Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecular design of protein-based materials – state of the art, opportunities and challenges at the interface between materials engineering and synthetic biology
Ebony
Shire
 ab,
André A. B.
Coimbra
ab,
André A. B.
Coimbra
 c,
Carlos
Barba Ostria
c,
Carlos
Barba Ostria
 de,
Leonardo
Rios-Solis
de,
Leonardo
Rios-Solis
 *c and
Diego
López Barreiro
*c and
Diego
López Barreiro
 *ab
*ab
aManufacturing Futures Lab, Department of Chemical Engineering, University College London, London, WC1E 7JE, UK. E-mail: d.lopezbarreiro@ucl.ac.uk
bCentre for Nature-Inspired Engineering, Department of Chemical Engineering, University College London, London, WC1E 7JE, UK
cDepartment of Biochemical Engineering, Bernard Katz Building, University College London, London, WC1E 6BT, UK. E-mail: leo.rios@ucl.ac.uk
dEscuela de Medicina, Colegio de Ciencias de la Salud Quito, Universidad San Francisco de Quito USFQ, Quito 170901, Ecuador
eInstituto de Microbiología, Universidad San Francisco de Quito USFQ, Quito 170901, Ecuador
First published on 19th September 2024
Abstract
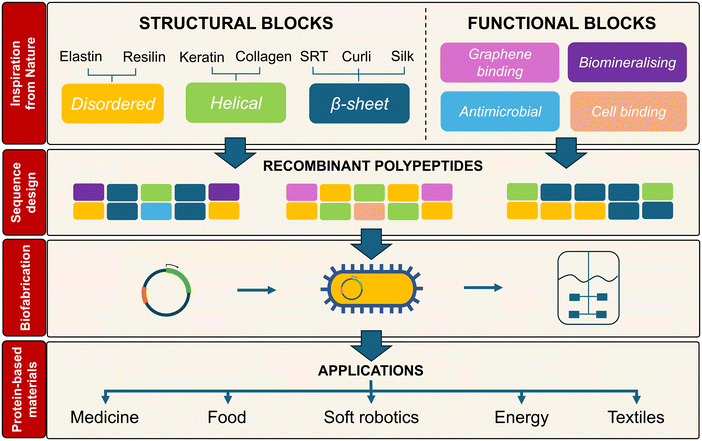
Structural proteins like silk, squid ring teeth, elastin, collagen, or resilin, among others, are inspiring the development of new sustainable biopolymeric materials for applications including healthcare, food, soft robotics, or textiles. Furthermore, advances in the fields of soft materials and synthetic biology have a joint great potential to guide the design of novel structural proteins, despite both fields progressing mostly in a separate fashion so far. Using recombinant DNA technologies and microbial fermentations, we can design new structural proteins with monomer-level sequence control and a dispersity of ca. 1.0, based on permutations of tandem repeats derived from natural structural proteins. However, the molecular design of recombinant and repetitive structural proteins is a nontrivial task that is generally approached using low-throughput trial-and-error experimentation. Here, we review recent progress in this area, in terms of structure–function relationships and DNA synthesis technologies. We also discuss experimental and computational advances towards the establishment of rapid prototyping pipelines for this family of biopolymers. Finally, we highlight future challenges to make protein-based materials a commercially viable alternative to current fossil-based polymers.
Design, System, ApplicationStructural proteins like elastin, resilin, collagen, or silk self-assemble into materials with a wide range of mechanical, structural, and dynamic properties. Inspired by this, scientists are increasingly researching structural proteins as sustainable biopolymers to manufacture materials with applications in healthcare, textiles or adhesives, to name a few. Thanks to recombinant DNA technology and synthetic biology developments, we can now generate with monomer-level precision new recombinant structural proteins that are inspired by nature, but that do not exist in nature. This allows us to create new multifunctional biopolymers that bring together dissimilar properties from natural proteins (e.g., the strength of silk, the stimuli-responsiveness of elastin, the stiffness of collagen) into a single new protein biopolymer. For these new recombinant proteins, it is hard to predict how the number and location of ordered/disordered, hydrophobic/hydrophilic, charged/uncharged, or structural/functional building blocks along their sequence affect the properties of the materials derived from them. Luckily, computational tools, such as molecular dynamics (MD) can help in unveiling sequence–property relationships, and thus accelerate the development of new recombinant proteins. |
1. Introduction
Many of our everyday materials are made of polymers derived from petroleum, using toxic solvents and/or harsh processing conditions via energy-intensive processes. Over 390 million metric tons of polymers are produced every year,1 accounting for 4–6% of the annual fossil oil use.2,3 Fossil-based polymers also generate a vast amount of waste that is at the core of an environmental plastic pollution disaster, affecting virtually all terrestrial and aquatic ecosystems.4 Additionally, the growing market for polymers (with a sales turnover of over £25 billion in the UK alone in 2022 (ref. 5)) challenges our ability to meet the demands for polymeric materials while minimising waste production. This is due to the persistence of plastic pollution in nature, with degradation times exceeding centuries for some materials. Therefore, there is an urgent unmet need for new sustainable polymers that fulfil the current role of plastics while enabling a bio-based and circular economy.To that end, nature is a great source of inspiration. Many natural proteins have a structural role that provides scaffolding or protection for the cells or organisms that produce those proteins. Structural proteins such as elastin, resilin, collagen, or silk can self-assemble into materials such as silkworm cocoons, the extracellular matrix, nails, squid ring teeth, or the adhesive threads of mussels.6–9 These materials span a wide range of mechanical and structural properties (e.g., from soft to stiff, from porous to densely packed, from dynamic to static) while being based on amide bonds, much like fossil-derived polymers such as polyurethanes or nylon.10 Inspired by this, scientists are increasingly researching structural proteins as sustainable biopolymers to manufacture materials with applications in food,11 medicine,12 adhesives,13 energy,14 textiles15 or separation processes,16 to name a few. Additional benefits of structural proteins include sustainability, lightweight, ease of processability, degradability, stimuli responsiveness and tuneable structural/mechanical properties.17 These features are determined by nano- and mesoscale motifs (e.g., β-sheets, helices, nanofibrils)18 that are encoded as short building blocks, also known as tandem repeats, in the amino acid sequence of those proteins.
Structural proteins are normally harvested from animal sources (e.g., silkworm cocoons, animal tissue), but these suffer from low yields, batch-to-batch variability, presence of contaminants, and cultural or religious concerns that limit their commercial use.18 Fortunately, advances in synthetic biology and bioprocess engineering allow us to overcome these issues and biofabricate new non-animal-derived structural proteins.19 Developments such as new DNA assembly techniques, new production hosts, or data-driven bioprocess optimisation are increasingly facilitating the biofabrication of new, high MW recombinant structural proteins with monomer-level sequence control and a dispersity of ca. 1.0.18 As a consequence, the yields of recombinant structural proteins (typically a few mg L−1) have attained productivities above 10 g L−1,20 bringing industrial production of recombinant structural proteins closer to economic feasibility. Furthermore, recombinant DNA technology allows us to explore new structural proteins beyond those selected for by evolution – it is now possible to design entirely new structural proteins with properties inspired by natural ones (e.g., the strength of silk, the stimuli-responsiveness of elastin, the stiffness of collagen), but that do not exist in Nature (Fig. 1). Nonetheless, this makes the molecular design space so vast as to become almost intractable via experimentation alone. Additionally, engineering proteins so that they self-assemble into materials with target mechanical (e.g., Young's modulus, toughness) or structural (e.g., mesh size, fibrillar networks) properties is a nontrivial task that is generally approached using low-throughput trial-and-error experimentation. Therefore, the design and engineering of protein-based materials would benefit from incorporating new computational techniques to accelerate the pace of development.
In this review, we aim to bring together two fields, synthetic biology and soft materials, that have mostly evolved independently. We first review the molecular design stage – the various protein building blocks available, their molecular-scale features, and how computational models aid in predicting in silico their behaviour. We then discuss synthetic biology aspects including DNA assembly methods, microbial hosts available, and the emerging field of engineered living materials (ELMs), in which the boundaries between materials and cells become blurred. Lastly, we provide an overview of the commercialisation landscape for recombinant protein biopolymers with materials applications. At the interface between biotechnology, bioprocess engineering, materials science, and computational modelling, this review aims to provide a comprehensive overview of the most recent developments in the field of microbially-produced protein-based materials.
2. Materials manufacturing from recombinant proteins
Many of the grand challenges that our society is facing, including healthcare, food and clean water supply, overuse of fossil fuels, or climate change, are intimately related to the materials that we chose to address those challenges. Fortunately, advances in engineering biology are increasingly enabling the design and engineering of new biopolymeric materials produced by microorganisms, using renewable feedstocks and sustainable biomanufacturing approaches. Recombinant synthesis of proteins is a good example of that. The genetic basis of sequence and length control in proteins, together with the manufacturing conditions applied (e.g., temperature, shear, solvent), allow us to control the self-assembly of structural protein biopolymers into macroscopic materials with long-range order and bespoke mechanical, structural, or biological properties that are hard to replicate in fossil-derived materials. In the following subsections, we will review common building blocks inspired by natural structural proteins, how they influence the properties of de novo recombinant structural proteins, and how computational tools can accelerate the prototyping of new materials.2.1. Nature-inspired motifs
The sequence of many natural structural proteins contains building blocks in the form of repetitive short peptides called minimal consensus repeats or tandem repeats. These repeats are usually represented by their constituent amino acids, which in this review will be noted using the single-letter code. In this section, we will review the main building blocks found in natural structural proteins, and how they determine the ability of proteins to adopt 3D conformations (i.e., helices, β-sheets, β-turns, coiled-coils, or disordered regions) (Table 1) or to display a dynamic behaviour, such as stimuli responsiveness. Beyond structural roles, many proteins display other functionalities, including electrical conductivity in the pili of some bacteria or the biomineralized skeletons of diatoms,21,22 that have promising applications in several engineering areas. Thus, by merging structural and functional properties of natural proteins into de novo recombinant proteins, we can engineer new protein-based materials with properties guided from the molecular scale up. However, we are still far from fully understanding how the macroscale properties of protein-based materials are linked to design features at the molecular scale (e.g., content in ordered/disordered, hydrophobic/hydrophilic, charged/uncharged, or structural/functional building blocks in the protein sequence) or the manufacturing routes used (i.e., solvent-based approaches, 3D printing, thermal moulding) – the so-called sequence–processing–property relationships. This explains why, to date, new structural recombinant proteins are mainly designed through low-throughput experimental trial-and-error permutations of a few building blocks.| Protein type | Structural block | Tandem repeat |
|---|---|---|
| Helical coiled coils | Collagen-like | GXY |
| Intrinsically disordered proteins | Elastin-like | VPGXG |
| Resilin-like | GGRPSDSYGAPGGGN or GGRPSSSYGAPGQGN | |
| β-Sheet-rich proteins | Curli | S(X)5QXGXGNXA(X)3Q |
| Silk-like (silkworm) | GAGAGS | |
| Silk-like (spider) | GAGAAAAAGGAGTS | |
| polyA | ||
| Squid-ring-teeth-like | PAAASVSTVHHP (hydrophobic) | |
| YGYGGLYGGLYGGLGY (hydrophilic) |
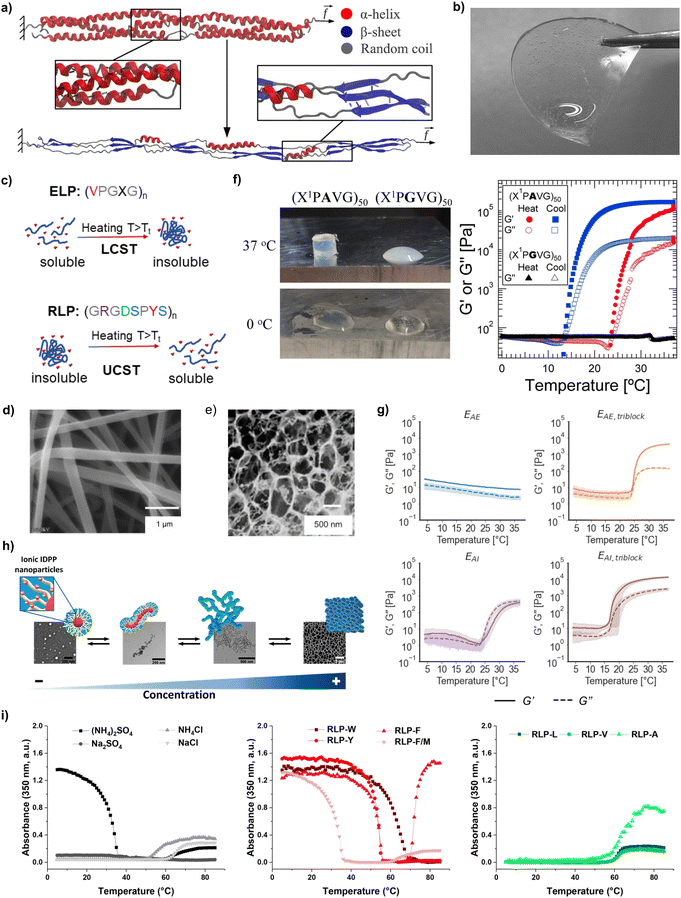 | ||
| Fig. 2 (a) Helical-to-β-sheet transition in a coiled-coil protein (reproduced and adapted with permission from ref. 26. Copyright 2012, American Chemical Society); (b) hydrogel produced from recombinant collagen-like polypeptides (reproduced and adapted with permission from ref. 28. Copyright 2021, American Chemical Society); (c) thermoreponsiveness of recombinant polypeptides with LCST behaviour (e.g., ELPs) and UCST behaviour (e.g., RLPs) (reproduced and adapted with permission from ref. 33. Copyright 2023, American Chemical Society); (d) fibres produced via electrospinning of aqueous solutions of SELPs (reproduced and adapted with permission from ref. 34. Copyright 2010, American Chemical Society); (e) porous films from BAB copolymeric ELPs after hydration in PBS (reproduced and adapted with permission from ref. 35. Copyright 2005, American Chemical Society); (f) photograph and temperature-dependent rheology (ω = 100 rad s−1, γo = 1%) of 20% (w/w) solutions ELPs containing plastic blocks (reproduced and adapted with permission from ref. 36. Copyright 2015, American Chemical Society); (g) rheological characterisation (storage modulus G′ and loss modulus G′′) as a function of triblock or alternate arrangements for neutral and positively charged ELP solutions in Milli-Q water (15 wt%) during a temperature sweep between 4 and 37 °C (reproduced and adapted with permission from ref. 37. Copyright 2022, American Chemical Society); (h) supramolecular assembly of ionic ELPs above Tt as a function of concentration (reproduced and adapted with permission from ref. 38. Copyright 2020, American Chemical Society); (i) UV-vis turbidity measurements of RLPs with different solvents and amino acid substitutions (reproduced and adapted with permission from ref. 39. Copyright 2021, American Chemical Society). | ||
One notable helical structure in Nature is collagen, the most abundant protein in the human body.40 Collagen self-assembles into a right-handed triple helix, and its tandem repeat is GXY. In mammalian collagen, proline (P) and 4-hydroxyproline (O) are the most common amino acids in the X and Y positions,41 respectively. 4-Hydroxyproline is a post-translational modification (PTM) of proline, thus requiring the additional co-expression of prolyl hydroxylases that are absent in bacterial cells.42 Thus, many researchers have focused on a simpler variant of collagen, namely unhydroxylated bacterial collagen-like polypeptides (CLPs) consisting of GPX tandem repeats28 (Fig. 2b). The extracellular secretion of CLPs has been demonstrated, which could lead to simple and more cost-effective purification and scale-up.43 The lack of O residues in bacterial CLPs does not impeded their self-assembly into triple helices,44 but comes at the expense of a reduced mechanical stability compared to hydroxylated collagen due to the formation of fewer hydrogen bonds between their constituent strands.45 Much like human collagen in bones, bacterial CLPs also showed the ability to biomineralise inorganic materials such as hydroxyapatite crystals, silver nanoparticles, and silica nanoparticles.46
Another common type of helix in Nature are α-helices in keratin.47 The helical structure of keratin is held together by hydrogen bonds between N–H groups and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, while intermolecular disulfide bonds render it insoluble.48 The sequence of keratin contains polar and nonpolar residues in a heptad pattern denoted as abcdefg, where a and d are always nonpolar.49 Additionally, keratin is often encountered as a higher-order quaternary structure called a coiled-coil, in which α-helices bundle together to create a stable superhelix50 that is the main structural component of hair, nails, skin and wool.51 The successful expression of recombinant keratin-like proteins has been reported, and their self-assembly into fibrils was confirmed.52 When compared against coatings produced from keratin extracted from human hair, recombinant keratin-like proteins led to more uniform coatings, which displayed increased biological activity. This was attributed to the increased homogeneity and the lack of contaminants in recombinant keratins. Moreover, the self-assembly of coiled-coil fibrils into higher order structures, such as hydrogels, can be modulated by pH, ionic strength, temperature, or sequence composition.53 As an example, Dooling and Tirrell54 synthesised seven different recombinant proteins with coiled-coil domains that differed by a single amino acid mutation. This allowed them to self-assemble hydrogels with a range of relaxation behaviours, spanning 5 orders of magnitude (from 0.22 ± 0.13 to 1608 ± 135 s).
O groups, while intermolecular disulfide bonds render it insoluble.48 The sequence of keratin contains polar and nonpolar residues in a heptad pattern denoted as abcdefg, where a and d are always nonpolar.49 Additionally, keratin is often encountered as a higher-order quaternary structure called a coiled-coil, in which α-helices bundle together to create a stable superhelix50 that is the main structural component of hair, nails, skin and wool.51 The successful expression of recombinant keratin-like proteins has been reported, and their self-assembly into fibrils was confirmed.52 When compared against coatings produced from keratin extracted from human hair, recombinant keratin-like proteins led to more uniform coatings, which displayed increased biological activity. This was attributed to the increased homogeneity and the lack of contaminants in recombinant keratins. Moreover, the self-assembly of coiled-coil fibrils into higher order structures, such as hydrogels, can be modulated by pH, ionic strength, temperature, or sequence composition.53 As an example, Dooling and Tirrell54 synthesised seven different recombinant proteins with coiled-coil domains that differed by a single amino acid mutation. This allowed them to self-assemble hydrogels with a range of relaxation behaviours, spanning 5 orders of magnitude (from 0.22 ± 0.13 to 1608 ± 135 s).
ELP tandem repeats are formed by the pentapeptide VPGXG, where X can be any amino acid excluding proline.69 The X amino acid directly impacts the Tt of ELPs, which arises from hydrophobic interactions between ELP chains. Below Tt, ELPs adopt random coil conformations due to the formation of an ordered hydrophobic hydration shell around the hydrophobic residues. Above Tt, the water molecules lose their ordered structure and ELP chains can self-assemble via intermolecular hydrophobic interactions.70 The value of Tt depends on the hydrophobicity of the amino acid residue in the X position:71 hydrophilic residues (e.g., glutamic or aspartic acid) increase Tt, whereas hydrophobic residues (e.g., as alanine, tryptophan, or isoleucine) reduce it.71 For instance, ELPs with a 46% ionic weight fraction displayed a Tt of 32 °C, while ELPs with a 45% non-ionic weight fraction had a Tt of 26 °C.38 The X amino acid also impacts the viscoelasticity of ELP hydrogels: ELP containing VPGIG (hydrophobic) blocks had a storage modulus (G′) 4.6× larger, and loss modulus (G′′) 29.5× larger than when the block was VPGEG (hydrophilic).37 Another common elastin-inspired building block widely used contains an alanine in the third position, and different variations of this tandem repeat have been proposed, such as VPAVG or IPAVG. Incorporating these blocks into the sequence of ELPs led to the formation of stiffer hydrogels with hysteresis between solvation and desolvation.37 To that end, Glassman et al.36 showed that 20 wt% solutions of (XPGVG)50 (where X was either 20% valine, 80% isoleucine, or 60% valine, 40% isoleucine) formed a turbid liquid at 37 °C. On the contrary, (XPAVG)50 formed a stiff hydrogel under the same conditions (Fig. 2f).
Additional methods to modulate the mechanical properties of ELP materials have been identified, mainly in studies related to the synthesis of hydrogels. The arrangement of blocks is one example. Hydrophilic–hydrophobic AB diblock ELPs formed micelles,72 whereas BAB triblock arrangements formed a hydrogel network.73 Triblock hydrophobic arrangements with IPAVG blocks and isoleucine as guest amino acid formed hydrogel networks with a storage modulus (G′ = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 1100 Pa) one order of magnitude larger than alternate arrangements with the same amino acid content and molecular weight (MW) (G′ = 1300 ± 400 Pa)37 (Fig. 2g). Higher protein molar concentration also promoted the formation of stiffer hydrogels for ELPs (Fig. 2h). As an example, the storage modulus of ELP hydrogels was found to increase from 2.9 kPa for a concentration of 2.5 mM, up to 5.8 kPa for a concentration of 2.8 mM.38 Similarly, higher MW also led to stiffer networks: hydrogels prepared from ELPs with 120 tandem repeats had a 5-fold increase in modulus (above 0.5 MPa) compared to sequences with only 70 repeats. The same study also underscored the effect of processing conditions (e.g., the ionic strength of the solvent) on the mechanical properties of ELP materials: hydrogels prepared in sodium phosphate buffer (pH 7.6) had 2- to 7-fold increases in high frequency moduli than those prepared in water.36 Mixing different types of ELPs is another strategy to control the properties of the resulting materials. Ternary mixtures of ELPs formed coacervates with different degrees of miscibility – from homogeneous mixtures to core–shell structures – depending on the guest amino acid (V, A, V50%/A50%, or V80%/A20%), the number of tandem repeats (from 40 to 160), or the concentration (volume fraction Φ from 0.001 to 0.1).74 The assembly of coacervates from ELP mixtures exhibited also a pathway-dependent assembly, enabling the synthesis of multiple assemblies from the same starting compounds and providing an additional handle to control the properties of ELP-based materials.75
000 ± 1100 Pa) one order of magnitude larger than alternate arrangements with the same amino acid content and molecular weight (MW) (G′ = 1300 ± 400 Pa)37 (Fig. 2g). Higher protein molar concentration also promoted the formation of stiffer hydrogels for ELPs (Fig. 2h). As an example, the storage modulus of ELP hydrogels was found to increase from 2.9 kPa for a concentration of 2.5 mM, up to 5.8 kPa for a concentration of 2.8 mM.38 Similarly, higher MW also led to stiffer networks: hydrogels prepared from ELPs with 120 tandem repeats had a 5-fold increase in modulus (above 0.5 MPa) compared to sequences with only 70 repeats. The same study also underscored the effect of processing conditions (e.g., the ionic strength of the solvent) on the mechanical properties of ELP materials: hydrogels prepared in sodium phosphate buffer (pH 7.6) had 2- to 7-fold increases in high frequency moduli than those prepared in water.36 Mixing different types of ELPs is another strategy to control the properties of the resulting materials. Ternary mixtures of ELPs formed coacervates with different degrees of miscibility – from homogeneous mixtures to core–shell structures – depending on the guest amino acid (V, A, V50%/A50%, or V80%/A20%), the number of tandem repeats (from 40 to 160), or the concentration (volume fraction Φ from 0.001 to 0.1).74 The assembly of coacervates from ELP mixtures exhibited also a pathway-dependent assembly, enabling the synthesis of multiple assemblies from the same starting compounds and providing an additional handle to control the properties of ELP-based materials.75
The other aforementioned thermoresponsive protein, resilin, is one of the most resilient materials known,76 and has thus inspired the development of RLPs for the manufacture of materials with low stiffness, high extensibility and outstanding resilience and fatigue life. RLPs are typically derived from variations of the tandem repeat GGRPSDSYGAPGGGN,77 typically by substituting the central tyrosine by amino acids such as phenylalanine, methionine, tryptophane or leucine. Similarly to ELPs, RLPs have also a high content of glycine and proline that promotes disorder and resilience. But conversely to ELPs, RLP's tandem repeat contains more hydrophilic amino acids, which enables a broader diversity of interchain interaction modes than in ELPs, such as electrostatic, hydrophobic, π–π stacking and hydrogen bonding interactions. The Tt and the material properties for RLP materials can be adjusted by tuning the MW, concentration, ionic strength, or amino acid substitutions.78–80 Amino acid mutations in RLP blocks affected the gelation propensity of RLP solutions, with aromatic residues (tryptophan, tyrosine, phenylalanine) promoting π–π or cation–π interactions that facilitated gelation, something that did not happen when aliphatic (leucine, valine, alanine) residues were used39,61 (Fig. 2i). To that end, Dzuricky et al.80 demonstrated that the interaction between cationic arginines and aromatic amino acids were the main drivers of RLP coacervation. Additionally, the Tt of RLPs was observed to increase with increasing MW. RLPs with a MW of 9.5 kDa phase separated at ca. 28 °C, whereas RLPs with a MW of 15.8 kDa phase separated at ca. 60 °C.81 The same study also showed that the Tt was lower for dilute RLP solutions.
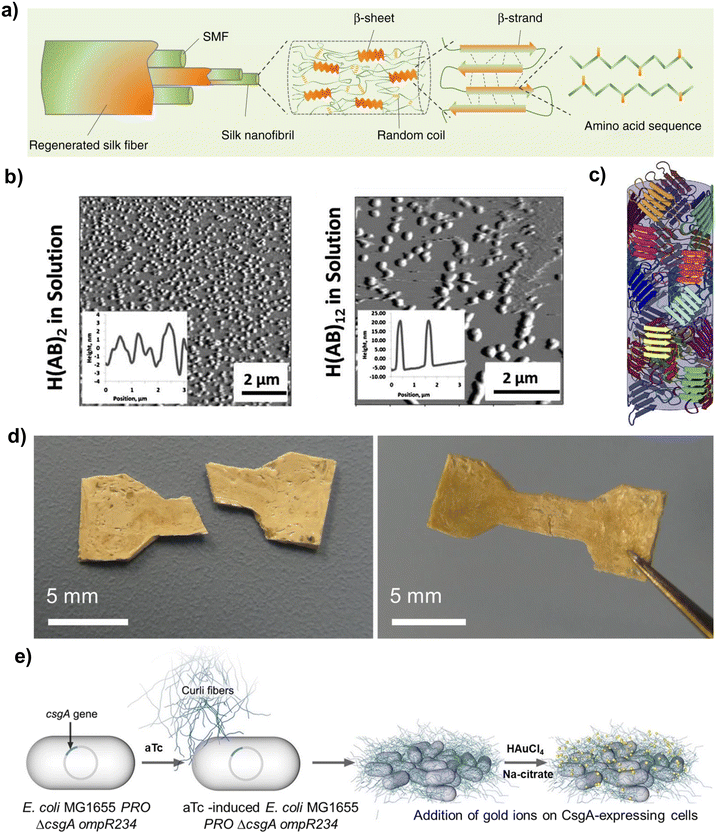 | ||
| Fig. 3 (a) Schematic representation of the alignment of β-sheet crystals in natural silk fibres (reproduced with permission from ref. 87. Copyright 2017, Nature Portfolio); (b) effect of MW on the self-assembly of silk-like polypeptides, with insets showing the spacing between self-assembled morphologies (reproduced and adapted with permission from ref. 88. Copyright 2014, Elsevier); (c) schematic representation of the isotropic distribution of β-sheet crystals in SRTs (reproduced with permission from ref. 89. Copyright 2017, Nature Portfolio); (d) self-healing properties of RLP-based materials (reproduced with permission from ref. 90. Copyright 2015, Nature Portfolio); (e) schematic representation of the secretion and self-assembly of recombinant curli fibrils that subsequently template the formation of gold nanoparticles (reproduced and adapted with permission from ref. 91. Copyright 2016, American Chemical Society). | ||
The archetypical tandem repeats used to promote the formation of β-sheets are the hexapeptide GAGAGS derived from the silkworm Bombyx mori,92 or polyalanine-rich polypeptides derived from the major ampullate spidroin of spider silk (such as GAGAAAAAGGAGTS).93,94 The degree of crystallinity (and hence, the strength) of silk-based materials directly correlates with the content in GAGAGS or polyalanine regions and with the manufacturing route used (e.g., casting, spinning). Furthermore, the balance between β-sheet-forming blocks and disordered blocks controls the microstructure of these materials: increasing the content of polyalanine blocks in silk-like polypeptides (SLPs) caused a segregation of hydrophobic blocks that changed the morphology of the self-assembled structures formed, from rod-like structures to micelles.95 The MW also influenced the self-assembly propensity, with higher MWs leading to the formation of larger aggregates and promoting the transition from disordered to lamellar structures88 (Fig. 3b).
SRT proteins (Fig. 3c) also contain a repetitive and alternating crystalline/disordered structure that can form antiparallel β-sheets with turns.90 The tandem repeats for SRT are typically formed by PAAASVSTVHHP (the β-sheet-forming block) and YGYGGLYGGLYGGLGY (the disordered block).96 The sequence within the β-sheet-forming domain alternates the position of H and A residues in neighbouring chains, leading to a compact structure, whereas the amorphous blocks show sequence repetition that may contribute to increased mechanical flexibility.97 SRTs have been proposed as a promising biopolymer for underwater adhesive applications, due to their high tensile (ca. 1.5 MPa) and shear strength (ca. 2.5 MPa) in wet conditions.83 The adhesive properties of SRTs were linked to their high content in β-sheets.98 Additionally, the stability of SRTs networks in wet environments was attributed to the high histidine content in their β-sheet-forming blocks,83 because of the versatility of the imidazole side chain in histidines to stabilise protein networks (e.g., via hydrogen bonding, metal coordination, or covalent crosslinking).
With respect to SRTs, Vural et al.99 demonstrated that the energy efficiency of SRT-based actuators with graphene oxide scaled inversely with the number of SRT tandem repeats. This was due to the ability of lower MWs to achieve higher curvatures, whereas SRTs with higher MWs were more restricted in their ability to bend due to the increased physical crosslinks and entanglements. SRTs were also used to develop films with tuneable thermal transport. Below <35% relative humidity, the thermal conductivity of SRT films was independent of the MW. In contrast, when the SRT films were hydrated, the thermal conductivity exhibited an almost linear dependency with the inverse of the number of tandem repeats in the SRT sequence. When analysing the mechanical properties of films, it was reported that an increasing MW did not alter the shear modulus of dry films. In turn, under hydrated conditions, SRTs with higher MW led to films with an increased shear modulus.100 SRTs have also been used to develop self-propelled microrobots,96 which could have applications in minimally invasive medical operations, removal of water contaminants, or targeted cargo delivery.
Curli proteins are another example of structural proteins rich in β-sheets. This protein class is extracellularly secreted and is the main component of bacterial biofilms from E. coli. Curli proteins self-assemble into very robust amyloid nanofibres with CsgA as the dominant protein and with a strength comparable to steel.101 The tandem repeat of curli proteins is S(X)5QXGXGNXA(X)3Q, and this is repeated five times to make up the amyloid core. S, Q and N residues align in stacks and hydrogen bonds stabilise the structure, which are reinforced by steric zipper interactions between side chain residues of neighbouring chains.18,102,103 The tendency of curli proteins to form amyloid fibres enabled the development of the biofilm-integrated nanofiber display (BIND) strategy. This strategy fused CsgA to other functional peptides (e.g., hydroxyapatite binding, gold binding) and used the secretory machinery of E. coli to secrete these fusion proteins and produce fibrillar meshes with the ability to i.e., template nanoparticle assembly, provide substrate adhesion, conduct electricity, or covalently immobilise enzymes.91,101
Recombinant proteins inspired by silk, SRT, or curli have been investigated for numerous applications, including tissue regeneration,94 engineering fibres,104 molecular motors96 or actuators.105 The non-covalent nature of β-sheet-rich networks allows them to exhibit fast self-healing kinetics after structural damage105–107 (Fig. 3d). It was also shown that this behaviour could be enhanced by applying localised heating to the damage site, due to the thermal mobility of proteins. For instance, damaged SRT-based pneumatic actuators exhibited 2–23 MPa strength after 1 s of healing under heat.105 In terms of microstructure, SRT-based materials exhibit an anisotropic packing of β-sheet crystals with a well-defined nanoscale size.89 In contrast, silk- and curli-like polypeptides tend to self-assemble into fibrillar structures. Additionally, it has been recently demonstrated that SLPs and SRT-based proteins can be thermally moulded from a melt into a solid.89,108 This technique is widely used for fossil-based polymers, and thus could facilitate the adoption of recombinant proteins for the manufacture of sustainable and high-performance bioplastics.
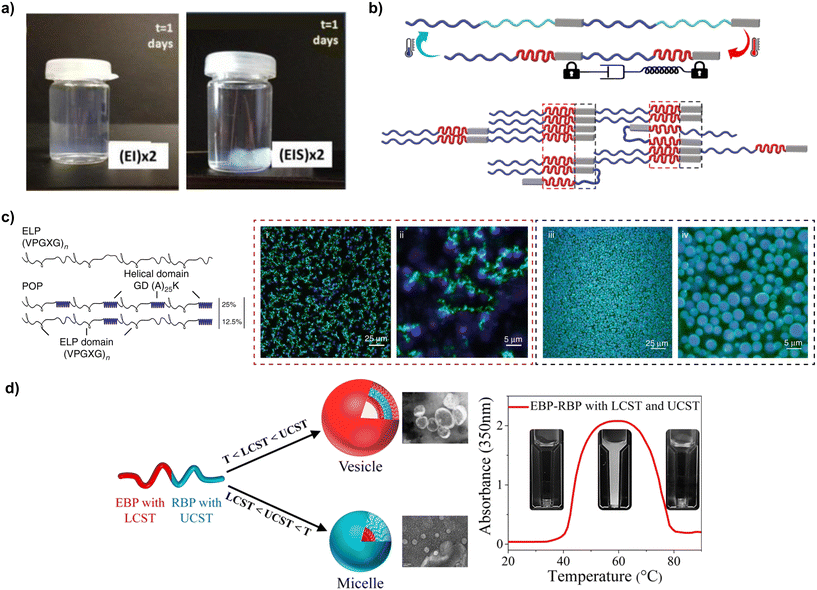 | ||
| Fig. 4 (a) Adding silk-like blocks to ELPs enhances the robustness and durability of hydrogels produced from those polypeptides (reproduced and adapted with permission from ref. 114. Copyright 2014, American Chemical Society); (b) representation of the elastin-like blocks (red regions) bring silk-like blocks in close proximity (grey regions) facilitating β-sheet formation (reproduced and adapted with permission from ref. 115. Copyright 2022, Elsevier); (c) schematic representation of the sequence and architecture for ELPs and ELP-POP fusions, and the different network architectures that they can form imaged using confocal microscopy (i–iv) (reproduced and adapted with permission from ref. 117. Copyright 2020, Nature Portfolio); (d) ELP–RLP fusions and their dual thermoresponsiveness (reproduced and adapted with permission from ref. 118. Copyright 2021, American Chemical Society). | ||
The properties of SELPs (and other fusion proteins) can be modulated using a variety of strategies. The viscoelastic properties are concentration dependent, with G′ and G′′ values of SELP increasing in a nonlinear fashion with the wt%.114 The ratio between blocks is another important parameter influencing gelation, with higher silk-to-elastin ratios increasing Tt.112,119 However, recent studies hint at the existence of a critical silk-to-elastin ratio above which the silk-like blocks begin to dominate the properties of the material, and the thermoresponsiveness of elastin blocks vanishes, making the gelation quasi-irreversible115 (Fig. 4b). Another relevant parameter for the self-assembly of SELPs is charge of the X amino acid in elastin-like blocks. Like in ELPs, SELPs with uncharged amino acids (e.g., phenylalanine or tyrosine) display a lower Tt than those with charged amino acids (e.g., glutamic acid or lysine).119 Charge also modulates the microstructure of SELP coacervates, promoting microphase separation.120 SRT, another β-sheet-forming polypeptide, was also fused to ELPs, thus forming SRT–ELP fusions in which SRT blocks enhance the mechanical properties of the resulting materials in a similar way to silk-like blocks in SELPs.121 To that end, 12mer, 24mer and 36mer SRT–ELP fusions were reported, ranging from 49.8 to 145.5 kDa. These fusion proteins were used to produce fibres, and the results indicated an enhancement of mechanical properties at higher MWs, with breaking strengths increasing from 303 ± 14 MPa (12mer) to 550 ± 20 MPa (36mer).
SLPs have also been fused to other natural structural proteins, such as mussel foot protein 5 (Mfp5), an IDP used for surface adhesion underwater. When Mfp5 was fused to the end termini of silk blocks, it increased the fibre tensile strength by up to 345% and toughness by up to 1970%.122 Golinska et al.123 fused SLPs with CLPs with the aim of merging two orthogonal self-assembly mechanisms: the fibrillation ability of SLPs and the tendency to form triple helices of CLPs. They showed that SLP–CLP fusions could be applied to synthesise self-healing and pH-responsiveness hydrogels with up to G′ = 1700 Pa (protein concentration of 2 wt%). In a subsequent study, Rombouts et al.27 demonstrated that the self-assembly of CLP trimers was thermoreversible, enabling the synthesis of hydrogels with switchable stiffness controlled by temperature (ca. 3 kPa at 40 °C and ca. 6 kPa at 20 °C).
ELPs have been fused to other types of blocks too. In a study by Roberts et al., ELP tandem repeats were fused with helical partially ordered polypeptides (POPs), forming porous viscoelastic networks (Fig. 4c). The Tt and microstructure of these networks were controlled by the ratio between ELP and POP regions. Increasing the percentage of POPs from 0–50% (whilst keeping the same ELP block sequence and MW) resulted in a 10 °C difference between solvation and desolvation.124 CLP is another helix-forming block that has been fused to ELPs. The tendency of CLP blocks to self-assemble into triple helices caused an increase of the concentration of ELPs at the edge of the helices formed that CLPs, which lowered their Tt.125 However, when the temperature increased beyond the melting temperature of CLPs, the crowding effect disappeared due to the disassembly of the CLP triple helix, leading to a re-dissolution of the fusion proteins.
Materials with dual thermoresponsiveness have also been developed by engineering RLP–ELP fusions. The resulting materials were fully miscible in water below LCST or above UCST but were immiscible between LCST and UCST118 (Fig. 4d). Different nanostructures (namely, disordered, cylindrical, and lamellar) were obtained, with their specific shape determined by temperature, RLP block length or protein concentration. The size of the nanostructures (lamellae, hexagonally packed cylinders, and disordered structures) increased as the RLP fraction increased, or as the concentration of the polymer decreased.126 Fusions of three different types of tandem repeats have also been reported. One example are RLP–ELP–CLP fusions, in which RLP and ELP blocks were used as elastomeric hydrophilic and hydrophobic blocks, respectively. The higher polarity of RLPs aided in the dissolution of RLP–ELP–CLP fusions, whereas the CLP region provided the material with the propensity towards self-assembling into fibrillar structures after incubation in water (0.5 mg mL−1) at 50 °C.127
2.2. Computer-aided protein design
The 3D conformation of recombinant structural proteins is tightly linked to the mechanical properties of the materials produced from them, but it is difficult to predict that conformation for de novo recombinant proteins. Computational modelling can aid in exploring in silico the vast design and conformational space for new recombinant protein sequences,128 which would reduce the number and cost of time-consuming design–build–test–learn cycles that are used to develop protein-based materials.129,130 Furthermore, the effect of manufacturing techniques (e.g., shear, temperature, ionic strength, solvent) can also be incorporated into simulations.131 There are several computational tools available to that end, depending on the desired level of resolution. At the most fundamental level are quantum mechanical methods, using e.g., density functional theory (DFT).132 DFT can aid in assessing molecular geometries, chemical bonding, bandgaps, or cohesive energies, but they are computationally expensive because they take into account the effects of electrons. Thus, they can only be used to investigate systems below ca. 5000 atoms for timescales in the ps range.133 Moving up in length- and timescales, the Rosetta macromolecular modelling tools134 enable the prediction of atomistic structures for protein sequences and have been applied to design nanoscale protein blocks with specific 2D or 3D assemblies.135The presence of highly mobile and disordered regions in many natural and recombinant structural proteins makes it also important to assess their dynamic behaviour. To that end, molecular dynamics (MD) simulations are a powerful methodology that can deliver structural, kinetic, and thermodynamic information for recombinant structural proteins. Fully atomistic MD simulations coalesce the influence of electrons with their nuclei and apply the Newtonian equations of motion to study the dynamics of systems of up to millions of atoms and for timescales in the microseconds range. Fully atomistic MD simulations have been successfully applied to many recombinant and natural structural proteins (Fig. 5a), including silk,136,137 elastin,138,139 collagen,140 resilin,141 and fusions thereof.112,130 These simulations use force fields that accurately calculate the forces and energies for these systems. Some examples of extensively validated force fields for biomolecular simulations include CHARMM,142 AMBER143 or OPLS.144 However, predicting the secondary structure for de novo recombinant proteins via fully atomistic MD simulations can be computationally costly, especially if solvent molecules are explicitly represented in the simulation. To overcome this issue, several advanced sampling techniques exist, among which replica exchange MD (REMD) simulations are a very common one145 that has been successfully applied study silk,146–148 elastin,149 collagen150 or silk–elastin112,151 protein-based materials. REMD can accelerate the prediction of the protein secondary structure by focusing on conformational sampling, but at the expense of losing dynamical information. At larger scales, coarse grained (CG) MD simulations can be applied152 (Fig. 5b). CG MD sacrifices atomistic resolution by lumping several atoms into one single bead, enabling the exploration of 10–100× larger length- and timescales than MD simulations.93,151–153
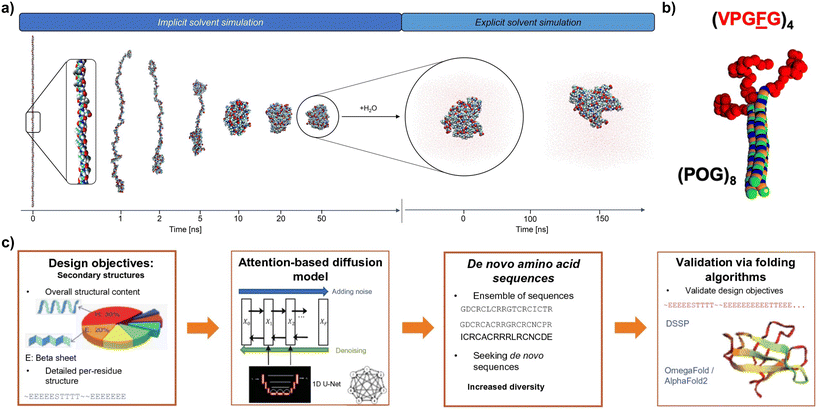 | ||
| Fig. 5 (a) Screenshots of the folding process (in implicit solvent) and dynamic run (in explicit solvent) of MD simulations for an ELP (reproduced and adapted with permission from ref. 37. Copyright 2022, American Chemical Society); (b) coarse grain model for ELP–CLP conjugates (ELPs in shown red) (reproduced and adapted with permission from ref. 152. Copyright 2019, American Chemical Society); (c) implementation of a workflow for the AI-assisted design de novo proteins with target mechanical properties using attention-based diffusion models (reproduced and adapted with permission from ref. 128. Copyright 2023, Springer). | ||
Finally, ML is emerging as a promising tool for high-throughput discovery of new protein-based materials (Fig. 5c). ML methods can be classified into supervised learning, unsupervised learning, reinforcement learning and semi-supervised learning.128 These methods have shown promising results in connecting protein sequences with their secondary structure (forward design), or in predicting amino acid sequences based on secondary structure design objectives (inverse design).148,154 ML methodologies are heavily dependent on the availability of reliable and annotated data, for which the establishment of comprehensive and accessible databases for protein-based materials is key.155
3. Synthetic biology for materials design and fabrication
3.1. DNA assembly methods for genes encoding protein biopolymers
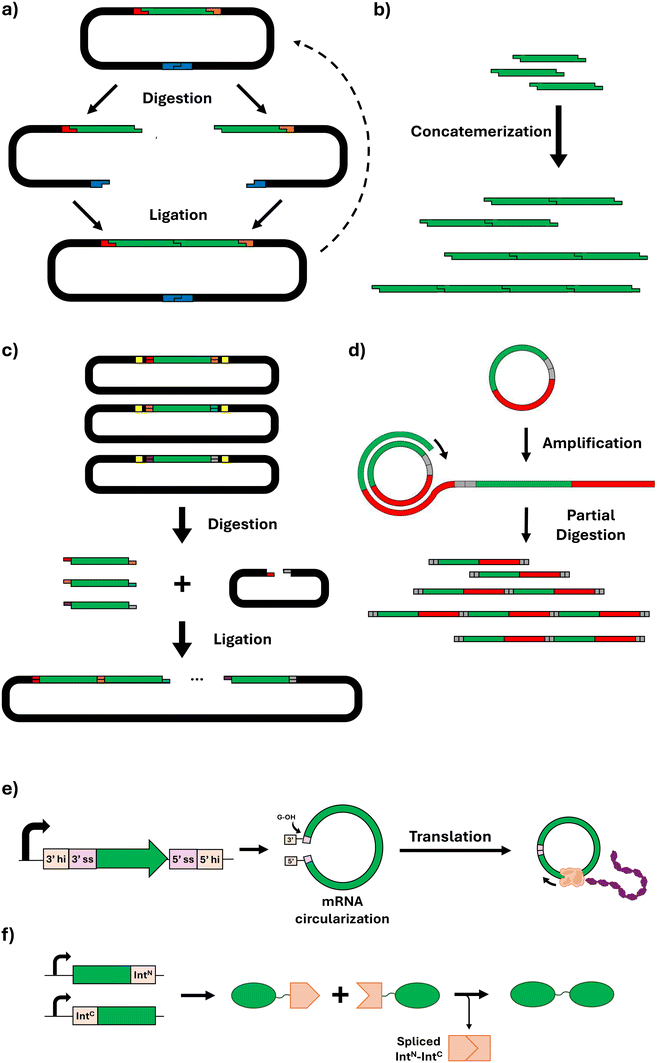
Essential to the biomanufacture of recombinant structural proteins is the strategic design of the DNA encoding for those proteins. The repetitive nature of many structural protein-based materials in nature presents certain challenges for their synthesis using microbial cells, including genetic instability due to increased recombination events (often with a high GC content);156 difficulties in proper protein expression and folding leading to misfolded proteins and the formation of inclusion bodies; and a significant metabolic burden on the host cells that can hinder cell growth and productivity. These features present significant challenges in assembling short DNA fragments into genes that faithfully encode the desired full-length protein biopolymers.To express structural proteins in microbial platforms, cloning is typically the first step, but genes encoding for repetitive proteins are unavoidably repetitive. This can make conventional PCR-based cloning methods fail, because non-specific binding can occur during primer annealing in repeated sequences. Other cloning strategies such as restriction-enzyme-based strategies can also be challenging, due to the presence of restriction sites in many repetitive sequences. Nonetheless, the rapid progression in synthetic biology's gene assembly methodologies has paved the way for the fabrication of multifunctional protein-based block copolymers.157 The concept of gene assembly allows for the consolidation of various functionalities into a singular protein entity, thereby enabling the stepwise creation of intricate protein substances which can be customised in their composition, quantity, and sequential structure of amino acid blocks. To that end, this section reviews the guiding principles behind several novel DNA assembly methods that have been reported to facilitate the construction of repetitive gene sequences. It also describes the diverse synthetic biology tools available for such gene construction, emphasising their advantages and disadvantages.
This method typically allows for reliable ligation of up to ten fragments simultaneously; beyond this number, the likelihood of successful assembly tends to decrease. For larger constructs, sequential rounds of the Golden Gate process may be employed, although this approach depends on the availability of distinct overhang sequences for each fragment.172 Alternatively, larger constructs can be generated by combining Golden Gate with other assembly methods/strategies. For instance, Tang and Chilkoti produced ELPs with 150 repeat units by using Golden Gate to join 30 repeat unit monomers generated by combinatorial codon scrambling.158 Additionally, due to its combinatorial and modular nature, Golden Gate assembly can also facilitate the production of fusion proteins.173,174
Inteins are protein segments that can excise themselves from a precursor protein and simultaneously ligate the remaining (extein) sequences with a peptide bond, without the need for external enzymes or energy sources.182 In this process known as protein splicing, the intein entity is absent from the ultimate protein sequence, thereby enabling a seamless peptide ligation.183 Split inteins (SIs), representing a variant where the intein is divided into two segments, have demonstrated significant utility in the assembly of very HMW proteins (Fig. 6f). This is exemplified in the construction of dragline spider silk proteins, such as major ampullate spidroin 1 (MaSp1). Through the application of SI ligation, two individually expressed segments comprising 96 repeats were successfully ligated, yielding a silk protein fusing 192 repeats with a resultant MW of 556 kDa.184 Beyond the synthesis of repetitive silk proteins, SI-mediated ligation has been employed in the biosynthesis of high-MW mussel foot proteins (Mfps) from Mytilus galloprovincialis.185 Additionally, further expanding the applications of SIs for protein-based materials, this approach was used to perform multiple consecutive ligation steps within microbial cells, achieving the production of protein biopolymers inspired by titin (a protein conferring muscle tissues with passive strength, damping capacity, and rapid mechanical recovery) with exceptionally high MWs of >5 MDa.186
3.2. Microbial host systems for protein biopolymer expression
Leveraging living organisms for material manufacturing takes advantage of their innate, evolved cellular mechanisms. The capability for organism engineering is tied to the knowledge of its metabolism and genome, with computational tools predicting genetic elements from genomic data. Optimal engineering frameworks require well-characterised genetic components and established genetic modification techniques. Consequently, foundational synthetic biology for materials has centred on well-understood unicellular organisms such as bacterial hosts Escherichia coli or Bacillus subtilis, yeast such as Saccharomyces cerevisiae, Komagataella phaffii, and recently also industrially relevant mammalian cell systems. Such organisms facilitate the swift development of genetic circuits, which must be carefully tailored to the host to prevent interference with cellular functions. Beyond unicellular hosts, there have also been advances in using complex organisms such as genetically modified plants, insects, and animals for synthesising protein biopolymers,164 though this is out of scope for this review. The following section primarily explores unicellular hosts used for protein synthesis with materials applications. Their advantages and disadvantages are summarised in Table 2.| Host | Advantages | Disadvantages |
|---|---|---|
| E. coli | • Genetic malleability | • Limited secretion ability |
| • Cost-effectiveness | • Limited PTMs for recombinant eukaryotic protein production | |
| • Rapid growth | • Premature stop codons and truncated proteins due to aminoacyl-tRNA shortages | |
| • Scalability | ||
| B. subtilis | • Well-characterised genetic background | • Limited PTMs for recombinant eukaryotic protein production |
| • Reliable genetic manipulation technologies | • Premature stop codons and truncated proteins due to aminoacyl-tRNA shortages | |
| • Superior secreting ability compared to E. coli | ||
| • Amenability to high-density fermentation | ||
| S. cerevisiae | • Very extensive and standardised synthetic biology tools; established genome engineering tools | • PTMs with high immunogenicity |
| • Can perform some eukaryotic PTMs | • Cannot perform very complex mammalian PTMs | |
| • Robustness under fermentation conditions | ||
| K. phaffii | • Standardised synthetic biology tools | • Requires methanol induction |
| • Superior secretion ability relative to S. cerevisiae | • Some PTMs differ from higher eukaryotes (e.g. N-glycosylations) | |
| • Glycosylation patterns similar to mammalian cells | ||
| • High protein titers | ||
| • Robustness under fermentation conditions | ||
| Mammalian cells | • Superior secretion ability | • Lower production levels |
| • Can perform complex and humanised PTMs | • High-cost fermentation | |
After synthesising protein biopolymers, their distribution within the host organism requires additional considerations. These newly formed biopolymers may be found in different cellular locations such as the cytosol, on the cell surface, or within the extracellular space. Recent observations suggest certain proteins, including SLPs or RLPs can undergo phase separation, leading to the formation of condensates specifically at the bacterial cell poles.192 Furthermore, it is possible to leverage the ability of E. coli to secrete curli proteins, thereby transforming the bacteria into continuous “living” factories.193,194
Meanwhile, the Gram-positive bacterium B. subtilis has also been widely used in industrial protein production, given its well characterised genetic background, its amenability to high-density fermentation, and the existence of reliable genetic manipulation technologies.195B. subtilis is particularly relevant as a chassis due to its high protein-secreting ability.196 Indeed, Xie et al. have engineered B. subtilis capable of secreting silk proteins TasA, which spontaneously assemble into fibres on the cell surface.197 In a different study, TasA fibres were functionalized with a mussel foot protein and an engineered hydrophobin-like protein, thus creating a material with underwater adhesion properties.198
As previously mentioned, the expression of repetitive and HMW recombinant structural proteins in microbial hosts can bring along some issues. Specifically, aminoacyl-tRNA shortages, for example, glycyl-tRNA during the production of glycine-rich proteins (such as collagen, elastin, silk), can lead to premature stop codons and truncated proteins.199 Boosting the levels of these tRNAs in host cells can therefore significantly improve protein biopolymer production. As an example, enhancing the internal pool of tRNAGly through the overexpression of glyVXY genes led to a 10- to 35-fold rise in yield for the silk proteins.200 Similarly, increasing both the tRNAAla and tRNAGly pools and supplementing with alanine and glycine elevated the production of MaSp2 proteins, although an excess of these amino acids impeded cell growth and protein synthesis.201,202
The production of recombinant eukaryotic proteins in microbial hosts present the additional challenge of replicating PTMs in microbial hosts. Nevertheless, researchers have been able to engineer an E. coli strain capable of producing a silica-mineralising silaffin R5 peptide, native to diatoms, complete with its necessary PTMs.203 Their method involved evaluating various PTM enzymes from diverse species to determine their effectiveness on the R5 peptide. At the same time, it should be noted that other researchers have been able to leverage the R5 peptide for silica biomineralisation without the need of PTMs, which poses the question of whether those are needed for the R5 function.94,204
One interesting aspect of yeasts as expression hosts is their secretion capacity to the culture medium. To that end, the methylotrophic yeast K. phaffii (Pichia pastoris) has emerged as an effective platform, due to its proficient secretion system210 that can drastically reduce the number of purification steps needed to isolate the target proteins. K. phaffii is also favoured for its ability to grow to high cell densities in cost-effective media. Consequently, K. phaffii has been effectively used to biosynthesise and secrete various protein biopolymers, such as spider silk proteins. To that end, Jansson et al.211 reported a comparable total yield of SLPs in K. phaffii (extracellular and intracellular) similar to that of E. coli. However, amount of secreted protein was about one third of the total protein. K. phaffii was also used to produce recombinant human collagen, reaching 2.33 g L−1 in a 5 L bioreactor.212
3.3. Engineered living materials
The desirable properties observed in many natural materials (e.g., self-assembled and hierarchically organised structures, dynamic responsiveness, autonomous patterning, or self-repair) are usually achieved thanks to the synergistic action of biopolymeric scaffolds and living cells embedded within them.216 Inspired by this, the emergent field of engineered living materials (ELMs) aims at hybridising genetically engineered cells and polymeric scaffolds to manufacture synthetic materials that replicate the sophistication of natural materials.217 ELMs blur the boundaries between cells and materials, and have been mostly researched for healthcare applications.218 Nonetheless, they are expected to impact other fields as well, such as biosensing,219 photosynthetic devices,220 agriculture,221 or patterned materials.222 Several technological hurdles need to be overcome before ELMs can become a commercially viable option. These hurdles include improving the lifespan and mechanical properties of ELMs, which are currently relatively weak compared to synthetic engineering materials,223,224 or accurately understanding the growth and spatial distribution of cellular populations within ELM scaffolds.225Current ELMs are generally made by combining fossil-based polymers (polycaprolactone, polyethylene oxide, polyvinyl alcohol, Pluronic F127, polydimethyl siloxane) or natural biopolymers (e.g., agarose, bacterial cellulose, alginate, gelatin, hyaluronic acid) with genetically engineered cells.218,226 Several microbial hosts have also been used227 including E. coli due to its aforementioned desirable characteristics; B. subtilis due to its protein secretion possibilities; K. rhaeticus because of its ability to secrete bacterial cellulose; the yeast S. cerevisiae; microalgae (e.g., Chlorella reinhardtii, Platymonas sp.); or the fungi mycelium,228 due to its ability to form bulk composite materials. Furthermore, mixed populations (e.g., K. rhaeticus and S. cerevisiae) have also been used to assemble ELMs.222 However, the ultimate goal of the ELMs field is to “grow” materials from simple growth media and an inoculum of engineered cells.157 To that end, recombinant structural proteins are promising scaffolding materials for that purpose: they offer a biocompatible environment for cells, and cells can be engineered to secrete those proteins as they grow. Some proof-of-principle studies have already been reported, using fusion recombinant proteins inspired in curli proteins,193,229,230 elastin,67,231 or mussel foot proteins232 that were able to self-assemble into soft materials. Additionally, the fusion of recombinant structural proteins with functional blocks has shown to confer scaffolds with properties beyond their structural role, such as the ability to bind to other biopolymers or to biomineralise.222,233 Here too, predictive computational tools for sequence–processing–properties in new recombinant structural proteins, as well as scaffold–cell interactions234 are expected to significantly contribute to the development of the ELMs field.
4. Scale-up and commercialisation
While the production of enzymes and biopharmaceuticals at industrial scale has been demonstrated already for decades, the use of microbial cell factories to produce materials is still lagging. Nonetheless, engineering biology to biofabricate materials has been recognised as a key player in sustainability and circular economy ambitions – some estimates indicate that 60% of the physical inputs to the global economy could be derived from biological systems.235 This has led to the creation of multiple companies active in the field of biopolymeric materials. The main efforts towards commercialisation have focused on β-sheet-rich polypeptides, including silk-like polypeptides (e.g., AMSilk,236 Bolt Threads,237 Spiber238) or squid-ring proteins SRT polypeptides (Tandem Repeat239). The company Modern Meadow has also reported the production of recombinant collagen using K. phaffii.240 This was facilitated by techniques such as codon optimisation to improve gene expression and the integration of these optimised genes into the yeast genome. Furthermore, the concurrent expression of specific hydroxylase enzymes supported necessary PTMs, increasing the production scale of recombinant collagen.These companies have demonstrated pilot-scale fermentation and purification, using mostly bacterial (mainly E. coli) and yeast (K. phaffii) hosts.9,239–241 The main areas of activity of these companies are the biomedical (tissue engineering, drug deliver) or personal care (e.g., cosmetics) sectors, but there are application examples also for leather alternatives for products like furniture, bags, and shoes. The main barrier to commercialisation for these materials remains cost.9 To that end, several strategies could facilitate the downstream purification processes and hence reduce costs. These include extracellular secretion of the target proteins,19 or incorporating thermoresponsive ELP or RLP blocks that allow for purification via thermal cycling.187
5. Outlook
Nature utilises structural proteins to manufacture complex materials with properties that we cannot currently recreate in synthetic materials, such as a self-assembled and hierarchically organised structure, dynamic responsiveness, autonomous patterning, and self-healing. Such materials span a wide range of mechanical and structural properties – from soft to stiff, from porous to densely packed, from dynamic to static. Inspired by this, scientists are increasingly using structural proteins to manufacture materials with applications including food, healthcare, adhesives, energy, or textiles. Furthermore, developments in gene synthesis have enabled us to explore beyond the protein sequences that evolution has selected for and create materials from entirely new protein sequences. However, despite the ample opportunity for protein-based materials, there are several challenges that must be addressed to harness their full potential.On the materials side, the design freedom for recombinant protein sequences makes it impossible to explore their design space via experimentation alone. Furthermore, we are still far from understanding how protein sequences and manufacturing conditions regulate the self-assembly of structural proteins into macroscopic materials, as well as the mechanical and structural properties of those materials. To that end, progress in computational simulation techniques and increased computational power (see section 2.2), as well as novel high-throughput characterisation techniques,242,243 will accelerate the exploration of the design space and contribute to uncover sequence–manufacturing–property relationships. High-throughput techniques will also enable the application of machine learning in the field of protein-based materials.244 However, a significant challenge will be the regulatory approval for these materials, especially for biomedical applications or materials containing living (genetically modified) cells, such as ELMs.
On the bioprocessing side, large scale fermentation has been successfully demonstrated for a few protein biopolymers (section 4), but there is still uncertainty about the economic feasibility of these biopolymers, especially for non-biomedical applications. One of the primary hurdles is the high cost of gene synthesis, particularly for large, repetitive protein sequences that are common in materials applications. Advances in synthetic biology and automation will be essential to making these materials economically competitive.245 Another challenge is the optimisation of microbial strains, which often struggle to efficiently produce HMW and complex proteins due to the metabolic burden they impose. Strain engineering efforts are needed to improve the expression and folding of these proteins while ensuring the host organism's viability and productivity.246 In addition, bioreactor design must be optimised to support stable and scalable fermentation processes that maintain product quality, consistency, and yield. Furthermore, downstream processing remains labour-intensive and expensive. This is another bottleneck that must be addressed through the development of more efficient purification methods, such as thermoresponsive blocks or simplified secretion strategies.247
Overall, protein-based materials is an exciting field of research at the intersection between materials engineering and synthetic biology. The on-going scale-up and commercialisation efforts taking place for several applications, be it biomedicine, textiles, or cosmetics, underscore the promise of these materials to address the urgent unmet need for new sustainable polymers. Nonetheless, the multifaceted challenges describe above demand continuing research efforts and multidisciplinary teams to tackle them and make protein-based materials an economically viable alternative to the well-established and cheaper manufacturing processes for fossil-derived polymers.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
DLB acknowledges funding from the Royal Society RG\R2\232238 Research Grant. AABC acknowledges funding from EPSRC UKRI EP/W524335/1.References
- P. Europe, Plastics - the Facts, 2022, https://plasticseurope.org/knowledge-hub/plastics-the-facts-2022/ (accessed 10th April, 2024) Search PubMed.
- W. E. Forum, The New Plastics Economy: Rethinking the future of plastics, 2016 Search PubMed.
- B. P. Federation, https://www.bpf.co.uk/press/Oil_Consumption.aspx (accessed 10th April, 2024).
- W. W. Y. Lau, Y. Shiran, R. M. Bailey, E. Cook, M. R. Stuchtey, J. Koskella, C. A. Velis, L. Godfrey, J. Boucher, M. B. Murphy, R. C. Thompson, E. Jankowska, A. Castillo Castillo, T. D. Pilditch, B. Dixon, L. Koerselman, E. Kosior, E. Favoino, J. Gutberlet, S. Baulch, M. E. Atreya, D. Fischer, K. K. He, M. M. Petit, U. R. Sumaila, E. Neil, M. V. Bernhofen, K. Lawrence and J. E. Palardy, Science, 2020, 369, 1455–1461 CrossRef CAS PubMed.
- B. P. Federation, https://www.bpf.co.uk/industry/Default.aspx (accessed 10th April, 2024).
- N. C. Abascal and L. Regan, Open Biol., 2018, 8, 180113 CrossRef CAS PubMed.
- I. R. Campbell, M.-Y. Lin, H. Iyer, M. Parker, J. L. Fredricks, K. Liao, A. M. Jimenez, P. Grandgeorge and E. Roumeli, Annu. Rev. Mater. Res., 2023, 53, 81–104 CrossRef CAS.
- M. J. Harrington and P. Fratzl, Prog. Mater. Sci., 2021, 120, 100767 CrossRef CAS.
- A. Miserez, J. Yu and P. Mohammadi, Chem. Rev., 2023, 123, 2049–2111 CrossRef CAS PubMed.
- Y. Cao and B. D. Olsen, Biomacromolecules, 2022, 23, 3286–3295 CrossRef CAS PubMed.
- D. Kim, Y. Cao, D. Mariappan, M. S. Bono Jr., A. J. Hart and B. Marelli, Adv. Funct. Mater., 2021, 31, 2005370 CrossRef CAS.
- A. Lee, A. R. Hudson, D. J. Shiwarski, J. W. Tashman, T. J. Hinton, S. Yerneni, J. M. Bliley, P. G. Campbell and A. W. Feinberg, Science, 2019, 365, 482–487 CrossRef CAS PubMed.
- K. A. Burke, D. C. Roberts and D. L. Kaplan, Biomacromolecules, 2016, 17, 237–245 CrossRef CAS PubMed.
- X. Jia, C. Wang, V. Ranganathan, B. Napier, C. Yu, Y. Chao, M. Forsyth, F. G. Omenetto, D. R. MacFarlane and G. G. Wallace, ACS Energy Lett., 2017, 2, 831–836 CrossRef CAS.
- G. Matzeu, L. Mogas-Soldevila, W. Li, A. Naidu, T. H. Turner, R. Gu, P. R. Blumeris, P. Song, D. G. Pascal, G. Guidetti, M. Li and F. G. Omenetto, Adv. Mater., 2020, 32, 2001258 CrossRef CAS PubMed.
- J. Guo, C. Li, S. Ling, W. Huang, Y. Chen and D. L. Kaplan, Biomaterials, 2017, 145, 44–55 CrossRef CAS PubMed.
- S. Ling, D. L. Kaplan and M. J. Buehler, Nat. Rev. Mater., 2018, 3, 18016 CrossRef CAS PubMed.
- Y. J. Yang, A. L. Holmberg and B. D. Olsen, Annu. Rev. Chem. Biomol. Eng., 2017, 8, 549–575 CrossRef CAS PubMed.
- M. W. T. Werten, G. Eggink, M. A. Cohen Stuart and F. A. de Wolf, Biotechnol. Adv., 2019, 37, 642–666 CrossRef CAS PubMed.
- M. Barroca, P. Rodrigues, R. Sobral, M. M. R. Costa, S. R. Chaves, R. Machado, M. Casal and T. Collins, Sci. Rep., 2016, 6, 39329 CrossRef CAS PubMed.
- K. Xiao, N. S. Malvankar, C. Shu, E. Martz, D. R. Lovley and X. Sun, Sci. Rep., 2016, 6, 23385 CrossRef CAS PubMed.
- J. Strobl, F. Kozak, M. Kamalov, D. Reichinger, D. Kurzbach and C. F. Becker, Adv. Mater., 2023, 35, 2207586 CrossRef CAS PubMed.
- M. J. Buehler, J. Mater. Res., 2006, 21, 1947–1961 CrossRef CAS.
- H. S. Rapoport and R. E. Shadwick, Biomacromolecules, 2002, 3, 42–50 CrossRef CAS PubMed.
- Z. Qin and M. J. Buehler, Phys. Rev. Lett., 2010, 104, 198304 CrossRef PubMed.
- A. Zhmurov, O. Kononova, R. I. Litvinov, R. I. Dima, V. Barsegov and J. W. Weisel, J. Am. Chem. Soc., 2012, 134, 20396–20402 CrossRef CAS PubMed.
- W. H. Rombouts, D. W. de Kort, T. T. H. Pham, C. P. M. van Mierlo, M. W. T. Werten, F. A. de Wolf and J. van der Gucht, Biomacromolecules, 2015, 16, 2506–2513 CrossRef CAS PubMed.
- K. Merrett, F. Wan, C.-J. Lee and J. L. Harden, ACS Biomater. Sci. Eng., 2021, 7, 1414–1427 CrossRef CAS PubMed.
- L. K. Hill, M. Meleties, P. Katyal, X. Xie, E. Delgado-Fukushima, T. Jihad, C.-F. Liu, S. O'Neill, R. S. Tu, P. D. Renfrew, R. Bonneau, Y. Z. Wadghiri and J. K. Montclare, Biomacromolecules, 2019, 20, 3340–3351 CrossRef CAS PubMed.
- F. A.-O. X. Wang, O. Gnewou, C. Modlin, L. C. Beltran, C. A.-O. Xu, Z. Su, P. Juneja, G. Grigoryan, E. A.-O. Egelman and V. A.-O. Conticello, Nat. Commun., 2021, 12, 407 CrossRef CAS PubMed.
- J. M. Fletcher, R. L. Harniman, F. R. H. Barnes, A. L. Boyle, A. Collins, J. Mantell, T. H. Sharp, M. Antognozzi, P. J. Booth, N. Linden, M. J. Miles, R. B. Sessions, P. Verkade and D. N. Woolfson, Science, 2013, 340, 595–599 CrossRef CAS PubMed.
- A. S. Cristie-David, P. Koldewey, B. A. Meinen, J. C. A. Bardwell and E. N. G. Marsh, Protein Sci., 2018, 27, 1893–1900 CrossRef CAS PubMed.
- R. L. Strader, Y. Shmidov and A. Chilkoti, Acc. Chem. Res., 2024, 57, 302–311 CrossRef CAS PubMed.
- K. Nagapudi, W. T. Brinkman, B. S. Thomas, J. O. Park, M. Srinivasarao, E. Wright, V. P. Conticello and E. L. Chaikof, Biomaterials, 2005, 26, 4695–4706 CrossRef CAS PubMed.
- K. Nagapudi, W. T. Brinkman, J. Leisen, B. S. Thomas, E. R. Wright, C. Haller, X. Wu, R. P. Apkarian, V. P. Conticello and E. L. Chaikof, Macromolecules, 2005, 38, 345–354 CrossRef CAS.
- M. J. Glassman and B. D. Olsen, Biomacromolecules, 2015, 16, 3762–3773 CrossRef CAS PubMed.
- D. López Barreiro, A. Folch-Fortuny, I. Muntz, J. C. Thies, C. M. J. Sagt and G. H. Koenderink, Biomacromolecules, 2022, 24(1), 489–501 CrossRef PubMed.
- S. Acosta, L. Poocza, L. Quintanilla-Sierra and J. C. Rodríguez-Cabello, Biomacromolecules, 2021, 22, 158–170 CrossRef CAS PubMed.
- C. Garcia Garcia, S. S. Patkar, N. Jovic, J. Mittal and K. L. Kiick, ACS Biomater. Sci. Eng., 2021, 7, 4244–4257 CrossRef CAS PubMed.
- M. D. Shoulders and R. T. Raines, Annu. Rev. Biochem., 2009, 78, 929–958 CrossRef CAS PubMed.
- A. Khedr, M. A. N. Soliman and M. A. Elsawy, in Peptide Bionanomaterials: From Design to Application, ed. M. A. Elsawy, Springer International Publishing, Cham, 2023, pp. 1–52, DOI:10.1007/978-3-031-29360-3_1.
- C. Rutschmann, S. Baumann, J. Cabalzar, K. B. Luther and T. Hennet, Appl. Microbiol. Biotechnol., 2014, 98, 4445–4455 CrossRef CAS PubMed.
- Z. Abdali, M. Renner-Rao, A. Chow, A. Cai, M. J. Harrington and N.-M. Dorval Courchesne, Biomacromolecules, 2022, 23, 1557–1568 CrossRef CAS PubMed.
- P. J. Kaur, R. Strawn, H. Bai, K. Xu, G. Ordas, H. Matsui and Y. Xu, J. Biol. Chem., 2015, 290, 9251–9261 CrossRef CAS PubMed.
- Y. Qiu, C. Zhai, L. Chen, X. Liu and J. Yeo, ACS Biomater. Sci. Eng., 2023, 9, 3778–3795 CrossRef CAS PubMed.
- Z. Abdali, M. Aminzare, A. Chow and N.-M. Dorval Courchesne, Biomed. Mater., 2023, 18, 015001 CrossRef CAS PubMed.
- L. Pauling, R. B. Corey and H. R. Branson, Proc. Natl. Acad. Sci. U. S. A., 1951, 37, 205–211 CrossRef CAS PubMed.
- H. H. Bragulla and D. G. Homberger, J. Anat., 2009, 214, 516–559 CrossRef CAS PubMed.
- D. N. Woolfson, in Advances in Protein Chemistry, Academic Press, 2005, vol. 70, pp. 79–112 Search PubMed.
- A. Lupas, Trends Biochem. Sci., 1996, 21, 375–382 CrossRef CAS PubMed.
- J. G. Rouse and M. E. Van Dyke, Materials, 2010, 3, 999–1014 CrossRef.
- R. N. Parker, A. Trent, K. L. Roth Stefaniak, M. E. Van Dyke and T. Z. Grove, Biomed. Mater., 2020, 15, 065006 CrossRef CAS PubMed.
- W. A. Petka, J. L. Harden, K. P. McGrath, D. Wirtz and D. A. Tirrell, Science, 1998, 281, 389–392 CrossRef CAS PubMed.
- L. J. Dooling and D. A. Tirrell, ACS Cent. Sci., 2016, 2, 812–819 CrossRef CAS PubMed.
- L. Faltova, A. M. Küffner, M. Hondele, K. Weis and P. Arosio, ACS Nano, 2018, 12, 9991–9999 CrossRef CAS PubMed.
- L. Malinovska, S. Kroschwald and S. Alberti, Biochim. Biophys. Acta, Proteins Proteomics, 2013, 1834, 918–931 CrossRef CAS PubMed.
- V. N. Uversky, Front. Phys., 2019, 7, 10 CrossRef.
- X. Jia and K. L. Kiick, Macromol. Biosci., 2009, 9, 140–156 CrossRef CAS PubMed.
- S. Lerch, R. Zuber, N. Gehring, Y. Wang, B. Eckel, K.-D. Klass, F.-O. Lehmann and B. Moussian, BMC Biol., 2020, 18, 195 CrossRef CAS PubMed.
- A. J. Bailey, J. Macmillan, P. R. Shrewry, A. S. Tatham, J. Gosline, M. Lillie, E. Carrington, P. Guerette, C. Ortlepp and K. Savage, Philos. Trans. R. Soc. London, Ser. B, 2002, 357, 121–132 CrossRef PubMed.
- S. S. Patkar, C. Garcia Garcia, L. L. Palmese and K. L. Kiick, Biomacromolecules, 2023, 24, 3729–3741 CrossRef CAS PubMed.
- X. H. To, N. Pebere, N. Pelaprat, B. Boutevin and Y. Hervaud, Corros. Sci., 1997, 39, 1925–1934 CrossRef CAS.
- B. Lim, J. Kim, M. S. Desai, W. Wu, I. Chae and S.-W. Lee, Biomacromolecules, 2023, 24, 118–131 CrossRef CAS PubMed.
- C. Ma, J. Sun, B. Li, Y. Feng, Y. Sun, L. Xiang, B. Wu, L. Xiao, B. Liu, V. S. Petrovskii, L. Bin, J. Zhang, Z. Wang, H. Li, L. Zhang, J. Li, F. Wang, R. Göstl, I. I. Potemkin, D. Chen, H. Zeng, H. Zhang, K. Liu and A. Herrmann, Nat. Commun., 2021, 12, 3613 CrossRef CAS PubMed.
- I. El Maachi, A. Loewen, S. Acosta, S. Rütten, J. C. Rodríguez-Cabello, S. Jockenhoevel and A. Fernández-Colino, Adv. Funct. Mater., 2024, 2313204 CrossRef CAS.
- R. Herrero-Vanrell, A. C. Rincón, M. Alonso, V. Reboto, I. T. Molina-Martinez and J. C. Rodríguez-Cabello, J. Controlled Release, 2005, 102, 113–122 CrossRef CAS PubMed.
- M. Dai, J.-P. Belaïdi, G. Fleury, E. Garanger, M. Rielland, X. Schultze and S. Lecommandoux, Biomacromolecules, 2021, 22, 4956–4966 CrossRef CAS PubMed.
- A. K. Varanko, J. C. Su and A. Chilkoti, Annu. Rev. Biomed. Eng., 2020, 22, 343–369 CrossRef CAS PubMed.
- S. Roberts, M. Dzuricky and A. Chilkoti, FEBS Lett., 2015, 589, 2477–2486 CrossRef CAS PubMed.
- D. W. Urry, M. M. Long, B. A. Cox, T. Ohnishi, L. W. Mitchell and M. Jacobs, Biochim. Biophys. Acta, Protein Struct., 1974, 371, 597–602 CrossRef CAS PubMed.
- D. W. Urry, D. C. Gowda, T. M. Parker, C.-H. Luan, M. C. Reid, C. M. Harris, A. Pattanaik and R. D. Harris, Biopolymers, 1992, 32, 1243–1250 CrossRef CAS PubMed.
- E. Garanger, S. R. MacEwan, O. Sandre, A. Brûlet, L. Bataille, A. Chilkoti and S. Lecommandoux, Macromolecules, 2015, 48, 6617–6627 CrossRef CAS.
- R. E. Sallach, W. Cui, J. Wen, A. Martinez, V. P. Conticello and E. L. Chaikof, Biomaterials, 2009, 30, 409–422 CrossRef CAS PubMed.
- J. R. Simon, N. J. Carroll, M. Rubinstein, A. Chilkoti and G. P. López, Nat. Chem., 2017, 9, 509–515 CrossRef CAS PubMed.
- J. Pille, A. Aloi, D. H. T. Le, I. Vialshin, N. van de Laar, K. Kevenaar, M. Merkx, I. K. Voets and J. C. M. van Hest, Small, 2021, 17, 2007234 CrossRef CAS PubMed.
- S. Mayavan, N. K. Dutta, N. R. Choudhury, M. Kim, C. M. Elvin and A. J. Hill, Biomaterials, 2011, 32, 2786–2796 CrossRef CAS PubMed.
- C. M. Elvin, A. G. Carr, M. G. Huson, J. M. Maxwell, R. D. Pearson, T. Vuocolo, N. E. Liyou, D. C. C. Wong, D. J. Merritt and N. E. Dixon, Nature, 2005, 437, 999–1002 CrossRef CAS PubMed.
- S. S. Patkar, Y. Tang, T. Zhang, A. M. Bisram, J. G. Saven, D. J. Pochan and K. L. Kiick, Biomacromolecules, 2024, 25, 2449–2461 CrossRef CAS PubMed.
- R. Balu, N. K. Dutta, A. K. Dutta and N. R. Choudhury, Nat. Commun., 2021, 12, 149 CrossRef CAS PubMed.
- M. Dzuricky, B. A. Rogers, A. Shahid, P. S. Cremer and A. Chilkoti, Nat. Chem., 2020, 12, 814–825 CrossRef CAS PubMed.
- F. G. Quiroz and A. Chilkoti, Nat. Mater., 2015, 14, 1164–1171 CrossRef CAS PubMed.
- L. C. Pauling, R. B. Corey and W. T. Astbury, Proc. R. Soc. London, Ser. B, 1997, 141, 21–33 Search PubMed.
- A. Pena-Francesch, B. Akgun, A. Miserez, W. Zhu, H. Gao and M. C. Demirel, Adv. Funct. Mater., 2014, 24, 6227–6233 CrossRef CAS.
- M. F. B. G. Gebbink, D. Claessen, B. Bouma, L. Dijkhuizen and H. A. B. Wösten, Nat. Rev. Microbiol., 2005, 3, 333–341 CrossRef CAS PubMed.
- Z. Xu and M. J. Buehler, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2010, 81, 061910 CrossRef PubMed.
- A. Ibáñez-Fonseca, D. Orbanic, F. J. Arias, M. Alonso, D. I. Zeugolis and J. C. Rodríguez-Cabello, Small, 2020, 16, 2001244 CrossRef PubMed.
- S. Ling, Z. Qin, C. Li, W. Huang, D. L. Kaplan and M. J. Buehler, Nat. Commun., 2017, 8, 1387 CrossRef PubMed.
- O. S. Tokareva, S. Lin, M. M. Jacobsen, W. Huang, D. Rizzo, D. Li, M. Simon, C. Staii, P. Cebe, J. Y. Wong, M. J. Buehler and D. L. Kaplan, J. Struct. Biol., 2014, 186, 412–419 CrossRef CAS PubMed.
- V. Latza, P. A. Guerette, D. Ding, S. Amini, A. Kumar, I. Schmidt, S. Keating, N. Oxman, J. C. Weaver, P. Fratzl, A. Miserez and A. Masic, Nat. Commun., 2015, 6, 8313 CrossRef CAS PubMed.
- V. Sariola, A. Pena-Francesch, H. Jung, M. Çetinkaya, C. Pacheco, M. Sitti and M. C. Demirel, Sci. Rep., 2015, 5, 13482 CrossRef PubMed.
- U. O. S. Seker, A. Y. Chen, R. J. Citorik and T. K. Lu, ACS Synth. Biol., 2017, 6, 266–275 CrossRef CAS PubMed.
- C.-Z. Zhou, F. Confalonieri, M. Jacquet, R. Perasso, Z.-G. Li and J. Janin, Proteins: Struct., Funct., Bioinf., 2001, 44, 119–122 CrossRef CAS PubMed.
- S. Lin, S. Ryu, O. Tokareva, G. Gronau, M. M. Jacobsen, W. Huang, D. J. Rizzo, D. Li, C. Staii, N. M. Pugno, J. Y. Wong, D. L. Kaplan and M. J. Buehler, Nat. Commun., 2015, 6, 6892 CrossRef PubMed.
- Z. Martín-Moldes, D. Ebrahimi, R. Plowright, N. Dinjaski, C. C. Perry, M. J. Buehler and D. L. Kaplan, Adv. Funct. Mater., 2018, 28, 1702570 CrossRef PubMed.
- O. S. Rabotyagova, P. Cebe and D. L. Kaplan, Biomacromolecules, 2009, 10, 229–236 CrossRef CAS PubMed.
- A. Pena-Francesch, J. Giltinan and M. Sitti, Nat. Commun., 2019, 10, 3188 CrossRef PubMed.
- H. Jung, A. Pena-Francesch, A. Saadat, A. Sebastian, D. H. Kim, R. F. Hamilton, I. Albert, B. D. Allen and M. C. Demirel, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 6478–6483 CrossRef CAS PubMed.
- K. Deepankumar, C. Lim, I. Polte, B. Zappone, C. Labate, M. P. De Santo, H. Mohanram, A. Palaniappan, D. S. Hwang and A. Miserez, Adv. Funct. Mater., 2020, 30, 1907534 CrossRef CAS.
- M. Vural, Y. Lei, A. Pena-Francesch, H. Jung, B. Allen, M. Terrones and M. C. Demirel, Carbon, 2017, 118, 404–412 CrossRef CAS.
- J. A. Tomko, A. Pena-Francesch, H. Jung, M. Tyagi, B. D. Allen, M. C. Demirel and P. E. Hopkins, Nat. Nanotechnol., 2018, 13, 959–964 CrossRef CAS PubMed.
- P. Q. Nguyen, Z. Botyanszki, P. K. R. Tay and N. S. Joshi, Nat. Commun., 2014, 5, 4945 CrossRef CAS PubMed.
- M. L. Evans and M. R. Chapman, Biochim. Biophys. Acta, Mol. Cell Res., 2014, 1843, 1551–1558 CrossRef CAS PubMed.
- E. P. DeBenedictis, D. Ma and S. Keten, RSC Adv., 2017, 7, 48102–48112 RSC.
- M. Andersson, Q. Jia, A. Abella, X.-Y. Lee, M. Landreh, P. Purhonen, H. Hebert, M. Tenje, C. V. Robinson, Q. Meng, G. R. Plaza, J. Johansson and A. Rising, Nat. Chem. Biol., 2017, 13, 262–264 CrossRef CAS PubMed.
- A. Pena-Francesch, H. Jung, M. C. Demirel and M. Sitti, Nat. Mater., 2020, 19, 1230–1235 CrossRef CAS PubMed.
- Q. Wang, S. Ling, X. Liang, H. Wang, H. Lu and Y. Zhang, Adv. Funct. Mater., 2019, 29, 1808695 CrossRef.
- Y. Kikuchi, A. Pena-Francesch, M. Vural and M. C. Demirel, ACS Appl. Bio Mater., 2020, 3, 2507–2515 CrossRef CAS PubMed.
- C. Guo, C. Li, H. V. Vu, P. Hanna, A. Lechtig, Y. Qiu, X. Mu, S. Ling, A. Nazarian, S. J. Lin and D. L. Kaplan, Nat. Mater., 2020, 19, 102–108 CrossRef CAS PubMed.
- A. Tarakanova, W. Huang, Z. Qin, D. L. Kaplan and M. J. Buehler, ACS Biomater. Sci. Eng., 2017, 3, 2889–2899 CrossRef CAS PubMed.
- L. Chambre, Z. Martín-Moldes, R. N. Parker and D. L. Kaplan, Adv. Drug Delivery Rev., 2020, 160, 186–198 CrossRef CAS PubMed.
- S. Salinas-Fernández, M. Santos, M. Alonso, L. Quintanilla and J. C. Rodríguez-Cabello, Appl. Mater. Today, 2020, 18, 100500 CrossRef.
- W. Huang, A. Tarakanova, N. Dinjaski, Q. Wang, X. Xia, Y. Chen, J. Y. Wong, M. J. Buehler and D. L. Kaplan, Adv. Funct. Mater., 2016, 26, 4113–4123 CrossRef CAS PubMed.
- Y. S. Wang, W. W. Huang, Y. Wang, X. Mu, S. J. Ling, H. P. Yu, W. S. Chen, C. C. Guo, M. C. Watson, Y. J. Yu, L. D. Black, M. Li, F. G. Omenetto, C. M. Li and D. L. Kaplan, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 14602–14608 CrossRef CAS PubMed.
- A. Fernández-Colino, F. J. Arias, M. Alonso and J. C. Rodríguez-Cabello, Biomacromolecules, 2014, 15, 3781–3793 CrossRef PubMed.
- C. Gonzalez-Obeso, J. C. Rodriguez-Cabello and D. L. Kaplan, Acta Biomater., 2022, 141, 14–23 CrossRef CAS PubMed.
- M. D. Golinska, T. T. H. Pham, M. W. T. Werten, F. A. de Wolf, M. A. Cohen Stuart and J. van der Gucht, Biomacromolecules, 2013, 14, 48–55 CrossRef CAS PubMed.
- S. Roberts, V. Miao, S. Costa, J. Simon, G. Kelly, T. Shah, S. Zauscher and A. Chilkoti, Nat. Commun., 2020, 11, 1342 CrossRef CAS PubMed.
- A. Basheer, S. Shahid, M. J. Kang, J. H. Lee, J. S. Lee and D. W. Lim, ACS Appl. Mater. Interfaces, 2021, 13, 24385–24400 CrossRef CAS PubMed.
- Q. Wang, X. Xia, W. Huang, Y. Lin, Q. Xu and D. L. Kaplan, Adv. Funct. Mater., 2014, 24, 4303–4310 CrossRef CAS PubMed.
- C. González-Obeso, M. González-Pérez, J. F. Mano, M. Alonso and J. C. Rodríguez-Cabello, Small, 2020, 16, 2005191 CrossRef PubMed.
- Y. Li, J. Li, J. Sun, H. He, B. Li, C. Ma, K. Liu and H. Zhang, Angew. Chem., Int. Ed., 2020, 59, 8148–8152 CrossRef CAS PubMed.
- J. Li, B. Jiang, X. Chang, H. Yu, Y. Han and F. Zhang, Nat. Commun., 2023, 14, 2127 CrossRef CAS PubMed.
- M. D. Golinska, M. K. Włodarczyk-Biegun, M. W. T. Werten, M. A. C. Stuart, F. A. de Wolf and R. de Vries, Biomacromolecules, 2014, 15, 699–706 CrossRef CAS PubMed.
- S. Roberts, T. S. Harmon, J. L. Schaal, V. Miao, K. Li, A. Hunt, Y. Wen, T. G. Oas, J. H. Collier, R. V. Pappu and A. Chilkoti, Nat. Mater., 2018, 17, 1154–1163 CrossRef CAS PubMed.
- T. Luo and K. L. Kiick, J. Am. Chem. Soc., 2015, 137, 15362–15365 CrossRef CAS PubMed.
- L. A. Navarro, J. J. Ryan, M. Dzuricky, M. Gradzielski, A. Chilkoti and S. Zauscher, Biomacromolecules, 2021, 22, 3827–3838 CrossRef CAS PubMed.
- A. Bracalello, V. Santopietro, M. Vassalli, G. Marletta, R. Del Gaudio, B. Bochicchio and A. Pepe, Biomacromolecules, 2011, 12, 2957–2965 CrossRef CAS PubMed.
- S. E. Arevalo and M. J. Buehler, MRS Bull., 2023, 48, 1140–1153 CrossRef.
- W. Huang, D. Ebrahimi, N. Dinjaski, A. Tarakanova, M. J. Buehler, J. Y. Wong and D. L. Kaplan, Acc. Chem. Res., 2017, 50, 866–876 CrossRef CAS PubMed.
- D. López Barreiro, A. Folch-Fortuny, I. Muntz, J. C. Thies, C. M. J. Sagt and G. H. Koenderink, Biomacromolecules, 2023, 24, 489–501 CrossRef PubMed.
- H. Shi, T. Ji, C. Zhai, J. Lu, W. Huang and J. Yeo, J. Mater. Chem. B, 2022, 10, 6133–6142 RSC.
- A. Paul and T. Birol, Annu. Rev. Mater. Res., 2019, 49, 31–52 CrossRef CAS.
- G. Gronau, S. T. Krishnaji, M. E. Kinahan, T. Giesa, J. Y. Wong, D. L. Kaplan and M. J. Buehler, Biomaterials, 2012, 33, 8240–8255 CrossRef CAS PubMed.
- A. Leaver-Fay, M. Tyka, S. M. Lewis, O. F. Lange, J. Thompson, R. Jacak, K. W. Kaufman, P. D. Renfrew, C. A. Smith, W. Sheffler, I. W. Davis, S. Cooper, A. Treuille, D. J. Mandell, F. Richter, Y.-E. A. Ban, S. J. Fleishman, J. E. Corn, D. E. Kim, S. Lyskov, M. Berrondo, S. Mentzer, Z. Popović, J. J. Havranek, J. Karanicolas, R. Das, J. Meiler, T. Kortemme, J. J. Gray, B. Kuhlman, D. Baker and P. Bradley, in Methods in Enzymology, ed. M. L. Johnson and L. Brand, Academic Press, 2011, vol. 487, pp. 545–574 Search PubMed.
- T. F. Huddy, Y. Hsia, R. D. Kibler, J. Xu, N. Bethel, D. Nagarajan, R. Redler, P. J. Y. Leung, C. Weidle, A. Courbet, E. C. Yang, A. K. Bera, N. Coudray, S. J. Calise, F. A. Davila-Hernandez, H. L. Han, K. D. Carr, Z. Li, R. McHugh, G. Reggiano, A. Kang, B. Sankaran, M. S. Dickinson, B. Coventry, T. J. Brunette, Y. Liu, J. Dauparas, A. J. Borst, D. Ekiert, J. M. Kollman, G. Bhabha and D. Baker, Nature, 2024, 627, 898–904 CrossRef CAS PubMed.
- S. Keten, Z. Xu, B. Ihle and M. J. Buehler, Nat. Mater., 2010, 9, 359–367 CrossRef CAS PubMed.
- G. Bratzel and M. J. Buehler, Biopolymers, 2012, 97, 408–417 CrossRef CAS PubMed.
- S. Rauscher and R. Pomès, eLife, 2017, 6, e26526 CrossRef PubMed.
- N. K. Li, S. Roberts, F. G. Quiroz, A. Chilkoti and Y. G. Yingling, Biomacromolecules, 2018, 19, 2496–2505 CrossRef CAS PubMed.
- A. Gautieri, S. Vesentini, A. Redaelli and M. J. Buehler, Nano Lett., 2011, 11, 757–766 CrossRef CAS PubMed.
- A. Pizzi, L. Sori, C. Pigliacelli, A. Gautieri, C. Andolina, G. Bergamaschi, A. Gori, P. Panine, A. M. Grande, M. B. Linder, F. Baldelli Bombelli, M. Soncini and P. Metrangolo, Small, 2022, 18, 2200807 CrossRef CAS PubMed.
- B. R. Brooks, C. L. Brooks III, A. D. Mackerell Jr., L. Nilsson, R. J. Petrella, B. Roux, Y. Won, G. Archontis, C. Bartels, S. Boresch, A. Caflisch, L. Caves, Q. Cui, A. R. Dinner, M. Feig, S. Fischer, J. Gao, M. Hodoscek, W. Im, K. Kuczera, T. Lazaridis, J. Ma, V. Ovchinnikov, E. Paci, R. W. Pastor, C. B. Post, J. Z. Pu, M. Schaefer, B. Tidor, R. M. Venable, H. L. Woodcock, X. Wu, W. Yang, D. M. York and M. Karplus, J. Comput. Chem., 2009, 30, 1545–1614 CrossRef CAS PubMed.
- R. Salomon-Ferrer, D. A. Case and R. C. Walker, WIREs Comput. Mol. Sci., 2013, 3, 198–210 CrossRef CAS.
- W. L. Jorgensen, D. S. Maxwell and J. Tirado-Rives, J. Am. Chem. Soc., 1996, 118, 11225–11236 CrossRef CAS.
- Y. Sugita and Y. Okamoto, Chem. Phys. Lett., 1999, 314, 141–151 CrossRef CAS.
- T. Shu, K. Zheng, Z. Zhang, J. Ren, Z. Wang, Y. Pei, J. Yeo, G. X. Gu and S. Ling, Biomacromolecules, 2021, 22, 1955–1965 CrossRef CAS PubMed.
- S. T. Krishnaji, G. Bratzel, M. E. Kinahan, J. A. Kluge, C. Staii, J. Y. Wong, M. J. Buehler and D. L. Kaplan, Adv. Funct. Mater., 2013, 23, 241–253 CrossRef CAS.
- C.-H. Yu, W. Chen, Y.-H. Chiang, K. Guo, Z. Martin Moldes, D. L. Kaplan and M. J. Buehler, ACS Biomater. Sci. Eng., 2022, 8, 1156–1165 CrossRef CAS PubMed.
- A. Tarakanova, W. Huang, A. S. Weiss, D. L. Kaplan and M. J. Buehler, Biomaterials, 2017, 127, 49–60 CrossRef CAS PubMed.
- J. Yeo, Y. Qiu, G. S. Jung, Y.-W. Zhang, M. J. Buehler and D. L. Kaplan, Biomaterials, 2020, 240, 119857 CrossRef CAS PubMed.
- J. Yeo, W. Huang, A. Tarakanova, Y.-W. Zhang, D. L. Kaplan and M. J. Buehler, J. Mater. Chem. B, 2018, 6, 3727–3734 RSC.
- A. Prhashanna, P. A. Taylor, J. Qin, K. L. Kiick and A. Jayaraman, Biomacromolecules, 2019, 20, 1178–1189 CrossRef CAS PubMed.
- J. E. Condon, T. B. Martin and A. Jayaraman, Soft Matter, 2017, 13, 2907–2918 RSC.
- B. Ni, D. L. Kaplan and M. J. Buehler, Chem, 2023, 9, 1828–1849 CAS.
- C. Zhai, T. Li, H. Shi and J. Yeo, J. Mater. Chem. B, 2020, 8, 6562–6587 RSC.
- W. V. Srubar, Trends Biotechnol., 2021, 39, 574–583 CrossRef CAS PubMed.
- P. Q. Nguyen, N.-M. D. Courchesne, A. Duraj-Thatte, P. Praveschotinunt and N. S. Joshi, Adv. Mater., 2018, 30, 1704847 CrossRef PubMed.
- N. C. Tang and A. Chilkoti, Nat. Mater., 2016, 15, 419–424 CrossRef CAS PubMed.
- N. Dinjaski, W. Huang and D. L. Kaplan, in Peptide Self-Assembly: Methods and Protocols, ed. B. L. Nilsson and T. M. Doran, Springer New York, New York, NY, 2018, pp. 181–192 Search PubMed.
- J. R. McDaniel, J. A. MacKay, F. G. Quiroz and A. Chilkoti, Biomacromolecules, 2010, 11, 944–952 CrossRef CAS PubMed.
- S.-H. Cheon, B.-Y. Seo, Y.-J. Lee, D. Sim, S.-B. Lee, P. Guruprasath, T. D. Singh, B.-H. Lee, V. Sarangthem and R.-W. Park, ACS Biomater. Sci. Eng., 2020, 6, 5024–5031 CrossRef CAS PubMed.
- T. Liu, A. Liang, Z. Liang, G. Li and F. Wang, 3 Biotech, 2018, 8, 252 CrossRef PubMed.
- R. E. Sallach, V. P. Conticello and E. L. Chaikof, Biotechnol. Prog., 2009, 25, 1810–1818 CrossRef CAS PubMed.
- K.-K. Tian, Z.-G. Qian and X.-X. Xia, Adv. Drug Delivery Rev., 2023, 194, 114728 CrossRef CAS PubMed.
- C. Engler, R. Gruetzner, R. Kandzia and S. Marillonnet, PLoS One, 2009, 4, e5553 CrossRef PubMed.
- E. Weber, C. Engler, R. Gruetzner, S. Werner and S. Marillonnet, PLoS One, 2011, 6, e16765 CrossRef CAS PubMed.
- J. M. Pryor, V. Potapov, K. Bilotti, N. Pokhrel and G. J. S. Lohman, ACS Synth. Biol., 2022, 11, 2036–2042 CrossRef CAS PubMed.
- A. Scior, S. Preissler, M. Koch and E. Deuerling, BMC Biotechnol., 2011, 11, 87 CrossRef CAS PubMed.
- M. T. A. Nguyen, M. V. Gobry, N. Sampedro Vallina, G. Pothoulakis and E. S. Andersen, ACS Synth. Biol., 2024, 13, 963–968 CrossRef CAS PubMed.
- D. Wu, A. Koscic, S. Schneider, R. C. A. Dubini, D. C. Rodriguez Camargo, S. Schneider and P. Rovó, Biomacromolecules, 2024, 25, 1759–1774 CrossRef CAS PubMed.
- E. Kim, J. Jeon, Y. Zhu, E. D. Hoppe, Y.-S. Jun, G. M. Genin and F. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 48457–48468 CrossRef CAS PubMed.
- B. Dai, C. J. Sargent, X. Gui, C. Liu and F. Zhang, Biomacromolecules, 2019, 20, 2015–2023 CrossRef CAS PubMed.
- X. Yang, J. Wei, Y. Wang, C. Yang, S. Zhao, C. Li, Y. Dong, K. Bai, Y. Li, H. Teng, D. Wang, N. Lyu, J. Li, X. Chang, X. Ning, Q. Ouyang, Y. Zhang and L. Qian, ACS Synth. Biol., 2018, 7, 2331–2339 CrossRef CAS PubMed.
- W. Ott, T. Nicolaus, H. E. Gaub and M. A. Nash, Biomacromolecules, 2016, 17, 1330–1338 CrossRef CAS PubMed.
- M. Amiram, F. G. Quiroz, D. J. Callahan and A. Chilkoti, Nat. Mater., 2011, 10, 141–148 CrossRef CAS PubMed.
- A. Fire and S. Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 4641–4645 CrossRef CAS PubMed.
- S. O. Lee, Q. Xie and S. D. Fried, ACS Cent. Sci., 2021, 7, 1736–1750 CrossRef CAS PubMed.
- N. Eger, L. Schoppe, S. Schuster, U. Laufs and J.-N. Boeckel, in Circular RNAs: Biogenesis and Functions, ed. J. Xiao, Springer Singapore, Singapore, 2018, pp. 41–52 Search PubMed.
- X. Liu, Y. Zhang, S. Zhou, L. Dain, L. Mei and G. Zhu, J. Controlled Release, 2022, 348, 84–94 CrossRef CAS PubMed.
- D.-F. Chen, L.-J. Zhang, K. Tan and Q. Jing, Biotechnol. Biotechnol. Equip., 2018, 32, 116–123 CrossRef CAS.
- L. Liu, P. Wang, D. Zhao, L. Zhu, J. Tang, W. Leng, J. Su, Y. Liu, C. Bi and X. Zhang, Appl. Environ. Microbiol., 2022, 88, e00028-22 CrossRef PubMed.
- A. Nanda, S. S. Nasker, A. Mehra, S. Panda and S. Nayak, Microorganisms, 2020, 8, 2004 CrossRef CAS PubMed.
- F. Pinto, E. L. Thornton and B. Wang, Nat. Commun., 2020, 11, 1529 CrossRef CAS PubMed.
- C. H. Bowen, B. Dai, C. J. Sargent, W. Bai, P. Ladiwala, H. Feng, W. Huang, D. L. Kaplan, J. M. Galazka and F. Zhang, Biomacromolecules, 2018, 19, 3853–3860 CrossRef CAS PubMed.
- E. Kim, B. Dai, J. B. Qiao, W. Li, J. D. Fortner and F. Zhang, ACS Appl. Mater. Interfaces, 2018, 10, 43003–43012 CrossRef CAS PubMed.
- C. H. Bowen, C. J. Sargent, A. Wang, Y. Zhu, X. Chang, J. Li, X. Mu, J. M. Galazka, Y.-S. Jun, S. Keten and F. Zhang, Nat. Commun., 2021, 12, 5182 CrossRef CAS PubMed.
- J. C. Rodríguez-Cabello, A. Girotti, A. Ribeiro and F. J. Arias, in Nanotechnology in Regenerative Medicine: Methods and Protocols, ed. M. Navarro and J. A. Planell, Humana Press, Totowa, NJ, 2012, pp. 17–38 Search PubMed.
- G. Bhattacharyya, P. Oliveira, S. T. Krishnaji, D. Chen, M. Hinman, B. Bell, T. I. Harris, A. Ghazitabatabaei, R. V. Lewis and J. A. Jones, Protein Expression Purif., 2021, 183, 105839 CrossRef CAS PubMed.
- M.-T. Chen, C.-F. Hu, H.-B. Huang, Z.-G. Qian and X.-X. Xia, Angew. Chem., Int. Ed., 2022, 61, e202214177 CrossRef CAS PubMed.
- Y. Y. Peng, L. Howell, V. Stoichevska, J. A. Werkmeister, G. J. Dumsday and J. A. M. Ramshaw, Microb. Cell Fact., 2012, 11, 146 CrossRef CAS PubMed.
- T. Collins, M. Barroca, F. Branca, J. Padrão, R. Machado and M. Casal, Biomacromolecules, 2014, 15, 2701–2708 CrossRef CAS PubMed.
- S.-P. Wei, Z.-G. Qian, C.-F. Hu, F. Pan, M.-T. Chen, S. Y. Lee and X.-X. Xia, Nat. Chem. Biol., 2020, 16, 1143–1148 CrossRef CAS PubMed.
- A. Y. Chen, Z. Deng, A. N. Billings, U. O. Seker, M. Y. Lu, R. J. Citorik, B. Zakeri and T. K. Lu, Nat. Mater., 2014, 13, 515–523 CrossRef CAS PubMed.
- Y. Cao, Y. Feng, M. D. Ryser, K. Zhu, G. Herschlag, C. Cao, K. Marusak, S. Zauscher and L. You, Nat. Biotechnol., 2017, 35, 1087–1093 CrossRef CAS PubMed.
- K. Zhang, L. Su and J. Wu, Annu. Rev. Food Sci. Technol., 2020, 11, 295–318 CrossRef CAS PubMed.
- J. van Dijl and M. Hecker, Microb. Cell Fact., 2013, 12, 3 CrossRef CAS PubMed.
- Q. Xie, S. On Lee, N. Vissamsetti, S. Guo, M. E. Johnson and S. D. Fried, Angew. Chem., Int. Ed., 2023, 62, e202305178 CrossRef CAS PubMed.
- C. Zhang, J. Huang, J. Zhang, S. Liu, M. Cui, B. An, X. Wang, J. Pu, T. Zhao, C. Fan, T. K. Lu and C. Zhong, Mater. Today, 2019, 28, 40–48 CrossRef.
- A. Heidebrecht and T. Scheibel, in Advances in Applied Microbiology, ed. S. Sariaslani and G. M. Gadd, Academic Press, 2013, vol. 82, pp. 115–153 Search PubMed.
- X.-X. Xia, Z.-G. Qian, C. S. Ki, Y. H. Park, D. L. Kaplan and S. Y. Lee, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14059–14063 CrossRef CAS PubMed.
- H. Cao, S. Parveen, D. Ding, H. Xu, T. Tan and L. Liu, Sci. Rep., 2017, 7, 11365 CrossRef PubMed.
- Y.-X. Yang, Z.-G. Qian, J.-J. Zhong and X.-X. Xia, Process Biochem., 2016, 51, 484–490 CrossRef CAS.
- A. K. Wallace, N. Chanut and C. A. Voigt, Adv. Funct. Mater., 2020, 30, 2000849 CrossRef CAS.
- Z. Martin-Moldes, D. L. Barreiro, M. J. Buehler and D. L. Kaplan, Acta Biomater., 2021, 120, 203–212 CrossRef CAS PubMed.
- K. Malcı, E. Watts, T. M. Roberts, J. Y. Auxillos, B. Nowrouzi, H. O. Boll, C. Z. S. D. Nascimento, A. Andreou, P. Vegh, S. Donovan, R. Fragkoudis, S. Panke, E. Wallace, A. Elfick and L. Rios-Solis, ACS Synth. Biol., 2022, 11, 2527–2547 CrossRef PubMed.
- K. Malcı, L. E. Walls and L. Rios-Solis, Front. Bioeng. Biotechnol., 2020, 8, 589468 CrossRef PubMed.
- J. Beal, A. Goñi-Moreno, C. Myers, A. Hecht, M. D. C. de Vicente, M. Parco, M. Schmidt, K. Timmis, G. Baldwin, S. Friedrichs, P. Freemont, D. Kiga, E. Ordozgoiti, M. Rennig, L. Rios, K. Tanner, V. de Lorenzo and M. Porcar, EMBO Rep., 2020, 21, e50521 CrossRef CAS PubMed.
- K. Malcı, N. Jonguitud-Borrego, H. van der Straten Waillet, U. Puodžiūnaitė, E. J. Johnston, S. J. Rosser and L. Rios-Solis, ACS Synth. Biol., 2022, 11, 3629–3643 CrossRef PubMed.
- K. V. Sidoruk, L. I. Davydova, D. G. Kozlov, D. G. Gubaidullin, A. V. Glazunov, V. G. Bogush and V. G. Debabov, Appl. Biochem. Microbiol., 2015, 51, 766–773 CrossRef CAS.
- Z. Yang and Z. Zhang, Biotechnol. Adv., 2018, 36, 182–195 CrossRef CAS PubMed.
- R. Jansson, C. H. Lau, T. Ishida, M. Ramström, M. Sandgren and M. Hedhammar, Biotechnol. J., 2016, 11, 687–699 CrossRef CAS PubMed.
- L. Ma, X. Liang, S. Yu and J. Zhou, Bioresour. Bioprocess, 2022, 9, 119 CrossRef PubMed.
- A. Lazaris, S. Arcidiacono, Y. Huang, J.-F. Zhou, F. Duguay, N. Chretien, E. A. Welsh, J. W. Soares and C. N. Karatzas, Science, 2002, 295, 472–476 CrossRef CAS PubMed.
- S. Frischholz, F. Beier, I. Girkontaite, K. Wagner, E. Pöschl, J. Turnay, U. Mayer and K. von der Mark, J. Biol. Chem., 1998, 273, 4547–4555 CrossRef CAS PubMed.
- S. Grip, A. Rising, H. Nimmervoll, E. Storckenfeldt, S. J. McQueen-Mason, N. Pouchkina-Stantcheva, F. Vollrath, W. Engström and A. Fernandez-Arias, Cancer Genomics Proteomics, 2006, 3, 83–87 CAS.
- F. Burla, Y. Mulla, B. E. Vos, A. Aufderhorst-Roberts and G. H. Koenderink, Nat. Rev. Phys., 2019, 1, 249–263 CrossRef.
- A. P. Liu, E. A. Appel, P. D. Ashby, B. M. Baker, E. Franco, L. Gu, K. Haynes, N. S. Joshi, A. M. Kloxin, P. H. J. Kouwer, J. Mittal, L. Morsut, V. Noireaux, S. Parekh, R. Schulman, S. K. Y. Tang, M. T. Valentine, S. L. Vega, W. Weber, N. Stephanopoulos and O. Chaudhuri, Nat. Mater., 2022, 21, 390–397 CrossRef CAS PubMed.
- A. Rodrigo-Navarro, S. Sankaran, M. J. Dalby, A. del Campo and M. Salmeron-Sanchez, Nat. Rev. Mater., 2021, 6, 1175–1190 CrossRef.
- T.-C. Tang, E. Tham, X. Liu, K. Yehl, A. J. Rovner, H. Yuk, C. de la Fuente-Nunez, F. J. Isaacs, X. Zhao and T. K. Lu, Nat. Chem. Biol., 2021, 17, 724–731 CrossRef CAS PubMed.
- S. Balasubramanian, K. Yu, A. S. Meyer, E. Karana and M.-E. Aubin-Tam, Adv. Funct. Mater., 2021, 31, 2011162 CrossRef CAS.
- A. T. Zvinavashe, E. Lim, H. Sun and B. Marelli, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 25555–25561 CrossRef CAS PubMed.
- C. Gilbert, T.-C. Tang, W. Ott, B. A. Dorr, W. M. Shaw, G. L. Sun, T. K. Lu and T. Ellis, Nat. Mater., 2021, 20, 691–700 CrossRef CAS PubMed.
- S. Molinari, R. F. Tesoriero and C. M. Ajo-Franklin, Matter, 2021, 4, 3095–3120 CrossRef CAS.
- C. M. Heveran and C. J. Hernandez, Matter, 2023, 6, 3705–3718 CrossRef CAS.
- J.-J. Oh, S. Ammu, V. D. Vriend, R. Kieffer, F. H. Kleiner, S. Balasubramanian, E. Karana, K. Masania and M.-E. Aubin-Tam, Adv. Mater., 2024, 36, 2305505 CrossRef CAS PubMed.
- B. An, Y. Wang, Y. Huang, X. Wang, Y. Liu, D. Xun, G. M. Church, Z. Dai, X. Yi, T.-C. Tang and C. Zhong, Chem. Rev., 2023, 123, 2349–2419 CrossRef CAS PubMed.
- T.-C. Tang, B. An, Y. Huang, S. Vasikaran, Y. Wang, X. Jiang, T. K. Lu and C. Zhong, Nat. Rev. Mater., 2021, 6, 332–350 CrossRef CAS.
- S. Gantenbein, E. Colucci, J. Käch, E. Trachsel, F. B. Coulter, P. A. Rühs, K. Masania and A. R. Studart, Nat. Mater., 2023, 22, 128–134 CrossRef CAS PubMed.
- J. Huang, S. Liu, C. Zhang, X. Wang, J. Pu, F. Ba, S. Xue, H. Ye, T. Zhao, K. Li, Y. Wang, J. Zhang, L. Wang, C. Fan, T. K. Lu and C. Zhong, Nat. Chem. Biol., 2019, 15, 34–41 CrossRef CAS PubMed.
- A. M. Duraj-Thatte, A. Manjula-Basavanna, J. Rutledge, J. Xia, S. Hassan, A. Sourlis, A. G. Rubio, A. Lesha, M. Zenkl, A. Kan, D. A. Weitz, Y. S. Zhang and N. S. Joshi, Nat. Commun., 2021, 12, 6600 CrossRef CAS PubMed.
- S. Molinari, R. F. Tesoriero, D. Li, S. Sridhar, R. Cai, J. Soman, K. R. Ryan, P. D. Ashby and C. M. Ajo-Franklin, Nat. Commun., 2022, 13, 5544 CrossRef CAS PubMed.
- Y. Wang, B. An, B. Xue, J. Pu, X. Zhang, Y. Huang, Y. Yu, Y. Cao and C. Zhong, Nat. Chem. Biol., 2021, 17, 351–359 CrossRef CAS PubMed.
- S.-Y. Kang, A. Pokhrel, S. Bratsch, J. J. Benson, S.-O. Seo, M. B. Quin, A. Aksan and C. Schmidt-Dannert, Nat. Commun., 2021, 12, 7133 CrossRef CAS PubMed.
- H. Zhai and J. Yeo, ACS Biomater. Sci. Eng., 2023, 9, 269–279 CrossRef CAS PubMed.
- Report on engineering biology: opportunities for the UK economy and national goals, https://www.gov.uk/government/publications/advice-on-engineering-biology/report-on-engineering-biology-opportunities-for-the-uk-economy-and-national-goals-html, (accessed 10th April, 2024) Search PubMed.
- AMSilk, https://www.amsilk.com/, (accessed 10th April, 2024).
- Bolt Threads, https://boltthreads.com/, (accessed 10th April, 2024).
- Spiber, https://spiber.inc/en/, (accessed 10th April, 2024).
- Tandem Repeat, https://tandemrepeat.com/, (accessed 10th April, 2024).
- K. R.-J. L. Dai and D. T. Williamson, US Pat., 11028146B2, 2021 Search PubMed.
- D. N. Breslauer, ACS Biomater. Sci. Eng., 2020, 6, 5980–5986 CrossRef CAS PubMed.
- M. Meleties, D. Britton, P. Katyal, B. Lin, R. L. Martineau, M. K. Gupta and J. K. Montclare, Macromolecules, 2022, 55, 1239–1247 CrossRef CAS.
- F. Burla, T. Sentjabrskaja, G. Pletikapic, J. van Beugen and G. H. Koenderink, Soft Matter, 2020, 16, 1366–1376 RSC.
- R. L. Martineau, A. V. Bayles, C.-S. Hung, K. G. Reyes, M. E. Helgeson and M. K. Gupta, Adv. Biol., 2022, 6, 2101070 CrossRef CAS PubMed.
- M. A. Morris, R. A. Bataglioli, D. J. Mai, Y. J. Yang, J. M. Paloni, C. E. Mills, Z. D. Schmitz, E. A. Ding, A. C. Huske and B. D. Olsen, Mol. Syst. Des. Eng., 2023, 8, 227–239 RSC.
- O. Gueta, O. Sheinenzon, R. Azulay, H. Shalit, D. S. Strugach, D. Hadar, S. Gelkop, A. Milo and M. Amiram, Front. Bioeng. Biotechnol., 2022, 10, 913057 CrossRef PubMed.
- J. Su, K. Zhao, Y. Ren, L. Zhao, B. Wei, B. Liu, Y. Zhang, F. Wang, J. Li, Y. Liu, K. Liu and H. Zhang, Angew. Chem., Int. Ed., 2022, 61, e202117538 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |