Open Access Article
Open Access ArticleOptimization of extraction conditions to synthesize green carbon nanodots using the Maillard reaction
Duyen H.H.
Nguyen
 *abc,
Arjun
Muthu
*abc,
Arjun
Muthu
 cd,
Hassan
El-Ramady
ae,
Lajos
Daróczi
f,
Lajos
Nagy
g,
Sándor
Kéki
cd,
Hassan
El-Ramady
ae,
Lajos
Daróczi
f,
Lajos
Nagy
g,
Sándor
Kéki
 g,
Áron
Béni
d,
Istvan
Csarnovics
g,
Áron
Béni
d,
Istvan
Csarnovics
 h and
József
Prokisch
a
h and
József
Prokisch
a
aInstitute of Animal Science, Faculty of Agricultural and Food Sciences and Environmental Management, Biotechnology and Nature Conservation, University of Debrecen, 138 Böszörményi Street, 4032 Debrecen, Hungary. E-mail: nguyen.huu.huong.duyen@agr.unideb.hu
bTay Nguyen Institute for Scientific Research, Vietnam Academy of Science and Technology, 118 Xo Viet Nghe Tinh Street, Da Lat, Vietnam
cDoctoral School of Nutrition and Food Science, University of Debrecen, Debrecen, Hungary
dInstitute of Agricultural Chemistry and Soil Science, Faculty of Agricultural and Food Sciences and Environmental Management, University of Debrecen, 138 Böszörményi Street, 4032 Debrecen, Hungary
eSoil and Water Department, Faculty of Agriculture, Kafrelsheikh University, Kafr El-Sheikh 33516, Egypt
fDepartment of Solid State Physics, University of Debrecen, PO Box 400, Debrecen, Hungary
gDepartment of Applied Chemistry, Faculty of Sciences and Technology, University of Debrecen, Egyetem tér 1, H-4032 Debrecen, Hungary
hDepartment of Experimental Physics, University of Debrecen, Debrecen, Hungary
First published on 1st March 2024
Abstract
Carbon nanodots (CNDs) are a class of nanoparticles with unique optical properties with broad applications in various fields. However, synthesizing CNDs with high fluorescence intensity and small size using a green solvent and low temperature remains challenging. In this study, we investigated the use of the Maillard reaction for synthesizing CNDs and optimized the reaction conditions at 120 °C for 12 h to achieve CNDs with desirable properties. The results showed that glycine was the most effective amino acid for CNDs formation when combined with sucrose. A molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for glycine
1 for glycine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sucrose resulted in the highest fluorescence intensity. The fluorescence intensity increased remarkably with 40% ethanol as the extraction solvent. However, a high ethanol concentration (above 60%) had an inverse relationship with CNDs’ fluorescence intensity, indicating that a high concentration of ethanol solution prevented the Schiff base formation. The purified CNDs (M-CNDs) were characterized using fluorescence spectrophotometry, UV-Vis spectroscopy, Raman spectroscopy, and TEM techniques. These findings provide a purification process of sustainable CNDs using the Maillard reaction and green solvents with optimized conditions and low temperatures.
sucrose resulted in the highest fluorescence intensity. The fluorescence intensity increased remarkably with 40% ethanol as the extraction solvent. However, a high ethanol concentration (above 60%) had an inverse relationship with CNDs’ fluorescence intensity, indicating that a high concentration of ethanol solution prevented the Schiff base formation. The purified CNDs (M-CNDs) were characterized using fluorescence spectrophotometry, UV-Vis spectroscopy, Raman spectroscopy, and TEM techniques. These findings provide a purification process of sustainable CNDs using the Maillard reaction and green solvents with optimized conditions and low temperatures.
Introduction
Carbon nanodots (CNDs) with sizes smaller than 10 nm have been reported for the first time since 2004.1 CNDs are attractive because of their intense fluorescent properties, high water solubility, stability, and ease of functionalization.1–3 Since their discovery, CNDs have garnered considerable attention across various sectors due to their highly fluorescent properties and potential applications in diverse fields, including biomedical,4 food safety,5 and the bio-electrical industry.6 In the food industry, CNDs have emerged as promising compounds for developing high-sensitivity probes to detect contaminants such as heavy metals,7,8 pesticides,9,10 and food additives,11,12 as well as packaging materials to detect spoilage of foods,13–15 and even detecting foodborne pathogens.16,17Notably, CNDs are low-toxicity compounds18 and have been used in biocompatible drug delivery and bioimaging applications.19,20 Studies show that nitrogen-doped CNDs with surface modifications do not harm human lung epithelial tissue in vitro and can exhibit antioxidant and anti-cancer properties.21,22 However, there is a research gap to prove that CNDs can be used without any health concerns. CNDs have low cytotoxicity depending on the dose, synthesis process, and precursors.23–25 Therefore, choosing safe precursors for the synthesis of CNDs is emerging as a promising strategy to enhance their safety for food industry applications.
The Maillard reaction, a complex series of reactions between reducing sugars and amino groups, is a crucial process in the food industry, affecting food's aesthetic, functional, and nutritional properties.26 This reaction shows a close relationship with the formation of CNDs.27,28 CNDs are a marker to control the Maillard reaction,28 directly related to the formation of melanoidins, brown pigments with high aromaticity.27
In this study, we investigated the use of amino acids and sucrose as a model system of the Maillard reaction for synthesizing CNDs and optimizing the reaction conditions to achieve CNDs with desirable properties. Using safe precursors (amino acids and sugars) from a fundamental food reaction is advantageous for the safety of CNDs. Furthermore, using lower temperatures and greener solvents, we aim to reduce the formation of toxins in the synthesis process and create safer CNDs with standardized quality. Purified CNDs were characterized using transmission electron microscopy (TEM), fluorescence, Raman, and UV-Vis spectroscopy techniques. The obtained CNDs were used as a standard to measure the concentration of CNDs forming in the breadmaking process.
Materials and methods
Materials
Amino acids (L-arginine, glycine, L-tyrosine, L-proline, L-glutamine, and L-phenylalanine) and sucrose were obtained from Vital-Trend Ltd (Budapest, Hungary). Hydrochloric acid (1.0 mol L−1), ethanol (≥ 99.5%), and sodium hydroxide solution (0.01 mol L−1) were used as-obtained (VWR International Ltd, Pennsylvania, United States). Ultrapure water was used in all the experiments. Fresh bread was bought from the ALDI supermarket on the experiment day.Characterization
The morphology and particle size of CNDs were measured using a transmission electron microscope (TEM) of JEM-2000FXII (JEOL Ltd, Tokyo, Japan). A Lambda 35 UV-Vis spectrophotometer (Norwalk, USA) was used to measure the ultraviolet-visible (UV-vis) spectra in the 200–600 nm range. Olympus Endosonic Ultrasonicator (Olympus Co., Tokyo, Japan) was used for ultrasound steps. Raman spectra were measured using a LabRAM HR Evolution Confocal Raman Microscope (Horiba, Ltd, Kyoto, Japan).29 The fluorescence spectra were measured using a Spectrofluorometer FP-8500, Jasco, Oklahoma, United States.30Experimental design
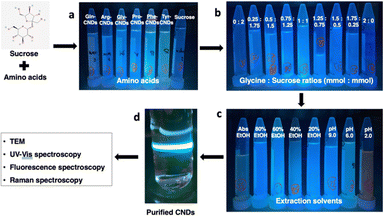
The synthesis method of CNDs from the Maillard reaction was based on the published procedure with modification.25 The optimal conditions for the Maillard reaction were investigated to achieve CNDs with high fluorescence intensity and small size following three experiments, which are illustrated in Fig. 1. (1) Initially, six amino acids—L-arginine (Arg), glycine (Gly), L-tyrosine (Tyr), L-proline (Pro), L-glutamine (Gln), and L-phenylalanine (Phe) were selected as precursors in combination with sucrose to form CNDs. (2) Following the selection of an amino acid that formed the highest quality CNDs, the ratio between amino acid and sucrose was examined using nine combinations, including 2 mmol: 0 mmol, 1.75 mmol: 0.25 mmol, 1.5 mmol: 0.5 mmol, 1.25 mmol: 0.75 mmol, 1 mmol: 1 mmol, 0.75 mmol: 1.25 mmol, 0.5 mmol: 1.5 mmol, 0.25 mmol: 1.75 mmol, and 0 mmol: 2 mmol. (3) Subsequently, various extraction solvents were assessed using nine different options: control (DI water), DI water adjusted to pH 2.0, DI water adjusted to pH 9.0, ethanol (EtOH) at 20%, EtOH 40%, EtOH 60%, EtOH 80%, and absolute EtOH. The optimal conditions were determined based on fluorescent spectra, and each treatment was performed in triplicate.To synthesize and purify CNDs, the solution was heated at 120 °C for 12 h and naturally cooled to room temperature. Cold ethanol was added to the product at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) ratio to remove residual amino acids and incubated for 30 minutes at −20 °C. Subsequently, the mixture was centrifuged at 10
1 (v/v) ratio to remove residual amino acids and incubated for 30 minutes at −20 °C. Subsequently, the mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min. The supernatant obtained was passed through a cellulose column, and the resulting solution was collected and filtered through a 0.22 μm filter. After freeze-drying, the powder of the Maillard reaction-derived carbon nanodots (MR-CNDs) was collected and stored for future use. The purified CNDs were characterized using TEM, and Raman, UV-Vis, and fluorescence spectroscopy techniques.
000 rpm for 30 min. The supernatant obtained was passed through a cellulose column, and the resulting solution was collected and filtered through a 0.22 μm filter. After freeze-drying, the powder of the Maillard reaction-derived carbon nanodots (MR-CNDs) was collected and stored for future use. The purified CNDs were characterized using TEM, and Raman, UV-Vis, and fluorescence spectroscopy techniques.
Statistical analysis
The statistical results were analyzed using IBM SPSS version 25.0 (New York, USA). TEM images and particle distribution were processed using ImageJ software version 1.54D, developed by the National Institutes of Health (New York, USA). All charts were illustrated using GraphPad Prism version 9.0 (California, USA).Results and discussion
Optimization for the synthesis of carbon nanodots using the Maillard reaction
To investigate the impact of the extraction conditions on the formation of carbon nanodots (CNDs) through the Maillard reaction, all parameters, except for the studied variable, were kept constant according to the following procedure. Initially, glycine (0.053 g, 1 mmol) and sucrose (0.180 g, 1 mmol) were dissolved in 20 mL of deionized (DI) water. The solution underwent heating at 120 °C for 12 h, followed by natural cooling to room temperature. Subsequently, it was mixed with cold ethanol in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (v/v), and the mixture was incubated in the freezer for 30 min. Afterward, centrifugation at 10
1 ratio (v/v), and the mixture was incubated in the freezer for 30 min. Afterward, centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min separated the supernatant, which was then passed through a cellulose column to eliminate impurities. The resulting liquid was filtered through a 0.22 μm filter and subjected to freeze-drying. The obtained powder represented the Maillard reaction-derived carbon nanodots (MR-CNDs) product. This method was modified at a lower temperature31 to prevent the formation of toxic compounds such as 5-HMF during the Maillard reactions.32
000 rpm for 30 min separated the supernatant, which was then passed through a cellulose column to eliminate impurities. The resulting liquid was filtered through a 0.22 μm filter and subjected to freeze-drying. The obtained powder represented the Maillard reaction-derived carbon nanodots (MR-CNDs) product. This method was modified at a lower temperature31 to prevent the formation of toxic compounds such as 5-HMF during the Maillard reactions.32
The fluorescence signal was measured using a fluorescence spectrophotometer with an excitation wavelength selected after scanning the excitation and emission wavelengths of standardized CNDs, as discussed in the subsequent section.
The effect of different amino acids on the formation of CNDs
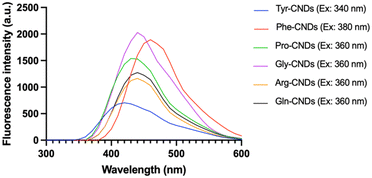
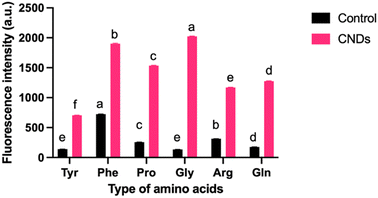
The Maillard reaction is a complex series of reactions between reducing sugars and amino acids.26 These reactions can lead to the formation of various new compounds, including CNDs. Furthermore, CNDs have been reported as a marker to control the Maillard reaction.28 In this study, the relationship between the Maillard reaction and the formation of CNDs was understood through a chosen study model between amino acids and sucrose. Fig. 2 illustrates the distinct fluorescent spectra of CNDs generated from sucrose and various amino acids, including tyrosine, proline, glycine, arginine, glutamine, and phenylalanine. The distinct excitation wavelengths for each amino acid are highlighted in the figure: 340 nm for tyrosine, 360 nm for proline, and 380 nm for glycine, arginine, glutamine, and phenylalanine. The fluorescence intensity of the different amino acids considerably varies (Fig. 2). Glycine-based carbon nanodots have the highest fluorescence intensity, followed by phenylalanine, proline, glutamate, arginine, and tyrosine, as shown in Fig. 2. It is likely due to glycine's enhanced reactivity in the Maillard reaction, which is attributed to its higher susceptibility for forming Schiff bases, intermediate products of the Maillard reaction. These Schiff bases can then react to form other fluorescent compounds, such as melanoidins and brown pigments. These melanoidins are essential precursors for forming CNDs.27The different chemical structures of the amino acids can explain this phenomenon. Tyrosine and phenylalanine contain aromatic rings known for fluorescence.33 Proline has a ring structure, but it is not aromatic. Glycine and arginine are both non-aromatic amino acids. Glutamine is an amino acid that can be converted to glutamine, a non-fluorescent amino acid.34 Fluorescence intensity depends on the structure of amino acids; the simpler the structure, the higher the formation of CNDs, and the higher the fluorescence intensity, as depicted in Fig. 3. Fig. 3 illustrates the fluorescence intensity of amino acids (without the synthesis process) and amino acid-based CNDs. Although phenylalanine resulted in the highest intensity (724.0 ± 4.0), the intensity of Phe-CNDs (1900.0 ± 9.1) was lower than that of Gly-CNDs (2024.0 ± 3.7), where glycine was the weakest in the fluorescence intensity at the beginning (134.9 ± 1.9). These findings suggest that glycine is a good precursor for synthesizing CNDs with high fluorescence intensity.
Effect of the ratio between glycine and sugars (mmol mmol−1) on the formation of CNDs
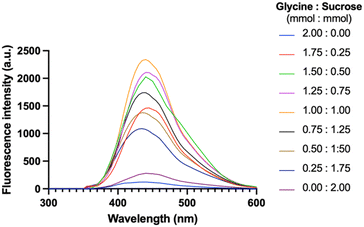
Based on glycine's selection as the precursor in the previous experiment, the excitation wavelength of 360 nm was readily adopted to synthesize CNDs. Fig. 4 shows the fluorescence emission spectra of CNDs formed from different ratios between glycine and sucrose (mmol mmol−1). The fluorescence intensity of the CNDs increased with increasing glycine content until the ratio of 1 mmol glycine to 1 mmol sucrose (Fig. 4). This is likely because glycine is more susceptible to forming Schiff bases. However, when the amount of glycine in the mixture continuously increased, the fluorescence intensity was remarkably reduced. This trend was attributed to the excess glycine interfering with the formation of Schiff bases, which are essential for CNDs formation. Overall, it suggests that the fluorescence intensity of CNDs can be controlled by adjusting the glycine: sucrose ratio. The ratio between glycine and sucrose was maintained at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mmol mmol−1). It exhibited the highest fluorescence intensity. This could be useful for developing CNDs with specific fluorescence properties for different applications.
1 (mmol mmol−1). It exhibited the highest fluorescence intensity. This could be useful for developing CNDs with specific fluorescence properties for different applications.
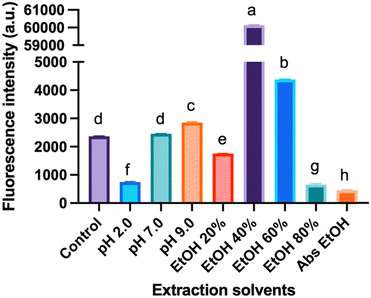
Effect of extraction solvents on the formation of CNDs
Fig. 5 depicts the fluorescence intensity of CNDs formed using various extraction solvents, with the control sample extracted using deionized (DI) water at pH 6.0. Comparing different pH extraction solvents, the higher fluorescence intensity of CNDs was obtained when extracted with DI water at pH 9.0. In contrast, pH 2.0 seems to destroy the presence of CNDs. The structure and fluorescence properties of CNDs could be entirely converted by changing the pH of solvents in the reaction.35 Notably, the highest fluorescence intensity of CNDs was achieved when extracted with 40% ethanol. These results were similar to the previous report,36 which mentioned that ethanolic solution can significantly enhance the reaction rate of the Maillard reaction, resulting in the browning at 420 nm.Furthermore, this phenomenon can be attributed to the favorable conditions for forming Schiff bases, crucial intermediates in the Maillard reaction. The Maillard reaction governs the complex interaction between reducing sugars and amino acids, providing strong evidence for the link between the reaction and CNDs formation. In contrast, an inverse relationship is observed between high ethanol concentration (60% – absolute ethanol) and fluorescence intensity of CNDs, with increasing ethanol concentrations hindering the Schiff base formation. These collective findings suggest a tunable control over the fluorescence intensity of CNDs by manipulating the pH of the extraction solvent and ethanol concentration. Specific observations highlight the maximization of fluorescence intensity at pH 9.0 and the inverse correlation with increasing ethanol concentration, reaching its lowest point with absolute ethanol extraction. These insights are valuable in tailoring CNDs for versatile applications, such as bioimaging or sensing, by optimizing fluorescence properties based on the extraction conditions.
Purification and characterization of CNDs prepared using the Maillard reaction
The isolation procedure was performed using 1 mmol of glycine and 1 mmol of sucrose mixed in 20 mL of 40% ethanol solution at 120 °C for 12 h, using a modified published procedure.25 The resulting solution turned dark brown and exhibited intense fluorescence, from colorless without fluorescence. To extract the M-CNDs further, the mixture was sonicated for 30 min. The obtained solution was then mixed with cold ethanol in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (v/v) to remove impurities at −20 °C for 30 min. The mixture was centrifuged at 10
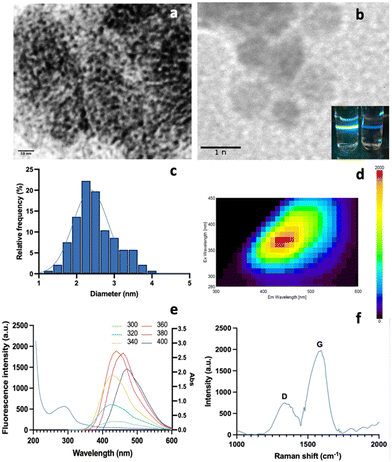
1 ratio (v/v) to remove impurities at −20 °C for 30 min. The mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min to obtain the purified M-CNDs. The supernatant was collected and filtered with a 0.22 μm syringe filter. The purified M-CNDs were characterized using TEM, fluorescence, Raman, and UV-Vis spectroscopy (Fig. 6).
000 rpm for 30 min to obtain the purified M-CNDs. The supernatant was collected and filtered with a 0.22 μm syringe filter. The purified M-CNDs were characterized using TEM, fluorescence, Raman, and UV-Vis spectroscopy (Fig. 6).
TEM images with 10 nm and 1 nm scales revealed the dark spherical morphology of the M-CNDs without any contaminated compounds. The size distribution of the M-CNDs ranged from 1.25 to 4 nm, with an average size of 2.5 ± 0.5 nm based on measurements of almost 300 particles in the TEM image with a scale of 10 nm (Fig. 6A). In Fig. 6E, the UV-vis spectrum of M-CNDs exhibited one peak at 275 nm, indicating the presence of the π–π* electronic transition in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C.37 Blue solid fluorescence of M-CNDs was detected under UV light (Fig. 6B) with a maximum emission wavelength of 438 nm and an excitation wavelength of 360 nm (Fig. 6D). These findings were consistent with those from the previous studies that used glucose and lysine or melatonin as starting materials.25,27
C.37 Blue solid fluorescence of M-CNDs was detected under UV light (Fig. 6B) with a maximum emission wavelength of 438 nm and an excitation wavelength of 360 nm (Fig. 6D). These findings were consistent with those from the previous studies that used glucose and lysine or melatonin as starting materials.25,27
Furthermore, Raman spectroscopy analysis of the synthesized M-CNDs revealed distinct D and G bands at 1350 and 1580 cm−1, respectively, as depicted in Fig. 6F. Consistent with established literature, the low-intensity ratio of the D to G band (ID < IG) underscores the moderate graphitic structure of the M-CNDs, affirming the absence of significant carbonaceous impurities, as corroborated by similar findings in the reference study.38 Overall, the results explain the successful isolation and characterization of M-CNDs with nanosized, intense fluorescence properties, which could be used as a safe carbon nanodot source for various applications.
Use of M-CNDs as a standard for detecting CNDs in food samples using a fluorescent spectrophotometer
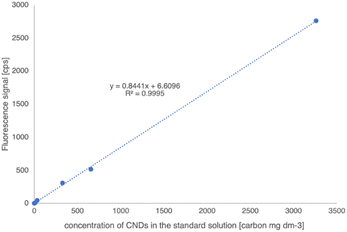
To evaluate the purity of M-CNDs and quantitatively determine the carbon content of M-CNDs stock solution, the purified M-CNDs were analyzed using an elemental analyzer, resulting in a total carbon content in the stock M-CNDs solution of 0.326%. Based on the carbon content, the concentration of the stock standard was 3260 mg dm−3. Standard curves were generated by measuring the intensity of the fluorescent spectrum at the excitation wavelength of 360 nm across a range of M-CNDs concentrations. The standard curve was prepared freshly on the measurement day. The resulting calibration curve for CNDs had a linear relationship for a five-point calibration curve in the concentration range from 0 to 3200 mg dm−3 (Fig. 7). The reproducibility of measurement was below 0.1%. The calculated detection limit of the method was 0.14 mg dm−3.Inter-day variation was also assessed, and a measurement was carried out with a standard addition method. The inter-day variation of the method was less than 0,1%. The calibration and the standard addition methods gave the same concentration result values.
Along with the obtained standard curve in Fig. 7, the concentration of CNDs in breadmaking, including dough, bread crust, and bread crumb, was measured. Note that 1 g of the sample was mixed with 10 mL of distilled water and then sonicated for 30 min. The mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The supernatant was collected and mixed with cold ethanol 40% in a 1
000 rpm for 10 min. The supernatant was collected and mixed with cold ethanol 40% in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) ratio at 4 °C in the freezer for 30 min and then centrifuged at 10
1 (v/v) ratio at 4 °C in the freezer for 30 min and then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min. The supernatant was collected and filtered with 0.22 μm and the content of CNDs was measured using fluorescence spectroscopy at the excitation wavelength of 360 nm. Our results, depicted in Table 1, revealed that no CNDs were detected in the dough, while the highest concentration of CNDs was detected on the bread crust with 512 ± 1 mg kg−1 DW. The difference in concentration of CNDs in breadmaking is supposed to be related to the temperature and heat treatment time, where the bread crust directly coming in contact with high temperature showed higher CNDs content than the bread crumb with 322 ± 4 mg kg−1 DW. The possible relationship between CNDs and heat treatment has been reported since 2012 when CNDs were found in bread and caramelized sugars for the first time.39
000 rpm for 30 min. The supernatant was collected and filtered with 0.22 μm and the content of CNDs was measured using fluorescence spectroscopy at the excitation wavelength of 360 nm. Our results, depicted in Table 1, revealed that no CNDs were detected in the dough, while the highest concentration of CNDs was detected on the bread crust with 512 ± 1 mg kg−1 DW. The difference in concentration of CNDs in breadmaking is supposed to be related to the temperature and heat treatment time, where the bread crust directly coming in contact with high temperature showed higher CNDs content than the bread crumb with 322 ± 4 mg kg−1 DW. The possible relationship between CNDs and heat treatment has been reported since 2012 when CNDs were found in bread and caramelized sugars for the first time.39
| Food Items | Concentration of CNDs | |||
|---|---|---|---|---|
| Dough | ND | |||
| Bread crumb | 322 | ± | 4 | mg kg−1 in DW a |
| Bread crust | 512 | ± | 1 | mg kg−1 in DW b |
The data are expressed as the mean ± standard deviation. Values within the same column with different letters (a–e) were significantly different (P < 0.05).
This method's precision, repeatability, day-to-day variation, and linearity were accessed according to the validation procedure of the European Medicines Agency and AOAC International.40,41 The standard curve was built with the regression line: y = 0,8441x + 6.61 with R2 = 0.9995 with a limit of detection (LOD) of 0.14 mg dm−3 and a limit of quantification (LOQ) of 0.47 mg dm−3. Considering the ten times dilution in the sample preparation, the detection limit calculated for the solid food sample is 1.4 mg kg−1. With all the assessments, this method was reliable for measuring CNDs in foods.
Conclusion
In conclusion, this study has demonstrated that the use of the Maillard reaction at low temperatures (120 °C) and long time (12 h) is a promising method for synthesizing carbon nanodots (CNDs) with high fluorescence intensity and small size (2.5 ± 0.5 nm). This study systematically explored the optimization of the synthesis of CNDs through the Maillard reaction, including (1) selecting an appropriate amino acid, (2) choosing the effective glycine-to-sucrose ratio, and (3) examining the influence of extraction solvents. Glycine enhanced the reactivity in the Maillard reaction, resulting in superior fluorescence intensity of CNDs compared to other amino acids. Moreover, the investigation into the glycine-to-sucrose ratio revealed that a 1 mmol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mmol ratio delivered the highest fluorescence intensity, offering a critical parameter for the synthesis of tailored CNDs. Expanding our exploration to extraction solvents, the crucial role of pH and ethanol concentration provided a valuable guide for controlling the fluorescence properties of CNDs, where 40% ethanol solution showed the most effective extraction to form CNDs with intense fluorescent intensity. The subsequent purification and characterization of Maillard reaction-derived carbon nanodots (M-CNDs) underscored the effectiveness of our isolation procedure. TEM imaging confirmed the nanosized, dark spherical morphology of M-CNDs, with an average size of 2.5 ± 0.5 nm. The UV-Vis spectrum and intense blue fluorescence of M-CNDs further validated their exceptional properties. Raman spectroscopy analysis supported the absence of significant carbonaceous impurities, confirming the moderate graphitic structure of M-CNDs. The obtained M-CNDs were used as a standard for quantifying the CNDs’ content in the breadmaking processing, where CNDs were only found in the bread crusts and crumbs after baking. It was shown that the presence of CNDs was closely related to the heat treatment process. The successful isolation and characterization of M-CNDs elevate the potential of these nanostructures in diverse applications, emphasizing their role as a promising carbon dot source. The results of this study have the potential to advance the development of carbon nanodots for bioimaging, sensing, and other applications.
1 mmol ratio delivered the highest fluorescence intensity, offering a critical parameter for the synthesis of tailored CNDs. Expanding our exploration to extraction solvents, the crucial role of pH and ethanol concentration provided a valuable guide for controlling the fluorescence properties of CNDs, where 40% ethanol solution showed the most effective extraction to form CNDs with intense fluorescent intensity. The subsequent purification and characterization of Maillard reaction-derived carbon nanodots (M-CNDs) underscored the effectiveness of our isolation procedure. TEM imaging confirmed the nanosized, dark spherical morphology of M-CNDs, with an average size of 2.5 ± 0.5 nm. The UV-Vis spectrum and intense blue fluorescence of M-CNDs further validated their exceptional properties. Raman spectroscopy analysis supported the absence of significant carbonaceous impurities, confirming the moderate graphitic structure of M-CNDs. The obtained M-CNDs were used as a standard for quantifying the CNDs’ content in the breadmaking processing, where CNDs were only found in the bread crusts and crumbs after baking. It was shown that the presence of CNDs was closely related to the heat treatment process. The successful isolation and characterization of M-CNDs elevate the potential of these nanostructures in diverse applications, emphasizing their role as a promising carbon dot source. The results of this study have the potential to advance the development of carbon nanodots for bioimaging, sensing, and other applications.
Author contributions
Conceptualization, J. P. and D. H. H. N.; methodology, D. H. H. N.; doing experiments, D. H. H. N., A. M., J. P., L. N., and L. D.; analyzing data, D. H. H. N, A. M, and J. P.; writing – original draft preparation, A. M., D. H. H. N., L. N; writing – discussion and conclusion, H. E.-R., D. H. H. N., visualization, D. H. H. N., supervision, J. P., H. E.-R. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are thankful for the support of the Stipendium Hungaricum Scholarship Program and the grant No. K-132685 from National Research, Development and Innovation Office, Hungary.References
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- H. Li, Z. Kang, Y. Liu and S.-T. Lee, J. Mater. Chem., 2012, 22, 24230 RSC.
- S. C. Ray, A. Saha, N. R. Jana and R. Sarkar, J. Phys. Chem. C, 2009, 113, 18546–18551 CrossRef CAS.
- S. Khan, A. Dunphy, M. S. Anike, S. Belperain, K. Patel, N. H. Chiu and Z. Jia, Int. J. Mol. Sci., 2021, 22, 6786 CrossRef CAS PubMed.
- W. Zhang, H. Zhong, P. Zhao, A. Shen, H. Li and X. Liu, Food Control, 2022, 133, 108591 CrossRef.
- D. Dhamodharan, H.-S. Byun, M. V. Shree, D. Veeman, L. Natrayan and B. Stalin, J. Ind. Eng. Chem., 2022, 110, 68–83 CrossRef CAS.
- M. Pajewska-Szmyt, B. Buszewski and R. Gadzała-Kopciuch, Spectrochim. Acta, Part A, 2020, 236, 118320 CrossRef CAS PubMed.
- S. C. Pandey, A. Kumar and S. K. Sahu, J. Photochem. Photobiol., A, 2020, 400, 112620 CrossRef CAS.
- C. Cao and W. Guo, Food Chem., 2024, 435, 137578 CrossRef CAS PubMed.
- F. Y. Vadia, S. Ghosh, V. N. Mehta, S. Jha, N. I. Malek, T. J. Park and S. K. Kailasa, Food Chem., 2023, 428, 136796 CrossRef CAS PubMed.
- B. J. Wang, Z. Y. Xu, Z. Sun, Z. Q. Li, Y. H. Luo, H. Q. Luo and N. B. Li, Anal. Bioanal. Chem., 2023, 415, 4639–4647 CrossRef CAS PubMed.
- M. A. Albalawi, H. Gomaa, M. A. El Hamd, M. A. S. Abourehab and M. A. Abdel-Lateef, Luminescence, 2023, 38, 92–98 CrossRef CAS PubMed.
- S. K. Tammina and J.-W. Rhim, Chemosphere, 2023, 313, 137627 CrossRef CAS PubMed.
- A. Khan, Z. Riahi, J. Tae Kim and J.-W. Rhim, Food Chem., 2024, 432, 137215 CrossRef CAS PubMed.
- S. Chen, Q. Zeng, X. Tan, M. Ye, Y. Zhang, L. Zou, S. Liu, Y. Yang, A. Liu, L. He and K. Hu, Carbohydr. Polym., 2023, 314, 120938 CrossRef CAS PubMed.
- X. Gao, H. Zhang, L. Liu, M. Jia, X. Li and J. Li, Food Chem., 2024, 432, 137144 CrossRef CAS PubMed.
- D. Zhao, Z. Zhang, X. Liu, R. Zhang and X. Xiao, Mater. Sci. Eng., C, 2021, 119, 111468 CrossRef CAS PubMed.
- K. Wang, Z. Gao, G. Gao, Y. Wo, Y. Wang, G. Shen and D. Cui, Nanoscale Res. Lett., 2013, 8, 122 CrossRef PubMed.
- J. A. Jaleel and K. Pramod, J. Controlled Release, 2018, 269, 302–321 CrossRef CAS PubMed.
- M. Tuerhong, Y. Xu and X.-B. Yin, Chin. J. Anal. Chem., 2017, 45, 139–150 Search PubMed.
- Z. Ji, Z. Yin, Z. Jia and J. Wei, Langmuir, 2020, 36, 8632–8640 CrossRef CAS PubMed.
- E. Durantie, H. Barosova, B. Drasler, L. Rodriguez-Lorenzo, D. A. Urban, D. Vanhecke, D. Septiadi, L. Hirschi-Ackermann, A. Petri-Fink and B. Rothen-Rutishauser, Biointerphases, 2018, 13, 06D404 CrossRef PubMed.
- K. Hola, Y. Zhang, Y. Wang, E. P. Giannelis, R. Zboril and A. L. Rogach, Nano Today, 2014, 9, 590–603 CrossRef CAS.
- S. Sahu, B. Behera, T. K. Maiti and S. Mohapatra, Chem. Commun., 2012, 48, 8835 RSC.
- D. Li, X. Na, H. Wang, Y. Xie, S. Cong, Y. Song, X. Xu, B.-W. Zhu and M. Tan, J. Agric. Food Chem., 2018, 66, 1569–1575 CrossRef CAS PubMed.
- M. Saltmarch and T. P. Labuza, Diabetes, 1982, 31, 29–36 CrossRef CAS.
- D. Li, Y. Xie, X. Na, Y. Li, C. Dai, Y. Li and M. Tan, Food Funct., 2019, 10, 4414–4422 RSC.
- S. Matiacevich and M. Pilarbuera, Food Chem., 2006, 95, 423–430 CrossRef CAS.
- A. C. Ferrari and J. Robertson, Philos. Trans. R. Soc. London, Ser. A, 2004, 362, 2477–2512 CrossRef CAS PubMed.
- Y.-M. Long, C.-H. Zhou, Z.-L. Zhang, Z.-Q. Tian, L. Bao, Y. Lin and D.-W. Pang, J. Mater. Chem., 2012, 22, 5917 RSC.
- D. Li, X. Na, H. Wang, Y. Xie, S. Cong, Y. Song, X. Xu, B.-W. Zhu and M. Tan, J. Agric. Food Chem., 2018, 66, 1569–1575 CrossRef CAS PubMed.
- Y. Chen, H. Lin, Y. Li, M. Lin and J. Chen, Ind. Crops Prod., 2019, 135, 146–152 CrossRef CAS.
- J. B. Alexander Ross, W. R. Laws, K. W. Rousslang and H. R. Wyssbrod, in Topics in Fluorescence Spectroscopy, ed. J. R. Lakowicz, Kluwer Academic Publishers, Boston, 2002, vol. 3, pp. 1–64 Search PubMed.
- R. D’Souza and J. Powell-Tuck, J. R. Soc. Med., 2004, 97, 425–427 CrossRef PubMed.
- J. Bai, Y. Ma, G. Yuan, X. Chen, J. Mei, L. Zhang and L. Ren, J. Mater. Chem. C, 2019, 7, 9709–9718 RSC.
- S. C. Shen and J. S. B. Wu, J. Food Sci., 2004, 69, FCT273–FCT279 CrossRef CAS.
- H. Ding, S.-B. Yu, J.-S. Wei and H.-M. Xiong, ACS Nano, 2016, 10, 484–491 CrossRef CAS PubMed.
- R. Atchudan, T. N. J. I. Edison and Y. R. Lee, J. Colloid Interface Sci., 2016, 482, 8–18 CrossRef CAS PubMed.
- M. P. Sk, A. Jaiswal, A. Paul, S. S. Ghosh and A. Chattopadhyay, Sci. Rep., 2012, 2, 383 CrossRef PubMed.
- I. ICH, Topic Q 2 (R1) Validation of Analytical Procedures: Text and Methodology. European Medicines Agency, CPMP/ICH/381/95, 1995.
- AOAC International, 2016.
| This journal is © The Royal Society of Chemistry 2024 |