Open Access Article
Open Access ArticleZeolitic imidazolate framework-8 encapsulated with Mo-based polyoxometalates as surfaces with antibacterial activity against Escherichia coli†
Mariam M.
Abdelkhalek
a,
Aya M.
Mohamed
b,
Rehab Z.
Abdallah
 c,
Ghada E.
Khedr
c,
Ghada E.
Khedr
 d,
Rania
Siam
c and
Nageh K.
Allam
d,
Rania
Siam
c and
Nageh K.
Allam
 *a
*a
aEnergy Materials Laboratory, Physics Department, School of Sciences and Engineering, The American University in Cairo, New Cairo 11835, Egypt. E-mail: nageh.allam@aucegypt.edu
bDepartment of Chemistry, Faculty of Science, Cairo University, Cairo 12613, Egypt
cDepartment of Biology, School of Sciences and Engineering, The American University in Cairo, New Cairo 11835, Egypt
dDepartment of Analysis and Evaluation, Egyptian Petroleum Research Institute, Cairo 11727, Egypt
First published on 2nd May 2024
Abstract
Bacterial infections represent a major global health concern, causing millions of deaths and a significant economic burden. The development of antibacterial nanoporous surfaces with potential mechano-bactericidal effects can revolutionize infection control practices. In this study, a hybrid material of zeolitic imidazolate framework-8 (ZIF-8) doped with phosphomolybdic acid (PMA) was synthesized and characterized by field emission scanning electron microscopy (FESEM), energy-dispersive X-ray spectroscopy (EDS), X-ray diffraction (XRD), Fourier-transform infrared spectroscopy (FTIR), and N2 sorption isotherms. PMA@ZIF-8 performance as an antibacterial agent against E. coli was superior to that of its individual constituents, suggesting a synergistic effect of PMA and ZIF-8. The incorporation of PMA into ZIF-8 significantly enhanced its antibacterial efficacy, as evidenced by a twofold reduction in MIC (375 μg mL−1vs. 750 μg mL−1) and a 4.35 times increase in the bactericidal kinetics rate constant. The time-kill curve experiment revealed that PMA@ZIF-8 achieved a 3-log reduction within 7 hours, whereas ZIF-8 required 24 hours to reach the same level of reduction. The density functional theory (DFT) calculated bandgap of PMA@ZIF-8 was significantly less than that of ZIF-8. Also, PMA@ZIF-8 has caused the elimination of 56.72% of the thiol group as detected by Ellman's assay. Accordingly, PMA@ZIF-8 can be both computationally and experimentally demonstrated as an oxidative nanozyme. PMA@ZIF-8's surface topology revealed nanorod protrusions, suggesting a potential mechano-bactericidal effect, which was confirmed by live/dead assay on PMA@ZIF-8-coated glass. This study highlights the potential of the PMA@ZIF-8 hybrid as a highly effective antibacterial agent, holding promise for creating multifunctional antibacterial surfaces.
1. Introduction
More than 7.7 million human deaths in 2019 were caused by bacterial infections.1 This makes bacterial-related deaths the second leading cause of mortality worldwide, being responsible for 13.6% of all global deaths in the same year.1 On the other hand, bacterial diseases cause a substantial economic burden around the world. It is difficult to estimate the exact total economic losses caused by bacterial infections. However, the USA spent as much as 5.6 billion dollars on the treatment of 7 drug-resistant bacteria in 2017.2 This enormous number may reflect the magnitude of the economic disaster of bacteria-caused infections and diseases. Disinfecting environmental surfaces is a critical aspect of infection control strategies to prevent the transmission of bacterial infections by indirect contact.3 Classically, several chemical and physical methods are implemented to disinfect and sterilize surfaces including ultraviolet (UV) irradiation, hydrogen peroxide systems, ethylene oxide gas, and liquid chemical disinfectants, including quaternary ammonium compounds and chlorine-based disinfectants.3 However, all these methods are limited by their short-term effectiveness; even after efficient disinfection, surfaces can rapidly get recontaminated.3Consequently, the development of antibacterial surfaces can significantly contribute to addressing the persistent challenge of surface contamination, which would revolutionize infection control practices.4,5 Antibacterial surfaces can exhibit their effect through either release-based systems with controlled elution of antibacterial agents or mechano-bactericidal surfaces. The latter induce antibacterial effects by mechanical stress and rupture of the bacterial cell because of the topological roughness and nanosized protrusions or pillars.6 The development of a highly porous nanomaterial that has topological roughness can be a promising candidate for the creation of antibacterial surfaces with combined release-based and mechano-bactericidal effects.7
In 1995, a breakthrough emerged in the field of nanoporous materials with the introduction of metal–organic frameworks (MOFs).8 MOFs are primarily composed of metal nodes or clusters attached by coordination bonds to organic linkers, constituting a highly customizable and adjustable porous structure and an exceptionally high surface area ranging from 1000 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 m2 g−1.8–10 Zeolite-imidazolate frameworks (ZIFs) are a subclass of MOFs that were first introduced in 2006.11,12 They are composed of metal nodes (M) tetrahedrally coordinated with 4 imidazolate-based organic linkers (IM), each acting as a bridge between two metal nodes, typically a transition metal (e.g., Co, Zn).11–13 The metal–linker–metal (M–IM–M) angle is topologically similar to the Si–O–Si angle in zeolites (145°). Hence, the name of that class of MOFs was coined as zeolite-imidazolate frameworks.14
000 m2 g−1.8–10 Zeolite-imidazolate frameworks (ZIFs) are a subclass of MOFs that were first introduced in 2006.11,12 They are composed of metal nodes (M) tetrahedrally coordinated with 4 imidazolate-based organic linkers (IM), each acting as a bridge between two metal nodes, typically a transition metal (e.g., Co, Zn).11–13 The metal–linker–metal (M–IM–M) angle is topologically similar to the Si–O–Si angle in zeolites (145°). Hence, the name of that class of MOFs was coined as zeolite-imidazolate frameworks.14
ZIF-8 is a Zn-based ZIF that was among the first introduced ZIFs in 2006 and immediately gained attention due to its high stability.12,15 Pristine ZIF-8 has been evidenced to have weak antibacterial activity, which is mediated mainly by Zn2+ cations which trigger the creation of reactive oxygen species (ROS).16,17 ZIF-8 has multiple characteristics that inspired researchers to use it in applications with infection control purposes. Its hydrophobic nature was employed in the synthesis of water-repellent sprayable coatings that resist the attachment of microorganisms.18 Being photoactive, ZIF-8 has been incorporated into air filtration systems with photocatalytic activity.19,20 Due to its unique porous structure, ZIF-8 can be utilized as a carrier for potent antibacterial agents, enabling controlled release systems such as the curcumin-release system for enhanced food packaging.21 Moreover, both pristine ZIF-8 and composites based on ZIF-8 have been employed to modify cotton fabrics, resulting in the production of antibacterial cotton textiles.5,15
In this study, we introduce a novel approach to enhance the antibacterial efficacy of ZIF-8 via doping with phosphomolybdic acid (PMA), a member of the broad family of polyoxometalates (POMs).22,23 POMs are polyatomic anionic clusters composed of transition metal ions (usually tungsten, molybdenum, or vanadium) in a high oxidation state, with oxygen ligands bridging in between, making a polymeric three-dimensional framework.22,23 The unique structure of POMs, with high affinity for electron acquisition, renders them “electron sponges” with remarkable redox activity.24
Although the first discovered POM (ammonium phosphomolybdate, [PMo12O40]3− anion) was reported as early as 1826, and the Keggin structure of phosphotungstate was characterized in 1934, the potential antibacterial properties of a POM were not described until 1993.25,26 After that, several POMs have shown promising independent and synergistic antibacterial actions in multiple Gram-negative and Gram-positive bacteria.26,27 POMs are water-soluble agents, that can cause their antibacterial activity by penetration of the outer membrane,27,28 causing oxidation of different electron carriers (e.g., NADH), leading to paralysis of the bacterial respiratory chain.26,29 POMs are also strong oxidizing agents which can create ROS.26,29
The recent literature has extensively studied the PMA and ZIF-8 antimicrobial properties,14,26 yet their combined synergistic effect has not been reported. Several factors have driven the interest in the current study to test the antibacterial properties of the PMA@ZIF-8 hybrid. Firstly, the antibacterial mechanisms of action of both ZIF-8 and PMA seem to complement each other, with potential multiple target shooting and subsequent synergy. Secondly, based on a recent finding that Mo@ZIF-8 acts as a nanozyme with strong capability of ROS generation,30 we expect that the PMA@ZIF-8 hybrid may also exhibit similar activity since PMA's structure contains Mo as the transition metal atom. In addition, since PMA has a strong anionic structure, the electrostatic attraction between Zn2+ cations and PMA anions during crystal growth is anticipated to induce topological changes and surface roughness in the hybrid structure,31 a characteristic that is known to enhance mechano-bactericidal effects.
