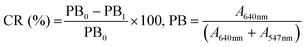Open Access Article
Open Access ArticleRapid detection of Salmonella using an aptamer-functionalized PDA liposome sensor with naked-eye colorimetric sensing†
Goeun
Lee
ab,
Byeongsung
Kim
a,
Inseung
Jang
a,
Moon
Il Kim
 c,
Seunghan
Shin
c,
Seunghan
Shin
 ad and
Kiok
Kwon
ad and
Kiok
Kwon
 *a
*a
aGreen and Sustainable Materials R&D Department, Korea Institute of Industrial Technology (KITECH), Republic of Korea. E-mail: kioks@kitech.re.kr
bDepartment of Chemical and Biomolecular Engineering, Yonsei University, Seodaemun-gu, Seoul, 03722, Republic of Korea
cDepartment of BioNano Technology, Gachon University, Seongnamdae-ro, Sujeong-gu, Seongnam, Gyeonggi 13120, Republic of Korea
dDepartment of Green Process and System Engineering, Korea University of Science & Technology (UST), Cheonan, Chungnam 31056, Republic of Korea
First published on 17th January 2024
Abstract
The increasing occurrence of Salmonella pathogens responsible for foodborne diseases is of mounting global concern within the sphere of public health and consumer welfare. Detecting Salmonella holds pivotal importance in averting these illnesses. The complex pretreatment process, time-consuming nature, and expensive equipment associated with existing detection methods have posed barriers to the development of sensors capable of rapidly and effortlessly detecting pathogens in real-life scenarios. Therefore, the development of a colorimetric sensor capable of rapid and facile pathogen detection, without the need for complex sample preparation or expensive detection equipment, is a crucial research direction for ensuring food safety. In this study, we successfully engineered a PDA-based liposome sensor decorated with an aptamer that specifically interacts with Salmonella typhimurium, enabling the rapid and accurate detection of Salmonella via a colorimetric response. This aptamer-functionalized PDA sensor exhibited a distinct color change (colorimetric response of 15.7% for 1 × 104 CFU mL−1Salmonella) within 15 minutes, accompanied by a highly linear increase in the CR values as the Salmonella concentration increased. In addition, the developed sensor materials can also effectively detect Salmonella in real food through a colorimetric reaction. We developed a colorimetric biosensor capable of detecting foodborne pathogens like Salmonella, with the naked eye in a simple and fast way, which demonstrates promising potential for sensor materials that could be extensively utilized in practical applications.
1. Introduction
Foodborne illnesses caused by pathogenic microorganisms like Salmonella, Listeria, Staphylococcus, and Escherichia coli pose a serious global threat to public health.1,2 Among these pathogens, Salmonella is particularly lethal and can lead to fatalities. Therefore, the urgent development of rapid and precise bacterial detection sensors is crucial to ensure food safety.The enzyme-linked immunosorbent assay (ELISA)3,4 and polymerase chain reaction (PCR)5,6 are traditional culture methods that provide high detection sensitivity and accuracy. They can be automated and used for large-scale testing with suitable equipment and software. However, they require specialized technical expertise and specific knowledge of genes or antibodies, potentially restricting their applicability to certain targets. Moreover, the analysis process is time-consuming, presenting challenges for obtaining real-time results. Hence, surface plasmon resonance (SPR),7–9 surface enhanced Raman scattering (SERS),10–12 and electrochemical methods like electrochemical impedance spectroscopy (EIS)13–15 or cyclic voltammetry (CV)16–18 have been employed to expedite detection. These methods provide rapid responses and high sensitivity, enabling the detection of low concentrations of target substances. However, akin to traditional methods, they are constrained by the requirement for specialized equipment and the elevated cost of result verification. However, similar to conventional techniques, their utility is restricted by the necessity for specialized equipment and substantial expense of result verification.
In order to address these issues, multiple ongoing studies are currently developing colorimetric biosensors that use polydiacetylene (PDA) to enable visual detection.19–21 PDA sensors display easily observable color changes, affordability, and versatile application potential, driving comprehensive research in this field. The polymerization of diacetylene (DA) to form PDA occurs through a photoreaction without the need for a chemical initiator, enabling the synthesis of high purity PDA without byproducts. The color characteristics of PDA arise from the π–π* absorption in the organized alternating ene–yne backbone.22 Because of their amphiphilic nature, DA monomers will spontaneously arrange in a water-based solution due to the van der Waals interactions occurring among the hydrophobic alkyl chains.23,24 Hence, when exposed to 254 nm UV irradiation, PDA supramolecules appear in blue. PDA, a conjugated polymer with distinct optical properties, is well-suited for creating color-changing and fluorescent sensors. It undergoes a chromatic transition from blue to red in response to external stimuli such as temperature,25 pH,26 light, organic solvents,27 and receptor binding.28 This color change is visually observable, cost-effective, and biocompatible. Interestingly, the PDA sensor can operate as a selective colorimetric sensor, detecting target substances with specificity.29 This is accomplished through the functionalization of receptor entities (e.g., ligands, lipids, aptamers, and antibodies) capable of recognizing specific substances, onto the surface of PDA liposomes. Among these, aptamers exhibit thermal stability and remarkable storage longevity due to their inherent chemical characteristics. Their cost-effectiveness and relative simplicity stem from their potential to chemically or enzymatically synthesize significant quantities of aptamers.30 These advantages have led to the development of numerous aptamer-based analysis systems (aptasensors) for diverse applications, including food safety, environmental monitoring, and disease diagnostics.31,32 Therefore, despite having a high detection limit, the PDA liposome–aptamer sensor demonstrates distinctiveness from existing Salmonella detection systems in that it allows for real-time confirmation of Salmonella concentrations at levels harmful to humans within a short period and visually. Moreover, it offers cost savings as it does not require specialized technical expertise and equipment.
In this study, we introduce a PDA-based liposome sensor capable of rapidly and accurately detecting Salmonella via a colorimetric response. The developed PDA liposome sensor is composed of a PDA liposome material surface-functionalized with an aptamer, a single-stranded DNA molecule exhibiting exceptional binding capability towards Salmonella. It is a notable achievement that the PDA liposome–aptamer sensor can detect Salmonella within less than 15 minutes through a colorimetric change observable to the naked eye. In contrast to previous intricate and time-intensive approaches, these results showcase a swift and straightforward real-time method for detecting foodborne pathogens like Salmonella, devoid of costly equipment or intricate preprocessing. These results demonstrate promising potential of sensor materials that can be extensively utilized in practical applications.
2. Experimental methods
2.1. Materials
10,12-Tricosadiynoic acid (TCDA, ≥98%), N-hydroxysuccinimide (NHS, 98%), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC-HCl), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), chloroform (ACS reagent, ≥99.8%), ethanolamine (≥98%) and potassium bromide (KBr, Fourier transform infrared (FTIR) grade, ≥99%) were purchased from Sigma-Aldrich. 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES, pH 7.0) was purchased from Biosolution. The ssDNA sequences (/5AmMC12/AGT AAT GCC CGG TAG TTA TTC AAA GAT GAG TAG GAA AAG A) were sourced from Integrated DNA Technologies (IDT).2.2. Synthesis of NHS-modified diacetylene monomers (TCDA-NHS)
TCDA-NHS monomers were synthesized following a previously reported method.33 10,12-Tricosadiynoic acid (TCDA, 0.72 mmol, 0.25 g), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC-HCl 1.35 mmol, 0.26 g) and N-hydroxysuccinimide (NHS, 1.07 mmol, 0.12 g) were dissolved in 4 mL of methylene chloride (Fig. S1, ESI†). The mixture was then stirred for 2 hours at room temperature. After the solution was evaporated using a rotary evaporator and the residue was purified by extraction with ethyl acetate, TCDA-NHS monomers were obtained as a white solid. The chemical structure of TCDA-NHS was confirmed by using 1H nuclear magnetic resonance (NMR, 300 MHz, chloroform-d, δ): ppm 2.85 (s, 4H), 2.61 (t, 2H), 2.25 (t, 4H), 1.79–1.26 (m, 28H), 0.89 (t, 3H).2.3. Preparation of PDA liposomes conjugated with aptamers
PDA liposomes conjugated with aptamers were synthesized using the established batch method.33 TCDA and TCDA-NHS were separately dissolved in chloroform in glass vials, and the solutions were combined in a 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of TCDA to TCDA-NHS. Following this, DMPC was added to the mixture solution at a 0.6 ratio and dissolved in chloroform to produce a 1 mM TCDA liposome solution. Next, the solution was filtered through a 0.2 μm syringe filter to eliminate any aggregated material or large particles. The solvent was then removed by nitrogen purging, resulting in a thin film on the flask's surface. Following this, 10 mL of deionized water was added to the flask. The solution was sonicated at 80 °C for 5 minutes until a translucent cloudy suspension was achieved. Subsequently, the solution was gradually cooled to room temperature and stored at 4 °C overnight. A 1% aptamer solution was conjugated to the liposome surface by 4-hour agitation. 0.13 mol of ethanolamine was added to inactivate the remaining unreacted NHS. To make an optically active PDA liposome, the PDA liposome–aptamer solution was exposed to UV light at 254 nm for approximately 30 minutes, during which the milky solution transformed into a blue colored solution by forming a conjugated backbone structure with an alternating ene–yne (C
1 molar ratio of TCDA to TCDA-NHS. Following this, DMPC was added to the mixture solution at a 0.6 ratio and dissolved in chloroform to produce a 1 mM TCDA liposome solution. Next, the solution was filtered through a 0.2 μm syringe filter to eliminate any aggregated material or large particles. The solvent was then removed by nitrogen purging, resulting in a thin film on the flask's surface. Following this, 10 mL of deionized water was added to the flask. The solution was sonicated at 80 °C for 5 minutes until a translucent cloudy suspension was achieved. Subsequently, the solution was gradually cooled to room temperature and stored at 4 °C overnight. A 1% aptamer solution was conjugated to the liposome surface by 4-hour agitation. 0.13 mol of ethanolamine was added to inactivate the remaining unreacted NHS. To make an optically active PDA liposome, the PDA liposome–aptamer solution was exposed to UV light at 254 nm for approximately 30 minutes, during which the milky solution transformed into a blue colored solution by forming a conjugated backbone structure with an alternating ene–yne (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C).
C).
2.4. Characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100). The FTIR spectra were acquired through transmission measurements across a scanning range of 3600–600 cm−1, with a resolution of 2 cm−1 and 16 scans.
100). The FTIR spectra were acquired through transmission measurements across a scanning range of 3600–600 cm−1, with a resolution of 2 cm−1 and 16 scans.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 minute and the supernatant was removed. For cell lysis, the cell pellet from each incubation was resuspended in 4 mL of B-PER® Bacterial Protein Extraction Reagent per 1 g of cell weight and left to react at room temperature for 10–15 minutes. Subsequently, the lysed bacteria were gradually diluted to a concentration range of 1 × 10 CFU mL−1 to 1 × 107 CFU mL−1 using 100 mM HEPES buffer. The final concentration of PDA liposomes was adjusted to 0.1 mM by mixing 300 μL of 1 mM PDA solution with 2700 μL of standard Salmonella solution diluted to the specified concentration. At a Salmonella concentration of 104 CFU mL−1, we monitored color changes and UV-visible spectra every 15 minutes for 60 minutes at 37 °C to track alterations in color and CR values over time. To assess the sensitivity and linearity of the PDA liposome–aptamer sensor, we varied the Salmonella concentration from 0 CFU mL−1 to 107 CFU mL−1. CR values for different Salmonella concentrations were determined with a 15-minute incubation time.
000 rpm for 1 minute and the supernatant was removed. For cell lysis, the cell pellet from each incubation was resuspended in 4 mL of B-PER® Bacterial Protein Extraction Reagent per 1 g of cell weight and left to react at room temperature for 10–15 minutes. Subsequently, the lysed bacteria were gradually diluted to a concentration range of 1 × 10 CFU mL−1 to 1 × 107 CFU mL−1 using 100 mM HEPES buffer. The final concentration of PDA liposomes was adjusted to 0.1 mM by mixing 300 μL of 1 mM PDA solution with 2700 μL of standard Salmonella solution diluted to the specified concentration. At a Salmonella concentration of 104 CFU mL−1, we monitored color changes and UV-visible spectra every 15 minutes for 60 minutes at 37 °C to track alterations in color and CR values over time. To assess the sensitivity and linearity of the PDA liposome–aptamer sensor, we varied the Salmonella concentration from 0 CFU mL−1 to 107 CFU mL−1. CR values for different Salmonella concentrations were determined with a 15-minute incubation time.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 minutes to eliminate any solid impurities in the egg white. Bacteria in the resulting supernatant were filtered using a 0.22 μm pore size filter. To examine the impact of the food matrix on the detection of S. typhimurium, a solution comprising 1 mL of the food matrix and 1 mL of the bacterial solution was prepared, and the volume was adjusted to 10 mL with a 100 mM HEPES buffer. Then, PDA liposome–aptamer sensor solutions were applied to capture S. typhimurium from real samples. The following operations were the same as those described in Section 2.4.6.
000 rpm for 10 minutes to eliminate any solid impurities in the egg white. Bacteria in the resulting supernatant were filtered using a 0.22 μm pore size filter. To examine the impact of the food matrix on the detection of S. typhimurium, a solution comprising 1 mL of the food matrix and 1 mL of the bacterial solution was prepared, and the volume was adjusted to 10 mL with a 100 mM HEPES buffer. Then, PDA liposome–aptamer sensor solutions were applied to capture S. typhimurium from real samples. The following operations were the same as those described in Section 2.4.6.
3. Results and discussion
3.1. Mechanism of colorimetric detection of Salmonella
Fig. 1 demonstrates the fabrication and sensing mechanism of the PDA liposome sensor material, enabling swift visual Salmonella detection. Polydiacetylene (PDA) monomers, bearing double and triple bonds, self-assemble into bilipid liposomes.35 UV or γ irradiation prompts the creation of an alternating ene–yne conjugated polymer backbone chain in PDA through 1,4-addition. As a consequence of the optical π–π* absorption of the polymer main chain, the resulting PDA exhibits a blue color with a maximum wavelength (λmax) of 640 nm.36,37 Different stimuli like temperature,25 pH,26 mechanical stress, or chemical bonding38 cause the blue PDA liposome to experience an energy gap increase between overlapping p-orbitals. This leads to an absorption spectrum expansion in the conjugated backbone, inducing a distinct color shift from blue to red with a maximum wavelength (λmax) of 550 nm.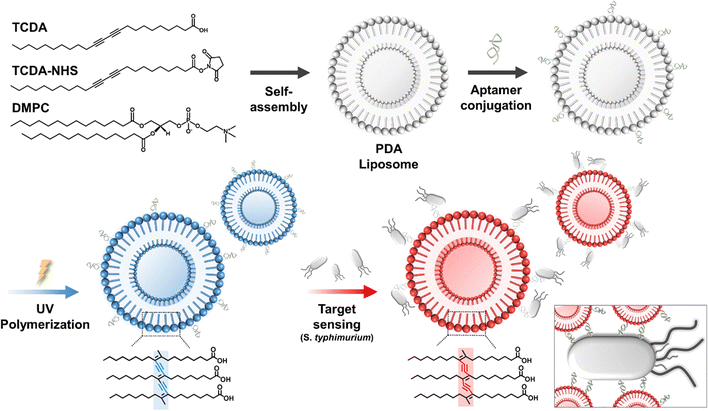 | ||
| Fig. 1 Schematic of the fabrication of the PDA liposome–aptamer sensor for Salmonella typhimurium detection. | ||
We engineered PDA-based liposomes with high-specificity aptamers to selectively bind to bacteria and pathogens. The nano-sized PDA vesicle was activated by N-hydroxysuccinimide (NHS) and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC-HCl) to form a reactive intermediate. The carboxyl group of the PDA vesicle and the amino group of the aptamer underwent peptide bonding, leading to the synthesis of PDA liposome–aptamer. After incorporating a Salmonella-specific single-stranded DNA aptamer onto the PDA liposomes, in an environment where bacteria are present, the bacteria specifically bind to the aptamer surface. The distorted conjugated backbone of the PDA liposome, induced by external stimuli, exhibits a visible color change from blue to red. Through this process, Salmonella can be easily and straightforwardly detected. The DNA sequence of the Salmonella-specific aptamer was AGT AAT GCC CGG TAG TTA TTC AAA GAT GAG TAG GAA AAG A, which was selected based on previous work.39 The aptamer sensor provides confirmation of Salmonella detection through molecular recognition and conformational changes in the PDA–liposome structure.
In this study, the PDA monomer used was 10,12-tricosadiynoic acid (TCDA). In addition, TCDA-NHS was employed as a co-monomer to achieve surface modification of liposomes with aptamers, wherein the NHS functional group of TCDA facilitated the formation of amide bonds with amine-terminated aptamers, thus enhancing the functionalization of the liposome surface. To enhance the structural flexibility of the liposomes and improve sensing sensitivity, the phospholipid 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was included. In the initial experimental phase, we investigated different DA monomers, phospholipid varieties and composition ratios. To simplify the experimental process, we chose liposome compositions based on assessing the thermal colorimetric response properties of various liposome components and ratios without incorporating aptamers (see Table S1, ESI†). The resulting optimal liposome composition was determined to be TCDA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TCDA-NHS
TCDA-NHS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMPC in a molar ratio of 0.9
DMPC in a molar ratio of 0.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1
0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.6 and was prepared using an established water bath method.33 These three components (TCDA, TCDA-NHS, and DMPC) self-assembled to form liposomes with a bilipid structure, as depicted in Fig. 1. The aminated aptamer was functionalized on the liposome surface by forming an amide bond with the NHS group of the TCDA-NHS. A blue PDA–liposome aptamer sensor was prepared successfully by sequentially conjugating the aptamer and undergoing UV polymerization processes.
0.6 and was prepared using an established water bath method.33 These three components (TCDA, TCDA-NHS, and DMPC) self-assembled to form liposomes with a bilipid structure, as depicted in Fig. 1. The aminated aptamer was functionalized on the liposome surface by forming an amide bond with the NHS group of the TCDA-NHS. A blue PDA–liposome aptamer sensor was prepared successfully by sequentially conjugating the aptamer and undergoing UV polymerization processes.
3.2. Characterization of the PDA liposome–aptamer sensor
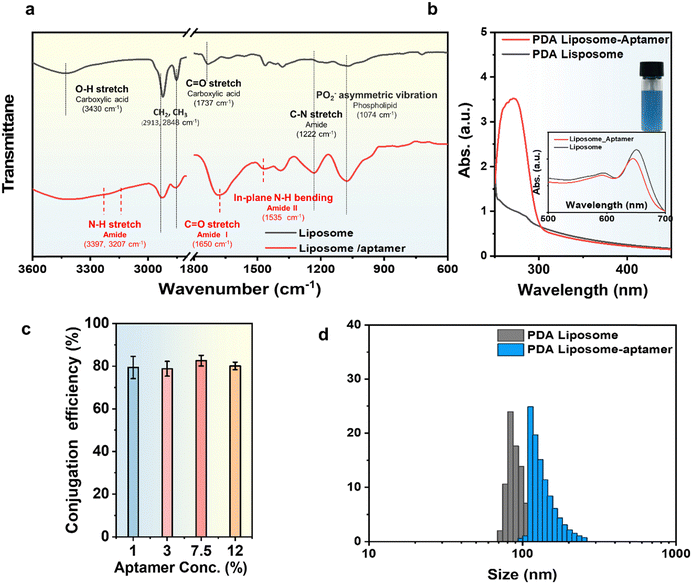
Fig. 2 shows the results of the structural analysis of the successfully prepared liposomes. To validate the successful conjugation of the aminated aptamer onto the liposome surface using the aforementioned method, we examined the UV-Vis and FT-IR spectra of liposome samples before and after aptamer attachment. Unconjugated aptamers were previously eliminated through dialysis.
Fig. 2a illustrates the FT-IR spectra of a liposome and a liposome conjugated with an aptamer. The characteristic peaks for the carboxylic acid of TCDA appeared at 3430 cm−1 (O–H stretching) and 1737 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching). The characteristic peak of the phospholipid was identified at 1074 cm−1 (PO2 asymmetric vibration). The amine group of the aptamer reacts with the carboxylic acid (–COOH) of TCDA, resulting in the formation of an amide bond. The presence of these amide bonds is confirmed by the characteristic peaks observed at 3397 cm−1 and 3207 cm−1 (N–H stretching), 1650 cm−1 (C
O stretching). The characteristic peak of the phospholipid was identified at 1074 cm−1 (PO2 asymmetric vibration). The amine group of the aptamer reacts with the carboxylic acid (–COOH) of TCDA, resulting in the formation of an amide bond. The presence of these amide bonds is confirmed by the characteristic peaks observed at 3397 cm−1 and 3207 cm−1 (N–H stretching), 1650 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), and 1535 cm−1 (in-plane N–H bending) in the FT-IR spectrum of the liposome–aptamer sample. These results demonstrate the successful chemical conjugation of the aptamer to the PDA–liposome.
O stretching), and 1535 cm−1 (in-plane N–H bending) in the FT-IR spectrum of the liposome–aptamer sample. These results demonstrate the successful chemical conjugation of the aptamer to the PDA–liposome.
In addition, the conjugation between liposomes and aptamers can also be confirmed by UV-Vis spectral investigation (Fig. 2b). The aptamer used in this study is a single-stranded DNA comprised of the four nucleotide bases: guanine (G), cytosine (C), adenine (A), and thymine (T). The aromatic rings within the purine and pyrimidine structures of guanine, cytosine, adenine, and thymine have absorbance in the 260 nm region (Fig. S2, ESI†).40 As depicted in Fig. 2b, aptamer-conjugated liposomes (PDA liposome–aptamer) have a high absorbance at 260 nm, a characteristic peak attributed to the aptamer nucleotide. The UV-Vis spectra were employed to quantitatively determine the conjugation efficiency of the aptamer to the liposome. This determination involves correlating the attached aptamer amount with the initially fed aptamer quantity by measuring the absorbance at 260 nm and comparing it to the aptamer concentration in the standard sample. Fig. 2c demonstrates the modulation of aptamer concentration in relation to the PDA monomer, ranging from 1 to 12 mol% (Fig. S3, ESI†). Notably, the aptamer consistently exhibited a chemical binding efficiency of approximately 80% relative to its initial introduced content in all instances. A PDA liposome–aptamer with 1% aptamer concentration was selected as the sensor material, as attaching aptamer concentrations exceeding 3% to PDA liposomes led to reduced polymerization efficiency. Fig. 2d presents the results of particle size analysis (PSA), validating the size of liposomes prepared using the mentioned bath method. The initial size of the PDA liposome was determined to be approximately 100 nm. After aptamer conjugation, the size of the liposome particles slightly increased to around 137 nm. This outcome is attributed to the presence of an extra hydrophilic molecule.
3.3. Feasibility of Salmonella detection
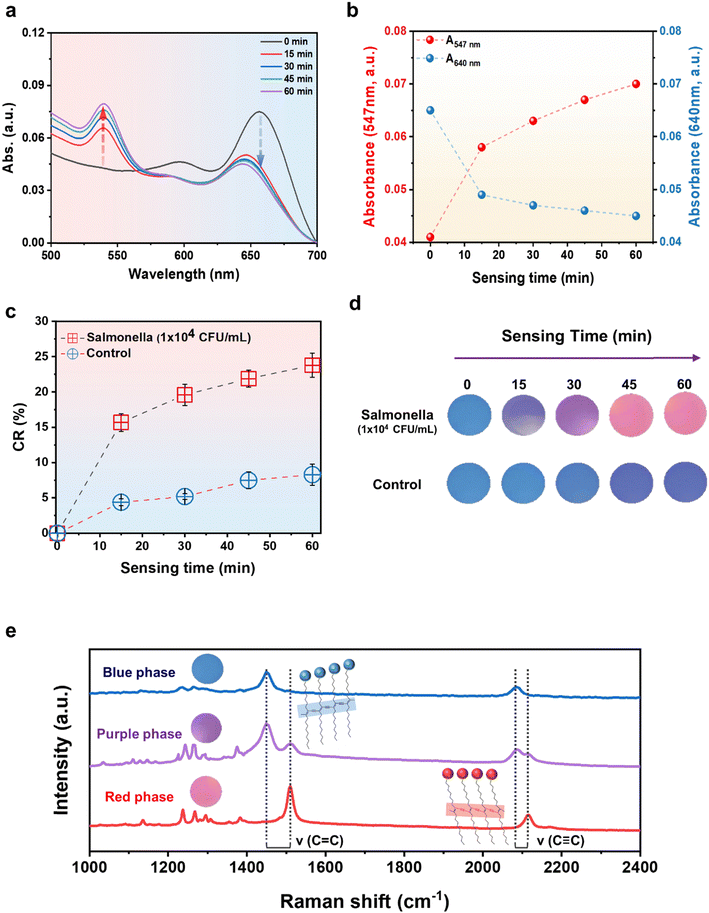
The developed PDA–aptamer liposomal sensor material possesses characteristics that enable the naked-eye detection of Salmonella within 15 minutes through a rapid and precise colorimetric reaction. Fig. 3 illustrates that the PDA liposome–aptamer sensor, loaded with Salmonella at a concentration of 1 × 104 CFU mL−1, undergoes a rapid visible color transition from blue to purple within 15 minutes. Furthermore, after 45 minutes, the sensor material transforms into a deep pink color. This change can be attributed to the fact that binding of a Salmonella-specific aptamer to Salmonella triggers surface interaction, leading to a conformational change in the PDA-conjugated chain and resulting in a color shift from blue to red.41UV-Vis spectrum measurements were performed over sensing time to investigate the alterations in optical properties within the PDA liposome–aptamer resulting from the introduction of Salmonella at a concentration of 1 × 104 CFU mL−1. Upon adding Salmonella to the prepared PDA liposome–aptamer solution, a gradual decrease in absorption in the 640 nm region and an increase in absorption in the 547 nm region were observed over time (Fig. 3a and b), indicating a typical blue-to-red transition of the PDA liposome–aptamer. As illustrated in Fig. 3b, during the initial phase (t < 15 min), the absorption at 640 nm diminished while the absorption at 547 nm simultaneously increased. A pronounced color alteration became apparent at the 15-minute mark. Additionally, at the 60-minute interval, the absorption peaks at 640 and 547 nm reached their zenith.
The quantitative assessment of changes in the absorbance spectrum can be determined by the colorimetric response (CR) value calculated as follows:
Fig. 3c and d demonstrates the clear color change of the sample, turning purple at 15 minutes and pink at 45 minutes, with corresponding CR values of 15.7% and 21.9%, respectively. A CR value of 7–8% or higher is commonly considered to be a value that can clearly distinguish the color change with the naked eye; the CR value of the PDA liposome–aptamer sensor exceeds 15.7% within 15 minutes, which indicates a high detection efficiency of Salmonella Interestingly, the control condition with a Salmonella concentration of 0 CFU mL−1 shows no significant color change in the PDA liposome–aptamer material after 30 minutes, maintaining its initial blue color, with color changes contributing to less than 6% CR value (Fig. 3d). However, for a more accurate comparison, it is recommended to consider color changes occurring within 30 minutes, as the CR value of the control sample reaches 8.3% at 60 minutes. These results hold significance as they allow for quick and easy confirmation of the presence of Salmonella using naked-eye observation. This approach contrasts with conventional detection methods that are known for their complexity and time-consuming nature.3
As shown in Fig. 3e, the blue-to-red chromatic transition of the PDA liposome was confirmed using Raman spectroscopy with a laser excitation wavelength of 633 nm. In the blue phase, two bands associated with the conjugated alkyne–alkene structure were observed at 2082 cm−1 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C) and 1450 cm−1 (C
C) and 1450 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) in the PDA liposome–aptamer. The Raman spectrum of the purple phase PDA liposome–aptamer material, after 15 minutes of inoculation with Salmonella (1 × 104 CFU mL−1), revealed a partial shift of the alkyne–alkene band to higher frequencies at 2122 cm−1 and 1817 cm−1, indicating a partial distortion of the conjugated yne–ene chain. However, the red phase, which was exposed to Salmonella for a longer period (45 minutes), displays a notable upward shift of the alkyne–alkene band to higher frequencies, which demonstrates that the increased binding of numerous Salmonella fragments to the aptamer leads to the full distortion of the conjugated yne–ene chain.
C) in the PDA liposome–aptamer. The Raman spectrum of the purple phase PDA liposome–aptamer material, after 15 minutes of inoculation with Salmonella (1 × 104 CFU mL−1), revealed a partial shift of the alkyne–alkene band to higher frequencies at 2122 cm−1 and 1817 cm−1, indicating a partial distortion of the conjugated yne–ene chain. However, the red phase, which was exposed to Salmonella for a longer period (45 minutes), displays a notable upward shift of the alkyne–alkene band to higher frequencies, which demonstrates that the increased binding of numerous Salmonella fragments to the aptamer leads to the full distortion of the conjugated yne–ene chain.
As mentioned earlier, we selected a Salmonella-specific aptamer (sequence: AGTAATGCC CGGTAGTTATTCAAAGATGAGTAGGAAAAGA) for this experiment based on previously reported papers.39 As shown in the above experiment, the PDA liposome–aptamer sensor confirmed a clear colorimetric reaction from blue to red of the PDA liposome material through the specific binding of the aptamer to Salmonella. Comparative experiments were performed to investigate the function of Salmonella-selective aptamers in PDA liposome sensors (see Fig. S4, ESI†). For the purpose of comparison, we categorized liposomal materials into three types. Type 1 liposomes, lacking aptamer functionalization, and type 3 liposomes, possessing non-Salmonella-specific aptamers, do not exhibit a specific colorimetric response to Salmonella. Conversely, type 2, equipped with Salmonella-specific aptamers, demonstrates a noticeable color alteration in the presence of Salmonella. These experimental findings reveal that the particular colorimetric reaction of the PDA liposome–aptamer sensor to Salmonella results from the specific interaction between the aptamer and Salmonella.
3.4. Sensitivity and limit of detection (LOD) range
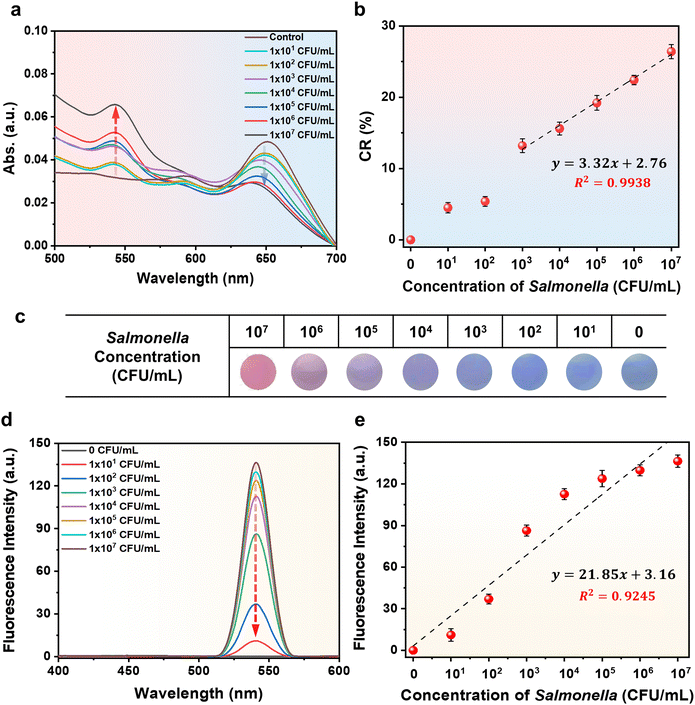
Next, we focused our attention towards evaluating the PDA liposome–aptamer and its chromatic transition properties in response to changing Salmonella concentrations. PDA liposome–aptamer solutions were exposed to different concentrations of S. typhimurium, ranging from 1 × 101 CFU mL−1 to 1 × 107 CFU mL−1, for a duration of 15 minutes at 37 °C. To monitor the blue-to-red transition upon addition of Salmonella, we recorded the UV-Vis spectrum of each sample (Fig. 4a). As anticipated, the introduction of Salmonella results in a characteristic blue-to-red color transition, which becomes more pronounced with higher concentrations of Salmonella. In Fig. 4b, an excellent linear correlation between CR% and Salmonella concentration is observed within the range of 1 × 103 CFU mL−1 to 1 × 107 CFU mL−1, with a linear response described by the equation y = 3.32x + 2.76 (R2 = 0.99). Based on this correlation curve, it is evident that quantitative analysis of an unknown Salmonella concentration is achievable.A photograph of a PDA liposome–aptamer solution exposed to different concentrations of Salmonella during a 15-minute incubation at 37 °C is shown in Fig. 4c. A noticeable color change, although subtle, in comparison to the control, is evident at Salmonella concentrations as low as 1 × 103 CFU mL−1. Furthermore, concentrations surpassing 1 × 104 CFU mL−1 display a distinct and substantial color change. These results indicate that the limit of detection (LOD) for Salmonella on the prepared PDA liposome–aptamer sensor is 1 × 103 CFU mL−1, which is comparable to the lowest value of the visual detection limit in a previously reported study on Salmonella detection sensors.42,43 The detection capability of the PDA liposome sensor is highly significant as it enables the naked-eye identification of Salmonella at concentrations above 1 × 103 CFU mL−1, which is reported as a human health hazard,44 within a rapid timeframe of 15 minutes.
The conjugated backbone of PDA liposomes, distorted by external stimuli, is known to exhibit strong fluorescent properties in addition to the well-known blue-to-red color change. We observed the fluorescence properties of PDA liposome–aptamer sensors exposed to various concentrations of Salmonella for a duration of 15 minutes at 37 °C (Fig. 4d).
As expected, the PDA liposome–aptamer emitted strong fluorescence at 550 nm with increasing Salmonella concentration. The fluorescence intensity of the PDA liposome–aptamer was also plotted against the Salmonella concentration (Fig. 4e). A linear correlation (R2 = 0.92) was obtained over the concentration range of 1 × 101–1 × 107 CFU mL−1.
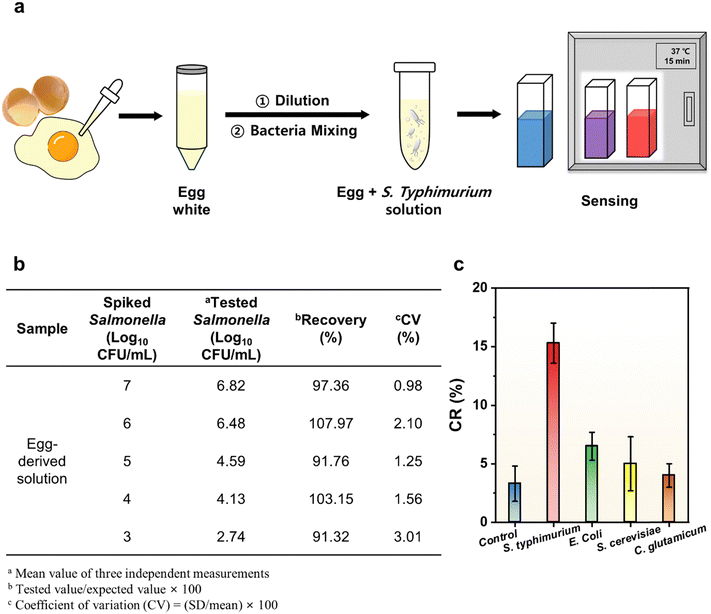
3.5. Detection of S. typhimurium in eggs and recovery
In order to evaluate the feasibility of the sensor, Salmonella in actual food samples was detected using a PDA liposome–aptamer sensor. Fig. 5a shows the general procedure for food sample treatment and the sensing procedure. An egg white served as the food matrix and various concentrations of Salmonella (103, 104, 105, 106, and 107 CFU mL−1) were spiked. The procedures for preparing the food matrix and bacterial spiking were adopted from a previous paper.34 Then, PDA liposome–aptamer sensor solutions were applied to capture S. typhimurium from real samples at 37 °C for 15 minutes and the color change was monitored. Fig. 5b shows the recovery and coefficient of variation (CV) of the PDA liposome–aptamer sensor in the food matrix. The recoveries from all samples range from 91.32 to 107.97%. The recoveries for each sample were achieved as follows: 91.32% for 103 CFU mL−1, 103.15% for 104 CFU mL−1, 91.76% for 105 CFU mL−1, 107.97% for 106 CFU mL−1, and 97.36% for 107 CFU mL−1Salmonella. The reproducibility of the PDA liposome–aptamer sensor was also evaluated by measuring its response to Salmonella concentrations ranging from 103 to 107 CFU mL−1. The repeatability of the sensor was measured three times independently under the same working conditions, and the coefficient of variation (CV) values for concentrations of 103, 104, 105, 106, and 107 CFU mL−1 were calculated to be 0.98%, 2.10%, 1.25%, 1.56% and 3.01%, respectively. The above results demonstrate the high reproducibility and repeatability of the sensor we have developed. In addition to the colorimetric response, strong fluorescence characteristics of the PDA liposome–aptamer sensor were also observed in authentic food samples (Fig. S5, ESI†).To assess the specificity of the PDA liposome–aptamer sensor for S. typhimurium, we examined its response to different strains, including S. typhimurium, E. coli, S. cerevisiae, and C. glutamicum. The sensing characteristics of the PDA liposome–aptamer for each strain were assessed by incubation at 37 °C for 15 minutes with a concentration of 1 × 104 CFU mL−1 and the CR values were determined using a previously described method. As depicted in Fig. 5c, the prepared PDA liposome–aptamer sensor demonstrates a markedly elevated CR value (15.3%) for S. typhimurium, in contrast to the other experimental groups with CR values below 7%. Notably, the resemblance of the CR values for S. cerevisiae and C. glutamicum to those of the control samples implies that the PDA liposome–aptamer sensors have a distinct color response to S. typhimurium. These results can be attributed to the unique ability of the aptamers to specifically bind to Salmonella. Furthermore, the PDA–liposome aptamer sensor material exhibits a discriminating and enhanced colorimetric reaction to Salmonella when compared to E. coli, a common bacterium that could be found in food samples. Remarkably, this specificity is visually confirmable, unequivocally validating the sensitive and precise Salmonella detection achieved by the PDA liposome–aptamer sensor.
Table 1 summarizes and contrasts the attributes of various Salmonella detection sensors, including our newly devised PDA sensor. The PDA liposome–aptamer sensor demonstrates a rapid detection time of 15 minutes, markedly shorter than those of traditional techniques like PCR5 and ELISA.3 Furthermore, distinct from SPR,7 SERS,10 and electrochemical methods13,14,16,17 requiring costly apparatus, our sensor uniquely detects Salmonella at concentrations surpassing 1 × 103 CFU mL−1, a level hazardous to humans, within a 15-minute timeframe through direct visual assessment, notwithstanding its relatively higher detection threshold. Our novel devised PDA liposome–aptamer sensor enables naked-eye Salmonella detection within a brief timeframe (15 minutes), contrasting with earlier colorimetric approaches (5 hours for a DNAzyme-decorated gold nanoparticle sensor45 and 48 hours for a PCDA/SPH/CHO/Lysine sensor46). Consequently, the Salmonella-specific PDA liposome–aptamer sensor holds significant potential for swift visual Salmonella detection.
| Detection techniques | Detection method | Detection time | Materials | Linear detection range (CFU mL−1) | Limit of detection (CFU mL−1) | Ref. |
|---|---|---|---|---|---|---|
| PCR | qPCR | Several days | — | — | 5 | |
| ELISA | Microplate reader | 24 hours | 1.4 × 105 | — | 3 | |
| SPR | SPR assay | 4 hours | 3.2 × 104–105 | 5.0 × 104 | 7 | |
| SERS | Raman | — | — | 1.0 × 102–107 | 15 | 10 |
| Electrochemical detecting | EIS, CV | 1 hour | 1.0 × 103–107 | 5.0 × 102 | 13 | |
| EIS | 2 hours | 1.0 × 102–106 | 80 | 14 | ||
| DPV | 1 hour | 1.0 × 10–108 | 10 | 16 | ||
| DPV | 2 hours | 9.6 × 1–10 | 8.1 | 17 | ||
| Colorimetric | Naked-eye | 5 hours | DNAzyme probe self-assembled GN | 3.0 × 103–106 | 3.0 × 103 | 40 |
| Naked-eye | 48 hours | PCDA/SPH/CHO/Lysine | 1.0 × 1–108 | 10 | 41 | |
| Naked-eye | 15 min | PDA liposome/aptamer | 1.0 × 103–107 | 1.0 × 103 | This work |
4. Conclusions
We have successfully engineered a colorimetric biosensor with the capability to rapidly detect Salmonella through direct visual observation. The material composition of this biosensor was meticulously formulated by creating liposomes via the combination of TCDA:TCDA-NHS:DMPC, followed by a process of aptamer surface functionalization and subsequent UV polymerization, resulting in an affinity specifically tailored for Salmonella recognition. The synthesized biosensor framework showcases multiple advantageous aspects compared to established Salmonella detection methodologies and alternative sensing paradigms.Foremost, the facile synthesis of both PDA liposomes and aptamers in considerable quantities presents a cost-effective and straightforward strategy for biosensor fabrication. Furthermore, the biosensor exhibited a distinct color transformation (CR: 25% for 1 × 104 CFU mL−1) within a rapid 15-minute window at a temperature of 37 °C. Additionally, a linear correlation of CR values was observed across a spectrum of Salmonella concentrations. Impressively, Salmonella concentrations surpassing 1 × 103 CFU mL−1 were visibly detectable without the necessity for sophisticated analytical instrumentation. Concurrently, we have successfully demonstrated the biosensor's efficacy in detecting Salmonella in authentic food samples, thus underscoring the sensor's adaptability and its potential relevance in everyday scenarios.
Collectively, these empirical observations substantiate the biosensor's effectiveness and accessibility in detecting the pathogenic bacterium Salmonella, with the prospective capacity to significantly impact public health. Subsequent research efforts could focus on refining and optimizing the Salmonella detection approach, potentially encompassing the development of paper-based sensor arrays or the conception of user-friendly film-based kit configurations.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Big Issue Program (PEO24030) of the Korea Institute of Industrial Technology (KITECH), Republic of Korea.References
- R. V. Tauxe, Emerging Infect. Dis., 1997, 3, 425–434 CrossRef CAS PubMed
.
- S. Finstad, C. A. O’Bryan, J. A. Marcy, P. G. Crandall and S. C. Ricke, Food Res. Int., 2012, 45, 789–794 CrossRef
.
- Y. He, Y. Ren, B. Guo, Y. Yang, Y. Ji, D. Zhang, J. Wang, Y. Wang and H. Wang, Food Chem., 2020, 25, 125942 CrossRef PubMed
.
- L. P. Mansfield and S. J. Forsythe, Food Microbiol., 2001, 18, 361–366 CrossRef CAS
.
- M. Siala, A. Barbana, S. Smaoui, S. Hachicha, C. Marouane, S. Kammoun, R. Gdoura and F. Messadi-Akrout, Front. Microbiol., 2017, 8, 1–10 Search PubMed
.
- D. De Medici, L. Croci, E. Delibato, S. Di Pasquale, E. Filetici and L. Toti, Appl. Environ. Microbiol., 2003, 69, 3456–3461 CrossRef CAS PubMed
.
- D. Bhandari, F.-C. Chen and R. C. Bridgman, Sensors, 2022, 22, 475 CrossRef CAS PubMed
.
- B. K. Oh, Y. K. Kim, K. W. Park, W. H. Lee and J. W. Choi, Biosens. Bioelectron., 2004, 19, 1497–1504 CrossRef CAS PubMed
.
- A. D. Taylor, J. Ladd, Q. Yu, S. Chen, J. Homola and S. Jiang, Biosens. Bioelectron., 2006, 22, 752–758 CrossRef CAS PubMed
.
- C. Wei, M. Li and X. Zhao, Front. Microbiol., 2018, 9, 1–9 CrossRef PubMed
.
- X. Ma, X. Xu, Y. Xia and Z. Wang, Food Control, 2018, 84, 232–237 CrossRef CAS
.
- Q. Yu, T. Wu, B. Tian, J. Li, Y. Liu, Z. Wu, X. Jin, C. Wang, C. Wang and B. Gu, Anal. Chim. Acta, 2023, 1286, 341931 CrossRef PubMed
.
- J. Dong, H. Zhao, M. Xu, Q. Maa and S. Ai, Food Chem., 2013, 141, 1980–1986 CrossRef CAS PubMed
.
- L. Wang, X. Huo, W. Qi, Z. Xia, Y. Li and J. Lin, Talanta, 2020, 211, 120715 CrossRef CAS PubMed
.
- P. Wang, B. Luo, K. Liu, C. Wang, H. Dong, X. Wang, P. Hou and A. Li, RSC Adv., 2022, 12, 27940–27947 RSC
.
- S. Muniandy, S. J. Teh, J. N. Appaturi, K. L. Thong, C. W. Lai, F. Ibrahim and B. F. Leo, Bioelectrochemistry, 2019, 127, 136–144 CrossRef CAS PubMed
.
- Y. Ye, W. Yan, Y. Liu, S. He, X. Cao, X. Xu, H. Zheng and S. Gunasekaran, Anal. Chim. Acta, 2019, 1074, 80–88 CrossRef CAS PubMed
.
- S. Muniandy, K. L. Thong, J. N. Appaturi, C. W. Lai and B. F. Leo, Sens. Diagn., 2022, 1, 1209–1217 RSC
.
- M. Weston, A. H. Pham, J. Tubman, Y. Gao, A. D. Tjandra and R. Chandrawati, Mater. Adv., 2022, 3, 4088–4102 RSC
.
- T. N. Nguyen, V. D. Phung and V. Van Tran, Biosensors, 2023, 13, 586 CrossRef CAS PubMed
.
- J. Chen, C. Wang, X. Qin, X. Yang, C. Yang, H. Nie, H. Chen and H. Li, Coord. Chem. Rev., 2023, 497, 215433 CrossRef CAS
.
- J. T. Wen, J. M. Roper and H. Tsutsui, Ind. Eng. Chem. Res., 2018, 57, 9037–9053 CrossRef CAS
.
- R. W. Carpick, D. Y. Sasaki, M. S. Marcus, M. A. Eriksson and A. R. Burns, J. Phys.: Condens. Matter, 2004, 16, 679–697 CrossRef
.
- D. C. Lee, S. K. Sahoo, A. L. Cholli and D. J. Sandman, Macromolecules, 2002, 35, 4347–4355 CrossRef CAS
.
- D. E. Wang, L. Zhao, M. Sen Yuan, S. W. Chen, T. Li and J. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 28231–28240 CrossRef CAS PubMed
.
- D. J. Ahn, E. H. Chae, G. S. Lee, H. Y. Shim, T. E. Chang, K. D. Ahn and J. M. Kim, J. Am. Chem. Soc., 2003, 125, 8976–8977 CrossRef CAS PubMed
.
- J. Yoon, S. K. Chae and J. M. Kim, J. Am. Chem. Soc., 2007, 129, 3038–3039 CrossRef CAS PubMed
.
- H. Shen, J. Wang, H. Liu, Z. Li, F. Jiang, F. B. Wang and Q. Yuan, ACS Appl. Mater. Interfaces, 2016, 8, 19371–19378 CrossRef CAS PubMed
.
- S. Hussain, R. Deb, S. Suklabaidya, D. Bhattacharjee and S. Arshad Hussain, Mater. Today: Proc., 2022, 65, 2765–2772 CAS
.
- S. Ni, Z. Zhuo, Y. Pan, Y. Yu, F. Li, J. Liu, L. Wang, X. Wu, D. Li, Y. Wan, L. Zhang, Z. Yang, B. T. Zhang, A. Lu and G. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 9500–9519 CrossRef CAS PubMed
.
- H. R. Jia, Z. Zhang, X. Fang, M. Jiang, M. Chen, S. Chen, K. Gu, Z. Luo, F. G. Wu and W. Tan, Mater. Today Nano, 2022, 18, 100188 CrossRef CAS
.
- J. Chen, C. Yang, H. Nie and H. Li, Spectrochim. Acta, Part A, 2023, 293, 122451 CrossRef CAS PubMed
.
- A. D. Tjandra, M. Weston, J. Tang, R. P. Kuchel and R. Chandrawati, Colloids Surf., A, 2021, 619, 126497 CrossRef CAS
.
- Y. Jiao, Z. Zhang, K. Wang, H. Zhang and J. Gao, Food Chem.: X, 2023, 19, 100798 CAS
.
- Y. K. Jung, T. W. Kim, J. Kim, J. M. Kim and H. G. Park, Adv. Funct. Mater., 2008, 18, 701–708 CrossRef CAS
.
- S. Lee, J. Y. Kim, X. Chen and J. Yoon, Chem. Commun., 2016, 52, 9178–9196 RSC
.
- X. Sun, T. Chen, S. Huang, L. Li and H. Peng, Chem. Soc. Rev., 2010, 39, 4244–4257 RSC
.
- J. Lee, H. J. Kim and J. Kim, J. Am. Chem. Soc., 2008, 130, 5010–5011 CrossRef CAS PubMed
.
- N. Duan, S. Wu, X. Chen, Y. Huang, Y. Xia, X. Ma and Z. Wang, J. Agric. Food Chem., 2013, 61, 3229–3234 CrossRef CAS PubMed
.
- C. Sester, J. A. J. McCone, A. Sen, J. Vorster, J. E. Harvey and J. M. Hodgkiss, Biophys. J., 2022, 121, 2193–2205 CrossRef CAS PubMed
.
- R. Joshi, H. Janagama, H. P. Dwivedi, T. M. A. Senthil Kumar, L. A. Jaykus, J. Schefers and S. Sreevatsan, Mol. Cell. Probes, 2009, 23, 20–28 CrossRef CAS PubMed
.
- Y. Fu, J. Wei, S. Yao, L. Zhang, M. Zhang, X. Zhuang, C. Zhao, J. Li and B. Pang, Microchim. Acta, 2022, 189, 218 CrossRef CAS PubMed
.
- S. Du, Z. Lu, L. Gao, Y. Ge, X. Xu and H. Zhang, Microchim. Acta, 2020, 187, 627 CrossRef CAS PubMed
.
- L. Zheng, G. Cai, W. Qi, S. Wang, M. Wang and J. Lin, ACS Sens., 2020, 5, 65–72 CrossRef CAS PubMed
.
- R. Luo, Y. Li, X. Lin, F. Dong, W. Zhang, L. Yan, W. Cheng, H. Ju and S. Ding, Sens. Actuators, B, 2014, 198, 87–93 CrossRef CAS
.
- T. V. De Oliveira, N. D. F. F. Soares, N. J. De Andrade, D. J. Silva, E. A. A. Medeiros and A. T. Badaró, Food Chem., 2015, 172, 428–432 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00840a |
| This journal is © The Royal Society of Chemistry 2024 |