Open Access Article
Open Access ArticleA comprehensive review of challenges and advances in exosome-based drug delivery systems
Sushesh Srivatsa
Palakurthi
,
Brijesh
Shah
,
Sumedha
Kapre
,
Nitin
Charbe
,
Susan
Immanuel
 ,
Sindhura
Pasham
,
Sindhura
Pasham
 ,
Maharshi
Thalla
,
Ankit
Jain
and
Srinath
Palakurthi
,
Maharshi
Thalla
,
Ankit
Jain
and
Srinath
Palakurthi
 *
*
Department of Pharmaceutical Sciences, Irma Lerma Rangel College of Pharmacy, Texas A&M University, Kingsville, TX 78363, USA. E-mail: palakurthi@tamu.edu; Tel: +1-361-221-0748
First published on 29th October 2024
Abstract
Exosomes or so-called natural nanoparticles have recently shown enormous potential for targeted drug delivery systems. Several studies have reported that exosomes as advanced drug delivery platforms offer efficient targeting of chemotherapeutics compared to individual polymeric nanoparticles or liposomes. Taking structural constituents of exosomes, viz., proteins, nucleic acids, and lipids, into consideration, exosomes are the most promising carriers as genetic messengers and for treating genetic deficiencies or tumor progression. Unfortunately, very little attention has been paid to the factors like source, scalability, stability, and validation that contribute to the quality attributes of exosome-based drug products. Some studies suggested that exosomes were stable at around −80 °C, which is impractical for storing pharmaceutical products. Currently, no reports on the shelf-life and in vivo stability of exosome formulations are available. Exosomes are quickly cleared from blood circulation, and their in vivo distribution depends on the source. Considering these challenges, further studies are necessary to address major limitations such as poor drug loading, reduced in vivo stability, a need for robust, economical, and scalable production methods, etc., which may unlock the potential of exosomes in clinical applications. A few reports based on hybrid exosomes involving hybridization between different cell/tumor/macrophage-derived exosomes with synthetic liposomes through membrane fusion have shown to overcome some limitations associated with natural or synthetic exosomes. Yet, sufficient evidence is indispensable to prove their stability and clinical efficacy.
1. Introduction
Exosomes are extracellular membrane bound bio-vesicles produced by most cell types. They are generated through an endocytic pathway that produces multivesicular body (MVB)/late endosomes and are subsequently released by fusion with the cell membrane. Exosomes are a subset of the large population of extracellular vesicles (EVs) with a size normally ranging from 30 to 120 nm in diameter.1–5 Their unique origin and properties like the ability to cross cytoplasmic membranes and the blood–brain barrier (BBB) make them suitable drug delivery systems. A variety of nanomaterials have been used for drug delivery. These include liposomes,6 polymers,7 mesoporous silica,8 graphene oxide,9 and iron oxide.10 Exosomes possess unique characteristics of nanocarriers in addition to low immunogenicity, biocompatibility due to the cellular origin, and enhanced barrier permeability.11,12 The advantages of exosomes over other nanomaterial drug delivery systems are highlighted in Table 1. Identifying the incidence of RNA in exosomes was the breakthrough that later demonstrated the function of exosomes in cell–cell interactions. Exosomes can attach to target cells and deliver payloads due to the occurrence of extensive surface adhesion proteins.4,13–15 Depending on the cell origin, exosomes have preferential homing targets. For example, exosomes from melanoma explicitly target sentinel lymph nodes to promote tumor metastasis.16 Therefore, cancer cell-secreted exosomes could be potentially used to target cancer. Exosomes were also reported to be good carriers of genes, proteins, and other biological materials with low toxicity.4,17,18 Due to their cellular origin, all exosomes carry diverse groups of proteins including membrane transport proteins and fusion proteins (GTPase, annexins, and flotillin), tetraspanins (CD9, CD63, and CD81), proteins involved in MVB biogenesis (TSG101 and Alix), heat shock proteins (Hsp70 and Hsp90), integrins, cell surface proteins, lipid-related proteins and phospholipases.19–21 Though limited data on the lipid composition of exosomes are available so far, some show that cholesterol, sphingomyelin, phosphatidyl serine, and phosphatidylcholine primarily constitute up to 80% of lipid content. Exosomes typically contain saturated or monounsaturated fatty acids20 along with ceramide, phosphatidylethanolamine, diacylglycerol, and hexosylceramides, which are present in small quantities.1,19–22 Exosomes have also been described to harbor sizeable quantities of miRNA, mRNA, and other non-coding RNAs suggesting their potential as outstanding gene carriers.4,17,20,21,23–25 Exosomes have also been demonstrated to protect payloads like catalase and curcumin from degradation when stored at 37 °C.26–28 In 2019, Wang et al. designed exosomes as a delivery vehicle for curcumin to prevent neuronal death and alleviate Alzheimer's disease (AD) symptoms. The exosomes substantially enhanced curcumin's solubility, bioavailability and drug permeation across the BBB and inhibited Tau phosphorylation, thereby acting as a potential source of drug carriers for targeted AD therapy.29 Various cannabinoids such as Δ9-tetrahydrocannabinol, cannabidiol, cannabigerol, Δ9-tetrahydrocannabivarin, and cannabichromene have also been loaded into EVs that showed superior anticancer effects.30 The autologous exosomes collected from the host could be beneficial, as these can evade clearance by the reticuloendothelial system (RES), thereby allowing them to have more circulation time to hit the target tissues.19 Site-specific delivery, nano-size, and acceptance by the body make exosomes unique carriers that are supposed to be involved in several biological, pathological, and diagnostic applications.31–33Recently, a concept of engineered and/or modified exosomes has emerged, allowing for the customization of their surface structure after isolation from the primary source to achieve specific characteristics like enhanced physiological barrier permeability, enhanced cell-specific targeting, in vivo tracking, improved stability, and higher efficacy.34,35 The surface functionalization of exosomes can be achieved by endogenous or exogenous strategies. The endogenous method involves the genetic modification of the exosome-secreting cell source.17,36 In this context, Alvarez-Erviti et al. engineered dendritic cells with a fusion gene to produce designer exosomes that carry neuron-specific rabies viral glycoprotein (RVG) and lysosome-associated membrane protein 2B (Lamp2B), an exosomal membrane protein. These proteins impart targeting abilities to the exosomes derived from the engineered dendritic cells, which specifically transport siRNA to the mouse brain by crossing the BBB.17 Although highly promising, endogenous exosome engineering can lead to inadvertent cell changes and may not be appropriate to produce unnatural molecules on the exosomal surface. In contrast, the exogenous strategy for exosome functionalization involves the chemical conjugation of secreted exosomes with molecules of interest to produce hybrid exosomes that can be used as a targeted delivery system. Recently, Wrobel et al. conducted a study exploring the interaction between two varieties of amphiphilic carbosilane dendritic structures, namely DDN-1 and DDN-2, with specific neutral and negatively charged lipid model membranes. Amphiphilic dendrons exhibited a high affinity to model lipid bilayers, making them a promising platform for decorating exosomes.37 The targeting ability of exosomes can be further augmented by functionalization with specific ligands, such as antibodies or aptamers.30,31
Exosome preparations may be synthetic, natural (derived from human or plant cells) or semi-synthetic wherein natural exosomes are modified before or after isolation for a specific purpose. However, low yield and modifications on a large scale are the limiting concerns for the clinical application of semi-synthetic exosomes. This led to the design of fully synthetic or so-called mimetic vesicles, holding huge therapeutic potential. Fully synthetic/artificial exosomes can be manufactured either by top-down or bottom-up approaches. The top-down method generates nano-sized vesicles by breaking larger and more complex units like cultured cells into nano-sized membrane fragments through extrusion or microfluidic devices. The bottom-up method uses specific lipids and/or proteins resembling plasma membranes, which further self-assemble into a bilayer structure and can be functionalized with selected proteins to mimic the desired exosomal functions. Microfluidic platforms offer high-precision processing and microscale liquid handling. Wang et al. (2021) reviewed microfluidic strategies to isolate exosomes depending on various characteristics of exosomes, including size, immunoaffinity, hydrodynamic and dielectrophoretic properties. The research concentrated on the label-free isolation of exosomes, aiming to maintain their active biological properties and intact morphological structures. It also introduced microfluidic techniques for the sensitive and specific detection of exosomal proteins and RNAs.38 The knowledge about exosomes has been systematically cataloged in various exosome databases, including Vesiclepedia (http://www.microvesicles.org/), ExoCarta (http://www.exocarta.org/), Exosomes.gene-quantification.info (https://www.gene-quantification.de/exosomes.html), GOA (https://www.ebi.ac.uk/GOA/EXOSOME), ExRNA (http://exrna-atlas.org/), and ExoRBase (http://www.exrobase.org/).39 While exosome-based delivery systems have been extensively investigated in the past decade, drug-loaded exosomes are still facing ample challenges in scale-up due to a lack of scalable isolation and purification techniques, a lack of reproducible protocols for producing well-characterized and homogeneous exosomes and limited data on exosome stability. This paper is an overview of current knowledge on engineered exosomes and the critical contributing factors/challenges that must be evaluated for the successful development of exosome-based drug products.
2. Exosome source, extraction methods, and scalability
2.1 Exosome sources
It is of utmost importance to pay attention while selecting a source for exosomal drug products. Factors like drug loading capacity, homing potential, membrane penetration immunomodulatory properties, etc., are directly dependent on the source of the exosomes and hence should be carefully considered.20,40 Although most cell types produce exosomes, the yield with each cell type is highly variable.14,41Though MSCs were reported to be a great source of exosomes,14,41–43 one of the major drawbacks is the finite expansion with the necessity of constantly deriving new batches of cells and the subsequent expensive and time-consuming processes of validation.44 Chen et al. used the transfection strategy to address the problem of MSC derivation using a lentivirus vector carrying the MYC gene and showed that this method could ensure the supply of exosomes in the milligram range.45 Pan et al. (2023) discussed exosome engineering techniques such as altering the culture conditions, cellular expansion through three-dimensional dynamic culture, etc., that subsequently modulated the biogenesis and secretion process of exosomes.46
Exosomes from various other sources like milk, plant extracts, saliva, etc. have also been reported (Fig. 1).40,41,47–56 However, only a few sources are suitable for producing exosomes in high quantities with homogeneity. Saliva, urine, or human blood seems to be only appropriate for isolating exosomes for diagnostic purposes.25,49–51,53,57 On the other hand, exosomes from bovine milk and colostrum have been investigated for their potential in delivering cancer therapeutics.46 Their plentiful availability, cost-effectiveness, scalability, capacity for high drug loading, ability to be functionalized for targeted delivery, and lack of toxicity make them promising candidates. Bovine milk exosomes (M-Exo) have been shown to maintain their physiochemical characteristics under harsh and denaturing environments of the gastrointestinal (GI) tract and recurring freeze–thaw cycles.58 Agrawal et al. designed a novel nano-oral delivery system of paclitaxel (PTX) using bovine M-Exos (ExoPac) and reported narrow particle size (∼108 nm) and size distribution (∼0.190) with improved stability in simulated GI fluids. ExoPac showed significant tumor growth inhibition (60%; p < 0.001) compared to intraperitoneal PTX (31%) against human lung tumor xenografts.59 Our recent studies revealed that M-Exos had relatively large sizes in a range of 100–200 nm (Fig. 2).
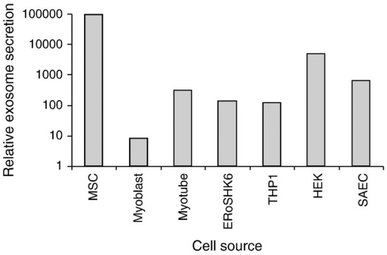 | ||
| Fig. 1 The relative quantity of exosomes secreted from different cell lines. Five different cells or cell lines were evaluated using culture media conditioned for three days by each of these cell lines. The cells or cell lines used were cmyc-transformed hESC-MSC (MSC), a mESC-derived insulin-producing cell line (ERoSHK6), the human acute monocytic leukemia cell line (THP-1), the human embryonic kidney cell line (HEK), a primary human small airway epithelial cell abbreviated as SAEC (Lonza, Cologne, Germany) and primary human myoblasts and myotubes differentiated from them (Lonza Cologne, Germany) (the figure has been adapted from ref. 41 with permission from Elsevier, Copyright 2013). | ||
Plant-derived EVs (PEVs) have been identified as promising bioactive nutraceutical molecules.60,61 Nanovesicles from grapefruits were extracted by Wang et al. and used as carriers for methotrexate (MTX) to treat dextran sulfate sodium (DSS)-induced colitis. These nanovesicles were shown to lower the toxicity of MTX and enhance its effects in colitis-induced mice.62 The significance of EVs from fruit sources has surged tremendously as human interaction with fruit juices is inevitable. Recently, Kilasoniya et al. (2023) evaluated the functional properties of PEVs sourced from grapefruit and tomato juices as antioxidants and delivery vehicles. Despite tomato PEVs being larger, grapefruit PEVs had a higher yield. The antioxidant activity of PEVs was comparatively lower than their juice sources, suggesting the restrictive role of PEVs in the juice's antioxidant properties. The grapefruit PEVs were more efficient in loading and delivery of cargo (Hsp70) than tomato PEVs. These results suggest that grapefruit PEVs hold greater promise as functional ingredients in juice and can deliver functional molecules to human cells.63 Exosomes/EVs derived from animal milk or plants, despite their wide range of applications, also have disadvantages. It should be noted that these exosomes could potentiate immunological reactions, especially upon intravenous (IV) administration. Long-term toxicity assessment is crucial for fully understanding and harnessing their therapeutic potential.64
2.2 Exosome extraction techniques and scalability
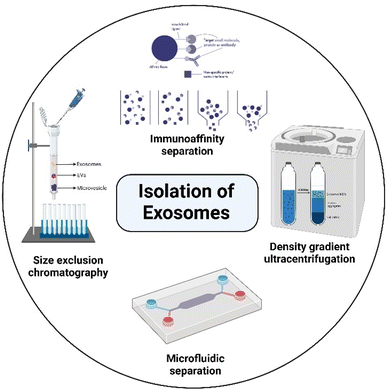
Exosome extraction and purification involve several different isolation methods, including differential ultracentrifugation (UC), solvent precipitation, size-exclusion chromatography (SEC), immunoaffinity techniques, density gradient separation, and microfluidic techniques, as shown in Fig. 3.19,65–68 Unfortunately, there is limited understanding of the relative efficiency of the isolation methods and the physicochemical properties of the isolated exosomes, such as size, size distribution, zeta potential, and dispersibility, even though those parameters have a profound impact on the characteristics and therapeutic potential of exosomes as drug carriers.69 The lack of scalability and poor validation of extraction methods are still major factors hindering the clinical translation of exosomes, which is extremely important in large-scale production. Differential UC remains the prevailing method for isolating exosomes from fluids. This technique involves a series of centrifugation cycles of variable centrifugal force and time, aimed at isolating exosomes based on their density and size.70 This method is cost-effective, has a low risk of contamination from separation reagents, and allows the collection of a large number of exosomes.36,71 However, exosomes isolated by this method commonly have lots of impurities and the yield is also relatively low.72–74 Yamashita et al. compared the properties of exosomes extracted from murine melanoma B16-BL6 by three UC-based methods: simple UC, UC with an iodixanol cushion, and UC on an iodixanol density gradient. The results showed that the type of extraction method had little effect on yield. However, the exosomes isolated by the density gradient method were highly dispersed over simple and cushioned UC.69 Another report by Jo et al. described the manufacturing of nanovesicles containing intracellular RNAs, intracellular proteins, and plasma membrane proteins using centrifugal force and a micron-sized filter. The nanovesicle isolation yield was found to be 250 times higher than that of exosome isolation (952 μg vs. 3.7 μg) from the same number of cells (1 × 108). Additionally, the size of nanovesicles was <100 nm with two times higher RNA and protein content compared to exosomes, which had a diameter >200 nm, and concluded that this method seems to be promising at a large-scale level.75 The SEC method can isolate exosomes with low contamination but usually results in a diluted sample and may require an additional concentration step. This method involves the use of a porous column packed with polymeric beads containing numerous pores through which macromolecules are sorted based on their size or diameter. A long run time and lower concentration are disadvantages of this method.76 However, the chromatography isolation method is widely accepted over centrifugation methods, since isolated exosomes are not impacted by shearing forces that can alter the vesicle structure.77 Immunoaffinity seems to be a better method for the extraction of exosomes when compared to differential UC in terms of reproducibility and efficiency.72 However, the utilization of immunoaffinity techniques is not a practical approach for generating large quantities of exosomes. Moreover, exosomes generated by this method are not cost-effective for clinical applications and there could be alterations or even destruction of the immunotherapeutic properties when exosomes are eluted from the beads.74 This method is also limited to the isolation of total exosomes,76 and it seems to be more favorable for diagnostic applications. Kalra et al. compared the quality of exosomes extracted from normal human plasma using 3 methods: differential centrifugation coupled with UC, immunoaffinity pull-down using epithelial cell adhesion molecules, and OptiPrep™ density gradient separation. Of the three, the OptiPrep™ density gradient method emerged as the best for isolating pure exosomes, free of plasma proteins.78 The microfluidic-based exosome isolation technique is still in the nascent phase of development. Although it holds great promise, low yield is the main limitation and further efforts are necessary to improve it.19 Considering large-scale production, Heinemann et al. developed a technique that is high-speed, automatable, scalable, and specific in isolating exosomes from complex biological systems. It was a 3-step filtration process that involved membranes with different pore sizes to specifically remove cell debris, proteins, and large vesicles from the body fluids or cell culture supernatant.79 Sunkara et al. (2023) have recently introduced a new method called ExoPRISM for precipitating EVs by altering the ionic strength. This method is a simple, cost-effective, and user-oriented approach for separating high yields of EVs without tampering with their functional characteristics. This method entails introducing an electrolyte solution into blood plasma, leading to the successive precipitation of proteins and EVs. Further on, a fractional separation of EVs is achieved through low-speed centrifugation. The co-precipitated electrolytes can be removed, and the entire process takes less than an hour. It has proven successful in separating EVs from various bio-sources, including culture medium, urine, plasma, and serum.80 In 2023, Williams et al. conducted a study to determine the impact of different isolation methods on EV purity and yield. The study isolated EVs through various methods such as SEC, UC, polyethylene glycol (PEG) precipitation, Total Exosome Isolation Reagent and an aqueous two-phase system with and without repeat washes. The study found that all isolation methods extracted EV-like particles, but the purity and the comparative expression of surface protein markers (Alix, CD9, CD63, CD81 and Annexin A2) varied. The choice of the characterization method significantly impacted the assessment of sample purity, as total particle counts and particle-to-protein (PtP) ratios frequently diverged from the quantitative analysis of surface markers evaluated using high-resolution nano-flow cytometry. The study revealed a preference for SEC and UC due to their overall efficiency. However, concerns arose about the scalability of these methods which could impede its application in therapeutics.81 Despite efforts, large-scale exosome production continues to be the major hurdle in the clinical application of exosome-based drug delivery systems.2.3 Exosomes for drug delivery
In the past two decades, exosomes from various sources have been loaded with drugs of interest to make them suitable carriers for various targeted therapies. Drug loaded M-Exos have been widely studied in cancer therapeutics as they enhance the human intestinal permeability of drugs like curcumin in vitro.82 Munagala et al. isolated exosomes from bovine raw milk and studied for the delivery of chemopreventive agents (curcumin, withaferin A, and bilberry-derived anthocyanidins) and chemotherapeutic drugs (PTX and docetaxel (DOX)) in tumor xenografts.48 Agrawal and group have stated that oral delivery of ExoPac showed remarkably lower systemic and immunogenic toxicity in C57BL/6 female mice compared to IV administration.59 Further, these exosomes should be conjugated with suitable ligands to target desired sites in vivo.Tumor-derived exosomes are being recognized as a potential biomarker for cancer diagnosis and therapy. Exosomes isolated from the A549 cell line could target tumors and deliver DTX, an anti-cancer drug. The exosomes facilitated the cellular uptake of the drug and displayed improved targeting and cytotoxicity to cancer cells compared to free DTX in mice.83 Anti-tumor drugs such as triptolide (TP), bleomycin (BLM), etc. have shown great potential against cancer cells. However, their clinical use is limited due to adverse side effects. A targeted approach with exosomes has opened up promising avenues for the delivery of these efficient drugs to specific cell types with minimal side effects. In vitro and in vivo studies have shown assuring results where the exosomes loaded with TP/BLM manifested tremendous anti-tumor activity with reduced toxicity compared to the free drugs.84,85 For treating melanoma, Gu et al. (2022) developed a bionic-targeted drug delivery system using exosomes derived from human umbilical cord MSCs (hUCMSCs). This system encapsulates TP using the cyclic peptide arginine–glycine–aspartate (cRGD) and achieves synergism and toxicity reduction.86 Fisher et al. (2022) studied EVs (enriched with CD63) derived from the prostate (PC3) and melanoma (M21) cancer cell lines. The two cell lines produced vesicles of variable size profiles, with a constant fraction of exosomes to EV content. The EVs had a non-spherical morphology and showed enhanced solubility for PTX in aqueous solution.87 Apart from the delivery of typical cancer drugs, EVs can also be loaded with potential immune therapy agents such as immune checkpoint inhibitors (ICIs), which instigate the immune system to target and attack cancer cells. ICIs have been explored for the treatment of melanoma skin cancer and lung cancer. These are antibodies raised against programmed cell death-1 (PD-1), programmed death-ligand 1 (PD-L1), and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4). Studies have shown that conjugating EVs with ICIs can improve efficacy and reduce side effects when administered systemically.88 Cui et al. (2022) considered the potential of co-delivering cisplatin (CDDP), a first-generation platinum-based drug and CD47 antibodies with MDA-MB-231 cell-derived exosome (CaCE) to treat lung cancer. Interestingly, the CDDP treated tumor xenograft model showed enhanced expression of CD47. The co-delivery of CD-47 antibodies led to the blockage of CD-47, which in turn magnified the macrophages' phagocytic activity on CDDP treated tumor cells. The administration of CDDP and anti-CD-47 within CaCE in a lung carcinoma model exhibited a powerful anticancer effect in terms of tumor weight and the survival rate of the animal model, which was not observed in the co-delivery of CDDP and anti-CD-47. The concentrations of IL-12p and IFN-γ cytokines increased, while that of TGF-β decreased, indicating an enhanced antitumor effect of CDDP. The study demonstrated the impact of exosomes, not just as a delivery vehicle but also for its therapeutic potential that can dramatically change the outcome of cancer treatment.89 Several other studies have also reported the therapeutic effect of exosomes. For example, Wei et al. (2022) developed a nanodrug called Exo-DOX from exosomes and doxorubicin. Exo-DOX was more effective against osteosarcoma cells than free DOX. The drug was targeted specifically at cancer cells and caused less toxicity in the heart tissue. The targeting capability of Exo-DOX stemmed from the chemotaxis of MSC-derived exosomes to osteosarcoma cells via the SDF1-CXCR4 axis.90
Natural exosomes, while possessing several advantages as delivery systems, also suffer from limitations like immunogenicity, toxicity and rapid clearance by the reticuloendothelial system, thereby reducing their circulation time. These problems can be addressed by engineering the surface of exosomes, which is described in the following section.
3. Modified exosomes
Customizing exosomes by surface modifications, attaching additional ligands, or loading essential proteins can greatly alter the functions of native exosomes and result in target-specific engineered exosomes with desired characteristics like surface charge and enhanced circulation at the therapeutic site.91–96 As described earlier, two strategies can be employed to engineer exosomes, namely endogenous and exogenous methods. Early studies on exosome engineering involved the endogenous method. For instance, Ohno et al. transfected HEK293 cells with the pDisplay vector that encodes GE11 or the epidermal growth factor (EGF). The exosomes produced subsequently had membrane-bound GE11 or EGF that can specifically target the EGF receptor (EGFR) expressing breast tumor cells and deliver therapeutic microRNA (miRNA) via an EGFR mediated endocytic mechanism.95 In an attempt to deliver DOX to αv integrin-positive breast cancer cells, Tian et al. produced exosomes with a membrane protein (Lamp2B) fused to an αv integrin-specific iRGD peptide by engineering mouse immature dendritic cells. The targeted DOX-loaded exosomes showed specific accumulation in tumor tissues and dramatically inhibited tumor growth after systemic injection into breast tumor-bearing mice.94 Consequently, Xin et al. (2021) loaded methioninase (rMETase) into iRGD-Exos by electroporation to create iRGD-Exos-rMETase, which was found to be an effective anticancer therapy that delivered rMETase to gastric tumor tissue. iRGD-Exos-rMETase had a strong tumor suppressive effect and significantly reduced tumor growth.97 Pullan et al. (2022) have developed a new treatment for triple-negative breast cancer called iDHRX. It involves bovine milk exosomes that can penetrate tumors and respond to hypoxia. The exosomes have been modified with an iRGD peptide and a hypoxia-responsive lipid to increase their effectiveness. In lab tests, iDHRX was able to target and kill triple-negative breast cancer cells in both monolayer and 3D spheroid models. These modified exosomes could be a promising new treatment for triple-negative breast cancer, as they effectively reduce the survival of cancer cells.98 In addition to chemotherapeutics, iRGD-Exos could also deliver siRNA that specifically silences Carnitine palmitoyltransferase 1A (CPT1A), a key enzyme of fatty acid oxidation that is involved in conferring resistance to colon cancer cells against oxaliplatin therapy.99 With the view to target the central nervous system (CNS), Lamp2B-expressing exosomes were fused to the CNS-specific rabies virus glycoprotein (RVG) peptide. This system was designed to carry siRNA/mRNA for precisely targeting different regions of the brain such as neurons and microglia by crossing the BBB92,96 following IV injection. Later, Yang et al. presented a similar approach for delivering miR-124 to the brain, promoting cortical neurogenesis by using RVG exosomes as the carriers.93 Iyaswamy et al. (2023) engineered exosomes derived from hippocampal neurons to overexpress Fe65, a protein that interacts with the amyloid-β precursor protein (APP) intracellular domain (AICD). The APP is normally overexpressed in the AD and therefore the engineered exosomes loaded with corynoxine-B (Cory-B) were directed toward neuronal cells in the brains of AD mice. This enabled APP-targeted delivery of Cory-B that subsequently blocked the usual interaction between Fe65 and APP. This led to the enrichment of cognitive decline and pathogenesis in AD mice. The study demonstrated the potential of an exosome-based targeted drug delivery system as a potential treatment for AD.100 Hou et al. have recently discovered that oxygen glucose deprivation (OGD)-pretreated astrocyte-derived exosomes (ADEVs) can alleviate the disruption of the BBB caused by intracerebral hemorrhage (ICH). This is achieved through the regulation of the miR-27a-3p/ARHGAP25/Wnt/β-catenin axis. OGD-activated astrocytes release EVs, which reduce hemin-induced endothelial hyper-permeability. OGD-activated astrocyte-derived EVs (ADEVs) have been shown to alleviate BBB dysfunction following ICH, both in vivo and in vitro. MicroRNA microarray analysis has revealed that miR-27a-3p is a major component that is highly expressed in OGD-pretreated ADEVs. The transfer of miR-27a-3p from OGD-ADEVs into bEnd.3 cells regulates the ARHGAP25/Wnt/β-catenin pathway, mitigating BBB injury and improving neurological deficits following ICH. The findings suggest that OGD-ADEVs may be a promising strategy for the treatment of ICH.101In recent times, endogenous methods are less preferred as they display various disadvantages like complexity of genetic engineering, off-target effects, scalability issues and heterogeneity of engineered exosomes. On the other hand, the exogenous method of exosome engineering offers several benefits including versatility in cargo loading, reduced off-target effects, minimized immunogenicity and opportunities to scale. From this perspective, Jang et al. developed a simple post-insertion technique that integrated folate (FA) into the exosomal membrane via hydrophobic interaction. Due to the elastic nature of the exosomes, a lipidic anchor such as phosphatidylethanolamine (PE) was used as a linker molecule for FA. The resulting hybrid exosomes were loaded with DOX and were selectively taken up by FA receptor overexpressed tumor cells via endocytosis.58 Pishavar et al., in 2023, conjugated aptamers with exosomes from mesenchymal stem cells (MSCs) derived from adipocyte tissue (ADSCs) to deliver SN38. Aptamers specific to Mucin-1 (MUC-1) were conjugated to SN-38 loaded exosomes by covalent interaction using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide (NHS) chemistry to produce SN38/Exo-Apt. SN38/Exo-Apt was effectively taken up by colon tumor cells overexpressing MUC-1 and demonstrated significant cytotoxicity, while exhibiting minimal toxicity towards normal cells (CHO cells).102
Limited loading efficiency is still one of the main obstacles to using nanomedicine for protein therapy. Exosome engineering was used to efficiently load and intracellularly deliver proteins of interest by integrating a reversible protein–protein interaction module modulated by blue light.103 By labeling Cre recombinase (a tyrosine recombinase enzyme) with WW domains of Nedd4 family ubiquitin ligase, Sterzenbach et al. demonstrated that WW-Cre proteins were recognized by the late domain (L-domain)-containing protein Ndfip1, showing efficient loading into exosomes. Interestingly, the engineered exosomes effectively transported the loaded proteins to recipient neurons in different brain regions after intranasal administration.104 Kojima et al. developed an exosomal device for RNA packaging that consists of archaeal ribosomal protein L7Ae conjugated to the C-terminus of the CD-63 biomarker and assessed the ability to deliver mRNA into the brain for Parkinson's disease.96 The authors implanted an RNA packaging device (CD63-L7Ae) into living mice. The device steadily delivered mRNA of interest to the brain, attenuating neurotoxicity and neuroinflammation in an in vivo model of Parkinson's disease.96 Manipulating the behavior of nanocarriers, the formation of protein corona (PC) can be challenged. Multifunctional exosome-mimetics (EM) have been developed and functionalized with angiopep-2 (Ang) to improve drug delivery for glioblastoma (GBM) by controlling the PC. DTX-loaded EM with Ang modification (DTX@Ang-EM) exhibited lower serum protein absorption and decreased macrophage phagocytosis. In addition, Ang-EM was able to penetrate the BBB effectively and target the GBM, eventually leading to the delivery of high doses of DTX to the tumor site with reduced systemic side effects.105
Numerous autologous proteins have shown tremendous potential in alleviating disorders. Yet their instability in the native form has restricted their use in therapeutics. Exosomes, when naturally packaged with such kinds of molecules, will effectively preserve the protein of interest and deliver it to the specific target. In this context, bone morphogenetic proteins-7 (BMP-7), a potential but unstable protein involved in cartilage homeostasis and repair, were over-expressed in synovial mesenchymal stem cells. The exosomes (SMSCs-exo) derived thereafter were investigated for their therapeutic potential against osteoarthritis (OA). The BMP-7-exo treatment promoted macrophage and chondrocyte proliferation, reduced inflammation, and increased polarization toward M2. BMP-7-exo also attenuated inflammation and cartilage injury, suggesting that it is a promising therapeutic option for OA.106
Exosomes are becoming the rising star in the area of the targeted delivery system. However, to address the unmet clinical needs, challenges and limitations like isolation source, poor loading, purification methods, immunogenicity, and scalability associated with natural or synthetic exosomes, hybrid technology can be the key to achieving desired functions and clinical efficacy. In this context, artificial exosome mimetic vesicles would be of great interest, whereby the fusion of synthetic lipidic carriers like nanoliposomes is performed with exosomes to form hybridosomes.107,108 During this fusion, cell membrane proteins from various cell sources interact with the phospholipid bilayer to design hybrid exosomes. This hybrid system will have the combined advantages of individual exosome and liposome systems and can overcome their respective shortcomings, such as modification flexibility, yield, and endogenous functionality, respectively.109 Sato et al. designed membrane-engineered exosomes by modifying the exosomal interface through direct membrane fusion between liposomes and exosomes using the freeze–thaw method and stated that such hybrid exosomes would have lower immunogenicity effects, higher colloidal stability, and improved biological half-life.110 The authors isolated exosomes from supernatants of mouse fibroblast sarcoma-derived Raw 264.7 cell cultures and fused them with fluorescently labeled liposomes by the freeze–thaw method. The lipid dilution ratio was evaluated as a function of fusion efficiency and was found to be 1.6-fold higher for the exosome–liposome system compared to the liposome–liposome system due to the interaction of liposomal lipids with membrane proteins on the exosomal interface. Exosomes isolated from CMS7 cells overexpressing the HER2 receptor were also fused with liposomes, which showed a higher fusion efficiency compared to the Raw 264.7 exosome-liposome system. The comparative cellular uptake efficiency of fluorescently labeled plain exosomes and hybrid exosomes in HeLa cells displayed a significantly increased uptake of hybrid exosomes, almost two-fold, compared to untreated exosomes (750 counts vs. 350 counts). The authors concluded that modifying the exogenous lipids could significantly impact the interaction between the exosomes and their target cells.110 In yet another study, Rayamajhi et al. designed DOX-loaded hybrid exosomes through membrane fusion between mouse macrophage (J774A.1) derived exosomes and synthetic liposomes (Fig. 4a) and exploited their potential for breast tumor-targeted delivery.109 In addition to the freeze–thaw method, hybrid exosomes can be designed by a membrane extrusion method that produces a slightly larger vesicle (177 nm) compared to exosomes (139 nm) and liposomes (131 nm), since exosomes were inserted into a lipidic bilayer (Fig. 4b). Protein analysis confirmed the presence of major protein markers like CD81, CD63, and CD9 and tumor susceptibility gene 101 protein in hybrid exosomes. DOX loading efficiency was found to be 99%. Cellular internalization in mouse breast cancer cells (4T1) using Rhodamine-B labeled liposomes and hybrid exosomes revealed higher internalization with hybrid exosomes after 3 h of incubation and suggested higher selectivity for the hybrid system compared to plain carriers towards tumor cells (Fig. 4c).109
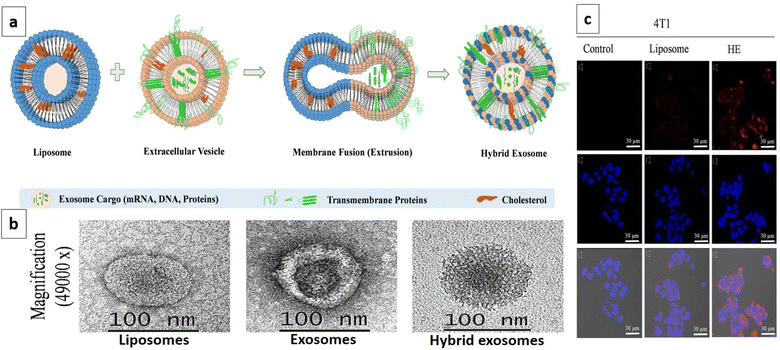 | ||
| Fig. 4 DOX loaded hybrid exosomes. (a) Schematic representation depicting the fabrication of hybrid exosomes by membrane fusion between mouse macrophage (J774A.1) derived exosomes and synthetic liposomes, (b) TEM images of liposomes, exosomes, and hybrid exosomes, and (c) confocal image showing cellular internalization in mouse breast cancer cells 4T1 with Rhodamine-B labeled liposomes and hybrid exosomes after 3 h of incubation, and untreated cells were used as a control. The highest internalization was observed with hybrid exosomes compared to liposomes (this figure has been reproduced from ref. 109 with permission from Elsevier, copyright 2019). | ||
Following cancer drug delivery, modified exosomes were also applied to liver diseases. Phillygenin (PHI) is a substance that protects the liver and can impede the progress of fibrosis by inhibiting the activation of hepatic stellate cells (HSCs). Unfortunately, its low aqueous solubility has created challenges for its application within a clinical environment. Gong and colleagues (2023) have developed a drug delivery system called PHI-HA-mEXO that uses milk-derived exosomes (mEXO) as nanocarriers. PHI is encapsulated in mEXO and then conjugated with hyaluronic acid (HA) that confers specificity to HPH-A-mEXO. This system can bind to CD44 (over-expressed on the HSC surface with the progression of liver fibrosis), which facilitates the delivery of PHI to activated HSCs (aHSCs) through endocytosis. Effective delivery of PHI results in the induction of aHSC death without affecting quiescent HSCs and hepatocytes. The PHI-HA-mEXO system shows encouraging results in alleviating liver fibrosis through an aHSC-targeted mechanism.111 Several other hybrid exosome-based investigations are listed in Table 2 along with their outcomes.
| Exosome origin | Synthetic carrier | Hybridization technique | Cargo/aim | Size (nm) | Marker proteins in a hybrid system | Pharmacokinetic/therapeutic implication | Ref. |
|---|---|---|---|---|---|---|---|
| a Abbreviations: CER: ceramide, CRISPR/Cas9: clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9, CT26 cells: mouse colon carcinoma cells, DPPC: dipalmitoylphosphatidylcholine, DiD: 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzene sulfonate salt, EMN: exosome mimetic nanosystems, HEK293FT: human embryonic kidney 293 cells, LCBM: lung cancer-bearing mice (inoculation of luciferase-expressing A549 lung carcinoma cells into the tail vein of nude mice), MSC: mesenchymal stem cells, mTHPC: (3,3′,3′′,3′′′-(2,3-dihydro porphyrin-5,10,15,20 tetrayl) tetraphenol) (antitumor photosensitizer), NR: not reported, PC: phosphatidylcholine, PE: phosphatidylethanolamine, PEG 8000: polyethylene glycol 8000, PLGA: poly(lactic-co-glycolic acid), and SM: sphingomyelin. | |||||||
| HUVECs (human umbilical vein endothelial cells) | Liposome: Foslip (liposomal form of mTHPC) | PEG mediated fusion: exosomes were fused with liposomes in the presence of 30% w/v PEG8000 for 2 h | mTHPC: to improve loading efficiency, minimize leakage, and compare the delivery of mTHPC in CT26 cells | <500 | NR | Compared to normal exosomes and liposomes, the hybrid system showed 20-fold higher encapsulation with 65% presence of the liposomal zone into hybrid exosomes and the absence of leakage. After 4 h of incubation with CT26 cells, hybrid exosomes showed 60–70% mTHPC transfer into cells against 10–20% transfer with Foslip and free mTHPC | 112 |
| A549 cells: integrin α6β4 tailored exosomes for lung organotropism | Liposome: PC, CH, SM, and CER composed liposome | Fusion method: exosomes and liposomes were fused to design functionalized EMN containing miR145 | miR145. EMN for targeted delivery of therapeutic oligonucleotides to lung adenocarcinoma cells | ∼100 | Alix, β-actin, N-cadherin, CD9, CD63, CD81 | Integrin α6β4 functionalized EMN showed significantly higher transfection of miR145 into A549 mouse lung cancer cells compared to normal exosomes. A biodistribution study in LCBM following intraperitoneal administration showed accumulation of miRNA in the lungs with localization of miR145 in the neighborhood of Ki67 positive proliferative tumor cells and significantly lower (p < 0.05) distribution in the kidneys and liver compared to plain exosomes | 113 |
| HEK293FT: exosomes were isolated from sgRNA expressing HEK293FT cells | Liposome: liposomes were transfected with the Cas9 expressing vector | Simple incubation for 12 h at 37 °C by mixing exosomes and liposomes and then adding it to murine MSC | CRISPR/Cas9. To deliver CRISPR/Cas9 in MSC for in vivo gene manipulation | <500 | Alix, TSG101, CD63, GAPDH | Compared with the control group (without the vector), the hybrid exosomes substantially increased both sgRNA and Cas9 mRNA expression into MSC and confirmed the presence of CRISPR/Cas9 in transfection-resistant cells | 114 |
| Mouse macrophage cells Raw264.7 | Amphiphilic cationic nanogel of pullulan (CNP) | Simple mixing of the exosome suspension and nanogel suspension in PBS for 30 min using ice | For effective intracellular delivery of biologics into cells using an exosome–nanogel hybrid complex | <200 | CD9, Hsc70 | Cellular uptake study in HeLa cells showed that after 30 min hybrids were adsorbed onto the cells' surface and internalized after 2 h, showing enhanced internalization compared to plain exosomes. A detailed internalization study using endocytic inhibitors revealed that exosome uptake was inhibited by every inhibitor. Contrastingly, exosome-nanogel hybrid complex uptake was significantly suppressed by the caveolae-mediated endocytosis inhibitor suggesting endocytosis being the primary mechanism behind this internalization. Internalized hybrids also showed fusion between the endosome and exosome | 115 |
| Human RBC membrane derived from whole blood | Human platelet membrane derived from platelet-rich plasma | Membrane fusion: fused RBC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) platelet (1 platelet (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) membrane was coated on PLGA cores to form RBC-PLT hybrid cores 1) membrane was coated on PLGA cores to form RBC-PLT hybrid cores |
To combine individual functions of the RBC membrane and platelet membrane into a single hybrid system to achieve dual-membrane enhanced functionality in imaging and targeting delivery | <150 | CD47, CD41, CD61, A antigen, CD235a | Platelets contain a large number of disease-specific binding markers on the surface, which were also present in RBC–PLT hybrid cores, since they bound firmly to the highly metastatic MDA-MB-231 human breast cancer cells and RBC alone did not show any binding affinity due to the absence of CD41 and CD61 adhesion markers. In vivo targeting in an atherosclerosis-induced mouse model after IV administration of DiD labeled RBC, PLT, and hybrid RBC–PLT cores revealed that after 24 h of administration, aortas from euthanized mice with a hybrid treated group showed enhanced fluorescence compared to individual RBC and PLT, thereby confirming higher binding because of the fused advantages of RBC and PLT together into a hybrid system | 116 |
| U937 monocytes (human macrophage cell line) | Liposomes: DPPC and CH-composed liposomes | Membrane fusion: lipid film was hydrated using U937 derived vesicle suspension to encapsulate exosomes into liposomes (EXOPLEX), followed by extrusion | Doxorubicin: to evaluate the potential of EXOPLEX for targeted delivery of chemotherapeutics | ∼200 | Alix, TSG101, CD63, CD9 | Cellular uptake study using DOX-loaded liposomes and EXOPLEX in HeLa cells indicated that at 6 h about 6.5% of HeLa cells had internalized the liposome, while 39.4% of cells had taken up DOX from EXOPLEX indicating the improved cellular uptake and better cell killing effect of cancerous HeLa cells. MTT assay on HeLa cells over 48 h showed the highest cytotoxicity of free DOX, while the cytotoxicity profile of EXOPLEX was reported to be better than that of liposomes | 117 |
Although exosomes from cancer cells have proved to be efficient in drug delivery, there is always a risk that their cargo (DNA, RNA, and protein) may produce specific outcomes on cancer progression involving angiogenesis, invasion and metastasis.118 In fact, recently, a patient-oriented therapy study conducted by Hyung et al. investigated the effects of exosomes on gastric cancer (GC) invasiveness and angiogenesis in an ex vivo 3D autologous tumor spheroid microfluidic system. The study used exosomes derived from malignant ascites (MET oncogene-amplified and non-amplified) to validate the role of exosomes in GC progression. The microfluidic system was intended to mimic the fluid interfaces observed in patients suffering from peritoneal disease. For individuals with MET amplification, there is a consistent and deliberate packaging of MET into the exosomes, which aligns with the findings of primary tumor genomics. The study demonstrated the uptake of exosomal MET by the tumor cells, leading to increased invasion and angiogenesis, while the MET depleted exosomes impaired the invasion and angiogenesis of MET-amplified GC. Thus, the study evidently showed that manipulating exosomal content, similar to MET depletion, may significantly impact the target cancer cells to be more sensitive towards MET-directed therapies, as shown in Fig. 5 and this strategy can be of therapeutic value in impeding cancer progression.119
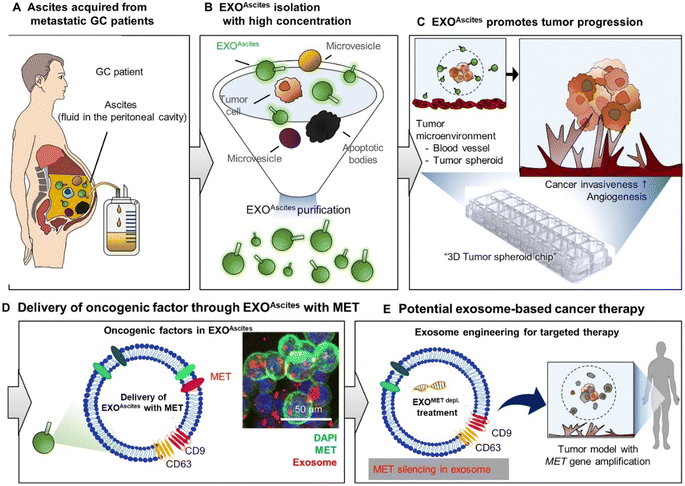 | ||
| Fig. 5 Applied exosome therapy as personalized medicine in gastric cancer. The schematic illustrates the potential use of exosomes as biomarkers and for predicting therapy response. EXOAscites isolated from cancer patients (A and B) induced cancer invasion and angiogenesis (C). EXOAscites from MET-amplified cancer cells contained MET, which drove tumor invasiveness and angiogenesis in a dose-dependent manner, regardless of the MET amplification status (D). The MET protein found in EXOAscites can serve as a useful biomarker for both the diagnosis and prognosis of gastric cancer (GC) (D). Additionally, a promising approach for precision medicine in GC could involve using engineered exosomes to remove MET or other relevant therapeutic proteins (E).119 (from ref. 119 © The Authors, some rights reserved; exclusive licensee AAAS. Distributed under a CC BY-NC 4.0 license http://creativecommons.org/licenses/by-nc/4.0/. Reprinted with permission from AAAS.119 with permission from AAAS). | ||
A new approach for mitigating the risks of tumorigenic exosomes is the use of reassembly-exosomes (R-EXO). These modified cancer cell-derived exosomes are prepared by a brief sonication process with intermittent breaks, followed by reassembly in an aqueous environment. The so-formed R-EXOs are devoid of internal contents and yet retain the cell membrane and other characteristic proteins that impart natural homologous targeting ability. In a recent study, R-EXOs were derived from homologous glioma cells and were loaded with temozolomide (TMZ) and dihydrotanshinone (DHT), a chemo-immune combination therapy that may alleviate the TMZ drug resistance exhibited by glioma cells. R-EXO-T/D nanoparticles have several advantages, including their small size, which allows them to penetrate the BBB more easily, their ability to accumulate in tumors and their improved anti-tumor activity. These advantages can help overcome resistance to TMZ and stimulate the immune system to fight cancer.120
4. Drug loading and release from exosomes
Efficient loading of active materials into exosomes without compromising the integrity and stability of a loaded drug is a colossal challenge. Sufficient drug loading is critical when developing exosomal drug products as it involves numerous factors such as the source of exosomes,36 the nature of the drug, drug loading methods, etc. Methods of loading active cargo into exosomes can be categorized into three methods, viz., exogenous loading (post-loading), endogenous loading (pre-loading), and a fusion method.121 The exogenous and fusion loading methods are more popular compared to endogenous methods in terms of loading efficiency and stability of exosomes. Exogenous loading can be achieved through several methods like freeze–thaw cycles, incubation, electroporation, sonication, extrusion, and saponin-mediated loading. A comparison of the loading efficiency (%) of catalase enzymes into RAW264.7 macrophage-derived exosomes is illustrated in Table 3.| Loading method | Loading efficiency (%) |
|---|---|
| Incubation at room temperature | 4.9 ± 0.5% |
| Incubation with saponin | 18.5 ± 1.3% |
| Freeze–thaw cycle | 14.7 ± 1.1% |
| Sonication | 26.1 ± 1.2% |
| Extrusion | 22.2 ± 3.1% |
The incubation method is the easiest method involving the mixing of isolated vesicles with the cargo. In this method, the drug permeates across the exosomal membrane by passive diffusion during incubation.26,27,122–126 Although simple, incubation is not often adopted due to the low loading efficiency.27,123 On the other hand, permeation enhancers could be used to increase the drug loading efficiency into exosomes.27,48,56,59,124 As shown in Fig. 6, the presence of saponin significantly improved the loading efficiency of catalase and porphyrin into exosomes and EVs without affecting their sizes.27,124 Organic solvents like ethanol, dimethyl sulfoxide (DMSO), or acetonitrile with concentrations up to 10% (w/v) may be used to enhance the exosomal membrane permeability, thereby enhancing the drug loading efficiency without significantly affecting their physicochemical properties.48,56,59 pH-gradients can also be used to increase the loading of drugs such as piceatannol into extracellular vesicles, which showed promising results.127 One should notice that all the studies using the incubation method have been performed at room temperature, which could adversely affect the stability and therapeutic effects of exosomes. Incubation at lower temperatures and increasing the time of incubation may be a suitable alternative to overcome these issues with stability.
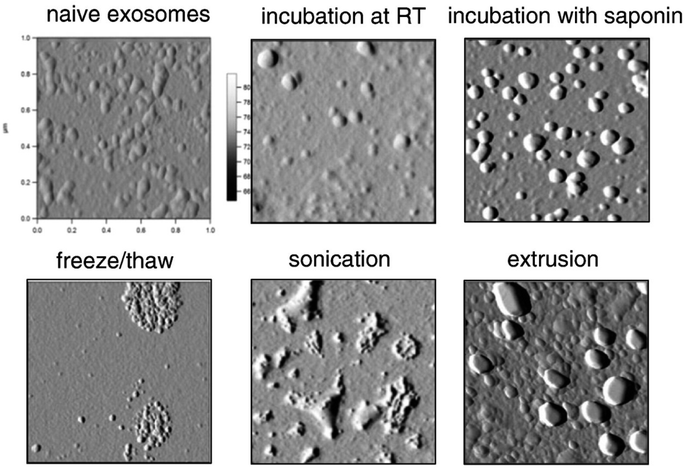 | ||
| Fig. 6 AFM morphology of catalase-loaded exosomes prepared by different methods (this figure has been reproduced from ref. 27 with permission from Elsevier, copyright 2015). | ||
Electroporation is an approach commonly employed for loading siRNA into exosomes.123,128–131 The technique uses an electric field to generate small pores in the phospholipid bilayer, thereby allowing passage of the cargo into the vesicles. However, a potential problem associated with electroporation is the membrane damage, fusion, and aggregation of exosomes.131 This was also confirmed by Kooijmans et al., where the disruption of the exosome integrity with electroporation was demonstrated.132 The study also highlighted other problems such as the formation of insoluble siRNA when loaded into EVs, which lead to the overassessment of the loaded siRNA and the potential by-products that may influence the delivery.132,133 The authors proposed strategies like using ethylene diamine tetraacetic acid (EDTA), cuvettes with conductive polymer electrodes, and acidic citrate electroporation buffer to reduce the aggregation. Nevertheless, siRNA encapsulated within the EVs was extremely low.132 The drug nanocarrier development in exosomes has faced challenges due to limited drug encapsulation efficiencies and complicated drug-loading procedures. A new acoustofluidic device has been reported that can perform drug loading and exosome encapsulation simultaneously. Using acoustic radiation force, microstreaming, and shear stresses in a rotating droplet, the device produces drug-loaded silica nanocarriers enclosed in an exosomal membrane. This acoustofluidic drug loading system is a facile drug loading method that enhances drug loading efficiency, with almost 30% of free drug molecules (such as DOX) being loaded into the nanocarriers.134 Active loading methods using energy were reported to enhance the transport of cargo into exosomes. Nevertheless, the applied energy could potentially damage the exosome structure and cause the aggregation of components like proteins. Mild sonication did not affect the proteins of exosomes and significantly enhanced the loading efficiency of drugs, yet it may lead to a significant increase in exosome size.27,123,135,136 Gholami Farashah et al. (2023) investigated the effects of two different methods of loading estradiol onto bone marrow mesenchymal stem cell (BMMSC)-derived exosomes, namely incubation and sonication methods, on the survival of BMMSCs. The researchers identified that the spherical morphology of the exosomes was preserved irrespective of the condition. On the other hand, the particle size was increased significantly when loaded by the sonication method.137 In another study, PTX was packaged into macrophage-derived exosomes using sonication, incubation, and electroporation methods and Kim et al. reported higher loading efficiency of PTX into exosomes with sonication over incubation and electroporation methods (28.29% vs. 1.44% vs. 5.3%). It should be noted that sonication did not alter the contents of the exosomes (protein and lipid) but the size of PTX-loaded exosomes was significantly higher (287.7 nm) compared to incubation (132.2 nm) and electroporation (145.3 nm) methods.123 This result was in line with the outcome reported by Haney et al., who also claimed the highest loading efficiency for catalase into RAW264.7 macrophage-derived exosomes with the sonication method (Table 3) over other post-loading approaches. Although not the highest, the loading efficiency of catalase through the extrusion method was comparable to that of sonication.27 The extrusion method involves the passing of the drug and vesicle mixture through a syringe-based lipid extruder containing different pore-size polycarbonate membranes ranging from 100 to 400 nm. Transient change and/or disruption in the vesicle membrane enabled the loading of drugs into the vesicles.138 Porphyrin was loaded into different cell-derived exosomes by the extrusion method using a hand-held mini-extruder and polycarbonate membranes of 400 nm pore size.124 A comparison of other loading methods like electroporation and saponin-mediated incubation concluded that the extrusion method showed variation in zeta potential over the other two methods, which could be attributed to the modulation of the vesicle membrane components since the vesicles were extruded 31 times. As a result, even porphyrin loading was lowest with the extrusion method and exosomes showed cytotoxicity, while the cytotoxicity level with the other two methods was significantly lower.124 Chemical transfection using cationic lipids is an alternative for loading siRNA into exosomes, although it is challenging to separate siRNA exosomes from the micelles used in the chemical transfection protocol.129,130 The freeze–thaw cycle, extrusion, and hypotonic dialysis are some uncommon methods used for loading drugs into exosomes. However, all of these methods were reported to cause an increase in exosome size.27,124 Like all other pharmaceutical processes, the drug loading methods must be scalable and have their validation established. To our current understanding, there are no reports on the use of exosomes as carriers for hydrophilic drugs, presumably due to the lack of a method for efficiently transporting hydrophilic drugs across the lipid membrane of exosomes without disrupting it. Our recent data showed that the loading efficiency of sulforhodamine B into M-Exos using passive incubation was extremely low. However, with a minimal amount of organic solvent, the entrapment could be improved without any damage to the exosome membrane.
The source of the exosome and release conditions have a huge impact on the release of active cargo from the exosome. PTX-loaded macrophage-derived exosomes showed a burst release within the first three hours in PBS pH 7.4, followed by a sustained release profile. However, the maximum release of PTX from exosomes was 70% in 150 h.123 In contrast, M-Exos showed complete release of PTX within 24 h in PBS pH 7.4 containing tween 80.48 The above differences in PTX release could be due to the differences in the source of exosomes and the release conditions. The pH could significantly impact the release rate of drugs from exosomes. For instance, a study was conducted to investigate the release of DOX from exosomes under two conditions: pH 7.4 and pH 5.0 representing the physiological environment and late endosomes. Burst release was observed under both pH conditions. The release of DOX at pH 7.4 was not complete and reached only 50% after 48 hours, with about 40% release in the first 5 hours. The results may be accounted for by the fact that DOX was not entirely entrapped into exosomes but was just bound to the surface. On the other hand, at pH 5.0 DOX was released slowly after 10 h and showed complete release in 48 h (Fig. 7).126 Similarly, Wang et al. also showed that the release of DOX from exosomes was higher under acidic conditions (pH 5.5) than at pH 7.4.139 Since exosomes are of endosomal origin, the mildly acidic conditions might not affect their structures, and therefore, the rapid release of DOX is probably the result of high DOX protonation.126,139 The release of unionized drugs like PTX was shown to be slightly impacted by the pH of the release medium. Agrawal et al. made a comparative study of the release of PTX from M-Exos in fed-state simulated gastric fluid (pH 5.0), intestinal fluid (pH 5.8) and PBS (pH 6.8). Interestingly, similar initial release kinetics were observed in all the simulated fluids suggesting the minimal effects of pH and enzymes on the PTX-loaded exosomes. More than 90% of PTX was released in PBS pH 6.8 in 48 hours.59
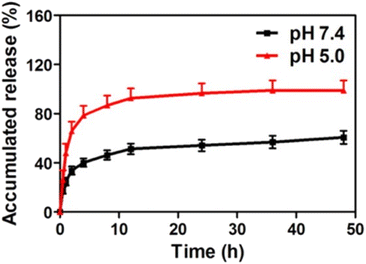 | ||
| Fig. 7 In vitro release profile of DOX-loaded exosomes in pH 5.0 acetate buffer and pH 7.4 phosphate buffer (this figure has been reproduced from ref. 126, with permission from American Chemical Society, Copyright 2016). | ||
5. Exosome stability and storage condition
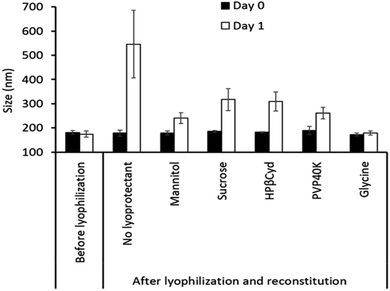
While exosomes, as drug delivery vehicles, have created significant interest, novel approaches for improving their stability have to be developed for realizing their translation into pharmaceutical products. Physicochemical properties and protein content are two main parameters commonly evaluated while studying exosome stability. Unfortunately, most of the studies only determine the effects of temperature on the exosome stability and the reported data are also controversial.140–142 Lee et al. reported the effect of storage conditions on the stability of exosomes extracted from HEK 293 cell cultures. The exosomes exposed to temperatures at and beyond RT were mostly degraded in 30 min. Under long-term storage conditions, more than 70% loss in CD63 was observed with incubation at 4 °C and RT, compared to −70 °C storage. At RT for 10 days, the exosome groups became well distributed compared to those stored at −70 °C and freshly isolated. The results collectively suggest that storing exosomes below −70 °C is a favorable condition for maintaining their cargo/packaging. However, studies beyond 10 days of storage would give us better insights into their long-term stability.140 In a related study, Agrawal et al. reported that M-Exos had excellent shelf stability at −80 °C.143 The storage conditions not only affect the cargo but also their size and integrity. Sokolova et al. observed the stability of human cell-derived exosomes at different temperatures (−20°, 4°, and 37 °C) and revealed that the sizes of exosomes stored at 4 °C and 37 °C reduced.142 In a contrasting study, exosomes collected from bronchoalveolar lavage fluid (BALF) stored at 4 °C and −80 °C showed an increase in diameter by 10% and 25%, respectively, in comparison to freshly isolated exosomes.144 Importantly, the authors also indicated that storage at −80 °C even had stronger adverse effects on the surface proteins, morphology, size, and zeta potential of those exosomes when compared to 4 °C. The stability of exosomes/EVs also relies on the source including cell culture medium or biological fluids. Various studies have shown that EVs isolated from milk, serum, plasma urine, etc. have retained the protein markers fairly well for 1 month of storage at −80 °C, provided that the freeze–thaw cycle was restricted to 1 or 2 cycles.142,145 Ruzycka-Ayoush and colleagues conducted a study to determine the optimal storage buffer for exosomes derived from lung cancer cells. The research revealed that PBS containing 25 mM trehalose emerged as the most effective cryoprotectant for maintaining the integrity of exosomes at −80 °C. Exosomes stored under these conditions exhibited consistent concentration, size distribution, zeta potential and total protein cargo level over time.146 Several studies have also suggested that the use of protease inhibitors like phenylmethylsulfonyl fluoride (PMSF) or the all-purpose protease inhibitor cocktail can preserve the exosomal proteins during storage.147,148 In addition to exosomes, it is also vital to assess the short-term and long-term stability and functionality of drug-loaded EVs/exosomes. DOX-loaded vesicles were stable when stored at 4 °C for one week and the authors reported that drug-loaded vesicles had similar pharmacological efficacy to kill H22-hepatocarcinoma tumor cells even after one week.141 Lőrincz et al. showed that 4 °C and −20 °C conditions significantly affected the size and antibacterial effects of neutrophilic granulocyte-derived EVs.149 Our group has investigated the stability of bovine M-Exos at 4 °C and RT for 10 days. The M-Exos were found to be stable in PBS and purified water in terms of particle size and size distribution (data not published). We have also evaluated the impact of temperature on the stability of SN-38 loaded exosomes and SN-38 solution for up to five days. The SN-38 content in exosomes was stable at RT and 4 °C, while its solution showed an almost 1.5-fold decline in the content after 5 days under both storage conditions, concluding that exosomes could be a promising carrier to protect and deliver antineoplastic drugs like SN-38. The studies on the stability of exosomes in physiological fluids are extremely critical since the design of exosomal drug formulations is significantly affected by these fluids. Unfortunately, not enough attention has been paid to this area and the reported data were also minimal. Agrawal et al. reported that the size of M-Exos had outstanding stability when exposed to simulated gastrointestinal fluid.59 A study by Wang et al. exhibited that nanovesicles from grapefruits were relatively stable in mouse gastrointestinal fluid in terms of surface charges and particle sizes.62 One can notice that exosomes used in stability tests were all in aqueous dispersion, so making lyophilized products of exosomes might be a good approach for overcoming the stability problems. Additionally, data on the stability of drug-loaded exosomes are very limited. It remains to be determined whether loaded drugs are retained in exosomes or drugs are gradually released from exosomes during storage. Our research group has been working on the stability of exosomes, which has been poorly defined so far. Limited stability studies on milk-based exosomes revealed that the loaded drug (coumarin-6) continuously released from the exosomes when stored in a liquid state even at 4 °C, as fluorescence intensity gradually decreased from day zero till the fourth day. In contrast, lyophilization improved the stability with constant fluorescence intensity for coumarin-6 from freeze-dried exosomes (0.5% w/v glycine) for up to four days, indicating the absence of leakage (data not published). Although several studies have indicated that the storage of exosomes at −80 °C is optimal, it may not be practical for general use. Appropriate storage conditions such as temperature, avoiding frequent freeze–thaw cycles, and using an appropriate cryoprotectant and a storage buffer would help to ensure the reliability and the usefulness of exosomes in a clinical setting.6. Exosome formulation and route of administration
The formulations of exosomes commonly tested in in vivo studies are simple aqueous dispersions without other excipients.17,26,27,48,125,150 Early studies in our laboratory indicated that lyophilization improved the stability and prevented the leakage of the drug, and a significant change in the size of lyophilized exosomes was observed 1 day after storage at 4 °C in the absence of cryoprotectants. However, the inclusion of cryoprotectants remarkably improved the exosome stability during lyophilization (Fig. 8). Further research on optimizing the optimal concentration of cryoprotectants and understanding the mechanism of the cryoprotectants is underway. In addition, our group is currently investigating the impacts of other manufacturing processes like filtration and sterilization on the stability and drug release of exosomal formulations.The administration route plays a pivotal role in defining the design of drug formulations. Whilst numerous studies have concentrated on drug delivery and targeting by IV and intraperitoneal routes, subcutaneous injection was also reported.17,26,48,150 Wassmer et al. investigated an intravitreal administration of exosome-associated adeno-associated virus (exo-AAV) for gene delivery into the murine retina. The authors encapsulated an AAV genome containing a green fluorescent protein (GFP) into an exo-AAV2 vector and injected it into adult mice intravitreally. After four weeks post-injection, exo-AAV2 demonstrated enhanced penetration into the retina, effectively reaching the inner nuclear and outer plexiform layers. This suggests that Exo-AAV2 can serve as a reliable gene delivery tool for the retina.151 No topical ocular formulations based on exosomes have been reported yet. However, topical application of exosomes has attracted significant attention in recent years for cosmetics152 and for treating skin-related disorders such as psoriasis.153 Tofacitinib (TFC) loaded keratinocyte exosomes were used for the treatment of psoriasis. The drug-loaded exosomes were mixed with a cold cream base and the anti-inflammatory properties were assessed on in vitro and in vivo disease models. The topical formulation of the TFC loaded exosomes targeted the immune cells that induce psoriasis inflammation and showed a higher therapeutic effect on animal models compared to free TFC.154 Intranasal administration provides a very convenient and rapid route for transporting drugs to the central nervous system (CNS). It was shown that drugs administered intranasally could traverse the single epithelial cell layer directly to the systemic bloodstream or along the olfactory nerve cells, eluding the BBB.154,155 Moreover, this approach is appealing, as it offers the potential for non-invasive, repetitive treatments with high levels of patient adherence. Interestingly, intranasal routes have been used in some studies to deliver exosome formulation to the brain. Curcumin exosomes were applied intranasally to treat brain inflammatory diseases.125 In a related study, Haney et al. used a Parkinson's disease mouse model and demonstrated a higher accumulation of catalase-loaded exosomes in the brain with intranasal administration in comparison to IV injection.27 When engineered exosomes were used to deliver Cre-recombinase proteins via the intranasal route in mice, exosomes were found predominantly distributed in recipient neurons in different brain regions like the olfactory bulb, cortex, striatum, hippocampus, and cerebellum, suggesting their capability to deliver protein molecules across the BBB.104 The potential of exosomal formulations for oral delivery has not been highly explored, except in a few studies, presumably due to their vulnerability to an acidic environment and the presence of proteolytic enzymes in the GI tract. Wang et al. isolated nanovesicles from grapefruits and used them as carriers for methotrexate (MTX) delivery to treat colitis mice.62 MTX-loaded nanovesicles showed significantly lower toxicity and enhanced therapeutic effects compared to free MTX, suggesting the potential of these nanovesicles in oral drug administration. Agrawal et al. used M-Exos for oral delivery of PTX and reported a significant reduction in tumor growth in a murine lung tumor xenograft model.59 Although the surface charge and particle size in simulated gastrointestinal fluids were used as the parameters to indicate exosome stability, it is not clear if the loaded drug was retained in the exosomes, as the impact on drug encapsulation and/or loading was not evaluated.59,62 Detailed stability studies on nanovesicles/exosomes in the gastrointestinal tract are warranted because the lipid and protein components of the vesicle are extremely vulnerable, thereby increasing the leakage of loaded drugs, resulting in stability problems.
7. Pre-clinical studies on exosomes
To develop exosomes as delivery vehicles, it is crucial to comprehend the mechanism and locations of exogenously administered exosome absorption, distribution, and elimination in vivo. The cellular uptake of bovine M-Exos was reported to be mediated by endocytosis via exosome surface glycoproteins.23 Although pharmacokinetic data are sparse, some suggest that exosomes undergo rapid clearance from the bloodstream and are dispersed to specific tissues depending on their source. For example, an in vivo mouse model showed that both intravenously and intraperitoneally administered exosomes rapidly cleared from the blood circulation and accumulated mainly in organs like the liver, spleen, and kidneys.26,56 In a related study, B16-BL6 murine melanoma cell-derived exosomes largely accumulated in the liver and lungs and to a lesser extent in the kidneys.156,157 Wiklander et al. analyzed the in vivo distribution of exosomes from different cell sources: B16F10 murine melanoma cells, C2C12 murine myoblast cells, bone marrow-derived DCs, and HEK293T human embryonic kidney cells. Size distribution analysis revealed that exosomes from different cell sources were within 100–150 nm. Although all exosomes were predominantly found in the liver, spleen, lungs, and GI tract after IV administration, distinct differences were also observed. C2C12 murine myoblast cell-derived exosomes displayed more substantial liver accumulation, B16F10 murine melanoma cell-EVs were more readily found in the GI tract, and bone marrow-derived DC-EVs showed increased accumulation in the spleen.158 The differences in the biodistribution of exosomes from different sources might account for the heterogeneity in exosomal surface biomarkers.159 Also, it is observed that the macrophages may have a vital role in the systemic clearance of B16-BL6 cell-derived exosomes.160A systematic in vivo study on the use of exosomes as a therapy to treat spinal cord injury was performed by Guo et al. Success in treating spinal cord injury is limited, leading to compromised cognitive and behavioral functions as well as permeant neurological deficits, which underscores the necessity to target the diminished neuronal and axonal regions, particularly phosphatase and tensin homolog (PTEN) levels. An MSC-derived exosome loaded with PTEN-siRNA (ExoPTEN) for intranasal delivery was designed to improve the expression of PTEN and to treat spinal cord injured rats. The study involved the administration of PTEN-siRNA and ExoPTEN via intralesional and intranasal routes into rats with complete spinal injury. The BBB locomotor score for each study group was observed up to 8 weeks. Intranasal ExoPTEN-treated rats led to significant locomotor recovery (p < 0.001) compared to other groups and 28.6% of this group rats showed a BBB locomotor score >14, illustrating steady plantar stepping, toe clearance, and stable forelimb–hindlimb coordination. In vivo MRI images (Fig. 9) confirmed the integrity of the spinal cord after intranasal ExoPTEN treatment. The results observed are very promising and can have huge therapeutic applications in neurological disorders.161
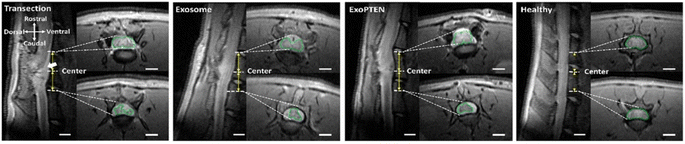 | ||
| Fig. 9 In vivo MRI imaging showing sagittal images with 4 mm caudal and rostral to the T10 epicenter in healthy rats or the injury epicenter in untreated (transection), exosome-treated, or ExoPTEN-treated rats. A large cyst (white arrow) is seen around the epicenter in the untreated spinal cord injured control rats. Exosome and ExoPTEN treated groups showing neural tissue regeneration and organization (scale bar: 2 mm) (this figure has been adapted from ref. 161, with permission from American Chemical Society, Copyright 2019). | ||
Recently, an in vivo study was conducted to assess the potential and the toxicity of IV administered hybrid exosomes. Parts of the exosomes that confer targeting ability (exosomal membrane) were coated over polymeric nanoparticles to improve their circulation time by evading the immune system. Exosomal membrane modified poly(lactic-co-glycolic acid) (PLGA) nanoparticles with AS1411 aptamers (AS-EP) were developed for better tumor targeting. AS-EP was assembled within 10 minutes using microfluidic sonication and cholesterol-modified aptamer functionalization. These AS-EPs exhibited enhanced in vivo circulation and high tumor-targeting efficiency through specific binding to nucleolin on tumor cell membranes. The IV administration of AS-EP to mice was safe and did not trigger any pathological anomalies. This study provided new insights for creating and functionalizing biomembrane-coated nanoparticles for targeted drug delivery.162 In a related study, chemotherapeutics were loaded into aptamer modified hybrid exosomes and their biodistribution was investigated post IV administration in a mouse colon adenocarcinoma model. DOX loaded MSC exosomes were developed for this purpose and were decorated with the MUC1 aptamer (DOX@exosome-apt), providing selective drug delivery. DOX@exosome-apt efficiently transported DOX to MUC1-positive cancer cells, significantly suppressing tumor growth in vivo compared to free DOX. Ex vivo fluorescent imaging verified DOX@exosome-apt's desirable biodistribution.163 In addition to intranasal and IV, in vivo studies were also performed on oral administration of hybrid exosomes. For instance, Han et al. investigated the effectiveness of treating colitis in mice using M-Exos and exosomes from HEK293T (H-Exos) loaded with TNF-α siRNA (M-Exo/siR) and labelled with Cy5.5. The organ distribution and pharmacokinetics of the Exos from both sources were observed in mice post oral administration. The distribution results showed intense fluorescent signals only within the GI tract of M-Exos fed mice (Fig. 10A). The presence of M-Exos in the GI tract is believed to be due to the unique composition of lipids in M-Exos. Further evaluation of the physicochemical properties of these Exos from a lipidomic standpoint (Fig. 10B) unveiled the presence of TAG (three fatty acids esterified to a glycerol backbone) constituting over half of the lipids in M-Exos.164
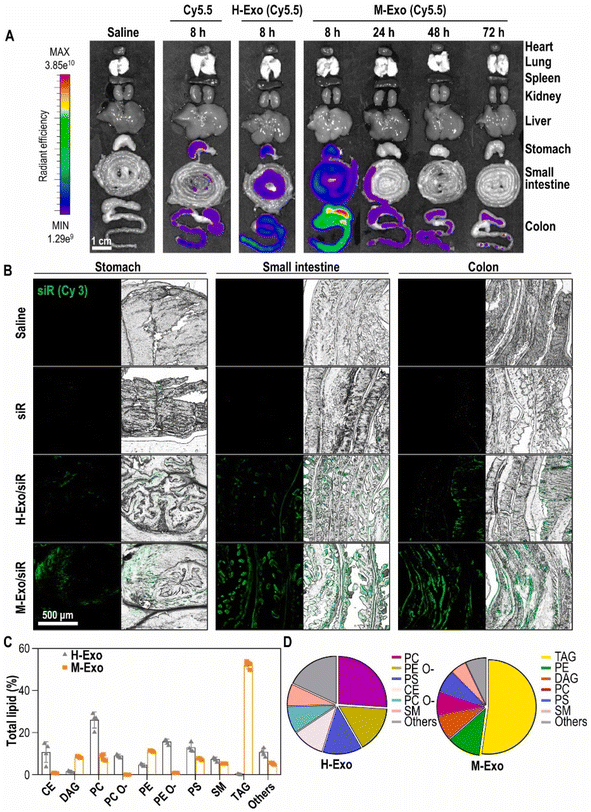 | ||
| Fig. 10 Stability and lipid composition analysis of M-Exos and H-Exos. (A) Bio-distribution of Cy5.5-labeled M-Exos. After oral administration of saline, cyanine 5.5, cyanine 5.5-labeled H-Exos, and cyanine 5.5-labeled M-Exos to 8 week-old female BALB/c. (B) Tissue images of Cy3-labeled siRNA absorbed into the stomach, small intestine, and colon. Green (cyanine 3-labeled TNF-α siRNA). (C) Lipidomics analysis of M-Exos compared to H-Exos [data are represented as mean ± SD (n = 4)]. (D) Lipid composition ratio constituting H-Exos and M-Exos (this figure has been adapted from ref. 164, with permission from Elsevier, Copyright 2024). | ||
From a therapeutic perspective of engineered/hybrid exosomes, Zhang et al. developed biomimetic artificial chimeric exosomes (ACEs) using RBC and MCF-7 cell membrane proteins in the form of artificial exosomes (ARE and AME) and fused them with liposomes. These hybrid exosomes were shown to have individual cell specific protein markers of RBC (CD47) and MCF-7 (EpCAM, galectin 3, and cadherin) and were loaded with DOX.165 The encapsulation of DOX in ACE (90%) was comparable with that of liposomes, ARE and AME (91–93%), which denotes that the inclusion of membrane proteins did not significantly alter the loading efficiency. An in vivo imaging study using indocyanine green fluorescent dye in MCF-7 tumor-bearing mice exhibited no fluorescence signals with liposomes up to 24 h and very little fluorescence with AME and ARE in the tumor region after 8 h, while the ACE treated group showed the highest fluorescence signals with enhanced tumor accumulation (Fig. 11a). Imaging data were in line with in vivo biodistribution, whereby the tumor showed the highest DOX concentration 24 h post-injection. Compared to liposomes, ACE showed more than three-fold higher DOX accumulation in tumors, while the liver and kidneys showed approximately 40% reduction in DOX accumulation, suggesting enhanced circulation and physiological stability of the developed hybrid ACE in cancer therapy (Fig. 11b).165 The authors also evaluated the antitumor efficacy in MCF-7 tumor xenograft models with an initial tumor volume of 80 mm3 and on the 18th day the tumor volume was ∼1260, ∼965, ∼721, and ∼307 mm3 after treatment with liposomes, ARE, AME and ACE, respectively, which showed the enhanced potential antitumor efficacy of ACE (Fig. 11c and d).165
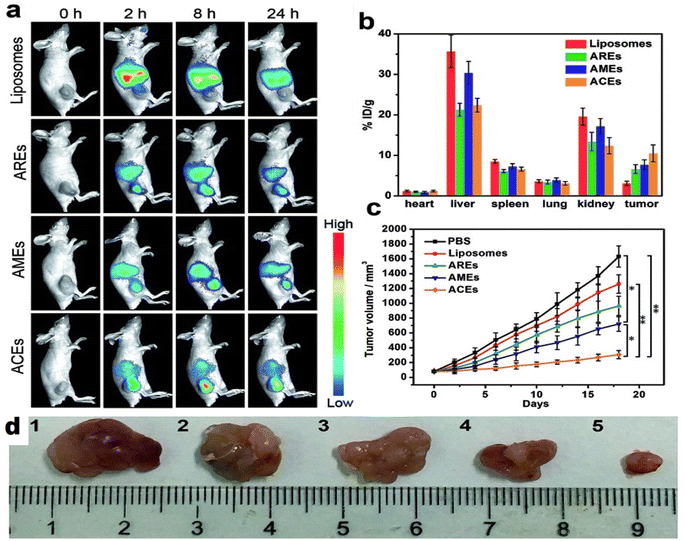 | ||
| Fig. 11 In vivo biodistribution, pharmacokinetics and antitumor efficacy of liposomes, AME, ARE and ACE after intravenous injection; (a) time-lapse fluorescence imaging in MCF-7 breast tumor-bearing nude mice showing the highest fluorescence signal with ACE after 8 h, (b) biodistribution of DOX at 24 h after intravenous administration at a dose of 6 mg kg−1 (means ± SD, n = 3) revealing the highest accumulation of DOX in tumor with ACE, (c) tumor growth curves of different groups after treatments up to 18 days (means ± SD, n = 5; t-test, *p < 0.05, and **p < 0.01) showing very slow increase in tumor growth with the lowest tumor volume after ACE treatment and (d) representative tumor photos indicating tumor volume from MCF-7 tumor-bearing mice after treatment with (1) PBS, (2) liposomes, (3) ARE, (4) AME and (5) ACE (this figure has been reproduced from ref. 165, with permission from Royal Society of Chemistry, Copyright 2010). | ||
8. Quality control and translation of exosomes
The standardization of the quality control parameters of exosomes for pharmaceutical applications is very crucial. Factors like size, size polydispersity, protein content, lipid content, and batch-to-batch variation should be taken into consideration.166 Like other types of nanoformulations, exosome size and polydispersity could potentially affect biodistribution, clearance, permeability across physiological barriers, and cellular uptake.167 In some cases, the surface protein content of exosomes significantly contributes to the targeting and the fusion-with-target cells.168,169 Several investigations have revealed that exosomes are relatively safe for application in humans,170–174 but immunogenicity could be a potential side effect due to the high protein content in exosomes. Additionally, RNA in exosomes could be a source of chronic adverse effects. Therefore, the toxicity standard should also be considered when developing drug products based on exosomes. With the ever-growing exosome database and the constant changes that are made with discoveries, it is challenging to develop standard protocols or manufacturing practices. Whilst several pharmaceutical companies are on the verge of advancement in exosome based medical interventions, the establishment of good manufacturing practice facilities for the preparation of exosomes remains crucial. With lyophilization being a useful technique to preserve the exosomes and transport them for over 36 months at 4 °C, the translation of therapeutic exosomes from bench to industry is within reach.Exosome application has been explored in several phase I clinical studies, mainly focusing on the immunotherapy of cancer. A phase I study using exosomes purified from autologous monocyte-derived dendritic cell cultures was accomplished in patients with melanoma stage IIIB-IV and tumor MAGE3 antigen positive. Patients were reported to tolerate the treatment well with no grade II toxicity, suggesting the safety of exosome administration.175 A similar strategy was presented in another phase I study among patients with progressed (stage IIIB-IV) non-small cell lung cancer and tumor expression of MAGE-A3 or A4. Leukapheresis was performed to generate dendritic cells that produced exosomes and was further loaded with several MAGE peptides. Patients well tolerated the exosomes and interestingly, some patients encountered enduring disease stability and activation of immune responses.176 Importantly, these two studies also suggest the viability of adhering to good manufacturing practices (GMPs) to produce dendritic cell-derived exosome vaccine.175,176 A phase I study of autologous ascites-derived exosomes in combination with granulocyte-macrophage colony-stimulating factor (GM-CSF) for colorectal cancer was also performed.177 It was found that ascites-derived exosomes, as a stand-alone treatment and in combination, were safe and well tolerated. However, the combination of ascites-derived exosomes with GM-CSF demonstrated immunological antitumor responses that were lacking in exosome-alone therapy.177 Currently, there are over 90 active trials on native/engineered exosomes for multiple organ failure, tumors, respiratory disease syndrome, diabetic retinopathy, etc. Since exosomes represent a new frontier in therapeutics, no specific guidelines exist for their manufacturing and quality assessment. Setting up suitable cell culture and exosome purification methodologies requires conscious decisions as the quality and functionality of exosomes are markedly affected by the cell source and the purification conditions. Developers need to collaborate closely with regulatory authorities to preemptively resolve any unforeseen issues. Comprehensive discussions with the industry should establish regulatory guidelines to prevent inappropriate use.178
9. Conclusion and future perspectives
The exploration of exosomes as next-generation drug delivery systems represents a promising frontier in pharmaceutical research, particularly in the context of targeted therapies for genetic disorders and cancer. Compared to traditional nanoparticulate systems like liposomes, exosomes are positioned as superior candidates due to their intrinsic properties derived from structural components such as proteins, nucleic acids, and lipids. The potential advantages of exosomes in achieving targeted delivery and reducing off-target effects underscore their significance in advancing therapeutic strategies. However, despite these promising attributes, several critical challenges must be addressed to transform exosomes into viable drug delivery systems for clinical practice.One major area of focus is enhanced production and scalability. Developing standardized protocols for the isolation, purification, and characterization of exosomes is crucial for ensuring reproducibility and quality control across different production batches. Additionally, investing in scalable production technologies, such as bioreactor systems and high-throughput isolation methods, may facilitate large-scale manufacturing, making exosome-based therapies more feasible for clinical use. Drug loading and release mechanisms must also be optimized. Innovations in drug loading techniques are needed to increase the payload capacity of exosomes without compromising their stability and functionality. Additionally, designing exosomes for controlled release of their therapeutic cargo in response to specific physiological conditions could improve efficacy and reduce side effects. Research into formulations that enhance the stability of exosomes under practical storage conditions is also essential. Potential strategies may include the use of stabilizing agents or advanced lyophilization techniques. Moreover, comprehensive studies to determine the shelf-life and long-term stability of exosome formulations under various conditions will be critical for their practical application.
In vivo performance also requires attention, particularly the need for an extended circulation time of exosomes in the bloodstream. Developing methods such as surface modifications or encapsulation technologies could improve their therapeutic efficacy. Furthermore, research on targeting mechanisms is necessary to optimize exosome delivery to specific tissues or cells, based on their origin and the disease being targeted, thereby enhancing therapeutic precision. The exploration of hybrid systems offers another avenue for advancement. Continued investigation into hybrid exosome systems that combine natural exosomes with synthetic elements or other nanoparticles may address current limitations such as stability and drug loading. However, rigorous testing and validation of these hybrid systems are essential to ensure their safety, efficacy, and compatibility with biological systems. In addition, regulatory and ethical considerations play a vital role in the approval of exosome-based therapies. Developing clear regulatory guidelines and frameworks for approval and commercialization will be important for widespread adoption. Additionally, addressing ethical concerns related to the source of exosomes, particularly those derived from human tissues, is necessary for responsible research and application. Finally, the success of exosome-based therapies hinges on well-designed preclinical and clinical trials to assess efficacy, safety, and potential side effects.
In conclusion, while exosomes hold great promise as advanced drug delivery vehicles, substantial research and development are required to address their current limitations. To advance exosomes as drug delivery systems, several key areas including production, stability, storage, in vivo performance, drug loading and release, and regulatory and ethical considerations need attention. Addressing these areas will be crucial for ensuring the translation of exosomes from experimental platforms to practical, reliable therapeutic solutions.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors have stated that they do not have any financial or non-financial interests that may affect the publication of this manuscript.References
- C. Thery, L. Zitvogel and S. Amigorena, Nat. Rev. Immunol., 2002, 2, 569–579 CrossRef CAS PubMed.
- M. Simons and G. Raposo, Curr. Opin. Cell Biol., 2009, 21, 575–581 CrossRef CAS PubMed.
- B. Fevrier and G. Raposo, Curr. Opin. Cell Biol., 2004, 16, 415–421 CrossRef CAS.
- H. Valadi, K. Ekström, A. Bossios, M. Sjöstrand, J. J. Lee and J. O. Lötvall, Nat. Cell Biol., 2007, 9, 654 CrossRef CAS PubMed.
- Q.-F. Han, W.-J. Li, K.-S. Hu, J. Gao, W.-L. Zhai, J.-H. Yang and S.-J. Zhang, Mol. Cancer, 2022, 21, 207 CrossRef.
- I. Hasan, S. Roy, E. Ehexige, R. Wu, Y. Chen, Z. Gao, B. Guo and C. Chang, Nanoscale, 2023, 15, 18108–18138 RSC.
- P. Das and N. R. Jana, ACS Appl. Polym. Mater., 2021, 3, 4791–4811 CrossRef CAS.
- P. Das, S. Pujals, L. M. Ali, M. Gary-Bobo, L. Albertazzi and J.-O. Durand, Nanoscale, 2023, 15, 12008–12024 RSC.
- S. Xiong, J. Luo, Q. Wang, Z. Li, J. Li, Q. Liu, L. Gao, S. Fang, Y. Li, H. Pan, H. Wang, Y. Zhang, Q. Wang, X. Chen and T. Chen, Biomater. Sci., 2021, 9, 1705–1715 RSC.
- S. Patri, N. T. K. Thanh and N. Kamaly, Nanoscale, 2024, 16, 15446–15464 RSC.
- J. E. Pullan, M. I. Confeld, J. K. Osborn, J. Kim, K. Sarkar and S. Mallik, Mol. Pharm., 2019, 16, 1789–1798 CrossRef CAS.
- C. Di, Q. Zhang, Y. Wang, F. Wang, Y. Chen, L. Gan, R. Zhou, C. Sun, H. Li, X. Zhang, H. Yang and H. Zhang, Artif. Cells, Nanomed., Biotechnol., 2018, 46, S564–S570 CrossRef.
- C. Théry, M. Ostrowski and E. Segura, Nat. Rev. Immunol., 2009, 9, 581 CrossRef PubMed.
- R. C. Lai, R. W. Yeo, K. H. Tan and S. K. Lim, Biotechnol. Adv., 2013, 31, 543–551 CrossRef CAS PubMed.
- F. André, N. Chaput, N. E. Schartz, C. Flament, N. Aubert, J. Bernard, F. Lemonnier, G. Raposo, B. Escudier and D.-H. Hsu, J. Immunol., 2004, 172, 2126–2136 CrossRef PubMed.
- J. L. Hood, R. S. San and S. A. Wickline, Cancer Res., 2011, 71, 3792–3801 CrossRef CAS PubMed.
- L. Alvarez-Erviti, Y. Seow, H. Yin, C. Betts, S. Lakhal and M. J. Wood, Nat. Biotechnol., 2011, 29, 341–345 CrossRef CAS PubMed.
- A. Tan, J. Rajadas and A. M. Seifalian, Adv. Drug Delivery Rev., 2013, 65, 357–367 CrossRef CAS.
- E. V. Batrakova and M. S. Kim, J. Controlled Release, 2015, 219, 396–405 CrossRef CAS.
- A. V. Vlassov, S. Magdaleno, R. Setterquist and R. Conrad, Biochim. Biophys. Acta, Gen. Subj., 2012, 1820, 940–948 CrossRef CAS PubMed.
- S. W. Ferguson and J. Nguyen, J. Controlled Release, 2016, 228, 179–190 CrossRef CAS PubMed.
- J. Kowal, M. Tkach and C. Thery, Curr. Opin. Cell Biol., 2014, 29, 116–125 CrossRef CAS.
- T. Wolf, S. R. Baier and J. J. T. Zempleni, J. Nutr., 2015, 145, 2201–2206 CrossRef CAS.
- Y. Lee, S. El Andaloussi and M. J. Wood, Hum. Mol. Genet., 2012, 21, R125–R134 CrossRef CAS PubMed.
- S. El-Andaloussi, Y. Lee, S. Lakhal-Littleton, J. Li, Y. Seow, C. Gardiner, L. Alvarez-Erviti, I. L. Sargent and M. J. Wood, Nat. Protoc., 2012, 7, 2112–2126 CrossRef CAS PubMed.
- D. Sun, X. Zhuang, X. Xiang, Y. Liu, S. Zhang, C. Liu, S. Barnes, W. Grizzle, D. Miller and H.-G. Zhang, Mol. Ther., 2010, 18, 1606–1614 CrossRef CAS PubMed.
- M. J. Haney, N. L. Klyachko, Y. Zhao, R. Gupta, E. G. Plotnikova, Z. He, T. Patel, A. Piroyan, M. Sokolsky and A. V. Kabanov, J. Controlled Release, 2015, 207, 18–30 CrossRef CAS.
- T. Tian, H.-X. Zhang, C.-P. He, S. Fan, Y.-L. Zhu, C. Qi, N.-P. Huang, Z.-D. Xiao, Z.-H. Lu and B. A. Tannous, Biomaterials, 2018, 150, 137–149 CrossRef CAS PubMed.
- H. Wang, H. Sui, Y. Zheng, Y. Jiang, Y. Shi, J. Liang and L. Zhao, Nanoscale, 2019, 11, 7481–7496 Search PubMed.
- S. Kaur, A. Nathani and M. Singh, Cancer Lett., 2023, 566, 216243 CrossRef CAS PubMed.
- M. P. Caby, D. Lankar, C. Vincendeau-Scherrer, G. Raposo and C. Bonnerot, Int. Immunol., 2005, 17, 879–887 CrossRef CAS.
- H. Zhou, T. Pisitkun, A. Aponte, P. S. Yuen, J. D. Hoffert, H. Yasuda, X. Hu, L. Chawla, R. F. Shen, M. A. Knepper and R. A. Star, Kidney Int., 2006, 70, 1847–1857 CrossRef CAS.
- C. Admyre, S. M. Johansson, K. R. Qazi, J. J. Filen, R. Lahesmaa, M. Norman, E. P. Neve, A. Scheynius and S. Gabrielsson, J. Immunol., 2007, 179, 1969–1978 Search PubMed.
- J. Mondal, S. Pillarisetti, V. Junnuthula, M. Saha, S. R. Hwang, I. K. Park and Y. K. Lee, J. Controlled Release, 2023, 353, 1127–1149 CrossRef CAS PubMed.
- Z. Zou, H. Li, G. Xu, Y. Hu, W. Zhang and K. Tian, Int. J. Nanomed., 2023, 18, 4751–4778 CrossRef CAS PubMed.
- S. Ohno, M. Takanashi, K. Sudo, S. Ueda, A. Ishikawa, N. Matsuyama, K. Fujita, T. Mizutani, T. Ohgi, T. Ochiya, N. Gotoh and M. Kuroda, Mol. Ther., 2013, 21, 185–191 CrossRef CAS PubMed.
- D. Wrobel, A. Edr, E. Zemanova, T. Strasak, A. Semeradtova and J. Maly, Chem. Phys. Lipids, 2023, 255, 105314 CrossRef CAS PubMed.
- J. Wang, P. Ma, D. H. Kim, B. F. Liu and U. Demirci, Nano Today, 2021, 37, 101066 CrossRef CAS PubMed.
- F. U. Rehman, Y. Liu, M. Zheng and B. Shi, Biomaterials, 2023, 293, 121949 CrossRef CAS PubMed.
- R. A. Haraszti, M. C. Didiot, E. Sapp, J. Leszyk, S. A. Shaffer, H. E. Rockwell, F. Gao, N. R. Narain, M. DiFiglia, M. A. Kiebish, N. Aronin and A. Khvorova, J. Extracell. Vesicles, 2016, 5, 32570 CrossRef PubMed.
- R. W. Yeo, R. C. Lai, B. Zhang, S. S. Tan, Y. Yin, B. J. Teh and S. K. Lim, Adv. Drug Delivery Rev., 2013, 65, 336–341 CrossRef CAS.
- R. C. Lai, T. S. Chen and S. K. Lim, Regener. Med., 2011, 6, 481–492 Search PubMed.
- S. Rani, A. E. Ryan, M. D. Griffin and T. Ritter, Mol. Ther., 2015, 23, 812–823 CrossRef CAS PubMed.
- K. B. Johnsen, J. M. Gudbergsson, M. N. Skov, L. Pilgaard, T. Moos and M. Duroux, Biochim. Biophys. Acta, Rev. Cancer, 2014, 1846, 75–87 CrossRef CAS PubMed.
- T. S. Chen, F. Arslan, Y. Yin, S. S. Tan, R. C. Lai, A. B. Choo, J. Padmanabhan, C. N. Lee, D. P. de Kleijn and S. K. Lim, J. Transl. Med., 2011, 9, 47 CrossRef CAS.
- W. Pan, H. Chen, A. Wang, F. Wang and X. Zhang, Life Sci., 2023, 319, 121524 CrossRef CAS PubMed.
- L. Pascucci, V. Coccè, A. Bonomi, D. Ami, P. Ceccarelli, E. Ciusani, L. Viganò, A. Locatelli, F. Sisto and S. M. Doglia, J. Controlled Release, 2014, 192, 262–270 Search PubMed.
- R. Munagala, F. Aqil, J. Jeyabalan and R. C. Gupta, Cancer Lett., 2016, 371, 48–61 CrossRef CAS PubMed.
- A. Michael, S. D. Bajracharya, P. S. Yuen, H. Zhou, R. A. Star, G. G. Illei and I. Alevizos, Oral Dis., 2010, 16, 34–38 Search PubMed.
- V. Palanisamy, S. Sharma, A. Deshpande, H. Zhou, J. Gimzewski and D. T. Wong, PLoS One, 2010, 5, e8577 Search PubMed.
- C. Lau, Y. Kim, D. Chia, N. Spielmann, G. Eibl, D. Elashoff, F. Wei, Y. L. Lin, A. Moro, T. Grogan, S. Chiang, E. Feinstein, C. Schafer, J. Farrell and D. T. Wong, J. Biol. Chem., 2013, 288, 26888–26897 CrossRef CAS.
- T. Pisitkun, R. F. Shen and M. A. Knepper, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 13368–13373 CrossRef CAS PubMed.
- J. Nilsson, J. Skog, A. Nordstrand, V. Baranov, L. Mincheva-Nilsson, X. O. Breakefield and A. Widmark, Br. J. Cancer, 2009, 100, 1603–1607 CrossRef CAS PubMed.
- A. Clayton, J. P. Mitchell, J. Court, M. D. Mason and Z. Tabi, Cancer Res., 2007, 67, 7458–7466 CrossRef CAS PubMed.
- C. Lasser, V. S. Alikhani, K. Ekstrom, M. Eldh, P. T. Paredes, A. Bossios, M. Sjostrand, S. Gabrielsson, J. Lotvall and H. Valadi, J. Transl. Med., 2011, 9, 9 CrossRef PubMed.
- T. Smyth, M. Kullberg, N. Malik, P. Smith-Jones, M. W. Graner and T. J. Anchordoquy, J. Controlled Release, 2015, 199, 145–155 CrossRef CAS.
- S. Alvarez, C. Suazo, A. Boltansky, M. Ursu, D. Carvajal, G. Innocenti, A. Vukusich, M. Hurtado, S. Villanueva, J. Carreno and A. Rogelio, Transplant. Proc., 2013, 45, 3719–3723 CrossRef CAS.
- H. Jang, H. Kim, E. H. Kim, G. Han, Y. Jang, Y. Kim, J. W. Lee, S. C. Shin, E. E. Kim, S. H. Kim and Y. Yang, Biomater. Res., 2023, 27, 124 CrossRef CAS PubMed.
- A. K. Agrawal, F. Aqil, J. Jeyabalan, W. A. Spencer, J. Beck, B. W. Gachuki, S. S. Alhakeem, K. Oben, R. Munagala, S. Bondada and R. C. Gupta, Nanomedicine, 2017, 13, 1627–1636 CrossRef CAS.
- M. Cao, N. Diao, X. Cai, X. Chen, Y. Xiao, C. Guo, D. Chen and X. Zhang, Mater. Horiz., 2023, 10, 3879–3894 RSC.
- S. Rahmati, H. Karimi, M. Alizadeh, A. H. Khazaei, A. C. Paiva-Santos, L. Rezakhani and E. Sharifi, Hum. Cell, 2024, 37, 121–138 CrossRef PubMed.
- B. Wang, X. Zhuang, Z. B. Deng, H. Jiang, J. Mu, Q. Wang, X. Xiang, H. Guo, L. Zhang, G. Dryden, J. Yan, D. Miller and H. G. Zhang, Mol. Ther., 2014, 22, 522–534 Search PubMed.
- A. Kilasoniya, L. Garaeva, T. Shtam, A. Spitsyna, E. Putevich, B. Moreno-Chamba, J. Salazar-Bermeo, E. Komarova, A. Malek, M. Valero and D. Saura, Antioxidants, 2023, 12, 943 Search PubMed.
- M. Wallen, F. Aqil, W. Spencer and R. C. Gupta, Cancer Lett., 2023, 561, 216141 Search PubMed.
- D. W. Greening, R. Xu, H. Ji, B. J. Tauro and R. J. Simpson, in Proteomic Profiling, Springer, 2015, pp. 179–209 Search PubMed.
- M. A. Livshits, E. Khomyakova, E. G. Evtushenko, V. N. Lazarev, N. A. Kulemin, S. E. Semina, E. V. Generozov and V. M. Govorun, Sci. Rep., 2015, 5, 17319 CrossRef PubMed.
- B. J. Tauro, D. W. Greening, R. A. Mathias, H. Ji, S. Mathivanan, A. M. Scott and R. J. Simpson, Methods, 2012, 56, 293–304 CrossRef CAS PubMed.
- S. S. Kanwar, C. J. Dunlay, D. M. Simeone and S. Nagrath, Lab Chip, 2014, 14, 1891–1900 RSC.
- T. Yamashita, Y. Takahashi, M. Nishikawa and Y. Takakura, Eur. J. Pharm. Biopharm., 2016, 98, 1–8 CrossRef CAS PubMed.
- P. Li, M. Kaslan, S. H. Lee, J. Yao and Z. Gao, Theranostics, 2017, 7, 789–804 CrossRef CAS PubMed.
- R. C. Lai, F. Arslan, M. M. Lee, N. S. Sze, A. Choo, T. S. Chen, M. Salto-Tellez, L. Timmers, C. N. Lee, R. M. El Oakley, G. Pasterkamp, D. P. de Kleijn and S. K. Lim, Stem Cell Res., 2010, 4, 214–222 CrossRef CAS PubMed.
- J. Caradec, G. Kharmate, E. Hosseini-Beheshti, H. Adomat, M. Gleave and E. Guns, Clin. Biochem., 2014, 47, 1286–1292 CrossRef CAS.
- I. Helwa, J. Cai, M. D. Drewry, A. Zimmerman, M. B. Dinkins, M. L. Khaled, M. Seremwe, W. M. Dismuke, E. Bieberich, W. D. Stamer, M. W. Hamrick and Y. Liu, PLoS One, 2017, 12, e0170628 CrossRef PubMed.
- H. G. Lamparski, A. Metha-Damani, J. Y. Yao, S. Patel, D. H. Hsu, C. Ruegg and J. B. Le Pecq, J. Immunol. Methods, 2002, 270, 211–226 CrossRef CAS PubMed.
- W. Jo, J. Kim, J. Yoon, D. Jeong, S. Cho, H. Jeong, Y. J. Yoon, S. C. Kim, Y. S. Gho and J. Park, Nanoscale, 2014, 6, 12056–12064 RSC.
- D. D. Taylor and S. Shah, Methods, 2015, 87, 3–10 CrossRef CAS PubMed.
- K. Yakimchuk, Mater. Methods, 2015, 5(5), 1450 Search PubMed.
- H. Kalra, C. G. Adda, M. Liem, C. S. Ang, A. Mechler, R. J. Simpson, M. D. Hulett and S. Mathivanan, Proteomics, 2013, 13, 3354–3364 CrossRef CAS.
- M. L. Heinemann, M. Ilmer, L. P. Silva, D. H. Hawke, A. Recio, M. A. Vorontsova, E. Alt and J. Vykoukal, J. Chromatogr. A, 2014, 1371, 125–135 CrossRef CAS PubMed.
- V. Sunkara, J. Park, J. Han, J. S. Del Rio, H. J. Cho, I. J. Oh and Y. K. Cho, ACS Appl. Mater. Interfaces, 2023, 15, 56807–56819 CAS.
- S. Williams, M. Fernandez-Rhodes, A. Law, B. Peacock, M. P. Lewis and O. G. Davies, J. Tissue Eng., 2023, 14, 20417314231174609 CrossRef PubMed.
- M. Vashisht, P. Rani, S. K. Onteru and D. Singh, Appl. Biochem. Biotechnol., 2017, 183, 993–1007 CrossRef CAS PubMed.
- Y. Wang, M. Guo, D. Lin, D. Liang, L. Zhao, R. Zhao and Y. Wang, Drug Delivery, 2021, 28, 1510–1523 CrossRef CAS PubMed.
- S. Shaikh, M. Younis, S. Yingying, T. Tanziela and L. Yuan, Life Sci., 2023, 330, 121977 CrossRef CAS PubMed.
- D. He, X. Xu, L. Li, C. Chen, K. Gong, Q. Guo, F. Liu, Y. Wang, Y. Duan and H. Li, J. Biomed. Nanotechnol., 2021, 17, 426–438 CrossRef CAS PubMed.
- Y. Gu, Y. Du, L. Jiang, X. Tang, A. Li, Y. Zhao, Y. Lang, X. Liu and J. Liu, J. Nanobiotechnol., 2022, 20, 384 CrossRef CAS PubMed.
- W. S. Fisher, C. Tchounwou, S. Wei, L. Roberts, K. K. Ewert and C. R. Safinya, Biochim. Biophys. Acta, Biomembr., 2022, 1864, 183841 CrossRef CAS PubMed.
- S. Najafi, J. Majidpoor and K. Mortezaee, Drug Delivery Transl. Res., 2023, 13, 2790–2806 CrossRef CAS.
- Z. Cui, Z. Ruan, J. Zeng, J. Sun, W. Ye, W. Xu, X. Guo, L. Zhang and L. Song, Thorac. Cancer, 2022, 13, 2723–2731 CrossRef CAS PubMed.
- H. Wei, F. Chen, J. Chen, H. Lin, S. Wang, Y. Wang, C. Wu, J. Lin and G. Zhong, Int. J. Nanomed., 2022, 17, 3483–3495 CrossRef.
- O. P. Wiklander, M. Á. Brennan, J. Lötvall, X. O. Breakefield and S. E. Andaloussi, Sci. Transl. Med., 2019, 11, eaav8521 CrossRef CAS.
- L. Alvarez-Erviti, Y. Seow, H. Yin, C. Betts, S. Lakhal and M. J. Wood, Nat. Biotechnol., 2011, 29, 341 CrossRef CAS.
- J. Yang, X. Zhang, X. Chen, L. Wang and G. Yang, Mol. Ther.–Nucleic Acids, 2017, 7, 278–287 CrossRef CAS PubMed.
- Y. Tian, S. Li, J. Song, T. Ji, M. Zhu, G. J. Anderson, J. Wei and G. Nie, Biomaterials, 2014, 35, 2383–2390 CrossRef CAS.
- S.-i. Ohno, M. Takanashi, K. Sudo, S. Ueda, A. Ishikawa, N. Matsuyama, K. Fujita, T. Mizutani, T. Ohgi and T. Ochiya, Mol. Ther., 2013, 21, 185–191 CrossRef CAS PubMed.
- R. Kojima, D. Bojar, G. Rizzi, G. Charpin-El Hamri, M. D. El-Baba, P. Saxena, S. Ausländer, K. R. Tan and M. Fussenegger, Nat. Commun., 2018, 9, 1305 CrossRef PubMed.
- L. Xin, Y. W. Yuan, C. Liu, L. Q. Zhou, L. Liu, Q. Zhou and S. H. Li, Dig. Dis. Sci., 2021, 66, 1045–1053 CrossRef CAS.
- J. Pullan, K. Dailey, S. Bhallamudi, L. Feng, L. Alhalhooly, J. Froberg, J. Osborn, K. Sarkar, T. Molden, V. Sathish, Y. Choi, A. Brooks and S. Mallik, ACS Appl. Bio Mater., 2022, 5, 2163–2175 CrossRef CAS PubMed.
- D. Lin, H. Zhang, R. Liu, T. Deng, T. Ning, M. Bai, Y. Yang, K. Zhu, J. Wang, J. Duan, S. Ge, B. Sun, G. Ying and Y. Ba, Mol. Oncol., 2021, 15, 3430–3446 CrossRef CAS.
- A. Iyaswamy, A. Thakur, X. J. Guan, S. Krishnamoorthi, T. Y. Fung, K. Lu, I. Gaurav, Z. Yang, C. F. Su, K. F. Lau, K. Zhang, R. C. Ng, Q. Lian, K. H. Cheung, K. Ye, H. J. Chen and M. Li, Signal Transduction Targeted Ther., 2023, 8, 404 CrossRef CAS PubMed.
- Y. Hou, Y. Xie, X. Liu, Y. Chen, F. Zhou and B. Yang, Fluids Barriers CNS, 2024, 21, 8 CrossRef CAS.
- E. Pishavar, R. Yazdian-Robati, K. Abnous, M. Hashemi, M. Ebrahimian, R. Feizpour, Z. Salmasi and S. M. Taghdisi, Iran. J. Basic Med. Sci., 2023, 26, 388–394 Search PubMed.
- N. Yim, S.-W. Ryu, K. Choi, K. R. Lee, S. Lee, H. Choi, J. Kim, M. R. Shaker, W. Sun and J.-H. Park, Nat. Commun., 2016, 7, 12277 CrossRef CAS PubMed.
- U. Sterzenbach, U. Putz, L.-H. Low, J. Silke, S.-S. Tan and J. Howitt, Mol. Ther., 2017, 25, 1269–1278 CrossRef CAS PubMed.
- J. Y. Wu, Y. J. Li, J. Wang, X. B. Hu, S. Huang, S. Luo and D. X. Xiang, J. Nanobiotechnol., 2021, 19, 405 CrossRef CAS PubMed.
- W. Sun, S. Qu, M. Ji, Y. Sun and B. Hu, Heliyon, 2023, 9, e19934 CrossRef CAS.
- D. Ha, N. Yang and V. Nadithe, Acta Pharm. Sin. B, 2016, 6, 287–296 CrossRef.
- A. Liu, G. Yang, Y. Liu and T. Liu, Front. Bioeng. Biotechnol., 2022, 10, 939441 CrossRef.
- S. Rayamajhi, T. D. T. Nguyen, R. Marasini and S. Aryal, Acta Biomater., 2019, 94, 482–494 CrossRef CAS PubMed.
- Y. T. Sato, K. Umezaki, S. Sawada, S. A. Mukai, Y. Sasaki, N. Harada, H. Shiku and K. Akiyoshi, Sci. Rep., 2016, 6, 21933 CrossRef CAS.
- L. Gong, H. Zhou, Y. Zhang, C. Wang, K. Fu, C. Ma and Y. Li, Mater. Today Bio, 2023, 23, 100804 CrossRef CAS.
- M. Piffoux, A. K. A. Silva, C. Wilhelm, F. Gazeau and D. Tareste, ACS Nano, 2018, 12, 6830–6842 CrossRef CAS PubMed.
- A. J. Vazquez-Rios, A. Molina-Crespo, B. L. Bouzo, R. Lopez-Lopez, G. Moreno-Bueno and M. de la Fuente, J. Nanobiotechnol., 2019, 17, 85 CrossRef.
- Y. Lin, J. Wu, W. Gu, Y. Huang, Z. Tong, L. Huang and J. Tan, Adv. Sci., 2018, 5, 1700611 CrossRef.
- S. I. Sawada, Y. T. Sato, R. Kawasaki, J. I. Yasuoka, R. Mizuta, Y. Sasaki and K. Akiyoshi, Biomater. Sci., 2020, 8, 619–630 RSC.
- D. Dehaini, X. Wei, R. H. Fang, S. Masson, P. Angsantikul, B. T. Luk, Y. Zhang, M. Ying, Y. Jiang, A. V. Kroll, W. Gao and L. Zhang, Adv. Mater., 2017, 29, 1606209 CrossRef.
- W. J. Goh, S. Zou, C. K. Lee, Y. H. Ou, J. W. Wang, B. Czarny and G. Pastorin, Biomacromolecules, 2018, 19, 22–30 CrossRef CAS PubMed.
- M. D. A. Paskeh, M. Entezari, S. Mirzaei, A. Zabolian, H. Saleki, M. J. Naghdi, S. Sabet, M. A. Khoshbakht, M. Hashemi, K. Hushmandi, G. Sethi, A. Zarrabi, A. P. Kumar, S. C. Tan, M. Papadakis, A. Alexiou, M. A. Islam, E. Mostafavi and M. Ashrafizadeh, J. Hematol. Oncol., 2022, 15, 83 CrossRef CAS PubMed.
- S. Hyung, J. Ko, Y. J. Heo, S. M. Blum, S. T. Kim, S. H. Park, J. O. Park, W. K. Kang, H. Y. Lim, S. J. Klempner and J. Lee, Sci. Adv., 2023, 9, eadk1098 CrossRef CAS PubMed.
- R. Wang, Q. Liang, X. Zhang, Z. Di, X. Wang and L. Di, Colloids Surf., B, 2022, 215, 112505 CrossRef CAS PubMed.
- X. Zhao, D. Wu, X. Ma, J. Wang, W. Hou and W. Zhang, Biomed. Pharmacother., 2020, 128, 110237 CrossRef CAS.
- T. Yang, P. Martin, B. Fogarty, A. Brown, K. Schurman, R. Phipps, V. P. Yin, P. Lockman and S. Bai, Pharm. Res., 2015, 32, 2003–2014 CrossRef CAS.
- M. S. Kim, M. J. Haney, Y. Zhao, V. Mahajan, I. Deygen, N. L. Klyachko, E. Inskoe, A. Piroyan, M. Sokolsky, O. Okolie, S. D. Hingtgen, A. V. Kabanov and E. V. Batrakova, Nanomedicine, 2016, 12, 655–664 CrossRef CAS.
- G. Fuhrmann, A. Serio, M. Mazo, R. Nair and M. M. Stevens, J. Controlled Release, 2015, 205, 35–44 CrossRef CAS PubMed.
- X. Zhuang, X. Xiang, W. Grizzle, D. Sun, S. Zhang, R. C. Axtell, S. Ju, J. Mu, L. Zhang, L. Steinman, D. Miller and H. G. Zhang, Mol. Ther., 2011, 19, 1769–1779 CrossRef CAS PubMed.
- H. Qi, C. Liu, L. Long, Y. Ren, S. Zhang, X. Chang, X. Qian, H. Jia, J. Zhao, J. Sun, X. Hou, X. Yuan and C. Kang, ACS Nano, 2016, 10, 3323–3333 CrossRef CAS PubMed.
- J. Gao, S. Wang and Z. Wang, Biomaterials, 2017, 135, 62–73 CrossRef CAS PubMed.
- Y. Tian, S. Li, J. Song, T. Ji, M. Zhu, G. J. Anderson, J. Wei and G. Nie, Biomaterials, 2014, 35, 2383–2390 CrossRef CAS.
- J. Wahlgren, T. D. L. Karlson, M. Brisslert, F. Vaziri Sani, E. Telemo, P. Sunnerhagen and H. Valadi, Nucleic Acids Res., 2012, 40, e130 CrossRef CAS PubMed.
- T. A. Shtam, R. A. Kovalev, E. Y. Varfolomeeva, E. M. Makarov, Y. V. Kil and M. V. Filatov, Cell Commun. Signaling, 2013, 11, 88 CrossRef CAS PubMed.
- A. B. Banizs, T. Huang, K. Dryden, S. S. Berr, J. R. Stone, R. K. Nakamoto, W. Shi and J. He, Int. J. Nanomed., 2014, 9, 4223–4230 CAS.
- S. A. Kooijmans, S. Stremersch, K. Braeckmans, S. C. de Smedt, A. Hendrix, M. J. Wood, R. M. Schiffelers, K. Raemdonck and P. Vader, J. Controlled Release, 2013, 172, 229–238 CrossRef CAS PubMed.
- G. Fuhrmann, I. K. Herrmann and M. M. Stevens, Nano Today, 2015, 10, 397–409 CrossRef CAS.
- Z. Wang, J. Rich, N. Hao, Y. Gu, C. Chen, S. Yang, P. Zhang and T. J. Huang, Microsyst. Nanoeng., 2022, 8, 45 CrossRef CAS PubMed.
- M. S. Kim, M. J. Haney, Y. Zhao, D. Yuan, I. Deygen, N. L. Klyachko, A. V. Kabanov and E. V. Batrakova, Nanomedicine, 2018, 14, 195–204 Search PubMed.
- G. Jia, Y. Han, Y. An, Y. Ding, C. He, X. Wang and Q. Tang, Biomaterials, 2018, 178, 302–316 CrossRef CAS.
- M. S. Gholami Farashah, M. Javadi, J. Soleimani Rad, S. K. Shakouri, S. Asnaashari, S. Dastmalchi, S. Nikzad and L. Roshangar, Adv. Pharm. Bull., 2023, 13, 736–746 CrossRef.
- S. G. Antimisiaris, S. Mourtas and A. Marazioti, Pharmaceutics, 2018, 10, 218 CrossRef CAS PubMed.
- J. Wang, Y. Dong, Y. Li, W. Li, K. Cheng, Y. Qian, G. Xu, X. Zhang, L. Hu and P. Chen, Adv. Funct. Mater., 2018, 28, 1707360 CrossRef.
- M. Lee, J.-J. Ban, W. Im and M. Kim, Biotechnol. Bioprocess Eng., 2016, 21, 299–304 CrossRef CAS.
- K. Tang, Y. Zhang, H. Zhang, P. Xu, J. Liu, J. Ma, M. Lv, D. Li, F. Katirai and G.-X. Shen, Nat. Commun., 2012, 3, 1282 CrossRef.
- V. Sokolova, A.-K. Ludwig, S. Hornung, O. Rotan, P. A. Horn, M. Epple and B. Giebel, Colloids Surf., B, 2011, 87, 146–150 CrossRef CAS.
- A. K. Agrawal, F. Aqil, J. Jeyabalan, W. A. Spencer, J. Beck, B. W. Gachuki, S. S. Alhakeem, K. Oben, R. Munagala and S. Bondada, Nanomed. Nanotechnol. Biol. Med., 2017, 13, 1627–1636 CrossRef CAS.
- R. Maroto, Y. Zhao, M. Jamaluddin, V. L. Popov, H. Wang, M. Kalubowilage, Y. Zhang, J. Luisi, H. Sun, C. T. Culbertson, S. H. Bossmann, M. Motamedi and A. R. Brasier, J. Extracell. Vesicles, 2017, 6, 1359478 CrossRef PubMed.
- A. Sivanantham and Y. Jin, Life, 2022, 12, 697 CrossRef CAS.
- M. Ruzycka-Ayoush, A. M. Nowicka, A. Kowalczyk, A. Gluchowska, A. Targonska, G. Mosieniak, K. Sobczak, M. Donten and I. P. Grudzinski, Eur. J. Pharm. Sci., 2023, 181, 106369 CrossRef CAS PubMed.
- H. Kalra, C. G. Adda, M. Liem, C. S. Ang, A. Mechler, R. J. Simpson, M. D. Hulett and S. Mathivanan, Proteomics, 2013, 13, 3354–3364 CrossRef CAS PubMed.
- H. Zhou, P. S. Yuen, T. Pisitkun, P. A. Gonzales, H. Yasuda, J. W. Dear, P. Gross, M. A. Knepper and R. A. Star, Kidney Int., 2006, 69, 1471–1476 Search PubMed.
- Á. M. Lőrincz, C. I. Timár, K. A. Marosvári, D. S. Veres, L. Otrokocsi, Á. Kittel and E. Ligeti, J. Extracell. Vesicles, 2014, 3, 25465 CrossRef PubMed.
- Y. Liu, D. Li, Z. Liu, Y. Zhou, D. Chu, X. Li, X. Jiang, D. Hou, X. Chen, Y. Chen, Z. Yang, L. Jin, W. Jiang, C. Tian, G. Zhou, K. Zen, J. Zhang, Y. Zhang, J. Li and C. Y. Zhang, Sci. Rep., 2015, 5, 17543 CrossRef CAS PubMed.
- S. J. Wassmer, L. S. Carvalho, B. György, L. H. Vandenberghe and C. A. Maguire, Sci. Rep., 2017, 7, 45329 CrossRef CAS PubMed.
- K. Zhang, L. Yu, F. R. Li, X. Li, Z. Wang, X. Zou, C. Zhang, K. Lv, B. Zhou, S. Mitragotri and M. Chen, Int. J. Nanomed., 2020, 15, 2859–2872 CrossRef CAS.
- P. Dehghani, J. Varshosaz, M. Mirian, M. Minaiyan, M. Kazemi and M. Bodaghi, Pharm. Res., 2024, 41, 263–279 CrossRef CAS PubMed.
- I. Kawikova and P. W. Askenase, Brain Res., 2015, 1617, 63–71 CrossRef CAS PubMed.
- L. Kozlovskaya, M. Abou-Kaoud and D. Stepensky, J. Controlled Release, 2014, 189, 133–140 CrossRef CAS.
- Y. Takahashi, M. Nishikawa, H. Shinotsuka, Y. Matsui, S. Ohara, T. Imai and Y. Takakura, J. Biotechnol., 2013, 165, 77–84 CrossRef CAS PubMed.
- M. Morishita, Y. Takahashi, M. Nishikawa, K. Sano, K. Kato, T. Yamashita, T. Imai, H. Saji and Y. Takakura, J. Pharm. Sci., 2015, 104, 705–713 CrossRef CAS PubMed.
- O. P. Wiklander, J. Z. Nordin, A. O'Loughlin, Y. Gustafsson, G. Corso, I. Mäger, P. Vader, Y. Lee, H. Sork and Y. Seow, J. Extracell. Vesicles, 2015, 4, 26316 CrossRef PubMed.
- M. Morishita, Y. Takahashi, M. Nishikawa and Y. Takakura, J. Pharm. Sci., 2017, 106, 2265–2269 CrossRef CAS PubMed.
- T. Imai, Y. Takahashi, M. Nishikawa, K. Kato, M. Morishita, T. Yamashita, A. Matsumoto, C. Charoenviriyakul and Y. Takakura, J. Extracell. Vesicles, 2015, 4, 26238 CrossRef PubMed.
- S. Guo, N. Perets, O. Betzer, S. Ben-Shaul, A. Sheinin, I. Michaelevski, R. Popovtzer, D. Offen and S. Levenberg, ACS Nano, 2019, 13, 10015–10028 CrossRef CAS PubMed.
- Z. Han, W. Lv, Y. Li, J. Chang, W. Zhang, C. Liu and J. Sun, ACS Appl. Bio Mater., 2020, 3, 2666–2673 CrossRef CAS.
- E. Bagheri, K. Abnous, S. A. Farzad, S. M. Taghdisi, M. Ramezani and M. Alibolandi, Life Sci., 2020, 261, 118369 CrossRef CAS.
- G. Han, H. Kim, H. Jang, E. S. Kim, S. H. Kim and Y. Yang, Bioact. Mater., 2024, 34, 138–149 CAS.
- K. L. Zhang, Y. J. Wang, J. Sun, J. Zhou, C. Xing, G. Huang, J. Li and H. Yang, Chem. Sci., 2019, 10, 1555–1561 RSC.
- E. Tzng, N. Bayardo and P. C. Yang, J. Extracell. Vesicles, 2023, 2, 100023 CrossRef.
- F. Caponnetto, I. Manini, M. Skrap, T. Palmai-Pallag, C. Di Loreto, A. P. Beltrami, D. Cesselli and E. Ferrari, Nanomedicine, 2017, 13, 1011–1020 CrossRef CAS.
- S. El Andaloussi, S. Lakhal, I. Mager and M. J. Wood, Adv. Drug Delivery Rev., 2013, 65, 391–397 Search PubMed.
- D. Xitong and Z. Xiaorong, Gene, 2016, 575, 377–384 CrossRef PubMed.
- J. Rezaie, M. Feghhi and T. Etemadi, Cell Commun. Signaling, 2022, 20, 145 CrossRef.
- F. Tan, X. Li, Z. Wang, J. Li, K. Shahzad and J. Zheng, Signal Transduction Targeted Ther., 2024, 9, 17 CrossRef PubMed.
- S. Dai, D. Wei, Z. Wu, X. Zhou, X. Wei, H. Huang and G. Li, Mol. Ther., 2008, 16, 782–790 CrossRef CAS PubMed.
- M. A. Morse, J. Garst, T. Osada, S. Khan, A. Hobeika, T. M. Clay, N. Valente, R. Shreeniwas, M. A. Sutton, A. Delcayre, D. H. Hsu, J. B. Le Pecq and H. K. Lyerly, J. Transl. Med., 2005, 3, 9 CrossRef.
- B. Escudier, T. Dorval, N. Chaput, F. Andre, M. P. Caby, S. Novault, C. Flament, C. Leboulaire, C. Borg, S. Amigorena, C. Boccaccio, C. Bonnerot, O. Dhellin, M. Movassagh, S. Piperno, C. Robert, V. Serra, N. Valente, J. B. Le Pecq, A. Spatz, O. Lantz, T. Tursz, E. Angevin and L. Zitvogel, J. Transl. Med., 2005, 3, 10 CrossRef.
- B. Escudier, T. Dorval, N. Chaput, F. André, M.-P. Caby, S. Novault, C. Flament, C. Leboulaire, C. Borg and S. Amigorena, J. Transl. Med., 2005, 3, 10 CrossRef PubMed.
- M. A. Morse, J. Garst, T. Osada, S. Khan, A. Hobeika, T. M. Clay, N. Valente, R. Shreeniwas, M. A. Sutton and A. Delcayre, J. Transl. Med., 2005, 3, 9 Search PubMed.
- S. Dai, D. Wei, Z. Wu, X. Zhou, X. Wei, H. Huang and G. Li, Mol. Ther., 2008, 16, 782–790 CrossRef CAS PubMed.
- S. H. Ahn, S. W. Ryu, H. Choi, S. You, J. Park and C. Choi, Mol. Cells, 2022, 45, 284–290 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |