Open Access Article
Open Access ArticleRecent progress in chemoenzymatic synthesis of human glycans
Shengzhou
Ma
ab,
Jinhua
Gao
a,
Yinping
Tian
a and
Liuqing
Wen
 *ab
*ab
aCarbohydrate-Based Drug Research Center, State Key Laboratory of Chemical Biology, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China. E-mail: lwen@simm.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 22nd August 2024
Abstract
Glycan is an essential cell component that usually exists in either a free form or a glycoconjugated form. Glycosylation affects the regulatory function of glycoconjugates in health and disease development, indicating the key role of glycan in organisms. Because of the complexity and diversity of glycan structures, it is challenging to prepare structurally well-defined glycans, which hinders the investigation of biological functions at the molecular level. Chemoenzymatic synthesis is an attractive approach for preparing complex glycans, because it avoids tedious protecting group manipulations in chemical synthesis and ensures high regio- and stereo-selectivity of glucosides during glycan assembly. Herein, enzymes, such as glycosyltransferases (GTs) and glycosidases (GHs), and sugar donors involved in the chemoenzymatic synthesis of human glycans are initially discussed. Many state-of-the-art chemoenzymatic methodologies are subsequently displayed and summarized to illustrate the development of synthetic human glycans, for example, N- and O-linked glycans, human milk oligosaccharides, and glycosaminoglycans. Thus, we provide an overview of recent chemoenzymatic synthetic designs and applications for synthesizing complex human glycans, along with insights into the limitations and perspectives of the current methods.
Introduction
Glycan is one of the most complicated and diverse molecules found in organisms, and it is free or covalently linked to other molecules to form glycoconjugates, such as glycoproteins, proteoglycans, glycolipids, and glycoRNAs.1,2 Glycosylation regulates various biological processes, including signal transduction, cell migration, inflammation, immune evasion, cancer metastasis, and bacterial and viral infection.3–6 However, the mechanism of action used by glycans in many biological events remains unclear. One substantial reason for this is the lack of structurally well-defined glycans. The complexity and diversity of glycan structures considerably hinder the synthesis of structurally well-defined glycans. In contrast to the synthesis of nucleic acids and proteins, the biosynthesis of glycans is a non-template-driven process. The expression levels of glycosyltransferases (GTs) and glycosidases (GHs)7 and the availability of sugar nucleotides can affect the formation of glycans in vivo.8–10Extraction from natural sources11 and artificial synthesis in vitro are the two main approaches for obtaining glycans. Because of the low abundance and micro-heterogeneity of endogenous glycans, it is difficult to obtain structurally well-defined glycans from nature for application in biological studies, despite using state-of-the-art analytical tools. In contrast, artificial synthesis is an attractive alternative method for preparing glycans. Chemical synthesis can be used to produce various natural or non-natural glycans and glycan derivatives.12–14 However, it is difficult and time-consuming to create scale-prepared complex glycans because of the tedious protection of group manipulations and challenging stereo-selective glycosylation that must be performed.
Enzymatic glycosylation is another synthetic method used to produce glycans.15,16 One of its advantages is its ability to produce highly regio- and stereo-selective glycosidic bonds without protecting group participation. However, one enzyme, for example one GT, can usually catalyze the formation of one glycosidic bond. Thus, the availability of enzymes limits the application of this method. To address these limitations, a chemoenzymatic synthesis strategy combining chemical synthesis and enzymatic glycosylation was developed and has been widely used in the past few decades to synthesize complex glycans.17,18 There is no need for tedious protecting group manipulations in chemical synthesis with this novel synthetic method, and it ensures high regio- and stereo-selectivity of glucosides during glycan assembly. As a result, it offers great potential for the efficient synthesis of highly complex branched glycans (Fig. 1). This mini-review provides an overview of the typical chemoenzymatic synthetic methods used to produce complex human glycans in recent years.
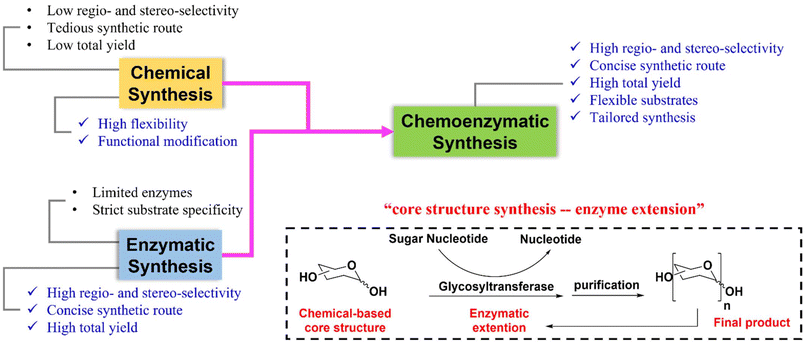 | ||
| Fig. 1 Comparison between chemical or enzymatic strategies and chemoenzymatic strategy, and a schematic illustration of the ‘Core structure synthesis – enzyme extension’ synthetic strategy. | ||
Enzymes and sugar donors
GTs and GHs are two major enzymes applied to the (chemo)enzymatic synthesis of glycans. GTs use nucleotide sugars as activated donors to natively catalyze the biosynthesis of glycans and glycoconjugates,19 and are the most widely used in glycan synthesis. The Carbohydrate-Active Enzymes (CAZy) database (https://www.cazy.org) groups more than 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 GTs into 135 families (as of 30th May 2024), with an additional 60
000 GTs into 135 families (as of 30th May 2024), with an additional 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 GT sequences remaining unclassified. Although hundreds of thousands of GTs have been discovered, the members successfully used in the chemoenzymatic synthesis of human glycans are mainly glucosyltransferases (GlcTs), N-acetylglucosaminyltransferases (GlcNAcTs), glucouronyltransferases (GlcATs), galactosyltransferases (GalTs), N-acetylgalactosaminyltransferases (GalNAcTs), mannosyltransferases (ManTs), sialyltransferases (SiaTs), and fucosyltransferases (FucTs).
000 GT sequences remaining unclassified. Although hundreds of thousands of GTs have been discovered, the members successfully used in the chemoenzymatic synthesis of human glycans are mainly glucosyltransferases (GlcTs), N-acetylglucosaminyltransferases (GlcNAcTs), glucouronyltransferases (GlcATs), galactosyltransferases (GalTs), N-acetylgalactosaminyltransferases (GalNAcTs), mannosyltransferases (ManTs), sialyltransferases (SiaTs), and fucosyltransferases (FucTs).
GTs used in chemoenzymatic synthesis should have the following ideal properties: high expression level, high stability during storage and application, high activity, high regio- and stereo-selectivity, minimal undesirable activities, and promiscuity towards modified substrate analogs.20 However, it is unrealistic to expect that all the ideal properties will exist for wild-type GTs. To address this, prokaryotic and eukaryotic expression systems21,22 have been constructed and combined with the enzyme engineering technique to generate GTs and GT mutants better suited for synthesis.
GHs, also called glycoside hydrolases, are enzymes that hydrolyze the glycosidic linkages in glycans and glycoconjugates.23 GHs are usually classified into exo-glycosidases and endo-glycosidases based on their cleavage sites. Exo-glycosidases individually hydrolyze nonreducing saccharide units, whereas endo-glycosidases hydrolyze internal glycosidic bonds to cleave more than one sugar residue.24 The hydrolysis feature of GHs has been widely utilized as a complementary tool in the chemoenzymatic synthesis of glycans. For example, a well-designed N-glycan precursor was treated with different GHs and chemical deprotection to provide a high mannose-type N-glycan library.25 Most importantly, the hydrolytic activity of GHs is reversible, which implies that GHs possess the potential to synthesize glycosides.
Many oligosaccharides and glucosides have been successfully synthesized utilizing GHs that intrinsically are able to perform transglycosylation capability.26–28 In addition, many mutational GHs presented many more highly efficient transglycosylations in the synthesis of glycans,29,30 glycoproteins,31 and the glycoengineering of antibodies.32 Mutational GHs have also been used for cell surface transglycosylation for the site-specific analysis of glycosylation of glycoproteins.33–35 Furthermore, some GHs can be engineered into mutants, called glycosynthases, which are used in glycan synthesis and prevent glycosidic bond hydrolysis.36 Many excellent reports provide a superb overview of the application of GHs and glycosynthases in biocatalytic glycoside synthesis.37–39 The current review focuses on the development of the biomimetic synthesis of human glycans using activated donors and GTs.
Monosaccharides need to be converted to the activated form (sugar donors) before being recognized and transferred by GTs to build glycans.40 Sugar donors usually include sugar nucleotides, dolichol phosphate sugars, and specific monosaccharide analogs. Sugar nucleotides are the most commonly used sugar donors in the biosynthesis and chemoenzymatic synthesis of glycans. Most GTs recognize sugar nucleotides as the donor for glycosylation,41 while a few GTs recognize dolichol phosphate sugar, which is present in the initial stages of glycan biosynthesis in the endoplasmic reticulum.42 In addition, phosphorylated sugars and fluoride sugars are often recognized by glycosynthases.43,44
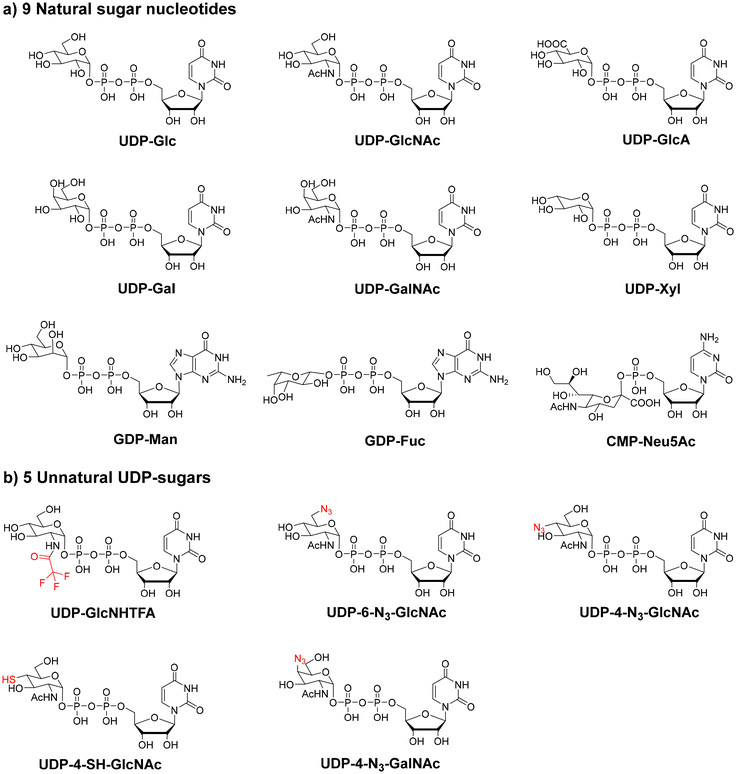
In humans, there are nine common sugar nucleotides, namely, UDP-Glc, UDP-GlcNAc, UDP-GlcA, UDP-Gal, UDP-GalNAc, UDP-Xyl, GDP-Man, GDP-Fuc, and CMP-Neu5Ac (Fig. 2a). The formation of sugar nucleotides in vivo is complicated and involves multiple pathways with multi-enzymes.45,46 Therefore, the biomimetic synthesis of sugar nucleotides in vitro is challenging. To achieve the large-scale preparation of sugar nucleotides, one-pot multi-enzyme strategies47,48 and in situ sugar nucleotide regeneration systems49,50 have been developed. Wen and coworkers reported an efficient cofactor-driven cascade conversion strategy for preparing several rare sugar nucleotides10,51,52 that are important synthetic precursors for many rare sugar-containing glycans.
Unnatural sugar nucleotides (structural analogs of sugar nucleotides) have been designed and prepared for glycan synthesis purposes. They can be recognized by GTs, which then incorporate unnatural sugar residues into glycan chains. Glycans containing unnatural sugar residues can be further manipulated to achieve precise structural control. For example, uridine 5′-diphosphate-N-trifluoroacetylglucosamine (UDP-GlcNHTFA) has been widely used as an analog of UDP-GlcNAc in glycan synthesis.53–55 In addition, UDP-6-N3-GlcNAc, UDP-4-N3-GlcNAc, UDP-4-SH-GlcNAc, and UDP-4-N3-GalNAc were also successfully prepared and used in the chemoenzymatic synthesis of glycans (Fig. 2b).48,56,57 Another application of unnatural sugar nucleotides is in situ synthesis of cell surface glycans to achieve glycan visualization and labeling.58–60
N-Linked glycans
Human N-glycans can be structurally divided into complex N-glycan, hybrid N-glycan, and high-mannose N-glycan, which have a common core pentasaccharide structure (Man3GlcNAc2) attached to the typical Asn-X-Ser/Thr sequon of protein with an N-glycosidic bond.5,61–64 Obtaining the core pentasaccharide is a prerequisite for the chemoenzymatic synthesis of N-glycans. The current chemoenzymatic methods involve the initial preparation of the core structure, and then its enzymatic extension.65–68 Following the concept of ‘core structure synthesis-enzyme extension’, numerous efficient chemoenzymatic synthetic methods have been developed for the synthesis of symmetric and asymmetric N-glycans.Wong and co-workers were among the first to elongate N-glycans using GTs. They later designed and developed a synthetic strategy called the modular assembly method.69,71,72 In this method, acceptor and donor modules are chemically synthesized. Thereafter, the coupling of both modules installs the core structure of N-glycans. Finally, enzymatic extension and modification are performed to achieve the production of targeted N-glycans (Fig. 3a). Although chemical manipulations are challenging, this method allows for the synthesis of many complex N-glycans. N-Glycans are usually asymmetrically substituted with a unique saccharide appendage at each branching point.5,73
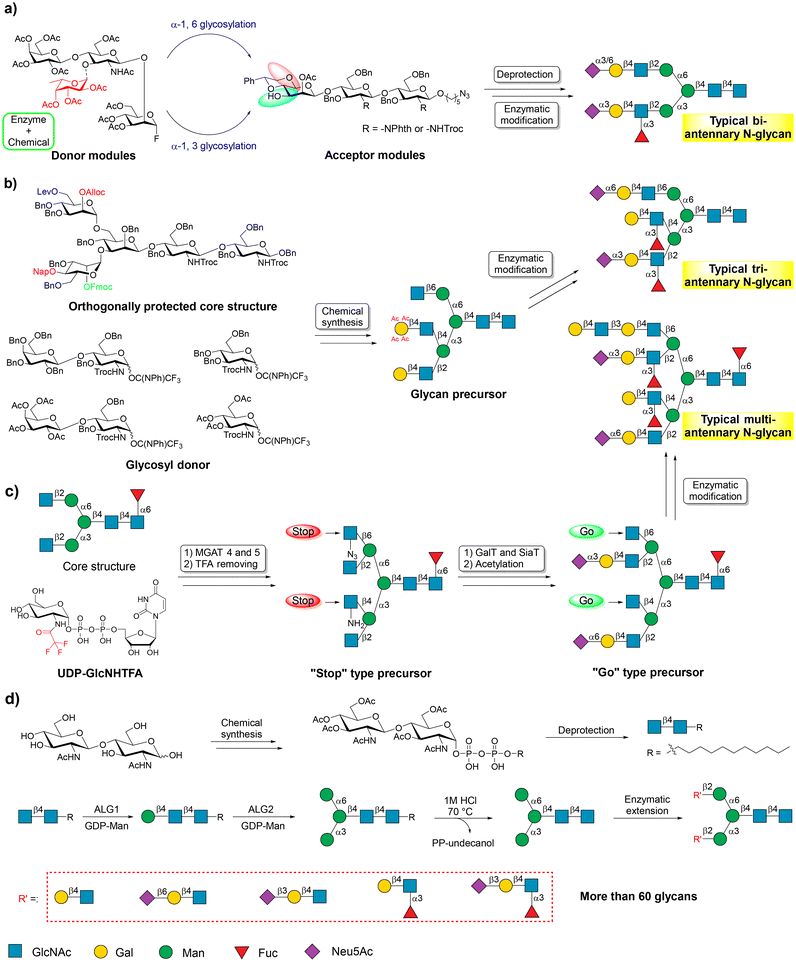 | ||
Fig. 3 Chemoenzymatic strategies for the synthesis of N-glycans. (a) Modular assembly strategy for the synthesis of N-glycans. Reproduced with permission from Shivatare et al.69 Copyright 2016, Nature Publishing Group. (b) Orthogonally protected synthetic strategy along with enzymatic modification for the synthesis of asymmetrically branched N-glycans. Reproduced with permission from Wang et al.65 Copyright 2013, American Association for the Advancement of Science. (c) The “stop and go” strategy for chemoenzymatic synthesis of complex N-glycans. Reproduced with permission from Liu et al.55 Copyright 2018, Nature Publishing Group. (d) Large-scale core structure-based concise chemoenzymatic synthesis of N-glycans.70 Copyright 2024, Elsevier Inc. The representation symbol of glucosamine should be “![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)  ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ” according to the guidance of Symbol Nomenclature for Glycans (SNFG). We use “ ” according to the guidance of Symbol Nomenclature for Glycans (SNFG). We use “ ” in place of “ ” in place of “![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)  ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ” in (c) to clearly show the conversion of functional groups. ” in (c) to clearly show the conversion of functional groups. | ||
Many efforts have been made to synthesize asymmetrical N-glycans, resulting in their easy access for experimentation.65,74,75 In 2013, Boons and co-workers reported a general strategy for the chemoenzymatic synthesis of asymmetrically branched N-glycans.65 In this work, a core pentasaccharide modified by orthogonal protecting groups, including levulinoyl (Lev), fluorenylmethyloxycarbonate (Fmoc), allyloxycarbonate (Alloc), and 2-naphthylmethyl (Nap) groups, at potential branching positions was synthesized as a common synthetic precursor. This allowed for the selective attachment of specific saccharide moieties by chemical glycosylation, followed by branch elongation by GTs to produce libraries of asymmetrical N-glycans (Fig. 3b). In the same year, Ito et al. reported a top-down chemoenzymatic strategy to produce a library of high mannose-type N-glycans.25 Similarly, other methods have been reported to synthesize various N-glycans.76–79
Trimming sialoglycopeptide (SGP) extracted from egg or egg yolk powder by GHs is another method used to obtain the core structure of N-glycans.80–82 Boons and co-workers developed a “stop and go” strategy, starting from SGP, to synthesize asymmetrical multi-antennary N-glycans.55 This method allows for the conversion of a bi-antennary heptasaccharide into multi-antennary N-glycans within ten or fewer chemical and enzymatic steps. In this work, UDP-GlcNHTFA was employed as an analog of UDP-GlcNAc to attach GlcNHTFA to the bi-antennary heptasaccharide from SGP through the catalysis of human mannosyl-glycoprotein N-acetylglucosaminyltransferases (MGAT4 and MGAT5), forming additional controllable arms on the core structure of N-glycan.
The GlcNHTFA can be converted to glucosamine (GlcNH2) under moderate basic conditions. GlcNH2 can further be converted to 2-deoxy-2-azido-glucose (GlcN3). The GlcNH2 and GlcN3 arms are not recognized by human β-1,4-galactosyltransferase (B4GalT1), thus allowing regio-selective galactosylation of GlcNAc. Finally, the GlcNH2 and GlcN3 residues can be easily converted back to GlcNAc, which is a suitable galactosylation site for B4GalT1 (Fig. 3c). Another study proved that the human N-acetylglucosaminyltransferases MGAT1 and MGAT2 can also use UDP-GlcNHTF as a donor. Thus, a new “stop and go” chemoenzymatic methodology was developed to prepare 32 asymmetrical bi-antennary N-glycans found in airway tissues.83
The large-scale production of core pentasaccharide, a key component for synthesizing various N-glycans, has been challenging due to difficulties in obtaining it through chemical synthesis or isolation from nature. Although many excellent studies have identified the catalytic roles of human mannosyltransferases Alg1 and Alg2, which are key enzymes in the biosynthesis of the core pentasaccharide, large-scale preparation of the core structure has been unsuccessful.84–86
To address this issue, Wen and coworkers recently developed a general platform for the concise chemoenzymatic synthesis of N-glycans.70 In this study, an inexpensive commercially available disaccharide GlcNAcβ1,4GlcNAc was chemically treated and coupled with an 11-carbon lipid tail (undecanol) to produce the common starting materials. This starter is well recognized as an acceptor by recombinant Alg1. Furthermore, the mannosylated starter is also recognized by recombinant Alg2, which is responsible for the installation of α-1,3- and α-1,6-mannose into the core pentasaccharide. Based on these, large-scale core pentasaccharides were efficiently synthesized using a few facile chemical and enzymatic steps. Starting from the synthetic core structure, more than 60 N-glycans were successfully synthesized (Fig. 3d).
In addition, many biologically related antibodies equipped with N-glycans have proven that N-glycosylation on antibodies exerts a significant role in their effector function.87–89 Therefore, chemoenzymatic methods have also been exploited to prepare a panel of N-glycans found on human serum immunoglobulin G (IgG). In 2020, Unverzagt and co-workers successfully galactosylated the bisecting GlcNAc of rare N-glycans identified in IgG.90 In this study, four multi-antennary N-glycans were treated with a bovine β-1,4-galactosyltransferase. Only bi-antennary bisected N-glycans can be galactosylated. Li and co-workers constructed a 64-membered IgG N-glycan library starting from SGP.91 The N-glycan remodeling of antibodies using enzymes, such as endo-β-N-acetylglucosaminidase mutants, for producing engineered antibodies with homogeneous N-glycan forms has been reviewed in recent years and is not discussed here.31,92,93
O-Linked glycans
O-GalNAc and O-mannose are two major O-linked glycan forms in humans.94,95 They are the focus of many chemoenzymatic strategies.96,97 A regioselective one-pot multienzyme (OPME) chemoenzymatic strategy was developed to systematically synthesize the sialyl Core 2 glycans.98 This study exploited the restricted acceptor specificity of sialyltransferases (Pasteurella multocida α-2,3-sialyltransferase 3) and the rational design of acceptors to successfully achieve the synthesis of regioselective sialylated Core 2 glycans (Fig. 4a).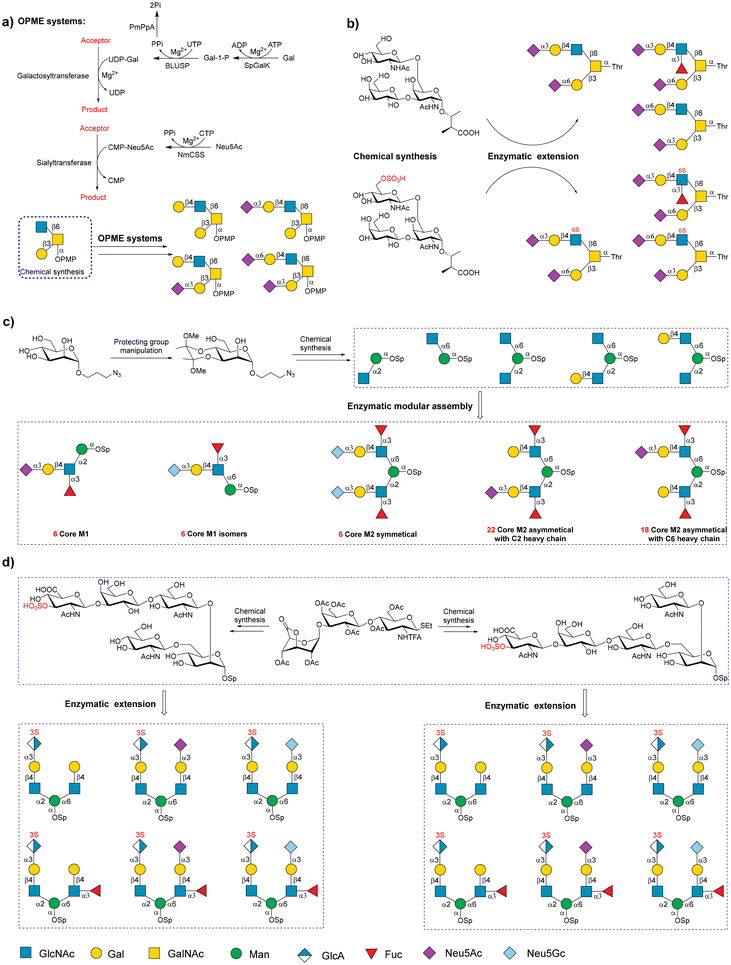 | ||
| Fig. 4 Chemoenzymatic strategies for the synthesis of O-GalNAc and O-mannose glycans. (a) Regioselective one-pot multienzyme (OPME) chemoenzymatic strategies for the synthesis of sialyl Core 2 glycans. SpGalK: Streptococcus pneumoniae TIGR4 galactokinase. BLUSP: UDP-sugar pyrophosphorylase from Bifidobacterium longum. PmPPA: pyrophosphatase from Pasteurella multocida. NmCSS: Neisseria meningitidis CMP-sialic acid synthetase. Reproduced with permission from Santra et al.98 Copyright 2018 American Chemical Society. (b) Diversity-oriented chemoenzymatic approach for the preparation of sulfated/nonsulfated Core 2 glycans. Reproduced with permission from Xu et al.99 Copyright 2021 American Chemical Society. (c) Chemoenzymatic assembly of mammalian O-mannosyl glycans. Reproduced with permission from Meng et al.101 Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (d) Chemoenzymatic synthesis of a sulfated HNK-1 epitope bearing O-mannosyl glycans. Reproduced with permission from Gao et al.102 Copyright 2019 American Chemical Society. | ||
In addition to the regioselective challenge, site-specific modification, such as sulfation of O-GalNAc glycans, complicates the synthesis process. Li and coworkers chemically synthesized sulfated and non-sulfated Core 2 trisaccharides as starters to enzymatically prepare over 30 structurally well-defined Core 2 glycans (Fig. 4b).99 In addition, other core structures of O-GalNAc were synthesized using a robust chemoenzymatic modular assembly strategy.100 This method focuses on exploiting three chemical building blocks to convergently assemble the O-GalNAc Core 1–4 and Core 6 structures.
Enzymatic decoration was then applied to create a series of O-GalNAc glycans. A total of 83 O-GalNAc glycans covering different natural glycan epitopes was synthesized in this study, and a glycan microarray was constructed to identify various lectins, anti-glycan antibodies, and serum samples from patients with colorectal cancer. Recently, three α-linked rare cores (Core 5, Core 7, and Core 8) and their sialylated forms were prepared by Li's group.103 The Photobacterium damselae α-2,6-sialyltransferase (Pd2,6ST) demonstrated restricted substrate specificity toward O-GalNAc glycan cores 5, 7, and 8, selectively installing α-2,6 sialic acid in the 6-OH of the internal GalNAc residue of Cores 5 and 8, and in the 6-OH of the terminal GalNAc residue of Core 7. These methods greatly facilitated the preparation of many structurally diverse O-GalNAc glycans, which further deepened the biological function study of O-GalNAc glycans.
The chemoenzymatic synthesis of O-mannosyl glycans is also performed using rational methodology designing. The O-mannosyl glycans are usually bi-antennary structures, in which two GlcNAc residues bearing further elaborations are each linked to core mannose with β-1,2- and β-1,6-glycosidic bonds.95 Achieving the regioselective chemoenzymatic synthesis of O-mannosyl glycans is challenging. Cao and coworkers successfully achieved the diversity-oriented assembly of 58 complex O-mannosyl glycans starting from five chemically synthesized core structures.101 Five synthesized core structures determined the regioselective sialylations and fucosylations of all of the complex glycans. Among these diverse glycans, 55 glycans were first synthesized (Fig. 4c).
Li and coworkers developed a scaffold synthesis/enzymatic extension strategy to efficiently synthesize 45 O-mannosyl glycans derived from human beings.104 Because muscular dystrophy-related α-dystroglycan (α-DG) is often mannosylated,105O-mannosyl α-DG is an attractive biomarker for disease in clinical settings.106 Peng and coworkers prepared several O-mannosylated glycopeptides using a microwave-assisted SPPS method.107 The human natural killer-1 (HNK-1) epitope, 3-S-GlcAβ1,3Galβ1,4GlcNAc trisaccharide, is a unique component of O-mannosyl glycans in brain tissues. Cao and coworkers first synthesized a series of HNK-1-bearing O-mannosyl glycans using a judicious chemoenzymatic synthesis approach.102 In this method, a chemically synthesized trisaccharide lactone donor was converted into three HNK-1-containing branched O-mannosyl precursors via chemical glycosylation, selective cleavage of lactone, and sulfation. These precursors were enzymatically elaborated, taking advantage of substrate promiscuities of bacterial GTs to produce various complex HNK-1-bearing O-mannosyl glycans (Fig. 4d).
Poly-LacNAc glycans
Poly-N-acetyl lactosamine (poly LacNAc) glycans consist of type II LacNAc repeating units and are common structural motifs of various complex human glycans, such as N- and O-linked glycans, and human milk oligosaccharides (HMOs). The fucosylation and sialylation of poly LacNAc glycans produce biologically related glycan epitopes, such as Lewis X or sialyl Lewis X. The synthesis of regioselective glycosylated poly LacNAc glycans is an attractive topic. Taking advantage of sequential enzymatic and chemical reactions, galactose oxidase, and reductive amination, Elling and coworkers chemoenzymatically synthesized a panel of branched LacNAc oligomers bearing LacNAc and/or N′,N′′-diacetyllactosamine (LacdiNAc) glycan epitopes (Fig. 5a).108 Oxidation by galactose oxidase converted LacNAc to 6′-aldehyde LacNAc, which can be recognized by Helicobacter pylori β-3-N-acetylglucosaminyltransferase (β3GlcNAcT) as the acceptor to elongate the LacNAc chain. The reserved aldehyde site was used for coupling with amine-linked glycans via reductive amination.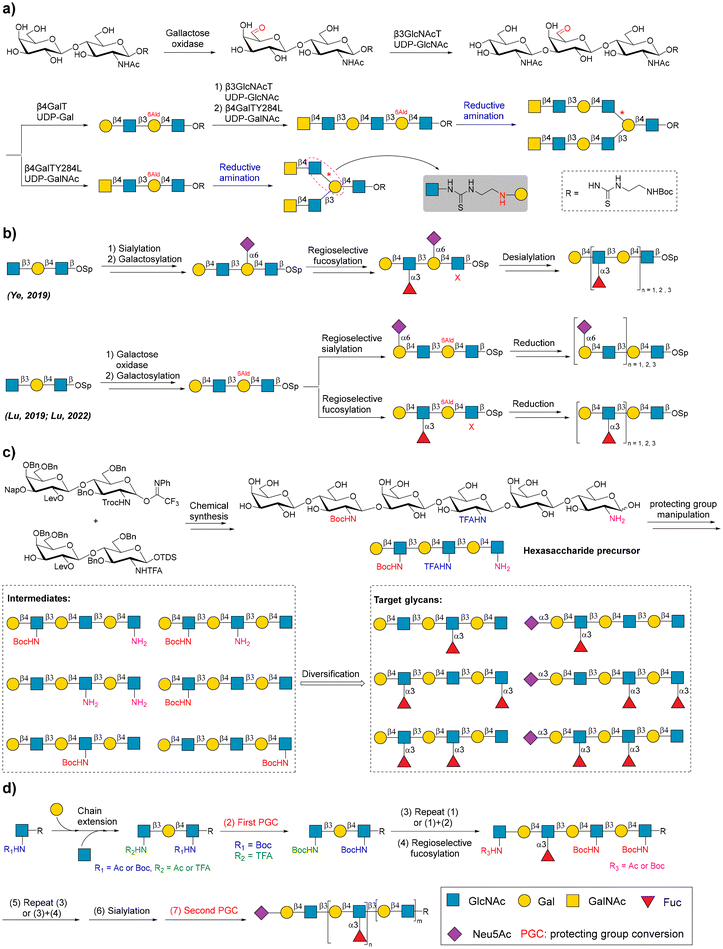 | ||
Fig. 5 Chemoenzymatic strategies for the synthesis of poly LacNAc glycans. (a) Chemoenzymatic synthesis of branched poly LacNAc glycans. Reproduced with permission from Laaf et al.108 Copyright 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) Glycosylation and GOase-controlled regioselective synthesis of poly LacNAc glycans. Reproduced with permission from Ye et al., Lu et al., and Lu et al.109–111 Copyright 2019, Nature Publishing Group. Copyright 2019 American Chemical Society. Copyright 2022 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (c) Unnatural substrate-controlled regioselective synthesis of poly LacNAc glycans. Reproduced with permission from Gagarinov et al.112 Copyright 2022 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (d) Acceptor-mediated regioselective fucosylation of poly LacNAc glycans. Reproduced with permission from Tseng et al.114 Copyright 2023 American Chemical Society. The representation symbol of glucosamine should be “![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)  ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ” according to the guidance of Symbol Nomenclature for Glycans (SNFG). We use “ ” according to the guidance of Symbol Nomenclature for Glycans (SNFG). We use “ ” in place of “ ” in place of “![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)  ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ” in (c) to clearly show the conversion of functional groups. ” in (c) to clearly show the conversion of functional groups. | ||
In 2019, Cao's group described a reprogrammed enzymatic assembly line for regioselective fucosylation of the poly LacNAc backbone.109 In this study, the α-2,6-linked sialic acid on LacNAc repeating units was used as the protecting group against fucosylation of LacNAc repeating units. Thus, the fucosylation of poly-LacNAc glycans was precisely controlled in a site-specific manner. Finally, the unwanted sialic acid can be removed by the use of sialidase. In the same year, they also reported a general redox-controlled site-specific sialylation strategy.110 This strategy used galactose oxidase to selectively oxidize the C-6-hydroxyl group of Gal, solving the problem of regioselective sialylation of poly LacNAc glycans. Recently, this redox-controlled glycosylation strategy was used for the site-specific enzymatic fucosylation of many N- and O-linked glycans (Fig. 5b).111
The exploitation of protecting group manipulation-based glycosyl donors also achieved regioselective sialylation and fucosylation of poly LacNAc glycans. Boons and coworkers developed a regioselective chemoenzymatic strategy that produced various fucosylated and sialylated poly LacNAc glycans.112 In this study, a hexasaccharide precursor containing GlcNH2, GlcNHTFA, and GlcNHBoc residues was chemically synthesized, in which unnatural GlcNH2, GlcNHTFA, and GlcNHBoc moieties were easily converted into natural GlcNAc. Recombinant fucosyl transferases Hp39-FT and FUT5 recognized LacNAc and LacNHTFA as acceptors for α-1,3 fucosylation, but not LacNH2 and LacNHBoc. By modifying the LacNH2 and LacNHBoc moieties before and after enzymatic fucosylation and sialylation, a library of structurally diversified poly LacNAc glycans was obtained (Fig. 5c).
Recently, Boons et al. found that the LacNHBoc moiety in linear poly LacNAc glycan can prevent the branched glycosylation of the proximal Gal residue by β-1,6-N-acetylglucosaminyltransferase 2 (GCNT2).113 This allows for the controlled synthesis of branched poly LacNAc glycans. GlcNHBoc can be galactosylated by bacterial galactosyltransferases, such as recombinant β-1,4-galactosyltransferase from Helicobacter pylori (HpGalT) and β-1,3-galactosyltransferase from Chromobacterium violaceum (CvGalT), and LacNHBoc cannot be recognized by fucosyltransferase. Lin and coworkers proposed a solution to bypass complex chemical glycosylations when preparing regioselective fucosylated poly LacNAc glycans (Fig. 5d).114
Human milk oligosaccharides (HMOs)
HMOs are usually used as metabolic substrates of probiotics to facilitate the formation of the neonatal gut microbiome.118,119 HMOs may also serve as soluble decoys for viruses and parasites to prevent infants from being infected.120,121 Because of their structural complexity, access to structurally well-defined HMOs is a major obstacle to elucidating their biological roles and achieving their scaled-up applications. The generation of complex HMOs begins with lactose, and the backbone is elongated by LacNAc, in which the terminal or internal LacNAc moieties are often produced by fucosylations or sialylations.119,122 Inspired by this repertoire, various HMOs have been synthesized via chemoenzymatic methods.123,124In 2015, Cao and coworkers reported the synthesis of lacto-N-tetrasaccharide and sialyl lacto-N-tetrasaccharides using the OPME method.125 In this study, the glycosyl acceptor lacto-N-tetrasaccharide-proN3 was first chemically synthesized. Thereafter, sialic acid was installed in the nonreducing end of lacto-N-tetrasaccharide-proN3 to form sialyl lacto-N-tetrasaccharides, leveraging the OPME method. Wang and coworkers developed a core synthesis/enzymatic extension strategy to synthesize diverse HMOs.115 Taking advantage of oligosaccharyl thioether and oligosaccharyl bromide donors, three core structures were chemically prepared using a convergent coupling strategy. Subsequently, these core structures were extended and further decorated by different GTs to create a glycan library of 31 HMOs (Fig. 6a). Despite the OPME strategy significantly simplifying the synthesis procedures, excessive chemical manipulations still limit the synthesis of HMOs. To address this issue, Chen's group attempted to enzymatically assemble lacto-N-tetrasaccharide-proN3 by recombinant GT. They identified a highly productive β-1,3-galactosyltransferase from C. violaceum, which exhibited high-efficiency catalysis toward lacto-N-triose.126
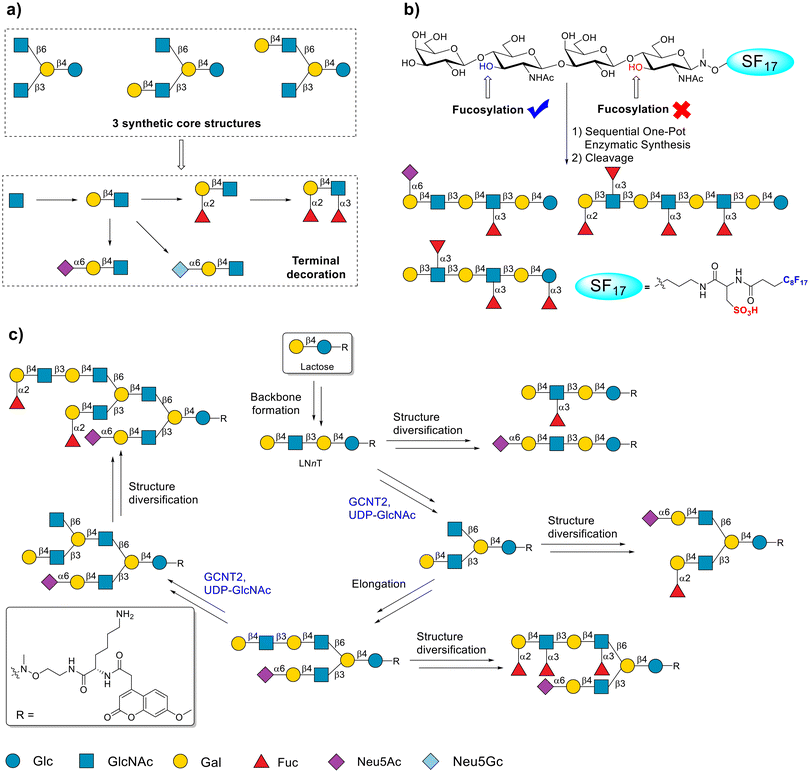 | ||
| Fig. 6 Chemoenzymatic strategies for the synthesis of HMOs. (a) The core synthesis/enzymatic extension (CSEE) method for synthesis of HMOs. Reproduced with permission from Xiao et al.115 Copyright 2016 American Chemical Society. (b) Acceptor-mediated regioselective synthesis of HMOs. Reproduced with permission from Huang et al.116 Copyright 2021 American Chemical Society. (c) Chemoenzymatic synthesis of asymmetrical multiantennary HMOs. Reproduced with permission from Prudden et al.117 Copyright 2017 Proceedings of the National Academy of Sciences. | ||
α-1,3/4-Fucosyltransferase from Helicobacter pylori DSM 6709 (FucTIII) was expressed and ingeniously applied by Yu and coworkers to prepare several fucosylated HMOs.127 In this study, FucTIII was incorporated into a sequential one-pot enzymatic strategy to produce a series of fucosylated HMOs, in which Lewis antigen structures, such as Lewis X, Lewis A, and sialyl Lewis X, were included. Furthermore, the enzyme kinetics assay indicated that FucTIII is preferred for the recognition of type II N-acetyl lactosamine (LacNAc) as the acceptor, and the catalytic efficiency of FucTIII can be increased by the addition of GlcNAc at the nonreducing end of the acceptor.
In 2021, Yu and coworkers proved that a sulfo-fluorous tag (SF17) added to the reducing end of lactose enables site-selective enzymatic fucosylation of the HMO core (Fig. 6b).116 SF17-tagged HMO acceptors have the following advantages: (1) enabling site-selective fucosylation by wild-type α-1,3/4-fucosyltransferase from Bacteroides fragilis (Bf13FT) and α-1,3/4-FucT from H. pylori UA948, (2) enabling selective α-2,6-sialylation on the terminal Gal of the acceptor by wild-type Pd2,6ST, which usually leads to multi-sialylation for the tag-freed or alkyl chain-tagged HMO acceptors, and (3) increasing the catalytic efficiency of β-1,3-galactosyltransferase (β1,3-GalT) from E. coli O55:H7 (WbgO), β-1,3-GlcNAcT, and Bf13FT. These strategies fully demonstrate that GTs from bacteria are powerful tools in the field of HMO synthesis.
Human GTs are also available to prepare HMOs. Boons and coworkers systematically described the synthesis of asymmetrical multi-antennary HMOs using a limited number of human GTs.117 In this study, lactose, bearing the hydrophobic coumarin moiety, acted as a starter to prepare tetrasaccharide LNnT using human β-1,3-N-acetylglucosaminyltransferase 2 and B4GalT1 in the presence of UDP-GlcNAc and UDP-Gal, respectively. Subsequently, the involvement of GCNT2 enabled the linear tetrasaccharide LNnT to produce a branching GlcNAc structure at the internal position, which is the first step in generating multi-antennary asymmetric HMOs. Thereafter, a panel of human GTs (galactoside α-1,2-fucosyltransferase 1, lactosamine α-1,3/4-fucosyltransferase 3, and α-2,6-sialyltransferase 1) was selected to further modify asymmetrical multi-antennary HMO backbones. Finally, a glycan library containing 60 asymmetric, multi-antennary HMOs was constructed and applied in a glycan microarray to probe the interaction of HMOs and several glycan-binding proteins (Fig. 6c). Recently, Fang and coworkers advantageously utilized the expression of GCNT2 in Pichia pastoris, and reported an enzymatic modular strategy that produced a library of branched HMOs.128
Glycosaminoglycans (GAGs)
Glycosaminoglycans (GAGs) are a family of common linear anionic polysaccharides with specific sulfation patterns. They are composed of alternating hexosamine and uronic acid or galactose residues.129–131 According to the disaccharide repeating units of each subtype of GAGs, they are classified into four species: hyaluronic acid (HA), which consists of -β4GlcA-β3GlcNAc1- repeating units; heparin/heparan sulfate (HP/HS), which contains either (-α4IdoA-α4GlcA1)n- or (-α4GlcA-α4IdoA)n-α4GlcNAc- with different sulfation patterns; chondroitin sulfate/dermatan sulfate (CS/DS), which is composed of -β4GlcA/IdoA-β3GalNAc1- repeating units with different sulfation patterns; and keratan sulfate (KS), which is poly-N-acetyllactosamine (-β3Gal-β4GlcNAc1)n- with sulfation at the 6-hydroxy positions.131,132Compared with other polysaccharides, GAGs exhibit substantial market value in medical and cosmetic drug development. For example, hyaluronan is used for arthritis and cosmetic applications,133 and heparin is a traditional anti-thrombosis agent.134 In particular, the discovery of the inhibitory effect of GAGs on severe acute respiratory syndrome coronavirus 2 stimulated the design and development of GAG-based antiviral drugs.135 However, limited access to GAGs, especially homogeneous GAGs, remains, which hinders the achievement of scaled production. The chemoenzymatic approach is thought to be a promising strategy for the large-scale production of GAGs.
A soluble microbial hyaluronan synthase from Pasteurella multocida (PmHAS) was used to catalyze GlcA and GlcNAc polymerization to form a hyaluronan chain. PmHAS contains two distinct GT domains that can recognize UDP-GlcNAc and UDP-GlcA as donors.136,137 Therefore, with the availability of UDP sugar precursors, the elongation of HA only requires one enzyme (PmHAS), facilitating the sequential synthesis of HA.138,139 Because of the uneconomical use of sugar nucleotide precursors, further studies focused on exploiting the OPME system to produce the desired HA.140,141 OPME systems require inexpensive monosaccharide modules (GlcNAc and Glc or GlcA) as substrates, rather than expensive sugar nucleotide precursors that provide the substrates. For example, using inexpensive sucrose and GlcNAc as substrates, Elling and coworkers described an economic OPME method to synthesize high-molecular-weight (HMw)-HA, in which the UDP-GlcA and UDP-GlcNAc can be in situ regenerated (Fig. 7a).141
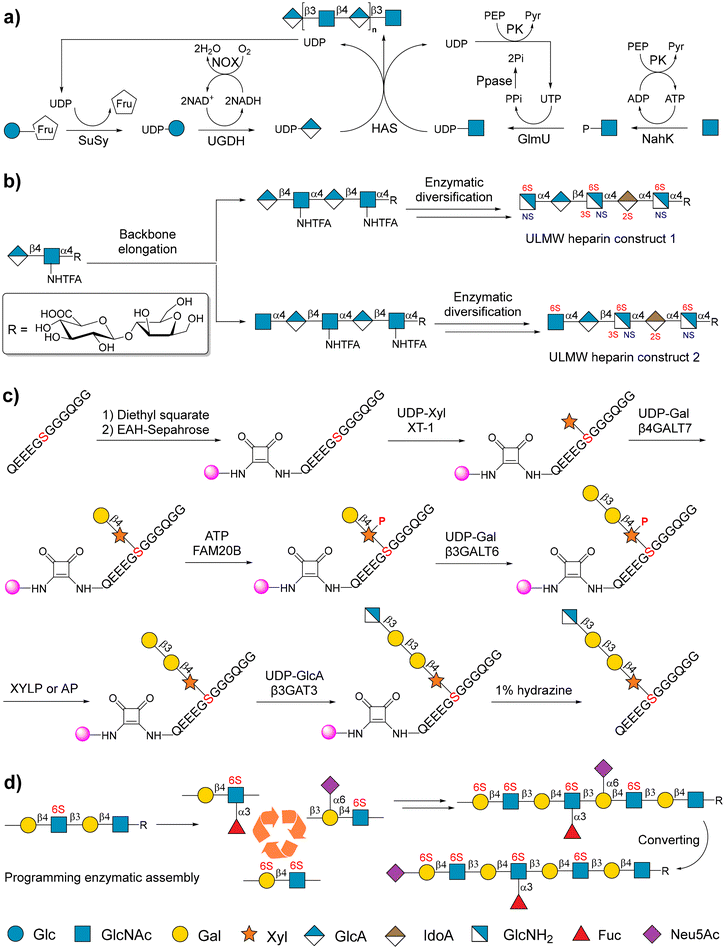 | ||
| Fig. 7 Chemoenzymatic strategies for the synthesis of GAGs. (a) OPME chemoenzymatic synthesis of HA. SuSy: sucrose synthase from potato. UGDH: UDP-glucose dehydrogenase from Streptococcus zooepidemicus. NOX: nicotinamide adenine dinucleotide (NADH) oxidase. NahK: N-acetylhexosamine-1-kinase from Bifidobacterium longum. PK: pyruvate kinase from the rabbit. GlmU: UDP-GlcNAc pyrophosphorylase Streptococcus zooepidemicus. PPase: pyrophosphatase from yeast. HAS: hyaluronan synthase from Pasteurella multocida. Reproduced with permission from Eisele et al.141 Copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) Chemoenzymatic synthesis of homogeneous ultralow molecular weight (ULMW) HP. Reproduced with permission from Xu et al.53 Copyright 2011, American Association for the Advancement of Science. (c) Solid-phase-supported chemoenzymatic synthesis of CSPG. Reproduced with permission from Lin et al.150 Copyright 2024 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (d) Programming the chemoenzymatic synthesis of KS. Reproduced with permission from Wu et al.151 Copyright 2024 American Chemical Society. | ||
The chemoenzymatic synthesis of HP/HS depends on the bi-functional enzymes from Pasteurella multocida PmHS1 and PmHS2,142 or a combination of KfiA and KfiC obtained from the E. coli K5 strain.143 A panel of sulfotransferases, including 6-O-sulfotransferase (6-OST), 3-O-sulfotransferase (3-OST), and N-sulfotransferase (NST), contributes to further diversity in the structure of HP/HS. Liu and coworkers reported a simple chemoenzymatic strategy for the tailored synthesis of two structurally homogeneous ULMW heparins (Fig. 7b).53 In this study, a disaccharide was used as a starting substrate, which was elongated using PmHS2 and KfiA. An unnatural glycosyl donor called UDP-GlcNHTFA was recognized by KfiA, and thus, GlcNHTFA was inserted into the backbone of the target molecule. The GlcNHTFA moiety was converted to GlcNH2 for further N-sulfation. Taking advantage of this property, synthetic hexasaccharide and heptasaccharide intermediates were further sulfated at specific sites using C5-epimerases and 2/3/6-OST to accomplish the preparation of two ULMW heparins. Many other efforts have resulted in the cost-effective, controlled, and even gram-scaled synthesis of structurally well-defined HP/HS,144–147 and HP/HS derivatives and mimetics.57,148,149
CS and DS have been widely investigated in chemoenzymatic synthesis for their promising anticoagulant properties.152,153 Using GT from the E. coli K4 strain (KfoC), which can transfer both GalNAc and GlcA from UDPGalNAc and UDP-GlcA to the acceptor to form CS, Liu and coworkers synthesized a library of 15 different CS glycans covering different sulfation pattern glycans, from trisaccharides to nona-saccharides.154 This group has reported the chemoenzymatic synthesis of seven structurally homogeneous HS and CS chimeras, in which a CS heptasaccharide domain on the reducing end was coupled with an HS pentasaccharide domain on the nonreducing end.155
Recently, Huang and coworkers developed a solid phase-supported strategy for the successful chemoenzymatic synthesis of CS proteoglycans (CSPG) for the first time.150 In this study, Sepharose beads were used as the solid phase support, and were attached to the terminal of peptide substrates using a cleavable linker. Subsequently, the first saccharide xylose was transferred to the peptide substrates using xylosyl transferase-1 to form the initial glycopeptides. The initial glycopeptides were sequentially elongated by β-1,4-galactosyl transferase 7, β-1,4-galactosyl transferase 6, and β-1,3-glucuronic acid transferase 3 (β3GAT3) to form tetra-saccharide-bearing glycopeptides. The phosphorylation of xylose 2-O in glycopeptides is required for the catalysis of β3GAT3 (Fig. 7c). Finally, enzymatic chain extension and sulfation result in the preparation of different CSPG glycopeptides. Different from CS, the chemoenzymatic synthesis of DS from chondroitin requires C5-epimerization by epimerase (DS-ep) and sulfation by 4-O sulfotransferase (D4ST1).156
KS consists of LacNAc repeating units with sulfation occurring on the 6-OH of GlcNAc and Gal. Although KS is involved in many biological events, poor efforts have been made to synthesize structurally well-defined KS glycans. One method for synthesizing KS glycans uses transglycosylation of mutant keratanase II to install the sulfated glycan moieties to produce KS glycans. Ohmae and coworkers efficiently synthesized several fucosylated and sialylated KS glycans using oxazoline donors and mutant keratanase II.30,157 Another approach involves the sulfation of poly LacNAc glycans by sulfotransferase to produce KS glycans.
Recently, Boons and coworkers reported the use of recombinant human GlcNAc-6-O-sulfotransferase (CHST2) and keratin sulfate Gal-6 sulfotransferase (CHST1) for chemoenzymatic synthesis of KS glycans.158 Their study indicated that CHST2 only sulfates the terminal GlcNAc moieties of LacNAc oligosaccharides, and CHST1 can sulfate the internal Gal moieties in LacNAc chains rather than the terminal Gal.158 Further work uncovered detailed substrate specificities of CHST1 and also confirmed that α-1,3-fucosylated LacNAc cannot be accepted by CHST1, and Gal-6-O sulfation of LacNAc hinders enzymatic α-1,3-fucosylation (Fig. 7d).151 Through these discoveries, a range of KS glycans with different sulfation and fucosylation patterns was synthesized.
Summary and outlook
In the past few decades, numerous critical physiological or pathological processes involving glycans have been revealed. These findings urgently require glycochemists to prepare structurally well-defined glycans for further exploration of their biological functions. The chemoenzymatic strategy is a powerful tool for preparing a range of biologically related complex glycans, and even glycoconjugates, such as glycopeptides, glycoproteins,31 and glycolipids.159,160 Enzymatic glycosylations combined with chemical handles will circumvent tedious protecting group manipulation, and will also increase the regio- and stereo-selectivity of glycosylation. However, at least two factors limit the development of this methodology.The first factor is the limited availability and poor understanding of GTs. To address this, different protein expression systems have been constructed to produce large-scale GTs, such as E. coli expression systems that can efficiently yield a series of bacterial GTs, and an insect expression system that overexpresses many mammalian GTs. Furthermore, crystal structure and mutagenesis studies are promising approaches to increase our understanding of GTs and provide the desired GTs for synthesis.
The second factor is the time-consuming and inefficient reaction post-processing procedures. For example, the synthesis of a deca-saccharide theoretically requires at least nine enzymatic reaction steps, and therefore, at least nine isolation and purification procedures need to be performed. To address this, machine-driven automated enzymatic synthesis has been developed to provide easy access to synthesize complex glycans.161,162 In addition, many state-of-the-art techniques and tools, such as ultra-performance liquid chromatography,163 high-resolution mass spectrometry,164 nanopore sequencing,165–167 and machine learning,168 are facilitating the identification and structural analysis of complex glycans, which will be helpful for the rational designing of chemoenzymatic strategies in glycan synthesis. In the future, general protocols will enable the synthesis of glycans that will be as easy as the PCR amplification of DNA.
Author contributions
All authors contributed equally to the manuscript.Data availability
Data availability is not applicable to this article.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (22007092 to L. W. and 22307127 to Y. T.), the Natural Science Foundation of Shanghai (22ZR1474000 to L. W.), and the Shanghai Municipal Science and Technology Major Project.References
- R. A. Flynn, K. Pedram, S. A. Malaker, P. J. Batista, B. A. H. Smith, A. G. Johnson, B. M. George, K. Majzoub, P. W. Villalta, J. E. Carette and C. R. Bertozzi, Small RNAs are modified with N-glycans and displayed on the surface of living cells, Cell, 2021, 184(12), 3109–3124 CrossRef CAS.
- S. S. Shivatare, V. S. Shivatare and C. H. Wong, Glycoconjugates: Synthesis, Functional Studies, and Therapeutic Developments, Chem. Rev., 2022, 122(20), 15603–15671 CrossRef CAS PubMed.
- A. Varki, Biological Roles of Oligosaccharides - All of the Theories Are Correct, Glycobiology, 1993, 3(2), 97–130 CrossRef CAS.
- K. Ohtsubo and J. D. Marth, Glycosylation in cellular mechanisms of health and disease, Cell, 2006, 126(5), 855–867 CrossRef CAS PubMed.
- K. W. Moremen, M. Tiemeyer and A. V. Nairn, Vertebrate protein glycosylation: diversity, synthesis and function, Nat. Rev. Mol. Cell Biol., 2012, 13(7), 448–462 CrossRef CAS.
- K. T. Schjoldager, Y. Narimatsu, H. J. Joshi and H. Clausen, Global view of human protein glycosylation pathways and functions, Nat. Rev. Mol. Cell Biol., 2020, 21(12), 729–749 CrossRef CAS.
- S. M. Hancock, M. D. Vaughan and S. G. Withers, Engineering of glycosidases and glycosyltransferases, Curr. Opin. Chem. Biol., 2006, 10(5), 509–519 CrossRef CAS.
- G. K. Wagner, T. Pesnot and R. A. Field, A survey of chemical methods for sugar-nucleotide synthesis, Nat. Prod. Rep., 2009, 26(9), 1172–1194 RSC.
- S. Sanz, G. Bandini, D. Ospina, M. Bernabeu, K. Marino, C. Fernandez-Becerra and L. Izquierdo, Biosynthesis of GDP-fucose and other sugar nucleotides in the blood stages of Plasmodium falciparum, J. Biol. Chem., 2013, 288(23), 16506–16517 CrossRef CAS.
- S. Wang, J. Zhang, F. Wei, W. Li and L. Wen, Facile Synthesis of Sugar Nucleotides from Common Sugars by the Cascade Conversion Strategy, J. Am. Chem. Soc., 2022, 144(22), 9980–9989 CrossRef CAS.
- X. Song, H. Ju, Y. Lasanajak, M. R. Kudelka, D. F. Smith and R. D. Cummings, Oxidative release of natural glycans for functional glycomics, Nat. Methods, 2016, 13(6), 528–534 CrossRef CAS PubMed.
- A. Pardo-Vargas, M. Delbianco and P. H. Seeberger, Automated glycan assembly as an enabling technology, Curr. Opin. Chem. Biol., 2018, 46, 48–55 CrossRef CAS PubMed.
- Q. Zhu, Z. Shen, F. Chiodo, S. Nicolardi, A. Molinaro, A. Silipo and B. Yu, Chemical synthesis of glycans up to a 128-mer relevant to the O-antigen of Bacteroides vulgatus, Nat. Commun., 2020, 11(1), 4142 CrossRef CAS.
- W. Yao, D.-C. Xiong, Y. Yang, C. Geng, Z. Cong, F. Li, B.-H. Li, X. Qin, L.-N. Wang, W.-Y. Xue, N. Yu, H. Zhang, X. Wu, M. Liu and X.-S. Ye, Automated solution-phase multiplicative synthesis of complex glycans up to a 1,080-mer, Nat. Synth., 2022, 1(11), 854–863 CrossRef.
- T. Rexer, D. Laaf, J. Gottschalk, H. Frohnmeyer, E. Rapp and L. Elling, Enzymatic Synthesis of Glycans and Glycoconjugates, Adv. Biochem. Eng./Biotechnol., 2021, 175, 231–280 CrossRef.
- C. C. Liu, J. Ye and H. Cao, Chemical Evolution of Enzyme-Catalyzed Glycosylation, Acc. Chem. Res., 2024, 514–522 Search PubMed.
- S. Muthana, H. Cao and X. Chen, Recent progress in chemical and chemoenzymatic synthesis of carbohydrates, Curr. Opin. Chem. Biol., 2009, 13(5–6), 573–581 CrossRef CAS PubMed.
- R. M. Schmaltz, S. R. Hanson and C. H. Wong, Enzymes in the synthesis of glycoconjugates, Chem. Rev., 2011, 111(7), 4259–4307 CrossRef CAS.
- M. Kumar, C. K. Bandi and S. P. Chundawat, High-throughput screening of glycosynthases using azido sugars for oligosaccharides synthesis, in Methods Enzymol, Elsevier, 2023, vol. 682, pp. 211–245 Search PubMed.
- J. B. McArthur and X. Chen, Glycosyltransferase engineering for carbohydrate synthesis, Biochem. Soc. Trans., 2016, 44(1), 129–142 CrossRef CAS.
- R. Chen, Bacterial expression systems for recombinant protein production: E. coli and beyond, Biotechnol. Adv., 2012, 30(5), 1102–1107 CrossRef CAS.
- K. W. Moremen, A. Ramiah, M. Stuart, J. Steel, L. Meng, F. Forouhar, H. A. Moniz, G. Gahlay, Z. Gao, D. Chapla, S. Wang, J. Y. Yang, P. K. Prabhakar, R. Johnson, M. D. Rosa, C. Geisler, A. V. Nairn, J. Seetharaman, S. C. Wu, L. Tong, H. J. Gilbert, J. LaBaer and D. L. Jarvis, Expression system for structural and functional studies of human glycosylation enzymes, Nat. Chem. Biol., 2018, 14(2), 156–162 CrossRef CAS.
- Y. Bourne and B. Henrissat, Glycoside hydrolases and glycosyltransferases: families and functional modules, Curr. Opin. Struct. Biol., 2001, 11(5), 593–600 CrossRef CAS PubMed.
- A. Kobata, Exo- and endoglycosidases revisited, Proc. Jpn. Acad., Ser. B, 2013, 89(3), 97–117 CrossRef CAS.
- A. Koizumi, I. Matsuo, M. Takatani, A. Seko, M. Hachisu, Y. Takeda and Y. Ito, Top-down chemoenzymatic approach to a high-mannose-type glycan library: synthesis of a common precursor and its enzymatic trimming, Angew. Chem., Int. Ed., 2013, 52(29), 7426–7431 CrossRef CAS PubMed.
- L. Guo, X. Chen, L. Xu, M. Xiao, L. Lu and H. Atomi, Enzymatic Synthesis of 6′-Sialyllactose, a Dominant Sialylated Human Milk Oligosaccharide, by a Novel exo-α-Sialidase from Bacteroides fragilis NCTC9343, Appl. Environ. Microbiol., 2018, 84(13), e00071–e00018 CrossRef CAS.
- H. T. T. Pham, G. A. T. Kate, L. Dijkhuizen and S. S. van Leeuwen, Synthesis and Characterization of Sialylated Lactose- and Lactulose-Derived Oligosaccharides by Trans-sialidase, J. Agric. Food Chem., 2019, 67(12), 3469–3479 CrossRef CAS.
- B. Zeuner and A. S. Meyer, Enzymatic transfucosylation for synthesis of human milk oligosaccharides, Carbohydr. Res., 2020, 493, 108029 CrossRef CAS PubMed.
- B. Zeuner, M. Vuillemin, J. Holck, J. Muschiol and A. S. Meyer, Loop engineering of an alpha-1,3/4 l-fucosidase for improved synthesis of human milk oligosaccharides, Enzyme Microb. Technol., 2018, 115, 37–44 CrossRef CAS.
- S. Yuge, A. Tateishi, K. Numata and M. Ohmae, Chemoenzymatic Synthesis of Sialyl Sulfo-Oligosaccharides as Potent Siglec-8 Ligands via Transglycosylation Catalyzed by Keratanase II, Biomacromolecules, 2022, 23(1), 316–325 CrossRef CAS.
- C. Li and L. X. Wang, Chemoenzymatic Methods for the Synthesis of Glycoproteins, Chem. Rev., 2018, 118(17), 8359–8413 CrossRef CAS.
- L. X. Wang, X. Tong, C. Li, J. P. Giddens and T. Li, Glycoengineering of Antibodies for Modulating Functions, Annu. Rev. Biochem., 2019, 88, 433–459 CrossRef CAS.
- Y. Tian, Y. Wang, H. Yin, Y. Luo, F. Wei, H. Zhou and L. Wen, A Sensitive and Reversible Labeling Strategy Enables Global Mapping of the Core–Fucosylated Glycoproteome on Cell Surfaces, Angew. Chem., Int. Ed. Engl., 2022, 134(49), e202206802 CrossRef.
- Y. Luo, Y. Wang, Y. Tian, H. Zhou and L. Wen, “Two Birds One Stone” Strategy for the Site-Specific Analysis of Core Fucosylation and O-GlcNAcylation, J. Am. Chem. Soc., 2023, 145(29), 15879–15887 CrossRef CAS PubMed.
- Y. Wang, R. Yuan, B. Liang, J. Zhang, Q. Wen, H. Chen, Y. Tian, L. Wen and H. Zhou, A “One-Step” Strategy for the Global Characterization of Core-Fucosylated Glycoproteome, JACS Au, 2024, 4(5), 2005–2018 CrossRef CAS PubMed.
- S. J. Williams and S. G. Withers, Glycosynthases: Mutant glycosidases for glycoside synthesis, Aust. J. Chem., 2002, 55(1–2), 3–12 CrossRef CAS.
- M. R. Hayes and J. Pietruszka, Synthesis of Glycosides by Glycosynthases, Molecules, 2017, 22(9), 1434 CrossRef.
- T. M. Gloster, Exploitation of carbohydrate processing enzymes in biocatalysis, Curr. Opin. Chem. Biol., 2020, 55, 180–188 CrossRef CAS PubMed.
- C. Moulis, D. Guieysse, S. Morel, E. Séverac and M. Remaud-Siméon, Natural and engineered transglycosylases: Green tools for the enzyme-based synthesis of glycoproducts, Curr. Opin. Chem. Biol., 2021, 61, 96–106 CrossRef CAS.
- J. P. Dolan, S. C. Cosgrove and G. J. Miller, Biocatalytic Approaches to Building Blocks for Enzymatic and Chemical Glycan Synthesis, JACS Au, 2023, 3(1), 47–61 CrossRef CAS.
- L. Mestrom, M. Przypis, D. Kowalczykiewicz, A. Pollender, A. Kumpf, S. R. Marsden, I. Bento, A. B. Jarzebski, K. Szymanska, A. Chrusciel, D. Tischler, R. Schoevaart, U. Hanefeld and P. L. Hagedoorn, Leloir Glycosyltransferases in Applied Biocatalysis: A Multidisciplinary Approach, Int. J. Mol. Sci., 2019, 20(21), 5263 CrossRef CAS.
- Y. Maeda and T. Kinoshita, Dolichol-phosphate mannose synthase: structure, function and regulation, Biochim. Biophys. Acta, 2008, 1780(6), 861–868 CrossRef CAS PubMed.
- H. Yu, V. Thon, K. Lau, L. Cai, Y. Chen, S. Mu, Y. Li, P. G. Wang and X. Chen, Highly efficient chemoenzymatic synthesis of beta1–3-linked galactosides, Chem. Commun., 2010, 46(40), 7507–7509 RSC.
- C. Li, S. Zhu, C. Ma and L. X. Wang, Designer alpha1,6-Fucosidase Mutants Enable Direct Core Fucosylation of Intact N-Glycopeptides and N-Glycoproteins, J. Am. Chem. Soc., 2017, 139(42), 15074–15087 CrossRef CAS PubMed.
- M. Bar-Peled and M. A. O'Neill, Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling, Annu. Rev. Plant Biol., 2011, 62, 127–155 CrossRef CAS PubMed.
- L. Cai, Recent Progress in Enzymatic Synthesis of Sugar Nucleotides, J. Carbohydr. Chem., 2012, 31(7), 535–552 CrossRef CAS.
- H. Yu, H. Yu, R. Karpel and X. Chen, Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases, Bioorg. Med. Chem., 2004, 12(24), 6427–6435 CrossRef CAS PubMed.
- Y. Chen, V. Thon, Y. Li, H. Yu, L. Ding, K. Lau, J. Qu, L. Hie and X. Chen, One-pot three-enzyme synthesis of UDP-GlcNAc derivatives, Chem. Commun., 2011, 47(38), 10815–10817 RSC.
- T. I. Tsai, H. Y. Lee, S. H. Chang, C. H. Wang, Y. C. Tu, Y. C. Lin, D. R. Hwang, C. Y. Wu and C. H. Wong, Effective sugar nucleotide regeneration for the large-scale enzymatic synthesis of Globo H and SSEA4, J. Am. Chem. Soc., 2013, 135(39), 14831–14839 CrossRef CAS PubMed.
- M. T. Anwar, S. K. Kawade, Y. R. Huo, A. K. Adak, D. Sridharan, Y. T. Kuo, C. Y. Fan, H. R. Wu, Y. S. Lee, T. Angata and C. C. Lin, Sugar nucleotide regeneration system for the synthesis of Bi- and triantennary-glycans and exploring their activities against siglecs, Eur. J. Med. Chem., 2022, 232, 114146 CrossRef CAS PubMed.
- Y. Zheng, J. Zhang, J. Meisner, W. Li, Y. Luo, F. Wei and L. Wen, Cofactor-Driven Cascade Reactions Enable the Efficient Preparation of Sugar Nucleotides, Angew. Chem., Int. Ed., 2022, 61(20), e202115696 CrossRef CAS.
- F. Wei, R. Yuan, Q. Wen and L. Wen, Systematic Enzymatic Synthesis of dTDP–Activated Sugar Nucleotides, Angew. Chem., 2023, 135(20), e202217894 CrossRef.
- Y. Xu, S. Masuko, M. Takieddin, H. Xu, R. Liu, J. Jing, S. A. Mousa, R. J. Linhardt and J. Liu, Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins, Science, 2011, 334(6055), 498–501 CrossRef CAS PubMed.
- Y. Xu, C. Cai, K. Chandarajoti, P. H. Hsieh, L. Li, T. Q. Pham, E. M. Sparkenbaugh, J. Sheng, N. S. Key, R. Pawlinski, E. N. Harris, R. J. Linhardt and J. Liu, Homogeneous low-molecular-weight heparins with reversible anticoagulant activity, Nat. Chem. Biol., 2014, 10(4), 248–250 CrossRef CAS PubMed.
- L. Liu, A. R. Prudden, C. J. Capicciotti, G. P. Bosman, J. Y. Yang, D. G. Chapla, K. W. Moremen and G. J. Boons, Streamlining the chemoenzymatic synthesis of complex N-glycans by a stop and go strategy, Nat. Chem., 2019, 11(2), 161–169 CrossRef CAS.
- X. Zhang, D. E. Green, V. L. Schultz, L. Lin, X. Han, R. Wang, A. Yaksic, S. Y. Kim, P. L. DeAngelis and R. J. Linhardt, Synthesis of 4-azido-N-acetylhexosamine uridine diphosphate donors: clickable glycosaminoglycans, J. Org. Chem., 2017, 82(18), 9910–9915 CrossRef CAS PubMed.
- P. He, X. Zhang, K. Xia, D. E. Green, S. Baytas, Y. M. Xu, T. Pham, J. Liu, F. M. Zhang, A. Almond, R. J. Linhardt and P. L. DeAngelis, Chemoenzymatic synthesis of sulfur-linked sugar polymers as heparanase inhibitors, Nat. Commun., 2022, 13(1), 7438 CrossRef CAS.
- L. Wen, D. Liu, Y. Zheng, K. Huang, X. Cao, J. Song and P. G. Wang, A One-Step Chemoenzymatic Labeling Strategy for Probing Sialylated Thomsen–Friedenreich Antigen, ACS Cent. Sci., 2018, 4(4), 451–457 CrossRef CAS PubMed.
- L. Q. Wen, M. R. Gadi, Y. Zheng, C. Gibbons, S. M. Kondengaden, J. B. Zhang and P. G. Wang, Chemoenzymatic Synthesis of Unnatural Nucleotide Sugars for Enzymatic Bioorthogonal Labeling, ACS Catal., 2018, 8(8), 7659–7666 CrossRef CAS.
- Y. P. Tian, S. Z. Ma and L. Q. Wen, Towards chemoenzymatic labeling strategies for profiling protein glycosylation, Curr. Opin. Chem. Biol., 2024, 80, 102460 CrossRef CAS.
- A. Helenius and M. Aebi, Intracellular functions of N-linked glycans, Science, 2001, 291(5512), 2364–2369 CrossRef CAS PubMed.
- L. Ellgaard and A. Helenius, Quality control in the endoplasmic reticulum, Nat. Rev. Mol. Cell Biol., 2003, 4(3), 181–191 CrossRef CAS.
- E. Weerapana and B. Imperiali, Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems, Glycobiology, 2006, 16(6), 91R–101R CrossRef CAS.
- F. Schwarz and M. Aebi, Mechanisms and principles of N-linked protein glycosylation, Curr. Opin. Struct. Biol., 2011, 21(5), 576–582 CrossRef CAS PubMed.
- Z. Wang, Z. S. Chinoy, S. G. Ambre, W. Peng, R. McBride, R. P. de Vries, J. Glushka, J. C. Paulson and G. J. Boons, A general strategy for the chemoenzymatic synthesis of asymmetrically branched N-glycans, Science, 2013, 341(6144), 379–383 CrossRef CAS.
- P. Wang, J. Zhu, Y. Yuan and S. J. Danishefsky, Total synthesis of the 2,6-sialylated immunoglobulin G glycopeptide fragment in homogeneous form, J. Am. Chem. Soc., 2009, 131(46), 16669–16671 CrossRef CAS.
- C. Gao, M. S. Hanes, L. A. Byrd-Leotis, M. Wei, N. Jia, R. J. Kardish, T. R. McKitrick, D. A. Steinhauer and R. D. Cummings, Unique Binding Specificities of Proteins toward Isomeric Asparagine-Linked Glycans, Cell Chem. Biol., 2019, 26(4), 535–547 CrossRef CAS PubMed.
- A. D. Srivastava, L. Unione, M. Bunyatov, I. A. Gagarinov, S. Delgado, N. G. A. Abrescia, A. Arda and G. J. Boons, Chemoenzymatic Synthesis of Complex N-Glycans of the Parasite S. mansoni to Examine the Importance of Epitope Presentation on DC-SIGN recognition, Angew. Chem., Int. Ed., 2021, 60(35), 19287–19296 CrossRef CAS PubMed.
- S. S. Shivatare, S. H. Chang, T. I. Tsai, S. Y. Tseng, V. S. Shivatare, Y. S. Lin, Y. Y. Cheng, C. T. Ren, C. C. Lee, S. Pawar, C. S. Tsai, H. W. Shih, Y. F. Zeng, C. H. Liang, P. D. Kwong, D. R. Burton, C. Y. Wu and C. H. Wong, Modular synthesis of N-glycans and arrays for the hetero-ligand binding analysis of HIV antibodies, Nat. Chem., 2016, 8(4), 338–346 CrossRef CAS.
- F. Wei, L. Zang, P. Zhang, J. Zhang and L. Wen, Concise chemoenzymatic synthesis of N-glycans, Chem, 2024 DOI:10.1016/j.chempr.2024.05.006.
- S. S. Shivatare, S. H. Chang, T. I. Tsai, C. T. Ren, H. Y. Chuang, L. Hsu, C. W. Lin, S. T. Li, C. Y. Wu and C. H. Wong, Efficient convergent synthesis of bi-, tri-, and tetra-antennary complex type N-glycans and their HIV-1 antigenicity, J. Am. Chem. Soc., 2013, 135(41), 15382–15391 CrossRef CAS PubMed.
- S. Pawar, L. Hsu, T. N. Reddy, M. Ravinder, C. T. Ren, Y. W. Lin, Y. Y. Cheng, T. W. Lin, T. L. Hsu, S. K. Wang, C. H. Wong and C. Y. Wu, Synthesis of Asymmetric Glycans as Common Core Substrates for Structural Diversification through Selective Enzymatic Glycosylation, ACS Chem. Biol., 2020, 15(9), 2382–2394 CrossRef CAS PubMed.
- S. J. North, P. G. Hitchen, S. M. Haslam and A. Dell, Mass spectrometry in the analysis of N-linked and O-linked glycans, Curr. Opin. Struct. Biol., 2009, 19(5), 498–506 CrossRef CAS PubMed.
- I. A. Gagarinov, T. Li, J. S. Torano, T. Caval, A. D. Srivastava, J. A. Kruijtzer, A. J. Heck and G. J. Boons, Chemoenzymatic Approach for the Preparation of Asymmetric Bi-, Tri-, and Tetra-Antennary N-Glycans from a Common Precursor, J. Am. Chem. Soc., 2017, 139(2), 1011–1018 CrossRef CAS PubMed.
- Z. S. Chinoy, F. Friscourt, C. J. Capicciotti, P. Chiu and G. J. Boons, Chemoenzymatic Synthesis of Asymmetrical Multi-Antennary N-Glycans to Dissect Glycan-Mediated Interactions between Human Sperm and Oocytes, Chemistry, 2018, 24(31), 7970–7975 CrossRef CAS.
- K. Fujikawa, A. Koizumi, M. Hachisu, A. Seko, Y. Takeda and Y. Ito, Construction of a high-mannose-type glycan library by a renewed top-down chemo-enzymatic approach, Chemistry, 2015, 21(8), 3224–3233 CrossRef CAS PubMed.
- L. Li, Y. Liu, C. Ma, J. Qu, A. D. Calderon, B. Wu, N. Wei, X. Wang, Y. Guo, Z. Xiao, J. Song, G. Sugiarto, Y. Li, H. Yu, X. Chen and P. G. Wang, Efficient Chemoenzymatic Synthesis of an N-glycan Isomer Library, Chem. Sci., 2015, 6(10), 5652–5661 RSC.
- Z. Wu, Y. Liu, C. Ma, L. Li, J. Bai, L. Byrd-Leotis, Y. Lasanajak, Y. Guo, L. Wen, H. Zhu, J. Song, Y. Li, D. A. Steinhauer, D. F. Smith, B. Zhao, X. Chen, W. Guan and P. G. Wang, Identification of the binding roles of terminal and internal glycan epitopes using enzymatically synthesized N-glycans containing tandem epitopes, Org. Biomol. Chem., 2016, 14(47), 11106–11116 RSC.
- A. D. Calderon, J. Zhou, W. Guan, Z. Wu, Y. Guo, J. Bai, Q. Li, P. G. Wang, J. Fang and L. Li, An enzymatic strategy to asymmetrically branched N-glycans, Org. Biomol. Chem., 2017, 15(35), 7258–7262 RSC.
- A. Seko, M. Koketsu, M. Nishizono, Y. Enoki, H. R. Ibrahim, L. R. Juneja, M. Kim and T. Yamamoto, Occurrence of a sialylglycopeptide and free sialylglycans in hen's egg yolk, Biochim. Biophys. Acta, Gen. Subj., 1997, 1335(1–2), 23–32 CrossRef CAS PubMed.
- B. Sun, W. Bao, X. Tian, M. Li, H. Liu, J. Dong and W. Huang, A simplified procedure for gram-scale production of sialylglycopeptide (SGP) from egg yolks and subsequent semi-synthesis of Man3GlcNAc oxazoline, Carbohydr. Res., 2014, 396, 62–69 CrossRef CAS.
- L. Liu, A. R. Prudden, G. P. Bosman and G. J. Boons, Improved isolation and characterization procedure of sialylglycopeptide from egg yolk powder, Carbohydr. Res., 2017, 452, 122–128 CrossRef CAS PubMed.
- S. Ma, L. Liu, D. Eggink, S. Herfst, R. A. M. Fouchier, R. P. de Vries and G.-J. Boons, Asymmetrical Biantennary Glycans Prepared by a Stop-and-Go Strategy Reveal Receptor Binding Evolution of Human Influenza A Viruses, JACS Au, 2024, 4(2), 607–618 CrossRef CAS PubMed.
- J. D. Valderrama-Rincon, A. C. Fisher, J. H. Merritt, Y. Y. Fan, C. A. Reading, K. Chhiba, C. Heiss, P. Azadi, M. Aebi and M. P. DeLisa, An engineered eukaryotic protein glycosylation pathway in Escherichia coli, Nat. Chem. Biol., 2012, 8(5), 434–436 CrossRef CAS PubMed.
- A. S. Ramirez, J. Boilevin, C. W. Lin, B. H. Gan, D. Janser, M. Aebi, T. Darbre, J. L. Reymond and K. P. Locher, Chemo-enzymatic synthesis of lipid-linked GlcNAc2Man5 oligosaccharides using recombinant Alg1, Alg2 and Alg11 proteins, Glycobiology, 2017, 27(8), 726–733 CrossRef CAS PubMed.
- M. H. Xiang, X. X. Xu, C. D. Wang, S. Chen, S. Xu, X. Y. Xu, N. Dean, N. Wang and X. D. Gao, Topological and enzymatic analysis of human Alg2 mannosyltransferase reveals its role in lipid-linked oligosaccharide biosynthetic pathway, Commun. Biol., 2022, 5(1), 117 CrossRef CAS PubMed.
- N. I. Majewska, M. L. Tejada, M. J. Betenbaugh and N. Agarwal, N-Glycosylation of IgG and IgG-Like Recombinant Therapeutic Proteins: Why Is It Important and How Can We Control It?, Annu. Rev. Chem. Biomol. Eng., 2020, 11, 311–338 CrossRef CAS PubMed.
- S. Malik, I. Grunert, M. F. von Roman, H. Walch, T. Dams, M. Thomann and R. Falkenstein, Implementation of in vitro glycoengineering of monoclonal antibodies into downstream processing of industrial production, Glycobiology, 2022, 32(2), 123–135 CrossRef CAS PubMed.
- N. Cohen Saban, A. Yalin, T. Landsberger, R. Salomon, A. Alva, T. Feferman, I. Amit and R. J. S. i. Dahan, Fc glycoengineering of a PD-L1 antibody harnesses Fcγ receptors for increased antitumor efficacy, Sci. Immunol., 2023, 8(81), eadd8005 CrossRef CAS PubMed.
- M. Weiss, D. Ott, T. Karagiannis, M. Weishaupt, M. Niemietz, S. Eller, M. Lott, M. Martinez-Orts, A. Canales, N. Razi, J. C. Paulson and C. Unverzagt, Efficient Chemoenzymatic Synthesis of N-Glycans with a beta1,4-Galactosylated Bisecting GlcNAc Motif, ChemBioChem, 2020, 21(22), 3212–3215 CrossRef CAS PubMed.
- W. Ma, Z. Xu, Y. Jiang, J. Liu, D. Xu, W. Huang and T. Li, Divergent Enzymatic Assembly of a Comprehensive 64-Membered IgG N-Glycan Library for Functional Glycomics, Adv. Sci., 2023, 10(30), e2303832 CrossRef.
- F. Tang, Y. Yang, Y. Tang, S. Tang, L. Yang, B. Sun, B. Jiang, J. Dong, H. Liu, M. Huang, M. Y. Geng and W. Huang, One-pot N-glycosylation remodeling of IgG with non-natural sialylglycopeptides enables glycosite-specific and dual-payload antibody-drug conjugates, Org. Biomol. Chem., 2016, 14(40), 9501–9518 RSC.
- A. J. Fairbanks, The ENGases: versatile biocatalysts for the production of homogeneous N-linked glycopeptides and glycoproteins, Chem. Soc. Rev., 2017, 46(16), 5128–5146 RSC.
- D. J. Gill, H. Clausen and F. Bard, Location, location, location: new insights into O-GalNAc protein glycosylation, Trends Cell Biol., 2011, 21(3), 149–158 CrossRef CAS PubMed.
- S. H. Stalnaker, R. Stuart and L. Wells, Mammalian O-mannosylation: unsolved questions of structure/function, Curr. Opin. Struct. Biol., 2011, 21(5), 603–609 CrossRef CAS.
- L. Na, R. Li and X. Chen, Recent progress in synthesis of carbohydrates with sugar nucleotide-dependent glycosyltransferases, Curr. Opin. Chem. Biol., 2021, 61, 81–95 CrossRef CAS PubMed.
- Z. F. Hu, K. Zhong and H. Cao, Recent advances in enzymatic and chemoenzymatic synthesis of N- and O-glycans, Curr. Opin. Chem. Biol., 2024, 78, 102417 CrossRef CAS.
- A. Santra, Y. Li, T. Ghosh, R. Li, H. Yu and X. Chen, Regioselective One-Pot Multienzyme (OPME) Chemoenzymatic Strategies for Systematic Synthesis of Sialyl Core 2 Glycans, ACS Catal., 2019, 9(1), 211–215 CrossRef CAS PubMed.
- Z. Xu, Y. Deng, Z. Zhang, W. Ma, W. Li, L. Wen and T. Li, Diversity-Oriented Chemoenzymatic Synthesis of Sulfated and Nonsulfated Core 2 O-GalNAc Glycans, J. Org. Chem., 2021, 86(15), 10819–10828 CrossRef CAS.
- S. Wang, C. Chen, M. R. Gadi, V. Saikam, D. Liu, H. Zhu, R. Bollag, K. Liu, X. Chen, F. Wang, P. G. Wang, P. Ling, W. Guan and L. Li, Chemoenzymatic modular assembly of O-GalNAc glycans for functional glycomics, Nat. Commun., 2021, 12(1), 3573 CrossRef CAS.
- C. Meng, A. Sasmal, Y. Zhang, T. Gao, C. C. Liu, N. Khan, A. Varki, F. Wang and H. Cao, Chemoenzymatic Assembly of Mammalian O-Mannose Glycans, Angew. Chem., Int. Ed., 2018, 57(29), 9003–9007 CrossRef CAS PubMed.
- T. Gao, J. Y. Yan, C. C. Liu, A. S. Palma, Z. M. Guo, M. Xiao, X. Chen, X. M. Liang, W. G. Chai and H. Z. Cao, Chemoenzymatic Synthesis of O-Mannose Glycans Containing Sulfated or Nonsulfated HNK-1 Epitope, J. Am. Chem. Soc., 2019, 141(49), 19351–19359 CrossRef CAS.
- M. R. Gadi, C. C. Chen, S. M. Bao, S. S. Wang, Y. X. Guo, J. H. Han, W. D. Xiao and L. Li, Convergent chemoenzymatic synthesis of GalNAc rare cores 5, 7, 8 and their sialylated forms, Chem. Sci., 2023, 14(7), 1837–1843 RSC.
- S. Wang, Q. Zhang, C. Chen, Y. Guo, M. R. Gadi, J. Yu, U. Westerlind, Y. Liu, X. Cao, P. G. Wang and L. Li, Facile Chemoenzymatic Synthesis of O-Mannosyl Glycans, Angew. Chem., Int. Ed., 2018, 57(30), 9268–9273 CrossRef CAS PubMed.
- M. O. Sheikh, S. M. Halmo and L. Wells, Recent advancements in understanding mammalian O-mannosylation, Glycobiology, 2017, 27(9), 806–819 CrossRef CAS PubMed.
- M. Van Scherpenzeel, E. Willems and D. J. Lefeber, Clinical diagnostics and therapy monitoring in the congenital disorders of glycosylation, Glycoconjugate J., 2016, 33(3), 345–358 CrossRef CAS.
- T. Li, Y. Zhang, T. Li, H. Zhuang, F. Wang, N. Wang, R. R. Schmidt and P. Peng, Divergent Synthesis of Core m1, Core m2 and Core m3 O–Mannosyl Glycopeptides via a Chemoenzymatic Approach, Chin. J. Chem., 2022, 40(13), 1571–1577 CrossRef CAS.
- D. Laaf, H. Steffens, H. Pelantová, P. Bojarová, V. Křen and L. Elling, Chemo–Enzymatic Synthesis of Branched N–Acetyllactosamine Glycan Oligomers for Galectin–3 Inhibition, Adv. Synth. Catal., 2017, 359(22), 4015–4024 CrossRef CAS.
- J. F. Ye, H. Xia, N. Sun, C. C. Liu, A. R. Sheng, L. L. Chi, X. W. Liu, G. F. Gu, S. Q. Wang, J. Zhao, P. Wang, M. Xiao, F. S. Wang and H. Z. Cao, Reprogramming the enzymatic assembly line for site-specific fucosylation, Nat. Catal., 2019, 2(6), 514–522 CrossRef CAS.
- N. Lu, J. Ye, J. Cheng, A. Sasmal, C. C. Liu, W. Yao, J. Yan, N. Khan, W. Yi, A. Varki and H. Cao, Redox-Controlled Site-Specific alpha2–6-Sialylation, J. Am. Chem. Soc., 2019, 141(11), 4547–4552 CrossRef CAS.
- N. Lu, Y. Li, H. Xia, K. Zhong, C. Jia, J. Ye, X. Liu, C. C. Liu and H. Cao, A Redox–Controlled Substrate Engineering Strategy for Site–Specific Enzymatic Fucosylation, Angew. Chem., 2022, 134(50), e202211032 CrossRef.
- I. A. Gagarinov, T. Li, N. Wei, J. S. Torano, R. P. de Vries, M. A. Wolfert and G. J. Boons, Protecting-Group-Controlled Enzymatic Glycosylation of Oligo-N-Acetyllactosamine Derivatives, Angew. Chem., Int. Ed., 2019, 58(31), 10547–10552 CrossRef CAS PubMed.
- G. M. Vos, Y. Wu, R. van der Woude, R. P. de Vries and G. J. Boons, Chemo-Enzymatic Synthesis of Isomeric I-branched Polylactosamines Using Traceless Blocking Groups, Chemistry, 2024, 30(5), e202302877 CrossRef CAS.
- H. K. Tseng, H. K. Wang, C. Y. Wu, C. K. Ni and C. C. Lin, Exploring Regioselective Fucosylation Catalyzed by Bacterial Glycosyltransferases through Substrate Promiscuity and Acceptor-Mediated Glycosylation, ACS Catal., 2023, 13(16), 10661–10671 CrossRef CAS.
- Z. Xiao, Y. Guo, Y. Liu, L. Li, Q. Zhang, L. Wen, X. Wang, S. M. Kondengaden, Z. Wu, J. Zhou, X. Cao, X. Li, C. Ma and P. G. Wang, Chemoenzymatic Synthesis of a Library of Human Milk Oligosaccharides, J. Org. Chem., 2016, 81(14), 5851–5865 CrossRef CAS PubMed.
- Y. T. Huang, Y. C. Su, H. R. Wu, H. H. Huang, E. C. Lin, T. W. Tsai, H. W. Tseng, J. L. Fang and C. C. Yu, Sulfo-Fluorous Tagging Strategy for Site-Selective Enzymatic Glycosylation of Human Milk Oligosaccharides, ACS Catal., 2021, 11(5), 2631–2643 CrossRef CAS.
- A. R. Prudden, L. Liu, C. J. Capicciotti, M. A. Wolfert, S. Wang, Z. Gao, L. Meng, K. W. Moremen and G. J. Boons, Synthesis of asymmetrical multiantennary human milk oligosaccharides, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(27), 6954–6959 CrossRef CAS.
- M. R. Charbonneau, L. V. Blanton, D. B. DiGiulio, D. A. Relman, C. B. Lebrilla, D. A. Mills and J. I. Gordon, A microbial perspective of human developmental biology, Nature, 2016, 535(7610), 48–55 CrossRef CAS PubMed.
- R. E. Moore, L. Y. L. Xu and S. D. Townsend, Prospecting Human Milk Oligosaccharides as a Defense Against Viral Infections, ACS Infect. Dis., 2021, 7(2), 254–263 CrossRef CAS.
- C. Kunz and S. Rudloff, Potential anti-inflammatory and anti-infectious effects of human milk oligosaccharides, Adv. Exp. Med. Biol., 2008, 606, 455–465 CrossRef CAS.
- S. Etzold and L. Bode, Glycan-dependent viral infection in infants and the role of human milk oligosaccharides, Curr. Opin. Virol., 2014, 7, 101–107 CrossRef CAS PubMed.
- M. Lu, I. Mosleh and A. Abbaspourrad, Engineered Microbial Routes for Human Milk Oligosaccharides Synthesis, ACS Synth. Biol., 2021, 10(5), 923–938 CrossRef CAS PubMed.
- X. Chen, Human Milk Oligosaccharides (HMOS): Structure, Function, and Enzyme-Catalyzed Synthesis, Adv. Carbohydr. Chem. Biochem., 2015, 72, 113–190 CrossRef PubMed.
- J. Zheng, H. Xu, J. Fang and X. Zhang, Enzymatic and chemoenzymatic synthesis of human milk oligosaccharides and derivatives, Carbohydr. Polym., 2022, 291, 119564 CrossRef CAS PubMed.
- W. Yao, J. Yan, X. Chen, F. Wang and H. Cao, Chemoenzymatic synthesis of lacto-N-tetrasaccharide and sialyl lacto-N-tetrasaccharides, Carbohydr. Res., 2015, 401, 5–10 CrossRef CAS PubMed.
- J. B. McArthur, H. Yu and X. Chen, A Bacterial beta1-3-Galactosyltransferase Enables Multigram-Scale Synthesis of Human Milk Lacto-N-tetraose (LNT) and Its Fucosides, ACS Catal., 2019, 9(12), 10721–10726 CrossRef CAS.
- T.-W. Tsai, J.-L. Fang, C.-Y. Liang, C.-J. Wang, Y.-T. Huang, Y.-J. Wang, J.-Y. Li and C.-C. Yu, Exploring the Synthetic Application of Helicobacter pylori α1,3/4-Fucosyltransferase FucTIII toward the Syntheses of Fucosylated Human Milk Glycans and Lewis Antigens, ACS Catal., 2019, 9(12), 10712–10720 CrossRef CAS.
- Y. Li, Y. Li, Y. Guo, C. Chen, L. Yang, Q. Jiang, P. Ling, S. Wang, L. Li and J. Fang, Enzymatic modular synthesis of asymmetrically branched human milk oligosaccharides, Carbohydr. Polym., 2024, 333, 121908 CrossRef CAS.
- U. Lindahl and M. Hook, Glycosaminoglycans and Their Binding to Biological Macromolecules, Annu. Rev. Biochem., 1978, 47(1), 385–417 CrossRef CAS.
- N. S. Gandhi and R. L. Mancera, The Structure of Glycosaminoglycans and their Interactions with Proteins, Chem. Biol. Drug Des., 2008, 72(6), 455–482 CrossRef CAS.
- X. Zhang, L. Lin, H. Huang and R. J. Linhardt, Chemoenzymatic Synthesis of Glycosaminoglycans, Acc. Chem. Res., 2020, 53(2), 335–346 CrossRef CAS PubMed.
- J. Gottschalk and L. Elling, Current state on the enzymatic synthesis of glycosaminoglycans, Curr. Opin. Chem. Biol., 2021, 61, 71–80 CrossRef CAS PubMed.
- G. Kogan, L. Soltes, R. Stern and P. Gemeiner, Hyaluronic acid: a natural biopolymer with a broad range of biomedical and industrial applications, Biotechnol. Lett., 2007, 29(1), 17–25 CrossRef CAS.
- L. A. Linkins, A. L. Dans, L. K. Moores, R. Bona, B. L. Davidson, S. Schulman and M. Crowther, Treatment and prevention of heparin-induced thrombocytopenia: Antithrombotic Therapy and Prevention of Thrombosis, 9th ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines, Chest, 2012, 141(2 Suppl), e495S–e530S CrossRef CAS PubMed.
- L. Liu, P. Chopra, X. Li, K. M. Bouwman, S. M. Tompkins, M. A. Wolfert, R. P. de Vries and G. J. Boons, Heparan Sulfate Proteoglycans as Attachment Factor for SARS-CoV-2, ACS Cent. Sci., 2021, 7(6), 1009–1018 CrossRef CAS.
- P. H. Weigel and P. L. DeAngelis, Hyaluronan synthases: a decade-plus of novel glycosyltransferases, J. Biol. Chem., 2007, 282(51), 36777–36781 CrossRef CAS PubMed.
- J. Mandawe, B. Infanzon, A. Eisele, H. Zaun, J. Kuballa, M. D. Davari, F. Jakob, L. Elling and U. Schwaneberg, Directed Evolution of Hyaluronic Acid Synthase from Pasteurella multocida towards High-Molecular-Weight Hyaluronic Acid, ChemBioChem, 2018, 19(13), 1414–1423 CrossRef CAS PubMed.
- W. Jing and P. L. DeAngelis, Synchronized chemoenzymatic synthesis of monodisperse hyaluronan polymers, J. Biol. Chem., 2004, 279(40), 42345–42349 CrossRef CAS PubMed.
- X. Fu, W. J. Shang, S. S. Wang, Y. P. Liu, J. Y. Qu, X. Chen, P. G. Wang and J. Q. Fang, A general strategy for the synthesis of homogeneous hyaluronan conjugates and their biological applications, Chem. Commun., 2017, 53(25), 3555–3558 RSC.
- S. Li, S. Wang, X. Fu, X. W. Liu, P. G. Wang and J. Fang, Sequential one-pot multienzyme synthesis of hyaluronan and its derivative, Carbohydr. Polym., 2017, 178, 221–227 CrossRef CAS PubMed.
- A. Eisele, H. Zaun, J. Kuballa and L. Elling, In Vitro One–Pot Enzymatic Synthesis of Hyaluronic Acid from Sucrose and N–Acetylglucosamine: Optimization of the Enzyme Module System and Nucleotide Sugar Regeneration, ChemCatChem, 2018, 10(14), 2969–2981 CrossRef CAS.
- A. E. Sismey-Ragatz, D. E. Green, N. J. Otto, M. Rejzek, R. A. Field and P. L. DeAngelis, Chemoenzymatic synthesis with distinct Pasteurella heparosan synthases: monodisperse polymers and unnatural structures, J. Biol. Chem., 2007, 282(39), 28321–28327 CrossRef CAS.
- N. Hodson, G. Griffiths, N. Cook, M. Pourhossein, E. Gottfridson, T. Lind, K. Lidholt and I. S. Roberts, Identification That KfiA, a Protein Essential for the Biosynthesis of the Escherichia coli K5 Capsular Polysaccharide, Is an α-UDP-GlcNAc Glycosyltransferase, J. Biol. Chem., 2000, 275(35), 27311–27315 CrossRef CAS.
- X. Zhang, V. Pagadala, H. M. Jester, A. M. Lim, T. Q. Pham, A. M. P. Goulas, J. Liu and R. J. Linhardt, Chemoenzymatic synthesis of heparan sulfate and heparin oligosaccharides and NMR analysis: paving the way to a diverse library for glycobiologists, Chem. Sci., 2017, 8(12), 7932–7940 RSC.
- Y. Yu, L. Fu, P. He, K. Xia, S. Varghese, H. Wang, F. Zhang, J. Dordick and R. J. Linhardt, Chemobiocatalytic Synthesis of a Low-Molecular-Weight Heparin, ACS Chem. Biol., 2022, 17(3), 637–646 CrossRef CAS.
- G. Zhang, K. Yang, L. Wang, Y. Cheng and C. Liu, Facile chemoenzymatic synthesis of unmodified anticoagulant ultra-low molecular weight heparin, Org. Biomol. Chem., 2022, 20(42), 8323–8330 RSC.
- S. Zhao, T. Zhang, Y. Kan, H. Li and J. P. Li, Overview of the current procedures in synthesis of heparin saccharides, Carbohydr. Polym., 2024, 339, 122220 CrossRef CAS PubMed.
- W. Lu, C. Zong, P. Chopra, L. E. Pepi, Y. Xu, I. J. Amster, J. Liu and G. J. Boons, Controlled Chemoenzymatic Synthesis of Heparan Sulfate Oligosaccharides, Angew. Chem., Int. Ed., 2018, 57(19), 5340–5344 CrossRef CAS PubMed.
- L. F. Sun, P. Chopra and G. J. Boons, Chemoenzymatic Synthesis of Heparan Sulfate Oligosaccharides having a Domain Structure, Angew. Chem., Int. Ed., 2022, 61(47), e202211112 CrossRef CAS.
- P.-H. Lin, Y. Xu, S. K. Bali, J. Kim, A. Gimeno, E. T. Roberts, D. James, N. M. S. Almeida, N. Loganathan, F. Fan, A. K. Wilson, I. J. Amster, K. W. Moremen, J. Liu, J. Jiménez-Barbero and X. Huang, Solid–Phase–Supported Chemoenzymatic Synthesis and Analysis of Chondroitin Sulfate Proteoglycan Glycopeptides, Angew. Chem., 2024, e202405671 CAS.
- Y. Wu, G. P. Bosman, D. Chapla, C. Huang, K. W. Moremen, R. P. de Vries and G. J. Boons, A Biomimetic Synthetic Strategy Can Provide Keratan Sulfate I and II Oligosaccharides with Diverse Fucosylation and Sulfation Patterns, J. Am. Chem. Soc., 2024, 146(13), 9230–9240 CrossRef CAS.
- S. G. Chen, C. H. Xue, L. A. Yin, Q. J. Tang, G. L. Yu and W. G. Chai, Comparison of structures and anticoagulant activities of fucosylated chondroitin sulfates from different sea cucumbers, Carbohydr. Polym., 2011, 83(2), 688–696 CrossRef CAS.
- E. Tykesson, M. Maccarana, H. Thorsson, J. Liu, A. Malmström, U. Ellervik and G. Westergren-Thorsson, Recombinant dermatan sulfate is a potent activator of heparin cofactor II-dependent inhibition of thrombin, Glycobiology, 2019, 29(6), 446–451 CrossRef CAS PubMed.
- J. Li, G. Su and J. Liu, Enzymatic Synthesis of Homogeneous Chondroitin Sulfate Oligosaccharides, Angew. Chem., Int. Ed., 2017, 56(39), 11784–11787 CrossRef CAS.
- E. Stancanelli, W. Liu, R. Wander, J. Li, Z. Wang, K. Arnold, G. Su, A. Kanack, T. Q. Pham, V. Pagadala, A. Padmanabhan, Y. Xu and J. Liu, Chemoenzymatic Synthesis of Homogeneous Heparan Sulfate and Chondroitin Sulfate Chimeras, ACS Chem. Biol., 2022, 17(5), 1207–1214 CrossRef CAS PubMed.
- E. Tykesson, A. Hassinen, K. Zielinska, M. A. Thelin, G. Frati, U. Ellervik, G. Westergren-Thorsson, A. Malmström, S. Kellokumpu and M. Maccarana, Dermatan sulfate epimerase 1 and dermatan 4–sulfotransferase 1 form complexes that generate long epimerized 4–sulfated blocks, J. Biol. Chem., 2018, 293(35), 13725–13735 CrossRef CAS.
- Y. Yamazaki, S. Kimura and M. Ohmae, Reaction specificity of keratanase II in the transglycosylation using the sugar oxazolines having keratan sulfate repeating units, Carbohydr. Res., 2018, 456, 61–68 CrossRef CAS PubMed.
- Y. Wu, G. M. Vos, C. Huang, D. Chapla, A. L. M. Kimpel, K. W. Moremen, R. P. de Vries and G.-J. Boons, Exploiting Substrate Specificities of 6-O-Sulfotransferases to Enzymatically Synthesize Keratan Sulfate Oligosaccharides, JACS Au, 2023, 3(11), 3155–3164 CrossRef CAS.
- R. C. R. Jala, S. Vudhgiri and C. G. Kumar, A comprehensive review on natural occurrence, synthesis and biological activities of glycolipids, Carbohydr. Res., 2022, 516, 108556 CrossRef CAS.
- H. Ando and N. Komura, Recent progress in the synthesis of glycosphingolipids, Curr. Opin. Chem. Biol., 2024, 78, 102423 CrossRef CAS.
- J. Zhang, C. Chen, M. R. Gadi, C. Gibbons, Y. Guo, X. Cao, G. Edmunds, S. Wang, D. Liu, J. Yu, L. Wen and P. G. Wang, Machine–Driven Enzymatic Oligosaccharide Synthesis by Using a Peptide Synthesizer, Angew. Chem., 2018, 130(51), 16880–16884 CrossRef.
- T. Li, L. Liu, N. Wei, J. Y. Yang, D. G. Chapla, K. W. Moremen and G. J. Boons, An automated platform for the enzyme-mediated assembly of complex oligosaccharides, Nat. Chem., 2019, 11(3), 229–236 CrossRef CAS PubMed.
- R. Basharat, V. Kotra, L. Y. Loong, A. Mathews, M. M. Kanakal, C. H. B. P. Dev, S. Nyamathulla, R. Varala, L. C. Ming, K. R. S. S. Rao, B. H. Babu and M. M. Alam, A Mini-review on Ultra Performance Liquid Chromatography, Orient. J. Chem., 2021, 37(4), 847–857 CAS.
- J. Q. Wang, J. Zhao, S. P. Nie, M. Y. Xie and S. P. Li, Mass spectrometry for structural elucidation and sequencing of carbohydrates, TrAC, Trends Anal. Chem., 2021, 144, 116436 CrossRef CAS.
- B. Xia, J. Fang, S. Ma, M. Ma, G. Yao, T. Li, X. Cheng, L. Wen and Z. Gao, Mapping the Acetylamino and Carboxyl Groups on Glycans by Engineered alpha-Hemolysin Nanopores, J. Am. Chem. Soc., 2023, 145(34), 18812–18824 CrossRef CAS.
- G. Yao, Y. Tian, W. Ke, J. Fang, S. Ma, T. Li, X. Cheng, B. Xia, L. Wen and Z. Gao, Direct Identification of Complex Glycans via a Highly Sensitive Engineered Nanopore, J. Am. Chem. Soc., 2024, 146(19), 13356–13366 CrossRef CAS PubMed.
- G. Yao, W. Ke, B. Xia and Z. Gao, Nanopore-based glycan sequencing: state of the art and future prospects, Chem. Sci., 2024, 15(17), 6229–6243 RSC.
- J. L. Abrahams, G. Taherzadeh, G. Jarvas, A. Guttman, Y. Zhou and M. P. Campbell, Recent advances in glycoinformatic platforms for glycomics and glycoproteomics, Curr. Opin. Struct. Biol., 2020, 62, 56–69 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |