DNA-templated fluorescent metal nanoclusters and their illuminating applications
Ashwin
Rajeev
 and
Dhiraj
Bhatia
and
Dhiraj
Bhatia
 *
*
Department of Biological Sciences and Engineering, Indian Institute of Technology Gandhinagar, Palaj, Gandhinagar, Gujarat–382355, India. E-mail: dhiraj.bhatia@iitgn.ac.in
First published on 10th September 2024
Abstract
After the discovery of DNA during the mid-20th century, a multitude of novel methodologies have surfaced which exploit DNA for its various properties. One such recently developed application of DNA is as a template in metal nanocluster formation. In the early years of the new millennium, a group of researchers found that DNA can be adopted as a template for the binding of metal nanoparticles that ultimately form nanoclusters. Three metal nanoclusters have been studied so far, including silver, gold, and copper, which have a plethora of biological applications. This review focuses on the synthesis, mechanisms, and novel applications of DNA-templated metal nanoclusters, including the therapies that have employed them for their wide range of fluorescent properties, and the future perspectives related to their development by exploiting machine learning algorithms and molecular dynamics simulation studies.
Introduction
Metal nanoclusters (NCs) have been exploited vastly for their biomedical applications, especially in biosensing and bioimaging, over the past half a century. However, the overall stability of these structures was often mediocre for the advanced applications for which they were exploited. Then, another strategy was adopted, wherein a template was introduced as a scaffold, which aids in the formation of clusters of desired sizes and shapes, along with the enhanced functionality of the metal NC as a whole.1 Soon, DNA was discovered to be a worthy candidate as a template, given its wealth of advantageous properties. DNA-templated fluorescent metal NCs have found immense applications over the past couple of decades, ranging from food safety detection to virus detection.2,3 Nevertheless, the focal point of their development dates back to the utilization of noble metal nanoclusters as a separate class of nanomaterials.4A typical metal NC comprises about 2 to 100 atoms, with peculiar properties that are dependent upon the cluster size, shape, and charge of these systems.5 These particles are smaller than plasmonic nanoparticles, with a size range in the order of sub-nanometres, which provides them quantum confinement effects that impart molecular properties such as considerable band gaps and high optical absorption.6 Since then, various surface ligands and templates, consisting of small organic ligands and synthetic polymers, for the nanoclusters have been used for overall stability and even endowing them with novel functionalities.7,8 Further research explored the nuances of biomolecules such as proteins and peptides,9 and DNA,10 and even polymers and dendrimers11 as templates for metal nanoclusters.12 The scope of generating assemblies of templates using proteins and DNA with wider functionalities and greater structural diversity for the incorporation of metal centers and nanomaterials has increased upon the emergence of ab initio modeling.13–15 Until a certain point in the scientific era, metal coordination was always associated with proteins, and this belief was shattered through certain Raman studies, as it was found that the nitrogen of nitrogenous bases could coordinate with metal ions.16
In the year 2004, it was reported for the first time that DNA could be used as a template for metal NCs, wherein the researchers used single-stranded DNA (ssDNA) as the template for silver (Ag) nanoclusters in aqueous solution, with a high degree of fluorescence.18 The unique structure and facile programmability of DNA aid in the formation of clusters with different metal nanoparticles. The repeating nucleotides, with each nucleotide consisting of a nitrogenous nucleobase, a deoxyribose sugar moiety, and a phosphate moiety, make up the long chain of DNA.19 The negative charge imparted to the DNA by the backbone made up of phosphate groups help in forming electrostatic and coordination bonds with different metal ions under physiological pH conditions.20 Each nucleotide of DNA has a specific metal-binding site, according to its respective nitrogenous nucleobases. The N7 atom of purines and the N3 atom of pyrimidines are involved in metal binding.21 It was found that the N3 atom of thymine (T) and the N3 atom of cytosine (C) have a strong interaction with Cu2+ and Ag2+, respectively (Fig. 1).22,23 Therefore, the inherent chemical functionality of DNA aided its ascent as an inorganic nanomaterial, which can be attained after the reduction of the DNA–metal complex, with the potential for expanding to various metal ions.21,24 Several properties of DNA came in handy for attaining this particular functionality of forming templates for metal NCs. DNA has an ability to form nanostructures through hairpin formation of ssDNA, formation of dsDNA through base-pairing, amplification of high-quality DNA segments through low automation methods such as polymerase chain reaction (PCR), manipulation of DNA structures with the applications of specific enzymes, and the adoption of aptamers which can bind to a wide range of analytes rather than just complementary nucleic acids.25–27 Due to the adaptability of this nanocluster nanomaterial, it has been proven to have multiple applications.2,20
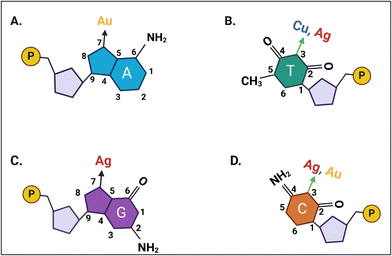 | ||
| Fig. 1 Simplified representation of the nitrogen-containing metal binding sites on the nucleobases of the DNA template, with A, B, C, and D representing adenine, thymine, guanine, and cytosine nucleobases respectively. Figure adapted from ref. 17. Created with BioRender. | ||
This review deals with the different synthesis methods of DNA-bound metal NCs, mechanisms of fluorescence, various types of metal ions adopted for NC formation, latest applications pertaining to these nanomaterials, and future perspectives for these materials, particularly by adopting machine learning algorithms and molecular simulation studies, thereby providing enhanced fluorescence properties and therefore advanced functionalities.
Importance of DNA as a template for nanoclusters
In recent years, numerous biomolecules have emerged as potential templates that are stable and already widely used for different applications.28,29 The hydrophilicity of DNA with solubility under physiological conditions and tuneable selectivity lead to the preference for its use over other biomolecules.30 For any application making use of fluorescence, its stability should be taken into account. Numerous factors, such as the sizes of NCs, templates used, properties of the solutions, and other related factors, determine the fluorescence properties of these NCs.31–33 NCs tend to aggregate given their high surface energy and extremely small size, which necessitates the use of stabilizers.20 In the study mentioned earlier pertaining to the first use of DNA as a template for metal NCs, the authors had used C-rich DNA as the template for the fluorescent silver NCs (Fig. 2),18 as a transition from the earlier used dendrimers.34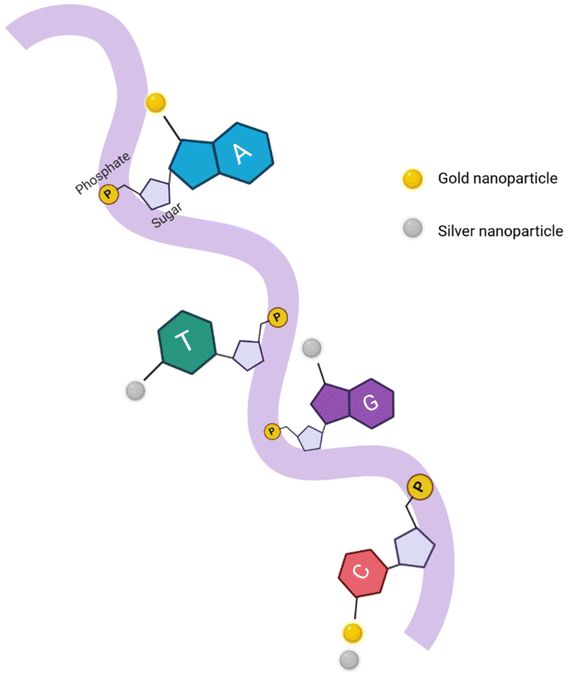 | ||
| Fig. 2 A simplified representation of a DNA strand with overhanging nitrogenous nucleobases attaching to gold and silver nanoparticles from their respective N7 and N3 positions on purines and pyrimidines. Figure adapted from ref. 6. Created with BioRender. | ||
Several reasons were attributed to the use of DNA in place of dendrimers, such as a higher quantum yield (QY) by manipulating the DNA sequence, which is difficult with other types of polymers, the ability of DNA aptamers to specifically bind to a variety of analytes, control over the NC size, the use of water as a solvent for the reaction rather than any organic solvents, and the cost-effectiveness of DNA compared to other synthetic templates.35 The emergence of DNA nanotechnology has propelled the applicability of fluorescent NCs as sensors, given its flexibility in positioning and tailoring the nanoparticles with a high level of precision.36 The controlled nanoparticle aggregation for the construction of explicit clusters and lattices using DNA is of significant importance.37 Clusters comprising varying geometries of the trimer, tetrahedral, octahedral, and 2D/3D lattices of plasmonic nanoparticles have been developed over the years, which upholds the feasibility of using DNA as templates for NCs.28–30,38–42 Given that there are numerous templates available for NCs in the form of peptides, proteins, and thiols, the selection of the ligand is based on the exact application, and compared to all these, DNA has an advantage of being highly cost-effective and tuneable, and choosing the appropriate sequence provides varying responses with the NCs.43,44 Moreover, the self-assembly capability through hydrogen bonding interactions and multifunctionality through the introduction of various functional groups ease the choice of DNA as the template over other biomolecules.45–48
Synthesis of some common metal nanoclusters
For the preparation of DNA-templated metal NCs, the metal clusters should be synthesized prior to mounting them on the DNA template. Although the synthesis of metal NCs can take up either a top-down or bottom-up approach, the latter is usually seen to be employed for this purpose in the solution phase using strong or weak reducing agents.49 The founding works in DNA-AgNCs used NaBH4 as the reductant and the 5′-AGGTCGCCCCC-3′ oligonucleotide as the ssDNA template, with 5 mM phosphate buffer as the solution phase at pH 7.5.18 The study strengthened the previous reports that Ag+ prefers binding to heterocyclic bases rather than phosphate groups.50 According to a previous study, the appropriate ratio of Ag+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA followed in the case of an ssDNA is 6
DNA followed in the case of an ssDNA is 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and if Ag+ is in excess, it may aid in the formation of plasmonic nanoparticles.51,52
1, and if Ag+ is in excess, it may aid in the formation of plasmonic nanoparticles.51,52
As opposed to AgNCs, for the preparation of gold (Au) NCs, the DNA strand used is abundant in the adenine nucleobase. The reaction solution consists of specified concentrations of chloroauric acid (HAuCl4), DNA oligonucleotide, and sodium citrate, with the pH of the final solution to be around 6 at room temperature. The solution is incubated for more than an hour.53 Contrary to both the previous studies of fluorescent metal NCs, copper (Cu) NCs employing ssDNA oligonucleotides rich in thymine residues as the template efficiently exhibited high fluorescence compared to sequences rich in the other three bases.54 Apart from this, poly(AT–TA) dsDNA had a higher efficiency as a template for CuNC formation in comparison with other dsDNA.54,55 The synthesis of the three different types of metal NCs is presented in Fig. 3.
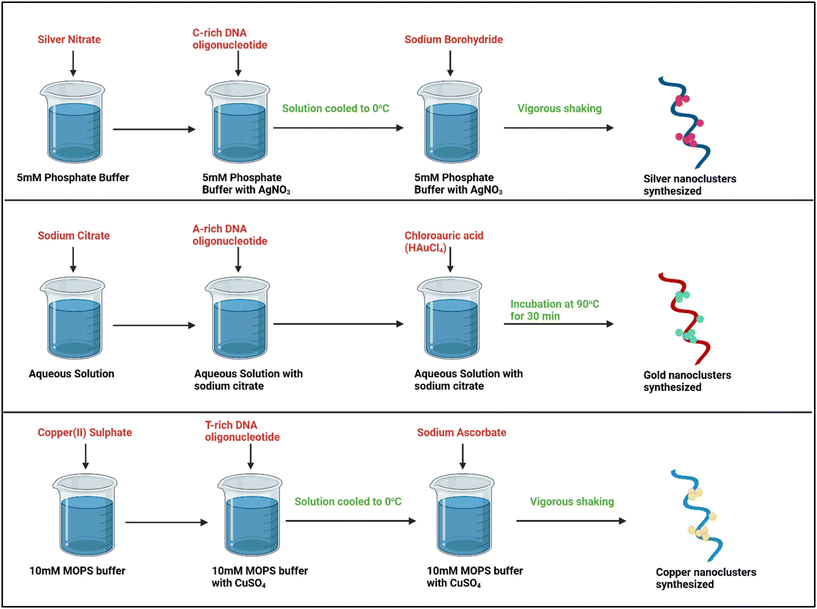 | ||
| Fig. 3 Schematic representation of the synthesis methods of the three different types of DNA-templated metal NCs, viz., silver, gold, and copper NCs, respectively. Created with BioRender. | ||
During the synthesis of DNA-templated fluorescent silver (Ag) NCs (F-AgNCs), the C-rich region of the template determines the aggregation of Ag0 on the DNA strand.56 In the case of cytosine, the N3 nitrogen does not possess a proton at neutral pH, which facilitates the facile binding of metal to the template at neutral pH of the reaction solution. In contrast, the N3 nitrogen of thymine is attached to a proton group at neutral pH, which necessitates the reaction solution to be maintained at an extremely basic pH for the easy binding of metal to this position on the template DNA.57 For adenine and guanine nucleosides, it is rather challenging to form fluorescent NCs, although the N7 nitrogen provides ample affinity for metal coordination. This adversity arises due to the combined effect of all other nitrogen on the purine ring, which makes the reduction process laborious to control and form F-AgNCs.58,59 Apart from ssDNA, numerous other forms of DNA such as dsDNA, aptamers, G-quadruplex DNA, and G-rich DNA and RNA have been reported to be used for various applications.60–62 An advantage related to AgNCs is their capability to exhibit fluorescence for a prolonged period, as observed by a research group, which found that around 32% of AgNC fluorescence remained after a period of 310 days at ambient room temperature.63
The limitation that arose from the use of F-AgNCs was their extreme susceptibility to photobleaching. This compelled the scientific community to search for another metal that could overcome this disadvantage. This ultimately resulted in the introduction of Au as a potential metal for NC formation on DNA.64,65 Following this, F-AuNCs were reported to be synthesized using a specific DNA sequence, with either dimethylamine borane as the reducing agent, at pH 7.0 and an NC size of 5 nm, or citrate as the reducing agent for the 30-mer DNA template.66–70 Over the years, the strategies for the synthesis of AuNCs have undergone change to include photoluminescence reduction and etching, in addition to chemical reduction.67,68 The bottom-up approach has been a common strategy for synthesis, with Xie et al. constructing red emitting AuNCs with bovine serum albumin (BSA) as the template, using simple methods.69 The top down approach involves etching of comparatively larger gold nanoparticles to form luminescent AuNCs. This was demonstrated by Yang et al., wherein they synthesized luminescent AuNCs using bis(phosphorothiolate) (ps) as the template for AuNPs and an acidic etchant.70 Apart from DNA, numerous biomolecules, such as proteins, peptides, and amino acids, have garnered attention as viable options for forming the template for AuNCs.71
In this scenario, F-CuNCs were introduced in view of their facile synthesis, tuneable fluorescence, and cost-effectiveness compared to their Au and Ag counterparts.72 In a particular study, it was found that dsDNA, and not ssDNA, was found to be efficient as the template for F-CuNCs.73 After a couple of years, it was reported that polyT-DNA ssDNA could be used as the template for the successful synthesis of F-CuNCs, but not with any other bases.74 Also, it was found that the emission intensity of these NCs was dependent on the length of the template.20 A drawback of CuNCs is the instability associated with fluorescence, which could be overcome through changes in the solution phase, as done by Kim et al., where they introduced fructose into the solution that increased the fluorescence lifetime by over 5 × 105 times.75
Biocompatibility of DNA-templated metal nanoclusters
In the case of Ag, its ions are toxic to humans as well as the environment at lethal concentrations.76 The confinement of electrons in molecular dimensions possess discrete energy levels, that can provide altered physical and chemical properties, including high photoluminescence and photostability, excellent biocompatibility, and sub-nanometer size.77 Various cytotoxicity tests conclude that DNA-AgNCs at a concentration of less than 1 μM have a minimal effect on cell viability,78–80 except for the micromolar concentration of AS1411 aptamer-functionalized AgNCs that could possibly induce cell death.81 AuNCs are also biocompatible as evidenced in the results of Wang et al., who tested them on human aortic endothelial cells and endothelial progenitor cells, and by various studies done earlier for inflammatory diseases adopting Au particles.82 Given that copper is a micronutrient in living beings, the nanomaterials created using it are comparatively more biocompatible than their heavy metal counterparts.83Mechanism of fluorescence from F-DNA metal NCs
From prior research spanning over several decades, it has been deduced that the NC size ranges around 2 nm and is somewhat midway between that of metal atoms and nanoparticles.84 The sporadic vibration of electrons of the metal atoms of NCs gives rise to the emission of bright light, which is exploited for its applications.1 The observed luminescence property is mainly because of the electronic transitions originating due to energy splitting, which includes intra-band transition (sp–sp) as well as inter-band transition (sp–d).84 Both these transitions can be grouped as electron transitions between the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) energy levels which result in the absorption and emission of light.85 The ligand-to-metal charge transfer provides luminescence of enhanced quality, compared to sole metal NCs, and this charge transfer can occur in two ways: (i) ligand-to-metal charge transfer (LCMT) and (ii) ligand-to-metal-metal charge transfer (LMMCT).86 Apart from this, it has been proven that luminescence from metal NCs is affected by their structures to some extent, which can be due to the differences in the number of free electrons or variation in the electronic transitions in the HOMO–LUMO energy levels.87,88Apart from the quantum confinement effects discussed above, numerous other factors have also been found to influence the photoemission properties such as size-dependent and independent emission that include the ligand effect, metal valence state correlated emission and self-assembly dependent emission.89 It was reported a decade ago that metal NCs dissolved in water show enhanced photoluminescence (PL) compared to those dissolved in organic solvents,90 indicating a role for the environment of the metal NCs. Adopting wet chemical synthesis strategies opened up opportunities for tunable and improved PL emission, through the synthesis of water-soluble metal nanoclusters of varying sizes.91,92 This correlation between the particle size and number could not solely explain the PL emissions since variations were observed between Au and Ag NCs with the same size and number. This called for a deeper and detailed analysis of further potential mechanisms behind the PL emissions, apart from the size-dependent effect of quantum confinement mechanics.89,93
Surface protective ligands have a profound effect on the stability of metal NCs, which would otherwise aggregate and result in a reduced emission rate. The ligand-dependent effect is well established for ligand-protected metal NCs, as in the case of DNA oligomer-templated AgNCs that provide extended emission from blue to the NIR region by manipulating the sequences.94 Another factor contributing to the luminescence of metal NCs is the valence state of the metal involved. This was observed in the case of AuNCs with glutathione as the ligand, in which 40 to 50% was made up of Au(I). The luminescence lifetime depended on the excitation wavelength. When excited at 420 nm, an orange color was emitted at 565 nm for several microseconds, while when excited at 530 nm, the same wavelength emission was observed, but for a short lifetime of around a few nanoseconds.95 Similar effects were observed by Zhu et al. in AuNCs by changing the oxidation states using oxidants such as O2, H2O2, and cesium sulfate.96,97 Yet another phenomenon determines photoemission, which is the surface-to-volume ratio, which is dependent on solvent molecules and oxygen. In such a scenario, aggregation induced emission (AIE) is an apt strategy to enhance luminescence.89
The theoretical analysis of the electronic structure and optical properties is propelled through density functional theory (DFT) calculations. This allows for the analysis of the ground state electronic structure and optical absorption, if the number and structure of the participating DNA strands are known beforehand.98 This can be done by assigning the charge of the clusters in aqueous solvents and analyzing the DNA-NC interactions, thereby the effect on the cluster charge.99 Apart from this, ligand fragmentation, damping, and basis set are important parameters for modelling the optical properties of DNA-AgNCs.100 Furthermore, simulation studies of electronic structures have revealed the existence of low-energy coupled excitonic states and the high-speed transport of energy between building blocks, and have given insights into the origin of fluorescence in coupled DNA-AgNCs.101,102
Characterization of F-DNA metal NCs20,57–61,63–66,73,74
Several characterization techniques have been employed for DNA-templated metal NCs according to the type of synthesis and requirement of the NC system. A few of the important techniques are described below, and an overview is presented in Fig. 4.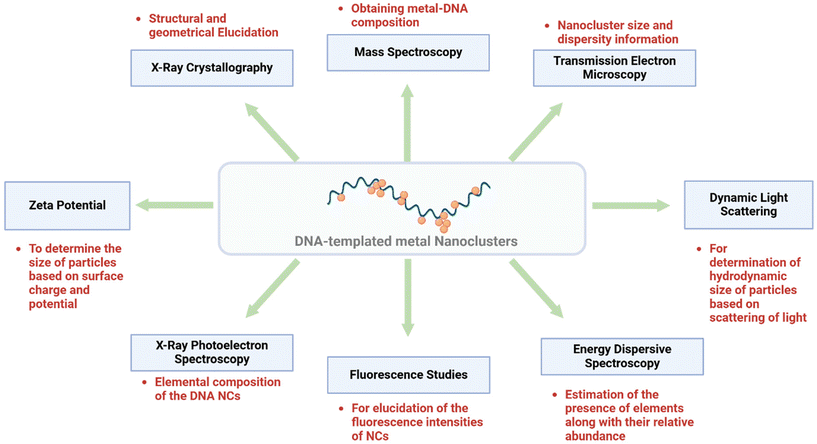 | ||
| Fig. 4 A representative overview of the above-described characterization techniques of DNA-templated metal NCs. Created with BioRender. | ||
Mass spectrometry (MS)
It was not until 2019 that the structural elucidation of NCs involved crystallographic studies.103,104 Until that point in time, the only source of information, even distantly related to its structure, could be obtained only from the correlation studies of experimentally determined absorption and excitation/emission for metal NCs of known composition using computational models. The depth of structural detailing provided by these methods is inferior compared to the recently available structural data of different crystals, although they provide all-inclusive structure–property relationships of DNA-bound metal NCs.103–106 Most of the above-mentioned studies have been conducted using AgNCs as a probe system and were used to determine the Ag-DNA composition. Advanced forms of MS analyses such as ESI-MS and ICP MS were used to determine the number of atoms of each metal in the DNA-Cu/Ag NCs, in which both the analyses pointed out the presence of two Ag atoms and one Cu atom per strand.107 Furthermore, studies conducted by Romolini et al. adopting the same ESI-MS and ICP-OES (inductively coupled plasma-optical emission spectrometry) have shown the presence of 21 silver atoms per 2 DNA strands, which is in agreement with founding works by Petty et al.108,109X-ray crystallography
Recently, crystallographic studies have been adopted to elucidate the structures of certain metal nanoclusters, especially of silver-bound DNA templates. These studies have reported the shape of the complex formed by the silver atoms along with the interatomic distances of metal atoms.5 This technique, along with advanced MS studies, is particularly useful in deciphering the chemical composition and crystal structure of metal NCs as opposed to larger nanoparticles with a multi-disperse size range and varying surface compositions since metal NCs are usually of smaller size.110 Crystallographic studies provide immense information regarding the bonds formed between different entities of the NC system, such as between the N3 of cytosines and Ag+ (in the case of the AgNC system), and also about metal–metal distances on the DNA template.5,111Transmission electron microscopy (TEM)
After the synthesis of the DNA-templated metal NCs, TEM characterization is performed, which allows for the estimation of the nanocluster size and dispersity. From a study involving F-AgNCs, it was found that DNA-AgNCs were spherical in shape with an average diameter of 2.09 nm, and these NCs were monodisperse on the DNA strand.112 In a study that included G-quadruplexes, they confirmed that the clusters are not aggregating, leading to the formation of large size nanoparticles, mainly through TEM analysis.113 Related to metal nanoclusters, TEM analyses are important given that the optical properties of these materials largely depend on the cluster sizes, and TEM gives account of the sizes of the particles involved, although subjected to errors due to the low resolution of TEM, paving the way for the optimized synthesis of metal NCs on DNA templates with enhanced fluorescence properties.60 The drawback associated with TEM is that, given the low resolution, smaller NCs might not be observed, and the images observed can be of aggregates of these smaller NCs (as observed by Teng et al. for AgNCs). To overcome this, these particles are subsequently subjected to atomic force microscopy (AFM) characterization to assess the morphology of the structures accurately.114X-ray photoelectron spectroscopy (XPS)
Since the XPS data provide information sensitive to the surface features of a material, in the nanocluster system as well, it is employed to gain insights into the atomic composition, chemical structures, and electronic structure of the concerned atoms.115,116 This characterization technique was used to determine the elemental composition of DNA-Cu/Ag NCs to extract information about the occurrence of copper and silver atoms on the DNA strands, especially the number of each of the atoms.107 In a similar study, this technique was employed to estimate the NC size and also analyze the transition bands of the 3d orbital of the Ag atom.117 XPS was used to address the questions concerning the actual structures of Ag-DNA complexes. In another study, to analyze the emissions from Ag as a whole, different forms of Ag were analyzed such as cations, fluorescent NCs in solution, and Ag clusters deposited on the DNA template.118Energy dispersive spectroscopy (EDS)
To assess the presence of elements along with their relative abundance, EDS has been employed in many studies, which are carried out in conjunction with scanning electron microscopy (SEM). This setup conveniently involves the use of the secondary electron, which moves out from the lower energy shell, as it is used in SEM for imaging, and the photon released as part of the electron transition (which has energy comparable to that of X-rays) to be captured by the EDS sensor.116 Especially related to DNA-templated metal nanoclusters, it has been used for the identification of the average number of metal atoms forming the nanocluster system on the ssDNA template, along with other structural information. In the case of F-DNA AgNCs, the fixed number of phosphate groups in these structures is exploited by calculating the relative atomic percentages of silver and phosphate atoms, which is, in turn, used for the estimation of silver atoms on the template.119,120Fluorescence studies
The fundamental property associated with DNA-templated metal NCs is their ability to fluoresce at different wavelengths of incident light depending on the composition of the material.121 UV-Vis absorption provides insights into the silver atom concentration in the solution through the molar absorption coefficient (ε).109The UV-Vis spectrum in a study based on DNA–AgNCs had two profound absorption peaks (Fig. 5C).122 The UV absorption peak in this study varied between 260 nm and 270 nm, while that of visible light was not fixed. In addition to that, the emission wavelength of UV excitation was at par with that of visible light but with the intensity increased by 2 to 4 times, which can be explained by the increased charge transfer from the cytosine of DNA to that of the bound Ag ions, upon UV excitation.123,124 Several prior studies have attributed to the fact that the emission properties of any nucleic acid-templated metal NCs depend on the size of both the template and the NCs, the compositional variations, and the overall geometry of the material. Optimizations in these aspects have led to the discovery of NCs with emission properties ranging from blue to the near-infrared region (NIR) range, some of them even surpassing 950 nm.125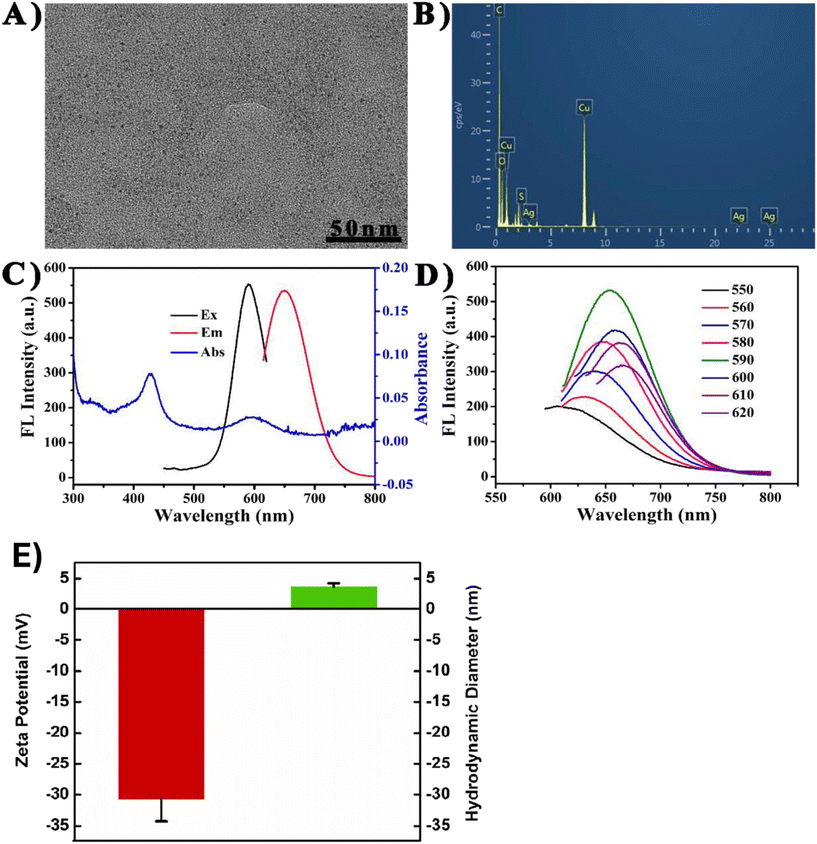 | ||
| Fig. 5 Representation of different characterization techniques employed for analyzing different types of metal NCs. (A) TEM image of DNA-AgNCs, (B) EDS spectrum of DNA-AgNCs, (C) optical spectrum indicating the excitation wavelength, emission wavelength, and absorbance of DNA-AgNCs, (D) fluorescence observed at different excitation wavelengths of DNA-AgNCs. Images A to D were reprinted with permission from ref. 135. Copyright 2021, Elsevier B.V. (E) Zeta potential (in red) and hydrodynamic diameter (in green) of CuNCs. Reprinted with permission from ref. 131. Copyright 2022, Elsevier B.V. | ||
For DNA-AgNCs, LMCT from the N atom of DNA to AgNCs is considered to be the initiation of photoluminescence.126,127 Liu et al. studied the changes in photoluminescence to the varying excitation wavelengths. When DNA1-AgNCs were excited at 350 or 555 nm, a distinct peak of emission was observed at 625 nm, whereas excitation at 435 nm did not yield any emission peak at 625 nm, which might be due to the surface plasmon absorption at 435 nm.128
Zeta potential
It deals with the surface potential difference between two layers associated with any material, particularly in colloidal solutions, and this charge potential determines the stability of these materials. In essence, the magnitude of this potential remains low in the case of aggregate formation, and vice versa.129 This aspect of the zeta potential is utilized as a primary measure to estimate whether or not the aggregation of nanoparticles has occurred. This feature has been exploited in numerous studies of metal NCs. In one study, zeta potential measurements were obtained to estimate the binding of carbon quantum dots onto DNA-templated CuNCs. Here, prior to the addition of quantum dots to the solution of CuNCs, the zeta potential was around −34 eV, which drastically changed to 2.3 eV with the addition of carbon quantum dots (CDs), which provides a preliminary confirmation that aggregation has taken place, which might be due to the binding of quantum dots to the CuNCs forming a complex of DNA-CD/CuNCs.130 In some cases, these studies are done in conjunction with dynamic light scattering studies in order to obtain the hydrodynamic size of the nanoparticles along with their surface charge (Fig. 5E).131Dynamic light scattering (DLS)
DLS is a technique employed to analyze the average hydrodynamic size of a particle, which is accomplished through the scattering of incident light on a particle while conserving the wavelength of the scattered light (Rayleigh scattering).132 This particular characteristic of DLS has been exploited to estimate nanoparticle sizes and as a preliminary confirmation for the aggregation of nanoparticles. Even in the case of metal NCs, this has been utilized to determine the formation of clusters of specific sizes, which in turn has unique properties of its own.133 Based on the results of this technique, in one study, it was estimated that the average hydrodynamic radius of DNA-templated AgNCs was around 2.5 nm.20 However, DLS is usually adopted in the case of metal nanoparticles, given their larger size, and for metal NCs, it might lead to errors, which eventually led to the development of DLS-based metallic nanobiosensors (nano-DLS) for point-of-care applications.134Applications of the different types of metal NCs
Over the past decade and a half, the number of metal nanoparticles that can be used for NC formation was heavily studied, with some fruitful discoveries opening avenues for the use of a multitude of metals. Most of the studies are undertaken using Au, Ag, and Cu nanoparticles, which have been used for diversified applications (Table 1).5,136,137| Metal nanocluster | Applications | Ref. |
|---|---|---|
| F-AgNCs | Heavy metal ion detection (Cu2+, Hg2+, and Pb2+) | 20, 138 and 139 |
| Detection of D-penicillamine | 135 | |
| Inhibit bacterial growth (while being non-toxic to mammalian cell lines) | 120 | |
| Food safety detection | 2, 45 and 140 | |
| Glucose level detection | 141 | |
| Detection of antibiotic resistance genes | 142 | |
| Detection of microRNA | 143 | |
| Detection of tetracycline (residues in food) | 144 | |
| β-Amyloid oligomer sensor | 145 | |
| Detection of thiram146 | 146 | |
| F-AuNCs | Cancer cell imaging and cancer treatment | 147 and 148 |
| Near-infrared (NIR) imaging of microRNAs and proteases | 149 | |
| Hg2+ sensing | 150 | |
| Sensing of specific nucleic acids in human serum | 151 | |
| Detection of sulfide ions | 152 | |
| Detection of deoxynivalenol | 153 | |
| F-CuNCs | Heavy metal ion detection (Cu2+, Hg2+, Pb2+, and Mn2+) | 20 and 154 |
| Detection of hydrogen peroxide release from live cells | 155 | |
| Detection of Hepatitis B Virus (HBV) DNA | 156 | |
| Screening of inorganic pyrophosphate activity and that of its inhibitor | 157 | |
| Enhanced electrochemiluminescence as a biosensor | 158 | |
| High-throughput logical analysis of biomarkers | 159 | |
| Detection of tetracycline in milk | 160 |
Apart from the applications mentioned above, these systems have been used in numerous other scenarios as well. These DNA-templated metal NCs have the potential to be used as fluorescent ‘turn-on’ probes, as the traditional molecular beacons used for the same purpose have limitations related to high background fluorescence and reduced sensitivity to different analytes. The novel nanocluster beacons (NCBs) overcome these limitations as they do not depend on Förster resonance energy transfer (FRET) and provide excellent detection of analytes, especially in the case of nucleic acid detection.161,162 DNA templated AgNCs have been employed for this application, where a weakly green fluorescent DNA templated AgNC is converted into a bright red emitting system in the presence of an enhancer sequence of DNA.163
Another application of these NCs comes in the form of protein detection. One of the hurdles in forming a protein detection system using NCs on DNA templates is the inability to use bioconjugates to bind to the target molecule since the system undergoes degradation during the bioconjugation step. In view of this, a DNA aptamer–AgNC system with intrinsic fluorescence and a target recognition motif was developed, with high specificity and sensitivity, simultaneously.164 This aptamer-based system was synthesized in a single step, without the requirement of successive attachment steps of the aptamer to a fluorophore, which ultimately reduces the cost and time. The efficiency of this system was assessed and confirmed by a study that explored α-thrombin as the protein of interest.6 Some of the common and latest applications employing DNA-templated metal NCs are mentioned in Table 1, and a few of these are discussed below.
Heavy metal ion detection
Heavy metal ion sensing is an application that spans all three types of metal NCs, i.e., Ag, Au, and Cu NCs. According to the current data, there are a plethora of methods available for monitoring heavy metal ions, out of which fluorescence-based methods are of importance due to their high sensitivity and specificity, facile synthesis methods, and affordability.165,166 In general, DNA-templated fluorescent metal NCs follow mechanisms that can be categorized into three: (i) turn-on fluorescence, (ii) turn-off fluorescence, and (iii) ratiometric fluorescence.20 Heavy metals can be either one or a combination of mostly Cu2+, Hg2+, and Pb2+. Cupric ions are usually present as trace elements in the human body,167 and even a vital component of hemocyanin.168 However, excessive exposure to copper might lead to disease conditions such as Wilson disease, amyotrophic lateral sclerosis, and Alzheimer's disease.169 Compared to Hg2+, the toxicity of Cu2+ is lower in water. Back in 2010, a method for copper ion detection that works based on the formation of a DNA-templated Cu/Ag alloy NC from a DNA-templated F-AgNC and based on the principle of turn-on fluorescence was developed, with increased emission of more than four times compared to normal AgNC fluorescence (detection limit of 10 nM).170 A year later, another method of detection based on turn-off fluorescence was developed, wherein the presence of Cu2+ was found to quench the fluorescence of F-AgNCs.171 A representation of both the turn-on and turn-off mechanism related to the sensing of Cu2+ by F-DNA AgNCs is given in Fig. 6.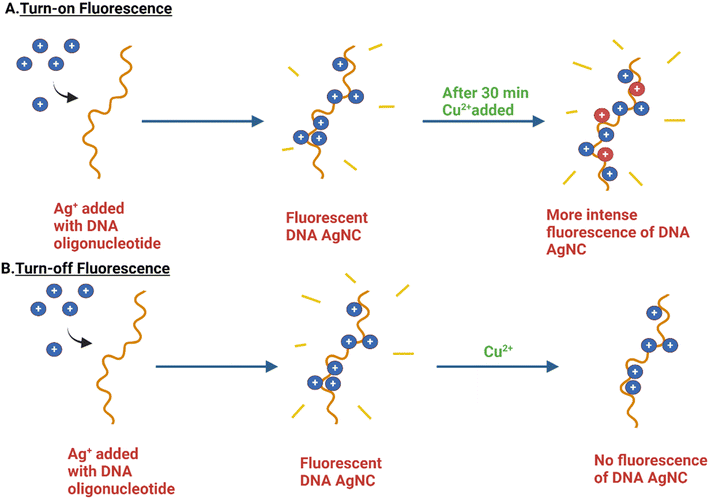 | ||
| Fig. 6 Representation of both the turn-on and turn-off mechanisms related to the sensing of Cu2+ by F-DNA AgNCs. (A) The turn-on fluorescence mechanism exploited for the detection of Cu2+ by DNA-AgNCs gives an extended, intense fluorescence in the presence of Cu2+ due to the formation of DNA-Ag/Cu NCs, as observed by Lan et al.170 (B) The turn-off fluorescence mechanism exploited for the detection of Cu2+ by DNA-AgNCs, which quenches fluorescence in the presence of Cu2+. Adapted from ref. 171. Created with BioRender. | ||
Hg2+ ion detection is of particular importance given its carcinogenic implications in the body and adverse effects on the environment in the long term.172,173 So, its detection from supposable sources such as drinking water should be efficiently carried out to minimize adverse epidemiological aftermaths. In the late 2000s, a group of researchers found that the DNA-templated F-AgNCs had the ability to detect Hg2+ in water using the turn-off fluorescence method. This turn-off was due to the static quenching of fluorescence by Hg2+, as it disrupted the bonding between DNA and AgNCs.174 Subsequently, several turn-on and ratiometric detection methods with improved detection levels were developed over the years by different research groups.175,176 Pb2+ is another ion of particular importance as it has severe implications on health, such as adverse effects on kidneys, cognition, and even muscles if consumed at toxic levels.165,177 In the early years after 2010, several researchers came up with fluorescence-based detection systems for lead ions from tap water, drinking water, and urine samples. Pb2+ quenches the intrinsic fluorescence of F-CuNCs by interacting with the copper ions of the cluster, thereby forming a turn-off detection system.178,179
Food safety detection
Determination of food safety using F-metal NCs has a close connection with the previously discussed heavy metal detection methods since contamination of food through heavy metal pollutants is a leading cause of morbidity among the population. Similar turn-on, turn-off, and ratiometric methods can be used to determine the level of different heavy metals in food preparations. Apart from this, multi-metal (Cu2+ and Hg2+) detection systems can be employed for higher throughput screening using F-DNA AgNCs.180 Another type of contaminant in food systems is the presence of microbial toxins. These toxins pose a great challenge as extremely minute detection levels are required to screen them. Zearalenone (ZEN) is a secondary metabolite of a typical fungus that was found to be hazardous to humans and is present in crops, which, when ingested, might lead to serious health complications, which can even affect the reproductive system.181 A detection system using aptamer-based AgNCs and metal–organic framework (MOF) based Fe3O4/carbon octahedra was developed as a potential biosensor based on FRET for microbial detection.182 A similar system was later used by another research group for the detection of T-2 toxin in wheat and corn.183 The detection systems are not just limited to metabolites but also include the detection of different pathogenic bacterial strains. Bacterial species such as Staphylococcus aureus, Listeria monocytogenes, and Escherichia coli are important players in food deterioration through toxins and other innate characteristics.184 Detection systems based on quenching and ratiometric sensing have been applied for bacterial detection.185,186Cancer cell imaging
Numerous techniques are being tested day by day to bring out novel methods of detecting and imaging cancer cells, given their worldwide notoriety. Metal NCs were employed for potential imaging capabilities and a better understanding of the dynamics of cancer cells, exploiting their extreme resistance to photobleaching and relatively less toxicity compared to numerous other imaging techniques.187 Recently, DNA nanoribbons (DNRs) were employed as the templates for AuNCs, which resulted in an enhancement in the intensity of intracellular fluorescence of the NCs, as compared to other templates or without DNR as a template. Here, the enhancement in the fluorescence intensity was attributed to the intracellular biosynthesis of AuNCs and the higher stability of these NCs in the lysosome.147NIR imaging of microRNAs and proteases
Biomarkers of tumors, such as microRNAs and proteases, are important entities to be considered for understanding the mechanism and pathway of tumorigenesis.188,189 Either DNA or peptide-functionalized fluorescent nanomaterials have been used for imaging purposes over the years,190,191 but not without their own set of limitations, such as poor retention and poor permeability into cells.192,193 To counteract these drawbacks, a novel material was sought after, that had size-tuneable properties.194 This culminated in the introduction of metal NCs, which had self-assembly and aggregation-induced emission (AIE).195,196 Later, optimization studies gradually led to the introduction of DNA as a template for these NCs, and finally, DNA-templated AuNCs were synthesized, which were successfully used for monitoring microRNA activity through a specific self-assembly approach.197,198 These advantages of AuNCs were exploited in a study involving the imaging of microRNAs and proteases in vivo.133Detection of hydrogen peroxide (H2O2) release from live cells
H2O2 is an important metabolite that is present in cancer cells in abundance compared to normal cells. Substantial generation of H2O2 can lead to numerous adverse health effects, as it causes oxidative stress, ultimately impacting cells adversely.199,200 Being an important biomarker of reactive oxygen species (ROS), it is crucial to devise appropriate detection systems to account for H2O2 release.199 In this arena, the concept of metal NCs was applied, which has brought fruitful results in the form of advanced detection systems. These NCs act as redox-active clusters, which in turn take part in the reaction as electron conductors and electron transfer mediators. Given the substantial cost of noble metals such as Ag, Au, and Pt, less expensive alternatives had to be searched for, which came in the form of Cu. Cu has widespread availability and is economically viable, which favors its use as an NC system for detection purposes.155 CuNCs have higher yields under moderate conditions, along with excellent water solubility, which makes them an appealing candidate for this application. Nevertheless, CuNCs have their own limitations, which propels the need to include noble metals for enhanced catalytic activity.201,202Detection of Hepatitis B virus (HBV) DNA
HBV is a circular double-stranded DNA virus that is spread primarily through body fluids, and the infection has been reported worldwide, with a quarter of a billion people infected already.203 Given this scenario, it is high time that an appropriate diagnostic method and treatment regime were designed. As of now, the detection methods consist of different types of immunoassays combined with the polymerase chain reaction (PCR), which, although they provide precise results, involve heavy machinery that is least suitable for developing countries to explore.156 To counter these shortcomings, cost-effective and reliable methods were developed over the years, one of which was the colorimetric method of detecting DNA, which is easier to visualize, as it can be observed with the naked eye.204,205In a study, this colorimetric method was utilized in the form of DNA-templated F-CuNCs to detect the presence of HBV from human sample specimens. In this application, a probe ssDNA is immobilized on a surface to which target ssDNA strands are added, and through reduction with CuSO4, the formation of dsDNA occurs, which then allows the profuse attachment of copper ions leading to the formation of CuNCs,206 which are successively liberated from the template during addition of nitric acid.207 These liberated copper ions will complex with the creatinine added thereafter, which in turn oxidizes ABTS,208 resulting in the formation of a green color solution. Different amounts of the target DNA will have different colors and serve as a quantitative technique above its qualitative capabilities.156 Recently, AgNCs have also been employed for the detection of HBV DNA by leveraging a concentration imbalance-driven DNA circuit (CIDDC) as an operational amplifier.209
Antibacterial activity of DNA-AgNCs
Compared to Ag nanoparticles (AgNPs), AgNCs have enhanced antibacterial efficiency given their higher stability and tuneability due to the template.210 Similar to AgNPs, reactive oxygen species (ROS) generation has been found to be the primary mechanism behind the antibacterial properties, along with some other mechanisms at play, which requires further studies for confirmation.211 The already identified positive correlation between the optical properties and antibacterial efficacy of these NCs can be put to effective use for bacterial sensing and visualization, simultaneously with bacterial growth inhibition.212 The antibacterial activity against the formation and growth of the biofilm from Pseudomonas aeruginosa has been demonstrated by Sengupta et al., with minimal risk of development of antibiotic resistance.213Detection of adenosine triphosphate (ATP) and cocaine
DNA-AgNCs have been mostly used for the detection of ATP by numerous groups. Zhang et al. used DNA-AgNCs to form an aptasensor for the detection of ATP with a detection limit of 5 μM that is based on turn-on fluorescence. This is due to the change in the conformation of the aptamer as ATP is introduced into the system, thereby altering the entire microenvironment.214 Zhang and Wei developed another turn-on fluorescence detection system for ATP and adenosine deaminase (ADA) using DNA-Cu/Ag NCs. The mechanism of detection is similar to that of the previous study, with a detection limit of 7.0 μM.215 Li et al. developed yet another turn-on fluorescent nanoprobe for the same application, with a detection limit of 3 μM.216 Li et al. proposed a Cu2+-mediated fluorescence biosensor for the detection of ATP using AgNCs, with a detection limit of 16 nM.217 Zhang et al. proposed a mechanism for the detection of cocaine using DNA-AgNCs by the nicking endonuclease assisted amplification method, with an excellent detection limit of 2 nM using a comparatively small volume of 5 μL.218 This method is cost-effective, simple, and easily detected using microplate readers.Future perspectives
From the initiation of the application of metal NCs on DNA templates, aspects related to synthesis, characterization, and applications have evolved at a rapid pace. Specifically in this regard, the synthesis of DNA templates has taken a high road since the adoption of machine learning (ML) models for designing specific DNA sequences for efficient metal–DNA binding and fluorescence properties.219 While designing the template DNA, it is always desirable to design in such a way that the final product will have time-dependent stability as well as enhanced fluorescence. From prior studies, it is evident that cytosine and guanine nucleobases are paramount for obtaining highly fluorescent AgNCs, while the presence of multiple nucleobases in the template stretch ensures the time stability of the system.220–222 Tweaking with the synthesis methods has been found to provide altered results,99 some of which are desirable while others may not be. In this regard, applying data mining and ML models for designing and synthesizing novel sequences might provide enhanced quality of NC systems, as compared to conventionally used ones.223Another related advancement that can be applied to DNA-templated nanoclusters is the design of DNA templates using DNA computing, which is mostly based on logic operators. Binary-encoded DNAs are used as inputs, and the outputs can be obtained in the form of DNA strands, their optical properties, or even electrochemical signals. This combination of Boolean operation with DNA can be applied to numerous bio-based smart applications spanning diagnostics to drug delivery.224–226 Using these approaches, especially ML algorithms, novel F-DNA AgNCs have been synthesized that exhibit fluorescence emission in the NIR region, which is extremely important in the analysis of biological tissues and fluids.227–229
Conclusion
Over the past few decades, DNA nanotechnology has developed and spread across a wide range of applications, from biosensing to drug delivery, and has been a forerunner for a multitude of novel detection systems. One such system is that of F-DNA metal NCs, which have numerous applications based on their fluorescence. Optimizing the synthesis methods of these NCs aids in different properties of the system as a whole, out of which some properties may be of interest based on the application. Most of the applications of these NCs are based on fluorescence turn-on or turn-off and are incredibly useful for detecting the presence of certain analytes in a particular environment. Numerous characterization techniques can be employed to obtain the physical and chemical properties of each of the novel systems developed. Additionally, the sensitivity and specificity of these systems can be enhanced by adopting novel techniques of pre-screening the systems through computational aids such as machine learning models. Thus, although the niche of DNA-templated metal NCs has progressed within a short span of time, there seems to be yet a gorge of untapped potential for advancement that can be attained in the near future.Data availability
No new data were generated for writing this review article.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We sincerely thank all the members of the DB group for critically reading the manuscript and for their valuable feedback. AR thanks IITGN and MHRD for the MTech fellowship. DB thanks the SERB, GoI, for the Core research grant, MoES for the STARS grant, IITGN for the start-up grant, and DBT-EMR, Gujcost-DST and GSBTM for research grants.References
- Y. Pan, Z. Han, S. Chen, K. Wei and X. Wei, Coord. Chem. Rev., 2023, 478, 214964 CrossRef CAS.
- B. Zhou, I. M. Khan, X. Ding, S. Niazi, Y. Zhang and Z. Wang, Talanta, 2024, 273, 125834 CrossRef CAS PubMed.
- Y. Guo, F. Cao, X. Lei, L. Mang, S. Cheng and J. Song, Nanoscale, 2016, 8, 4852–4863 RSC.
- R. Jin, C. Zeng, M. Zhou and Y. Chen, Chem. Rev., 2016, 116, 10346–10413 CrossRef CAS.
- A. Gonzàlez-Rosell, C. Cerretani, P. Mastracco, T. Vosch and S. M. Copp, Nanoscale Adv., 2021, 3, 1230–1260 RSC.
- Y. Chen, M. L. Phipps, J. H. Werner, S. Chakraborty and J. S. Martinez, Acc. Chem. Res., 2018, 51, 2756–2763 CrossRef CAS.
- Y. Bao, H. C. Yeh, C. Zhong, S. A. Ivanov, J. K. Sharma, M. L. Neidig, D. M. Vu, A. P. Shreve, R. B. Dyer, J. H. Werner and J. S. Martinez, J. Phys. Chem. C, 2010, 114, 15879–15882 CrossRef CAS.
- A. Desireddy, B. E. Conn, J. Guo, B. Yoon, R. N. Barnett, B. M. Monahan, K. Kirschbaum, W. P. Griffith, R. L. Whetten, U. Landman and T. P. Bigioni, Nature, 2013, 501, 399–402 CrossRef CAS.
- Y. Hu, W. Guo and H. Wei, Isr. J. Chem., 2015, 55, 682–697 CrossRef CAS.
- Y.-S. Borghei, M. Hosseini and M. R. Ganjali, Microchimica Acta., 2017, 184, 2671–2677 CrossRef CAS.
- J. Benavides, I. Quijada-Garrido and O. García, Nanoscale, 2020, 12, 944–955 RSC.
- L. Yang, C. Yao, F. Li, Y. Dong, Z. Zhang and D. Yang, Small, 2018, 14, 1800185 CrossRef.
- D. Yang, M. R. Hartman, T. L. Derrien, S. Hamada, D. An, K. G. Yancey, R. Cheng, M. Ma and D. Luo, Acc. Chem. Res., 2014, 47, 1902–1911 CrossRef CAS.
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Science, 2015, 347(6224), 1260901 CrossRef.
- E. N. Salgado, R. J. Radford and F. A. Tezcan, Acc. Chem. Res., 2010, 43, 661–672 CrossRef CAS.
- J. Duguid, V. A. Bloomfield, J. Benevides and G. J. Thomas, Biophys. J., 1993, 65, 1916–1928 CrossRef CAS PubMed.
- J. Liu, TrAC, Trends Anal. Chem., 2014, 58, 99–111 CrossRef CAS.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 5207–5212 CrossRef CAS PubMed.
- L. Zhu, Z. Qing, L. Hou, S. Yang, Z. Zou, Z. Cao and R. Yang, ACS Sens., 2017, 2, 1198–1204 CrossRef CAS.
- C. Song, J. Xu, Y. Chen, L. Zhang, Y. Lu and Z. Qing, Molecules, 2019, 24(22), 4189 CrossRef CAS.
- L. Berti and G. A. Burley, Nat. Nanotechnol., 2008, 3, 81–87 CrossRef CAS.
- A. Ono, S. Cao, H. Togashi, M. Tashiro, T. Fujimoto, T. MacHinami, S. Oda, Y. Miyake, I. Okamoto and Y. Tanaka, Chem. Commun., 2008, 4825–4827 RSC.
- A. Ono, H. Torigoe, Y. Tanaka and I. Okamoto, Chem. Soc. Rev., 2011, 40, 5855–5866 RSC.
- P. P. Pompa, L. Martiradonna, A. Della Torre, F. Della Sala, L. Manna, M. de Vittorio, F. Calabi, R. Cinagolani and R. Rinaldi, Nat. Nanotechnol., 2006, 1, 126–130 CrossRef CAS PubMed.
- Z. Qing, J. Xu, J. Hu, J. Zheng, L. He, Z. Zou, S. Yang, W. Tan and R. Yang, Angew. Chem., Int. Ed., 2019, 58, 11574–11585 CrossRef CAS PubMed.
- Z. Qing, L. Zhu, L. Hou, Z. Zou, S. Yang and R. Yang, Sci. China: Chem., 2018, 61, 1630–1636 CrossRef CAS.
- Z. Peng and H. Liu, Chem. Mater., 2016, 28, 1012–1021 CrossRef CAS.
- Y. Yu, B. Y. L. Mok, X. J. Loh and Y. N. Tan, Adv. Healthc. Mater., 2016, 5, 1844–1859 CrossRef CAS PubMed.
- S. C. L. Tan, Z. He, G. Wang, Y. Yu and L. Yang, Molecules, 2023, 28, 5531 CrossRef CAS PubMed.
- J. Qiu, F. Ahmad, J. Ma, Y. Sun, Y. Liu, Y. Xiao, L. Xu, T. Shu and X. Zhang, Anal. Bioanal. Chem., 2024, 416, 3923–3944 CrossRef CAS PubMed.
- Z. Qing, T. Qing, Z. Mao, X. He, K. Wang, Z. Zou, H. Shi and D. He, Chem. Commun., 2014, 50, 12746–12748 RSC.
- N. Schaeffer, B. Tan, C. Dickinson, M. J. Rosseinsky, A. Laromaine, D. W. McComb, M. M. Stevens, Y. Wang, L. Petit, C. Barentin, D. G. Spiller, A. I. Cooper and R. Lévy, Chem. Commun., 2008, 3986–3988 RSC.
- L. Y. Chen, C. W. Wang, Z. Yuan and H. T. Chang, Anal. Chem., 2015, 87, 216–229 CrossRef CAS PubMed.
- J. Zheng and R. M. Dickson, J. Am. Chem. Soc., 2002, 124, 13982–13983 CrossRef CAS PubMed.
- A. Pandya, A. N. Lad, S. P. Singh and R. Shanker, RSC Adv., 2016, 6, 113095–113114 RSC.
- J. A. Fan, Y. He, K. Bao, C. Wu, J. Bao, N. B. Schade, V. N. Manoharan, G. Shvets, P. Nordlander, D. R. Liu and F. Capasso, Nano Lett., 2011, 11, 4859–4864 CrossRef CAS PubMed.
- G. Chen, I. Roy, C. Yang and P. N. Prasad, Chem. Rev., 2016, 116, 2826–2885 CrossRef CAS PubMed.
- D. Nykypanchuk, M. M. Maye, D. Van Der Lelie and O. Gang, Nature, 2008, 451, 549–552 CrossRef CAS PubMed.
- M. A. Boles, M. Engel and D. V. Talapin, Chem. Rev., 2016, 116, 11220–11289 CrossRef CAS PubMed.
- A. M. Hung, C. M. Micheel, L. D. Bozano, L. W. Osterbur, G. M. Wallraff and J. N. Cha, Nat. Nanotechnol., 2010, 5, 121–126 CrossRef CAS PubMed.
- C. M. Soto, A. Srinivasan and B. R. Ratna, J. Am. Chem. Soc., 2002, 124, 8508–8509 CrossRef CAS PubMed.
- A. Shahrokhtash and D. S. Sutherland, ACS Appl. Mater. Interfaces, 2024, 16, 21534–21545 CrossRef CAS PubMed.
- Y. Zhang, Y. Cai, Z. Qi, L. Lu and Y. Qian, Anal. Chem., 2013, 85, 8455–8461 CrossRef CAS PubMed.
- Z. Huang, F. Pu, Y. Lin, J. Ren and X. Qu, Chem. Commun., 2011, 47, 3487–3489 RSC.
- X. Tang, M. Lu, J. Wang, S. Man, W. Peng and L. Ma, J. Agric. Food Chem., 2024, 72, 5542–5554 CrossRef CAS PubMed.
- A. Gonzàlez-Rosell and S. M. Copp, Acc. Chem. Res., 2024, 57, 2117 CrossRef PubMed.
- A. A. Buglak and M. T. Nguyen, Biophys. Rev., 2024 DOI:10.1007/S12551-024-01200-X.
- Y. Han, Q. Xu, H. Liu, F. Ma and C. Y. Zhang, Coord. Chem. Rev., 2024, 509, 215801 CrossRef CAS.
- Y. Lu and W. Chen, Chem. Soc. Rev., 2012, 41, 3594–3623 RSC.
- A. C. Schwartz, Z. Allg. Mikrobiol., 1975, 15, 388–388 CrossRef.
- H. Liu, Y. Wang, L. Zhang, Y. Shao and B. Zheng, Mater. Lett., 2015, 139, 265–267 CrossRef CAS.
- A. Nandy, S. Chakraborty, U. Pramanik, S. Nandi and S. Mukherjee, J. Phys. Chem. Lett., 2020, 11(7), 2436–2442 CrossRef.
- H. B. Wang, Y. Li, H. Y. Bai and Y. M. Liu, Sens. Actuators, B, 2018, 259, 204–210 CrossRef CAS.
- Z. Qing, X. He, D. He, K. Wang, F. Xu, T. Qing and X. Yang, Angew. Chem., Int. Ed., 2013, 52, 9719–9722 CrossRef CAS PubMed.
- T. Qing, Z. Qing, Z. Mao, X. He, F. Xu, L. Wen, D. He, H. Shi and K. Wang, RSC Adv., 2014, 4, 61092–61095 RSC.
- Z. Liasi, A. E. Hillers-Bendtsen, L. Jensen and K. V. Mikkelsen, J. Phys. Chem. Lett., 2023, 14, 5727–5733 CrossRef CAS.
- B. Sengupta, C. M. Ritchie, J. G. Buckman, K. R. Johnsen, P. M. Goodwin and J. T. Petty, J. Phys. Chem. C, 2008, 112, 18776–18782 CrossRef CAS.
- J. A. R. Navarro and B. Lippert, Coord. Chem. Rev., 1999, 185–186, 653–667 CrossRef CAS.
- S. Verma, A. K. Mishra and J. Kumar, Acc. Chem. Res., 2010, 43, 79–91 CrossRef CAS.
- J. Ai, W. Guo, B. Li, T. Li, D. Li and E. Wang, Talanta, 2012, 88, 450–455 CrossRef CAS.
- D. Schultz and E. Gwinn, Chem. Commun., 2011, 47, 4715–4717 RSC.
- H. Liu, X. Yang, B. Huang and H. Liu, Spectrochim. Acta, Part A, 2023, 297, 122740 CrossRef CAS.
- J. Sharma, R. C. Rocha, M. L. Phipps, H. C. Yeh, K. A. Balatsky, D. M. Vu, A. P. Shreve, J. H. Werner and J. S. Martinez, Nanoscale, 2012, 4, 4107–4110 RSC.
- G. Liu, Y. Shao, K. Ma, Q. Cui, F. Wu and S. Xu, Gold Bull., 2012, 45, 69–74 CrossRef CAS.
- T. A. C. Kennedy, J. L. MacLean and J. Liu, Chem. Commun., 2012, 48, 6845–6847 RSC.
- G. Liu, Y. Shao, F. Wu, S. Xu, J. Peng and L. Liu, Nanotechnology, 2012, 24, 015503 CrossRef PubMed.
- S. K. Kailasa, S. Borse, J. R. Koduru and Z. V. P. Murthy, Trends Environ. Anal. Chem., 2021, 32, e00140 CrossRef CAS.
- L. Chen, M. Gharib, Y. Zeng, S. Roy, C. K. Nandi and I. Chakraborty, Mater. Today Chem., 2023, 29, 101460 CrossRef CAS.
- J. Xie, Y. Zheng and J. Y. Ying, J. Am. Chem. Soc., 2009, 131, 888–889 CrossRef CAS.
- L. Yang, J. Chen, T. Huang, L. Huang, Z. Sun, Y. Jiang, T. Yao and S. Wei, J. Mater. Chem. C, 2017, 5, 4448–4454 RSC.
- G. Yang, Z. Wang, F. Du, F. Jiang, X. Yuan and J. Y. Ying, J. Am. Chem. Soc., 2023, 145, 11879–11898 CrossRef CAS.
- C. Wei, H. Lin and H. Bai, Microchim. Acta, 2023, 190, 1–10 CrossRef.
- A. Rotaru, S. Dutta, E. Jentzsch, K. Gothelf and A. Mokhir, Angew. Chem., Int. Ed., 2010, 49, 5665–5667 CrossRef CAS PubMed.
- G. Liu, Y. Shao, J. Peng, W. Dai, L. Liu, S. Xu, F. Wu and X. Wu, Nanotechnology, 2013, 24, 345502 CrossRef PubMed.
- S. Kim, E. S. Lee, B. S. Cha and K. S. Park, Nano Lett., 2022, 22, 6121–6127 CrossRef CAS PubMed.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- L. Zhang and E. Wang, Nano Today, 2014, 9, 132–157 CrossRef CAS.
- G. M. Han, Z. Z. Jia, Y. J. Zhu, J. J. Jiao, D. M. Kong and X. Z. Feng, Anal. Chem., 2016, 88, 10800–10804 CrossRef CAS PubMed.
- Y. Cao, Y. Dai, H. Chen, Y. Tang, X. Chen, Y. Wang, J. Zhao and X. Zhu, Biosens. Bioelectron., 2019, 130, 132–138 CrossRef PubMed.
- J. Li, Y. Dai, S. Wang, C. Han and K. Xu, Sens. Actuators, B, 2016, 232, 1–8 CrossRef CAS.
- Y. J. Zhu, W. J. Li, Z. Y. Hong, A. N. Tang and D. M. Kong, J. Mater. Chem. B, 2017, 5, 9229–9237 RSC.
- H. H. Wang, C. A. J. Lin, C. H. Lee, Y. C. Lin, Y. M. Tseng, C. L. Hsieh, C. H. Chen, C. H. Tsai, C. T. Hsieh, J. L. Shen, W. H. Chan, W. H. Chang and H. I. Yeh, ACS Nano, 2011, 5, 4337–4344 CrossRef CAS PubMed.
- Z. Qing, A. Bai, S. Xing, Z. Zou, X. He, K. Wang and R. Yang, Biosens. Bioelectron., 2019, 137, 96–109 CrossRef CAS PubMed.
- Y. Su, T. Xue, Y. Liu, J. Qi, R. Jin and Z. Lin, Nano Res., 2019, 12, 1251–1265 CrossRef CAS.
- A. Fernando, K. L. D. M. Weerawardene, N. V. Karimova and C. M. Aikens, Chem. Rev., 2015, 115, 6112–6216 CrossRef CAS PubMed.
- Z. Liu, Z. Wu, Q. Yao, Y. Cao, O. J. H. Chai and J. Xie, Nano Today, 2021, 36, 101053 CrossRef CAS.
- L. Shang, S. Dong and G. U. Nienhaus, Nano Today, 2011, 6, 401–418 CrossRef CAS.
- M. Yang, X. Chen, Y. Su, H. Liu, H. Zhang, X. Li and W. Xu, Front. Chem., 2020, 8, 601621 CrossRef CAS PubMed.
- T. Q. Yang, B. Peng, B. Q. Shan, Y. X. Zong, J. G. Jiang, P. Wu and K. Zhang, Nanomaterials, 2020, 10(2), 261 CrossRef CAS PubMed.
- J. Kong, W. Zhang, Y. Wu and M. Zhou, Aggregate, 2022, 3, e207 CrossRef CAS.
- R. Jin, H. Qian, Z. Wu, Y. Zhu, M. Zhu, A. Mohanty and N. Garg, J. Phys. Chem. Lett., 2010, 1, 2903–2910 CrossRef CAS.
- L. A. Angel, L. T. Majors, A. C. Dharmaratne and A. Dass, ACS Nano, 2010, 4, 4691–4700 CrossRef CAS PubMed.
- T. Udaya Bhaskara Rao and T. Pradeep, Angew. Chem., Int. Ed., 2010, 49, 3925–3929 CrossRef CAS PubMed.
- C. I. Richards, S. Choi, J. C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y. L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038 CrossRef CAS PubMed.
- C. Zhou, C. Sun, M. Yu, Y. Qin, J. Wang, M. Kim and J. Zheng, J. Phys. Chem. C, 2010, 114, 7727 CrossRef CAS PubMed.
- L. Luo, Z. Liu, X. Du and R. Jin, Commun. Chem., 2023, 6, 1–6 CrossRef PubMed.
- M. Zhu, W. T. Eckenhoff, T. Pintauer and R. Jin, J. Phys. Chem. C, 2008, 112, 14221–14224 CrossRef CAS.
- S. Malola, M. F. Matus and H. Häkkinen, J. Phys. Chem. C, 2023, 127, 16553–16559 CrossRef CAS.
- Z. Liasi, L. Jensen and K. V. Mikkelsen, J. Chem. Theory Comput., 2024, 20, 937–945 CrossRef CAS PubMed.
- V. Bonac, M. Perić and Ž Sanader, J. Phys. Chem. Lett., 2018, 9, 2584–2589 CrossRef PubMed.
- P. G. Lisinetskaya and R. Mitrić, J. Phys. Chem. Lett., 2019, 10, 7884–7889 CrossRef CAS PubMed.
- Z. V. Reveguk, V. A. Pomogaev, M. A. Kapitonova, A. A. Buglak and A. I. Kononov, J. Phys. Chem. C, 2021, 125, 3542–3552 CrossRef CAS.
- D. J. E. Huard, A. Demissie, D. Kim, D. Lewis, R. M. Dickson, J. T. Petty and R. L. Lieberman, J. Am. Chem. Soc., 2019, 141, 11465–11470 CrossRef CAS PubMed.
- C. Cerretani, H. Kanazawa, T. Vosch and J. Kondo, Angew. Chem., Int. Ed., 2019, 58, 17153–17157 CrossRef CAS PubMed.
- C. Cerretani, J. Kondo and T. Vosch, RSC Adv., 2020, 10, 23854–23860 RSC.
- C. Cerretani, J. Kondo and T. Vosch, CrystEngComm, 2020, 22, 8136–8141 RSC.
- G. Y. Lan, W. Y. Chen and H. T. Chang, Analyst, 2011, 136, 3623–3628 RSC.
- J. T. Petty, C. Fan, S. P. Story, B. Sengupta, A. St. John Iyer, Z. Prudowsky and R. M. Dickson, J. Phys. Chem. Lett., 2010, 1, 2524–2529 CrossRef CAS PubMed.
- G. Romolini, C. Cerretani, V. Rück, M. B. Liisberg, C. B. Mollerup and T. Vosch, Nanoscale, 2024, 16, 12559–12566 RSC.
- Y. Zhong, Z. Wu, X. Bai, Y. Zhang and J. Xie, Mater. Today, 2024, 76, 72–93 CrossRef CAS.
- A. Gonzàlez-Rosell, C. Cerretani, P. Mastracco, T. Vosch and S. M. Copp, Nanoscale Adv., 2021, 3, 1230–1260 RSC.
- Y. T. Su, G. Y. Lan, W. Y. Chen and H. T. Chang, Anal. Chem., 2010, 82, 8566–8572 CrossRef CAS PubMed.
- I. Díez, M. Pusa, S. Kulmala, H. Jiang, A. Walther, A. S. Goldmann, A. H. E. Müller, O. Ikkala and R. H. A. Ras, Angew. Chem., Int. Ed., 2009, 48, 2122–2125 CrossRef PubMed.
- Y. Teng, X. Yang, L. Han and E. Wang, Chem. – Eur. J., 2014, 20, 1111–1115 CrossRef CAS PubMed.
- A. A. Makarova, E. V. Grachova, V. S. Neudachina, L. V. Yashina, A. Blüher, S. L. Molodtsov, M. Mertig, H. Ehrlich, V. K. Adamchuk, C. Laubschat and D. V. Vyalikh, Sci. Rep., 2015, 5, 8710 CrossRef CAS PubMed.
- A. Avula, A. Galor, P. Blackwelder, M. Carballosa-Gautam, A. S. Hackam, B. Jeng and N. Kumar, Cornea, 2017, 36, 752 CrossRef PubMed.
- Y.-X. Lin and C.-W. Chang, RSC Adv., 2019, 9, 26061–26066 RSC.
- I. L. Volkov, A. Smirnova, A. A. Makarova, Z. V. Reveguk, R. R. Ramazanov, D. Y. Usachov, V. K. Adamchuk and A. I. Kononov, J. Phys. Chem. B, 2017, 121, 2400–2406 CrossRef CAS PubMed.
- D. Beasock and K. A. Afonin, Methods Mol. Biol., 2023, 2709, 163 CrossRef CAS PubMed.
- L. Rolband, L. Yourston, M. Chandler, D. Beasock, L. Danai, S. Kozlov, N. Marshall, O. Shevchenko, A. V. Krasnoslobodtsev and K. A. Afonin, Molecules, 2021, 26(13), 4045 CrossRef CAS PubMed.
- M. Zhou, T. Higaki, G. Hu, M. Y. Sfeir, Y. Chen, D. E. Jiang and R. Jin, Science, 2019, 364, 279–282 CrossRef CAS PubMed.
- M. Berdakin, M. Taccone, K. J. Julian, G. Pino and C. G. Sánchez, J. Phys. Chem. C, 2016, 120, 24409–24416 CrossRef CAS.
- P. R. Oneill, E. G. Gwinn and D. K. Fygenson, J. Phys. Chem. C, 2011, 115, 24061–24066 CrossRef CAS.
- M. Yang, L. Zhu, W. Yang and W. Xu, Coord. Chem. Rev., 2023, 491, 215247 CrossRef CAS.
- S. M. Swasey, S. M. Copp, H. C. Nicholson, A. Gorovits, P. Bogdanov and E. G. Gwinn, Nanoscale, 2018, 10, 19701–19705 RSC.
- Y. Chen, T. Yang, H. Pan, Y. Yuan, L. Chen, M. Liu, K. Zhang, S. Zhang, P. Wu and J. Xu, J. Am. Chem. Soc., 2014, 136, 1686–1689 CrossRef CAS PubMed.
- I. Díez, R. H. A. Ras, M. I. Kanyuk and A. P. Demchenko, Phys. Chem. Chem. Phys., 2012, 15, 979–985 RSC.
- X. Liu, R. Hu, Z. Gao and N. Shao, Langmuir, 2015, 31, 5859–5867 CrossRef CAS PubMed.
- J. D. Clogston and A. K. Patri, Methods Mol. Biol., 2011, 697, 63–70 CrossRef CAS PubMed.
- X. Bu, Y. Fu, X. Jiang, H. Jin and R. Gui, Microchim. Acta, 2020, 187, 1–10 CrossRef.
- Y. Tao, K. Yi, H. Wang, K. Li and M. Li, Sens. Actuators, B, 2022, 361, 131711 CrossRef CAS.
- T. Zheng, S. Bott and Q. Huo, ACS Appl. Mater. Interfaces, 2016, 8, 21585–21594 CrossRef CAS.
- T. Wang, K. Jiang, Y. Wang, L. Xu, Y. Liu, S. Zhang, W. Xiong, Y. Wang, F. Zheng and J. J. Zhu, Chem. Sci., 2024, 15, 1829–1839 RSC.
- M. Tavakkoli Yaraki and Y. N. Tan, Sens. Int., 2020, 1, 100049 CrossRef.
- L. Liu, Q. Zhang, F. Li, M. Wang, J. Sun and S. Zhu, Spectrochim. Acta, Part A, 2020, 1, 100049 Search PubMed.
- S. M. van de Looij, E. R. Hebels, M. Viola, M. Hembury, S. Oliveira and T. Vermonden, Bioconjugate Chem., 2022, 33, 4–23 CrossRef CAS.
- X. Ouyang, M. Wang, L. Guo, C. Cui, T. Liu, Y. Ren, Y. Zhao, Z. Ge, X. Guo, G. Xie, J. Li, C. Fan and L. Wang, Angew. Chem., 2020, 132, 11934–11942 CrossRef.
- Y. Zhang, Y. Liao, X. Yin, Y. Zhang, Z. Yang, H. Wang, W. Yang and P. Pang, Microchem. J., 2023, 189, 108544 CrossRef CAS.
- J. Li, M. Chen, Q. Jiang, W. Zhang, Y. Lan, M. M. Ahmed, C. Ma, J. Huang and Q. Xu, Anal. Chem., 2024, 96, 9209–9217 CrossRef CAS PubMed.
- D. Sabarinathan, A. S. Sharma, M. Murugavelu, B. Kirubasankar, I. Balusamy, Z. Han, H. Li and Q. Chen, Heliyon, 2023, 9, e15655 CrossRef CAS.
- Z. Qiao, Y. Yan and S. Bi, Sens. Actuators, B, 2022, 352, 131073 CrossRef CAS.
- N. Chen, C. Gong and H. Zhao, Sci. Total Environ., 2023, 882, 163559 CrossRef CAS PubMed.
- L. Ning, Y. Li, Z. Zhang, Y. Zhou, L. Yang, Q. Yu, F. Yu and Z. Tong, Appl. Biochem. Biotechnol., 2023, 195, 6334–6344 CrossRef CAS.
- S. Yang, C. Li, H. Zhan, R. Liu, W. Chen, X. Wang and K. Xu, J. Nanobiotechnol., 2023, 21, 22 CrossRef CAS.
- C. Yan, L. Mu, M. Mei, Y. Wang, G. She and W. Shi, Anal. Chem., 2023, 95, 6915–6922 CrossRef CAS PubMed.
- Z. Yang, L. Hu, K. Ning, Y. Wu and J. Liang, Food Chem., 2023, 413, 135428 CrossRef CAS PubMed.
- X. Ouyang, N. Jia, J. Luo, L. Li, J. Xue, H. Bu, G. Xie and Y. Wan, JACS Au, 2023, 3, 2566–2577 CrossRef CAS PubMed.
- D. Kim, S. J. Kim, J. Jeong, S. Han, H. Kim, S. Lee, I. Choi, J. Hong, J. O. Jin and J. B. Lee, ACS Nano, 2024, 18, 1744–1755 CrossRef CAS PubMed.
- T. Wang, K. Jiang, Y. Wang, L. Xu, Y. Liu, S. Zhang, W. Xiong, Y. Wang, F. Zheng and J. J. Zhu, Chem. Sci., 2023, 15, 1829–1839 RSC.
- T. Qing, X. He, D. He, Z. Qing, K. Wang, Y. Lei, T. Liu, P. Tang and Y. Li, Talanta, 2016, 161, 170–176 CrossRef CAS PubMed.
- Z. Y. Li, Y. T. Wu and W. L. Tseng, ACS Appl. Mater. Interfaces, 2015, 7, 23708–23716 CrossRef CAS PubMed.
- W. Y. Chen, G. Y. Lan and H. T. Chang, Anal. Chem., 2011, 83, 9450–9455 CrossRef CAS PubMed.
- W. Yu, X. Lin, N. Duan, Z. Wang and S. Wu, Anal. Chim. Acta, 2023, 1244, 340846 CrossRef CAS PubMed.
- S. Mayuri and N. S. Jha, Microchim. Acta, 2023, 190, 1–13 CrossRef PubMed.
- L. Luo, Y. Xing, Y. Fu, L. Li, X. Yang, Y. Xue, J. Luo, H. Bu, F. Chen and X. Ouyang, J. Colloid Interface Sci., 2024, 660, 1–9 CrossRef CAS PubMed.
- X. Mao, S. Liu, C. Yang, F. Liu, K. Wang and G. Chen, Anal. Chim. Acta, 2016, 909, 101–108 CrossRef CAS PubMed.
- J. Pang, Y. Lu, X. Gao, L. He, J. Sun, F. Yang and Y. Liu, Mikrochim. Acta, 2020, 187(12), 672 CrossRef CAS PubMed.
- X. Ouyang, Y. Wu, L. Guo, L. Li, M. Zhou, X. Li, T. Liu, Y. Ding, H. Bu, G. Xie, J. Shen, C. Fan and L. Wang, Angew. Chem., Int. Ed., 2023, 62(21), e202300893 CrossRef CAS PubMed.
- C. Zhang, M. Wu, S. Hu, S. Shi, Y. Duan, W. Hu and Y. Li, Anal. Chem., 2023, 95, 11978–11987 CrossRef CAS PubMed.
- N. N. Wu, L. G. Chen and H. B. Wang, Appl. Spectrosc., 2023, 77, 1206–1213 CrossRef CAS PubMed.
- H. C. Yeh, J. Sharma, I. M. Shih, D. M. Vu, J. S. Martinez and J. H. Werner, J. Am. Chem. Soc., 2012, 134, 11550–11558 CrossRef CAS PubMed.
- H. C. Yeh, J. Sharma, J. J. Han, J. S. Martinez and J. H. Werner, Nano Lett., 2010, 10, 3106–3110 CrossRef CAS PubMed.
- H. C. Yeh, J. Sharma, J. J. Han, J. S. Martinez and J. H. Werner, IEEE Nanotechnol. Mag., 2011, 5, 28–33 Search PubMed.
- J. Sharma, H. C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2011, 47, 2294–2296 RSC.
- Y. Guo, L. Zhang, S. Zhang, Y. Yang, X. Chen and M. Zhang, Biosens. Bioelectron., 2015, 63, 61–71 CrossRef CAS PubMed.
- J. Huang, X. Su and Z. Li, Biosens. Bioelectron., 2017, 96, 127–139 CrossRef CAS PubMed.
- Q. Lou, F. Lai, J. Li, K. Mao, H. Wan and Y. He, Apoptosis, 2024, 2024, 1–26 Search PubMed.
- C. J. Coates and E. M. Costa-Paiva, Subcell. Biochem., 2020, 94, 233–250 CAS.
- H. Deng, S. Zhu, H. Yang, H. Cui, H. Guo, J. Deng, Z. Ren, Y. Geng, P. Ouyang, Z. Xu, Y. Deng and Y. Zhu, Biol. Trace Elem. Res., 2011, 1, 3 Search PubMed.
- G. Y. Lan, C. C. Huang and H. T. Chang, Chem. Commun., 2010, 46, 1257–1259 RSC.
- M. Zhang and B. C. Ye, Analyst, 2011, 136, 5139–5142 RSC.
- M. R. Knecht and M. Sethi, Anal. Bioanal. Chem., 2009, 394, 33–46 CrossRef CAS PubMed.
- Z. Qing, L. Zhu, X. Li, S. Yang, Z. Zou, J. Guo, Z. Cao and R. Yang, Environ. Sci. Technol., 2017, 51, 11884–11890 CrossRef CAS PubMed.
- W. Guo, J. Yuan and E. Wang, Chem. Commun., 2009, 3395–3397 RSC.
- J. L. MacLean, K. Morishita and J. Liu, Biosens. Bioelectron., 2013, 48, 82–86 CrossRef CAS PubMed.
- J. Yin, X. He, X. Jia, K. Wang and F. Xu, Analyst, 2013, 138, 2350–2356 RSC.
- Y. Guo, Z. Wang, W. Qu, H. Shao and X. Jiang, Biosens. Bioelectron., 2011, 26, 4064–4069 CrossRef CAS PubMed.
- L. J. Ou, X. Y. Li, H. W. Liu, L. J. Li and X. Chu, Anal. Sci., 2014, 30, 723–727 CrossRef CAS PubMed.
- J. Chen, J. Liu, Z. Fang and L. Zeng, Chem. Commun., 2012, 48, 1057–1059 RSC.
- S. Li, W. Cao, A. Kumar, S. Jin, Y. Zhao, C. Zhang, G. Zou, P. C. Wang, F. Li and X. J. Liang, New J. Chem., 2014, 38, 1546–1550 RSC.
- W. Zheng, N. Feng, Y. Wang, L. Noll, S. Xu, X. Liu, N. Lu, H. Zou, J. Gu, Y. Yuan, X. Liu, G. Zhu, J. Bian, J. Bai and Z. Liu, Food Chem. Toxicol., 2019, 126, 262–276 CrossRef CAS PubMed.
- Y. Sun, Y. Zhang and Z. Wang, Sens. Actuators, B, 2021, 347, 130661 CrossRef CAS.
- I. M. Khan, S. Zhao, S. Niazi, A. Mohsin, M. Shoaib, N. Duan, S. Wu and Z. Wang, Sens. Actuators, B, 2018, 277, 328–335 CrossRef CAS.
- M. Schirone, P. Visciano, R. Tofalo and G. Suzzi, Front. Microbiol., 2019, 10, 481870 Search PubMed.
- L. Ma, J. Wang, Y. Li, D. Liao, W. Zhang, X. Han and S. Man, J. Hazard. Mater., 2023, 443, 130234 CrossRef CAS PubMed.
- M. Yang, X. Chen, L. Zhu, S. Lin, C. Li, X. Li, K. Huang and W. Xu, ACS Appl. Mater. Interfaces, 2021, 13, 38647–38655 CrossRef CAS PubMed.
- W. Hou, F. Xia, G. Alfranca, H. Yan, X. Zhi, Y. Liu, C. Peng, C. Zhang, J. M. de la Fuente and D. Cui, Biomaterials, 2017, 120, 103–114 CrossRef CAS PubMed.
- S. Yu, Y. Zhou, Y. Sun, S. Wu, T. Xu, Y. C. Chang, S. Bi, L. P. Jiang and J. J. Zhu, Angew. Chem., Int. Ed., 2021, 60, 5948–5958 CrossRef CAS PubMed.
- J. Sun, K. Jiang, Y. Wang, Y. Liu, T. Wang, S. Ding, X. Zhang, W. Xiong, F. Zheng, H. Yang and J. J. Zhu, Adv. Healthc. Mater., 2023, 12, 2302016 CrossRef CAS PubMed.
- D. Jiang, Y. Pan, H. Yao, J. Sun, W. Xiong, L. Li, F. Zheng, S. Sun and J. J. Zhu, Anal. Chem., 2022, 94, 9074–9080 CrossRef CAS PubMed.
- F. Zheng, T. Meng, D. Jiang, J. Sun, H. Yao, J. J. Zhu and Q. Min, Angew. Chem., Int. Ed., 2021, 60, 21565–21574 CrossRef CAS PubMed.
- W. Jiang, B. Y. S. Kim, J. T. Rutka and W. C. W. Chan, Nat. Nanotechnol., 2008, 3, 145–150 CrossRef CAS PubMed.
- E. A. Sykes, J. Chen, G. Zheng and W. C. W. Chan, ACS Nano, 2014, 8, 5696–5706 CrossRef CAS PubMed.
- W. Yu, R. Liu, Y. Zhou and H. Gao, ACS Cent. Sci., 2020, 6, 100–116 CrossRef CAS PubMed.
- F. Semcheddine, N. El Islem Guissi, W. Liu, Tayyaba, L. Gang, H. Jiang and X. Wang, Mater. Horiz., 2021, 8, 2771–2784 RSC.
- Z. Wu, Q. Yao, O. J. H. Chai, N. Ding, W. Xu, S. Zang and J. Xie, Angew. Chem., Int. Ed., 2020, 59, 9934–9939 CrossRef CAS.
- X. Ran, Z. Wang, F. Pu, E. Ju, J. Ren and X. Qu, Mater. Horiz., 2021, 8, 1769–1775 RSC.
- W. Shu, X. Zhang, H. Tang, L. Wang, M. Cheng, J. Xu, R. Li and X. Ran, Anal. Chim. Acta, 2023, 1268, 341372 CrossRef CAS.
- Z. Zhou, Y. Li, W. Su, B. Gu, H. Xu, C. Wu, P. Yin, H. Li and Y. Zhang, Sens. Actuators, B, 2019, 280, 120–128 CrossRef CAS.
- E. Dervisevic, N. H. Voelcker, G. Risbridger, K. L. Tuck and V. J. Cadarso, Anal. Chem., 2022, 94, 1726–1732 CrossRef CAS.
- H. Li, H. Zhao, Z. Wang, F. Zhou and M. Lan, Microchem. J., 2022, 172, 106972 CrossRef CAS.
- G. Li, Y. Chen, F. Liu, W. Bi, C. Wang, D. Lu and D. Wen, Microsyst. Nanoeng., 2023, 9, 1–10 CrossRef.
- J. Vimali, Y. K. Yong, A. Murugesan, K. Vishnupriya, R. Ashwin, E. A. Daniel, P. Balakrishnan, S. Raju, M. Rosmawati, V. Velu, M. Larsson and E. M. Shankar, Front. Med., 2022, 9 DOI:10.3389/FMED.2022.1019230.
- J.-S. Lee, M. S. Han and C. A. Mirkin, Angew. Chem., 2007, 119, 4171–4174 CrossRef.
- J. Hu, T. Wang, J. Kim, C. Shannon and C. J. Easley, J. Am. Chem. Soc., 2012, 134, 7066–7072 CrossRef CAS.
- X. Yang, Y. Zhuo, S. Zhu, Y. Luo, Y. Feng and Y. Xu, Biosens. Bioelectron., 2015, 64, 345–351 CrossRef CAS.
- S. Paul, R. Clérac, N. G. R. Hearns and D. Ray, Cryst. Growth Des., 2009, 9, 4032–4040 CrossRef CAS.
- A. Singh, S. Patra, J. A. Lee, K. H. Park and H. Yang, Biosens. Bioelectron., 2011, 26, 4798–4803 CrossRef CAS.
- S. Lv, Q. Yao, J. Yi, J. Si, Y. Gao, S. Su and C. Zhu, JACS Au, 2024, 4, 2323–2334 CrossRef CAS PubMed.
- S. Javani, R. Lorca, A. Latorre, C. Flors, A. L. Cortajarena and Á. Somoza, ACS Appl. Mater. Interfaces, 2016, 8, 10147–10154 CrossRef CAS.
- N. Joshi, B. T. Ngwenya, I. B. Butler and C. E. French, J. Hazard. Mater., 2015, 287, 51–58 CrossRef CAS PubMed.
- M. Yang, X. Chen, L. Zhu, S. Lin, C. Li, X. Li, K. Huang and W. Xu, ACS Appl. Mater. Interfaces, 2021, 13, 38647–38655 CrossRef CAS.
- B. Sengupta, P. Adhikari, E. Mallet, R. Havner and P. Pradhan, Molecules, 2020, 25(16), 3631 CrossRef CAS.
- B. Zhang, Z. Yang, Y. Li, L. Ma, F. Li, X. Lv and G. Wen, RSC Adv., 2022, 12, 30024–30029 RSC.
- B. Zhang and C. Wei, Anal. Bioanal. Chem., 2020, 412, 2529–2536 CrossRef CAS PubMed.
- Y. Li, Z. Meng, Y. Liu and B. Zhang, RSC Adv., 2024, 14, 5594–5599 RSC.
- J. Li, G. Peng, Y. Yu, B. Lin, L. Zhang, M. Guo, Y. Cao and Y. Wang, Microchim. Acta, 2023, 190, 1–9 CrossRef PubMed.
- K. Zhang, K. Wang, X. Zhu, J. Zhang, L. Xu, B. Huang and M. Xie, Chem. Commun., 2013, 50, 180–182 RSC.
- P. Mastracco and S. M. Copp, Chem. Commun., 2023, 59, 10360–10375 RSC.
- E. Gwinn, D. Schultz, S. M. Copp and S. Swasey, Nanomaterials, 2015, 5, 180–207 CrossRef PubMed.
- C. I. Richards, S. Choi, J. C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y. L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038–5039 CrossRef CAS PubMed.
- D. Schultz and E. G. Gwinn, Chem. Commun., 2012, 48, 5748–5750 RSC.
- W. Zhang, Z. Sun, Y. Tian, Y. Mou, Y. Guo, X. Sun and F. Li, Sens. Actuators, B, 2024, 406, 135427 CrossRef CAS.
- Y. J. Chen, B. Groves, R. A. Muscat and G. Seelig, Nat. Nanotechnol., 2015, 10, 748–760 CrossRef CAS PubMed.
- S. Auslände, D. Ausländer, M. Müller, M. Wieland and M. Fussenegger, Nature, 2012, 487, 123–127 CrossRef PubMed.
- J. Yin, J. Wang, R. Niu, S. Ren, D. Wang and J. Chao, Chem. Res. Chin. Univ., 2020, 36, 219–226 CrossRef CAS.
- G. Hong, A. L. Antaris and H. Dai, Nat. Biomed. Eng., 2017, 1, 1–22 CrossRef.
- P. Mastracco, A. Gonzàlez-Rosell, J. Evans, P. Bogdanov and S. M. Copp, ACS Nano, 2022, 16, 16322–16331 CrossRef CAS PubMed.
- W. Phanchai, J. Thonghlueng and T. Puangmali, ACS Appl. Nano Mater., 2024, 7, 5554–5563 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |
