Advances in lipid nanoparticle mRNA therapeutics beyond COVID-19 vaccines
Yeung
Wu
a,
Sinuo
Yu
a and
Irene
de Lázaro
 *abc
*abc
aDepartment of Biomedical Engineering, NYU Tandon School of Engineering, New York University, USA. E-mail: irene.delazaro@nyu.edu
bCardiovascular Research Center, Division of Cardiology, Department of Medicine, NYU Grossman School of Medicine, New York University, USA
cHarvard John A. Paulson School of Engineering and Applied Science, Harvard University, USA
First published on 13th March 2024
Abstract
The remarkable success of two lipid nanoparticle-mRNA vaccines against coronavirus disease (COVID-19) has placed the therapeutic and prophylactic potential of messenger RNA (mRNA) in the spotlight. It has also drawn attention to the indispensable role of lipid nanoparticles in enabling the effects of this nucleic acid. To date, lipid nanoparticles are the most clinically advanced non-viral platforms for mRNA delivery. This is thanks to their favorable safety profile and efficiency in protecting the nucleic acid from degradation and allowing its cellular uptake and cytoplasmic release upon endosomal escape. Moreover, the development of lipid nanoparticle-mRNA therapeutics was already a very active area of research even before the COVID-19 pandemic, which has likely only begun to bear its fruits. In this Review, we first discuss key aspects of the development of lipid nanoparticles as mRNA carriers. We then highlight promising preclinical and clinical studies involving lipid nanoparticle-mRNA formulations against infectious diseases and cancer, and to enable protein replacement or supplementation and genome editing. Finally, we elaborate on the challenges in advancing lipid nanoparticle-mRNA technology to widespread therapeutic use.
1. Introduction
Messenger RNA (mRNA) is a transient nucleic acid intermediator that transfers genetic information from DNA to protein synthesis in the cytoplasm.1 By delivering mRNAs that encode antigens of disease-causing microorganisms or cancerous cells, genome-editing tools, or other relevant proteins, mRNA technology has shown its therapeutic potential. This recently became a reality with the regulatory approval of the Pfizer/BioNTech and Moderna vaccines against SARS-CoV-2 in 2020, which spared millions of lives during the COVID-19 pandemic. As a therapeutic, the advantages of mRNA over other nucleic acids revolve around its mechanism and site of action. Unlike DNA, mRNA poses no risk of insertional mutagenesis in the host's genome and does not need to overcome the nuclear membrane barrier. Its transient expression, which decays in ∼48 hours, can be advantageous or a limitation depending on the desired application. Initial roadblocks related to the instability and immunogenicity of mRNA have been largely mitigated thanks to new strategies implemented during in vitro transcription (IVT) such as capping,2,3 the inclusion of a poly-A tail,4 and nucleoside substitution.5However, mRNA cannot realize its therapeutic potential alone. It is subject of rapid degradation by nucleases in the bloodstream and tissue compartments, and its large molecular size and negative charge severely limit cellular internalization.6 The development of nanoparticle formulations that encapsulate mRNA, protecting it from enzymatic degradation and facilitating its translocation to the cytoplasm, has proved as equally crucial for achieving the desired biological effect as the development of functional and stable mRNA molecules itself. So far, a variety of nanoscale delivery vectors have been developed for mRNA delivery, including liposomes, lipid nanoparticles, micelles, polymer and protein-based nanoparticles.7–10 However, many of them suffer from low encapsulation efficiencies,11 poor cellular internalization12–14 and/or limited endosomal escape,15 among other issues. To date, lipid nanoparticles face no close competition in mRNA encapsulation and transfection efficiency.
Here, we first provide a general overview of the formulation and preparation of lipid nanoparticles, and of the mRNA delivery mechanism that they enable. We then highlight recent advances in lipid nanoparticle-mRNA therapeutics that are being developed for diseases other than COVID-19. The latter have been the focus of various comprehensive reviews by other colleagues and are therefore not the scope of our work.16–19 For conciseness, we only discuss those formulations that have been at least tested in preclinical animal models. Those that have entered clinical trials are summarized in Tables 1–4. We also discuss challenges and limitations of lipid nanoparticle-mRNA delivery systems that need still to be overcome to achieve widespread clinical presence.
| mRNA | Indication | Administration route | Study type, phase | Clinical trial ID | Status | Sponsor |
|---|---|---|---|---|---|---|
| HA: hemagglutinin; RSV: respiratory syncytial virus; hMPV: human metapneumovirus; RABV-G: Rabies virus glycoprotein; CHIKV: Chikungunya virus; gB: glycoprotein B; PC: pentameric complex; PV3: parainfluenza virus type 3; prME: pre-membrane and envelope structural proteins; HIV: human immunodeficiency virus. The information summarized in this table was obtained by searching the ClinicalTrials.gov database with the following keywords: [mRNA, lipid nanoparticle], [mRNA, LNP], [messenger RNA, lipid nanoparticle], [messenger RNA, LNP], [messenger ribonucleic acid, lipid nanoparticle] and [LNP, vaccine] between August and December 2023. | ||||||
| H3 mRNA/LNP mRNA encoding full-length HA of influenza virus | Influenza H3N2 | Intramuscular | Interventional, phase 1 | NCT05829356 | Recruiting | Sanofi Pasteur |
| VRC-FLUNPF099-00-VP mRNA encoding Vaccine Research Center(VRC) H1ssF 3928 | Influenza | Intramuscular | Interventional, phase 1 | NCT05755620 | Recruiting | National Institute of Allergy and Infectious Diseases (NIAID) |
| DCVC H1 HA mRNA encoding full-length H1 HA of influenza A/California/07/2009 (H1N1) | Influenza H | Intramuscular | Interventional, phase 1 | NCT05945485 | Recruiting | National Institute of Allergy and Infectious Diseases (NIAID) |
| mRNA-1851 (VAL-339851) H7N9 antigen mRNA | Influenza | Intramuscular | Interventional, phase 1 | NCT03345043 | Completed | ModernaTX, Inc. |
| mRNA-1440 (VAL-506440) H10N8 antigen mRNA | Influenza | Intramuscular | Interventional, phase 1 | NCT03076385 | Completed | ModernaTX, Inc. |
| mRNA-1010 mRNA encoding membrane-bound HA surface glycoproteins of four influenza strains (A/H1N1, A/H3N2, B/Victoria, and B/Yamagata) | Influenza | Intramuscular | Interventional, phase 3 | NCT05827978 | Recruiting | ModernaTX, Inc. |
| mIRB, bIRB, qIRB one, two of four mRNAs encoding influenza antigens | Influenza | Intramuscular | Interventional, phase 1 & 2 | NCT05052697 | Completed | Pfizer |
| CL-0059 or CL-0137 RSV mRNA | Respiratory syncytial virus | Intramuscular | Interventional, phase 1 & 2 | NCT05639894 | Active, not recruiting | Sanofi Pasteur |
| mRNA-1345 mRNA encoding RSV stabilized prefusion F glycoprotein | Respiratory syncytial virus | Intramuscular | Interventional, phase 1 | NCT04528719 | Active, not recruiting | ModernaTX, Inc. |
| Intramuscular | Interventional, phase 3 | NCT06067230 | Not yet recruiting | ModernaTX, Inc. | ||
| N/A | Observational | NCT05572658 | Active, not recruiting | ModernaTX, Inc. | ||
| Intramuscular | Interventional, phase 3 | NCT06060457 | Not yet recruiting | ModernaTX, Inc. | ||
| Intramuscular | Interventional, phase 2 & 3 | NCT05127434 | Active, not recruiting | ModernaTX, Inc. | ||
| mRNA-1345 mRNA encoding RSV stabilized prefusion F glycoprotein & mRNA-1365 mRNAs encoding RSV stabilized prefusion F glycoprotein and hMPV F protein | Respiratory syncytial virus & human metapneumovirus | Intramuscular | Interventional, phase 1 | NCT05743881 | Recruiting | ModernaTX, Inc. |
| CV7202 RABV-G mRNA | Rabies virus | Intramuscular | Interventional, phase 1 | NCT03713086 | Completed | CureVac |
| mRNA-1944 mRNA encoding the heavy and light chains of a CHIKV-specific monoclonal neutralizing antibody, CHKV-24 | Chikungunya virus | Intramuscular | Interventional, phase 1 | NCT03829384 | Completed | ModernaTX, Inc. |
| mRNA-1388 mRNA encoding full CHIKV structural polyprotein from CHIKV West African strain 37,997 | Chikungunya virus | Intramuscular | Interventional, phase 1 | NCT03325075 | Completed | ModernaTX, Inc. |
| mRNA-1647 gB and PC mRNAs | Cytomegalovirus | Intramuscular | Interventional, phase 2 | NCT04232280 | Completed | ModernaTX, Inc. |
| mRNA-1653 mRNAs encoding full-length membrane-bound fusion proteins of MPV and PIV3 | Metapneumovirus and parainfluenza virus type 3 | Intramuscular | Interventional, phase 1 | NCT03392389 | Completed | ModernaTX, Inc. |
| mRNA-1325 prME mRNA from Micronesia 2007 Zika virus isolate | Zika virus | Intramuscular | Interventional, phase 1 | NCT03014089 | Completed | ModernaTX, Inc. |
| mRNA-1893 prME mRNA from RIO-U1 Zika virus | Zika virus | Intramuscular | Interventional, phase 1 | NCT04064905 | Completed | ModernaTX, Inc. |
| V3G CH848 mRNA-Tr2 Native-like HIV-1 Envelope Trimer mRNA | HIV | Not specified | Interventional, phase 1 | NCT05903339 | Recruiting | National Institute of Allergy and Infectious Diseases (NIAID) |
| Drug name/mRNA | Indication | Administration route | Study type, phase | Clinical Trial ID | Status | Sponsor |
|---|---|---|---|---|---|---|
| HNSCC: head and neck squamous cell carcinoma; NSCLC: non-small cell lung cancer. The information summarized in this table was obtained by searching the ClinicalTrials.gov database with the following keywords: [mRNA, lipid nanoparticle], [mRNA, LNP], [messenger RNA, lipid nanoparticle], [messenger RNA, LNP], [messenger ribonucleic acid, lipid nanoparticle] and [LNP, vaccine] between August and December 2023. | ||||||
| OTX-2002 MYC epigenetic controller mRNA | Hepatocellular carcinoma | Intravenous | Interventional, phase 1 & 2 | NCT05497453 | Recruiting | Omega Therapeutics |
| mRNA-5671 (V941) G12D, G12V, G13D, and G12C KRAS mutations mRNAs | Carcinoma, non-small-cell lung & pancreatic neoplasms, colorectal neoplasms | Intramuscular | Interventional, phase 1 | NCT03948763 | Completed | Merck Sharp & Dohme LLC |
| mRNA-4157 (V940) mRNA encoding up to 34 patient-specific neoantigens | NSCLC | Intramuscular | Interventional, phase 3 | NCT06077760 | Recruiting | Merck Sharp & Dohme LLC |
| Melanoma | Intramuscular | Interventional, phase 3 | NCT05933577 | Recruiting | Merck Sharp & Dohme LLC | |
| Intramuscular | Interventional, phase 2 | NCT03897881 | Recruiting | ModernaTX, Inc. | ||
| Solid tumors | Intramuscular | Interventional, phase 1 | NCT03313778 | Active, not recruiting | ModernaTX, Inc. | |
| mRNA-4650 mRNA backbone encoding 20 patient-specific neoantigens | Melanoma, colon cancer, gastrointestinal cancer, genitourinary cancer, hepatocellular cancer | Intramuscular | Interventional, phase 1 & 2 | NCT03480152 | Terminated (slow accrual) | National Cancer Institute (NCI) |
| mRNA-2752 OX40L, IL-23 and IL-36γ mRNAs | Relapsed/refractory solid tumor malignancies, lymphoma, triple negative breast cancer, HNSCC, non-hodgkins, urothelial cancer, immune checkpoint refractory melanoma, and NSCLC lymphoma | Intratumoral | Interventional, phase 1 | NCT03739931 | Active, not recruiting | ModernaTX, Inc. |
| mRNA-2416 OX40L mRNA | Ovarian cancer, relapsed/refractory solid tumor malignancies, lymphoma | Intratumoral | Interventional, phase 1 & 2 | NCT03323398 | Terminated (efficacy endpoints not met) | ModernaTX, Inc |
| MEDI1191 IL-12 mRNA | Advanced solid tumors | Intratumoral | Interventional, phase 1 | NCT03946800 | Completed | MedImmune LLC |
| Drug name/mRNA | Indication | Admin. route | Study type, phase | Clinical Trial ID | Status | Sponsor |
|---|---|---|---|---|---|---|
| MUT: methylmalonil-CoA mutase; PCCA: propionyl-CoA carboxylase, subunit A; PCCB: propionyl-CoA carboxylase, subunit B; CFTR: cystic fibrosis transmembrane conductance regulator; OTC: ornithine transcarbamylase. The information summarized in this table was obtained by searching the ClinicalTrials.gov database with the following keywords: [mRNA, lipid nanoparticle], [mRNA, LNP], [messenger RNA, lipid nanoparticle], [messenger RNA, LNP], [messenger ribonucleic acid, lipid nanoparticle] and [LNP, vaccine] between August and December 2023. | ||||||
| mRNA-3704 MUT mRNA | Methylmalonic acidemia (MMA) | Intravenous | Interventional, phase 1 & 2 | NCT03810690 | Withdrawn (business decision) | ModernaTX, Inc. |
| mRNA-3927 PCCA & PCCB mRNAs | Propionic acidemia | Intravenous | Interventional, phase 1 & 2 | NCT04159103 | Recruiting | ModernaTX, Inc. |
| MRT5005 CFTR mRNA | Cystic fibrosis | Inhalation | Interventional, phase 1 & 2 | NCT03375047 | Unknown state | Translate Bio, Inc. |
| ARCT-032 CFTR mRNA | Cystic fibrosis | Inhalation | Interventional, phase 1 | NCT05712538 | Recruiting | Arcturus Therapeutics, Inc |
| ARCT-810 OTC mRNA | Ornithine transcarbamylase deficiency | Intravenous | Interventional, phase 2 | NCT05526066 | Recruiting | Arcturus Therapeutics, Inc |
| Intravenous | Interventional, phase 1b | NCT04442347 | Active, not recruiting | Arcturus Therapeutics, Inc. | ||
| Intravenous | Interventional, phase 1 | NCT04416126 | Completed | Arcturus Therapeutics, Inc. | ||
| MRT5201 OTC mRNA | Ornithine transcarbamylase deficiency | Intravenous | Interventional, phase 1 & 2 | NCT03767270 | Withdrawn (Program was discontinued) | Translate Bio, Inc. |
| Drug name/mRNA | Indication | Admin. route | Study type, phase | Clinical Trial ID | Status | Sponsor |
|---|---|---|---|---|---|---|
| sgRNA: single-guide RNA, TTR: transthyretin, Cas9: CRISPR associated protein 9. The information summarized in this table was obtained by searching the ClinicalTrials.gov database with the following keywords: [mRNA, lipid nanoparticle], [mRNA, LNP], [messenger RNA, lipid nanoparticle], [messenger RNA, LNP], [messenger ribonucleic acid, lipid nanoparticle] and [LNP, vaccine] between August and December 2023. | ||||||
| NTLA-2001 (sgRNA against the TTR gene and Cas9 mRNA) | Transthyretin-related (ATTR) familial amyloid polyneuropathy, transthyretin-related (ATTR) familial amyloid cardiomyopathy, wild-type transthyretin cardiac amyloidosis | N/A | Observational | NCT05697861 | Recruiting | Intellia Therapeutics |
| Intravenous | Interventional, phase 3 | NCT06128629 | Recruiting | Intellia Therapeutics | ||
| Intravenous | Interventional, phase 1 | NCT04601051 | Active, not recruiting | Intellia Therapeutics | ||
2. Lipid nanoparticles: components, formulation and mRNA delivery mechanism
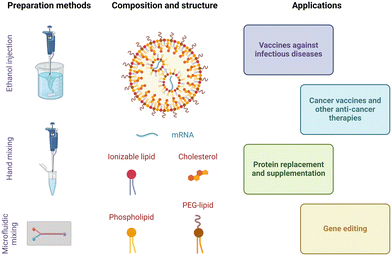
Lipid nanoparticles are spherical and nanoscale delivery systems (typically, 80–200 nm in diameter) composed of several distinct types of lipids. The groundwork for this colloidal formulation was laid in Pieter Cullis’ laboratory decades ago.20 His group's basic research on lipid asymmetry enabled them to develop an ‘ionizable cationic lipid’ that can exist in either a neutral or protonated form depending on pH. This turned out to be extremely useful for gene therapy, because these lipids are able to efficiently encapsulate highly negatively charged nucleic acids and to promote endosomal escape when positively charged, at low pH, while reducing in vivo toxicity thanks to their close to neutral charge at physiological pH. Today, lipid nanoparticles formulated with ionizable cationic lipids constitute the most clinically mature delivery vectors for nucleic acid therapeutics. They are the formulations behind Patisiran (Onpattro®, siRNA drug) and the Moderna and Pfizer/BioNTech mRNA vaccines against SARS-CoV-2. Schematic representations of a typical lipid nanoparticle mRNA formulation, together with manufacturing methods and biomedical applications, are provided in Fig. 1.Composition
Lipid nanoparticles are typically composed of an ionizable lipid, a phospholipid, cholesterol, and a lipid conjugated to polyethylene glycol (PEG-lipid). These components are crucial for efficiently protecting and delivering mRNA into cells.The chemical identity of the ionizable lipid has also been seen to impact the biodistribution profile of lipid nanoparticles.36–40 Escalona-Rayo et al. demonstrated in vivo biodistribution of DiD-labeled lipid nanoparticles containing clinically approved ionizable cationic lipids (MC3, SM-102, and ALC-0315). They discovered that 24 hours after injection, these lipid nanoparticles had accumulated in endothelial cells and the extravascular space throughout the zebrafish embryo body. Notably, ALC-0315-based lipid nanoparticles accumulated the most in the extravascular space.36 Lam et al. compared intramuscular administration of firefly luciferase mRNA encapsulated in lipid nanoparticles containing lipid benchmarks (MC3, SM-102, and ALC-0315), in mice. Six hours post-injection, ALC-0315 and SM-102 nanoparticles provided comparable luciferase expression levels at the injection site, which outperformed those observed with MC3 lipids. Additionally, ALC-0315 nanoparticles showed high luciferase expression in the liver, unlike the other formulations.37 When systemically administered, MC3-containing lipid nanoparticles have been consistently shown to accumulate in the liver preferentially.38–40 However, nanoparticles incorporating ionizable lipids with quaternary ammoniums have shown preferential mRNA delivery to the lungs.41 New ionizable lipids continue to be synthesized, and combinatorial design and screenings of large lipid libraries have been implemented to identify formulations that preferentially accumulate in specific organs.23 For example, using combinatorial design and synthesis together with high-throughput in vitro screening, Chen et al. identified the ionizable lipid Iso-A11B5C1 that enables efficient muscle-targeted mRNA delivery with minimal off-target effects, in mice.42 A remarkable advance has been the implementation of a high throughput DNA barcoding system, first demonstrated by Dahlman and colleagues, to simultaneously investigate the biodistribution of a library of tens of different lipid nanoparticles in vivo.43
Formulation
Lipid nanoparticles can be prepared by several methods. All of them share the rapid mixing of an ethanolic lipid mixture with an aqueous mRNA solution, at low pH, to produce lipid nanoparticles via self-assembly.59 Ethanol injection and hand mixing are bulk processes that do not require specialized equipment and are accessible to all laboratories.60,61 Ethanol injection is performed by mixing the ethanolic and aqueous phases with the aid of a magnetic stir plate. Hand-mixing simply involves rapidly pipetting the ethanolic lipid mixture into the aqueous nucleic acid solution. However, due to a lack of precise control over the mixing speed, both processes tend to yield heterogeneous and larger particles (>100 nm in diameter) with low encapsulation efficiency and high batch-to-batch variability. This is important because particle size at least partially dictates the pharmacokinetics and transfection efficiency of lipid nanoparticles in vivo.62 Alternatively, microfluidic mixing has been designed to minimize this variability and achieve better control over particle size.63,64 In this method, streams of the ethanol and aqueous phases are forced to mix rapidly within a microfluidic channel. The lipids begin to precipitate and self-assemble into nanoparticles due to reciprocal diffusion of ethanol and water over the ethanol/water interface. Particle size is predominantly determined by the mixing time (t) and solvent diffusion coefficient (D).65 The architecture and dimensions of the microfluidic channel also regulate the hydrodynamic-focusing flow behaviour and the mixing process can be regulated by adjusting the width of the focused stream (w) in eqn (1).66 Overall, this process allows precise control of particle size, more homogeneous size distribution, higher encapsulation efficiency, and batch-to-batch consistency than bulk processes.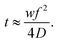 | (1) |
mRNA encapsulation and delivery mechanism
During lipid nanoparticle-mRNA preparation, regardless of the method of choice, the ionizable lipid becomes protonated in contact with the acidic environment provided by the aqueous mRNA phase (generally, a buffer with pH below 5.0).67 Electrostatic interactions between the positively charged lipids and negatively charged mRNA molecules, together with lipid self-assembly, enable encapsulation of the nucleic acid.68 Then, the lipid nanoparticle-mRNA suspension is dialyzed to neutral pH, so ionizable lipids become uncharged and thus less toxic.44Cellular uptake of lipid nanoparticles is mediated by endocytosis.69 However, unlike other nanoscale delivery vectors, ionizable lipid-based nanoparticles can efficiently escape the endosome largely thanks to the membrane-destabilizing effect of the ionizable lipid,20,70 it is hypothesized. In brief, ionizable lipids become again protonated in the acidic environment of the endosome and interact with naturally occurring anionic phospholipids in the endosomal membrane to form cone-shaped ion pairs that are incompatible with a bilayer.71 The cationic–anionic lipid pair drives the transition from the bilayer structure to the inverted hexagonal HII phase, which promotes membrane destabilization and endosomal escape of lipid nanoparticles.23 Ultimately, the mRNA cargo is released into the cytoplasm, where it can be translated into protein in the ribosomes.
3. Lipid nanoparticle-mRNA vaccines for infectious diseases
To date, successful vaccination campaigns have eradicated more than 20 life-threatening diseases, and the World Health Organization estimates that immunization saves 3.5 to 5 million lives yearly that would otherwise be lost to diphtheria, tetanus, pertussis, influenza and measles.72 Furthermore, it is estimated that in the first year of the COVID-19 vaccination program, successful vaccine development avoided 14.4 million fatalities worldwide.73 The outbreak of the COVID-19 pandemic expedited the fastest vaccine development in global history, with nucleic acid vaccines based on mRNA at the forefront of those achievements. When compared to conventional vaccines that use inactivated, live attenuated, and recombinant microbes, which require lengthy, often several years long development, the rapid manufacturing process of mRNA makes this class of vaccines a promising candidate to bridge the time gap between pathogen emergence and vaccine licensure. Additionally, large-scale production of mRNA can be achieved via a one or two-step in vitro cell-free synthesis followed by relatively simple purification74 and the lack of infectivity and genomic integration capabilities of mRNA add extra layers of safety. Vaccines against infectious diseases based on lipid nanoparticle mRNA delivery that are currently undergoing clinical trials are summarized in Table 1. These include mRNA-based immunizations against Influenza, Zika, HIV, Rabies, Cytomegalovirus, Chikungunya and Respiratory Syncytial Virus. Below, we provide more information on the development of some of these promising candidates.Influenza viruses
Mortality owing to influenza viruses is estimated at 290![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–650
000–650![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 yearly deaths worldwide.75 Influenza viruses are single-stranded RNA viruses classified into four types: A, B, C, and D, with type A viruses causing seasonal epidemic or pandemic outbreaks in animals and humans, including the possibility of animal-to-human and human-to-animal transmission.76 Current vaccination technology primarily targets hemagglutinin (HA) glycoproteins in the viral surface, which promote viral entry.77 Neuraminidase (NA), an enzyme essential for the release and spread of progeny virions is also often targeted.78 However, because of the diversity of endemic influenza strains, as well as virus evolution by point mutations in HA and NA (denoted as “antigenic drift” and “antigenic shift”, respectively), annual vaccine efficacy ranges from 10 to 60% only.79 As a result, influenza vaccines must be reviewed and reformulated on a yearly basis to ensure vaccine efficacy. Another caveat is that most influenza antigens, such as those included in intramuscular inactivated vaccines and intranasal live-attenuated vaccines, are prepared in chicken eggs. These take a long time to produce and purify, and viral mutations in chicken eggs can render them ineffective in humans. For example, the loss of a glycosylation site in the HA of egg-adapted H3N2 vaccine strain resulted in poor efficacy during the 2016–2017 influenza season.80 Alternate antigen targets and production methods are in high demand and in vitro transcribed synthetic mRNAs stand as promising candidates thanks to ease of antigen modification and rapid large-scale production if an entirely new influenza strain emerges globally. In 2013, eight days after the sequences coding for the HA and NA genes of the H7N9 influenza virus were available, a lipid nanoparticle-based self-amplifying mRNA vaccine candidate was formulated.81 At the time, progress to a Phase I trial was halted due to lack of GMP facilities for mRNA manufacturing, but this case still exemplifies how fully synthetic vaccine technology could enable remarkable speed in responding to the initial wave of pandemic influenza viruses.
000 yearly deaths worldwide.75 Influenza viruses are single-stranded RNA viruses classified into four types: A, B, C, and D, with type A viruses causing seasonal epidemic or pandemic outbreaks in animals and humans, including the possibility of animal-to-human and human-to-animal transmission.76 Current vaccination technology primarily targets hemagglutinin (HA) glycoproteins in the viral surface, which promote viral entry.77 Neuraminidase (NA), an enzyme essential for the release and spread of progeny virions is also often targeted.78 However, because of the diversity of endemic influenza strains, as well as virus evolution by point mutations in HA and NA (denoted as “antigenic drift” and “antigenic shift”, respectively), annual vaccine efficacy ranges from 10 to 60% only.79 As a result, influenza vaccines must be reviewed and reformulated on a yearly basis to ensure vaccine efficacy. Another caveat is that most influenza antigens, such as those included in intramuscular inactivated vaccines and intranasal live-attenuated vaccines, are prepared in chicken eggs. These take a long time to produce and purify, and viral mutations in chicken eggs can render them ineffective in humans. For example, the loss of a glycosylation site in the HA of egg-adapted H3N2 vaccine strain resulted in poor efficacy during the 2016–2017 influenza season.80 Alternate antigen targets and production methods are in high demand and in vitro transcribed synthetic mRNAs stand as promising candidates thanks to ease of antigen modification and rapid large-scale production if an entirely new influenza strain emerges globally. In 2013, eight days after the sequences coding for the HA and NA genes of the H7N9 influenza virus were available, a lipid nanoparticle-based self-amplifying mRNA vaccine candidate was formulated.81 At the time, progress to a Phase I trial was halted due to lack of GMP facilities for mRNA manufacturing, but this case still exemplifies how fully synthetic vaccine technology could enable remarkable speed in responding to the initial wave of pandemic influenza viruses.
Another key advantage of lipid nanoparticle-mRNA technology is the possibility to co-administer several antigens in one immunization. This directly resonates with long-pursued efforts to develop a “universal influenza virus vaccine” that would improve current suboptimal vaccine effectiveness by providing immunity against a variety of influenza strains and subtypes. For example, intradermal vaccination of mice with mRNA encoding full-length HA of influenza A/PuertoRico/8/1934 demonstrated long-lived and protective activity against influenza A virus infection (H1N1, H3N2, and H5N1 strains).82 A single immunization with 30 μg of full-length HA-encoding mRNA in lipid nanoparticles (A/California/07/2009 HA lipid nanoparticle-mRNAs) induced antibody responses against both homologous H1N1 and heterologous H1N1 viruses in mice, and protected them from a lethal dose of H5N1 viruses.83 Another study in 2022 employed a lipid nanoparticle-mRNA vaccine encoding separate hemagglutinin antigens from all 20 known influenza A virus subtypes and influenza B virus lineages.84 This multivalent vaccine elicited substantial levels of cross-reactive and subtype-specific antibodies and provided protection against matched and mismatched viral strains in both immunized mice and ferrets. Currently, lipid nanoparticle mRNA influenza vaccines dominate the clinical trial scene of this technology, only after those against SARS-CoV2, and may soon be widely available. Moderna's lead candidate seasonal flu vaccine (mRNA-1010) is currently in Phase 3 clinical trials, while Pfizer has begun a Phase 3 trial of a quadrivalent mRNA influenza vaccine candidate in the United States.85
Zika virus
The Zika virus (ZIKV) is a mosquito-borne virus that was first discovered in 1947, with evidence of infection and illnesses in humans emerging in the 1950s. The majority of individuals infected by ZIKV do not develop symptoms. However, ZIKV emerged as a global health threat when the causal link between ZIKV and congenital malformations was revealed.86 There is currently no treatment or prophylactic intervention for ZIKV infection or illness, but a neutralization study with ZIKV-immune convalescent sera in 2016 revealed a single serotype, implying that vaccination against an antigen from contemporary South American, Asian, and early African ZIKV strains can elicit broadly neutralizing antibodies against multiple strains.87 The flavivirus envelope (E) protein is the primary target of neutralizing antibodies.88 A proof-of-concept DNA vaccine was first developed using the coding sequences of the viral membrane and envelop protein (prM-E) against ZIKV and tested in mice89 and rhesus monkeys.90 It generated high-levels of ZIKV-specific neutralizing antibodies and protected both species from ZIKV challenges.89,90 A year later, a single low-dose (30 μg in mice or 50 μg in non-human primates) intradermal immunization with a lipid nanoparticle-mRNA vaccine encoding prM-E elicited rapid and lasting protective immunity. The anti-ZIKV neutralizing antibodies produced by this mRNA vaccine were orders of magnitude higher than those produced by DNA vaccines in mice and rhesus macaques.91 Furthermore, a lipid nanoparticle modified mRNA prM-E subunit vaccine has been optimized to avoid the generation of cross-reacting antibodies that could enhance Dengue virus infection.92 This vaccine was tested in pregnant mice, which showed diminished viral levels, and most fetuses presented no evidence of infection.93 This technology completed Phase 1 clinical trials in healthy adults in 2019 (NCT03014089, NCT04064905).94HIV
HIV/AIDS has been one of the world's deadliest and most persistent epidemics since the first cases were recorded in 1981. Globally, 39 million people were living with HIV at the end of 2022.95 In contrast to the development of vaccines against SARS-CoV2, accomplished in record time, no effective HIV immunization has been achieved after 40 years of research and the testing of more than 200 vaccine candidates. Developing an effective HIV vaccine has been an uphill struggle, primarily owing to the antigenic diversity of the HIV envelope protein, the extensive ‘glycan shield’ that conceals critical envelope protein epitopes, and the ability of retroviruses to integrate in the host DNA.96 Now, recent advances in mRNA vaccines could help overcome this roadblock and several studies have already utilized lipid nanoparticles towards this goal. In 2019, Blakney et al. reported a systematic comparison of lipid nanoparticles carrying self-amplifying RNA encoding HIV-1 Env gp140 on the particle's interior versus the exterior, providing an alternative way to the RNA encapsulation paradigm.97 Two years later, Zhang et al. showed that a lipid nanoparticle vaccine co-encapsulating mRNAs encoding for membrane-anchored HIV-1 envelop and simian immunodeficiency virus Gag proteins resulted in a 79% per-exposure risk reduction from SHIV AD8 in vaccinated macaques.98 However, although this mRNA platform offers a promising approach for developing a preventative HIV-1 vaccination, it requires seven or more sequential immunizations, which would be challenging to implement in humans.Generally, the most effective, clinically used vaccines elicit neutralizing antibodies as their primary mode of protection.99 Thus, the induction of antibodies that broadly protect against heterologous HIV-1 strains is also a topmost objective of HIV-1 vaccine development. Broadly neutralizing antibodies (bNbs), particularly VRC01, that targets the HIV-1 envelope glycoprotein are an innovative HIV vaccination approach. In 2016, Pardi et al. showed that humanized mice were protected against an intravenous HIV-1 challenge by a single administration of a lipid nanoparticle-mRNA encoding VRC01.83 A combination regime of ferritin and lipid nanoparticles delivering mRNAs that encode for HIV neutralizing antibodies has recently entered Phase 1 clinical trials (NCT05903339).
4. Lipid nanoparticle-mRNA cancer vaccines and other anti-cancer therapies
Responsible for nearly one in six demises per year, cancer remains a leading cause of death worldwide.100 However, in the last decades, fundamental advancements in cancer immunology have unveiled new strategies to harness the power of the immune system to fight cancer. Synthetic mRNA serves as a template for the translation of virtually any given protein, so it can be a versatile tool to participate in many of such strategies. For example, mRNA can encode tumor-associated antigens,101 so it can stimulate antigen-specific anti-tumor immune responses, chimeric antigen receptors,102 to engineer CAR-T cells, cytokines,103 to modify the immune-suppressive tumor microenvironment, and anti-tumor antibodies,104 such as anti-HER2. Another advantage of lipid nanoparticle mRNA technologies in the context of cancer immunotherapy is that these delivery vectors tend to accumulate in immune-rich organs such as the liver and spleen and can be internalized by antigen-presenting cells.105 What has been a long-standing caveat in the cancer nanomedicine field is now an opportunity for cancer nano-immunotherapy. The most relevant clinical trials involving cancer vaccines and other anti-cancer therapies based on lipid nanoparticle mRNA technology are summarized in Table 2.mRNA cancer vaccines
Prophylactic and therapeutic cancer vaccines aim to strengthen the patient's adaptive immune system against specific tumor antigens, providing immunological memory and ensuring robust CD8+ T cell activation to suppress tumor growth through direct killing. These vaccines typically involve exogenous delivery of tumor-specific antigens coupled with adjuvants, in free form or carried by nanoparticles, that are processed by and activate antigen presenting cells. Therefore, identification of tumor-specific antigens is a key aspect in the successful development of anti-cancer vaccines, and can be challenging due to high heterogeneity among cancers and individuals.106A common strategy is to utilize proteins that are mutated in cancer cells as antigens. This ensures that the immune response targets tumor cells specifically and minimizes off-target effects. A lipid nanoparticle mRNA vaccine encoding for four mutations of KRAS (G12D, G12V, G13D, and G12C) has recently completed Phase 1 clinical trials (NCT03948763), including patients with non-small-cell lung cancer (NSCLC), pancreatic and colorectal neoplasms. However, not all cancers express conserved mutations across different patients and, in the last few years, personalized cancer immunotherapy and, especially, the search for patient-specific neoantigens, have gained traction thanks to advances in next-generation sequencing,107 computational algorithms,108 and machine learning.109 Moreover, the deployment of the neoantigen paradigm may also benefit from the rapid mRNA production process and the facile co-administration of multiple sequences enabled by lipid nanoparticles. For example, the lipid nanoparticle-mRNA formulation mRNA-4157 encodes up to 34 patient-specific tumour neoantigens that are administered intramuscularly. This vaccine is now being tested in Phase I trials in patients with unresectable solid tumors (NCT03313778). A total of 79 patients received at least one dose of mRNA-4157, and preliminary data concluded that this vaccine has an acceptable safety profile in combination with the anti-PD-1 antibody pembrolizumab, with only low-grade and reversible treatment-related adverse events such as fatigue, injection-site soreness, colitis, and myalgia reported.110 In parallel, Phase 2 and 3 clinical trials (NCT03897881 and NCT05933577, respectively) are testing this therapy in patients with high-risk melanoma111 and a Phase 3 trial involving NSCLC is also underway (NCT06077760). Similarly, mRNA-4650, developed by the National Cancer Institute (NCI), involved an mRNA backbone encoding for 20 neoantigens. Unfortunately, a Phase 1 and 2 trial of this technology was terminated due to low patient accrual (NCT03480152).
Cancer treatment with other lipid nanoparticle-mRNA therapeutics
Lipid nanoparticle-mRNA formulations encoding cytokines aim to reprogram the immunosuppressive tumor microenvironment,112 can be administered either as monotherapies or in combination with other immunotherapies, and through systemic or local routes depending on the anatomical location of the tumor.6,113 This strategy can mitigate the toxicity associated with recombinant cytokine therapies in their free form114 thanks to preferential accumulation of the particles at the tumor site, effectively transforming into a cytokine production center,6,112 and minimizing off-target effects. An example is the administration of lipid nanoparticle-mRNA encoding Interleukin-12 (IL-12) for the treatment of MYC-driven hepatocellular carcinoma.115 After intravenous administration, the nanoparticles accumulate in the liver and local IL-12 production triggers a substantial influx of CD4+ T helper cells into the tumor and surrounding liver tissue, increasing IFN-γ production and significantly extending survival in mice. Lipid nanoparticles encapsulating IL-12 mRNA have also been administered intratumorally to promote local IL-12 production and enhance T cell response with improved tolerability compared with systemic administration for extrahepatic tumors.116 The combination of these particles (MEDI1191) with the anti-PD-L1 antibody durvalumab successfully concluded an open-label Phase 1 trial for the treatment of advanced solid tumors (NCT03946800).117 The combination proved to be safe and feasible in patients, showing preliminary evidence of clinical benefit, with 29.0% of patients exhibiting partial responses or maintaining stable disease as their best overall response at 12 weeks. Indeed, local administration is more suitable when the tumor is localized at a site where lipid nanoparticles do not tend to accumulate upon systemic administration. For example, Hewitt et al. intratumorally injected a cocktail of mRNA-encoding cytokines (IL-23, IL-36γ, and OX40L) formulated in lipid nanoparticles in various tumor models in mice.118 This local therapy not only led to the clearance of distal untreated tumors but also ignited immune memory to prevent cancer recurrence, and the expressed triplet mRNA therapy induced a broader immune response compared to treatment with mRNA coding for any of these cytokines individually. Moderna is currently leading a Phase 1 clinical trial based on the intratumoral co-administration of these three mRNAs (NCT03739931) after the administration of OX40L mRNA alone did not meet its efficacy endpoints (NCT03323398).CAR-T cell therapy is another anti-cancer treatment that may be improved via lipid nanoparticle-mRNA technology. Engineering lipid nanoparticles able to target and transfect T cells with CAR receptors in their mRNA form in vivo has been demonstrated by including targeting antibodies, and may bypass the complications and high costs associated with ex vivo CAR-T cell production.38 Moreover, the inherent transiency of mRNA may also ameliorate CAR-T cell induce cytotoxic effects.
5. Lipid nanoparticle-mRNA for protein replacement or supplementation
Lipid nanoparticle-mRNA formulations have also been investigated to provide proteins that alleviate genetic or acquired diseases. Delivery of the mRNA form transforms the cell into its own protein factory and offers certain advantages including easier delivery (especially when working with high molecular weight proteins), facile mRNA and thus protein modification, and enhanced stability thanks to lipid nanoparticle encapsulation.119 However, a caveat to consider, particularly for genetic diseases that may require chronic protein supplementation, is the short half-life of mRNA, that typically decays in 48 hours.In terms of protein replacement in the context of genetic diseases, clinical trials of lipid nanoparticle-mRNA therapeutics have mainly focused on metabolic disorders, including methylmalonic acidaemia (NCT03810690), propionic acidaemia (NCT04159103), and ornithine transcarbamylase deficiency (NCT03767270). The supplement of deficient enzymes in their mRNA forms can slow down disease progression. Two clinical trials for cystic fibrosis have also been registered (NCT05712538, NCT03375047). All of these trials are summarized in Table 3. In addition, mRNA-based protein replacement therapies have been applied to phenylketonuria,120 arginase I deficiency121 and haemophilia A122 and B,123 in preclinical studies.
Others have also deployed lipid nanoparticle-mRNA drugs to deliver proteins over specific periods of time, to re-establish tissue function of induce regeneration, although all of these are still at the preclinical stage. For example, Swingle et al. screened different ionizable lipids to engineer lipid nanoparticles with preferential accumulation in the mouse placenta. They used this platform to deliver VEGF mRNA and promote placental vasodilation during pregnancy, with a better safety profile than benchmark C12–200 lipid nanoparticles. This approach could be developed into a therapeutic strategy against pre-eclampsia.124 Interestingly, lipid nanoparticles formulated with specific ionizable lipids have also demonstrated in utero mRNA delivery that in turns triggers the production of therapeutic proteins in the fetus’ liver.125 This technology could be further developed to achieve in utero protein replacement or gene editing.
6. Lipid nanoparticle-mRNA for genome editing
Gene editing mediated by regularly interspaced short palindromic repeat (CRISPR)/CRISPR associated protein (Cas) has emerged as the third generation of gene manipulation technology following zinc finger and TALEN. The UK MHRA and US FDA have recently approved the first ever ex vivo gene editing therapy for the treatment of sickle cell anemia, CRISPR-enabled Exa-cel®, so we may see other similar therapies reaching clinical practice in the near future. CRISPR gene editing is broadly classified into three types: non-homologous end joining (NHEJ), homology-directed repair (HDR), and base editing. NHEJ and HDR primarily aim to interrupt the expression of disease-causing genes or completely replace them, whereas base editing makes targeted changes to the gene sequences and normalizes protein production.126–128 CRISPR/Cas gene editing typically requires the delivery of – at least – a type of Cas enzyme (e.g., Cas9), responsible for cutting DNA at specific sites of the genome, and a single-guide RNA (sgRNA) that guides Cas9 to that specific site.129 However, because of the large size of Cas9 (160 kDa, 4300 bases) and concerns of increased off-target effects if the expression of the editing system is sustained for an extended period of time, efficient and safe delivery of CRISPR-Cas9 for gene editing is difficult to achieve with conventional viral and nonviral delivery systems.130 Lipid nanoparticles co-encapsulating sgRNA and Cas9 in its mRNA form could help bypass these caveats. Indeed, in vivo NHEJ editing of the Polo-like Kinase 1 (Plk1) gene was achieved with efficiency as high as 70% with a system using a novel ionizable amino lipid with dilinoleyl chain and tertiary amine head group. Plk1 editing caused tumor cell apoptosis, inhibited tumor growth by 50%, and improved survival by 30% in glioblastoma-bearing mice, after direct intracranial injection.131 HDR requires additional delivery of donor DNA as repair template. There have been very few reports of HDR using lipid nanoparticles, and editing efficiencies have been disappointing, likely due to limited nuclear translocation of the DNA.132 However, a recent report described two improvements to augment the efficiency of HDR ex vivo, in primary cells. First, the HDR template can be engineered to interact with Cas9 ribonucleoproteins (RNPs) that shuttle DNA to the nucleus, increasing HDR efficiency by two- to fourfold. Second, stabilizing Cas9 RNPs into nanoparticles with polyglutamic acid further enhances editing efficiency.133In vivo cytidine base editing using lipid nanoparticles as delivery vectors has also been demonstrated and corrected phenylketonuria in a transgenic mouse model.134,135 Moreover, prime editing (PE), a groundbreaking gene-editing technique introduced in 2019, offers a more precise method for rewriting new genetic information into the genome compared to earlier techniques.136 PE facilitates the direct introduction of new genetic information into a specific DNA site, allowing for precise modification of the DNA without causing double-strand breaks. The PE system involves a catalytically impaired Cas9 endonuclease fused with an engineered reverse transcriptase, guided by a prime editing guide RNA, to directly modify specific genetic sequences within the genome with enhanced precision and minimal off-target effects.136 Chen et al. recently demonstrated lipid nanoparticle-mediated simultaneous delivery of chemically modified pegRNA and PE mRNA, which enabled gene editing in wild-type mice.137 Currently, the biotech company Prime Medicine is developing an extensive pipeline using PE for the treatment of diseases as diverse as cystic fibrosis, metabolic diseases, and hematologic disorders, among many others, with several of their candidates exploiting lipid nanoparticle technology. However, at the time of writing this article none of them have reached clinical investigation.138 Clinical trials involving genome editing based on lipid nanoparticle mRNA technology are summarized in Table 4.7. Challenges in the clinical translation of lipid nanoparticle-mRNA therapeutics
Despite the recent success and widespread adoption of lipid nanoparticle-mRNA COVID-19 vaccines and other remarkable preclinical and clinical advances highlighted in this article, there are still key areas of improvement and challenges ahead in increasing the presence of this type of therapy in the clinical practice.Tissue and cell targeting
Clinical translation of lipid nanoparticle-mRNA therapeutics has been limited in practice due to poor understanding of how to target specific organs or cell types, especially when local administration is not an option. A common caveat in nanomedicine, systemic administration via intravenous injection typically results in predominant accumulation in liver and spleen.105 Strategies to target other organs have been developed and remain an important area or research. For example, large screenings of ionizable lipids have been performed, as discussed in Section 2, to identify those that skew nanoparticle biodistribution to certain tissues and organs.41,42,139 A different approach, Selective ORgan Targeting (SORT), has shown excellent capabilities to shift biodistribution from liver to spleen and lung.140 SORT consists in the addition of a fifth component – typically a permanently cationic or anionic lipid – to the lipid nanoparticle formulation. By changing the internal charge of the formulation, changes in biodistribution and organ targeting are achieved. Active targeting of lipid nanoparticles by including cell-specific ligands has also been explored. For example, CD4+ T cells in the spleen have been successfully targeted by conjugating a CD4 antibody on the lipid nanoparticle surface.141 However, it is to be mentioned that this strategy has not yet contributed any FDA approved formulations, even in the case of more established nanoparticles.105 A detailed review of current targeting technologies for lipid nanoparticles has been recently published by Dilliard and Siegwart.142Biocompatibility and safety
Lipid nanoparticles have demonstrated an overall favorable safety profile, thanks to the absence of positive charges at neutral pH and the biodegradability of most if not all of their components. However, some concerns that warrant further research are related to the use of PEG. Anti-PEG antibodies – that may be pre-existing or newly formed – have been detected is some patients administered with PEG-containing lipid nanoparticle mRNA vaccines. These antibodies can induce hypersensitivity reactions and rapid blood clearance of the nanoparticles,143,144 which is not only a health risk but could also decrease the therapeutic efficacy of the treatment. As for the lipidic components, the current trend aims to move towards the use of biodegradable lipids. Advances in lipid synthesis are also enabling the development of ever-increasing ionizable lipid libraries. The safety profile of every newly synthesized lipid should be thoroughly investigated. Lastly, as new formulations enable targeting of tissues other than commonly targeted liver, spleen and muscle, their biocompatibility in such organs must be also investigated.The main known safety risk related to synthetic mRNA is its immunogenicity, which could in turn cause endothelial damage, edema, hypercoagulation, and thromboembolic events among other problems.145 To abate the immunogenicity of mRNA, two approaches are commonly used. First, substitution of uridine with pseudouridine (ψ) or N1-methylpseudouridine (m1ψ), among other non-immunogenic modified nucleosides, in the mRNA sequence, prevents innate immune activation.5,146 Second, chromatographic purification can reduce immune reactions triggered by double-stranded RNAs and other IVT byproducts.147 Nevertheless, considering the relatively novelty of the use of synthetic mRNA therapeutically, longer-term studies will still likely add useful information regarding its safety profile.
Storage and stability
Despite advances in IVT technology that have enhanced mRNA stability, the overall stability of lipid nanoparticle mRNA formulations remains suboptimal and requires cold storage conditions that may not be available in every healthcare setup. Currently, Pfizer/BioNTech BNT162b2 requires ultra-cold storage (−60 °C to −80 °C) while Moderna mRNA-1273 requires less stringent conditions (−15 °C to −25 °C), and both of them require sucrose as cryoprotectant.10 Significant efforts are being made at finding a lyophilized formulation that could be stored at room temperature or under 4 °C refrigeration. However, this will likely impact nanoparticle size and could affect mRNA expression and thus must be thoroughly tested. For example, Muramatsu et al. developed a lyophilization procedure that ensures that physiochemical properties and bioactivity of lyophilized lipid nanoparticle-mRNA vaccines remain stable for 12 and 24 weeks after storage at room temperature and 4 °C, respectively. This was achieved by freezing samples at −45 °C for 3 hours, followed by an 84-hour primary dry cycle at 25 °C per 20 mTorr and a 5-hour secondary dry cycle at 25 °C per 20 mTorr. Finally, nitrogen was added to the vials for storage.148 In addition, Meulewaeter et al. investigated a continuous freeze-drying technique based on spin-freezing to enable storage of lipid nanoparticle-mRNA formulations at room- and even higher temperatures. They confirmed that lyophilized lipid nanoparticle-mRNA formulations preserved their functionality when stored at 4 °C, 22 °C and even at 37 °C for a period of 12 weeks.149 Advances on these technologies are expected to facilitate the deployment of lipid nanoparticle mRNA therapeutics, including in areas where cold storage is not readily available.8. Conclusions
Fast development and extraordinary success of lipid nanoparticle mRNA COVID-19 vaccines has shown great promise in the combination of this delivery system and new class of therapeutics. Fuelled by this encouragement, and standing on decades of extensive research in both mRNA and lipid-based nanoparticles synthesis, a growing number of lipid nanoparticle mRNA-based therapies are expected to populate the clinical space in the near future. Although challenges still revolve around targeting capabilities, long-term safety profiles and storage and deployment logistics, this technology is expected to make significant impact in areas as diverse as infectious diseases, cancer immunotherapy, protein replacement, regenerative medicine, and gene-editing.Author contributions
YW: writing – original draft, writing – review and editing, visualization. SY: data curation, visualization. IdL: conceptualization, writing – original draft, writing – review and editing, visualization, supervision.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
YW and SY are both recipients of Graduate Fellowships from the NYU Tandon School of Engineering. IdL acknowledges startup funding from the NYU Tandon School of Engineering.References
- F. Hia and O. Takeuchi, Cell. Mol. Life Sci., 2021, 78, 1909–1928 CrossRef CAS PubMed.
- A. N. Kuhn, M. Diken, S. Kreiter, A. Selmi, J. Kowalska, J. Jemielity, E. Darzynkiewicz, C. Huber, Ö. Türeci and U. Sahin, Gene Ther., 2010, 17, 961–971 CrossRef CAS.
- M. Ishikawa, R. Murai, H. Hagiwara, T. Hoshino and K. Suyama, Nucleic Acids Symp. Ser., 2009, 53, 129–130 CrossRef CAS PubMed.
- H. Fuke and M. Ohno, Nucleic Acids Res., 2008, 36, 1037–1049 CrossRef CAS PubMed.
- K. Karikó, M. Buckstein, H. Ni and D. Weissman, Immunity, 2005, 23, 165–175 CrossRef PubMed.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Nat. Rev. Mater., 2021, 6, 1078–1094 CrossRef CAS PubMed.
- K. A. Hajj and K. A. Whitehead, Nat. Rev. Mater., 2017, 2, 1–17 Search PubMed.
- S. Guan and J. Rosenecker, Gene Ther., 2017, 24, 133–143 CrossRef CAS PubMed.
- M. S. Gebre, L. A. Brito, L. H. Tostanoski, D. K. Edwards, A. Carfi and D. H. Barouch, Cell, 2021, 184, 1589–1603 CrossRef CAS PubMed.
- J. Kim, Y. Eygeris, M. Gupta and G. Sahay, Adv. Drug Delivery Rev., 2021, 170, 83–112 CrossRef CAS PubMed.
- F. Martinon, S. Krishnan, G. Lenzen, R. Magne, E. Gomard, J. G. Guillet, J. P. Levy and P. Meulien, Eur. J. Immunol., 1993, 23, 1719–1722 CrossRef CAS.
- Y. Yoshizaki and T. Konno, Molecules, 2023, 28, 4479 CrossRef CAS.
- D. Yohan, C. Cruje, X. Lu and D. B. Chithrani, Nano-Micro Lett., 2016, 8, 44–53 CrossRef.
- B. M. Rothen-Rutishauser, S. Schürch, B. Haenni, N. Kapp and P. Gehr, Environ. Sci. Technol., 2006, 40, 4353–4359 CrossRef CAS PubMed.
- J. Gilleron, W. Querbes, A. Zeigerer, A. Borodovsky, G. Marsico, U. Schubert, K. Manygoats, S. Seifert, C. Andree, M. Stöter, H. Epstein-Barash, L. Zhang, V. Koteliansky, K. Fitzgerald, E. Fava, M. Bickle, Y. Kalaidzidis, A. Akinc, M. Maier and M. Zerial, Nat. Biotechnol., 2013, 31, 638–646 CrossRef CAS PubMed.
- T. Kisby, A. Yilmazer and K. Kostarelos, Nat. Nanotechnol., 2021, 16, 843–850 CrossRef CAS.
- G. Guerrini, D. Magrì, S. Gioria, D. Medaglini and L. Calzolai, Nat. Nanotechnol., 2022, 17, 570–576 CrossRef CAS.
- Y. Suzuki and H. Ishihara, Drug Metab. Pharmacokinet., 2021, 41, 100424 CrossRef CAS.
- B. Wilson and K. M. Geetha, J. Drug Delivery Sci. Technol., 2022, 74, 103553 CrossRef CAS.
- C. Horejs, Nat. Rev. Mater., 2021, 6, 1075–1076 CrossRef CAS.
- P. L. Felgner, T. R. Gadek, M. Holm, R. Roman, H. W. Chan, M. Wenz, J. P. Northrop, G. M. Ringold and M. Danielsen, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 7413–7417 CrossRef CAS PubMed.
- J. D. Tousignant, A. L. Gates, L. A. Ingram, C. L. Johnson, J. B. Nietupski, S. H. Cheng, S. J. Eastman and R. K. Scheule, Hum. Gene Ther., 2000, 11, 2493–2513 CrossRef CAS PubMed.
- X. Han, H. Zhang, K. Butowska, K. L. Swingle, M. G. Alameh, D. Weissman and M. J. Mitchell, Nat. Commun., 2021, 12, 8–13 CrossRef PubMed.
- P. S. Kowalski, A. Rudra, L. Miao and D. G. Anderson, Mol. Ther., 2019, 27, 710–728 CrossRef CAS PubMed.
- G. Tilstra, J. Couture-Senécal, Y. M. A. Lau, A. M. Manning, D. S. M. Wong, W. W. Janaeska, T. A. Wuraola, J. Pang and O. F. Khan, J. Am. Chem. Soc., 2023, 145, 2294–2304 CrossRef CAS PubMed.
- C. Y. Meng, Z. Chen, G. Li, T. Welte and H. F. Shen, Adv. Ther., 2021, 4, 2000099 CrossRef CAS.
- E. Samaridou, J. Heyes and P. Lutwyche, Adv. Drug Delivery Rev., 2020, 154–155, 37–63 CrossRef CAS.
- A. Akinc, M. A. Maier, M. Manoharan, K. Fitzgerald, M. Jayaraman, S. Barros, S. Ansell, X. Du, M. J. Hope, T. D. Madden, B. L. Mui, S. C. Semple, Y. K. Tam, M. Ciufolini, D. Witzigmann, J. A. Kulkarni, R. van der Meel and P. R. Cullis, Nat. Nanotechnol., 2019, 14, 1084–1087 CrossRef CAS PubMed.
- S. Patel, R. C. Ryals, K. K. Weller, M. E. Pennesi and G. Sahay, J. Controlled Release, 2019, 303, 91–100 CrossRef CAS PubMed.
- M. Zhang, J. Sun, M. Li and X. Jin, Mol. Ther.–Methods Clin. Dev., 2020, 18, 702–712 CrossRef CAS PubMed.
- E. Robinson, K. D. MacDonald, K. Slaughter, M. McKinney, S. Patel, C. Sun and G. Sahay, Mol. Ther., 2018, 26, 2034–2046 CrossRef CAS.
- J. F. Nabhan, K. M. Wood, V. P. Rao, J. Morin, S. Bhamidipaty, T. P. Labranche, R. L. Gooch, F. Bozal, C. E. Bulawa and B. C. Guild, Sci. Rep., 2016, 6, 1–10 CrossRef PubMed.
- M. A. Maier, M. Jayaraman, S. Matsuda, J. Liu, S. Barros, W. Querbes, Y. K. Tam, S. M. Ansell, V. Kumar, J. Qin, X. Zhang, Q. Wang, S. Panesar, R. Hutabarat, M. Carioto, J. Hettinger, P. Kandasamy, D. Butler, K. G. Rajeev, B. Pang, K. Charisse, K. Fitzgerald, B. L. Mui, X. Du, P. Cullis, T. D. Madden, M. J. Hope, M. Manoharan and A. Akinc, Mol. Ther., 2013, 21, 1570–1578 CrossRef CAS PubMed.
- L. Zhang, K. R. More, A. Ojha, C. B. Jackson, B. D. Quinlan, H. Li, W. He, M. Farzan, N. Pardi and H. Choe, npj Vaccines, 2023, 8, 156 CrossRef CAS PubMed.
- M. Paloncýová, P. Čechová, M. Šrejber, P. Kührová and M. Otyepka, J. Phys. Chem. Lett., 2021, 12, 11199–11205 CrossRef PubMed.
- O. Escalona-Rayo, Y. Zeng, R. A. Knol, T. J. F. Kock, D. Aschmann, B. Slütter and A. Kros, Biomed. Pharmacother., 2023, 165, 115065 CrossRef CAS PubMed.
- K. Lam, A. Leung, A. Martin, M. Wood, P. Schreiner, L. Palmer, O. Daly, W. Zhao, K. McClintock and J. Heyes, Adv. Mater., 2023, 35(15), e2209624 CrossRef.
- M. M. Billingsley, N. Gong, A. J. Mukalel, A. S. Thatte, R. El-Mayta, S. K. Patel, A. E. Metzloff, K. L. Swingle, X. Han, L. Xue, A. G. Hamilton, H. C. Safford, M. G. Alameh, T. E. Papp, H. Parhiz, D. Weissman and M. J. Mitchell, Small, 2023, 2304378, 1–14 Search PubMed.
- B. L. Mui, Y. K. Tam, M. Jayaraman, S. M. Ansell, X. Du, Y. Y. C. Tam, P. J. Lin, S. Chen, J. K. Narayanannair, K. G. Rajeev, M. Manoharan, A. Akinc, M. A. Maier, P. Cullis, T. D. Madden and M. J. Hope, Mol. Ther.–Nucleic Acids, 2013, 2, e139 CrossRef CAS PubMed.
- N. Davies, D. Hovdal, N. Edmunds, P. Nordberg, A. Dahlén, A. Dabkowska, M. Y. Arteta, A. Radulescu, T. Kjellman, A. Höijer, F. Seeliger, E. Holmedal, E. Andihn, N. Bergenhem, A.-S. Sandinge, C. Johansson, L. Hultin, M. Johansson, J. Lindqvist, L. Björsson, Y. Jing, S. Bartesaghi, L. Lindfors and S. Andersson, Mol. Ther.–Nucleic Acids, 2021, 24, 369–384 CrossRef CAS.
- M. Qiu, Y. Tang, J. Chen, R. Muriph, Z. Ye, C. Huang, J. Evans, E. P. Henske and Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2116271119 CrossRef CAS.
- J. Chen, Y. Xu, M. Zhou, S. Xu, A. J. Varley, A. Golubovic, R. X. Z. Lu, K. C. Wang, M. Yeganeh, D. Vosoughi and B. Li, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2309472120 CrossRef CAS.
- J. E. Dahlman, K. J. Kauffman, Y. Xing, T. E. Shaw, F. F. Mir, C. C. Dlott, R. Langer, D. G. Anderson and E. T. Wang, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 2060–2065 CrossRef CAS PubMed.
- C. Hald Albertsen, J. A. Kulkarni, D. Witzigmann, M. Lind, K. Petersson and J. B. Simonsen, Adv. Drug Delivery Rev., 2022, 188, 114416 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, J. Leung, Y. Y. C. Tam and P. R. Cullis, Nanoscale, 2019, 11, 21733–21739 RSC.
- A. R. Ferhan, S. Park, H. Park, H. Tae, J. A. Jackman and N. J. Cho, Adv. Funct. Mater., 2022, 2203669, 1–38 Search PubMed.
- B. K. Kim, G. B. Hwang, Y. B. Seu, J. S. Choi, K. S. Jin and K. O. Doh, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 1996–2001 CrossRef CAS PubMed.
- X. Cheng and R. J. Lee, Adv. Drug Delivery Rev., 2016, 99, 129–137 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, J. Leung, Y. Y. C. Tam and P. R. Cullis, Nanoscale, 2019, 11, 21733–21739 RSC.
- S. C. Semple, A. Chonn and P. R. Cullis, Biochemistry, 1996, 35, 2521–2525 CrossRef CAS PubMed.
- Y. Eygeris, S. Patel, A. Jozic and G. Sahay, Nano Lett., 2020, 20, 4543–4549 CrossRef CAS.
- K. Paunovska, A. J. Da Silva Sanchez, C. D. Sago, Z. Gan, M. P. Lokugamage, F. Z. Islam, S. Kalathoor, B. R. Krupczak and J. E. Dahlman, Adv. Mater., 2019, 31, 1–7 Search PubMed.
- S. K. Patel, M. M. Billingsley, C. Frazee, X. Han, K. L. Swingle, J. Qin, M. G. Alameh, K. Wang, D. Weissman and M. J. Mitchell, J. Controlled Release, 2022, 347, 521–532 CrossRef CAS PubMed.
- S. Patel, N. Ashwanikumar, E. Robinson, Y. Xia, C. Mihai, J. P. Griffith, S. Hou, A. A. Esposito, T. Ketova, K. Welsher, J. L. Joyal, Ö. Almarsson and G. Sahay, Nat. Commun., 2020, 11, 1–13 CrossRef.
- J. S. Suk, Q. Xu, N. Kim, J. Hanes and L. M. Ensign, Adv. Drug Delivery Rev., 2016, 99, 28–51 CrossRef CAS PubMed.
- K. Knop, R. Hoogenboom, D. Fischer and U. S. Schubert, Angew. Chem., Int. Ed., 2010, 49, 6288–6308 CrossRef CAS PubMed.
- T. T. H. Thi, E. H. Pilkington, D. H. Nguyen, J. S. Lee, K. D. Park and N. P. Truong, Polymers, 2020, 12, 298 CrossRef CAS PubMed.
- S. S. Nogueira, A. Schlegel, K. Maxeiner, B. Weber, M. Barz, M. A. Schroer, C. E. Blanchet, D. I. Svergun, S. Ramishetti, D. Peer, P. Langguth, U. Sahin and H. Haas, ACS Appl. Nano Mater., 2020, 3, 10634–10645 CrossRef CAS.
- S. J. Shepherd, D. Issadore and M. J. Mitchell, Biomaterials, 2021, 274, 120826 CrossRef CAS PubMed.
- D. M. Strelkova Petersen, N. Chaudhary, M. L. Arral, R. M. Weiss and K. A. Whitehead, Eur. J. Pharm. Biopharm., 2023, 192, 126–135 CrossRef CAS PubMed.
- N. Maurer, K. F. Wong, H. Stark, L. Louie, D. McIntosh, T. Wong, P. Scherrer, S. C. Semple and P. R. Cullis, Biophys. J., 2001, 80, 2310–2326 CrossRef CAS PubMed.
- N. Hoshyar, S. Gray, H. Han and G. Bao, Nanomedicine, 2016, 11, 673–692 CrossRef CAS PubMed.
- N. M. Belliveau, J. Huft, P. J. Lin, S. Chen, A. K. Leung, T. J. Leaver, A. W. Wild, J. B. Lee, R. J. Taylor, Y. K. Tam, C. L. Hansen and P. R. Cullis, Mol. Ther.–Nucleic Acids, 2012, 1, e37 CrossRef PubMed.
- P. M. Valencia, O. C. Farokhzad, R. Karnik and R. Langer, Nat. Nanotechnol., 2012, 7, 623–629 CrossRef CAS PubMed.
- M. Lu, A. Ozcelik, C. L. Grigsby, Y. Zhao, F. Guo, K. W. Leong and T. J. Huang, Nano Today, 2016, 11, 778–792 CrossRef CAS PubMed.
- M. C. Operti, Y. Dölen, J. Keulen, E. A. W. van Dinther, C. G. Figdor and O. Tagit, Pharmaceutics, 2019, 11, 1–17 CrossRef PubMed.
- E. Fang, X. Liu, M. Li, Z. Zhang, L. Song, B. Zhu, X. Wu, J. Liu, D. Zhao and Y. Li, Signal Transduction Targeted Ther., 2022, 7, 1–31 CrossRef PubMed.
- J. Kim, Y. Eygeris, M. Gupta and G. Sahay, Adv. Drug Delivery Rev., 2021, 170, 83–112 CrossRef CAS PubMed.
- M. Maugeri, M. Nawaz, A. Papadimitriou, A. Angerfors, A. Camponeschi, M. Na, M. Hölttä, P. Skantze, S. Johansson, M. Sundqvist, J. Lindquist, T. Kjellman, I. L. Mårtensson, T. Jin, P. Sunnerhagen, S. Östman, L. Lindfors and H. Valadi, Nat. Commun., 2019, 10, 4333 CrossRef CAS PubMed.
- I. M. S. Degors, C. Wang, Z. U. Rehman and I. S. Zuhorn, Acc. Chem. Res., 2019, 52, 1750–1760 CrossRef CAS PubMed.
- P. R. Cullis and M. J. Hope, Mol. Ther., 2017, 25, 1467–1475 CrossRef CAS PubMed.
- World Health Organization, Vaccines and Immunization, https://www.who.int/health-topics/vaccines-and-immunization#tab=tab_1, (accessed 30 December, 2023).
- O. J. Watson, G. Barnsley, J. Toor, A. B. Hogan, P. Winskill and A. C. Ghani, Lancet Infect. Dis., 2022, 22, 1293–1302 CrossRef CAS PubMed.
- S. S. Rosa, D. M. F. Prazeres, A. M. Azevedo and M. P. C. Marques, Vaccine, 2021, 39, 2190–2200 CrossRef CAS PubMed.
- A. D. Iuliano, K. M. Roguski, H. H. Chang, D. J. Muscatello, R. Palekar, S. Tempia, C. Cohen, J. M. Gran, D. Schanzer, B. J. Cowling, P. Wu, J. Kyncl, L. W. Ang, M. Park, M. Redlberger-Fritz, H. Yu, L. Espenhain, A. Krishnan, G. Emukule, L. van Asten, S. Pereira da Silva, S. Aungkulanon, U. Buchholz, M. A. Widdowson, J. S. Bresee, E. Azziz-Baumgartner, P. Y. Cheng, F. Dawood, I. Foppa, S. Olsen, M. Haber, C. Jeffers, C. R. MacIntyre, A. T. Newall, J. G. Wood, M. Kundi, T. Popow-Kraupp, M. Ahmed, M. Rahman, F. Marinho, C. V. Sotomayor Proschle, N. Vergara Mallegas, F. Luzhao, L. Sa, J. Barbosa-Ramírez, D. M. Sanchez, L. A. Gomez, X. B. Vargas, a. B. Acosta Herrera, M. J. Llanés, T. K. Fischer, T. G. Krause, K. Mølbak, J. Nielsen, R. Trebbien, A. Bruno, J. Ojeda, H. Ramos, M. an der Heiden, L. del Carmen Castillo Signor, C. E. Serrano, R. Bhardwaj, M. Chadha, V. Narayan, S. Kosen, M. Bromberg, A. Glatman-Freedman, Z. Kaufman, Y. Arima, K. Oishi, S. Chaves, B. Nyawanda, R. A. Al-Jarallah, P. A. Kuri-Morales, C. R. Matus, M. E. J. Corona, A. Burmaa, O. Darmaa, M. Obtel, I. Cherkaoui, C. C. van den Wijngaard, W. van der Hoek, M. Baker, D. Bandaranayake, A. Bissielo, S. Huang, L. Lopez, C. Newbern, E. Flem, G. M. Grøneng, S. Hauge, F. G. de Cosío, Y. de Moltó, L. M. Castillo, M. A. Cabello, M. von Horoch, J. Medina Osis, A. Machado, B. Nunes, A. P. Rodrigues, E. Rodrigues, C. Calomfirescu, E. Lupulescu, R. Popescu, O. Popovici, D. Bogdanovic, M. Kostic, K. Lazarevic, Z. Milosevic, B. Tiodorovic, M. Chen, J. Cutter, V. Lee, R. Lin, S. Ma, A. L. Cohen, F. Treurnicht, W. J. Kim, C. Delgado-Sanz, S. de mateo Ontañón, A. Larrauri, I. L. León, F. Vallejo, R. Born, C. Junker, D. Koch, J. H. Chuang, W. T. Huang, H. W. Kuo, Y. C. Tsai, K. Bundhamcharoen, M. Chittaganpitch, H. K. Green, R. Pebody, N. Goñi, H. Chiparelli, L. Brammer and D. Mustaquim, Lancet, 2018, 391, 1285–1300 CrossRef PubMed.
- J. Piret and G. Boivin, Front. Microbiol., 2021, 11, 631736 CrossRef PubMed.
- N. Chaudhary, D. Weissman and K. A. Whitehead, Nat. Rev. Drug Discovery, 2021, 20, 817–838 CrossRef CAS PubMed.
- M. N. Matrosovich, T. Y. Matrosovich, T. Gray, N. A. Roberts and H.-D. Klenk, J. Virol., 2004, 78, 12665–12667 CrossRef CAS PubMed.
- W. C. Liu, R. Nachbagauer, D. Stadlbauer, A. Solórzano, F. Berlanda-Scorza, A. García-Sastre, P. Palese, F. Krammer and R. A. Albrecht, Front. Immunol., 2019, 10, 1–18 CrossRef PubMed.
- S. J. Zost, K. Parkhouse, M. E. Gumina, K. Kim, S. D. Perez, P. C. Wilson, J. J. Treanor, A. J. Sant, S. Cobey and S. E. Hensley, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 12578–12583 CrossRef CAS PubMed.
- A. Hekele, S. Bertholet, J. Archer, D. G. Gibson, G. Palladino, L. A. Brito, G. R. Otten, M. Brazzoli, S. Buccato, A. Bonci, D. Casini, D. Maione, Z. Q. Qi, J. E. Gill, N. C. Caiazza, J. Urano, B. Hubby, G. F. Gao, Y. Shu, E. De Gregorio, C. W. Mandl, P. W. Mason, E. C. Settembre, J. B. Ulmer, J. Craig Venter, P. R. Dormitzer, R. Rappuoli and A. J. Geall, Emerging Microbes Infect., 2013, 2, 1–7 CrossRef PubMed.
- B. Petsch, M. Schnee, A. B. Vogel, E. Lange, B. Hoffmann, D. Voss, T. Schlake, A. Thess, K. J. Kallen, L. Stitz and T. Kramps, Nat. Biotechnol., 2012, 30, 1210–1216 CrossRef CAS PubMed.
- N. Pardi, K. Parkhouse, E. Kirkpatrick, M. McMahon, S. J. Zost, B. L. Mui, Y. K. Tam, K. Karikó, C. J. Barbosa, T. D. Madden, M. J. Hope, F. Krammer, S. E. Hensley and D. Weissman, Nat. Commun., 2018, 9, 1–12 CrossRef CAS PubMed.
- C. P. Arevalo, M. J. Bolton, V. Le Sage, N. Ye, C. Furey, H. Muramatsu, M. G. Alameh, N. Pardi, E. M. Drapeau, K. Parkhouse, T. Garretson, J. S. Morris, L. H. Moncla, Y. K. Tam, S. H. Y. Fan, S. S. Lakdawala, D. Weissman and S. E. Hensley, Science, 2022, 378, 899–904 CrossRef CAS PubMed.
- Pfizer, Pfizer Initiates Phase 3 Study of mRNA-Based Influenza Vaccine, https://www.pfizer.com/news/press-release/press-release-detail/pfizer-initiates-phase-3-study-mrna-based-influenza-vaccine, (accessed 30 December, 2023).
- J. Lessler, L. H. Chaisson, L. M. Kucirka, Q. Bi, K. Grantz, H. Salje, A. C. Carcelen, C. T. Ott, J. S. Sheffield, N. M. Ferguson, D. A. T. Cummings, C. J. E. Metcalf and I. Rodriguez-Barraquer, Science, 2016, 353, aaf8160 CrossRef PubMed.
- K. A. Dowd, C. R. DeMaso, R. S. Pelc, S. D. Speer, A. R. Y. Smith, L. Goo, D. J. Platt, J. R. Mascola, B. S. Graham, M. J. Mulligan, M. S. Diamond, J. E. Ledgerwood and T. C. Pierson, Cell Rep., 2016, 16, 1485–1491 CrossRef CAS PubMed.
- F. X. Heinz and K. Stiasny, Microbiol. Mol. Biol. Rev., 2017, 81, 1–27 CrossRef PubMed.
- R. A. Larocca, P. Abbink, J. P. S. Peron, P. M. D. A. Zanotto, M. J. Iampietro, A. Badamchi-Zadeh, M. Boyd, D. Ng'ang'a, M. Kirilova, R. Nityanandam, N. B. Mercado, Z. Li, E. T. Moseley, C. A. Bricault, E. N. Borducchi, P. B. Giglio, D. Jetton, G. Neubauer, J. P. Nkolola, L. F. Maxfield, R. A. De La Barrera, R. G. Jarman, K. H. Eckels, N. L. Michael, S. J. Thomas and D. H. Barouch, Nature, 2016, 536, 474–478 CrossRef CAS.
- P. Abbink, R. A. Larocca, R. A. De La Barrera, C. A. Bricault, E. T. Moseley, M. Boyd, M. Kirilova, Z. Li, D. Ng'ang'a, O. Nanayakkara, R. Nityanandam, N. B. Mercado, E. N. Borducchi, A. Agarwal, A. L. Brinkman, C. Cabral, A. Chandrashekar, P. B. Giglio, D. Jetton, J. Jimenez, B. C. Lee, S. Mojta, K. Molloy, M. Shetty, G. H. Neubauer, K. E. Stephenson, J. P. S. Peron, P. M. A. De Zanotto, J. Misamore, B. Finneyfrock, M. G. Lewis, G. Alter, K. Modjarrad, R. G. Jarman, K. H. Eckels, N. L. Michael, S. J. Thomas and D. H. Barouch, Science, 2016, 353, 1129–1132 CrossRef CAS PubMed.
- N. Pardi, M. J. Hogan, R. S. Pelc, H. Muramatsu, H. Andersen, C. R. DeMaso, K. A. Dowd, L. L. Sutherland, R. M. Scearce, R. Parks, W. Wagner, A. Granados, J. Greenhouse, M. Walker, E. Willis, J. S. Yu, C. E. McGee, G. D. Sempowski, B. L. Mui, Y. K. Tam, Y. J. Huang, D. Vanlandingham, V. M. Holmes, H. Balachandran, S. Sahu, M. Lifton, S. Higgs, S. E. Hensley, T. D. Madden, M. J. Hope, K. Karikó, S. Santra, B. S. Graham, M. G. Lewis, T. C. Pierson, B. F. Haynes and D. Weissman, Nature, 2017, 543, 248–251 CrossRef CAS PubMed.
- J. M. Richner, S. Himansu, K. A. Dowd, S. L. Butler, V. Salazar, J. M. Fox, J. G. Julander, W. W. Tang, S. Shresta, T. C. Pierson, G. Ciaramella and M. S. Diamond, Cell, 2017, 168, 1114–1125 CrossRef CAS PubMed.
- J. M. Richner, B. W. Jagger, C. Shan, C. R. Fontes, K. A. Dowd, B. Cao, S. Himansu, E. A. Caine, B. T. D. Nunes, D. B. A. Medeiros, A. E. Muruato, B. M. Foreman, H. Luo, T. Wang, A. D. Barrett, S. C. Weaver, P. F. C. Vasconcelos, S. L. Rossi, G. Ciaramella, I. U. Mysorekar, T. C. Pierson, P. Y. Shi and M. S. Diamond, Cell, 2017, 170, 273–283 CrossRef CAS PubMed.
- B. Essink, L. Chu, W. Seger, E. Barranco, N. Le Cam, H. Bennett, V. Faughnan, R. Pajon, Y. D. Paila, B. Bollman, S. Wang, J. Dooley, S. Kalidindi and B. Leav, Lancet Infect. Dis., 2023, 23, 621–633 CrossRef CAS.
- A. Vass, Br. Med. J., 2001, 323, 1271 CrossRef CAS.
- D. R. Burton and J. P. Moore, Nat. Med., 1998, 4, 495–498 CrossRef CAS.
- A. K. Blakney, P. F. McKay, B. I. Yus, Y. Aldon and R. J. Shattock, Gene Ther., 2019, 26, 363–372 CrossRef CAS.
- P. Zhang, E. Narayanan, Q. Liu, Y. Tsybovsky, K. Boswell, S. Ding, Z. Hu, D. Follmann, Y. Lin, H. Miao, H. Schmeisser, D. Rogers, S. Falcone, S. M. Elbashir, V. Presnyak, K. Bahl, M. Prabhakaran, X. Chen, E. K. Sarfo, D. R. Ambrozak, R. Gautam, M. A. Martin, J. Swerczek, R. Herbert, D. Weiss, J. Misamore, G. Ciaramella, S. Himansu, G. Stewart-Jones, A. McDermott, R. A. Koup, J. R. Mascola, A. Finzi, A. Carfi, A. S. Fauci and P. Lusso, Nat. Med., 2021, 27, 2234–2245 CrossRef CAS PubMed.
- B. F. Haynes, K. Wiehe, P. Borrow, K. O. Saunders, B. Korber, K. Wagh, A. J. McMichael, G. Kelsoe, B. H. Hahn, F. Alt and G. M. Shaw, Nat. Rev. Immunol., 2023, 23, 142–158 CrossRef CAS PubMed.
- N. Sobhani, B. Scaggiante, R. Morris, D. Chai, M. Catalano, D. R. Tardiel-Cyril, P. Neeli, G. Roviello, G. Mondani and Y. Li, Cancer Treat. Rev., 2022, 109, 102429 CrossRef CAS.
- X. Sun, L. Zeng and Y. Huang, J. Gene Med., 2019, 21, 1–14 Search PubMed.
- M. M. Billingsley, N. Singh, P. Ravikumar, R. Zhang, C. H. June and M. J. Mitchell, Nano Lett., 2020, 20, 1578–1589 CrossRef CAS PubMed.
- J. Q. Liu, C. Zhang, X. Zhang, J. Yan, C. Zeng, F. Talebian, K. Lynch, W. Zhao, X. Hou, S. Du, D. D. Kang, B. Deng, D. W. McComb, X. F. Bai and Y. Dong, J. Controlled Release, 2022, 345, 306–313 CrossRef CAS PubMed.
- Y. Rybakova, P. S. Kowalski, Y. Huang, J. T. Gonzalez, M. W. Heartlein, F. DeRosa, D. Delcassian and D. G. Anderson, Mol. Ther., 2019, 27, 1415–1423 CrossRef CAS PubMed.
- I. de Lázaro and D. J. Mooney, Nat. Mater., 2021, 20, 1469–1479 CrossRef.
- T. N. Yamamoto, R. J. Kishton and N. P. Restifo, Nat. Med., 2019, 25, 1488–1499 CrossRef CAS.
- R. Kamps, R. D. Brandão, B. J. van den Bosch, A. D. C. Paulussen, S. Xanthoulea, M. J. Blok and A. Romano, Int. J. Mol. Sci., 2017, 18, 308 CrossRef PubMed.
- G. Fotakis, Z. Trajanoski and D. Rieder, Immunol.-Oncol. Technol., 2021, 12, 100052 CrossRef CAS PubMed.
- J. Herrera-Bravo, L. Herrera Belén, J. G. Farias and J. F. Beltrán, Comput. Biol. Chem., 2021, 19, 107452 CrossRef PubMed.
- Moderna, United States Securities and Exchange Commission Form 10-K, https://www.sec.gov/Archives/edgar/data/1682852/000168285220000006/moderna10-k12312019.htm, (accessed 30 December, 2023).
- A. Khattak, J. S. Weber, T. Meniawy, M. H. Taylor, G. Ansstas, K. B. Kim, M. McKean, G. V. Long, R. J. Sullivan, M. B. Faries, T. Tran, C. L. Cowey, T. M. Medina, J. M. Segar, V. Atkinson, G. T. Gibney, J. J. Luke, E. I. Buchbinder, R. S. Meehan and M. S. Carlino, J. Clin. Oncol., 2023, 41, LBA9503–LBA9503 CrossRef.
- J. Deckers, T. Anbergen, A. M. Hokke, A. de Dreu, D. P. Schrijver, K. de Bruin, Y. C. Toner, T. J. Beldman, J. B. Spangler, T. F. A. de Greef, F. Grisoni, R. van der Meel, L. A. B. Joosten, M. Merkx, M. G. Netea and W. J. M. Mulder, Nat. Rev. Bioeng., 2023, 1, 286–303 CrossRef PubMed.
- K. J. Hassett, K. E. Benenato, E. Jacquinet, A. Lee, A. Woods, O. Yuzhakov, S. Himansu, J. Deterling, B. M. Geilich, T. Ketova, C. Mihai, A. Lynn, I. McFadyen, M. J. Moore, J. J. Senn, M. G. Stanton, Ö. Almarsson, G. Ciaramella and L. A. Brito, Mol. Ther.–Nucleic Acids, 2019, 15, 1–11 CrossRef CAS PubMed.
- J. D. Beck, D. Reidenbach, N. Salomon, U. Sahin, Ö. Türeci, M. Vormehr and L. M. Kranz, Mol. Cancer, 2021, 20, 1–24 CrossRef PubMed.
- I. Lai, S. Swaminathan, V. Baylot, A. Mosley, R. Dhanasekaran, M. Gabay and D. W. Felsher, J. ImmunoTher. Cancer, 2018, 6, 125 CrossRef PubMed.
- S. L. Hewitt, D. Bailey, J. Zielinski, A. Apte, F. Musenge, R. Karp, S. Burke, F. Garcon, A. Mishra, S. Gurumurthy, A. Watkins, K. Arnold, J. Moynihan, E. Clancy-Thompson, K. Mulgrew, G. Adjei, K. Deschler, D. Potz, G. Moody, D. A. Leinster, S. Novick, M. Sulikowski, C. Bagnall, P. Martin, J. M. Lapointe, H. Si, C. Morehouse, M. Sedic, R. W. Wilkinson, R. Herbst, J. P. Frederick and N. Luheshi, Clin. Cancer Res., 2020, 26, 6284–6298 CrossRef CAS PubMed.
- B. A. Carneiro, D. Zamarin, T. Marron, M. Inderjit, S. P. Patel, V. Subbiah, A. El-khoueiry, D. Grand, K. Garcia-reyes, S. Goel, P. Martin, J. Wang, Y. Wu, S. Eck, B. Ridgway, N. Elgeioushi, J. Eyles, N. Durham, A. Azaro and O. Hamid, First-in-human study of MEDI1191 (mRNA encoding IL-12) plus durvalumab in patients with advanced solid tumors, https://s29.q4cdn.com/435878511/files/doc_presentations/2022/04/AACR2022_CT183_Carneiro-AACR-2022-MEDI1191-Poster_Final.pdf, (accessed 15 March, 2024).
- S. L. Hewitt, A. Bai, D. Bailey, K. Ichikawa, J. Zielinski, R. Karp, A. Apte, K. Arnold, S. J. Zacharek, M. S. Iliou, K. Bhatt, M. Garnaas, F. Musenge, A. Davis, N. Khatwani, S. V. Su, G. MacLean, S. J. Farlow, K. Burke and J. P. Frederick, Sci. Transl. Med., 2019, 11, eaat9143 CrossRef CAS.
- S. Qin, X. Tang, Y. Chen, K. Chen, N. Fan, W. Xiao, Q. Zheng, G. Li, Y. Teng, M. Wu and X. Song, Signal Transduction Targeted Ther., 2022, 7, 166 CrossRef CAS.
- M. L. Cacicedo, C. Weinl-Tenbruck, D. Frank, M. J. Limeres, S. Wirsching, K. Hilbert, M. A. Pasha Famian, N. Horscroft, J. B. Hennermann, F. Zepp, F. Chevessier-Tünnesen and S. Gehring, Front. Bioeng. Biotechnol., 2022, 10, 1–14 Search PubMed.
- K. H. Asrani, L. Cheng, C. J. Cheng and R. R. Subramanian, RNA Biol., 2018, 15, 914–922 CrossRef PubMed.
- C. Y. Chen, D. M. Tran, A. Cavedon, X. Cai, R. Rajendran, M. J. Lyle, P. G. V. Martini and C. H. Miao, Mol. Ther.–Nucleic Acids, 2020, 20, 534–544 CrossRef CAS PubMed.
- S. Ramaswamy, N. Tonnu, K. Tachikawa, P. Limphong, J. B. Vega, P. P. Karmali, P. Chivukula and I. M. Verma, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E1941–E1950 CrossRef CAS PubMed.
- K. L. Swingle, H. C. Safford, H. C. Geisler, A. G. Hamilton, A. S. Thatte, M. M. Billingsley, R. A. Joseph, K. Mrksich, M. S. Padilla, A. A. Ghalsasi, M. G. Alameh, D. Weissman and M. J. Mitchell, J. Am. Chem. Soc., 2023, 145, 4691–4706 CrossRef CAS PubMed.
- R. S. Riley, M. V. Kashyap, M. M. Billingsley, B. White, M.-G. Alameh, S. K. Bose, P. W. Zoltick, H. Li, R. Zhang, A. Y. Cheng, D. Weissman, W. H. Peranteau and M. J. Mitchell, Sci. Adv., 2021, 7, eaba1028 CrossRef CAS PubMed.
- Y. Xu and Z. Li, Comput. Struct. Biotechnol. J., 2020, 18, 2401–2415 CrossRef CAS PubMed.
- F. Ran, P. Hsu, J. Wright, V. Agarwala and D. A. Scott, Nat. Protoc., 2006, 8, 1–10 Search PubMed.
- E. M. Porto, A. C. Komor, I. M. Slaymaker and G. W. Yeo, Nat. Rev. Drug Discovery, 2020, 19, 839–859 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- H. Yin, K. J. Kauffman and D. G. Anderson, Nat. Rev. Drug Discovery, 2017, 16, 387–399 CrossRef CAS PubMed.
- D. Rosenblum, A. Gutkin, R. Kedmi, S. Ramishetti, N. Veiga, A. M. Jacobi, M. S. Schubert, D. Friedmann-Morvinski, Z. R. Cohen, M. A. Behlke, J. Lieberman and D. Peer, Sci. Adv., 2020, 6, eabc9450 CrossRef CAS PubMed.
- L. Farbiak, Q. Cheng, T. Wei, E. Álvarez-Benedicto, L. T. Johnson, S. Lee and D. J. Siegwart, Adv. Mater., 2021, 33, 1–8 CrossRef PubMed.
- D. N. Nguyen, T. L. Roth, P. J. Li, P. A. Chen, R. Apathy, M. R. Mamedov, L. T. Vo, V. R. Tobin, D. Goodman, E. Shifrut, J. A. Bluestone, J. M. Puck, F. C. Szoka and A. Marson, Nat. Biotechnol., 2020, 38, 44–49 CrossRef CAS PubMed.
- L. Villiger, T. Rothgangl, D. Witzigmann, R. Oka, P. J. C. Lin, W. Qi, S. Janjuha, C. Berk, F. Ringnalda, M. B. Beattie, M. Stoffel, B. Thöny, J. Hall, H. Rehrauer, R. van Boxtel, Y. K. Tam and G. Schwank, Nat. Biomed. Eng., 2021, 5, 179–189 CrossRef CAS PubMed.
- L. Villiger, H. M. Grisch-Chan, H. Lindsay, F. Ringnalda, C. B. Pogliano, G. Allegri, R. Fingerhut, J. Häberle, J. Matos, M. D. Robinson, B. Thöny and G. Schwank, Nat. Med., 2018, 24, 1519–1525 CrossRef CAS PubMed.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby, A. Raguram and D. R. Liu, Nature, 2019, 576, 149–157 CrossRef CAS PubMed.
- Z. Chen, K. Kelly, H. Cheng, X. Dong, A. K. Hedger, L. Li, E. J. Sontheimer and J. K. Watts, GEN Biotechnol., 2023, 2, 490–502 CrossRef CAS.
- Pipeline | Prime Medicine, https://primemedicine.com/pipeline/, (accessed March 2, 2024).
- P. P. G. Guimaraes, R. Zhang, R. Spektor, M. Tan, A. Chung, M. M. Billingsley, R. El-Mayta, R. S. Riley, L. Wang, J. M. Wilson and M. J. Mitchell, J. Controlled Release, 2019, 316, 404–417 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS PubMed.
- I. Tombácz, D. Laczkó, H. Shahnawaz, H. Muramatsu, A. Natesan, A. Yadegari, T. E. Papp, M. G. Alameh, V. Shuvaev, B. L. Mui, Y. K. Tam, V. Muzykantov, N. Pardi, D. Weissman and H. Parhiz, Mol. Ther., 2021, 29, 3293–3304 CrossRef PubMed.
- S. A. Dilliard and D. J. Siegwart, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- Y. Ju, J. M. Carreño, V. Simon, K. Dawson, F. Krammer and S. J. Kent, Nat. Rev. Immunol., 2023, 23, 135–136 CrossRef CAS PubMed.
- M. Estapé Senti, C. A. de Jongh, K. Dijkxhoorn, J. J. F. Verhoef, J. Szebeni, G. Storm, C. E. Hack, R. M. Schiffelers, M. H. Fens and P. Boross, J. Controlled Release, 2022, 341, 475–486 CrossRef PubMed.
- N. Pardi, M. J. Hogan, F. W. Porter and D. Weissman, Nat. Rev. Drug Discovery, 2018, 17, 261–279 CrossRef CAS PubMed.
- K. Karikó, H. Muramatsu, F. A. Welsh, J. Ludwig, H. Kato, S. Akira and D. Weissman, Mol. Ther., 2008, 16, 1833–1840 CrossRef PubMed.
- J. Lee, M. C. Woodruff, E. H. Kim and J. H. Nam, Exp. Mol. Med., 2023, 55, 1305–1313 CrossRef CAS PubMed.
- H. Muramatsu, K. Lam, C. Bajusz, D. Laczkó, K. Karikó, P. Schreiner, A. Martin, P. Lutwyche, J. Heyes and N. Pardi, Mol. Ther., 2022, 30, 1941–1951 CrossRef CAS PubMed.
- S. Meulewaeter, G. Nuytten, M. H. Y. Cheng, S. C. De Smedt, P. R. Cullis, T. De Beer, I. Lentacker and R. Verbeke, J. Controlled Release, 2023, 357, 149–160 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |