Open Access Article
Open Access ArticlePorous microwell scaffolds for 3D culture of pancreatic beta cells to promote cell aggregation and insulin secretion
Huajian
Chen
 a,
Tianjiao
Zeng
ab,
Toru
Yoshitomi
a,
Tianjiao
Zeng
ab,
Toru
Yoshitomi
 a,
Naoki
Kawazoe
a,
Hirotake
Komatsu
c,
Yingnan
Yang
d and
Guoping
Chen
a,
Naoki
Kawazoe
a,
Hirotake
Komatsu
c,
Yingnan
Yang
d and
Guoping
Chen
 *ab
*ab
aResearch Center for Macromolecules and Biomaterials, National Institute for Materials Science, 1-1 Namiki, Tsukuba, Ibaraki 305-0044, Japan. E-mail: Guoping.CHEN@nims.go.jp; Fax: 81-29-860-4673; Tel: 81-29-860-4496
bGraduate School of Science and Technology, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8577, Japan
cDepartment of Translational Research & Cellular Therapeutics, Arthur Riggs Diabetes & Metabolism Research Institute of City of Hope, 1500 E. Duarte Rd., Duarte, CA 91010, USA
dGraduate School of Life and Environmental Science, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8572, Japan
First published on 15th January 2024
Abstract
Malfunctioning of the pancreas leads to insufficient insulin secretion from pancreatic beta cells in type 1 diabetes mellitus patients. Even though transplantation of pancreatic beta cells is making progress in diabetic treatment, further promoting insulin secretion of pancreatic beta cells remains a challenge. In this study, gelatin–PLGA mesh based porous microwell scaffolds were prepared for three-dimensional (3D) culture of pancreatic beta cells to promote their aggregation and insulin secretion functions. The microwell structure was fabricated on a biodegradable polymer mesh by using an ice particulate template method. The size of microwells could be controlled by the dimension of ice particulates. The microwells had a concave morphology surrounded by dense ultra-small pores. RIN-5F cells cultured in the porous microwell scaffolds had high viability and formed grape-like aggregates. More importantly, insulin secretion of RIN-5F cells was enhanced by culturing in the porous microwell scaffolds compared to culturing in flat scaffold and normal cell culture plates. The results suggested that the porous microwell scaffolds promoted aggregation formation and insulin secretion of pancreatic beta cells. The porous microwell scaffolds hold high potential as a novel 3D culture model for diabetes mellitus.
Introduction
Type 1 diabetes mellitus (T1DM) is a chronic disease with insufficient insulin secretion of pancreas that leads to dysregulation of blood glucose homeostasis and other serious complications.1,2 Insulin injection is the most common strategy that clinically maintains glucose homeostasis and relieves complications in T1DM patients. However, lifetime injection of insulin is accompanied by tremendous pains and inconveniences.3,4 Therefore, cell therapy has emerged as an alternative and prospective strategy for treatment of T1DM.5–8 Pancreatic islets are the functional units in pancreas regulating blood glucose homeostasis.9 Pancreatic islets consist of different types of endocrine cells. Among them, pancreatic beta cells are in charge of insulin secretion.10,11 Thus, transplantation of beta cells is a superior way of insulin administration compared to conventional insulin injection for T1DM treatment.12–14 Pancreatic beta cells should be maintained at high viability and insulin secretion functionality for cell therapy. Therefore, it is pivotal to develop an optimal culture strategy to meet the requirements and achieve the expected therapeutic effect of transplantation of pancreatic beta cells.Conventional 2D monolayer culture of pancreatic beta cells shows lower insulin secretion, which is inadequate for cell therapy of T1DM.15 Many studies have indicated that the spheroid structure of islets is crucial to keep functions of endocrine cells.16–18 Thus, some cell culture methods have been developed to promote formation of a spheroid structure and insulin secretion function of beta cells.19–21 For instance, beta cells have been cultured in ultra-low-attachment plates under constant shaking to form islet-like aggregates.22,23 Although insulin secretion of beta cells is improved due to the formation of cell aggregates, the size of beta cell aggregates has a wide distribution and large size of aggregates affects cell viability. Hydrogels and concave microwells have also been studied for cell aggregate formation to promote insulin secretion.24–31 Hydrogels such as thiol–ene hydrogels have been used for culture of pancreatic beta cells for the formation and retrieval of cell spheroids.24 The pancreatic beta cells encapsulated in the hydrogels have higher insulin secretion capacity than the cells cultured in 2D culture systems.24,25 Concave microwells prepared from polydimethylsiloxane (PDMS) and poly(ethylene glycol) (PEG) hydrogel have been used for 3D culture of beta cells to mimic the pancreatic structure.26–30
These previous studies suggest that concave microwell structure has a promotive effect on cell aggregation and spheroid formation.27–32 However, PDMS and PEG used for the microwell preparation are non-degradable materials. Biodegradable biomaterials are desirable for the fabrication of microwell structures that can be directly implanted after the formation of pancreatic beta cell aggregates. Moreover, porous structures should be considered to benefit the diffusion of nutrients and oxygen to maintain high viability of transplanted pancreatic beta cells.33
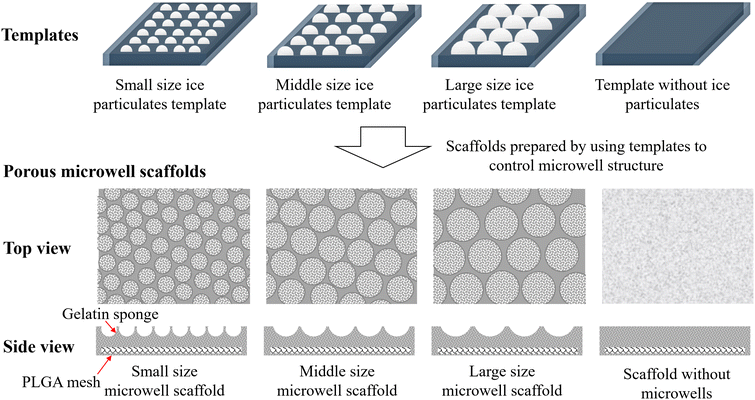
In this research, porous microwell scaffolds of biodegradable polymers of gelatin and poly(lactic-co-glycolic acid) (PLGA) were prepared by using ice particulates (Scheme 1). Gelatin porous microwells were fabricated on a PLGA knitted mesh to construct gelatin–PLGA porous microwell scaffolds. The porous microwell scaffolds facilitated the aggregate formation of pancreatic beta cells and promoted insulin secretion. The scaffolds should be a good platform to control the functions of pancreatic beta cells for cell therapy of T1DM.
Materials and methods
Preparation and characterization of gelatin–PLGA porous microwell scaffolds
Gelatin–PLGA porous microwell scaffolds were prepared by using an ice particulate template method.34–36 At first, a copper plate (8 cm × 8 cm) was covered with a Teflon film and placed in a sealed chamber. A moist environment was created by constantly filling chamber with pure water vapor generated by a vaporizer. A water droplet array was formed on the plate under exposure to the moist environment. Moisture exposure time of the Teflon film-covered copper plate was controlled at 10, 15 and 20 min to prepare different sizes of water droplets. Then, the water droplet array was frozen in a −80 °C freezer for 30 min to form the ice particulate template. In the meantime, a PLGA mesh (Vicryl knitted mesh made of copolymer of glycolic acid and lactic acid at a ratio of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, Ethicon Inc., USA) was placed on a glass plate wrapped with a plastic film. The surrounding of the PLGA mesh was covered by a silicone frame (1 mm in thickness). 5% (wt%) gelatin aqueous solution was poured on the PLGA mesh in the silicone frame and the gelatin solution surface was flattened (1 mm in thickness) at room temperature. The construct was transferred into a low temperature chamber at −2 °C for 2 min to cool the construct without freezing. The ice particulate template was transferred from the freezer to the low temperature chamber and immediately placed on the top of the pre-cooled construct to allow the ice particulates facing the gelatin solution. The construct was immediately transferred to the −80 °C freezer and frozen for 3 h, followed by 48 h of freeze-drying. After freeze-drying, the scaffolds were immersed in ethanol and cross-linked with 50 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 20 mM N-hydroxysuccinimide dissolved in a mixture solution of water and ethanol at a series ratio of 5/95, 10/90 and 15/85 (v/v) with 0.1% 4-morpholineethanesulfonic acid, each for 8 h. After cross-linking, the porous microwell scaffolds were immersed in 0.1 M glycine aqueous solution overnight and washed by pure water before the second freeze-drying. Three types of microwell scaffolds were prepared by using different sizes of ice particulates as templates that were prepared with different moisture exposure times of 10, 15 and 20 min, which were referred to as small, middle and large microwell scaffolds, respectively. As a control, gelatin–PLGA porous scaffold without microwell structure was prepared using the same procedures without the ice particulate template. The control gelatin–PLGA porous scaffold was referred as a flat scaffold. The porous structure of the scaffolds was observed by a scanning electron microscope (SEM, Hitachi S-4800, Japan). The size of each microwell in the porous microwell scaffolds and the size of small pores in walls of microwells were analyzed by imageJ software. Five samples of each type of scaffold and 50 microwells and 50 small pores of each microwell were used for the analysis.
10, Ethicon Inc., USA) was placed on a glass plate wrapped with a plastic film. The surrounding of the PLGA mesh was covered by a silicone frame (1 mm in thickness). 5% (wt%) gelatin aqueous solution was poured on the PLGA mesh in the silicone frame and the gelatin solution surface was flattened (1 mm in thickness) at room temperature. The construct was transferred into a low temperature chamber at −2 °C for 2 min to cool the construct without freezing. The ice particulate template was transferred from the freezer to the low temperature chamber and immediately placed on the top of the pre-cooled construct to allow the ice particulates facing the gelatin solution. The construct was immediately transferred to the −80 °C freezer and frozen for 3 h, followed by 48 h of freeze-drying. After freeze-drying, the scaffolds were immersed in ethanol and cross-linked with 50 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 20 mM N-hydroxysuccinimide dissolved in a mixture solution of water and ethanol at a series ratio of 5/95, 10/90 and 15/85 (v/v) with 0.1% 4-morpholineethanesulfonic acid, each for 8 h. After cross-linking, the porous microwell scaffolds were immersed in 0.1 M glycine aqueous solution overnight and washed by pure water before the second freeze-drying. Three types of microwell scaffolds were prepared by using different sizes of ice particulates as templates that were prepared with different moisture exposure times of 10, 15 and 20 min, which were referred to as small, middle and large microwell scaffolds, respectively. As a control, gelatin–PLGA porous scaffold without microwell structure was prepared using the same procedures without the ice particulate template. The control gelatin–PLGA porous scaffold was referred as a flat scaffold. The porous structure of the scaffolds was observed by a scanning electron microscope (SEM, Hitachi S-4800, Japan). The size of each microwell in the porous microwell scaffolds and the size of small pores in walls of microwells were analyzed by imageJ software. Five samples of each type of scaffold and 50 microwells and 50 small pores of each microwell were used for the analysis.
Cell culture, cell viability and morphology
RIN-5F cells were cultured in a conventional cell culture flask with RPMI-1640 containing 10% fetal bovine serum before seeding in the scaffolds. Trypsin/EDTA solution was used to detach the subcultured RIN-5F cells. The harvested RIN-5F cells were resuspended in RPMI 1640 serum medium at a density of 2 × 107 cells per mL. The flat and microwell porous scaffolds were punched into discs with a diameter of 10 mm and sterilized with 70% ethanol. A cell strainer with 70 μm pores was used during cell seeding. To control cell suspension solution passing through the scaffold discs without peripheral leakage, a silicon frame with an inner hole of 10 mm was placed in the cell strainer to hold the scaffold disc. Scaffold disc was placed in the silicon frame on the cell strainer. After washing twice with PBS and twice with serum medium, the cell suspension solution was dropwise added to the scaffold discs. 2 mL of cell suspension solution was added to each scaffold disc for cell seeding. After cell seeding, the scaffold discs were transferred to cell culture dishes and incubated in a CO2 incubator at 37 °C for 1 day and 7 days. Half of the medium was changed every day during culture. Samples were used in triplicate for cell seeding and following investigation.After culture for 1 day and 7 days, RIN-5F cells were stained by a live/dead staining kit. 4 μL calcein-AM and 6 μL propidium iodide were directly added to the culture medium to stain live and dead cells for 10 min. After staining, the cell/scaffold constructs were directly observed by fluorescence microscopy. The morphology of aggregates in the flat and porous microwell scaffolds after 7 days of culture was further observed by confocal laser scanning microscopy.
DNA quantification
The proliferation of beta cells in the scaffolds was quantified by measuring the DNA amount in the cell/scaffold constructs after 1 day and 7 days of culture. At first, the cell/scaffold constructs were carefully washed with PBS 3 times and pure water once, followed by freeze-drying. Then, the freeze-dried cell/scaffold constructs were digested in 400 μg mL−1 papain solution containing 5 mM EDTA and 5 mM L-cysteine in 0.1 M phosphate buffer (pH 6.0). Finally, the DNA amount in each sample was measured by using a Hoechst 33258 dye (Sigma Aldrich, USA) and a fluorescence spectrometer (FP8500, JASCO, Japan). Samples were used in triplicate for the quantification.Evaluation of insulin secretion
The secreted insulin from the cell/scaffold constructs after 1 day and 7 days of culture was measured using an ELISA kit. At first, the cell/scaffold discs were transferred to 5 mL of fresh RPMI 1640 culture medium and incubated for 3 h to release insulin. Subsequently, 500 μL of medium from each sample was taken and stored at −80 °C. The insulin amount in all samples was measured using a rat insulin ELISA kit based on the protocol. In brief, insulin standards and the collected samples were diluted by a dilution buffer. A sandwich ELISA system where the insulin antibody was pre-embedded in a 96 well plate was used. After washing 4 times with buffer, the standards and samples were added to wells one by one. After incubation for 2 h, the wells were washed once. The second antibody with horseradish peroxidase (HRP) was added to each well and incubated for 0.5 h. After washing, the colorimetric reaction was applied by adding o-phenylenediamine dihydrochloride (OPD) solution to each well. A microplate reader was used to read the absorbance of each sample after adding sulfuric acid (1 M) to block the reaction. Absorbance at 492 nm with a reference wavelength of 650 nm was measured. The secretion amount of insulin from each sample was calculated according to the standard curve of insulin. The results were normalized with DNA amount. Samples of each group were used in triplicate for the measurement to calculate the means and standard deviations.Statistical analysis
All the quantitative experiments were repeated in triplicate (n = 3). All the results are expressed as means ± standard deviations. Statistical analysis of the experimental data was performed by using one-way analysis of variance (ANOVA) with Tukey's post hoc test in Origin Pro software (8.0). The p value was used to determine the level of significance: *p < 0.05, **p < 0.01, and ***p < 0.001.Results
Preparation and characterization of gelatin–PLGA porous microwell scaffolds
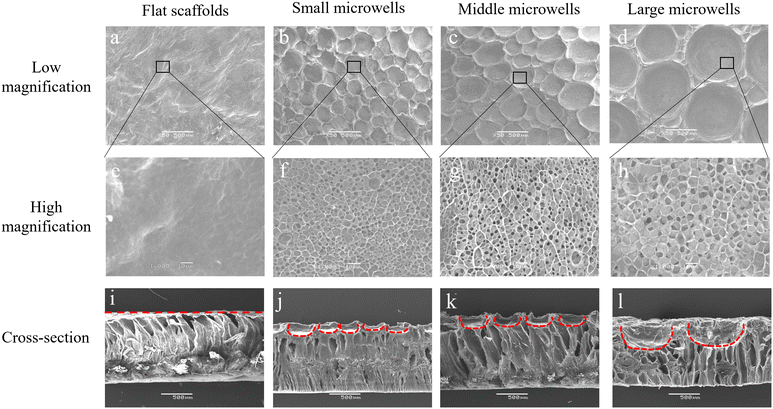
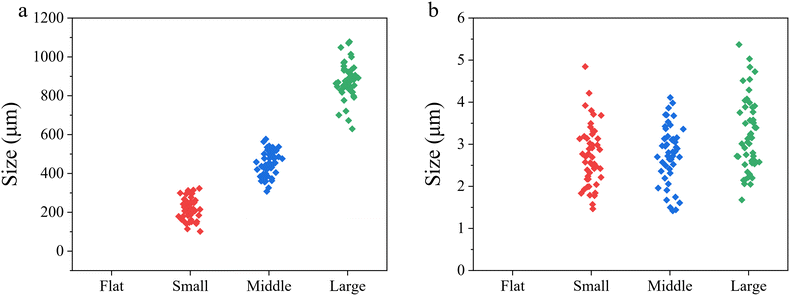
The gelatin–PLGA porous microwell scaffolds were prepared by using an ice particulate template method (Scheme 1). The PLGA mesh was covered with gelatin aqueous solution during the preparation process, resulting in formation of a gelatin porous structure on the PLGA mesh. Through changing the ice particulate templates, porous microwells of different sizes were formed on the PLGA mesh. SEM observation showed that there was no microwell structure in the control flat scaffold (Fig. 1a and e). On the other hand, microwells of different sizes were formed on the surface of the small, middle and large microwell scaffolds (Fig. 1b–d). The microwells had a concave morphology separated by thin ridges. The size of microwells in the small size, middle size and large size microwell scaffolds was 221.1 ± 34.8, 453.1 ± 64.0 and 874.3 ± 89.4 μm, respectively (Fig. 2a). Observation at a high magnification of SEM showed that the walls of the microwells had dense small pores (Fig. 1f–h). The size of the small pores on the walls of the small size, middle size and large size microwell scaffolds was 2.7 ± 0.7, 2.8 ± 0.7, and 3.2 ± 0.9 μm, respectively (Fig. 2b). The cross-sectional view of the scaffolds also showed microwell structures of the gelatin–PLGA porous microwell scaffolds (Fig. 1j–l). However, the control scaffold had a flat surface (Fig. 1i). The PLGA mesh was located on one side of the flat scaffold and the microwell scaffolds while the gelatin areas showed porous structures. The average depth of microwells in the small size, middle size and large size microwell scaffolds was around 148 μm, 232 μm and 410 μm, respectively.Cell viability and proliferation
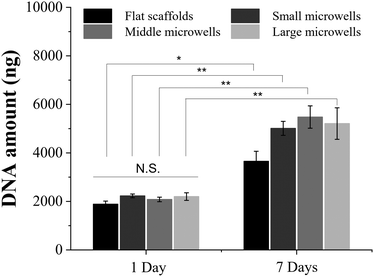
RIN-5F cells were seeded in the scaffolds by dropping cell suspension solution on the scaffolds. RIN-5F cells would be trapped in the porous microwells while the medium penetrated through scaffolds. Cell/scaffold constructs were cultured in serum medium for 1 day and 7 days. Half of the culture medium was carefully removed and fresh medium was added slowly on each day after cell seeding to avoid loss of cells during medium changing. Live/dead staining results showed high viability of RIN-5F cells in the porous microwell scaffolds after 1 day and 7 days of culture (Fig. 3). No dead cells were observed in either scaffold.Proliferation of RIN-5F cells in the scaffolds was investigated by measuring DNA amount after 1 day and 7 days of culture (Fig. 4). The DNA amount after 1 day of culture in the flat scaffold and porous microwell scaffolds had no significant difference. After 7 days of culture, the DNA amount in the porous microwell scaffolds showed significant increase compared to that after 1 day of culture. The DNA amount after 7 days of culture in the flat, small size microwell, middle size microwell and large size microwell scaffolds increased to 1.9, 2.3, 2.6, and 2.4 times higher than that after 1 day of culture. Cell proliferation in the porous microwell scaffolds was higher than that in the flat scaffold.
Formation of cell aggregates
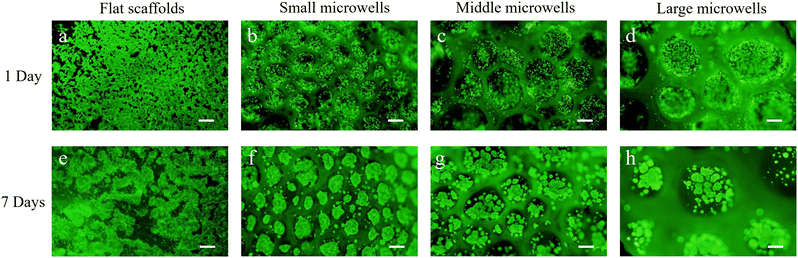
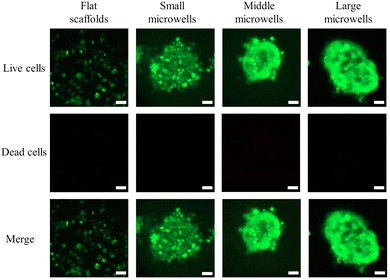
Cell distribution in all scaffolds was observed by fluorescence microscopy after live/dead staining (Fig. 3). The cell distribution in the flat scaffold was different from that in the porous microwell scaffolds. In the flat scaffold, the cells adhered to the flat surface of the scaffold (Fig. 3a). In contrast, the cells in the porous microwell scaffolds were predominantly trapped in the microwells (Fig. 3b–d). The cells attached along the wall of each microwell. After 7 days of culture, the cells in the flat scaffold formed flake-like cell clusters (Fig. 3e). On the other hand, RIN-5F cells in the porous microwell scaffolds formed grape-like aggregates (Fig. 3f–h). The cells formed small grape-like aggregates that were further connected to form large aggregates. In the small size microwell scaffold, the small grape-like aggregates were connected to form one big aggregate in each microwell. However, in the middle size and large size microwell scaffolds, the small grape-like aggregates were connected to form a few large grape-like aggregates. The small grape-like aggregates in all the porous microwell scaffolds were only loosely connected. The loose connection among the small grape-like aggregates could facilitate diffusion of nutrient and oxygen into each of the small aggregates, therefore maintaining high cell viability.Confocal laser scanning microscopy was used to observe the flake-like cell clusters in the flat scaffold and the middle section of grape-like aggregates in the porous microwell scaffolds at a high magnification (Fig. 5). In the flat scaffolds, the cells in the flake-like clusters were sparsely distributed. On the other side, the cells in the grape-like aggregates interacted well with each other to form spheroidal aggregates where tight junctions might be established among cells. Furthermore, no red fluorescence in the middle of the aggregates indicated that the size of the aggregates was reasonable to maintain high cell viability in the center of aggregates.
Insulin secretion
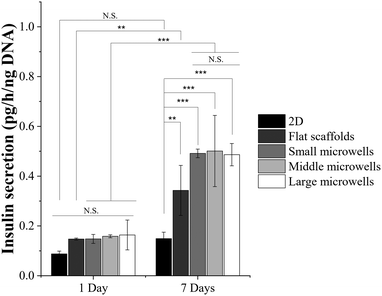
Insulin secretion of RIN-5F cells cultured in the scaffolds and 2D plates was investigated by an ELISA kit after 1 day and 7 days of culture (Fig. 6). The results were normalized with the DNA amount for evaluating the insulin secretion function of RIN-5F cells. After 1 day of culture, RIN-5F cells in the scaffolds showed higher insulin secretion than that in the 2D plates. However, the increase was not significant. After 7 days of culture, insulin secretion of RIN-5F cells cultured in 2D plates did not increase significantly. On the other hand, insulin secretion of RIN-5F cells in the scaffolds increased significantly. Insulin secretion in the scaffolds was significantly higher than that in the 2D plates. Culture in the porous microwell scaffolds further promoted insulin secretion of RIN-5F cells compared to the flat scaffold. Insulin secretion in the small, middle and large microwell scaffolds was similar.Discussion
Gelatin–PLGA porous microwell scaffolds were prepared by using an ice particulate template for 3D culture of pancreatic beta cells to promote cell aggregation and enhance insulin secretion. At first, an ice particulate template was prepared by a moisture method in which atomized water was deposited on a hydrophobic surface to form water droplets. The size of water droplets could be adjusted by controlling the moisture exposure time of the hydrophobic substrate. And then, porous microwells of gelatin were constructed on a PLGA knitted mesh. The PLGA knitted mesh was used to support the microwell structure because it has high mechanical strength and good biocompatibility.37–39 Microwell structure with a concave morphology was formed and independently separated from each other (Fig. 1). To investigate the size influence, microwells of three different sizes (221.1, 453.1 and 874.3 μm) were prepared because the size of normal human islets has a range of 50–500 μm.40 Two sizes were in the range of human islet size while one was larger. Dense ultra-small pores were observed in the wall of microwells. The dense ultra-small pores were formed during rapid freezing in a −80 °C freezer. Rapid freezing could trigger micro-phase separation of polymer aqueous solution.41,42 The porous structure of a microwell scaffold could allow free permeation of oxygen and nutrients that are important for maintaining cell viability by alleviating hypoxia stress, importing glucose and secreting insulin.43,44RIN-5F cells were cultured in the porous microwell scaffolds for investigation of the influence of microwell structures on cell aggregation formation and insulin secretion. RIN-5F cells are derived from rat pancreas and are usually used for investigating 3D culture of beta cells.21 It has been reported that non-adhesive materials, such as PDMS, can be used for 3D culture of beta cells.45 Beta cells would be forced to aggregate at the bottom of non-adhesive microwells as a single cluster through centrifugation. However, such microwell scaffolds are lack of biodegradability and require complicated procedures for transplantation. It is more attractive to form cell aggregates in transplantable devices or scaffolds for direct implantation after cell culture. The gelatin–PLGA porous microwell scaffolds should be a good candidate for such applications. The gelatin–PLGA porous microwell scaffolds could maintain the high viability of RIN-5F cells (Fig. 3 and 4). This should be attributed to the porous structure that could facilitate nutrient diffusion throughout the scaffolds. RIN-5F cells cultured in the gelatin–PLGA porous microwell scaffolds could interact with each other and form grape-like aggregates. The small aggregates were connected to form large aggregates like a bunch of grapes. The connection between the small grape-like aggregates was loose so that the viability of cells inside aggregates could be maintained due to the diffusion of oxygen and nutrients (Fig. 5).
The major function of pancreatic beta cells used for T1DM treatment is the insulin secretion.46 The gelatin–PLGA porous microwell scaffolds promoted insulin secretion of RIN-5F cells. RIN-5F cells cultured in the small, middle and large microwell scaffolds showed the same level of insulin secretion (Fig. 6). The microwell size did not affect the insulin secretion capacity because all the porous microwells of different sizes facilitated the formation of small grape-like cell aggregates. It has been widely studied that insulin secretion of beta cells is highly related to the cell–cell interaction.47 The small grape-like cell aggregates had similar size. The cell–cell interaction in the small grape-like aggregates was strong. Although large microwells could accommodate more cells in each microwell and form a few large aggregates, the insulin secretion should be dominated by the small aggregates. The interaction between the small grape-like cell aggregates was not strong, and therefore did not affect insulin secretion significantly. The cells cultured in the flat scaffolds showed a loose structure of flake-like cell clusters that cell–cell interaction might be weakened, resulting in deficient insulin secretion. Therefore, the porous microwell scaffolds could accommodate more cells in the porous microwells to facilitate cell aggregation, maintain cell viability and promote insulin secretion. The cell aggregates that attached to the porous microwells could be important for transplantation together with the scaffolds. In this study, only one cell type, pancreatic beta cells, was cultured in the porous microwell scaffolds. In the future, co-culture of different cell types should be considered because primary pancreatic islets consist of different types of cells. Angiogenesis should also be considered to mimic the natural pancreatic structure. The gelatin–PLGA porous microwell scaffolds should provide a useful tool for 3D culture of not only pancreatic beta cells but also co-culture of different cell types for T1DM treatment.
Conclusions
Gelatin–PLGA porous microwell scaffolds were prepared for 3D culture of RIN-5F cells to promote formation of cell aggregates and promote insulin secretion. The porous microwell scaffolds had a controllable microwell size. RIN-5F cells cultured in the porous microwell scaffolds showed high viability and formed grape-like cell aggregates. The cells in the porous microwell scaffolds had high insulin secretion. The porous microwell scaffolds showed a high potential for serving as a 3D culture model for T1DM treatment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by JSPS KAKENHI Grant number 19H04475, 21H03830 and 22K19926.References
- L. DiMeglio, C. Evans-Molina and R. Oram, Lancet, 2018, 391(10138), 2449–2462 CrossRef PubMed.
- M. Lotfy, J. Adeghate, H. Kalasz, J. Singh and E. Adeghate, Curr. Diabetes Rev., 2017, 13(1), 3–10 CrossRef CAS PubMed.
- A. Janež, C. Guja, A. Mitrakou, T. Tankova, L. Czupryniak, A. Tabák, M. Prazny, E. Martinka and L. Smircic-Duvnjak, Diabetes Ther., 2020, 11, 387–409 CrossRef PubMed.
- S. Roze, J. Smith-Palmer, W. Valentine, S. de Portu, K. Nørgaard and J. Pickup, Diabetic Med., 2015, 32(11), 1415–1424 CrossRef CAS PubMed.
- Y. Aghazadeh and C. Nostro, Curr. Diabetes Rep., 2017, 17, 1–9 CrossRef CAS PubMed.
- C. Aguayo-Mazzucato and S. Bonner-Weir, Nat. Rev. Endocrinol., 2010, 6(3), 139–148 CrossRef PubMed.
- M. Bellin and T. Dunn, Diabetologia, 2020, 63, 2049–2056 CrossRef PubMed.
- A. Shapiro, M. Pokrywczynska and C. Ricordi, Nat. Rev. Endocrinol., 2017, 13(5), 268–277 CrossRef CAS PubMed.
- M. A. Atkinson, G. S. Eisenbarth and A. W. Michels, Lancet, 2014, 383(9911), 69–82 CrossRef PubMed.
- D. Steiner, A. Kim, K. Miller and M. Hara, Islets, 2010, 2(3), 135–145 CrossRef PubMed.
- G. Rutter, T. Pullen, D. Hodson and A. Martinez-Sanchez, Biochem. J., 2015, 466(2), 203–218 CrossRef CAS PubMed.
- M. Narushima, N. Kobayashi, T. Okitsu, Y. Tanaka, S. Li, Y. Chen, A. Miki, K. Tanaka, S. Nakaji, K. Takei, A. Gutierrez, J. Rivas-Carrillo, N. Navarro-Alvarez, H. Jun, K. Westerman, H. Noguchi, J. Lakey, P. Leboulch, N. Tanaka and J. Yoon, Nat. Biotechnol., 2005, 23(10), 1274–1282 CrossRef CAS PubMed.
- M. Bellin and T. Dunn, Diabetologia, 2020, 63(10), 2049–2056 CrossRef PubMed.
- M. Vantyghem, E. de Koning, F. Pattou and M. Rickels, Lancet, 2019, 394(10205), 1274–1285 CrossRef CAS PubMed.
- J. Kim, I. K. Shim, D. G. Hwang, Y. N. Lee, M. Kim, H. Kim, S. W. Kim, S. Lee, S. C. Kim, D. W. Cho and J. Jang, J. Mater. Chem. B, 2019, 7(10), 1773–1781 RSC.
- O. Cabrera, D. M. Berman, N. S. Kenyon, C. Ricordi, P. O. Berggren and A. Caicedo, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(7), 2334–2339 CrossRef CAS PubMed.
- Y. Jun, A. R. Kang, J. S. Lee, G. S. Jeong, J. Ju, D. Y. Lee and S. H. Lee, Biomaterials, 2013, 34(15), 3784–3794 CrossRef CAS PubMed.
- K. W. Lee, S. K. Lee, J. W. Joh, S. J. Kim, B. B. Lee, K. W. Kim and K. U. Lee, Tissue Eng., 2004, 10(7–8), 965–977 CAS.
- H. Tanaka, S. Tanaka, K. Sekine, S. Kita, A. Okamura, T. Takebe, Y. W. Zheng, Y. Ueno, J. Tanaka and H. Taniguchi, Biomaterials, 2013, 34(23), 5785–5791 CrossRef CAS PubMed.
- M. Zhang, S. Yan, X. Xu, T. Yu, Z. Guo, M. Ma, Y. Zhang, Z. Gu, Y. Feng, C. Du, M. Wan, K. Hu, X. Han and N. Gu, Biomaterials, 2021, 270, 120687 CrossRef CAS PubMed.
- N. Kawazoe, X. Lin, T. Tateishi and G. Chen, J. Bioact. Compat. Polym., 2009, 24(1), 25–42 CrossRef CAS.
- J. Hilderink, S. Spijker, F. Carlotti, L. Lange, M. Engelse, C. van Blitterswijk, E. de Koning, M. Karperien and A. van Apeldoorn, J. Cell. Mol. Med., 2015, 19(8), 1836–1846 CrossRef CAS PubMed.
- X. Liu, F. Yan, H. Yao, M. Chang, J. Qin, Y. Li, Y. Wang and X. Pei, Cell Tissue Res., 2014, 358(2), 359–369 CrossRef CAS PubMed.
- C. Lin, A. Raza and H. Shih, Biomaterials, 2011, 32(36), 9685–9695 CrossRef CAS PubMed.
- A. Ghasemi, E. Akbari and R. Imani, Front. Bioeng. Biotechnol., 2021, 9, 662084 CrossRef PubMed.
- J. Hwang, B. Lee, M. Jung, H. Jung, Y. Hwang, M. Kim, S. Lee and D. Lee, Macromol. Res., 2011, 19, 1320–1326 CrossRef CAS.
- Y. N. Lee, H. J. Yi, H. Goh, J. Y. Park, S. Ferber, I. K. Shim and S. C. Kim, Cells, 2020, 9(12), 2551 CrossRef CAS PubMed.
- A. Bernard, C. Lin and K. Anseth, Tissue Eng., Part C, 2012, 18(8), 583–592 CrossRef CAS PubMed.
- C. K. Huang, G. J. Paylaga, S. Bupphathong and K. H. Lin, Biofabrication, 2020, 12(2), 025016 CrossRef CAS PubMed.
- Y. Pang, Y. Horimoto, S. Sutoko, K. Montagne, M. Shinohara, D. Mathiue, K. Komori, M. Anzai, T. Niino and Y. Sakai, Biofabrication, 2016, 8(3), 035016 CrossRef CAS PubMed.
- M. Buitinga, F. Assen, M. Hanegraaf, P. Wieringa, J. Hilderink, L. Moroni, R. Truckenmüller, C. van Blitterswijk, G. W. Römer, F. Carlotti, E. de Koning, M. Karperien and A. van Apeldoorn, Biomaterials, 2017, 135, 10–22 CrossRef CAS PubMed.
- E. Hadavi, J. Leijten, M. Engelse, E. de Koning, P. Jonkheijm, M. Karperien and A. van Apeldoorn, Tissue Eng., Part C, 2019, 25(2), 71–81 CrossRef CAS PubMed.
- F. Tokito, M. Shinohara, M. Maruyama, K. Inamura, M. Nishikawa and Y. Sakai, J. Biosci. Bioeng., 2021, 131(5), 543–548 CrossRef CAS PubMed.
- S. Chen, T. Nakamoto, N. Kawazoe and G. Chen, Biomaterials, 2015, 73, 23–31 CrossRef CAS PubMed.
- Y. Chen, S. Chen, N. Kawazoe and G. Chen, Sci. Rep., 2018, 8(1), 14143 CrossRef PubMed.
- S. Chen, N. Kawazoe and G. Chen, Tissue Eng., Part C, 2017, 23(6), 367–376 CrossRef CAS PubMed.
- Y. Chen, K. Lee, Y. Yang, N. Kawazoe and G. Chen, Biofabrication, 2020, 12(2), 025027 CrossRef CAS PubMed.
- N. Kawazoe, C. Inoue, T. Tateishi and G. Chen, Biotechnol. Prog., 2010, 26(3), 819–826 CrossRef CAS PubMed.
- G. Chen, T. Sato, H. Ohgushi, T. Ushida, T. Tateishi and J. Tanaka, Biomaterials, 2005, 26(15), 2559–2566 CrossRef CAS PubMed.
- C. Ricordi, Pancreas, 1991, 6(2), 242–244 CrossRef CAS PubMed.
- R. Bansil, J. Phys. IV, 1993, 3(C1), C1-225 CrossRef.
- A. Kumar, R. Mishra, Y. Reinwald and S. Bhat, Mater. Today, 2010, 13(11), 42–44 CrossRef CAS.
- D. Rojas-Canales, M. Waibel, A. Forget, D. Penko, J. Nitschke, F. Harding, B. Delalat, A. Blencowe, T. Loudovaris, S. Grey, H. Thomas, T. Kay, C. Drogemuller, N. Voelcker and P. Coates, Endocr. Connect., 2018, 7(3), 490–503 CAS.
- Y. Jun, J. Lee, S. Choi, J. Yang, M. Sander, S. Chung and S. Lee, Sci. Adv., 2019, 5(11), eaax4520 CrossRef CAS PubMed.
- M. Shinohara, H. Kimura, K. Montagne, K. Komori, T. Fujii and Y. Sakai, Biotechnol. Prog., 2014, 30(1), 178–187 CrossRef CAS PubMed.
- L. M. Weber, K. N. Hayda and K. S. Anseth, Tissue Eng., Part A, 2008, 14(12), 1959–1968 CrossRef CAS PubMed.
- E. S. O'Sullivan, A. S. Johnson, A. Omer, J. Hollister-Lock, S. Bonner-Weir, C. K. Colton and G. C. Weir, Diabetologia, 2010, 53, 937–945 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |