Open Access Article
Open Access ArticleOvercoming microbial resuscitation using stable ultrafine gold nanosystems†
Anindita
Thakur
a,
Pranay Amruth
Maroju
a,
Ramakrishnan
Ganesan
 *b and
Jayati
Ray Dutta
*b and
Jayati
Ray Dutta
 *a
*a
aDepartment of Biological Sciences, Birla Institute of Technology and Science (BITS), Pilani, Hyderabad Campus, Jawahar Nagar, Kapra Mandal, Medchal District, Hyderabad, Telangana – 500078, India. E-mail: jayati@hyderabad.bits-pilani.ac.in
bDepartment of Chemistry, Birla Institute of Technology and Science (BITS), Pilani, Hyderabad Campus, Jawahar Nagar, Kapra Mandal, Medchal District, Hyderabad, Telangana – 500078, India. E-mail: ram.ganesan@hyderabad.bits-pilani.ac.in
First published on 7th March 2024
Abstract
While ultrafine gold nanosystems (UGNs) are being extensively studied for their antimicrobial activities, hitherto, no report is available on their propensity towards mitigating bacterial resuscitation—a potential factor contributing to the antimicrobial resistance. The investigations herein with two categories of gold nanosystems—modulated for their stability and surface accessibility through glutathione capping—have provided insights into overcoming resuscitation. Additionally, the study cautions that even moderate resistance development in bacteria exposed to nanosystems can result in significant cross-resistance against conventional antibiotics.
Gold nanosystems (nanoparticles (NPs) and nanoclusters (NCs)) have recently emerged as high-potential wide-spectrum antimicrobials, which could possibly be a clinical reality in the near future for combating microbe-borne infections, as witnessed by the feasibility of oral administration, facile renal excretion and selective ability to target prokaryotic over eukaryotic cells.1–9 Recent reports have highlighted the influence of surface capping agents on the antimicrobial performance of gold nanosystems.10–15 Besides, nano-antimicrobials have been shown to function differently from traditional antibiotics and hence it is possible that they may differ in mechanisms of resistance development compared to conventional counterparts.16–21 Nevertheless, extensive studies are required to understand the microbial resistance against nanosystems. Another vital aspect that could potentially contribute to resistance is resuscitation—a revival phenomenon from an apparently dead or quiescent phase—which is mainly elicited by dormant phenotypes like persister and viable-but-nonculturable cells (VBNCs).22 A review article by Li et al., describes the characteristics and significance of VBNCs in human bacterial pathogens.23 However, to the best of our knowledge, no report is available to date on the microbial resuscitation propensity of metal-based nanosystems.
Recently, our group has reported the synthesis of weakly-capped infant Au NCs through a citric acid-mediated pseudo-solid-phase approach, wherein citric acid acts as a reducing agent and a weak protectant (Fig. S1†).24 When added to water, the citric acid matrix dissolves instantaneously, facilitating maximum surface accessibility of NCs, leading to record-high catalytic activity. However, the high surface energy of these weakly-capped NCs makes them prone to aggregation within minutes, which significantly affects the catalytic activity at later stages. Hence it is reasonable to term such systems as burst-active but labile. On the contrary, nanosystems stabilized using non-toxic strong capping agents may mask the gold surface and decrease the activity but would impart stability against aggregation, providing longevity.10,25
In the current work, the surface accessibility and stability of these weakly-capped Au NCs were modulated using glutathione (GSH) as the capping agent via a ligand-displacement approach.26–28 This protectant plays a dual role of concentration-dependent blocking of Au NCs' active sites and presenting a microbe-friendly environment due to its non-toxic nature.25 We developed two categories of gold nanosystems, namely, (i) non-GSH-capped and (ii) (partially or fully) GSH-capped, to study their antibacterial activity against a representative Gram-negative bacterium Escherichia coli K-12 (DH5α). It is well-known that Gram-negative microbes have a higher propensity to develop resistance than Gram-positive ones—a factor majorly attributed to the thinner outer membrane as well as extensive efflux pumps and porin channels in the former.29
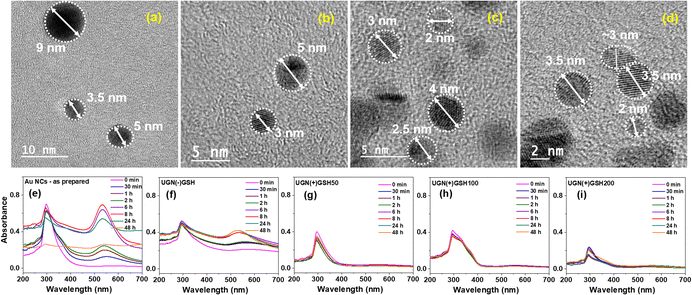
It can be noted that the matrix of our as-synthesized Au NCs was predominantly composed of citric acid (CA), with an estimated composition of 96%. Therefore, prior to modulating the stability using GSH, the effect of pristine CA on the antibacterial property was initially explored by resazurin assay. The study revealed significant toxicity exerted by pristine CA in the concentration range of ∼50–500 μg mL−1, which was attributed to the acid stress caused by the pH value of <4 (Fig. S2a†). To nullify this, CA in the Au NCs was neutralized with a stoichiometric quantity of NaOH to render the ensuing sodium citrate matrix microbe-friendly, which was ascertained through a control experiment devoid of Au NCs (Fig. S2b†). This approach paved the path for understanding the antimicrobial efficacy of the NCs exclusively. The neutralized solution having a pH of 6.5–7.0 was termed as UGN(−)GSH, while those added with different amounts of GSH, such as 50%, 100% and 200% with respect to gold atoms, were termed UGN(+)GSH50, UGN(+)GSH100 and UGN(+)GSH200, respectively. In the case of UGN(−)GSH, the high-resolution transmission electron microscopy (HR-TEM) analysis revealed the particle sizes majorly in the range of 4–9 nm. However, in GSH-capped gold nanosystems, the particle size was mostly found to be less than 4 or 5 nm (Fig. 1). Since this size indicates the formation of NPs, additional study was required to confirm any plasmonic signature (vide infra). It can be noted that the aggregation during the sampling could also have resulted in a slightly higher size. Our earlier work on weakly-capped Au NCs required a customized solid-phase sampling procedure over a holey carbon grid to correctly identify the diameter as sub-2 nm, as otherwise, the conventional liquid phase sampling over Cu grid resulted in particle aggregation/growth.
Initially, the binding of GSH over the surface-active sites of Au NCs was assessed using liquid chromatography (LC; Fig. S3†) and 1H NMR spectroscopy (Fig. S4†), following which its effect on the modulation of stability was probed using electronic spectroscopy (Fig. S5† and 1). The as-synthesized Au NCs exhibited the signature peak of NCs at ∼300 nm in solution, whose intensity, upon incubation, was found to decrease with time, accompanied with a concomitant plasmonic peak appearance at ∼520 nm due to the growth of NCs into NPs.24 Such a plasmonic peak formation was also observed with UGN(−)GSH from the 6th hour of incubation; however, the overall particle growth was found to be slower. On the contrary, no such growth was observed with the GSH-capped NCs, starting from 50% GSH content, even till 48 h. Thus, correlating with the HR-TEM analyses, the slight increase in the particle size in TEM measurements could be attributed to the drying step in the sample preparation process (vide supra). Nevertheless, these particles may fall within the size range that overlaps NCs and NPs, so they can be deemed ultrafine gold nanosystems (UGNs).
Resazurin assay was followed to assess the inhibitory concentration-50 (IC50) of the UGNs by incubating them for 6 h with the bacterial cultures.30,31 Both UGN(−)GSH and UGN(+)GSH50 exhibited an identical IC50 of 8.3 μg mL−1. On the other hand, the IC50 increased to 55 μg mL−1 for UGN(+)GSH100, whereas, even at this elevated concentration, UGN(+)GSH200 did not show any significant bactericidal activity (Fig. 2a). These findings indicate a reduced bactericidal performance with increased GSH capping, likely due to decreased gold surface availability. Minimum inhibitory concentration (MIC) using the conventional two-fold dilution method was also evaluated for UGN(−)GSH and UGN(+)GSH50, which revealed their MIC values as 8.3 μg mL−1 (equal to IC50) and 16.6 μg mL−1 (2 × IC50), respectively (Table S2†). As a control antibiotic, ampicillin demonstrated an IC50 and MIC of 1 μM (0.35 μg mL−1) and 2 μM (0.7 μg mL−1), respectively (Fig. 2b and Table S2†).
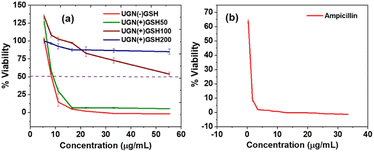 | ||
| Fig. 2 The bacterial viability studies in triplicate using resazurin assay with the (a) GSH-capped and non-GSH-capped UGNs and (b) ampicillin. | ||
The minimum bactericidal concentration (MBC) of these nano-antimicrobials was then assessed using the standard plating method. As the treatment duration in the resazurin assay was fixed as 6 h, in MBC studies, the bacterial cultures were treated with UGN(−)GSH and UGN(+)GSH50 for a similar duration, followed by agar plate incubation for 24 h (Fig. S6†).32 UGN(−)GSH exhibited an MBC of 8 × IC50, while MBC was not reached with UGN(+)GSH50, even at 16 × IC50, indicating the decreased bactericidal activity due to GSH capping of surface gold atoms. It is noteworthy that the resazurin assay quantifies the metabolically active state and does not account for persister/VBNC cells, which can resuscitate in a nutrient-rich medium.22,23 Therefore, the higher bacterial growth in the GSH-capped UGNs can be attributed to two factors: (i) decreased availability of surface gold atoms with GSH capping, leading to decreased activity and (ii) increased stability imparted by the non-toxic protectant, resulting in slowing down the initial stronger burst attack.
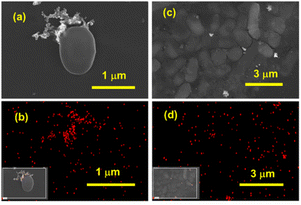
Field-emission scanning electron microscopy (FE-SEM) imaging and elemental mapping of gold were performed on cultures treated with 2 × IC50 concentrations of UGN(−)GSH and UGN(+)GSH50 (Fig. 3). With UGN(−)GSH, a small amount of pellet was obtained and as a consequence only a few isolated bacteria were observed under FE-SEM. These UGNs were found to be adsorbed and aggregated on the cell surface of these bacteria. On the other hand, a significant bacterial colonization, with a relatively even distribution of UGNs was observed with the UGN(+)GSH50.
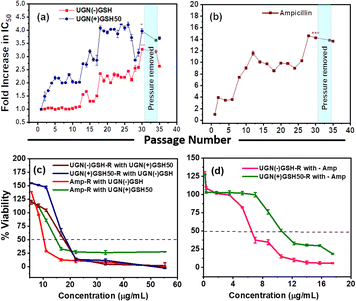
To assess resistance development upon repeated exposure, the bacterial cultures were subjected to 30 cycles of passaging (∼5400 generations) by periodic exposure to IC50 values of UGN(−)GSH and UGN(+)GSH50 (Fig. 4).32,33 A control study with no antimicrobial treatment was also conducted to eliminate any influence from environmental factors. Following 30 passages with ampicillin, a 14-fold increase in IC50 and a 16-fold increase in MIC were observed. In comparison to the ancestor population (not subjected to passaging), the bacteria started developing resistance against ampicillin and UGN(+)GSH50 from passage-2 onwards. On the contrary, no resistance development was observed until 12 passages with UGN(−)GSH, after which resistance gradually emerged. We speculate that the initial burst activity leading to a stronger attack by the UGNs likely contributed to delaying resistance development. Nevertheless, after 30 passages, the bacteria developed ∼3.5–4-fold resistance against both UGN(−)GSH and UGN(+)GSH50. The resistance persisted even after temporarily lifting the antimicrobial pressure for three cycles and then reinstating it, indicating horizontal and vertical transfer of the resistance gene(s), signifying heritability. The studies pertaining to the gold uptake and oxidative stress are presented in the ESI,† aiming to illuminate potential interaction mechanisms (Fig. S7 and Table S3†). The population resistant to one antimicrobial agent was then studied for any cross-resistance or collateral sensitivity (enhanced sensitivity) against the remaining two. The cultures passaged with UGN(−)GSH showed ∼2-fold cross-resistance against UGN(+)GSH50 and vice versa. This ∼2-fold cross-resistance, instead of the expected ∼3.5–4-fold, may be considered a manifestation of collateral sensitivity, given the similarities in chemical nature between both UGN categories, apart from partial surface coverage with GSH. Conversely, the ampicillin-resistant strain showed a cross-resistance of 1.5–2-fold relative change in the IC50 against the UGNs. However, the cultures resistant to the UGNs exhibited significant cross-resistance (∼20–30-fold) against ampicillin, possibly due to the activation of multi-modal resistance pathways.
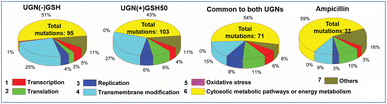
To derive preliminary insights into the genetic level changes underlying bacterial resistance, we employed a total-population passaging strategy to uncover the potential mechanistic pathways adopted by the microbes. The investigation was carried out using next-generation sequencing (NGS) over the ancestor, control and resistant populations. Significantly, with both types of UGNs, hundreds (∼160–200) of single nucleotide polymorphisms (SNPs) and a few insertions/deletions (in-dels) were found in the coding regions of the resistant populations' genomes. On the other hand, only 32 mutations were observed in the ampicillin-resistant population. The mutations were categorized based on the event's occurrence specific to non-GSH-capped or GSH-capped UGNs and those in common. Gene Ontology (GO) analyses, specifically examining biological processes, cellular components and molecular functions, revealed UGNs to have a higher enrichment score and a greater impact on processes compared to ampicillin (Fig. S8†). Processes were further classified based on gene function and location, and the results are presented in a pie–chart diagram, highlighting a profound impact on genes involved in cytosolic metabolic and transmembrane modification processes (Fig. 5). Notably, the percentage of transmembrane modification was significantly higher with UGNs compared to ampicillin, indicating the active role of membrane components (proteins, porins, efflux pumps, etc.) in resistance evolution. These observations indicate a severe and multi-faceted attack by the UGNs. For further insights into the specific pathways impacted by mutations, additional analysis was conducted using the STRING software tool (Fig. S9†). Only mutations common to both UGN categories were processed by the software and prioritized by the false discovery rate (FDR) (Table S4†). The major pathways impacted were metabolic, oxidative phosphorylation, and RNA degradation.
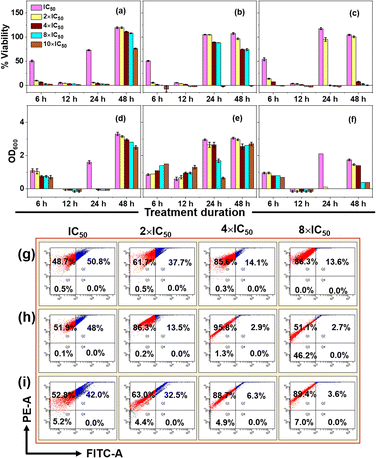
To investigate bacterial resuscitation dynamics, extended time-point studies with all the formulations up to 48 h in the concentration range of IC50 to 10 × IC50 were conducted and the results are presented in Fig. 6. For this, absorbance at 600 nm (OD600) and resazurin assay were performed to differentiate the aggregation of the UGNs and cell viability. An increase in OD600 values may result from cell growth, particle aggregation, or a combination of both factors (Fig. S10†). Cell viability decreased with increasing antimicrobial concentration at 6 h. By 12 h, all concentrations inhibited bacterial growth. Notably, the resazurin assay showed gradual cell resuscitation at 24 and 48 h with lower antimicrobial concentrations, while higher concentrations maintained complete inhibition. In particular, a clear distinction between the GSH-capped and non-GSH-capped UGNs was observed, wherein UGN(−)GSH exhibited an initial burst activity, but the same did not sustain later time points due to aggregation, as indicated by the higher OD600 values due to light scattering by aggregated particles. Complete resuscitation of cells against UGN(−)GSH was observed up to 8 × IC50 from 24 h onwards, with no viability at 10 × IC50 until 48 h. In contrast, UGN(+)GSH50 and UGN(+)GSH100 elicited modest activity up to 6 h, but exhibited sustained activity due to their stability, ensuring a persistent antimicrobial threat. Only a small degree of resuscitation was observed in these GSH-capped cases, even with concentrations as low as 4 × IC50 at 48 h. It can be noted that the control experiment with ampicillin showed a similar trend till 24 h, but at 48 h, complete resuscitation was observed with all the employed concentrations (up to 10 × IC50).
To gain insights into the concentration of the antimicrobials and the degree of cell rupture, fluorescence-activated cell sorter (FACS) analysis was conducted on microbial cultures treated with ampicillin, UGN(−)GSH and UGN(+)GSH50 in the concentration range of IC50 to 8 × IC50, beyond which cell pellets were insufficient for analysis (Fig. 6g–i). The sorted cells were divided into Q1 (dead) and Q2 (viable, including lethally injured, persister and VBNC) categories.32 In general, the population of dead cells increased with higher antimicrobial concentrations. At 8 × IC50, ampicillin exhibited a cell population of ∼14–15% in Q2. In contrast, the UGNs resulted in less than 5% cell population in this quadrant, indicating a significantly higher degree of cell rupture. For UGN(−)GSH at 8 × IC50, nearly half the population was observed in Q3 (unstained region), plausibly due to the high concentration of labile nanosystems aggregated over the cell surface and thereby interfering with the sorting process. Nevertheless, it is evident from the study that at appropriate concentrations of UGNs, the cell damage was severe to the extent that they could not be resuscitated, while such a degree of cell damage was not achieved with ampicillin at similar IC50 levels.
To further validate the time-point studies presented in Fig. 6, the MBC of UGN(−)GSH and UGN(+)GSH50 was determined by incubating them with the bacterial culture for 6, 12, 24 and 48 h followed by plating (Table S5†). The results mirrored the resazurin assay trends for both categories of UGNs. At 16 × IC50, UGN(−)GSH completely failed to inhibit the bacterial growth from 24 h onwards, while UGN(+)GSH50 effectively prevented resuscitation even at a lower concentration of 12 × IC50 in the 24–48 h range. These observations highlight the steady-but-sustained activity of UGN(+)GSH50 as its key strength in preventing resuscitation.
Conclusions
In summary, two categories of UGNs were employed, whose stability against aggregation was modulated through GSH capping. Their antimicrobial performance exhibited distinct trends: burst activity with the less stable ones and steady-but-sustained activity with the more stable ones. Passaging studies revealed the development of modest but heritable resistance against both categories of UGNs upon periodic exposure to IC50 levels in 30 days. Importantly, in a direct comparison with the conventional antibiotic, the lineages with 3–4-fold resistance to UGNs exhibited ∼20–30-fold cross-resistance against ampicillin (an observation that prompts additional studies on the collateral impact). Even with moderate resistance development against UGNs, such a profound effect was attributed to the multi-modal counter-response by microbes, as revealed by the hundreds of SNPs and several in-dels in the gene sequencing analyses over the resistant populations. Resuscitation studies showed the failure of the conventional antibiotic in resisting the bacterial revival after 24 h, even at 10 × IC50, whereas stable UGNs effectively resisted bacterial resuscitation at much lower IC50 values, revealing the persistent antimicrobial activity. Overall, these studies revealed that the appropriate nature, treatment duration and concentration of UGNs could play a vital role in countering microbial resistance. Future works replacing the non-toxic GSH with potent antimicrobial substances might be an interesting strategy for enhancing the multi-modal attack.4,34 The need for deeper insight into the intricate relationship between bacteria and UGNs has been emphasized in the study, with the aim of devising more effective approaches to overcome bacterial resistance and resuscitation. Further research is imperative to fully comprehend this complex interplay.Author contributions
AT: investigation, formal analysis, validation. PAM: formal analysis, validation. JRD: conceptualization, methodology, resources, supervision, writing – review and editing. RG: conceptualization, methodology, resources, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the Department of Science and Technology (DST) for the financial support (File No. SPG/2021/004518). The authors thank Prof. K. N. Mohan and Ms. Anuhya Anne from the Department of Biological Sciences, BITS Pilani Hyderabad Campus (BPHC) for their useful assistance with the Partek software for gene analysis. The XPS and FE-SEM facilities of the Central Analytical Laboratory of BPHC are greatly acknowledged.Notes and references
- Z. Pang, et al. , Chem. Sci., 2021, 12, 14871–14882 RSC.
- K. Zheng, et al. , ACS Nano, 2017, 11, 6904–6910 CrossRef CAS PubMed.
- G. B. Hwang, et al. , Nat. Commun., 2020, 11, 1207 CrossRef CAS PubMed.
- H. Tang, et al. , Angew. Chem., Int. Ed., 2021, 60, 13829–13834 CrossRef CAS PubMed.
- P. J. Weldick, et al. , Nanoscale, 2022, 14, 4018–4041 RSC.
- Z. Cao, et al. , Nanoscale, 2022, 14, 10016–10032 RSC.
- Z. Cao, et al. , ACS Appl. Bio Mater., 2020, 3, 5275–5286 CrossRef CAS PubMed.
- E. M. Higbee-Dempsey, et al. , J. Am. Chem. Soc., 2020, 142, 7783–7794 CrossRef CAS PubMed.
- R. Chowdhury, et al. , Nanoscale, 2017, 9, 14074–14093 RSC.
- K. Zheng, et al. , Chem. Mater., 2018, 30, 2800–2808 CrossRef CAS.
- Y. Zheng, et al. , Bioconjugate Chem., 2018, 29, 3094–3103 CrossRef CAS PubMed.
- D. Pranantyo, et al. , Biomacromolecules, 2019, 20, 2922–2933 CrossRef CAS PubMed.
- S. Zhu, et al. , ACS Appl. Mater. Interfaces, 2020, 12, 11063–11071 CrossRef CAS PubMed.
- W. Ndugire, et al. , Angew. Chem., Int. Ed., 2023, 62, e202214086 CrossRef CAS PubMed.
- C. Y. Tay, et al. , Nano Res., 2014, 7, 805–815 CrossRef CAS.
- M. Miethke, et al. , Nat. Rev. Chem, 2021, 5, 726–749 CrossRef CAS PubMed.
- E. Valentin, et al. , Nanoscale, 2020, 12, 2384–2392 RSC.
- H. Wang, et al. , Nat. Commun., 2021, 12, 3331 CrossRef CAS PubMed.
- M. Xie, et al. , Angew. Chem., Int. Ed., 2023, e202217345 CAS.
- H. Mude, et al. , Adv. Ther., 2023, 6, 2300015 CrossRef CAS.
- Y. Xie, et al. , Angew. Chem., Int. Ed., 2018, 57, 3958–3962 CrossRef CAS PubMed.
- M. Ayrapetyan, et al. , Infect. Immun., 2015, 83, 4194–4203 CrossRef CAS PubMed.
- L. Li, et al. , Front. Microbiol., 2014, 5, 258 Search PubMed.
- D. Patra, et al. , Nanoscale Adv., 2020, 2, 5384–5395 RSC.
- K. Zheng, et al. , Nano Res., 2021, 14, 1026–1033 CrossRef CAS.
- W. Suzuki, et al. , J. Am. Chem. Soc., 2022, 144, 12310–12320 CrossRef CAS PubMed.
- Y. Cao, et al. , Nat. Commun., 2020, 11, 5498 CrossRef CAS PubMed.
- L. Chen, et al. , Nano Res., 2023, 16, 7770–7776 CrossRef CAS.
- S. J. Lam, et al. , Nat. Microbiol., 2016, 1, 1–11 Search PubMed.
- H. Mude, et al. , Food Chem., 2022, 374, 131830 CrossRef CAS PubMed.
- H. Mude, et al. , ACS Food Sci. Technol., 2022, 2, 368–377 CrossRef CAS.
- H. Mude, et al. , ACS Appl. Bio Mater., 2021, 4, 8396–8406 CrossRef CAS PubMed.
- W. Zheng, et al. , Nano Lett., 2021, 21, 1992–2000 CrossRef CAS PubMed.
- G. Giunta, et al. , ACS Nano, 2023, 17, 23391–23404 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00046c |
| This journal is © The Royal Society of Chemistry 2024 |