Open Access Article
Open Access ArticleUnveiling the functional components and anti-Alzheimer's activity of Koelreuteria elegans (Seem.) A.C. Sm. using UHPLC-MS/MS and molecular networking†
Mohamed S.
Demerdash‡
a,
Reem T.
Attia‡
b,
Moshera M.
El-Sherei
a,
Wafaa M.
Aziz
a,
Sherif Ashraf
Fahmy
 *c and
Marwa Y.
Issa
*c and
Marwa Y.
Issa
 *a
*a
aDepartment of Pharmacognosy, Faculty of Pharmacy, Cairo University, Cairo 11562, Egypt. E-mail: marwa.issa@pharma.cu.edu.eg
bDepartment of Pharmacology, Toxicology, and Biochemistry, Faculty of Pharmacy, Future University in Egypt, Cairo 11865, Egypt
cDepartment of Chemistry, School of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, R5 New Garden City, New Administrative Capital, AL109AB, Cairo 11835, Egypt. E-mail: sheriffahmy@aucegypt.edu
First published on 12th March 2024
Abstract
The use of plant extracts and their phytochemicals as candidates for treating Alzheimer's disease (AD) has been increasingly demanded lately. AD is a progressive neurodegenerative disorder, assumed to be associated with the formation of Aβ plaques and neurofibrillary tangles as well as with neuroinflammation, mediated by cytokines. The metabolomic profiles of Koelreuteria elegans (Seem.) A.C. Sm. leaf and fruit methanol extracts (KEL and KEF, respectively) were explored using UHPLC-MS/MS analysis aided by molecular networking in negative and positive modes for the first time. A total of 139 metabolites of different classes were tentatively identified. The molecular networking (MN) reflected high levels of phenolics and flavonoids. KEL and KEF showed great effects on memory function and spatial learning in behavioral experiments of the injured streptozotocin (STZ)-treated mice. The plant extracts led to pronounced improvement in the histopathological profile of the cerebral cortex of the injured STZ-treated mice. The effect of extracts on the levels of neuroinflammatory mediators TNF-α, NF-κB and IL-1β in AD-induced mice was assessed. Both extracts reduced all these markers of inflammation and neurodegeneration in AD.
1. Introduction
Alzheimer's disease (AD) is the most common progressive neurodegenerative disorder, afflicting the health of the elderly. It primarily affects the hippocampal and cerebral cortex regions, causing deficits in memory, learning, thinking, and spatial orientation skills, as well as cognitive dysfunctions and personality alterations. The main pathological changes in the brain include the formation of extracellular senile plaques and neurofibrillary tangles and loss of neurons, leading to atrophy of the cerebral cortex, which is accompanied by a defect in the psychic and physical abilities.1,2 The chronic inflammatory processes accompanying AD played a key role in the development of neurodegeneration in AD. Cytokines are products of the immune system and affect a wide range of biological functions, including immunity, inflammation and repair. Although most cytokines are produced at low levels in a healthy brain, neuroinflammation can be detected years before neuronal apoptosis. Pro-inflammatory cytokines represent immunoregulatory molecules that promote inflammation. The anti-inflammatory cytokines control the pro-inflammatory cytokine response. Inflammation is characterized by a co-ordination between pro- and anti-inflammatory cytokines and their imbalance may be an essential factor in AD.2Tumor necrosis factor alpha (TNF-α), the most studied pro-inflammatory cytokine in the pathophysiology of AD, plays an important role in the cytokine cascade during an inflammatory response. The concentration of TNF-α increases in blood and the cerebrospinal fluid of AD patients as reported by many clinical and animal studies, indicating a link between the elevation of TNF-α levels in the brain and AD progression.3 Chronic neuronal TNF-α production causes synaptic dysfunction and severe neuronal death, leading to the evolution of AD and cognitive decline.4
Nuclear factor kappa B (NF-κB) is an inflammatory transcription factor that fuels neurodegeneration. Upon exposure to pro-inflammatory mediators such as cytokines, NF-κB target genes are activated and expressed, and consequent elevation of cytokines and chemokines in microglia results in the chronic neuroinflammation observed in AD. Moreover, the elevation of NF-κB levels in the cerebral cortex of AD patients is correlated with the formation of amyloid fibrils, which consequently aggregate into amyloid plaques.5,6
Interleukin-1 (IL-1) is a pleiotropic cytokine family comprising a network of eleven pro-inflammatory cytokines capable of regulating acute and chronic inflammatory responses. Studies suggested the possible role of IL-1 in the immune processes in chronic neurodegenerative diseases, such as AD. The first members of the IL-1 family to be identified were IL-1α and IL-1β.2 The pro-inflammatory cytokine IL-1β has a critical modulatory effect in the pathogenesis of AD. Studies in human beings have demonstrated that an increase in IL-1β expression has been associated with AD brain pathology. Experimental models showed that elevation of serum levels of IL-1β is directly implicated in neurodegenerative injury and neural loss.7,8
Koelreuteria elegans (Seem.) A.C. Sm. (K. formosana Hayata or K. henryi Dumm.) is a deciduous, ornamental landscape tree belonging to the family Sapindaceae, native to Taiwan and Fiji and also cultivated in South America, Australia and some Asian countries.9 It is a fast-growing species and tolerant of various environmental conditions.9 The plant species have been used in traditional Taiwanese medicine; its roots, bark, twigs, and leaves have been used to treat diarrhea, urethritis, and malaria and improve liver functions. Moreover the seeds of K. elegans were used as insecticides and the leaves as anti-fungal and anti-bacterial agents, besides being used as a black hair dye.10–15 Previous phytochemical studies of this species led to the identification of phenolic compounds, flavonoids, lignans, sterols, tocopherols and triterpenes.11,14–21 The metabolites of K. elegans form the basis for the determination of its biological activities. A great suppressive effect on dihydrodiol dehydrogenase expression has been demonstrated.20 Protein-tyrosine kinase (PTK) was inhibited by kaempferol and quercetin and their glycosides that were isolated from the leaves and twigs;16 in addition, the antiproliferative activities of different fractions of isolated compounds against various human tumor cell lines were reported.11,14,17,22 Antioxidant and ROS scavenging activities of different fractions of leaf extract of K. elegans,21,23,24 aqueous extract of its flowers,13 and 1,3,4,5-tetra-O-galloylquinic acid isolated from the leaves19 and extracts of aerial plant parts12 were documented. Furthermore, El Naggar demonstrated the antimicrobial activity of the aqueous methanolic extract of K. elegans leaves and its pure compounds, 1,3,4,5-tetra-O-galloylquinic acid butyl ester and methyl gallate, against Geotrichum candidum, Staphylococcus aureus, Bacillus subtilis, Enterococcus faecalis, Salmonella typhimurium and Escherichia coli, along with their hepatoprotective effect.15
Concerning current literature, nothing was found dealing with the identification and elucidation of the metabolite profiles of K. elegans leaf or fruit crude extracts or their anti-AD activity. This activity in genus Kolreuteria was only studied for K. paniculata Laxm.1
Our study aimed to analyze the chemical profiles of K. elegans leaf and fruit methanol extracts by LC MS/MS-based molecular networking in both negative and positive high-resolution electrospray ionization (ESI) modes to characterize their bioactive metabolites and find possible metabolomic differences. In addition, the effects of the tested samples on memory function and spatial learning in behavioral experiments and on the histopathological changes of the injured tissue induced by streptozotocin (STZ) in the cerebral cortex of the tested mice, as well as on the levels of elevated neuroinflammatory mediators TNF-α, NF-KB and IL-1β in the STZ-induced AD mouse model, were investigated.
2. Materials and methods
2.1. Plant materials
Fresh leaves and fruits of Koelreuteria elegans were collected in May 2021 from the Agriculture Research Centre, Giza, Egypt. The plant was kindly identified by Prof. Dr Abdel Halim Abdel Mojali, Head of the Department of Flora Researches and Plant Taxonomy at the Agriculture Research Centre, Giza, Egypt and was verified by Prof. Dr Rim Samir Hamdy, Professor of Plant Taxonomy and Flora at the Department of Botany, Faculty of Science, Cairo University. The plant materials were air-dried before being ground into a powder and kept in a closed container. A voucher specimen was deposited in the Herbarium of the Pharmacognosy Department at Cairo University under registration number 18.12.23.2.2. Chemicals and reagents
For LC-MS/MS analysis, formic acid (≥95.0%), water, acetonitrile, and methanol were of LC-MS grade and supplied by Merck (Darmstadt, Germany). The hematoxylin–eosin (H&E) staining kit (500 mL) was purchased from TissuePRO, catalogue #: 90888820. Thiopental sodium was obtained from EIPICO, 10th of Ramadan City, Egypt and formalin from the Gomhouria Company, Egypt, Catalogue #: L24810. TNF-α and IL-1β ELISA kits were bought from CUSA-BIO Inc., Houston, TX, USA, Catalogue #: CSB-E11987r and the NF-KB ELISA kit was purchased from EiAAB, Wuhan, China, Catalogue #: E1824r. All other solvents were of analytical grade.2.3. Plant extraction
Four kilograms and 600 grams of air-dried powdered leaves and fruits, respectively, were separately extracted by cold maceration using methanol successively until exhaustion. Each extract was individually collected, evaporated to dryness at temperature not exceeding 40 °C under vacuum to obtain a dark green solid extract of leaves (KEL, 535 g) and a dark reddish solid extract of fruits (KEF, 40 g). The dried extracts were kept in a refrigerator at 4 °C for phytochemical and biological studies.2.4. Sample preparation for UPLC-Orbitrap HRMS analysis
KEL and KEF, 100 mg each, were mixed with 5 mL of 100% MeOH containing 10 μg mL−1 umbelliferone as the internal standard, using a Turrax mixer (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) for five 20-s periods at 1 min intervals separating each period to prevent heating. The extracts were vortexed vigorously and centrifuged at 3000 rpm for 30 min to remove debris and filtered using a 22 μm pore size filter. An aliquot of 500 μL was placed on a C18 cartridge (500 mg) preconditioned with MeOH and H2O. Samples were then eluted with 5 mL of 100% MeOH, the eluent was evaporated under nitrogen stream, and the collected dry residue was resuspended in 500 μL of MeOH. Three microlitres of the supernatant were used for UPLC-MS analysis.
000 rpm) for five 20-s periods at 1 min intervals separating each period to prevent heating. The extracts were vortexed vigorously and centrifuged at 3000 rpm for 30 min to remove debris and filtered using a 22 μm pore size filter. An aliquot of 500 μL was placed on a C18 cartridge (500 mg) preconditioned with MeOH and H2O. Samples were then eluted with 5 mL of 100% MeOH, the eluent was evaporated under nitrogen stream, and the collected dry residue was resuspended in 500 μL of MeOH. Three microlitres of the supernatant were used for UPLC-MS analysis.
2.5. UPLC-Orbitrap HRMS analysis
Both negative and positive high-resolution ESI modes and collision-induced dissociation (CID) MS spectra were obtained using an Orbitrap Elite mass spectrometer (Thermo Fischer Scientific, Darmstadt, Germany) equipped with a heated electrospray ion source adjusted at 3 kV and 4 kV in negative and positive modes, respectively, with a capillary temperature of 300 °C, a source heater temperature of 250 °C, and an FTMS resolution of 30.000. The MS spectrometer was coupled to a UHPLC system (Dionex UltiMate 3000, Thermo Fischer Scientific), equipped with an RP-18 column (30 mm × 2.1 mm × 1.8 μm), Acquity HSS T3, H2O, column temperature: 40 °C, DAD (220–600 nm, Thermo Fischer Scientific). The mobile phase consisted of H2O (A) and acetonitrile (B) provided with 0.1% formic acid. The following elution gradient was used: at 0–1 min 5% (B), followed by linear increase to reach 100% B until 11 min, then from 11 to 19 min 100% (B) was used, and finally from 19 to 30 min (B) was reduced to 5%. The flow rate used was 150 μL min−1 and the injection volume was 2 μL. The CID mass spectra were recorded using a normalized collision energy (NCE) of 35%. Calibration of the instrument was performed externally by using Pierce ESI negative ion calibration solution (Product no. 88324) and Pierce ESI positive ion calibration solution (Product no. 88323) from Thermo Fisher Scientific. The data were evaluated using Mass Hunter software version B.06.00.2.6. LC-MS/MS data preparation
UPLC-MS files were converted into the mzXML format using the ProteoWizard tool MSConvert.25 The converted data were processed using the free software MZmine 2.37 for peak picking, deconvolution, deisotoping, alignment, and formula prediction.2.7. Feature-based molecular networking
The processed mzXML files were uploaded to the Global Natural Product Social Molecular Networking (GNPS) using the Winscp cross-platform. A feature-based molecular network (FBMN) in GNPS could integrate the spectral data to generate the molecular network, which compares and clusters the dataset. In the case of negative mode data, the setup parameters of the molecular network are as follows – network's basic options: precursor ion mass tolerance and fragment ion mass tolerance were set to 0.02 Da; advanced network options: minimum pairs cosine: 0.65; network TopK: 10; minimum matched fragment ions: 6; maximum connected component size: 100; maximum shift between precursors: 500 Da. All the other parameters were set to their default values. The same parameters processed the positive mode data; however, the minimum pairs cosine was set to 0.55 and the minimum matched fragment ions were set to 3. The generated networks were imported to the open-source software platform, Cytoscape 3.10.0 software, for visualization.2.8. Metabolites and molecular formula identification
The bioactive metabolites were identified in the extracts after processing UPLC-MS files using MZmine 2.37 software. The structural interpretation was achieved by extensively examining the high-resolution mass spectra. The CSI:FingerID interface of Sirius 5.6.3.0 software based on the high-resolution mass was used to identify or annotate the metabolites. Metabolite identification was further supported by MN exploration and the proposed GNPS spectral library search. The parameters of GNPS spectral library search were set to a precursor ion mass tolerance of 0.02 Da with minimum matched peaks of 4, a fragment ion mass tolerance of 0.02 Da and a score threshold of 0.65. According to the accurate mass and fragmentation pattern, the most likely molecular formula of the metabolite was selected and confirmed. PubChem, HMDB, Reaxys, LIPID MAPS and COCONUT were also used.2.9. Neuroprotective activity of KEL and KEF
The study was approved and all methods were performed in agreement with the appropriate guidelines and protocols of the Ethics Committee for Animal Experimentation of Faculty of Pharmacy, Cairo University (Permit Number: MP 3324) and conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 2011).
The Morris water maze (MWM; probe test) and object recognition tests were performed 24 h following the last administration of either extract. In order to reduce variability resulting from circadian rhythms, the tests were consistently conducted at approximately the same time each day. Following the behavioral assessments, each group was further divided into two subgroups (with n = 3 in each), and the mice were euthanized using an overdose injection of thiopental (IP 200 mg kg−1). The brains were rapidly extracted and separated. One set of whole brains were preserved in 10% formalin in saline for subsequent histopathological and immunohistochemical examination, while the brains of the other set were designated for biochemical analyses.
3. Results
3.1. LC-MS/MS metabolomic profiling aided by molecular networking
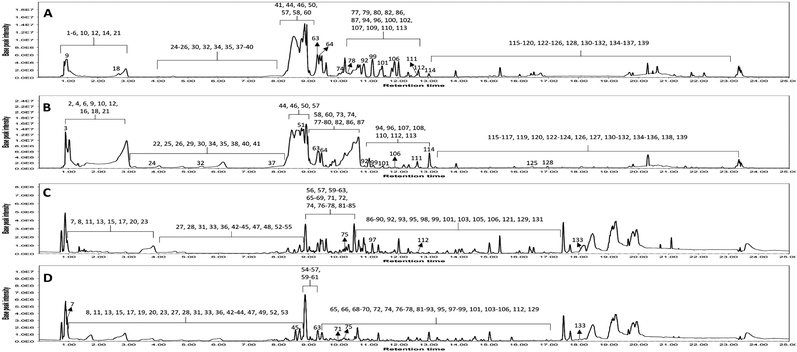
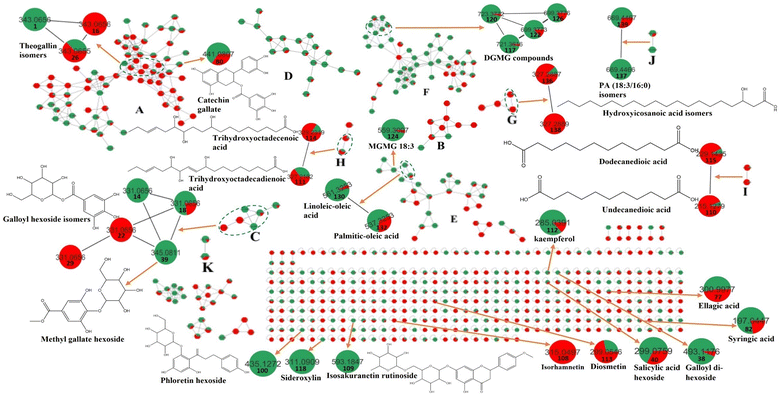
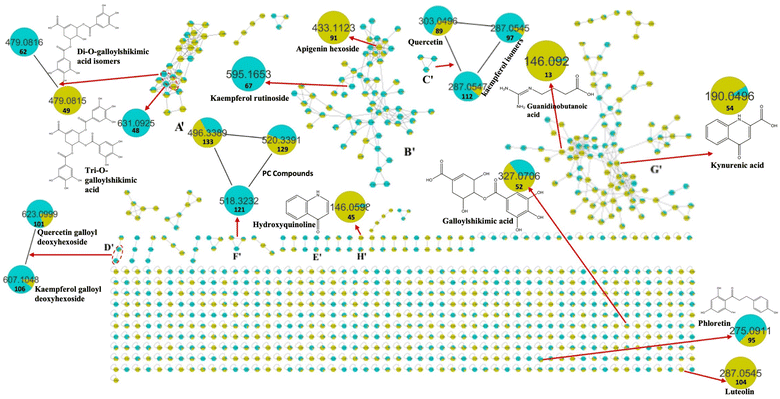
Metabolomic profiling conducted using UHPLC-MS/MS analysis aided by MN in negative and positive modes was performed to assess the metabolite composition of K. elegans leaf and fruit extracts. The analysis led to the characterization of several known compounds that were detected before in the Koelreuteria genus or discovered for the first time. Metabolomic assessments were made by comparing retention times, distribution, accurate masses and fragmentation patterns, which were further supported by MN exploration, together with the proposed GNPS spectral library search, followed by literature data comparison for confirmation. The metabolites along with their peak numbers, observed m/z of the detected molecular ions, errors (ppm), molecular formulae, product ions (MS/MS), compound classes, and retention times are presented in Table 1. The representative base peak chromatograms of the two extracts along with detailed fragmentation labelling of some identified compounds are displayed in Fig. 1 and Fig. S1–S21 (ESI†). Putative structures of representative groups of metabolites are shown in Fig. 2. A total of 139 metabolites were identified after overlapping both negative and positive data of KEL and KEF. The chief identified secondary metabolites were phenolics and flavonoids (a total of ninety-four), as confirmed by previous reports.14–16,18,20–22 Negative and positive MNs were established (Fig. 3 and 4). They allowed us to investigate UPLC-MS/MS data and observe metabolite distribution among the two samples in both modes. Depending on MS/MS fragmentation pattern similarities, they classified molecules into clusters, where metabolites with fragmentation patterns close enough to each other were connected, while those with dissimilar fragmentation patterns were separated,33 based on controlled parameters described before. Nodes were displayed as a pie chart to describe the semi-relative abundance of the detected molecular ions in the two extracts, while the edges indicated the mass differences between the connected nodes. Nodes were also colored by the sample type (i.e., KEL and KEF) and labeled with their precursor m/z values. The established MN of the negative mode was composed of 590 nodes connected in twenty-six clusters (a minimum of two connected nodes) and 350 self-looped nodes. The detected metabolites are identified as follows: phenolic acids: clusters A, B, and C; flavonoids: cluster D; cluster E was mostly formed by the assigned lipids; glycolipids were arranged in cluster F; most of the determined fatty acids were present in clusters G, H, and I; two phosphatidic acids were formed in cluster J; and cluster K comprised one dihydrochalcone. Regarding self-looped nodes, they mostly corresponded to some flavonoids and phenolic acids, among other metabolites. The constructed MN of positive mode consisted of 833 nodes comprising thirty-six clusters (a minimum of two connected nodes) and 561 self-looped nodes. Cluster A′ encompassed phenolic acids, while clusters B′, C′, D′ and E′ represented flavonoids. Phosphocholines were arranged in cluster F′. The identified hydroxyquinoline occurred in cluster H′ and cluster G′ contained amino acids, organic acids, phenols, hydroxycoumarins and an isocoumarin. Some flavonoids and phenolic acids besides a dihydrochalcone were arranged in single-looped nodes.| Peak | RT | Assignment | Precursor ion (m/z) | Error (ppm) | Molecular formula | Product ions MS/MS | Class | KEL | KEF |
|---|---|---|---|---|---|---|---|---|---|
| Note: DGMG, digalactosylmonoacylglycerol; MGMG, monogalactosylmonoacylglycerol; PE, phosphoethanolamine; PC, phosphocholine; PA, phosphatidic acid and PI, phosphoinositol. | |||||||||
| 1 | 0.90 | Theogallin isomer I | 343.0656[M − H]− | −2.62 | C14H16O10 | 191, 169, 125 | Phenolic acid | + | − |
| 2 | 0.90 | Methyl-O-galloyl hexoside isomer I | 345.0812[M − H]− | −2.90 | C14H18O10 | 345, 169, 124, 151, 161, 191, 85 | Phenolic acid | + | + |
| 3 | 0.90 | Quinic acid | 191.0553[M − H]− | −1.57 | C7H12O6 | 85, 191, 59, 93, 127, 109, 111, 173 | Organic acid | + | + |
| 4 | 0.92 | Di-O-galloylquinic acid isomer I | 495.0757[M − H]− | −3.64 | C21H20O14 | 169, 343, 325, 495 | Phenolic acid | + | + |
| 5 | 0.93 | Gallic acid | 169.0136[M − H]− | −0.59 | C7H6O5 | 125, 79, 69, 81, 97, 107, 53 | Phenolic acid | + | − |
| 6 | 0.94 | Tri-O-galloylquinic acid isomer I | 647.0862[M − H]− | −3.34 | C28H24O18 | 495, 647, 343, 477, 325, 169, 191 | Phenolic acid | + | + |
| 7 | 0.95 | Proline betaine | 144.1017[M + H]+ | −5.55 | C7H13NO2 | 144, 58, 84, 102 | Amino acid betaine | + | + |
| 8 | 0.95 | Methyl-O-galloyl hexoside isomer I | 347.0963[M + H]+ | −4.32 | C14H18O10 | 153, 109, 125, 347, 81, 155, 145 | Phenolic acid | + | + |
| 9 | 0.97 | Tetra-O-galloylquinic acid isomer I | 799.0965[M − H]− | −3.63 | C35H28O22 | 601, 629, 477, 169, 647, 495, 343, 191 | Phenolic acid | + | + |
| 10 | 0.98 | Digallic acid isomer I | 321.0238[M − H]− | −2.80 | C14H10O9 | 169, 125 | Phenolic acid | + | + |
| 11 | 1.13 | Betaine | 118.0860[M + H]+ | −6.77 | C5H11NO2 | 59, 58, 118 | Modified amino acid | + | + |
| 12 | 1.31 | Shikimic acid | 173.0448[M − H]− | −1.16 | C7H10O5 | 93, 73, 83, 137 | Organic acid | + | + |
| 13 | 1.43 | Guanidinobutanoic acid | 146.0920[M + H]+ | −6.16 | C5H11N3O2 | 87, 86, 60, 69, 83, 146, 56, 111 | Organic acid | + | + |
| 14 | 1.59 | Galloyl hexoside isomer I | 331.0656[M − H]− | −2.72 | C13H16O10 | 169, 59, 331, 151, 211, 125, 271, 89 | Phenolic acid | + | − |
| 15 | 1.63 | Theogallin isomer I | 345.0811[M + H]+ | −3.19 | C14H16O10 | 153, 125, 79, 81, 171, 229, 85 | Phenolic acid | + | + |
| 16 | 1.73 | Theogallin isomer II | 343.0656[M − H]− | −2.62 | C14H16O10 | 169, 191, 125 | Phenolic acid | − | + |
| 17 | 2.03 | Isoleucine | 132.1017[M + H]+ | −6.06 | C6H13NO2 | 86, 69, 58, 57, 91 | Amino acid | + | + |
| 18 | 2.70 | Galloyl hexoside isomer II | 331.0656[M − H]− | −2.72 | C13H16O10 | 169, 331, 151, 59, 123, 211, 271, 89 | Phenolic acid | + | + |
| 19 | 2.86 | Pyrogallol | 127.0387[M + H]+ | −6.30 | C6H6O3 | 81, 53, 109, 79, 67 | Phenol | − | + |
| 20 | 2.89 | Gallic acid | 171.0285[M + H]+ | −5.26 | C7H6O5 | 81, 107, 109, 125, 153, 53, 79, 69, 97 | Phenolic acid | + | + |
| 21 | 2.94 | Pyrogallol | 125.0239[M − H]− | 0 | C6H6O3 | 125, 79, 51, 69, 97, 81, 107 | Phenol | + | + |
| 22 | 3.38 | Galloyl hexoside isomer III | 331.0656[M − H]− | −2.72 | C13H16O10 | 169, 211, 271, 125, 331, 59, 89, 151 | Phenolic acid | − | + |
| 23 | 3.72 | Methyl-O-galloyl hexoside isomer II | 347.0966[M + H]+ | −3.46 | C14H18O10 | 153, 127, 109, 349, 125, 155, 81, 174 | Phenolic acid | + | + |
| 24 | 3.80 | Methyl-O-galloyl hexoside isomer II | 345.0811[M − H]− | −3.19 | C14H18O10 | 345, 169, 124, 151, 193, 161 | Phenolic acid | + | + |
| 25 | 3.97 | Protocatechuic acid isomer I | 153.0187[M − H]− | −0.65 | C7H6O4 | 109, 81, 53 | Phenolic acid | + | + |
| 26 | 4.10 | Theogallin isomer II | 345.0812[M + H]+ | −2.90 | C14H16O10 | 153, 171, 111, 125, 51, 327, 109 | Phenolic acid | + | + |
| 27 | 4.67 | Phenylalanine | 166.0861[M + H]+ | −4.21 | C9H11NO2 | 120, 103, 84, 93, 77, 91 | Amino acid | + | + |
| 28 | 4.75 | Galloyl hexoside isomer IV | 331.0656[M − H]− | −2.72 | C13H16O10 | 271, 169, 211, 125, 59, 151 | Phenolic acid | − | + |
| 29 | 5.31 | Protocatechuic acid hexoside isomer I | 315.0708[M − H]− | −2.54 | C13H16O9 | 153, 109, 108, 152 | Phenolic acid | + | + |
| 30 | 5.35 | Methyl-O-galloyl hexoside isomer III | 347.0967[M + H]+ | −3.17 | C14H18O10 | 153, 109, 127, 79, 81, 171, 53, 329, 141 | Phenolic acid | + | + |
| 31 | 5.44 | Methyl-O-galloyl hexoside isomer III | 345.0812[M − H]− | −2.90 | C14H18O10 | 345, 124, 169, 151, 161, 85 | Phenolic acid | + | + |
| 32 | 5.97 | Theogallin isomer III | 343.0655[M − H]− | −2.91 | C14H16O10 | 191, 169, 125 | Phenolic acid | + | + |
| 33 | 6.13 | Theogallin isomer III | 345.0813[M + H]+ | −2.61 | C14H16O10 | 153, 125, 345, 171, 285, 107, 285 | Phenolic acid | + | + |
| 34 | 6.15 | Protocatechuic acid isomer II | 153.0187[M − H]− | −0.65 | C7H6O4 | 109, 81, 53, 91 | Phenolic acid | + | + |
| 35 | 6.65 | Protocatechuic acid hexoside isomer II | 315.0706[M − H]− | −3.17 | C13H16O9 | 152, 108, 153, 315, 109 | Phenolic acid | + | + |
| 36 | 7.75 | Galloylglycerol | 245.0649[M + H]+ | −4.90 | C10H12O7 | 153, 125, 107, 140, 79 | Phenolic acid | + | + |
| 37 | 7.79 | Galloylglycerol | 243.0498[M − H]− | −2.88 | C10H12O7 | 124, 169, 243, 59, 151, 89, 91 | Phenolic acid | + | + |
| 38 | 7.81 | Galloyl di-hexoside | 493.1176[M − H]− | −3.65 | C19H26O15 | 493, 313, 169, 271, 191, 331, 125 | Phenolic acid | + | + |
| 39 | 7.93 | Methyl gallate hexoside | 345.0811[M − H]− | −3.19 | C14H18O10 | 183, 59, 225, 89, 285, 71, 169, 124 | Phenolic acid | + | − |
| 40 | 7.97 | Salicylic acid hexoside | 299.0759[M − H]− | −2.67 | C13H16O8 | 137, 59, 89, 101, 93, 119 | Phenolic acid | + | + |
| 41 | 8.07 | Galloylshikimic acid | 325.0550[M − H]− | −3.08 | C14H14O9 | 169, 125, 325, 173, 93, 111 | Phenolic acid | + | + |
| 42 | 8.17 | Galloylshikimic acid isomer I | 327.0706[M + H]+ | −3.06 | C14H14O9 | 153, 95, 109, 125, 171, 139, 214 | Phenolic acid | + | + |
| 43 | 8.41 | Syringic acid hexoside | 383.0945[M + Na]+ | −2.35 | C15H20O10 | 383, 221, 185, 251, 253 | Phenolic acid | + | + |
| 44 | 8.47 | Hydroxybenzoic acid | 137.0239[M − H]/139.0387[M + H]+ | 0/−5.75 | C7H6O3 | 93, 137, 108, 119/77, 121, 95, 65, 56 | Phenolic acid | + | + |
| 45 | 8.56 | Hydroxyquinoline | 146.0598[M + H]+ | −5.48 | C9H7NO | 146, 77, 91, 118, 104, 128 | Hydroquinolone | + | + |
| 46 | 8.66 | Tri-O-galloylquinic acid isomer II | 647.0861[M − H]− | −3.55 | C28H24O18 | 477, 647, 495, 343, 169, 449, 325, 191 | Phenolic acid | + | + |
| 47 | 8.67 | Methyl galloylquinic acid | 359.0967[M + H]+ | −3.06 | C15H18O10 | 153, 111, 93, 171, 127, 359, 143, 167 | Phenolic acid | + | + |
| 48 | 8.69 | Tri-O-galloylshikimic acid | 631.0925[M + H]+ | −1.58 | C28H22O17 | 153, 461, 631 | Phenolic acid | + | − |
| 49 | 8.70 | Di-O-galloylshikimic acid isomer I | 479.0815[M + H]+ | −2.30 | C21H18O13 | 153, 309, 479, 171, 95 | Phenolic acid | − | + |
| 50 | 8.72 | Di-O-galloylquinic acid isomer II | 495.0758[M − H]− | −3.43 | C21H20O14 | 343, 169, 495, 325, 191 | Phenolic acid | + | + |
| 51 | 8.76 | Methyl gallate | 183.0293[M − H]− | −0.55 | C8H8O5 | 124, 183, 78, 168 | Phenolic acid | − | + |
| 52 | 8.81 | Galloylshikimic acid isomer II | 327.0706[M + H]+ | −3.06 | C14H14O9 | 153, 95, 139, 143, 255, 279, 111 | Phenolic acid | + | + |
| 53 | 8.81 | Di-O-galloylquinic acid | 497.0920[M + H]+ | −2.21 | C21H20O14 | 153, 309, 327, 479, 328, 171, 125 | Phenolic acid | + | + |
| 54 | 8.83 | Kynurenic acid | 190.0496[M + H]+ | −4.21 | C10H7NO3 | 144, 116, 113, 89, 162, 59 | Organic acid | + | + |
| 55 | 8.88 | Methyl gallate | 185.0443[M + H]+ | −3.78 | C8H8O5 | 126, 153, 107, 125, 59, 67, 95, 79 | Phenolic acid | + | + |
| 56 | 8.99 | Digallic acid | 323.0394[M + H]+ | −2.79 | C14H10O9 | 153, 125, 79 | Phenolic acid | + | + |
| 57 | 9 | Catechin/Epicatechin | 289.0707[M − H]−/291.0858[M + H]+ | −1.73\-3.78 | C15H14O6 | 247, 109, 203, 191, 123, 219, 179, 151\139, 123, 147, 207, 140, 177, 162 | Flavonoid | + | + |
| 58 | 9.02 | Digallic acid isomer II | 321.0239[M − H]− | −2.49 | C14H10O9 | 169, 125 | Phenolic acid | + | + |
| 59 | 9.13 | Scopoletin isomer I | 193.0491[M + H]+ | −5.18 | C10H8O4 | 133, 178, 193, 137, 105, 194, 122, 109, 80, 149, 76 | Hydroxycoumarin | + | + |
| 60 | 9.14 | Brevifolin carboxylic acid | 291.0134[M − H]−/293.0288[M + H]+ | −2.41\−3.07 | C13H8O8 | 247, 191, 219, 147, 229/219, 191, 247, 293, 220, 205, 163, 179 | Isocoumarin | + | + |
| 61 | 9.21 | Trimethoxyphenol | 185.0805[M + H]+ | −4.86 | C9H12O4 | 125, 153, 110, 139, 127, 59, 95, 107, 79 | Phenol | + | + |
| 62 | 9.30 | Di-O-galloylshikimic acid isomer II | 479.0816[M + H]+ | −2.09 | C21H18O13 | 153, 309, 479, 171, 95 | Phenolic acid | + | − |
| 63 | 9.30 | Tri-O-galloylquinic acid isomer III | 647.0863[M − H]−/649.1027[M + H]+ | −3.25\−2.16 | C28H24O18 | 495, 647, 343, 477, 325, 169, 191/153, 479, 305, 263, 309, 631, 461, 281, 171 | Phenolic acid | + | + |
| 64 | 9.41 | Tetra-O-galloylquinic acid isomer II | 799.0966[M − H]− | −3.50 | C35H28O22 | 799, 601, 629, 477, 169, 647, 495, 343, 191 | Phenolic acid | + | + |
| 65 | 9.44 | Tetra-O-galloyllapiitol isomer I | 783.1029[M + Na]+ | 1.02 | C33H28O21 | 153, 783, 305, 461, 263, 613, 433 | Phenolic acid | + | + |
| 66 | 9.53 | 3-Methoxy-4-hydroxyphenol-1-O-β-d-(6'-O-galloyl)-glucoside isomer I | 493.0972[M + Na]+ | 2.84 | C20H22O13 | 153, 323, 493 | Phenolic acid | + | + |
| 67 | 9.62 | Kaempferol rutinoside isomer I | 595.1653[M + H]+ | −1.68 | C27H30O15 | 287, 449, 71, 85, 243, 147, 153 | Flavonoid | + | − |
| 68 | 9.77 | Galloyl-(epi)gallocatechin (epi)gallocatechin | 801.1133[M + K]+ | 7.99 | C37H30O18 | 153, 305, 631, 479, 263, 783, 433, 457, 461, 111, 171, 327, 586, 281, 309, 613, 291 | Flavonoid | + | + |
| 69 | 9.81 | Calycosin hexoside | 447.1280[M + H]+ | −2.46 | C22H22O10 | 285, 153, 447, 309, 270 | Flavonoid | + | + |
| 70 | 9.84 | Tetra-O-galloyllapiitol isomer II | 783.1026[M + Na]+ | 0.63 | C33H28O21 | 153, 305, 783, 461, 309, 613, 631 | Phenolic acid | − | + |
| 71 | 9.98 | Cirsimaritin hexoside | 477.1385[M + H]+ | −2.51 | C23H24O11 | 315, 477, 153 | Flavonoid | + | + |
| 72 | 10.05 | Vaniline | 153.0544[M + H]+ | −4.57 | C8H8O3 | 65, 93, 110, 125, 79 | Phenol | + | + |
| 73 | 10.08 | Quercetin di-deoxyhexoside | 593.1485[M − H]− | −3.71 | C27H30O15 | 593, 447, 284, 301, 183, 299, 271 | Flavonoid | − | + |
| 74 | 10.13 | Quercetin rutinoside | 609.1433[M − H]−/611.1600[M + H]+/633.1417[M + Na]+ | −3.76/−2.00/−2.37 | C27H30O16 | 609, 300, 301/303, 85, 129, 71, 465/633, 331, 325, 153 | Flavonoid | + | + |
| 75 | 10.20 | 3-Methoxy-4-hydroxyphenol-1-O-β-D-(6'-O-galloyl)-glucoside isomer II | 493.097[M + Na]+ | 2.43 | C20H22O13 | 153, 493, 323 | Phenolic acid | + | + |
| 76 | 10.20 | 1'-O-galloyl-3,4,5-trihydroxybenzyl alcohol 4-O-β-D-(6′′-O-galloyl)-glucopyranoside | 645.1078[M + Na]+ | 1.55 | C27H26O17 | 153, 475, 645, 305, 323, 273 | Phenolic acid | + | + |
| 77 | 10.26 | Ellagic acid | 300.9977[M − H]−/303.0132[M + H]+ | −2.33/−2.64 | C14H6O8 | 301/303, 285, 275, 165, 257, 137, 153 | Phenolic acid | + | + |
| 78 | 10.26 | Quercetin hexoside | 463.0862[M − H]−/465.1021[M + H]+/487.084[M + Na]+ | −3.24/−2.58/−2.67 | C21H20O12 | 300, 463, 271, 178, 151, 255/303, 91, 85, 61, 305, 97, 73, 127/487, 185, 325 | Flavonoid | + | + |
| 79 | 10.27 | Kaempferol rutinoside | 593.1484[M − H]− | −3.88 | C27H30O15 | 593, 285 | Flavonoid | + | + |
| 80 | 10.28 | Catechin gallate | 441.0807[M − H]− | −3.40 | C22H18O10 | 169, 289, 245, 125 | Flavonoid | + | + |
| 81 | 10.32 | Kaempferol hexoside | 449.1073[M + H]+ | −2.45 | C21H20O11 | 287 | Flavonoid | + | + |
| 82 | 10.37 | Syringic acid | 197.0447[M − H]−/199.0599[M + H]+ | −1.52/−4.02 | C9H10O5 | 123, 182, 97, 167/140, 107, 167, 59, 123, 67, 95 | Phenolic acid | + | + |
| 83 | 10.45 | Scopoletin isomer II | 193.0492[M + H]+ | −4.66 | C10H8O4 | 178, 133, 193, 137, 149, 122, 194, 105, 165 150, 117, 94 | Hydroxycoumarin | + | + |
| 84 | 10.45 | Kaempferol rutinoside isomer II | 595.1650[M + H]+ | −2.18 | C27H30O15 | 287, 85, 71, 129, 449 | Flavonoid | + | + |
| 85 | 10.54 | Kaempferol hexoside | 471.0893[M + Na]+ | −2.12 | C21H20O11 | 471, 185, 309 | Flavonoid | + | + |
| 86 | 10.55 | Quercetin pentoside | 433.0757[M − H]−/435.0918[M + H]+/457.0736[M + Na]+ | −3.23/−2.07/−2.41 | C20H18O11 | 300, 433, 271, 178, 151/303, 73, 305, 61, 115/475, 325, 155 | Flavonoid | + | + |
| 87 | 10.65 | Methyl digallate | 335.0395[M − H]−/337.0550[M + H]+ | −2.39/−3.00 | C15H12O9 | 183, 124, 168, 78/153, 125, 185 | Phenolic acid | + | + |
| 88 | 10.66 | Diosmetin | 301.0703[M + H]+ | −3.00 | C16H12O6 | 301, 286, 258, 153 | Flavonoid | + | + |
| 89 | 10.70 | Quercetin isomer I | 303.0496[M + H]+ | −3.00 | C15H10O7 | 303, 153, 229, 285, 165 | Flavonoid | + | + |
| 90 | 10.75 | Isorhamnetin hexoside | 501.0995[M + Na]+ | −2.79 | C22H22O12 | 501, 339, 185, 317 | Flavonoid | + | + |
| 91 | 10.76 | Apigenin hexoside isomer I | 433.1123[M + H]+ | -2.77 | C21H20O10 | 271 | Flavonoid | − | + |
| 92 | 10.85 | Kaempferol pentoside | 417.0809[M − H]−/441.0792[M + Na]+ | −3.12/−1.36 | C20H18O10 | 284, 417, 255, 227, 151/441, 309, 155 | Flavonoid | + | + |
| 93 | 10.90 | Diosmetin hexoside | 463.1231[M + H]+ | −1.94 | C22H22O11 | 301, 286, 258 | Flavonoid | + | + |
| 94 | 10.92 | Kaempferide hexoside | 461.1069[M − H]− | −3.25 | C22H22O11 | 461, 446, 283, 298, 255, 269, 315 | Flavonoid | + | + |
| 95 | 10.96 | Phloretin | 275.0911[M + H]+ | −3.27 | C15H14O5 | 107, 169, 77 | Dihydrochalcone | + | + |
| 96 | 10.99 | Phloretin hexoside isomer I | 435.1275[M − H]− | −3.68 | C21H24O10 | 273, 167, 341, 391, 125 | Dihydrochalcone | + | + |
| 97 | 11.12 | Kaempferol isomer I | 287.0545[M + H]+ | −3.83 | C15H10O6 | 287, 153, 121, 165 | Flavonoid | + | + |
| 98 | 11.12 | Apigenin hexoside isomer II | 433.1124[M + H]+ | −2.54 | C21H20O10 | 271 | Flavonoid | + | + |
| 99 | 11.13 | Kaempferol deoxyhexoside | 431.0964[M − H]−/455.0943[M + Na]+ | −3.25/−2.42 | C21H20O10 | 285, 431, 255, 227/309, 455, 169, 310, 85, 71 | Flavonoid | + | + |
| 100 | 11.28 | Phloretin hexoside isomer II | 435.1272[M − H]− | −4.37 | C21H24O10 | 273, 167, 221, 191, 315 | Dihydrochalcone | + | − |
| 101 | 11.45 | Quercetin galloyl deoxyhexoside | 599.1016[M − H]−/601.1180[M + H]+/623.0999[M + Na]+ | −3.51/−2.16/−2.25 | C28H24O15 | 599, 301, 297, 169/153, 299, 154, 303, 300/321, 623, 303, 325, 175, 281, 153, 69 | Flavonoid | + | + |
| 102 | 11.54 | Dihydrokaempferol | 287.0549[M − H]− | −2.44 | C15H12O6 | 135, 123, 151, 183, 223 | Flavonoid | + | − |
| 103 | 11.80 | Daidzein hexoside | 417.1174[M + H]+ | −2.64 | C21H20O9 | 255 | Flavonoid | + | + |
| 104 | 11.85 | Luteolin | 287.0545[M + H]+ | −3.83 | C15H10O6 | 287, 197, 257, 152, 269 | Flavonoid | − | + |
| 105 | 11.87 | Quercetin isomer II | 303.0495[M + H]+ | -3.30 | C15H10O7 | 303, 285, 165, 257, 229, 153 | Flavonoid | + | + |
| 106 | 11.87 | Kaempferol galloyl deoxyhexoside | 583.1065[M − H]−/607.1048[M + Na]+ | −3.94/−2.64 | C28H24O14 | 285, 583, 297, 169/321, 607, 303, 309, 281, 153, 175 | Flavonoid | + | + |
| 107 | 11.89 | Quercetin | 301.0340[M − H]− | −2.66 | C15H10O7 | 151, 301, 179, 121, 107, 65 | Flavonoid | + | + |
| 108 | 11.94 | Isorhamnetin | 315.0497[M − H]− | −2.54 | C16H12O7 | 300, 125, 315, 112, 187, 71 | Flavonoid | − | + |
| 109 | 12.52 | Isosakuranetin rutinoside | 593.1847[M − H]− | −3.88 | C28H34O14 | 285 | Flavonoid | + | − |
| 110 | 12.57 | Undecanedioic acid | 215.1279[M − H]− | −1.86 | C11H20O4 | 197, 153, 215 | Fatty acid | + | + |
| 111 | 12.62 | Trihydroxyoctadecadienoic acid | 327.2162[M − H]− | −3.06 | C18H32O5 | 327, 211, 229, 171, 85, 97, 291 | Fatty acid | + | + |
| 112 | 12.66 | Kaempferol isomer II | 285.0391[M − H]−/287.0547[M + H]+ | −2.81/−3.14 | C15H10O6 | 285/287, 153, 121, 165 | Flavonoid | + | + |
| 113 | 12.76 | Diosmetin | 299.0546[M − H]− | −3.34 | C16H12O6 | 284, 299, 256, 79 | Flavonoid | + | + |
| 114 | 13.02 | Trihydroxyoctadecenoic acid | 329.2319[M − H]− | −2.73 | C18H34O5 | 329, 211, 171, 229, 139, 99 | Fatty acid | + | + |
| 115 | 13.30 | Dodecanedioic acid | 229.1435[M − H]− | −2.18 | C12H22O4 | 211, 167, 229 | Fatty acid | + | + |
| 116 | 14.22 | Chrysin | 253.0495[M − H]− | −2.37 | C15H10O4 | 253, 158, 177, 209, 63, 143, 79 | Flavonoid | + | + |
| 117 | 15.39 | DGMG (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
721.3616[M + HCOO]− | −4.30 | C33H56O14 | 397, 675, 415, 277, 235, 721, 253 | Glycolipid | + | + |
| 118 | 15.76 | Sideroxylin | 311.0909[M − H]− | −3.54 | C18H16O5 | 296, 311 | Flavonoid | + | − |
| 119 | 15.94 | PI (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/0 2/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
595.2859[M − H]− | −4.03 | C27H49O12P | 595, 279, 153, 315, 241, 415, 79, 259 | Phosphoinositol | + | + |
| 120 | 16.04 | DGMG (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
723.3772[M + HCOO]− | −4.29 | C33H58O14 | 397, 677, 415, 279, 89, 235, 305, 723, 253 | Glycolipid | + | + |
| 121 | 16.14 | PC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3/0 3/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
518.3232[M + H]+ | −2.89 | C26H48NO7P | 184, 104, 518, 500, 86, 258, 60, 125 | Phosphocholine | + | − |
| 122 | 16.23 | DGMG (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) isomer I 0) isomer I |
699.3773[M + HCOO]− | −4.29 | C31H58O14 | 397, 653, 415, 255, 235, 699, 89, 253 | Glycolipid | + | + |
| 123 | 16.38 | PE (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/0 2/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
476.2759[M − H]− | −3.78 | C23H44NO7P | 279, 476, 196, 214, 79, 140, 153 | Phosphoethanolamine | + | + |
| 124 | 16.42 | MGMG 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
559.3097[M + HCOO]− | −3.75 | C27H46O9 | 277, 253, 235, 101, 513, 559 | Glycolipid | + | + |
| 125 | 16.46 | PI (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
571.2863[M − H]− | −3.50 | C25H49O12P | 571, 255, 153, 241, 315, 393, 79, 259 | Phosphoinositol | + | + |
| 126 | 16.52 | DGMG (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) isomer II 0) isomer II |
699.3776[M + HCOO]− | −3.86 | C31H58O14 | 397, 653, 415, 255, 235, 89, 699, 253 | Glycolipid | + | + |
| 127 | 16.66 | PE (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) isomer I 0) isomer I |
452.2760[M − H]− | −3.76 | C21H44NO7P | 255, 113, 452, 181, 153, 79, 214, 140 | Phosphoethanolamine | − | + |
| 128 | 16.96 | PE (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) isomer II 0) isomer II |
452.2759[M − H]− | −3.98 | C21H44NO7P | 255, 452, 196, 140, 79, 214, 153 | Phosphoethanolamine | + | + |
| 129 | 17.07 | PC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
520.3391[M + H]+ | −2.31 | C26H50NO7P | 184, 104, 520, 502, 86, 337, 60, 258 | Phosphocholine | + | + |
| 130 | 17.19 | Linoleic–oleic acid | 561.3253[M − H]− | −3.92 | C28H50O11 | 279, 253, 504, 235 | Fatty acid | + | + |
| 131 | 17.41 | PA (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/0 2/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
433.2339[M − H]− | −3.69 | C21H39O7P | 153, 79, 433, 171, 97, 279 | Phosphatidic acid | + | + |
| 132 | 17.78 | Palmitic–oleic acid | 537.3253[M − H]− | −4.09 | C26H50O11 | 255, 253, 235 | Fatty acid | + | + |
| 133 | 18.03 | PC (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
496.3389[M + H]+ | −2.82 | C24H50NO7P | 104, 184, 496, 478, 86, 258, 313 | Phosphocholine | + | + |
| 134 | 18.20 | PA (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16 0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
409.2341[M − H]− | −3.42 | C19H39O7P | 153, 79, 409, 255, 97 | Phosphatidic acid | + | + |
| 135 | 18.67 | PA (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
435.2495[M − H]− | −3.91 | C21H41O7P | 153, 79, 97, 171, 281 | Phosphatidic acid | + | + |
| 136 | 20.3 | Hydroxyicosanoic acid isomer I | 327.2887[M − H]− | −3.67 | C20H40O3 | 59, 255, 101, 327 | Fatty acid | + | + |
| 137 | 20.30 | PA (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3/16 3/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) isomer I 0) isomer I |
669.4466[M − H]− | −4.33 | C37H67O8P | 391, 669, 255, 409, 153, 79, 277, 413, 97 | Phosphatidic acid | + | − |
| 138 | 20.88 | Hydroxyicosanoic acid isomer II | 327.2889[M − H]− | −3.06 | C20H40O3 | 59, 327, 101 | Fatty acid | − | + |
| 139 | 23.33 | PA (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3/16 3/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) isomer II 0) isomer II |
669.4467[M − H]− | −4.18 | C37H67O8P | 391, 669, 255, 79, 153, 409, 413, 277, 97 | Phosphatidic acid | + | + |
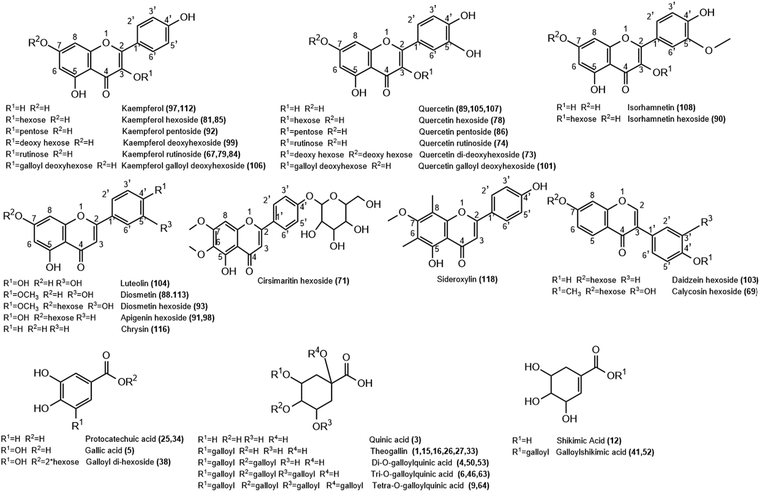 | ||
| Fig. 2 Structure of representative groups of metabolites identified in the K. elegans leaf methanol extract (KEL) and K. elegans fruit methanol extract (KEF). The carbon numbering system for each compound is based on analogy rather than on IUPAC rules. Metabolite numbers are listed in Table 1. | ||
3.1.2.1. Flavonols. Twenty-two flavonols were detected in K. elegans methanolic extracts, including 12 kaempferols: two of them were isomers of kaempferol aglycone (peaks 97 and 112), one as dihydrokaempferol (peak 102) and nine as glycosides (peaks 67, 79, 81, 84, 85, 92, 94, 99 and 106), 8 quercetin isomers; 3 of them as aglycones (peaks 89, 105 and 107) and 5 as glycosides (peaks 73, 74, 78, 86 and 101) and 2 isorhamnetin isomers (peaks 90 and 108). Furthermore, peaks 67 and 102 were observed only in KEL, while peaks 73 and 108 were noticed only in KEF. Indeed, most of the displayed flavonols were represented by O-glycosides that showed a higher abundant ion [Aglycone − H]−/[Aglycone + H]+, derived from a homolytic cleavage, at m/z 285/287 (kaempferol), and 301/303 (quercetin) relative to the corresponding ion [Aglycone]−, derived from a heterolytic cleavage, at m/z 284 and 300, respectively. Specifically, rutinosyl glycosides were observed in compounds 74 and 79, which showed [M − H]− at m/z 609.1433 and 593.1484, respectively. After losing the rutinosyl moiety, the fragments ions 301 and 285 were produced, indicating quercetin and kaempferol aglycones, respectively. Aglycone hexosides were detected as peaks 78, 85 and 90 with [M + Na]+ at m/z 487.084, 471.0893 and 501.0995, respectively. They revealed the [Aglycone + Na]+ ion peaks at m/z 325 (quercetin), 309 (kaempferol) and 339 (isorhamnetin), respectively, after loss of the hexose moiety. Compounds 86 (m/z 433.0757) and 92 (m/z 417.0809) revealed aglycone product ions at m/z 300 and 284, respectively due to loss of the pentose sugar unit. Compounds 73 and 99 (m/z 593.1485 and 431.0964, respectively) were identified as quercetin di-deoxyhexoside and kaempferol deoxy hexoside based on the relative abundances of the aglycone ion after the loss of di-deoxy (−292 Da) and deoxy (−146 Da) hexose moieties, respectively. Peak 94 was putatively identified as kaempferide hexoside with m/z 461.1069[M − H]−. It exhibited product ions at m/z 446 and 283 due to successive loss of methyl and hexose moieties, in addition to the fragment ion at m/z 298 due to direct loss of the hexose unit from the parent mass corresponding to kaempferide aglycone. Methyl ether of quercetin or isorhamnetin (peak 108) showed a molecular ion peak at m/z 315.0497 and a fragment ion peak at 300 m/z due to loss of the methyl group (−15 Da). Finally, compounds 101 and 106 showed conjugation between sugar and gallyl moieties. They were annotated as quercetin galloyl deoxy hexoside (m/z 599.1016) and kaempferol galloyl deoxy hexoside (m/z 583.1065), respectively. They showed fragment ions at m/z 169 and 297 corresponding to galloyl and dehydrated galloyl deoxy hexose moieties, respectively.
3.1.2.2. Flavones and isoflavones. Nine flavones (peaks 71, 88, 91, 93, 98, 104, 113, 116 and 118) were tentatively detected in both leaf and fruit extracts. However, peaks 91 and 104 could not be identified in KEL, while peak 118 was absent in KEF. They were present as aglycones aside from peaks 71, 91, 93 and 98, which were present in the form of glycosides. Compound 71 showed m/z 477.1385 [M + H]+ and was identified as cirsimaritin hexoside. It had a base peak at m/z 315 due to loss of the hexose unit. Similarly, peaks 91 and 98 revealed an aglycone abundant fragment ion at 271 m/z belonging to apigenin after losing the sugar moiety. Their [M + H]+ species were nearly similar: m/z 433.1123 and 433.1124, respectively, and they were considered as isomers of apigenin hexoside. Luteolin-containing compounds are listed in Table 1 as peaks 88, 93, 104 and 113 based on the relative abundances of the aglycone ion. Compound 104 showed m/z 287.0545 [M + H]+ corresponding to luteolin aglycone. Compounds 88 and 113 were tentatively identified as diosmetin, while 93 was their glycosidic form (m/z 463.1231 [M + H]+) due to loss of hexose sugar (ms2 301 m/z). Diosmetin produced product ions at m/z 286 and 284 due to loss of the methyl group (−15 Da) in positive (peaks 88 and 93) and negative (peak 113) analyses, respectively. Peak 116 was identified as chrysin with [M − H]− at m/z 253.0495. Peak 118 was identified as sideroxylin (311.0909 m/z) with a fragment ion at m/z 296 due to demethylation. Isoflavones were represented as peaks 69 [m/z 447.1280] and 103 [m/z 417.1174], which were identified as calycosin and daidzein hexosides, respectively. They showed [Aglycone + H]+ product ions at m/z 285 and 255, respectively, due to loss of sugar unit. Furthermore, peak 69 revealed a fragment ion at m/z 270 corresponding to demethylation of calycosin aglycone.
3.1.2.3. Flavanols and flavanones. Flavan-3-ols or catechins were detected as peaks 57, 68 and 80 and are listed in Table 1. They were tentatively characterized as catechin/epicatechin, galloyl-(epi)gallocatechin-(epi)gallocatechin and catechin gallate, respectively. Compound 57 revealed m/z at 289.0707 [M − H]− and 291.0858 [M + H]+, while 68 showed [M + K]+ at m/z 801.1133. Their fragmentation patterns were in agreement with the reported literature.38 Peak 80 showed a conjugation between galloyl and catechin units with fragment ions at m/z 289 and 169 due to loss of galloyl ions and gallic acid, respectively. Finally, only one flavanone was observed in KEL and was identified as isosakuranetin rutinoside, peak 109. It revealed m/z 593.1847 [M − H]− with abundant [Aglycone − H]− at m/z 285 due to loss of the rutinosyl moiety.
It is worth noting that the established MN was capable of discriminating ions from several flavonoid analogues as observed for the negative MN. Custer D was considered the main cluster for flavonoid glycosides. Catechin gallate was separated from the main flavonoid cluster and presented in cluster A. This might be due to the presence of galloyl moiety, which was the main part of cluster A. Sideroxylin, isorhamnetin, diosmetin, and kaempferol aglycones appeared as self-looped nodes, due to the absence of sugar moieties. In the positive MN, cluster B′ included flavonoid glycosides, cluster C′ contained aglycones only and D′ included galloyl flavonol glycosides.
A total of eighteen lipids were examined in the K. elegans extracts, which could be differentiated into two classes (i.e., phospholipids and glycolipids). Among phospholipids, metabolites ascribable to phosphoinositols (PI), phosphoethanolamines (PE), phosphocholines (PC), and phosphatidic acids (PA) were tentatively identified. In particular, peak 119 [m/z 595.2859 (C27H48O12P−)], an example of PI, exhibited diagnostic fragment ions at m/z 315 and 241, due to dehydrated glycerophosphoinositol (C9H16O10P−) and inositol-phosphate (C6H10O8P−) ions, respectively. The ions at m/z 415 and 279 are related to the fatty acid-glycerophosphate (C21H36O6P−) and the 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 fatty acid carboxylate anion (C18H31O2−), respectively. However, the appearance of daughter ions at m/z 259, 153 and 79 led to their characterization as inositol phosphate (C6H12O9P−), dehydrated glycerol phosphate (C3H6O5P−) and phosphate (PO3−) ions, respectively. Compound 119 was identified as octadecadienoyl-glycero-phospho-myo-inositol (PI (18
2 fatty acid carboxylate anion (C18H31O2−), respectively. However, the appearance of daughter ions at m/z 259, 153 and 79 led to their characterization as inositol phosphate (C6H12O9P−), dehydrated glycerol phosphate (C3H6O5P−) and phosphate (PO3−) ions, respectively. Compound 119 was identified as octadecadienoyl-glycero-phospho-myo-inositol (PI (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/0
2/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0)). The analysis of peak 127 MS/MS spectrum [m/z 452.2760 (C21H43NO7P−)], as an example of PE, showed the characteristic ions of phospholipids (m/z 153 and 79) and a base peak at m/z 255 corresponding to the 16
0)). The analysis of peak 127 MS/MS spectrum [m/z 452.2760 (C21H43NO7P−)], as an example of PE, showed the characteristic ions of phospholipids (m/z 153 and 79) and a base peak at m/z 255 corresponding to the 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 fatty acid carboxylate anion (C16H31O2−). The ions at m/z 214 and 140 represented glycerophosphoethanolamine (C5H13NO6P−) and phosphoethanolamine (C2H7NO4P−), respectively. Therefore, it was identified as hexadecanoyl-sn-glycero-phosphoethanolamine (PE (16
0 fatty acid carboxylate anion (C16H31O2−). The ions at m/z 214 and 140 represented glycerophosphoethanolamine (C5H13NO6P−) and phosphoethanolamine (C2H7NO4P−), respectively. Therefore, it was identified as hexadecanoyl-sn-glycero-phosphoethanolamine (PE (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0
0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0)). PA was putatively identified as compound 137 [m/z 669.4466, C37H66O8P−]. It contained the diagnostic ions of phospholipids at m/z 153, 79 and 97 (H2PO4−). Its major ions were observed at m/z 277 and 255 corresponding to the 18
0)). PA was putatively identified as compound 137 [m/z 669.4466, C37H66O8P−]. It contained the diagnostic ions of phospholipids at m/z 153, 79 and 97 (H2PO4−). Its major ions were observed at m/z 277 and 255 corresponding to the 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (C18H29O2−) and 16
3 (C18H29O2−) and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 fatty acid carboxylate anions, respectively. Compound 137 was assigned as octadecatrienoyl-hexadecanoyl-glycero-phosphate (PA (18
0 fatty acid carboxylate anions, respectively. Compound 137 was assigned as octadecatrienoyl-hexadecanoyl-glycero-phosphate (PA (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3/16
3/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0)). Compound 129 [m/z 520.3391, C26H51NO7P+], as a representative of PC, revealed product ion peaks at m/z 258, 184 and 104, corresponding to glycerolphosphocholine [C8H20NO6P + H]+, phosphocholine [C5H14NO4P + H]+ and choline [C5H13NO + H]+ ions, respectively. In addition, a specific ion at m/z 337, corresponding to dehydrated glycerol conjugated with 18
0)). Compound 129 [m/z 520.3391, C26H51NO7P+], as a representative of PC, revealed product ion peaks at m/z 258, 184 and 104, corresponding to glycerolphosphocholine [C8H20NO6P + H]+, phosphocholine [C5H14NO4P + H]+ and choline [C5H13NO + H]+ ions, respectively. In addition, a specific ion at m/z 337, corresponding to dehydrated glycerol conjugated with 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 fatty acid (C21H37O3+), was observed. Accordingly, compound 129 was identified as octadecadienoyl-sn-glycero-phosphocholine (PC (18
2 fatty acid (C21H37O3+), was observed. Accordingly, compound 129 was identified as octadecadienoyl-sn-glycero-phosphocholine (PC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2)). The MS2 spectra of glycolipids showed the typical product ions at m/z 253 and 235 attributed to the glyceryl hexoside anion (C9H17O8−) followed by dehydration, respectively. Compound 124 contained one hexose unit and it was identified as MGMG in cluster E. Cluster F contained peaks 117, 120, 122 and 126, which exhibited an extra hexose moiety, and they showed fragment ions at m/z 415 and 397, corresponding to the glyceryl di-hexoside ion (C15H27O13−) and successive loss of water molecules, respectively. All glycolipids showed sharp peaks due to the involved fatty acid.
2)). The MS2 spectra of glycolipids showed the typical product ions at m/z 253 and 235 attributed to the glyceryl hexoside anion (C9H17O8−) followed by dehydration, respectively. Compound 124 contained one hexose unit and it was identified as MGMG in cluster E. Cluster F contained peaks 117, 120, 122 and 126, which exhibited an extra hexose moiety, and they showed fragment ions at m/z 415 and 397, corresponding to the glyceryl di-hexoside ion (C15H27O13−) and successive loss of water molecules, respectively. All glycolipids showed sharp peaks due to the involved fatty acid.
A total of four organic acids were detected. Quinic and shikimic acids appeared mainly as scattered nodes in the negative MN, while guanidinobutanoic and kynurenic acids were present in the same cluster of amino acids, due to their nitrogen containment. The MS/MS spectra displayed abundant ions due to the loss of H2O, CO2, CO and CH2 groups, in addition to loss of nitrogen groups in the case of nitrogen-containing organic acid. The assessment of organic acids was based on their accurate masses, MS/MS fragmentation behaviors, and previous studies.41–43
Hydroxycoumarins and isocoumarins are listed in Table 1 as peaks 59, 83 (isomers of scopoletin) and 60 (brevifolin carboxylic acid). They showed the characteristic fragmentation patterns of their classes in agreement with reference data.45 In addition, specific product ions of 59 and 83 were mainly due to dehydration, while 60 corresponded to decarboxylation and dehydration.
Regarding the four examined phenols, peaks 19 [m/z 127.0387 (C6H7O3+)] and 21 [m/z 125.0239 (C6H5O3−)] were identified as pyrogallol,46 while peaks 61 and 72 were identified as trimethoxyphenol [m/z 185.0805 (C9H13O4+)] and vaniline [m/z 153.0544 (C8H9O3+)], respectively. Compounds 61 and 72 showed fragment ions mainly due to the loss of methoxy and carbonyl groups. Finally, peak 45 was identified as hydroxyquinoline,47 with [M + H] + at m/z 146.0598.
3.2. Toxicity study
Screening of the toxic effect of increased oral doses of KEL and KEF revealed that they were non-toxic up to 2000 mg kg−1.3.3. Morris water maze test
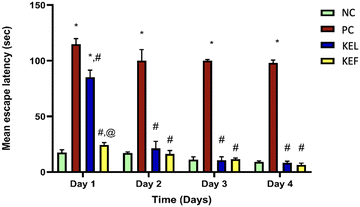
During the 4-day memory acquisition trial, the latency to find the hidden platform profoundly declined in all mice treated with STZ, with a profound deterioration in their cognitive functions in the second, third, and fourth training days, respectively, as compared to the NC group. The effect of the treatment with leaf or fruit extracts quickly normalized the mice memory from the second day (Table 2 and Fig. 5).| Day/latency time (s) | Normal control | Positive control | KEL | KEF |
|---|---|---|---|---|
| KEL, Koelreuteria elegans leaf methanol extract and KEF, Koelreuteria elegans fruit methanol extract. | ||||
| Day 1 | 17.5 ± 2.5 | 114.9 ± 5 | 85.2 ± 6.4 | 24.3 ± 2.1 |
| Day 2 | 17 ± 1 | 100 ± 10 | 21.2 ± 3.2 | 16.1 ± 3.2 |
| Day 3 | 11.1 ± 3.2 | 100 ± 7.2 | 10.5 ± 2.1 | 11.6 ± 1.5 |
| Day 4 | 9 ± 1 | 98 ± 2.6 | 8.2 ± 1.5 | 6.5 ± 1.5 |
3.4. Histopathological examination
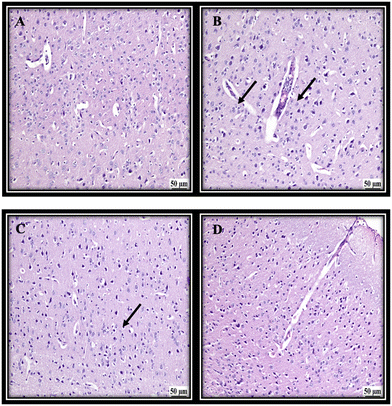
Due to the involvement of the cerebral cortex in the memory function and being a primary target of Alzheimer's disease, it was chosen to assess the effect of different treatments. From the histopathology images it was apparent that the first group (NC) revealed a normal histological structure of the cerebral cortex. The untreated group (PC) showed various degenerated neurons in the cerebral cortex, which suggested a decline in the memory function. The group treated with the KEL showed a decline in the number of degenerated neurons with a moderate number of dark degenerated neurons in the cerebral cortex. Finally, the group treated with the KEF showed better modification of the degenerated neurons with only a few dark degenerated neurons in the cerebral cortex, suggesting a better outcome of treatment (Fig. 6).3.5. The effect of K. elegans on TNF-α
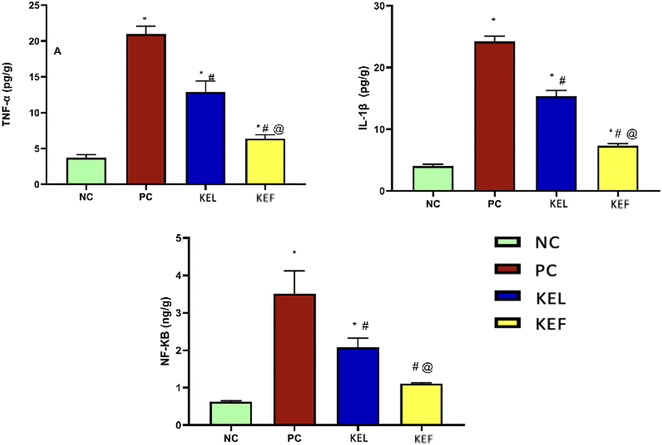
The PC group showed a 4.6-fold increase in pro-inflammatory TNF-α; this rise was controlled significantly by 38.5% upon the administration of KEL, with 70% decrease in the KEF treated group. Notably, the treatment with the fruit extract showed a 51% significant decline in the level of TNF-α compared to the group treated with the leaf extract (Table 3 and Fig. 7).| Parameter/group | Normal control | Positive control | KEL | KEF |
|---|---|---|---|---|
| TNF-α, tumor necrosis factor alpha; NF-κB, nuclear factor kappa B; IL-1β, interleukin-1 beta; KEL, Koelreuteria elegans leaf methanol extract; and KEF, Koelreuteria elegans fruit methanol extract. | ||||
| TNF-α | 3.73 ± 0.41 | 21 ± 1 | 12.9 ± 1.5 | 6.3 ± 0.5 |
| NF-KB | 0.62 ± 0.03 | 3.5 ± 0.6 | 2.07 ± 0.24 | 1.1 ± 0.02 |
| IL-1β | 4.03 ± 0.3 | 24.2 ± 0.85 | 15.3 ± 0.95 | 7.3 ± 0.33 |
3.6. The effect of K. elegans on NF-κB
The transcription factor NF-κB was increased by 82% in the PC group. The administration of KEL significantly controlled the level of NF-κB, with 41% decline in its level. The KEF treated group exhibited 68.5% decline in the tissue NF-κB level (Table 3 and Fig. 7).3.7. The effect of K. elegans on IL-1β
The increase in the IL-1β level of the PC group (83.3%) was controlled significantly by the administration of KEL and KEF, with 37% and 69.8% decline in its level in the tissues, respectively (Table 3 and Fig. 7).4. Discussion
Previous phytochemical studies of K. elegans led to the detection of phenolic compounds, flavonoids, lignans, sterols, tocopherols and triterpenes.11,14–21 Despite its potential as a producer of bioactive natural compounds, no studies are available on the anti-AD activity of K. elegans. The only study that discussed the anti-AD potential was conducted on Koelreuteria paniculata,1 with the isolation and elucidation of five barrigenol-type triterpenoid compounds from the seeds of K. paniculata. These compounds were evaluated for their anti-AD activity in okadaic acid (OA)-induced learning and memory impaired mice. The results revealed that two of these metabolites could improve the learning and memory deficits induced by OA, along with the attenuation of the provoked tau hyperphosphorylation by regulating the levels of GSK-3β and PP2A.1 Our study identified secondary metabolites, such as phenolics, flavonoids and fatty acids. Phenolic acids have been demonstrated to attenuate the aggregation of proteins contributing to the pathogenesis of various neurodegenerative disorders characterized by cognitive deterioration, including Alzheimer's disease.48,49 Gallic, protocatechuic, ellagic and syringic acids detected in KEL and KEF have been previously evaluated for their potential to protect neurons from Aβ-induced neurotoxicity. They showed improvement of memory deficits and synaptic dysfunction by suppressing the release of the pro-inflammatory cytokines: TNF-α, NF-κB, and IL-1β.48 Several studies evaluated the biological activities of flavonoids as neuronal antioxidants, exhibiting anti-amyloidogenic and anti-inflammatory potential and neuroprotection and improving cognition.50–55K. elegans leaves and fruits are rich in flavonoids, which have potential against AD by regulating several important physiological responses. Quercetin, catechin-3-gallate, apigenin, apigenin hexoside, kaempferol and luteolin are detected in KEL and KEF. They have been perceived in several reports as anti-neuroinflammatory by blocking the release of cytokines (TNF-α and IL-1β), inhibiting NF-κB expression, and reducing intracellular Aβ and the hyperphosphorylation of tau.50–53 The results of the study displayed that the fruit extract of K. elegans has a more potent anti-inflammatory effect than the leaf extract in a mice model of AD. The fruit extract reduced TNF-α, NF-κB, and IL-1β levels more than the leaf extract, which are all markers of inflammation and neurodegeneration in AD. This is consistent with some recent studies revealing that natural plant extracts can have beneficial effects on AD by modulating the immune system and clearing amyloid-β plaques.56 The PC group, which represents an Alzheimer's disease model, showed sky-rocketed pro-inflammatory cytokine TNF-α levels. This is consistent with the well-documented involvement of TNF-α in neuroinflammation and its association with Alzheimer's disease. Elevated TNF-α levels are often observed in the brains of AD patients and are thought to contribute to neuroinflammatory processes.4 Oxidative stress is known to play a significant role in the pathogenesis of neurodegenerative diseases, including Alzheimer's disease.The capability of antioxidant drugs to shield neurons from amyloid-induced neurodegeneration is based on their ability to counteract oxidative stress and its detrimental effects on neuronal health.57 In light of this, El Naggar et al. reported that K. elegans had a hepatoprotective effect due to its ability to increase the enzymatic levels of superoxide dismutase and glutathione, indicative of its antioxidant properties. Building on this, it is reasonable to propose that K. elegans extracts can exhibit neuroprotective effects.15 Furthermore, Kumari et al. demonstrated the antioxidant activity of K. elegans leaf extract using the DPPH method, and Waleed et al. supported these findings by confirming the strong radical scavenging properties of K. elegans. These studies collectively suggest the potential neuroprotective effects of K. elegans through its antioxidant mechanisms.12 In addition, research has shown that antioxidant treatment can attenuate neuronal loss, improve cognitive function, and reduce the accumulation of amyloid β plaques in the brain.58 These findings provide additional support for the notion that KE extracts may hold promise in slowing down the neurodegeneration process.
The administration of the leaf extract of K. elegans resulted in the decline of the levels of TNF-α, while the fruit extract of K. elegans led to an even more substantial diminution. This suggested that both extracts have anti-inflammatory effects, with the fruit extract being more effective. Affecting TNF-α triggers a trajectory that activates NF-κB in the brain tissues.59 The PC group exhibited an amplification in the transcription factor NF-κB. Treatment with the leaf extract of K. elegans resulted in a reduction in NF-κB levels, while the fruit extract showed a more extensive effect. In light of the previous parameters, the untreated mice group showed an upsurge in the pro-inflammatory cytokine IL-1β, which is another key player in neuroinflammation associated with Alzheimer's disease.7,8 The effect of the fruit extract was more pronounced than that of the leaf extract in decreasing IL-1β. These results indicated that both extracts have anti-inflammatory effects, with the fruit extract showing stronger activity. In the context of Alzheimer's disease research, these findings are promising. Since the antioxidant activity of the K. elegans extract was previously discussed,12 this study sheds light on the anti-inflammatory capabilities of the plant extract. Neuroinflammation is increasingly recognized as a contributing factor to the progression of AD.2 Hence, reducing pro-inflammatory markers like TNF-α, NF-KB, and IL-1β could have therapeutic potential. Further research, including clinical trials, is needed to determine the efficacy and safety of these extracts in humans with Alzheimer's disease.
5. Conclusion
The current study provides the first comprehensive metabolite profiles of leaf and fruit extracts of K. elegans using UHPLC-MS in both negative and positive modes assisted by MN. A total of 139 metabolites were tentatively detected after a detailed interpretation of the data using the CSI:FingerID interface of Sirius 5.6.3.0 software, and the metabolite identification was further supported by MN exploration, together with the proposed GNPS spectral library search. The identified compounds belonged to various classes encompassing fifty-seven phenolic acids, thirty-seven flavonoids, four amino acids, four organic acids, four phenols, two hydroxycoumarins, one isocoumarin, three dihydrochalcones, and one hydroquinolone. Additionally, twenty-six lipids of different classes were characterized, including eight fatty acids, two phosphoinositols, three phosphoethanolamines, three phosphocholines, five glycolipids and five phosphatidic acids. The analysis of the data showed the compositional similarities and differences in the metabolites among the leaf and fruit extracts. Remarkably, KEL could ameliorate the learning and memory deficits induced by STZ in behavioral experiments, besides improving the histopathological profile of the cerebral cortex of the injured mice, while KEF led to an even more potent effect. Based on the in vivo experiments, the fruit extract reduced TNF-α, NF-κB, and IL-1β levels more than the leaf extract, which are all markers of inflammation and neurodegeneration in AD. Altogether, these findings provide support for further research with the raw leaf and fruit extracts or after fractionation and purification of specific compounds. More detailed and conclusive in vivo and clinical studies are highly recommended to exploit the potential of K. elegans in treating patients with Alzheimer's disease.Abbreviations
| AD | Alzheimer's disease |
| Aβ | Amyloid beta |
| CID | Collision induced dissociation |
| DAD | Diode-array detection |
| DDH | Dihydrodiol dehydrogenase |
| DGMG | Digalactosylmonoacylglycerol |
| ESI | Electrospray ionization |
| FBMN | Feature-based molecular network |
| GNPS | Global natural product social molecular networking |
| HRMS | High resolution mass spectrometry |
| ICV | Intracerebroventricular injection |
| IL-1β | Interleukin-1 beta |
| IP | Intraperitoneal |
| KEF | Koelreuteria elegans fruit methanol extract |
| KEL | Koelreuteria elegans leaf methanol extract |
| MEL | Mean escape latency |
| MGMG | Monogalactosylmonoacylglycerol |
| MN | Molecular network |
| MWM | Morris water maze |
| NC | Normal control |
| NF-κB | Nuclear factor kappa B |
| OECD | Organization for economic development |
| PA | Phosphatidic acid |
| PC | Positive control |
| PC | Phosphocholine |
| PE | Phosphoethanolamine |
| PI | Phosphoinositol |
| PTK | Protein-tyrosine kinase |
| ROS | Reactive oxygen species |
| STZ | Streptozotocin |
| TNF-α | Tumor necrosis factor alpha |
| UHPLC | Ultra high performance liquid chromatography |
Conflicts of interest
There are no conflicts to declare.References
- X. Lu, L. Sun, Y. Zhang and W. Li, J. Funct. Foods, 2019, 61, 103459 CrossRef CAS.
- M. Culjak, M. N. Perkovic, S. Uzun, D. S. Strac, G. N. Erjavec, M. B. Leko, G. Simic, L. Tudor, M. Konjevod, O. Kozumplik, N. Mimica and N. Pivac, Curr. Alzheimer Res., 2020, 17, 972–984 CrossRef CAS PubMed.
- F. Brosseron, M. Krauthausen, M. Kummer and M. T. Heneka, Mol. Neurobiol., 2014, 50, 534–544 CrossRef CAS PubMed.
- M. V. Lourenco, J. R. Clarke, R. L. Frozza, T. R. Bomfim, L. Forny-Germano, A. F. Batista, L. B. Sathler, J. Brito-Moreira, O. B. Amaral, C. A. Silva, L. Freitas-Correa, S. Espírito-Santo, P. Campello-Costa, J. C. Houzel, W. L. Klein, C. Holscher, J. B. Carvalheira, A. M. Silva, L. A. Velloso, D. P. Munoz, S. T. Ferreira and F. G. De Felice, Cell Metab., 2013, 18, 831–843 CrossRef CAS PubMed.
- E. Sun, A. Motolani, L. Campos and T. Lu, Int. J. Mol. Sci., 2022, 23 Search PubMed.
- Y. Tang, D. Zhang, X. Gong and J. Zheng, Adv. Funct. Mater., 2022, 32, 2208022 CrossRef CAS.
- D. Di Bona, A. Plaia, S. Vasto, L. Cavallone, F. Lescai, C. Franceschi, F. Licastro, G. Colonna-Romano, D. Lio, G. Candore and C. Caruso, Brain Res. Rev., 2008, 59, 155–163 CrossRef CAS PubMed.
- L. Xie, Y. Lai, F. Lei, S. Liu, R. Liu and T. Wang, Mol. Med. Rep., 2015, 11, 3219–3228 CrossRef CAS PubMed.
- F. Meyer, J. Arnold Arbor., 1976, 57, 129–166 CrossRef.
- Chinese medicinal plants, ed. Y. C. Jeng, Publisher's Reader’s Digest Association Far East Ltd., Hong Kong, 1994, vol. 207 Search PubMed.
- C. C. Wu, K. F. Huang, T. Y. Yang, Y. L. Li, C. L. Wen, S. L. Hsu and T. H. Chen, PLoS One, 2015, 10, e0132052 CrossRef PubMed.
- P. Kumari, S. Nehra and M. Deen, J. Pharmacogn. Phytochem., 2019, 8, 1724–1728 CAS.
- W.-C. Tsai, H.-C. Chang, H.-Y. Yin, M.-C. Huang, D. C. Agrawal and H.-W. Wen, Electron. J. Biotechnol., 2020, 47, 89–99 CrossRef CAS.
- F. Abo-Elghiet, M. Ibrahim and A. A. Sleem, Al-Azhar J. Pharm. Sci., 2021, 1, 23–29 Search PubMed.
- D. El Naggar, Al-Azhar J. Pharm. Sci., 2022, 65, 16–39 CrossRef.
- M. Abou-Shoer, G. E. Ma, X. H. Li, N. M. Koonchanok, R. L. Geahlen and C. J. Chang, J. Nat. Prod., 1993, 56, 967–969 CrossRef CAS PubMed.
- Y. N. Song, H. L. Zhang, C. J. Chang and D. M. Bollag, J. Nat. Prod., 1994, 57, 1670–1674 CrossRef CAS PubMed.
- T. H. Lee, Y. H. Chiang, C. H. Chen, P. Y. Chen and C. K. Lee, J. Nat. Med., 2009, 63, 209–214 CrossRef CAS PubMed.
- C. H. Chen, P. Y. Chen, K. C. Wang and C. K. Lee, J. Chin. Chem. Soc., 2010, 57, 404–410 CrossRef CAS.
- Y. Y. Chiang, S. L. Wang, C. L. Yang, H. Y. Yang, H. C. Yang, J. N. Sudhakar, C. K. Lee, H. W. Huang, C. M. Chen, S. H. Chiou, S. F. Chiang, H. Y. Fang, C. Y. Chen, S. H. Shieh and K. C. Chow, Int. J. Mol. Med., 2013, 32, 577–584 CrossRef CAS PubMed.
- C. Y. Lin, P. N. Chen, Y. S. Hsieh and S. C. Chu, Food Chem., 2014, 146, 299–307 CrossRef CAS PubMed.
- C. Y. Lin, P. N. Chen, L. S. Hsu, D. Y. Kuo, S. C. Chu and Y. S. Hsieh, Mol. Med. Rep., 2014, 10, 3334–3342 CrossRef CAS PubMed.
- M. H. Lee, C. B. Jiang, S. H. Juan, R. D. Lin and W. C. Hou, Fitoterapia, 2006, 77, 109–115 CrossRef PubMed.
- C. H. Chen, H. C. Chan, Y. T. Chu, H. Y. Ho, P. Y. Chen, T. H. Lee and C. K. Lee, Molecules, 2009, 14, 2947–2958 CrossRef CAS PubMed.
- M. C. Chambers, B. Maclean, R. Burke, D. Amodei, D. L. Ruderman, S. Neumann, L. Gatto, B. Fischer, B. Pratt, J. Egertson, K. Hoff, D. Kessner, N. Tasman, N. Shulman, B. Frewen, T. A. Baker, M. Y. Brusniak, C. Paulse, D. Creasy, L. Flashner, K. Kani, C. Moulding, S. L. Seymour, L. M. Nuwaysir, B. Lefebvre, F. Kuhlmann, J. Roark, P. Rainer, S. Detlev, T. Hemenway, A. Huhmer, J. Langridge, B. Connolly, T. Chadick, K. Holly, J. Eckels, E. W. Deutsch, R. L. Moritz, J. E. Katz, D. B. Agus, M. MacCoss, D. L. Tabb and P. Mallick, Nat. Biotechnol., 2012, 30, 918–920 CrossRef CAS PubMed.
- C. J. Ugwah-Oguejiofor, C. O. Okoli, M. O. Ugwah, M. L. Umaru, C. S. Ogbulie, H. E. Mshelia, M. Umar and A. A. Njan, Heliyon, 2019, 5, e01179 CrossRef PubMed.
- OECD, Test No. 425: Acute Oral Toxicity: Up-and-Down Procedure, 2022.
- M. A. Pelleymounter, M. Joppa, M. Carmouche, M. J. Cullen, B. Brown, B. Murphy, D. E. Grigoriadis, N. Ling and A. C. Foster, J. Pharmacol. Exp. Ther., 2000, 293, 799–806 CAS.
- M. A. Pelleymounter, M. Joppa, N. Ling and A. C. Foster, J. Pharmacol. Exp. Ther., 2002, 302, 145–152 CrossRef CAS PubMed.
- G. Warnock, 2007.
- M. E. Sorial and N. El Sayed, Naunyn Schmiedebergs Arch. Pharmacol., 2017, 390, 581–593 CrossRef CAS PubMed.
- B. Singh, B. Sharma, A. S. Jaggi and N. Singh, J. Renin Angiotensin Aldosterone Syst., 2013, 14, 124–136 CrossRef CAS PubMed.
- T. M. M. Jouaneh, N. Motta, C. Wu, C. Coffey, C. W. Via, R. D. Kirk and M. J. Bertin, Fitoterapia, 2022, 159, 105200 CrossRef CAS PubMed.
- N. Kumar and N. Goel, Biotechnol. Rep., 2019, 24, e00370 CrossRef PubMed.
- W. Xu, M. Huang, H. Li, X. Chen, Y. Zhang, J. Liu, W. Xu, K. Chu and L. Chen, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2015, 986–987, 69–84 CrossRef CAS PubMed.
- M. d F. R. de Lima, L. A. Cavalcante, E. C. T. de Araújo Costa, B. O. de Veras, M. V. da Silva, L. N. Cavalcanti and R. M. Araújo, Phytochem. Lett., 2021, 41, 186–192 CrossRef CAS.
- A. N. Panche, A. D. Diwan and S. R. Chandra, J. Nutr. Sci., 2016, 5, e47 CrossRef CAS PubMed.
- A. Singh, S. Kumar and B. Kumar, Nat. Prod. Commun., 2018, 13(5) DOI:10.1177/1934578X1801300511.
- R. M. Ibrahim, G. F. Elmasry, R. H. Refaey and R. A. El-Shiekh, ACS omega, 2022, 7, 17339–17357 CrossRef CAS PubMed.
- J. T. Pierson, G. R. Monteith, S. J. Roberts-Thomson, R. G. Dietzgen, M. J. Gidley and P. N. Shaw, Food Chem., 2014, 149, 253–263 CrossRef CAS PubMed.
- T. Guo, C. Tang, H. Song, Y. Dong and Q. Ma, Food Chem., 2021, 353, 129446 CrossRef CAS PubMed.
- S. Uysal, G. Zengin, K. I. Sinan, G. Ak, R. Ceylan, M. F. Mahomoodally, A. Uysal, N. B. Sadeer, J. Jekő, Z. Cziáky, M. J. Rodrigues, E. Yıldıztugay, F. Elbasan and L. Custodio, RSC Adv., 2021, 11, 5295–5310 RSC.
- L. Chen, Z. Dai, C. Ge, D. Huang, X. Zhou, K. Pan, W. Xu, J. Fu and J. L. Du, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2022, 1203, 123260 CrossRef CAS PubMed.
- A. R. Jesus, A. P. Marques and A. P. Rauter, Pure Appl. Chem., 2016, 88, 349–361 CrossRef CAS.
- L. Cissé, A. Tine, L. Kaboré and A. Saba, Spectrosc. Lett., 2009, 42, 95–99 CrossRef PubMed.
- I. E. Sallam, U. Rolle-Kampczyk, S. S. Schäpe, S. S. Zaghloul, R. S. El-Dine, P. Shao, M. V. Bergen and M. A. Farag, Molecules, 2022, 27 Search PubMed.
- A. Ermakov, V. Voronin, A. Sorokin, N. Épshtein, I. Muravskaya, V. Chistyakov and A. Zuev, Chem. Heterocycl. Compd., 1984, 20, 637–642 CrossRef.
- G. Caruso, J. Godos, A. Privitera, G. Lanza, S. Castellano, A. Chillemi, O. Bruni, R. Ferri, F. Caraci and G. Grosso, Nutrients, 2022, 14 Search PubMed.
- Y. Tang, D. Zhang, X. Gong and J. Zheng, Biophysical Chemistry, 2022, 281, 106735 CrossRef CAS PubMed.
- A. Calderaro, G. T. Patanè, E. Tellone, D. Barreca, S. Ficarra, F. Misiti and G. Laganà, Int. J. Mol. Sci., 2022, 23 Search PubMed.
- P. Bellavite, Antioxidants, 2023, 12, 280 CrossRef CAS PubMed.
- R. A. Arias-Sánchez, L. Torner and B. Fenton Navarro, Molecules, 2023, 28, 5415 CrossRef PubMed.
- F. Hadrich, M. Chamkha and S. Sayadi, Food Chem. Toxicol., 2022, 159, 112752 CrossRef CAS PubMed.
- P. H. Nguyen, A. Ramamoorthy, B. R. Sahoo, J. Zheng, P. Faller, J. E. Straub, L. Dominguez, J.-E. Shea, N. V. Dokholyan and A. De Simone, Chemical reviews, 2021, 121, 2545–2647 CrossRef CAS PubMed.
- Y. Tang, D. Zhang and J. Zheng, ACS Chem. Neurosci., 2023, 14, 3143–3155 CrossRef CAS PubMed.
- A. Halle, V. Hornung, G. C. Petzold, C. R. Stewart, B. G. Monks, T. Reinheckel, K. A. Fitzgerald, E. Latz, K. J. Moore and D. T. Golenbock, Nat. Immunol., 2008, 9, 857–865 CrossRef CAS PubMed.
- R. Dhapola, S. K. Beura, P. Sharma, S. K. Singh and D. HariKrishnaReddy, Mol. Biol. Rep., 2024, 51, 48 CrossRef CAS PubMed.
- S. Dubey and E. Singh, Inflammopharmacology, 2023, 31, 717–730 CrossRef CAS PubMed.
- B. Kaltschmidt and C. Kaltschmidt, Cold Spring Harb. Perspect. Biol., 2009, 1, a001271 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma00007b |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |