Open Access Article
Open Access ArticleImmunomagnetic particles exhibiting programmable hierarchical flower-like nanostructures for enhanced separation of tumor cells†
Na
He
a,
Han
Bao
bc,
Jingxin
Meng
 bc,
Yongyang
Song
bc,
Yongyang
Song
 *bc,
Li-Ping
Xu
*bc,
Li-Ping
Xu
 *a and
Shutao
Wang
*a and
Shutao
Wang
 bc
bc
aBeijing Key Laboratory for Bioengineering and Sensing Technology, School of Chemistry and Biological Engineering, University of Science and Technology Beijing, Beijing 100083, P. R. China. E-mail: xuliping@ustb.edu.cn
bCAS Key Laboratory of Bio-inspired Materials and Interfacial Science, Technical Institute of Physics and Chemistry, Chinese Academy of Sciences, Beijing, 100190, P. R. China. E-mail: yysong@mail.ipc.ac.cn
cUniversity of Chinese Academy of Sciences, Beijing, 100049, P. R. China
First published on 18th September 2024
Abstract
Immunomagnetic particles are extensively used for the separation of biological molecules and particles, and have exhibited great potential in many fields including biosensors, disease diagnosis and biomedical engineering. However, most immunomagnetic particles exhibit a smooth surface, resulting in a limited separation efficiency for biological particles featuring enormous surface nanostructures, such as tumor cells. Here we report flower-like immunomagnetic particles (FIMPs) prepared by streptavidin (SA)-assisted biomineralization and one-step antibody modification, and demonstrate their superior capability for highly efficient and selective separation of circulating tumor cells (CTCs). SA can link inorganic nanosheets and magnetic nanoparticles together to obtain FIMPs with programmable hierarchical flower-like nanostructures and provide enormous binding sites for post-antibody modification. The synergetic effect of nano-sized petals and micro-sized particles in the hierarchical nanostructure enhances the interaction between the cells and the matrix, thus enabling FIMPs to separate CTCs with high selectivity and high efficiency. Our study provides a promising platform for the selective separation of trace biological molecules and particles from complex samples and shows great potential for downstream detection and diagnosis.
1. Introduction
Cancer is one of the most devastating diseases, causing millions of deaths each year.1,2 With the rising cancer incidence, early and accurate diagnosis of cancer is vital since it provides valuable information to improve the therapeutic efficiency and survival rate of cancer patients.3,4 Circulating tumor cells (CTCs), escaping from the primary tumor site into the peripheral blood, are considered to be one of the important prognostic biomarkers for tumor metastasis,5,6 cancer diagnosis7,8 and cancer treatment.9,10 The low abundance of CTCs in human biofluids11,12 requires high-sensitivity platforms for CTC isolation.13,14Due to an unmet need for the detection of CTCs in the medical community, much endeavor has been invested in CTC enrichment, using techniques such as density gradient centrifugation,15,16 flow cytometry,17 microfluidic microarrays,18,19 size filtration20,21 and immunomagnetic separation.22,23 Among them, immunomagnetic separation is one of the most widely used techniques due to its simplicity of operation, large surface-to-volume ratio, fast kinetics, rapid magnetic response, and high recovery efficiency.24,25 The conventional separation of CTCs merely relies on specific molecular interaction between antigens in cells and recognition molecules on the surface of immunomagnetic particles.26,27 These immunomagnetic particles often encounter separation efficiency bottlenecks due to undesired antibody modification efficiency and neglect of the surface structure. On the one hand, the binding of these specific molecules always requires tedious reaction steps.28,29 For example, CTC capture was achieved using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) for activation and conjugated with an antibody to epithelial cell adhesion molecule (anti-EpCAM).30,31 To modify the surface of Fe3O4 nanoparticles with an antibody to achieve CTC capture, procedures including a complex layer-by-layer assembly (LBL) technique using quantum dots and mercaptan and the binding of anti-EpCAM were used, while GSH-mediated disulfide bond cleavage was used for further biofriendly recovery of CTCs.32 Multiple reaction steps may influence the modification efficiency for specific molecules and thereafter result in undesired CTC separation efficiency.33,34 On the other hand, most existing immunomagnetic particles exhibit smooth surface structures.35,36 However, the surface nanostructure can significantly improve the capture efficiency of CTCs due to topological interaction between the surface and the CTCs.37,38 Therefore, there is an increasing demand for developing immunomagnetic particles with efficient specific molecule modification and programmable surface nanostructures.
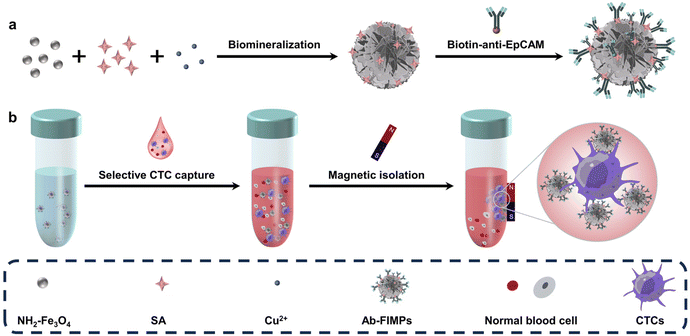
Mimicking the biomineralization process in nature, many wonderful organic–inorganic hybrid materials have been created with unique nanostructures.39–41 Here we report flower-like immunomagnetic particles (FIMPs) with efficient antibody modification and programmable nanostructures prepared by SA-assisted biomineralization and one-step antibody modification. Cu2+, magnetic nanoparticles, and SA were mixed for biomineralization, and the magnetic particles were modified with anti-EpCAM via a one-step reaction to obtain Ab-FIMPs (Fig. 1a). SA can assist the nanostructure formation of the magnetic particles with programmable hierarchical flower-like nanostructures, and provides enormous binding sites for post-antibody modification. The obtained Ab-FIMPs can selectively separate CTCs with high selectivity and high efficiency from biofluids with normal cells (Fig. 1b), providing an important tool for the rapid capture of CTCs. Compared with conventional immunomagnetic particles, the Ab-FIMPs enhance topographic interaction with the cells and significantly improve the cell capture efficiency, which provides new insight into the fabrication of immunomagnetic particles and holds great promise for reliable and sensitive CTC capture in the clinical setting.
2. Materials and methods
2.1. Chemicals and reagents
SA, CuSO4·5H2O (≥98%), and ethanol (≥99.8%, GR) were purchased from Sigma-Aldrich. Glutaraldehyde and hexamethyl disilylamine (HMDS) were purchased from Aladdin Chemistry Co. Ltd. Red blood cell lysis buffer, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (Dil), the Calcein/PI Cell Viability/Cytotoxicity Assay Kit, and 3,3-dioctadecyloxacarbocyanine perchlorate (DiO) were purchased from Sigma-Aldrich. Trypsin-EDTA (0.25%), Gibco Ham's F-12K (Kaighn's), Dulbecco's modified Eagle's medium (DMEM), and Roswell Park Memorial Institute 1640 (RPMI 1640) were obtained from Invitrogen. Fetal bovine serum (FBS) and penicillin–streptomycin (PS) were purchased from Thermo Fisher Scientific. Biotinylated anti-EpCAM was purchased from R&D Systems (USA). The prostate cancer cell line (PC-3, EpCAM-positive), human hepatocellular carcinomas (HepG2, EpCAM-positive), T lymphocyte cell line (Jurkat, EpCAM-negative), human breast cancer cell line (MCF-7, EpCAM-positive), and lymphoma cell line (Daudi, EpCAM-negative) were purchased from Beijing Xiehe Hospital. All other reagents were of analytical grade and used as received without further purification. Ultrapure water (Milli-Q, 18.2 MΩ cm) was used in all experiments.2.2. Apparatus
Scanning electron microscope (SEM) (SU8010, Hitachi, US) and transmission electron microscope (TEM) (HT7700, Hitachi, US) were employed to characterize the synthesis of the FIMPs. A Nicolet 400 Fourier transform infrared spectrometer (Madison, WI), an AXIS ULTRADLD spectrometer (Kratos, Japan), and an X-ray photoelectron spectrometer (Thermo Scientific K-Alpha+) were employed to characterize the composition of the FIMPs. The images of the cells adsorbed on the Ab-FIMPs were monitored using a fluorescence microscope (Nikon, TE2000).2.3. Preparation of Fe3O4–NH2 microparticles
Fe3O4 microparticles were synthesized according to a previous work.42 0.82 g of FeCl3·6H2O and 0.6 g of FeCl2·4H2O were dissolved in 10 mL of ultrapure water at a temperature of 30 °C and stirred well for 20 min. Then, 2.5 mL of 28% ammonia was added drop by drop, and stirred at 30 °C for 40 min. Then the precipitate was washed three to five times with ultrapure water and then transferred to a centrifuge tube, placed into a vacuum freeze dryer at 60 °C, dried for 48 h.The obtained Fe3O4 (20 mg) was mixed with 20 mL of anhydrous methanol, and 1 mL of 3-aminopropyltriethoxysilane (APTES), followed by sonication for 5 min and stirred for 7 h at room temperature to obtain amino-modified Fe3O4. The NH2–Fe3O4 was washed with ultrapure water and repeated three to five times. The washed NH2–Fe3O4 was transferred to a centrifuge tube and 4 mL of ultrapure water was added to prepare a suspension of NH2–Fe3O4 (5 mg mL−1), which was kept at 4 °C.
2.4. Preparation of flower-like immunomagnetic particles (FIMPs)
The FIMPs were prepared according to our previous work with a slight modification.43 In brief, SA solution (20 μL, 5 mg mL−1), NH2–Fe3O4 solution (15 μL, 5 mg mL−1), and CuSO4 solution (17.5 μL, 120 mM) were added to 1 mL of PBS buffer (100 mM, pH = 7.4). After reaction at room temperature for 48 h, the solution was centrifuged at 4000 rpm for 5 min, and the obtained FIMPs were washed three times with ultrapure water to remove excess CuSO4, NH2–Fe3O4, and SA. The FIMPs were suspended in 100 μL of PBS buffer (10 mM) for further use.2.5. Preparation of FIMPs modified with anti-EpCAM (Ab-FIMPs)
The FIMPs were incubated with biotin-anti-EpCAM for 45–90 min. The supernatant was removed after magnet adsorption and washed three times repeatedly with PBS. The following cell capture experiments were then performed.2.6. Cell culture
The culture medium was supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. MCF-7 and HepG2 were cultured in DMEM. PC-3 was cultured in Ham's F-12K medium, and other cells (Jurkat, Daudi) were cultured in RPMI-1640 medium. All cells were incubated at 37 °C in an incubator containing 5% CO2. When the cells had grown for 2 days to reach the exponential phase and pass through at approximately 90% fusion, the culture medium was removed and the cells were washed with PBS. The cells were digested with 0.25% trypsin solution for 3 min and then twice the amount of fresh culture medium was added to inhibit trypsin activity. The cells were isolated from the medium by centrifugation at 1000 rpm for 3 min, resuspended and stored in fresh medium, then counted using a cell counting plate.2.7. Capture of CTCs
Taking target PC-3 cells as an example, the Ab-FIMPs were added to the PC-3 cells solution (1 mL, 105 cells per mL) for cell capture. The mixture was incubated in a cell culture incubator at 37 °C with 0.5% CO2 for 45 min with gentle shaking. Several slight shakes at 5 min intervals were required to allow full contact of the Ab-FIMPs with the PC-3 cells. The captured PC-3 were collected by magnetic separation and washed three times. The capture efficiency of the captured PC-3 cells was calculated by dividing the number of captured cells by the initial number of cells before separation. Jurkat cells and Daudi cells were incubated with the Ab-FIMPs as a negative control, and MCF-7 cells and HepG2 cells were used as a positive control.2.8. SEM characterization of the morphology of the captured cells
The morphology of the cells captured by the Ab-FIMPs was observed. The captured cells were fixed with 2.5% glutaraldehyde solution at room temperature overnight. Subsequently, the cells were dehydrated using different concentrations of ethanol (30%, 50%, 70%, 90%, 95%, and 100%). Afterwards, the cells were treated with 50% and 100% HMDS in anhydrous ethanol for 15 min each time, respectively. Then the cells were dried in a vacuum freeze dryer for 12 h and the morphology of the captured cells was observed by SEM.2.9. Cell staining
PC-3 and Daudi cells were stained with cellular dyes (DiO and Dil, 2 μg mL−1). After incubation in an incubator for 20 min, the cell suspensions were centrifuged at 1000 rpm for 3 min and the cells were washed twice with PBS for capture experiments. The results were observed by fluorescence microscope.2.10. Cell viability analysis
The collected tumor cells were stained with calcein (AM) and propidium iodide (PI) and analyzed for viability. Suspensions of isolated tumor cells were prepared in a culture medium (Ham's F-12K), then the cellular dyes AM and PI (2 μg mL−1) were added and mixed well. After incubation in an incubator for 20 min, the cell suspension was collected by magnetoadsorption and washed twice with PBS. The results were visualized by fluorescence microscope.2.11. Preparation and treatment of lysed blood samples from healthy volunteers
The red blood cells were removed from blood samples according to the manufacturer's instructions by adding 10 mL of red blood cell lysis buffer to 1.0 mL of blood sample, and incubated at room temperature for 12 min, then centrifuged at 400g for 5 min, and the supernatant was discarded. PBS was added to wash the precipitate 2–3 times, and the precipitate was resuspended in DMEM to perform the CTC capture experiment.2.12. Immunocytochemistry identification of the captured CTCs
To mimic patient blood samples, 10, 20, 50, and 100 PC-3 cells (pre-stained with Dil) were spiked into 1 mL of whole blood sample, respectively. The simulated samples were incubated with magnetic beads under optimized cell capture conditions. The captured cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 20 min, blocked with 2% BSA for 1 h, stained with PE-labeled anti-CK (CK, a protein marker for epithelial cells) for 2 h, stained with FITC-labeled anti-CD45 (CD45, a leukocyte marker) for 12 h, and nuclear staining with DAPI was performed for 15 min. The cells with CK19 positive and DAPI positive but CD45 negative phenotypes were classified as CTCs using fluorescence microscope.3. Results and discussion
3.1. Characterization of the nanostructure and morphology of FIMPs
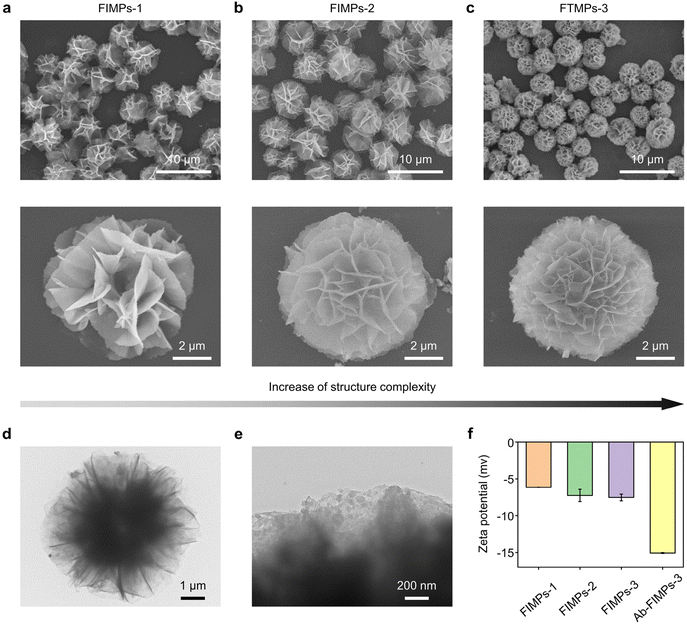
FIMPs were synthesized by SA-assisted biomineralization. In the preparation of FIMPs, SA and NH2–Fe3O4 were used as precursors. When SA, NH2–Fe3O4, and CuSO4 were added to the PBS solution, the coordination between Cu2+ and the amino groups in SA and NH2–Fe3O4 induced the formation of a copper phosphate primary nucleus, and it was used as a nucleation site for the formation of larger copper phosphate nanoflowers. As the incubation time increases, more nano-petals combine to form large aggregates and begin to produce flower-like nanostructures. After a few hours, the flower nanostructure is fully formed. Three different FIMPs with controllable nanostructures were obtained at different biomineralization times (12 h, 24 h, and 48 h) (Fig. 2), i.e. FIMPs-1, FIMPs-2, and FIMPs-3. The morphology of the FIMPs was characterized using SEM and TEM. As shown in Fig. 2(a–c), the FIMPs have a relatively homogeneous flower-like morphology, consisting of nanosheets assembled in interlocking combinations with good mono-dispersity. The high-resolution SEM images show that the FIMPs have a hierarchical nanostructure assembled by hundreds of nano-petals. The size of the obtained FIMPs does not vary at different biomineralization times, with sizes of FIMPs-1, FIMPs-2, and FIMPs-3 for 5.25 ± 0.02, 5.34 ± 0.12, and 5.35 ± 0.03, respectively. However, their hierarchical nanostructure becomes gradually obvious, and the density of the petals gradually increases, which leads to a more complex nanostructure. The petal nanostructure of the FIMPs can be seen more clearly from the TEM image (Fig. 2d). The high-resolution TEM (Fig. 2e) image further shows that the Fe3O4 nanoparticles are uniformly embedded in the petals of the FIMPs. The zeta potential and fluorescence intensity were used to characterize the amount of antibody on the Ab-FIMPs. The decrease of zeta potential indicates more modification of anti-EpCAM (Fig. 2f and Fig. S1a†). It was also shown that the negative potentials of FIMPs-1, FIMPs-2, and FIMPs-3 gradually increased with the prolongation of biomineralization time, which might be due to more SA being encapsulated in the fabricated FIMPs. The antibody loading rates were obtained according to the fluorescence intensity. As shown in Fig. S1b,† the anti-EpCAM load of FITC-Ab-FIMPs-3 is higher than those of FITC-Ab-FIMPs-1 and FITC-Ab-FIMPs-2, indicating more antibody modification. As shown in Fig. S1(c and d),† the anti-EpCAM loading rates of FIMPs-1, FIMPs-2, and FIMPs-3 are 82.9% ± 0.3%, 87.3% ± 0.4%, and 95.5% ± 1.5%, respectively. The high antibody loading rate makes the fabricated FIMPs very promising in biosensing. Moreover, with increasing biomineralization time and hierarchical nanostructures of the FIMPs, the loading rate of anti-EpCAM is higher. The FIMPs can be separated using commercial magnets within 15 s (Fig. S2†), indicating their potential for magnetic separation. The analysis of the FIMPs was also carried out using XRD, XPS, and FT-IR (Fig. S3†). The Ab-FIMPs can be stably stored at 4 °C for 7 days, and the capture efficiency of CTCs is little changed (Fig. S4†).3.2. The ability of Ab-FIMPs to capture circulating tumor cells
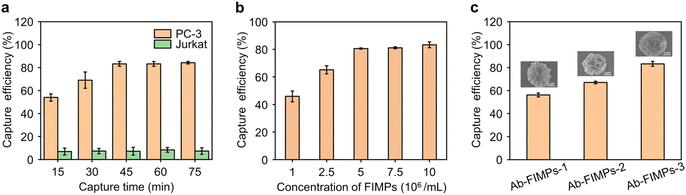
The influence of capture time on capture efficiency was evaluated. In our study, PC-3 cells with rich EpCAM expression were selected as positive target cells, while Jurkat T lymphocyte cells were selected as negative control cells. At the same concentrations of Ab-FIMPs and cells, the capture efficiency of PC-3 cells increases significantly with the incubation time and reaches a plateau after 45 min. For the negative control cells, the capture efficiency of Jurkat cells is lower than 11% (Fig. 3a). In order to obtain the highest capture specificity, 45 min was chosen as the optimal cell capture time.The Ab-FIMP concentration was also optimized to get higher cell capture efficiency. The cell capture efficiency increases with the increase of Ab-FIMP concentration and reaches a plateau at an Ab-FIMP concentration of about 5 × 106 mL−1 (Fig. 3b). Thus the subsequent cell capture experiment was performed at an Ab-FIMP concentration of 5 × 106 mL−1.
3.3. Enhanced specific recognition from the hierarchical nanostructures of Ab-FIMPs
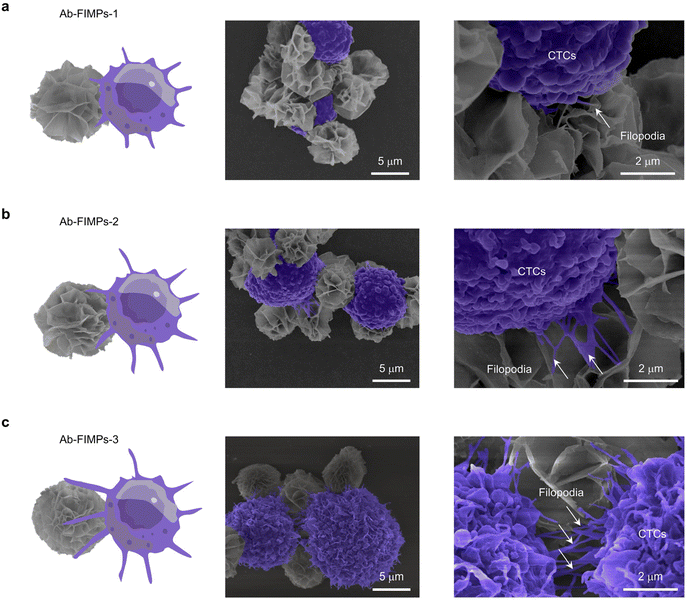
Considering the significant effect of magnetic bead morphology on capture efficiency, three kinds of Ab-FIMPs with different hierarchical nanostructures were obtained at various biomineralization times and employed to capture the target PC-3 cells (Fig. 3c). Ab-FIMPs with different hierarchical nanostructures (Ab-FIMPs-1, Ab-FIMPs-2, Ab-FIMPs-3) show cell capture efficiency of 56.2% ± 1.7%, 67.1% ± 1.2%, and 83.3% ± 2.1%, respectively. These results suggest that the increased nanostructure complexity of Ab-FIMPs can improve the cell capture efficiency significantly.To reveal the relationship between the capture efficiency and the nanostructure of Ab-FIMPs, the morphology of captured cells was characterized by SEM (Fig. 4(a–c)). After cell capture by using Ab-FIMPs-1 with fewer petals, PC-3 cells were surrounded by magnetic particles, indicating that CTC capture can be achieved using the fabricated magnetic particles. Few and short filopodia could be observed. For Ab-FIMPs-2, more and longer filopodia of the cells appeared. After cell capture using Ab-FIMPs-3, we observed the emergence of large lamellar pseudopods that extended into more and longer protruding filopodia and grasped the nano-petals on Ab-FIMPs-3. The enhanced capture efficiency could be attributed to the increased nanostructure complexity by integrating nano-sized petals and micro-sized particles. As the biomineralization time increases, the densities of the nano-sized petals and recognition probe also increase, forming a morphological gradient. These results suggest that the topological effect between the Ab-FIMPs-3 and CTCs favors their interaction and improves their cell capture efficiency.
3.4. Capture performance analysis
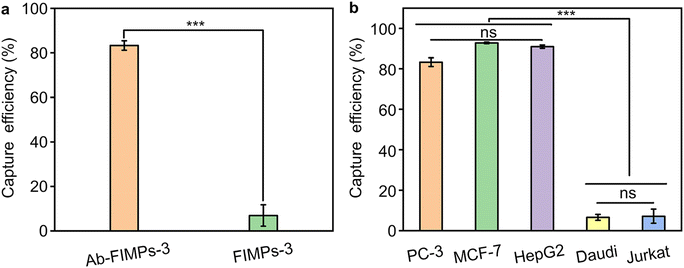
The cell capture specificity of the Ab-FIMPs was studied by comparing the capture efficiency of Ab-FIMPs-3 and FIMPs-3. Compared with Ab-FIMPs-3, FIMPs-3 exhibits a much lower capture efficiency for PC-3 with a significant difference (Fig. 5a), suggesting that the specific CTC capture of FIMPs-3 depends on the anti-EpCAM modification. The cell capture specificity of the Ab-FIMPs was further evaluated by employing three EpCAM-positive cell lines (including PC-3, MCF-7, and HepG2) and two unspecific cell lines (Daudi and Jurkat). The different cell lines (PC-3, MCF-7, HepG2, Daudi, and Jurkat) show cell capture efficiency of 84.3% ± 0.9%, 92.83% ± 0.4%, 90.97% ± 0.8%, 6.6% ± 1.5%, and 6.8% ± 4.9%, respectively. The capture efficiency for the EpCAM-positive cell lines of Ab-FIMPs-3 is higher than that of nonspecific cell lines with a significant difference (Fig. 5b). We have tested the capture efficiency of different cell lines (PC-3, MCF-7, and HepG2) at a lower tumor cell concentration (103 cells per mL). The cell capture efficiencies of PC-3, MCF-7, and HepG2 at a tumor cell concentration of 103 cells per mL are 84.2 ± 0.9%, 91.64% ± 0.84%, and 90.67 ± 0.72%, respectively (Fig. S5†). Thus, there is no significant change in the capture efficiency when the cell concentration is reduced. These results demonstrate that the Ab-FIMPs are highly efficient and specific for recognizing EpCAM-positive cell lines. We also tested the performance of the Ab-FIMPs in recognizing and isolating target CTCs from the cell mixture, and the results confirm that the fabricated Ab-FIMPs can identify and capture rare CTCs from the cell mixture selectively (Fig. S6†).To deeply understand the effect of the capture process on cell activity, changes in the PC-3 cell viability before and after capture were studied by a well-established AM/PI staining method. The results show that the cell viability of the captured CTCs is comparable to the pre-capture results (Fig. S7†). Therefore, the fabricated Ab-FIMPs not only capture CTCs efficiently and specifically, but also do not affect the survival of the CTCs, which is important for further single-cell analysis.
3.5. Capture sensitivity of Ab-FIMPs-3 for rare CTCs in the mimic sample
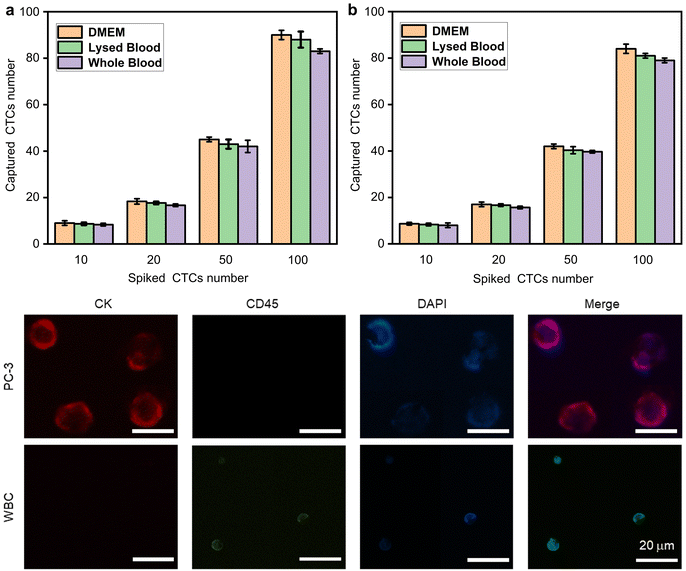
We further explored the performance of Ab-FIMPs-3 in capturing rare CTCs from the simulated patient blood samples. Samples spiked with MCF-7 cells and PC-3 cells, respectively, were tested under optimal cell capture conditions. MCF-7 cells and PC-3 cells (stained with Dil) were added to 1 mL of DMEM, lysed blood, and whole blood samples from healthy volunteers, respectively. The counting results (Fig. 6(a and b)) show that the capture efficiency of MCF-7 cells in DMEM, lysed blood, and whole blood are 91.65% ± 2.4%, 86.4% ± 2.3%, and 83.4% ± 0.4%, respectively. The capture efficiency of PC-3 cells in DMEM, lysed blood, and whole blood are 84.93% ± 1.3%, 82.05% ± 1.5%, and 79.15% ± 0.7%, respectively. Thus, the great potential of Ab-FIMPs for efficient and specific detection of CTCs in actual clinical samples is confirmed.PC-3 cells (FITC-labeled anti-CK) captured on the Ab-FIMPs can also be identified from non-specifically attached white blood cells (WBCs, PE-labeled anti-CD45) by the commonly used three-color immunocytochemistry method. The intensity of DAPI and the expression levels of CK and CD45 in individual cells were quantified using a fluorescent inverted microscope. As shown in Fig. 6c, CTCs have a strong CK signal and a weak CD45 signal. In contrast, WBCs exhibit high CD45 and low CK expression levels. PC-3 cells (CK+/CD45−/DAPI+), WBCs (CK−/CD45+/DAPI+), and cellular debris were distinguished using combinatorial information. These results demonstrate that the Ab-FIMPs could convincingly overcome the interference of the complex matrix from whole blood, exhibit the potential for utilization in further clinical diagnosis.
4. Conclusions
In summary, a facile biomineralization-inspired method has been proposed to fabricate immunomagnetic particles for efficient and specific CTC capture. SA assists the fabrication of immunomagnetic particles with programmable hierarchical flower-like nanostructures and provides a large number of binding sites for anti-EpCAM modification. The nanostructure of the obtained immunomagnetic particles can be controlled. The high capture efficiency of Ab-FIMPs-3 reveals the synergetic effect of the nano-sized petals and micro-sized particles in the hierarchical nanostructure enhance the interaction between the cells and the matrix, which may provide new insight into the fabrication of cell chips. Moreover, the capture efficiency for CTCs with the Ab-FIMPs is comparable to or even better than that of most existing materials (Table S1†) and the obtained Ab-FIMPs can also achieve efficient separation of CTCs from whole blood samples, which indicates its promising potential for early cancer screening in clinical preoperative tests.Author contributions
Na He: conceptualization, methodology, data curation, formal analysis, investigation, and writing – original draft. Han Bao: conceptualization, methodology, and formal analysis. Jingxin Meng: writing – review and editing. Yongyang Song: supervision, project administration, data curation, and writing – review and editing. Li-Ping Xu: supervision, validation, project administration, funding acquisition, and writing – review and editing. Shutao Wang: supervision, validation, project administration, funding acquisition, and writing – review and editing.Data availability
Additional data are made available in the ESI† of this manuscript. The authors will supply the relevant data in response to reasonable requests.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge funding from the National Key R&D Program of China (2022YFA1206900, 2019YFA0709300, and 2023YFF0714401), the National Natural Science Foundation of China (22375016), and the Beijing Municipal Science and Technology Commission (2182036).References
- B. Wang, S. Zhang, J. Meng, L. Min, J. Luo, Z. Zhu, H. Bao, R. Zang, S. Deng, F. Zhang, L. Ma and S. Wang, Adv. Mater., 2021, 33, 2103999 CrossRef CAS PubMed.
- S. Li, K. Wang, S. Hao, F. Dang, Z.-Q. Zhang and J. Zhang, Anal. Chem., 2022, 94, 6754–6759 CrossRef CAS PubMed.
- W.-J. Zheng, P.-X. Wang, Y.-F. Sun, J.-W. Cheng, Y.-C. Zhong, Y. Xu, W. Guo, B. Hu, J. Zhou, J. Fan, X. Chen and X.-R. Yang, ACS Appl. Mater. Interfaces, 2022, 14, 36425–36437 CrossRef CAS PubMed.
- X. Xu, J. Lin, Y. Guo, X. Wu, Y. Xu, D. Zhang, X. Zhang, X. Yujiao, J. Wang, C. Yao, J. Yao, J. Xing, Y. Cao, Y. Li, W. Ren, T. Chen, Y. Ren and A. Wu, Biosens. Bioelectron., 2022, 210, 114305 CrossRef CAS PubMed.
- H. Huang, B. Zou, S. Zhu, X. Zhang, J. Huang, J. Wang, X. Li and T. Chen, Nano Res., 2023, 17, 4350–4358 CrossRef.
- J. Li, X. Li, Y. Zhang, K. Jin, Y. Yuan, R. Ming, Y. Yang and T. Chen, Nano Res., 2022, 16, 873–881 CrossRef.
- Y. Gong, Y. Yao, W. Cheng, C. Duan, D. Li, Z. Wang and Y. Xiang, Anal. Chem., 2022, 94, 11767–11772 CrossRef CAS PubMed.
- B. Lu, Y. Deng, Y. Peng, Y. Huang, J. Ma and G. Li, Anal. Chem., 2022, 94, 12822–12827 CrossRef CAS PubMed.
- C. Li, R. Li, X. Wu, Y. Zuo, G. Xiong, M. Huang, Y. Sun, R. Liao, Y. Xiao, L. Hu, C. Gao and Y. Yu, Anal. Chem., 2022, 94, 15240–15249 CrossRef CAS PubMed.
- X. Jiang, X. Zhang, C. Guo, Y. Yu, B. Ma, Z. Liu, Y. Chai, L. Wang, Y. Du, B. Wang, N. Li, D. Dong, Y. Li, X. Huang and L. Ou, Nanoscale, 2022, 14, 8474–8483 RSC.
- J. Li, C. Dong, H. Gan, X. Gu, J. Zhang, Y. Zhu, J. Xiong, C. Song and L. Wang, Biosens. Bioelectron., 2023, 231, 115273 CrossRef CAS PubMed.
- J. Xiong, C. Dong, J. Zhang, X. Fang, J. Ni, H. Gan, J. Li and C. Song, Biosens. Bioelectron., 2022, 213, 114442 CrossRef CAS PubMed.
- M. G. Ahmed, M. F. Abate, Y. Song, Z. Zhu, F. Yan, Y. Xu, X. Wang, Q. Li and C. Yang, Angew. Chem., Int. Ed., 2017, 56, 10681–10685 CrossRef CAS PubMed.
- S. Gao, S. Chen and Q. Lu, Biomater. Sci., 2019, 7, 4027–4035 RSC.
- D. Bang, T. Lee, J. Park, G. Lee, S. Haam and J. Park, BioChip J., 2018, 12, 38–45 CrossRef CAS.
- Q. Huang, F.-B. Wang, C.-H. Yuan, Z. He, L. Rao, B. Cai, B. Chen, S. Jiang, Z. Li, J. Chen, W. Liu, F. Guo, Z. Ao, S. Chen and X.-Z. Zhao, Theranostics, 2018, 8, 1624–1635 CrossRef CAS PubMed.
- A. L. Allan, S. A. Vantyghem, A. B. Tuck, A. F. Chambers, I. H. Chin-Yee and M. Keeney, Cytometry, Part A, 2005, 65, 4–14 CrossRef PubMed.
- Y. Liu, Z. Lin, Z. Zheng, Y. Zhang and L. Shui, ACS Sens., 2022, 7, 666–673 CrossRef CAS PubMed.
- L. L. Wu, Z. L. Zhang, M. Tang, D. L. Zhu, X. J. Dong, J. Hu, C. B. Qi, H. W. Tang and D. W. Pang, Angew. Chem., Int. Ed., 2020, 59, 11240–11244 CrossRef CAS PubMed.
- J. Bu, Y. J. Kim, Y.-T. Kang, T. H. Lee, J. Kim, Y.-H. Cho and S.-W. Han, Biomaterials, 2017, 125, 1–11 CrossRef CAS PubMed.
- M. L. Jørgensen, C. Müller, M. Sikkersoq, M. Nadzieja, Z. Zhang, Y. Su, J. Just, K. L. Garm Spindler and M. Chen, Mater. Today Bio, 2020, 6, 100052 CrossRef PubMed.
- C. Huang, G. Yang, Q. Ha, J. Meng and S. Wang, Adv. Mater., 2014, 27, 310–313 CrossRef PubMed.
- A. Mishra, T. D. Dubash, J. F. Edd, M. K. Jewett, S. G. Garre, N. M. Karabacak, D. C. Rabe, B. R. Mutlu, J. R. Walsh, R. Kapur, S. L. Stott, S. Maheswaran, D. A. Haber and M. Toner, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 16839–16847 CrossRef CAS PubMed.
- S. Sun, S. Yang, X. Hu, C. Zheng, H. Song, L. Wang, Z. Shen and Z.-S. Wu, ACS Sens., 2020, 5, 3870–3878 CrossRef CAS PubMed.
- Y. Zhang, F. Zhang, Y. Song, X. Shen, F. Bu, D. Su, C. Luo, L. Ge, S. Deng, Z. Wu, Z. Zhang, P. Duan, N. Li, L. Min, S. Zhang and S. Wang, Adv. Mater., 2023, 35, 2303821 CrossRef CAS PubMed.
- C. Ding, C. Zhang, S. Cheng and Y. Xian, Adv. Funct. Mater., 2020, 30, 1909781 CrossRef CAS.
- C. Ding, C. Zhang, X. Yin, X. Cao, M. Cai and Y. Xian, Anal. Chem., 2018, 90, 6702–6709 CrossRef CAS PubMed.
- H. Cui, R. Li, J. Du, Q.-F. Meng, Y. Wang, Z.-X. Wang, F.-F. Chen, W.-F. Dong, J. Cao, L.-L. Yang and S.-S. Guo, Talanta, 2019, 202, 230–236 CrossRef CAS PubMed.
- C. Song, C. Gao, J. Zhao and Z. Wang, J. Biomater. Appl., 2020, 35, 947–957 CrossRef PubMed.
- Z.-M. Chang, H. Zhou, C. Yang, R. Zhang, Q. You, R. Yan, L. Li, M. Ge, Y. Tang, W.-F. Dong and Z. Wang, J. Mater. Chem. B, 2020, 8, 5019–5025 RSC.
- P. Xu, Y. Yu, T. Li, H. Chen, Q. Wang, M. Wang, M. Wan and C. Mao, Anal. Chim. Acta, 2020, 1129, 60–68 CrossRef CAS PubMed.
- X. Zhou, B. Luo, K. Kang, S. Ma, X. Sun, F. Lan, Q. Yi and Y. Wu, J. Mater. Chem. B, 2019, 7, 393–400 RSC.
- Z.-M. Chang, Z. Wang, D. Shao, J. Yue, H. Xing, L. Li, M. Ge, M. Li, H. Yan, H. Hu, Q. Xu and W.-F. Dong, ACS Appl. Mater. Interfaces, 2018, 10, 10656–10663 CrossRef CAS PubMed.
- H. Ruan, X. Wu, C. Yang, Z. Li, Y. Xia, T. Xue, Z. Shen and A. Wu, ACS Biomater. Sci. Eng., 2018, 4, 1073–1082 CrossRef CAS PubMed.
- H.-X. Cao, P.-F. Liu, L. Wang, Z.-J. Liu, S.-Y. Ye and G.-X. Liang, Sens. Actuators, B, 2020, 318, 128287 CrossRef CAS.
- Y. Liu, X. Wang, Y. Zhou, G. Yang, J. Hou and S. Zhou, Nanoscale, 2021, 13, 16923–16931 RSC.
- H. Cui, B. Wang, W. Wang, Y. Hao, C. Liu, K. Song, S. Zhang and S. Wang, ACS Appl. Mater. Interfaces, 2018, 10, 19545–19553 CrossRef CAS PubMed.
- W. Wang, H. Cui, P. Zhang, J. Meng, F. Zhang and S. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 10537–10543 CrossRef CAS PubMed.
- J. C. Munyemana, H. He, S. Ding, J. Yin, P. Xi and J. Xiao, New J. Chem., 2018, 42, 12824–12829 RSC.
- Y. Liu, J. Chen, M. Du, X. Wang, X. Ji and Z. He, Biosens. Bioelectron., 2017, 92, 68–73 CrossRef CAS PubMed.
- Z. Li, H. Liu, R. Wang, C. Ji, Y. Wei, M. Shi, Y. Wang, Y. Du, Y. Zhang, Q. Yuan and C. Yan, ACS Nano, 2020, 14, 16085–16095 CrossRef CAS PubMed.
- G. Prabha and V. Raj, J. Iran. Chem. Soc., 2018, 15, 871–884 CrossRef CAS.
- T. Yan, S. Zhang, Y. Yang, Y. Li and L.-P. Xu, Microchim. Acta, 2022, 189, 260 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr02929a |
| This journal is © The Royal Society of Chemistry 2024 |