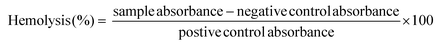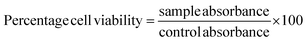Open Access Article
Open Access ArticleDoxorubicin-Polysorbate 80 conjugates: targeting effective and sustained delivery to the brain†
S.
Ram Prasad‡
 *a,
Sruthi Sudheendran
Leena
b,
Ani
Deepthi
*a,
Sruthi Sudheendran
Leena
b,
Ani
Deepthi
 b,
A. N.
Resmi
c,
Ramapurath S.
Jayasree
b,
A. N.
Resmi
c,
Ramapurath S.
Jayasree
 c,
K. S.
Sandhya
b and
A.
Jayakrishnan§
*ab
c,
K. S.
Sandhya
b and
A.
Jayakrishnan§
*ab
aDivision of Chemical Biology, Rajiv Gandhi Centre for Biotechnology, Jagathy, Trivandrum 695 014, Kerala, India. E-mail: jayakrishnana@rgcb.res.in; jayakrishnan1953@gmail.com (A.J); dr.rps1789@gmail.com (S.R.P)
bDepartment of Chemistry, University of Kerala, Kariavattom, Trivandrum 695 581, Kerala, India
cBiophotonics and Imaging Laboratory, Biomedical Technology Wing, Sree Chitra Tirunal Institute for Medical Sciences and Technology, Trivandrum 695 012, Kerala, India
First published on 10th May 2024
Abstract
Targeting therapeutic agents to the brain to treat central nervous system (CNS) diseases is a major challenge due to the blood–brain barrier (BBB). In this study, an attempt was made to deliver a model drug such as doxorubicin (DOX), to the brain in a mouse model through DOX-Polysorbate 80 (DOX-PS80) conjugates. DOX was successfully conjugated with the non-ionic surfactant Polysorbate 80 (PS80) by carbamate linkage and the conjugate was characterized by different spectroscopic techniques, such as FTIR, UV-Visible and NMR. The DOX conjugation efficacy was found to be 43.69 ± 4.72%. The in vitro cumulative release of DOX from the conjugates was found to be 4.9 ± 0.8% in PBS of pH 7.3 and 3.9 ± 0.6% in simulated cerebrospinal fluid (CSF) of pH 7.3 at the end of 10 days. An in vitro BBB permeability assay was carried out using bEnd.3 cells and DOX-PS80 conjugate showed a 3-fold increase in BBB permeability compared with controls. In vitro cytotoxicity assay using U251 human glioblastoma cells showed an IC50 value of 38.10 μg mL−1 for DOX-PS80. Cell uptake studies revealed that DOX-PS80 was effectively taken up (90%) by the bEnd.3 and U251 cells and localized in cytoplasm at the end of 24 h. Pharmacokinetic parameters for DOX-PS80 were evaluated using in silico studies. Tumor spheroid assay and in vivo experiments in Swiss albino mouse demonstrated the possibility of DOX-PS80 conjugate crossing the BBB and delivering the drug molecules to the target site for treating CNS disorders.
1. Introduction
Targeting therapeutic agents to the brain to treat central nervous system (CNS) diseases is a major challenge due to the blood–brain barrier (BBB), the major role of which is to restrict the entry of pathogenic organisms and unwanted substances into the brain.1–3 In spite of the prodigious developments in our understanding of the molecular structure and the physiology of the BBB as well as the advances in the therapeutic approaches to treat CNS diseases, effective delivery of many potential therapeutic agents to treat CNS disorders and diseases is still in its infancy. Any attempt to transport drugs across the BBB is against the natural function of BBB.4 The main anatomical and functional site of the BBB is the brain endothelium.5 The presence of tight junctions (TJ) between adjacent endothelial cells, formed by an intricate complex of transmembrane proteins, impedes the drug transport into the brain via the blood circulation.3–5In order to overcome the constraints in delivering drugs to the brain or CNS, it is important to understand the physiology of the BBB, the response of the BBB to physical and chemical stimuli, the different transporter receptors present there, and its permeability under different pathological or disease conditions, and to have knowledge about the drug delivery technologies that could overcome the barrier constraints.4–7 Currently various approaches and methodologies are attempted for transporting drugs across the BBB, such as opening the TJ using chemical stimulants like sodium dodecyl sulphate, transport system-mediated delivery (adsorptive-mediated transcytosis (AMT), cationic proteins, and cell-penetrating peptides (CPP) capable of entering cells) and nanocarrier-based delivery using liposomes and polymeric nanoparticles.8–20
Among various systems, considerable research has been reported over the years on the use of polymeric/liposomal/inorganic nanoparticulate drug delivery systems for crossing the BBB to deliver a number of therapeutic agents.21–28 While the nanoparticle (NP) approach is interesting from the point view of examining the possibility of delivering drugs across the BBB, there are many problems associated with nanocarriers, especially when targeting the brain.26–30 NPs coated with polysorbate (PS)-80, a non-ionic surfactant, have been shown to cross the BBB and deliver a number of drugs to the brain.31–33 Despite the fact that PS-coated NPs displayed BBB permeability, there are still some issues to be resolved, such as the toxicity of polycyanoacrylates or inorganic/metals used as NPs and their toxic byproducts. There is the possibility of accumulation of NPs as well as their degradation products in the brain capillaries. The drug cargo in the NPs is often only a very small percentage of the weight of the NPs, and fabrication and incorporation of drugs in the NPs usually involve organic solvents, traces of which are often difficult to remove completely. This is the case even when more biocompatible and biodegradable NPs from poly(lactic acid) or poly(glycolic acid) are used. The encapsulation process also results in wasting a significant portion of the drug, as encapsulation efficiencies are rather low in most encapsulation processes. In the work described wherein NPs are coated with polysorbate-80,27–39 the surfactant is physically adsorbed onto the NPs and one has little control on the extent of adsorption of the surfactant.
If polysorbates are responsible for the transport of NPs across the BBB as has been reported for many drugs, and the mechanism of transport is believed to be the adsorption of ApoE or ApoB on the NPs to facilitate their transport,31,38,39 then drugs that are conjugated to polysorbates should obviously have the ability to cross the BBB. Polysorbates are surfactants based on poly(oxyethylene) sorbitan esters and have been approved by the US FDA as additives in pharmaceutical preparations (e.g., for Tween-80, Tween-20). In a recent study,40 we have shown that amphotericin B (AmB) conjugated to Tween 20 through hydrolysable carbamate linkages increases the solubility of an otherwise water-insoluble AmB dramatically, is able to release the drug when incubated in phosphate buffer, exhibited negligible hemolytic potential even at a concentration of 1000 μM, and was not toxic against human embryonic kidney cell line (HEK293T) at similar concentrations.40 Various NP carriers, including gold, iron oxide, and PLGA NPs, as well as polysorbate-modified NPs, were reported to be used to deliver drugs to the brain.40–48
The current study is based on the hypothesis that a polymeric prodrug like DOX-PS80 should be able to pass the BBB in sustained manner if the polycyanoacrylate/albumin/silica/polymer NPs coated with PS80 are able to do so, as previously reported in numerous studies.29–48 Here, the drug DOX was coupled to the surfactant polysorbate-80 via a hydrolysable carbamate linkage to test the hypothesis, thereby allowing precise control over the prodrug's structure and composition and its solubility in aqueous vehicles for injection without the burden of the toxicity of NPs as carriers, their insolubility, aggregation, and degradation. DOX-PS80 conjugates displayed significant BBB permeation and prolonged DOX release in both in vitro and in vivo investigations. We are aware that DOX is not the standard drug for treatment for brain tumors; the objective of this investigation is to demonstrate that polysorbate-based drug conjugates will be an effective approach for crossing the BBB and delivering small molecules to the brain to treat CNS disorders.
2. Materials and methods
2.1. Materials
Polysorbate 80 (PS80) was purchased from S. D. Fine Chemicals, Mumbai, India. It was dried by azeotropic distillation with toluene and stored in a desiccator. p-Nitrophenyl chloroformate (PNPC) and 4-dimethylamino pyridine (DMAP) were obtained from Spectrochem Ltd, Mumbai, India. Tetrahydrofuran (THF) was dried over sodium/benzophenone and distilled prior to use. Dry dimethyl formamide (DMF) was prepared by drying the solvent overnight over barium oxide, followed by distillation in a vacuum. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT), 4′,6-diamidino-2-phenylindole (DAPI) and Dulbecco's Modified Eagle's medium (DMEM) media were from Himedia labs, India. Potassium bromide (KBr) was from Himedia Labs, Mumbai, India. Chloroform D (D, 99.8%) (CDCl3) was from Cambridge Isotope Laboratories, Inc. (USA). Brain endothelial cells (bEnd.3) were procured from NCCS, Pune and U251 glioblastoma cells were procured from Cancer cell repository, Rajiv Gandhi Center for Biotechnology (RGCB), Trivandrum. Distilled water was used throughout. All other solvents such as chloroform, methanol, hexane, ethyl acetate etc., were of analytical grade. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), India. RGCB CPCSEA registration number: 326/GO/ReBiBt/S/2001/CPCSEA. The experimental protocols were approved by the Institutional Animal Ethics Committee of Animal Research Facility (IAEC-ARF), RGCB, Trivandrum India. In vivo biodistribution animal studies IAEC-ARF approval number: IAEC/856,877/RAM/2021-2022.2.2. Instrumentation
1H NMR spectra were recorded using a Bruker instrument (Bruker AV III 400 MHz FT NMR, USA). Fourier-transform infrared (FTIR) spectra were taken using KBr pellets using a PerkinElmer FT-IR spectrophotometer (Spectrum Two Model, USA). UV-visible spectra were recorded using a UV-Visible spectrometer (PerkinElmer, Lambda 25, USA). The Elisa microplate reader used was BIORAD, iMark, India. Confocal microscopy was used to capture cell imaging (Leica SP8 WLL, Germany). A Varioskan™ LUX multimode microplate reader was used to measure the fluorescence intensity. The quantification of drug uptake in cells was performed by flow cytometry (BD FACSVerseTM, BD Biosciences, USA).2.3. Synthesis of PS80-PNPC
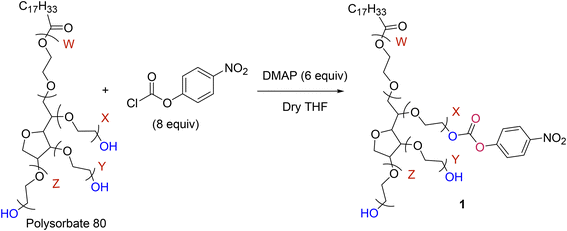
PS80 was activated using PNPC using the following procedure.40 PS80 (0.381 mmol) was dissolved in dry THF (5 mL) and 279 mg (2.29 mmol) of DMAP was added, and the contents were stirred for 10 min. Then, PNPC (614 mg, 3.05 mmol) was added, and the mixture was stirred at room temperature overnight. The product formation was identified by thin layer chromatography (Silica gel 60 F254 aluminium sheet) every 2 h. After product formation, the precipitate was filtered using Whatman filter paper grade 1 (150 mm) and the crude product was collected in a round-bottom flask and concentrated using a rotavapor. The crude product was purified by column chromatography on silica gel (100–200 mesh) using ethyl acetate/hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) followed by chloroform/methanol (9.5
1) followed by chloroform/methanol (9.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5) to give a yellow-coloured viscous liquid which was characterized and named PS80-PNPC. Yield: 60 to 80% (Scheme 1).
0.5) to give a yellow-coloured viscous liquid which was characterized and named PS80-PNPC. Yield: 60 to 80% (Scheme 1).
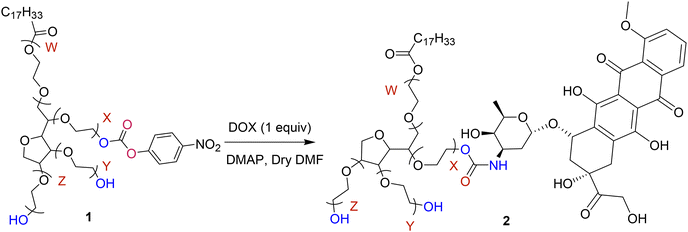
2.4. Synthesis of DOX-PS80 conjugate
The bioconjugate of DOX with PS80-PNPC was synthesized using the following procedure. PS80-PNPC (100 mg, 0.0677 mmol) and DMAP (8.27 mg, 0.0677 mmol) were dissolved in dry DMF (3 mL), and the solution was treated with a suspension of doxorubicin (36.79 mg, 0.0677 mmol) in DMF (2 mL) for 24 h at room temperature (Scheme 2). After the reaction, the clear solution was poured into cold ether (50 mL) and the product was collected by filtration on a sintered glass filter under nitrogen and was washed with ether (3 × 20 mL). The residue was then dissolved in methanol (5 mL), precipitated with cold ether (25 mL), filtered and washed with hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ether (70
ether (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) until the free DOX and other impurities were removed. The washing procedure was repeated (5×) to ensure complete removal of free DOX. The precipitate was finally washed with hexane and dried in a vacuum desiccator (Yield 80%). The compound is named as DOX-PS80 Conjugate.
30) until the free DOX and other impurities were removed. The washing procedure was repeated (5×) to ensure complete removal of free DOX. The precipitate was finally washed with hexane and dried in a vacuum desiccator (Yield 80%). The compound is named as DOX-PS80 Conjugate.
2.5. Estimation of drug content
The amount of DOX conjugated in the PS80 was calculated by using UV-visible spectroscopy. DOX-PS80 (5 mg) was dissolved in 200 μL methanol and made up to 5 mL using phosphate buffered saline (PBS) to achieve 1 mg mL−1 concentration. Then samples were serially diluted, and the absorbance was measured at 480 nm. The amount of DOX conjugated to the PS80 was calculated using a standard curve of DOX in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 methanol:PBS and measuring the absorbance at 480 nm. Measurements were done in triplicate and averaged.
9 methanol:PBS and measuring the absorbance at 480 nm. Measurements were done in triplicate and averaged.
2.6. In vitro drug release studies
The in vitro drug release studies were performed using phosphate buffered saline (PBS) pH 7.3 and simulated cerebrospinal fluid (CSF) pH 7.3. DOX-PS80 conjugate (5 mg) was dissolved in 5 ml of buffer and transferred into 3.5 kDa dialysis tubing. The dialysis tubing was kept in 100 mL PBS pH 7.3 or CSF pH 7.3 in 250 mL beakers in a constant temperature bath maintained at 37 °C. At regular intervals, 1 mL of supernatant from the samples was withdrawn and replaced with fresh buffer. The samples were withdrawn for 10 days at regular time points. The amount of drug released from the conjugates was estimated by UV-visible spectrophotometry by measuring the absorbance at 480 nm.2.7. Hemolysis
Hemolysis assay was performed using the protocol of Francis et al.47 Briefly, human blood was obtained from the blood bank in EDTA-coated vacutainers. Red blood cells (RBCs) were separated from the whole blood by centrifuging at 1500 rpm for 5 min. The sediment of RBC pellets was separated, and supernatant was discarded. The centrifuged RBCs were washed with 150 mM of normal saline (3×). Then the concentrated RBCs were dispersed in PBS (pH 7.4) and diluted to 10-fold from initial concentration. DOX-PS80 and DOX were prepared at different concentrations of 20 to 100 μg ml−1 and transferred to 2 mL Eppendorf tubes and sterilized with UV radiation for 2 h in a bio-safety cabinet. The diluted RBCs (200 μL) were added to the Eppendorf tubes and the volume was adjusted to 1 mL using PBS (pH 7.4). The control samples were prepared without drug. RBCs dispersed in PBS and water were used as negative and positive control, respectively. All the samples were incubated for 1 h at 37 °C. Then the Eppendorf tubes were centrifuged for 5 min at 1500 rpm. The supernatant solution was separated and absorbance was measured at 540 nm using a microplate reader (BIORAD, iMark, India). The hemolysis percentage was calculated by following formula,2.8. Cell culture studies
2.9. Ex vivo tumor spheroids assay
The tumor spheroids formation protocol was adapted from Piyush et al.48 Briefly, U251 cells were grown in T25 flasks using DMEM with 10% FBS and 1% antibiotic-antimitotic solution and incubated at 37 °C/5% CO2 until reaching 90% confluence. Then, 96-well plates were coated with 2% agar and sterilized under UV for 2 h. Once the cells were confluent and healthy, cells were trypsinized and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded per well in 200 μL DMEM media and incubated at 37 °C. The plates were regularly monitored until the cells were in the shape of uniform spheroids. At the end of 48 h, uniform spheroids were formed, and images were captured. Then, the spheroids were treated with free DOX (5 μg ml−1), DOX-PS80 (10 μg ml−1) and free PS80 (5 μg ml−1) and incubated at 37 °C. Control was without any sample. Every 24 h, the spheroids’ morphology was observed, and images were captured using a phase contrast microscope (Nikon Ti eclipse, USA) at 4× magnification and compared with control spheroids. The average diameter of untreated and sample treated tumor spheroids was calculated using Image J software (U. S. National Institutes of Health, Bethesda, Maryland, USA).
000 cells were seeded per well in 200 μL DMEM media and incubated at 37 °C. The plates were regularly monitored until the cells were in the shape of uniform spheroids. At the end of 48 h, uniform spheroids were formed, and images were captured. Then, the spheroids were treated with free DOX (5 μg ml−1), DOX-PS80 (10 μg ml−1) and free PS80 (5 μg ml−1) and incubated at 37 °C. Control was without any sample. Every 24 h, the spheroids’ morphology was observed, and images were captured using a phase contrast microscope (Nikon Ti eclipse, USA) at 4× magnification and compared with control spheroids. The average diameter of untreated and sample treated tumor spheroids was calculated using Image J software (U. S. National Institutes of Health, Bethesda, Maryland, USA).
2.10. Biodistribution studies by IVIS imaging
The transportation of the drug conjugates to the brain was analyzed using IVIS animal imaging (Xenogen IVIS Spectrum, PerkinElmer, USA). Healthy adult Swiss albino mice (male) in the age group of 8–10 weeks (19–22 g) were selected for the study. The experiments were performed with Institute (RGCB) animal ethical committee approval (No.: IAEC/856,877/RAM/2021-2022). The animals were divided into 3 groups, namely DOX, DOX-PS80 and untreated control. These preparations were injected i.v. into the tail vein using the following dose regimens: 2.5 mg ml−1 for DOX-PS80 conjugate and 1.25 mg ml−1 of DOX (equivalent to DOX in conjugate). The whole animal imaging was carried out after the animals were anaesthetized with isoflurane at different time points, i.e., 1 h and 24 h. The images were captured by fixing DOX excitation wavelength (480 nm) and emission filters with background subtraction. Tissue/blood auto-fluorescence was excluded by setting GFP excitation/emission filter from untreated mice.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phosphate buffer pH 3
phosphate buffer pH 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (25
methanol (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) and the mixture was centrifuged at 8000 rpm for 15 min and supernatant was separated. The supernatant samples were syringe filtered (0.45 μm) and analyzed using fluorescence spectroscopy by fixing the excitation and emission wavelength at 480 nm and 555 nm.49,50
30) and the mixture was centrifuged at 8000 rpm for 15 min and supernatant was separated. The supernatant samples were syringe filtered (0.45 μm) and analyzed using fluorescence spectroscopy by fixing the excitation and emission wavelength at 480 nm and 555 nm.49,50
2.11. In silico studies: analysis of pharmacokinetic parameters
Computational modeling studies are gold standard techniques to evaluate the drug likeness and pharmacokinetic profiles of new chemical entities.60,61 Among different computational resources, artificial intelligence (AI) and machine learning (ML)-based prediction software packages have recently been the focus for predicting the absorption (A), distribution (D), metabolism (M), excretion (E) and toxicity (T) (ADMET) properties for new therapeutic molecules.61,62 In our study, to acquire a deeper understanding of the ADMET properties of DOX, PS80, and DOX-PS80 conjugates, a variety of AI-based ADMET tools were utilized, including ADMET-AI,62 ADMETlab 2.0,63 PreADMET,64 admetSAR65 and SwissADME.66 The physicochemical parameters and ADMET score of all the three compounds were analyzed in different ADMET tools and best outcomes were reported.2.12. Statistical analysis
For all the experiments one-way and two-way ANOVA was performed using Graph Pad Prism (version 5.0) software and the values are expressed as mean ± SD. The differences were considered significant if p < 0.05.3. Results and discussion
PS80 was activated by para nitrophenyl chloroformate (PNPC) in dry tetrahydrofuran (THF) leading to a stable carbonate intermediate as previously reported from our laboratory40 (Scheme 1). Briefly, the activation of the PS80's hydroxyl group was achieved by employing the 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
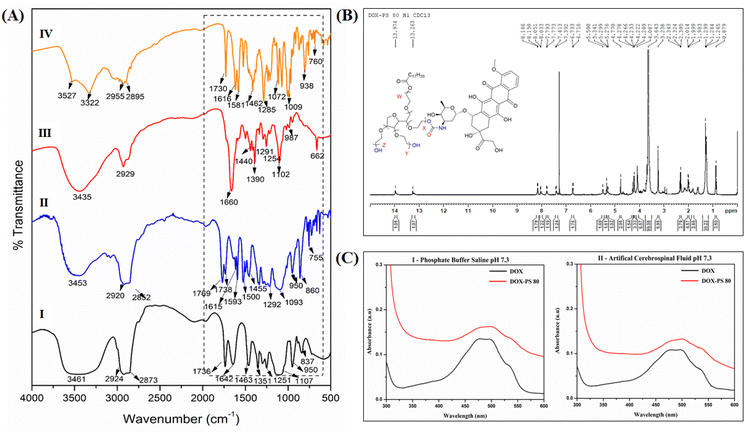
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 equivalent dimethyl amino pyridine (DMAP) and PNPC. The activation of hydroxyls was confirmed by both FTIR and 1H NMR spectroscopy. PS80 displays characteristic absorption peaks at 3461 cm−1 (OH stretching), 2924 cm−1 (CH2 stretching), 1736 cm−1 (ester carbonyl) and ether linkages of PEG at 1104 cm−1. When PS80 was activated by PNPC through a carbonate bond, a characteristic absorption peak of carbonate was observed in the region of 1769 cm−1, along with the strong absorption of nitro group in the region of 1217 cm−1 confirming the activation of the hydroxyl group of PS80 (Fig. 1A-I).
8 equivalent dimethyl amino pyridine (DMAP) and PNPC. The activation of hydroxyls was confirmed by both FTIR and 1H NMR spectroscopy. PS80 displays characteristic absorption peaks at 3461 cm−1 (OH stretching), 2924 cm−1 (CH2 stretching), 1736 cm−1 (ester carbonyl) and ether linkages of PEG at 1104 cm−1. When PS80 was activated by PNPC through a carbonate bond, a characteristic absorption peak of carbonate was observed in the region of 1769 cm−1, along with the strong absorption of nitro group in the region of 1217 cm−1 confirming the activation of the hydroxyl group of PS80 (Fig. 1A-I).
A comparison of the FTIR spectra of PS80 and PS80-(4-nitrophenyl carbonate) was done as follows. PS80 showed characteristic IR absorptions (Fig. 1A-I) at 3461 cm−1 (OH stretch), 2924 cm−1 (C–H stretch), 1736 cm−1 (ester CO stretch) and 1107 cm−1 (ethereal C–O stretch) (Fig. 1A-I). Activation of PS80 by PNPC is through a carbonate bond and its characteristic stretching frequencies are from the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, O–C–O and O–C–C vibrations. The IR spectrum of activated PS80 (Fig. 1A-II) clearly showed the strong peaks at 1769 cm−1 (carbonate C
O, O–C–O and O–C–C vibrations. The IR spectrum of activated PS80 (Fig. 1A-II) clearly showed the strong peaks at 1769 cm−1 (carbonate C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch), 1738 cm−1 (ester C
O stretch), 1738 cm−1 (ester C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch), 1217 cm−1 (O–C–O stretch) and 1093 cm−1 (C–O stretch). Additionally, the strong peak at 1593 cm−1 can be attributed to the N–O stretching of the nitro group. All this evidence pointed out the successful activation of PS80 by PNPC resulting in PS80-PNPC.
O stretch), 1217 cm−1 (O–C–O stretch) and 1093 cm−1 (C–O stretch). Additionally, the strong peak at 1593 cm−1 can be attributed to the N–O stretching of the nitro group. All this evidence pointed out the successful activation of PS80 by PNPC resulting in PS80-PNPC.
Furthermore, the activation of PS80 by PNPC was confirmed by NMR spectroscopy. PS80 (CDCl3, δ ppm) 1H NMR spectrum showed peaks at 5.33 ppm (CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 4.2 ppm (OCOCH2) and 3.6 ppm (CH2) corresponding to olefic proton, 2.3 ppm (CH2OCO), 1.9 ppm (CH2CH2OCO) and peaks at 1.2–1.3 ppm (CH2), 0.8 ppm (CH3) corresponding to lauric acid residues (Fig. S1†). The 13C NMR of PS80 is shown in Fig. S2.† In addition to the polysorbate peaks, the appearance of new signals at 8.29, 8.27 ppm and 7.41, 7.39 ppm was attributed to the aromatic protons from PNPC (Fig. S3†). The peak signals of sorbitan polyethoxylates (SPEs) series i.e., O–CH2–CH2–O of PS80 at 5.3 ppm and aromatic proton at 8.2 ppm from PNPC confirmed the activation of polysorbate. The 13C NMR spectrum also confirmed the activation of PS80 by PNPC (Fig. S4†). The aromatic carbonyl carbon of PNPC was observed at δ 155 ppm, 145 ppm, 121 ppm and 125 ppm and the carbonate carbonyl carbon was observed at δ 171.1 ppm (Fig. S4†). NMR spectra of PS80-PNPC were further analysed, 1H NMR (400 MHz, CDCl3, δ ppm) showed 8.27 ppm (d, J = 9.2 Hz, 2H), 7.39 ppm (d, J = 9.2 Hz, 2H), 5.3 ppm (m, 1H), 4.4 ppm (d, J = 9.2 Hz, 2H), 4.25 (t, J = 4.5 Hz, 2H), 3.84–3.78 ppm (m, 1H), 3.69 ppm (q, J = 0 Hz, 4H), 3.65 ppm (dd, J = 7.6 Hz, 59H), 2.33 ppm (t, J = 7.6 Hz, 2H), 2.04–1.96 ppm (m, 1H), 1.37–1.21 ppm (m, 22 H) and 0.87 ppm (t, J = 7.0 Hz, 3H).
CH), 4.2 ppm (OCOCH2) and 3.6 ppm (CH2) corresponding to olefic proton, 2.3 ppm (CH2OCO), 1.9 ppm (CH2CH2OCO) and peaks at 1.2–1.3 ppm (CH2), 0.8 ppm (CH3) corresponding to lauric acid residues (Fig. S1†). The 13C NMR of PS80 is shown in Fig. S2.† In addition to the polysorbate peaks, the appearance of new signals at 8.29, 8.27 ppm and 7.41, 7.39 ppm was attributed to the aromatic protons from PNPC (Fig. S3†). The peak signals of sorbitan polyethoxylates (SPEs) series i.e., O–CH2–CH2–O of PS80 at 5.3 ppm and aromatic proton at 8.2 ppm from PNPC confirmed the activation of polysorbate. The 13C NMR spectrum also confirmed the activation of PS80 by PNPC (Fig. S4†). The aromatic carbonyl carbon of PNPC was observed at δ 155 ppm, 145 ppm, 121 ppm and 125 ppm and the carbonate carbonyl carbon was observed at δ 171.1 ppm (Fig. S4†). NMR spectra of PS80-PNPC were further analysed, 1H NMR (400 MHz, CDCl3, δ ppm) showed 8.27 ppm (d, J = 9.2 Hz, 2H), 7.39 ppm (d, J = 9.2 Hz, 2H), 5.3 ppm (m, 1H), 4.4 ppm (d, J = 9.2 Hz, 2H), 4.25 (t, J = 4.5 Hz, 2H), 3.84–3.78 ppm (m, 1H), 3.69 ppm (q, J = 0 Hz, 4H), 3.65 ppm (dd, J = 7.6 Hz, 59H), 2.33 ppm (t, J = 7.6 Hz, 2H), 2.04–1.96 ppm (m, 1H), 1.37–1.21 ppm (m, 22 H) and 0.87 ppm (t, J = 7.0 Hz, 3H).
The activated PS80-PNPC was conjugated with DOX using DMAP in dry DMF (Scheme 2) with a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The conjugation of DOX with PS80 was characterized by FTIR, NMR and UV-Visible spectroscopy. FTIR spectrum of DOX-PS80 (Fig. 1A-III) showed a sharp peak at 1660 cm−1 corresponding to the C
1. The conjugation of DOX with PS80 was characterized by FTIR, NMR and UV-Visible spectroscopy. FTIR spectrum of DOX-PS80 (Fig. 1A-III) showed a sharp peak at 1660 cm−1 corresponding to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch of the carbamate group and the absence of the carbonate signal, strongly suggesting the formation of the DOX-PS80 conjugate. Furthermore, 1H NMR spectrum of DOX-PS80 showed signals at δ 8.01, 7.79 and 7.74 ppm, ascribed to the aromatic protons from DOX, while the peak at δ 8.16 ppm corresponded to the N–H proton and peaks at δ 13.96 and 13.26 ppm corresponded to the phenolic hydroxyl protons (Fig. 1B). The 13C NMR spectrum showed the disappearance of carbonyl carbon of PNPC at δ 155, 145, 121, 125, 171.1 ppm and the appearance of DOX carbonyl peaks at δ 56, 110, 117, 19, 129, 135, 137,160, 213 ppm, confirming the conjugation of DOX with polysorbate (Fig. S5†).
O stretch of the carbamate group and the absence of the carbonate signal, strongly suggesting the formation of the DOX-PS80 conjugate. Furthermore, 1H NMR spectrum of DOX-PS80 showed signals at δ 8.01, 7.79 and 7.74 ppm, ascribed to the aromatic protons from DOX, while the peak at δ 8.16 ppm corresponded to the N–H proton and peaks at δ 13.96 and 13.26 ppm corresponded to the phenolic hydroxyl protons (Fig. 1B). The 13C NMR spectrum showed the disappearance of carbonyl carbon of PNPC at δ 155, 145, 121, 125, 171.1 ppm and the appearance of DOX carbonyl peaks at δ 56, 110, 117, 19, 129, 135, 137,160, 213 ppm, confirming the conjugation of DOX with polysorbate (Fig. S5†).
DOX-PS80 1H NMR (400 MHz, CDCl3δ ppm): 13.97 ppm (d, J = 9.28 Hz, 1H), 13.52–12.94 ppm (m, 1H), 8.16 ppm (d, J = 6.52 Hz, 2H), 8.10–7.95 ppm (m, 1H), 7.81–7.77 ppm (m, 1H), 7.41–7.39 ppm (m, 1H), 6.72 ppm (d, J = 6.8 Hz, 2H), 5.50 ppm (s, 2H), 5.40–5.32 ppm (m, 1H), 5.31–5.22 ppm (m, 1H), 4.77 ppm (s, 2H), 4.32–4.25 ppm (m, 1H), 4.22 ppm (dd, J = 5.7, 3.9 Hz, 3H), 4.08 ppm (d, J = 3.4 Hz, 5H), 3.64 ppm (q, J = 3.5 Hz, 99 H), 3.24 ppm (s, 6H), 2.32 ppm (t, J = 7.5 Hz, 4H), 2.16 ppm (dd, J = 15.3, 3.3 Hz, 4H), 2.06–1.96 ppm (m, 4H), 1.50–1.10 ppm (m, 30H), 0.89 ppm (t, J = 6.9 Hz, 3H).
The conjugation of DOX with PS80 was also confirmed by UV visible spectroscopy. DOX-PS80 conjugate exhibited characteristic absorptions peaks of DOX at 480, 496 and 538 nm, confirming the DOX coupling onto the PS80. The absorption spectra of conjugate in PBS (pH 7.3) and CSF (pH 7.3) are shown in Fig. 1C. The amount of DOX in the conjugates was calculated by measuring the absorption of the DOX-PS80 conjugate at 480 nm using a standard plot of DOX constructed at different concentrations. The amount of drug conjugated onto the PS80 was found to be 43.69 ± 4.72%.
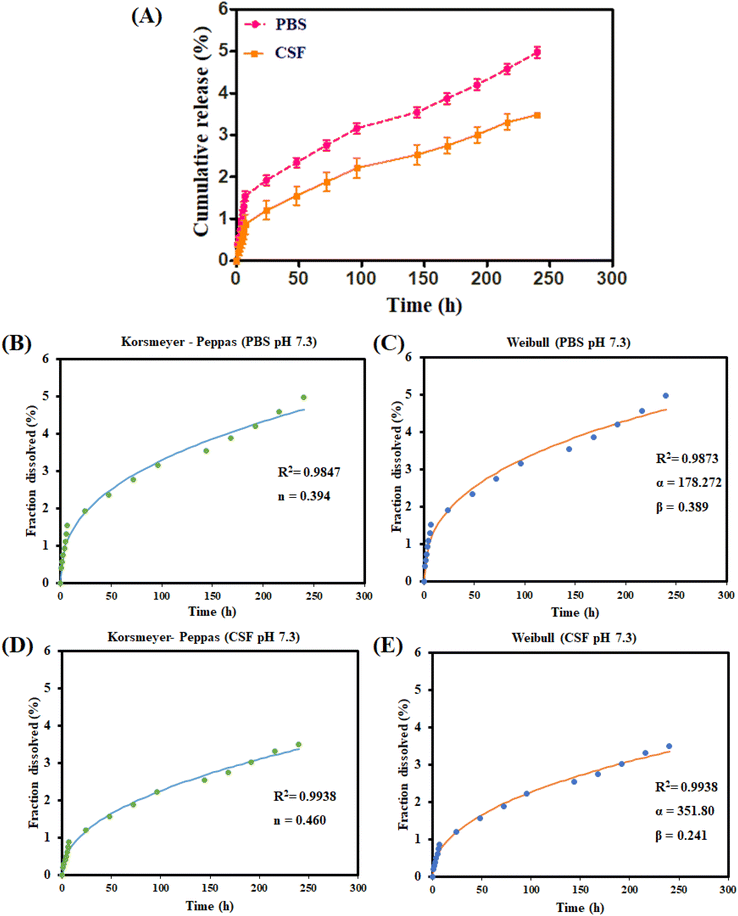
3.1. In vitro drug release and kinetic studies
The drug release studies of DOX-PS80 conjugate were performed using PBS pH 7.3 and CSF pH 7.3 using the dialysis method. At the end of 10 days, the cumulative drug released from DOX-PS80 was found to be 4.9 ± 0.8% in PBS and 3.9 ± 0.6% in CSF (Fig. 2A). No burst release effect was observed in either PBS or CSF buffers. Sustained release of DOX was observed over the period of time. In a previous study from our laboratory40 the prodrug of AmB with Tween 20 with a carbamate linkage showed 6% release at the end of 48 h. According to a previous report by Conover et al., drug release via carbamate ester linkage-based systems was less than 5% in PBS, while the pattern for release in rat plasma was completely different38,39 (50%). A wide range of mathematical models were established to fit the drug dissolution data and most of them were non-linear equations (Zhang et al., 2010).51 In order to understand the drug release mechanism from DOX-PS80 conjugate, the drug release profiles of DOX were fitted with different kinetic models. The mechanism of drug release was analyzed using the correlation coefficient (R2) and other release parameters. The (R2) values of each model with other parameters are compared and tabulated as shown in Table 1.| Kinetic model | Equation | Drug release parameters | Release from DOX-PS80 | |
|---|---|---|---|---|
| PBS pH 7.3 | CSF pH 7.3 | |||
| Zero order | F = k0 × t | R 2 | 0.7057 | 0.8161 |
| K 0 | 0.023 | 0.017 | ||
| First order | F = 100 × [1 − exp(−k1 × t)] | R 2 | 0.7123 | 0.8199 |
| K 1 | 0.001 | 0.001 | ||
| Higuchi | F = kH × t0.5 | R 2 | 0.9652 | 0.9913 |
| K H | 0.317 | 0.222 | ||
| Korsmeyer Peppas | F = kKP × tn | R 2 | 0.9847 | 0.9938 |
| KK p | 0.535 | 0.271 | ||
| n | 0.394 | 0.460 | ||
| Hixson Crowell | F = 100 × [1 − (1 − kHC × t)3] | R 2 | 0.7101 | 0.8186 |
| K HC | 0.001 | 0.001 | ||
| Hopfenberg | F = 100 × [1 − (1 − kHB × t)n] | R 2 | 0.7121 | 0.8189 |
| n | 54.068 | 38.909 | ||
| Weibull | F = 100 × {1 − exp[−(tβ)/α]} | R 2 | 0.9873 | 0.9938 |
| α | 178.272 | 351.80 | ||
| β | 0.389 | 0.241 | ||
Korsmeyer Peppas and Weibull models showed highest the correlation coefficient values (R2) in the range of 0.96 to 0.99 compared with the other kinetic models. The Peppas model is a simple and semi-empirical relationship which describes drug release mechanism from the polymeric and matrix systems.52,53 Drug release profiles of DOX in both PBS and CSF showed n values less than 1 (PBS n = 0.394 and CSF, n = 0.460) which demonstrated a lower Fickian diffusion release mechanism. The Peppas model fit curve for DOX from DOX-PS80 is shown in Fig. 2(B) and (D).
Weibull is an empirical model and widely applied for extended-release dosage forms as well as matrix system.52,53 It can be used for all types of dosage forms and drug release mechanisms, such as diffusion, dissolution, and dissolution–diffusion rate. The equation of Weibull is F = 100 × {1 − e[−(tβ)/α]}; in this equation, F is fraction of drug dissolved as a function of time t, α defines a scale parameter of geometry, and β defines a shape parameter which explains the drug dissolution curve. In the case of β = 1, the shape of the curve corresponds to exponential, β > 1 defines the sigmoid S-shaped upward curve, and β < 1 denotes a parabolic with initial high slope followed by consistent exponential. Release profiles of DOX from PBS and CSF both showed β value less than 1, i.e., 0.389 (PBS) and 0.241 (CSF), which denotes a parabolic curve with high initial slope. Scale parameter (α) defines the release rate of dissolved drug molecules; if the scale parameter is high, the drug release rate will be slower. CSF buffer showed higher α values (α = 351.80) compared with the PBS (α = 178.27) at alkali environment (pH 7.3) which shows the steady and sustained release of DOX from the conjugates. The Weibull fit curve for DOX from the DOX-PS80 conjugate is shown in Fig. 2(C) and (E).
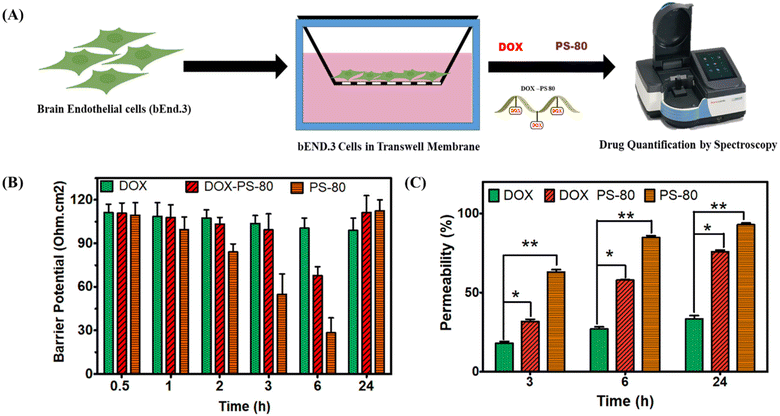
3.2. In vitro barrier potential and BBB permeability assay
To evaluate the effect of BBB permeability of DOX-PS80, bEnd 3 cells were grown on a transwell milli-insert with 0.4 μm pore size (Millipore) (Fig. 3). The barrier potential of the bEnd.3 cells was regularly monitored to ensure the formation of monolayer cells which mimic the BBB. When the potential of the cells reached 110 Ohm cm−2 on the 6th day, the samples were exposed, and the barrier potential was monitored. Once the samples were added, barrier potential of PS80 dropped to 90 Ohm cm−2 at the end of 3 h and 30 Ohm cm−2 at 6 h, but again the membrane potential was regained to 110 Ohm cm−2 at the end of 24 h. In contrast, the potential of both DOX and DOX-PS80 was stable up to 3 h, followed by a decrease at 6 h and again a regain of the potential at the end of 24 h (Fig. 3B). The drug molecules which crossed the transwell barrier, i.e., passing through the membrane, were quantified using UV visible spectroscopy at regular intervals. The concentration-dependent permeability was observed from 3 h to 24 h incubation (Fig. 3C). At the end of 3 h, PS80 showed more than 60% permeability, while DOX and DOX-PS80 showed 30% permeability. At the end of 24 h incubation, DOX-PS80 showed more than 70% permeability, whereas DOX showed only less than 40% and PS80 showed a high 90% permeability. In comparison with DOX, DOX-PS 80 conjugate showed two-fold increased drug permeability at the end of 24 h (Fig. 3C).3.3. Biocompatibility and cell uptake by bEnd3 cells
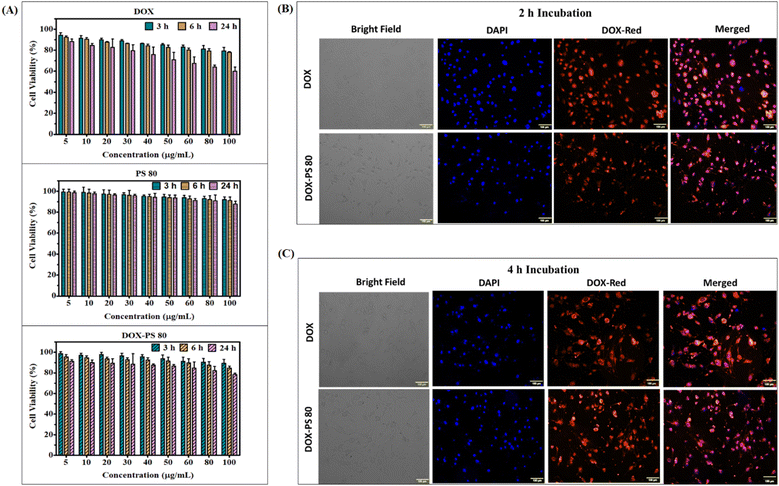
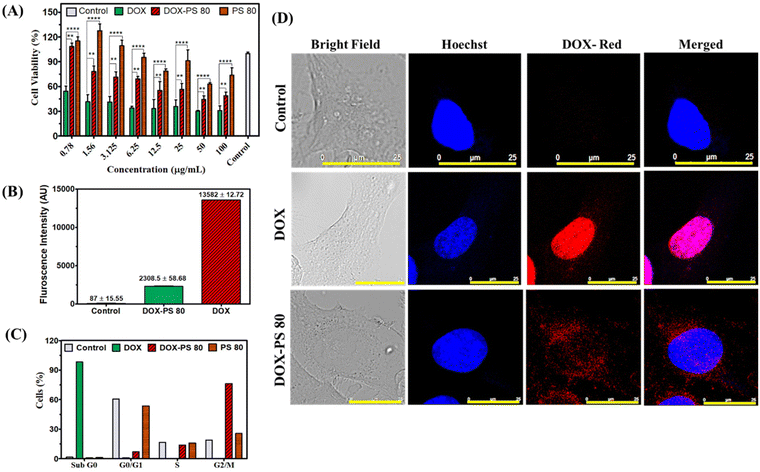
The biocompatibility of synthesized DOX-PS80 was evaluated in brain endothelial cells (bEnd3) by incubating samples for different periods of time with controls (PS80 and DOX). All the samples showed more than 90% viability at 3 and 6 h incubation. DOX showed concentration-dependent toxicity at 24 h incubation with more than 60% viability even at a concentration of 100 μg ml−1 (Fig. 4A). Meanwhile, DOX-PS80 showed more than 80% viability for all the concentrations at 24 h incubation. The cell viability assay showed that the synthesised DOX-PS80 was highly biocompatible with the brain endothelial cells. The intracellular uptake of DOX-PS80 and DOX was analyzed using bEnd3 cells by fluorescence imaging. The samples were incubated at 2 h and 4 h followed by fixing the cells using 4% paraformaldehyde. Then the cells were mounted on the glass slide and imaged at 10× magnification. Both DOX-PS80 and DOX showed intracellular uptake within 2 h. The red fluorescence of drug molecules was observed in cytoplasm as well in the nucleus, which confirmed the internalization of drug molecules throughout the cells (Fig. 4B).3.4. Cytotoxicity, intracellular uptake and cell cycle analysis in U251 cells
The cytotoxicity of DOX-PS80, DOX and PS80 was tested in human glioblastoma U251 cells. PS80 showed more than 60% cell viability at 48 h incubation and did not show significant cytotoxicity compared with the control cells. In the case of DOX and DOX-PS80, both showed concentration-dependent toxicity at 48 h incubation (Fig. 5A). The minimum inhibitory concentration (IC50) for DOX and DOX-PS80 was calculated at the end of 48 h. The IC50 of DOX-PS80 was found to be 38.10 μg mL−1 and DOX showed 1 μg mL−1. Our results are in agreement with the previous results of Stan et al.54 who reported the IC50 of free DOX as 1 μg mL−1 in U 87 MG and U373 glioma cells. DOX-PS80 showed higher inhibitory concentration compared with free DOX, which could be due to sustained release of DOX from the prodrug. Comparing the drug release data with the cytotoxicity assay, DOX-PS80 in PBS pH 7.4 showed around 2% DOX release at the end of 48 h. Though the drug release from the conjugate is slow and controlled, there is not much difference in the tumor inhibitory effect of DOX with the glioma cells.The cellular uptake behaviour of DOX-PS80 and DOX was studied in glioblastoma U251 cells at 2, 4 and 24 h incubation. The nucleus was stained with Hoechst, which will give blue fluorescence and DOX will emit red fluorescence. After drug treatment, incubation and staining the dyes, the cells were fixed with paraformaldehyde. The localization of drug molecules in the cells at different time points was observed under confocal microscope at 20× and 60× magnification. The single cell image at 60× magnification (24 h time point) is shown in Fig. 5D and 20× magnification for 2 h, 4 h and 24 h shown in Fig. S6.† Confocal images clearly demonstrated that DOX-PS80 localized mainly in the cytoplasm and around the nucleus, but in case of DOX, it entered the nucleus within 2 h incubation. Further increases in incubation time to 4 and 24 h also showed a similar effect with increased fluorescence intensity. Compared with the control cells, DOX and DOX-PS80 both showed a similar effect and time-dependent drug uptake at all time points. During confocal imaging, it was observed that conjugation of DOX with PS led to a decrease in the auto fluorescence property of DOX.
The internalization of DOX-PS80 and DOX was quantified using flow cytometry at different time points of 2, 4 and 24 h incubation. The data for cell uptake at 24 h are shown in Fig. 5B. The mean fluorescence intensity (MFI) for DOX and DOX-PS80 was calculated, and the results were compared and correlated (Fig. S7†). The mean fluorescence intensity of DOX-PS80 was found to be 837.5 ± 19.10 (2 h), 1124 ± 28.22 (4 h), 2308.5 ± 58.68 (24 h) and for DOX was 2959 ± 4.24 (2 h), 5761 ± 1.41 (4 h), 13582 ± 12.7 (24 h), respectively (Fig. S7†). The drug uptake results showed 80 to 90% uptake at the end of 2 h, while 4 h and 24 h incubation showed more than 95% uptake for both DOX and DOX-PS80 (Fig. S8†). Both DOX and DOX-PS80 showed increase in fluorescence intensity with increasing drug incubation time. While comparing the MFI of DOX-PS80 with DOX, DOX showed higher fluorescence intensity. The reason for the lower MFI for DOX-PS80 may be the quenching effect of auto fluorescence of DOX when conjugated with PS80. The results are in conformity with the previous confocal imaging results. Also, the higher MFI for free DOX may be due to passive diffusion of small molecules through the cell membrane; meanwhile, the mechanism of DOX-PS80 intracellular uptake remains to be explored.
To check the effect of cell cycle phases in U251 cells, PS80, DOX and DOX-PS80 conjugate were treated for 24 h and analyzed by flow cytometry (Fig. 5C). Control cells and PS80 showed the same cell cycle pattern, with more than 50% cells in G1 phase (growth phase), meanwhile DOX showed majority of cells in Sub G0 phase (98.2%) which may be due to the higher concentration of DOX resulting in apoptosis. Apoptotic cells, cells that had already lost their DNA by the release of apoptotic bodies, cellular fragments containing fragments of chromatin, damaged nuclei, chromosomes, and cellular debris made up the sub-G0/G1 peak.55,56 A higher proportion of cells (76.3%) were arrested in the G2/M phase of mitosis as a result of DOX-PS80 conjugate treatment, along with 13.8% of the population of cells in the S phase (DNA synthesis), and 6.9% of cells were retained in the G1 phase. The percentages of cell cycle phases for all the samples are shown in Fig. S9.†
3.5. Ex vivo tumor spheroid assay
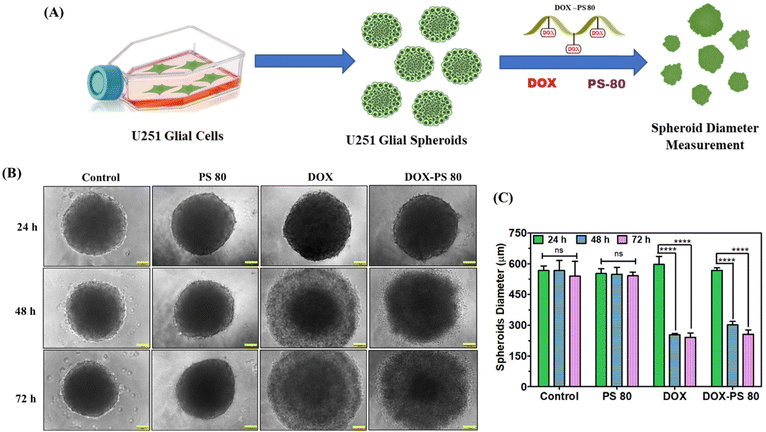
The tumor spheroid assay is the best ex vivo model, and closely resembles the solid tumor microenvironment for testing the anticancer efficacy of synthesised compounds.48,57 In our study, the tumor inhibitory efficacy of DOX-PS80 was assessed using a glioblastoma U251 cell spheroid model with controls (Fig. 6A). The morphology of tumor spheroids was captured at the end of 24, 48 and 72 h using phase contrast microscopy. The morphology of drug-treated tumor spheroids was compared with the control. PS80 does not show any morphological changes in spheres until 72 h. Meanwhile both DOX-PS80 and free DOX-treated spheroids showed slight morphological changes after 24 h (Fig. 6B). Comparatively, DOX-treated spheres showed stepwise degradation of spheres followed by cell death from 24 to 72 h. More interestingly, DOX-PS80-treated samples showed disruption of cells slowly from spheres followed by degradation. Polysorbate has a solubilizing property, and due to this property DOX-PS80 samples showed disruption followed by cell death. PS80-treated samples also showed slight cell disruption behaviour (Fig. 6B). The average diameter of the tumor spheroids was calculated and compared with the control (Fig. 6C). At the end of 72 h, control spheroids showed average diameter of 539.76 ± 71.53 μm and spheroids treated with PS80, DOX and DOX-PS80 showed average diameter of 541.62 ± 17.44 μm, 240.62 ± 22.08 μm and 256.57 ± 20.89 μm, respectively.The morphological changes as well the average diameter of spheroids showed that both DOX and DOX-PS80 exhibited a very similar tumor inhibitory effect. Due to sustained release of DOX from the DOX-PS80 conjugate, the tumor inhibitory effect happened in a slow fashion. This may be due to PS80 solubilizing and disrupting the tumor spheroids’ morphology and DOX degrading the cells slowly followed by apoptosis. Control and PS80 did not show tumor inhibitory effects and there was no change in spheroids’ diameter. At 24 h incubation, there were no changes in the test group compared with untreated spheroids. Compared with the control, DOX and DOX-PS80 conjugate showed significant difference at 48 and 72 h incubation, demonstrating the anti-tumor efficacy of the drug molecules (Fig. 6B and C).
3.6. Hemolysis
The hemolytic effect of the synthesized DOX-PS80 was assessed using human RBCs. Different concentrations of PS80, DOX and DOX-PS80 were prepared using PBS pH 7.4 and the assay was performed according to Francis et al.47 The percentages of hemolysis for all the concentrations are shown in Fig. S10.† PS80 did not show any hemolytic activity. DOX-conjugated PS80 samples showed less hemolytic activity compared with the pure DOX. At higher concentrations such as 100 μg ml−1, DOX-PS80 conjugates showed 5.16 ± 1.4% and free DOX showed around 14.71 ± 0.7% hemolysis.3.7. Biodistribution, toxicology, and histopathology studies in Swiss albino mice
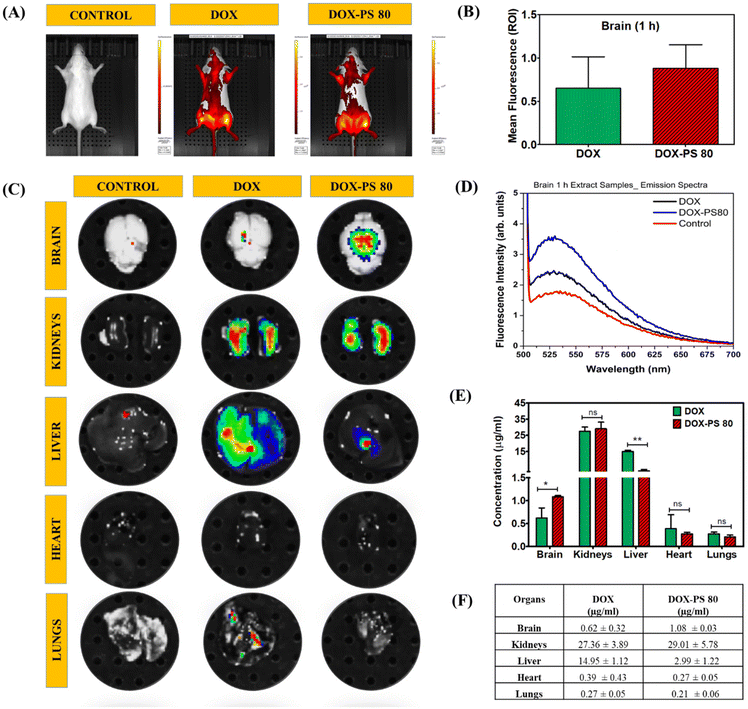
Biodistribution studies were performed to evaluate the transportation of DOX-PS80 conjugate in different organs.58,59 Through IVIS live animal imaging, the whole animal and dissected organs were imaged and compared with the control (Fig. 7A and C). Whole live animal imaging was unable to provide a clear understanding of the transportation of drug conjugates into the brain because DOX fluorescence interfered with the fluorescence of the animal's blood and fur. To evaluate the drug concentration, all the organs, namely the brain, kidneys, liver, lungs, and heart, were dissected and imaged by IVIS (Fig. 7C). DOX-PS80 showed higher fluorescence compared with the DOX in IVIS imaging and drug quantification. Fluorescence signal (ROI) for brain was measured separately before digestion and results for DOX and DOX-PS80 were compared (Fig. 7B). To further quantify the amount of drug in the brain and other organs, all the tissues were ground and digested with buffer [acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phosphate buffer pH 3
phosphate buffer pH 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (25
methanol (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30)] and the amount of drug in each tissue was measured using fluorescence spectroscopy at 590 nm (Fig. 7D–F).
30)] and the amount of drug in each tissue was measured using fluorescence spectroscopy at 590 nm (Fig. 7D–F).
When the concentrations of free DOX and DOX-PS80 conjugate in the brain were compared, DOX-PS80 showed a significant difference, demonstrating the effective permeation of drug conjugate across the BBB. The concentration of DOX in other organs was quantified. Within 1 h, kidneys and liver showed maximum drug, followed by other organs, namely brain, heart, and lungs. On comparing the amount of free DOX and DOX-PS80 conjugate in the brain alone, DOX-PS80 showed around 1.08 ± 0.03 μg ml−1 and DOX showed 0.62 ± 0.32 μg ml−1. The concentrations of drug molecules of different organs were quantified and tabulated, and are shown in Fig. 7F.
The drug biodistribution analysis was also performed for 24 h drug treatment and results were compared with respective controls. IVIS spectrum of whole animal and different organ analysis data for 24 h are shown in Fig. S11 and Table S1.† All the experiments were subjected to statistical analysis and the average results are reported. For histopathology, organs were dissected and stained with hematoxylin and eosin after 24 h of drug treatment. Sections were photographed at 10× magnification and any morphological or pathological changes were investigated. Histopathological investigation of the sections indicated no appreciable changes in any of the dissected organs. The results were compared and correlated with the control group (Fig. S12†).
3.8. In silico studies: analysis of pharmacokinetic parameters
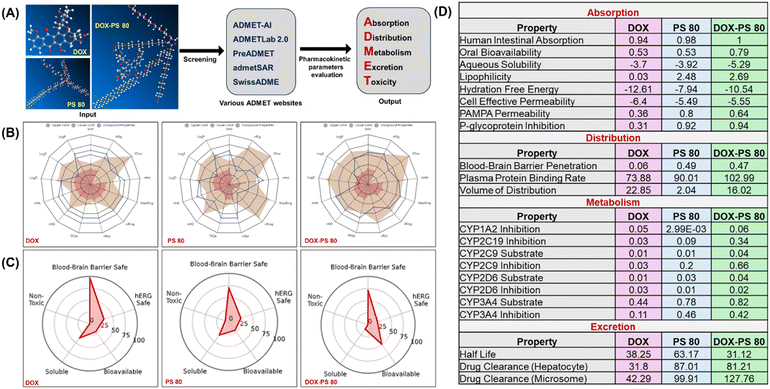
ADMET properties for DOX, PS80 and DOX-PS 80 were comprehensively analyzed through different web server-based platforms (Fig. 8A). The physicochemical and ADMET properties of all the three molecules were analyzed using ADMET-AI, ADMETlab 2.0, Pre ADME, admetSAR, and SwissADME.60–65 Every tool forecasts the ADMET characteristics using a distinct source and approach; all the predictions for DOX, PS80 and DOX-PS80 were subsequently assessed and verified. Among those, machine learning platform ADMET-AI web server (https://admet.ai.greenstonebio.com/) provided fast and accurate ADMET predictions and holds the highest rank on the therapeutic data commons leaderboard.62 In addition to ADMET-AI, the web-based ADMETlab 2.0 (https://adMETmesh.scbdd.com/) demonstrated precise and similar characteristics to the predictions generated by ADMET-AI.Even though we investigated ADMET predictions from a variety of programs, we have opted to present the data from ADMET-AI and ADMETlab2, as these programs produced the most accurate projections. In Fig. 8B, a radar spider plot illustrates the comparison of the physicochemical parameters of DOX, PS80, and DOX-PS80 from ADMETlab 2.0 predictions. All three compounds showed optimal values with reference range; however, DOX-PS80 showed a higher polar surface area (478.97 Å) followed by PS80 (280.82 Å) and DOX (206.07 Å) based on Veber rules. The logarithmic aqueous solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S) of DOX-PS80, PS80 and DOX showed −4.751, −4.562 and −3.419 mol L−1, which is the optimum range. The predicted log
S) of DOX-PS80, PS80 and DOX showed −4.751, −4.562 and −3.419 mol L−1, which is the optimum range. The predicted log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of DOX-PS80 showed 4.667 mol L−1, which represents higher lipophilic behavior of the compound compared with DOX (2.012 mol L−1) and PS80 (2.508 mol L−1). Interestingly, the distribution coefficient (log
P value of DOX-PS80 showed 4.667 mol L−1, which represents higher lipophilic behavior of the compound compared with DOX (2.012 mol L−1) and PS80 (2.508 mol L−1). Interestingly, the distribution coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D) values of PS80 and DOX-PS80 showed optimal values of 2.713 and 2.534 mol L−1, respectively. However, DOX showed a log
D) values of PS80 and DOX-PS80 showed optimal values of 2.713 and 2.534 mol L−1, respectively. However, DOX showed a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D value less than 1 (0.738) which is considered as out of range. Both log
D value less than 1 (0.738) which is considered as out of range. Both log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and log
P and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D values are direct measurements of lipophilicity, which is an important requirement for blood–brain barrier membrane permeability. Detailed descriptions of all the physicochemical parameters are available on the ADMETlab 2.0 website (https://admetmesh.scbdd.com/explanation/index).
D values are direct measurements of lipophilicity, which is an important requirement for blood–brain barrier membrane permeability. Detailed descriptions of all the physicochemical parameters are available on the ADMETlab 2.0 website (https://admetmesh.scbdd.com/explanation/index).
The radial plot from ADMET-AI (Fig. 8C) summarizes the significant ADMET properties, i.e., solubility, bioavailability, toxicity, BBB and hERG safety. The ADMET AI prediction clearly indicated that all three compounds are non-toxic and BBB-safe. It is interesting to note that DOX-PS80 showed a better bioavailability value of 0.79 compared with DOX (0.53) and PS80 (0.53). The human ether-a-go-go related gene (hERG) potassium channel prediction data values for DOX (0.61), PS80 (0.74) and DOX-PS80 (0.81) showed that the values are within the safe range.
The ADMET properties of DOX, PS80 and DOX-PS80 from ADMET-AI software were compared and tabulated (Fig. 8D). In terms of absorption factors such bioavailability, solubility, lipophilicity, and permeability, DOX-PS80 outperformed DOX with respect to predicted values. The BBB penetration scores clearly demonstrated that DOX-PS80 had superior BBB penetration compared with free DOX. Whereas free DOX showed a value of 0.06, the DOX-PS80 and PS80 displayed values of 0.47 and 0.49. This exemplifies the significance of conjugating PS80 with hydrophilic dug moieties such as DOX. These findings are corroborated with in vitro BBB permeability results (Fig. 3C) and in vivo data (Fig. 7). The human cytochrome (CYP) P450 family enzymes and isozymes play a major role in the drug metabolism process. The CYP substrate and the inhibition prediction values for DOX, PS80, and DOX-PS80 exhibited values less than 1 (within range). The half-life (t1/2) is an important parameter which correlates the volume of distribution and clearance. PS80 exhibited a maximum half-life of 63.17 h, while DOX and DOX-PS80 displayed 38.25 and 31.12 h, respectively. Both DOX-PS80 and PS80 showed the highest clearance values compared with free DOX (Fig. 8D).
4. Conclusions
Doxorubicin was successfully conjugated with PS80 by carbamate linkage and the conjugation was confirmed by different spectroscopic techniques. The amount of DOX conjugated to the PS80 was found to be 43.69 ± 4.72%. The percentage cumulative release of DOX from the conjugates was found to be 4.9 ± 0.8% (PBS pH 7.3) and 3.9 ± 0.6% (CSF pH 7.3) at the end of 10 days. In vitro biocompatibility testing in bEnd.3 cells revealed that the synthesized DOX-PS80 conjugate was biocompatible and non-toxic. BBB permeability assay by in vitro method revealed the effective permeation of DOX-PS80 conjugates compared with DOX. The cytotoxic effect of DOX-PS80 was analyzed using glioblastoma U251 cells and the IC50 value was found to be 38.10 μg ml−1. Intra-cellular uptake studies revealed that the synthesized DOX-PS80 was able to localize in cytoplasm and nucleus at different time intervals in both brain endothelial bEnd.3 as well in glioblastoma U251 cells. Cell cycle analysis of DOX-PS80 conjugate showed G2/M phase arrest and apoptosis. Tumor spheroids assay in U251 cells showed a significant reduction in tumor size in treated cells compared with control. IVIS live animal imaging and fluorescence spectroscopy results revealed that most of the drug molecules were transported to the liver and kidneys, followed by other organs. Brain sections showed a higher amount of DOX-PS80 conjugate compared with DOX, showing the enhanced permeation of DOX-PS80 through BBB. In silico pharmacokinetic parameters were screened using ADMET web-based platforms. These preliminary findings show that drugs conjugated to PS80 have the potential to cross the BBB for treating CNS disorders.Conflicts of interest
The authors declared no conflict of interest.Acknowledgements
This work was supported by the Department of Atomic Energy, Government of India. A. J. thanks the Department of Atomic Energy, Government of India for the Raja Ramanna Fellowship. S. R. P. thanks the Department of Atomic Energy, Government of India for a Research Asssociateship. The authors acknowledge the Animal Research Facility, FACS and Confocal Imaging facilities at Rajiv Gandhi Center for Biotechnology for technical support.References
- A. Achar, R. Myers and C. Ghosh, Drug delivery challenges in brain disorders across the blood–brain barrier: novel methods and future considerations for improved therapy, Biomedicines, 2021, 9, 1834, DOI:10.3390/biomedicines9121834.
- I. G. Tucker, Drug delivery to the brain via the blood–brain barrier: a review of the literature and some recent patent disclosures, Ther. Delivery, 2011, 2(3), 311–327, DOI:10.4155/tde.11.3.
- W. M. Pardridge, The blood-brain barrier: bottleneck in brain drug development, NeuroRx, 2005, 2, 3–14, DOI:10.1602/neurorx.2.1.3.
- Drug Delivery to the Brain: Physiological Concepts, Methodologies and Approaches, ed. M. Hammarlund-Udenaes, E. C. M. de Lange and R. G. Thorne, Springer, New York, 2014. https://link.springer.com/book/10.1007/978-1-4614-9105-7 Search PubMed.
- H. Kadry, B. Noorani and L. Cucullo, A blood–brain barrier overview on structure, function, impairment, and biomarkers of integrity, Fluids Barriers CNS, 2020, 17, 1–24, DOI:10.1186/s12987-020-00230-3.
- J. Rip, G. J. Schenk and A. G. de Boer, Differential receptor-mediated drug targeting to the diseased brain, Expert Opin. Drug Delivery, 2009, 6, 227–237, DOI:10.1517/17425240902806383.
- N. J. Abbott, L. Ronnback and E. Hansson, Astrocyte-endothelial interactions at the blood–brain barrier, Nat. Rev. Neurosci., 2006, 7, 41–53, DOI:10.1038/nrn1824.
- A. Saija, P. Princi, D. Trombetta, M. Lanza and A. De Pasquale, Changes in the permeability of the blood–brain barrier following sodium dodecyl sulphate administration in the rat, Exp. Brain Res., 1997, 115, 546–551, DOI:10.1007/PL00005725.
- K. Hynynen, Ultrasound for drug and gene delivery to the brain, Adv. Drug Delivery Rev., 2008, 60, 1209–1217, DOI:10.1016/j.addr.2008.03.010.
- R. Stam, Electromagnetic fields and the blood–brain barrier, Brain Res. Rev., 2010, 65, 80–97, DOI:10.1016/j.brainresrev.2010.06.001.
- F. Herve, N. Ghinea and J. M. Scherrmann, CNS delivery via adsorptive transcytosis, AAPS J., 2008, 10, 455–472, DOI:10.1208/s12248-008-9055-2.
- M. Zorko and U. Langel, Cell-penetrating peptides: mechanism and kinetics of cargo delivery, Adv. Drug Delivery Rev., 2005, 57, 529–545, DOI:10.1016/j.addr.2004.10.010.
- C. Rousselle, P. Clair, J. M. Lefauconnier, M. Kaczorek, J. M. Scherrmann and J. Temsamani, New advances in the transport of doxorubicin through the blood–brain barrier by a peptide vector-mediated strategy, Mol. Pharmacol., 2000, 57, 679–686, DOI:10.1124/mol.57.4.679.
- A. G. de Boer and P. J. Gaillard, Drug targeting to the brain, Annu. Rev. Pharmacol. Toxicol., 2007, 47, 323–355, DOI:10.1146/annurev.pharmtox.47.120505.105237.
- J. F. Poduslo and G. L. Curran, Polyamine modification increases the permeability of proteins at the blood–nerve and blood–brain barriers, J. Neurochem., 1996, 66, 1599–1609, DOI:10.1046/j.1471-4159.1996.66041599.x.
- W. J. Rogers and P. Basu, Factors regulating macrophage endocytosis of nanoparticles: implications for targeted magnetic resonance plaque imaging, Atherosclerosis, 2005, 178, 67–73, DOI:10.1016/j.atherosclerosis.2004.08.017.
- J. Panyam and V. Labhasetwar, Dynamics of endocytosis and exocytosis of poly(D, Llactideco-glycolide) nanoparticles in vascular smooth muscle cells, Pharm. Res., 2003, 20, 212–220, DOI:10.1023/A:1022219003551.
- D. L. Daleke, K. Hong and D. Papahadjopoulos, Endocytosis of liposomes by macrophages: binding, acidification and leakage of liposomes monitored by a new fluorescence assay, Biochim. Biophys. Acta, 1990, 1024, 352–366, DOI:10.1016/0005-2736(90)90365-U.
- S. Jain, V. Mishra, P. Singh, P. K. Dubey, D. K. Saraf and S. P. Vyas, RGD-anchored magnetic liposomes for monocytes/neutrophils-mediated brain targeting, Int. J. Pharm., 2003, 261, 43–55, DOI:10.1016/S0378-5173(03)00269-2.
- Y.-E. L. Koo, G. R. Reddy, M. Bhojani, R. Schneider, M. A. Philbert, A. Rehemtulla, B. D. Ross and R. Kopelman, Brain cancer diagnosis and therapy with nanoplatforms, Adv. Drug Delivery Rev., 2006, 58, 1556–1577, DOI:10.1016/j.addr.2013.09.006.
- S. Bhaskar, F. Tian, T. Stoeger, W. Kreyling, J. M. de la Fuente, V. Grazu, P. Borm, G. Estrada, V. Ntziachristos and D. Razansky, Multifunctional Nanocarriers for diagnostics, drug delivery and targeted treatment across blood–brain barrier: perspectives on tracking and neuroimaging, Part. Fibre Toxicol., 2010, 7, 3, DOI:10.1186/1743-8977-7-3.
- E. Garcia-Garcia, K. Andrieux, S. Gil and P. Couvreur, Colloidal carriers and blood-brain barrier (BBB) translocation: a way to deliver drugs to the brain?, Int. J. Pharm., 2005, 298, 274–292, DOI:10.1016/j.ijpharm.2005.03.031.
- R. Pahuja, K. Seth and A. Shukla, et al., Trans-blood brain barrier delivery of dopamine-loaded nanoparticles reverses functional deficits in parkinsonian rats, ACS Nano, 2015, 9, 4850–4871, DOI:10.1021/nn506408v.
- I. Brigger, C. Dubernet and P. Couvreur, Nanoparticles in cancer therapy and diagnosis, Adv. Drug Delivery Rev., 2002, 54, 631–651, DOI:10.1016/S0169-409X(02)00044-3.
- S. Santra, H. Yang, P. H. Holloway, J. T. Stanley and R. A. Mericle, Synthesis of water dispersible fluorescent, radio-opaque, and paramagnetic CdS:Mn/ZnS quantum dots: a multifunctional probe for bioimaging, J. Am. Chem. Soc., 2005, 127, 1656–1657, DOI:10.1021/ja0464140.
- A. Poustforoosh, M. H. Nematollahi, H. Hashemipour and H. A. Pardakhty, Recent advances in Bio-conjugated nanocarriers for crossing the Blood-Brain Barrier in (pre-) clinical studies with an emphasis on vesicles, J. Controlled Release, 2022, 17, 343, DOI:10.1016/j.jconrel.2022.02.015 , 777–797.
- U. Schröder and B. A. Sabel, Nanoparticles, a drug carrier system to pass the blood-brain barrier, permit central analgesic effects of i.v. dalargin injections, Brain Res., 1996, 710, 121–124, DOI:10.1016/0006-8993(95)01375-X.
- P. Blasi, S. Giovagnoli, A. Schoubben, M. Ricci and C. Rossi, Solid lipid nanoparticles for targeted brain drug delivery, Adv. Drug Delivery Rev., 2007, 59, 454–477, DOI:10.1016/j.addr.2007.04.011.
- J. Kreuter, Mechanism of polymeric nanoparticle-based drug transport across the blood brain barrier (BBB), J. Microencapsulation, 2013, 30, 49–54, DOI:10.3109/02652048.2012.692491.
- J. Kreuter, Transport of drugs across the blood-brain barrier by nanoparticles, Curr. Med. Chem.: Cent. Nerv. Syst. Agents, 2002, 2, 241–249, DOI:10.1016/j.jconrel.2011.08.017.
- J. Kreuter, P. Ramge, V. Petrov, S. Hamm, E. Svetlana, G. B. Engelhardt, A. H. Briesen and D. J. Begley, Direct evidence that polysorbate-80-coated poly (butylcyanoacrylate) nanoparticles deliver drugs to the CNS via specific mechanisms requiring prior binding of drug to the nanoparticles, Pharm. Res., 2003, 20, 409–416, DOI:10.1023/A:1022604120952.
- A.E Gulyaev, S.E Gelperina, I.N Skidan, A.S Antropov, G.Y Kivman and J. Kreuter, Significant transport of doxorubicin into the brain with polysorbate 80-coated nanoparticles, Pharm. Res., 1999, 16, 1564–1569, DOI:10.1023/A:1018983904537.
- J. Kreuter, V. E. Petrov, D. A. Kharkevich and R. N. Alyautdin, Influence of the type of surfactant on the analgesic effects induced by the peptide dalargin after its delivery across the blood–brain barrier using surfactant-coated nanoparticles, J. Controlled Release, 1997, 49, 81–87, DOI:10.1016/S0168-3659(97)00061-8.
- J. Kreuter, Nanoparticulate systems for brain delivery of drugs, Adv. Drug Delivery Rev., 2001, 23, 65–81, DOI:10.1016/S0169-409X(00)00122-8.
- S. F. Basharzad, M. Hamidi, A. Maleki, Z. Karami, H. Mohamadpour and M. R. S. Zanjani, Polysorbate-coated mesoporous silica nanoparticles as an efficient carrier for improved rivastigmine brain delivery, Brain Res., 2022, 1781, 147786, DOI:10.1016/j.brainres.2022.147786.
- X. Tao, S. Mao, Q. Zhang, H. Yu, Y. Li, X. He, S. Yang, Z. Zhang, Z. Yi, Y. Song and X. Feng, Brain-targeted polysorbate 80-emulsified donepezil drug-loaded nanoparticles for neuroprotection, Nanoscale Res. Lett., 2021, 16, 1–15, DOI:10.1186/s11671-021-03584-1.
- S. Sharma and S. Dang, Polysorbate 80 Surface Modified PLGA Nanoparticles: An In vitro Evaluation of Cellular Uptake & Cytotoxicity on Neuro-2a Cells, J. Microencapsulation, 2023, 40, 1–17, DOI:10.1080/02652048.2023.2244095.
- A. Zensi, D. Begley, C. Pontikis, C.L. Legros, L. Mihoreanu, S. Wagner, C. Buchel, H. von Briesen and J. Kreuter, Albumin nanoparticles targeted with Apo E enter the CNS by transcytosis and are delivered to neurones, J. Controlled Release, 2009, 137, 78–86, DOI:10.1016/j.jconrel.2009.03.002.
- V. Ravichandran, M. Lee, T. G. Nguyen Cao and M. S. Shim, Polysorbate-based drug formulations for brain-targeted drug delivery and anticancer therapy, Appl. Sci., 2021, 11(19), 9336, DOI:10.3390/app11199336.
- V. Ravichandran, V. Kesavan, S. Cojean, P. M. Loiseau and A. Jayakrishnan, Polysorbate surfactants as drug carriers: Tween 20-amphotericin B conjugates as anti-fungal and antileishmanial agents, Curr. Drug Delivery, 2018, 15, 1028–1037, DOI:10.2174/1567201815666180503122829.
- C. D. Conover, H. Zhao, C. B. Longley, K. L. Shum and R. B. Greenwald, Utility of poly (ethylene glycol) conjugation to create prodrugs of amphotericin B, Bioconjugate Chem., 2003, 14(3), 661–666, DOI:10.1021/bc0256594.
- L. V. Nair, R. V. Nair, S. J. Shenoy, A. Thekkuveettil and R. S. Jayasree, Blood brain barrier permeable gold nanocluster for targeted brain imaging and therapy: an in vitro and in vivo study, J. Mater. Chem. B, 2017, 5, 8314–8321, 10.1039/C7TB02247F.
- S. C. Steiniger, J. Kreuter, A. S. Khalansky, I. N. Skidan, A. I. Bobruskin, Z. S. Smirnova, S. E. Severin, R. Uhl, M. Kock, K. D. Geiger and S. E. Gelperina, Chemotherapy of glioblastoma in rats using doxorubicin–loaded nanoparticles, Int. J. Cancer, 2004, 109, 759–767, DOI:10.1002/ijc.20048.
- Y. Li, M. Wu, N. Zhang, C. Tang, P. Jiang, X. Liu, F. Yan and H. Zheng, Mechanisms of enhanced antiglioma efficacy of polysorbate 80−modified paclitaxel–loaded PLGA nanoparticles by focused ultrasound, J. Cell. Mol. Med., 2018, 22(9), 4171–4182, DOI:10.1111/jcmm.13695.
- C. Zhang, J. Song, L. Lou, X. Qi, L. Zhao, B. Fan, G. Sun, Z. Lv, Z. Fan, B. Jiao and J. Yang, Doxorubicin–loaded nanoparticle coated with endothelial cells–derived exosomes for immunogenic chemotherapy of glioblastoma, Bioeng. Transl. Med., 2020, e10203, DOI:10.1002/btm2.10203.
- M. Norouzi, V. Yathindranath, J. A. Thliveris, B. M. Kopec, T. J. Siahaan and D. W. Miller, Doxorubicin-loaded iron oxide nanoparticles for glioblastoma therapy: A combinational approach for enhanced delivery of nanoparticles, Sci. Rep., 2020, 10, 1–8 CrossRef PubMed.
- A. P. Francis, S. Gurudevan and A. Jayakrishnan, Synthetic polymannose as a drug carrier: synthesis, toxicity and anti-fungal activity of polymannose-amphotericin B conjugates, J. Biomater. Sci., Polym. Ed., 2018, 29, 1529–1548, DOI:10.1080/09205063.2018.1469186.
- P. K. Gupta, S. K. Tripathi, S. Pappuru, S. C. Chabattula, K. Govarthanan, S. Gupta, B. K. Biswal, D. Chakraborty and R. S. Verma, Metal-free semi-aromatic polyester as nanodrug carrier: A novel tumor targeting drug delivery vehicle for potential clinical application, Mater. Sci. Eng., C, 2020, 107, 110285, DOI:10.1016/j.msec.2019.110285.
- A. K. Verma, A. Chanchal and K. Chutani, Augmentation of anti–tumour activity of cisplatin by pectin nano–conjugates in B–16 mouse model: pharmacokinetics and in–vivo biodistribution of radio–labelled, hydrophilic nano–conjugates, Int. J. Nanotechnol., 2012, 10, 872–886, DOI:10.1504/IJNT.2012.049452.
- A. K. Verma, A. Leekha, V. Kumar, I. Moin and S. Kumar, Biodistribution and In vivo efficacy of doxorubicin loaded chitosan nanoparticles in Ehrlich Ascites Carcinoma (EAC) bearing Balb/C mice, J. Nanomed. Nanotechnol., 2018, 9, 510, DOI:10.4172/2157-7439.1000510.
- Y. Zhang, M. Huo, J. Zhou, A. Zou, W. Li, C. Yao and S. Xie, DDSolver: an add-in program for modeling and comparison of drug dissolution profiles, AAPS J., 2010, 12, 263–271, DOI:10.1208/s12248-010-9185-1.
- P. Costa and J. M. S. Lobo, Modeling and comparison of dissolution profiles, Eur. J. Pharm. Sci., 2001, 13, 123–133, DOI:10.1016/S0928-0987(01)00095-1.
- R. Y. Basha, T. S. S. Kumar and M. Doble, Dual delivery of tuberculosis drugs via cyclodextrin conjugated curdlan nanoparticles to infected macrophages, Carbohydr. Polym., 2019, 218, 53–62, DOI:10.1016/j.carbpol.2019.04.056.
- A. C. Stan, S. Casares, D. Radu, G. F. Walter and T. D. Brumeanu, Doxorubicin-induced cell death in highly invasive human gliomas, Anticancer Res., 1999, 19, 941–950 CAS . PMID: 10368637.
- H. S. Kim, Y.S Lee and D. K. Kim, Doxorubicin exerts cytotoxic effects through cell cycle arrest and Fas-mediated cell death, Pharmacology, 2009, 84, 300–309, DOI:10.1159/000245937.
- M. Levi, R. Salaroli, F. Parenti, R. De Maria, A. Zannoni, C. Bernardini, C. Gola, A. Brocco, A. Marangio, C. Benazzi and L. V. Muscatello, Doxorubicin treatment modulates chemoresistance and affects the cell cycle in two canine mammary tumour cell lines, BMC Vet. Res., 2021, 17, 1–5, DOI:10.1186/s12917-020-02709-5.
- M. Millard, I. Yakavets, V. Zorin, A. Kulmukhamedova, S. Marchal and L. Bezdetnaya, Drug delivery to solid tumors: The predictive value of the multicellular tumor spheroid model for nanomedicine screening, Int. J. Nanomed., 2017, 12, 7993, DOI:10.2147/IJN.S146927.
- S. E. Gelperina, A. S. Khalansky and I. N. Skidan, et al., Toxicological studies of doxorubicin bound to polysorbate 80-coated poly(butyl cyanoacrylate) nanoparticles in healthy rats and rats with intracranial glioblastoma, Toxicol. Lett., 2002, 126, 131–141, DOI:10.1016/S0378-4274(01)00456-8.
- A. E. Gulyaev, S. E. Gelperina, I. N. Skidan, A. S. Antropov, G. Y. Kivman and J. Kreuter, Significant transport of doxorubicin into brain with polysorbate-80 coated nanoparticles, Pharm. Res., 1999, 16, 1564–1569, DOI:10.1023/A:1018983904537.
- H. Komura, R. Watanabe and K. Mizuguchi, The trends and future prospective of in silico models from the viewpoint of ADME evaluation in drug discovery, Pharmaceutics, 2023, 15(11), 2619, DOI:10.3390/pharmaceutics15112619.
- J. A. Pradeepkiran, S. B. Sainath and K. V. L. Shrikanya, In silico validation and ADMET analysis for the best lead molecules, in Brucella Melitensis, Academic Press, 2021, pp. 133–176. DOI:10.1016/B978-0-323-85681-2.00008-2.
- K. Swanson, P. Walther, J. Leitz, S. Mukherjee, J. C. Wu, R. V. Shivnaraine and J. Zou, ADMET-AI: A machine learning ADMET platform for evaluation of large-scale chemical libraries, bioRxiv, 2023, 12. DOI:10.1101/2023.12.28.573531.
- G. Xiong, Z. Wu, J. Yi, L. Fu, Z. Yang, C. Hsieh, M. Yin, X. Zeng, C. Wu, A. Lu and X. Chen, ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties, Nucleic Acids Res., 2021, 49, W5–W14, DOI:10.1093/nar/gkab255.
- S. K. Lee, I. H. Lee, H. J. Kim, G. S. Chang, J. E. Chung and K. T. No, The PreADME Approach: Web-based program for rapid prediction of physico-chemical, drug absorption and drug-like properties, EuroQSAR 2002 Designing Drugs and Crop Protectants: processes, problems and solutions, 2003, pp. 418–420.
- H. Yang, C. Lou, L. Sun, J. Li, Y. Cai, Z. Wang, W. Li, G. Liu and Y. Tang, admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties, Bioinformatics, 2019, 35(6), 1067–1069, DOI:10.1093/bioinformatics/bty707.
- Z. Y. U. Ibrahim, A. Uzairu, G. Shallangwa and S. Abechi, Molecular docking studies, drug-likeness and in-silico ADMET prediction of some novel β-Amino alcohol grafted 1, 4, 5-trisubstituted 1, 2, 3-triazoles derivatives as elevators of p53 protein levels, Sci. Afr., 2020, 10, e00570, DOI:10.1016/j.sciaf.2020.e00570.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4pm00053f |
| ‡ Present address: Department of Chemical & Biological Engineering, Colorado School of Mines, 1613 Illinois St, Golden, Colorado 80401, USA. |
| § Present address: Department of Chemistry, University of Kerala, Kariavattom Campus, Thiruvananthapuram 695 581 Kerala, India. |
| This journal is © The Royal Society of Chemistry 2024 |