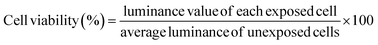Open Access Article
Open Access ArticleInfluence of albumin concentration on surface characteristics and cellular responses in the pre-incubation of multi-walled carbon nanotubes†
Sun Young
Lee‡
 a,
Jae Won
Choi‡
ab,
Tae Geol
Lee
*a,
Min Beom
Heo
*a and
Jin Gyeong
Son
a,
Jae Won
Choi‡
ab,
Tae Geol
Lee
*a,
Min Beom
Heo
*a and
Jin Gyeong
Son
 *a
*a
aNanobio Measurement Group, Division of Biomedical Metrology, Korea Research Institute of Standards and Science, 267 Gajeong-ro, Yuseong-gu, Daejeon 34113, Republic of Korea. E-mail: tglee@kriss.re.kr; mbheo@kriss.re.kr; yeskyoung@kriss.re.kr
bDepartment of Biomedical Science and Technology, Graduate School, Kyung Hee University, Seoul 02447, Republic of Korea
First published on 10th October 2024
Abstract
Reliable characterization of protein coronas (PCs) that form when nanomaterials are introduced into biological fluids is a critical step in the development of safe and efficient nanomedicine. We observed that bovine serum albumin (BSA)-coated multi-walled carbon nanotubes (MWCNTs) do not induce cytotoxicity, but have different cellular uptake rates depending on the BSA pretreatment concentration. To determine how these slight differences affect A549 cell responses and intracellular changes, we conducted spectroscopic (circular dichroism and Fourier-transform infrared) and spectrometric (nanoflow liquid chromatography–electrospray ionization–tandem mass spectrometry) analyses. The various characterization techniques conducted in this study reveal the following. (i) The composition ratio of PCs on MWCNTs differs depending on the BSA concentration. (ii) Analysis of the secondary structure of the proteins revealed that the α-helix structure increased with increasing BSA concentration. (iii) Proteomic analysis showed that different biological pathways were activated at levels higher and lower than 5 mg mL−1. Such combined spectroscopic and spectrometric approaches provide an integrated understanding of PC composition as well as how nano/bio-interface states are linked to cellular-level responses. Our results can support reliable and practical applications of nanomedicine development.
Introduction
Multi-walled carbon nanotubes (MWCNTs), consisting of multiple layers of cylindrical graphene sheets, are applied as nanocarriers because their small diameters enable them to penetrate cells and tissues.1–3 They are widely used in biomedical science due to their ability to prevent drug degradation, which increases drug stability and treatment efficiency.4,5 However, MWCNTs in their original form easily aggregate and generate non-specific binding within a cell or organism, leading to undesired results.6,7 To improve the biocompatibility of MWCNTs, their surfaces should be modified to ensure homogeneous dispersion, stability in biological fluids, and inhibition of non-specific binding.8,9The most common method of preventing the aggregation of nanomaterials (NMs) to increase their dispersibility is surface functionalization.9–12 The surfaces of NMs can be functionalized by modifying the surface functional groups that are prone to aggregation using chemical reagents or by modifying the surface properties using proteins. In particular, protein surface modification is the preferred method for biological research due to its high biocompatibility and absence of in vitro and in vivo toxic effects.13–15 The most commonly used protein for surface modification is bovine serum albumin (BSA) as it offers the advantages of high water solubility, abundance, and low cost.16 NMs, including MWCNTs, are coated with BSA to prevent aggregation and increase dispersion in biological environments,17,18 for which one strategy is the pre-incubation of NMs with BSA.13,14,16,19–21 In addition to surface modification, ultrasonication is frequently utilised to facilitate the dispersion of MWCNTs. The application of ultrasonication disrupts the aggregates, thus enabling the nanotubes to disperse on an individual basis. The combination of BSA coating and ultrasonication is of great importance for the improvement of dispersion uniformity and the maintenance of the mechanical performance of MWCNTs in biological environments.22–25
When NMs are introduced into a biological environment, regardless of whether pre-coated with proteins or not, they inevitably interact and bind with the surrounding proteins in the exposed environment. A layer called the protein corona (PC) quickly forms around the NMs, altering their surface properties.3,26,27 For example, Cedervall et al. demonstrated that the formation of the protein corona is highly dependent on the nanoparticles' surface characteristics, affecting protein binding affinity and exchange rates.28 It is known that the shape, size, and molecular composition of the NMs determine the proteins that make up the PCs.29–31 The PCs formed on NMs give them a new biological identity, differing from that of the intrinsic NMs, which can significantly affect their bioactivity.11 For example, Zhang et al. reported that using BSA and immunoglobulin G (IgG) on the same NMs to form PCs in different ways resulted in different cellular responses.20 Similarly, Fleischer et al. reported that the BSA coating of NMs with different surface charges can result in secondary structure variations in the proteins composing the PCs, which can affect their interaction with cells.32,33 Recent advances in in situ assays have revealed that the soft corona, the outer layer of the protein corona, plays an important role in determining the biological identity of nanomaterials and undergoes dynamic evolution even within minutes of exposure.34,35 Furthermore, it has been reported that using different concentrations of the same protein to coat NMs results in conformational changes in the proteins that make up the PCs and also differential cellular uptake.13 Such reports suggest that the composition and conformation of PCs generated on the surfaces of NMs play36 a more crucial role in their interaction with cells than the NMs themselves. Therefore, in order to understand the interaction between PCs generated on NMs and the surrounding environment, it is essential to understand not only the identity of the proteins adsorbed on NMs, but also their conformation.37 From this perspective, previous studies on MWCNTs have reported differences in their properties,38–40 PC composition influenced by the surrounding environment and the protein used in pre-coating,12–14,17,19,20 and responses of MWCNTs under in vivo and in vitro conditions.1,6,41,42 However, there is a lack of studies on how the concentration of BSA used in pre-incubation affects PC formation and cellular response to BSA-coated MWCNTs.
The present study examined the impact of varying concentrations of BSA on the surface modification of MWCNTs, with a particular focus on the alterations induced in PC compositions and cellular responses. In our experimental model, the pre-incubation and ultrasonication treatment of BSA resulted in the formation of a hard corona on MWCNTs that was sufficiently stabilised. Subsequently, an additional layer of the protein corona, designated as the soft corona, formed on the MWCNTs that had been pre-incubated with BSA in a 10% FBS cell culture medium. This model provides an appropriate framework for investigating the manner in which the behaviour of newly formed nanomaterials within a biological environment is regulated by the type of hard corona that forms on their surface. It was observed that while altered protein coronas did not affect the viability of A549 cells, they significantly reduced the uptake of MWCNTs, particularly at BSA concentrations of 5 and 10 mg mL−1. The use of circular dichroism (CD) and Fourier-transform infrared (FT-IR) spectroscopy revealed an increase in α-helix structures within the PCs at higher BSA concentrations. Proteomic analysis using nanoflow liquid chromatography-electrospray ionisation-tandem mass spectrometry (nLC-ESI-MS/MS)36,40 demonstrated that MWCNTs pre-incubated with 0 and 1 mg mL−1 BSA exhibited downregulation of proteins associated with the ribosome and oxidative phosphorylation pathways, whereas 5 and 10 mg mL−1 BSA resulted in downregulation of proteins involved in the mRNA surveillance pathway. These findings suggest that the concentration of BSA influences the secondary structure and composition of the PCs, resulting in disparate cellular responses at concentrations exceeding and falling below 5 mg mL−1. The findings of this study offer valuable insights into the potential for enhancing the biocompatibility of MWCNTs through protein pre-incubation.
Results and discussion
Characterization of BSA-coated MWCNTs
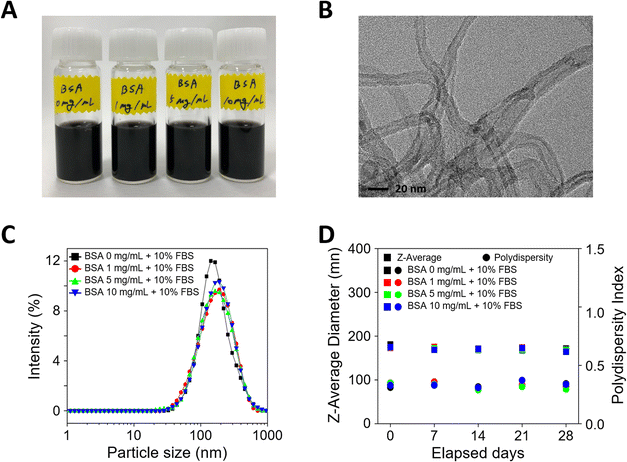
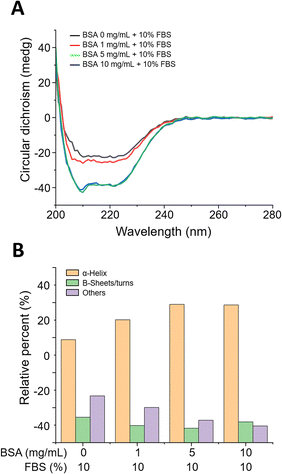
When MWCNTs are introduced into cell culture, aggregation occurs due to surface charge imbalance. To increase dispersion stability, MWCNTs pre-incubated at different BSA concentrations (0, 1, 5, and 10 mg mL−1) were prepared and evaluated for dispersion stability. First, the cell culture media dispersed with MWCNTs prepared after pre-incubation with various concentrations of BSA appeared black due to the uniformly dispersed MWCNTs, as shown in Fig. 1A. To verify the morphology of the MWCNTs, transmission electron microscopy (TEM) measurements were performed, which confirmed that they consisted of multiple layers of carbon and an elongated cylindrical shape, as shown in Fig. 1B. Next, DLS measurements were performed to evaluate the dispersion stability of the MWCNTs in solution. In Fig. 1C, the particle size according to the concentration of BSA showed a similar and consistent range, measuring about 170–180 nm, with no significant differences between samples. Additionally, the polydispersity index (PI), a measure of sample heterogeneity based on size, sustained at about 0.3–0.4 over a 28 day experimental period, confirming that the samples did not aggregate during the analysis (Fig. 1D).The surface functionality of BSA-coated MWCNTs can be evaluated through protein structure investigation. The interaction of proteins with NMs can lead to secondary structure modifications of the proteins that compose the PCs formed on NM surfaces.38 Since these changes can determine the function and biological activity of proteins, it is important to evaluate protein conformational changes upon interaction between proteins and MWCNTs. The secondary structure of proteins can be determined by circular dichroism (CD) spectroscopy, a widely used method to measure conformational changes in proteins. In Fig. 2A, CD spectral analysis of the four types of BSA-coated MWCNTs reveals two minima at 209 nm and 222 nm with increasing concentration of the BSA coating, which was attributed to the α-helix structure. As the concentration of the pre-coated BSA solution increases, the CD signal becomes more negative, indicating an increase in the proportion of α-helix structures. The β-sheet structure typically appears in the 210–230 nm range, with a peak around 215 nm. As illustrated in Fig. 2A, the CD measurements also reveal that the spectrum at 215 nm becomes more negative with increasing BSA pre-coating concentration, indicative of a more pronounced β-sheet structure. The relative contribution of each secondary structural element was calculated as shown in Fig. 2B. The extent of the α-helix structures was determined using the 209 nm and 222 nm minima, from which the α-helix structure of the MWCNTs without BSA was found to be at the 60% level. For MWCNTs in the presence of BSA at 1 mg mL−1, 5 mg mL−1, and 10 mg mL−1, the respective α-helicity values were around 70%, 80%, and 80%; the α-helix structure increased with the concentration of BSA, saturating at 5 mg mL−1. On the other hand, β-sheets and β-turn structures were the highest without BSA pre-incubation, while the percentage of these structures decreased with BSA pre-incubation. Overall, an increase in the concentration of the BSA pre-coating results in a more negative CD signal, indicating an enhancement in the secondary structures of the protein, namely the α-helix and β-sheet. This suggests that the protein structure is stabilised and that specific secondary structures are reinforced as the concentration of the BSA pre-coating increases.
Another approach for determining the secondary structure is to use infrared (IR) spectroscopy because structural arrangements in the protein chain are associated with specific vibration bands. We investigated the differences in the secondary structure of BSA-coated MWCNTs using FT-IR; the spectra are presented in Fig. S1 (ESI†). The BSA structural analysis was performed in the amide I vibration band (1700–1600 cm−1) and the amide II region (1600–1480 cm−1).43 The 1655–1650 cm−1 region is attributed to the α-helix structure, and the peak in this region was found to increase significantly with increasing BSA concentration. In the absence of the BSA pre-coating, the peak observed near 1654.69 cm−1 exhibited a broad and diffuse character. However, as the concentration of the BSA pre-coating increased to 1, 5, and 10 mg mL−1, the peak became sharper and more defined, indicating an increase in the α-helix content within the protein. This structural alteration suggests that the quantity of BSA adsorbed on the MWCNT surface rises in conjunction with elevated BSA pre-coating concentrations. The main peak near 1540 cm−1 in the amide II region is associated with β-sheet structures. As the concentration of the BSA pre-coating solution increases, the peak in the spectrum becomes more pronounced and sharper. In particular, the peak shifts from 1549.58 cm−1 under conditions without the BSA pre-coating and with a 1 mg mL−1 BSA pre-coating to 1546.69 cm−1 at 5 mg mL−1 and further to 1545.24 cm−1 at 10 mg mL−1. This shift suggests that as the protein structure undergoes alteration, the proportion of β-sheet structures may decrease, or other secondary structural elements may increase. This trend is consistent with the CD measurement results, which also indicate a reduction in the β-sheet content.
The integration of the CD and IR data reveals a clear enhancement in the signals associated with the α-helix structure as the concentration of the BSA pre-coating increases. In the case of the β-sheet structure, while the intensity of the structure increases with increasing BSA concentrations, the data also indicate that structural changes occur. This suggests that when BSA is exposed to MWCNTs at high concentrations initially, without competition from other proteins, it can adsorb in a well-aligned manner on the surface. Considering the impact of BSA pre-incubation concentration on the secondary structure of proteins, it is likely that the initial pre-incubation of BSA contributes to structural changes in the protein. At higher pre-incubation concentrations, BSA may strongly adsorb onto the MWCNT surface, maintaining its structural stability. The higher the concentration of BSA during pre-incubation, the more likely it is that the pre-adsorbed BSA will hinder or limit the adsorption of other proteins. This could result in the BSA structure being more distinctly preserved, suggesting that BSA may act as a barrier against dynamic surface changes that could occur in the medium. Conversely, if the BSA pre-incubation concentration is low, other proteins in the 10% FBS medium may more easily adsorb onto the surface and compete with BSA. The changes in the secondary structure observed in the IR and CD data can be understood as a result of interactions that occur when BSA-pre-incubated MWCNTs are introduced into the 10% FBS medium. The analysis indicates that α-helical and β-sheet structures are more stably maintained or even enhanced at higher BSA pre-incubation concentrations. At lower pre-incubation concentrations, structural changes may occur more readily, as evidenced by shifts in peak positions and intensities in the IR and CD spectra. This reflects the competitive adsorption between BSA and other proteins in the 10% FBS medium. In conclusion, the amount of BSA stably adsorbed onto the MWCNT surface serves as a primary driving force influencing the formation of the secondary protein layer on the MWCNTs, acting as a barrier to the subsequent adsorption of other proteins and potentially playing a crucial role in stabilizing the MWCNTs.
Protein coronas adsorbed on BSA-coated MWCNTs
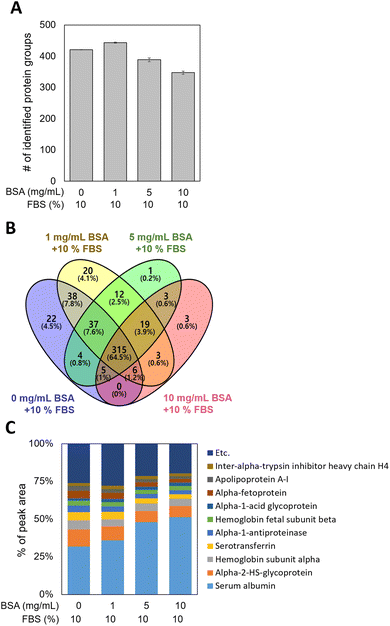
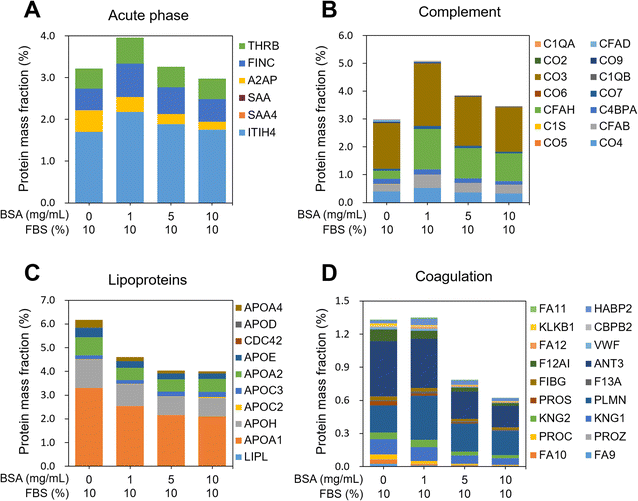
MWCNTs were pre-incubated with 0, 1, 5, and 10 mg mL−1 of BSA (0-, 1-, 5-, and 10-BSA) and stored in DMEM medium with 10% fetal bovine serum (FBS), during which PCs formed on the MWCNT surfaces. Since the behavior of MWCNTs depends on their surface properties,42,44 it is important to investigate the composition and amount of the proteins of the PC, which can define the surface properties of the MWCNTs. To extract the PCs from the MWCNT surfaces, non-specifically bound proteins were first eliminated using several PBS washings. Then the PCs were extracted from the washed MWCNT pellets, and digested samples were introduced into the nLC-ESI-MS/MS system to identify and quantify the PCs adsorbed on the BSA-coated MWCNTs. PC proteomic analysis identified approximately 400 proteins in the four groups, regardless of the concentration of BSA used in the pre-incubation, as shown in Fig. 3A. As displayed in the Venn diagram in Fig. 3B, 315 common proteins (64.5%) were identified in the four groups. Interestingly, 22 and 20 unique proteins were measured in the 0- and 1-BSA groups, respectively, while 1 and 3 unique proteins were measured in the 5- and 10-BSA groups, respectively. In addition, by examining the percentage of the peak area values of the identified proteins, the peak area of serum albumin was found to be higher at the high concentrations of BSA for the MWCNT coating, while those of other proteins were lower (Fig. 3C). These results suggest that BSA, which has a higher affinity for hydrophobic MWCNT surfaces compared to other proteins in serum,45,46 was used in the pre-incubation in a concentration-dependent manner, reducing the opportunity for other proteins to adsorb onto the MWCNT surface, thus affecting the PC composition.In order to characterize the PCs adsorbed on the MWCNTs, their molecular weight (MW), isoelectric point (pI), and biological functions were investigated. The distributions of the MW and pI of the PCs were similar regardless of the concentration of BSA (Fig. S2A and B in the ESI†). In addition, classification according to four physiological functions of the serum proteins was investigated, namely acute phase, complement, lipoproteins, and coagulation. The protein mass fraction (%) was calculated with reference to a previously reported study.47 First, the protein mass fraction for six proteins related to the acute phase was highest in 1-BSA (Fig. 4A). The protein mass fractions at other concentrations were similar, but the proportions of individual proteins were different. Second, in complement, as in the acute phase, the protein mass fraction was highest in the 1-BSA group, and the protein mass fraction decreased with increasing BSA concentration (Fig. 4B). Unusually, cytokine inducible SH2 protein (CIS) was the only protein measured in 0-BSA, unlike the other groups. On the other hand, the protein mass fraction of complement factor H (CFAH) increased significantly after coating with BSA. Third, lipoproteins showed the highest protein mass fraction in 0-BSA, and the protein mass fraction decreased with increasing BSA concentrations (Fig. 4C). The primary lipoproteins that showed a significant reduction were Apolipoprotein A1 (APOA1) and Apolipoprotein H (APOH). APOA1 is the major protein component of high-density lipoprotein (HDL) and plays a crucial role in the reverse cholesterol transport (RCT) process. It possesses anti-inflammatory properties, which help suppress inflammatory responses.48 APOA1 can inhibit the production of pro-inflammatory cytokines, reduce oxidative stress, and decrease vascular inflammation, thereby potentially mitigating the progression of atherosclerosis.49 APOH, also known as beta-2-glycoprotein I, is primarily involved in the blood coagulation process by binding to lipids and phospholipids in plasma and modulating coagulation.50 Finally, the proteins including coagulation were found to be similarly high for 0-BSA and 1-BSA, and the protein mass fraction decreased significantly with increasing concentration (Fig. 4D). Among the coagulation factors, Antithrombin III (ANT3) exhibited the most significant decrease. ANT3 is a key protein that inhibits blood coagulation by binding to and inactivating thrombin and other activated coagulation factors.51 Through its anti-inflammatory and anticoagulant actions, ANT3 can suppress vascular inflammatory responses.52 The protein analysis results of Fig. 4C and D indicate that when BSA-coated MWCNTs were exposed to 10% FBS media, there was a significant decrease in the adsorption of lipoproteins and coagulation factors as the BSA pre-coating concentration increased.
The comprehensive examination of the PC components indicated that the adsorption of specific protein groups (lipoproteins and coagulation factors) diminished with elevated BSA pre-incubation concentrations. This observation implies that BSA effectively hinders the adsorption of other proteins. Consequently, BSA pre-coating can be employed to enhance the biocompatibility of nanoparticles or to mitigate non-specific interactions with particular proteins. This ultimately allows for the regulation of how MWCNTs interact with other proteins when exposed to biological environments.
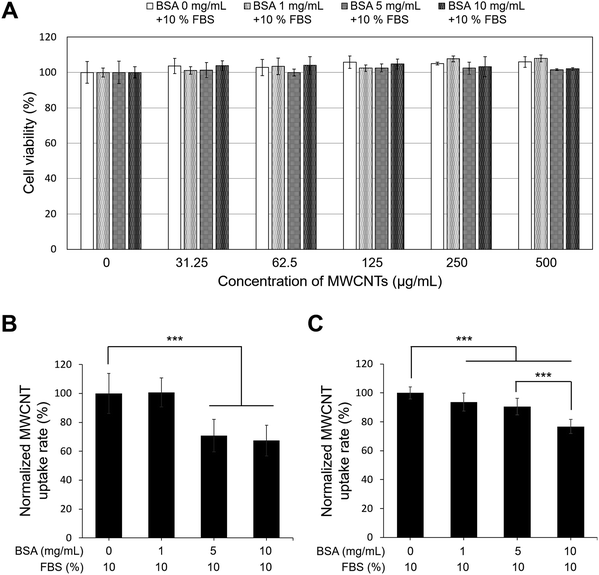
Cellular viability of A549 cells with BSA-coated MWCNTs
In this study, A549 cells were exposed to MWCNTs coated with BSA at various concentrations for 24 h, and toxicity was evaluated using the ATP assay, a method based on mitochondrial activity. As shown in Fig. 5A, no discernible toxicity related to the concentrations of BSA and MWCNTs was observed. Therefore, regardless of the BSA concentration, it was found that the MWCNTs did not affect intracellular mitochondrial activity. This is consistent with other reports that BSA-coated MWCNTs exhibit lower cytotoxicity.14,20 Then A549 cells were treated with 50 μg mL−1 of BSA-coated MWCNTs for 4 and 24 h to quantify cellular uptake over time. The quantity of MWCNTs taken up by cells was calculated with reference to a previously reported study.53Fig. 5B and C compare the uptake of BSA-coated MWCNTs in the A549 cells based on different BSA concentrations for 4 and 24 h of exposure, respectively. After 4 h of exposure, the A549 cells treated with BSA concentrations of 5 mg mL−1 and 10 mg mL−1 exhibited uptake rates of 70.9 ± 11.2% and 67.4 ± 10.7%, respectively, which were lower than those treated with 0 mg mL−1 and 1 mg mL−1 BSA, indicating minimal uptake at higher concentrations. However, after 24 h of exposure, the cell absorption rate decreased in a concentration-dependent manner compared to 0 mg mL−1, and at 10 mg mL−1, the absorption rate was 76.9 ± 4.5%, indicating that the higher the BSA concentration, the lower the cell absorption efficiency. The observed decrease in the cellular uptake of MWCNTs in our study may be attributed to variations in the protein composition, quantity, and secondary structure of the PC formed around MWCNTs, depending on the concentration of BSA. Changes in the composition and structure of the PC can interfere with the recognition of MWCNTs by cellular receptors, leading to reduced cellular internalization. This finding is consistent with previous studies that observed a decrease in the cellular uptake of BSA-coated MWCNTs in A549 cells.54 However, our results appear to contradict macrophage research indicating higher cellular uptake of BSA-coated MWCNTs compared to pure MWCNTs.19,20 This discrepancy may arise from the fact that nanoparticle–cell interactions are influenced not only by surface properties and size but also by the specific cell type. Different cell types present distinct sets of biomolecules on their surfaces,14,55 which can significantly impact the interaction and uptake of nanoparticles.Proteomic analysis of cellular responses in A549 cells with BSA-coated MWCNTs
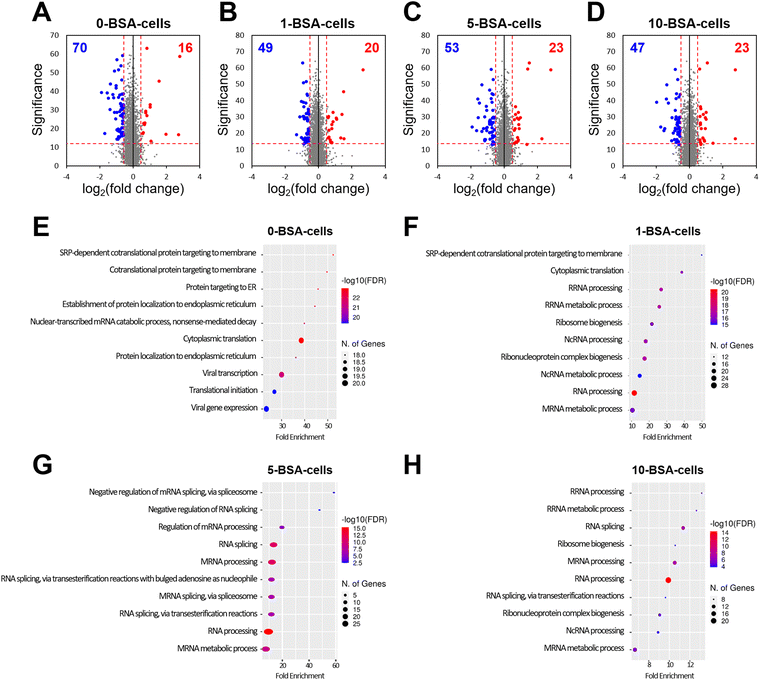
Through a cell viability assay, we found that the exposure of A549 cells to BSA-coated MWCNTs did not affect their viability, but their cellular uptake decreased as the concentration of BSA used in the pre-incubation increased. Therefore, to investigate the alteration of proteins in the cells based on the pre-incubated BSA concentration, we carried out a quantitative analysis of proteins from A549 cells exposed to four concentrations of BSA using nLC-ESI-MS/MS.To measure the alterations of protein expression in cells exposed to MWCNTs according to BSA concentration in pre-incubation, A549 cells treated with the four different BSA concentrations (0-, 1-, 5-, and 10-BSA cells) were compared with a control unexposed to MWCNTs. A total of 3348 proteins were quantified from the four different groups. Compared to the control, the number of downregulated proteins slightly differed in the four groups (70, 49, 53, and 47 proteins in 0-, 1-, 5-, and 10-BSA cells, respectively, Fig. 6A–D), whereas the number of upregulated proteins was similar (16, 20, 23, and 23 proteins in 0-, 1-, 5-, and 10-BSA cells). Biological processes that can provide detailed information on proteins were investigated using differentially expressed proteins (DEPs) measured from the four groups. No biological process was identified for the upregulated proteins. The downregulated proteins measured in the 0-BSA cell group were associated with protein targeting, protein localization, and translation (Fig. 6E). In the 1-BSA cells group, proteins involved in RNA processing, the RNA metabolic process, and ribonucleoprotein complex biogenesis were downregulated, unlike in the 0-BSA cell group (Fig. 6F). The proteins downregulated in the 5-BSA cell group were similar to those in the 1-BSA cell group, and the pathways involved in RNA splicing via either transesterification with bulged adenosine as a nucleophile or the spliceosome were also identified (Fig. 6G). Proteins involved in overall RNA processing, RNA metabolism, and RNA splicing were found to be downregulated in the 10-BSA cells (Fig. 6H). From the biological process results, MWCNTs without BSA coating, whose PCs were formed from the FBS, are mainly involved in protein localization and cytoplasmic translation in A549 cells. However, MWCNTs pre-incubated with BSA showed a different composition of PCs that resulted in altered surface functionality, and this downregulated the pathways involved in RNA processing in cells. In addition, the higher the concentration used for BSA coating, the greater the downregulation of the proteins involved in RNA splicing.
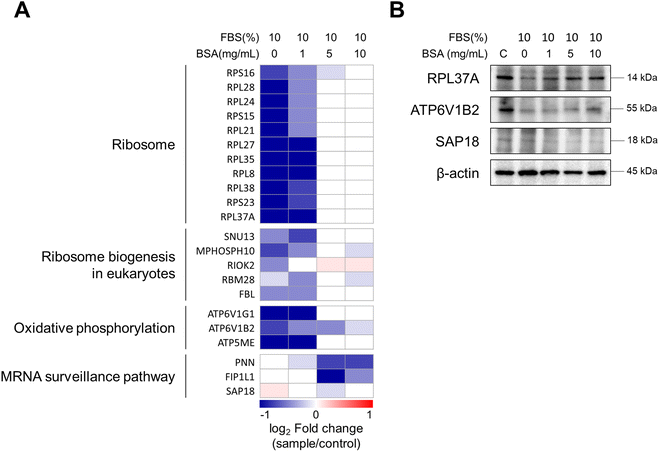
Compared to the control, the expression pattern of DEPs was similar between the 0- and 1-BSA cell groups (low concentration groups) (Fig. S3A in the ESI†). On the other hand, the protein expression levels of the high concentration groups (5- and 10-BSA cells) showed different patterns from the low concentration groups. By comparing the low and high concentration groups, we identified the three most differentiated clusters and investigated the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Fig. 3B in the ESI†). The proteins in cluster 1, which were significantly downregulated in the low concentration groups, were involved in pathways related to the ribosome, oxidative phosphorylation, and ribosome biogenesis in eukaryotes. The proteins in cluster 2, which were significantly downregulated in the high concentration groups, were involved in the nucleocytoplasmic transport and mRNA surveillance pathways. Finally, the proteins in cluster 3, which were downregulated in the low concentration groups and upregulated in the 10-BSA cell group, were associated with the pathway of ribosome biogenesis in eukaryotes.
Based on the above results, the expression levels of the proteins in four of these pathways are described in Fig. 7A. In the low concentration groups, the proteins related to the ribosome, ribosome biogenesis in eukaryotes, and oxidative phosphorylation pathways were significantly downregulated. In the high concentration groups, only proteins involved in the mRNA surveillance pathway were downregulated, which correlates with the RNA splicing pathway described in the analysis in Fig. 6. The response of RNA splicing in the high concentration group (5- and 10-BSA cells), as shown in Fig. 6G and H, may explain the downregulation of the mRNA surveillance pathway. Heatmap results are also consistent with the western blot data. As shown in Fig. 7B and S4 (ESI†), 60S ribosomal protein L37a (RPL37A) and V-type proton ATPase subunit B brain isoform (ATP6V1B2), which were downregulated at 0 and 1 mg mL−1 BSA concentrations, were also downregulated in the western blot results and showed similar expression levels to the control at higher concentrations. In contrast, histone deacetylase complex subunit SAP18 (SAP18) was similar to the control at low BSA concentrations (0 and 1 mg mL−1) and decreased in expression at high BSA concentrations (5 and 10 mg mL−1). The extent of internalization of MWCNTs and its impact on biological processes in A549 cells can be attributed to varying levels of cellular stress, which depend on the concentration of pre-coated BSA. When cells are exposed to stressors such as MWCNTs, it is known that RNA splicing can be disrupted.56 In addition, previous research reported that long-term exposure to MWCNTs downregulated ribosomal function and oxidative phosphorylation in lung cells of mice.57 The results we observed in vitro with A549 cells are similar to the chronic toxicity results seen in vivo with mice. Our findings demonstrate that the chronic toxic effects observed at the tissue level in mouse models can indeed be reflected at the cellular level, thereby underscoring the importance of pathway-specific studies in the field of nanotoxicology. This further substantiates the utility of our model in elucidating the mechanisms through which nanomaterials, even those devoid of acute toxicity, can influence cellular processes. Additionally, it is noteworthy that when MWCNTs were pre-incubated with BSA at concentrations of 5 and 10 mg mL−1, no reduction in protein synthesis pathways was observed. This suggests that adequately passivated MWCNTs with BSA exhibit reduced interactions with cells, thereby preventing significant intracellular changes. This variation appears to be linked to changes in the secondary structure of the proteins composing the PCs of MWCNTs. The secondary structure of the PC proteins is an important factor in determining receptor recognition upon entry into the cell,33 and it is known that protein–NM complexes are maintained upon entry into the cell.32 Therefore, BSA-coated MWCNTs with PCs that have altered the secondary structures of proteins can change the biological pathways within the cell. Overall, we demonstrated through proteomic analysis of PCs and cells that the intracellular response to MWCNTs is highly dependent on the concentration of BSA used in the pre-incubation.
Conclusions
In this study, we sought to correlate PCs formed on MWCNTs and their effects on cells according to pre-incubation BSA concentration. Analysis using CD and FT-IR showed that the high concentration group (5 and 10 mg mL−1 BSA) had increased α-helix structures in the PCs on the MWCNT surfaces, which affected their surface functionalization. In addition, BSA-coated MWCNTs did not significantly affect the viability of cells, but the cellular uptake of the MWCNTs decreased with increasing concentration of BSA. Through nLC-ESI-MS/MS-based proteomic analysis, we found that the composition ratio of PC proteins adsorbed on MWCNTs differs depending on the concentration of BSA in pre-incubation. Additionally, the mass fraction of the proteins composing the PCs was also affected by the BSA concentration. In terms of biological processes, A549 cells treated with pre-coated MWCNTs at low BSA concentrations (0 and 1 mg mL−1 BSA cells) had inhibited ribosome and oxidative phosphorylation pathways, while the high concentration group (5 and 10 mg mL−1 BSA) exhibited a suppressed mRNA surveillance pathway. Such results indicate that cellular responses to BSA-coated MWCNTs can differ depending on the concentration of BSA. We found that even for the same MWCNTs, the composition of the PCs is affected by the protein concentration in the biological environment, and that cells respond differently to the secondary structures of the surface-adsorbed proteins, rather than to the MWCNT properties. Based on this multifaceted analysis of the composition and secondary structure of protein coronas on MWCNTs along with cellular responses, we conclude that BSA-concentration-dependent surface functionality may provide important clues for modifying related methods to improve the biocompatibility of nanomaterials.Experimental
Materials
Tris–HCl buffer (pH 8.0), sodium chloride (NaCl), phosphate buffered saline (PBS), ammonium bicarbonate (AmBic), dithiothreitol (DTT), iodoacetamide (IAA), L-cysteine, and formic acid (FA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile (ACN) and water containing LC-MS grade 0.1% FA, n-dodecyl beta-D-maltoside (DDM), trypsin, and bicinchoninic acid (BCA) assay reagent were purchased from Thermo Fisher Scientific (Rockford, IL, USA). The protease inhibitor cocktail was purchased from Roche Diagnostic GmbH (Mannheim, Germany). To purify protein digests, HLB cartridges were purchased from Waters (MA, USA) and HPLC grade water and ACN were purchased from Honeywell (MI, USA).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The diluted MWCNT dispersions were prepared in three parallel samples. The absorbance values of the MWCNM dispersions in cuvettes at 750 nm were measured using a UV-Vis-NIR spectrometer. A calibration line was generated by plotting the average value of absorbance versus concentration. Three 6-well plates were used for three groups (two for the control and one for the MWCNT test). The cell dispersion (approximately 3 × 105 cells/3 mL) was seeded in each well of 6-well plates and incubated for 24 h. The cells were incubated for 24 h, after which the medium was replaced with MWCNT-containing medium (50 μg mL−1) in the plate for MWCNTs or fresh medium in the two plates for the control group. The cells were incubated for a further 4 h and 24 h. One plate was used for cell counting. After washing with PBS for cell counting, cells were harvested by treating with 0.25% trypsin–EDTA. The number of cells in each well of the control plate was counted using a cell counter. Another control plate and a plate for the MWCNT test were washed twice with PBS after removing the supernatant, and each well was treated with 0.5 mL of cell lysis reagent for 30 min at room temperature. After pipetting and transferring to a new e-tube, 0.5 mL of SDBS was added. The cell dispersions were sonicated for 10 s with a horn-type sonicator. The concentration of MWCNTs in the cell lysates was estimated based on the corresponding calibration curves from the absorbance at 750 nm.
1). The diluted MWCNT dispersions were prepared in three parallel samples. The absorbance values of the MWCNM dispersions in cuvettes at 750 nm were measured using a UV-Vis-NIR spectrometer. A calibration line was generated by plotting the average value of absorbance versus concentration. Three 6-well plates were used for three groups (two for the control and one for the MWCNT test). The cell dispersion (approximately 3 × 105 cells/3 mL) was seeded in each well of 6-well plates and incubated for 24 h. The cells were incubated for 24 h, after which the medium was replaced with MWCNT-containing medium (50 μg mL−1) in the plate for MWCNTs or fresh medium in the two plates for the control group. The cells were incubated for a further 4 h and 24 h. One plate was used for cell counting. After washing with PBS for cell counting, cells were harvested by treating with 0.25% trypsin–EDTA. The number of cells in each well of the control plate was counted using a cell counter. Another control plate and a plate for the MWCNT test were washed twice with PBS after removing the supernatant, and each well was treated with 0.5 mL of cell lysis reagent for 30 min at room temperature. After pipetting and transferring to a new e-tube, 0.5 mL of SDBS was added. The cell dispersions were sonicated for 10 s with a horn-type sonicator. The concentration of MWCNTs in the cell lysates was estimated based on the corresponding calibration curves from the absorbance at 750 nm.
Sample preparation for proteomic analysis
For the protein coronas analysis, 1000 μL of MWCNTs distributed in cell culture medium was introduced into new tubes to analyze the PCs adsorbed on the MWCNTs. The solution was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C, after which the supernatants were removed. For the elimination of the remaining cell culture medium, the pellets were gently rinsed using PBS solution, and the solution was removed after centrifugation. This washing process was repeated three times. The washed MWCNT pellets were added to 50 μL of lysis solution consisting of 0.2% DDM, 150 mM NaCl, 50 mM Tris–HCl, and one tablet of protease inhibitor cocktail, and reacted at 95 °C for 10 min in a thermomixer to isolate the proteins from the MWCNTs. Then the solution was centrifuged at 20
000g for 30 min at 4 °C, after which the supernatants were removed. For the elimination of the remaining cell culture medium, the pellets were gently rinsed using PBS solution, and the solution was removed after centrifugation. This washing process was repeated three times. The washed MWCNT pellets were added to 50 μL of lysis solution consisting of 0.2% DDM, 150 mM NaCl, 50 mM Tris–HCl, and one tablet of protease inhibitor cocktail, and reacted at 95 °C for 10 min in a thermomixer to isolate the proteins from the MWCNTs. Then the solution was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C to remove the MWCNTs. The total protein concentration of the extracted proteins was measured using the BCA assay. For protein digestion, 20 μg of proteins was reduced using 50 mM AmBic solution containing 10 mM DDT. To prevent re-folding of the disulfide bonds, the solution was alkylated with IAA at a final concentration of 20 mM and reacted at room temperature for 30 min. The alkylated proteins in a tube were digested by adding trypsin (protein
000g for 30 min at 4 °C to remove the MWCNTs. The total protein concentration of the extracted proteins was measured using the BCA assay. For protein digestion, 20 μg of proteins was reduced using 50 mM AmBic solution containing 10 mM DDT. To prevent re-folding of the disulfide bonds, the solution was alkylated with IAA at a final concentration of 20 mM and reacted at room temperature for 30 min. The alkylated proteins in a tube were digested by adding trypsin (protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) enzyme = 20
enzyme = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/w) at 37 °C for 18 h to form peptides. After purification of the peptide mixture with an HLB cartridge, the solution was dried under vacuum conditions. The dried sample was stored up to −20 °C prior to analysis by nLC-ESI-MS/MS.
1, w/w) at 37 °C for 18 h to form peptides. After purification of the peptide mixture with an HLB cartridge, the solution was dried under vacuum conditions. The dried sample was stored up to −20 °C prior to analysis by nLC-ESI-MS/MS.
For the A549 cell effect analysis, cell harvests treated with BSA-coated MWCNTs incubated for 24 hours were added to 500 μL of lysis solution and mixed vigorously at 95 °C for 10 min. To remove cell debris, the solution was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C. The concentration of the extracted proteins was detected through the BCA assay. Then 25 μg of proteins was used for proteolytic digestion. All processes for protein digestion with trypsin were the same as the digestive method for the PCs. The dried peptides were stored at −20 °C prior to nLC-ESI-MS/MS analysis.
000g for 15 min at 4 °C. The concentration of the extracted proteins was detected through the BCA assay. Then 25 μg of proteins was used for proteolytic digestion. All processes for protein digestion with trypsin were the same as the digestive method for the PCs. The dried peptides were stored at −20 °C prior to nLC-ESI-MS/MS analysis.
nLC-ESI-MS/MS analysis
The dried samples were reconstituted with an aqueous solution containing LC-MS grade 2% ACN and 0.1% FA, and then 250 ng of the peptide mixture was introduced into a NanoElute LC system connected to a hybrid trapped ion mobility spectrometry-quadrupole time-of-flight mass spectrometer (timsTOF Pro, Bruker Daltonics, Bremen, Germany) equipped with a modified nano-electrospray ion source (CaptiveSpray, Bruker Daltonics). In an in-house column (75 μm inner diameter and 250 mm length) packed with C18 resins (1.9 μm, 120 Å, Dr. Maisch), the peptide mixture was separated at 50 °C with a constant flow of 400 nL min−1, followed by elution using the following binary gradient of mobile phases A (0.1% FA in water) and B (0.1% FA in ACN): 2% to 17% B for 45.0 min, 17% to 25% for 22.5 min, 25% to 37% for 7.5 min, and 37% to 80% for 5.0 min, and then this level was maintained for 10 min to rinse the analytical column. The timsTOF Pro system was operated in parallel accumulation serial fragmentation (PASEF) acquisition mode using Bruker Compass Hystar 5.0.37.1. The settings for the MS and MS/MS scans were as follows: mass range of 100 to 1700 m/z, 1/K0 start at 0.6 V s cm−2 and end at 1.6 V s cm−2, capillary voltage of 1500 V, dry gas flow rate of 3 L min−1, and dry temperature of 180 °C; PASEF mode: 10 MS/MS scans (total cycle time 1.16 s), charge range of 0 to 5, active exclusion for 0.4 min, scheduling target intensity of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and intensity threshold of 2500, depending on the precursor mass and charge.
000, and intensity threshold of 2500, depending on the precursor mass and charge.
Data analysis
The obtained spectral data were submitted to PEAKS Studio 10.5 (Bioinformatics Solutions, Waterloo, Canada) for identification and label-free quantification (LFQ) searches of PCs and cell-derived proteins against the SwissProt database of Bos taurus (bovin, UP00000913, downloaded 20/12/2019, 6008 entries) and Homo sapiens (human, UP00000564, downloaded 22/11/2019, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 379 entries) from Uniprot (http://www.uniprot.org/) with a false discovery rate of 0.01. The parameters for protein identification were as follows: (a) trypsin as the specific enzyme, with two missed cleavages allowed; (b) fixed modification: carbamidomethylation of cysteine and variable modification: oxidation of methionine and acetylation of protein N-term, allowing for three variable PTMs per peptide; (c) precursor mass error tolerance of 20.0 ppm; (d) fragment mass error tolerance of 0.05 Da. Following the completion of protein identification, LFQ was performed based on the analyzed PEAKS dataset. The analysis of variance (ANOVA) method was used to conduct LFQ analysis, and significance thresholds were set at two unique peptides, a data filter in at least two samples per group, a significance of 20 (p-value = 0.01), and a 1.5-fold change. Total ion chromatography (TIC) was used to perform data normalization. GO-term enrichment of proteins was carried out using the ShinyGO program (version 0.77) after the data were exported to Microsoft Excel.
379 entries) from Uniprot (http://www.uniprot.org/) with a false discovery rate of 0.01. The parameters for protein identification were as follows: (a) trypsin as the specific enzyme, with two missed cleavages allowed; (b) fixed modification: carbamidomethylation of cysteine and variable modification: oxidation of methionine and acetylation of protein N-term, allowing for three variable PTMs per peptide; (c) precursor mass error tolerance of 20.0 ppm; (d) fragment mass error tolerance of 0.05 Da. Following the completion of protein identification, LFQ was performed based on the analyzed PEAKS dataset. The analysis of variance (ANOVA) method was used to conduct LFQ analysis, and significance thresholds were set at two unique peptides, a data filter in at least two samples per group, a significance of 20 (p-value = 0.01), and a 1.5-fold change. Total ion chromatography (TIC) was used to perform data normalization. GO-term enrichment of proteins was carried out using the ShinyGO program (version 0.77) after the data were exported to Microsoft Excel.
Western blot
A549 cells (90–95% confluency) were treated with different concentrations of BSA (0, 1, 5, and 10 mg mL−1) in the presence of 50 μg mL−1 MWCNTs for 24 h. Proteins in the cell lysates were quantified using Bradford analysis (5X SDS-PAGE sample buffer, TLP-102.1), and an equal amount of protein was separated by sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) electrophoresis. Subsequently, the proteins were transferred to a PVDF membrane (polyvinylidene fluoride, 0.45 μm pore size) at 385 mA for 1 h and then blocked with 3% BSA dissolved in phosphate buffered saline with Tween detergent (0.05%) for 1 h. Afterward, the membrane was incubated overnight at 4 °C with primary antibodies: β-actin (mouse monoclonal, sc-47778, Santa Cruz Biotechnology), ATP6V1B2 (rabbit polyclonal, PA5-52518, Invitrogen), RPL37A (rabbit polyclonal, PA5-75866, Invitrogen), and SAP18 (rabbit polyclonal, PA5-76167, Invitrogen). The membrane was washed three times with PBST (0.05%) for 10 min each time and then incubated with a secondary antibody (mouse anti-rabbit IgG-HRP, sc-2357, Santa Cruz Biotechnology) at room temperature for 2 h. Finally, the blotting was visualized using a ChemiDoc XRS+ system (Bio-Rad Laboratories, Hercules, CA, USA).Author contributions
Conceptualization: J. G. S., M. B. H., and T. G. L.; data curation: J. G. S., M. B. H., S. Y. L., and J. W. C.; formal analysis: S. Y. L. and J. W. C.; funding acquisition: J. G. S., M. B. H., and T. G. L.; investigation: S. Y. L. and J. W. C.; methodology: S. Y. L. and J. W. C.; project administration: P. R. and G. P.; resources: O. M., P. R., and G. P.; supervision: J. G. S., M. B. H., and T. G. L.; validation: S. Y. L. and J. W. C.; visualization: S. Y. L. and J. W. C.; writing – original draft: S. Y. L. and J. W. C.; writing –review & editing: J. G. S., M. B. H., and T. G. L.Data availability
Proteomic data for this study are available in the PRIDE database under accession number PXD052226.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Development of Measurement Standards and Technology for Biomaterials and Medical Convergence funded by the Korea Research Institute of Standards and Science (No. KRISS-2024-GP2024-0007), the Basic Science Research Program (No. 2021RC1C1011598), and the Nano & Material Technology Development Program (RS-2024-00452934) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT, Republic of Korea.References
- G. M. Hilton, A. J. Taylor, C. D. McClure, G. N. Parsons, J. C. Bonner and M. S. Bereman, Toxicology, 2015, 329, 80–87 CrossRef CAS
.
- Y. Ando, X. Zhao, H. Shimoyama, G. Sakai and K. Kaneto, Int. J. Inorg. Mater., 1999, 1, 77–82 CrossRef CAS
.
- N. Lu, Y. Sui, R. Tian and Y. Y. Peng, Chem. Res. Toxicol., 2018, 31, 1061–1068 Search PubMed
.
- K. H. Ahn, S. M. Kim and I. J. Yu, Saf. Health Work, 2011, 2, 65–69 Search PubMed
.
- W. Bai, A. Raghavendra, R. Podila and J. M. Brown, Int. J. Nanomed., 2016, 11, 4357–4371 Search PubMed
.
- M. Allegri, D. K. Perivoliotis, M. G. Bianchi, M. Chiu, A. Pagliaro, M. A. Koklioti, A. A. Trompeta, E. Bergamaschi, O. Bussolati and C. A. Charitidis, Toxicol Rep, 2016, 3, 230–243 Search PubMed
.
- B. Koh and W. Cheng, Langmuir, 2014, 30, 10899–10909 CrossRef CAS PubMed
.
- J. W. Lee, Y. C. Choi, R. Kim and S. K. Lee, Biomed. Res. Int., 2015, 2015, 485343 Search PubMed
.
- M. Nicoletti, C. Gambarotti and E. Fasoli, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2021, 1163, 122504 CrossRef CAS PubMed
.
-
F. N. A. M. Sabri, M. R. Zakaria and H. M. Akil, Presented in Part at the 3rd International Postgraduate Conference on Materials, Minerals & Polymer (Mamip), 2020 Search PubMed
.
- C. H. Z. Martins, F. Côa, G. H. Da Silva, J. Bettini, M. A. De Farias, R. V. Portugal, G. d. A. Umbuzeiro, O. L. Alves and D. S. T. Martinez, Environ. Sci.: Nano, 2022, 9, 2887–2905 RSC
.
- M. Nicoletti, C. Gambarotti and E. Fasoli, Biotechnol. Appl. Biochem., 2021, 68, 1003–1013 CrossRef CAS PubMed
.
- M. T. Huynh, C. Mikoryak, P. Pantano and R. Draper, Nanomaterials, 2021, 11(2), 539 CrossRef CAS PubMed
.
- J. Long, X. Li, Y. Kang, Y. Ding, Z. Gu and Y. Cao, RSC Adv., 2018, 8, 9253–9260 RSC
.
- M. Kucki, J.-P. Kaiser, M. Clift, B. Rothen-Rutishauser, A. Petri-Fink and P. Wick, Fibers, 2014, 2, 187–210 CrossRef CAS
.
- F. Coa, F. S. Delite, M. Strauss and D. S. T. Martinez, NanoImpact, 2022, 27, 100413 CrossRef CAS
.
- S. H. De Paoli, L. L. Diduch, T. Z. Tegegn, M. Orecna, M. B. Strader, E. Karnaukhova, J. E. Bonevich, K. Holada and J. Simak, Biomaterials, 2014, 35, 6182–6194 CrossRef CAS
.
- K. Nagaraju, R. Reddy and N. Reddy, J. Appl. Biomater. Funct. Mater., 2015, 13, e301–e312 CAS
.
- W. Bai, Z. Wu, S. Mitra and J. M. Brown, J. Nanomater., 2016, 2016, 2159537 Search PubMed
.
- T. Zhang, M. Tang, Y. Yao, Y. Ma and Y. Pu, Int. J. Nanomed., 2019, 14, 993–1009 CrossRef CAS
.
- D. Elgrabli, S. Abella-Gallart, O. Aguerre-Chariol, F. Robidel, F. Rogerieux, J. Boczkowski and G. Lacroix, Nanotoxicology, 2009, 1, 266–278 CrossRef
.
- A. J. Blanch, C. E. Lenehan and J. S. Quinton, Carbon, 2011, 49, 5213–5228 CrossRef CAS
.
- S. Alrekabi, A. B. Cundy, A. Lampropoulos, R. L. D. Whitby and I. Savina, Mater. Des., 2017, 136, 223–237 CrossRef CAS
.
- S. Alrekabi, A. Cundy, A. Lampropoulos and I. Savina, Int. Schol. Sci. Res. Innov., 2016, 10, 268–274 Search PubMed
.
- J. Yu, N. Grossiord, C. E. Koning and J. Loos, Carbon, 2007, 45, 618–623 CrossRef CAS
.
- L. Garcia-Hevia, M. Saramiforoshani, J. Monge, N. Iturrioz-Rodriguez, E. Padin-Gonzalez, F. Gonzalez, L. Gonzalez-Legarreta, J. Gonzalez and M. L. Fanarraga, J. Nanobiotechnol., 2021, 19, 129 CrossRef CAS PubMed
.
- Z. Gu, Z. Yang, Y. Chong, C. Ge, J. K. Weber, D. R. Bell and R. Zhou, Sci. Rep., 2015, 5, 10886 CrossRef CAS
.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS
.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS PubMed
.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS
.
- Y. Cong, R. Qiao, X. Wang, Y. Ji, J. Yang, D. Baimanov, S. Yu, R. Cai, Y. Zhao, X. Wu, C. Chen and L. Wang, J. Am. Chem. Soc., 2024, 146, 10478–10488 CrossRef CAS
.
- C. C. Fleischer and C. K. Payne, Acc. Chem. Res., 2014, 47, 2651–2659 CrossRef CAS PubMed
.
- C. C. Fleischer and C. K. Payne, J. Phys. Chem. B, 2014, 118, 14017–14026 CrossRef CAS PubMed
.
- H. Mohammad-Beigi, Y. Hayashi, C. M. Zeuthen, H. Eskandari, C. Scavenius, K. Juul-Madsen, T. Vorup-Jensen, J. J. Enghild and D. S. Sutherland, Nat. Commun., 2020, 11, 4535 CrossRef CAS PubMed
.
- D. Baimanov, J. Wang, J. Zhang, K. Liu, Y. Cong, X. Shi, X. Zhang, Y. Li, X. Li, R. Qiao, Y. Zhao, Y. Zhou, L. Wang and C. Chen, Nat. Commun., 2022, 13, 5389 CrossRef CAS PubMed
.
- L. Digiacomo, F. Giulimondi, A. L. Capriotti, S. Piovesana, C. M. Montone, R. Z. Chiozzi, A. Lagana, M. Mahmoudi, D. Pozzi and G. Caracciolo, Nanoscale Adv., 2021, 3, 3824–3834 RSC
.
- G. Bashiri, M. S. Padilla, K. L. Swingle, S. J. Shepherd, M. J. Mitchell and K. Wang, Lab Chip, 2023, 23, 1432–1466 RSC
.
- A. G. Kenry, Y. Liu, K. P. Loh and C. Teck Lim, NPG Asia Mater., 2017, 9, e422 CrossRef CAS
.
- S. S. Poulsen, K. B. Knudsen, P. Jackson, I. E. Weydahl, A. T. Saber, H. Wallin and U. Vogel, PLoS One, 2017, 12, e0174167 CrossRef
.
- H. Haniu, Y. Matsuda, K. Takeuchi, Y. A. Kim, T. Hayashi and M. Endo, Toxicol. Appl. Pharmacol., 2010, 242, 256–262 CrossRef CAS PubMed
.
- S. Phuyal, M. Kasem, O. Knittelfelder, A. Sharma, D. M. Fonseca, V. Vebraite, S. Shaposhnikov, G. Slupphaug, V. Skaug and S. Zienolddiny, Nanotoxicology, 2018, 12, 138–152 CrossRef CAS
.
- L. Ju, G. Zhang, X. Zhang, Z. Jia, X. Gao, Y. Jiang, C. Yan, P. J. Duerksen-Hughes, F. F. Chen, H. Li, X. Zhu and J. Yang, PLoS One, 2014, 9, e84974 CrossRef
.
- A. Y. Gerasimenko, G. N. Ten, D. I. Ryabkin, N. E. Shcherbakova, E. A. Morozova and L. P. Ichkitidze, Spectrochim. Acta, Part A, 2020, 227, 117682 CrossRef CAS
.
- S. Chortarea, F. Zerimariam, H. Barosova, D. Septiadi, M. J. D. Clift, A. Petri-Fink and B. Rothen-Rutishauser, Applied In Vitro, Toxicology, 2019, 5, 47–61 CAS
.
- C. Ge, J. Du, L. Zhao, L. Wang, Y. Liu, D. Li, Y. Yang, R. Zhou, Y. Zhao, Z. Chai and C. Chen, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16968–16973 CrossRef CAS
.
- D. Dutta, S. K. Sundaram, J. G. Teeguarden, B. J. Riley, L. S. Fifield, J. M. Jacobs, S. R. Addleman, G. A. Kaysen, B. M. Moudgil and T. J. Weber, Toxicol. Sci., 2007, 100, 303–315 CrossRef CAS
.
- S. Y. Lee, J. G. Son, J. H. Moon, S. Joh and T. G. Lee, Biointerphases, 2020, 15, 061002 CrossRef CAS PubMed
.
- A. S. Bhale and K. Venkataraman, Biomed. Pharmacother., 2022, 154, 113634 CrossRef CAS PubMed
.
- S. Patel, B. A. Di Bartolo, S. Nakhla, A. K. Heather, T. W. Mitchell, W. Jessup, D. S. Celermajer, P. J. Barter and K. A. Rye, Atherosclerosis, 2010, 212, 392–397 CrossRef CAS PubMed
.
- T. McDonnell, C. Wincup, I. Buchholz, C. Pericleous, I. Giles, V. Ripoll, H. Cohen, M. Delcea and A. Rahman, Blood Rev., 2020, 39, 100610 CrossRef CAS
.
- G. M. Rodgers and A. Mahajerin, Clin. Appl. Thromb./Hemostasis, 2023, 29, 10760296231205279 CrossRef
.
- A. R. Rezaie and H. Giri, J. Thromb. Haemostasis, 2020, 18, 528–533 CrossRef
.
- M. Zhang, M. Yang, T. Okazaki and M. Yudasaka, e-J. Surf. Sci. Nanotechnol., 2018, 16, 93–96 CrossRef CAS
.
- V. H. Nguyen, N. M. Meghani, H. H. Amin, T. T. D. Tran, P. H. L. Tran, C. Park and B. J. Lee, Colloids Surf., B, 2018, 170, 179–186 CrossRef CAS PubMed
.
- R. Purohit, N. V. S. Vallabani, R. K. Shukla, A. Kumar and S. Singh, Bioinspired, Biomimetic Nanobiomater., 2016, 5, 121–131 CrossRef
.
- M. Bhadra, P. Howell, S. Dutta, C. Heintz and W. B. Mair, Hum. Genet., 2020, 139, 357–369 CrossRef PubMed
.
- J. M. Dymacek, B. N. Snyder-Talkington, R. Raese, C. Dong, S. Singh, D. W. Porter, B. Ducatman, M. G. Wolfarth, M. E. Andrew, L. Battelli, V. Castranova, Y. Qian and N. L. Guo, Int. J. Toxicol., 2018, 37, 276–284 CrossRef CAS PubMed
.
- ISO, ISO/TS 23034:2021 Nanotechnologies — Method to estimate cellular uptake of carbon nanomaterials using optical absorption, International Organization for Standardization, 2021, Available at: https://www.iso.org/standard/79657.html, accessed 5 September 2024 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00743c |
| ‡ These authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2024 |