Therefore, this study investigated the antibacterial activity of PMA@ZIF-8 on the model organism Escherichia coli (E. coli) and compared it to that of the individual compounds using CFU counting, measuring the minimum inhibitory concentration, and bacterial death kinetics. Additionally, we computationally predicted the potential oxidative nanozyme effect of the hybrid PMA@ZIF-8 using density functional theory (DFT). DFT was proven to be a robust tool for investigating electronic properties32–35 and drug design.36 Our study presents PMA@ZIF-8 as a promising material suitable for incorporation into antibacterial surfaces applicable across healthcare facilities, and public and household spaces.
2. Experimental section
2.1. Utilized reagents and bacteria
Phosphomolybdic acid (H3PMo12O40, PMA), Zn(NO3)2·6H2O (zinc nitrate hexahydrate), 2-methylimidazole (2-MeIM), Luria–Bertani (LB) broth (Difco), LB agar (Difco), resazurin dye (Loba) acridine orange (Loba), propidium iodide (Loba), DTNB (YB Pharmacy), N-acetyl cysteine (Sedico) and physiological saline (NaCl 0.9%) were used. TOP10 chemically competent E. coli was used as a model organism.2.2. Synthesis of PMA@ZIF-8
A simple co-precipitation reaction was conducted to synthesize the PMA@ZIF-8 MOF material. Typically, 0.3 g of Zn(NO3)2·6H2O and 0.055 g of PMA were dissolved in 5 mL deionized water (DI) H2O to form solution A. Afterwards, 2.75 g of 2-MeIM was dissolved in 20 mL DI H2O via magnetic stirring to form solution B. Subsequently, solution B was added dropwise to solution A. After the homogeneous mixing of solutions A & B under magnetic stirring, a colloidal white precipitate was formed and was left to stand for 1 hour at room temperature without further stirring. Eventually, the white precipitate was collected and separated using centrifugation and it was washed with DI H2O several times to discard any unreacted residues. Finally, the as-prepared (PMA@ZIF-8) nanocomposite was dried at 60 °C overnight. For comparison, ZIF-8 was prepared through the same steps except for the addition of PMA.2.3. Materials characterization
The surface morphology of the prepared ZIF-8 and PMA@ZIF-8 hybrid was assessed via a Zeiss Ultra 60 field emission scanning electron microscope (FESEM). An X-ray diffractometer X'Pert PRO MRD with Cu kα radiation (λ = 0.15406 nm) was utilized to inspect the crystal structure. The structural functional groups were verified from the executed FTIR absorption curves in the range of 400–4000 cm−1via an ATI Unicam (Mattson 936) benchtop spectrometer using pressed KBr pellets. To gain more insights into the surface texture of the prepared materials, the N2 physisorption measurements were conducted at 77 K using a Microtrac BELSORP surface area and pore size distribution analyzer. Before conducting the measurements, the materials were degassed under vacuum at 150 °C overnight. The specific surface area (m2 g−1) was estimated using the BET model, whereas the pore size distribution was assessed from the desorption branch of the isotherm according to the BJH method.2.4. Antibacterial assays
2.5. Preparation of bacterial inocula
An E. coli suspension was prepared using the growth method as per CLSI recommendations.37 A single distinct E. coli colony was suspended in LB broth and incubated overnight at 37 °C. Following this, OD600 was measured and calibrated as required.2.6. PMA, ZIF-8, and PMA@ZIF-8 antibacterial activity screening
1000 μg mL−1 of PMA, ZIF-8, and PMA@ZIF-8 suspensions were mixed with 1–5 × 105 CFU mL−1E. coli. Positive control tubes with the same concentrations of the bacteria without the NPs were also prepared. Sterility control tubes containing 1000 μg mL−1 of NPs in LB broth and negative control tubes containing only LB broth were additionally prepared. All tubes were then incubated in a shaking incubator at 180 rpm for 24 hours at 26 ± 1 °C. Thereafter, serial dilutions of the tubes were spread on LB agar plates. The plates were incubated for 24 hours at 37 °C, and colony counts were then recorded. The CFU of the initial bacterial inoculum was also enumerated to set a reference point for calculating the reduction percentage. All experiments were performed in triplicate and mean values and standard deviations (SDs) were calculated. The reduction percentage and the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 inhibition were calculated using eqn (1) and (2), respectively.
10 inhibition were calculated using eqn (1) and (2), respectively. | (1) |
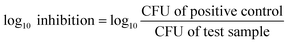 | (2) |
2.7. Minimum inhibitory concentrations (MICs) of the nanomaterials
MICs of ZIF-8 and PMA@ZIF-8 against E. coli were determined by the plate microdilution method in 96-well plates. Serial dilutions of ZIF-8 and PMA@ZIF-8 were prepared in physiological saline, and different concentrations of ZIF-8 and PMA@ZIF-8 were pipetted into wells along with the E. coli inoculum. Negative controls consisting of only LB broth and positive controls containing bacterial inocula were included. The concentration of E. coli in all tests and the positive controls were adjusted to 1–5 × 105 CFU mL−1. In addition, sterility controls were prepared, containing equal volumes of serial dilutions of NPs and LB broth. The bacterial inocula and the NP suspensions were vortexed before addition to the wells. Plates were incubated at 180 rpm in a shaking incubator for 18 hours at 37 °C. All experiments were performed in triplicate, and mean values and standard deviations (SDs) were calculated. In the case of MIC determination of the NPs the resazurin-based assay was favored over the optical density measurements (OD600) since it is less prone to interference by the optical density of nanoparticles.38 Viable bacterial cells reduce blue-colored, non-fluorescent resazurin to pink-colored, highly fluorescent resorufin.39 Resazurin-based assay was conducted according to the previously established protocol.39 Resazurin dye was dissolved in sterile physiological saline to a final concentration of 0.1 mg mL−1. Then, the solution was sterilized by syringe filtration. 50 μL of the resazurin solution was pipetted into each well and incubated for an additional 75 minutes. The fluorescence of the 96-well plate was measured with excitation at 570 nm and emission at 590 nm using a Tecan i-control (Infinite 200 PRO) plate reader. The fluorimetric MIC was determined as the concentration of the well showing no significant increase in fluorescence intensity as compared to the sterility control wells.2.8. Evaluation of bactericidal kinetics
To evaluate the time-dependent bactericidal effectiveness of PMA@ZIF-8 and ZIF-8 NPs against E. coli, NP suspensions were challenged using E. coli at a concentration of 1–5 × 105 CFU mL−1. The experiment was conducted at four times the MIC (1500 μg mL−1 PMA@ZIF-8 and 3000 μg mL−1 for ZIF-8). Sterility control tubes containing the same concentration of the NPs in the absence of bacteria were additionally prepared along with positive and negative control tubes. All tubes were incubated in a shaking incubator at 180 rpm at 26 ± 1 °C. At each time point, samples were drawn from the tubes, serially diluted, and spread on LB agar plates. The plates were incubated for 24 hours at 37 °C. Colony counts were then recorded. Experiments were carried out in triplicate, and the data were used to calculate the means and standard deviations.2.9. Visualization of bactericidal activity by field emission scanning electron microscopy (FESEM)
106 bacterial cells were treated with double MIC values of ZIF-8 and PMA@ZIF-8 for 3 hours. The cells were separated from the culture medium via centrifugation, washed twice with physiological saline, and fixed in 2.5% glutaraldehyde. Dehydration was carried out using a gradient of ethanol (30%, 50%, 70%, 90%, and 100%). The samples were placed on glass slides and gold-sputtered for 4 minutes at 15 microamperes.2.10. Live/dead cell viability assay
PMA@ZIF-8 and ZIF-8 coated slides were prepared using a homogeneous solution, consisting of 250 μL of 5% Nafion as a binder, 5 mL of isopropanol as a solvent, and 25 mg of either PMA@ZIF-8 or ZIF-8. Subsequently, this solution was spin-coated onto the surface of glass slides for 30 seconds at a speed of 500 rpm. Then, the slides were left to dry. Next, a suspension of 20 μL of E. coli (107 CFU mL−1) was seeded onto the slides' surfaces. The slides were then incubated for 3 hours at a temperature of 26 ± 1 °C. Following incubation, they were submerged in the dye solution for 20 minutes in darkness. Finally, the stained samples were visualized using a Leica DMI8 microscope.2.11. Computational details
The Dmol3 code as implemented in Materials Studio 2017 software with the generalized gradient approximation (GGA) was used. The Perdew–Burke–Ernzerhof functional and double numeric basis set (DNP) was utilized. DFT semi-core pseudopotentials (DSPPs) were utilized to include the core electrons. To ensure accuracy in the simulations, the geometries were optimized, and the energies were determined under the following convergence criteria, including a maximum force tolerance of 0.002 Ha Å−1, an energy tolerance of 1.0 × 10−5 Ha, and a maximum displacement tolerance of 0.005 Å. Additionally, an empirical dispersion-corrected density functional theory (DFT-D) method, sophisticated by Grimme, was utilized to address the inadequate representation of Haggard interactions in the standard PBE functional.2.12 N-Acetyl cysteine oxidation
The oxidation of N-acetyl cysteine (NAC) by PMA@ZIF-8 NPs was assessed through Ellman's assay. Initially, a 50 mM NAC solution was prepared and mixed with a PMA@ZIF-8 solution in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, resulting in a final concentration of PMA@ZIF-8 at 1.5 mg mL−1. Subsequently, 1 mL of this mixture was combined with 1.8 mL of Tris–HCl buffer (pH = 8), and 40 μL of DTNB (100 mM) was added. The solution was thoroughly mixed using a vortex shaker. To remove any PMA@ZIF-8 particles, the mixture was passed through a 0.22 μm filter. A negative control experiment (without nanoparticles) and a positive control experiment treated with 3% H2O2 were also conducted for comparison. The absorption value at 412 nm was then measured. The percentage of oxidation of NAC was measured using the percentage of loss of the thiol group using the following equation:
1 ratio, resulting in a final concentration of PMA@ZIF-8 at 1.5 mg mL−1. Subsequently, 1 mL of this mixture was combined with 1.8 mL of Tris–HCl buffer (pH = 8), and 40 μL of DTNB (100 mM) was added. The solution was thoroughly mixed using a vortex shaker. To remove any PMA@ZIF-8 particles, the mixture was passed through a 0.22 μm filter. A negative control experiment (without nanoparticles) and a positive control experiment treated with 3% H2O2 were also conducted for comparison. The absorption value at 412 nm was then measured. The percentage of oxidation of NAC was measured using the percentage of loss of the thiol group using the following equation: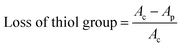 | (3) |
2.13. Statistical analysis
Statistical analysis was conducted using GraphPad Prism software. An unpaired t-test with Welch correction was used to evaluate the differences in CFU count between samples in the CFU counting test. Analysis of variance between different time points in the time-kill curve of PMA@ZIF-8 and ZIF-8 was conducted using a two-way ANOVA with multiple comparison tests corrected with the Sidak method. P-values less than 0.05 were considered statistically significant.3. Results and discussion
3.1. Characterization of the PMA@ZIF-8 hybrid
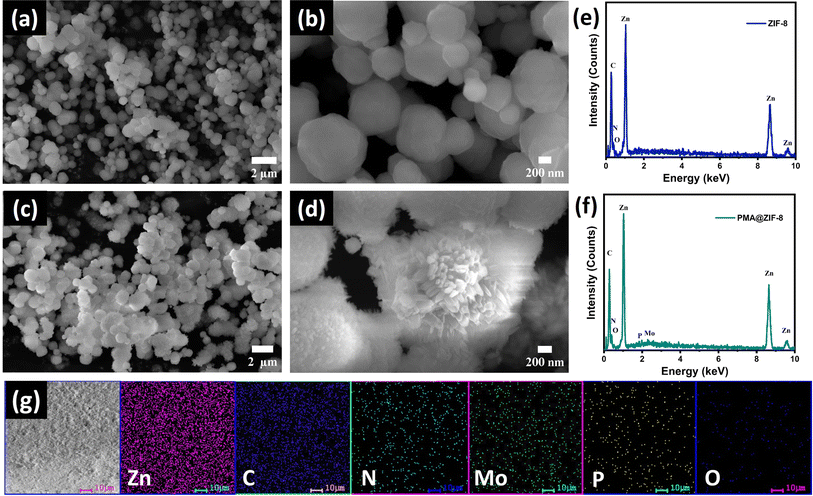
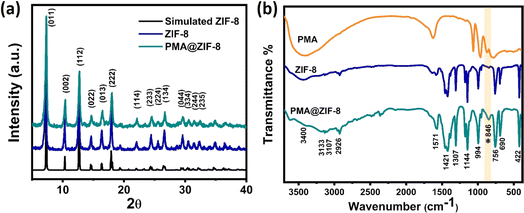
ZIF-8 and PMA@ZIF-8 nanocomposites were synthesized via a simple one-step co-precipitation reaction, where the fabrication was conducted at room temperature, Scheme S1.† Throughout the synthesis process, the ZIF-8 framework is self-assembled around the PMA particles encapsulating them within its cavities, thus preventing their dissolution. The morphology of the as-synthesized ZIF materials could be assessed from the top-view images shown in Fig. 1. The FESEM images illustrate a distorted polyhedron morphology displayed by the bare MOF material with an average size of ∼550 nm, Fig. 1a and b. The high magnification power image reveals the assembly of a smooth surface, Fig. 1b. On the other hand, the PMA@ZIF-8 sample exhibited a rough surface due to the development of nanorod-like structures from the polyhedron units, Fig. 1c and d. The deviation from the polyhedron morphology of the parent MOF is most probably ascribed to the electrostatic attraction between the Zn2+ cations and PMA anions during the crystal development.31 Moreover, it could be attributed to the sensitivity of ZIF-8 to the acidity of phosphomolybdic acid.40 Furthermore, the existence of Mo, P, and O from the PMA in the PMA@ZIF-8 sample along with Zn, C, and N from ZIF-8 was revealed via the EDS analysis of the synthesized PMA@ZIF-8 nanocomposite, see Fig. 1c and d. Besides, the elemental mapping confirms the homogeneous distribution of PMA within the structure of ZIF-8.To scrutinize the crystal structure of the prepared MOF, the X-ray diffraction (XRD) analysis was performed. Fig. 2a demonstrates the XRD patterns of ZIF-8 and the hybrid PMA@ZIF-8. The parent ZIF-8 MOF displayed diffraction peaks at 2θ of 7.27°, 10.32°, 12.68°, 14.62°, 16.37°, 17.99°, 22.01°, 24.41°, 25.53°, 26.68°, 29.68°,30.43°, 31.36°, and 32.35°, which are indexed to the respective symmetry planes of (011), (002), (112), (022), (013), (222), (114), (233), (224), (134), (044), (334), (244), and (235) of pure ZIF-8. The XRD peaks agree perfectly with the previously reported data confirming the assembly of a pure phase of ZIF-8 MOF.41,42 Furthermore, the XRD of the PMA@ZIF-8 hybrid revealed that encapsulating the PMA nanoparticles within the ZIF-8 cavities does not alter the original crystal structure of the parent ZIF-8 MOF. This could be evidenced by the absence of the diffraction peaks related to PMA, which signifies the uniform distribution of the amorphous PMA nanoparticle within the cavities of ZIF-8.24,43
The FT-IR spectra of the ZIF-8 and PMA@ZIF-8 hybrid are displayed in Fig. 2b. The spectrum of ZIF-8 MOF demonstrates a sharp peak at 422 cm−1 corresponding to the Zn–N stretching vibration of the coordination between the N atom of the 2-MIM linker and Zn,44,45 whereas the absorption peaks located at 756 and 690 cm−1 are attributed to the out-of-plane bending of the imidazole ring.46,47 Moreover, the bending and stretching vibrations of the C–N bond in the 1,3-diazole ring are identified from the absorption bands at 994 and 1144 cm−1, respectively.47 Furthermore, the peaks observed at 1307 and 1421 cm−1 are assigned to the entire elongation of the imidazole ring.41 Besides, the band detected at 1571 cm−1 is ascribed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibration,46 whereas in the higher frequency region, the aliphatic and aromatic C–H stretch of 1,3-diazole is situated at 2926 and 3107 cm−1, respectively.46 Also, the band at 3133 cm−1 is endorsed by the vibrational stretching of the C–C bond in the imidazole ring.41 However, the broad band located at around ∼3400 cm−1 is attributed to the O–H vibration in the bonded water.48,49 On the other hand, the FTIR spectra of the PMA@ZIF-8 hybrid demonstrated absorption bands similar to those of pure MOF along with the emergence of a new band located at 846 cm−1, corresponding to the Mo–O–Mo vertex bond in the PMA anions,50 confirming its successful encapsulation within the MOF cavities without altering their structure.
N stretching vibration,46 whereas in the higher frequency region, the aliphatic and aromatic C–H stretch of 1,3-diazole is situated at 2926 and 3107 cm−1, respectively.46 Also, the band at 3133 cm−1 is endorsed by the vibrational stretching of the C–C bond in the imidazole ring.41 However, the broad band located at around ∼3400 cm−1 is attributed to the O–H vibration in the bonded water.48,49 On the other hand, the FTIR spectra of the PMA@ZIF-8 hybrid demonstrated absorption bands similar to those of pure MOF along with the emergence of a new band located at 846 cm−1, corresponding to the Mo–O–Mo vertex bond in the PMA anions,50 confirming its successful encapsulation within the MOF cavities without altering their structure.
To gain more insights into the textural features of the as-synthesized ZIF-8 and PMA@ZIF-8 materials, N2 adsorption–desorption isotherms were recorded at 77 K. Upon applying low relative pressures, the adsorption branch of the isotherm for the pristine ZIF-8 sample displayed large N2 uptake, revealing the dominance of a microporous surface as evidenced by a type I isotherm, Fig. 3a. The parent MOF sample exhibited an enormous specific surface area (SSA) of 1120 m2 g−1 with a total pore volume of 0.69 cm3 g−1. Remarkably, encapsulating the Keggin PMA units within ZIF-8 cavities not only preserved the microporous feature of the isotherm, Fig. 3b, but also maintained a high SSA of 862 m2 g−1. Besides, a slight reduction in the total pore volume down to 0.52 cm3 g−1 was observed confirming the successful encapsulation of the PMA units within the ZIF-8 voids.24,43 Moreover, the pore size distributions were assessed from the desorption branch of the isotherms according to the BJH model. As noted from the insets in Fig. 3a and b, the composite PMA@ZIF-8 demonstrated a wider pore size distribution with a hierarchical micro/meso porous structure, consistent with the different morphologies depicted in the FESEM images. This hierarchical porous structure could be beneficial for offering various active antibacterial sites with different features.51
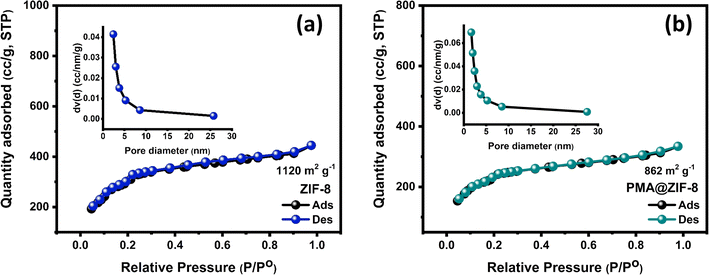 | ||
| Fig. 3 N2 adsorption–desorption isotherms of the fabricated (a) ZIF-8 and (b) PMA@ZIF-8 samples with the insets showing the corresponding pore size distribution. | ||
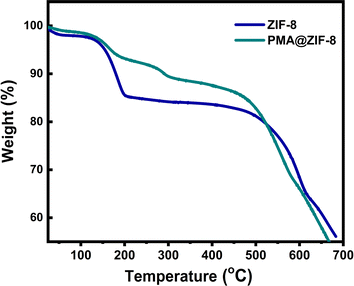
To investigate the thermal stability of the prepared materials, thermal gravimetric analysis (TGA) measurements were conducted under ambient air pressure as demonstrated in Fig. 4. The samples were heated in air from 25 to 700 °C at a heating rate of 10 °C min−1. At a temperature lower than 150 °C, both samples showed minimal weight loss ∼4%, which is ascribed to the removal of the residual solvent water molecules.42,52 Upon heating the samples from 150 to 450 °C, gradual weight losses of ∼17.2% and 13.5% were observed for ZIF-8 and PMA@ZIF-8 materials, respectively. This weight loss was ascribed to the removal of the unreacted species on the surface of the material as well as the guest molecules from the MOF cavities.42,53 It is remarked that the weight loss experienced by PMA@ZIF-8, due to the evaporation of solvent molecules and the unreacted surface species, is lower than that in the case of bare ZIF-8, which is in agreement with the N2 adsorption–desorption measurements results and with the literature.54 Sharp weight losses were demonstrated between 470 and 700 °C corresponding to the decomposition of the organic imidazolate species and the collapse of the framework into its derived metal oxide.53 Thus, it can be concluded that our prepared composite demonstrates an outstanding thermal stability.
3.2. Experimental determination of antibacterial activity
The antibacterial properties of PMA@ZIF-8 were experimentally evaluated in comparison to ZIF-8 using CFU counting, MIC values, and bactericidal kinetics.3.3. Microbial CFU screening assay
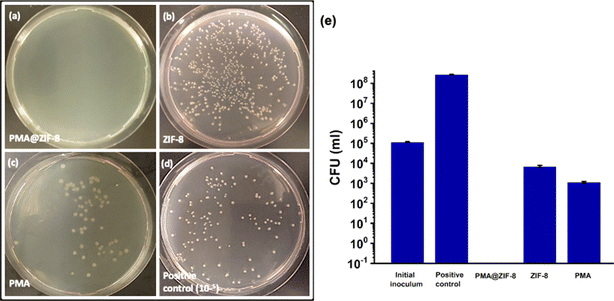
In the screening assay, the bactericidal activities of PMA, ZIF-8, and PMA@ZIF-8 were evaluated by measuring the reduction percentage in the initial inoculum. All three materials caused significant inhibition of bacterial growth as compared to the positive control (P-value < 0.001). Under the described testing conditions PMA@ZIF-8 achieved complete (100%) elimination of the bacterial cells, PMA exhibited a slightly lower reduction (99.01%), and ZIF-8 displayed the lowest reduction percentage (93.93%), Fig. 5. The CFU count in the case of the PMA@ZIF-8 sample was significantly less than that of PMA (P-value = 0.003) and ZIF-8 (p-value = 0.004).Additionally, the log inhibition values were gauged relative to the positive control to provide insights into the extent of inhibition of bacterial growth achieved using each material. While PMA@ZIF-8 resulted in complete growth inhibition, PMA exhibited a log inhibition value of 5.38, while ZIF-8's value was 4.59, which was in alignment with previously reported values (4.07).11 These results highlighted that PMA@ZIF-8 has a more potent antibacterial activity as compared to its individual constituents, which reflects experimental evidence of the synergism. It is to be noted that CFU counting was selected for screening instead of the widely used agar diffusion method as the latter is not a favorable option in our experimental setting because the zone of inhibition in the agar diffusion method is influenced by the diffusion rate and solubility of the material in agar.55 ZIF-8's unique characteristics, such as high porosity coupled with low specific gravity17 and its solubility in organic media16 render it unsuitable for direct agar diffusion comparison.
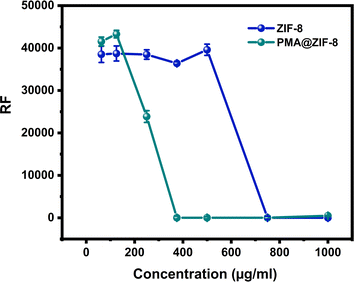
3.4. Determination of minimum inhibitory concentration (MIC)
To achieve a more objective understanding of the enhanced antibacterial activity of ZIF-8 upon doping with PMA a MIC analysis was conducted. This analysis aimed to provide quantitative insights into the extent of antibacterial activity improvement upon the incorporation of PMA into ZIF-8. The resazurin assay fluoroscopic analysis revealed that the MIC of PMA@ZIF-8 was 375 μg mL−1, while that of ZIF-8 alone was 750 μg mL−1, Fig. 6. These findings clearly demonstrate a significant enhancement in the antibacterial effectiveness of the composite, showcasing a noteworthy two-fold reduction in the MIC when compared to ZIF-8 alone.The MIC of ZIF-8 detected in the current study (750 μg mL−1) closely aligns with previously reported values, such as 768 μg mL−1 (ref. 56) and >512 μg mL−1.57 However, it is worth noting that Ahmad et al. reported higher MIC values for ZIF-8 against E. coli (>10 gm/L) in two distinct studies.17,58 This discrepancy in MIC values is probably attributed to the relatively shorter incubation period (30 minutes) in their experiments.17,58 This short incubation period may underestimate the ZIF-8 MIC due to the relatively slow antibacterial kinetics of ZIF-8, as revealed in the time-kill curve experiment of the current study. On the other hand, Taheri et al. reported a relatively low minimum bactericidal concentration (MBC) value of 250 mg L−1 for ZIF-8.16 By definition, the MBC value must be equal to or greater than the MIC value, indicating a potentially lower MIC value in their study. However, Taheri et al. determined the MBC value in phosphate-buffered saline (PBS) rather than LB broth. PBS is a buffer solution that preserves bacterial cells at a constant number without providing nutrients to promote growth. Conversely, LB broth promotes bacterial flourishing and growth by supplying essential nutrients, with a subsequent need for more antibacterial material to exert its bacteriostatic and/or bactericidal effects.16 This underscores the substantial impact of experimental conditions and media composition on the determination of MIC and MBC values for ZIF-8.
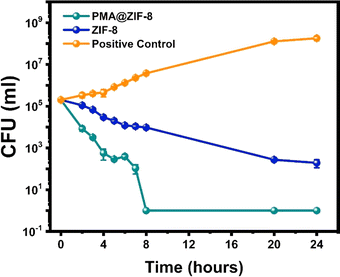
3.5. Time-kill kinetics analysis
To comprehensively understand the bactericidal potential of both PMA@ZIF-8 and ZIF-8, a time-kill curve analysis was undertaken. This involved the monitoring of CFU at various time points, Fig. 7. Furthermore, the extent of CFU reduction relative to the initial inoculum was quantified as percentages at various time points, and these values were tabulated for reference, Table 1.| Time | Percentage of CFU reductiona of the ZIF-8-treated sample | Percentage of CFU reductiona of the PMA@ZIF-8-treated sample | P-valueb |
|---|---|---|---|
| a Percentage of CFU reduction was calculated based on the CFU count of the initial inoculum. b P-values were calculated by comparing the CFU counts between the three replicas of ZIF-8 and PMA@ZIF-8. | |||
| 2 hours | 45.17% | 95.76% | <0.0001 |
| 3 hours | 66.42% | 98.41% | <0.0001 |
| 4 hours | 85.39% | 99.72% | <0.0001 |
| 5 hours | 90.06% | 99.86% | <0.0001 |
| 6 hours | 94.02% | 99.80% | 0.0045 |
| 7 hours | 94.59% | 99.95% | 0.0101 |
| 8 hours | 95.26% | 100.00% | 0.0315 |
| 20 hours | 99.86% | 100.00% | >0.9999 |
| 24 hours | 99.90% | 100.00% | >0.9999 |
The time-dependent antibacterial activity of ZIF-8 was rather slow, with only 45.17% eradication of the initial bacterial inocula after 2 hours. A single logarithmic reduction (90.06%) was achieved after 5 hours, whereas a 3-log reduction was not attained before 24 hours. On the other hand, PMA@ZIF-8 demonstrated significantly faster bactericidal kinetics manifesting a 95.76% bacterial death within the initial 2 hour time point, 2-log reduction after 4 hours, 3-log reduction after 7 hours, and complete eradication by the 8-hour time point. The CFU count difference between PMA@ZIF-8 and ZIF-8 was significantly lower for PMA@ZIF-8 from the 2-hour to the 8-hour time points (p-values < 0.05). However, this difference ceased to be statistically significant at the 20-hour time point.
For a quantitative depiction of bacterial death kinetics, an exponential decay equation was derived through regression analysis based on the natural logarithmic values of CFU counts. The derived equations for the PMA@ZIF-8 and ZIF-8 samples are:
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFUp = −1.2612t + 12.015 CFUp = −1.2612t + 12.015 |
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFUz = −0.2901t + 11.679 CFUz = −0.2901t + 11.679 |
It is important to note that the bacterial kinetics experiment was conducted at 4 times the MIC, which amounted to 3000 μg mL−1 for ZIF-8 and 1500 μg mL−1 for PMA@ZIF-8. Remarkably, at half the concentration, PMA@ZIF-8 exhibited significantly better bactericidal kinetics and overall antibacterial behavior as compared to ZIF-8 alone. This finding highlights the superior synergistic effect of doping ZIF-8 with PMA, with subsequent enhanced efficacy and cost-effectiveness of PMA@ZIF-8 as an antibacterial agent.
3.6. PMA@ZIF-8 induced phenotypic bacterial changes observed by FESEM
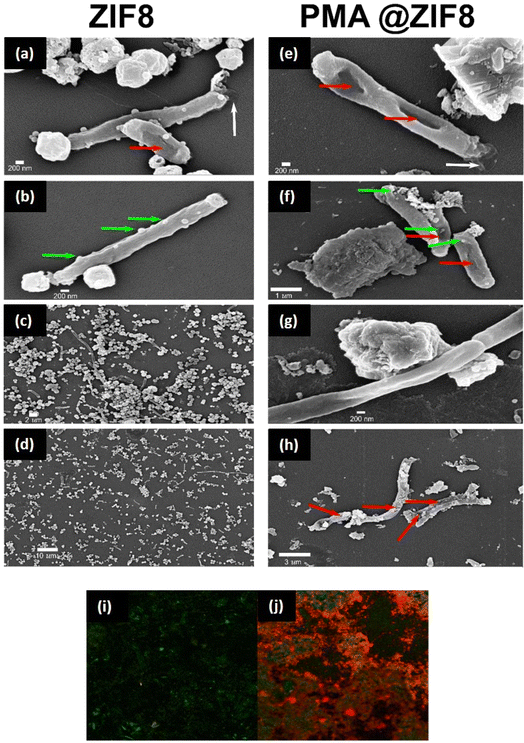
To gain insights into the consequence of contact between the bacteria and nanoparticles, FESEM was employed to visualize subsequent phenotypic changes, Fig. 8. Both ZIF-8 and PMA@ZIF-8 could attach to the bacterial cells, causing different stress responses. In the case of ZIF-8 treated cells, the most prominent sign was filamentation of the cells reached up to 20 μm, Fig. 8c and d. Filamentation was observed to a lesser extent in the case of PMA@ZIF-8 (Fig. 8g); however, other signs became more obvious and frequent such as digging of the nanoparticles into the cell membranes (Fig. 8f), membrane tunneling (Fig. 8e and f), and cytoplasmic leakage (Fig. 8h). Ghost cells and cell debris were also observed (Fig. 8h). These FESEM observations collectively suggest that while both ZIF-8 and PMA@ZIF-8 nanoparticles interact with bacterial cells, PMA@ZIF-8 exhibits a more pronounced impact although it was tested at half the concentration of ZIF-8 (750 μg mL−1 for PMA@ZIF-8 vs. 1500 μg mL−1 for ZIF-8).3.7. Bactericidal activity of PMA@ZIF-8 reported by live/dead cell viability assay
To assess the PMA@ZIF-8 coated surface's capability to disrupt bacterial cell membranes, a live/dead cell viability assay utilizing acridine orange/propidium iodide staining was performed. Acridine orange is a potent dye that can infiltrate intact cell membranes of live cells, while propidium iodide can only penetrate damaged cell membranes of dead cells.59 In the case of ZIF-8, the absolute majority of the bacterial cells had intact membranes, Fig. 8i. On the other hand, PMA@ZIF-8 coated slides showed a dramatic increase in the number of cells with damaged membranes, Fig. 8j. This observed difference seems to be related to the mechano-bactericidal activity of PMA@ZIF-8, attributed to its surface roughness and the presence of nanorods protruding from its surface. These unique structural features have probably contributed to the effective disruption of bacterial cell membranes, which was not observed with the smooth topology of ZIF-8.3.8. Molecular modeling
To explore the effect of addition of the primitive cell of ZIF-8, its 3D cubic cell and PMA@ZIF-8 were constructed as shown in Fig. 9a–c for subsequent calculations. The density of states (DOS) besides the band structures of ZIF-8 and PMA@ZIF-8 was calculated. Pristine ZIF-8 exhibits a bandgap value of 4.89 eV, indicating its semiconductor nature, which is well-established through electrochemical background studies.60 However, it was observed that ZIF-8 displays insufficient electronic conductivity despite its semiconductor properties. Its conductivity was increased by implementing PMA. The densities of states of bare ZIF-8 and PMA@ZIF-8 are illustrated in Fig. 9d and e. The implementation of PMA reduces the bandgap significantly. It allows electrons to jump from the valence band to the conduction band, creating ROS, which attack the carbon atom of the carbonyl group in the peptide linkages, leading to the oxidation and degradation of bacterial proteins with subsequent bacterial death.24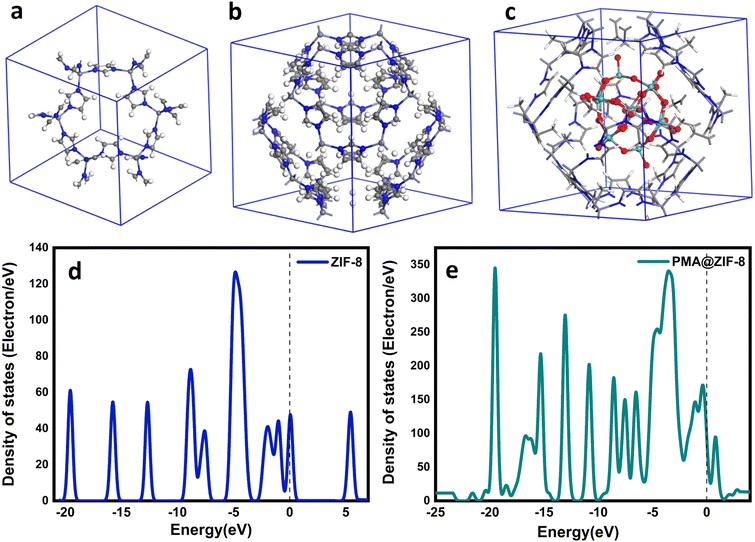 | ||
| Fig. 9 Side view of (a) ZIF-8 primitive cell, (b) ZIF-8-unit cell, and (c) PMA@ ZIF-8-unit cell, and DOS for (d) pristine ZIF-8 and (e) PMA@ZIF-8. | ||
3.9. N-Acetyl cysteine oxidation
To experimentally validate the capability of PMA@ZIF-8 to catalyze oxidative reactions, Ellman's assay with NAC as the substrate was used. NAC is an acetylated form of the amino acid L-cysteine.61 Similar to many thiols (R–SH), NAC can be oxidized by free radicals and oxidizing agents to N-acetyl cystine (R–S–S–R).62 According to the spectroscopic measurements, the positive control reduced the thiol group by 93.88% while PMA@ZIF-8 caused a 56.72% reduction, which reflects the oxidation of NAC. This provides experimental evidence of the oxidative nanozyme effect of PMA@ZIF-8.3.10. Mechanism of synergism between PMA and ZIF-8
Multiple factors have played roles in the experimentally observed synergy between ZIF-8 and PMA in the hybrid structure. The first is the significant decrease in the band gap of ZIF-8 upon doping with PMA, with subsequent powerful oxidative capability and a subsequent antibacterial effect. According to this finding, the PMA@ZIF-8 hybrid can be behave as an oxidative nanozyme. This finding was experimentally confirmed by the NAC oxidation experiment.Secondly, the synergistic mechanism of PMA and ZIF-8 can be understood in the context of the complementary mechanisms of action. Besides the oxidative effect of PMA@ZIF-8, the release of PMA from the composite may aggravate the oxidative stress. POMs can penetrate the outer membrane through porins, causing oxidation of the electron carriers with subsequent decreased ATP production.26,28,29 Additionally, they can decrease the reduced glutathione pole, sensitizing the bacterial cells to oxidative stress.26,29,63 The observed synergism between PMA and ZIF-8 can be attributed to their distinct but complementary abilities to target multiple mechanisms. Hence, further research is needed to explore the potential bacterial protein targets of PMA@ZIF-8, which could shed light on the specific molecular pathways involved in its enhanced antibacterial activity. The third potential factor that contributed to the observed synergism between PMA and ZIF-8 in the hybrid PMA@ZIF-8 system is the surface roughness and nanorods emerging from the hybrid structure's surface, Fig. 1d. These morphological properties can lead to mechanical stress by piercing or disrupting the bacterial cell membrane resulting in the so-called morphology-related antibacterial effect,64,65 which was consistent with the results of the live/dead assay. This is the cornerstone feature in the choice of materials for the development of mechano-bactericidal surfaces, which are designed to mimic the naturally occurring nanopillars on the surface of cicada wings and shark skin.6,66
Interestingly, a previously reported study demonstrated the development of nano-dagger arrays for use as mechano-bactericidal surfaces by using ZIF-L, because it is distinguished by a feathery shape with sharp angles.66 The shift of the ZIF-8 shape in the current study, from the classic polyhedron shape with a smooth surface to a rough surface with nanorod protrusions, may provide exciting opportunities for its testing and application in similar mechano-bactericidal surface designs. This novel surface topology of our new hybrid could potentially enhance the antibacterial activity by providing an additional mechanism for the mechanical disruption of bacterial cells. One limitation of the current study is that the PMA doping was tested at a single preliminary concentration in the hybrid PMA@ZIF-8 material. Therefore, future studies should be conducted to explore a broader range of PMA concentrations with subsequent determination of the optimum concentration achieving the highest synergistic antimicrobial activity in the PMA@ZIF-8 system.
4. Conclusion
In this study, a PMA@ZIF-8 hybrid was successfully synthesized by a simple co-precipitation method. The antibacterial performance of PMA@ZIF-8 was much better than that of the pristine MOF, clearly demonstrated by the twofold reduction in the MIC and elevating the rate constant of bactericidal kinetics by an impressive factor of 4.35. Moreover, the hybrid material showed oxidative nanozyme capability as predicted using DFT calculations and confirmed by the NAC oxidation experiment. Furthermore, the addition of PMA influenced ZIF-8's surface topology with the acquisition of surface nanorods and protrusions, suggesting potential morphology-related bactericidal activity, which was confirmed by live/dead assay. On the other hand, PMA's addition caused just a minor decrease in the SSA of ZIF-8, with plenty of roomy residual SSA, allowing for potential loading of other antibacterial agents. The enhanced efficacy, kinetics, and nanozyme activity with the improved surface topology and the preserved high porosity render PMA@ZIF-8 a very promising candidate to be used in antibacterial surfaces with combined release-base and mechano-bactericidal effects, and it is hence a promising candidate for a wide range of infection control applications.Conflicts of interest
There are no conflicts to declare.References
- K. S. Ikuta, et al , Lancet, 2022, 400, 2221–2248 CrossRef PubMed.
- G. J. Buckley and G. H. Palmer, Combating Antimicrobial Resistance and Protecting the Miracle of Modern Medicine, 2022 Search PubMed.
- W. A. Rutala and D. J. Weber, Am. J. Infect. Control, 2019, 47, A96–A105 CrossRef PubMed.
- W. A. Abbas, I. H. Abdullah, B. A. Ali, N. Ahmed, A. M. Mohamed, M. Y. Rezk, N. Ismail, M. A. Mohamed and N. K. Allam, Nanoscale Adv., 2019, 1, 2801–2816 RSC.
- C. Mu, J. Ren, H. Chen, Y. Wu, Q. Xu, X. Sun and K. Yan, ACS Appl. Nano Mater., 2021, 4, 10634–10644 CrossRef CAS.
- D. P. Linklater, V. A. Baulin, S. Juodkazis, R. J. Crawford, P. Stoodley and E. P. Ivanova, Nat. Rev. Microbiol., 2021, 19, 8–22 CrossRef CAS PubMed.
- A. Lewis, P. Hazelton, F. S. Butt, N. A. Mazlan, X. Wei, N. Radacsi, X. Chen, Y. Yang, S. Yang and Y. Huang, ACS Appl. Nano Mater., 2022, 5, 16250–16263 CrossRef CAS.
- S. M. Abdou, A. Moustafa and N. K. Allam, ACS Appl. Bio Mater., 2024, 7, 2569–2581 CrossRef CAS PubMed.
- J. Li, W. Ye and C. Chen, Interface Sci. Technol., 2019, 29, 217–279 CrossRef CAS.
- A. M. Mohamed, M. Ramadan and N. K. Allam, J. Energy Storage, 2021, 34, 102195 CrossRef.
- X. C. Huang, Y. Y. Lin, J. P. Zhang and X. M. Chen, Angew. Chem., Int. Ed., 2006, 45, 1557–1559 CrossRef CAS PubMed.
- K. S. Park, Z. Ni, A. P. Côté, J. Y. Choi, R. Huang, F. J. Uribe-Romo, H. K. Chae, M. O'Keeffe and O. M. Yaghi, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10186–10191 CrossRef CAS PubMed.
- M. Bergaoui, M. Khalfaoui, A. Awadallah-F and S. Al-Muhtaseb, J. Nat. Gas Sci. Eng., 2021, 96, 104289 CrossRef CAS.
- H. N. Abdelhamid, Curr. Med. Chem., 2021, 28, 7023–7075 CAS.
- W. L. Teo, J. Liu, W. Zhou and Y. Zhao, ACS Appl. Nano Mater., 2021, 4, 6562–6567 CrossRef CAS.
- M. Taheri, D. Ashok, T. Sen, T. G. Enge, N. K. Verma, A. Tricoli, A. Lowe, D. R. Nisbet and T. Tsuzuki, Chem. Eng. J., 2021, 413, 127511 CrossRef CAS.
- N. Ahmad, N. A. H. M. Nordin, J. Jaafar, A. F. Ismail and M. K. N. B. Ramli, J. Environ. Chem. Eng., 2021, 9, 105887 CrossRef CAS.
- D. Ashok, M. Taheri, P. Garg, D. Webb, P. Parajuli, Y. Wang, B. Funnell, B. Taylor, D. C. Tscharke, T. Tsuzuki, N. K. Verma, A. Tricoli and D. R. Nisbet, Adv. Sci., 2022, 9, 1–15 Search PubMed.
- R. Ni, H. Xu, J. Ma, Q. Lu, Y. Hu, C. Huang, Q. Ke and Y. Zhao, Mater. Today Chem., 2022, 23, 100689 CrossRef CAS.
- P. Li, J. Li, X. Feng, J. Li, Y. Hao, J. Zhang, H. Wang, A. Yin, J. Zhou, X. Ma and B. Wang, Nat. Commun., 2019, 10, 1–10 CrossRef PubMed.
- Y. Cai, J. Guan, W. Wang, L. Wang, J. Su and L. Fang, J. Food Sci., 2021, 86, 3550–3562 CrossRef CAS PubMed.
- M. T. Pope, Heteropoly and Isopoly Oxometalates, Springer, Berlin, Heidelberg, 1983, vol. 8 Search PubMed.
- B. Hasenknopf, Front. Biosci., 2005, 10, 275–287 CrossRef CAS PubMed.
- A. M. Mohamed, W. A. Abbas, G. E. Khedr, W. Abass and N. K. Allam, Sci. Rep., 2022, 12, 15989 CrossRef CAS PubMed.
- J. F. Keggin, Nature, 1933, 131, 908–909 CrossRef CAS.
- A. Bijelic, M. Aureliano and A. Rompel, Chem. Commun., 2018, 54, 1153–1169 RSC.
- L. Hao, Q. Zheng, M. Guan, M. Zhou, Z. Yin, H. Chen, H. Zhou and X. Zhou, ACS Appl. Nano Mater., 2023, 6, 4754–4769 CrossRef CAS.
- M. Inoue, K. Segawa, S. Matsunaga, N. Matsumoto, M. Oda and T. Yamase, J. Inorg. Biochem., 2005, 99, 1023–1031 CrossRef CAS PubMed.
- J. Chang, M. Li, J. Du, M. Ma, C. Xing, L. Sun and P. Ma, Int. J. Mol. Sci., 2022, 23, 9651 CrossRef CAS PubMed.
- Z. Lian, C. Lu, J. Zhu, X. Zhang, T. Wu, Y. Xiong, Z. Sun and R. Yang, Front. Chem., 2022, 10, 1–8 Search PubMed.
- L. Zhang, T. Mi, M. A. Ziaee, L. Liang and R. Wang, J. Mater. Chem. A, 2018, 6, 1639–1647 RSC.
- M. M. Hasan, G. E. Khedr and N. K. Allam, ACS Appl. Nano Mater., 2022, 5, 15457–15464 CrossRef CAS.
- S. M. Fawzy, G. E. Khedr and N. K. Allam, Int. J. Hydrogen Energy, 2023, 48, 33111–33118 CrossRef CAS.
- I. M. Badawy, G. E. Khedr, A. M. Hafez, E. A. Ashour and N. Allam, Chem. Commun., 2023, 59, 7974–7977 RSC.
- M. M. Hasan, A. K. Gomaa, G. E. Khedr, K. E. Salem, B. S. Shaheen and N. K. Allam, Energy Fuels, 2022, 36, 14371–14381 CrossRef CAS.
- A. M. Asran, M. A. Mohamed, G. E. Khedr, G. M. G. Eldin, A. M. Yehia, R. K. Mishra and N. K. Allam, J. Therm. Anal. Calorim., 2022, 147, 5779–5786 CrossRef CAS.
- CLSI, Performance Standards for Antimicrobial Susceptability Tests, Clin. Lab. Standars Inst., 2015, 32, pp. 18 Search PubMed.
- M. Vukomanovic and E. Torrents, J. Nanobiotechnol., 2019, 17, 1–11 CrossRef PubMed.
- S. Foerster, V. Desilvestro, L. J. Hathaway, C. L. Althaus and M. Unemo, J. Antimicrob. Chemother., 2017, 72, 1961–1968 CrossRef CAS PubMed.
- Z. Huang, Z. Yang, M. Z. Hussain, B. Chen, Q. Jia, Y. Zhu and Y. Xia, Electrochim. Acta, 2020, 330, 135335 CrossRef CAS.
- B. A. Costa, M. P. Abuçafy, T. W. L. Barbosa, B. L. da Silva, R. B. Fulindi, G. Isquibola, P. I. da Costa and L. A. Chiavacci, Pharmaceutics, 2023, 15, 1–14 Search PubMed.
- H. Kaur, G. C. Mohanta, V. Gupta, D. Kukkar and S. Tyagi, J. Drug Deliv. Sci. Technol., 2017, 41, 106–112 CrossRef CAS.
- A. M. Mohamed, M. Ramadan, N. Ahmed, A. O. A. ElNaga, H. H. Alalawy, T. Zaki, S. A. Shaban, H. B. Hassan and N. K. Allam, J. Energy Storage, 2020, 28, 101292 CrossRef.
- K. Zhou, B. Mousavi, Z. Luo, S. Phatanasri, S. Chaemchuen and F. Verpoort, J. Mater. Chem. A, 2017, 5, 952–957 RSC.
- A. M. Mohamed, D. M. Sayed and N. K. Allam, ACS Appl. Mater. Interfaces, 2023, 15, 16755–16767 CrossRef CAS PubMed.
- M. Ammar, S. Jiang and S. Ji, J. Solid State Chem., 2016, 233, 303–310 CrossRef CAS.
- R. Ediati, P. Elfianuar, E. Santoso, D. O. Sulistiono and M. Nadjib, in Mesoporous Materials - Properties and Applications, 2019, pp. 1–14 Search PubMed.
- A. M. Mohamed, A. O. A. E. Naga, T. Zaki, H. B. Hassan and N. K. Allam, ACS Appl. Energy Mater., 2020, 3, 8064–8074 CrossRef CAS.
- H. M. El Sharkawy, A. M. Mohamed, M. Ramadan and N. K. Allam, J. Energy Storage, 2022, 54, 105272 CrossRef.
- L. Lu and Y. Xie, New J. Chem., 2016, 41, 335–346 RSC.
- H. M. El Sharkawy, A. M. Mohamed and N. K. Allam, Electrochim. Acta, 2023, 471, 143368 CrossRef.
- S. Goyal, M. S. Shaharun, C. F. Kait and B. Abdullah, J. Phys.: Conf. Ser., 2018, 1123, 012062 CrossRef.
- F. Chu, Y. Zheng, B. Wen, L. Zhou, J. Yan and Y. Chen, RSC Adv., 2018, 8, 2426–2432 RSC.
- C. Wang, A. R. Li and Y. L. Ma, Fuel Process. Technol., 2021, 212, 106629 CrossRef CAS.
- M. Moghayedi, E. K. Goharshadi, K. Ghazvini, H. Ahmadzadeh, R. Ludwig and M. Namayandeh-Jorabchi, Mater. Chem. Phys., 2017, 188, 58–67 CrossRef CAS.
- C. Zhang, Z. Shu, H. Sun, L. Yan, C. Peng, Z. Dai, L. Yang, L. Fan and Y. Chu, Appl. Surf. Sci., 2023, 611, 155599 CrossRef CAS.
- Y. Zhang, L. Lai, Y. Liu, B. Chen, J. Yao, P. Zheng, Q. Pan and W. Zhu, ACS Appl. Mater. Interfaces, 2022, 14, 6453–6464 CrossRef CAS PubMed.
- N. Ahmad, N. A. H. M. Nordin, J. Jaafar, N. A. N. N. Malek, A. F. Ismail, M. N. F. Yahya, S. A. M. Hanim and M. S. Abdullah, Particuology, 2020, 49, 24–32 CrossRef CAS.
- X. Wang, J. Qiao, F. Yuan, R. Hang, X. Huang and B. Tang, Surf. Coat. Technol., 2014, 240, 167–178 CrossRef CAS.
- A. Baghban, S. Habibzadeh and F. Z. Ashtiani, RSC Adv., 2020, 10, 22929–22938 RSC.
- M. Zafarullah, W. Q. Li, J. Sylvester and M. Ahmad, Cell. Mol. Life Sci., 2003, 60, 6–20 CrossRef CAS PubMed.
- Z. Abedinzadeh, J. Arroub and M. Gardes-Albert, Can. J. Chem., 1994, 72, 2102–2107 CrossRef CAS.
- Y. Fang, C. Xing, S. Zhan, M. Zhao, M. Li and H. Liu, J. Mater. Chem. B, 2019, 7, 1933–1944 RSC.
- E. Jeong, C. U. Kim, J. Byun, J. Lee, H. E. Kim, E. J. Kim, K. J. Choi and S. W. Hong, Sci. Total Environ., 2020, 712, 136574 CrossRef CAS PubMed.
- R. Gandotra, Y. R. Chen, T. Murugesan, T. W. Chang, H. Y. Chang and H. N. Lin, J. Alloys Compd., 2021, 873, 159769 CrossRef CAS.
- Y. Yuan and Y. Zhang, Nanomedicine, 2017, 13, 2199–2207 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00142g |
| This journal is © The Royal Society of Chemistry 2024 |