Open Access Article
Open Access ArticlePExM: polyplex expansion microscopy for cell trafficking studies†
María
Navalón-López
 a,
Pau
Castells-Colldeforns
a,
Pau
Castells-Colldeforns
 a,
Silvia
Pujals
a,
Silvia
Pujals
 *b,
Cristina
Fornaguera
*b,
Cristina
Fornaguera
 *a and
Salvador
Borrós
*a and
Salvador
Borrós
 a
a
aGrup d'Enginyeria de Materials (Gemat), Institut Químic de Sarrià (IQS), Universitat Ramon Llull (URL), Via Augusta 390, 08017, Barcelona, Spain. E-mail: cristina.fornagura@iqs.url.edu
bInstitute of Advanced Chemistry of Catalonia (IQAC-CSIC), Carrer Jordi Girona 18-24, 08034, Barcelona, Spain. E-mail: silvia.pujals@iqac.csic.es
First published on 3rd June 2024
Abstract
Nanomedicine is a field at the intersection of nanotechnology and medicine, promising due to its potential to revolutionize healthcare. Despite its long trajectory, there is still a long road ahead for its full development, and smart design of nanomedicines is still a challenge. Among other problems, this is due to the scarcity of tools available for the precise visualization and comprehension of nano–bio interactions, impeding progress towards the clinical phase. One of the developed tools that stands out to be a strong nanoscopy technique for studying nano-delivery systems within cellular environments is expansion microscopy (ExM). This technique was used for tissue and cell expansion and most recently for lipid molecule expansion inside cells. Herein, we present for the first time polyplex expansion microscopy (PExM); a comprehensive examination of ExM as an already developed technique, but adapted for expanding polymer based nanocarriers, in particular polyplexes within cells, allowing the analysis of their trafficking. With our method set up, PExM will be extensively used for the study of polyplex nanoparticle cell trafficking, becoming a high-resolution technique which can also be applied to primary amine containing polymeric nanoparticles without requiring expensive super-resolution microscopes.
Introduction
The intersection of nanotechnology with biomedical and pharmaceutical sciences has given birth to nanomedicine, an emerging field that encompasses drug delivery, nanoimaging, and theragnostics.1–3 Nanocarriers, typically nanoparticles (NPs), are utilized for encapsulating and delivering (bio)therapeutics, owing to their distinct advantages over conventional medicines. Key characteristics of nanocarriers include enhanced pharmaceutical properties (such as improved solubility and stability, prolonged drug half-life, and increased accumulation in tumors),2,4–6 improvement of the therapeutic effectiveness and/or reduction of toxicity and targeted delivery to specific cells or tissues.3,6,7 The majority of these attributes, especially the pharmacokinetic ones, are achieved through polymeric carriers capable of biodegrading and undergoing chemical modifications, allowing for additional targeting.8 To achieve precise site-specific delivery, the design of engineered NPs must be meticulously tailored to surmount various challenges upon entering the systemic circulation, including interactions with plasma proteins, immune system detection, and overcoming various intracellular barriers.2 Despite polymeric carriers being considered gold standards in the field of delivery systems and having shown some hints of success, due to all the aforementioned reasons,9,10 they have failed to progress beyond clinical trials. One of the factors hindering the transference of these nanoparticles is the lack of nanoscale characterization techniques for their thorough comprehension and control. To beat this limitation, there is a need to develop imaging tools and techniques that overcome the nanometric resolution barrier to be capable to analyze and develop safer and more potent nanomedicines.11 Specifically, the study of their localization in different organelles and cell compartments is of utmost importance to control their trafficking, and consequently, study their cargo release and their fate, key issues to not only enable the rational design of efficient formulations but also to ensure their safe use. One interesting possibility is expansion microscopy (ExM), a powerful and accessible high-resolution imaging strategy where biomolecules are anchored to a swellable hydrogel that enables samples to be expanded by a factor of four to twenty,12 providing high resolutions with a conventional microscope.13,14 ExM was first proposed in 2015, by Chen, Tillberg, and Boyden, devoted to the expansion of biological molecules.15 Further work in 2022, by White and coworkers described the adaptation of this technique for the study of lipid molecules.13 However, to date, no studies exist reporting the possibility of using ExM for the nanoscopic study of polyplex interactions with cells. Thus, the main objective of this communication is to shed light on polymeric nanoparticle (pNP) trafficking within cells by developing an ExM technique specific for polyplex-based carriers that can also be carried out generally with any polymeric nanoparticles that contain primary amines or by attaching them into any carrier-type. Therefore, these carriers’ amplification and visualization within the different cellular compartments through different labelings enables an affordable and straightforward methodology. To achieve this, we have devised a technique, termed polyplex expansion microscopy (PExM), which facilitates the examination of spatial and temporal localization of pNPs at both the cellular and subcellular levels using ExM. Using our proprietary poly(beta aminoester) – pBAE – NPs as proof-of-concept biomaterials, for the first time, pNPs within cells can be rigorously studied with this new PExM technique. pBAEs are an interesting choice due to their sought after characteristics such as low toxicity, biodegradability, biocompatibility, the ability to compact genetic material, and their easy synthesis and chemical modification.16,17 In this work, we demonstrate that the obtention of enough resolution allows nanoparticle–cell interactions to be guided with the intention of controlling their fate and regulating target localization and degradation, and finally provides a deep understanding of pNP interactions with cells.18 Our communication is expected to guide researchers in the characterization of PExM interactions with cells for the development of other pNPs for biomedical uses.Protocol development
Here, we report a reexamination of the traditional protein retention ExM19 detaining not only endogenous amino-containing macromolecules in cultured cells but also synthetic molecules such as polyplexes on a swellable gel. Following the ExM principle of anchoring primary amines to a hydrogel using a commercially available cross-linking molecule known as the succinimidyl ester of 6-((acryloyl)amino)hexanoic acid (AcX)15 we can colocalize NPs inside cells. In this communication we will proceed to explain step by step the developed protocol to achieve it, starting from the anchoring of the fluorophores to the nanosystem and to the hydrogel, gelation of the polymer chains that form the network which traps and immobilizes the cells and molecules, digestion of random chains of the polymer network to allow swelling of the hydrogel, and expansion of the hydrogel to finish with the microscopy visualization.Process preparation
To perform conventional immunostaining after seeding, we first created a PDMS scaffold (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a 35 mm diameter that fits a Petri dish, but with a 5 mm diameter glass bottom inside where the cells will be seeded (Fig. 1A). This allows to have the cells in a localized region. Once the hydrogel is formed, the scaffold is removed, enabling gel expansion along the Petri dish without the need to manipulate the gel, thus avoiding potential breakage as seen in conventional ExM protocols.20 Once the scaffold was ready and sterilized with UV light for 30 minutes, we proceeded with the seeding of the cells of interest. In our case, this has been done with A549 cells, human lung adenocarcinoma cells, as a proof of concept that could be extrapolated to any cell line growing attached to the surface. It is worth noting that to achieve proper confluence after the entire expansion process, it is necessary to seed around 250
1) with a 35 mm diameter that fits a Petri dish, but with a 5 mm diameter glass bottom inside where the cells will be seeded (Fig. 1A). This allows to have the cells in a localized region. Once the hydrogel is formed, the scaffold is removed, enabling gel expansion along the Petri dish without the need to manipulate the gel, thus avoiding potential breakage as seen in conventional ExM protocols.20 Once the scaffold was ready and sterilized with UV light for 30 minutes, we proceeded with the seeding of the cells of interest. In our case, this has been done with A549 cells, human lung adenocarcinoma cells, as a proof of concept that could be extrapolated to any cell line growing attached to the surface. It is worth noting that to achieve proper confluence after the entire expansion process, it is necessary to seed around 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL (10
000 cells per mL (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mm2), a higher density than recommended in standard protocols (Fig. 1B).21 Subsequently, transfection can be performed with any polymeric system with the presence of amine groups in its structure to anchor the NPs to the hydrogel. We used our polymeric poly(β-aminoester) (pBAE) NPs as a proof of concept, a family of biodegradable cationic polymers which are an interesting choice for nucleic acid delivery.17 In addition, the properties of pBAEs can be boosted by adding some modifications to the end-capping groups of the backbone molecule. In particular, our proprietary adjustments22 are based on using different positive peptide moieties,23 such as arginine (C6CR3) and histidine (C6CH3) oligopeptides (Fig. 1C and ESI Fig. S1†). Namely, in the presented case, R-pBAE were labeled with Atto647N in a 1
000 cells per mm2), a higher density than recommended in standard protocols (Fig. 1B).21 Subsequently, transfection can be performed with any polymeric system with the presence of amine groups in its structure to anchor the NPs to the hydrogel. We used our polymeric poly(β-aminoester) (pBAE) NPs as a proof of concept, a family of biodegradable cationic polymers which are an interesting choice for nucleic acid delivery.17 In addition, the properties of pBAEs can be boosted by adding some modifications to the end-capping groups of the backbone molecule. In particular, our proprietary adjustments22 are based on using different positive peptide moieties,23 such as arginine (C6CR3) and histidine (C6CH3) oligopeptides (Fig. 1C and ESI Fig. S1†). Namely, in the presented case, R-pBAE were labeled with Atto647N in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer
1 polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dye molar ratio. To do so, NPs were prepared containing a mixture of 60% C6CR3 and 40% C6CH3 (simplified as RH NPs).23–25 To delineate the cell structure, DAPI and Orange CellMask were used for the cell nuclei and cytoplasmic membrane staining, respectively. AF488 was used as the secondary antibody of Rab5, an early endosome marker, thus allowing colocalization studies with our pBAE NPs to understand their internalization process. Crosstalk between the selected fluorophores was checked, for more detailed information see the ESI, Fig. S2.†
dye molar ratio. To do so, NPs were prepared containing a mixture of 60% C6CR3 and 40% C6CH3 (simplified as RH NPs).23–25 To delineate the cell structure, DAPI and Orange CellMask were used for the cell nuclei and cytoplasmic membrane staining, respectively. AF488 was used as the secondary antibody of Rab5, an early endosome marker, thus allowing colocalization studies with our pBAE NPs to understand their internalization process. Crosstalk between the selected fluorophores was checked, for more detailed information see the ESI, Fig. S2.†
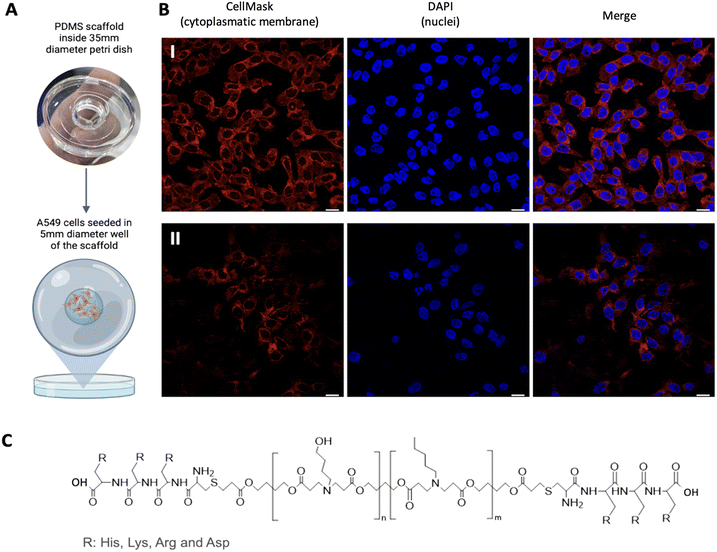 | ||
Fig. 1 Pre-expansion. (A) Scaffold of PDMS (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with a 35 mm diameter and a 5 mm diameter hole in the center for cell seeding. (B) A549 cells stained with DAPI and CellMask (I) seeded at 250 1) with a 35 mm diameter and a 5 mm diameter hole in the center for cell seeding. (B) A549 cells stained with DAPI and CellMask (I) seeded at 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL confluency and B (II) seeded at 150 000 cells per mL confluency and B (II) seeded at 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL confluency. (C) Oligo-modified poly-β(amino ester) backbone and the possible end terminal tripeptides; for more information see ESI Fig. S1.† Scale bar is 20 μm. 000 cells per mL confluency. (C) Oligo-modified poly-β(amino ester) backbone and the possible end terminal tripeptides; for more information see ESI Fig. S1.† Scale bar is 20 μm. | ||
Labeling
For an optimum observation of cells with the microscope being able to differentiate their subcellular compartments, labeling selection is a key issue. To achieve optimal results, various fluorophores had to be fine-tuned, both in terms of concentration and the pre- or post-expansion application method. Given the necessity to modify the entire conventional protocol for NP visualization, fluorophore concentrations have also been an affected factor. For nuclear staining, DAPI was used at a concentration of 0.5 μg ml−1, while CellMask Orange was employed at 20 μg mL−1 for cell membrane delineation. To detect endosomes, Rab5 was utilized as the primary antibody at a concentration of 5 μg mL−1, with AF488 as the secondary antibody at a concentration of 5 μg mL−1, as indicated in ESI Table S1.† The used concentrations in PExM do not differ in the order of magnitude from the classic ExM protocol, nonetheless, the labeling incubations in our protocol are shorter. Two staining approaches were carried out, a pre-expansion and a post-expansion labeling, but the second one led to a significant loss in image quality. There exist additional fluorophores that could be used;26 however, among these, the aforementioned ones align most suitably with our system's requirements and are readily available for our specific overlapping needs.Gelation, homogenization and expansion
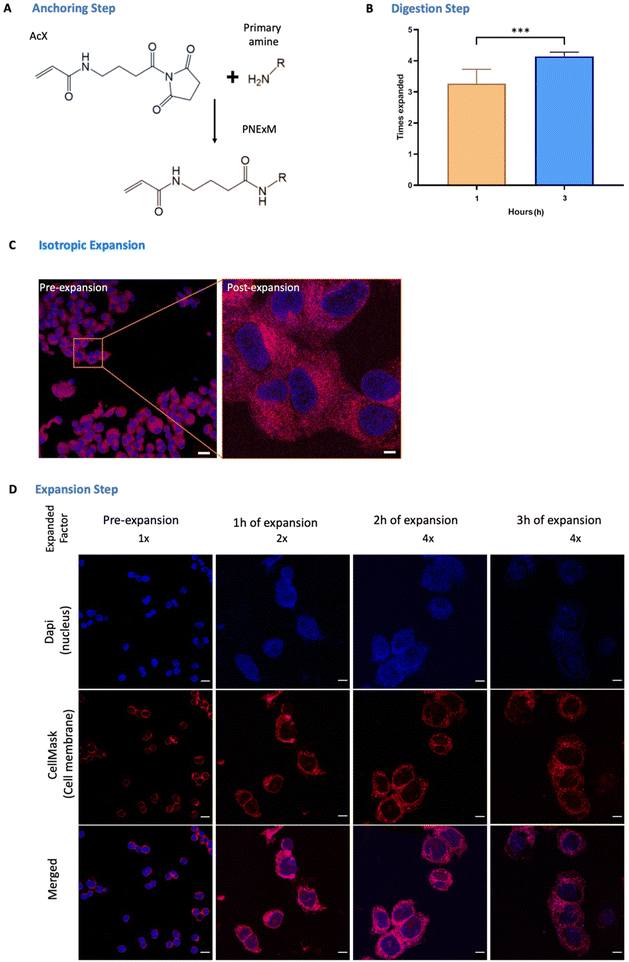
Once the cells have been seeded and the NPs of interest have been labeled, we have to anchor the large amount of primary amines in the system to the terminal NHS groups of the succinimidyl ester of 6-((acryloyl)amino)hexanoic acid (AcX) molecules (Fig. 2A), to allow the retention of our NPs and cell structure in the gel (anchoring step). In our case, as we need to visualize not only the proteins and cell membranes that classic ExM protocols do but also the NPs, we functionalized the maximum possible quantity of primary amines, including the ones present in the carriers. To do so, AcX was incubated overnight in the dark at room temperature to enhance this mentioned reaction of AcX with the NH2 groups of the polymeric carriers allowing the NPs’ attachment to the hydrogel. Protocols that only involve the retention of lipids and proteins only require a minimum of 3 h, using a 0.1 mg ml−1 concentration in all cases. In contrast, this anchoring step enables our system (cells and NPs) to crosslink with the polymeric network formed by an acrylamide/bisacrylamide gel, which is the reason why an overnight incubation is needed otherwise there was no NP retention nor expansion (ESI Fig. S3†). The next step is the gelation process in which the gel is formed by mixing the monomer (Stock X), TEMED, and APS solutions in a 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volumetric ratio, as in the standard way. This solution was vortexed for 10 seconds and immediately placed in ice to avoid premature polymerization. Then, the AcX solution was aspirated and the gelation solution was applied to the sample and transferred to the incubator at 37 °C for 1 hour in the dark. The digestion step, performed with proteinase K, is required to enable the swelling of the hydrogel by the cleavage of some peptide bonds formed in the gelation process. An excessive duration of proteinase K incubation, as indicated in some ExM protocols, could cause the breakage of all peptide bonds and no sample remaining in the gel. In contrast, short incubation times could produce the opposite effect, not enough space for the hydrogel to swell. Therefore, different timings were tested, and a better ratio of resolution
1 volumetric ratio, as in the standard way. This solution was vortexed for 10 seconds and immediately placed in ice to avoid premature polymerization. Then, the AcX solution was aspirated and the gelation solution was applied to the sample and transferred to the incubator at 37 °C for 1 hour in the dark. The digestion step, performed with proteinase K, is required to enable the swelling of the hydrogel by the cleavage of some peptide bonds formed in the gelation process. An excessive duration of proteinase K incubation, as indicated in some ExM protocols, could cause the breakage of all peptide bonds and no sample remaining in the gel. In contrast, short incubation times could produce the opposite effect, not enough space for the hydrogel to swell. Therefore, different timings were tested, and a better ratio of resolution![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) expansion was obtained with 3 h incubation in the dark and at room temperature in an orbital shaker at 60 rpm (Fig. 2B). Finally, to perform gel expansion, the ProK solution was aspirated, the scaffold was removed and the gel was unhooked from the dish surface, allowing a homogeneous expansion as shown in Fig. 2C. Comparison of pre-and post-PExM images revealed minimal sample distortion, with a root-mean-square (RMS) error of <3% for distances of up to 10 μm2, demonstrating that the PExM protocol expands isotropically (ESI Fig. S4†). Additionally, Fig. 2C demonstrates that, thanks to this stablished protocol, a higher ressolution of a 4.6× post-expanded image, in comparison to the pre-expanded image, can be obtained. This swelling step will be determined by the previous steps of anchoring and digestion, and finding the optimum balance between them leads to success. In Fig. 2D we see that immersing the gel in Milli-Q water in the dark and at room temperature for 3 h resulted in fluorophore loss, and no more than 4× of expansion was achieved. In contrast, with 1 h of swelling just 3× of expansion was obtained though the fluorophores were still intact. Therefore, 2 h of gel swelling with a 30 minute water replacement (this was done in all cases) leads, as seen in Fig. 2D, to an optimum ratio between visualization and expansion.
expansion was obtained with 3 h incubation in the dark and at room temperature in an orbital shaker at 60 rpm (Fig. 2B). Finally, to perform gel expansion, the ProK solution was aspirated, the scaffold was removed and the gel was unhooked from the dish surface, allowing a homogeneous expansion as shown in Fig. 2C. Comparison of pre-and post-PExM images revealed minimal sample distortion, with a root-mean-square (RMS) error of <3% for distances of up to 10 μm2, demonstrating that the PExM protocol expands isotropically (ESI Fig. S4†). Additionally, Fig. 2C demonstrates that, thanks to this stablished protocol, a higher ressolution of a 4.6× post-expanded image, in comparison to the pre-expanded image, can be obtained. This swelling step will be determined by the previous steps of anchoring and digestion, and finding the optimum balance between them leads to success. In Fig. 2D we see that immersing the gel in Milli-Q water in the dark and at room temperature for 3 h resulted in fluorophore loss, and no more than 4× of expansion was achieved. In contrast, with 1 h of swelling just 3× of expansion was obtained though the fluorophores were still intact. Therefore, 2 h of gel swelling with a 30 minute water replacement (this was done in all cases) leads, as seen in Fig. 2D, to an optimum ratio between visualization and expansion.
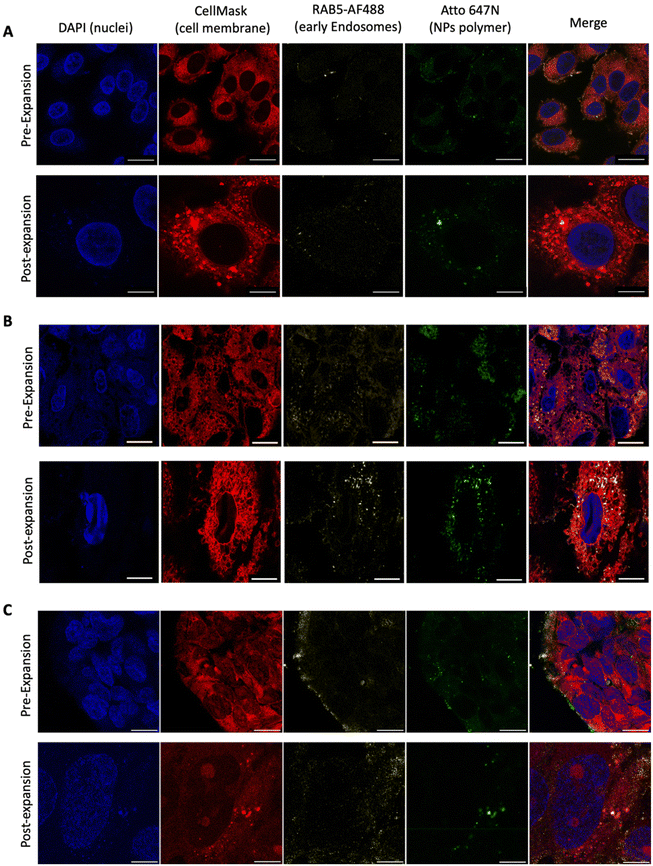
Proof of concept results
Having performed all the steps of the NP expansion protocol, we achieved a significantly enhanced resolution in visualizing the NPs compared to the pre-expanded state, as depicted in Fig. 3A. This enhancement enabled individual NP observation using a simple 63× objective. To validate the developed PExM protocol, as mentioned above, we used our pBAE particle system, and the magnification obtained was up to 4.6 times. The protocol was also demonstrated to be useful for other NPs such as poly(ethylene imine) – PEI – NPs as another example of the utility of the protocol in Fig. 3B and additionally it has been tested with a non-tumorigenic cell line from human bronchial epithelium named BEAS 2B, taken as an example of epithelial cells as shown in Fig. 3C.Thus, this technique enables the differentiation of our NPs in various cell line compartments. For instance, in the case illustrated in Fig. 4A, we aimed to investigate the internalization route of our NPs. Therefore, following the fluorophore section, we employed RAB5 conjugation to study primary endosomes. In the initial observation after 2 h of transfection, a majority of the NPs were colocalized with primary endosomes, predominantly on the cell membrane surface. However, after 4 h of nanoparticle transfection, the same NPs no longer coincided with primary endosomes, having migrated to the perinuclear region. By using the Fiji JACoP plugin as a colocalization tool, we obtained the overlap coefficient of NPs and early endosomes at 2 h as 34.3 ± 1.5% and at 4 h, 28.6 ± 1.1%, these two values being significantly different (p < 0.01), therefore corroborating our previous hypothesis (ESI Fig. S5†).
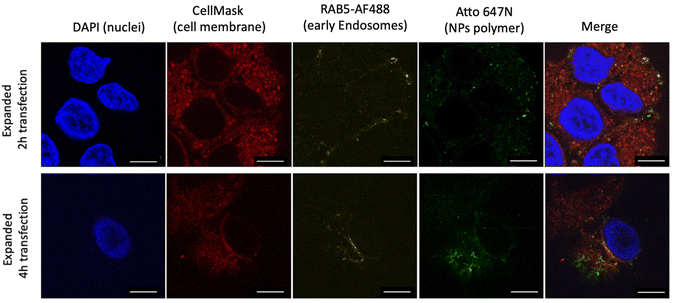 | ||
| Fig. 4 Expanded images of A549 cells with different pBAE NP transfection times. Nanoparticles were transfected for 2 h and 4 h with the PExM protocol. Scale bars are 93 μm. | ||
Conclusions
We present here a novel protocol we developed: Polyplex Expansion Microscopy (PExM). This is a revised approach to the conventional protein retention method in Expansion Microscopy (ExM), which not only retains endogenous proteins within cultured cells but also accommodates synthetic molecules such as polymer nanoparticles within a swelling gel matrix. The PExM protocol enhancements encompass the sequential stages from affixing the fluorophores to the NPs and hydrogel, polymer chain gelation constituting the network that captures and immobilizes cells and molecules, enzymatic digestion of random polymer chains to enable hydrogel swelling, subsequent expansion of the hydrogel and the ultimate visualization using conventional microscopes. This capability allows us to comprehend the cell trafficking of NPs, enabling the study of their internalization, as we demonstrated for pBAE, proprietary polymers used as a proof-of-concept to validate the protocol. In conclusion, our protocol could be an appealing tool for the fine characterization of other polymeric NP interactions with cells that will boost their translation to clinical use.Author contributions
Conceptualization: SB, CF and SP. Data curation: MNL, PCC. Formal analysis: MNL, PCC. Funding acquisition: SB, CF and SP. Investigation: MNL, PCC. Methodology: MNL, PCC. Project administration: SB, CF and SP. Resources: SB, CF and SP. Software: MNL, PCC. Supervision: SB, CF and SP. Validation: SB, CF and SP. Visualization: SB, CF and SP. Writing – original draft: MNL. Writing – review & editing: SB, CF and SP.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support received from PID2021-125910OB-I00 and PID2019-109450RB-I00 funded by MCIN/AEI /10.13039/501100011033/FEDER, UE; co-funding by the Institute of Health Carlos III (ISCIII) (AC22/00042), Award no. TRNSC213882FORN by FCAECC and within the TRANSCAN framework and by the Joint Transnational Initiative ERA-NET TRANSCAN-3, European Commission. This publication is based upon work from COST Action CA21135 supported by COST (European Cooperation in Science and Technology). Yvette Rabadà is acknowledged for help with secondary antibodies and Nil Gonzalez and Pau Jorba are acknowledged for polymer synthesis.References
- C. Lee Ventola, Progress in Nanomedicine: Approved and Investigational Nanodrugs, 2017, 42(12) Search PubMed.
- D. Bobo, K. J. Robinson, J. Islam, K. J. Thurecht and S. R. Corrie, Nanoparticle-Based Medicines: A Review of FDA-Approved Materials and Clinical Trials to Date, Pharm. Res., 2016, 33(10), 2373–2387, DOI:10.1007/S11095-016-1958-5.
- V. Sainz, J. Conniot, A. I. Matos, C. Peres, E. Zupančič, L. Moura, L. C. Silva, H. F. Florindo and R. S. Gaspar, Regulatory Aspects on Nanomedicines, Biochem. Biophys. Res. Commun., 2015, 468(3), 504–510, DOI:10.1016/J.BBRC.2015.08.023.
- S. Kumar, N. Dilbaghi, R. Saharan and G. Bhanjana, Nanotechnology as Emerging Tool for Enhancing Solubility of Poorly Water-Soluble Drugs, Bionanoscience, 2012, 4(2), 227–250, DOI:10.1007/S12668-012-0060-7.
- H. Havel, G. Finch, P. Strode, M. Wolfgang, S. Zale, I. Bobe, H. Youssoufian, M. Peterson and M. Liu, Nanomedicines: From Bench to Bedside and Beyond, 2016, 18, 1373–1378, DOI:10.1208/s12248-016-9961-7.
- J. Wolfram, M. Zhu, Y. Yang, J. Shen, E. Gentile, D. Paolino, M. Fresta, G. Nie, C. Chen, H. Shen, M. Ferrari and Y. Zhao, Safety of Nanoparticles in Medicine, Curr. Drug Targets, 2015, 16(14), 1671, DOI:10.2174/1389450115666140804124808.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Engineering Precision Nanoparticles for Drug Delivery, Nat. Rev. Drug Discovery, 2021, 20, 101–124, DOI:10.1038/s41573-020-0090-8.
- D. M. Valcourt, M. N. Dang, M. A. Scully and E. S. Day, Nanoparticle-Mediated Co-Delivery of Notch-1 Antibodies and ABT-737 as a Potent Treatment Strategy for Triple-Negative Breast Cancer, ACS Nano, 2020, 14, 3388, DOI:10.1021/acsnano.9b09263.
- E. Pérez-Herrero and A. Fernández-Medarde, Advanced Targeted Therapies in Cancer: Drug Nanocarriers, the Future of Chemotherapy, Eur. J. Pharm. Biopharm., 2015, 93, 52–79, DOI:10.1016/J.EJPB.2015.03.018.
- B. Li, Q. Li, J. Mo and H. Dai, Drug-Loaded Polymeric Nanoparticles for Cancer Stem Cell Targeting, Front. Pharmacol., 2017, 8, 239124, DOI:10.3389/FPHAR.2017.00051/BIBTEX.
- S. Hua, M. B. C. de Matos, J. M. Metselaar and G. Storm, Current Trends and Challenges in the Clinical Translation of Nanoparticulate Nanomedicines: Pathways for Translational Development and Commercialization, Front. Pharmacol., 2018, 9, 790, DOI:10.3389/fphar.2018.00790.
- J.-B. Chang, F. Chen, Y.-G. Yoon, E. E. Jung, H. Babcock, J. S. Kang, S. Asano, H.-J. Suk, N. Pak, P. W. Tillberg, A. T. Wassie, D. Cai and E. S. Boyden, Iterative Expansion Microscopy, Nat. Methods, 2017, 14, 593–599, DOI:10.1038/nmeth.4261.
- B. M. White, P. Kumar, A. N. Conwell, K. Wu and J. M. Baskin, Lipid Expansion Microscopy, J. Am. Chem. Soc., 2022, 2022, 18212–18217, DOI:10.1021/jacs.2c03743.
- V. Sheth, X. Chen, E. M. Mettenbrink, W. Yang, M. A. Jones, O. M'Saad, A. G. Thomas, R. S. Newport, E. Francek, L. Wang, A. N. Frickenstein, N. D. Donahue, A. Holden, N. F. Mjema, D. E. Green, P. L. DeAngelis, J. Bewersdorf and S. Wilhelm, Quantifying Intracellular Nanoparticle Distributions with Three-Dimensional Super-Resolution Microscopy, ACS Nano, 2023, 17(9), 8376–8392, DOI:10.1021/acsnano.2c12808.
- F. Chen, P. W. Tillberg and E. S. Boyden, Expansion Microscopy, Science, 2015, 347(6221), 543–548, DOI:10.1126/science.1260088.
- J. Karlsson, K. R. Rhodes, J. J. Green and S. Y. Tzeng, Poly(Beta-Amino Ester)s as Gene Delivery Vehicles: Challenges and Opportunities, Expert Opin. Drug Delivery, 2020, 17(10), 1395–1410, DOI:10.1080/17425247.2020.1796628.
- N. Segovia, P. Dosta, A. Cascante, V. Ramos and S. Borrós, Oligopeptide-Terminated Poly(b-Amino Ester)s for Highly Efficient Gene Delivery and Intracellular Localization, Acta Biomater., 2014, 10(5), 2147–2158, DOI:10.1016/j.actbio.2013.12.054.
- P. Dosta, C. Demos, V. Ramos, D. W. Kang, S. Kumar, H. Jo and S. Borrós, Delivery of SiRNA to Endothelial Cells In Vivo Using Lysine/Histidine Oligopeptide-Modified Poly(b-Amino Ester) Nanoparticles, Cardiovasc. Eng. Technol., 2021, 12, 114–125, DOI:10.1007/s13239-021-00518-x.
- A. Klimas, B. Gallagher and Y. Zhao, Basics of Expansion Microscopy, Curr. Protoc. Cytom., 2019, 91(1), 1–16, DOI:10.1002/cpcy.67.
- C. Zhang, J. S. Kang, S. M. Asano, R. Gao and E. S. Boyden, Expansion Microscopy for Beginners: Visualizing Microtubules in Expanded Cultured HeLa Cells, Curr. Protoc. Neurosci., 2020, 92(1), e96, DOI:10.1002/cpns.96.
- Human wild-type A549 cell line (ab275463) | Abcam https://www.abcam.com/products/cell-lines/human-wild-type-a549-cell-line-ab275463.html# (accessed 2023-11-03).
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Surface charge tunability as a powerful strategy to control electrostatic interaction for high efficiency silencing using tailored oligopeptide-modified poly (beta-amino ester) s (PBAEs), Acta Biomater., 2015, 20, 82–93, DOI:10.1016/j.actbio.2015.03.029.
- M. Navalón-López, A. Dols-Perez, S. Grijalvo, C. Fornaguera and S. Borrós, Unravelling the Role of Individual Components in PBAE/Polynucleotide Polyplexes in the Synthesis of Tailored Carriers for Specific Applications: On the Road to Rational Formulations, Nanoscale Adv., 2023, 5(6), 1611–1623, 10.1039/d2na00800a.
- R. Riera, N. Feiner-Gracia, C. Fornaguera, A. Cascante, S. Borrós and L. Albertazzi, Tracking the DNA Complexation State of PBAE Polyplexes in Cells with Super Resolution Microscopy, Nanoscale, 2019, 11(38), 17869–17877, 10.1039/c9nr02858g.
- R. Riera, J. Tauler, N. Feiner-Gracia, S. Borrós, C. Fornaguera and L. Albertazzi, Complex PBAE Nanoparticle Cell Trafficking: Tracking Both Position and Composition Using Super Resolution Microscopy, ChemMedChem, 2022, 17(13), e202100633, DOI:10.1002/cmdc.202100633.
- A. T. Wassie, Y. Zhao and E. S. Boyden, Expansion Microscopy: Principles and Uses in Biological Research, Nat. Methods, 2019, 16(1), 33–41, DOI:10.1038/s41592-018-0219-4.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr00917g |
| This journal is © The Royal Society of Chemistry 2024 |