Open Access Article
Open Access ArticleSynthetic graphene–copper nanocomposites interact with the hACE-2 enzyme and inhibit its biochemical activity
Shoukath
Sulthana†
a,
Abeera
Bhatti†
b,
Elza
Mathew
 b,
Sohel H.
Quazi
ac,
Natasha N.
Gaudreault
*b,
Robert
DeLong
b,
Sohel H.
Quazi
ac,
Natasha N.
Gaudreault
*b,
Robert
DeLong
 *d and
Santosh
Aryal
*d and
Santosh
Aryal
 *a
*a
aDepartment of Pharmaceutical Sciences and Health Outcomes, The Ben and Maytee Fisch College of Pharmacy, University of Texas at Tyler, Tyler, TX 75799, USA. E-mail: santosharyal@uttyler.edu
bDepartment of Diagnostic Medicine/Pathobiology, College of Veterinary Medicine, Kansas State University, Manhattan, KS 66506, USA. E-mail: nng5757@vet.k-state.edu
cDepartment of Biology, Division of Natural and Computational Sciences, Texas College, Tyler, TX 75702, USA
dLandmark Bio, Innovation Development Laboratory, Watertown, MA 02472, USA. E-mail: rdelong@landmarkbio.com
First published on 10th November 2023
Abstract
This study demonstrates the copper nanocomposite-induced enzymatic inhibition of human angiotensin I-converting enzyme-2 (hACE-2) by complex stabilization through the formation of the enzyme nanocomposite. The immediate application of this work is related to ACE-2 as a mechanism of SARS-CoV-2 entry into cells. Moreover, ACE-2 enzyme regulation is a potential therapeutic strategy in hypertension and cardiovascular disease, diabetes, lung injury, and fibrotic disorders. Thus, inhibition of ACE-2 with nanocomposite therapy, may have pharmacologic application with regard to infectious and non-infectious diseases. Synthesized copper nanocomposites described here alone with a commercially available compound, were tested for their potential to inhibit hACE-2 activities. Following wet chemical synthesis, Cu/CuO nanoparticles and graphene–copper (GO–Cu) complexes were synthesized and characterized for their chemical integrity. Cu/CuO formed well-dispersed clusters of 390 ± 100 nm, that when complexed with the hACE-2 enzyme exhibited larger clusters of 506 ± 56 nm. The formation of the Cu/CuO and hACE-2 enzyme complex was monitored by analyzing the zeta potential, which reflected the surface charge distribution of the complex. A negatively charged Cu/CuO nanocomposite nearly becomes neutral when complexed with hACE-2 further assuring the complex formation. Formation of this complex and its inactivation of hACE-2 was evaluated using a standardized protocal for enzymatic activity. Similarly, carboxylate-functionalized graphene was complexed with copper, and its inhibitory effect was studied. Each step in the GO–Cu composite formation was monitored by characterizing its surface electrical properties, resulting in a decrease in its zeta potential and conductivity when complexed with copper. The interaction of the nanocomposites with hACE-2 was confirmed by 2D-FDS and gel electrophoresis analysis. GO–Cu was a rapid and efficacious inhibitor compared to Cu–CuO, especially at lower concentrations (2 μg ml−1). Considering the environmental friendliness of copper and graphene and their use in industries as surface coating materials, we anticipate that use of these composites once proven effective, may have future antimicrobial application. Utility of nanocomposites as antimicrobials, either as a surface antimicrobial or as an in vivo therapeutic, could be invisioned for use against current unknown and/or emergent pathogens.
Introduction
A nanoparticulate drug delivery biomaterial, broadly defined as nanomedicine, has been developed with some approved candidates for clinical use by the US FDA.1–4 These nanomedicines are also in continuous development and preclinical research with the goal of overcoming the biological barriers that otherwise are not possible with free small molecule or large macromolecular drugs.5–17 Currently in the clinics, many organic and inorganic nanoparticles (NPs) are being used for the diagnosis and therapy of cancer. Example of these includes, DOXIL® for ovarian cancer (secondary to platinum-based therapies)18 and HIV-associated Kaposi's sarcoma (secondary to chemotherapy), multiple myeloma (secondary); Abraxane® for advanced non-small cell lung cancer, metastatic breast cancer, and pancreatic cancer, VYXEOS® for acute myeloid leukemia, Hensify® for locally-advanced squamous cell carcinoma, Resovist® as a magnetic resonance imaging contrast agent and the list further increases with the total of more than 50 approved nanomedicine for clinical use.15 While significant efforts have been dedicated to engineering synthetic and biomimetic nanomedicine against cancer,5,7–9,16,19–27 the studies related to the effect of NPs on enzyme receptor biochemical activity have been given less attention to envisioning its possible impact on infectious disease and immune modulation.Recently, efforts have been put forth toward the development of nanomedicine-based agents capable of inhibiting the activity of human angiotensin I-converting enzyme-2 (hACE-2), which is the receptor for the Spike (S) protein of the SARS-CoV and SARS-CoV-2 coronaviruses, facilitating the viral entry into the target cells.28–30 A general example of the use of nanomedicine against coronaviruses is the COVID-19 vaccine that use as lipid NPs and adjuvant and delivery vehicle for mRNA encoding the spike protein to induce a protective immune response against SARS-CoV-2. Adaptive mutations in the SARS-CoV-2 genome can alter viral transmission, pathogenesis and immune protection. Therefore, the development of efficacious diagnostics, treatments, and vaccines against COVID-19 remains important; metal-based nanocomposites are of great interest as a potential application towards preventing virus infection and transmission. In the recent report presented by Sportelli et al.,31 coronaviruses are inactivated by copper surfaces due to ROS created by the release of copper ions. Similarly, the antiviral activity of CuI NPs against the influenza A virus (H1N1), inactivating the virus through the generation of hydroxide radicals capable of degrading viral proteins is reported by Fujimori et al.32 An exponential improvement in the antimicrobial properties was reviewed by Ingle et al., with Cu/CuO NPs, where authors focused on the bioactivities and cytotoxicity and pointed out many opportunities with Cu-based nanomaterials.33 Owing to the high surface-to-volume ratio of NPs, they are associated with a wide surface area, which assists in improving their interaction with microbes, thereby allowing better opportunity for a broad spectrum of antimicrobial activities.
Copper and copper-based NPs are earth-abundant, inexpensive, and particularly easier to manufacture as compared to other NPs. They are prone to oxidation under atmospheric conditions, but they still play a vital role because of their various uses.34 Cu plays a vital role in the functions of proteins and the emerging use of NPs has caused an increased number of applications of Cu and Cu-based NPs in various ways because of their antimicrobial activities.35 They have a strong ability to inhibit pathogenic bacteria, fungi, and viruses and have been used as a coating on portable devices, such as mobile phones to prevent the spread and accumulation of microbes.
In addition to Cu and its composites, graphene nanomaterials have also been found to exhibit antiviral and antimicrobial properties at the nanoscale along with outstanding optical and electrical properties that warrant it as a potential candidate for designing and developing high-performance components and devices required for the COVID-19 pandemic and other future disasters.36 Sametband et al.37 reported the antiviral properties of graphene oxide against Herpes Simplex Virus Type-1 (HSV-1) through a competitive inhibition mechanism. Graphene oxide-based nanomaterials have been used as label-free detection and disinfection kits against environmental viruses such as Enterovirus 71 (EV71) and endemic gastrointestinal avian influenza A virus (H9N2) that have great environmental stability and low sensitivity for organic disinfectants and soaps.36,38 Compared to other surfaces such as plastic and steel, graphene and copper surfaces were found to be unfavorable to the virus, which either denatures the viral protein or creates a chemical environment that significantly reduces the viral viability.
While the number of SARS-CoV-2 vaccines, including, mRNA, viral vector, inactivated virus, and live attenuated virus vaccines have been reported, some SARS-CoV-2 variants have challenged the efficacy of these vaccines. Also, there are no available specific medications, approved by FDA against COVID-19, except some monoclonal antibody therapies. Therefore, finding new solutions and revisiting existing strategies are very important in order to combat this rapidly changing virus, SARS-CoV-2. Considering the opportunities available in nanotechnology, we herein demonstrate the synthesis and characterization of Cu/CuO and graphene–Cu nanomaterials and study their potential for inhibiting ACE-2, a vital receptor that is widely expressed in alveolar epithelial cells of lungs.39 Based on the antiviral activity of the composite biomaterials against other viral pathogens, we hypothesize that the nanostructured copper composites, such as Cu/CuO NPs and GO–copper complexes can be a potential effective tool in the fight against COVID-19 and future pandemics. In this regard, one of the potential applications is coating the nanocomposites on commonly touched surfaces in public, high human traffic, spaces to potentially mitigate pathogen transmission.
Materials and methods
Materials
The substrate Mca-APK(Dnp) TFA salt (Mca = 7-Methoxycoumarin-4-yl; Dnp = 2,4-dinitrophenyl, APK = Ala-Pro-Lys) was purchased from Sigma-Aldrich (product number: SML2948) and used as received for assessing the enzymatic activity of ACE-2 with regard to functionality of the proline-lystine cleavage function of the enzyme. Enzyme hACE-2 was purchased from Sigma-Aldrich (product number: 176872) and used as received throughout the experiments. The Cu/CuO nanocomposite was purchased from PlasmaScience and stored at 4 °C. Graphene oxide was purchased from TCI and was used as a control and a substrate. Copper sulfate pentahydrate was purchased from Thermo Scientific and was used as a critial reagent for nanocomposite synthesis.Synthesis of the graphene–copper complex
The graphene–copper complex was synthesized in two steps. Firstly, carboxylation of graphene oxide (GO) was carried out in piranha solution to develop metal ion chelators. In the second step, Cu complexation was performed at ambient temperature. Briefly, 10 mg of GO powder was treated with 6 ml of piranha solution (H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O2) and stirred overnight at 50 °C. The GO–COOH product was collected and washed three times with ultrapure water. After purification, samples were lyophilized to powder and kept at −80 °C for further use. For Cu complexation, the GO–COOH product (5 mg) was treated with 5 ml of 25 gm L−1 of CuSO4 for 24 h at 70 °C. The final graphene–copper complex was collected and washed several times with water and freeze-dried to obtain the powder.
H2O2) and stirred overnight at 50 °C. The GO–COOH product was collected and washed three times with ultrapure water. After purification, samples were lyophilized to powder and kept at −80 °C for further use. For Cu complexation, the GO–COOH product (5 mg) was treated with 5 ml of 25 gm L−1 of CuSO4 for 24 h at 70 °C. The final graphene–copper complex was collected and washed several times with water and freeze-dried to obtain the powder.
Synthesis of Cu/CuO nanoparticles
Cu/CuO NP was synthesized by a chemical reduction process using copper(II) sulfate pentahydrate (CuSO4·5H2O) as the precursor salt and ascorbic acid as a mild reducing agent.40 Briefly, 120 ml of 0.1 M CuSO4·5H2O was prepared in deionized water and placed in a round bottom flask. Next, 50 ml of 0.2 M ascorbic acid was added slowly under continuous rapid stirring. Then, 30 ml of 1 M NaOH was added slowly with constant stirring and heating at 80 °C for 2 h. As the reduction process continued, the color of the solution slowly became ocher. After completion, the solution was left overnight for sedimentation, and the supernatant was carefully removed from the round bottom flask. The ocher-colored Cu/CuO NPs were collected in 50 ml centrifuging tubes and washed two times with water and once with ethanol following centrifugation at 2000g for 5 min. After washing, Cu/CuO NPs were dried under a vacuum and stored at 4 °C for future use.Biochemical enzyme assays
A working stock of the hACE-2 enzyme was prepared by dissolving 0.1 units of the enzyme in 75 μl of 1X PBS buffer. The Mca-APK (Dnp) TFA salt substrate stock solution was also prepared by dissolving 1 mg of the substrate in 75 μl of 1X PBS buffer. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution of the substrate was prepared by adding 2 μl of the substrate in 398 μl of 1X PBS. A 1
200 dilution of the substrate was prepared by adding 2 μl of the substrate in 398 μl of 1X PBS. A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution of the enzyme was prepared by adding 4 μl of the enzyme in 196 μl of 1X PBS. 4.4 mg ml−1 of the stock solution of Cu/CuO NPs (lab-made), followed by a 9 mg ml−1 stock solution of Cu/CuO NPs (plasmaChem) was also prepared. 2 μl of the fluorometric substrate and 4 μl of hACE-2 enzyme were added into the BRAND 96-well black with clear flat bottom plates. 50 μl of 400 μg ml−1, 200 μg ml−1, 20 μg ml−1, and 2 μg ml−1 dilutions of both types of Cu/CuO NPs were added separately in the same wells with enzyme and substrate. Separate wells containing 2 μl of the substrate and 4 μl of the enzyme (with a total volume of 56 μl) with no NP were used as controls. The same enzyme assay was repeated using different NPs to compare the inhibition of hACE-2 enzymes by Cu/CuO NPs with graphene carboxylate and graphene Cu-complex. 9 mg ml−1 of the stock solution Cu/CuO NPs (PlasmaChem), followed by 1.5 mg ml−1 stock solution of graphene carboxylate complex and 1.4 mg ml−1 stock solution of Graphene Cu-Complex was also prepared. 2 μl of a fluorometric substrate and 4 μl of hACE-2 enzyme were added into the wells. 50 μl of 400 μg ml−1, 200 μg ml−1, 20 μg ml−1, and 2 μg ml−1 dilutions of all types of NPs were added separately in the same wells with the enzyme and the substrate. Separate wells of the 2 μl substrate and 4 μl of the enzyme only (with a total volume of 56 μl) with no NP were used as controls. Fluorometric readings were taken on the Spectramax i3X (molecular devices) with the settings of the fluorescence spectrum, fixed excitation of 320 nm, and emission 405 to 425 nm in 5 nm steps while the temperature was 37 °C. The area under the curve (AUC) was calculated for enzyme + substrate, and samples containing 400 μg ml−1, 200 μg ml−1, 20 μg ml−1, and 2 μg ml−1 dilutions of Cu/CuO NPs using the formula, (Y1 + Y2)/2 × (X2 − X1) where X1 = 405 wavelength, X2 = 410 wavelength, Y1 is the RFU at 405 wavelength and, Y2 is the RFU at 410 wavelengths. The same process was repeated for each wavelength and the corresponding RFU to obtain each AUC. All the AUCs were then added to obtain the total area under the curve for each concentration and their standard deviations were calculated.
50 dilution of the enzyme was prepared by adding 4 μl of the enzyme in 196 μl of 1X PBS. 4.4 mg ml−1 of the stock solution of Cu/CuO NPs (lab-made), followed by a 9 mg ml−1 stock solution of Cu/CuO NPs (plasmaChem) was also prepared. 2 μl of the fluorometric substrate and 4 μl of hACE-2 enzyme were added into the BRAND 96-well black with clear flat bottom plates. 50 μl of 400 μg ml−1, 200 μg ml−1, 20 μg ml−1, and 2 μg ml−1 dilutions of both types of Cu/CuO NPs were added separately in the same wells with enzyme and substrate. Separate wells containing 2 μl of the substrate and 4 μl of the enzyme (with a total volume of 56 μl) with no NP were used as controls. The same enzyme assay was repeated using different NPs to compare the inhibition of hACE-2 enzymes by Cu/CuO NPs with graphene carboxylate and graphene Cu-complex. 9 mg ml−1 of the stock solution Cu/CuO NPs (PlasmaChem), followed by 1.5 mg ml−1 stock solution of graphene carboxylate complex and 1.4 mg ml−1 stock solution of Graphene Cu-Complex was also prepared. 2 μl of a fluorometric substrate and 4 μl of hACE-2 enzyme were added into the wells. 50 μl of 400 μg ml−1, 200 μg ml−1, 20 μg ml−1, and 2 μg ml−1 dilutions of all types of NPs were added separately in the same wells with the enzyme and the substrate. Separate wells of the 2 μl substrate and 4 μl of the enzyme only (with a total volume of 56 μl) with no NP were used as controls. Fluorometric readings were taken on the Spectramax i3X (molecular devices) with the settings of the fluorescence spectrum, fixed excitation of 320 nm, and emission 405 to 425 nm in 5 nm steps while the temperature was 37 °C. The area under the curve (AUC) was calculated for enzyme + substrate, and samples containing 400 μg ml−1, 200 μg ml−1, 20 μg ml−1, and 2 μg ml−1 dilutions of Cu/CuO NPs using the formula, (Y1 + Y2)/2 × (X2 − X1) where X1 = 405 wavelength, X2 = 410 wavelength, Y1 is the RFU at 405 wavelength and, Y2 is the RFU at 410 wavelengths. The same process was repeated for each wavelength and the corresponding RFU to obtain each AUC. All the AUCs were then added to obtain the total area under the curve for each concentration and their standard deviations were calculated.
2D FDS using Cu/CuO NPs
The working stock of the hACE-2 enzyme was prepared by dissolving 0.1 units of the enzyme in 75 μl of 1X PBS buffer. The Mca-APK (Dnp) TFA salt substrate stock solution was also prepared by dissolving 1 mg of the substrate in 75 μl of 1X PBS buffer. AP dye 647 Hydrazide (AXISPharm) solution was also prepared by dissolving 1 mg of dye in 100 μl 1X PBS. Then, Cu/CuO NPs (PlasmaChem) solution was prepared by dissolving 2 mg of Cu/CuO NPs in 1000 μl of 1X PBS. 2 μl hACE-2 solutions were added in 96-well plates, followed by 2 μl of the dye and 100 μl of 1X PBS and fluorometric readings were taken on the Spectramax i3X (Molecular Devices) instrument with the settings for the fluorescence spectrum, and unknown fixed excitation and emission, to determine the fixed excitation and emission using the optimization wizard function while the temperature was set to 37 °C. The same procedure was repeated, using 2 μl of the substrate along with enzyme and dye, and readings were taken. Then, 50 μl of Cu/CuO NPs were added in a separate well of 96-well plates, along with the same quantities of dye, enzyme, substrate, and 50 μl of 1X PBS, while fluorometric readings were taken at each of the steps. The same procedure was repeated using the same quantities of enzyme, substrate, and AP dye, along with a 50 μl solution of 1 mg ml−1 of graphene carboxylate, graphene copper complex, and graphene oxide.Colloidal and electrical properties
Zeta potential, size, and polydispersity indices were measured for Cu/CuO NP and Cu/CuO NP + hACE-2. Both the treatments contained 20 μg of Cu/CuO NP, 0.1 μg of hACE-2, and 970 μl of RNase-free water. Samples were incubated for 15 minutes at 150 rpm on an orbital shaker (MIDSCI) and were dispersed in RNase-free water. Samples were analyzed at a volume of 1 ml in a capillary cell cuvette on Malvern Zetasizer Nano Range Dynamic Light Scattering (DLS) equipment.Gel electrophoresis
A stock solution containing 5 mg ml−1 of Cu/CuO NPs was prepared and four dilutions from the stock solution were prepared with the concentrations of 400 μg ml−1, 200 μg ml−1, 20 μg ml−1, and 2 μg ml−1 6 samples were prepared by adding 2 μl of hACE-2 enzyme and 8 μl of 1X PBS for the first one; 2 μl of hACE-2 enzyme, 1 μl of the substrate, and 7 μl of 1X PBS, for the second one; 2 μl of hACE-2 enzyme, 1 μl of the substrate and 7 μl of 400 μg ml−1 Cu/CuO NP solution, for the third one; 2 μl of hACE-2 enzyme, 1 μl of the substrate and 7 μl of 200 μg ml−1 Cu NP solution, for the fourth one; 2 μl of hACE-2 enzyme, 1 μl of the substrate and 7 μl of 20 μg ml−1 Cu NP solution, for the fifth one; and 2 μl of hACE-2 enzyme, 1 μl of the substrate and 7 μl of 2 μg ml−1 Cu NP solution for the sixth one. After adding loading buffer (10 μl each), the samples were heated at 100 °C for 5 minutes. The samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 1X Tris-Glycine buffer at 100 V for 45 minutes. The Bullseye pre-stained protein ladder (10–180 kDa) was used as the reference. The protein bands were visualized using GelCode™ Blue Safe Protein Stain. The gel was imaged using the Gel Doc™ XR + gel documentation system and analyzed using the Image Lab™ software.Results and discussion
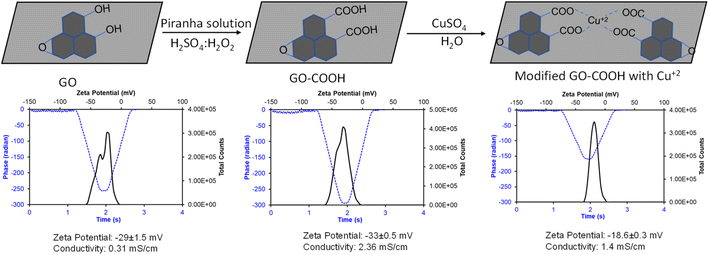
The graphene–Cu complex has widespread application in biology and electronics due to its ease of functionalization using redox chemistry,36,38,41,42 and is widely used in contact areas. The goal of this study was to understand the inhibitory efficiency of the graphene–Cu complex on hACE-2 enzymatic activity. Starting with commercially available graphene oxide (GO), we synthesized the GO–Cu complex in a two-step reaction (Fig. 1).In the first step, GO was further oxidized into its carboxylate (GO–COO−) form, followed by complexation with Cu++ in the second step (Fig. 1). The chemistry of each step was monitored by measuring the zeta potential to study the change in surface charge with the change in chemistry. In the zeta potential, an electrophoretic light scattering intensity measures the movement of a charged particle relative to the liquid under the influence of an applied electric field. The data is captured in the form of charge and the intensity of scattering light. The phase plot of the zeta potential measurement abstracts the difference in phase between the measured frequency and a reference frequency as a function of time, which gives the overall conductivity of the material in solution. GO obtained from the market shows −29 ± 1.5 mV, which further decreased with oxidation to carboxylate to −33 ± 0.5 mV and increased with copper complexation to −18.6 ± O.5 mV. The formation of the GO–Cu complex and the integrity of each chemical step were further confirmed by measuring the solution conductivity. The composite solution conductivity of GO–COO− increased from (GO) 0.31 mS cm−1 to 2.36 mS cm−1, which decreased to 1.4 mS cm−1 with Cu complexation. The decrease in the solution conductivity of the GO–Cu complex as compared to GO–COO– is possibly due to the charge neutralization with copper complexation, which also decreases the overall mass of GO–COO− with the addition of Cu++, because Cu has a single valence electron, while graphene pi-bond electrons are free to move and increase the conductivity of the graphene.
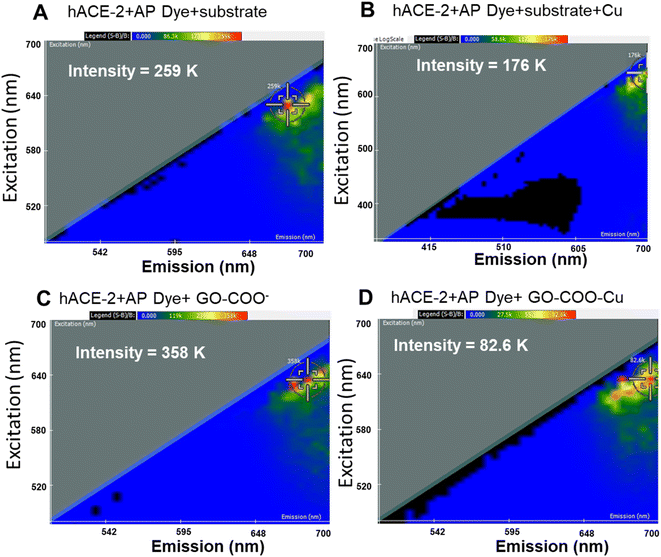
Thus, the synthesized GO–Cu complexes were further characterized for their inhibitory effect using 2D-FDS spectroscopy (Fig. 2A–D). As discussed above, we observed significant fluorescence quenching when hACE-2 is treated with the GO–Cu complex. We observed the intensity of hACE-2 with AP dye and the substrate (Fig. 2A) decreased from 259 to 176 in the presence of Cu (Fig. 2B). Similarly, the fluorescence intensity in the presence of GO–COO– (Fig. 2C) also decreased from 358 K to 82.6 K (Fig. 2D) when Cu was complexed. From these results, we can affirm that the presence of Cu in the system is an essential component for hACE-2 inhibition. In addition to the presence of Cu, the composite surface charge also plays a significant role, for example, GO-COO− has a high degree of negative zeta potential (−33 ± 0.5 mV) that could propel the enzyme far from its surface, resulting in a decrease in enzyme inhibitory response, whereas, with the copper complexation, zeta potential moved towards more positive value (−18.6 ± 0.5 mV). Interactions of the viral attachment proteins with protein receptor molecules are mostly determined by complementarity in surface charge distribution, hydrophobic interactions, and geometry.43 A model study suggested that hydrophobic residues at the ACE-2 surface were found in close proximity to the charged ridges, which could contribute to binding.43 Therefore, although the mechanism of the nanoparticle inhibition is unknown at present, it seems likely that the Cu and especially the Cu–graphene composite could alter biomolecular interactions at this interface.
To firmly analyze the properties of copper, colloidal Cu/CuO NPs were synthesized in the absence of graphene using ascorbic acid as a mild reducing and capping agent in a wet chemistry laboratory. Synthesized Cu/CuO NPs were characterized for their colloidal properties by monitoring the size, surface charge, and polydispersity (Table 1). These nanoaggregates exhibited a cluster with a hydrodynamic size of 390 ± 100 nm with a polydispersity index of 0.49 ± 0.11. At this stage, the Cu/CuO cluster has a negative zeta potential of −16.7 ± 4.7, which is presumably due to the oxide-rich surface, as discussed previously.34,44,45 These NPs when complexed with the hACE-2 protein, a dramatic change in the zeta potential to −4.25 ± 5.5, nearly neutral, was observed, confirming the surface modification with the hACE-2 protein. At this stage, the hydrodynamic size of the cluster increases to 506 ± 56 nm, which can be attributed to the surface protein and the increased hydration layer due to the protein. However, the polydispersity index (PDI) is not significantly altered, further confirming that the overall cluster remains the same before and after the hACE-2 modification.
| Type of nanomaterials | Zeta potential (mV) | Size (nm) | PDl |
|---|---|---|---|
| Cu/CuO NP | −16.7 ± 4.7 | 390 ± 100 | 0.49 ± 0.12 |
| Cu/CuO NP-hACE-2 | −4.25 ± 5.5 | 506 ± 56 | 0.52 ± 0.16 |
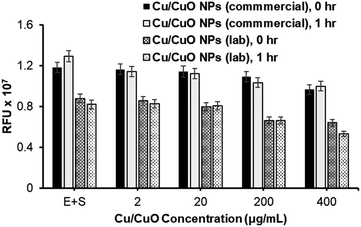
With the successful synthesis and characterization of Cu/CuO nanocomposites, we proceeded to study their effect on hACE-2 protein enzymatic activity and compared these activities with those of Cu/CuO NPs obtained from PlasmaChem company (Fig. 3). Both types of nanocomposites are capable of inhibiting the hACE-2 enzyme in a concentration-dependent manner (Fig. 3). However, significant differences in the inhibition properties were not observed, which allowed us to refine and proceed with one type of Cu/CuO nanocomposites in order to minimize efforts and maximize uniformity in the synthesized composite. A commercially available Cu/CuO nanocomposite from PlasmaChem's were used to conduct our experiments.
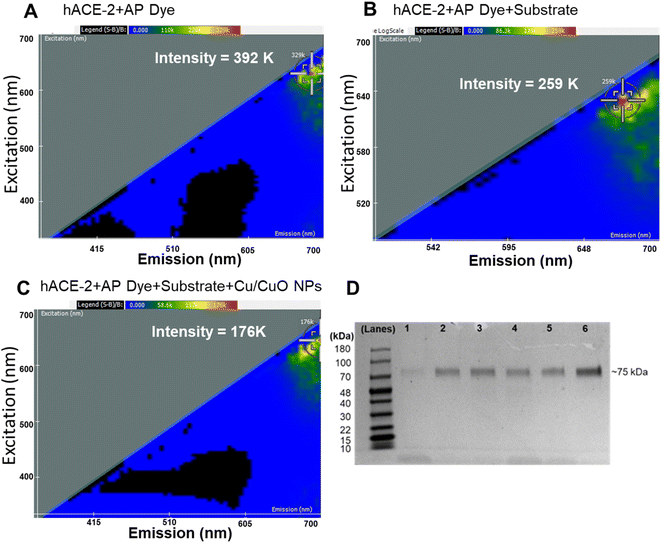
We employed two-dimensional fluorescence difference spectroscopy (2D FDS) that produces a unique spectral signature of metal composites that changes its signature when interacting with physiologically important proteins, for example, herein, hACE-2 (Fig. 4). Nanomaterials with their high surface area to volume ratio offer docking of multiple molecules, small molecules, or large macromolecules, which in turn alter fluorescent properties that can be easily tracked using 2D FDS spectroscopy. In Fig. 4, the excitation and emission spectra of the Cu/CuO composite were routinely compared to the AP dye and substrate used. The y-axis represents the excitation wavelength in nm, the x-axis represents the emission wavelength in nm, and the scale above the 2-D FDS plot represents the intensity in relative fluorescence units (RFU). Each spectral signature is distinct to the nanomaterial's chemistry, which shows a significant decrease in the overall intensity from 392 K to 176 K when binding to the hACE-2 protein. Fluorescent quenching was observed upon interaction with the Cu/CuO nanocomposite, diminishing the intensity by over 216![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 light units. In addition, we observed a red shift in the emission spectrum, which could be beneficial in biological experiments as tissue and blood-associated interferences are minimal in the near-infrared region.
000 light units. In addition, we observed a red shift in the emission spectrum, which could be beneficial in biological experiments as tissue and blood-associated interferences are minimal in the near-infrared region.
We further studied the binding efficiency of hACE-2 and the Cu/CuO nanocomposite using gel electrophoresis (Fig. 4). Our observation showed that a minimum amount of nanocomposite is sufficient to capture this protein.
Finally, the synthesized GO–Cu complex was routinely compared with Cu/CuO NPs for their potential for inhibiting the hACE-2 protein (Fig. 5A and B). Results show that the inhibition is concentration dependent, inhibition started in the beginning (time zero) of the nanocomposite treatment, suggesting the rapid binding of nanocomposite with the enzyme–substrate complex. Over the time of the treatment (post 1 h treatment), we observed a slight decrease in inhibition in all cases, which could be attributed to the production of the enzyme–substrate complex.
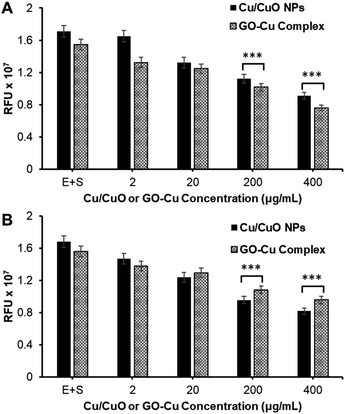 | ||
| Fig. 5 Study of nanocomposite-mediated hACE-2 enzyme inhibition. (A) At 0 h and (B) at 1 h of treatment. (*** = two-tailed P value < 0.0001, extremely significant, unpaired t-test). | ||
Conclusions and outlook
In this study, graphene–Cu and copper oxide nanocomposites were synthesized using a wet chemical procedure. The resulting composite nanomaterials along with a commercial product demonstrated an enhanced inhibitory effect on the hACE-2 enzyme. ACE-2 is the receptor for SAR-CoV-2 entry into cells. Among the synthesized nanocomposites, the GO–Cu complex exhibited a significantly high degree of hACE-2 inhibition demonstrated using a standardized assay for ACE-2 enzymatic activity. This basic scientific research has potential application as an inhibitor of infection or as a therapeutic. Moreover, given the functionality in the use of graphene and copper as coating materials, coating exposed surfaces with GO–Cu nanocomposite as an antimicrobial application could be used as a mitigation strategy to reduce SAR-CoV-2, once proven. Further research is focused on the extention of ACE-2 inhibition of SARS-CoV-2 in cells and its application on surfaces. The extentions of this work is intended to demonstrate the utility and application of this methodology against SARS-CoV-2 and other existing and emergent pathogens.Conflicts of interest
There are no conflicts to declare.References
- Research, C. for D. E. and FDA Approves Liposome-Encapsulated Combination of Daunorubicin-Cytarabine for Adults with Some Types of Poor Prognosis AML, FDA, 2019 Search PubMed.
- Drugs@FDA: FDA-Approved Drugs, https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process%26ApplNo=050718, accessed 2022-06-15.
- R. C. Kane, W. D. McGuinn, R. Dagher, R. Justice and R. Pazdur, Dexrazoxane (Totect™): FDA Review and Approval for the Treatment of Accidental Extravasation Following Intravenous Anthracycline Chemotherapy, Oncologist, 2008, 13(4), 445–450, DOI:10.1634/theoncologist.2007-0247.
- W. J. Gradishar, Albumin-Bound Paclitaxel: A next-Generation Taxane, Expert Opin. Pharmacother., 2006, 7(8), 1041–1053, DOI:10.1517/14656566.7.8.1041.
- A. Pitchaimani, T. D. T. Nguyen and S. Aryal, Natural Killer Cell Membrane Infused Biomimetic Liposomes for Targeted Tumor Therapy, Biomaterials, 2018, 160, 124–137, DOI:10.1016/j.biomaterials.2018.01.018.
- T. D. T. Nguyen, A. Pitchaimani, C. Ferrel, R. Thakkar and S. Aryal, Nano-Confinement-Driven Enhanced Magnetic Relaxivity of SPIONs for Targeted Tumor Bioimaging, Nanoscale, 2017, 10(1), 284–294, 10.1039/c7nr07035g.
- S. Rayamajhi, T. D. T. Nguyen, R. Marasini and S. Aryal, Macrophage-Derived Exosome-Mimetic Hybrid Vesicles for Tumor Targeted Drug Delivery, Acta Biomater., 2019, 94, 482–494, DOI:10.1016/j.actbio.2019.05.054.
- T. D. T. Nguyen, R. Marasini, S. Rayamajhi, C. Aparicio, D. Biller and S. Aryal, Erythrocyte Membrane Concealed Paramagnetic Polymeric Nanoparticle for Contrast-Enhanced Magnetic Resonance Imaging, Nanoscale, 2020, 12(6), 4137–4149, 10.1039/D0NR00039F.
- A. Pitchaimani, T. D. T. Nguyen, R. Marasini, A. Eliyapura, T. Azizi, M. Jaberi-Douraki and S. Aryal, Biomimetic Natural Killer Membrane Camouflaged Polymeric Nanoparticle for Targeted Bioimaging, Adv. Funct. Mater., 2019, 29(4), 1806817, DOI:10.1002/adfm.201806817.
- J. Abello, T. D. T. Nguyen, R. Marasini, S. Aryal and M. L. Weiss, Biodistribution of Gadolinium- and near Infrared-Labeled Human Umbilical Cord Mesenchymal Stromal Cell-Derived Exosomes in Tumor Bearing Mice, Theranostics, 2019, 9(8), 2325–2345, DOI:10.7150/thno.30030.
- R. H. Fang, A. V. Kroll, W. Gao and L. Zhang, Cell Membrane Coating Nanotechnology, Adv. Mater., 2018, 30(23), 1706759, DOI:10.1002/adma.201706759.
- B. T. Luk and L. Zhang, Cell Membrane-Camouflaged Nanoparticles for Drug Delivery, J. Control. Release, 2015, 220(Pt B), 600–607, DOI:10.1016/j.jconrel.2015.07.019.
- C.-M. J. Hu, R. H. Fang, K.-C. Wang, B. T. Luk, S. Thamphiwatana, D. Dehaini, P. Nguyen, P. Angsantikul, C. H. Wen, A. V. Kroll, C. Carpenter, M. Ramesh, V. Qu, S. H. Patel, J. Zhu, W. Shi, F. M. Hofman, T. C. Chen, W. Gao, K. Zhang, S. Chien and L. Zhang, Nanoparticle Biointerfacing by Platelet Membrane Cloaking, Nature, 2015, 526(7571), 118–121, DOI:10.1038/nature15373.
- S. Aryal, C.-M. J. Hu and L. Zhang, Polymer−Cisplatin Conjugate Nanoparticles for Acid-Responsive Drug Delivery, ACS Nano, 2010, 4(1), 251–258, DOI:10.1021/nn9014032.
- A. C. Anselmo and S. Mitragotri, Nanoparticles in the Clinic: An Update, Bioeng. transl. med., 2019, 4(3), e10143, DOI:10.1002/btm2.10143.
- N. I. Millagaha Gedara, X. Xu, R. DeLong, S. Aryal and M. Jaberi-Douraki, Global Trends in Cancer Nanotechnology: A Qualitative Scientific Mapping Using Content-Based and Bibliometric Features for Machine Learning Text Classification, Cancers, 2021, 13(17), 4417, DOI:10.3390/cancers13174417.
- R. Juliano, Nanomedicine: Is the Wave Cresting?, Nat. Rev. Drug Discovery, 2013, 12(3), 171–172, DOI:10.1038/nrd3958.
- K. Johnson-Arbor and R. Dubey, Doxorubicin, in StatPearls, StatPearls Publishing, Treasure Island (FL), 2022 Search PubMed.
- S. Rayamajhi, S. Wilson, S. Aryal and R. DeLong, Biocompatible FePO4 Nanoparticles: Drug Delivery, RNA Stabilization, and Functional Activity, Nanoscale Res. Lett., 2021, 16(1), 169, DOI:10.1186/s11671-021-03626-8.
- R. Marasini, T. D. Thanh Nguyen, S. Rayamajhi and S. Aryal, Synthesis and Characterization of a Tumor-Seeking LyP-1 Peptide Integrated Lipid–Polymer Composite Nanoparticle, Adv. Mater., 2020, 1(3), 469–480, 10.1039/D0MA00203H.
- S. Rayamajhi, R. Marasini, T. D. T. Nguyen, B. L. Plattner, D. Biller and S. Aryal, Strategic Reconstruction of Macrophage-Derived Extracellular Vesicles as a Magnetic Resonance Imaging Contrast Agent, Biomater. Sci., 2020, 8(10), 2887–2904, 10.1039/D0BM00128G.
- S. Rayamajhi, J. Marchitto, T. D. T. Nguyen, R. Marasini, C. Celia and S. Aryal, PH-Responsive Cationic Liposome for Endosomal Escape Mediated Drug Delivery, Colloids Surf., B, 2020, 188, 110804, DOI:10.1016/j.colsurfb.2020.110804.
- T. D. T. Nguyen, A. Pitchaimani and S. Aryal, Engineered Nanomedicine with Alendronic Acid Corona Improves Targeting to Osteosarcoma, Sci. Rep., 2016, 6, 36707, DOI:10.1038/srep36707.
- L. Zhang, J. M. Chan, F. X. Gu, J.-W. Rhee, A. Z. Wang, A. F. Radovic-Moreno, F. Alexis, R. Langer and O. C. Farokhzad, Self-Assembled Lipid–Polymer Hybrid Nanoparticles: A Robust Drug Delivery Platform, ACS Nano, 2008, 2(8), 1696–1702, DOI:10.1021/nn800275r.
- A. V. Kroll, R. H. Fang and L. Zhang, Biointerfacing and Applications of Cell Membrane-Coated Nanoparticles, Bioconjug. Chem., 2017, 28(1), 23–32, DOI:10.1021/acs.bioconjchem.6b00569.
- Cancer nanomedicine: progress, challenges and opportunities|Nature Reviews Cancer, https://www.nature.com/articles/nrc.2016.108, accessed 2022-06-14.
- P. Chakravarty, W. Qian, M. A. El-Sayed and M. R. Prausnitz, Delivery of Molecules into Cells Using Carbon Nanoparticles Activated by Femtosecond Laser Pulses, Nat. Nanotechnol., 2010, 5(8), 607–611, DOI:10.1038/nnano.2010.126.
- P. Zhou, X.-L. Yang, X.-G. Wang, B. Hu, L. Zhang, W. Zhang, H.-R. Si, Y. Zhu, B. Li, C.-L. Huang, H.-D. Chen, J. Chen, Y. Luo, H. Guo, R.-D. Jiang, M.-Q. Liu, Y. Chen, X.-R. Shen, X. Wang, X.-S. Zheng, K. Zhao, Q.-J. Chen, F. Deng, L.-L. Liu, B. Yan, F.-X. Zhan, Y.-Y. Wang, G.-F. Xiao and Z.-L. Shi, A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin, Nature, 2020, 579(7798), 270–273, DOI:10.1038/s41586-020-2012-7.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N.-H. Wu, A. Nitsche, M. A. Müller, C. Drosten and S. Pöhlmann, SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, 2020, 181(2), 271–280.e8, DOI:10.1016/j.cell.2020.02.052.
- W. Li, M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe and M. Farzan, Angiotensin-Converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus, Nature, 2003, 426(6965), 450–454, DOI:10.1038/nature02145.
- M. C. Sportelli, M. Izzi, E. A. Kukushkina, S. I. Hossain, R. A. Picca, N. Ditaranto and N. Cioffi, Can Nanotechnology and Materials Science Help the Fight against SARS-CoV-2?, Nanomater, 2020, 10(4), 802, DOI:10.3390/nano10040802.
- Y. Fujimori, T. Sato, T. Hayata, T. Nagao, M. Nakayama, T. Nakayama, R. Sugamata and K. Suzuki, Novel Antiviral Characteristics of Nanosized Copper(I) Iodide Particles Showing Inactivation Activity against 2009 Pandemic H1N1 Influenza Virus, Appl. Environ. Microbiol., 2012, 78(4), 951–955, DOI:10.1128/AEM.06284-11.
- A. P. Ingle, N. Duran and M. Rai, Bioactivity, Mechanism of Action, and Cytotoxicity of Copper-Based Nanoparticles: A Review, Appl. Microbiol. Biotechnol., 2014, 98(3), 1001–1009, DOI:10.1007/s00253-013-5422-8.
- M. B. Gawande, A. Goswami, F.-X. Felpin, T. Asefa, X. Huang, R. Silva, X. Zou, R. Zboril and R. S. Varma, Cu and Cu-Based Nanoparticles: Synthesis and Applications in Catalysis, Chem. Rev., 2016, 116(6), 3722–3811, DOI:10.1021/acs.chemrev.5b00482.
- H. L. Karlsson, P. Cronholm, Y. Hedberg, M. Tornberg, L. De Battice, S. Svedhem and I. O. Wallinder, Cell Membrane Damage and Protein Interaction Induced by Copper Containing Nanoparticles–Importance of the Metal Release Process, Toxicology, 2013, 313(1), 59–69, DOI:10.1016/j.tox.2013.07.012.
- A. K. Srivastava, N. Dwivedi, C. Dhand, R. Khan, N. Sathish, M. K. Gupta, R. Kumar and S. Kumar, Potential of Graphene-Based Materials to Combat COVID-19: Properties, Perspectives, and Prospects, Mater. Today Chem., 2020, 18, 100385, DOI:10.1016/j.mtchem.2020.100385.
- M. Sametband, I. Kalt, A. Gedanken and R. Sarid, Herpes simplex virus type-1 attachment inhibition by functionalized graphene oxide, ACS Appl. Mater. Interfaces, 2014, 6(2), 1228–1235, DOI:10.1021/am405040z.
- Z. Song, X. Wang, G. Zhu, Q. Nian, H. Zhou, D. Yang, C. Qin and R. Tang, Virus Capture and Destruction by Label-Free Graphene Oxide for Detection and Disinfection Applications, Small, 2015, 11(9–10), 1171–1176, DOI:10.1002/smll.201401706.
- A. Pouremamali, A. Babaei, S. S. Malekshahi, A. Abbasi and N. Rafiee, Understanding the Pivotal Roles of ACE2 in SARS-CoV-2 Infection: From Structure/Function to Therapeutic Implication, Egypt. J. Med. Hum. Genet., 2022, 23(1), 103, DOI:10.1186/s43042-022-00314-9.
- A. Khan, A. Rashid, R. Younas and R. Chong, A Chemical Reduction Approach to the Synthesis of Copper Nanoparticles, Int. Nano Lett., 2016, 6(1), 21–26, DOI:10.1007/s40089-015-0163-6.
- R. Cassano, F. Curcio, M. L. Di Gioia and S. Trombino, Chapter 16 - Copper Nanoparticles-Based Stimuli-Responsive Approaches, in Stimuli-Responsive Nanocarriers, ed. Gajbhiye, V., Gajbhiye, K. R. and Hong, S., Academic Press, 2022, pp. 413–428, DOI:10.1016/B978-0-12-824456-2.00015-1.
- S. Tabassum, R. Kumar and L. Dong, Nanopatterned Optical Fiber Tip for Guided Mode Resonance and Application to Gas Sensing, IEEE Sens. J., 2017, 17(22), 7262–7272, DOI:10.1109/JSEN.2017.2748593.
- P. Prabakaran, X. Xiao and D. S. Dimitrov, A Model of the ACE2 Structure and Function as a SARS-CoV Receptor, Biochem. Biophys. Res. Commun., 2004, 314(1), 235–241, DOI:10.1016/j.bbrc.2003.12.081.
- C. Peng, C. Shen, S. Zheng, W. Yang, H. Hu, J. Liu and J. Shi, Transformation of CuO Nanoparticles in the Aquatic Environment: Influence of PH, Electrolytes and Natural Organic Matter, Nanomaterials, 2017, 7(10), 326, DOI:10.3390/nano7100326.
- Y. Hokita, M. Kanzaki, T. Sugiyama, R. Arakawa and H. Kawasaki, High-Concentration Synthesis of Sub-10-Nm Copper Nanoparticles for Application to Conductive Nanoinks, ACS Appl. Mater. Interfaces, 2015, 7(34), 19382–19389, DOI:10.1021/acsami.5b05542.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |