Open Access Article
Open Access ArticleBiosensors for detection of airborne pathogenic fungal spores: a review
Roomia
Memon
,
Javed H.
Niazi
 * and
Anjum
Qureshi
* and
Anjum
Qureshi
 *
*
Sabanci University, SUNUM Nanotechnology Research and Application Center, Orta Mah. Tuzla 34956, Istanbul, Turkey. E-mail: anjum@sabanciuniv.edu; javed@sabanciuniv.edu; Tel: +90 216 483 9879/2441
First published on 19th July 2024
Abstract
The excessive presence of airborne fungal spores presents major concerns with potential adverse impacts on public health and food safety. These spores are recognized as pathogens and allergens prevalent in both outdoor and indoor environments, particularly in public spaces such as hospitals, schools, offices and hotels. Indoor environments pose a heightened risk of pulmonary diseases due to continuous exposure to airborne fungal spore particles through constant inhalation, especially in those individuals with weakened immunity and immunocompromised conditions. Detection methods for airborne fungal spores are often expensive, time-consuming, and lack sensitivity, making them unsuitable for indoor/outdoor monitoring. However, the emergence of micro–nano biosensor systems offers promising solutions with miniaturized designs, nanomaterial integration, and microfluidic systems. This review provides a comprehensive overview of recent advancements in bio-nano-sensor system technology for detecting airborne fungal spores, while also discussing future trends in biosensor device development aimed at achieving rapid and selective identification of pathogenic airborne fungi.
1. Introduction
Since the industrial revolution, global concern over environmental pollution has grown exponentially.1 The deleterious ramifications of air pollution, water pollution, and soil degradation are now recognized as major environmental challenges attributable to a diverse array of emerging contaminants.2 Particularly concerning is the escalating problem of environmental contamination stemming from newly identified harmful agents, with the discharge of hazardous substances such as toxins, toxic heavy metals, pesticides, pharmaceutical residues and bioaerosols into the ecosystem raising significant worldwide concerns.3,4 Among these pollutants, bioaerosols are notable for their detrimental impact on both human health and the environment.5 Furthermore, extensive research has consistently revealed that the inhalation of bioaerosol-airborne biological particles can result in a wide spectrum of detrimental health consequences.6 Bio-aerosols are airborne particles that have varying sizes in nano-to-micron ranges. These particles vary in composition and size, encompassing living organisms such as bacteria (approximately 0.5–10 μm), fungi (around 5–100 μm), viruses (approximately 50–200 nm), as well as smoke particles (approximately 50 nm to 1 μm) and pollen. In recent years, there has been significant attention directed towards the monitoring and sampling analysis of airborne pathogenic microorganisms. This heightened focus is largely attributed to the presence of indoor airborne microbial/mold contamination and bioaerosols, which constitute significant contributors, accounting for approximately 5–34% of indoor air pollution.7,8 Surveillance and identification of airborne microbial particles represent a significant concern owing to their presence and the resulting health implications, spreading of infectious diseases, acute toxic effects, allergies, cancer and the potential danger of bioterrorism.9,10 Among these airborne microbial particles, fungi are recognized as a significant contributor to infectious and allergic diseases.11,12 Public spaces, such as hospitals, schools, offices, and hotels, are among the more susceptible places where fungal particle exposure is at higher risk. Individuals with weak immunity or immunocompromised conditions are the most vulnerable to infectious pulmonary diseases caused by the constant inhalation of numerous airborne fungal particles.Among the airborne microbial fungal particles, Aspergillus fumigatus has emerged as a highly prevalent airborne fungal pathogen. It is responsible for dangerous and invasive diseases that are frequently fatal, especially in immunocompromised hosts in developed nations.13 Over the past 12 years, there has been a quadruple rise in invasive aspergillosis (IA). IA accounted for around 30% of fungal infections in cancer patients, with an estimated occurrence of 10 to 25% in leukemia patients. Among leukemia patients, the mortality rate remains high at 80 to 90%, even after treatment.13,14 In leukemia treatment centers, solid-organ transplantation facilities, and bone marrow transplantation (BMT), invasive aspergillosis (IA) has been identified as a major cause of mortality.14 Alveolitis, allergic sinusitis, and asthma are examples of conditions that can result from recurrent exposure to Aspergillus sp. conidia or antigens, even in the absence of mycelial colonization. This information is based on a survey by the World Health Organization (WHO) regarding indoor air quality.15 Therefore, it is imperative to develop innovative methods for early monitoring and accurate detection of such pathogenic airborne fungal particles in contaminated air/water/aerosol environments, which may prevent the disease progression. The conventional fungi detection methods include microscopy, histopathology, growth in pure culture and morphologic study of the reproductive structures, which are challenging due to non-specificity and the requirement of additional time for the radiological imaging (CT scan) method.16 Currently, there is no tool available that can directly allow the detection of fungal pathogens from air/aerosols. Existing detection methods heavily rely on microscopic examination of air/aerosol samples that are qualitative and often inaccurate with false-positive/-negative results. Various assays have been developed for the potential detection of fungal spores in immunocompromised host systems, specifically from liquid phases (blood/serum) or biopsy tissue samples. These assays encompass the polymerase chain reaction (PCR), DNA-based methods,17 enzyme-linked immunosorbent assay (ELISA),18 and spectroscopic quantification,19 all utilized for the detection of pathogenic fungal species.20–25 These techniques are rather more complex, expensive, and time-consuming and often face challenges of false positives due to the high probability of cross-reactivity. Therefore, these methods are practically challenging for in situ real-time detection of pathogenic fungal particles from aerosols. Within this context, biosensors have emerged as invaluable tools for detecting minuscule concentrations of bioaerosols. These biosensors, characterized by compact analytical devices, bring forth a host of advantages, including simplicity, portability, ease of use, exceptional sensitivity, and selectivity. They have emerged as powerful solutions for detecting and analyzing a wide spectrum of bioaerosol pollutants across diverse environmental contexts.26,27 Biosensors can serve as early warning systems and provide active measurements by which precautionary measures can be taken to control the presence and levels of pathogenic fungal spores/particles in air/aerosol contaminants. In recent years, biosensors have particularly excelled as highly effective platforms for analyzing airborne pathogens.28 Simultaneously, the realm of microfluidic bioassays has witnessed significant advancements, facilitating both containment and rapid detection of airborne microbes. Nevertheless, despite these strides, the availability of commercially proven devices for real-world sample analysis remains limited.29 The challenges encompass factors such as low microbe concentrations in the air, the presence of interfering agents, and the intricate task of seamlessly integrating sampling chambers with sensing devices, thereby impeding the success of diagnostic applications.30
Nevertheless, significant advancements in this domain have been propelled by nanotechnology, leveraging the unique characteristics of nanomaterials, particularly their heightened surface-area-to-volume ratio. This has led to the development of highly efficient and sensitive detection methodologies.31 The use of nanomaterials has facilitated the miniaturization of sensing devices, leading to the creation of swift, portable, and sensitive diagnostic systems for pathogens.32 These systems can identify airborne pathogens in various settings, ranging from hospitals and ventilation systems to airplanes.33 Furthermore, they hold promise in preempting and identifying bioterrorism threats in public spaces, thereby bolstering overall security and public health.
In the realm of fungal pathogenicity and environmental health, the detection and characterization of Aspergillus fumigatus spores have emerged as a critical focal point. As a ubiquitous airborne fungal species, Aspergillus fumigatus presents both a formidable challenge and a compelling opportunity for research and diagnostics. Understanding the intricacies of its ecological role, while also mitigating potential health risks is essential for developing effective strategies to manage and control this fungus. Among the pathogenic aspergilli, Aspergillus fumigatus holds the distinction of being the most widespread in the environment and serves as the primary causative agent of disease. Subsequent in order of importance are Aspergillus niger, Aspergillus flavus, Aspergillus terreus, Aspergillus nidulans, and several species belonging to the fumigati family. These organisms exhibit comparable morphological characteristics to Aspergillus fumigatus. In this review, we delve into the latest advancements in biosensor technologies (Scheme 1), focusing on fabrication processes, materials, and biosensing methodologies in the realm of Aspergillus fumigatus spore detection, shedding light on the state-of-the-art techniques and their applications in various fields, from clinical diagnostics to environmental monitoring. Through this exploration, we aim to unravel the complexities surrounding Aspergillus spores and provide insights into the evolving landscape of detection strategies with sustainable biosensor technologies to revolutionize healthcare diagnostics and contribute to a greener and more environmentally conscious future.
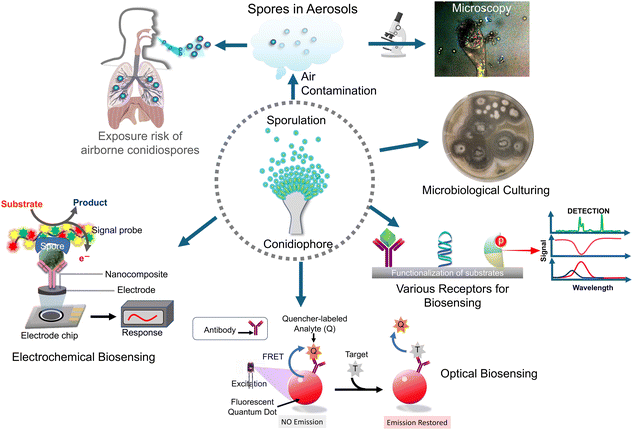 | ||
| Scheme 1 The schematic diagram showing an overview of biosensors for airborne pathogenic fungal spore detection. | ||
2. Conventional approaches to airborne pathogenic fungal spore detection
Evaluating the presence and quantity of airborne pathogenic fungal spores, notably those belonging to Aspergillus fumigatus, holds paramount importance in gauging the risk of respiratory infections and assessing indoor air quality. Over time, various conventional techniques have been employed to detect these fungal spores, including cell-culturing, molecular techniques involving the amplification of specific genes for identification by PCR, and detection for the presence and quantification of biomarker proteins by enzyme-linked immunosorbent assay (ELISA). These techniques are detailed in the following sections.2.1 Cell culture-based assay
Traditional culture-based methods are extensively employed for the isolation, cultivation, and quantification of airborne fungal spores, owing to their ease of use, cost-effectiveness, and operational simplicity. This technique involves incubating bioaerosol samples collected on appropriate nutritional agar surfaces or in buffer solutions at an appropriate temperature for 24 h to facilitate the growth and proliferation of fungal spores. The primary sampler widely employed for this approach is the Andersen sampler. This sampler operates as a multi-stage (cascade) impactor, allowing for a separate collection of spores of varying sizes. It has found application in recent studies conducted both indoors and outdoors.34,35 The resulting colonies (colony-forming units, CFUs) on the agar plates are counted to determine their concentration in bioaerosols followed by identification techniques for cultured microorganisms.36 Microscopic observations are pivotal in identifying fungal pathogens through the examination of morphological features, which are employed for taxonomic classification. These features include sporulation production, spore morphology, and the characteristics of sporulating structures that give rise to both asexual and sexual spore forms.37,38 Although culture-based methods are regarded as the benchmark for microbial testing, they do come with specific limitations. These include a high labour requirement, long detection times and incompatibility with non-cultural pathogens. Moreover, the culture-based method fails to provide accurate information about the total number of airborne microorganisms, limiting its suitability for on-site applications. In a research investigation by Zengmin Miao et al., the enumeration of airborne Aspergillus fumigatus fungal spores in a rabbit house was achieved through both culture-based counting (CCM) and the polymerase chain reaction (PCR). In this study, air samples were gathered and subsequently cultured on specific media to facilitate the growth of viable fungal spores. Following incubation, the count of colony-forming units (CFUs) was determined, representing the quantity of viable fungal spores in air. The results showed that the concentrations of Aspergillus fumigatus spores, as determined by CCM, were 2.5 × 102, 2.8 × 102 and 1.1 × 102 CFU per m3 of air in three different houses (A, B and C), respectively.39–422.2 Polymerase chain reaction (PCR)
PCR stands for the polymerase chain reaction, a highly effective molecular biological approach utilized to amplify and identify DNA sequences. For the detection of fungal spores, including Aspergillus fumigatus, it has been extensively employed. PCR is dependent on the synthesis of new DNA strands that are complementary to a target DNA sequence or a gene by DNA polymerase enzymes. The process involves repeated cycles of heating, cooling, and enzymatic reactions to amplify a specific DNA region of interest, a process similar to the popular COVID-19 RT-PCR method, except that it does not use the enzyme reverse transcriptase (RT) and labelled oligonucleotide probes used in RT-PCR for detection. Furthermore, a typical PCR comprises the following constituents. (a) Template DNA: this involves using a sample containing the target DNA, such as fungal spore DNA, as the template for PCR, (b) primers: these are brief DNA sequences crafted to complement the regions bordering the target DNA sequences. They are purposefully designed to bind to the target DNA or a gene, serving as initial points for DNA synthesis. (c) DNA polymerase: utilizing a heat-stable DNA polymerase enzyme, like Taq DNA polymerase, which can synthesize new DNA strands based on the template DNA and the primers. (d) Nucleotides, comprising the essential building blocks (A, T, C, and G), are crucial for DNA synthesis. The PCR procedure involves three main stages, with the first being (e) denaturation: the reaction mixture undergoes high-temperature heating (typically at 94–98 °C), inducing the separation or denaturation of DNA strands, leading to the formation of single-stranded DNA molecules; (f) annealing: after denaturation, the reaction mixture undergoes cooling to a temperature that enables the primers to selectively bind to their complementary sequences on the single-stranded template DNA. Typically, this step occurs at a temperature around 50–65 °C; (g) extension: following annealing, the reaction mixture is then heated to an optimal temperature, usually around 72 °C. The synthesis of new DNA strands by DNA polymerase enzymes is facilitated at this temperature through the addition of nucleotides to the primers. Thus, complementary copies of the target DNA sequences are generated. These three steps are repeated through multiple cycles, typically 25–35 cycles, leading to the exponential amplification of the target DNA, if that specific sequence is present in the sample. Significantly, each cycle doubles the amount of DNA, resulting in a substantial increase in the quantity of the target DNA sequence. In another study, the researchers developed and validated a PCR assay targeting the Aspergillus fumigatus gene encoding the mitochondrial small subunit rRNA. The PCR assay successfully detected Aspergillus fumigatus DNA in various clinical samples including the bronchoalveolar lavage fluid, sputum, and lung tissues which confirms its utility for diagnosing Aspergillosis, a fungal infection caused by Aspergillus species.42,43 Another study that employed real-time PCR (also reverse transcriptase PCR), a molecular technique utilized for the real-time amplification and quantification of specific fungal DNA sequences, has been documented. Here, identification and measurement of the genetic material of Aspergillus fumigatus spores in air were carried out by collecting samples from three rabbit houses. The results indicated that the concentrations of ambient Aspergillus fumigatus spores in three distinct residences (A, B, and C) were 3 × 103–5 × 103 spores per m3 respectively, as determined by real-time PCR. After comparing both techniques, the real-time PCR method consistently yielded higher spore concentrations compared to the CCM method. The former concentrations were reported to be 12–14 times higher than the latter concentrations in all three rabbit houses.2.3 Enzyme-linked immunosorbent assay (ELISA)
The enzyme-linked immunosorbent assay (ELISA) is a commonly used immunoassay method utilized for the detection and quantification of proteins, biomarkers or other target molecules. It utilizes the principles of antigen–antibody interactions and an enzymatic detection system to provide a measurable signal. In ELISA, the fungal spores coated on the surface, together with the primary antibody, create an antibody–antigen complex. Subsequently, this complex is identified through a secondary antibody linked to an enzyme, usually horseradish peroxidase (HRP), in the presence of H2O2 and a redox-sensitive chromogenic substrate. Here, the enzyme initiates a reaction with a substrate, resulting in the production of a detectable signal directly proportional to the presence or concentration of Aspergillus fumigatus spores in the aerosol samples. The signal is quantified using a spectrophotometer or a plate reader. For example, a study by Stetzenbach et al. (2004) demonstrated the application of ELISA in detecting fungal spores from air samples. The researchers coated microplate wells with fungal spores and used specific primary antibodies against the target spores. They then added a secondary antibody conjugated to alkaline phosphatase, which is another enzyme used in ELISA that catalyzes the hydrolysis of a colorless substrate, such as a p-nitrophenyl phosphate (pNPP) into yellow colored p-nitrophenol (pNP). The resulting colour change is measured spectrophotometrically to quantify the fungal spores present in the aerosol samples collected using an impactor.44,45Conventional techniques for detecting airborne fungi, including culture-based methods, PCR, ELISA, and microscopy, have both merits and demerits. While they offer sensitivity, specificity, and the ability to isolate and identify viable fungal spores, they are also time-consuming, labour-intensive, and may not detect non-viable spores or applicable to all fungal species. Additionally, sampling limitations, laboratory-based analysis, and delayed results (taking days to weeks) can hinder prompt interventions. These limitations highlight the need for alternative, rapid, and on-site detection methods, such as biosensors, to complement conventional techniques and improve airborne fungal detection and monitoring. Despite these drawbacks, traditional techniques are cost-effective and accessible, requiring minimal specialized equipment and expertise. Incorporating modern molecular methods can improve sensitivity and accuracy, offering a more comprehensive assessment of airborne fungal diversity and potential health risks.
