Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Discovery of highly potent SARS-CoV-2 nsp14 methyltransferase inhibitors based on adenosine 5′-carboxamides†‡
Hugo
Kocek
 ab,
Dominika
Chalupská
a,
Milan
Dejmek
ab,
Dominika
Chalupská
a,
Milan
Dejmek
 a,
Alexandra
Dvořáková
a,
Michala
Zgarbová
ac,
Michal
Šála
a,
Alexandra
Dvořáková
a,
Michala
Zgarbová
ac,
Michal
Šála
 a,
Karel
Chalupský
a,
Petra
Krafčíková
a,
Tomáš
Otava
ad,
Matúš
Drexler
a,
Karel
Chalupský
a,
Petra
Krafčíková
a,
Tomáš
Otava
ad,
Matúš
Drexler
 a,
Eliška
Procházková
a,
Eliška
Procházková
 a,
Blanka
Klepetářová
a,
Milan
Štefek
a,
Blanka
Klepetářová
a,
Milan
Štefek
 ae,
Ján
Kozic
ae,
Ján
Kozic
 a,
Helena
Mertlíková-Kaiserová
a,
Evzen
Boura
a,
Jan
Weber
a,
Helena
Mertlíková-Kaiserová
a,
Evzen
Boura
a,
Jan
Weber
 a and
Radim
Nencka
a and
Radim
Nencka
 *a
*a
aInstitute of Organic Chemistry, and Biochemistry of the Czech Academy of Sciences, Prague, Czech Republic. E-mail: nencka@uochb.cas.cz
bFaculty of Chemical Technology, University of Chemistry and Technology, Prague, Czech Republic
cDepartment of Genetics and Microbiology, Faculty of Science, Charles University, Prague, Czech Republic
dFaculty of Food and Biochemical Technology, University of Chemistry and Technology, Prague, Czech Republic
eDepartment of Organic Chemistry, Faculty of Science, Charles University, Prague, Czech Republic
First published on 1st August 2024
Abstract
The emergence of SARS-CoV-2, the causative agent of COVID-19, has highlighted the need for advanced antiviral strategies. Targeting the coronaviral methyltransferase nsp14, which is essential for RNA capping, offers a promising approach for the development of small-molecule inhibitors. We designed and synthesized a series of adenosine 5′-carboxamide derivatives as potential nsp14 inhibitors and identified coumarin analogs to be particularly effective. Structural modifications revealed the importance of the 5′-carboxyl moiety for the inhibitory activity, showing superior efficacy compared to other modifications. Notably, compound 18l (HK370) demonstrated high selectivity and favorable in vitro pharmacokinetic properties and exhibited moderate antiviral activity in cell-based assays. These findings provide a robust foundation for developing targeted nsp14 inhibitors as a potential treatment for COVID-19 and related diseases.
Introduction
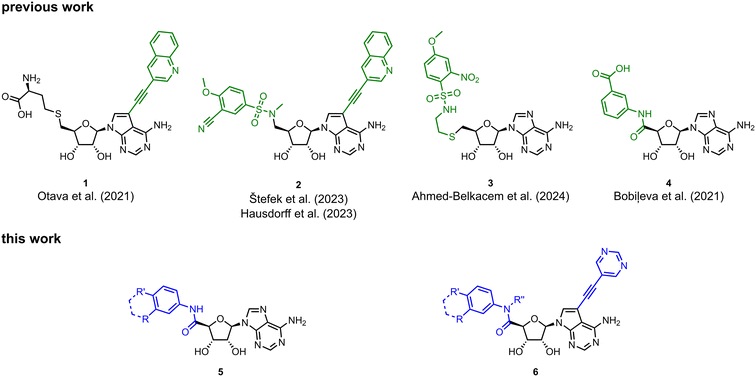
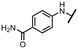
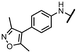
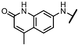
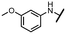
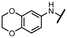
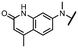
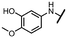
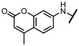
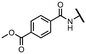
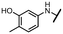
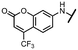
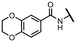
Viruses belonging to the family Coronaviridae (order Nidovirales) pose a significant pandemic threat, as was demonstrated by SARS-CoV, MERS-CoV and, most notably, by SARS-CoV-2 in 2002, 2012 and 2019, respectively.1 The genetic information of SARS-CoV-2 consists of a large (∼30 kb) positive-sense, single-stranded RNA (+ssRNA), which encodes 4 structural proteins, 16 non-structural proteins and several accessory factors.2 The virus replicates in double-membrane vesicles (DMVs) derived from the host endoplasmic reticulum and, therefore, does not have access to the host's mRNA capping machinery.3 The 5′ end of the eukaryotic mRNA is equipped with a cap-1 or cap-2 structure, which is important for several cellular processes including translation and self-recognition. The absence of this cap structure triggers the activation of cytosolic sensors (e.g., IFIT1 or MDA5) and subsequently initiates an immune response.4,5 To mimic the host's mRNA cap, SARS-CoV-2 utilizes its own capping enzymes, including two MTases – nsp14 (N7 methylation; cap-0) and nsp16 (2′O methylation; cap-1). Both MTases are SAM-dependent, which makes them a suitable target for small-molecule inhibitors.6Coronaviral MTases are a focal point of medicinal chemistry research since the beginning of the SARS-CoV-2 pandemic.7–21 Otava et al. described SAH analogs with a modified nucleobase (1) targeting the lateral cavity above the SAM-binding site7 and various replacements of the amino acid moiety on the 5′ end were explored by several groups.11,22–26 A significant portion of this research has focused on arylsulfonamides derived from 5′-aminoadenosine (2, 3), with the sulfonamide moiety being crucial for the inhibitory activity due to its specific geometry.24–28 Derivatives of adenosine-5′-carboxylic acid represent an under-explored area in the search for coronaviral nsp14 inhibitors. To date, only one such compound (4) has been described in the literature11 and is considered inferior due to its poor inhibitory activity against SARS-CoV-2 nsp14 (IC50 = 12 μM).
In this work, we synthesized a series of amides derived from adenosine-5′-carboxylic acid leading to the development of nanomolar inhibitors with general structures 5 and 6. We further examined the importance of the amidic moiety for inhibitory activity and tested our compounds in a cell-based assay (Fig. 1).
Results and discussion
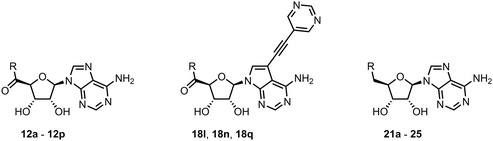
Synthesis
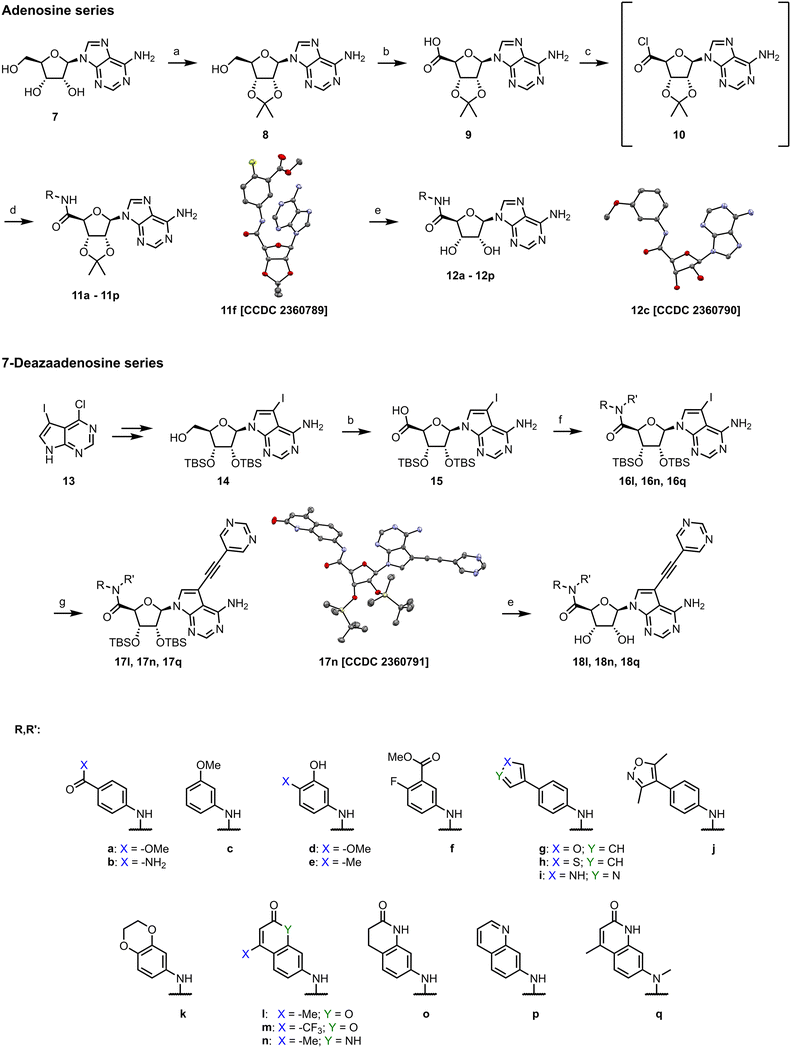
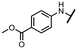
We started the synthesis from adenosine (7), which was isopropylidene-protected to afford 8.29 Subsequent oxidation of the 5′ carbon using TEMPO/PhI(OAc)2 yielded adenosine 5′-carboxylic acid 9.30 Treatment of 9 with SOCl2 afforded highly reactive intermediate 10, which was used without any purification in amidic coupling with a selected amine leading to amides 11. Final removal of the isopropylidene protecting group was achieved using 80% formic acid yielding 12.Synthesis of analogs with a modified nucleobase started from 2′,3′-protected 7-iodotubercidine 14, which was prepared according to a published procedure.31 Oxidation of the 5′ carbon was again achieved with TEMPO/PhI(OAc)2 yielding 15.30 A different approach was used for synthesis of 16, as some amines reacted poorly with acyl chloride 10. Propanephosphonic acid anhydride (T3P) mediated peptide coupling between the amine of choice and the acid 15 smoothly afforded products in 2 to 24 hours in moderate to good yields. Installation of the 5-ethynylpyrimidine moiety in position 7 of the nucleobase was achieved via the Sonogashira cross-coupling,7 leading to 17 which was subsequently treated with 80% formic acid to afford final compounds 18 (Scheme 1).
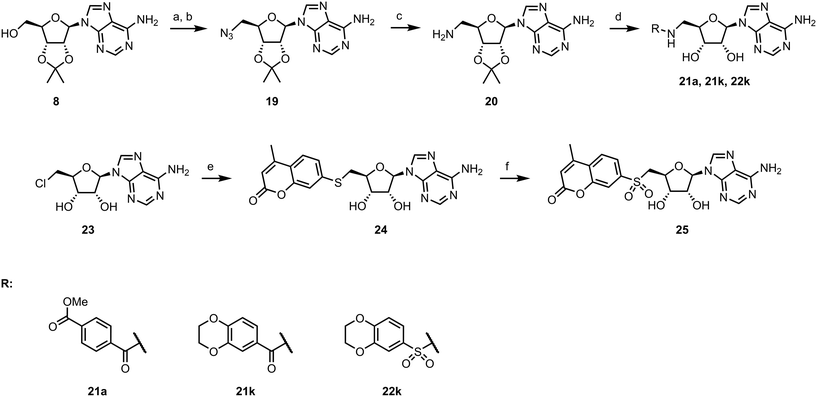
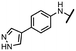
To explore other linkers, we mesylated compound 8 and, via nucleophilic substitution with NaN3, prepared compound 19. Subsequent hydrogenation yielded compound 20, which was then used for the preparation of amides (21a, 21k) and sulfonamide (22k).32 Finally, commercially available compound 23 was treated with 7-mercapto-4-methylcoumarin to afford compound 24. Oxidation of the sulfur linker using Oxone® yielded sulfone 25 (Scheme 2).33
Inhibition of SARS-CoV-2 MTase nsp14
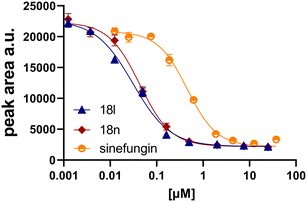
Initially, we focused on the synthesis of adenosine 5′-carboxamides using simple mono- and disubstituted anilines leading to 12a–f; however, these compounds exhibited only negligible activity. We then attempted to replace the ester moiety of 12a by decorating the para position of the aniline with various 5-membered heterocycles (compounds 12g–12j), but this resulted in inactive molecules. Introduction of a bicyclic benzo-1,4-dioxane core (compound 12k) led to an intriguing 1.8 μM inhibitory activity. Based on this result, we decided to explore further bicyclic cores via a scaffold hopping approach. This led to a series of coumarin-based inhibitors with exciting submicromolar potency (12l–12n).In recent studies,7,8 we identified several aromatic C-7 substituents that enhanced the biological activity of our ligands. Derivatives with the 5-ethynylpyrimidine substituent demonstrated very good inhibitory properties in all cases and our recent studies further revealed their superior solubility and metabolic stability properties (unpublished data). Consequently, we selected this substituent as our model and prepared compounds 18l and 18n, which demonstrated a 10-fold enhancement in activity in an enzymatic assay, with IC50 values of 30 nM and 43 nM, respectively (Fig. 2).
We also introduced a methyl group on the amidic nitrogen (18q), which has previously been shown to be highly beneficial in the case of sulfonamide 2.8,25,28 However, in this particular case, this modification proved to be unfavorable, resulting in a 20-fold decrease in inhibitory activity.
To better understand the role of the amidic linker, analogs bearing a reversed amidic linker, as well as sulfonamide, sulfide, and sulfone linkers, were prepared (21a, 21k, 22k, 24, 25). In all cases, this led to a significant decrease or complete loss of the inhibitory activity and clearly demonstrated the importance of the 5′-carboxyl moiety (Table 1).
Human cells rely on several MTases; therefore, it is necessary to assess the possible off-target effect. Compounds 12l, 12n, 18l, 18n, and 18q were tested against human RNMT which is an analog to coronaviral nsp14 as they both methylate the cap at position N7.34 None of the tested compounds exhibited any inhibitory activity against RNMT at 25 μM concentration of an inhibitor.
Docking study
To understand the binding mode of our inhibitors, we performed extensive docking experiments using the crystal structure of SARS-CoV-2 nsp14 in complex with SAH (PDB: 7R2V).35 Results from Glide36 docking show that our inhibitors target both the SAM-binding site and the RNA-binding site as the aromatic moiety on the 5′ end extends to the cap-site and interacts with phenylalanine 426 via π–π stacking interaction. This amino acid residue, F426, provides one of the key interactions with the RNA cap and F426A mutants lack MTase activity.37 In the case of the C7-modified analogs (series 18), the alkynyl moiety extends into a lateral cavity above the SAM-binding site as described by Otava et al.7 The results of docking experiments (GlideScore XP; Fig. 3) for 12k–n, 18l, and 18n generally correlate with the measured IC50 values, except for compound 12m. This discrepancy might be due to the presence of fluorine atoms, as we have previously experienced that fluorinated compounds can have their GlideScore values falsely overestimated. This phenomenon has been reported by other groups as well.38,39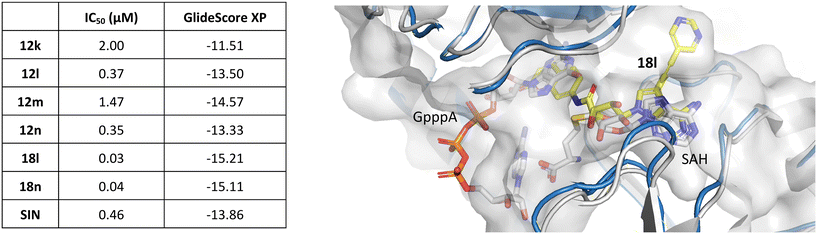 | ||
| Fig. 3 Correlation between IC50 values and GlideScore XP (left). Docking pose of 18l in nsp14 (PDB: 7R2V;35 protein in blue, inhibitor in yellow) overlaid with nsp14 with a cap analog (GpppA, gray) and SAH (PDB: 5C8S;40 gray) (right). | ||
Pharmacology
Enzymatic screening allowed us to advance our inhibitors in terms of their activity; however, pharmacological evaluation is a crucial step towards compounds capable of inhibiting viral replication in cells. Compounds 12l, 12m, 12n, 18l, 18n and 18q were evaluated for their stability in both human and mouse plasma and liver microsomes (Table 2). All tested compounds were stable in liver microsomes and only 12m and 18n were metabolized in plasma. Inhibitor 12m was shown to be unstable in both human and mouse plasma, potentially due to the strong electron-withdrawing effect of –CF3 in conjugation with the lactone moiety. In the case of 18n, metabolic instability was observed only in mouse plasma with a half-life of 94 minutes (see the ESI‡ for graphs, Fig. S2).| Plasma stability % of the parent compound after 120 min | Microsomal stability % of the parent compound after 45 min | Caco-2 permeability | |||||
|---|---|---|---|---|---|---|---|
| Human | Mouse | Human | Mouse | Papp (cm s−1) A–B × 10−6 | Papp (cm s−1) B–A × 10−6 | Efflux ratio | |
| 12l | 103 ± 3.3 | 93 ± 2.7 | 104 ± 4.1 | 95 ± 5.0 | 3.1 ± 1.3 | 10.7 ± 2.5 | 3.4 |
| 12m | 20 ± 1.4 | 67 ± 3.5 | 90 ± 9.1 | 93 ± 9.4 | 8.0 ± 2.9 | 32.9 ± 5.3 | 4.1 |
| 12n | 97 ± 2.0 | 78 ± 0.3 | 95 ± 0.0 | 90 ± 2.9 | 4.4 ± 1.8 | 2.2 ± 0.4 | 0.5 |
| 18l | 74 ± 0.2 | 89 ± 1.1 | 90 ± 4.2 | 84 ± 2.5 | 10.1 ± 4.0 | 8.3 ± 0.5 | 0.8 |
| 18n | 93 ± 0.4 | 41 ± 0.7 | 90 ± 2.0 | 85 ± 0.6 | 11.3 ± 3.7 | 7.3 ± 1.3 | 0.6 |
| 18q | 80 ± 1.1 | 86 ± 0.9 | 107 ± 6.6 | 117 ± 10.9 | 2.0 ± 0.3 | 2.1 ± 0.4 | 1.0 |
| Propantheline | 0.1 ± 0.0 | 9 ± 0.2 | N.D. | N.D. | N.D. | N.D. | N.D. |
| Verapamil | N.D. | N.D. | 32 ± 1.1 | 5 ± 0.2 | N.D. | N.D. | N.D. |
| Digoxin | N.D. | N.D. | N.D. | N.D. | 1.7 ± 0.2 | 13.8 ± 0.9 | 8.5 |
Monolayers of differentiated Caco-2 epithelial cells were used to simulate the intestinal absorption41,42 of selected inhibitors and all tested compounds exhibited reasonable transepithelial permeability. Two compounds exhibited an efflux ratio (ER) above 2, 12l (ER: 3.4) and 12m (ER: 4.1), suggesting their interaction with cellular efflux pumps (Table 2).43
Inhibition of SARS-CoV-2 in vitro
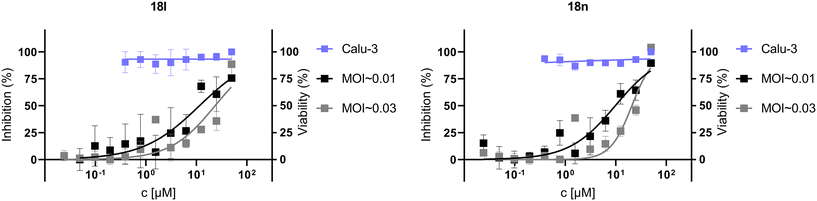
We tested the cytotoxicity and the anti-SARS-CoV-2 activity of compounds 12l, 12n, 18l, 18n, and 18q in two types of cell cultures – Calu-3 and Vero E6. The compounds were prepared in 2-fold serial dilutions, starting from a 50 μM solution. Cells were pretreated with the inhibitors two hours prior to the infection with the SARS-CoV-2 strain hCoV-19/Czech Republic/NRL_6632_2/2020 at two different multiplicities of infection (MOI ∼0.03 and MOI ∼0.01). Simultaneously, the same experimental setup without virus was used to determine the compounds' cytotoxicity. Cells were incubated for three days at 37 °C in 5% CO2. Cell viability was then assessed using the XTT assay.44 We detected an antiviral effect for compounds 18l and 18n in the Calu-3 cell line, with EC50 values of 12 ± 6 μM and 10 ± 3 μM, respectively, at an MOI ∼0.01, without any observed cytotoxicity (CC50 > 50 μM; Fig. 4). For compounds 12l, 12n, and 18q, the EC50 values were determined to be above 50 μM in the Calu-3 cell line (see the ESI‡ for graphs, Fig. S4). No antiviral effect was observed in the Vero E6 cell line for any of the five inhibitors which may be explained by the defective interferon response of this cell line.45Conclusion
In this work, we describe the discovery of SARS-CoV-2 nsp14 inhibitors with novel structural motifs based on adenosine 5′-carboxamides derived from bicyclic amines. Through scaffold hopping, we found out that coumarin analogs are particularly effective. We also explored the importance of the 5′-carboxyl moiety for inhibitory activity. Analogs bearing a methylated amide, reversed amidic linker, sulfonamide, sulfide, or sulfone showed a significant decrease or complete loss of the inhibitory activity. Compound 18l (HK370), with an IC50 value of 31 nM, exhibited high selectivity for nsp14 over human RNMT (IC50 (RNMT) > 25 μM; CC50 > 50 μM), a favorable in vitro metabolic profile and good transepithelial permeability. Notably, our inhibitors show moderate efficacy in a cell-based assay (EC50: 12 ± 6 μM, Calu-3 cell line). Overall, this work provides a strong foundation for the development of targeted nsp14 inhibitors as potential treatments for COVID-19 and other coronavirus-related diseases.Abbreviations
| SARS-CoV | Severe acute respiratory syndrome-related coronavirus |
| MERS-CoV | Middle East respiratory syndrome coronavirus |
| COVID-19 | Coronavirus disease 2019 |
| +ssRNA | Single-stranded positive-sense RNA |
| MTase | Methyltransferase |
| SAM | S-Adenosyl-L-methionine |
| SAH | S-Adenosyl-L-homocysteine |
| ER | Efflux ratio |
| MOI | Multiplicity of infection |
Data availability
The data supporting this article have been included as part of the ESI.‡ Crystallographic data for 11f, 12c, and 17n have been deposited at the CCDC under 2360789, 2360790, and 2360791.Author contributions
H. K., M. D., M. Š., T. O., and M. Š.: synthesis of the compounds; D. C., A. D., K. C., P. K., M. D., and J. K.: protein purification and biochemical assays; A. D. and M. Z.: cell-based assays; E. P.: compound characterization by NMR spectroscopy; B. K.: SC-XRD; H. K. and M. D.: writing – original draft; H. M.-K., E. B., J. W., and R. N.: supervision, funding acquisition, project administration, and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was supported by the Academy of Sciences of the Czech Republic as part of the Strategy AV 21 Virology and Antiviral Therapy programme and the Czech Academy of Sciences (RVO: 61388963). Also, this research was funded by the National Institute Virology and Bacteriology (Programme EXCELES, project no. LX22NPO5103) – funded by the European Union – Next Generation EU. We thank IOCB's Mass Spectrometry Core Facility for measuring HR-MS spectra, namely Kvetoslava Kertisová, Edita Kofroňová, and Kateřina Nováková. We also thank IOCB's Analytical Laboratory Core Facility for determining optical rotation, namely Lucie Holasová and Michaela Gazdurová. We used PyMOL software46 to prepare the figures both in the main text and ESI.‡References
- F. Wu, S. Zhao, B. Yu, Y.-M. Chen, W. Wang, Z.-G. Song, Y. Hu, Z.-W. Tao, J.-H. Tian and Y.-Y. Pei, Nature, 2020, 579, 265–269 CrossRef CAS PubMed.
- R. Lu, X. Zhao, J. Li, P. Niu, B. Yang, H. Wu, W. Wang, H. Song, B. Huang and N. Zhu, Lancet, 2020, 395, 565–574 CrossRef CAS PubMed.
- S. Klein, M. Cortese, S. L. Winter, M. Wachsmuth-Melm, C. J. Neufeldt, B. Cerikan, M. L. Stanifer, S. Boulant, R. Bartenschlager and P. Chlanda, Nat. Commun., 2020, 11, 5885 CrossRef CAS PubMed.
- R. Züst, L. Cervantes-Barragan, M. Habjan, R. Maier, B. W. Neuman, J. Ziebuhr, K. J. Szretter, S. C. Baker, W. Barchet and M. S. Diamond, Nat. Immunol., 2011, 12, 137–143 CrossRef PubMed.
- A. Pichlmair and C. R. e Sousa, Immunity, 2007, 27, 370–383 CrossRef CAS PubMed.
- R. Nencka, J. Silhan, M. Klima, T. Otava, H. Kocek, P. Krafcikova and E. Boura, Nucleic Acids Res., 2022, 50, 635–650 CrossRef CAS PubMed.
- T. Otava, M. Sala, F. Li, J. Fanfrlik, K. Devkota, S. Perveen, I. Chau, P. Pakarian, P. Hobza and M. Vedadi, ACS Infect. Dis., 2021, 7, 2214–2220 CrossRef CAS PubMed.
- M. Štefek, D. Chalupská, K. Chalupský, M. Zgarbová, A. Dvořáková, P. Krafčíková, A. S. M. Li, M. Šála, M. Dejmek and T. Otava, ACS Omega, 2023, 8, 27410–27418 CrossRef PubMed.
- I. Singh, F. Li, E. A. Fink, I. Chau, A. Li, A. Rodriguez-Hernández, I. Glenn, F. J. Zapatero-Belinchón, M. L. Rodriguez and K. Devkota, J. Med. Chem., 2023, 66, 7785–7803 CrossRef CAS PubMed.
- R. Amador, A. Delpal, B. Canard, J.-J. Vasseur, E. Decroly, F. Debart, G. Clavé and M. Smietana, Org. Biomol. Chem., 2022, 20, 7582–7586 RSC.
- O. Bobileva, R. Bobrovs, I. Kanepe, L. Patetko, G. Kalnins, M. Sisovs, A. L. Bula, S. Grinberga, M. R. S. BoroduŠķis and A. Ramata-Stunda, ACS Med. Chem. Lett., 2021, 12, 1102–1107 CrossRef CAS PubMed.
- H. Hassan, J. Chiavaralli, A. Hassan, L. Bedda, T. Krischuns, K.-Y. Chen, A. S. M. Li, A. Delpal, E. Decroly and M. Vedadi, RSC Med. Chem., 2023, 14, 507–519 RSC.
- M. Zmudzinski, W. Rut, K. Olech, J. Granda, M. Giurg, M. Burda-Grabowska, R. Kaleta, M. Zgarbova, R. Kasprzyk and L. Zhang, Sci. Rep., 2023, 13, 9161 CrossRef CAS PubMed.
- A. Nigam, M. F. D. Hurley, F. Li, E. Konkoľová, M. Klima, J. Trylčová, R. Pollice, S. S. Çinaroǧlu, R. Levin-Konigsberg, J. Handjaya, M. Schapira, I. Chau, S. Perveen, H.-L. Ng, H. Ü. Kaniskan, Y. Han, S. Singh, C. Gorgulla, A. Kundaje, J. Jin, V. A. Voelz, J. Weber, R. Nencka, E. Boura, M. Vedadi and A. Aspuru-Guzik, Digital Discovery, 2024, 3, 1327–1341 RSC.
- K. Devkota, M. Schapira, S. Perveen, A. Khalili Yazdi, F. Li, I. Chau, P. Ghiabi, T. Hajian, P. Loppnau and A. Bolotokova, SLAS Discovery, 2021, 26, 1200–1211 CrossRef CAS PubMed.
- J. Chen, Y. Zhou, X. Wei, X. Xu, Z. Qin, C. P. Ong, Z. W. Ye, D. Y. Jin, B. Boitrel, S. Yuan, J. F. Chan, H. Li and H. Sun, ACS Infect. Dis., 2024, 10, 858–869 CrossRef CAS PubMed.
- N. L. Inniss, J. Kozic, F. Li, M. Rosas-Lemus, G. Minasov, J. I. Rybáček, Y. Zhu, R. Pohl, L. Shuvalova and L. R. RulíŠek, ACS Infect. Dis., 2023, 9, 1918–1931 CrossRef CAS PubMed.
- F. Li, P. Ghiabi, T. Hajian, M. Klima, A. S. M. Li, A. K. Yazdi, I. Chau, P. Loppnau, M. Kutera and A. Seitova, Biochim. Biophys. Acta, Gen. Subj., 2023, 1867, 130319 CrossRef CAS PubMed.
- M. Zgarbová, T. Otava, J. Silhan, R. Nencka, J. Weber and E. Boura, Antiviral Res., 2023, 218, 105714 CrossRef PubMed.
- J. Silhan, M. Klima, T. Otava, P. Skvara, D. Chalupska, K. Chalupsky, J. Kozic, R. Nencka and E. Boura, Nat. Commun., 2023, 14, 2259 CrossRef CAS PubMed.
- P. Krafcikova, J. Silhan, R. Nencka and E. Boura, Nat. Commun., 2020, 11, 3717 CrossRef CAS PubMed.
- O. Bobileva, R. Bobrovs, E. E. Sirma, I. Kanepe, A. L. Bula, L. Patetko, A. Ramata-Stunda, S. Grinberga, A. Jirgensons and K. Jaudzems, Molecules, 2023, 28, 768 CrossRef CAS PubMed.
- R. Bobrovs, I. Kanepe, N. Narvaiss, L. Patetko, G. Kalnins, M. Sisovs, A. L. Bula, S. Grinberga, M. Boroduskis and A. Ramata-Stunda, Pharmaceuticals, 2021, 14, 1243 CrossRef CAS PubMed.
- E. Jung, R. Soto-Acosta, J. Xie, D. J. Wilson, C. D. Dreis, R. Majima, T. C. Edwards, R. J. Geraghty and L. Chen, ACS Med. Chem. Lett., 2022, 13, 1477–1484 CrossRef CAS PubMed.
- R. Ahmed-Belkacem, M. Hausdorff, A. Delpal, P. Sutto-Ortiz, A. M. Colmant, F. Touret, N. S. Ogando, E. J. Snijder, B. Canard and B. Coutard, J. Med. Chem., 2022, 65, 6231–6249 CrossRef CAS PubMed.
- R. Ahmed-Belkacem, P. Sutto-Ortiz, M. Guiraud, B. Canard, J.-J. Vasseur, E. Decroly and F. Debart, Eur. J. Med. Chem., 2020, 201, 112557 CrossRef CAS PubMed.
- R. Ahmed-Belkacem, J. Troussier, A. Delpal, B. Canard, J.-J. Vasseur, E. Decroly and F. Debart, RSC Med. Chem., 2024, 15, 839–847 RSC.
- M. Hausdorff, A. Delpal, S. Barelier, L. Nicollet, B. Canard, F. Touret, A. Colmant, B. Coutard, J.-J. Vasseur and E. Decroly, Eur. J. Med. Chem., 2023, 256, 115474 CrossRef CAS PubMed.
- R. Shah, A. Strom, A. Zhou, K. M. Maize, B. C. Finzel and C. R. Wagner, ACS Med. Chem. Lett., 2016, 7, 780–784 CrossRef CAS PubMed.
- J. B. Epp and T. S. Widlanski, J. Org. Chem., 1999, 64, 293–295 CrossRef CAS PubMed.
- S. De Ornellas, J. M. Slattery, R. M. Edkins, A. Beeby, C. G. Baumann and I. J. Fairlamb, Org. Biomol. Chem., 2015, 13, 68–72 RSC.
- Y. Luan, L. L. Blazer, H. Hu, T. Hajian, J. Zhang, H. Wu, S. Houliston, C. H. Arrowsmith, M. Vedadi and Y. G. Zheng, Org. Biomol. Chem., 2016, 14, 631–638 RSC.
- B. Yu, A.-H. Liu, L.-N. He, B. Li, Z.-F. Diao and Y.-N. Li, Green Chem., 2012, 14, 957–962 RSC.
- M. J. Ensinger and B. Moss, J. Biol. Chem., 1976, 251, 5283–5291 CrossRef CAS PubMed.
- A. Czarna, J. Plewka, L. Kresik, A. Matsuda, A. Karim, C. Robinson, S. O'Byrne, F. Cunningham, I. Georgiou and P. Wilk, Structure, 2022, 30, 1050–1054 CrossRef CAS PubMed e1052.
- R. A. Friesner, J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley and J. K. Perry, J. Med. Chem., 2004, 47, 1739–1749 CrossRef CAS PubMed.
- N. S. Ogando, P. El Kazzi, J. C. Zevenhoven-Dobbe, B. W. Bontes, A. Decombe, C. C. Posthuma, V. Thiel, B. Canard, F. Ferron and E. Decroly, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2108709118 CrossRef CAS PubMed.
- F. Fratev, T. Steinbrecher and S. Ó. Jónsdóttir, ACS Omega, 2018, 3, 4357–4371 CrossRef CAS PubMed.
- M. Ashram, A. Y. Habashneh, S. Bardaweel and M. O. Taha, Med. Chem. Res., 2023, 32, 271–287 CrossRef CAS.
- Y. Ma, L. Wu, N. Shaw, Y. Gao, J. Wang, Y. Sun, Z. Lou, L. Yan, R. Zhang and Z. Rao, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 9436–9441 CrossRef CAS PubMed.
- P. Artursson and J. Karlsson, Biochem. Biophys. Res. Commun., 1991, 175, 880–885 CrossRef CAS PubMed.
- A. R. Hilgers, R. A. Conradi and P. S. Burton, Pharm. Res., 1990, 7, 902–910 CrossRef CAS PubMed.
- M. Yazdanian, K. Briggs, C. Jankovsky and A. Hawi, Pharm. Res., 2004, 21, 293–299 CrossRef CAS PubMed.
- N. W. Roehm, G. H. Rodgers, S. M. Hatfield and A. L. Glasebrook, J. Immunol. Methods, 1991, 142, 257–265 CrossRef CAS PubMed.
- J. M. Emeny and M. J. Morgan, J. Gen. Virol., 1979, 43, 247–252 CrossRef CAS PubMed.
- L. Schrödinger and W. DeLano, URL https://www.pymol.org/pymol, 2022.
Footnotes |
| † This article is dedicated to honoring and celebrating the retirement of Professor Christian Leumann. |
| ‡ Electronic supplementary information (ESI) available. CCDC 2360789–2360791. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4md00422a |
| This journal is © The Royal Society of Chemistry 2024 |