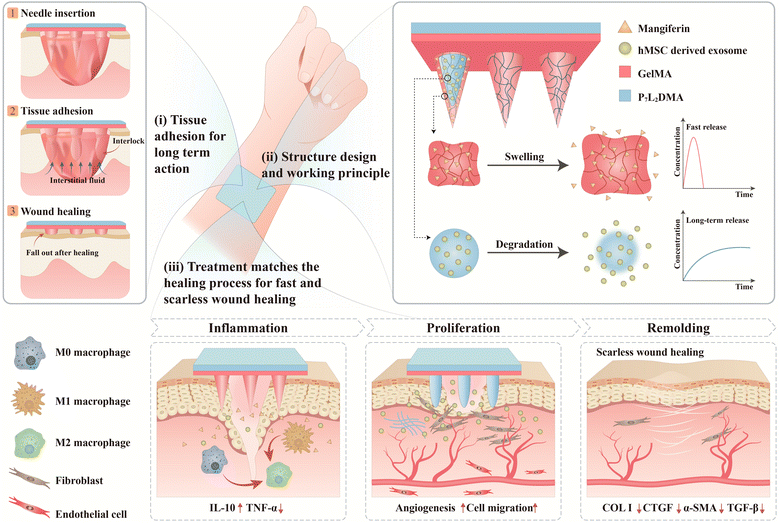
A differential-targeting core–shell microneedle patch with coordinated and prolonged release of mangiferin and MSC-derived exosomes for scarless skin regeneration†
Shang
Lyu
abc,
Qi
Liu
ab,
Ho-Yin
Yuen
ab,
Huizhi
Xie
d,
Yuhe
Yang
abe,
Kelvin Wai-Kwok
Yeung
 d,
Chak-yin
Tang
d,
Chak-yin
Tang
 f,
Shuqi
Wang
f,
Shuqi
Wang
 gh,
Yaxiong
Liu
i,
Bin
Li
gh,
Yaxiong
Liu
i,
Bin
Li
 *jk,
Yong
He
*jk,
Yong
He
 *c and
Xin
Zhao
*c and
Xin
Zhao
 *abe
*abe
aDepartment of Biomedical Engineering, The Hong Kong Polytechnic University, Hung Hom, Hong Kong SAR 999077, China
bDepartment of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hung Hom, Hong Kong SAR 999077, China. E-mail: xin.zhao@polyu.edu.hk
cState Key Laboratory of Fluid Power and Mechatronic Systems, School of Mechanical Engineering, Zhejiang University, Hangzhou, Zhejiang 310027, China. E-mail: yongqin@zju.edu.cn
dDepartment of Orthopaedics and Traumatology, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR 999077, China
eThe Hong Kong Polytechnic University Shenzhen Research Institute, Shenzhen, Guangdong 518057, China
fDepartment of Industrial & Systems Engineering, The Hong Kong Polytechnic University, Hung Hom, Hong Kong SAR 999077, China
gTianfu Jincheng Laboratory, City of Future Medicine, Chengdu 641400, China
hClinical Research Center for Respiratory Disease, West China Hospital, Sichuan University, Chengdu 610065, China
iJihua Laboratory, Foshan, Guangdong 528000, China
jMedical 3D Printing Center, Orthopedic Institute, Department of Orthopaedic Surgery, The First Affiliated Hospital, School of Biology & Basic Medical Sciences, Suzhou Medical College, Soochow University, Suzhou, Jiangsu 215006, China. E-mail: binli@suda.edu.cn
kCollaborative Innovation Center of Hematology, Soochow University, Suzhou, Jiangsu 215006, China
First published on 11th April 2024
Abstract
Microneedles for skin regeneration are conventionally restricted by uncontrollable multi-drug release, limited types of drugs, and poor wound adhesion. Here, a novel core–shell microneedle patch is developed for scarless skin repair, where the shell is composed of hydrophilic gelatin methacryloyl (GelMA) loaded with mangiferin, an anti-inflammatory small molecule, and the core is composed of hydrophobic poly (lactide-co-propylene glycol-co-lactide) dimethacrylates (PGLADMA) loaded with bioactive macromolecule and human mesenchymal stromal cell (hMSC)-derived exosomes. This material choice provides several benefits: the GelMA shell provides a swelling interface for tissue interlocking and rapid release of mangiferin at an early wound healing stage for anti-inflammation, whereas the PGLADMA core offers long-term encapsulation and release of exosomes (30% release in 3 weeks), promoting sustained angiogenesis and anti-inflammation. Our results demonstrate that the core–shell microneedle possesses anti-inflammatory properties and can induce angiogenesis both in vitro in terms of macrophage polarization and tube formation of human umbilical vein endothelial cells (HUVECs), and in vivo in terms of anti-inflammation, re-epithelization, and vessel formation. Importantly, we also observe reduced scar formation in vivo. Altogether, the degradation dynamics of our hydrophilic/hydrophobic materials enable the design of a core–shell microneedle for differential and prolonged release, promoting scarless skin regeneration, with potential for other therapies of long-term exosome release.
New conceptsWe propose a novel microneedle strategy (first-described in the literature) of coupling a hydrophilic hydrogel and hydrophobic polymer for anti-inflammation and long-term exosome release, not only offering a promising skin wound dressing for clinical treatment, but also providing a universal strategy for constructing next-generation multi-functional microneedles for diverse tissue regeneration. Specifically, we develop a core–shell microneedle using GelMA as the outer shell for mangiferin (an anti-inflammatory small molecule) loading and poly(lactide-co-propylene glycol-co-lactide) dimethacrylates (PGLADMA) as the inner core for human mesenchymal stromal cell (hMSC)-derived exosome loading. Such a hierarchical material strategy addresses the paradoxical relationship between drug-loading and its efficacy while preserving the benefits of microneedle design: on one hand, the GelMA shell swells and releases mangiferin rapidly, resulting in tissue-interlocking adhesion and early-stage anti-inflammation; on the other hand, the PGLADMA polymer can be photocrosslinked rapidly with low heat generation for efficient exosome loading, and prevents rapid diffusion of exosomes for sustained release along with material degradation. This sustained exosome release ability elevates its intrinsic properties for wound healing, that is, anti-inflammation via M2 polarization, improved tissue regeneration systematically, and enhanced angiogenesis and tube formation. Phenotypically, its sustained release can also promote angiogenesis and cell migration at the later proliferation/remodeling phase of wound healing. |
1. Introduction
The utilization of hydrogels as the base material of microneedles (e.g., gelatin methacryloyl (GelMA),1–3 hyaluronic acid methacrylate (HAMA),4–7 and chitosan8,9) has attracted widespread attention for tissue regeneration and wound healing due to their excellent biocompatibility and drug-loading capability for delivery of a wide range of cargos, including small molecules, growth factors,6,8 and extracellular vesicles.2,10–12 Despite the newfound capability of bioactive small or macromolecule encapsulation, the intrinsic properties of hydrogels, that is, their lossy molecular network, lead to paradoxically a fast degradation rate and eventually the burst release of encapsulated drugs (∼3 days),2,10,11,13,14 crippling their function as a delivery vehicle especially in the scenarios of chronic wounds where the healing process takes a course of usually several weeks or even months.15 Microneedle materials with a denser and more rigid polymer such as poly(lactic-co-glycolic acid) (PLGA), polycaprolactone (PCL), polystyrene, and polyglycolic acid16–21 are therefore developed for sustained drug release. Yet, these candidates cannot effectively load bioactive macromolecules with their harsh fabrication and drug-loading conditions, that is, the involvement of high temperature, organic solvents, and aspects that disrupt the bioactivity of the macromolecules.20,21 Although workaround solutions attempted to coat bioactive macromolecules on the surface of the needle tips17,22 or to enhance the porosity of microneedles for larger surface area and better drug loading capacity,19,23 these approaches again suffer from the excessively rapid drug release. In other words, the choice of the polymer (between a rigid polymer and hydrogel) is paradoxically tied to their drug-loading ability and drug efficacy, with better drug-loading capability being detrimental to drug efficacy, and vice versa. It is therefore imperative to develop a better material choice or strategy.In this study, we develop a core–shell microneedle patch using GelMA as the outer shell for mangiferin (an anti-inflammatory small drug molecule) loading and poly (lactide-co-propylene glycol-co-lactide) dimethacrylates (PGLADMA) as the inner core for bioactive macromolecule human mesenchymal stromal cell (hMSC)-derived exosome loading (Fig. 1). This hierarchical material strategy addressed the paradoxical relationship between drug-loading and its efficacy while preserving the benefits of microneedle design: on one hand, the GelMA shell swells and releases mangiferin rapidly, resulting in a tissue-interlocking adhesion and an early-stage anti-inflammation; on the other hand, the dense PGLADMA polymer can be photocrosslinked rapidly with low heat generation for efficient exosome loading, and can prevent rapid diffusion of exosomes for sustained release along with the material degradation. This sustained exosome release ability elevated its intrinsic beneficial properties for wound healing, that is, anti-inflammation via M2 polarization, improved tissue regeneration systematically, and enhanced angiogenesis and tube formation. Phenotypically, its sustained release can also promote angiogenesis and cell migration at the later proliferation/remodeling phase of wound healing. Under such core–shell design and therapeutic choices, the differential degradation as well as the encapsulation property of GelMA and PGLADMA in synergy allows a coordinated treatment of different wound healing phases in a single microneedle patch. We believe this novel microneedle strategy (first-described in the literature) of coupling a hydrophilic hydrogel and hydrophobic polymer for anti-inflammation and long-term exosome release not only offers a promising skin wound dressing for clinical treatment, as demonstrated in our experiments, but also provides a universal strategy for constructing next-generation multi-functional microneedles for diverse tissue regeneration.
2. Results and discussion
2.1. Synthesis of PGLADMA
We synthesized the PGLADMA as P7L2DMA where P7 denotes seven repeating units of propylene glycol (PPG) and L2 denotes two repeating units of lactide (LA) attached to either end of PPG (please see the Experimental section for protocol details). We previously demonstrated that this configuration possesses excellent in vitro and in vivo biocompatibility, capability to encapsulate and release bioactive macromolecules in the long term, and appropriate mechanical properties (with a compressive modulus of 50 MPa).24–28 The synthesized P7L2DMA was characterized by 1H NMR, which is similar to the previous data, and confirmed as P7L1.951.8MA (data not shown).2.2. Fabrication and characterization of the core–shell microneedle patch
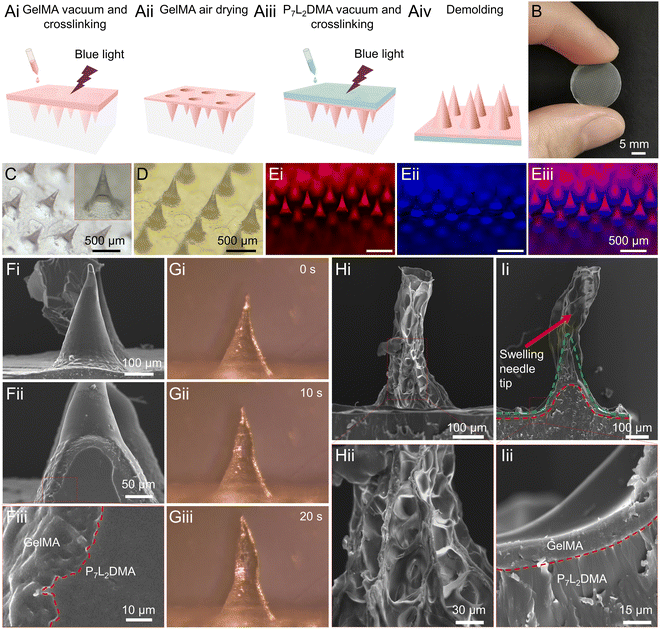
For the proposed core–shell microneedle system (Fig. 2A), we used PGLADMA as the core (inner-layer) material of the microneedle, and GelMA as the shell (outer-layer) material. As described in the previous section, this realized a PGLADMA inner-layer for slow release of exosomes and an adhesive GelMA outer-layer with the burst release property.29–32 We utilized a negative microneedle mold for the fabrication involving four steps: solidifying the GelMA in the mold by vacuum and photocrosslinking (Fig. 2Ai), air-drying the GelMA to form the outer-layer structure (Fig. 2Aii), solidifying the PGLADMA-in-GelMA by vacuuming and photocrosslinking (Fig. 2Aiii), and finally demolding the microneedle product (Fig. 2Aiv).We first examined the structure of the fabricated core–shell microneedle patch. Fig. 2B, C and D show the macroscopic image of the complete microneedle, the GelMA part of the microneedle under a microscope, and the full microneedle (with P7L2DMA) under a microscope, respectively. We measured each single needle having an average needle height of 403 ± 12 μm and a base diameter of 270 ± 0.40 μm. To visualize the internal structure of the microneedle, we stained the GelMA and P7L2DMA layer in red and blue, respectively. As shown in Fig. 2Ei–iii, the GelMA and P7L2DMA were clearly distinguished with a core–shell structure. Then, we examined the transitory interface between GelMA and P7L2DMA with a scanning electron microscope (SEM). As shown in Fig. 2F, the outer (GelMA) and inner (P7L2DMA) layers demonstrated a distinctive morphology, with the GelMA layer being rougher and more porous, and the P7L2DMA layer being more homogeneous. We believe that the irregularity of the GelMA layer facilitated its integration and binding with P7L2DMA in the two following aspects. Firstly, this allowed the P7L2DMA precursor to infiltrate into the GelMA shell during the vacuuming step (Fig. 2A), so that the two layers were interlocked during the crosslinking step. Secondly, after the demolding, the GelMA shell would continue to shrink due to the evaporation of residual water, and this led to shrinkage that tightly wrapped the P7L2DMA material. We also characterized the material distribution at the z-level by capturing a stained single needle tip at different heights (from 50 to 200 μm, Fig. S1, ESI†) using a confocal microscope. The GelMA shell (red) showed a perfect circular distribution around the P7L2DMA core (blue): as the section height increased and approached the needle point, the diameter of the core and shell decreased.
Since the morphological changes of the GelMA (shell) layer during swelling are important for both the tissue interlocking and the core–shell layer interlocking, we dipped the microneedle into water and examined the structural and morphological changes of a single needle tip under a microscope. Fig. 2Gi–iii show the changes of the general appearance of a single needle after absorbing water for different periods (i.e., 0, 10, and 20 s). The tip of the needle was greatly swollen within 20 s, while the base of the needle, which was mainly composed of P7L2DMA with a thin GelMA outer layer, was less swollen. Using SEM, we examined the swollen needle tip in Fig. 2H, and we further noted an emerging porous structure and volume expansion of the GelMA shell, which differs from the smooth structure in Fig. 2F. The dissected needle tip in Fig. 2I illustrates more details on the swelling behavior of the GelMA layer. Using the GelMA-P7L2DMA interface as reference (indicated by the red dashed line) and approximately the extent of the original GelMA layer (indicated by the green dashed line), we observed a significant swelling in the tip portion, while the base portion showed no obvious deformation, with the inner surface of the GelMA shell still adhered to the P7L2DMA firmly. Fig. 2Iii also illustrates the intact and tight bonding between the GelMA shell and the P7L2DMA core. These altogether indicate that the microneedle could swell at the tip of the needle for tissue interlocking and keep a tight bonding between the GelMA shell and the P7L2DMA core in the base, demonstrating successful fabrication of a functional core–shell structure for the goal of an adhesive and multi-step drug-releasing patch.
To assess the core–shell integrity of the microneedle after wound insertion, we conducted tissue insertion tests using chicken tissue as a soft and moist mimic of wound tissue (Fig. S2, ESI†). In Fig. S2A (ESI†), the core–shell microneedle was inserted into the chicken tissue and left in place for 1 min before removal. Fig. S2B (ESI†) shows slight swelling of the needle tips, with the water-absorbing component primarily concentrated at the tips that directly contact the tissue. Fig. S2Cii (ESI†) demonstrates no delamination at the interface between P7L2DMA and GelMA. Given that the water-absorbing segment is predominantly situated at the upper part of the needle tip, it does not impact the adhesion between the P7L2DMA core and the GelMA shell.
2.3. Mechanical and tissue adhesion properties of the core–shell microneedle patch
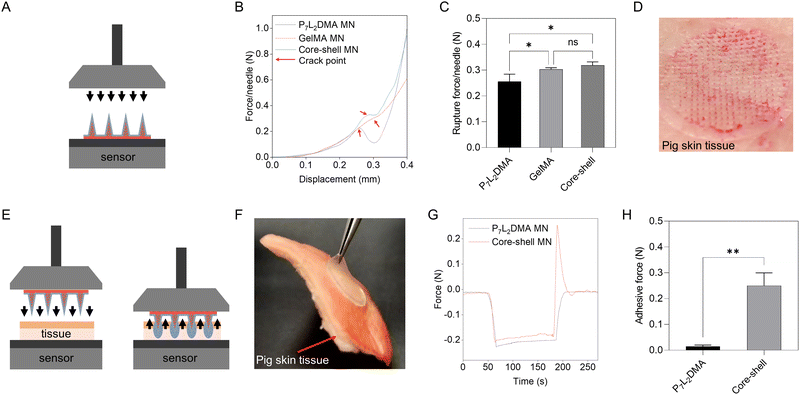
As the proposed microneedle system is used to insert into skin wounds (known to be softer and less elastic than the stratum corneum), a needle tip that is too stiff could easily cause secondary trauma to wounds.4 GelMA in its swollen state is sufficiently soft for tissue penetration, and also its dehydration can enforce a packing effect of its polymer network to increase its stiffness. Therefore, we opted for the use of the dehydrated GelMA shell to facilitate the wound insertion.33,34 After insertion, GelMA would first absorb the interstitial fluid, swell, and become softer when interlocking with the wound tissues, providing suitable tissue adhesive force and biocompatibility of the microneedles for the surrounding tissues. Fig. 3A illustrates our microneedle compression setup of the above properties. As shown in Fig. 3B, microneedles in all groups cracked at a similar displacement (∼280 μm), where the average rupture force of the pure P7L2DMA microneedle, the pure GelMA microneedle and the core–shell microneedle is 0.26 N per needle, 0.30 N per needle and 0.32 N per needle, respectively (Fig. 3C). In addition, the rupture force of the GelMA and core–shell microneedles had no significant difference, which indicates GelMA being the primary component that provided the insertion force and thus tissue-interlocking for our microneedle. We noted that the rupture force above is actually far beyond the required force to penetrate porcine skin (which is around 0.05 N per needle and it indicates sufficient wound insertion35), and indeed this was as demonstrated in Fig. 3D with an array of red dots. To further verify the dynamic tissue-interlocking mechanism of GelMA, we used a setup involving the following three steps: the insertion, stabilization, and pulling of the microneedle against a pork skin (Fig. 3E illustrates our setup). The microneedle was first inserted into the pork skin for 2 min to be attached to the skin surface. As shown in Fig. 3G, before the microneedle was pulled from the pork skin (0–180 s), the curve in the pure P7L2DMA and the core–shell groups fitted very well, showing a rapid loading increase to around 0.20 N during the insertion (0–60 s) and a steady loading value during the stabilization (60–180 s). However, during the pulling process (180–200 s), the core–shell microneedle showed a drastic rise of adhesive force in the loading curve, while the pure P7L2DMA microneedle showed no significant adhesion. Fig. 3H also shows that the average adhesive force of the core–shell microneedle was 0.25 N, which was ten times higher than that of the pure P7L2DMA microneedle (0.02 N). This strong interlocking force is attributed to the swelling GelMA shell layer that, without the P7L2DMA core, a pure GelMA microneedle would simply be too soft and fall apart during the pulling process (data not shown). This showed that our core–shell microneedle patch has sufficient tissue adhesive force for long-term drug delivery.2.4. In vitro drug release from the core–shell microneedle patch
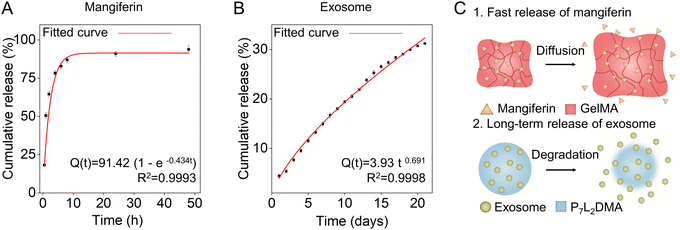
To fabricate the drug-loaded microneedles, we loaded exosomes into P7L2DMA solution and mangiferin into GelMA solution following the same process in Section 2.2 (Fig. 2Ai–iv), see the Experimental section for details. The drug encapsulation did not significantly influence the mechanical and tissue adhesion properties of the microneedle patch (data not shown). We found that 86.94 ± 2.25% of total mangiferin was released from GelMA rapidly within the initial 8 h and the cumulative percentage reached 93.84 ± 2.94% within 48 h (Fig. 4A). We chose Weibull function to fit the release data: Q(t) = Q∞ (1 – exp (−atb)), where Q(t) is the percentage of drug released at time t, Q∞ is the percentage of drug released at infinite time, a is the scale factor, and b is the shape factor. After the curve fitting, the release profile can be described by the equation Q(t) = 91.42 (1 – exp (−0.434t)), where b equals 1, suggesting the mangiferin release from GelMA obeying Fick's first law of diffusion.36–39 This rapid mangiferin release is expected to meet the requirement for an early stage of anti-inflammation, which usually lasts for 2–3 days.40For the exosome release, we started with characterizing our yield and we used a differential centrifugation method to extract the hMSC-derived exosomes (Fig. S3A, ESI†), see the protocol details in the Experimental section. After successful isolation, a nanoparticle tracking analyzer (NTA) was used to confirm the particle size distribution of the exosome samples (Fig. S3B, ESI†). The exosomes were found to have a modal diameter of 146.6 nm with a normal size distribution. They presented a distorted cup-shaped morphology (Fig. S3C, ESI†). The exosome samples were also positive for exosome marker proteins CD9 and ALIX while negative for GM130 (a Golgi marker exists only in cells but not in exosomes) (Fig. S3D, ESI†), indicating the high purity of the exosomes. These results supported that the exosomes were prepared successfully with exosomal molecular features and morphology. Then, we loaded the exosomes into P7L2DMA to prepare the core–shell microneedle patch (as illustrated in Fig. 2A) to characterize the release profile. Fig. 4B shows that the release rate of exosomes was found to be slower compared to mangiferin release, where the cumulative release of exosomes from P7L2DMA was 25.24 ± 0.66% in 14 days and 31.19 ± 0.27% in 21 days. Such slower exosome release is advantageous as the proliferation phase of wound healing usually lasts for 4–21 days.41 After the curve fitting, the release profile can be described by the equation Q(t) = 3.93 t0.691, where Q(t) is the amount of cumulative drug release at time t, following the Ritger–Peppas model with the release exponent n equaling 0.691. This implied that the exosomes were released from P7L2DMA by anomalous transport, based on a combination of diffusion and erosion events.42,43 In addition, the exosomes appeared intact after 21-day release from the microneedle patch, indicating that the P7L2DMA can preserve the structure of exosomes for long-term delivery (Fig. S3C and E compare the morphology of exosomes prior to encapsulation and after release, ESI†). To further verify the exosome release mechanisms, and confirm the relationship between exosome release kinetics and P7L2DMA degradation, the mass loss of P7L2DMA during degradation in PBS was measured. As shown in Fig. S4A (ESI†), 16.68 ± 0.51% and 21.90 ± 0.29% of P7L2DMA was eroded within 21 days and 56 days respectively. Compared with the exosome release rate of 31.19 ± 0.27%, which is higher than the mass loss of P7L2DMA, it further confirms that the release kinetics of exosome from P7L2DMA was based on a combination of diffusion and erosion. To further validate the hypothesis, we conducted tests to measure the release of lactate, which is presumed to be the primary hydrolysis product of P7L2DMA based on the established hydrolysis mechanism (Fig. S4B, ESI†).44 The results demonstrate a consistent release of lactate throughout the entire testing period, with the concentration of lactate in the degradation solution on day 56 recorded at 0.72 mg mL−1. This finding confirms the occurrence of material erosion during the water immersion of P7L2DMA, with lactate identified as one of the key degradation products. The above results suggested that our microneedle system could achieve a programmatic therapeutic delivery matching the wound healing process by promoting initial anti-inflammation and long-term angiogenesis and cell migration. In particular, this long-term exosome release is a breakthrough in using microneedles for biomacromolecule (e.g., proteins and cytokines) delivery, which usually lasts for 3 days due to the low-density polymer network of the microneedle patch materials.
2.5. In vitro anti-inflammation of the core–shell microneedle patch
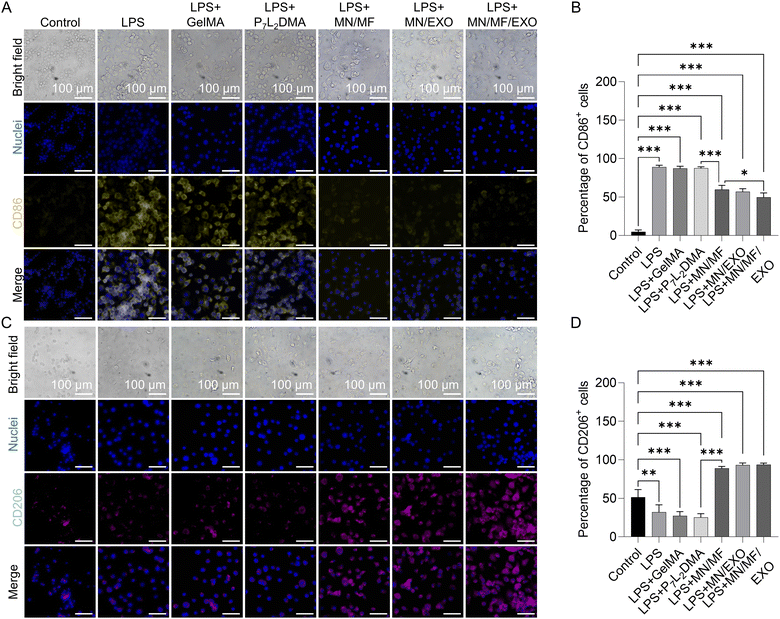
We next characterized the microneedle drug efficacy in vitro. Our target would be the prolonged inflammation of wound sites, which usually impedes the wound healing process and leads to extracellular matrix (ECM) degradation or even chronic wounds. Proper immunoregulation for anti-inflammation is not only beneficial to the angiogenesis that follows, but also to scar reduction.45–47 As noted in the previous section, we load mangiferin into the GelMA shell for its anti-oxidant, anti-inflammatory, and anti-microbial properties,48 all of which are beneficial at the early stage of wound healing; hMSC-derived exosomes are loaded into P7L2DMA for long-term anti-inflammation. Since macrophages are primary mediators of inflammation after injury, we first studied the immunomodulatory and anti-inflammatory effect in vitro using RAW 264.7 cells, namely by immunofluorescence staining of the M1 phenotype marker protein CD86, M2 phenotype marker protein CD206, anti-inflammatory factor IL-10, and proinflammatory factor TNF-α. Regarding determining the concentration of mangiferin and exosomes, we mainly refer to previous studies. For mangiferin, a concentration ranging from 20 to 200 μg mL−1 is usually used in the in vitro test of mangiferin.49 Many articles utilize mangiferin with 100 μg mL−1 and achieve a good anti-inflammatory and cytoprotective effect.50–53 Therefore, 100 μg mL−1 was chosen as the working concentration in this study. For exosome, most articles investigating exosomes choose 100–500 μg mL−1 as the working concentration.54–57 However, most of them were directly applied to the wound or encapsulated in hydrogels with fast release rate. Considering that the exosomes loaded in P7L2DMA would be slowly released, 1 mg mL−1 was chosen as the working concentration in this study. In summary, 100 μg mL−1 mangiferin and 1 mg mL−1 exosome were used in the following in vitro cell experiments.To investigate the anti-inflammatory effect of different components in microneedles, extraction solutions of different groups including control, LPS, P7L2DMA, GelMA, MN/MF (100 μg mL−1 mangiferin loaded microneedle), MN/EXO (1 mg mL−1 exosome loaded microneedle), and MN/MF/EXO (100 μg mL−1 mangiferin and 1 mg mL−1 exosome loaded microneedle) were used in the experiment (see Table S1 for details of the samples, ESI†). All extraction solutions were supplemented with 100 ng mL−1 LPS and were taken to treat the macrophages for 48 h. From the bright field images of Fig. 5A and C, macrophages in all treatment groups showed a larger cell area than the control groups, preliminarily indicating a phenotypic transformation. Fig. 5B shows the quantitative analysis of the percentage of CD86+ cells in different groups, where the LPS group (88.95 ± 2.15%) has a significantly higher percentage of CD86+ cells compared with the control group (4.99 ± 2.01%), proving a successful induction of the M1 transition by LPS. GelMA (87.33 ± 2.4%) and P7L2DMA (87.56 ± 1.63%) groups showed a similar CD86 level compared with the LPS group, indicating no obvious effect on immunoregulation. However, the MN/MF (59.77 ± 4.68%), MN/EXO (56.97 ± 3.48%), and especially the MN/MF/EXO (49.55 ± 5.06%) groups significantly decreased the proportion of CD86+ cells, suggesting that both mangiferin and exosomes can promote the M1-M2 transition of the macrophages. Similarly in Fig. 5D, GelMA (27.21 ± 4.74%) and P7L2DMA (25.22 ± 4.24%) groups showed a similar CD206 level with the LPS (31.94 ± 8.36%) group, indicating no obvious effect on immunoregulation, while the MN/MF (89 ± 2.04%), MN/EXO (93.13 ± 2.41%), and MN/MF/EXO (93.53 ± 1.93%) groups significantly increased the proportion of CD206+ cells compared with the control (51.44 ± 8.63%) and LPS group, indicating good immunoregulatory effects of the drugs. Fig. S5 (ESI†) also shows IL-10 and TNF-α immunostaining results to further confirm the anti-inflammatory effect of the core–shell microneedle. In brief, Fig. S5B (ESI†) shows the quantitative analysis of IL-10+ cells in different groups, where GelMA (23.16 ± 3.72%) and P7L2DMA (23.62 ± 3.72%) groups showed a similar IL-10 level compared with the LPS (23.48 ± 4.28%) group, indicating no obvious effect on anti-inflammation. However, the MN/MF (90.23 ± 1.84%), MN/EXO (92.83 ± 2.48%), and MN/MF/EXO (93.09 ± 2.56%) groups had higher level of IL-10 expression than the LPS group, proving good anti-inflammatory effect of these two drugs (mangiferin and exosomes). Similarly for TNF-α expression (Fig. S5D, ESI†), GelMA (70.04 ± 5.22%) and P7L2DMA (71.16 ± 5.62%) groups showed a similar TNF-α level with the LPS (69.48 ± 6.87%) group, indicating no obvious effect on anti-inflammation, while the MN/MF (19.20 ± 2.33%), MN/EXO (20.74 ± 2.63%), and especially MN/MF/EXO (10.65 ± 2.70%) groups significantly downregulated the TNF-α expression compared with the LPS group, indicating good anti-inflammatory effects of the drugs.
These results suggested that both the released mangiferin and exosomes showed good immunoregulatory effect and inhibited inflammation in vitro. More importantly, these results showed that P7L2DMA preserved the activity of exosomes, which is crucial for long-term treatment of wound healing. We thus believe the core–shell microneedle patch with programmed release of mangiferin and exosomes would achieve an effective and long-term anti-inflammation efficacy in our in vivo experiment in later sections.
2.6. In vitro angiogenesis and wound healing of the core–shell microneedle patch
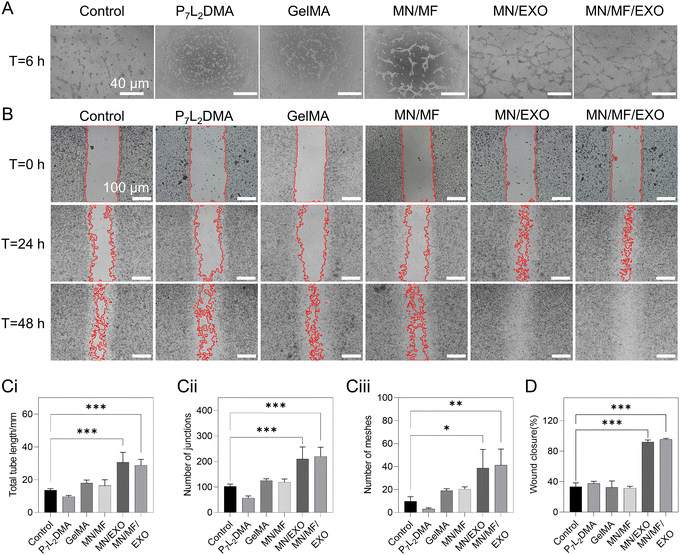
Angiogenesis and cell migration capacity are also critical in the wound healing process. For these two aspects, the tube formation of HUVECs and migration of L929 fibroblasts and HUVECs were investigated (Fig. 6 and Fig. S6, ESI†). Extraction solutions of six groups, including control, P7L2DMA, GelMA, MN/MF, MN/EXO, and MF/MF/EXO were used in the tests (see details of the samples in Table S2, ESI†). We cultured the extraction solutions for 6 h with HUVECs and first examined their tube formation. As shown in Fig. 6A, the MN/EXO group and MN/MF/EXO group both showed apparent tube formation in most areas; as for the control, GelMA, and MN/MF groups, only few short cell connections were observed. For quantitative measurement, we evaluated the total tube length, number of junctions, and number of meshes (Fig. 6Ci–iii). Consistent with our observations, all the three indexes were significantly higher in the MN/EXO group and MN/MF/EXO group compared to the rest of the groups, suggesting the pivotal role of exosomes in tube formation and angiogenesis. For cell migration, Fig. 6B shows the wound closure rate of different groups at 0, 24, and 48 h and Fig. 6D shows the quantitative analysis of the closure rate at 24 h. Similarly, for the cell migration experiment of the fibroblasts, the MN/EXO and MN/MF/EXO groups showed accelerated wound closure with average closure rates of 91.88 ± 2.34% and 95.40 ± 1.18%, respectively; the control, P7L2DMA, GelMA, and MN/MF groups showed no significant differences with wound closure rates of 33.32 ± 4.21%, 37.65 ± 2.27%, 32.46 ± 6.82%, and 31.47 ± 1.89%, respectively. In Fig. S6 (ESI†), the cell migration experiment of HUVECs shows similar results, with MN/EXO and MN/MF/EXO groups exhibiting the fastest wound closure rate among all groups. This indicates that the released exosomes were critical for promoting cell migration. All the above results suggested that the core–shell microneedle patch improved angiogenesis and migration of fibroblasts and HUVECs in vitro. The current drug loading concentration will also be used in the following in vivo tests.2.7. In vivo therapeutic effect of the core–shell microneedle patch
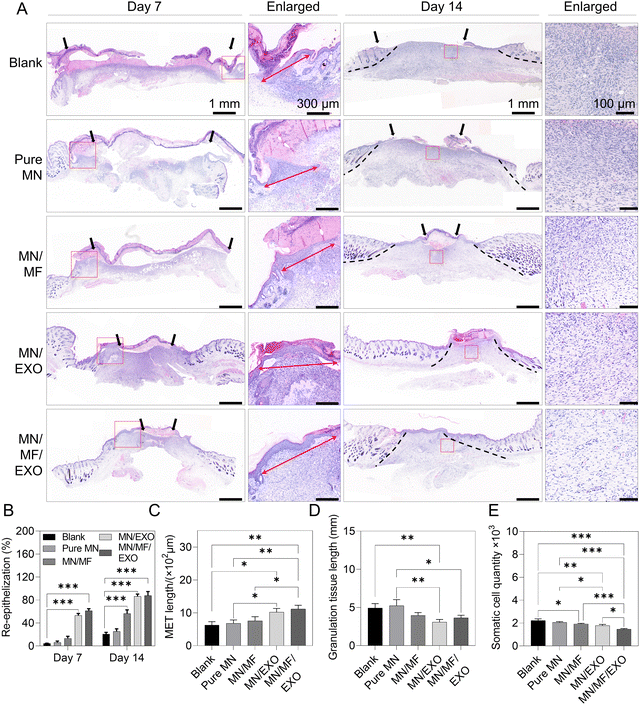
The excisional wound model is one of the common mouse models to investigate skin regeneration and scarless wound healing.58 For our experiment, we created an excisional full thickness wound on mouse dorsal skin. Then, pure MN, MN/MF, MN/EXO, or MN/MF/EXO groups were directly applied to the wound bed (the blank group had no treatment applied). Please see the Experimental section and Table S2 (ESI†) for the preparation and details of the samples. Wound images were taken on days 0, 7, and 14, and the wound areas were abstracted and are drawn in Fig. S7A and B (ESI†) to illustrate how the wounds change with time. We evaluated wound closure rate using the wound area on days 7 and 14 relative to the original wound area on day 0 (Fig. S7C, ESI†), and the resulting wound closure rate was as follows (lower value indicates quicker healing): 75.26 ± 2.00% and 10.32 ± 0.83%, respectively, in the blank group; 62.43 ± 2.09% and 7.07 ± 0.48%, respectively, in the pure MN group; 49.10 ± 1.38% and 4.11 ± 0.49%, respectively, in the MN/MF group; 31.57 ± 1.54% and 1.83 ± 0.26%, respectively, in the MN/EXO group; and 23.66 ± 1.50% and 0.49 ± 0.16%, respectively, in the MN/MF/EXO group. From the results, the MN/MF, MN/EXO, and MN/MF/EXO groups recovered over 50% on day 7 and over 95% on day 14, which was significantly faster than the blank and pure MN groups, indicating a critical effect of either exosomes or mangiferin, as well as when they were used together. Interestingly, the pure MN group also showed a better recovery than the blank group on both day 7 (62.43% healed in the blank group compared to 75.26% healed in the MN group) and day 14 (7.07% healed in the blank group compared to 10.32% in the MN group). This can be attributed to the hydration effect of the GelMA and the water retention effect of the P7L2DMA patch on the wound area (data not shown). Hydration is a crucial external factor essential for optimal healing, known as moist wound healing, which has been demonstrated to enhance epithelialization rate and reduce scarring.59,60 GelMA, an extensively studied biological hydrogel material, is recognized for its excellent hydration properties attributed to its swelling characteristics and high porosity.61 It efficiently absorbs interstitial fluid and exudate upon application to the wound, preventing rapid loss of wound moisture. Various forms of GelMA, such as hydrogel blocks,62,63 electrospun membranes,64 and microneedles,65,66 have been proven to accelerate wound closure. In our system, GelMA needle tips rapidly absorb wound interstitial fluid, providing localized moisturization. Moreover, the P7L2DMA in the microneedle substrate, which covers the entire wound, exhibits significant hydrophobicity as demonstrated in our previous research.24,27 When crosslinked, it forms a dense polymer network with a low water vapor transmission rate, effectively preserving the wound moisture and complementing the hydrating properties of GelMA. In conclusion, the combination of the GelMA shell and the P7L2DMA substrate design synergistically maintains wound moisture, resulting in a beneficial hydration effect.The wound healing effect of different groups was further analyzed through histological evaluation. Fig. 7A shows representative H&E staining images on days 7 and 14 of the wound tissues of different groups. Low-magnification images were used to observe the general re-epithelialization, inflammation, and granulation tissue morphology. Quantitative re-epithelialization ratio was also measured in Fig. 7B. On day 7, the MN/MF/EXO group (61.30 ± 3.51%) and MN/EXO group (53.00 ± 3.61%) demonstrated significantly more re-epithelialization than the other groups (blank 4.33 ± 0.58%; pure MN 6.00 ± 2.65%; MN/MF 13.33 ± 3.51%); on day 14, three groups (MN/MF/EXO 87.67 ± 7.10%; MN/EXO 86.33 ± 4.04%; MN/MF 56.33 ± 6.51%) showed significantly more re-epithelialization than the other groups (blank 20.67 ± 3.06%; pure MN 25.33 ± 4.51%). In addition, the MET length, which also represents the re-epithelialization capacity, was marked in the enlarged view of images on day 7 and quantitatively analyzed in Fig. 7C. According to the re-epithelialization data, MN/MF/EXO and MN/EXO groups showed longer MET length among all the groups, while the other three groups showed no significant difference on day 7. These results proved that mangiferin and especially exosomes greatly promoted cell migration in the wound bed, leading to a faster re-epithelialization. After 14 days of surgery, the granulation tissue maturation was observed in the MN/MF, MN/EXO, and especially MN/MF/EXO groups, with the presence of paler scars and absence of inflammatory cells from the enlarged view. Moreover, hair follicles were generated in the MN/MF/EXO group, suggesting a successful recovery of functional and healthy skin tissues in the MN/MF/EXO group, which is consistent with the systemic regenerative effect of exosomes reported in the literature.67 As the scar formation is mainly due to the excessive matrix deposition of somatic cells like fibroblasts and myofibroblasts, somatic cell quantity was adopted as a preliminary indicator for assessing scar formation.68,69 Quantitative results indicate that significant decrease in granulation tissue length and somatic cell quantity occurred in the MN/MF, MN/EXO, and MN/MF/EXO groups (Fig. 7D and E), indicating a trend in reducing scar formation by these treatments. Overall, our results demonstrated that mangiferin and exosomes greatly facilitated wound closure and re-epithelization of skin wounds.
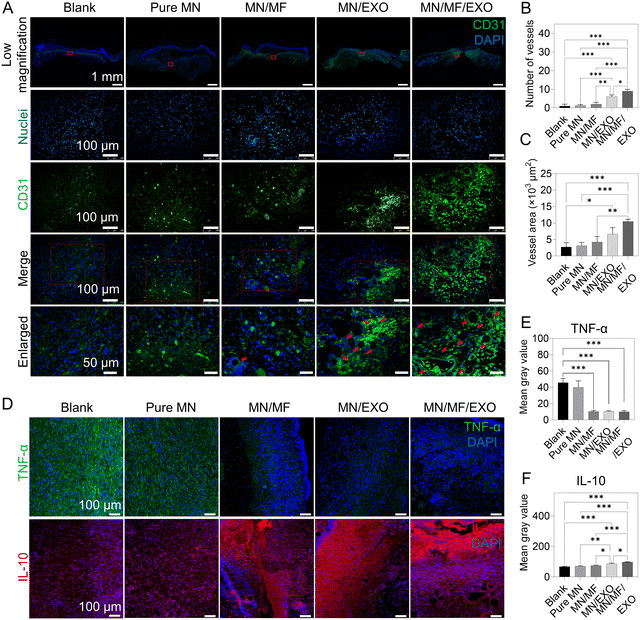
To elucidate the underlying mechanism of accelerated wound healing, we examined the signs of early inflammation and angiogenesis in tissue sections on day 7 using CD31 (angiogenesis marker), IL-10 (anti-inflammatory marker), and TNF-α (pro-inflammatory marker) immunostaining. Fig. 8A shows the representative images of CD31 in different groups, and the blood vessel number and vessel area were analyzed in Fig. 8B and C. Similar to our other in vivo results, the exosomes provided excellent systemic regeneration effect, making the MN/MF/EXO group having the most blood vessel number and vessel area compared to the other groups, followed by the MN/EXO group. The remaining groups showed no significant difference in the CD31 expression. For anti-inflammation, Fig. 8D shows the representative images of IL-10 and TNF-α in different groups and average fluorescence intensity was quantified in Fig. 8E and F. We found that mangiferin and exosomes significantly decreased the TNF-α expression and elevated the IL-10 expression, indicating the presence of anti-inflammatory response. The combination of mangiferin and exosomes had a stronger anti-inflammatory effect compared to either of the individual treatments, indicating a synergetic long-term anti-inflammatory effect when the two treatments were combined. Altogether, our core–shell microneedles successfully demonstrated the ability to inhibit inflammation and promote angiogenesis in vivo.
We further verified the anti-inflammatory and angiogenesis effect via real-time quantitative polymerase chain reaction (RT-qPCR) and western blot by choosing several typical markers (Fig. S8, ESI†). This includes vascular endothelial growth factor (VEGF), a classic marker for neovascularization, platelet derived growth factor (PDGF), a classic marker for stability of neovascularization, and nuclear factor-kappa B (NF-κB), a key mediator of its signaling pathway, associated with exosome-induced angiogenesis in endothelial cells. From the results (Fig. S8, ESI†), the expression of VEGF, PDGF, and NF-κB was significantly higher in the MN/MF/EXO and MN/EXO groups than the other three groups, indicating that our MSC-derived exosomes released from the core–shell microneedles could elevate the VEGF and PDGF expression and activate the NF-κB signaling pathway for promoting angiogenesis. This was consistent with the immunostaining results, which demonstrated the angiogenesis activity of the released exosome. We also evaluated the two inflammatory markers, IL-10 and TNF-α, to further support our previous data (Fig. 8). As shown in Fig. S8 (ESI†), IL-10 showed higher expression and TNF-α showed lower expression only in the MN/MF/EXO group, indicating that our mangiferin and MSC-derived exosomes released from the core–shell microneedle patch can inhibit inflammation in vivo.
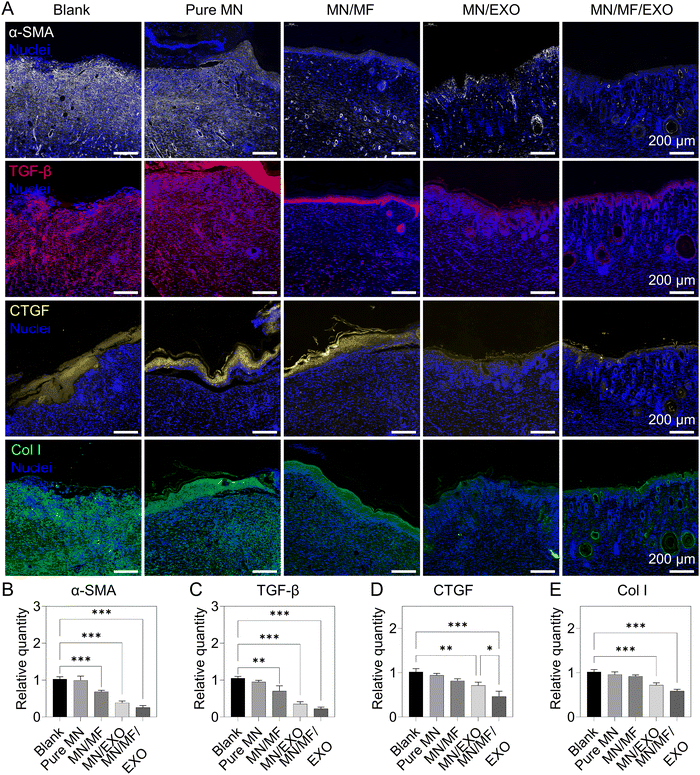
Apart from a restoration of regulatory processes like anti-inflammation and angiogenesis, scarless skin wound healing is also considered as another important indication of functional recovery of regenerated skin. We hypothesized that exosomes and mangiferin combined can inhibit myofibroblast activation, regulate ECM deposition and its microenvironment for less scar formation.70–72 Therefore, we evaluated the scar formation in the regenerated wounds of different groups after 14 days. The following scar-related proteins were selected for immunostaining: α-smooth muscle actin (α-SMA), representing myofibroblasts that secret ECM and are responsible for wound contraction and scar formation;73 transforming growth factor-β (TGF-β), representing ECM deposition and the associated stress environment in the wound area; connective tissue growth factor (CTGF), a key mediator for pathways involving myofibroblast activation, ECM deposition, and fibrosis, as well as a key marker for hypertrophic scars and keloids;74–76 and collagen type I (Col I), a major component of scar in general. Fig. 9 illustrates the resulting wound site of different microneedle groups. As shown in Fig. 9A and the quantitative results in Fig. 9B–E, the MN/MF/EXO group showed significantly lower expression in all markers compared to the other groups, following the MN/EXO group, and then the MN/MF group. Several structural differences can also be observed among different microneedle groups. For instance, TGF-β was not observed in the MN/MF/EXO group, persisted slightly in the newly formed epidermis of MN/EXO and MN/MF groups, and distributed abundantly and evenly in the granulation tissue of the blank and pure MN groups. Similarly, CTGF was only present as a thin layer in the epidermis of the MN/MF/EXO group, which was otherwise thicker in the other groups in the following order: the MN/EXO group, the MN/MF group, the pure MN group, and the blank group. In addition, the MN/EXO and MN/MF/EXO groups had a sparse distribution of Col I also with signs of hair follicles (round or oval-shaped structures) appearing in the wound area, an indication of functional recovery. On the contrary, there was an abundance of Col I in the granulation tissue (and hence scarring) of the blank, pure MN, and MN/MF groups. All these results suggested that our core–shell microneedle patch could greatly reduce the scar formation by regulating several key cytokines, including α-SMA, TGF-β, and CTGF. In particular, the hMSC-derived exosomes played a vital role during this process with systemic improvement and regulation of scar-related cytokines, and with mangiferin primarily affecting the α-SMA and TGF-β expression for effective anti-inflammation.
Altogether, the combination of exosomes, mangiferin, and the core–shell microneedle patch greatly enhanced wound healing by promoting anti-inflammation, angiogenesis, cell migration, and scar reduction. Attributed to the system design and material selection of our core–shell microneedle patch, these two drugs could be delivered programmatically to match the two different phases of wound healing: the inflammatory phase and the regenerative phase. Importantly, our choice of the core material, P7L2DMA, can encapsulate and release biomacromolecules in a sustained way without losing their activity, enabling the long-term effect of exosomes for promoting anti-inflammation, angiogenesis, cell migration, and ECM reorganization at later stages of wound healing (i.e., proliferation, and remodeling). This is a breakthrough to the current hydrogel-based microneedle systems which can only release encapsulated therapeutics in a short term and cannot address the full process of wound healing.
3. Conclusions
In summary, we have fabricated a PGLADMA/GelMA core–shell microneedle patch with mangiferin loaded in the shell and hMSC-derived exosomes loaded in the core to achieve scarless skin wound healing. The benefit of this core–shell structure is the rapid release of mangiferin from the GelMA shell for early anti-inflammation, and prolonged release of exosomes from the PGLADMA core for angiogenesis and cell proliferation. Our results showed that such a differential release is achieved successfully, including a significant effect on immunoregulation at the wound site in vivo with the suppression of the pro-inflammatory cytokine TNF-α and anti-inflammatory cytokine IL-10, enhanced cell migration and tube formation of HUVECs in vitro, and neovascularization with markers of the NF-κB signaling pathway (α-SMA, TGF-β, CTGF, and Col I) in vivo. Altogether, our core–shell microneedle not only realized a scarless skin regeneration strategy utilizing the release dynamics of the material properties and microneedle design, but also demonstrated for the first time a hydrogel strategy for prolonged encapsulation release of exosomes for therapeutic purposes. This provides an important guideline for future biomaterial studies taking advantage of the long-term benefits of exosome treatments, especially using hydrogel microneedles, for multi-phase tissue regeneration. However, there are still limitations to the current research. While the 14-day in vivo results have demonstrated the anti-scar efficacy of our core–shell microneedle, future animal experiments with a longer duration, such as 28 days, will be conducted to better showcase the scar reduction effect.4. Experimental section
4.1. Materials
GelMA (EFL-GM-60) was bought from Suzhou Intelligent Manufacturing Research Institute (Suzhou, China). Mangiferin (M3547), lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) and propylene glycol (PPG) were bought from Sigma-Aldrich (Hong Kong). Lactide (LA), methacryloyl chloride (MAC), trimethylamine (TEA), stannous octoate, trimethylamine (TEA), Irgacure 819, and 2-hydroxyethyl methacrylate (HEMA) were purchased from Macklin Reagent (Shanghai, China). Dichloromethane (DCM) and diethyl ether were purchased from Duksan (Hong Kong). Phosphatic buffer solution (PBS), Dulbecco's modified Eagle medium (DMEM), minimum essential medium α (MEM α), fetal bovine serum (FBS), and penicillin/streptomycin (PS) were bought from Gibco (Hong Kong). Exosome depleted fetal bovine serum (FBS) was purchased from Vivacell (Shanghai, China). Lipopolysaccharide (LPS), 4′,6-diamidino-2-phenylindole (DAPI), and Nile red dye were purchased from Thermo Fisher (Hong Kong).4.2. Synthesis of P7L2DMA
The synthetic process has been presented in our previous work.27 In brief, under nitrogen protection, 34.4 g PPG and 23 g LA underwent ring-opening polymerization using stannous octoate as a catalyst for 6 h at 150 °C. Then 4.22 g MAC and 4.05 g TEA were added in drops at 0 °C to conjugate methacrylate groups to the end of the polymer chain. Both MAC and TEA were diluted in DCM. To remove TEA·HCL generated during this process, the product (mixture of P7L2DMA and TEA·HCL) was dissolved in diethyl ether followed by filtration. Finally, to eliminate the residual solvent in the oil solvent phase, 2 h of rotary evaporation was used and pure P7L2DMA was obtained. To form photocrosslinkable P7L2DMA precursor, P7L2DMA was mixed with a HEMA solution containing Irgacure 819 (10 wt% of Irgacure 819 in HEMA) at a ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w).
1 (w/w).
4.3. Isolation of exosomes
Human mesenchymal stem cells (hMSCs) were chosen as the cell source of exosome extraction. hMSCs were cultured in MEM α containing 10% (v/v) FBS and 1% PS. All cells were cultured under humidified conditions at 37 °C in a 5% CO2 incubator and culture medium was replaced every two days. After the cells reached 80% confluence in the culture flask, the culture medium was changed to conditioned medium consisting of MEM α, 10% (v/v) exosome depleted FBS, and 1% (v/v) PS. The cells were incubated in the conditioned medium for another 48 h. The conditioned medium was then collected and subjected to centrifugation at 4 °C and 800g for 5 min, followed by 2000g for 20 min. The resulting supernatant was then filtered using a 0.22 μm pore-sized filter to obtain the clarified conditioned medium. Finally, the clarified conditioned medium underwent two rounds of centrifugation at 4 °C and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min each to obtain the pure exosomes.
000g for 70 min each to obtain the pure exosomes.
4.4. Nanoparticle tracking analyzer (NTA)
Exosome size profile and concentration were assessed using a NanoSight NS300HSBF (Malvern Panalytical GmbH, Germany) instrument featuring a CMOS camera and a 405 nm laser source. Samples were diluted using PBS to a concentration of 107–108 vesicles mL−1, with 50–200 vesicles observed per frame. A 60-second video was recorded using a NanoSight device. Data were analyzed using NanoSight NTA 3.1 software with a detection threshold of 5.4.5. Transmission electron microscope (TEM)
TEM was employed to examine the morphology of exosomes. 2 μL exosome solution was placed on carbon-coated grids. Subsequently, 1% (v/v) glutaraldehyde solution was used to fix the exosomes on the grid for 5 min. Double-distilled water was then used to wash the fixed exosomes several times. Excess water was dried under ambient conditions. To stain the samples, uranyl oxalate at pH 7 was applied for 5 min, followed by incubation in a mixture of 4% v/v uranyl acetate (UA) and 2% v/v methylcellulose on ice for 10 min. Excess liquid was dried in air. The fixed samples were observed using a TEM (JEOL, JEM-2010).4.6. Fabrication process of microneedles
The whole microneedle system was fabricated by a multistep micromolding technique. Firstly, to prepare mangiferin loaded GelMA solution for shell construction, 10% (w/v) GelMA solution was prepared by dissolving GelMA in PBS containing 0.5% (w/v) LAP. Then mangiferin was added into 10% (w/v) GelMA solution to achieve a final concentration of 2 mg mL−1. Secondly, to prepare exosome loaded P7L2DMA solution for core construction, P7L2DMA precursor and 10% (w/v) Irgacure 819 in hydroxyethyl methacrylate (HEMA, Sigma Aldrich, Hong Kong) solution were mixed at a ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w), forming the photocrosslinkable P7L2DMA solution. Then exosome solution was poured into the photocrosslinkable P7L2DMA solution (500 μg exosome in 1 mL P7L2DMA solution) and put into a freeze dryer to remove the water overnight. Thirdly, 200 μL mangiferin loaded GelMA solution was added into a PDMS mold (Y61, Weixinyiyao Technology, China) and put into a vacuum chamber to remove the bubbles between the mold and the solution for 3 min. Then a 405 nm blue light was used to crosslink the mangiferin loaded GelMA solution. The crosslinked GelMA solution along with the mold was put into a thermostat to dehydrate the GelMA at 40 °C for about 1 h, forming a layer of drug loaded GelMA shell on the bottom of the mold. Fourthly, 200 μL exosome loaded P7L2DMA solution was poured into the mold and put into a vacuum chamber to remove the bubbles for over 30 min. A 405 nm blue light was also used to crosslink P7L2DMA. Finally, the core–shell microneedle patch was demolded mildly from the mold.
1 (w/w), forming the photocrosslinkable P7L2DMA solution. Then exosome solution was poured into the photocrosslinkable P7L2DMA solution (500 μg exosome in 1 mL P7L2DMA solution) and put into a freeze dryer to remove the water overnight. Thirdly, 200 μL mangiferin loaded GelMA solution was added into a PDMS mold (Y61, Weixinyiyao Technology, China) and put into a vacuum chamber to remove the bubbles between the mold and the solution for 3 min. Then a 405 nm blue light was used to crosslink the mangiferin loaded GelMA solution. The crosslinked GelMA solution along with the mold was put into a thermostat to dehydrate the GelMA at 40 °C for about 1 h, forming a layer of drug loaded GelMA shell on the bottom of the mold. Fourthly, 200 μL exosome loaded P7L2DMA solution was poured into the mold and put into a vacuum chamber to remove the bubbles for over 30 min. A 405 nm blue light was also used to crosslink P7L2DMA. Finally, the core–shell microneedle patch was demolded mildly from the mold.
4.7. Microneedle morphology characterization
The general morphology of the microneedles was observed using an optical microscope (Nikon, Japan) and scanning electron microscope (SEM, Tescan VEGA3, Czech Republic) after gold sputtering using a sputter coater. To observe the core–shell structure of the microneedle, a fluorescence microscope (Nikon, Japan) and a confocal microscope (TCS SPE, Leica) were used. For shell staining, DAPI was added into GelMA solution during the fabrication process. For core staining, Nile red dye was mixed in the P7L2DMA solution during the fabrication process.4.8. In vitro drug release test
For testing the mangiferin release from GelMA, a standard curve which determined the concentration of mangiferin in PBS was obtained at first. A series of mangiferin solutions, with concentrations of 500, 250, 125, 62.5, 31.25, 15.625, 7.8, 3.9, 1.95, and 0.976 μg mL−1, were prepared by the serial dilution method in a 96-well plate. The optical density value of these solutions was detected using a microplate reader (Thermo Fisher, Hong Kong). The standard curve was drawn by Origin software, reflecting the relationship between the concentrations and optical density values. Subsequently, 2 mg mL−1 mangiferin/GelMA solution was prepared as mentioned above. Each sample was prepared by dehydrating 200 μL crosslinked mangiferin/GelMA solution. Finally, samples were immersed in 5 mL PBS for the drug release test. The PBS was fully refreshed at each time point (30 min, 1 h, 2 h, 4 h, 6 h, 8 h, 10 h, 12 h, 24 h, 48 h, 72 h, and 96 h) and the extracted water solution was used for optical density value measurement at 370 nm and calculation of the quantity of the released mangiferin. The cumulative release rate at each time point = cumulative release quantity/total quantity (0.4 mg). For testing the exosome release from P7L2DMA, 500 μg mL−1 exosome/P7L2DMA mixture was prepared. Each sample was prepared by crosslinking 200 μL exosome/P7L2DMA mixture and subsequent immersion in 5 mL PBS. PBS was fully refreshed at each time point (from day 1 to day 21) and the extracted water solution was used for concentration measurement using an exosome ELISA kit (Mlbio, China) according to the user manual. The cumulative release rate at each time point = cumulative release quantity/total quantity (100 μg).4.9. In vitro degradation test
For P7L2DMA degradation test, each sample was made into a disc shape with a radius of 8 mm and a thickness of 5 mm using a mold. The samples were firstly weighed using an analytical balance for initial weight, and then immersed in a 50 mL centrifuge tube containing 10 mL of PBS. After incubation for different time periods (7, 14, 21, 28, 35, 42, and 56 days) at 37 °C, the samples were taken out from the solution, and dried in a vacuum drier at room temperature, and weighed using an analytical balance. The mass loss (ML) at each time point was calculated according to the following equation: ML (%) = (M0 − Mt)/M0 × 100, where M0 is the initial weight of the samples, and Mt is the dry weight of samples at different time points. As low-polymerized lactic acid is the primary degradable chain segment in P7L2DMA, samples of the degradation solution were collected at various time points over 56 days for lactate detection using the lactate assay kit (ab65330, Abcam). The cumulative concentration of lactate at each time point was determined following the instructions provided in the user manual.4.10. Mechanical test
A mechanical testing system (ElectroForce 3200, Bose, America) was used for the compression test and tissue adhesion test of the microneedles. For the compression test, a piece of microneedle sample was attached to the stainless-steel platform (needles facing up) using glue, and the sensor probe approached the microneedle at a velocity of 5 mm min−1. The initial distance between the probe and the needle tips was 1 cm. All data were treated and analyzed by Origin software. For the tissue adhesion test, a piece of microneedle sample was attached to the sensor probe (needles facing down), and a piece of pig skin was attached to the stainless-steel platform using glue. The sensor probe was programmed to move down for 1 cm initially at a velocity of 5 mm min−1. Then the sensor probe was set to hold the position for 2 min for stabilization and finally be pulled up at a velocity of 5 mm min−1. The initial distance between the needle tips and the pork skin was 0.4 cm. All data were analyzed by Origin software. An insertion test was conducted using a pork skin tissue. The needle tip was firstly stained with red dye and then inserted into the pork skin directly.4.11. In vitro anti-inflammatory assay
RAW 264.7 macrophages were cultured using complete DMEM. The cells were incubated under humidified conditions at 37 °C and 5% CO2 in an incubator; the culture medium was refreshed every two days. Seven groups, including control (PBS), LPS, LPS + GelMA, LPS + P7L2DMA, LPS + MN/MF, LPS + MN/EXO, and LPS + MN/MF/EXO, were involved in the test. The concentration of loaded mangiferin and exosomes was determined based on the release kinetics of P7L2DMA and GelMA. Except the control and the LPS group, all groups used extraction solutions, made by immersing the samples in full DMEM for two weeks, for investigation. Then, except the control group, LPS was added to all groups at a concentration of 100 ng mL−1 as working medium. Finally, RAW 264.7 macrophages were pre-cultured for 24 h in full DMEM, followed by the medium changing to the working medium of the seven groups. The cells were cultured for another 48 h and prepared for immunostaining. 4% paraformaldehyde was used to fix cells for 30 min, then permeabilized with 0.5% Triton X-100 for 10 min and blocked for 1 h under room temperature. Subsequently, samples were incubated with primary antibodies, including CD86 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, ab239075, Abcam), CD206 (1
500, ab239075, Abcam), CD206 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900, ab64693, Abcam), TNF-α (1
900, ab64693, Abcam), TNF-α (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, ab183218, Abcam) and IL-10 (1
500, ab183218, Abcam) and IL-10 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, ab189392, Abcam), overnight at 4 °C followed by incubation with secondary antibodies, Alexa Fluor® 488 (1
500, ab189392, Abcam), overnight at 4 °C followed by incubation with secondary antibodies, Alexa Fluor® 488 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, ab150157, Abcam), for 1 h at room temperature. The samples were washed three times with PBS and then examined using a fluorescence microscope (Nikon, Japan).
500, ab150157, Abcam), for 1 h at room temperature. The samples were washed three times with PBS and then examined using a fluorescence microscope (Nikon, Japan).
4.12. Tube formation assay
Human umbilical vein endothelial cells (HUVECs) were cultured in full DMEM containing 10% (v/v) FBS and 1% PS. The cells were incubated under humidified conditions at 37 °C in a 5% CO2 incubator and culture medium was replaced every two days. Six groups, including control (PBS), P7L2DMA, GelMA, MN/MF, MN/EXO, and MN/MF/EXO, were involved in the test. Except the control group, all groups used extraction solutions, made by immersing the samples in full DMEM for two weeks, for investigation. HUVECs were plated at a density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 on a 24-well plate that had been precoated with 250 μL Matrigel and grown to confluency. Then the culture medium in each group was replaced by the solution prepared as mentioned above. The tube formation was imaged using an inverted microscope (Nikon, Japan) at three time points (4, 6, and 18 h) and analyzed with ImageJ software applying the angiogenesis analyzer plugin.
000 cells per cm2 on a 24-well plate that had been precoated with 250 μL Matrigel and grown to confluency. Then the culture medium in each group was replaced by the solution prepared as mentioned above. The tube formation was imaged using an inverted microscope (Nikon, Japan) at three time points (4, 6, and 18 h) and analyzed with ImageJ software applying the angiogenesis analyzer plugin.
4.13. Scratch wound healing assay
L929 fibroblasts and HUVECs were both cultured in full DMEM containing 10% (v/v) FBS and 1% PS. The cells were incubated under humidified conditions at 37 °C in a 5% CO2 incubator and culture medium was replaced every two days. Six groups, including control (PBS), P7L2DMA, GelMA, MN/MF, MN/EXO, and MN/MF/EXO, were involved in the test. Except the control group, all groups used extraction solutions which were made by immersing the samples in full DMEM for two weeks, for investigation. L929 fibroblasts were grown at a density of 1 × 105 cells per cm2 in a 24-well plate and grown to confluency. Then wounds were scraped in each well using a 200 μL pipette tip. The wound area was imaged using a microscope (Nikon, Japan) at five time points (0, 4, 12, 24, and 48 h) and analyzed with ImageJ software. HUVECs were grown at a density of 1 × 105 cells per cm2 in a 6-well plate and grown to confluency. Then wounds were scraped in each well using a 200 μL pipette tip. The wound area was imaged using a microscope (Nikon, Japan) at four different time points (0, 4, 12, and 24 h) and analyzed with ImageJ software.4.14. In vivo therapeutic test
The Animal Subjects Ethics Sub-committee of the Hong Kong Polytechnic University (Approval No. 21-22/83-BME-R-STUDENT) granted ethical approval for all animal experiments conducted in this study. Male C57BL/6J mice weighing approximately 20 g and aged 7 weeks were employed to establish the excisional full thickness wound model. Prior to the procedure, the mice were administered anesthesia using 1.5% isoflurane in 100% O2 gas anesthesia. A circular incision with a diameter of 18 mm was then made along the midline of the mouse dorsal skin, encompassing the epidermis, dermis, subcutis, and muscularis layers. A total of 80 mice were utilized and allocated into five groups. In the blank group, the wound bed was treated with 100 μL PBS. For the pure MN group, microneedles with the GelMA shell and the P7L2DMA core were applied. For the MN/MF group, microneedles with the mangiferin (100 μg mL−1) loaded GelMA shell and P7L2DMA core were applied. For the MN/EXO group, microneedles with the GelMA shell and exosome (1 mg mL−1) loaded P7L2DMA core were applied. For the MN/MF/EXO group, microneedles with the mangiferin loaded GelMA shell and exosome loaded P7L2DMA core were applied. Mice were sacrificed for histological analysis on days 7 and 14 post operation.4.15. Histology and wound healing evaluation
Wound images were taken on days 0, 7, and 14. Wound area was measured using ImageJ software by recognizing the border of the wound. Diagrams of wound areas at different time points were also generated using ImageJ software by setting the wound border as the region of interest (ROI). The tissue samples were subjected to hematoxylin–eosin (H&E) staining to visualize the newly generated skin tissue. Quantitative data, including the migrating epidermal tongue (MET) length, granulation tissue length, re-epithelialization ratio, and somatic cell quantity, were measured or counted using ImageJ. MET is the newly formed epithelium. Re-epithelialization ratio (%) = distance covered by the newly formed epithelium/wound width × 100%. TNF-α, IL-10, CD31, TGF-β, CTGF, Col I, and Col III immunostaining (Servicebio, China) was conducted to evaluate the angiogenesis, inflammation, and scar formation.4.16. Western blot analysis
Isolated exosomes, mouse skin homogenates and cell lysates were quantitated by bicinchoninic acid protein (BCA) assay. The samples were then separated on an 8–12% sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore). The protein loaded PVDF membranes were blocked for 30 min at room temperature using 5% skim milk. Following blocking, the membranes were incubated with primary antibodies, including CD9 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, ab223052, Abcam), ALIX (1
1000, ab223052, Abcam), ALIX (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, ab275377, Abcam), GM130 (1
1000, ab275377, Abcam), GM130 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, PA5-95727, Invitrogen), ACTIN (1
500, PA5-95727, Invitrogen), ACTIN (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000, GB15001, Servicebio), VEGFR1 (1
2000, GB15001, Servicebio), VEGFR1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, GB114200, Servicebio), PDGF B (1
1000, GB114200, Servicebio), PDGF B (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, GB11261, Servicebio), FGF2 (1
1000, GB11261, Servicebio), FGF2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, GB113969, Servicebio), P65 (1
1000, GB113969, Servicebio), P65 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, GB11997, Servicebio), IL-10 (1
1000, GB11997, Servicebio), IL-10 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, GB11108, Servicebio), and TNF-α (1
1000, GB11108, Servicebio), and TNF-α (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, GB11188, Servicebio), overnight at 4 °C. The PVDF membranes were washed three times for 5 min each with TBST and then incubated with the horseradish peroxidase conjugated secondary antibodies at room temperature for 1 h. The labeled proteins were observed using a luminescent image analyzer.
1000, GB11188, Servicebio), overnight at 4 °C. The PVDF membranes were washed three times for 5 min each with TBST and then incubated with the horseradish peroxidase conjugated secondary antibodies at room temperature for 1 h. The labeled proteins were observed using a luminescent image analyzer.
4.17. RT-qPCR
RNA extraction and reverse transcription process were performed referring to the instructions of the kit. mRNA expression levels of angiogenesis- and inflammation-related proteins were calculated relative to GAPDH (internal control gene) using primers specific for VEGF, PDGF, NF-κB, IL-10, TNF-α, and FGF-2 (Table S3, ESI†).4.18. Statistical analysis
GraphPad software was used for statistical analysis. Unless otherwise stated, all tests were repeated a minimum of three times. The results are shown as mean ± standard deviation (SD). A normality test was conducted for each set of data. One-way ANOVA was used in comparing two different groups. P-Value ≤ 0.05 was considered statistically significant (*p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001).Author contributions
Shang Lyu: writing – original draft, data curation, formal analysis, investigation, methodology, visualization. Qi Liu: data curation, visualization. Ho-Yin Yuen: writing – review & editing. Huizhi Xie: data curation, visualization. Yuhe Yang, Kelvin Wai-Kwok Yeung, Chak-yin Tang, Shuqi Wang and Yaxiong Liu: methodology, resources. Bin Li, Yong He and Xin Zhao: supervision, project administration, funding acquisition, resources, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the grant from the Guangdong Basic and Applied Basic Research Foundation (2020B1515130002), Collaborative Research with World-leading Research Groups from The Hong Kong Polytechnic University (P0039523), National Natural Science Foundation of China (82122002 and 52235007), the Science Fund for Creative Research Groups of the National Natural Science Foundation of China (No. T2121004), the funding support to the State Key Laboratories in Hong Kong from the Innovation and Technology Commission of the Government of the Hong Kong Special Administrative Region of China and The Hong Kong Polytechnic University (Project Code: BBX2), the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, Organ-on-a-Chip Innovation Base, Central Government-Guided Special Project for Local Scientific and Technological Development in Sichuan Province (2023ZYD0166), and 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYYC21010).References
- X. Zhang, G. Chen, Y. Liu, L. Sun, L. Sun and Y. Zhao, ACS Nano, 2020, 14, 5901–5908 CrossRef CAS PubMed.
- J. Gan, X. Zhang, W. Ma, Y. Zhao and L. Sun, Nano Today, 2022, 47, 101630 CrossRef CAS.
- Z. Guo, H. Liu, Z. Shi, L. Lin, Y. Li, M. Wang, G. Pan, Y. Lei and L. Xue, J. Mater. Chem. B, 2022, 10, 3501–3511 RSC.
- Y. Xu, X. Wu, X. Zhang, Y. Zu, Q. Tan and Y. Zhao, Adv. Funct. Mater., 2023, 33, 2209986 CrossRef CAS.
- Q. Lei, D. He, L. Ding, F. Kong, P. He, J. Huang, J. Guo, C. J. Brinker, G. Luo and W. Zhu, Adv. Funct. Mater., 2022, 2113269 CrossRef CAS.
- L. Sun, Y. Wang, L. Fan and Y. Zhao, Chem. Eng. J., 2023, 457, 141206 CrossRef CAS.
- Y. Wang, A. a Sheng, X. Jiang, S. Yang, L. Lin, M. Yang, F. Zhu, Y. Hu, J. Li and L. Chang, Bio-Des. Manuf., 2023, 6, 255–267 CrossRef CAS.
- J. Chi, X. Zhang, C. Chen, C. Shao, Y. Zhao and Y. Wang, Bioact. Mater., 2020, 5, 253–259 Search PubMed.
- T. Ning, F. Yang, D. Chen, Z. Jia, R. Yuan, Z. Du, S. Liu, Y. Yu, X. Dai, X. Niu and Y. Fan, Adv. Healthcare Mater., 2022, 11, 2102180 CrossRef CAS PubMed.
- M. Yuan, K. Liu, T. Jiang, S. Li, J. Chen, Z. Wu, W. Li, R. Tan, W. Wei, X. Yang, H. Dai and Z. Chen, J. Nanobiotechnol., 2022, 20, 147 CrossRef CAS PubMed.
- W. Ma, X. Zhang, Y. Liu, L. Fan, J. Gan, W. Liu, Y. Zhao and L. Sun, Adv. Sci., 2022, 9, 2103317 CrossRef CAS PubMed.
- A. Fang, Y. Wang, N. Guan, Y. Zuo, L. Lin, B. Guo, A. Mo, Y. Wu, X. Lin, W. Cai, X. Chen, J. Ye, Z. Abdelrahman, X. Li, H. Zheng, Z. Wu, S. Jin, K. Xu, Y. Huang, X. Gu, B. Yu and X. Wang, Nat. Commun., 2023, 14, 4011 CrossRef CAS PubMed.
- D. Deng, X. Li, J.-J. Zhang, Y. Yin, Y. Tian, D. Gan, R. Wu, J. Wang, B.-M. Tian, F.-M. Chen and X.-T. He, ACS Nano, 2023, 17, 8530–8550 CrossRef CAS PubMed.
- X. Tong, Y. Xu, T. Zhang, C. Deng, J. Xun, D. Sun and D. Xu, J. Orthop. Trans., 2023, 39, 100–112 Search PubMed.
- A. Przekora, Cells, 2020, 9, 1622 CrossRef CAS PubMed.
- A. Malek-Khatabi, M. Sadat Razavi, A. Abdollahi, M. Rahimzadeghan, F. Moammeri, M. Sheikhi, M. Tavakoli, M. Rad-Malekshahi and Z. Faraji Rad, Biomater. Sci., 2023, 11, 5390–5409 RSC.
- S. W. T. Chew, Y. Zeng, M. Cui, H. Chang, M. Zheng, S. Wei, W. Zhao and C. Xu, SLAS Technol., 2019, 24, 181–187 CrossRef CAS PubMed.
- R. D. Boehm, J. Daniels, S. Stafslien, A. Nasir, J. Lefebvre and R. J. Narayan, Biointerphases, 2015, 10 Search PubMed.
- H. Abe, Y. Matsui, N. Kimura and M. Nishizawa, Macromol. Mater. Eng., 2021, 306, 2100171 CrossRef CAS.
- A. D. Permana, M. Mir, E. Utomo and R. F. Donnelly, Int. J. Pharm.: X, 2020, 2, 100047 CAS.
- Y. Wu, L. K. Vora, Y. Wang, M. F. Adrianto, I. A. Tekko, D. Waite, R. F. Donnelly and R. R. S. Thakur, Eur. J. Pharm. Biopharm., 2021, 165, 306–318 CrossRef CAS PubMed.
- R. D. Boehm, J. Daniels, S. Stafslien, A. Nasir, J. Lefebvre and R. J. Narayan, Biointerphases, 2015, 10, 011004 CrossRef PubMed.
- Y. Wang, H. Lu, M. Guo, J. Chu, B. Gao and B. He, Adv. Healthcare Mater., 2022, 11, 2101659 CrossRef CAS PubMed.
- T. Xu, Y. Yang, E. H.-L. Yeung, Q. Chen, H.-P. Bei, Q. Yang, M. Yang, Y. Hao, B. Li and X. Zhao, Adv. Funct. Mater., 2023, 33, 2213428 CrossRef CAS.
- J. Fang, H. Liu, W. Qiao, T. Xu, Y. Yang, H. Xie, C.-H. Lam, K. W. K. Yeung and X. Zhao, Adv. Healthcare Mater., 2023, 12, 2201220 CrossRef CAS PubMed.
- Y. Yang, T. Xu, H. P. Bei, Y. Zhao and X. Zhao, Adv. Funct. Mater., 2021, 31, 2104636 CrossRef CAS.
- Y. Yang, Q. Zhang, T. Xu, H. Zhang, M. Zhang, L. Lu, Y. Hao, J. H. Fuh and X. Zhao, Biomaterials, 2020, 263, 120378 CrossRef CAS PubMed.
- X. Zhao, I. Olsen, H. Li, K. Gellynck, P. G. Buxton, J. C. Knowles, V. Salih and A. M. Young, Acta Biomater., 2010, 6, 845–855 CrossRef CAS PubMed.
- Y. Piao, H. You, T. Xu, H.-P. Bei, I. Z. Piwko, Y. Y. Kwan and X. Zhao, Eng. Regener., 2021, 2, 47–56 Search PubMed.
- X. Zhao, S. Liu, L. Yildirimer, H. Zhao, R. Ding, H. Wang, W. Cui and D. Weitz, Adv. Funct. Mater., 2016, 26, 2809–2819 CrossRef CAS.
- X. Zhao, Q. Lang, L. Yildirimer, Z. Y. Lin, W. Cui, N. Annabi, K. W. Ng, M. R. Dokmeci, A. M. Ghaemmaghami and A. Khademhosseini, Adv. Healthcare Mater., 2016, 5, 108–118 CrossRef CAS PubMed.
- H. Chen, L. Guo, J. Wicks, C. Ling, X. Zhao, Y. Yan, J. Qi, W. Cui and L. Deng, J. Mater. Chem. B, 2016, 4, 3770–3781 RSC.
- Z. Zeng, G. Jiang, T. Liu, G. Song, Y. Sun, X. Zhang, Y. Jing, M. Feng and Y. Shi, Bio-Des. Manuf., 2021, 4, 902–911 CrossRef CAS.
- M. Qu, H.-J. Kim, X. Zhou, C. Wang, X. Jiang, J. Zhu, Y. Xue, P. Tebon, S. A. Sarabi, S. Ahadian, M. R. Dokmeci, S. Zhu, Z. Gu, W. Sun and A. Khademhosseini, Nanoscale, 2020, 12, 16724–16729 RSC.
- W. C. Oliver and G. M. Pharr, J. Mater. Res., 1992, 7, 1564–1583 CrossRef CAS.
- M.-L. Laracuente, M. H. Yu and K. J. McHugh, J. Controlled Release, 2020, 327, 834–856 CrossRef CAS PubMed.
- S. Chavoshi, M. Rabiee, M. Rafizadeh, N. Rabiee, A. S. Shamsabadi, M. Bagherzadeh, R. Salarian, M. Tahriri and L. Tayebi, Bio-Des. Manuf., 2019, 2, 96–107 CrossRef CAS.
- E. Adams, D. Coomans, J. Smeyers-Verbeke and D. L. Massart, Int. J. Pharm., 2002, 240, 37–53 CrossRef CAS PubMed.
- V. Papadopoulou, K. Kosmidis, M. Vlachou and P. Macheras, Int. J. Pharm., 2006, 309, 44–50 CrossRef CAS PubMed.
- A. C. Gonzalez, T. F. Costa, Z. A. Andrade and A. R. Medrado, An. Bras. Dermatol., 2016, 91, 614–620 CrossRef PubMed.
- O. S. Manoukian, A. Ahmad, C. Marin, R. James, A. D. Mazzocca and S. G. Kumbar, in Wound Healing Biomaterials, ed. M. S. Ågren, Woodhead Publishing, 2016, pp. 451–481 Search PubMed.
- in Strategies to Modify the Drug Release from Pharmaceutical Systems, ed. M. L. Bruschi, Woodhead Publishing, 2015, pp. 63–86 Search PubMed.
- F. Yang, J. Wang, X. Li, Z. Jia, Q. Wang, D. Yu, J. Li and X. Niu, Bio-Des. Manuf., 2022, 5, 305–317 CrossRef CAS.
- S. J. de Jong, E. R. Arias, D. T. S. Rijkers, C. F. van Nostrum, J. J. Kettenes-van den Bosch and W. E. Hennink, Polymer, 2001, 42, 2795–2802 CrossRef CAS.
- H. S. Kim, X. Sun, J.-H. Lee, H.-W. Kim, X. Fu and K. W. Leong, Adv. Drug Delivery Rev., 2019, 146, 209–239 CrossRef CAS PubMed.
- H. Pratsinis, E. Mavrogonatou and D. Kletsas, Adv. Drug Delivery Rev., 2019, 146, 325–343 CrossRef CAS PubMed.
- P. Wu, Y. Liang and G. Sun, Biomater Trans., 2021, 2, 61–71 Search PubMed.
- S. Saha, P. Sadhukhan and P. C. Sil, BioFactors, 2016, 42, 459–474 CrossRef CAS PubMed.
- A. J. Núñez Selles, M. Daglia and L. Rastrelli, Biofactors, 2016, 42, 475–491 CrossRef PubMed.
- Z. Wei, L. Yan, Y. Chen, C. Bao, J. Deng and J. Deng, Mol. Med. Rep., 2016, 14, 1091–1098 CrossRef CAS PubMed.
- Q. J. Zhang and L. Yue, Afr. J. Tradit., Complementary Altern. Med., 2017, 14, 263–271 CrossRef CAS PubMed.
- J. Rodríguez, D. Di Pierro, M. Gioia, S. Monaco, R. Delgado, M. Coletta and S. Marini, Biochim. Biophys. Acta, Gen. Subj., 2006, 1760, 1333–1342 CrossRef PubMed.
- I. Rodeiro, M. T. Donato, I. Martínez, I. Hernández, G. Garrido, J. A. González-Lavaut, R. Menéndez, A. Laguna, J. V. Castell and M. J. Gómez-Lechón, Toxicol. In Vitro, 2008, 22, 1242–1249 CrossRef CAS PubMed.
- M. Li, T. Wang, H. Tian, G. Wei, L. Zhao and Y. Shi, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 3793–3803 CrossRef CAS PubMed.
- B. F. Hettich, M. Ben-Yehuda Greenwald, S. Werner and J. C. Leroux, Adv. Sci., 2020, 7, 2002596 CrossRef CAS PubMed.
- J. D. Anderson, H. J. Johansson, C. S. Graham, M. Vesterlund, M. T. Pham, C. S. Bramlett, E. N. Montgomery, M. S. Mellema, R. L. Bardini and Z. Contreras, Stem Cells, 2016, 34, 601–613 CrossRef CAS PubMed.
- J. Zhang, J. Guan, X. Niu, G. Hu, S. Guo, Q. Li, Z. Xie, C. Zhang and Y. Wang, J. Transl. Med., 2015, 13, 1–14 CrossRef CAS PubMed.
- M. Isakson, C. de Blacam, D. Whelan, A. McArdle and A. J. P. Clover, Stem Cells Int., 2015, 831095 CAS.
- K. Ousey, K. F. Cutting, A. A. Rogers and M. G. Rippon, J. Wound Care, 2016, 25, 122–130 CrossRef CAS PubMed.
- M. B. Dreifke, A. A. Jayasuriya and A. C. Jayasuriya, Mater. Sci. Eng., C, 2015, 48, 651–662 CrossRef CAS PubMed.
- S. Ding, S. He, K. Ye, X. Shao, Q. Yang and G. Yang, Int. J. Biol. Macromol., 2023, 253, 127151 CrossRef CAS PubMed.
- B. Jiang, A. Shi, Y. Xu, Y. Zhang, Y. Chen, X. Jiang, H. Liu and L. Zhang, Mater. Today Commun., 2023, 34, 105152 CrossRef CAS.
- T. Kushibiki, Y. Mayumi, E. Nakayama, R. Azuma, K. Ojima, A. Horiguchi and M. Ishihara, Sci. Rep., 2021, 11, 23094 CrossRef CAS PubMed.
- X. Sun, Q. Lang, H. Zhang, L. Cheng, Y. Zhang, G. Pan, X. Zhao, H. Yang, Y. Zhang, H. A. Santos and W. Cui, Adv. Funct. Mater., 2017, 27, 1604617 CrossRef.
- G. Wang, W. Wang, Z. Chen, T. Hu, L. Tu, X. Wang, W. Hu, S. Li and Z. Wang, Chem. Eng. J., 2024, 482, 148938 CrossRef CAS.
- Y. Yi, Z. Yang, C. Zhou, Y. Yang, Y. Wu and Q. Zhang, Nano TransMed, 2024, 3, 100030 CrossRef.
- A. K. Gupta, T. Wang and J. A. Rapaport, J. Cosmet. Dermatol., 2023, 22, 2424–2433 CrossRef PubMed.
- V. Sarrazy, F. Billet, L. Micallef, B. Coulomb and A. Desmoulière, Wound Repair Regen., 2011, 19, 10–15 CrossRef PubMed.
- Q. Zhang, L. Shi, H. He, X. Liu, Y. Huang, D. Xu, M. Yao, N. Zhang, Y. Guo, Y. Lu, H. Li, J. Zhou, J. Tan, M. Xing and G. Luo, ACS Nano, 2022, 16, 10163–10178 CrossRef CAS PubMed.
- H.-P. Bei, T. Xu, J. Zhou, Z. Dong, Y. Wang, K.-Y. Wong, H. Wang and X. Zhao, Appl. Mater. Today, 2022, 27, 101463 CrossRef.
- B. H. Pan, Q. Zhang, C. H. Lam, H. Y. Yuen, S. Kuang and X. Zhao, Drug Discovery Today, 2022, 27, 857–865 CrossRef CAS PubMed.
- L. Cheng, X. Sun, X. Zhao, L. Wang, J. Yu, G. Pan, B. Li, H. Yang, Y. Zhang and W. Cui, Biomaterials, 2016, 83, 169–181 CrossRef CAS PubMed.
- A. V. Shinde, C. Humeres and N. G. Frangogiannis, Biochim. Biophys. Acta, Mol. Basis Dis., 2017, 1863, 298–309 CrossRef CAS PubMed.
- C.-C. Tsai, S.-B. Wu, H.-C. Kau and Y.-H. Wei, Sci. Rep., 2018, 8, 7276 CrossRef PubMed.
- D. C. Yeo, C. Wiraja, A. S. Paller, C. A. Mirkin and C. Xu, Nat. Biomed. Eng., 2018, 2, 227–238 CrossRef CAS PubMed.
- K. E. Lipson, C. Wong, Y. Teng and S. Spong, Fibrog. Tissue Repair, 2012, 5, S24 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S17 and Tables S1–S3. See DOI: https://doi.org/10.1039/d3mh01910a |
| This journal is © The Royal Society of Chemistry 2024 |