3. Biosensors
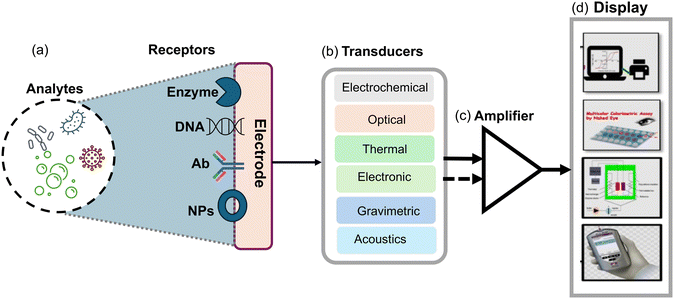
Biosensors are analytical devices that detect and quantify specific substances by combining a transducer with a biological recognition component. These devices are composed of three fundamental components: the biological recognition element, the transducer, and the signal processing system as shown in (Fig. 1). The biological recognition component is typically a biomolecule, such as an enzyme,46 antibody39,47 or nucleic acid48–50 which interacts specifically with the target analyte. This interaction generates a signal or response that is transduced by the transducer into a measurable output. The transducer converts the biochemical or biophysical signal into an electrical, optical and physical signal which can be quantitatively measured. This transduction process is achieved through various mechanisms such as electrochemical reactions, optical absorption or emission, piezoelectric changes, and thermal effects. Moreover, the signal processing system interprets and analyzes the transduced signal and provides quantitative information about the target analyte's concentration or presence compared to other types of sensors.51 Biosensors offer distinct advantages, such as they exhibit high selectivity and specificity, as the biological recognition element is designed to interact only with the target analyte, including fungal spores. This allows for accurate detection and minimizes interference from other substances. Furthermore, biosensors can offer measurements in real-time, allowing for continuous monitoring and prompt response to fluctuations in analyte concentration. Besides, biosensors also offer excellent sensitivity, often detecting analytes at low concentrations which is crucial in applications such as medical diagnostics and environmental monitoring. Biosensors can be designed for miniaturization, enabling portable and point-of-care testing. This portability and ease of use make biosensors highly suitable for on-site analysis and remote field applications.52 Biosensors possess numerous advantages; but besides this, they also have some limitations. For instance, the stability and shelf-life of biological recognition elements can be a challenge, requiring appropriate storage conditions and periodic replacement. Biosensors may also be susceptible to interference from complex sample matrices and necessitate sample preparation steps. Additionally, the development and optimization of biosensors for specific analytes may require significant expertise and time, as compared to other sensors, such as chemical sensors or physical sensors; biosensors offer superior specificity and sensitivity due to the specific interactions between the biological recognition element and the target analyte. Chemical sensors, for example, often rely on non-selective chemical reactions, leading to potential cross-reactivity with interfering substances. On the other hand, physical sensors may lack the specificity necessary for precise analyte detection. Therefore, biosensors with their biological recognition elements provide an elegant solution to overcome these limitations.44,53–55Moreover, biosensors provide rapid and on-site detection capabilities, complementing conventional techniques and improving airborne fungal detection and monitoring. These modern methods enhance sensitivity and accuracy, offering a more comprehensive assessment of airborne fungal diversity and potential health risks. Incorporating biosensors into detection strategies can significantly improve efficiency and effectiveness in identifying and responding to fungal contamination events. In the following section, we explore a range of optical, electrochemical, and microfluidic platforms that utilized nanomaterials with special emphasis on detecting airborne Aspergillus spores.
3.1 Optical biosensors
Optical sensors have a high sensitivity for detecting optical signals produced by analytes binding to probes. Their function is associated with light–matter interactions, which give rise to distinct sensing modes such as colorimetric, plasmonic, and energy transfer sensing methods. Among these, colorimetric sensing is the most employed due to its simplicity and user-friendliness. In a colorimetric biosensor, the signal manifests as a colour change that is easily perceptible and detectable to the naked eye. This eliminates the necessity for intricate equipment and reduces the time-consuming intermediate steps. This approach to detecting infections, pathogens, or endotoxins is extremely effective and trustworthy. Optical biosensors are constructed using fundamental optical principles, including reflection, refraction, absorption, and dispersion (Fig. 2a and b). Optical biosensors have been developed and advanced through the utilization of various transducers, including optical fiber, Raman spectroscopy, and surface plasmonic resonance (SPR). In the case of optical fiber, emitted light traverses through the tapered optical fiber on the detection surface and is subsequently detected. Light-spreading optical biosensors that detect airborne particles and endotoxins have been developed as a result of these methods.56 Reflectance spectroscopy, facilitated by SPR, is employed to identify pathogens and endotoxins.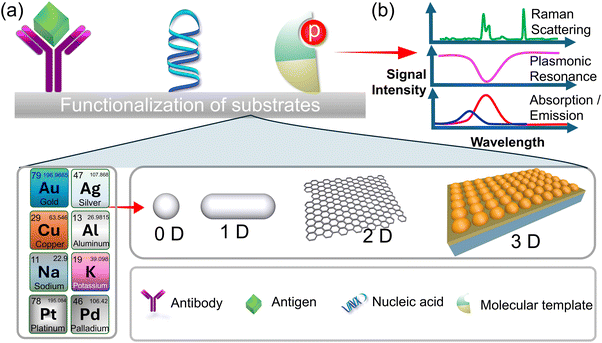 | ||
| Fig. 2 (a) The functionalization process of the substrate with various optical materials and the attachment of specific receptors for enhancement of sensitivity and specificity. (b) The signal detection mechanism, involving the selection of modes such as surface plasmon resonance (SPR), absorbance, transmission or Raman scattering, enabling precise analyte detection and analysis.93 | ||
Kattke et al.47 conducted a study to develop a quantum dot-based immunoassay for detecting A. amstelodami using FRET. The study focuses on developing a quantum dot (QD)-based immunoassay using Förster resonance energy transfer (FRET) for the rapid and specific detection of A. amstelodami. The authors emphasize the importance of optimizing the quenching efficiency and addressing challenges like mold autofluorescence to enhance sensitivity and signal consistency. To construct a complex, QDs were attached to antibodies along with the quencher BHQ-3-labeled mold analyte. This mold analyte had a lower affinity for the antibody than A. amstelodami. The characterization of QD–antibody conjugates included production and purification assessments concerning concentration and antibody-to-QD ratio, serving as quality control measures. To mitigate challenges arising from mold autofluorescence, adjustments were made to the excitation wavelength, and a QD with optimal signal-to-noise ratios was strategically selected. Additionally, quenching efficiency required optimization by choosing a suitable FRET donor–acceptor pair with maximal spectral overlap. A. fumigatus and A. amstelodami were identified as quencher and target analytes, respectively, for the ensuing FRET complex, as depicted in (Fig. 3a and b). It took the optimized displacement immunoassay no more than five minutes to find A. amstelodami concentrations as low as 103 spores per mL for identification. This observation was made while the baseline fluorescence stayed the same even though a negative control of 105 CFU per mL of heat-killed E. coli O157:H7 was present. Scientists did some research and found that they could make detection more sensitive and signal variability less noticeable by creating a more reliable way to attach QDs to antibodies and construct FRET complexes. Optimizing the quenching efficiency and regulating the number of antibodies present on QDs were both contemplated as potential strategies to attain reduced detection limits and consistent signal generation.
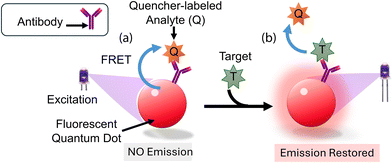 | ||
| Fig. 3 The mechanism of mold detection. (a) Antigen-binding of the QD-conjugated antibody to the quencher-labelled analyte results in the formation of the initial biosensor complex. The QD transmits its energy to the adjacent quencher molecules via FRET upon excitation. (b) FRET is disrupted and an increased emission signal from the QD donor results from the displacement of quencher-labeled analytes caused by the introduction of target analytes.47 Reprinted with permission from ref. 47 (copyright open access © 2011 MDPI). | ||
In general, the FRET-based immunoassay has rapidly progressed which provides accuracy and specificity in identifying the fungus species. The FRET mechanism occurs more commonly in fluorescent compounds and in some nanomaterials, particularly in QDs, which have been utilized in the field of pathogen detection coupled with immunoassays and offer more sensitivity but face drawbacks including high complexity and cost, stability issues, potential toxicity, and interference from autofluorescence. These challenges can complicate their use in pathogen detection, making alternative approaches like gold nanoparticles (AuNPs) for colorimetric sensing more appealing.
Building on the advancements in fungal spore detection technology, Lee et al.48 presented a highly selective and sensitive colorimetric sensor based on AuNPs modified with a rodlet-binding peptide from Aspergillus niger spores. The authors highlighted the efficiency of receptor-modified AuNPs for real-time detection and their potential for mobile applications. Here, a colorimetric sensor was developed which utilizes gold nanoparticles (AuNPs) with a rodlet-binding peptide from Aspergillus niger spore-binding peptide (ASBP), as illustrated in Fig. 4a. To identify the peptide, the scientists used phage display screening and found that a 12-mer peptide known as Aspergillus niger spore-binding peptide (ASBP) had a high affinity for binding to A. niger spores. They then investigated the use of ASBP-modified AuNPs for the real-time colorimetric detection of fungal spores. The research revealed that a substantial quantity of AuNPs modified with ASBP-based peptides efficiently adhered to particles measuring 3 to 5 μm Aspergillus niger spore particle size, causing noticeable color intensity variations in the AuNP suspension as spores settled. This result demonstrated the strong performance of AuNPs modified with ASBP as efficient colorimetric sensors for A. niger spore detection, as shown in Fig. 4b. The sensor system could detect as few as 50 spores in just 10 min and demonstrated an outstanding detection range of 50 to 100 spores, facilitated through a mobile device-specific application. These findings suggest that ASBP-modified AuNPs could be a practical and efficient method for detecting A. niger spores, with potential applications in mobile technology. The study contributes to broader efforts in developing portable and user-friendly biosensors for environmental and health monitoring, showing the potential of peptide-modified nanoparticles in creating efficient colorimetric sensors for pathogen detection.
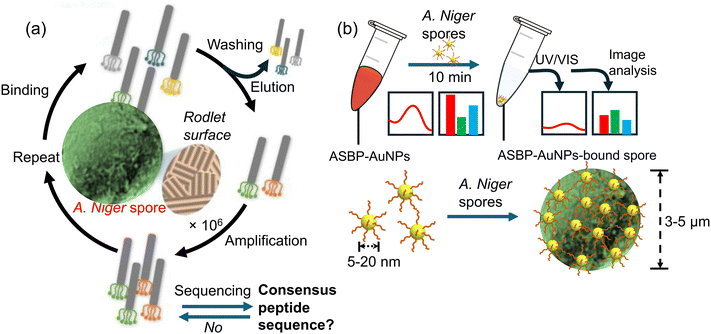 | ||
| Fig. 4 The process of identifying spore-binding peptide ligands by Aspergillus niger and its use in detecting fungal spores. (a) Firstly, the Aspergillus niger spore-binding peptide was discovered via screening of a phage display peptide library; (b) gold nanoparticles were modified with the Aspergillus niger spore-binding peptide (ASBP-AuNPs) to enable rapid colorimetric detection of Aspergillus niger spores. The change in the colour of the Aspergillus niger spore-binding peptide–gold nanoparticle (ASBP-AuNP) dispersion was analyzed by using UV/Vis spectroscopy.48 Reprinted with permission from ref. 48 (copyright © 2021Published by Elsevier B.V.). | ||
In line with advancement of mobile technology shifting away from bulky equipment for measurement and detection of signals, Ling Shan Yu et al.57 developed a portable and cost-effective lab-on-a-chip sensor using nucleic acid-based techniques for detecting azole-resistant Aspergillus fumigatus alleles. They emphasize the platform's compatibility with standard laboratory devices and its potential for early infection detection. The lab-on-a-chip platform utilizes a combination of both loop-mediated isothermal amplification (a slightly similar method to PCR amplication feature discussed above) and ion-sensitive field-effect transistors (FETs) to detect azole-resistant alleles in Aspergillus fumigatus. Here, the authors chose two promoter regions with TR34 and TR46 mutations for detection because these mutations are associated with the ubiquitous mechanism of Aspergillus fumigatus. The ion-sensitive FETs fabricated using unmodified complementary metal–oxide semiconductor (CMOS) technology enable electrochemical sensing without the need for bulky and complex optical equipment. The loop-mediated isothermal amplification method amplifies specific DNA sequences through isothermal incubation, eliminating the requirement for thermocycling and reducing hardware complexity, power consumption, and diagnostic platform size. This device can accurately detect TR34 and TR46 alleles, responsible for azole resistance, with high sensitivity and specificity within 30 min.
Additionally, the AndroidOS mobile application facilitates data acquisition, visualization, and cloud connectivity, making it a powerful tool for point-of-care and environmental surveillance, as illustrated in Fig. 5(a–d). The efficacy of the detection method is confirmed by testing samples from various countries, showcasing a notable sensitivity with a detection limit of 10 genomic copies per reaction within 30 min. This work contributed to the field of point-of-care diagnostics and environmental surveillance by integrating nucleic acid amplification and CMOS technology. It aligns with global health initiatives aimed at combating antimicrobial resistance.
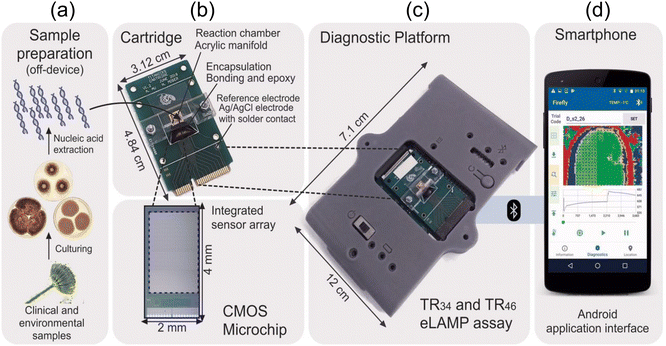 | ||
| Fig. 5 (a) The diagnostic technique for identifying TR34 and TR46 using the lab-on-a-chip platform. After cultivating clinical and environmental samples, nucleic acids are extracted in an off-device manner. Following this, (b–d) the nucleic acids that were extracted are merged with TR-LAMP reagents and transferred to a disposable cartridge that contains a CMOS microchip featuring 785x ISFET sensors. The LoC diagnostic platform establishes a Bluetooth connection with a smartphone upon receiving the inserted cartridge. Reprinted with permission from ref. 57 (copyright open access © 2020 Published by American Society for Microbiology). | ||
Furthermore, other studies58 introduced a novel nanoengineered plasmonic chip that utilizes the colorimetric principle with localized surface plasmonic resonance (LSPR) to modulate bright-field colors for detecting Aspergillus fumigatus and develop portable and smartphone-based detection systems. This approach departs from traditional dark-field imaging. The alternative strategy is well-suited for portable applications and can be integrated into smartphone-based detection systems, as illustrated in Fig. 6. For biological specificity of the above sensor system, researchers employed a unique approach by dip-pen nanolithography (DPN) to functionalize bioreceptors. This method systematically prints biomarkers on a microscale, which facilitates high-throughput processes, streamlining the detection process for large-scale screening and simultaneous detection of multiple analytes. The nanofabrication process combines three technologies, namely dip pin nanolithography, electron beam lithography and molecularly active self-assembly. This combination of techniques enabled the creation of Au-nanodisks (Au-NDs) as solid binding sites, which were subsequently modified with a specific probe such as single-stranded DNA (ssDNA) for detecting fungal infections. The surfaces of these modified Au-ND binding sites were coated with self-assemblies of silver nanoparticle (Ag-NP)-labelled target ssDNA. After the successful hybridization of the ssDNA (sandwiching), a color change occurs at the locations where the self-assembly of Ag-NPs took place. As a result, the electric field localization in the hotspots between the Au-NDs and Ag-NPs enhanced the Raman signals, allowing for surface-enhanced Raman scattering (SERS) mapping.
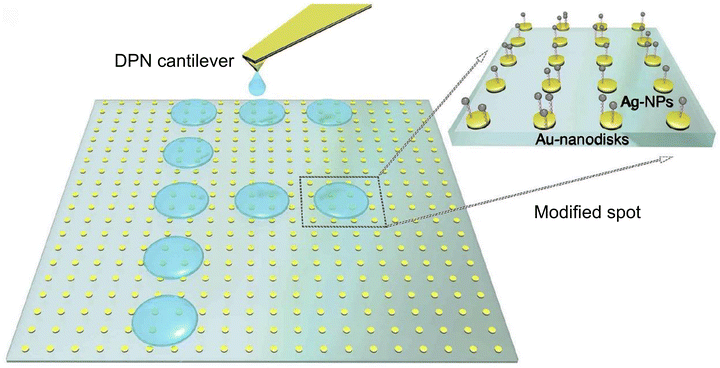 | ||
| Fig. 6 Schematic illustration of the fabrication process of plasmonic nanostructures. The bright-field image of Au-NDs is presented within a square surrounded by gold markers and a green-blue colour scheme. DPN is employed to immobilize and pattern the probe ssDNA onto the binding sites of Au-NDs. The utilization of this method produces the letter F on the left, which indicates an infection with Aspergillus fumigatus. A zoomed-in region on the right illustrates the process by which Ag-NPs self-assemble onto the Au-NDs-binding sites to the hybridization of the target ssDNA, which was initially modified by DPN. Reprinted with permission from ref. 58 (copyright © 2019 Published by Elsevier B.V.). | ||
The simplest plasmonic sensor system for detecting fungal spores is to mix clinical samples suspected of spore-contamination with receptor-labeled AuNPs. Tobiloba Sojinrin et al.59 employed this method for detecting fungal hyphae and spores, including human cutaneous fungal infections. The sensitivity and rapid detection capabilities of AuNPs allowed for distinguishing between infected and uninfected samples. Leveraging the localized surface plasmon resonance properties of gold nanoparticles, researchers have devised a swift and sensitive method for detecting fungal hyphae and spores. Gold nanoparticles with a diameter of 40 nm were functionalized with a peptide sequence designed to bind to fungal cell-wall components. These nanoparticles were then exposed to various fungi, including Candida albicans and Aspergillus fumigatus. The LSPR peak shift observed indicated the binding of the nanoparticles to the fungal cells as illustrated in Fig. 7. Researchers have also evaluated the nanoparticles on human skin samples infected with Trichophyton rubrum, a common cause of cutaneous fungal infections. These nanoparticles demonstrated the capability to differentiate between infected and uninfected skin samples with the aid of the LSPR peak shift phenomenon. This pioneering technology marks a significant advancement in the realm of fungal infection detection. This research builds on the application of plasmonic nanoparticles specifically for fungal infection detection, an advancement in biomedical diagnostics. It highlights the potential for clinical use and the importance of early diagnosis in preventing the spread of infections. However, several challenges remain, such as those identifying and harnessing the ability of natural biological receptors (host proteins, cell-wall/membrane structures, or nucleic acid probes that are derived from the analyte of interest, e.g. fungal spores) or synthetic molecules that serve as receptors, and effective ways to labelling them on nanoparticles, while preserving the nanoparticles’ colloidal forms.
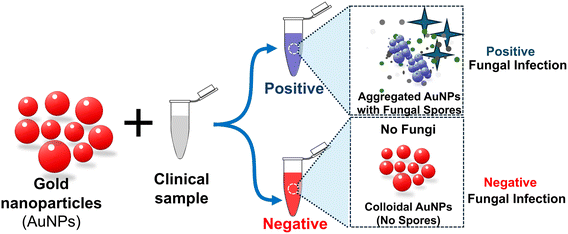 | ||
| Fig. 7 Schematic illustration of the gold nanoparticle system for plasmonic detection of fungi to facilitate self-diagnosis and hygiene control of clinical fungal infections. The progression in hue from red to pink to purple to blue signifies an escalation in the concentration of fungi present in a given clinical sample.59 | ||
Other nanoparticles that have non-LSPR features have been utilized for detecting fungal spores. For instance, Gaikwad and colleagues60 developed an economical method for synthesizing carbon dot (CD) films for detecting Aspergillus niger spores. This method relies on the interaction between CDs and spores, leading to fluorescence quenching and potential applications in real-world settings. Here, CD films were synthesized using a flame/combustion technique at the liquid–liquid interface, and these CD films were deposited on quartz plates via the Blodgett process, as shown in Fig. 8a and b.
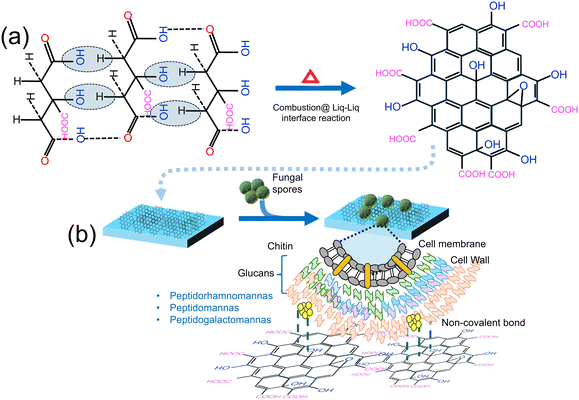 | ||
| Fig. 8 (a) Synthesis of the CD-based sensor film using flame/combustion at the liquid–liquid interface. (b) Schematic representation of fungal spore detection using the CD-based sensor film. Reprinted with permission from ref. 60 (copyright © 2019 Published by American Chemical Society). | ||
The sensitivity of the CD film sensor towards Aspergillus niger spores was evaluated by measuring the fluorescence intensity against varying spore concentrations while maintaining a constant CD concentration. The resulting gradual decrease in the emission intensity of the CDs corresponded to the increasing concentrations of A. niger spores. To further assess the sensor films’ ability to detect Aspergillus spores, a controlled environment was simulated in a closed wooden box, mimicking typical spore-rich conditions. Twelve sensor films were placed at different locations within the box. It was found that under moist environment conditions, the CD films tend to exhibit a significant change in emission properties, while no change was observed with the dry sensor films. This system responds efficiently under moist conditions that promote the interaction between the CDs and the fungal spores, conditions ideal for fungal propagation. The sensing mechanism operates by exploiting the increased accessibility of CDs’ surface area under moist conditions, facilitating bonding with –OH and –COOH groups on the surface through van der Waals and hydrogen bonds. This interaction leads to fluorescence quenching of CDs primarily due to the inner-filter effect (IFT), where overlapping absorption regions hinder electron relaxation, resulting in quenching. To test the practical application of the sensor films, they were exposed to real-world settings such as near a dustbin, in food and fruit storage areas, and hospitals. The quenching effect observed in these environments mirrored the simulated tests, confirming the sensor films’ effectiveness. This study contributes to the advancement of fluorescent sensors for environmental monitoring and food safety, while demonstrating the practicality of carbon-based nanomaterials in detecting fungal spores, aligning with efforts to create cost-effective and efficient biosensors. However, a major challenge with this approach is associated with specificity and selectivity, which can be overcome by introducing the bio-/chemical receptors.
Jin Woo Seo et al.49 employed cell-SELEX and spore-SELEX techniques to isolate aptamers specific to Aspergillus spores. The study highlights the high affinity and specificity of the identified aptamer, Asp-3, and its potential integration into sensors for detecting toxic Aspergillus species. Here, cell-SELEX and spore-SELEX techniques were employed to isolate aptamers capable of binding to the spores of three Aspergillus species, namely A. fumigatus, A. flavus, and A. niger (Fig. 9a and b). These fungal species are known to release airborne spores that can cause allergic reactions and severe invasive fungal infections (IFIs). After 12 successive iterative rounds of SELEX to identify aptamer candidates with high affinity and specificity for Aspergillus spores, the most efficient aptamer, Asp-3, emerged as a candidate for application as a bioreceptor (Fig. 9c). The dissociation constant (Kd) values for Asp-3 towards A. fumigatus, A. flavus, and A. niger were determined to be 80.12 nM, 35.17 nM, and 101.19 nM, respectively. It has also been found that Asp-3 binds to certain cell surface proteins on the spore surface. Asp-3 demonstrated high specificity towards Aspergillus species compared to other fungal species. Furthermore, the aptamer exhibited a quantitative linear relationship between itself and the concentrations of Aspergillus spores. These findings suggest that Asp-3 is a promising candidate for integration with sensors to detect the spores of three toxic Aspergillus species. The robust binding affinity, specificity, and quantitative relationships of Asp-3 position it as an asset in the advancement of sensor-based systems for the accurate detection of the perilous fungal spores. The research connects to the broader field of aptamer-based biosensors, emphasizing the potential of aptamers as selective recognition elements in diagnostic tools. It also supports ongoing efforts to developing sensitive and specific methods for fungal spore detection (Table 1).
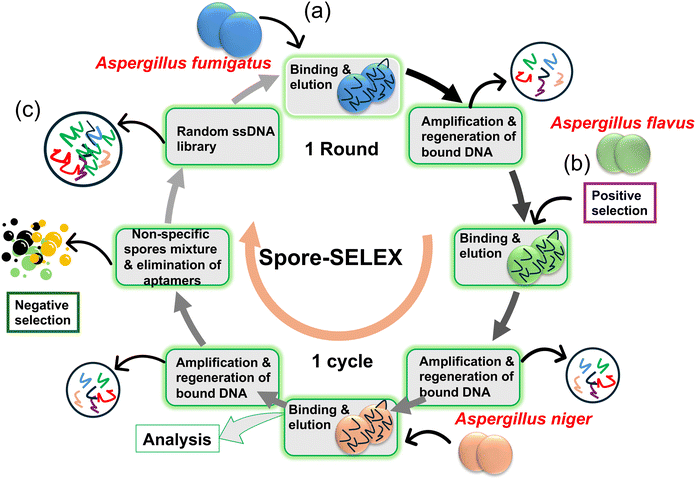 | ||
| Fig. 9 (a) Overview of the methods used to select aptamers that bind to the spores of three toxic Aspergillus species, starting with a random ssDNA library and undergoing four cycles of three rounds (a total of twelve rounds). (b) Positive selection was performed for A. fumigatus, A. flavus, and A. niger. (c) To exclude non-specific aptamers, negative selections were conducted at rounds 4 and 7 using non-specific spore mixtures49 (copyright open access © 2021 Published by RSC). | ||
| Species | Dynamic range | LOD | Time | Ref. |
|---|---|---|---|---|
| Aspergillus niger | 50–1500 spores | 50 spores | 10 min | 48 |
| TR34 and TR46 mutations associated with Aspergillus fumigatus | 5.6 × 103–2.9 × 105 copies per reaction | 10 genome copies per reaction | 20 min | 57 |
| A. amstelodami | 102–105 spores per mL | 103 spores per mL | Less than 5 min | 47 |
| Aspergillus niger | 1–16 CFU per mL | 10 CFU per mL | 2 min | 59 |
| Aspergillus niger | 1.2 × 107 spores per mL | ND | 3 h | 60 |
| A. fumigatus, A. flavus, & A. niger | 5 × 105, 106, 1.5 × 106, 2 × 106, 5 × 106, 1 × 107 spores per mL | ND | 30 min | 49 |
3.2 Electrochemical biosensors
An electrochemical biosensor serves as a tool for detecting and quantifying biological substances (such as biomolecules, cells and pathogens) by transforming their interactions with a biological recognition element into electrical signals. These sensors find extensive applications in diverse fields, including medical diagnostics, environmental monitoring, food safety and biotechnology.56,61,62Fig. 10a–d depict the operational principle of an electrochemical biosensor comprising three key components. (a) Biological recognition element: this element is designed to be specific to the target analyte (the biological substance under detection). It could be an enzyme, antibody, DNA/RNA aptamer, or any other biologically active molecule that selectively interacts with the target. The binding of the target molecule to the recognition element initiates a biological reaction. (b) Transducer: the transducer is responsible for converting the biochemical signal generated by the biological recognition element as a result of interaction with a target analyte, into an electrical, optical, or mechanical signal that can be measured and analyzed. The electric/electrochemical transducer operates based on various electrochemical transduction techniques, such as amperometry, potentiometry, or conductometry.63,64 Electrochemical measurement involves the interaction of the target analyte with the biological recognition element, leading to a change in the electrical properties of the sensor and the generation of an electrical signal. The magnitude of this signal is directly proportional to the concentration of the target analyte present in the sample. The primary types of electrochemical biosensors include amperometric biosensors; these biosensors quantify the current produced during a redox reaction. The presence of the target analyte triggers an oxidation or reduction reaction at the electrode surface, resulting in a current that is proportional to the analyte concentration. Potentiometric biosensors assess the potential difference between two electrodes. The binding of the target analyte to the recognition element induces a change in the surface charge, resulting in a detectable potential difference.62,65,66 Conductometric biosensors quantify alterations in the electrical conductivity of the medium between two electrodes. The recognition or binding of the target analyte to the element modifies the conductivity of the solution.62,67,68 The advantages of electrochemical biosensors include their high sensitivity, rapid response times and potential for miniaturization and integration with electronic devices. They have significantly contributed to advancements in healthcare, biotechnology, and environmental monitoring by providing fast and accurate detection of various biological substances.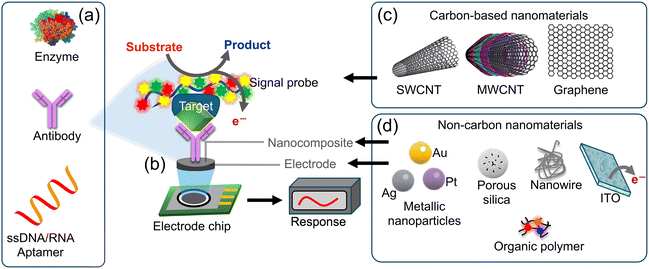 | ||
| Fig. 10 Schematic diagram of an electrochemical sensor: (a) biological recognition elements such as those but not limited to antibodies, enzymes, or DNA are designed for target specificity; (b) these elements are applied onto the working electrode, (c and d) materials that enhance the electrochemical signals have been used to modify the working electrodes such as carbon and non-carbon materials such as SWCNTs, MWCNTs, graphene, NPs, polymers, or nanowires. The transducer converts biochemical signals into electrical signals, facilitating accurate measurement and analysis.69 | ||
The following section explores recent advancements in biosensing technologies for detecting fungal contamination or spores, including electrochemical biosensors.
Identifying specific bioreceptors for fungal infections is challenging for two reasons, not all fungal species are invasive in nature, and their invasion is largely dependent on host's susceptibility, resistance, and immune response, particularly in individuals with compromised immune systems. Therefore, targeting the specific virulent gene as a genetic biomarker for fungi that cause infections, such as invasive aspergillosis (IA) proves to be an effective approach to biosensing. For instance, gliP gene of a few Aspergillus species is linked to the cause of IA disease. An oligonucleotide probe for this gliP gene (glip-T) has been utilized as a genetic biomarker for electrochemical biosensing of virulent Aspergillus species. The biosensor's sensitivity and specificity marking the first direct detection of glip-T using a biosensor has been reported.70 This biosensor was constructed by immobilizing glip-P onto chitosan-stabilized gold nanoparticles (AuNPs) attached to a 1,6-hexanedithiol (HDT) self-assembled monolayer (SAM) prepared on a gold electrode (Au-electrode). Detection of glip-T was accomplished through a hybridization reaction followed by intercalation of toluidine blue, yielding the analytical signal, as illustrated in Fig. 11a and b. The researchers optimized the analytical parameters for the biosensor, determined the detection limit of glip-T in both standard buffer and a real sample matrix, and assessed interference caused by random nucleotides and non-specific biochemicals. One of the advantages of utilizing oligo-probes on sensors is their ability to regenerate by simple chemical or physical denaturation, followed by their renaturation by bringing them back to normal conditions by washing or annealing. The detection limit as determined for glip-T by the above method was 0.32 ± 0.01 × 10−14 (RSD <5.2%), with a dynamic range between 1 × 10−14 and 1 × 10−2 M concentrations of glip-T.70 It marks the first direct detection of glip-T from the fungal strain using a biosensor for IA diagnosis advancing the field of electrochemical biosensors for fungal infection diagnosis. It aligns with the need for rapid and sensitive diagnostic tools for life-threatening infections, contributing to improved patient outcomes.
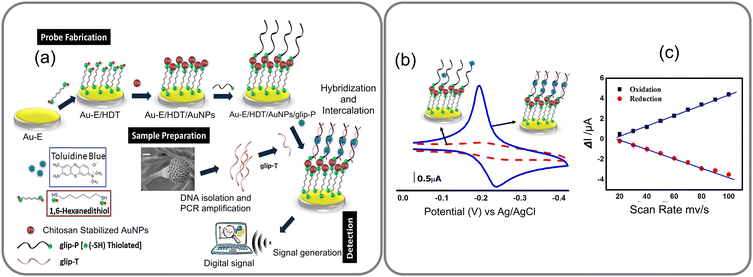 | ||
| Fig. 11 (a) The GLIP biosensor is fabricated by depositing a layer of gold nanoparticles on a gold electrode functionalized with hexane dithiol and immobilizing the GLIP protein on the surface of the AuNPs through covalent bonding. The GLIP biosensor detects target molecules based on the redox properties of the GLIP protein, which has three heme groups enabling it to participate in redox reactions with various electron acceptors. (b) CVs for Au-E/HDT/AuNPs/glip-P (red dotted curve) and Au-E/HDT/AuNPs/glip-P/glip-T (blue curve) after interacting with toluidine blue and (c) plots of the redox peak currents vs. scan rates for Au-E/HDT/AuNPs/glip-P/glip-T interacted with toluidine blue. Reprinted with permission from ref. 70 (copyright © 2018 Published by Elsevier B.V.). | ||
A highly sensitive aptamer-based biosensor was introduced by Xin Ma et al.71 for detecting gliotoxin using a dual-signal detection mechanism involving AIE and the CRISPR Cas12a-mediated cleavage reaction. Gliotoxin (GT) is an epidithiodioxopiperazine metabolite synthesized by several pathogenic fungi, such as Aspergillus fumigatus, Eurotium chevalieri, Gliocladium fimbriatum, and various Trichoderma and Penicillium species. The biosensor was operated through a dual-signal detection mechanism, by initially activating the AIE effect through target-induced stimulation and subsequently initiating CRISPR Cas12a (LbCpf1)-mediated cleavage (Fig. 12a and b). The aptamer-based biosensor involves two key stages: first, the interaction of the target with the aptamers results in the rapid breakdown of DNA building blocks, releasing Ac1 and forming ETTC–dsDNA, which aggregates, creating a fluorescence signal through the AIE effect. Second, the release of Ac2 triggers LbCpf1-crRNA, significantly enhancing ssDNA–Fc cleavage for signal amplification and achieving ultrasensitive target detection (Fig. 12c). This approach successfully overcomes the challenge of weak electrochemical signals observed in previous gliotoxin sensors. The breakthrough technique not only reduces the detection time by up to 55 minutes, but also achieves an impressive low detection limit of 2.4 femtomolar. The biosensor exhibits a highly satisfactory linear range of 50 femtomolar to 1 nanomolar for gliotoxin detection, showcasing a more efficient and effective method for gliotoxin detection. This research contributes to the development of innovative biosensors combining aptamer recognition and CRISPR technology. It supports the trend towards creating highly sensitive and specific diagnostic tools for mycotoxin detection.
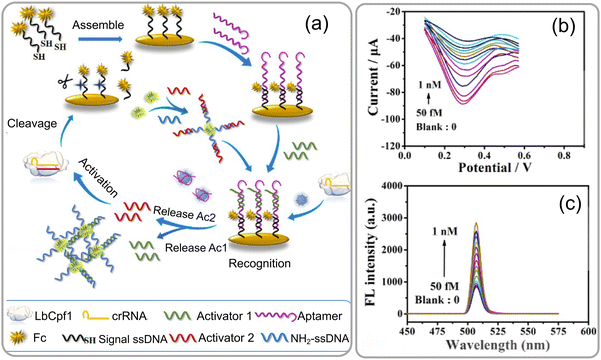 | ||
| Fig. 12 (a) Schematic representation of an ultrasensitive electrochemical-fluorescent-biosensing method. (b) The response of differential pulse voltage (DPV) was assessed at various concentrations of GT, and (c) variation in the fluorescence intensity of the biosensor at distinct concentrations of GT, as well as the calibration curve of −ΔI% against the logarithm. The inset illustrates the calibration curve of ΔFL% as a function of the logarithm. Reprinted with permission from ref. 71 (copyright © 2023 Published by American Chemical Society). | ||
Another most efficient, rapid and cost-effective approach for biosensor transducers is using a field-effect transistor (FET) that detects binding of charged molecules, such as protein receptors, on analyte surfaces to the gate of the FET. When these analytes bind, they change the charge distribution of the underlying semiconductor material, resulting in a change in the conductance of the FET channel. FET-based biosensors offer advantages such as intrinsic amplification, small size, and compatibility with microfluidic devices for lab-on-chip applications. FET-based biosensors have been employed for fungal allergen detection. For instance, Joon-Hyung Jin et al.39 presented a sensor designed for the swift detection of Aspergillus niger, a well-known allergenic fungal species. The study involves the use of single-walled carbon nanotube (SWNT) field-effect transistors (FETs) that were functionalized with pentameric antibodies specifically binding to Aspergillus species. By utilizing the electrostatic gating effect of the Aspergillus fungus, this approach facilitated real-time, remarkable sensitivity, and selectivity in detection of Aspergillus sp. SWNT-FETs were initially functionalized with mouse anti-Aspergillus species-specific primary antibodies (WF-AF-1 Abs) by covalent attachment on the SWNT channel using EDC/NHS chemistry, as depicted in Fig. 13a and b. The sensor exhibited a detection range of 0.5–10 mg mL−1 with a detection limit of 0.3 pg mL−1, representing a threefold improvement in sensitivity and a hundred-fold expansion in the detection range compared to any previously reported sensor (Fig. 13c–h). This research advances the application of carbon nanotube-based FETs in biosensing. It aligns with ongoing efforts to create highly sensitive and selective electrical biosensors for fungal pathogen detection, contributing to improved environmental and health monitoring.
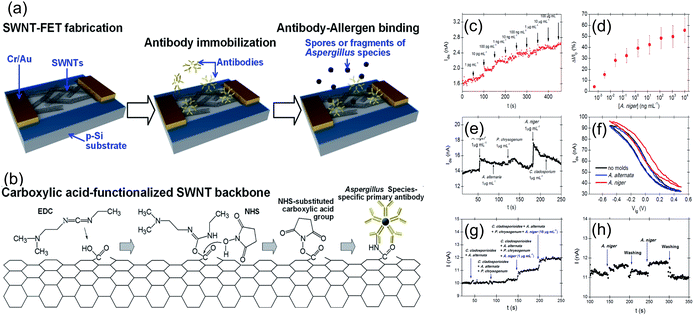 | ||
| Fig. 13 (a) Illustration of the antibody–allergen binding and the fabrication of fungi sensors based on SWNT-FETs for binding of allergens. (b) EDC/NHS chemistry was utilized to immobilize WF-AF-1 Abs directly onto the carboxylic acid-functionalized SWNT surface. (c)The real-time response of an SWNT-FET device immobilized with WF-AF-1 Abs to a series of injections of varying concentrations of A. niger-suspended PBS solution. (d) Corresponding calibration curve. (e) Response of the functionalized SWNT-FET device in real-time to a random succession of administrations of various mould-suspended PBS solutions. (f) Sensor possesses a liquid gating effect that is specific to target and non-target molds. (g) Injection of four distinct mixtures of fungal solutions sequentially. The initial two mixtures do not contain A. niger, whereas the final two mixtures do contain A. niger in ten-fold different concentrations. (h) Water and PBS buffer are used to thoroughly rinse the sensor to revive it. Reprinted with permission from ref. 39 (copyright © 2015 Published by RSC advance). | ||
CNTs play a crucial role in FET biosensors because of their high surface-to-volume ratios and excellent electrical properties, biocompatibility and faster response times. Such FET biosensors can be integrated with a microfluidic system, which enables extensive washing of the sensor device, sensor regeneration, extends the system operation time, and allows for removal of interference from contaminants, especially from bioaerosols that contain micron-sized particles, such as microplastics. Junhyup Kim et al.74 addressed the above issues and introduced the bioaerosol auto-monitoring instrument (BAMI), integrated with a bioaerosol sampler and two-channel carbon nanotube field-effect transistors (CNT-FETs), enabling real-time monitoring of airborne fungal particles (Fig. 14a and b).
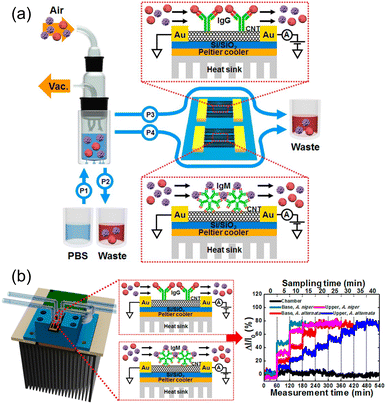 | ||
| Fig. 14 (a) Schematic diagram of the bioaerosol auto-monitoring instrument (BAMI) integrated with a bioaerosol sampler and two-channel carbon nanotube field-effect transistors (CNT-FETs) for real-time monitoring of airborne fungal particles. (b) Illustration of CNT-FET functionalization with specific antibodies for capturing airborne Aspergillus niger and A. alternata particles, resulting in changes in electrical conductivity for immediate analysis. Reprinted with permission from ref. 74 (copyright © 2016 Published by American Chemical Society). | ||
CNT-FETs functionalized with specific antibodies captured airborne Aspergillus niger and A. alternata fungal particles, resulting in detectable changes in the electrical conductivity for immediate analysis. This application in residential settings is crucial for maintaining indoor air quality and preventing health issues associated with fungal exposure. The integration of CNT-FETs with the bioaerosol sampler in a real-world environment demonstrated the system's capability to detect and differentiate between various airborne fungal species, specifically Aspergillus niger and Alternaria alternata. This enhanced both selectivity and sensitivity for environmental monitoring applications. By effectively identifying harmful fungal particles in homes, the BAMI system helps mitigate potential health risks associated with bioaerosols and ensure a safer living environment.
3.3 Miscellaneous sensor studies
Additionally, this section covers the use of biomarkers like glip-T for the biosensing of invasive aspergillosis, electromagnetic biosensing techniques that detect changes in magnetoresistance in response to applied oscillating magnetic fields, and the detection of fungal spores or contamination using cantilever-based atomic force microscopy (AFM).Magnetic biosensors offer rapid and direct signal generation from the ligand–receptor interactions on the sensor surfaces. Giant magnetoresistance type of transducer platforms generate a signal based on the dependence of electron scattering on spin orientation, observed in multilayer structures composed of alternating ferromagnetic and non-magnetic conductive layers. GMR-based biosensors work by detecting changes in the magnetic field caused by the presence of magnetic nanoparticles or magnetic-labels attached to biological receptors. A notable example of such an approach was reported recently,72 where a magneto-nanosensor biochip immunoassay method is employed for detecting Aspergillus fumigatus allergen Asp f using GMR spin-valve sensors. The study highlights the sensitivity and specificity of the detection process. GMR spin-valve sensors quantify the magnetic nanoparticles selectively bound to the sensor surface, as depicted in Fig. 15a–d. In the initial steps, antibodies are covalently immobilized on the sensor surfaces. These antibodies are designed to specifically bind to the Asp allergens. Upon capturing the Asp of allergens, detection antibodies bind, forming a sandwich-like structure. This configuration enhances both the specificity and sensitivity of the detection process. The detection antibodies used are pre-biotinylated and subsequently labelled with nanometer-diameter streptavidin-coated magnetic nanoparticles. During the measurement process, the magnetic nanoparticles generate a stray magnetic field in response to an applied oscillating external magnetic field. This magnetic field induces changes in the resistance of the spin-valve sensor due to the GMR effect. These changes in resistance correspond to the specific biological signal being measured. The study contributes to the development of magneto-nanosensors for allergens, supporting the trend towards integrating advanced sensor technologies in immunoassays and enhancing their application in environmental and clinical diagnostics.
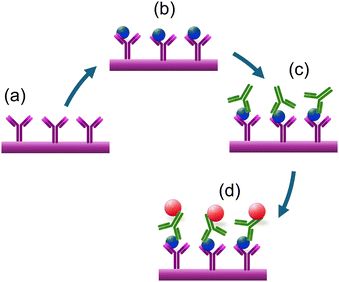 | ||
| Fig. 15 The schematic representation of the magneto-nanosensor biochip immunoassay involves several steps. (a and b) Immobilization of antibodies on the surface of the working electrode. (c) Sandwich structures are formed through the addition of biotinylated detection antibodies. (d) Magnetic nanoparticles bound to the detection antibodies generate a stray magnetic field that results in a change in sensor resistance due to the GMR effect.72 | ||
Other rapid and direct biosensing approaches include cantilever-based atomic force microscopy (AFM) biosensors. This type of biosensor device utilizes the deflection of a microfabricated cantilever to detect and measure biomolecular interactions. Cantilever probes that go into this method can be chemically or biologically modified to tailor them to bind the desired target analyte or biomarker on a given surface or medium. Nugaeva's research73 explored a similar technique for detecting individual fungal spores on cantilever surfaces. The authors emphasize the rapid detection capabilities and the potential for assessing spore contamination levels. In this study, two major fungal spores were chosen as a target sample to investigate a novel method for growth and detection using a cantilever array. The cantilever surfaces were functionalized with a variety of proteins. The researchers leveraged the specific biomolecular interactions of grafted proteins with the molecular structure on the fungal cell surface. It was observed that these proteins exhibited different affinities and efficiencies for binding to the spores. This discovery underscored the diverse and selective nature of the interactions between the proteins and the spore surfaces, as depicted on the immunoglobulin-G functionalized cantilever surface in Fig. 16. This rapid detection stood in stark contrast to conventional techniques that typically required several days. The biosensor demonstrated the capability to detect the target fungi in the range of 103–106 CFU per mL. The measured shift's magnitude was directly proportional to the mass of individual fungal spores, facilitating the assessment of spore contamination levels. This study connects to the broader field of AFM-based biosensors, demonstrating their potential for rapid and sensitive pathogen detection. It supports efforts to develop high-resolution and selective biosensors for environmental monitoring (Table 2).
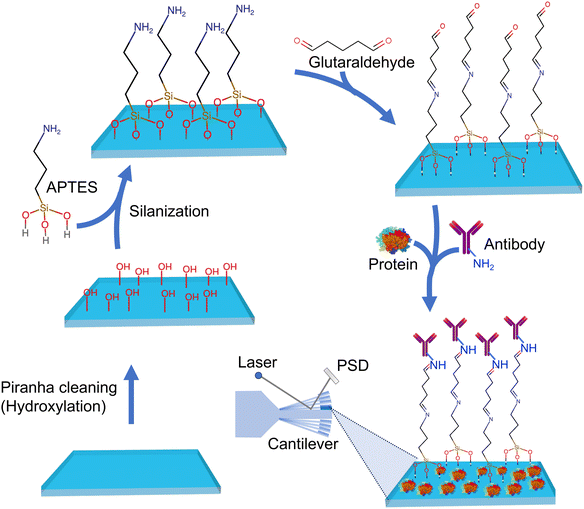 | ||
| Fig. 16 Scheme showing silicon microfabricated cantilever functionalization by treating the surface with APTES, glutaraldehyde followed by colvaent attachment of anti-Aspergillus niger polyclonal antibodies ∼IgG designed to capture fungal spores.73 Binding of spores to the functionalized cantilever induces its deflection of the laser beam falling on it which is sensitively detected by the position sensitive detector (PSD). | ||
| Species | Dynamic range | LOD | Time | Ref. |
|---|---|---|---|---|
| Glip-T | 1 × 10–14–10−2 M | 0.32 × 10−14 M | 30 min | 70 |
| Aspergillus niger mold | 0.5 pg mL−1–10 μg mL−1 weighed amount of spores | 0.3 pg mL−1 | NM | 39 |
| Gliotoxin | 50 fM–1 nM | 2.4 fM | 55 min | 71 |
| Aspergillus niger and A. alternata | 10 pg mL−1 to 1 ng mL−1 and 10 pg mL−1 to 1 μg mL−1 weighed amount of spores | 10 pg mL−1 | 5 min | 74 |
| Miscellaneous sensor studies | ||||
| Magnetic biosensor-Asp f 1 allergen | ∼0.1 to ∼10 ng mL−1 | ∼0.1 ng mL−1 | NM | 72 |
| Cantilever deflection biosensor-Aspergillus niger | Used concentration 17 × 103 CFU per mL | 103 CFU per mL | 4 h | 73 |
3.4 Micro-fluidic based biosensors
Microfluidic biosensors are small pieces of equipment or systems that incorporate elements for signal transduction directly into microchips.75,76 Fluid regulation, target identification, signal transmission, and output are all requirements of these biosensors. The critical factors that considerably impact the performance of microfluidic biosensors are fluid control specifically via microchannels, microstructures, and diverse fluid actuation methods.77–79 Design of microfluidic chips with a focus on appropriate materials and structures holds paramount importance. The evolution of the microfluidic chip concept has led to the development of 2D and 3D materials specifically designed for handling micro-volumes of fluid samples within the microfluidic chip modules. These materials include silicon,80 glass,81 quartz,82 polymethyl methacrylate (PMMA),83 and polydimethylsiloxane (PDMS).77,84,85 Biosensors integrated with microfluidics allow substantial reduction in detection times due to their advantageous characteristics, which include minimal sample requirements and reagent usage, adaptable liquid manipulation, and process integration. These biosensors can then be incorporated with a variety of signal transduction modes including electrical, magnetic, and optical, enabling rapid identification and detection of desired analytes, such as fungal spores within minutes. Microfluidics-based biosensors, with their inherent features, have gained significant attention for detecting pathogenic fungi in various fields, including clinical diagnostics, food safety, and environmental analysis. The term “lab-on-a-chip” coined in 1979 refers to the imparted microfluidic chip's ability of biosensors to scale down fluid flow, heat, and mass transfer to the micrometre or nanometer scale. The microfluidics platform in conjunction with a bio-transducer (biosensor) can be successfully miniaturized to perform complex operations such as specimen preparation (sampling and capture), reagent manipulation, biological recognition, and detection (Fig. 17a–c).86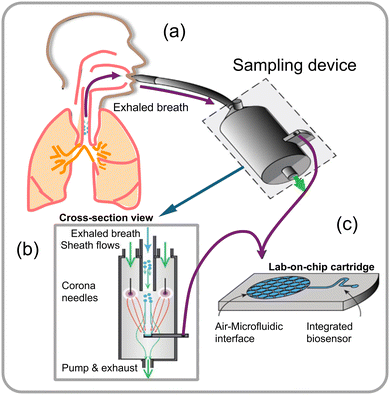 | ||
| Fig. 17 (a) Schematic illustration of a point-of-care (PoC) system designed for sampling aerosol from a high volume of air directly into a microfluidic device that captures exhaled breath condensate, (b) sampling device cross-sectional view integrated with a microfluidic detector, and (c) a lab-on-chip cartridge with a microfluidic interface that allows diagnosis from an exhaled-breath aerosol.86 | ||
These operations, which are otherwise multi-step tedious processes involving higher costs and lengthy procedures, can be streamlined using this microfluidics technology. The continuous advancements in microfluidics-based biosensor technology highlight its potential for revolutionizing analyte detection applications across diverse domains. Microfluidic systems offer high-throughput screening (HTS) capabilities suitable for screening many samples with speed and sensitivity. Researchers have developed a nanoliter-range droplet-based microfluidic system specifically adapted for HTS of large filamentous fungi libraries for secreted enzyme activities.
Portable microfluidic chips are highly useful for conducting multiplexed reactions and detecting airborne fungal spores through sensing fungus-specific nucleic acids. The detection of fungal spores in the environment is challenging using conventional methods, largely due to their dispersion in a large volume area. Researchers have integrated a portable microfluidic chip device that automates the process of nucleic acid detection. This device mechanically lyses fungal cells or spores, treats the samples with reagents, and releases fungal DNA required to perform loop-mediated isothermal amplification (LAMP) reactions. LAMP is a rapid and efficient DNA amplification technique that generates visible color change signals, making it a more advanced alternative to traditional PCR.50 The integrated microfluidic chip device is designed with multiple reaction chambers, each capable of performing independent LAMP reactions. These chambers are connected through microchannels, with a single inlet and three air vents (as shown in Fig. 18a–e). For the LAMP reaction to occur, the device requires the target DNA sequence from fungal cells or spores in aerosol, a set of 4–6 specially designed primers (short fragments that stick to ends of the target DNA), a DNA polymerase enzyme that copies the target DNA as a template with the primers and building blocks (deoxynucleotide triphosphates, dNTPs) to finally amplify at 60–65 °C. Fluorescent dyes in the reaction are added to intercalate the amplified double-stranded DNA to monitor and detect.
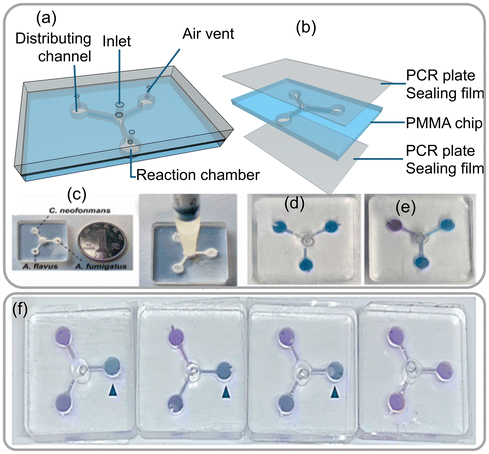 | ||
| Fig. 18 (a) Description of the apparatus. (b) Assembly of the chip. (c) Three sets of primers were preloaded with 0.5% low melt agarose in the reaction chamber. (d) The chip's adapter. (e) Verification of the possibility and cross-contamination of LAMP assays implemented on the microfluidic chips. (f) Outcomes of the colorimetric LAMP assays conducted on the chips at 4 × 106, 4 × 105, 4 × 104 and 4 × 103 spores per sample. Reprinted with permission from ref. 50 (copyright © 2022 Published by Elsevier B.V.). | ||
The integrated microfluidic device enables quick and efficient detection of various fungal species, such as Aspergillus fumigatus, Aspergillus flavus, and Cryptococcus spores making it a valuable tool for environmental monitoring and diagnosis. To validate the device's performance, a simulated spore-aerosol of A. fumigatus was generated and collected using a wet cyclone sampler into 4 mL 1 M sorbitol containing 0.5% Tween 20. After air sample collection, 100 μL of the sample was counted using Petri film and mold count Mold™ to determine the A. fumigatus spore quantity in the simulated air samples. The conventional real-time fluorescence LAMP assay carried out in Eppendorf tubes was able to detect spore samples ranging from 4 × 103 to 4 × 106 spores, while the same assay performed using the microfluidic chip device showed consistent signals confirming for 4 × 103 to 4 × 106 spores per sample (as shown in Fig. 18f). The above study demonstrated that the integrated microfluidic chip device has the potential to revolutionize the detection of airborne fungi by providing a rapid and efficient method to detect fungal spores.50 By providing accurate and reliable results, this innovative technology could help protect people's health and safety by identifying potential fungal hazards in the air.
Other portable and automated microfluidic devices that are made of different materials, designs, detection methods have been tested for detecting fungi. Li et al.87 developed a microfluidic platform for detecting Aspergillus niger spores using immunoassay reactions on carboxyl-modified microspheres with a size of 20.38 μm that were utilized to capture specific spores (Fig. 19a). Microspheres were immobilized with polyclonal rabbit antibodies and introduced into the reaction channels of a microfluidic device for spore detection (Fig. 19b). Detection of spores is carried out by sandwiching with a FITC-labeled anti-mouse secondary antibody injected into the reaction channel and the detection of a signal from the sandwiched complex can be observed under a microscope, such as a Nikon Eclipse Ti fluorescence microscope (Japan) (Fig. 19c and d). This microfluidic chip device comprised 16 identically designed channels (0.04 mm high × 0.6 mm wide × 40 mm long), arranged symmetrically from the center with a shared airflow outlet. Each channel had an inlet for air/liquid and a joint hole for liquid flow with a flow rate of 1.3–1.45 μL min−1 (80–90 mL min−1 for each of the 16 channels) (Fig. 19e and f). The sensor's detection limit was determined to be 300 spores per m3, which is equivalent to the limit of detection (LOD) of as low as approximately 20 spores. This research advances the application of microfluidic platforms in fungal spore detection. It supports ongoing efforts to develop sensitive and specific biosensors for environmental and health monitoring.
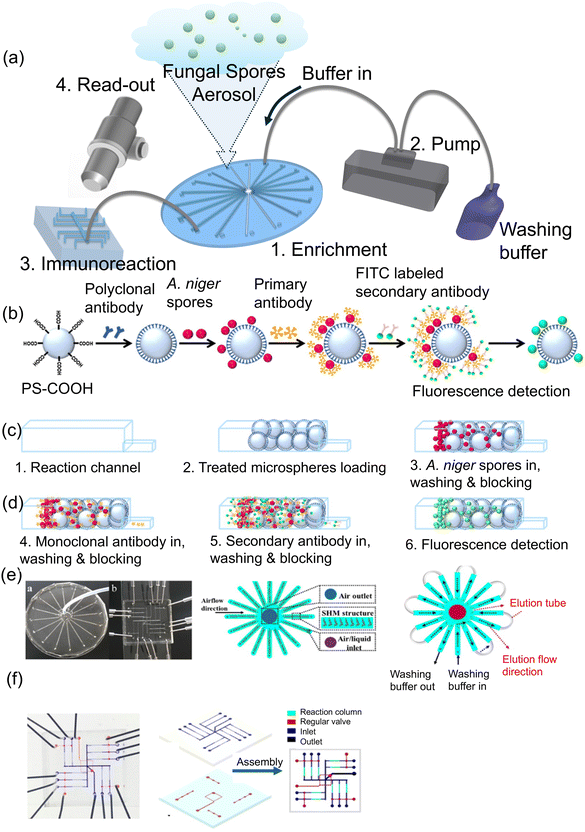 | ||
| Fig. 19 (a) Diagram showing the design of a microfluidic device. (b–d) Steps involved in conducting immunoassay reactions for detecting A. niger spores on microspheres. (e) Illustration of the open and closed inlet and outlets of a microfluidic system used for enriching and detecting airborne spore aerosols during the A. niger spore analysis on the detection chip. (f) Snapshot of the detection unit, displaying the fluid layer and control layer. Reprinted with permission from ref. 87 (copyright © 2018 Published by American Chemical Society). | ||
Other detection schemes can be integrated with microfluidics to generate the signal, such as those reported for fungal enzyme activity measurements.88 For instance, a microfluidic platform was integrated for systematically screening for Aspergillus niger strains by probing the enhanced α-amylase activity from fungi. Well-designed microfluidic devices were employed to generate droplets with a volume of 18 nL containing a fluorogenic substrate for enzyme α-amylase activity, which promoted encapsulation of individual spores and enabled measurement of enzyme activity (Fig. 20a). The droplets generated by this device-setup were composed of a fluorinated oil phase and a fluoro surfactant (KryJeffD900), which facilitate efficient gas delivery to the spores during the incubation period. The spore density within the droplets followed a Poisson distribution, maintaining an average number of spores per drop (λ) ranging from 0.1 to 0.3. Over the course of a 24 h incubation at 30 °C, the spores underwent germination, hyphal growth, and α-amylase secretion. The α-amylase activity within the droplets was accurately evaluated using a fluorogenic substrate, and a sorting device with cross-side configuration electrodes enabled the sorting of droplets based on their α-amylase activity (Fig. 20b). The sorted fungi were then subsequently retrieved, sporulated, and subjected to further characterization to assess their α-amylase production. This innovative microfluidic platform demonstrated its efficacy in screening large whole-genome mutated libraries of Aspergillus niger strains, leading to the identification of strains exhibiting significantly enhanced α-amylase overproduction. Notably, researchers applied this approach to the strain O58, renowned for its reference production of Soufflet α-amylase, exposing it to a chemical agent during the screening process, thereby proving to be a successful strategy. Furthermore, a robotic microfluidic platform is utilized to screen a sorted fungal population on a solid medium derived from rapeseed meals. After analyzing 1272 sorted fungi with α-amylase activity, 98.9% of the sorted fungi displayed α-amylase activity, with most displaying activity levels similar to O58. These findings demonstrate the efficiency of the microfluidic platform in identifying strains of Aspergillus niger with enhanced α-amylase production potential. This approach connects to the broader field of microfluidic screening platforms, demonstrating their potential not only in industrial biotechnology applications, but also in detecting fungi in environmental and clinical settings.
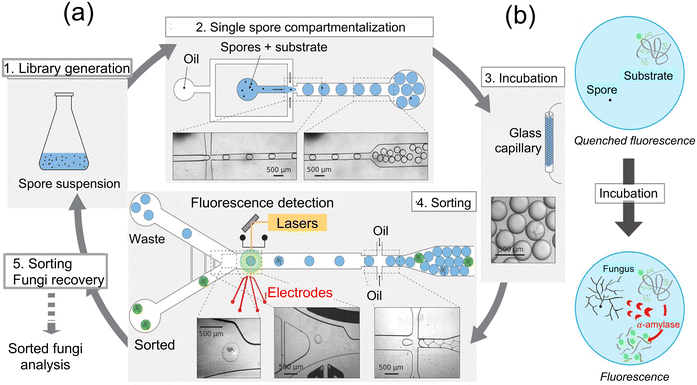 | ||
| Fig. 20 (a) Schematic illustration of sorting fungal spores using microfluidic devices and encapsulation of droplet containing labeled enzyme's substrate. 1. Spore suspension from a whole genome mutated library of fungi. 2. A suitable spore-to-droplet ratio is utilized to compartmentalize the spores into 10 nL droplets containing a fluorogenic enzyme substrate through emulsion. 3. For fungi growth, enzyme secretion, and substrate digestion within the particles, the emulsion was incubated off-chip at 30 °C by passing through a glass capillary tube for 24 h. 4. Enzymatic activity-based sorting of droplets is achieved by reintroducing them into a microfluidic sorting device and assessed their fluorescence intensity. 5. Fungi are extracted from the sorted droplets and subsequently undergo another round of mutagenesis and/or selection, characterization, or α-amylase fluorogenic assay. (b) Substrate is composed of a backbone composed of starch and numerous neutralized (quenched) BODIPY®FL fluorophores. Once the starch backbone is hydrolyzed by α-amylase, the quenched fluorophore unquenches leading to the generation of a fluorescence signal88 (copyright open access © 2016 Published by Springer Nature). | ||
Further developments in microfluidic devices used for detecting fungi can be found in a study89 that combined nanoelectrode activation and microwell arrays to achieve highly efficient capture and quantification of the airborne fungal pathogen Sclerotinia sclerotiorum. The nanoelectrodes serve to attract and immobilize the fungal spores, enabling their efficient capture within the microwells for subsequent analysis as shown in Fig. 21a and b. The microwell array provides a high-throughput platform for capturing multiple fungal spores simultaneously, allowing for efficient quantification of the pathogen in the air sample. By exploiting the unique properties of nanoelectrodes and microwells, this system offers improved sensitivity and specificity in detecting Sclerotinia sclerotiorum compared to traditional methods. This innovative approach holds promise for advancing the field of airborne pathogen detection and could have significant implications for disease surveillance and control strategies.89Table 3 summarizes reported performance parameters for microfluidic sensors for detection of fungal spores.
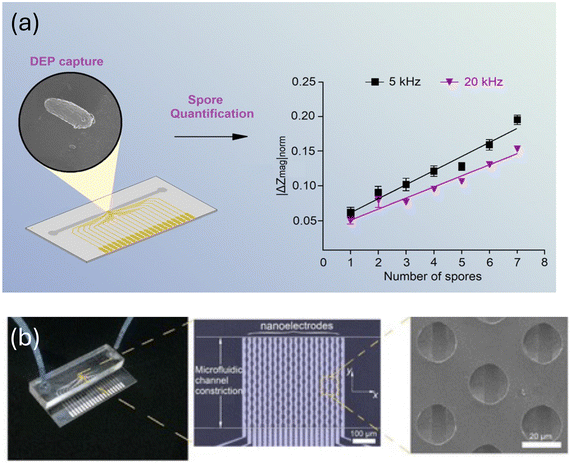 | ||
| Fig. 21 (a and b) A microfluidic device comprising 190 microwells and 20 aluminium nanoelectrodes adapted for performing dielectrophoresis (DEP) to capture spores, and the results of individual electrochemical impedance spectroscopy (EIS) measurements are shown in the line plot on right, enabling precise quantification. Reprinted with permission from ref. 89 (copyright open access © 2021 Published by American Chemical Society). | ||
| Species | Dynamic range | LOD | Time | Ref. |
|---|---|---|---|---|
| Aspergillus fumigatus, Aspergillus flavus, and Cryptococcus | 4 × 103–106 spores per sample | 4 × 104 spores | 90 min | 50 |
| Aspergillus niger | 1.1 × 106 spores per mL | ∼20 spores | 1 h | 87 |
| Aspergillus niger | 104 clone library | Screening of clones ∼7000 h−1 | 90 min | 88 |
| Sclerotinia sclerotiorum | ∼2 × 104 spores per mL | Single spore detection range | ∼20 s | 89 |
The approaches described above for optical, electrochemical and microfluidic based sensors present distinct strengths and weaknesses. Optical sensors, particularly FRET-based assays, demonstrated rapid detection times and high sensitivity for specific fungal species. Colorimetric sensors using AuNPs showed impressive sensitivity and selectivity, with potential for mobile applications suitable for remote settings. Lab-on-a-chip platforms provide portability and a cost-effective solution with high specificity integrating well with standard laboratory devices. Plasmonic chip-based methods offer innovative approaches for portable and smartphone-based detection systems, emphasizing the versatility of optical sensors in various settings.
Electrochemical sensors demonstrate exceptional sensitivity and specificity, often utilizing aptamers or antibodies for targeted detection. Advanced techniques such as magneto-nanosensors and AFM-based methods provided robust and innovative solutions for real-time and high-sensitivity fungal detection. These sensors showed potential for clinical and environmental applications, offering rapid and accurate detection capabilities.
Microfluidic biosensors excel in providing rapid, efficient, and high-throughput detection options suitable for screening a large number of samples with speed. These platforms enable the integration of advanced detection techniques such as LAMP and immunoassays in a compact and portable format. The ability to perform real-time monitoring and high-throughput screening makes microfluidic biosensors highly suitable for both environmental surveillance and industrial applications. The versatility and scalability of these platforms highlighted their potential for widespread adoption in various fields.
Each approach offers distinct advantages in fungal detection. The choice of method however depends on the specific application requirements, including sensitivity, specificity, detection speed, and practical deployment scenarios, ensuring that the most appropriate technology is used for each unique situation.
4. Commercially available technology/products for airborne fungal spores
Monitoring airborne fungi and other bio-aerosols in public places and hospitals is crucial for maintaining indoor air quality and mitigating potential health risks. Various bioaerosol sampling instruments have been developed, each exhibiting distinct mechanisms, performance characteristics, and analysis methods. This section provides an in-depth examination of commercially available technologies employed in public places and hospitals, emphasizing their fundamental principles.Portable and cost-effective lab-on-chip platforms have been developed for the detection of infectious diseases using nucleic acid-based techniques. For example, Myconostica Ltd is designed for the specific detection of genomic DNA from 15 distinct Aspergillus species. This assay particularly targets the four most prevalent pathogenic species: A. fumigatus, A. flavus, A. terreus, and A. niger. The foundational principle of this diagnostic test relies on molecular beacon real-time polymerase chain reaction (RT-PCR) technology, with the multicopy 18S ribosomal RNA (rRNA) gene as its target. To ensure result accuracy, the assay incorporates an internal amplification control (IAC) sequence from plant sources, crucial for identifying inhibitory factors that may impede the PCR process in the test sample. In the experimental procedure, PCRs are carried out on the Cepheid Smart Cycler, using 10 microliters of DNA template in a final reaction volume of 25 microliters, following manufacturer's guidelines. The determination of PCR positivity is based on a predetermined threshold of 36 amplification cycles, corresponding to an analytical sensitivity capable of detecting around 50 copies of the 18S rRNA gene. Notably, this sensitivity is approximately equivalent to the genomic content of a single A. fumigatus microorganism.90 The above example method for specific detection of a fungus is largely similar to the classical method that has been used for popular COVID-19 tests that are not only conducted in centralized laboratories, but are also slow, labor intensive, and expensive. However, they provide clues to the current challenges and researchers are continuously attempting to advance detection technologies that are highlighted in the above sections. A few other classical tools for detecting fungi that have been commercialized are described in the following sections.
4.1 MycoGenie (A. fumigatus; TR34/L98H)
Ademtech in Pessac, France developed MycoGenie assay, which is intricately crafted for the detection of A. fumigatus, specifically targeting the 28S rDNA and the TR34/L98H mutation. This versatile assay accommodates a range of clinical specimens, including biopsies, respiratory samples, and serum samples, and is compatible with analytical platforms such as CFX96, ABI 7500, LightCycler 480, SmartCycler, MX3000, and the Rotor-Gene. A comprehensive validation study conducted by Dannaoui et al. in 2017 underscores the robust performance of the MycoGenie R assay. Utilizing 88 respiratory samples (59 culture-positive) and 69 serum samples from individuals with proven or probable aspergillosis, the assay exhibited an exceptional sensitivity of 92.9% for respiratory samples and a noteworthy specificity of 90.1%. Similarly, serum samples displayed 100% sensitivity and 84.6% specificity. In a broad investigation led by Guegan et al. in 2018, involving 387 subjects across hematological and non-hematological contexts, the MycoGenie assay underwent a rigorous comparative evaluation against the AspergGenius assay and two distinct in-house assays. Analyzing bronchoalveolar lavage (BAL) samples, the MycoGenie assay demonstrated nuanced sensitivity profiles. Specifically, in patients with hematological conditions, a sensitivity of 53.7% was observed, highlighting its proficiency in detecting Aspergillus DNA in this cohort. Conversely, among non-hematological cases, the assay exhibited increased sensitivity at 75%, emphasizing its accuracy in detecting Aspergillus DNA. In this context, the AspergGenius R assay showed comparatively diminished sensitivities, highlighting the superior performance of the MycoGenie assay in this specialized diagnostic domain. Notably, sensitivities diminished in patients subjected to antifungal prophylaxis or treatment, suggesting the potential impact of therapeutic interventions on assay outcomes. Furthermore, the MycoGenie assay found meaningful application in the molecular detection of Aspergillus in French patients with fungal rhinosinusitis, alongside positive microscopy (N = 137). Impressively, the assay yielded positive results in 77.4% of cases, a marked contrast to the 32.1% culture positivity rate. Another investigation by Denis et al. in 2018 positioned the MycoGenie R assay in a comparative context with the A. fumigatus Bio-Evolution R assay. Both assays maintained 100% specificity, while sensitivities stood at 71% (MycoGenie) and 81% (A. fumigatus Bio-Evolution). Notably, sensitivities remained robust against antifungal treatment, and no resistance markers were identified, demonstrating the assay's reliability.55 In summary, the MycoGenie assay emerged as a promising diagnostic tool, particularly for its capability to identify a prominent azole-resistant mutation. While its diagnostic prowess is evident, further comprehensive data accumulation remains crucial to ascertain its definitive utility in clinical routine applications. The nuanced sensitivity modulation in the presence of therapeutic interventions underscores the intricate dynamics inherent in accurate assessment and interpretation in the realm of Aspergillus detection.4.2 RenDx Fungiplex assay
The RenDX, Fungiplex, assay was developed by Renshaw-diagnostic Limited in Glasgow, UK which is based on a polymerase chain reaction method, targeting specific sequences within the 18S rDNA and 28S rDNA regions. Following PCR amplification, the assay incorporates surface-enhanced Raman scattering for the selective and sensitive detection of targeted pathogenic oligonucleotides. Utilizing an Aspergillus probe alongside an extensive-range Saccharomycetales probe, the assay enables simultaneous detection of Aspergillus and Candida spp. In a validation study by White et al. in 2014, plasma samples from 14 predominantly haemato-oncological patients such as chronic leukemia, lymphoma, myeloma, and other disorders with proven/probable invasive aspergillosis (IA) and 80 patients without IA were examined. The assay demonstrated a sensitivity of 82.2% and a specificity of 87.5%. Notably, the RenDX Fungiplex R assay transitioned to RT-PCR technology, prompting a reformatting of its operational framework.91 Regarding the Fungiplex Aspergillus PCR assay developed by Bruker Daltonik GmbH in Bremen, researchers conducted a study to develop a quantum dot-based immunoassay for detecting A. amstelodami using FRET. Analytical performance in detecting DNA from A. fumigatus, Aspergillus flavus, Aspergillus niger, and Aspergillus terreus was reported by Green and Dougan in 2017.92Fundamentally, the RenDX Fungiplex assay demonstrates a sophisticated convergence of PCR and SERS technologies, designed for effective and accurate detection of pathogen-specific oligonucleotides with high sensitivity and specificity. While the assay holds promise in diagnostic capabilities and has embraced the transition to real-time PCR, a comprehensive assessment of its clinical utility, in comparison with the Fungiplex Aspergillus PCR assay, is still pending. Early assessments, nonetheless, highlight the assay's robust analytical proficiency in identifying diverse Aspergillus species, as evident from preliminary studies involving artificially introduced DNA samples.
4.3 AspergGenius assay
PathoNostics (Maastricht, Netherlands) devised the AspergGenius assay which is a multiplex real-time PCR that detects invasive aspergillosis (IA). It is compatible with the LightCycler 480, Rotor-Gene 6000 (Corbett), and Rotor-Gene. Q (Qiagen). This assay provides two respiratory secretions PCR assays; (a) identification and distinction of Aspergillus spp. using the AsperGenius species multiplex, which targets the 28Sr DNA. A probe for the A. fumigatus complex is incorporated, which provides coverage for multiple species, such as A. fumigatus., A. lentulus, A. udagawae and A. viridinutans. Another probe is available to detect a different pool of fungi, such as Aspergillus spp., which include A. fumigatus complex, A. terreus, A. flavus, and A. niger, and (b) identification of four azole resistance markers (L98H, TR34, T289A, Y121F) within the Cyp51A gene of A. fumigatus using AsperGenius resistance multiplex R. This assay provides good clinical performance with the added ability to detect azole resistance directly from noninvasive serum samples. While the available fungal burden may limit its application, it represents a significant classical diagnosis and management of aspergillosis.5. Conclusion and future perspectives
In this comprehensive review, we have explored various technologies and assays specifically developed for the detection of airborne fungal spores present in the air, with a particular emphasis on Aspergillus species. The integration of innovative platforms, including optical, electrochemical, microfluidics and molecular diagnostics, has significantly enhanced the precision and efficiency of fungal spore detection. The advancements discussed in this review underscore the importance of continuous innovation in the field of fungal spore detection, crucial for maintaining indoor air quality and preventing associated health risks. These technologies offer valuable insights into the prevalence characteristic of airborne fungal spores, aiding in timely clinical interventions.Future endeavors in biosensing of airborne pathogenic fungal spores should focus on the development of breath samplers integrated with nanosensors, enabling rapid and in situ detection. Large-scale production of nanosensors with excellent reproducibility and repeatability, adopting formats like LFIA, FET biosensors, or microfluidic assays, are crucial for broader commercial utilization. Progress is imperative to establish an efficient and proficient biosensing system encompassing automated rapid sampling, heightened sensitivity, affordability, stability, and real-time, on-site analyte detection.
Looking ahead, several avenues present exciting opportunities for further exploration:
Technological refinement: ongoing advancements in microfluidics, PCR methodologies, and other detection technologies are anticipated. Continuous refinement and integration of these technologies will likely yield more sophisticated, user-friendly, and cost-effective diagnostic platforms.
Multiplexing and specificity: future assays are expected to incorporate enhanced multiplexing capabilities for simultaneous detection of multiple fungal species and specific resistance markers. This will provide a more comprehensive understanding of the fungal landscape.
Point-of-care development: the development of point-of-care assays remains a key focus. Portable, rapid, and accessible diagnostic tools are crucial for timely interventions, especially in resource-limited settings.
Big data and AI integration: the integration of big data analytics and artificial intelligence will play a pivotal role in data interpretation. These technologies can offer nuanced insights into spore prevalence, geographical variations, and potential outbreaks.
Clinical validation and standardization: rigorous clinical validation studies across diverse patient populations and settings will be imperative. Standardization of methodologies and assays will enhance their reliability and facilitate their integration into routine clinical practices.
Moreover, interdisciplinary collaboration remains key for fostering knowledge exchange and advancing biosensor platforms. Advancements in miniaturization and microfabrication technologies, along with the integration of novel bio-recognition molecules and nanomaterials, contribute to the progress of biosensors. The successful development of biosensor devices involves critical factors such as studying interactions between bio-recognition elements and analytes, immobilization techniques, anti-fouling surface chemistries, device design, fabrication, and integration of biology with devices.
As research continues, the future holds promising prospects for biosensors in revolutionizing pathogen detection, especially in the context of airborne fungal spores, contributing to improved public health outcomes and a better understanding of indoor air quality.
Data availability
This review article was prepared based on findings from previously published studies. All data analyzed during this study are included in the published articles cited within the review. No new data were created or analyzed in this study. For specific datasets or further information, readers can refer to the original publications listed in the References section.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge the support received from the TÜBİTAK 1004 NANOSİS, project number 20AG004 (ASPİRA, LOC-SENS).References
- J. Gong, J. Qi, E. Beibei, Y. Yin and D. Gao, Concentration, viability and size distribution of bacteria in atmospheric bioaerosols under different types of pollution, Environ. Pollut., 2020, 257, 113485 CrossRef CAS PubMed.
- S. Khan, M. Naushad, M. Govarthanan, J. Iqbal and S. M. Alfadul, Emerging contaminants of high concern for the environment: Current trends and future research, Environ. Res., 2022, 207, 112609 CrossRef CAS PubMed.
- H. Harms, D. Schlosser and L. Y. Wick, Untapped potential: exploiting fungi in bioremediation of hazardous chemicals, Nat. Rev. Microbiol., 2011, 9(3), 177–192 CrossRef CAS PubMed.
- S. K. Brar, M. Verma, R. Surampalli, K. Misra, R. Tyagi and N. Meunier, et al. Bioremediation of hazardous wastes—a review, Pract. Period. Hazard., Toxic, Radioact. Waste Manage., 2006, 10(2), 59–72 CrossRef CAS.
- A. Grinn-Gofroń and B. Bosiacka, Effects of meteorological factors on the composition of selected fungal spores in the air, Aerobiologia, 2015, 31, 63–72 CrossRef PubMed.
- W. Xie, Y. Li, W. Bai, J. Hou, T. Ma and X. Zeng, et al. The source and transport of bioaerosols in the air: A review, Front. Environ. Sci. Eng., 2021, 15, 1–19 CrossRef.
- P. Srikanth, S. Sudharsanam and R. Steinberg, Bio-aerosols in indoor environment: composition, health effects and analysis, Indian J. Med. Microbiol., 2008, 26(4), 302–312 CrossRef PubMed.
- Pollution IA., Indoor air pollution [available from: https://www.pollutionissues.com/Ho-Li/Indoor-Air-Pollution.html].
- L. D. Stetzenbach, M. P. Buttner and P. Cruz, Detection and enumeration of airborne biocontaminants, Curr. Opin. Biotechnol., 2004, 15(3), 170–174 CrossRef CAS PubMed.
- T. G. O'Riordan and G. C. Smaldone, Respiratory medical societies and the threat of bioterrorism, Thorax, 2004, 59(3), 265–267 CrossRef PubMed.
- M. J. Mendell, A. G. Mirer, K. Cheung, M. Tong and J. Douwes, Respiratory and Allergic Health Effects of Dampness, Mold, and Dampness-Related Agents: A Review of the Epidemiologic Evidence, Environ. Health Perspect., 2011, 119(6), 748–756 CrossRef CAS PubMed.
- M. Breitenbach, The spectrum of fungal allergy, Int. Arch. Allergy Immunol., 2008, 145(1), 58–86 CrossRef PubMed.
- J. P. Latge, Aspergillus fumigatus and aspergillosis, Clin. Microbiol. Rev., 1999, 12(2), 310–350 CrossRef CAS PubMed.
- N. V. Sipsas and D. P. Kontoyiannis, Invasive fungal infections in patients with cancer in the Intensive Care Unit, Int. J. Antimicrob. Agents, 2012, 39(6), 464–471 CrossRef CAS PubMed.
- E. Heseltine and J. Rosen, WHO guidelines for indoor air quality: dampness and mould, WHO Regional Office Europe, 2009 Search PubMed.
- L. Senn, J. O. Robinson, S. Schmidt, M. Knaup, N. Asahi and S. Satomura, et al., 1,3-beta-D-glucan antigenemia for early diagnosis of invasive fungal infections in neutropenic patients with acute leukemia, Clin. Infect. Dis., 2008, 46(6), 878–885 CrossRef CAS PubMed.
- C. Lass-Florl, W. Mutschlechner, M. Aigner, K. Grif, C. Marth and M. Girschikofsky, et al., Utility of PCR in Diagnosis of Invasive Fungal Infections: Real-Life Data from a Multicenter Study, J. Clin. Microbiol., 2013, 51(3), 863–868 CrossRef PubMed.
- B. T. Fisher, The Role of Biomarkers for Diagnosis of and Therapeutic Decisions Related to Invasive Aspergillosis in Children, Curr. Fungal Infect. Rep., 2013, 7(1), 7–14 CrossRef PubMed.
- E. C. Y. Chan, K. K. Pasikanti and J. K. Nicholson, Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry, Nat. Protoc., 2011, 6(10), 1483–1499 CrossRef CAS PubMed.
- A. P. Snyder, A. Tripathi, W. M. Maswadeh, J. Ho and M. Spence, Field detection and identification of a bioaerosol suite by pyrolysis–gas chromatography–ion mobility spectrometry, Field Anal. Chem. Technol., 2001, 5(4), 190–204 CrossRef CAS.
- C. E. Enyoh, A. W. Verla, W. Qingyue, F. O. Ohiagu, A. H. Chowdhury and E. C. Enyoh, et al., An overview of emerging pollutants in air: Method of analysis and potential public health concern from human environmental exposure, Trends Environ. Anal. Chem., 2020, 28, e00107 CrossRef CAS.
- H. J. Tobias, M. P. Schafer, M. Pitesky, D. P. Fergenson, J. Horn and M. Frank, et al., Bioaerosol mass spectrometry for rapid detection of individual airborne Mycobacterium tuberculosis H37Ra particles, Appl. Environ. Microbiol., 2005, 71(10), 6086–6095 CrossRef CAS PubMed.
- C. Calderon, E. Ward, J. Freeman and A. McCartney, Detection of airborne fungal spores sampled by rotating-arm and Hirst-type spore traps using polymerase chain reaction assays, J. Aerosol Sci., 2002, 33(2), 283–296 CrossRef CAS.
- K. Miyajima, Y. Suzuki, D. Miki, M. Arai, T. Arakawa and H. Shimomura, et al., Direct analysis of airborne mite allergen (Der f1) in the residential atmosphere by chemifluorescent immunoassay using bioaerosol sampler, Talanta, 2014, 123, 241–246 CrossRef CAS PubMed.
- R. Urbano, B. Palenik, C. Gaston and K. Prather, Detection and phylogenetic analysis of coastal bioaerosols using culture dependent and independent techniques, Biogeosciences, 2011, 8(2), 301–309 CrossRef CAS.
- G. Qiu, Y. Yue, J. Tang, Y.-B. Zhao and J. Wang, Total bioaerosol detection by a Succinimidyl-Ester-functionalized Plasmonic biosensor to reveal different characteristics at three locations in Switzerland, Environ. Sci. Technol., 2020, 54(3), 1353–1362 CrossRef CAS PubMed.
- J. Ma, G. Jiang, Q. Ma, H. Wang, M. Du and C. Wang, et al., Rapid detection of airborne protein from Mycobacterium tuberculosis using a biosensor detection system, Analyst, 2022, 147(4), 614–624 RSC.
- Real-time bioaerosol detecting via combination of cyclone based collecting system and SiNW biosensor, ed. G. Woo, D. H. Lee, B. Y. Lee and T. Kim, IEEE, 2023, IEEE International Symposium on Medical Measurements and Applications (MeMeA), 2023 Search PubMed.
- P. Wang, R. Zhang, Y. Wu, Y. Chang and M. Liu, An Electrochemical Aptasensor Integrating Zeolitic Imidazolate Framework for Highly Selective Detection of Bioaerosols, Biosensors, 2022, 12(9), 725 CrossRef CAS PubMed.
- X. Su, L. Sutarlie and X. J. Loh, Sensors and analytical technologies for air quality: particulate matters and bioaerosols, Chem. – Asian J., 2020, 15(24), 4241–4255 CrossRef CAS PubMed.
- R. K. Ibrahim, M. Hayyan, M. A. AlSaadi, A. Hayyan and S. Ibrahim, Environmental application of nanotechnology: air, soil, and water, Environ. Sci. Pollut. Res., 2016, 23, 13754–13788 CrossRef CAS PubMed.
- N. Kumar, M. Kumari and R. K. Arun, Development and implementation of portable biosensors in microfluidic point-of-care devices for pathogen detection. Miniaturized Biosensing Devices: Fabrication and Applications, Springer, 2022, pp. 99–122 Search PubMed.
- P. L. Kashyap, P. Rai, S. Sharma, H. Chakdar, S. Kumar and K. Pandiyan, et al., Nanotechnology for the detection and diagnosis of plant pathogens, in Nanoscience in Food and Agriculture 2, 2016, pp. 253–276 Search PubMed.
- S.-Y. Cho, J.-P. Myong, W.-B. Kim, C. Park, S. J. Lee and S. H. Lee, et al., Profiles of environmental mold: indoor and outdoor air sampling in a hematology hospital in Seoul, South Korea, Int. J. Environ. Res. Public Health, 2018, 15(11), 2560 CrossRef CAS PubMed.
- Experimental study of fungal release characteristics in central air, ed. Y. Lv, G. Hu, W. Yuan, Y. Li, T. Liu, P. He, et al., Atlantis Press, 2018, 2018 7th International Conference on Energy, Environment and Sustainable Development (ICEESD 2018) Search PubMed.
- L. Liu and A. Corma, Metal catalysts for heterogeneous catalysis: from single atoms to nanoclusters and nanoparticles, Chem. Rev., 2018, 118(10), 4981–5079 CrossRef CAS PubMed.
- S. C. Ortiz, K. Pennington, D. D. Thomson and M. Bertuzzi, Novel insights into Aspergillus fumigatus pathogenesis and host response from state-of-the-art imaging of host–pathogen interactions during infection, J. Fungi, 2022, 8(3), 264 CrossRef CAS PubMed.
- T. Lee, S. A. Grinshpun, D. Martuzevicius, A. Adhikari, C. M. Crawford and T. Reponen, Culturability and concentration of indoor and outdoor airborne fungi in six single-family homes, Atmos. Environ., 2006, 40(16), 2902–2910 CrossRef CAS PubMed.
- J.-H. Jin, J. Kim, T. Jeon, S.-K. Shin, J.-R. Sohn and H. Yi, et al., Real-time selective monitoring of allergenic Aspergillus molds using pentameric antibody-immobilized single-walled carbon nanotube-field effect transistors, RSC Adv., 2015, 5(20), 15728–15735 RSC.
- C. F. Fronczek and J.-Y. Yoon, Biosensors for monitoring airborne pathogens, J. Lab. Autom., 2015, 20(4), 390–410 CrossRef CAS PubMed.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi and K. B. Ku, et al., Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor, ACS Nano, 2020, 14(4), 5135–5142 CrossRef CAS PubMed.
- Z. Miao, Y. Cai, J. Liu, D. Li and T. Chai, Quantification of airborne Aspergillus fumigatus spores in rabbit houses by real-time polymerase chain reaction, Aerobiologia, 2012, 28, 291–294 CrossRef.
- S. A. Balajee, A. Imhof, J. L. Gribskov and K. A. Marr, Determination of antifungal drug susceptibilities of Aspergillus species by a fluorescence-based microplate assay, J. Antimicrob. Chemother., 2004, 55(1), 102–105 CrossRef PubMed.
- L. D. Stetzenbach, M. P. Buttner and P. Cruz, Detection and enumeration of airborne biocontaminants, Curr. Opin. Biotechnol., 2004, 15(3), 170–174 CrossRef CAS PubMed.
- D. Méheust, P. Le Cann, G. Reboux, L. Millon and J.-P. Gangneux, Indoor fungal contamination: health risks and measurement methods in hospitals, homes and workplaces, Crit. Rev. Microbiol., 2014, 40(3), 248–260 CrossRef PubMed.
- J. S. West and R. Kimber, Innovations in air sampling to detect plant pathogens, Ann. Appl. Biol., 2015, 166(1), 4–17 CrossRef PubMed.
- M. D. Kattke, E. J. Gao, K. E. Sapsford, L. D. Stephenson and A. Kumar, FRET-based quantum dot immunoassay for rapid and sensitive detection of Aspergillus amstelodami, Sensors, 2011, 11(6), 6396–6410 CrossRef CAS PubMed.
- J. I. Lee, S. C. Jang, J. Chung, W.-K. Choi, C. Hong and G. R. Ahn, et al., Colorimetric allergenic fungal spore detection using peptide-modified gold nanoparticles, Sens. Actuators, B, 2021, 327, 128894 Search PubMed.
- J.-W. Seo, J. Y. Kim, J.-J. Oh, Y. J. Kim and G.-H. Kim, Selection and characterization of toxic Aspergillus spore-specific DNA aptamer using spore-SELEX, RSC Adv., 2021, 11(5), 2608–2615 RSC.
- H. Lu, J. Zhu, T. Zhang, X. Zhang, X. Chen and W. Zhao, et al., A rapid multiplex nucleic acid detection system of airborne fungi by an integrated DNA release device and microfluidic chip, Talanta, 2022, 246, 123467 CrossRef CAS.
- Z. Fattahi and M. Hasanzadeh, Nanotechnology-assisted microfluidic systems for chemical sensing, biosensing, and bioanalysis, TrAC, Trends Anal. Chem., 2022, 152, 116637 CrossRef CAS.
- H. Kholafazad-Kordasht, M. Hasanzadeh and F. Seidi, Smartphone based immunosensors as next generation of healthcare tools: Technical and analytical overview towards improvement of personalized medicine, TrAC, Trends Anal. Chem., 2021, 145, 116455 CrossRef CAS.
- A. P. Turner and S. J. Knechtle, Induction immunosuppression in liver transplantation: a review, Transplant Int., 2013, 26(7), 673–683 CrossRef CAS PubMed.
- K. E. Sapsford, T. Pons, I. L. Medintz and H. Mattoussi, Biosensing with luminescent semiconductor quantum dots, Sensors, 2006, 6(8), 925–953 CrossRef CAS.
- M. Asal, Ö. Özen, M. Şahinler, H. T. Baysal and İ. Polatoğlu, An overview of biomolecules, immobilization methods and support materials of biosensors, Sens. Rev., 2018, 39(3), 377–386 CrossRef.
- V. Naresh and N. Lee, A review on biosensors and recent development of nanostructured materials-enabled biosensors, Sensors, 2021, 21(4), 1109 CrossRef CAS PubMed.
- L.-S. Yu, J. Rodriguez-Manzano, N. Moser, A. Moniri, K. Malpartida-Cardenas and N. Miscourides, et al., Rapid detection of azole-resistant Aspergillus fumigatus in clinical and environmental isolates by use of a lab-on-a-chip diagnostic system, J. Clin. Microbiol., 2020, 58(11), e00843–20 CrossRef CAS PubMed.
- E. Heydari, Nanoplasmonic biodetection based on bright-field imaging of resonantly coupled gold-silver nanoparticles, in Photonics and Nanostructures-Fundamentals and Applications, 2019, vol. 36, p. 100708 Search PubMed.
- T. Sojinrin, J. Conde, K. Liu, J. Curtin, H. J. Byrne and D. Cui, et al., Plasmonic gold nanoparticles for detection of fungi and human cutaneous fungal infections, Anal. Bioanal. Chem., 2017, 409, 4647–4658 CrossRef CAS PubMed.
- A. Gaikwad, M. Joshi, K. Patil, S. Sathaye and C. Rode, Fluorescent carbon-dots thin film for fungal detection and bio-labeling applications, ACS Appl. Bio Mater., 2019, 2(12), 5829–5840 CrossRef CAS PubMed.
- W. R. Heineman, W. B. Jensen and L. C. Clark Jr., Biosens. Bioelectron., 2006, 21(8), 1403–1404 Search PubMed.
- A. Chaubey and B. Malhotra, Mediated biosensors, Biosens. Bioelectron., 2002, 17(6–7), 441–456 CrossRef CAS PubMed.
- B. D. Malhotra and M. A. Ali, Nanomaterials in biosensors: Fundamentals and applications, in Nanomaterials for Biosensors, 2018, p. 1 Search PubMed.
- A. Shanker, K. Lee and J. Kim, Synthetic hybrid biosensors. Encyclopedia of molecular cell biology and molecular medicine, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2014 Search PubMed.
- A. Shanker, K. Lee and J. Kim, Synthetic hybrid biosensors. 2014 Search PubMed.
- D. Grieshaber, R. MacKenzie, J. Vörös and E. Reimhult, Electrochemical biosensors-sensor principles and architectures, Sensors, 2008, 8(3), 1400–1458 CrossRef CAS PubMed.
- M. S. Alaejos and F. J. Garcia Montelongo, Application of amperometric biosensors to the determination of vitamins and α-amino acids, Chem. Rev., 2004, 104(7), 3239–3266 Search PubMed.
- R. C. Alkire, D. M. Kolb and J. Lipkowski, Advances in Electrochemical Science and Engineering: Bioelectrochemistry, Wiley-VCH, 2011 Search PubMed.
- I.-H. Cho, D. H. Kim and S. Park, Electrochemical biosensors: Perspective on functional nanomaterials for on-site analysis, Biomater. Res., 2020, 24(1), 6 Search PubMed.
- I. Bhatnagar, K. Mahato, K. K. R. Ealla, A. Asthana and P. Chandra, Chitosan stabilized gold nanoparticle mediated self-assembled gliP nanobiosensor for diagnosis of Invasive Aspergillosis, Int. J. Biol. Macromol., 2018, 110, 449–456 CrossRef CAS PubMed.
- X. Ma, Y. Zhang, X. Qiao, Y. Yuan, Q. Sheng and T. Yue, Target-Induced AIE Effect Coupled with CRISPR/Cas12a System Dual-Signal Biosensing for the Ultrasensitive Detection of Gliotoxin, Anal. Chem., 2023, 95(31), 11723–11731 Search PubMed.
- D. Kim and S. X. Wang, A magneto-nanosensor immunoassay for sensitive detection of Aspergillus fumigatus allergen Asp f 1, IEEE Trans. Magn., 2012, 48(11), 3266–3268 Search PubMed.
- N. Nugaeva, K. Y. Gfeller, N. Backmann, M. Düggelin, H. P. Lang and H.-J. Güntherodt, et al., An antibody-sensitized microfabricated cantilever for the growth detection of Aspergillus niger spores, Microsc. Microanal., 2007, 13(1), 13–17 Search PubMed.
- J. Kim, J.-H. Jin, H. S. Kim, W. Song, S.-K. Shin and H. Yi, et al., Fully automated field-deployable bioaerosol monitoring system using carbon nanotube-based biosensors, Environ. Sci. Technol., 2016, 50(10), 5163–5171 Search PubMed.
- Y. Song, B. Lin, T. Tian, X. Xu, W. Wang and Q. Ruan, et al., Recent progress in microfluidics-based biosensing, Anal. Chem., 2018, 91(1), 388–404 CrossRef PubMed.
- Y. Liu, H. Shen, X. Yang, S. Kang, L. Cai and T. Tian, et al., Recent progress in microfluidic biosensors with different driving forces, TrAC, Trends Anal. Chem., 2022, 116894 Search PubMed.
- G. Xing, W. Zhang, N. Li, Q. Pu and J.-M. Lin, Recent progress on microfluidic biosensors for rapid detection of pathogenic bacteria, Chin. Chem. Lett., 2022, 33(4), 1743–1751 CrossRef CAS.
- K. Chen, P. Dopico, J. Varillas, J. Zhang, T. J. George and Z. H. Fan, Integration of Lateral Filter Arrays with Immunoaffinity for Circulating–Tumor–Cell Isolation, Angew. Chem., 2019, 131(23), 7688–7692 CrossRef.
- Q. Ruan, W. Ruan, X. Lin, Y. Wang, F. Zou and L. Zhou, et al., Digital-WGS: Automated, highly efficient whole-genome sequencing of single cells by digital microfluidics, Sci. Adv., 2020, 6(50), eabd6454 CrossRef CAS PubMed.
- J. Yakovleva, R. Davidsson, A. Lobanova, M. Bengtsson, S. Eremin and T. Laurell, et al., Microfluidic enzyme immunoassay using silicon microchip with immobilized antibodies and chemiluminescence detection, Anal. Chem., 2002, 74(13), 2994–3004 CrossRef CAS PubMed.
- Y. Yalikun, Y. Hosokawa, T. Iino and Y. Tanaka, An all-glass 12 μm ultra-thin and flexible micro-fluidic chip fabricated by femtosecond laser processing, Lab Chip, 2016, 16(13), 2427–2433 RSC.
- S. Dochow, C. Beleites, T. Henkel, G. Mayer, J. Albert and J. Clement, et al., Quartz microfluidic chip for tumour cell identification by Raman spectroscopy in combination with optical traps, Anal. Bioanal. Chem., 2013, 405, 2743–2746 CrossRef CAS PubMed.
- H.-J. Kim and S.-J. Choi, Rapid single-cell detection of pathogenic bacteria for in situ determination of food safety, Anal. Methods, 2020, 12(46), 5621–5627 RSC.
- L. Zhou, S. Mao, Q. Huang, X. He and J.-M. Lin, Inhibition of anaerobic probiotics on colorectal cancer cells using intestinal microfluidic systems, Sci. China: Chem., 2018, 61, 1034–1042 CrossRef CAS.
- Y. Zheng, Z. Wu, J.-M. Lin and L. Lin, Imitation of drug metabolism in cell co-culture microcapsule model using a microfluidic chip platform coupled to mass spectrometry, Chin. Chem. Lett., 2020, 31(2), 451–454 CrossRef CAS.
- G. Pardon, L. Ladhani, N. Sandström, M. Ettori, G. Lobov and W. van der Wijngaart, Aerosol sampling using an electrostatic precipitator integrated with a microfluidic interface, Sens. Actuators, B, 2015, 212, 344–352 CrossRef CAS.
- X. Li, X. Zhang, Q. Liu, W. Zhao, S. Liu and G. Sui, Microfluidic system for rapid detection of airborne pathogenic fungal spores, ACS Sens., 2018, 3(10), 2095–2103 CrossRef CAS PubMed.
- T. Beneyton, I. Wijaya, P. Postros, M. Najah, P. Leblond and A. Couvent, et al., High-throughput screening of filamentous fungi using nanoliter-range droplet-based microfluidics, Sci. Rep., 2016, 6(1), 1–10 CrossRef PubMed.
- P. A. Duarte, L. Menze, L. Shoute, J. Zeng, O. Savchenko and J. Lyu, et al., Highly efficient capture and quantification of the airborne fungal pathogen Sclerotinia sclerotiorum employing a nanoelectrode-activated microwell array, ACS Omega, 2021, 7(1), 459–468 CrossRef PubMed.
- P.-M. Rath and J. Steinmann, Overview of commercially available PCR assays for the detection of Aspergillus spp. DNA in patient samples, Front. Microbiol., 2018, 9, 740 CrossRef PubMed.
- P. L. White, S. J. Hibbitts, M. D. Perry, J. Green, E. Stirling and L. Woodford, et al., Evaluation of a commercially developed semiautomated PCR–surface-enhanced Raman scattering assay for diagnosis of invasive fungal disease, J. Clin. Microbiol., 2014, 52(10), 3536–3543 CrossRef PubMed.
- The evaluation of a real time PCR assay for aiding the diagnosis of Invasive Candidiasis: Fungiplex Candida, ed. J. Green and J. Dougan, MYCOSES, WILEY 111 RIVER ST, HOBOKEN 07030-5774, NJ USA, 2017 Search PubMed.
- J. Liu, M. Jalali, S. Mahshid and S. Wachsmann-Hogiu, Are plasmonic optical biosensors ready for use in point-of-need applications?, Analyst, 2020, 145(2), 364–384 RSC.
| This journal is © The Royal Society of Chemistry 2024 |