Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Single-cell analysis of innate immune cell mechanics: an application to cancer immunology
Tom M.J.
Evers
ab,
Antoinette
van Weverwijk
cd,
Karin E.
de Visser
cde and
Alireza
Mashaghi
 *ab
*ab
aMedical Systems Biophysics and Bioengineering, Leiden Academic Centre for Drug Research, Faculty of Science, Leiden University, The Netherlands. E-mail: a.mashaghi.tabari@lacdr.leidenuniv.nl
bLaboratory for Interdisciplinary Medical Innovations, Centre for Interdisciplinary Genome Research, Faculty of Science, Leiden University, The Netherlands
cDivision of Tumor Biology & Immunology, Netherlands Cancer Institute, Amsterdam, The Netherlands
dOncode Institute, Utrecht, The Netherlands
eDepartment of Immunology, Leiden University Medical Center, Leiden, The Netherlands
First published on 4th March 2024
Abstract
The innate immune system forms a crucial defense mechanism of the human body against infections and foreign objects, orchestrates wound healing, and restores tissue homeostasis after resolving the insult. Furthermore, innate immune cells launch adaptive immune responses against tumors and mediate phagocytosis of cancer cells and cytotoxic tumor killing. They may also be exploited by cancer cells to promote angiogenesis and metastasis. Although the life cycle and function of innate immune cells require a continuous remodeling of their mechanical properties, data on the biomechanics of different innate immune cell subtypes are scarce. Recent advances in single-cell force spectroscopy have facilitated the exploration of immune cell mechanics at phyiological temperature, aiding in filling this knowledge gap. In this article, we discuss the existing data on the mechanical properties of innate immune cells and provide the first single-cell mechanical characterization of tumor-associated macrophages. The presented data on tumor-associated macrophages showcase the new technical possibilities for decoding immune cell mechanics in cancer, further contributing to our understanding of innate immune cell behavior in disease contexts.
1. Innate immunity
Cells of the innate immune system arise from hematopoietic stem cells in the bone marrow. Once matured, they are released from the bone marrow to the bloodstream, from where they can home to tissues until the body mobilizes them to cope with infection, trauma, or cancer.1–3 Innate immune cells detect molecular cues, such as changes induced by microbial infections and inflammatory mediators like cytokines and chemokines, and then launch adaptive immune responses while mounting their effector responses, such as phagocytosis for macrophages, and cytotoxicity for natural killer cells.3 Research into immune cell signal transduction by applying the tools of biochemistry, molecular biology, and genetics has identified networks of secreted ligands, cell surface receptors, intracellular signaling pathways, and transcriptional regulators that coordinate various aspects of cell behavior. There is a growing realization that the mechanical properties of a cell add an extra layer of regulation to innate immune cells.4It has become increasingly evident that the mechanical properties of a cell can offer intrinsic biophysical cues for determining physiological cell state transitions. The link between mechanical properties and cell function is particularly obvious in innate immune cells as they need to undergo extensive morphological changes during endothelial transmigration and phagocytosis, display rapid movement through blood vessels and tight interstitial spaces, strongly adhere under shear flow, and form highly dynamic interfaces with other cells.4–9 How mechanical forces are involved in regulating essential cellular processes such as cell-surface receptor activation, cell migration, intracellular signaling, and intercellular communication, in various immune cell types, has been the focus of numerous excellent reviews.4,7 Although the link between the mechanical properties of an innate immune cell and its function might seem obvious, data on the basic mechanical properties of various innate immune cell subtypes (at physiological temperature) are scarce and for diseased states lacking almost entirely. Several important early studies have described the mechanical behavior of an innate immune cell (mainly neutrophil) as a “fluid-filled bag with an elastic cortical envelope”.10,11 Recent advancements in single-cell force spectroscopy have played a pivotal role in unraveling immune cell mechanics and investigating the “cortical shell-liquid core” concept in greater depth. In this perspective, we discuss the existing data on the mechanics of innate immune cells, in particular monocytes and macrophages, and explore how current technology can be applied to study immune cells associated with cancer.
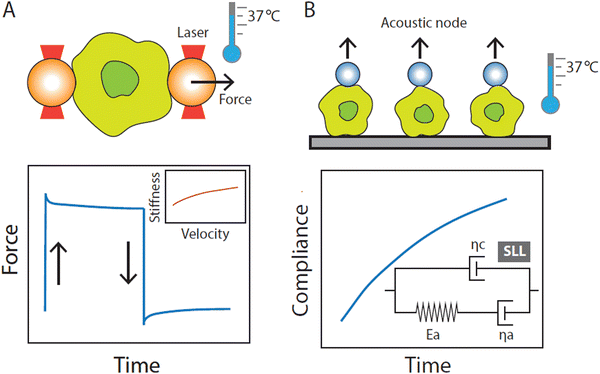
2. Advances in single-cell force spectroscopy
Progress in single-cell force spectroscopy (SCFS) techniques enables substantial advances in our understanding of the tight relationship between mechanics and function in innate immune cells. This, in turn, contributes to the emergence of the field of mechanoimmunology or immunomechanics. When performing SCFC experiments, biophysicists generally consider two readouts: cell elasticity, referring to the stretchiness of cells, and viscosity, referring to the fluidity of cells. Together, these parameters determine the mechanical phenotype of a cell. On one hand, the application of mechanical probes commonly employed in cell biophysics is relatively new to immunologists, and, on the other hand, biophysicists have focused mostly on the mechanics of the simplest blood cell, the red blood cells (RBCs). RBCs are therefore among the best-studied cells in the field of cell mechanics, and the values of their mechanical properties are supported by a large body of experiments by different SCFS methods like optical tweezers (OT), acoustic force spectroscopy (AFS), atomic force microscopy (AFM), micropipette aspiration (MA), and other techniques, all giving consistent results.12–19 Brief descriptions of commonly employed SCFS techniques are provided in Box 1. Despite the fact that the application of SCFS techniques in RBC studies has led to many important findings in hemorheology and improvement in understanding disease,20–28 data on the mechanical behavior of RBCs at biomedically relevant temperatures was lacking till very recently, mainly due to technical limitations. Motivated by this gap in knowledge, we have recently initiated a single-cell mechanics research program to characterize the mechanics of red and white blood cells by utilizing several complementary SCFS techniques, including AFS, OT, and AFM, in a temperature controlled system to mimic physiological and medically relevant conditions. Indeed, using RBCs as a model, we demonstrated that cellular mechanics are highly temperature-sensitive,29 emphasizing the need to probe cell mechanics in temperature-controlled environments. The temperature dependency of the mechanical properties has also been suggested for neutrophils, with a demonstrated decrease in apparent viscosity as temperature increases.10 Inspired by these observations, we recently moved into the direction of innate immune cell mechanics, specifically monocytes and macrophages, on which we will focus below.Box 1
Single-cell force spectroscopy (SCFS) techniques
Cell mechanics rely on technical approaches that enable the quantification of mechanical properties at the single-cell level. Several of the most commonly used strategies are described briefly below.3. Innate immunity from a biophysical perspective
The life of an innate immune cell is intensely physical, requiring a continuous remodeling of its mechanical properties. To illustrate this, we follow the physically perilous journey of a monocyte along its way to an inflamed tissue (Fig. 2). Inflamed tissues release potent inflammatory mediators that drive monocytes out of the bone marrow (or other reservoirs such as the spleen) and into the circulation to be rapidly transported to their destination.30 An example of such a chemical signal is the chemokine CCL2, a strong chemoattractant for monocytes and a powerful initiator of inflammation.31 Using a combination of AFS and OT (see Fig. 1 for experimental setup), we have recently shown that CCL2 exposure in vitro causes primary human monocytes to stiffen, as well as induces a more fluid-like phenotype.32 These very finely tuned chemokine-induced alterations in the mechanical properties of monocytes may help their switch to a migration-competent state and potentially their accumulation at sites of infection. During their transport in the blood, monocytes have to strongly deform to pass through the narrowest capillaries and bifurcations. This process depends on a finely tweaked cortical tension combined with a high cell viscosity that governs the fast transit through the vascular network while preserving cell integrity.33 While approaching the site of inflammation, monocytes move closer to the endothelium, establishing contact and eventually creating adhesions when they reach the low-flow postcapillary venule environment. Initially, these adhesions are transient and weak, mediated by selectins, causing the monocytes to roll along the endothelial surface.34,35 However, they eventually come to a halt and firmly attach by forming stronger integrin bonds in regions where activated endothelial cells exhibit high concentrations of chemokines on their luminal surface. Subsequently, monocytes extend cellular protrusions and commence crawling atop the endothelial cells, prompting them to find a path through the endothelium and into the underlying tissue.35–37 Extravasating monocytes undergo extensive mechanical alterations, helping them to squeeze through the narrow gaps in the endothelial barrier. Inside the affected tissue, monocytes differentiate into macrophages. It has previously been shown that during the course of differentiation, macrophages exhibit distinct mechanical properties compared to their precursor cells.38 Irrespective of the macrophage type, macrophages stiffen and behave less fluid-like than monocytes. However, monocyte differentiation into macrophages occurs complementary with the acquisition of a functional phenotype that depends on microenvironmental signals.39 Traditionally, macrophages have been categorized into two major polarized phenotypes, M1 and M2.40In vitro activation using the Th1-type cytokine interferon-gamma (IFNγ), granulocyte-macrophage colony-stimulating factor (GM-CSF), lipopolysaccharide (LPS) and/or Toll-like receptor 4 (TLR4) engagement gives rise to pro-inflammatory M1-like macrophages, whereas stimulation with macrophage colony-stimulating factor (M-CSF) or the Th2 cytokines interleukin 4 (IL-4), IL-10, or IL-13 generates the anti-inflammatory M2-like macrophages.41,42 It is important to note that in vivo macrophage polarization is not binary, but has a spectrum-like level of activation that cannot be defined by two extremes.39,40,43,44 Since macrophages are highly plastic cells that are able to acquire different functional phenotypes, it is tempting to speculate that they may also adopt different mechanical phenotypes to accommodate these different and sometimes opposing functions.45 To address this, we recently employed several single-cell force spectroscopy techniques to mechanically characterize human (in vitro GM-CSF-induced) M1-like and (in vitro M-CSF-induced) M2-like macrophages.46 Indeed, M2-like macrophages exhibit a higher overall viscosity and appear to be stiffer than M1-like macrophages. M2-like macrophages have been shown to exhibit higher phagocytic capacity than M1-like macrophages and phagocytosis can lead to increased stiffness caused by elevated production of reactive oxygen species.47 This stiffness hampers the ingestion of additional particles, as phagocytosis requires deformability as a prerequisite.47–49 Consequently, there seems to be a threshold for phagocytosis that can be reached either by engulfing numerous small particles or a few larger ones.50 In addition, the higher viscosity of M2-like macrophages presumably facilitates their migration in porous interstitial-like environments,38 supporting the major function of anti-inflammatory macrophages; phagocytosis followed by efflux from the tissue.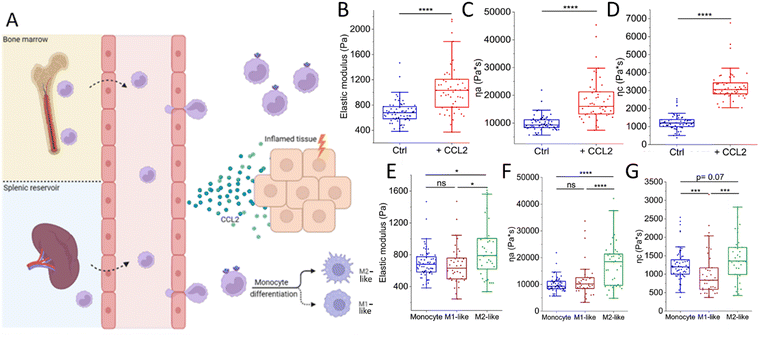 | ||
| Fig. 2 The intense physical life of a monocyte on its way to an inflamed tissue. (A) Inflamed tissues release chemokines such as CCL2 that drive monocytes out of the bone marrow and spleen into the circulation. Upon binding CCL2, monocytes switch to a migration-competent state, reflected by an increase in (B) stiffness (elastic modulus) and viscosity (C) ηa and (D) ηc. Then, monocytes transmigrate through the endothelium during which they undergo extensive (squeezing) mechanical alterations. Upon entering the tissue, monocytes differentiate into either M1-like or M2-like macrophages. M2-like macrophages are (E) stiffer and (F) and (G) less fluid-like than their precursors, whereas M1-like macrophages and monocytes exhibit similar mechanical properties at physiological temperature (37 °C). Macrophage experiments included 38 M1-like and 38 M2-like macrophages collected from 4 donors. Monocytes (ctrl: n = 63, +CCL2: n = 51) were collected from 3 donors. Graphs (B)–(D) are re-plotted from ref. 32 and graphs (E)–(G) are re-plotted from ref. 32 and 46. | ||
In summary, it has become increasingly clear that monocytes and macrophage subtypes exhibit distinct mechanical phenotypes, which correlate with their diverse functions, and that immune triggers such as the chemokine CCL2 and cytokines GM-CSF or M-CSF can modulate this mechanical phenotype at physiological temperature. Besides monocytes and macrophages, a significant body of evidence exists on the mechanical properties of human primary neutrophils in physiological and diseased states (Table 1). However, single-cell mechanical data for other human innate immune cells, particularly natural killer cells, mast cells, eosinophils, and basophils are scarce. Table 1 presents an overview of studies that employed direct single-cell measurements to mechanically characterize primary innate immune cells, or indirectly by measuring morphological properties as a proxy for mechanics.
| Innate immune cell type | SCFS technique | Experimental model | Conclusion |
|---|---|---|---|
| Neutrophil | OS | Human primary neutrophils (isolated from blood) | Complement product C5a increases neutrophil deformability87 |
| Human primary neutrophils (isolated from blood) | Mechanical deformation induces priming and rapid depolarization of neutrophils88 | ||
| MA | Human primary immature myeloblasts, promyelocytes, myelocytes, and neutrophils (isolated from bone-marrow) | The mature neutrophil is becoming progressively softer than its progenitors89 | |
| Human primary neutrophils (isolated from blood) | ICAM-1-mediated priming causes neutrophils to stiffen90 | ||
| Human primary neutrophils from ND-CRF and HD patients (isolated from blood) | Neutrophil deformability decreases in patients with CRF91 | ||
| Human primary neutrophils (isolated from blood) | The first model for the passive rheological behavior of neutrophils92 | ||
| Cell poker | Human primary neutrophils (isolated from blood) | fMLP-mediated priming causes neutrophils to stiffen93 | |
| AFM | Human primary neutrophils (isolated from blood) | Human primary neutrophils are six times softer than their precursors (HL60 cell-line)94 | |
| Rat neutrophils (isolated from blood) | Adherence-mediated priming causes neutrophils to stiffen95 | ||
| RT-DC | Human primary neutrophils (isolated from blood) | Neutrophil priming induces stiffening in the first 1–5 min, followed by softening within 15–30 min post-priming96 | |
| Sepsis patient-derived neutrophils (isolated from blood) | Neutrophils from patients with sepsis possessed an increased deformability97 | ||
| Psoriasis patient-derived neutrophils(isolated from blood) | CXCL16 and IL-8 soften neutrophils facilitating infiltration into skin98 | ||
| Micro-fluidics | Human primary neutrophils (isolated from blood) | Neutrophils exposed to serum from ARDS patients rapidly stiffen99 | |
| MR | Human primary neutrophils (isolated from blood) | Neutrophils stiffen during phagocytosis. The observed stiffening is dependent on the size of the engulfed particles100 | |
| Monocyte | AFS, OT | Human primary monocytes (isolated from blood) | Characterization of monocyte mechanics at physiological temperature. The work reports elastic and viscous parameters, strain dependency of stiffness, and resolves the effect of CCL232 |
| MR | Human primary monocytes (isolated from blood) | Characterization of elastic and viscous properties of monocytes, adhered to a fibronectin coated surface101 | |
| MF | Human primary monocytes (isolated from blood) | Monocytes exposed to serum from ARDS patients rapidly stiffen99 | |
| RT-DC | Systemic sclerosis (SSc) patient-derived monocytes (isolated from blood) | All three subpopulations of monocytes identified had higher deformation and cross-sectional area in SSc patients as compared to healthy controls102 | |
| Cell poker | Human and rabbit primary monocytes (isolated from blood) | LPS exposure induced monocyte stiffening103 | |
| Dendritic cell | RT-DC | Human primary monocyte-derived dendritic cells (isolated from blood) | Maturation of dendritic cells leads to increased cellular stiffness and higher membrane fluidity104 |
| MR | Human primary monocyte-derived dendritic cells (isolated from blood) | Characterization of elastic and viscous properties of dendritic cells. Exposure to IFNγ or LPS stiffens dendritic cells. The opposite occurs for a TNFα + PGE2 cocktail101 | |
| AFM | Mouse bone marrow-derived dendritic cells (isolated from bone marrow) | Dendritic cells stiffen during maturation, which enhances their ability to prime T-cells105 | |
| Macrophage | AFS, OT | Human primary monocyte-derived macrophages (M-CSF and GM-CSF stimulated) (isolated from blood) | Characterization of macrophage mechanics at physiological temperature. The work reports elastic and viscous parameters, strain dependency of stiffness, and resolves the difference between M1 and M246 |
| MR | Human primary monocyte-derived macrophages (M-CSF stimulated) (isolated from blood) | Characterization of elastic and viscous properties of macrophages, bound to a fibronectin coated surface. Exposure to IFNγ or LPS stiffens macrophages. The opposite occurs for a TNFα + PGE2 cocktail101 | |
| MTC | Human alveolar macrophages (isolated from lung tissue) | Macrophages stiffen upon exposure to LPS and IFN-γ. Macrophages stiffen when cultured on a rigid substrate and soften when cultured on a less rigid substrate49 | |
| ATC | Mouse peritoneal macrophages (isolated from peritoneal cavity) | Macrophages with rod-shaped crystal-like drug inclusions were softer than macrophages that engulfed solid polyethylene microbeads48 | |
| Eosinophil | RT-DC | Human primary eosinophils from atopic and non-atopic subjects (isolated from blood) | Eosinophils from atopic subjects show a higher total area ratio than eosinophils from non-atopic subjects, comparable with basal priming. Eosinophil deformability and area were unchanged between the atopic and non-atopic eosinophils106 |
| Natural killer (NK) cell | MA | Human primary NK-cells (isolated from blood) | IL-2-activated NK-cells show a reduction in rigidity after thioglycolate treatment107 |
| Human primary NK-cells (isolated from blood) | Reducing NK cell rigidity by thioglycolate facilitates NK infiltration into tissues, but compromises its immunotherapeutic efficacy108 | ||
| Human primary NK-cells (isolated from blood) | IL2 exposure increases NK cell rigidity, which was maintained for up to 96h after IL2 withdrawal109 |
4. Resolving immune cell mechanics in cancer
The physical microenvironment also plays an important role in the modulation of mechanical properties. An example of such a microenvironment is the one surrounding a growing tumor. In the following section, we will explore the effects of the tumor microenvironment on the mechanical properties of macrophages therein.Cancers evolve as complex ecosystems that consist of malignant tumor cells and numerous non-cancerous cells, such as immune cells, cancer-associated fibroblasts, endothelial cells, pericytes, and various additional tissue-resident cells that are embedded in a complex network of fibers and proteins known as the extracellular matrix (ECM).51 The ECM provides a physical scaffold for cells and helps to regulate their behavior.52 In the case of cancer, changes in the communication between cells and the ECM play a key role in tumor growth and spread. Cancer cells can alter the stiffness and composition of the ECM, creating a positive tumorigenic feedback loop that alters cancer cell mechanics and promotes their survival, proliferation, and invasiveness.53 Altered mechanics have therefore been considered a hallmark of disseminating tumor cells: more than two-thirds of metastatic cells exhibit lower levels of stiffness.54
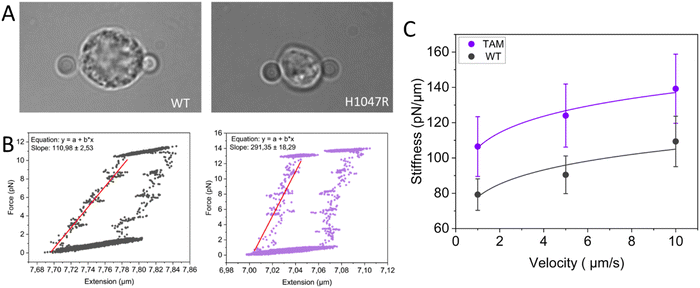
Tumor-associated macrophages (TAMs) are the most abundant innate immune cell type in solid tumors. TAMs are often recruited from bone marrow-derived monocytes via the tumor cell-derived CCL2-CCR2 chemokine signaling pathway, but can also differentiate from resident macrophages.55–57 Within the tumor microenvironment, tumor, and host cells are known to trigger macrophage polarization and hijack their inflammatory and wound-healing capacities to favor tumor progression.58,59 In many solid cancer types, anti-inflammatory TAMs with M2-like properties are the predominant macrophage population, which has been directly correlated with poor prognosis and increased metastatic potential.60–62 Whereas the alterations of cancer cell mechanics within the tumor microenvironment have been extensively studied,63–65 they remain to be elucidated for the macrophages and other immune cells present therein.66 This major shortcoming has mainly been due to the lack of methodologies to probe TAM mechanics. Here, we report preliminary data on the first single-cell mechanical characterization of TAMs at physiological temperature and discuss the technical possibilities offered by state-of-the-art technology and the challenges involved. We used single-cell optical tweezers to measure stiffness profiles of resident F4/80+ macrophages freshly isolated from healthy mammary gland tissue from littermate control FVB mice and TAMs isolated from spontaneous mammary tumors from the WapCre;Cdh1F/F;PIK3CA-H1047R genetically engineered mouse tumor model (FVB background), driven by the loss of E-cadherin and mutant PIK3CA.
In order to probe the mechanical properties of macrophages from healthy mammary glands and TAMs from mammary tumors, we performed multiple stressing–destressing cycles, in which the cells were pulled and relaxed at a constant strain rate at different values of velocity (1, 5, and 10 μm s−1). Examples of hysteresis loops of WT macrophages and TAMs at a stretching velocity of 10 μm s−1 are shown in Fig. 3B. From the slope of the linear ascending part of the stress–strain curve, the stiffness of the macrophages at each stretching velocity can be determined. Notably, only cells showing a distinguished hysteresis loop were included in the analysis. Possible reasons for not obtaining the characteristic loops are the improper attachment of the beads to the cells, cells that are in poor condition (i.e., entered apoptotic phase), and off-centered beads during stretching. Average stiffness values for WT macrophages and TAMs at each stretching velocity are presented in Fig. 3C. Our data suggest that, for this particular tumor model, TAMs are stiffer than the corresponding tissue-resident macrophages. It has previously been shown that YES-associated protein (YAP), a mature nuclear transcription coactivator that responds to a variety of mechanical changes, senses the hardness of the surrounding matrix and stimulates macrophages to transform into the immunosuppressive M2 type.67 In addition, in vivo and in vitro studies have shown that macrophages adjust their polarization state in response to surrounding mechanical stimuli.68 As such, the involvement of YAP in the polarization of macrophages towards the immunosuppressive type would be interesting for future studies.
These experiments are technically difficult, in particular, due to the limited number of immune cells that can be assessed, and their inherent heterogeneity requiring high resolution to detect small changes in mechanical properties. Nevertheless, the presented data demonstrate that sensitive measurements of immune cell mechanics are achievable thanks to technological advancements.
5. Future perspectives
Over the last decade, a conceptual framework has emerged for measuring and interpreting the mechanical properties of immune cells. However, the field is still in its infancy. Although various studies have provided a useful approximation of the mechanical behavior of innate immune cells by utilizing in vitro cell lines, they do not fully recapitulate in vivo immune cell physiology and functionality.38,69–78 We have started to unravel the mechanical properties of monocytes and macrophages and the potential link to their immune functions under physiological conditions and in cancer. These studies can help us better understand the function of innate immune cells in health and disease. It has recently been shown that mechanical markers can be used for tracking disease progression and treatment in various tissues.79,80 Such approaches may also be utilized as future possibilities in the field of mechanoimmunology. For instance, the mechanical properties of immune cells may provide us with insights into how these cells cope with challenging circumstances, such as the aberrant deposition and organization of ECM protein that results in increased matrix stiffness, which is often observed in the mammary tumor microenvironment.The mechanical characterization of innate immune cells could enable the development of mechanical biomarkers that can be used to monitor innate immune activity in both healthy and diseased states. To achieve this goal, a step in the right direction was taken by Toepfner and colleagues, who were able to detect human disease conditions by single-cell morpho-rheological phenotyping of blood.81 Real-time deformability cytometry, a novel high-throughput method for the continuous mechanical characterization of single cells, allowed them to identify all major blood cells in a single drop of blood and characterize their mechanical alterations in several disease conditions.
It must be noted here that macrophages, orchestrators of virtually all major diseases, do not circulate in the blood. The approach introduced by our lab, therefore, brings us a step closer to mechanically characterizing all innate immune cells (both circulating and tissue-resident) that impact disease progression, as we have shown for TAMs (Fig. 3). Future studies using a combination of organ-on-chip modeling and SCFS could circumvent the need for specifically isolating the relevant cells of interest, preventing the potential of inadvertent cell change.82,83
In addition to diagnostics, we hypothesize that mechanical phenotyping opens avenues for mechanics-targeting therapies in which external stimuli could be employed to program innate immune cells toward desired mechanical phenotypes. For instance, if we could solely change the mechanical properties of cells with minimally altering any other process, then we could soften and reprogram otherwise rigid M2-like TAMs to M1-like macrophages and see whether that halts tumor growth through enhanced tumor cell killing. Undoubtedly, cell mechanics is intricately linked with numerous other processes, making it a challenging task to identify specific factors that solely influence mechanical changes. In fact, mechanical cues experienced by immune cells are translated into biochemical signals through the process of mechanotransduction, which, through multiple signaling pathways, impacts crucial cellular functions including metabolism.4,7 We have recently described how mechanical cues, sensed through the cytoskeleton or distortion of the cell and organelles, can induce metabolic changes in the cell and how the resulting alterations in metabolism, in turn, can feed back to regulate the mechanical properties of cells and tissues.84 Advances in analytical technologies are enabling studying metabolism at the single-cell level,85 and have been recently applied to innate immune cells too.86 Future endeavors should therefore incorporate correlating mechanical information from a cell to other data including multi-omics.
Conflicts of interest
K.E.d.V. reports research funding from Roche/Genentech and is consultant for Macomics, outside the scope of this work.Acknowledgements
K.E.d.V. acknowledges funding from the Netherlands Organization for Scientific Research (grant NWO-VICI 91819616).References
- J. S. Marshall, R. Warrington, W. Watson and H. L. Kim, An introduction to immunology and immunopathology, Allergy, Asthma, Clin. Immunol., 2018, 14(Suppl 2), 49, DOI:10.1186/s13223-018-0278-1.
- M. Huber-Lang, J. D. Lambris and P. A. Ward, Innate immune responses to trauma review-article, Nat. Immunol., 2018, 19, 327–341, DOI:10.1038/s41590-018-0064-8.
- S. R. Woo, L. Corrales and T. F. Gajewski, Innate immune recognition of cancer, Annu. Rev. Immunol., 2015, 33, 445–474 CrossRef CAS PubMed.
- M. Huse, Mechanical forces in the immune system, Nat. Rev. Immunol, 2017, 17, 679–690, DOI:10.1038/nri.2017.74.
- F. Y. McWhorter, C. T. Davis and W. F. Liu, Physical and mechanical regulation of macrophage phenotype and function, Cell. Mol. Life Sci., 2015, 72, 1303–1316, DOI:10.1007/s00018-014-1796-8.
- N. Jain, J. Moeller and V. Vogel, Mechanobiology of Macrophages: How Physical Factors Coregulate Macrophage Plasticity and Phagocytosis, Annu. Rev. Biomed. Eng., 2019, 21, 267–297, DOI:10.1146/annurev-bioeng-062117.
- H. Du, et al., Tuning immunity through tissue mechanotransduction, Nat. Rev. Immunol., 2023, 23, 174–188, DOI:10.1038/s41577-022-00761-w.
- A. E. Ekpenyong, N. Toepfner, E. R. Chilvers and J. Guck, Mechanotransduction in neutrophil activation and deactivation, Biochim. Biophys. Acta, Mol. Cell Res., 2015, 3105–3116 CrossRef CAS PubMed.
- C. Zhu, W. Chen, J. Lou, W. Rittase and K. Li, Mechanosensing through immunoreceptors, Nat. Immunol., 2019, 10, 1269–1278 CrossRef PubMed.
- E. Evans and A. Yeung, Apparent viscosity and cortical tension of blood granulocytes determined by micropipet aspiration, Biophys. J., 1989, 56, 151–160 CrossRef CAS PubMed.
- R. M. Hochmuth, Micropipette Aspiration of Living Cells, J. Biomech., 2000, 33, 15–22 CrossRef CAS PubMed.
- A. Sinha, T. T. T. Chu, M. Dao and R. Chandramohanadas, Single-cell evaluation of red blood cell bio-mechanical and nano-structural alterations upon chemically induced oxidative stress, Sci. Rep., 2015, 5, 9768 CrossRef CAS PubMed.
- M. Dao, C. T. Lim and S. Suresh, Mechanics of the human red blood cell deformed by optical tweezers, J. Mech. Phys. Solids, 2003, 51, 2259–2280 CrossRef.
- R. Zhu, T. Avsievich, A. Popov and I. Meglinski, Optical Tweezers in Studies of Red Blood Cells, Cells, 2020, 9(3), 545 CrossRef CAS PubMed.
- M. A. S. de Oliveira, D. S. Moura, A. Fontes and R. E. de Araujo, Damage induced in red blood cells by infrared optical trapping: an evaluation based on elasticity measurements, J. Biomed. Opt., 2016, 21, 075012 CrossRef PubMed.
- H. Turlier, et al., Equilibrium physics breakdown reveals the active nature of red blood cell flickering, Nat. Phys., 2016, 12, 513–519 Search PubMed.
- G. Lenormand, S. Hénon, A. Richert, J. Siméon and F. Gallet, Direct measurement of the area expansion and shear moduli of the human red blood cell membrane skeleton, Biophys. J., 2001, 81, 43–56 CrossRef CAS PubMed.
- S. Hénon, G. Lenormand, A. Richert and F. Gallet, A new determination of the shear modulus of the human erythrocyte membrane using optical tweezers, Biophys. J., 1999, 76, 1145–1151 CrossRef PubMed.
- K. Svoboda, C. F. Schmidt, D. Branton and S. M. Block, Conformation and elasticity of the isolated red blood cell membrane skeleton, Biophys. J., 1992, 63, 784–793 CrossRef CAS PubMed.
- J. Sleep, D. Wilson, R. Simmons and W. Gratzer, Elasticity of the red cell membrane and its relation to hemolytic disorders: An optical tweezers study, Biophys. J., 1999, 77, 3085–3095 CrossRef CAS PubMed.
- S. Suresh, et al., Reprint of: Connections between single-cell biomechanics and human disease states: Gastrointestinal cancer and malaria, Acta Biomater., 2015, 23, S3–S15 CrossRef PubMed.
- S. Suresh, et al., Connections between single-cell biomechanics and human disease states: Gastrointestinal cancer and malaria, Acta Biomater., 2005, 1, 15–30 CrossRef CAS PubMed.
- S. K. Mohanty, A. Uppal and P. K. Gupta, Self-Rotation of Red Blood Cells in Optical Tweezers: Prospects for High Throughput Malaria Diagnosis, Biotechnol. Lett., 2004, 26, 971–974 CrossRef CAS PubMed.
- H. S. Byun, et al., Optical measurement of biomechanical properties of individual erythrocytes from a sickle cell patient, Acta Biomater., 2012, 8, 4130–4138 CrossRef PubMed.
- M. M. Brandão, et al., Optical tweezers for measuring red blood cell elasticity: Application to the study of drug response in sickle cell disease, Eur. J. Haematol., 2003, 70, 207–211 CrossRef PubMed.
- R. Agrawal, et al., Assessment of red blood cell deformability in type 2 diabetes mellitus and diabetic retinopathy by dual optical tweezers stretching technique, Sci. Rep., 2016, 6, 15873 CrossRef CAS PubMed.
- C. N. Lima, et al., Evaluating viscoelastic properties and membrane electrical charges of red blood cells with optical tweezers and cationic quantum dots – applications to β-thalassemia intermedia hemoglobinopathy, Colloids Surf., B, 2020, 186, 110671 CrossRef CAS PubMed.
- K. Fraczkowska, et al., Alterations of biomechanics in cancer and normal cells induced by doxorubicin, Biomed. Pharmacother., 2018, 97, 1195–1203 CrossRef CAS PubMed.
- V. Sheikhhassani, T. M. J. Evers, S. Lamba, F. Shokri and A. Mashaghi, Single cell force spectroscopy of erythrocytes at physiological and febrile temperatures reveals mechano-modulatory effects of atorvastatin, Soft Matter, 2022, 18, 2143–2148 RSC.
- C. Shi and E. G. Pamer, Monocyte recruitment during infection and inflammation, Nat. Rev. Immunol, 2011, 11, 762–774, DOI:10.1038/nri3070.
- M. Gschwandtner, R. Derler and K. S. Midwood, More Than Just Attractive: How CCL2 Influences Myeloid Cell Behavior Beyond Chemotaxis, Front. Immunol., 2019, 10, 2759, DOI:10.3389/fimmu.2019.02759.
- T. M. J. Evers, et al., Single-cell analysis reveals chemokine-mediated differential regulation of monocyte mechanics, iScience, 2022, 25(1), 103555 CrossRef CAS PubMed.
- J. Dupire, P. H. Puech, E. Helfer and A. Viallat, Mechanical adaptation of monocytes in model lung capillary networks, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 14798–14804 CrossRef CAS PubMed.
- K. D. Rinker, V. Prabhakar and G. A. Truskey, Effect of Contact Time and Force on Monocyte Adhesion to Vascular Endothelium, Biophys. J., 2001, 80(4), 1722–1732 CrossRef CAS PubMed.
- K. Ley, C. Laudanna, M. I. Cybulsky and S. Nourshargh, Getting to the site of inflammation: The leukocyte adhesion cascade updated, Nat. Rev. Immunol, 2007, 7, 678–689, DOI:10.1038/nri2156.
- S. Nourshargh, P. L. Hordijk and M. Sixt, Breaching multiple barriers: Leukocyte motility through venular walls and the interstitium, Nat. Rev. Mol. Cell Biol., 2010, 11, 366–378, DOI:10.1038/nrm2889.
- S. Nourshargh and R. Alon, Leukocyte Migration into Inflamed Tissues, Immunity, 2014, 41, 694–707, DOI:10.1016/j.immuni.2014.10.008.
- A. E. Ekpenyong, et al., Viscoelastic Properties of Differentiating Blood Cells Are Fate- and Function-Dependent, PLoS One, 2012, 7(9), e45237 CrossRef CAS PubMed.
- P. Italiani and D. Boraschi, From monocytes to M1/M2 macrophages: Phenotypical vs. functional differentiation, Front. Immunol., 2014, 5, 514, DOI:10.3389/fimmu.2014.00514.
- C. Yunna, H. Mengru, W. Lei and C. Weidong, Macrophage M1/M2 polarization, Eur. J. Pharmacol., 2020, 877, 173090, DOI:10.1016/j.ejphar.2020.173090.
- F. A. W. Verreck, T. de Boer, D. M. L. Langenberg, L. van der Zanden and T. H. M. Ottenhoff, Phenotypic and functional profiling of human proinflammatory type-1 and anti-inflammatory type-2 macrophages in response to microbial antigens and IFN-γ- and CD40L-mediated costimulation, J. Leukocyte Biol., 2006, 79, 285–293 CrossRef CAS PubMed.
- F. A. W. Verreck, et al., Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4560–4565 CrossRef CAS PubMed.
- S. K. Biswas and A. Mantovani, Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm, Nat. Immunol., 2010, 11, 889–896, DOI:10.1038/ni.1937.
- P. J. Murray, Macrophage Polarization, Annu. Rev. Physiol., 2017, 79, 541–566, DOI:10.1146/annurev-physiol-022516-034339.
- N. Jain, J. Moeller and V. Vogel, Mechanobiology of Macrophages: How Physical Factors Coregulate Macrophage Plasticity and Phagocytosis, Annu. Rev. Biomed. Eng., 2019, 21, 267–297, DOI:10.1146/annurev-bioeng-062117.
- T. M. J. Evers, et al., Single-Cell Mechanical Characterization of Human Macrophages, Adv. NanoBiomed Res., 2022, 2, 2100133 CrossRef CAS.
- M. Agarwal, P. Biswas, A. Bhattacharya and D. K. Sinha, Reactive oxygen species-mediated cytoplasmic stiffening impairs the phagocytic ability of the macrophage, J. Cell Sci., 2020, 133(5), jcs236471 CrossRef PubMed.
- X. Hong, et al., Acoustic tweezing cytometry for mechanical phenotyping of macrophages and mechanopharmaceutical cytotripsy, Sci. Rep., 2019, 9, 5702 CrossRef PubMed.
- N. R. Patel, et al., Cell Elasticity Determines Macrophage Function, PLoS One, 2012, 7, e41024 CrossRef CAS PubMed.
- P. Pacheco, D. White and T. Sulchek, Effects of Microparticle Size and Fc Density on Macrophage Phagocytosis, PLoS One, 2013, 8(4), e60989 CrossRef CAS PubMed.
- K. E. de Visser and J. A. Joyce, The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth, Cancer Cell, 2023, 41, 374–403 CrossRef CAS PubMed.
- J. C. Valdoz, et al., The ECM: To scaffold, or not to scaffold, that is the question, Int. J. Mol. Sci., 2021, 22(23), 12690, DOI:10.3390/ijms222312690.
- F. B. Kai, A. P. Drain and V. M. Weaver, The Extracellular Matrix Modulates the Metastatic Journey, Dev. Cell, 2019, 49, 332–346, DOI:10.1016/j.devcel.2019.03.026.
- S. E. Cross, Y. S. Jin, J. Rao and J. K. Gimzewski, Nanomechanical analysis of cells from cancer patients, Nat. Nanotechnol., 2007, 2, 780–783 CrossRef CAS PubMed.
- J. Jin, et al., CCL2: An Important Mediator Between Tumor Cells and Host Cells in Tumor Microenvironment, Front. Oncol., 2021, 11, 1–14, DOI:10.3389/fonc.2021.722916.
- R. N. Hanna, et al., Patrolling monocytes control tumor metastasis to the lung, Science, 1979, 350(985–990), 2015 Search PubMed.
- B. Z. Qian, et al., CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis, Nature, 2011, 475, 222–225 CrossRef CAS PubMed.
- C. Salvagno, et al., Therapeutic targeting of macrophages enhances chemotherapy efficacy by unleashing type I interferon response, Nat. Cell Biol., 2019, 21, 511–521 CrossRef CAS PubMed.
- D. J. Kloosterman and L. Akkari, Macrophages at the interface of the co-evolving cancer ecosystem, Cell, 2023, 186(8), 1627–1651, DOI:10.1016/j.cell.2023.02.020.
- C. E. Lewis and J. W. Pollard, Distinct Role of Macrophages in Different Tumor Microenvironments, Cancer Res., 2006, 66, 605–612 CrossRef CAS PubMed.
- M. Yang, D. McKay, J. W. Pollard and C. E. Lewis, Diverse Functions of Macrophages in Different Tumor Microenvironments, Cancer Res., 2018, 78, 5492–5503 CrossRef CAS PubMed.
- G. Luthria, et al., In vivo microscopy reveals macrophage polarization locally promotes coherent microtubule dynamics in migrating cancer cells, Nat. Commun., 2020, 11, 3521 CrossRef CAS PubMed.
- P. Katira, M. H. Zaman and R. T. Bonnecaze, How changes in cell mechanical properties induce cancerous behavior, Phys. Rev. Lett., 2012, 108(2), 028103 CrossRef PubMed.
- D. Wirtz, K. Konstantopoulos and P. C. Searson, The physics of cancer: The role of physical interactions and mechanical forces in metastasis, Nat. Rev. Cancer, 2011, 11, 512–522, DOI:10.1038/nrc3080.
- V. Swaminathan, et al., Mechanical Stiffness grades metastatic potential in patient tumor cells and in cancer cell lines, Cancer Res., 2011, 71, 5075–5080 CrossRef CAS PubMed.
- E. J. Hoffmann and S. M. Ponik, Biomechanical Contributions to Macrophage Activation in the Tumor Microenvironment, Front. Oncol., 2020, 10, 787, DOI:10.3389/fonc.2020.00787.
- V. S. Meli, et al., YAP-Mediated Mechanotransduction Tunes the Macrophage Inflammatory Response, Sci. Adv., 2020, 6(49), eabb8471 CrossRef CAS PubMed.
- S. Adams, L. M. Wuescher, R. Worth and E. Yildirim-Ayan, Mechano-Immunomodulation: Mechanoresponsive Changes in Macrophage Activity and Polarization, Ann. Biomed. Eng., 2019, 47, 2213–2231, DOI:10.1007/s10439-019-02302-4.
- F. Lautenschläger, et al., The regulatory role of cell mechanics for migration of differentiating myeloid cells, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(37), 15696–15701, DOI:10.1073/pnas.0811261106.
- M. A. Tsai, R. E. Waugh and P. C. Keng, Changes In Hl-60 Cell Deformability During Differentiation Induced By Dmso, Biorheology, 1996, 33, 1–15 CAS.
- K. D. Rinker, et al., Linoleic acid increases monocyte deformation and adhesion to endothelium, Atherosclerosis, 2004, 177, 275–285 CrossRef CAS PubMed.
- F. Richelme, A.-M. Benoliel and P. Bongrand, Dynamic Study of Cell Mechanical and Structural Responses to Rapid Changes of Calcium Level, Cytoskeleton, 2000, 45(2), 93–105 CrossRef CAS.
- A. Ravetto, H. M. Wyss, P. D. Anderson, J. M. J. Den Toonder and C. V. C. Bouten, Monocytic cells become less compressible but more deformable upon activation, PLoS One, 2014, 9(3), e92814 CrossRef PubMed.
- A. Menachery, et al., Dielectrophoretic characterization of dendritic cell deformability upon maturation, Biotechniques, 2020, 70, 29–36 CrossRef PubMed.
- Y. Zhao, G. Mahajan, C. R. Kothapalli and X.-L. Sun, Sialylation Status and Mechanical Properties of THP-1 Macrophages upon LPS Stimulation, Biochem. Biophys. Res. Commun., 2019, 518(3), 573–578 CrossRef CAS PubMed.
- J. Lam, M. Herant, M. Dembo and V. Heinrich, Baseline mechanical characterization of J774 macrophages, Biophys. J., 2009, 96, 248–254 CrossRef CAS PubMed.
- M. A. Tsai and D. A. Hammert, Rheology of Rat Basophilic Leukemia Cells, Ann. Biomed. Eng., 1997, 25, 62–68 CrossRef CAS PubMed.
- Z.-Y. Liu, J.-I. Young and E. L. Elson, Rat Basophilic Leukemia Cells Stiffen When They Secrete, J. Cell. Biol., 1987, 105(6 Pt 2), 2933–2943 CrossRef CAS PubMed.
- Z. Chang, L. Zhang, J. T. Hang, W. Liu and G. K. Xu, Viscoelastic Multiscale Mechanical Indexes for Assessing Liver Fibrosis and Treatment Outcomes, Nano Lett., 2023, 23, 9618–9625 CrossRef PubMed.
- Z. Chang, J. Zhang, Y. Liu, H. Gao and G. K. Xu, New Mechanical Markers for Tracking the Progression of Myocardial Infarction, Nano Lett., 2023, 23, 7350–7357 CrossRef CAS PubMed.
- N. Toepfner, et al., Detection of human disease conditions by single-cell morpho-rheological phenotyping of blood, eLife, 2018, 7, e29213, DOI:10.7554/eLife.29213.001.
- H. Tang, et al., Human Organs-on-Chips for Virology, Trends Microbiol., 2020, 28, 934–946, DOI:10.1016/j.tim.2020.06.005.
- D. E. Ingber, From tensegrity to human organs-on-chips: implications for mechanobiology and mechanotherapeutics, Biochem. J., 2023, 480, 243–257 CrossRef CAS PubMed.
- T. M. J. Evers, L. J. Holt, S. Alberti and A. Mashaghi, Reciprocal regulation of cellular mechanics and metabolism, Nat. Metab., 2021, 3, 456–468 CrossRef PubMed.
- T. M. J. Evers, et al., Deciphering Metabolic Heterogeneity by Single-Cell Analysis, Anal. Chem., 2019, 91, 13314–13323 CrossRef CAS PubMed.
- H. Tang, et al., Random forest and live single-cell metabolomics reveal metabolic profiles of human macrophages upon polarization, Biotechnol. Bioeng., 2023, 120, 2314–2325 CrossRef CAS PubMed.
- S. Denk, et al., Complement C5a-Induced Changes in Neutrophil Morphology During Inflammation, Scand. J. Immunol., 2017, 86, 143–155 CrossRef CAS PubMed.
- A. E. Ekpenyong, et al., Mechanical Deformation Induces Depolarization of Neutrophils, Sci. Adv., 2017, 3(6), e1602536 CrossRef PubMed.
- M. A. Lichtman, Cellular deformability during maturation of the myeloblast, N. Engl. J. Med., 1970, 283, 943–948 CrossRef CAS PubMed.
- A. Pai, P. Sundd and D. F. J. Tees, In situ microrheological determination of neutrophil stiffening following adhesion in a model capillary, Ann. Biomed. Eng., 2008, 36, 596–603 CrossRef PubMed.
- A. T. Skoutelis1, et al., Nephrology Dialysis Transplantation Polymorphonuclear leukocyte rigidity is defective in patients with chronic renal failure, Nephrol., Dial., Transplant., 2000, 15(11), 1788–1793 CrossRef PubMed.
- E. Evans and B. Kukan, Passive Material Behavior of Granulocytes Based on Large Deformation and Recovery After Deformation Tests, Blood, 1984, 64(5), 1028–1035 CrossRef CAS PubMed.
- G. S. Worthen, B. Schwab, E. L. Elson and G. P. Downey, Mechanics of stimulated neutrophils: cell stiffening induces retention in capillaries, Science, 1989, 245, 183–186 CrossRef CAS PubMed.
- M. J. Rosenbluth, W. A. Lam and D. A. Fletcher, Force microscopy of nonadherent cells: A comparison of leukemia cell deformability, Biophys. J., 2006, 90, 2994–3003 CrossRef CAS PubMed.
- P. Roca-Cusachs, et al., Rheology of passive and adhesion-activated neutrophils probed by atomic force microscopy, Biophys. J., 2006, 91, 3508–3518 CrossRef CAS PubMed.
- K. R. Bashant, et al., Real-time deformability cytometry reveals sequential contraction and expansion during neutrophil priming, J. Leukocyte Biol., 2019, 105, 1143–1153 CrossRef CAS PubMed.
- K. Crawford, et al., Rapid biophysical analysis of host immune cell variations associated with sepsis, Am. J. Respir. Crit. Care Med., 2018, 198, 280–282, DOI:10.1164/rccm.201710-2077LE.
- S. Steffen, et al., Toll-Like Receptor-Mediated Upregulation of CXCL16 in Psoriasis Orchestrates Neutrophil Activation, J. Invest. Dermatol., 2018, 138, 344–354 CrossRef CAS PubMed.
- P. Preira, et al., The leukocyte-stiffening property of plasma in early acute respiratory distress syndrome (ARDS) revealed by a microfluidic single-cell study: The role of cytokines and protection with antibodies, Crit. Care, 2016, 20, 8 CrossRef PubMed.
- A. Zak, et al., Distinct timing of neutrophil spreading and stiffening during phagocytosis, Biophys. J., 2022, 121, 1381–1394 CrossRef CAS PubMed.
- N. Bufi, et al., Human primary immune cells exhibit distinct mechanical properties that are modified by inflammation, Biophys. J., 2015, 108, 2181–2190 CrossRef CAS PubMed.
- A. Matei, et al., Biophysical phenotyping of circulating immune cells identifies a distinct monocyte-driven signature in systemic sclerosis, Arthritis Rheum., 2022, 75(5), 768–781, DOI:10.1002/art.42394.
- D. E. Doherty and G. S. Worthen, Lipopolysaccharide-induced Monocyte Retention in the Lungs of Rabbits, Chest, 1994, 105, 108S CAS.
- J. J. Lühr, et al., Maturation of Monocyte-Derived DCs Leads to Increased Cellular Stiffness, Higher Membrane Fluidity, and Changed Lipid Composition, Front. Immunol., 2020, 11, 590121 CrossRef PubMed.
- D. Blumenthal, V. Chandra, L. Avery and J. K. Burkhardt, Mouse t cell priming is enhanced by maturation-dependent stiffening of the dendritic cell cortex, eLife, 2020, 9, 1–44 CrossRef PubMed.
- L. Porter, et al., Metabolic profiling of human eosinophils, Front. Immunol., 2018, 9, 1404 CrossRef PubMed.
- R. J. Melder and R. K. Jain, Reduction of Rigidity in Human Activated Natural Killer Cells by Thioglycollate Treatment, J. Immunol. Methods, 1994, 175(1), 69–77 CrossRef CAS PubMed.
- R. J. Melder, C. A. Kristensen, L. L. Munn and R. K. Jain, Modulation of A-NK Cell Rigidity: In Vitro Characterization and in Vivo Implications for Cell Delivery, Biorheology, 2001, 38, 151–159 CAS.
- R. J. Melder and R. K. Jain, Kinetics of Interleukin-2 Induced Changes in Rigidity of Human Natural Killer, Cells., 1992, 20(2–3), 161–176 CAS.
| This journal is © The Royal Society of Chemistry 2024 |