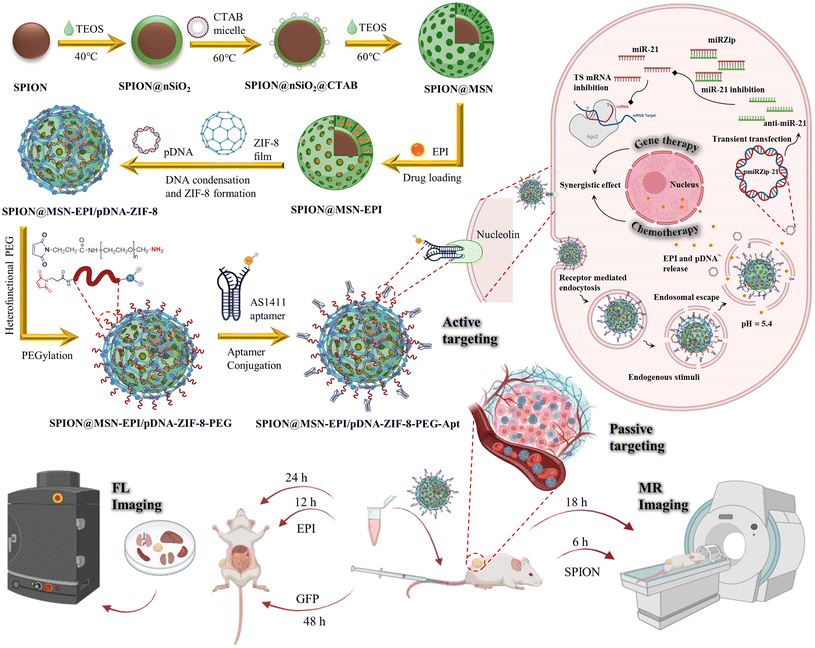
Enhanced theranostic efficacy of epirubicin-loaded SPION@MSN through co-delivery of an anti-miR-21-expressing plasmid and ZIF-8 hybridization to target colon adenocarcinoma†
Amir
Abrishami
a,
Ahmad Reza
Bahrami
ab,
Amir Sh.
Saljooghi
 *cd and
Maryam M.
Matin
*cd and
Maryam M.
Matin
 *ad
*ad
aDepartment of Biology, Faculty of Science, Ferdowsi University of Mashhad, Mashhad, Iran. E-mail: matin@um.ac.ir
bIndustrial Biotechnology Research Group, Institute of Biotechnology, Ferdowsi University of Mashhad, Mashhad, Iran
cDepartment of Chemistry, Faculty of Science, Ferdowsi University of Mashhad, Mashhad, Iran
dNovel Diagnostics and Therapeutics Research Group, Institute of Biotechnology, Ferdowsi University of Mashhad, Mashhad, Iran. E-mail: saljooghi@um.ac.ir
First published on 19th February 2024
Abstract
Using targeted drug delivery systems has emerged as a promising approach to increase the efficacy of chemotherapy, particularly in combination with gene therapy. The overexpression of miR-21 plays a crucial role in colorectal cancer (CRC) progression, and targeted inhibition of miR-21 offers significant potential for enhancing CRC chemotherapy outcomes. In this study, a theranostic system based on mesoporous silica and superparamagnetic iron oxide nanoparticles (SPION@MSNs) was synthesized as a core–shell structure. After loading epirubicin (EPI) in the open pores of MSN, the plasmid expressing anti-miR-21 (pDNA) covered the outer surface with the help of a ZIF-8 (zeolitic imidazolate framework-8) film. Afterward, polyethylene glycol (PEG) and AS1411 aptamer were conjugated to the surface to improve the protective, biocompatibility, and targeting abilities of the nanocarrier. Moreover, the physicochemical characteristics as well as the loading capacity and release profile of EPI and pDNA were fully evaluated. The uptake of the nanoparticles by CRC and normal cell lines in addition to the anticancer effects related to targeted combinational therapy were investigated in vitro. Finally, in vivo tests were performed on BALB/c mice bearing colorectal tumors to evaluate the effectiveness of the targeted nanoparticles, their possible side effects, and also their application in fluorescence and magnetic imaging in vivo. The successful synthesis of SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt nanoparticles (∼68 nm) and good loading efficiency and controlled release of EPI and pDNA were confirmed. Moreover, hemolysis and gel retardation assays demonstrated the biocompatibility and plasmid protection. Cellular uptake and expression of copGFP illustrated selective entry and transient transfection of targeted nanoparticles, consistent with the cytotoxicity results that indicated the synergistic effects of chemo–gene therapy. The results of animal studies proved the high antitumor efficiency of targeted nanoparticles with minimal tissue damage, which was in line with fluorescence and magnetic imaging results. The novel synthesized nanoparticles containing SPION@MSN-ZIF-8 were suitable for CRC theranostics, and the combined approach of chemo–gene therapy suppressed the tumor more effectively.
1. Introduction
The challenges in colorectal cancer (CRC) management due to limitations in traditional chemotherapy result in increased recurrence and mortality rates.1 Issues include the detrimental effects of anti-cancer drugs on normal tissues due to inadequate tumor targeting, the development of multidrug resistance (MDR) to standard chemotherapeutic agents, and the aggressive metastasis of CRC cells.2–4 To tackle these challenges, the integration of innovative therapeutic strategies has emerged as a promising solution.5 Gene therapy, in particular, offers several advantages such as targeted cancer cell destruction, disruption of multiple signaling pathways, and personalized treatment options based on individual genetic profiles.6 However, the widespread implementation of gene therapy is hindered by challenges related to effective gene delivery, long-term safety concerns, and the complex regulation of genes and immune response.7 Despite these obstacles, the combination of chemotherapy and gene therapy has garnered significant attention for its potential to synergistically enhance anti-tumor efficacy while minimizing the side effects.8 This powerful approach represents a noteworthy advancement in the field of cancer therapeutics, holding great potential in the fight against CRC.9The investigation of gene therapy via microRNA (miRNA, miR) targeting has shown promise in the field of cancer treatment. Researchers have explored two main approaches in miR-based gene therapy: (1) activation of tumor suppressor miRNAs and (2) inhibition of oncogenic miRNAs.10 Various blocking methods, including anti-miRNAs, miRNA sponges, miRNA masking, and small enabler molecules, have been used effectively to hinder the function of cancer-causing miRNAs like miR-21.11 Among the various strategies employed in miRNA-based cancer therapy, using lentiviral plasmids expressing anti-miRNAs has gained recognition as an effective approach.12 However, limited research has explored the potential of pmiRZIP-21, a lentiviral plasmid expressing anti-miR-21, in combination with co-delivery of chemotherapeutic drugs. Downregulation of miR-21 has been found to play a role in inducing apoptosis and inhibiting critical processes including drug resistance, proliferation, invasion, and migration through the targeting of important signaling pathways.13 Furthermore, miR-21 has demonstrated its capacity to inhibit tumor suppressor (TS) mRNAs, exerting precise control over their translation to TS proteins, underscoring its relevance to cancer progression.14,15 Nevertheless, a comprehensive understanding of the therapeutic potential of miRZIP-21 plasmid and its synergistic effects with chemotherapeutic drugs in CRC treatment remains necessary.
Despite advancements in miRNA-based therapies, limitations such as inefficient cellular transfer, poor stability, toxicity, and lack of targeted delivery have hindered their widespread applications. Therefore, the development of reliable delivery systems that fulfill these three properties is crucial: (1) protecting the genes from degradation by nucleases in the bloodstream and intracellular matrices, (2) facilitating the gene passage through the plasma membrane, and (3) ensuring that the delivery system is non-toxic. Recently, mesoporous nanoparticles have been widely used for gene therapy applications as non-viral or synthetic vectors.16
In recent years, there has been increasing interest in the development of theranostic systems with superparamagnetic iron oxide nanoparticles (SPIONs) through core–shell structures.17 SPIONs have emerged as valuable contrast agents for magnetic resonance imaging (MRI) due to their adjustable size (1–100 nm), biodegradability, and low toxicity. However, the half-life of SPIONs is influenced by surface interactions, which can be improved through their incorporation into mesoporous silica nanoparticles (MSNs).18 MSNs have emerged as a versatile drug delivery system for transferring various molecules, including anticancer drugs and genetic materials. Due to their safety, biodegradability, potential biocompatibility, and well-developed surface chemistry, MSNs are highly effective in encapsulating molecules and facilitating targeted delivery.18
Furthermore, zeolitic imidazolate frameworks (ZIFs), specifically ZIF-8, have shown promise in gene delivery applications.19 Using a biomimetic mineralization and co-precipitation process, ZIF-8 takes advantage of the biological affinity of macromolecules to metal ions and organic ligands in situ, allowing for efficient plasmid loading within its porous structure. ZIF-8 maintains the functionality of the biomolecule while safeguarding it within its narrow pore size distribution.20,21 Moreover, the ease of synthesis at room temperature facilitates efficient optimization of plasmid adsorption, and the pH-sensitive nature of ZIF-8 helps the controlled release of the cargo.22 By combining the unique capabilities of MSNs and ZIF-8, researchers have explored the development of MSN@ZIF-8 core–shell structures.23,24 This hybrid system allows for the integration of ZIF-8 as a pH-sensitive cap, enhancing the versatility and functionality of the formula in drug and gene delivery systems.
Nanoparticle-based drug delivery systems possess important features including the particles’ stability in the bloodstream, their impact on red blood cells, and the ability to evade recognition by the immune system, which account for their successful applications in vivo. To maximize the therapeutic impact of nanocarriers and their accumulation at tumor sites, it is crucial to specifically target cancer cells within the tumor microenvironment (TME).25 Polyethylene glycol (PEG) is a synthetic polymer that exhibits excellent suitability for biomedical applications due to its high solubility, biocompatibility, and tolerance.26 PEGs can be directly attached to drugs or loaded onto nanomaterials, to enhance the stability, solubility, and efficiency of the drug delivery systems.27 Additionally, a new generation of nanocarriers has been developed using ligands like aptamers. AS1411 aptamer, as a 26-nucleotide DNA aptamer with a G-quadruplex structure, specifically targets nucleolin (C23), a protein present in the nucleus and also distributed in the cytoplasm and on the cell surface.28 Nucleolin is often overexpressed and has a distinct location in rapidly multiplying cancerous cells, making it a promising target for AS1411 aptamer-mediated targeted drug delivery and cancer treatment.29–31
In this study, a multipurpose nanocarrier, SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt, was developed for targeted delivery of chemo–gene therapy and imaging agents. The nanocarrier was designed by combining an anticancer drug (epirubicin, EPI), a gene therapy agent (anti-miR-21 expressing plasmid), and a smart-release system (ZIF-8 cap) on MSNs containing SPIONs in their core. To enhance the biocompatibility and active targeting to CRC cells, PEG and AS1411 aptamer were functionalized onto the nanocarrier, respectively. Extensive characterizations were conducted to determine the size, morphology, and composition of the nanocarriers. The loading and release profiles of EPI and pDNA were assessed, along with the protective capabilities of the nanocarrier. Cellular uptake, gene transfection efficiency, and synergistic anti-cancer effects were evaluated in vitro using CRC and normal cell lines. In vivo experiments were performed on BALB/c mice bearing CRC tumors to investigate the effectiveness of the combined chemo–gene therapy approach, while fluorescence (FL) and magnetic resonance (MR) imaging were utilized to track the accumulation of nanoparticles in the tumor area (Scheme 1).
2. Experimental section
2.1. Materials
Fe3O4, tetraethylorthosilicate (TEOS), n-cetyl trimethyl ammonium bromide (CTAB), ammonium hydroxide (NH4OH), epirubicin (EPI), 2-methylimidazole (2-MIM), zinc nitrate hexahydrate (Zn(NO3)2·6H2O) and 4′,6-diamidino-2-phenylindole (DAPI) were obtained from Sigma-Aldrich. Heterobifunctional PEG polymer with amine, and maleimide terminal groups (H2N-PEG-MAL, MW: 2500 Da) was purchased from JenKem (USA). Fetal bovine serum (FBS), Dulbecco's modified Eagle's medium (DMEM), Roswell Park Memorial Institute 1640 (RPMI 1640) medium, and penicillin/streptomycin were purchased from Gibco (Scotland). Trypsin and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were obtained from Tinab Shimi (Iran). Fluorescein isothiocyanate (FITC)-annexin V with propidium iodide (PI) apoptosis detection kit was purchased from BioLegend (USA). Anti-miR-21-expressing plasmid (pmiRZip-21) was prepared from System Bioscience (SBI, USA), and was propagated in DH5α Escherichia coli and extracted using a HiPure Plasmid Maxiprep Kit (Invitrogen, Thermo Fisher Scientific, USA). The 26 mer AS1411 DNA aptamer [SH-5′-(GGTGGTGGTGGTTGTGGTGGTGGTGG)-3′] was synthesized by MicroSynth (Switzerland). Agarose powder, tris-acetate-EDTA (TAE) buffer, and DNA markers (50 bp and 1 kb) were purchased from DENAzist Asia (Iran). Ethidium bromide was obtained from SinaClon (Iran). Chemical reagents, absolute ethanol, and other solvents were purchased from Merck (Germany). Murine colon adenocarcinoma cells (C26) and mouse fibroblast cells (NIH/3T3) were obtained from Ferdowsi University of Mashhad, Iran, and cultured respectively in RPMI 1640 and DMEM medium supplemented with 10% FBS, at 37 °C in a 5% CO2 humidified incubator.2.2. Synthesis procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min), and the supernatant containing free EPI was collected. The amount of unloaded EPI was evaluated by assessing the absorbance of the supernatant at λ = 480 nm using a spectrophotometer (Eppendorf, Germany), and the corresponding concentration was calculated with the help of the standard curve equation of the drug. Eqn (1) and (2) were used to calculate drug encapsulation efficiency (EE%) and loading capacity (LC%).33,34
000g for 10 min), and the supernatant containing free EPI was collected. The amount of unloaded EPI was evaluated by assessing the absorbance of the supernatant at λ = 480 nm using a spectrophotometer (Eppendorf, Germany), and the corresponding concentration was calculated with the help of the standard curve equation of the drug. Eqn (1) and (2) were used to calculate drug encapsulation efficiency (EE%) and loading capacity (LC%).33,34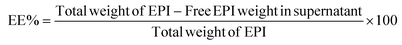 | (1) |
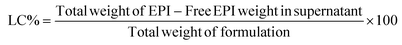 | (2) |
2.3. Physical characterization
Dynamic light scattering (DLS; Cordouan Technologies, France) and electrophoretic light scattering (ELS; CAD Instruments, France) methods were used to determine the particle size and zeta potential of nanocarriers in each step. Transmission electron microscopy (TEM; Philips EM208S 100KV, Netherland), field emission-scanning electron microscopy (FE-SEM; TESCAN MIRA, Czech Republic), and atomic force microscopy (AFM; BRUKER, USA) images were used to evaluate the size, structure and morphology of nanoparticles. The magnetic and optical properties of synthesized nanoparticles were analyzed using a vibrating sample magnetometer (VSM; Lake Shore Cryotronics, USA), and MR and FL imaging techniques. Furthermore, in order to collect the X-ray diffraction (XRD) patterns of the synthesized nanoparticles, a Multipurpose-Theta/Theta-high resolution diffractometer (PHILIPS_PW1730, Netherlands) was used. Specific surface areas, pore size distribution, and pore volume of SPION@MSN and SPION@MSN-EPI/pDNA-ZIF-8 were investigated at 77 K by Brunauer–Emmett–Teller (BET) and Barrett–Joyner–Halenda (BJH) methods, using a BELSORP Mini II instrument. Thermogravimetric analysis (TGA; Q600, USA) with a heating rate of 20 °C min−1 in the air was used to determine the thermal properties of SPION, SPION@MSN, and SPION@MSN-EPI/pDNA-ZIF-8-PEG. Electrophoresis was carried out to validate the attachment of the aptamer to the surface of nanoparticles. To this aim, each sample, namely DNA ladder, standalone aptamer, SPION@MSN-ZIF-8-PEG, and SPION@MSN-ZIF-8-PEG-Apt, was scrutinized using a 2% agarose gel that had an ethidium bromide concentration of 0.3 μg mL−1. The elemental compositions of each step of synthesis (Si, Fe, Zn, C, O, N, and P) were evaluated by energy dispersive X-ray spectroscopy (EDX; TESCAN MIRA, Czech Republic). Finally, Fourier-transform infrared (FT-IR) spectra for each step of the nanocarriers synthesis were analyzed to confirm the presence of functional groups using the AVATAR 370 FT-IR spectrometer (Therma Nicolet spectrometer, USA).2.4. In vitro cargo release
2.5. Binding affinity and protection for pDNA
In the next step, the protection of the pmiRZip-21 plasmid from DNase I degradation was evaluated.22 Each of the SPION@MSN-pDNA-ZIF-8-PEG-Apt, SPION@MSN-pDNA-ZIF-8-PEG, and SPION@MSN-pDNA-ZIF-8 nanocarriers (containing 0.5 μg plasmid) was dispersed in 25 μL PBS solution. Then 1 μL DNase I (2 U μL−1) was added to each solution, followed by incubation at 37 °C for 30 min. To inactivate DNase I, 1 μL EDTA (100 mM) was added, and then centrifugation (6000g for 5 min) was performed. The collected sediments were resuspended in 10 μL nuclease-free aqueous solution for agarose gel electrophoresis. In this regard, the plasmid extraction method with the help of heparin was used in order to evaluate the protective quality of DNase I-treated nanocarriers.8,21 For this purpose, similar to the previous step, the sediment collected after treatment with DNase I was resuspended in 25 μL PBS. Then 10 μL EDTA (100 mM) was added to the mixture to stop the enzyme activity and dissolve ZIF-8, completely. Next, 8 μL of sodium heparin solution (10 μg μL−1) was added to the mixture of nanocarriers for plasmid extraction and kept for 2 h at room temperature in the dark. Finally, 10 μL of samples were loaded in each well of 0.8% w/v agarose gel, and electrophoresis (100 V for 40 min) was performed. It should be noted that the protected plasmids in nanocarriers were treated and evaluated in the same conditions as the free plasmids.
2.6. Hemolysis assay
In order to investigate the biocompatibility of the prepared nanocarriers through their destructive effect in the vicinity of red blood cells (RBCs), the hemolysis assay was performed. To achieve this, first, a human blood sample from a healthy donor was stabilized with heparin, and the red blood cells were separated by centrifugation (1500g for 10 min at 4 °C). Then the resulting cell sediment was washed and diluted with cold PBS (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10). Afterward, 0.1 mL of diluted cells was mixed with 0.9 mL of SPION@MSN, SPION@MSN-EPI-ZIF-8, and SPION@MSN-EPI-ZIF-8-PEG at equivalent concentrations of SPION@MSN ranging from 25–400 μg mL−1. Moreover, 0.9 mL of PBS/DIW was added to diluted cells (0.1 mL) as negative/positive control, respectively. Finally, all the mixtures were kept at 37 °C in a shaker incubator for 4, 12, and 24 h at a speed of 100 rpm followed by centrifugation (2500g for 1 min) after the specified time periods. The absorption of released hemoglobin at λ = 541 nm was evaluated by a UV/Vis spectrophotometer and eqn (3) was used to calculate the percentage of hemolysis.43
10). Afterward, 0.1 mL of diluted cells was mixed with 0.9 mL of SPION@MSN, SPION@MSN-EPI-ZIF-8, and SPION@MSN-EPI-ZIF-8-PEG at equivalent concentrations of SPION@MSN ranging from 25–400 μg mL−1. Moreover, 0.9 mL of PBS/DIW was added to diluted cells (0.1 mL) as negative/positive control, respectively. Finally, all the mixtures were kept at 37 °C in a shaker incubator for 4, 12, and 24 h at a speed of 100 rpm followed by centrifugation (2500g for 1 min) after the specified time periods. The absorption of released hemoglobin at λ = 541 nm was evaluated by a UV/Vis spectrophotometer and eqn (3) was used to calculate the percentage of hemolysis.43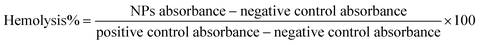 | (3) |
2.7. In vitro cellular uptake
Comparison of nanoparticles uptake into cancer and normal cells through the fluorescence property of EPI was investigated by two quantitative and qualitative methods. In the first step, equal numbers (2 × 105 cells per well) of C26 and NIH/3T3 cells were cultured in 6-well plates. After 24 h of incubation and cell network formation, cells were treated with free drug (Free EPI), non-targeted (SPION@MSN-EPI/pDNA-ZIF-8-PEG) and targeted (SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt) nanocarriers with the same concentration (equivalent to 6.25 μg mL−1 EPI, 2.5 μg mL−1 pDNA and 25 μg mL−1 NPs) for 6 h. In order to quantitatively check the entry of nanoparticles by flow cytometry, the medium was removed and cells were washed with PBS. After collecting the cells with trypsin–EDTA solution, centrifugation (400g for 15 min) was performed; the supernatant was carefully removed and cells were resuspended in 300 μL of cold PBS. Finally, the fluorescence emission of cells in the FL2 channel was evaluated using the BD FACSCalibur flow cytometer and the results were analyzed with FlowJo V10 software. Furthermore, cellular uptake was evaluated qualitatively by using a fluorescence microscope. In the same procedure, after 6 h of treating the cells with the mentioned concentrations and washing with PBS, cells were fixed with formalin (10%) for 20 min at 4 °C and stained with DAPI solution (1.5 μg mL−1) in the dark for 10 min. Finally, after washing the cells with PBS, the fluorescence emissions were examined using a fluorescence microscope (Olympus BX51, Japan).2.8. In vitro transfection
Transient transfection in eukaryotic cells is a way to introduce a gene into a cell without entering the host's genome which can lead to high and temporary expression of the target gene in a short period of time. Since the use of targeted nanocarriers can greatly facilitate the entry of the gene into the cell, in this study, the efficiency of the targeted nanocarriers containing plasmid in uptake and expression of the gene was evaluated by means of the copGFP fluorescent marker. First, two cell lines, C26 and NIH/3T3, were cultured in 24-well plates with a density of 5 × 104 cells per well and incubated for 24 h. When the cell network formed, the cells were treated with non-targeted (SPION@MSN-pDNA-ZIF-8-PEG) and targeted (SPION@MSN-pDNA-ZIF-8-PEG-Apt) nanoparticles containing plasmid (equivalent concentration to 2.5 μg mL−1 pDNA and 25 μg mL−1 NPs) for 6 h. After the medium exchange, the cells were incubated for another 18 h to check the transient transfection via copGFP expression.44 Finally, cells were evaluated qualitatively and quantitatively via green fluorescence emission of copGFP by using a fluorescence microscope and FL1 channel of the BD FACSCalibur flow cytometer, respectively.2.9. MTT assay
In order to investigate the synergistic anti-cancer effects of nanoparticles containing EPI and anti-miR-21-expressing plasmid, the MTT assay was performed. First, two cell lines, C26 as cancer cells with high expression of nucleolin and NIH/3T3 as normal cells with low expression of nucleolin, were seeded at a density of 6 × 103 and 8 × 103 cells per well in 96-well plates, respectively. After 24 h incubation, when the confluency reached about 70%, cells were treated in five groups, namely Free EPI, SPION@MSN-pDNA-ZIF-8-PEG, SPION@MSN-EPI-ZIF-8-PEG, SPION@MSN-EPI/pDNA-ZIF-8-PEG and SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt at equivalent concentrations of EPI (1.56 to 50 μg mL−1) and pDNA (0.62 to 20 μg mL−1) for 6 h. At the end of the treatment period, the medium was changed and the cells were incubated for 24, 48, and 72 h post-treatment. After each time interval, 20 μL of MTT (5 mg mL−1) was added to each well and incubated in the dark for 4 h. In order to dissolve the formazan crystals produced by MTT reduction, after draining the medium, 150 μL of DMSO was added to each well. Finally, the absorption of purple solutions was determined by an ELISA reader (Awareness Technology, Inc., USA) at 540 nm, and the half-maximal inhibitory concentration (IC50) and the corresponding graphs for both cell lines were analyzed using GraphPad Prism version 9.3.0 software.2.10. Studying cell death mechanism
Flow cytometry with the help of the FITC-annexin V/PI apoptosis detection kit was used to investigate the effects of targeted nanocarriers containing EPI drug and anti-miR-21-expressing plasmid on increasing programmed cell death (apoptosis). Since C26 cancer cells over-express both nucleolin as a receptor for AS1411 aptamer and miR-21 as an oncogene involved in drug treatment pathways, these cells were used to evaluate the effect of prepared nanocarriers in chemo–gene therapy. After 24 h of cell seeding (2 × 105 cells per well in 6-well plates), the cells were treated in five groups, namely Free EPI, SPION@MSN-pDNA-ZIF-8-PEG, SPION@MSN-EPI-ZIF-8-PEG, SPION@MSN-EPI/pDNA-ZIF-8-PEG and SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt at equivalent concentrations of EPI (6.25 μg mL−1) and pDNA (2.5 μg mL−1) for 6 h. Subsequently, 24 h post-treatment cells were collected and stained according to the protocol of the FITC-annexin V/PI kit. Finally, the cell death mechanism was assessed by flow cytometry and the results were analyzed using FlowJo V10 software.2.11. In vivo anti-tumor efficacy and biosafety
In order to induce colon subcutaneous tumors from C26 cells, female BALB/c mice (5–7 weeks old and inbred in an animal house at FUM) weighing about 20 g were used in accordance with the guidelines approved by the Animal Ethics Committee of Ferdowsi University of Mashhad (IR.UM.REC.1401.040). Then, 1 × 106 C26 cells per mouse were suspended in 100 μL PBS and injected into the right flank of mice, subcutaneously. When the induced tumors reached approximately 100 mm3 in volume, mice were randomly divided into 6 groups (n = 5 per each group). The treatment groups, namely PBS, Free EPI, SPION@MSN-pDNA-ZIF-8-PEG, SPION@MSN-EPI-ZIF-8-PEG, SPION@MSN-EPI/pDNA-ZIF-8-PEG and SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt (EPI and pDNA dosage were set as 5 mg kg−1 and 2 mg kg−1, respectively) were injected intravenously through tail vein in two doses at an interval of 3 days. Tumor volume was measured every other day for 15 days post-treatment based on (length × width × height)/2 formula by measuring the diameters of the tumor with a digital caliper (Mitutoyo, Japan). Furthermore, to evaluate biosafety and possible side effects in treated mice, body weight and liver weight were measured during and at the end of the study, respectively. Finally, mice were euthanized and the major organs including heart, lung, spleen, kidney, and liver, and also tumor tissues, were isolated and pathologically evaluated through H&E staining. The biosafety of PEGylated nanoparticles (SPION@MSN-EPI/pDNA-ZIF-8-PEG) was evaluated through biochemical assays and compared with Free EPI (administered at a dose of 5 mg kg−1). Three days after intravenous administration of PBS (control group), Free EPI, and SPION@MSN-EPI/pDNA-ZIF-8-PEG to healthy mice, blood samples were drawn from the hearts of animals (n = 3). The plasma samples were prepared after 20 min centrifugation at 4000g, and the levels of albumin (ALB), alkaline phosphatase (ALP), and blood urea nitrogen (BUN) were determined using biochemical assay kits.452.12. Ex vivo FL imaging
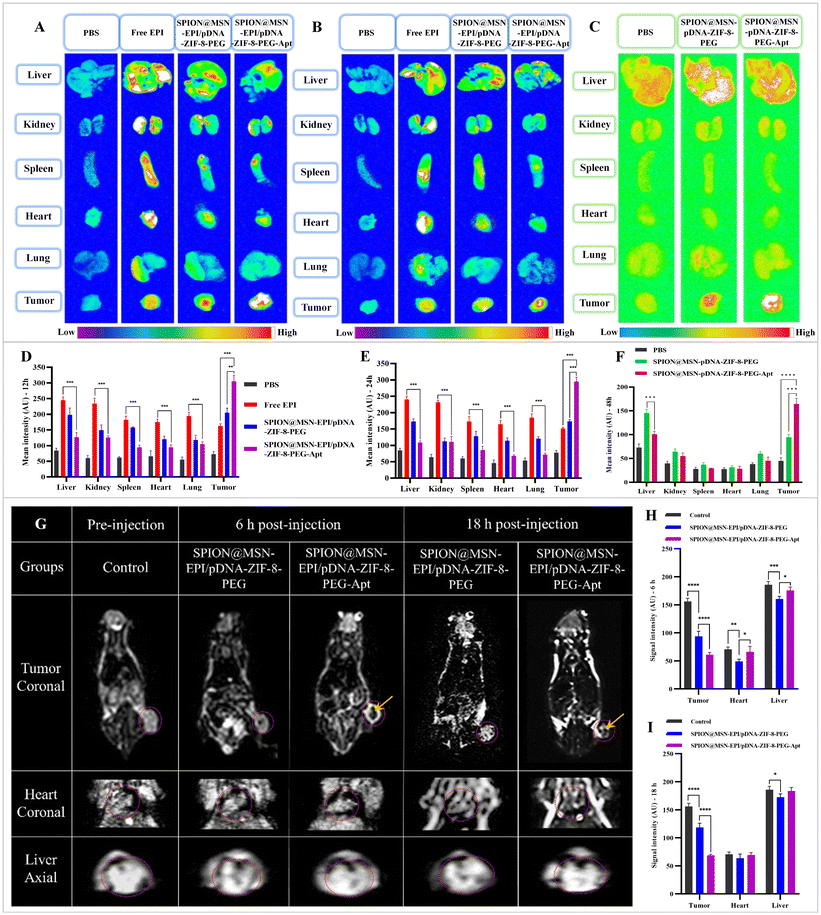
To investigate the biodistribution of free drug, targeted and non-targeted nanoparticles in the body of mice, the main organs and tumor were isolated, and associated fluorescence emissions were investigated in both qualitative and quantitative manners. To this aim, when the tumor volume reached about 200 mm3, food restriction was implemented for 12 h to reduce the effect of food fluorescence. Then, each of the PBS, Free EPI, SPION@MSN-EPI/pDNA-ZIF-8-PEG, and SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt groups was treated with one dose of intravenous injection (equivalent dose of EPI was set as 5 mg kg−1). After 12 and 24 h of injection, the mice were euthanized with CO2, and the main organs and tumor tissues were dissected. Finally, the mean fluorescence intensity (MFI) was qualitatively and quantitatively evaluated using the KODAK IS in vivo imaging system (λexcitation = 470 nm and λemission = 600 nm).Similarly, 48 h post-injection of PBS, SPION@MSN-pDNA-ZIF-8-PEG and SPION@MSN-pDNA-ZIF-8-PEG-Apt groups (equivalent to 2 mg kg−1 pDNA), ex vivo FLI was measured to check the expression of copGFP associated with delivered plasmid. Subsequently, the MFI of the tumor and various organs was evaluated qualitatively and quantitatively using the KODAK IS in vivo imaging system (λexcitation = 470 nm and λemission = 520 nm).
2.13. In vivo MR imaging
The magnetic efficacy of SPION (in the core of the nanocarrier) was evaluated in vivo by magnetic resonance imaging (MRI). For this purpose, BALB/c mice bearing C26 tumors with a volume of 200 mm3 were prepared and all animals in separate groups related to targeted and non-targeted nanoparticles were anesthetized with ketamine-xylazine, fixed with medical tapes, and imaged with MRI to minimize false results in the final images. Then, during two periods of 6 and 18 h after the intravenous injection of nanoparticles (SPION@MSN-EPI/pDNA-ZIF-8-PEG and SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt) with a dose of 20 mg kg−1, MRI was performed based on T2 contrast, similar to the first procedure. Finally, the differential intensity in the tumor area was qualitatively and quantitatively analyzed using DICOM viewer software (Medixant. RadiAnt DICOM Viewer [Software], Version 2020.2. Jul 19, 2020. https://www.radiantviewer.com). To obtain MR T2-weighted coronal and axial images under a 1.5 T MRI scanner (Ingenia CX; Philips, Netherlands), the following imaging parameters were used: protocol = turbo field echo (TSE); repetition time (TR) = 2800 ms; echo time (TE) = 34 ms; and slice thickness = 0.5 mm; resolution = 256 pixels; Number of Averages = 8; Scanning Sequence = SE; Flip Angle = 90; Columns = 240; Pixel Bandwidth = 957; Rows = 240; Echo Number (s) = 1; and Echo Train Length = 141.2.14. Statistical analysis
Statistical data analysis was carried out with GraphPad Prism version 9.3.0 (GraphPad Software, San Diego, CA). The results were reported as mean ± SD or SEM and considered to be statistically significant at p < 0.05.3. Results
3.1. Successful synthesis of NPs was demonstrated by characterization results
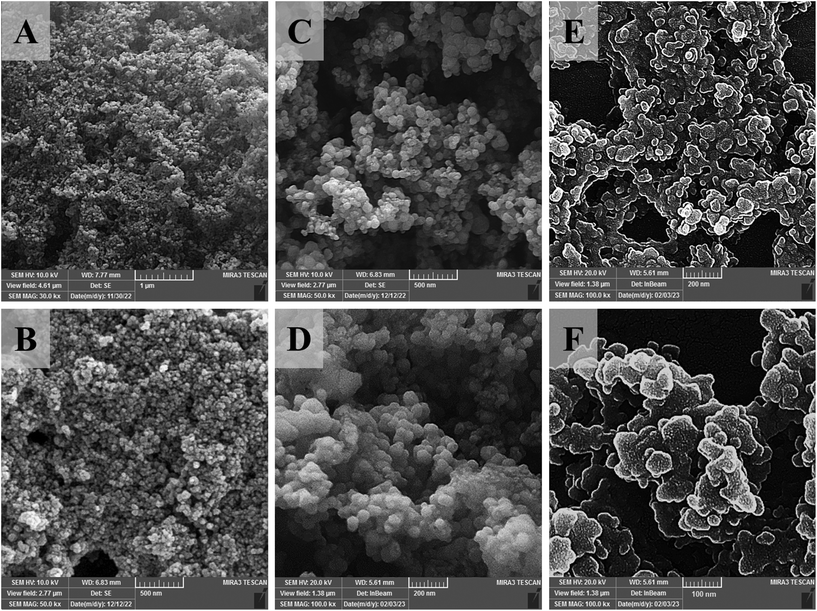
The structural characteristics of nanoparticles, including average particle size and zeta potential, were calculated in each step of synthesis by DLS and ELS methods (Table 1), and the corresponding results indicate the expected alterations. In addition, the spherical structure of SPION, SPION@MSN, and SPION@MSN-EPI/pDNA-ZIF-8 nanoparticles was estimated to be about 15, 25, and 45 nm on average by TEM microscopic images (Fig. S1A–F†). Furthermore, the spherical structure and uniform morphology of SPION@MSN, SPION@MSN-EPI/pDNA-ZIF-8, and SPION@MSN-EPI/pDNA-ZIF-8-PEG nanoparticles were investigated using FE-SEM (Fig. 1A–F). Besides the particle size growth of nanoparticles, the surface modifications made by ZIF-8 and PEG are also evident in the AFM results (Fig. S2A and B†). The size distribution map generated by DLS accurately indicates the intuitive size of nanoparticles (Fig. S3†).| Samples | Particle size (nm) | Polydispersity index (PDI) | Zeta potential (mV) |
|---|---|---|---|
| Particle size was measured by dynamic light scattering (DLS). Data are expressed as mean ± SD. Abbreviations: SPION, superparamagnetic iron oxide nanoparticle; MSN, mesoporous silica nanoparticle; EPI, epirubicin; pDNA, pmiRZip-21 plasmid; ZIF-8, zeolitic imidazolate framework-8; PEG, polyethylene glycol; Apt, Aptamer. | |||
| SPION | 15.26 ± 1.63 | 0.7 ± 0.32 | −20.21 ± 1.27 |
| SPION@MSN | 25.35 ± 1.21 | 0.4 ± 0.22 | −19.52 ± 2.12 |
| SPION@MSN-EPI | 27.43 ± 1.45 | 0.54 ± 0.30 | −9.23 ± 2.84 |
| SPION@MSN-EPI-ZIF-8 | 45.54 ± 2.31 | 0.34 ± 0.20 | +7.14 ± 4.32 |
| SPION@MSN-EPI/pDNA-ZIF-8 | 46.03 ± 2.74 | 0.3 ± 0.23 | +6.27 ± 3.64 |
| SPION@MSN-EPI/pDNA-ZIF-8-PEG | 61.24 ± 4.71 | 0.4 ± 0.14 | −13.45 ± 3.50 |
| SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt | 68.34 ± 4.29 | 0.3 ± 0.08 | −21.16 ± 4.73 |
The magnetic properties of the backbone nanoparticle (SPION@MSN) were confirmed in the presence of a magnetic field and also by MR imaging (Fig. S4A†). As shown in Fig. S4B,† the comparison between the magnetic properties of the backbone (SPION@MSN) and coated nanoparticles (SPION@MSN-ZIF-8-PEG), performed by using the VSM method, revealed a decrease in the magnetic saturation value (−5.03 emu g−1), potentially due to the increased size of the nanoparticles. Moreover, the paramagnetic properties of SPION@MSN and SPION@MSN-ZIF-8-PEG were clearly confirmed through an increase in their saturation magnetization (Ms) under a strong magnetic field. The efficacy of nanoparticles containing Fe and their ability as contrast agents in MRI were confirmed through the introduction of equivalent serial dilutions of nanoparticles. The impact of nanoparticle concentration on MRI was depicted using T2 weighted imaging, qualitatively and quantitatively (Fig. S4A and C†). The R2 relaxation or the inverse relaxation time (1/T2 (s−1)) demonstrated a direct relationship with all three nanoparticle concentrations as shown by the slope of the graph (Fig. S4C†). According to these results, hybridized nanoparticles containing Fe can be used as a differentiating factor in MR imaging. Fluorescence imaging (Fig. S4D†) was utilized to evaluate the radiation efficiency of EPI, SPION@MSN-EPI, and SPION@MSN-EPI-ZIF-8 fluorescence compounds. The findings confirmed that the fluorescence properties of these compounds were concentration-dependent. Quantitative analysis of FLI illustrated a direct relationship between the intensity of radiation fluorescence and the concentration of the compounds, which is further supported by the linear relationship with varying slopes (Fig. S4E†).
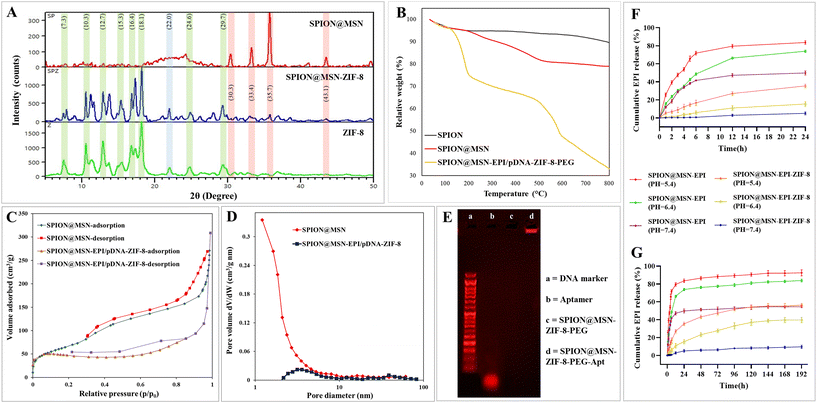
In the XRD analysis of the SPION@MSN-ZIF-8 hybrid nanoparticles shown in Fig. 2A, distinct peaks at 2θ values of 7.3°, 10.3°, 12.7°, 15.3°, 16.4°, 18.1°, 24.6°, and 29.5°, which are not present in the SPION@MSN pattern, correspond to the crystallographic planes specific to ZIF-8. The absence of these peaks in the SPION@MSN spectrum supports the successful coating of MSNs with ZIF-8, confirmed by the referenced literature.23,35 Meanwhile, the broad peak at 2θ = 22.6° in the SPION@MSN spectrum is attributed to the amorphous nature of silica within the MSN component, indicating its lack of crystalline order. Furthermore, the shared peaks at 2θ values of 30.3°, 33.4°, 35.7°, and 43.1° in both the SPION@MSN and SPION@MSN-ZIF-8 spectra are assigned to the iron oxide Fe3O4 component, confirming the presence of a magnetite core in the nanoparticles after coating with ZIF-8.37
Thermogravimetric analysis (TGA) illustrated the thermal stability, the amount of conjugated organic groups, and the thermal fracture pattern of nanoparticles (Fig. 2B). The highest weight loss (∼67%) corresponds to SPION@MSN-EPI/pDNA-ZIF-8-PEG, which has the highest organic composition compared with SPION (∼10%) and SPION@MSN (∼20%). In addition, the weight loss of SPION@MSN-EPI/pDNA-ZIF-8-PEG in the range of 500 °C is related to the decomposition of the imidazole groups of the ZIF-8 layer.35 As shown in Fig. 2C, N2 adsorption/desorption isotherms confirmed the isotherm type IV and II (H1 and H3 hysteresis loop) of SPION@MSN and SPION@MSN-EPI/pDNA-ZIF-8 and their mesoporous structures, respectively. Additionally, based on BET and BJH results, surface area, total pore volume, and pore diameter were calculated as 538.65 m2 g−1, 4.25 cm3 g−1, and 2.84 nm for SPION@MSN and 174.44 m2 g−1, 0.47 cm3 g−1, and 2.13 nm for SPION@MSN-EPI/pDNA-ZIF-8, respectively (Fig. 2C and D). As shown in Fig. 2E, the bright band of SPION@MSN-ZIF-8-PEG-Apt in the well compared with SPION@MSN-ZIF-8-PEG demonstrated the successful aptamer conjugation on the nanoparticle surface, while the bright band of free aptamer (with a length of 26 bp) below 50 bp size marker showed the expected molecular weight of the AS1411 aptamer. The encapsulation efficiency (EE%) and drug loading capacity (LC%) were calculated as 67% ± 1.71 and 25% ± 1.32, respectively. Furthermore, the intelligent EPI release was indicated in acidic pH environments. A rapid EPI release in the first hours even at neutral pH was shown in the drug release pattern of uncoated nanoparticles, while coated nanoparticles had minimal drug leakage (∼8%) at neutral pH after 8 days (Fig. 2F and G).
EDX spectra illuminated the elemental composition and chemical purity of nanoparticles in each step of the synthesis (Fig. S5A†) as the weight percentage (W%) of main elements (Si, Fe, Zn, C, O, N, and P), and the results are summarized in Table 2. The analysis of the elements and their distribution in SPION@MSN-EPI/pDNA-ZIF-8 nanoparticles by mapping method confirmed the existence and uniform distribution of the main elements in the structure (Fig. S5B†).
| Samples | Si | Fe | Zn | C | O | N | P |
|---|---|---|---|---|---|---|---|
| The data are represented as weight percentage (W%). Abbreviations: SPION, superparamagnetic iron oxide nanoparticle; MSN, mesoporous silica nanoparticle; EPI, epirubicin; pDNA, pmiRZip-21 plasmid; ZIF-8, zeolitic imidazolate framework-8; PEG, polyethylene glycol. | |||||||
| SPION@MSN | 35.21 | 2.58 | 0 | 6.63 | 55.58 | 0 | 0 |
| SPION@MSN-EPI-ZIF-8 | 2.41 | 0.15 | 9.11 | 39.64 | 12.61 | 36.08 | 0 |
| SPION@MSN-EPI/pDNA-ZIF-8 | 5 | 0.31 | 11.14 | 38.51 | 14.94 | 29.96 | 0.14 |
| SPION@MSN-EPI/pDNA-ZIF-8-PEG | 3.21 | 0.52 | 7.70 | 37.73 | 25.12 | 25.28 | 0.45 |
FT-IR spectroscopy was used to investigate and compare the functional groups of nanoparticles at each stage of synthesis (Fig. S6†). The obtained spectrum of SPION@MSNs represented the characteristic peaks related to Fe–O, Si–O–Si, and Si–O bonds located at 570, 460, 800, and 1080 cm−1, respectively.46,47 In addition, the presence of peaks at 1630 and 3410 cm−1 was ascribed to the vibrational band of O–H on the surface of SPION@MSNs.48,49 After loading the EPI drug in SPION@MSNs, the stretching vibration of the CH2 group at 2944 cm−1 and its bending vibration at 1400 cm−1 are evident.50 In addition, more absorption peaks related to hydroxyl (O–H) and carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) groups as well as the presence of NH2 bending vibration appeared in the region of 3410, 1619, and 1580 cm−1 of the SPION@MSN-EPI spectrum, respectively.50,51 After coating with the ZIF-8 layer, the peaks located in the region of 420 cm−1 were attributed to the stretching vibration of Zn–N groups. Moreover, the stretching vibrations of the C–N related to imidazole groups (2-MIM) appeared at 1145 and 995 cm−1, which indicated the successful incorporation of the ZIF-8 layer on the surface of the backbone.52 In this regard, the presence of 1307 and 1400 cm−1 vibrations was attributed to amine and methyl groups in the imidazole ring, respectively.53 In addition, the appearance of a new peak in the area of 3410 cm−1 in nanoparticles containing plasmid can be related to N–H or O–H groups of nucleic acid. The successful binding of heterofunctional PEG (H2N-PEG-MAL) to the surface of SPION@MSN-EPI/pDNA-ZIF-8 was confirmed by the presence of peaks at 1600 and 3400 cm−1 ascribed to N–H or NH2 groups and new peaks at 2929 and 2869 cm−1 corresponding to CH2 groups of PEG.54
O) groups as well as the presence of NH2 bending vibration appeared in the region of 3410, 1619, and 1580 cm−1 of the SPION@MSN-EPI spectrum, respectively.50,51 After coating with the ZIF-8 layer, the peaks located in the region of 420 cm−1 were attributed to the stretching vibration of Zn–N groups. Moreover, the stretching vibrations of the C–N related to imidazole groups (2-MIM) appeared at 1145 and 995 cm−1, which indicated the successful incorporation of the ZIF-8 layer on the surface of the backbone.52 In this regard, the presence of 1307 and 1400 cm−1 vibrations was attributed to amine and methyl groups in the imidazole ring, respectively.53 In addition, the appearance of a new peak in the area of 3410 cm−1 in nanoparticles containing plasmid can be related to N–H or O–H groups of nucleic acid. The successful binding of heterofunctional PEG (H2N-PEG-MAL) to the surface of SPION@MSN-EPI/pDNA-ZIF-8 was confirmed by the presence of peaks at 1600 and 3400 cm−1 ascribed to N–H or NH2 groups and new peaks at 2929 and 2869 cm−1 corresponding to CH2 groups of PEG.54
3.2. pDNA was well encapsulated in NPs
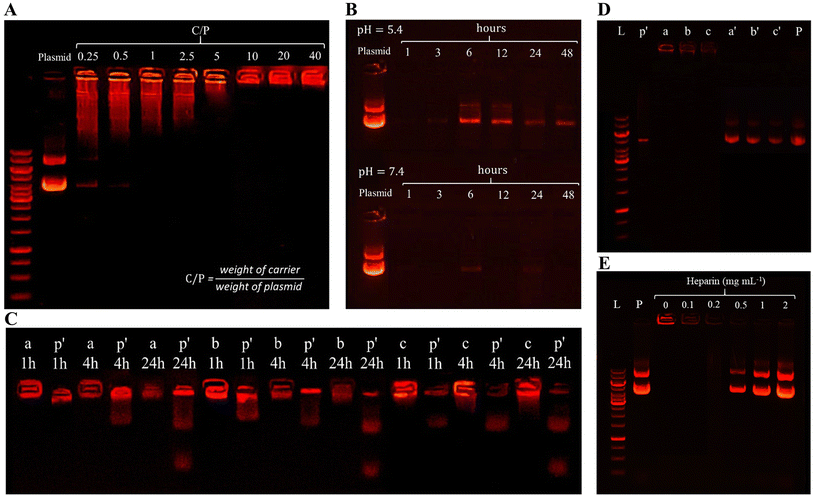
Preserving the integrity of plasmid DNA against enzymatic degradation is a critical aspect of gene delivery. To assess the stability of the pmiRZip-21 plasmid loaded in various nanocarriers (SPION@MSN-pDNA-ZIF-8-PEG-Apt, SPION@MSN-pDNA-ZIF-8-PEG, and SPION@MSN-pDNA-ZIF-8), an electrophoresis method was employed to evaluate DNase I degradation. The initial results of DNase I treatment, as depicted in Fig. 3D, revealed a distinct light band corresponding to the nanocarriers (denoted as a, b, and c) in their respective wells. The presence of this band suggests the protection of encapsulated plasmids from enzymatic digestion, while the weaker band corresponding to the free plasmid (p) exhibits a high degree of digestion. The pattern of plasmid migration, following extraction from the nanoparticles using heparin (represented as a′, b′, and c′), closely resembles the plasmid pmiRZip-21 band (P), indicating the quality of the loaded plasmids after treatment. Furthermore, the electrophoretic analysis of the SPION@MSN-pDNA-ZIF-8 nanocarrier treated with varying concentrations of sodium heparin demonstrated that a minimum concentration of 0.5 mg mL−1 of sodium heparin was sufficient for complete isolation of the plasmid from the nanocarrier, consistent with the migration pattern of the free plasmid (P) (Fig. 3E).
3.3. Coating with ZIF-8 and PEG resulted in increased biocompatibility
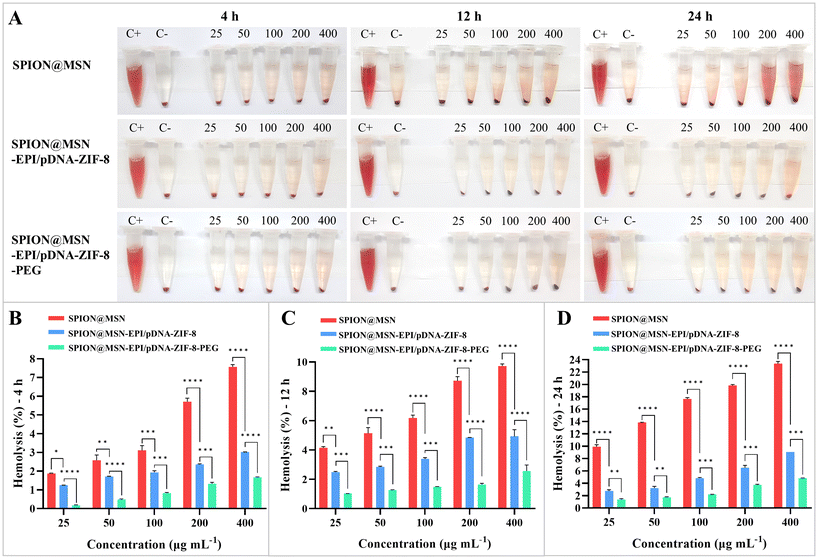
The biocompatibility of the nanocarriers coated with ZIF-8 and PEG, at equivalent concentrations based on SPION@MSNs (25–400 μg mL−1), was investigated by exposing SPION@MSN, SPION@MSN-EPI-ZIF-8, and SPION@MSN-EPI-ZIF-8-PEG to blood cells. The results depicted in Fig. 4A–D demonstrate a noticeable decrease in the detrimental effects of the coated nanocarriers at all three observed time periods (4, 12, and 24 h). Notably, the hemolysis results highlighted differences in the impact of different concentrations (ranging from 25 to 400 μg mL−1) of SPION@MSN and SPION@MSN-EPI-ZIF-8 at different time intervals. However, the percentage of hemolysis exhibited by PEG-coated nanoparticles was the lowest (1.6, 2.5, and 4.8% at 4, 12, and 24 h, respectively) and less than the hemolysis threshold (10%).3.4. Targeted NPs had higher uptake and transfection efficiency in CRC cells
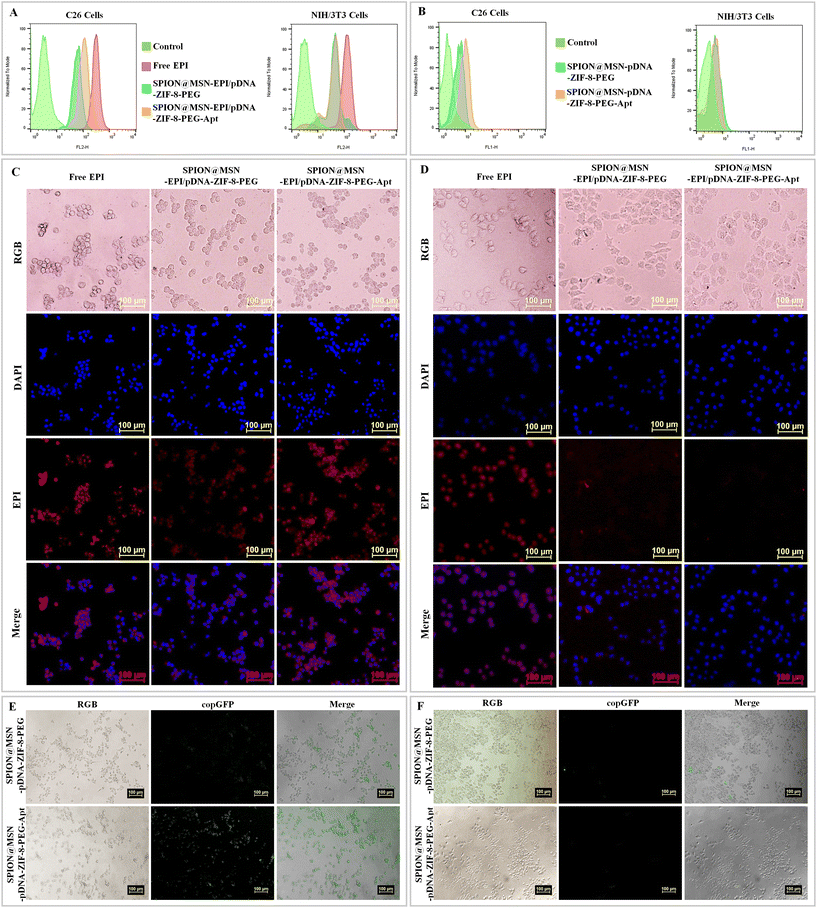
In this study, the first cellular assessment focused on investigating the cellular uptake of AS1411 aptamer-targeted nanoparticles by C26 (nucleolin positive) and NIH/3T3 (nucleolin negative) cells. The results of treating cells with free drug (Free EPI), non-targeted nanoparticles (SPION@MSN-EPI/pDNA-ZIF-8-PEG), and targeted nanoparticles (SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt) were evaluated using both quantitative (flow cytometry) and qualitative (fluorescence microscopy) methods. The flow cytometry results indicated the enhanced cellular uptake of targeted nanoparticles rather than non-targeted ones by C26 cells, as indicated by the fluorescence emission of the EPI drug (Fig. 5A). In contrast, there was no significant difference between these groups in NIH/3T3 cells. Moreover, the excess entrance of Free EPI was observed in both cancer and normal cells, suggesting the non-selective entry of the drug. Fluorescence microscopy results supported these findings, depicting greater red fluorescence in the nucleus and cytoplasm of C26 cells compared with NIH/3T3 cells after treatment with targeted nanoparticles (Fig. 5C and D). Based on these observations, the active internalization of targeted nanoparticles into C26 cancer cells was attributed to the interaction between the nucleolin receptor on these cells and the AS1411 aptamer attached to the surface of the targeted nanoparticles.To investigate and compare the efficiency of the targeted nanocarrier for plasmid delivery into cells, we evaluated the transient transfection via the expression of copGFP as a fluorescence marker with both quantitative and qualitative methods. Fig. 5B illuminates the flow cytometry results, indicating that C26 cells treated with targeted nanoparticles (SPION@MSN-pDNA-ZIF-8-PEG-Apt) show higher fluorescence emission compared with non-targeted nanoparticles. However, no significant difference in fluorescence emission was observed in NIH/3T3 cells treated with targeted and non-targeted nanoparticles. Furthermore, fluorescence microscopy results revealed a significant increase in green fluorescence attributed to copGFP in C26 cells treated with targeted nanoparticles compared with non-targeted ones (Fig. 5E). Conversely, NIH/3T3 cells exhibited minimal fluorescence emission upon treatment with targeted nanoparticles (Fig. 5F). The results not only confirm the successful expression of the copGFP gene in C26 cells through green fluorescence emission but also demonstrate the selective entry of targeted nanoparticles into C26 cancer cells via the active interaction between the AS1411 aptamer and the nucleolin receptor on these cells.
3.5. Synergistic cytotoxicity of targeted NPs was confirmed by MTT assay
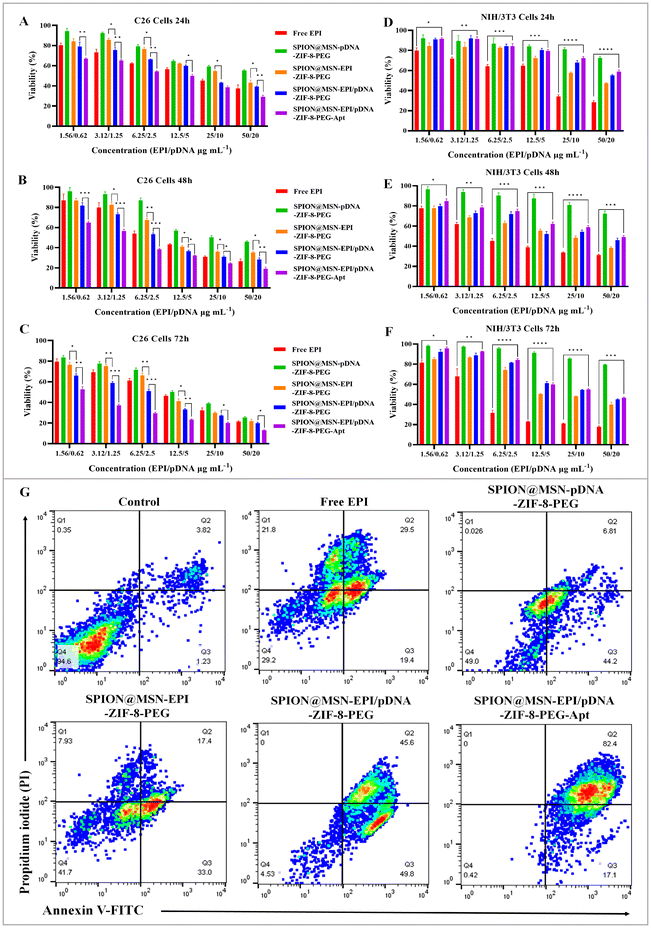
The synergistic toxicity of the EPI drug and the plasmid expressing anti-miR-21 was evaluated using the MTT assay. The results of treatment with non-targeted and targeted nanoparticles containing the drug and plasmid on C26 cells demonstrated higher cytotoxicity of SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt nanoparticles compared with nanoparticles without the aptamer, indicating enhanced and selective uptake of targeted nanoparticles into these cells (Fig. 6A–C). Conversely, targeted nanoparticles exhibited significantly (p < 0.0001) lower toxicity compared with Free EPI in nucleolin-negative NIH/3T3 cells (Fig. 6D–E). Hybrid nanoparticles carrying both chemo–gene therapy agents (SPION@MSN-EPI/pDNA-ZIF-8-PEG) demonstrated greater toxicity on C26 cancer cells compared with nanoparticles containing only one of these agents (either the drug or plasmid). Notably, the observed increase in toxicity over time suggests the controlled release of the drug and plasmid from the nanoparticles upon entry into cancer cells, thereby exerting prolonged effects. The IC50 values of the groups treated based on equivalent concentrations of EPI (μg mL−1) at three time points are summarized in Table 3. It is worth mentioning that nanoparticles lacking the drug and plasmid (SPION@MSN-ZIF-8-PEG) did not exhibit significant toxicity in either the C26 or NIH/3T3 cell lines (results not reported).| Treatments | IC50 (μg mL−1) ± SD (C26 cells) | IC50 (μg mL−1) ± SD (NIH/3T3 cells) | ||||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| Data are expressed as mean ± SD, n = 3. Abbreviations: SPION, superparamagnetic iron oxide nanoparticle; MSN, mesoporous silica nanoparticle; EPI, epirubicin; pDNA, pmiRZip-21 plasmid; ZIF-8, zeolitic imidazolate framework-8; PEG, polyethylene glycol; Apt, aptamer. | ||||||
| Free EPI | 6.90 ± 1.41 | 6.83 ± 1.11 | 7.19 ± 1.45 | 10.71 ± 1.45 | 5.87 ± 0.52 | 4.28 ± 0.74 |
| SPION@MSN-pDNA-ZIF-8-PEG | 21.61 ± 2.75 | 19.70 ± 2.14 | 10.96 ± 1.92 | 176.40 ± 1.78 | 156.80 ± 1.44 | 234.10 ± 0.38 |
| SPION@MSN-EPI-ZIF-8-PEG | 13.85 ± 1.53 | 8.86 ± 1.22 | 7.81 ± 2.70 | 29.85 ± 0.81 | 13.25 ± 0.94 | 18.05 ± 0.53 |
| SPION@MSN-EPI/pDNA-ZIF-8-PEG | 8.26 ± 1.82 | 5.82 ± 1.46 | 4.57 ± 1.15 | 48.61 ± 1.25 | 18.93 ± 1.95 | 26.99 ± 1.43 |
| SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt | 4.20 ± 2.62 | 3.18 ± 1.57 | 2.17 ± 1.43 | 62.78 ± 0.73 | 28.41 ± 1.10 | 28.11 ± 1.17 |
3.6. Synergistic cytotoxicity of targeted NPs led to a significant increase in apoptosis
To assess the synergistic lethal effects of targeted nanoparticles in chemo–gene therapy, the rate of cell death was evaluated in C26 cells using PI and annexin V staining. As anticipated, the highest rate of apoptotic cell death was observed in cells treated with targeted nanoparticles carrying both EPI and anti-miR-21-expressing plasmid (Fig. 6G). Notably, there was a 36.8% increase in the level of late apoptosis induced by targeted nanoparticles compared with similar non-targeted nanoparticles (SPION@MSN-EPI/pDNA-ZIF-8-PEG), indicating faster, enhanced, and selective entry of nanoparticles containing the aptamer into cells expressing the nucleolin receptor (C26). Furthermore, SPION@MSN-EPI/pDNA-ZIF-8-PEG nanoparticles exhibited an increase in late apoptosis of up to 28.2% and 38.8% compared with nanoparticles containing only one therapeutic agent: SPION@MSN-EPI-ZIF-8-PEG and SPION@MSN-pDNA-ZIF-8-PEG, respectively. This finding confirms the synergistic effects of the drug and plasmid. In contrast, only ∼30% of cells treated with the free drug (at the same concentration) underwent late apoptosis.3.7. Promising and selective anti-tumor effects of targeted NPs via the combined chemo–gene therapy approach
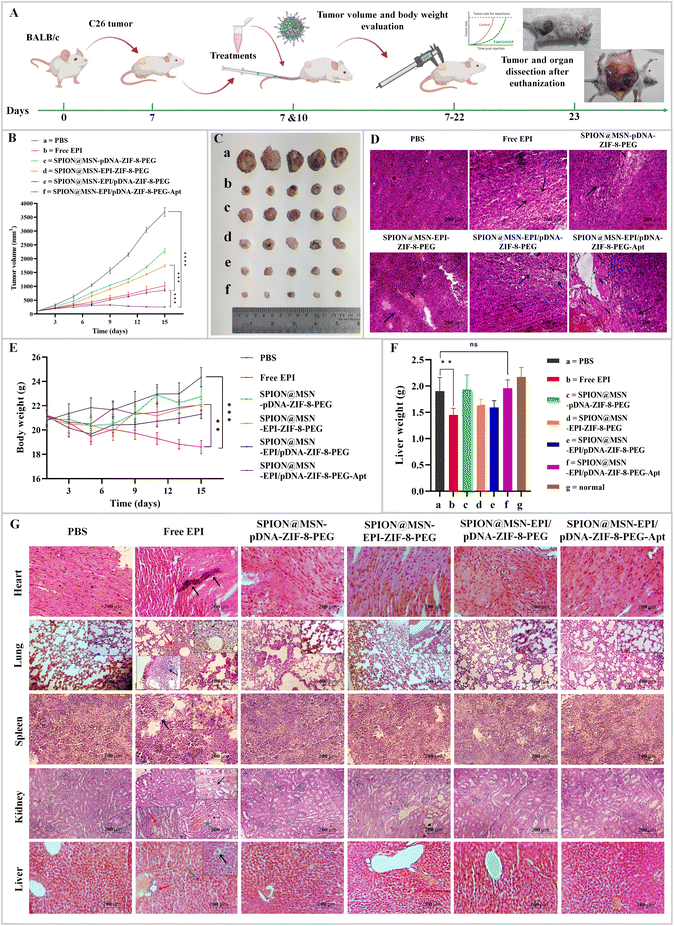
The effectiveness of manufactured nanoparticles on tumor growth was assessed in animal models of colon cancer after two injections across six treatment groups (Fig. 7A). Notably, Fig. 7B and C demonstrate the lowest tumor growth in the group treated with targeted nanoparticles containing both the drug and plasmid (SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt). This particular group exhibited remarkable efficacy in suppressing tumor growth (p < 0.001) compared with non-targeted nanoparticles and Free EPI. Furthermore, nanoparticles encapsulating both therapeutic agents demonstrated superior potency in suppressing tumor growth compared with nanoparticles containing only one agent or the free drug. Additionally, the extent of tissue damage resulting from various treatments was evaluated using hematoxylin and eosin (H&E) staining after the completion of the treatment period in different groups. Fig. 7D reveals a significant increase in apoptosis and necrosis within the tumor tissue in the group treated with targeted nanoparticles (black arrows and “N”, respectively). Non-targeted nanoparticles containing both therapeutic agents also induced tissue necrosis in comparison with the control group. These results show that SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt nanocarriers are highly efficient in actively targeting tumor cells through aptamer-based mechanisms. Moreover, they facilitate the specific release of the EPI drug and anti-miR-21-expressing plasmid within the tumor cells.3.8. Biosafety of prepared NPs was confirmed in vivo
In order to investigate possible side effects caused by the injection of the studied compounds in mouse models, the body weight of all mice was measured every other day until the 15th day after injection. The results indicated a significant decrease (p < 0.001) in the weight of mice treated with Free EPI compared with the control group (PBS) (Fig. 7E). After 15 days from the first injection, heart, lung, spleen, kidneys, and liver organs were dissected and the liver weight of mice treated with Free EPI showed a significant decrease (p < 0.01) compared with the control group (Fig. 7F).Histological evaluations were performed after the preparation of tissue sections and staining with H&E, and the representative images of the samples are shown in Fig. 7G. In light microscopy observations of the heart tissue, most cardiomyocytes related to various studied groups, except for Free EPI, were transparent and without congestion, bleeding, inflammation, or necrosis. Conversely, in the group receiving the free drug, multifocal necrosis along with calcification (black arrows) was observed. The pathological evaluation of the lung tissue related to the free drug group showed severe diffuse alveolar damage, intra-alveolar hemorrhage, and peribronchial mononuclear cell inflammation (black arrows) associated with hemosiderin-laden macrophages (red arrows). In contrast, no significant inflammation, edema, or necrosis was observed in the lung tissue of other treatment groups. Evaluation of the spleen tissue from the free drug group revealed hemosiderin pigment deposition related to hemosiderophages of the red pulp (red arrows), as well as severe degeneration and necrosis of both the white and red pulps (black arrows). Conversely, the white and red pulps, as well as the sinusoids, in other groups exhibited a relatively normal structure. Although the kidney tissue from most groups displayed a normal appearance with intact Bowman's capsule and healthy renal tubules, the kidney tissue from the free drug group exhibited degenerative and necrotic changes in the epithelium of the renal tubules (black arrows), along with infiltration of mononuclear inflammatory cells (red arrows) and kidney bleeding (blue arrows). In terms of liver pathology associated with the free drug group, necrosis (black arrows) and degeneration (blue arrows) of liver cells, along with focal accumulation of inflammatory cells (particularly in the portal area, indicated by red arrows), were observed. While mild degeneration was detected in the liver tissue related to non-targeted nanoparticles, the liver tissue from the targeted nanoparticles group did not exhibit liver cell degeneration, necrosis, or inflammation when compared with the control group. Moreover, a detailed assessment of nanoparticle biosafety markers involving liver and kidney function like ALB, ALP, and BUN revealed significant differences between Free EPI and PBS. In contrast, the PEGylated nanoparticles showed less variation in the levels of these biochemical markers compared with the PBS control group (Fig. S7†).
3.9. Ex vivo fluorescence imaging successfully tracked the NPs with high efficiency
The biodistribution analysis of Free EPI, non-targeted, and targeted nanoparticles was conducted in mice bearing C26 tumors using EPI and copGFP fluorescence emission. The KODAK IS in vivo imaging device was utilized to evaluate the fluorescence intensity in the liver, kidneys, spleen, heart, lungs, and tumor. As presented in Fig. 8, the MFI of targeted nanoparticles in the major organs was significantly lower compared with free drug (p < 0.001). This observation indicates effective drug delivery to the tumor site, demonstrating targeted drug delivery and minimal unwanted dispersion of the drug in other organs. Notably, the distribution of nanoparticles in various organs decreased during the 24 h period following injection in comparison with the 12 h, implying their gradual elimination from the body (Fig. 8D and E). Furthermore, the accumulation of targeted nanoparticles in the tumor tissue, as indicated by the high fluorescence emission of the EPI drug and the expression of the copGFP gene (present in the plasmid), signifies successful targeted drug delivery and gene therapy by aptamer-containing nanoparticles. Moreover, the SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt nanoparticles exhibited minimal biodistribution in the major organs at 12, 24, and 48 h post-injection when compared with SPION@MSN-EPI/pDNA-ZIF-8-PEG. The results obtained from fluorescence imaging demonstrate the active targeting of tumor tissue by targeted nanoparticles containing the AS1411 aptamer. This active targeting is achieved through the selective interaction of the aptamer with the nucleolin receptor on the surface of C26 tumor cells. These findings underscore the successful delivery of drugs and genes by these nanoparticles, establishing their potential for effective drug–gene delivery in cancer therapy.3.10. In vivo MR imaging demonstrated excellent traceability of the NPs
To investigate the biodistribution and accumulation of nanoparticles in tumor tissue, magnetic resonance imaging (MRI) was employed for the detection of nanoparticles featuring a SPION core as a contrast agent. T2-Weighted MR images revealed enhanced accumulation of targeted nanoparticles (SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt) compared with non-targeted nanoparticles (SPION@MSN-EPI/pDNA-ZIF-8-PEG) at the tumor site, as indicated by decreased signal intensity. This quantitative and qualitative difference within this region was demonstrated for both 6 and 18 h post-injection time points compared with pre-injection status (Fig. 8G–I). Additionally, targeted nanoparticles exhibited significantly lower biodistribution in the heart and liver compared with non-targeted nanoparticles.The findings from MRI, in alignment with the results obtained from fluorescence imaging, confirm the active targeting and delivery of therapeutic agents to the tumor tissue through the specific interaction of the AS1411 aptamer. These observations provide further evidence of the successful targeted delivery of therapeutic agents to tumor tissue and their suitability for theranostic applications.
4. Discussion
Improving the efficacy of cancer chemotherapy involves overcoming crucial challenges such as the toxic impact on healthy tissues, MDR, and aggressive cancer cell metastasis. To this aim, researchers have introduced synergistic therapeutic approaches that involve multiple modalities with diverse molecular mechanisms.55 In recent years, the combination of chemotherapy and gene therapy approaches has gained prominence, offering advantages such as synergistic anti-tumor effects and the ability to overcome the limitations of single-agent treatments.56 Studies have demonstrated that the inhibition of miR-21 can reverse the suppression of cell death and enhance the response to chemotherapeutic drugs.11,57 Therefore, a practical approach for synergistic cancer treatment could involve the co-delivery of chemotherapeutic drugs like EPI along with anti-miR-21-expressing plasmids via biocompatible nanoparticles such as MSNs.55,58In this research, the characteristics of SPION@MSN-based nanoparticles were investigated using physical and chemical methods. The fluorescence imaging of nanoparticles containing the EPI fluorescence drug demonstrated the remarkable optical properties of SPION@MSN-EPI-ZIF-8 nanoparticles both quantitatively and qualitatively. Additionally, the presence of SPION in the core of the nanoparticles provided magnetic properties, as evaluated by VSM analysis and MRI imaging, indicating their dual-purpose capability for imaging. These findings align with previous studies conducted by Chetty et al., that investigated the quantitative and qualitative fluorescence and magnetism of nanoparticles.59 Microscopic methods, such as TEM, AFM, and FE-SEM, confirmed the spherical structure and uniform morphology of the nanoparticles. The results of other characterizations, including zeta potential and particle size alterations at different stages, further confirmed the successful synthesis of the particles. Similar studies have shown that after coating with ZIF-8, the zeta potential becomes positive.23,60 The XRD spectroscopy of SPION@MSN-ZIF-8 hybrid nanoparticles revealed the presence of specific peaks related to ZIF-8, indicating the successful synthesis of the ZIF-8 layer, which is consistent with previous studies by Pan et al. and Sorribas et al.23,35,36
The theranostic nanoparticles for a combined chemo–gene therapy approach were developed by simultaneous loading of EPI drug and anti-miR-21-expressing plasmid into the pores of the SPION@MSN and ZIF-8 layer, respectively. The encapsulation efficiency (EE%) and drug loading capacity (LC%) of the EPI drug were found to be 67% ± 1.71 and 25% ± 1.32, respectively, indicating satisfactory entrapment of the drug in SPION@MSN pores. In comparison, previous studies utilizing MSN nanoparticles achieved EE% and LC% of approximately 47% and 16% for EPI, respectively.61,62 The second therapeutic agent, a DNA plasmid, was loaded onto the nanocarriers with an experimental loading efficiency determined by the ratio of the nanocarrier/plasmid (C/P = 10). Similar to previous studies, the optimal C/P ratio was determined through agarose gel electrophoresis.63,64 For instance, Babaei et al. condensed the plasmid expressing the survivin short hairpin RNA (shRNA) on the nanoparticle surface and found the optimal C/P ratio to be 6.9
The hybridization process of SPION@MSNs with the ZIF-8 layer is facilitated by the coordination bond between the OH groups on the silica surface, Zn2+ ions, and 2-MIM, leading to the accumulation of the ZIF-8 coating layer.23,40,60 In this study, the optimal loading of the plasmid was achieved through the formation of the ZIF-8 layer. A review by Ni et al. summarized that loading nucleic acids into metal–organic frameworks (MOFs) can be efficiently performed using various methods, offering protection against enzymatic degradation.21 Based on reference articles, the design and construction of the SPION@MSN-ZIF-8 hybrid nanocarrier were carried out by varying Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2-MIM
2-MIM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O ratios alongside a fixed amount of pDNA and SPION@MSN-EPI, allowing for investigation of nanoparticle size and the efficiency of drug and plasmid loading.23,36,40 For instance, Yang et al. employed a similar synthesis method to integrate Pd@Au nanoparticles, along with doxorubicin (DOX) drug, into a ZIF-8 coating, leveraging the combined capability of the nanocarrier for drug delivery and light-heat therapy to suppress liver cancer cells (SMMC-7721).38
H2O ratios alongside a fixed amount of pDNA and SPION@MSN-EPI, allowing for investigation of nanoparticle size and the efficiency of drug and plasmid loading.23,36,40 For instance, Yang et al. employed a similar synthesis method to integrate Pd@Au nanoparticles, along with doxorubicin (DOX) drug, into a ZIF-8 coating, leveraging the combined capability of the nanocarrier for drug delivery and light-heat therapy to suppress liver cancer cells (SMMC-7721).38
In addition to the successful loading of the drug and plasmid into the nanocarrier, achieving optimal and intelligent release of these cargos in cancer cells is a crucial factor in successful drug delivery. In this study, pH-dependent release was investigated under laboratory conditions, and the cumulative drug release after 8 days from nanoparticles capped with ZIF-8 demonstrated approximately 58% and 8% release in acidic conditions and neutral conditions, respectively (Fig. 2F and G). Similarly, Li et al. observed a slow release pattern of DOX over a 10-day period and the release of pDNA from MSN nanocarriers incorporating imidazole at various pH values (5.4, 6.4, and 7.4), which aligns with the release pattern observed in our study.41 As previously demonstrated by Zou et al., ZIF-8, a well-known MOF, is responsive to external stimuli, including pH changes.60 These nanoparticles, formed by the coordination of zinc ions with 2-methylimidazole, remain stable under physiological conditions but disintegrate under acidic conditions. This unique property of ZIF-8 nanoparticles can be harnessed for the intelligent release of therapeutic or diagnostic cargos in the TME or cancer cell endosomes.40
To enhance the biocompatibility properties of SPION@MSN-EPI/pDNA-ZIF-8 hybrid nanoparticles, PEG polymer was utilized, and its successful binding to the nanoparticle surface was confirmed through various characterization techniques, including DLS, TGA, FT-IR, and EDX. The increase in nanoparticle size (∼15 nm) and the decrease in positive zeta potential (∼20 mV) indicated the surface modifications resulting from the PEG coating. Moreover, the weight loss patterns observed in the TGA analysis confirmed the presence of PEG polymer within the nanoparticles’ composition. These findings align with previous studies, such as the work of Wu et al., who employed PEG functionalized with an amine group (NH2) to bind to the surface of ZIF-8 nanoparticles.54 In this approach, the PEG-NH2 polymer covers the external surface of the nanoparticles through coordination bonds between PEG amine groups and Zn2+ centers in the ZIF-8 structure, while minimizing permeability to the structure pores.52 In addition to the stabilizing role played by PEG in physiological environments, its coating on nanoparticles also improves blood circulation half-life and biocompatibility.65 The potential destructive effects of PEGylated nanoparticles in the presence of red blood cells were evaluated, and the results demonstrated a low rate of hemolysis (<5%) at multiple time points. Furthermore, the coating of SPION@MSNs with a biocompatible ZIF-8 layer significantly reduced the hemolysis rate, as supported by studies conducted by Zou et al.60 Although the final step of making the targeted nanocarrier was checked and confirmed by changes in particle size and zeta potential via DLS and ELS characterization, the electrophoresis method was also used in order to more closely examine the binding of aptamer to the surface of the nanocarrier. The electrophoresis results showed a bright band in the well of SPION@MSN-ZIF-8-PEG-Apt nanoparticles compared with non-targeted nanoparticles, which indicates the presence of AS1411 aptamer on the surface of nanoparticles (Fig. 2E). This finding is consistent with a study conducted by Sakhtianchi et al., who demonstrated the binding of the AS1411 aptamer to the surface of PEGylated nanoparticles using electrophoresis.66 Furthermore, Li et al. and Babaei et al. utilized a similar method, employing a thiol-functionalized aptamer (AS1411) covalently attached to PEG containing a maleimide group.9,67
In order to investigate the protective properties of the nanocarrier for the loaded plasmid, the coated nanoparticles at various stages were subjected to solutions containing serum and DNase I enzyme. Electrophoresis was then performed to evaluate the extent of pDNA degradation. Results demonstrated that nanoparticles coated with ZIF-8, PEG, and aptamer provided an effective protection against enzymatic degradation (Fig. 3C and D). Similarly, Mahmoodi et al. conducted a study exposing nanoparticles with different functional groups, containing plasmid, to DNase I enzyme, which corroborates our findings.64 The presence of the intense band in the agarose gel well and the corresponding bright band representing the extracted plasmid indicate protection against the degradative effects of the enzyme, highlighting the favorable quality of the plasmid after extraction. Similar electrophoresis patterns before and after plasmid extraction using sodium heparin were reported in other studies, consistent with our findings.8,21,68 In a study by Li et al., pEGFP-C1@ZIF-8-polymer nanoparticles loaded with plasmid were also exposed to DNase I enzyme before and after polymer coating, demonstrating up to ∼80% protection rate for the loaded plasmid.22 These findings further support the ability of the nanocarrier to safeguard the integrity of loaded plasmids.
In the initial cellular test, we assessed the uptake of targeted and non-targeted nanocarriers into C26 (nucleolin positive) and NIH/3T3 (nucleolin negative) cells compared with Free EPI by flow cytometry and fluorescence microscopy. The findings demonstrated a selective uptake of targeted nanoparticles by C26 cells, as indicated by a significantly higher emission of EPI drug fluorescence in the targeted nanoparticle group compared with the non-targeted group (Fig. 5A and C). In previous studies conducted by Yazdian-Robati et al. and Taghdisi et al., the entry of nanocarriers containing EPI, and specifically targeted with AS1411 aptamer, into C26 cells was similarly investigated by examining the fluorescence emission of the EPI drug.69,70 Furthermore, other research has consistently demonstrated the high expression of the nucleolin receptor on the surface of colorectal cancer cells, including the C26 cell line. Hence, AS1411 aptamer has been identified as a suitable ligand for specifically targeting these cells.31,71,72 The phenomenon of rapid absorption of a nanocarrier equipped with a specific ligand by the receptor on the target cell is due to the ligand–receptor interaction, which leads to the entry of nanoparticles through endocytosis.73
After facilitating the entry of targeted nanoparticles into cancer cells through receptor-mediated endocytosis, the release of nanocarrier cargoes, including drugs and plasmids, occurs within the acidic conditions of the endosome. One crucial aspect in evaluating the efficiency of gene therapy is the successful expression of the gene within the desired target cell. To assess this, the efficacy of the nanocarrier in transient cell transfection was evaluated by assessing the copGFP expression, which served as a fluorescence marker. The results of flow cytometry and fluorescence microscopy demonstrated the expression of the plasmid, as evidenced by the significant emission of green fluorescence light from C26 cells following treatment with targeted nanoparticles (Fig. 5B and E). In the same context, Babaei et al., in a study similar to Sanati et al., evaluated the expression of shSur-EGFP plasmid after subjecting C26 cells to targeted nanoparticles equipped with AS1411 aptamer for 24 h.9,42 Their results further support our findings, indicating enhanced expression of the GFP gene in cells treated with targeted nanoparticles, as observed through the presence of more green dots in the fluorescence microscope and FL1 channel of flow cytometry.
To investigate the anticancer effects of the targeted nanocarrier (SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt) and evaluate the performance of the drug and loaded plasmid in a combined chemo–gene therapy approach, in vitro tests were conducted using the MTT and FITC-annexin V/PI methods. Key findings from the MTT assay are as follows: (a) the nanoparticles containing both the drug and anti-miR-21-expressing plasmid exhibited synergistic toxicity on cancerous cells compared with nanoparticles containing only one of these therapeutic agents. (b) Nanoparticles containing the aptamer demonstrated stronger anticancer effects compared with those without it, on the C26 CRC cells. (c) The survival of cancer cells decreased with increasing time and dose. (d) The targeted nanoparticles did not cause significant toxicity on NIH/3T3 normal cells, while the free drug exhibited high toxicity on these cells. According to these results and similar studies, nanoparticles targeted with AS1411 aptamer exert their anticancer effects by specifically entering cancer cells through ligand–receptor interaction with nucleolin on the surface of these cells.31 For instance, Li et al. conducted an MTT test using the MSNs-anti-miR-155@PDA-Apt nanocarrier equipped with an anti-miR-155 gene therapy agent and AS1411 aptamer, showing a stronger inhibition of CRC cells (SW480).74 Additionally, the results from the PI and annexin V staining for investigating the mechanism of cell death demonstrated a high rate of late apoptosis (∼82%) in the group treated with targeted nanoparticles containing the EPI drug and anti-miR-21-expressing plasmid. This finding is corroborated by similar studies and supports the synergistic effects observed in the combined approach.8,9,75 The increased cell death rate can be attributed to the enhanced effects of the EPI drug, after the inhibition of the oncogenic miR-21 through the SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt nanocarrier. Rui et al. demonstrated similar synergistic anticancer effects of anti-miR-21 in combination with DOX in a DOX-resistant breast cancer cell line (MCF7/ADR).58 Similarly, Bahreyni et al. studied the co-delivery of EPI and anti-miR-21 to escalate the cytotoxicity for MCF-7 and C26 cell lines as target cells.76
In the second part of the biological studies, BALB/c mice bearing colorectal tumors were utilized as an animal model. Results from measuring the tumor volume over a 15-day period revealed a significant decrease in tumor growth rate in mice treated with the combined therapeutic agents of the drug and plasmid. While non-targeted SPION@MSN-EPI/pDNA-ZIF-8-PEG and Free EPI nanoparticles were able to significantly (p < 0.0001) reduce tumor growth rate, the targeted SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt nanoparticles exhibited a more pronounced reduction in tumor volume, even reaching levels lower than the initial volume of 100 mm3 by day 15, indicating effective suppression of tumor growth. Histological analysis of the tumors and major organs of the treated mice demonstrated extensive tissue destruction caused by the free drug, whereas the PEGylated nanoparticles did not cause any significant side effects. These in vivo results are in line with previous studies that have shown the most synergistic anti-tumor effects with nanoparticles containing both a chemotherapy drug and a plasmid expressing a gene therapy agent.8,41 For example, Yu et al. effectively inhibited liver tumor growth by simultaneously delivering PTX drug and a plasmid expressing survivin shRNA using a chitosan nanocarrier targeted with folic acid.77 Moreover, the efficacy of AS1411 aptamer-targeted nanocarriers in suppressing colorectal tumor growth has been summarized and supported by Yazdian-Robati et al.31
The presence of both fluorescence (EPI drug and copGFP-expressing plasmid) and magnetism (SPION core) agents, in the theranostic nanocarrier SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt, make it suitable for fluorescence imaging (FLI) and magnetic resonance imaging (MRI). Ex vivo studies conducted using a fluorescence imaging device at different time intervals illuminated a higher accumulation of targeted nanoparticles in the tumor area compared with non-targeted nanoparticles. In contrast, the scattered distribution of the red fluorescence emitted by EPI in other organs indicated the potential for widespread tissue damage in the mice treated with Free EPI. Additionally, the detection of green fluorescence resulting from copGFP expression in the tumor of mice treated with targeted nanoparticles was significantly higher than that observed in animals treated with non-targeted nanoparticles (p < 0.001). These findings align with previous studies using FLI that have compared the effects of the enhanced permeability and retention (EPR) phenomenon in passive targeting with the active targeting effects achieved through the use of AS1411 aptamer.9,42
Furthermore, MRI results indicated a higher accumulation of targeted nanoparticles at the tumor site compared with non-targeted ones, both at 6 and 18 h post-injection. The increased waves emitted from the tumor, heart, and liver at 18 h, in comparison with the 6 h period, suggest the gradual elimination of nanoparticles from the body, as evidenced by the decrease in differential intensity compared with the control group. These magnetic imaging results align with studies conducted on similar nanoparticles containing iron oxide, such as SPION, that have been targeted using various ligands.78,79 For instance, Sakhtianchi et al. introduced mesoporous silica nanoparticles with a SPION core as a theranostic nanoparticle capable of being traced in T2 magnetic imaging.66 In another study, Nosrati et al. developed a theranostic nanocarrier, SPION/PVD/MUC1Apt/DOX, targeted with MUC1 aptamer. This nanocarrier, which incorporated both magnetic (SPION) and fluorescence (PVD and DOX) components, could be efficiently detected by both MRI and FLI methods, and its accumulation in C26 tumor cells was confirmed.80 To the best of our knowledge, this research represents the first multi-purpose nanocarrier that combines the magnetic properties of SPION and MSN to load the EPI chemotherapy drug and anti-miR-21-expressing plasmid. The nanocarrier, integrated with ZIF-8 coating, PEG, and AS1411 aptamer, exhibited an intelligent and biocompatible structure with targeted release capabilities.
5. Conclusion
The core–shell nanoparticles composed of SPION@MSN were optimized to carry epirubicin and the anti-miR-21-expressing plasmid with ZIF-8 coating. The successful synthesis of the theranostic nanocarriers was confirmed through a comprehensive examination of their physicochemical characteristics. Subsequently, their performance was evaluated in both in vitro and in vivo conditions. Our research findings demonstrated the synergistic effectiveness of applying EPI along with suppression of miR-21, on CRC cells in a combinational chemo–gene therapy approach. The specific binding between the AS1411 aptamer on the surface of nanocarriers and nucleolin on C26 cancer cells resulted in the targeted delivery of the particles and induced a high cytotoxicity through apoptosis. Furthermore, the ZIF-8 and PEG-coated nanocarriers exhibited excellent biocompatibility in the vicinity of RBCs. In this regard, the SPION@MSN-EPI/pDNA-ZIF-8-PEG-Apt nanocarrier demonstrated a remarkable efficacy in suppressing colorectal cancer cells without causing too much damage to normal cells in both in vitro and in vivo studies. Moreover, the traceability of these nanoparticles in the body through FLI and MRI emphasized their theranostic capabilities to confirm passive and active targeting. Consequently, the introduced multimodal nanoparticles have the potential to serve as high-performance platforms for chemo–gene combinational therapy in targeted treatment approaches for patients with colorectal cancer. Further investigation is required to examine the biocompatibility, potential side effects, and the impact on cancer resistance and recurrence post-treatment, to clarify the clinical suitability of these nanocarriers.Abbreviations
| AFM | Atomic force microscopy |
| ALP | Alkaline phosphatase |
| ALB | Albumin |
| AU | Arbitrary unit |
| BET | Brunauer–Emmett–Teller |
| BJH | Barrett–Joyner–Halenda |
| BUN | Blood urea nitrogen |
| CRC | Colorectal cancer |
| CTAB | n-Cetyl trimethyl ammonium bromide |
| °C | Degree Celsius |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| DIW | Deionized water |
| DLS | Dynamic light scattering |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DMSO | Dimethyl sulfoxide |
| E. coli | Escherichia coli |
| ELISA | Enzyme-linked immunosorbent assay |
| EPI | Epirubicin |
| EPR | Enhanced permeability and retention |
| FBS | Fetal bovine serum |
| FE-SEM | Field emission-scanning electron microscopy |
| FL | Fluorescence |
| FT-IR | Fourier transform infrared |
| g | Gravity |
| GFP | Green fluorescent protein |
| H&E | Hematoxylin and eosin |
| IC50 | The half-maximal inhibitory concentration |
| MFI | Mean fluorescence intensity |
| mg kg−1 | Milligram per kilogram |
| mg mL−1 | Milligram per milliliter |
| miR-21 | microRNA-21 |
| mM | Millimolar |
| mm3 | Cubic millimeter |
| MOFs | Metal organic frameworks |
| MRI | Magnetic resonance imaging |
| MSN | Mesoporous silica nanoparticle |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| 2-MIM | 2-Methylimidazole |
| nm | Nanometer |
| NP | Nanoparticle |
| PBS | Phosphate-buffered saline |
| PEG | Polyethylene glycol |
| PI | Propidium iodide |
| pmiRZip-21 | miRZip-21 plasmid |
| RBC | Red blood cell |
| RPMI 1640 | Roswell Park Memorial Institute 1640 |
| S.D. | Standard deviation |
| SPION | Superparamagnetic iron oxide nanoparticle |
| TEM | Transmission electron microscopy |
| TEOS | Tetraethylorthosilicate |
| TGA | Thermogravimetric analysis |
| TME | Tumor microenvironment |
| U mL−1 | Unit per milliliter |
| VSM | Vibrating sample magnetometer |
| XRD | X-ray diffraction |
| ZIF-8 | Zeolitic imidazolate framework-8 |
| μg | Microgram |
| μg mL−1 | Microgram per milliliter |
| μg mL−1 | Microgram per microliter |
| μL | Microliter |
Author contributions
AA: methodology, formal analysis, data curation, investigation, resources, software, visualization, writing original draft. ARB: supervision, validation, funding acquisition. ASS: supervision, validation, funding acquisition. MMM: supervision, validation, funding acquisition, review & editing.Ethics approval and consent to participate
The animal experiments were performed in accordance with the guidelines approved by the Animal Ethics Committee of Ferdowsi University of Mashhad (IR.UM.REC.1401.040).Consent for publication
All authors consent for the manuscript to be published.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors would like to thank Dr Shaterzadeh, Miss Maryam Dolatshahi, and Dr Sonia Iranpour for their excellent support. This work was supported by Ferdowsi University of Mashhad, grant number: 57588. The schematic figures were created with BioRender.com.References
- R. Siegel, N. Wagle, A. Cercek, R. Smith and A. Jemal, Colorectal cancer statistics, 2023, CA-Cancer J. Clin., 2023, 73, 233–254, DOI:10.3322/caac.21772.
- T. Yan, S. Zhu, W. Hui, J. He, Z. Liu and J. Cheng, Chitosan based pH-responsive polymeric prodrug vector for enhanced tumor targeted co-delivery of doxorubicin and siRNA, Carbohydr. Polym., 2020, 250, 116781, DOI:10.1016/j.carbpol.2020.116781.
- Z. Han, M. Gao, Z. Wang, L. Peng, Y. Zhao and L. Sun, pH/NIR-responsive nanocarriers based on mesoporous polydopamine encapsulated gold nanorods for drug delivery and thermo-chemotherapy, J. Drug Delivery Sci. Technol., 2022, 75, 103610, DOI:10.1016/j.jddst.2022.103610.
- J. Shi, P. W. Kantoff, R. Wooster and O. C. Farokhzad, Cancer nanomedicine: progress, challenges and opportunities, Nat. Rev. Cancer, 2017, 17, 20–37, DOI:10.1038/nrc.2016.108.
- W. Wang, Y. Zhang, Y. Jian, S. He, J. Liu, Y. Cheng, S. Zheng, Z. Qian, X. Gao and X. Wang, Sensitizing chemotherapy for glioma with fisetin mediated by a microenvironment-responsive nano-drug delivery system, Nanoscale, 2023, 16, 97–109, 10.1039/d3nr05195a.
- M. P. Patel, M. S. Shah, M. N. Athalye and J. K. Patel, Gene therapy for cancer treatment: recent trends and clinical evidence, Gene Delivery Syst., 2022, 163–180 Search PubMed.
- F. Uddin, C. M. Rudin and T. Sen, CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future, Front. Oncol., 2020, 10, 1384, DOI:10.3389/fonc.2020.01387.
- Q. Li, X. Lv, C. Tang and C. Yin, Co-delivery of doxorubicin and CRISPR/Cas9 or RNAi-expressing plasmid by chitosan-based nanoparticle for cancer therapy, Carbohydr. Polym., 2022, 287, 119315, DOI:10.1016/j.carbpol.2022.119315.
- M. Babaei, K. Abnous, S. M. Taghdisi, S. Taghavi, A. S. Saljooghi, M. Ramezani and M. Alibolandi, Targeted rod-shaped mesoporous silica nanoparticles for the co-delivery of camptothecin and survivin shRNA in to colon adenocarcinoma in vitro and in vivo, Eur. J. Pharm. Biopharm., 2020, 156, 84–96, DOI:10.1016/j.ejpb.2020.08.026.
- E. Van Rooij and S. Kauppinen, Development of micro RNA therapeutics is coming of age, EMBO Mol. Med., 2014, 6, 851–864, DOI:10.15252/emmm.201100899.
- T. Akhtarkhavari, A. R. Bahrami and M. M. Matin, Downregulation of miR-21 as a promising strategy to overcome drug resistance in cancer, Eur. J. Pharmacol., 2022, 932, 175233, DOI:10.1016/j.ejphar.2022.175233.
- X. Guo, S. Guo, L. Pan, L. I. Ruan, Y. Gu and J. Lai, Anti-microRNA-21/221 and microRNA-199a transfected by ultrasound microbubbles induces the apoptosis of human hepatoma HepG2 cells, Oncol. Lett., 2017, 13, 3669–3675, DOI:10.3892/ol.2017.5910.
- L. X. Yan, Q. N. Wu, Y. Zhang, Y. Y. Li, D. Z. Liao, J. H. Hou, J. Fu, M. S. Zeng, J. P. Yun, Q. L. Wu, Y. X. Zeng and J. Y. Shao, Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth, Breast Cancer Res., 2011, 13, 1–14, DOI:10.1186/bcr2803.
- D. Bhere, N. Arghiani, E. R. Lechtich, Y. Yao, S. Alsaab, F. Bei, M. M. Matin and K. Shah, Simultaneous downregulation of miR-21 and upregulation of miR-7 has anti-tumor efficacy, Sci. Rep., 2020, 10, 1779, DOI:10.1038/s41598-020-58072-w.
- N. Arghiani and M. M. Matin, MiR-21: A key small molecule with great effects in combination cancer therapy, Nucleic Acid Ther., 2021, 31, 271–283, DOI:10.1089/nat.2020.0914.
- M. H. Butt, M. Zaman, A. Ahmad, R. Khan, T. H. Mallhi, M. M. Hasan, Y. H. Khan, S. Hafeez, E. E. S. Massoud, M. H. Rahman and S. Cavalu, Appraisal for the Potential of Viral and Nonviral Vectors in Gene Therapy: A Review, Genes, 2022, 13, 1370, DOI:10.3390/genes13081370.
- G. A. Marcelo, C. Lodeiro, J. L. Capelo, J. Lorenzo and E. Oliveira, Magnetic, fluorescent and hybrid nanoparticles: From synthesis to application in biosystems, Mater. Sci. Eng., C, 2020, 106, 110104, DOI:10.1016/j.msec.2019.110104.
- Y. Wang and H. Gu, Core-shell-type magnetic mesoporous silica nanocomposites for bioimaging and therapeutic agent delivery, Adv. Mater., 2015, 27, 576–585, DOI:10.1002/adma.201401124.
- M. Saeinasab, S. Iranpour, N. Hosseini-Giv, A. Sh. Saljooghi and M. M. Matin, Tumor-targeted delivery of SNHG15 siRNA using a ZIF-8 nanoplatform: Towards a more effective prostate cancer therapy, Int. J. Biol. Macromol., 2024, 259, 129233, DOI:10.1016/j.ijbiomac.2024.129233.
- S. Iranpour, A. R. Bahrami, M. Dayyani, A. Sh. Saljooghi and M. M. Matin, A potent multifunctional ZIF-8 nanoplatform developed for colorectal cancer therapy by triple-delivery of chemo/radio/targeted therapy agents, J. Mater. Chem. B, 2024, 12, 1096–1114, 10.1039/D3TB02571C.
- D. Ni, J. Lin, N. Zhang, S. Li, Y. Xue, Z. Wang, Q. Liu, K. Liu, H. Zhang, Y. Zhao, C. Chen and Y. Liu, Combinational application of metal–organic frameworks–based nanozyme and nucleic acid delivery in cancer therapy, WIREs Nanomed. Nanobiotechnol., 2022, 14, 1–22, DOI:10.1002/wnan.1773.
- Y. Li, K. Zhang, P. Liu, M. Chen, Y. Zhong, Q. Ye, M. Q. Wei, H. Zhao and Z. Tang, Encapsulation of plasmid DNA by nanoscale metal–organic frameworks for efficient gene transportation and expression, Adv. Mater., 2019, 31, 1901570, DOI:10.1002/adma.201901570.
- Q. S. Pan, T. T. Chen, C. P. Nie, J. T. Yi, C. Liu, Y. L. Hu and X. Chu, In situ synthesis of ultrathin ZIF-8 film-coated MSNs for co-delivering Bcl-2 siRNA and doxorubicin to enhance chemotherapeutic efficacy in drug-resistant cancer cells, ACS Appl. Mater. Interfaces, 2018, 10, 33070–33077, DOI:10.1021/acsami.8b13393.
- J. Sun, L. Zhang, X. Liu, A. Zhao, C. Hu, T. Gan and Y. Liu, Rational design of a mesoporous silica@ZIF-8 based molecularly imprinted electrochemical sensor with high sensitivity and selectivity for atropine monitoring, J. Electroanal. Chem., 2021, 903, 115843, DOI:10.1016/j.jelechem.2021.115843.
- Z. Fu and J. Xiang, Aptamer-functionalized nanoparticles in targeted delivery and cancer therapy, Int. J. Mol. Sci., 2020, 21, 1–39, DOI:10.3390/ijms21239123.
- T. Thanh, H. Thi, E. H. Pilkington, D. H. Nguyen and J. S. Lee, The importance of poly (ethylene glycol) alternatives for overcoming PEG immunogenicity in drug delivery and bioconjugation, Polymers, 2020, 12, 298, DOI:10.3390/polym12020298.
- D. Yadav and H. K. Dewangan, PEGYLATION: an important approach for novel drug delivery system, J. Biomater. Sci., Polym. Ed., 2021, 32, 266–280, DOI:10.1080/09205063.2020.1825304.
- P. J. Bates, D. A. Laber, D. M. Miller, S. D. Thomas and J. O. Trent, Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer, Exp. Mol. Pathol., 2009, 86, 151–164, DOI:10.1016/j.yexmp.2009.01.004.
- Z. Chen and X. Xu, Roles of nucleolin: Focus on cancer and anti-cancer therapy, Saudi Med. J., 2016, 37, 1312–1318, DOI:10.15537/smj.2016.12.15972.
- J. Liu, T. Wei, J. Zhao, Y. Huang, H. Deng, A. Kumar, C. Wang, Z. Liang, X. Ma and X.-J. Liang, Multifunctional aptamer-based nanoparticles for targeted drug delivery to circumvent cancer resistance, Biomaterials, 2016, 91, 44–56, DOI:10.1016/j.biomaterials.2016.03.013.
- R. Yazdian-Robati, P. Bayat, F. Oroojalian, M. Zargari, M. Ramezani, S. M. Taghdisi and K. Abnous, Therapeutic applications of AS1411 aptamer, an update review, Int. J. Biol. Macromol., 2020, 155, 1420–1431, DOI:10.1016/j.ijbiomac.2019.11.118.
- P. Yang, Z. Quan, Z. Hou, C. Li, X. Kang, Z. Cheng and J. Lin, A magnetic, luminescent and mesoporous core-shell structured composite material as drug carrier, Biomaterials, 2009, 30, 4786–4795, DOI:10.1016/j.biomaterials.2009.05.038.
- R. Pignatello, D. Amico, S. Chiechio, C. Spadaro, G. Puglisi and P. Giunchedi, Preparation and analgesic activity of eudragit RS100® microparticles containing diflunisal, Drug Delivery, 2001, 8, 35–45, DOI:10.1080/107175401300002748.
- M. Liu, A. Dasgupta, P. Koczera and S. Schipper, Drug loading in poly (butyl cyanoacrylate)-based polymeric microbubbles, Mol. Pharmaceutics, 2020, 17, 2840–2848, DOI:10.1021/acs.molpharmaceut.0c00242.
- S. Sorribas, B. Zornoza, C. Téllez and J. Coronas, Ordered mesoporous silica-(ZIF-8) core-shell spheres, Chem. Commun., 2012, 48, 9388–9390, 10.1039/c2cc34893d.
- S. Sorribas, B. Zornoza, C. Téllez and J. Coronas, Mixed matrix membranes comprising silica-(ZIF-8) core-shell spheres with ordered meso-microporosity for natural- and bio-gas upgrading, J. Membr. Sci., 2014, 452, 184–192, DOI:10.1016/j.memsci.2013.10.043.
- H. Liu, L. Chen and J. Ding, A core-shell magnetic metal organic framework of type Fe3O4@ZIF-8 for the extraction of tetracycline antibiotics from water samples followed by ultra-HPLC-MS analysis, Microchim. Acta, 2017, 184, 4091–4098, DOI:10.1007/s00604-017-2442-6.
- X. Yang, L. Li, D. He, L. Hai, J. Tang, H. Li, X. He and K. Wang, A metal-organic framework based nanocomposite with co-encapsulation of Pd@Au nanoparticles and doxorubicin for pH- and NIR-triggered synergistic chemo-photothermal treatment of cancer cells, J. Mater. Chem. B, 2017, 5, 4648–4659, 10.1039/c7tb00715a.
- H. Zheng, Y. Zhang, L. Liu, W. Wan, P. Guo, A. M. Nyström and X. Zou, One-pot synthesis of metal-organic frameworks with encapsulated target molecules and their applications for controlled drug delivery, J. Am. Chem. Soc., 2016, 138, 962–968, DOI:10.1021/jacs.5b11720.
- C. Zheng, Y. Wang, S. Zeng, F. Phua, W. Q. Lim and Y. Zhao, ZnO-DOX@ZIF-8 core-shell nanoparticles for pH-responsive drug delivery, ACS Biomater. Sci. Eng., 2017, 10, 2223–2229, DOI:10.1021/acsbiomaterials.7b00435.
- Z. Li, L. Zhang, C. Tang and C. Yin, Co-delivery of doxorubicin and survivin shRNA-expressing plasmid via microenvironment-responsive dendritic mesoporous silica nanoparticles for synergistic cancer therapy, Pharm. Res., 2017, 34, 2829–2841, DOI:10.1007/s11095-017-2264-6.
- S. Sanati, S. Taghavi, K. Abnous, S. M. Taghdisi, M. Babaei, M. Ramezani and M. Alibolandi, Fabrication of anionic dextran-coated micelles for aptamer targeted delivery of camptothecin and survivin-shRNA to colon adenocarcinoma, Gene Ther., 2021, 26, 55–68, DOI:10.1038/s41434-021-00234-0.
- I. P. Sæbø, M. Bjørås, H. Franzyk, E. Helgesen and J. A. Booth, Optimization of the hemolysis assay for the assessment of cytotoxicity, Int. J. Mol. Sci., 2023, 24, 2914, DOI:10.3390/ijms24032914.
- Y. Yang, J. Wang, X. Li, L. Lin and X. Yue, A near infrared fluorescent/ultrasonic bimodal contrast agent for imaging guided pDNA delivery via ultrasound targeted microbubble destruction, RSC Adv., 2015, 5, 8404–8414, 10.1039/c4ra15066j.
- F. S. Aljohani, M. T. Hamed, B. A. Bakr, Y. H. Shahin, M. M. Abu-Serie, A. K. Awaad, H. El-Kady and B. H. Elwakil, In vivo bio-distribution and acute toxicity evaluation of greenly synthesized ultra-small gold nanoparticles with different biological activities, Sci. Rep., 2022, 12, 1–20, DOI:10.1038/s41598-022-10251-7.
- R. Y. Hong, J. H. Li, S. Z. Zhang, H. Z. Li, Y. Zheng, J. min Ding and D. G. Wei, Preparation and characterization of silica-coated Fe3O4 nanoparticles used as precursor of ferrofluids, Appl. Surf. Sci., 2009, 255, 3485–3492, DOI:10.1016/j.apsusc.2008.09.071.
- A. Lapresta-Fernández, T. Doussineau, S. Dutz, F. Steiniger, A. J. Moro and G. J. Mohr, Magnetic and fluorescent core-shell nanoparticles for ratiometric pH sensing, Nanotechnology, 2011, 22, 415501, DOI:10.1088/0957-4484/22/41/415501.
- W. Premaratne, W. Priyadarshana, S. H. P. Gunawardena and A. A. P. De Alwis, Synthesis of nanosilica from paddy husk ash and their surface functionalization, J. Sci. Univ. Kelaniya, 2013, 8, 33–48, DOI:10.4038/josuk.v8i0.7238.
- L. Zhu, L. Zhou, N. Huang, W. Cui, Z. Liu, K. Xiao and Z. Zhou, Efficient preparation of enantiopure D-phenylalanine through asymmetric resolution using immobilized phenylalanine ammonia-lyase from rhodotorula glutinis JN-1 in a recirculating packed-bed reactor, PLoS One, 2014, 9, 108586, DOI:10.1371/journal.pone.0108586.
- J. Ding, G. Chen, G. Chen and M. Guo, One-pot synthesis of epirubicin-capped silver nanoparticles and their anticancer activity against hep G2 cells, Pharmaceutics, 2019, 11, 123, DOI:10.3390/pharmaceutics11030123.
- S. Iranpour, A. R. Bahrami, S. Nekooei, A. S. Saljooghi and M. M. Matin, Improving anti-cancer drug delivery performance of magnetic mesoporous silica nanocarriers for more efficient colorectal cancer therapy, J. Nanobiotechnol., 2021, 1–23, DOI:10.1186/s12951-021-01056-3.
- R. Ettlinger, N. Moreno, N. Ziółkowska, A. Ullrich, H. A. Krug von Nidda, D. Jirák, K. Kerl and H. Bunzen, In Vitro Studies of Fe3O4-ZIF-8 Core–Shell Nanoparticles Designed as Potential Theragnostics, Part. Part. Syst. Charact., 2020, 37, 2000185, DOI:10.1002/ppsc.202000185.
- A. Poddar, J. J. Conesa, K. Liang, S. Dhakal, P. Reineck, G. Bryant, E. Pereiro, R. Ricco, H. Amenitsch, C. Doonan, X. Mulet, C. M. Doherty, P. Falcaro and R. Shukla, Encapsulation, visualization and expression of genes with biomimetically mineralized zeolitic imidazolate framework-8 (ZIF-8), Small, 2019, 15, 1902268, DOI:10.1002/smll.201902268.
- M. Wu, X. Guo, F. Zhao and B. Zeng, A poly(ethylenglycol) functionalized ZIF-8 membrane prepared by coordination-based post-synthetic strategy for the enhanced adsorption of phenolic endocrine disruptors from water, Sci. Rep., 2017, 7, 1–11, DOI:10.1038/s41598-017-09364-1.
- Q. He and J. Shi, MSN anti-cancer nanomedicines: Chemotherapy enhancement, overcoming of drug resistance, and metastasis inhibition, Adv. Mater., 2014, 26, 391–411, DOI:10.1002/adma.201303123.
- G. Lin, R. A. Revia and M. Zhang, Inorganic nanomaterial-mediated gene therapy in combination with other antitumor treatment modalities, Adv. Funct. Mater., 2021, 31, 2007096, DOI:10.1002/adfm.202007096.
- E. Jung, J. Choi, J. S. Kim and T. S. Han, Microrna-based therapeutics for drug-resistant colorectal cancer, Pharmaceuticals, 2021, 14, 1–19, DOI:10.3390/ph14020136.
- M. Rui, Y. Qu, T. Gao, Y. Ge, C. Feng and X. Xu, Simultaneous delivery of anti-miR21 with doxorubicin prodrug by mimetic lipoprotein nanoparticles for synergistic effect against drug resistance in cancer cells, Int. J. Nanomed., 2017, 12, 217–237, DOI:10.2147/IJN.S122171.
- S. S. Chetty, S. Praneetha, A. Vadivel Murugan, K. Govarthanan and R. S. Verma, Human umbilical cord Wharton's jelly-derived mesenchymal stem cells labeled with Mn2+ and Gd3+ co-doped CuInS2-ZnS nanocrystals for multimodality imaging in a tumor mice model, ACS Appl. Mater. Interfaces, 2020, 12, 3415–3429, DOI:10.1021/acsami.9b19054.
- Z. Zou, S. Li, D. He, X. He, K. Wang, L. Li, X. Yang and H. Li, A versatile stimulus-responsive metal-organic framework for size/morphology tunable hollow mesoporous silica and pH-triggered drug delivery, J. Mater. Chem. B, 2017, 5, 2126–2132, 10.1039/c6tb03379b.
- L. Ansari, M. R. Jaafari, T. R. Bastami and B. Malaekeh-Nikouei, Improved anticancer efficacy of epirubicin by magnetic mesoporous silica nanoparticles: in vitro and in vivo studies, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 594–606, DOI:10.1080/21691401.2018.1464461.
- M. Yahya Hanafi-Bojd, M. Reza Jaafari, N. Ramezanian, K. Abnous and B. Malaekeh-Nikouei, Co-delivery of epirubicin and siRNA using functionalized mesoporous silica nanoparticles enhances in vitro and in vivo drug efficacy, Curr. Drug Delivery, 2015, 13, 1176–1182, DOI:10.2174/1567201813666151231094056.
- C. S. Cheng, T. P. Liu, F. C. Chien, C. Y. Mou, S. H. Wu and Y. P. Chen, Codelivery of plasmid and curcumin with mesoporous silica nanoparticles for promoting neurite outgrowth, ACS Appl. Mater. Interfaces, 2019, 11, 15322–15331, DOI:10.1021/acsami.9b02797.
- M. Mahmoodi, A. Behzad-Behbahani, S. Sharifzadeh, S. S. Abolmaali and A. M. Tamaddon, Co-condensation synthesis of well-defined mesoporous silica nanoparticles: Effect of surface chemical modification on plasmid DNA condensation and transfection, IET Nanobiotechnol., 2017, 11, 995–1004, DOI:10.1049/iet-nbt.2017.0078.
- M. Xie, H. Shi, Z. Li, H. Shen, K. Ma, B. Li, S. Shen and Y. Jin, A multifunctional mesoporous silica nanocomposite for targeted delivery, controlled release of doxorubicin and bioimaging, Colloids Surf., B, 2013, 110, 138–147, DOI:10.1016/j.colsurfb.2013.04.009.
- R. Sakhtianchi, B. Darvishi, Z. Mirzaie, F. Dorkoosh, S. Shanehsazzadeh and R. Dinarvand, Pegylated magnetic mesoporous silica nanoparticles decorated with AS1411 Aptamer as a targeting delivery system for cytotoxic agents., Pharm. Dev. Technol., 2019, 24, 1063–1075, DOI:10.1080/10837450.2019.1569678.
- L. Li, J. Hou, X. Liu, Y. Guo, Y. Wu, L. Zhang and Z. Yang, Nucleolin-targeting liposomes guided by aptamer AS1411 for the delivery of siRNA for the treatment of malignant melanomas, Biomaterials, 2014, 35, 3840–3850, DOI:10.1016/j.biomaterials.2014.01.019.
- S. Dong, Q. Chen, W. Li, Z. Jiang, J. Ma and H. Gao, A dendritic catiomer with an MOF motif for the construction of safe and efficient gene delivery systems, J. Mater. Chem. B, 2017, 5, 8322–8329, 10.1039/c7tb01966a.
- R. Yazdian-Robati, M. Ramezani, S. H. Jalalian, K. Abnous and S. M. Taghdisi, Targeted delivery of epirubicin to cancer cells by polyvalent aptamer system in vitro and in vivo, Pharm. Res., 2016, 33, 2289–2297, DOI:10.1007/s11095-016-1967-4.
- S. M. Taghdisi, N. M. Danesh, M. Ramezani, P. Lavaee, S. H. Jalalian, R. Y. Robati and K. Abnous, Double targeting and aptamer-assisted controlled release delivery of epirubicin to cancer cells by aptamers-based dendrimer in vitro and in vivo., Eur. J. Pharm. Biopharm., 2016, 102, 152–158, DOI:10.1016/j.ejpb.2016.03.013.
- F. Khatami, M. M. Matin, N. M. Danesh, A. R. Bahrami, K. Abnous and S. M. Taghdisi, Targeted delivery system using silica nanoparticles coated with chitosan and AS1411 for combination therapy of doxorubicin and antimiR-21, Carbohydr. Polym., 2021, 266, 118111, DOI:10.1016/j.carbpol.2021.118111.
- J. Mosafer, K. Abnous, M. Tafaghodi, A. Mokhtarzadeh and M. Ramezani, In vitro and in vivo evaluation of anti-nucleolin-targeted magnetic PLGA nanoparticles loaded with doxorubicin as a theranostic agent for enhanced targeted cancer imaging and therapy, Eur. J. Pharm. Biopharm., 2017, 113, 60–74, DOI:10.1016/j.ejpb.2016.12.009.
- H. Gao, J. Qian, S. Cao, Z. Yang, Z. Pang, S. Pan, L. Fan, Z. Xi, X. Jiang and Q. Zhang, Precise glioma targeting of and penetration by aptamer and peptide dual-functioned nanoparticles, Biomaterials, 2012, 33, 5115–5123, DOI:10.1016/j.biomaterials.2012.03.058.
- Y. Li, Y. Duo, P. Zhai, L. He, K. Zhong, Y. Zhang, K. Huang, J. Luo, H. Zhang and X. Yu, Targeted delivery of anti-miR-155 by functionalized mesoporous silica nanoparticles for colorectal cancer therapy, Nanomedicine, 2018, 13, 1753–1772, DOI:10.2217/nnm-2017-0353.
- Z. Li, L. Zhang, C. Tang and C. Yin, Co-delivery of doxorubicin and survivin shRNA-expressing plasmid via microenvironment-responsive dendritic mesoporous silica nanoparticles for synergistic cancer therapy, Pharm. Res., 2017, 34, 2829–2841, DOI:10.1007/S11095-017-2264-6.
- A. Bahreyni, M. Alibolandi, M. Ramezani, A. Sarafan Sadeghi, K. Abnous and S. M. Taghdisi, A novel MUC1 aptamer-modified PLGA-epirubicin-PβAE-antimir-21 nanocomplex platform for targeted co-delivery of anticancer agents in vitro and in vivo, Colloids Surf., B, 2019, 175, 231–238, DOI:10.1016/j.colsurfb.2018.12.006.
- B. Yu, C. Tang and C. Yin, Enhanced antitumor efficacy of folate modified amphiphilic nanoparticles through co-delivery of chemotherapeutic drugs and genes, Biomaterials, 2014, 35, 6369–6378, DOI:10.1016/j.biomaterials.2014.04.095.
- L. Zhu, Z. Zhou, H. Mao and L. Yang, Magnetic nanoparticles for precision oncology: Theranostic magnetic iron oxide nanoparticles for image-guided and targeted cancer therapy, Nanomedicine, 2017, 12, 73–87, DOI:10.2217/nnm-2016-0316.
- S. Sabale, P. Kandesar, V. Jadhav, R. Komorek, R. K. Motkuri and X. Y. Yu, Recent developments in the synthesis, properties, and biomedical applications of core/shell superparamagnetic iron oxide nanoparticles with gold, Biomater. Sci., 2017, 5, 2212–2225, 10.1039/c7bm00723j.
- R. Nosrati, K. Abnous, M. Alibolandi, J. Mosafer, S. Dehghani, S. M. Taghdisi and M. Ramezani, Targeted SPION siderophore conjugate loaded with doxorubicin as a theranostic agent for imaging and treatment of colon carcinoma, Sci. Rep., 2021, 11, 1–15, DOI:10.1038/s41598-021-92391-w.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr06642h |
| This journal is © The Royal Society of Chemistry 2024 |