% under acidic conditions (pH 5). The MIP demonstrated superior selectivity over the NIP, as evidenced by a selectivity coefficient of 7.7 towards ketoprofen while structurally similar substances were present.150
4.2. Pesticides
Another area of focus pertains to the identification and measurement of pest-controlling substances employed in agricultural crops. Pesticides, insecticides, herbicides, and similar agents have been extensively utilized to manage pests and enhance crop yield. Currently, there is increased attention towards detecting pesticides due to the reduction of maximum residue limits (MRLs) by both domestic and foreign experts. Consequently, there is a need for more efficient pre-treatment extraction techniques to enable trace-level pesticide detection.151 In Table 4, various pesticides detection methods utilizing MIPs-based sensors are outlined.| Pesticides | Sensor | Type of polymerization | Sample | Instrument | Recovery | LOD | Ref. |
|---|---|---|---|---|---|---|---|
| Pirimicarb | Quartz crystal microbalance sensor | — | Tomato sample and aqueous solutions. | LC–MS/MS | 91 and 94%. | 0.028 nM | 154 |
| Pesticides | Surface plasmon resonance sensor | Photopolymerization | Environmental water samples | LC/MS-IT-TOF | Between 90 and 95% | 8.3 ng L−1 | 153 |
| Dimethoate | 7.1 ng L−1 | ||||||
| Carbofuran | |||||||
| Malathion | Electrochemical | — | Contaminated olive oil and fruit samples | DPV | 87.9% | 0.06 pg mL−1. | 155 |
| Carbofuran | Electrochemical nanocomposite sensor | Bulk polymerization | Tap and river water | UV-vis | Between 94%–97% | 3 × ∼10−10 | 156 |
| Profenofos | Amperometric | — | Vegetable sample (spring onion, tomato, Chinese cabbage, cabbage, green and chili peppers). | HPLC | — | 0.002 μM | 152 |
A novel and straightforward approach for selectively measuring profenofos (PFF) involves employing a MIP-coated carbon nanotube (3D-CNTs@MIP) amperometric sensor, as suggested by Amatatongchai et al. The sensor was constructed by coating a glassy carbon electrode the fabricated material and subsequently extracting the imprinting template through. Characterization tests demonstrated that the integration of MIP onto the CNT surface notably increased the selective surface area, resulting in a greater number of imprinting locations. The produced MIP sensor exhibited rapid response and demonstrated excellent recognition capabilities.152
SPR sensor chip nanofilms were synthesized using molecular imprinting technology. To assess the selectivity of the pesticide-imprinted nanofilms, comparative adsorption experiments were conducted utilizing SPR sensors. The imprinted nanofilms displayed greater sensitivity and selectivity compared to the non-imprinted counterparts for pesticide detection. The findings provided evidence that SPR sensors offer enhanced selectivity, sensitivity, and lower detection limits.153
5. Future directions and challenges
MIPs have been extensively used in numerous domains such as extraction, sensors, catalysis, drug delivery, and more, serving as effective mimics of natural receptors.157 Nanoimprinting techniques have greatly enhanced the number of functional recognition sites, leading to significant improvements in imprinting capacity. Highly selective and sensitive MIP reliant fluorescent sensors can be developed by taking advantage the sensitivity of fluorescence. Incorporating magnetic materials and catalytic micromotors into MIP synthesis allows for the creation of magnetic imprinted micromotor microsensors, offering not only insights into target imprinting but also avenues for multi-functionalization and intelligent nano-devices.158 Within the realm of biosensing, MIPs have primarily centered around imprinting individual target species, subsequently optimizing their performance and assessing selectivity in the presence of structurally similar species. However, early detection of diseases often requires the synchronous recognition of multiple biomarkers present in the same medium. Therefore, it is imperative to create and advance multiplexed sensing platforms or arrays that can identify biomarkers relevant to diseases, thus broadening the clinical importance of MIP biosensors.In alignment with the increasing significance of green chemistry principles in various domains, molecular imprinting is also anticipated to embrace these principles. Presently, there is limited research focused on the utilization of less toxic and environmentally friendly chemicals that ensure user safety and minimize harm to the environment. By embracing green synthesis techniques, such as utilizing renewable reagents, minimizing the overall quantity of necessary reagents, and adopting safer analytical procedures, it is possible to implement approaches that prioritize sustainability and establish a “non-toxic” environment. This dedication to sustainability enhances the welfare of both healthcare and the ecosystem.159
Recently, MIPs have expanded beyond their traditional applications in sensors and analytical purposes, finding new potential in the field of nanomedicine. Significant advancements have been made in synthesis techniques to enhance biocompatibility, control size, and even manipulate imprint orientation. Compared to antibodies, MIP nanoparticles offer advantages such as cost-effectiveness, improved stability, and scalability, rendering them an attractive alternative for targeted applications in bioimaging and therapy. These developments have sparked interest in utilizing MIPs for nanomedicine.160 However, when using MIP particles in vivo, it is crucial to consider the formation of a protein corona, which can diminish their ability to identify the target molecules by obstructing access to the internal imprint within the polymer. This phenomenon has been investigated by Lagana et al. in 2020, emphasizing the need for careful consideration in the future development of MIPs for nanomedicine.161
Additionally, to impart new and desirable properties, MIPs can be integrated with inorganic materials like gold, silver or iron oxide nanoparticles, which display responsiveness to external physical stimuli. For instance, Asadi et al. demonstrated drug release triggered by polymer degradation in their Fe3O4-modified MIP nanoparticles containing olanzapine (Fig. 7). The release of the drug was observed at pH levels similar to the physiological environment, as the fructose-based polymer underwent degradation.162 These approaches showcase the potential of MIPs in nanomedicine and highlight the possibilities for tailored applications and enhanced functionality. Molecularly imprinted polymer (MIP)-based sensors have been deemed a valuable tool for the specific detection of diverse pathogenic bacteria and viruses.163
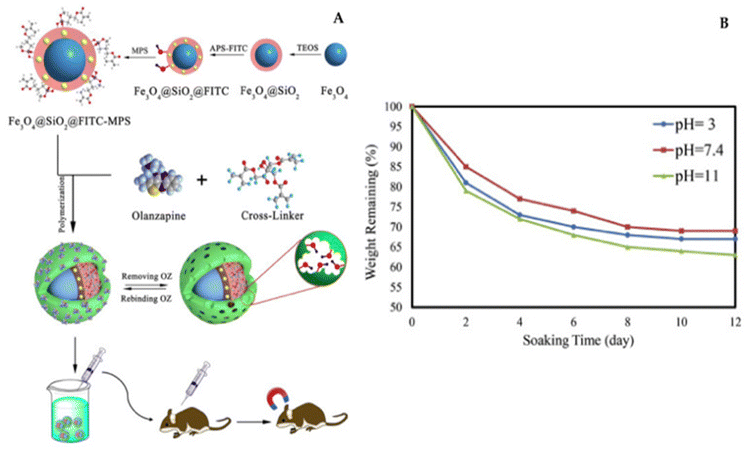 | ||
| Fig. 7 (A) A silica-coated iron oxide core is employed as the substrate, which is then functionalized with a fructose polymer imprinted specifically with olanzapine, an anti-psychotic medication. (B) In vivo, the natural degradation of the fructose polymer over the course of several days triggers the release of the drug, enabling drug release to occur across a wide pH range within the body. Reproduced from ref. 162 with permission from Elsevier, copyright 2023. | ||
Lu et al. conducted a study on the fabrication of bio-imprinting sensors targeting HIV-1-linked glycoprotein 41 (gp41), as depicted in Fig. 8(1).164 Particularly, magnetic MIPs have demonstrated significant advantages when integrated with immunoassays, providing a sensitive, straightforward, and cost-effective approach for the preliminary evaluation of HIV carriers, as illustrated in Fig. 8(2).165
 | ||
| Fig. 8 (1) The procedure illustrates the implementation of epitope-imprinting, where a MIP-coated quartz crystal microbalance (QCM) was utilized for precise detection of the target protein and template peptide. Reproduced from ref. 164, with permission from Elsevier, copyright 2023. (2) (A to C) The advancement of an electrochemiluminescence (ECL) immunosensor for accurate and selective quantification of type 1 HIV antibodies. Reproduced from ref. 165 with permission from Elsevier, copyright 2023. | ||
While molecularly imprinted polymers (MIPs) offer numerous advantages, their synthesis is influenced by various factors such as the properties of the solvent, functional monomer, and cross-linker, which can limit their application in biosensing. One key challenge in MIP synthesis relates to the nature of the monomer, as it significantly affects the polymer imprinting process. An additional obstacle arises from the insolubility of hydrophilic substances in organic solvents, leading to inadequate imprinting. However, this issue can be addressed by functionalizing the hydrophilic substance with alkyl chains, thereby converting it into a hydrophobic form.166
6. Conclusions
Molecularly imprinted polymers technology is experiencing rapid growth in multiple scientific disciplines. This review has compiled the fundamental principles of MIPs and their diverse applications. The monomers utilized in MIPs for non-covalent molecular imprinting procedures include MAA, 2-VP, dopamine, and styrene. Additionally, a crosslinker is employed to establish non-covalent interactions, with EGDMA being the most commonly used crosslinker. The choice of non-covalent molecular imprinting is driven by the simplicity of the method and the ease of template removal. The majority of porogens utilized in MIP synthesis is acetonitrile. The exceptional selectivity and sensitivity offered by MIPs have proven advantageous in numerous areas. To further enhance their properties, there is significant potential for introducing innovative approaches in the synthesis of MIPs by using diverse monomers and alternative solvents instead of the typical ones. However, further research is required to address the synthetic mechanisms and advance the design of MIPs. Moreover, careful consideration must be given to controlling solvent residues when organic solvents are employed in the productions of MIPs. The potential applications of MIPs span a variety of fields. The solid-phase approach in MIP preparation, along with the interaction between MIPs and imprinting targets, presents an innovative avenue for coating soluble polymers onto nanoparticles. As we look towards the future, a higher demand for the utilization of green synthesis methods alongside MIP sample preparation techniques is anticipated. This approach aligns with the principles of waste reduction, cost-effectiveness, ecological compatibility, production safety and conformance with sustainable chemistry. It is crucial to establish techniques that are both friendly to the environment and efficient for supervising food safety, addressing pollution and health concerns. The implementation of extraction systems based on MIPs will support the creation of analytical approaches that are eco-friendly, cost-effective, and fast by minimizing solvent usage. Ongoing research focuses on creating nano- or magnetic materials that can enhance the efficiency and capacity of analyte sorption. The ultimate goal is to create intelligent platforms appropriate for analysis in diverse research areas, including healthcare and environmental sciences.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The financial support received for this study from the Greek Ministry of Development and Investments (General Secretariat for Research and Technology) through the research project “Intergovernmental International Scientific and Technological Innovation-Cooperation. Joint declaration of Science and Technology Cooperation between China and Greece” with the topic “Development of monitoring and removal strategies of emerging micro-pollutants in wastewaters” (grant No. T7ΔKI-00220) and the support is gratefully acknowledged.References
- J. J. BelBruno, Chem. Rev., 2019, 119, 94–119 CrossRef CAS PubMed.
- S. He, L. Zhang, S. Bai, H. Yang, Z. Cui, X. Zhang and Y. Li, Eur. Polym. J., 2021, 143, 110179 CrossRef CAS.
- D. Qureshi, S. K. Nayak, S. Maji, A. Anis, D. Kim and K. Pal, Eur. Polym. J., 2019, 120, 109220 CrossRef CAS.
- C. Dong, H. Shi, Y. Han, Y. Yang, R. Wang and J. Men, Eur. Polym. J., 2021, 145, 110231 CrossRef CAS.
- G. Wulff and A. Sarhan, Angew. Chem., Int. Ed., 1972, 341–344 CAS.
- D. Çimen, N. Bereli and A. Denizli, Biomed. Mater. Devices, 2022 DOI:10.1007/s44174-022-00022-3.
- A. Araz, Turk. J. Chem., 2022, 46, 487–498 CrossRef CAS.
- Y. Suwanwong and S. Boonpangrak, Eur. Polym. J., 2021, 161, 110835 CrossRef CAS.
- H. Zhong and J. Deng, Polym. Rev., 2022, 62, 826–859 CrossRef CAS.
- S. Han, Y. Song, S. Liu, L. Zhao and R. Sun, Eur. Polym. J., 2022, 171, 111219 CrossRef CAS.
- S. Bhogal, I. Mohiuddin, K. Kaur, J. Lee, R. J. C. Brown, A. K. Malik and K.-H. Kim, Environ. Pollut., 2021, 275, 116613 CrossRef CAS PubMed.
- G. Pilvenyte, V. Ratautaite, R. Boguzaite, A. Ramanavicius, R. Viter and S. Ramanavicius, Int. J. Mater. Sci., 2023, 24, 4105 CAS.
- L. Andersson, B. Sellergren and K. Mosbach, Tetrahedron Lett., 1984, 25, 5211–5214 CrossRef CAS.
- G. Wulff, D. Oberkobusch and M. Minárik, React. Polym., Ion Exch., Sorbents, 1985, 3, 261–275 CrossRef CAS.
- A. G. Strikovsky, D. Kasper, M. Grün, B. S. Green, J. Hradil and G. Wulff, J. Am. Chem. Soc., 2000, 122, 6295–6296 CrossRef CAS.
- M. G. Metwally, A. H. Benhawy, R. M. Khalifa, R. M. El Nashar and M. Trojanowicz, Molecules, 2021, 26, 6515 CrossRef CAS PubMed.
- G. Wulff and J. Liu, Acc. Chem. Res., 2012, 45, 239–247 CrossRef CAS PubMed.
- W. Chen, Z. Meng, M. Xue and K. J. Shea, Mol. Imprinting, 2016, 4, 1–12 CAS.
- Molecular imprinting for nanosensors and other sensing applications, ed. A. Denizli, Elsevier, Amsterdam, 2021 Search PubMed.
- X. Li, J. Zhou, L. Tian, W. Li, Z. Ali, N. Ali, B. Zhang, H. Zhang and Q. Zhang, Sens. Actuators, B, 2016, 225, 436–445 CrossRef CAS.
- G. N. Wang, K. Yang, H. Z. Liu, M. X. Feng and J. P. Wang, Anal. Methods, 2016, 8, 5511–5518 RSC.
- A. R. Fareghi, P. N. Moghadam, J. Khalafy, M. Bahram and M. Moghtader, J. Appl. Polym. Sci., 2017, 134, 45581 CrossRef.
- W. Zheng, H. Wu, Y. Jiang, J. Xu, X. Li, W. Zhang and F. Qiu, Anal. Biochem., 2018, 555, 42–49 CrossRef CAS PubMed.
- W. Huang, P. Jiang, X. Yin, L. Zhang, S. Zhao, H. Zhou, X. Ni and W. Xu, J. Sep. Sci., 2021, 44, 513–520 CrossRef CAS PubMed.
- Ö. Erdem, Y. Saylan, N. Cihangir and A. Denizli, Biosens. Bioelectron., 2019, 126, 608–614 CrossRef PubMed.
- S. Ramanavicius and A. Ramanavicius, Adv. Colloid Interface Sci., 2022, 305, 102693 CrossRef CAS PubMed.
- T. Zhou, L. Ding, G. Che, W. Jiang and L. Sang, TrAC, Trends Anal. Chem., 2019, 114, 11–28 CrossRef CAS.
- F. Shahhoseini, A. Azizi and C. S. Bottaro, SSRN J., 2022, 156, 116695 CrossRef CAS.
- L. D. De León-Martínez, M. Rodríguez-Aguilar, R. Ocampo-Pérez, J. M. Gutiérrez-Hernández, F. Díaz-Barriga, L. Batres-Esquivel and R. Flores-Ramírez, Bull. Environ. Contam. Toxicol., 2018, 100, 395–401 CrossRef PubMed.
- M. Zhao, H. Shao, J. Ma, H. Li, Y. He, M. Wang, F. Jin, J. Wang, A. M. Abd El-Aty, A. Hacımüftüoğlu, F. Yan, Y. Wang and Y. She, J. Chromatogr., A, 2020, 1615, 460751 CrossRef CAS PubMed.
- K. Banan, D. Hatamabadi, H. Afsharara, B. Mostafiz, H. Sadeghi, S. Rashidi, A. D. Beirami, M.-A. Shahbazi, R. Keçili, C. M. Hussain and F. Ghorbani-Bidkorbeh, Trends Food Sci. Technol., 2022, 119, 164–178 CrossRef CAS.
- M. Arabi, A. Ostovan, J. Li, X. Wang, Z. Zhang, J. Choo and L. Chen, Adv. Mater., 2021, 33, 2100543 CrossRef CAS PubMed.
- L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- G. Liu, X. Huang, L. Li, X. Xu, Y. Zhang, J. Lv and D. Xu, Nanomaterials, 2019, 9, 1030 CrossRef CAS PubMed.
- R. Garcia, M. D. R. Gomes Da Silva and M. J. Cabrita, Talanta, 2018, 176, 479–484 CrossRef CAS PubMed.
- M. Zhao, H. Shao, J. Ma, H. Li, Y. He, M. Wang, F. Jin, J. Wang, A. M. Abd El-Aty, A. Hacımüftüoğlu, F. Yan, Y. Wang and Y. She, J. Chromatogr., A, 2020, 1615, 460751 CrossRef CAS PubMed.
- M. Parlapiano, Ç. Akyol, A. Foglia, M. Pisani, P. Astolfi, A. L. Eusebi and F. Fatone, J. Environ. Chem. Eng., 2021, 9, 105051 CrossRef CAS.
- H. Wu, G. Lin, C. Liu, S. Chu, C. Mo and X. Liu, Trends Environ. Anal. Chem., 2022, 36, e00178 CrossRef CAS.
- H. Kamyab, S. Chelliapan, O. Tavakkoli, M. Mesbah, J. K. Bhutto, T. Khademi, I. Kirpichnikova, A. Ahmad and A. A. Aljohani, Chemosphere, 2022, 308, 136471 CrossRef CAS PubMed.
- A. J. Kadhem, G. J. Gentile and M. M. Fidalgo De Cortalezzi, Molecules, 2021, 26, 6233 CrossRef CAS PubMed.
- Y. Han, J. Tao, N. Ali, A. Khan, S. Malik, H. Khan, C. Yu, Y. Yang, M. Bilal and A. A. Mohamed, Eur. Polym. J., 2022, 179, 111582 CrossRef CAS.
- D. S. Villarreal-Lucio, K. X. Vargas-Berrones, L. Díaz De León-Martínez and R. Flores-Ramíez, Environ. Sci. Pollut. Res., 2022, 29, 89923–89942 CrossRef CAS PubMed.
- S. I. Kaya, A. Cetinkaya and S. A. Ozkan, Trends Environ. Anal. Chem., 2023, 37, e00193 CrossRef CAS.
- S. E. Elugoke, A. S. Adekunle, O. E. Fayemi, E. D. Akpan, B. B. Mamba, E. M. Sherif and E. E. Ebenso, Electrochem. Sci. Adv., 2021, 1(2), e2000026 CrossRef CAS.
- M. Gao, Y. Gao, G. Chen, X. Huang, X. Xu, J. Lv, J. Wang, D. Xu and G. Liu, Front. Chem., 2020, 8, 616326 CrossRef CAS PubMed.
- Y. Yang and X. Shen, Molecules, 2022, 27, 7355 CrossRef CAS PubMed.
- G. Wulff, in Frontiers in Biosensorics I, ed. F. W. Scheller, F. Schubert and J. Fedrowitz, Birkhäuser Basel, Basel, 1997, vol. 80, pp. 13–26 Search PubMed.
- K. Mosbach and O. Ramström, Nat. Biotechnol., 1996, 14, 163–170 CrossRef CAS.
- A. Lusina and M. Cegłowski, Polymers, 2022, 14, 640 CrossRef CAS PubMed.
- T. Sajini and B. Mathew, Talanta Open, 2021, 4, 100072 CrossRef.
- T. Takeuchi, K. Akeda, S. Murakami, H. Shinmori, S. Inoue, W.-S. Lee and T. Hishiya, Org. Biomol. Chem., 2007, 5, 2368 RSC.
- F. Varenne, P. Kadhirvel, P. Bosman, L. Renault, A. Combès and V. Pichon, Anal. Bioanal. Chem., 2022, 414, 451–463 CrossRef CAS PubMed.
- L. M. Madikizela, N. T. Tavengwa, H. Tutu and L. Chimuka, Trends in Environmental Analytical Chemistry, 2018, 17, 14–22 CrossRef CAS.
- A. Beltran, F. Borrull, R. M. Marcé and P. A. G. Cormack, TrAC, Trends Anal. Chem., 2010, 29, 1363–1375 CrossRef CAS.
- J. Ashley, M.-A. Shahbazi, K. Kant, V. A. Chidambara, A. Wolff, D. D. Bang and Y. Sun, Biosens. Bioelectron., 2017, 91, 606–615 CrossRef CAS PubMed.
- A. Speltini, A. Scalabrini, F. Maraschi, M. Sturini and A. Profumo, Anal. Chim. Acta, 2017, 974, 1–26 CrossRef CAS PubMed.
- Y. A. Olcer, M. Demirkurt, M. M. Demir and A. E. Eroglu, RSC Adv., 2017, 7, 31441–31447 RSC.
- A. G. Mayes and K. Mosbach, Anal. Chem., 1996, 68, 3769–3774 CrossRef CAS PubMed.
- J. Haginaka, J. Chromatogr. B, 2008, 866, 3–13 CrossRef CAS PubMed.
- N. Pérez-Moral and A. G. Mayes, Anal. Chim. Acta, 2004, 504, 15–21 CrossRef.
- Y. Cui, J. Ding, Y. Su and L. Ding, Chem. Eng. J., 2023, 452, 139291 CrossRef CAS.
- H. Yan and K. Row, Int. J. Mater. Sci., 2006, 7, 155–178 CAS.
- K. Haupt, P. X. Medina Rangel and B. T. S. Bui, Chem. Rev., 2020, 120, 9554–9582 CrossRef CAS PubMed.
- M. S. Kang, E. Cho, H. E. Choi, C. Amri, J.-H. Lee and K. S. Kim, Biomater. Res., 2023, 27, 45 CrossRef CAS PubMed.
- Y. Zhang, G. Zhao, K. Han, D. Sun, N. Zhou, Z. Song, H. Liu, J. Li and G. Li, Molecules, 2022, 28, 301 CrossRef PubMed.
- Y. Chen, Z. Xie, L. Zhang and X. Hu, J. Biomater. Sci., Polym. Ed., 2020, 31, 954–968 CrossRef CAS PubMed.
- M. Azimi, M. Ahmadi Golsefidi, A. Varasteh Moradi, M. Ebadii and R. Zafar Mehrabian, J. Anal. Methods Chem., 2020, 2020, 1–9 CrossRef PubMed.
- J.-W. Zhang, J.-Y. He, C.-Z. Wang, F.-Q. Yang, L.-D. Zhou, Q.-H. Zhang, Z.-N. Xia and C.-S. Yuan, Talanta, 2020, 219, 121283 CrossRef CAS PubMed.
- Z.-F. Jiang, Q. Li, Q.-Y. Li, H.-X. Xu, J.-Y. He, C.-Z. Wang, L.-D. Zhou, Q.-H. Zhang, L. Luo and C.-S. Yuan, Microchem. J., 2022, 179, 107545 CrossRef CAS.
- Y. Wang, J. Li, L. Wang, J. Qi and L. Chen, Se Pu, 2021, 39, 134–141 CAS.
- A. M. Mostafa, S. J. Barton, S. P. Wren and J. Barker, TrAC, Trends Anal. Chem., 2021, 144, 116431 CrossRef CAS.
- N. Murdaya, A. L. Triadenda, D. Rahayu and A. N. Hasanah, Polymers, 2022, 14, 4441 CrossRef CAS PubMed.
- S. A. Mohajeri, G. Karimi, J. Aghamohammadian and M. R. Khansari, J. Appl. Polym. Sci., 2011, 121, 3590–3595 CrossRef CAS.
- W. Zhang, X. She, L. Wang, H. Fan, Q. Zhou, X. Huang and J. Tang, Materials, 2017, 10, 475 CrossRef CAS PubMed.
- Y. Liu, K. Hoshina and J. Haginaka, Talanta, 2010, 80, 1713–1718 CrossRef CAS PubMed.
- N. Ali, B. Zhang, H. Zhang, W. Zaman, S. Ali, Z. Ali, W. Li and Q. Zhang, J. Chin. Chem. Soc., 2015, 62, 695–702 CrossRef CAS.
- G. Dvorakova, R. Haschick, K. Chiad, M. Klapper, K. Müllen and A. Biffis, Macromol. Rapid Commun., 2010, 31, 2035–2040 CrossRef CAS PubMed.
- I. Veloz Martínez, J. I. Ek, E. C. Ahn and A. O. Sustaita, RSC Adv., 2022, 12, 9186–9201 RSC.
- H. Hang, C. Li, J. Pan, L. Li, J. Dai, X. Dai, P. Yu and Y. Feng, J. Sep. Sci., 2013, 36, 3285–3294 CrossRef CAS PubMed.
- F. Zhu, L. Li, N. Li, W. Liu, X. Liu and S. He, Microchem. J., 2021, 164, 106060 CrossRef CAS.
- K. Ramajayam, S. Ganesan, P. Ramesh, M. Beena, T. Kokulnathan and A. Palaniappan, Biomimetics, 2023, 8, 245 CrossRef CAS PubMed.
- M. M. Moein, A. Abdel-Rehim and M. Abdel-Rehim, Molecules, 2019, 24, 2889 CrossRef CAS PubMed.
- L. Fang, M. Jia, H. Zhao, L. Kang, L. Shi, L. Zhou and W. Kong, Trends Food Sci. Technol., 2021, 116, 387–404 CrossRef CAS.
- L. T. Hou, J. J. Chen, H. J. Fu and X. L. Fu, Acc. Mater. Res., 2014, 884–885, 33–36 Search PubMed.
- R. Gao, X. Mu, Y. Hao, L. Zhang, J. Zhang and Y. Tang, J. Mater. Chem. B, 2014, 2, 1733–1741 RSC.
- S. Beyazit, B. Tse Sum Bui, K. Haupt and C. Gonzato, Prog. Polym. Sci., 2016, 62, 1–21 CrossRef CAS.
- L. A. Camacho-Cruz, M. A. Velazco-Medel and E. Bucio, in Green Sustainable Process for Chemical and Environmental Engineering and Science, Elsevier, 2020, pp. 275–318 Search PubMed.
- M. Szwarc, Nature, 1956, 178, 1168–1169 CrossRef CAS.
- T. Otsu, J. Polym. Sci., Part A: Polym. Chem., 2000, 38, 2121–2136 CrossRef CAS.
- T. Otsu and M. Yoshida, Makromol. Chem., Rapid Commun., 1982, 3, 127–132 CrossRef CAS.
- T. E. Orowitz, P. P. A. A. Ana Sombo, D. Rahayu and A. N. Hasanah, Molecules, 2020, 25, 3256 CrossRef CAS PubMed.
- M. Fizir, A. Richa, H. He, S. Touil, M. Brada and L. Fizir, Rev. Environ. Sci. Biotechnol., 2020, 19, 241–258 CrossRef.
- H. Zhang, Polymer, 2014, 55, 699–714 CrossRef CAS.
- A. N. Hasanah, N. Safitri, A. Zulfa, N. Neli and D. Rahayu, Molecules, 2021, 26, 5612 CrossRef CAS PubMed.
- M. Marć and P. P. Wieczorek, Comprehensive Analytical Chemistry, Elsevier, 2019, vol. 86, pp. 1–15 Search PubMed.
- C. Nasraoui, N. Jaoued-Grayaa, L. Vanoye, Y. Chevalier and S. Hbaieb, Eur. Polym. J., 2022, 177, 111441 CrossRef CAS.
- X. Xie, Y. Bu and S. Wang, Rev. Anal. Chem., 2016, 35, 87–97 CrossRef CAS.
- P. Sikiti, T. A. Msagati, B. B. Mamba and A. K. Mishra, J. Environ. Health Sci. Eng., 2014, 12, 82 CrossRef PubMed.
- S. Bakhtiar, S. A. Bhawani and S. R. Shafqat, Chem. Biol. Technol. Agric., 2019, 6, 15 CrossRef.
- X. Sun, M. Wang, J. Peng, L. Yang, X. Wang, F. Wang, X. Zhang, Q. Wu, R. Chen and J. Chen, Talanta, 2019, 196, 47–53 CrossRef CAS PubMed.
- N. Amaly, G. Istamboulie, A. Y. El-Moghazy and T. Noguer, J. Chem. Res., 2021, 45, 102–110 CrossRef CAS.
- X. Tian, H. Song, Y. Wang, X. Tian, Y. Tang, R. Gao and C. Zhang, Talanta, 2020, 220, 121367 CrossRef CAS PubMed.
- Z. E. Mbhele, S. Ncube and L. M. Madikizela, Environ. Sci. Pollut. Res., 2018, 25, 36724–36735 CrossRef CAS PubMed.
- D. Yuan, D. Fu and C. Wang, Sep. Sci. Technol., 2021, 56, 233–241 CrossRef CAS.
- J. Meléndez-Marmolejo, L. Díaz De León-Martínez, V. Galván-Romero, S. Villarreal-Lucio, R. Ocampo-Pérez, N. A. Medellín-Castillo, E. Padilla-Ortega, I. Rodríguez-Torres and R. Flores-Ramírez, Environ. Sci. Pollut. Res., 2022, 29, 45885–45902 CrossRef PubMed.
- H. Li, J. Chen, L. Tan and J. Wang, Anal. Lett., 2019, 52, 2896–2913 CrossRef CAS.
- B. Sellergren and L. I. Andersson, Methods, 2000, 22, 92–106 CrossRef CAS PubMed.
- G. Wulff, Angew. Chem., Int. Ed. Engl., 1995, 34, 1812–1832 CrossRef CAS.
- R. Simon, M. E. Collins and D. A. Spivak, Anal. Chim. Acta, 2007, 591, 7–16 CrossRef CAS PubMed.
- D. A. Spivak, R. Simon and J. Campbell, Anal. Chim. Acta, 2004, 504, 23–30 CrossRef CAS.
- R. Pratiwi, S. Megantara, D. Rahayu, I. Pitaloka and A. N. Hasanah, J. Young Pharm., 2018, 11, 12–16 CrossRef.
- C. Lafarge, M. Bitar, L. El Hosry, P. Cayot and E. Bou-Maroun, Mater. Today Commun., 2020, 24, 101157 CrossRef CAS.
- Y. Hu, J. Pan, K. Zhang, H. Lian and G. Li, TrAC, Trends Anal. Chem., 2013, 43, 37–52 CrossRef CAS.
- B. B. Prasad, A. Prasad, M. P. Tiwari and R. Madhuri, Biosens. Bioelectron., 2013, 45, 114–122 CrossRef CAS PubMed.
- R. C. Advincula, Korean J. Chem. Eng., 2011, 28, 1313–1321 CrossRef CAS.
- H. J. Lim, T. Saha, B. T. Tey, W. S. Tan and C. W. Ooi, Biosens. Bioelectron., 2020, 168, 112513 CrossRef CAS PubMed.
- G. Ozcelikay, S. I. Kaya, E. Ozkan, A. Cetinkaya, E. Nemutlu, S. Kır and S. A. Ozkan, TrAC, Trends Anal. Chem., 2022, 146, 116487 CrossRef CAS.
- K. S. Verma and K. Xia, J. AOAC Int., 2010, 93, 1313–1321 CrossRef CAS PubMed.
- G. Vasapollo, R. D. Sole, L. Mergola, M. R. Lazzoi, A. Scardino, S. Scorrano and G. Mele, Int. J. Mater. Sci., 2011, 12, 5908–5945 CAS.
- P. Lucci, O. Núñez and M. T. Galceran, J. Chromatogr., A, 2011, 1218, 4828–4833 CrossRef CAS PubMed.
- N. Y. Mlunguza, S. Ncube, P. Nokwethemba Mahlambi, L. Chimuka and L. M. Madikizela, J. Environ. Chem. Eng., 2019, 7, 103142 CrossRef CAS.
- P. Manesiotis, A. J. Hall, J. Courtois, K. Irgum and B. Sellergren, Angew. Chem., Int. Ed., 2005, 44, 3902–3906 CrossRef CAS PubMed.
- M. Włoch and J. Datta, Comprehensive Analytical Chemistry, Elsevier, 2019, vol. 86, pp. 17–40 Search PubMed.
- J.-D. Marty, M. Tizra, M. Mauzac, I. Rico-Lattes and A. Lattes, Macromolecules, 1999, 32, 8674–8677 CrossRef CAS.
- X. Song, J. Li, J. Wang and L. Chen, Talanta, 2009, 80, 694–702 CrossRef CAS PubMed.
- N. Holland, J. Frisby, E. Owens, H. Hughes, P. Duggan and P. McLoughlin, Polymer, 2010, 51, 1578–1584 CrossRef CAS.
- A. Rosengren, B. Karlsson and I. Nicholls, Int. J. Mater. Sci., 2013, 14, 1207–1217 CAS.
- I. Gavrilović, K. Mitchell, A. D. Brailsford, D. A. Cowan, A. T. Kicman and R. J. Ansell, Steroids, 2011, 76, 478–483 CrossRef PubMed.
- A. Mueller, Molecules, 2021, 26, 5139 CrossRef CAS PubMed.
- A. Guyot and M. Bartholin, Prog. Polym. Sci., 1982, 8, 277–331 CrossRef CAS.
- D. Horák, F. Švec, M. Bleha and J. Kálal, Angew. Makromol. Chem., 1981, 95, 109–115 CrossRef.
- B. Sellergren and K. J. Shea, J. Chromatogr., A, 1993, 635, 31–49 CrossRef CAS.
- D. Spivak, Adv. Drug Delivery Rev., 2005, 57, 1779–1794 CrossRef CAS PubMed.
- P. A. G. Cormack and A. Z. Elorza, J. Chromatogr. B, 2004, 804, 173–182 CrossRef CAS PubMed.
- S. A. Piletsky, E. V. Piletskaya, T. L. Panasyuk, A. V. El'skaya, R. Levi, I. Karube and G. Wulff, Macromolecules, 1998, 31, 2137–2140 CrossRef CAS.
- J. Ma, M. Yan, G. Feng, Y. Ying, G. Chen, Y. Shao, Y. She, M. Wang, J. Sun, L. Zheng, J. Wang and A. M. Abd El-Aty, Talanta, 2021, 225, 122031 CrossRef CAS PubMed.
- E. Turan, A. Zengin, Z. Suludere, N. Ö. Kalkan and U. Tamer, Talanta, 2022, 237, 122926 CrossRef CAS PubMed.
- B. Bräuer, F. Thier, M. Bittermann, D. Baurecht and P. A. Lieberzeit, ACS Appl. Bio Mater., 2022, 5, 160–171 CrossRef PubMed.
- S. G. Surya, S. Khatoon, A. Ait Lahcen, A. T. H. Nguyen, B. B. Dzantiev, N. Tarannum and K. N. Salama, RSC Adv., 2020, 10, 12823–12832 RSC.
- X. Wei, L. Lv, Z. Zhang and W. Guan, J. Appl. Polym. Sci., 2020, 137, 49126 CrossRef CAS.
- A. Shaheen, A. Taj, F. Jameel, M. A. Tahir, A. Mujahid, F. K. Butt, W. S. Khan and S. Z. Bajwa, Appl. Nanosci., 2022, 12, 139–150 CrossRef CAS.
- L. Zhang, C. Zhu, C. Chen, S. Zhu, J. Zhou, M. Wang and P. Shang, Food Chem., 2018, 266, 170–174 CrossRef CAS PubMed.
- A. Azizi and C. S. Bottaro, J. Chromatogr., A, 2020, 1614, 460603 CrossRef CAS PubMed.
- J. S. Mankar, M. D. Sharma and R. J. Krupadam, J. Mater. Sci., 2020, 55, 6810–6825 CrossRef CAS.
- B. H. Fumes, M. R. Silva, F. N. Andrade, C. E. D. Nazario and F. M. Lanças, TrAC, Trends Anal. Chem., 2015, 71, 9–25 CrossRef CAS.
- X. Wang, P. Huang, X. Ma, X. Du and X. Lu, Talanta, 2019, 194, 7–13 CrossRef CAS PubMed.
- S. Cui, H. Cao, Y. Wang and Y. Su, Hum. Ecol. Risk Assess.: Int. J., 2020, 26, 1108–1123 CrossRef CAS.
- M. M. Gaho, G. Z. Memon, J. U. R. Memon, J. B. Arain, A. J. Arain, A. Shah and M. Q. Samejo, Chin. J. Anal. Chem., 2022, 50, 100079 Search PubMed.
- T. Gornik, S. Shinde, L. Lamovsek, M. Koblar, E. Heath, B. Sellergren and T. Kosjek, Polymers, 2020, 13, 120 CrossRef PubMed.
- L. M. Madikizela, S. S. Zunngu, N. Y. Mlunguza, N. T. Tavengwa, P. S. Mdluli and L. Chimuka, Water SA, 2018, 44 DOI:10.4314/wsa.v44i3.08.
- S. Farooq, J. Nie, Y. Cheng, Z. Yan, J. Li, S. A. S. Bacha, A. Mushtaq and H. Zhang, Analyst, 2018, 143, 3971–3989 RSC.
- M. Amatatongchai, W. Sroysee, P. Sodkrathok, N. Kesangam, S. Chairam and P. Jarujamrus, Anal. Chim. Acta, 2019, 1076, 64–72 CrossRef CAS PubMed.
- O. Çakır and Z. Baysal, Sens. Actuators, B, 2019, 297, 126764 CrossRef.
- O. Cakir, J. Mol. Recognit., 2019, 32(9), e2785 CrossRef PubMed.
- Y. Aghoutane, A. Diouf, L. Österlund, B. Bouchikhi and N. El Bari, Bioelectrochemistry, 2020, 132, 107404 CrossRef CAS PubMed.
- M. Khadem, F. Faridbod, P. Norouzi, A. R. Foroushani, M. R. Ganjali, R. Yarahmadi and S. J. Shahtaheri, J. Anal. Chem., 2020, 75, 669–678 CrossRef.
- J. Pan, W. Chen, Y. Ma and G. Pan, Chem. Soc. Rev., 2018, 47, 5574–5587 RSC.
- L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- Y. L. Mustafa, A. Keirouz and H. S. Leese, J. Mater. Chem. B, 2022, 10, 7418–7449 RSC.
- M. Garnier, M. Sabbah, C. Ménager and N. Griffete, Nanomaterials, 2021, 11, 3091 CrossRef CAS PubMed.
- A. Capriotti, S. Piovesana, R. Zenezini Chiozzi, C. M. Montone, A. M. Bossi and A. Laganà, J. Proteomics, 2020, 219, 103736 CrossRef CAS PubMed.
- E. Asadi, M. Abdouss, R. M. Leblanc, N. Ezzati, J. N. Wilson and D. Kordestani, Polymer, 2016, 97, 226–237 CrossRef CAS.
- F. Cui, Z. Zhou and H. S. Zhou, Sensors, 2020, 20, 996 CrossRef CAS PubMed.
- C.-H. Lu, Y. Zhang, S.-F. Tang, Z.-B. Fang, H.-H. Yang, X. Chen and G.-N. Chen, Biosens. Bioelectron., 2012, 31, 439–444 CrossRef CAS PubMed.
- J. Zhou, N. Gan, T. Li, F. Hu, X. Li, L. Wang and L. Zheng, Biosens. Bioelectron., 2014, 54, 199–206 CrossRef CAS PubMed.
- A. K. Yadav, D. Verma, N. Dalal, A. Kumar and P. R. Solanki, Biosens. Bioelectron.: X, 2022, 12, 100257 CAS.
| This journal is © The Royal Society of Chemistry 2024 |