Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Antimicrobial coatings from gramicidin D nanoparticles and polymers
Livia Cestaro de Souza
Camargo
,
Bianca Reche
Bazan
,
Rodrigo Tadeu
Ribeiro
 ,
Giovanna Maruyama
Quinto
,
Andrea Caroline Bazzan
Muniz
and
Ana Maria
Carmona-Ribeiro
,
Giovanna Maruyama
Quinto
,
Andrea Caroline Bazzan
Muniz
and
Ana Maria
Carmona-Ribeiro
 *
*
Biocolloids Laboratory, Departamento de Bioquímica, Instituto de Química, Universidade de São Paulo, Avenida Professor Lineu Prestes, 748, Butantan, São Paulo 05508-000, Brazil. E-mail: amcr@usp.br; Tel: +55 11 30912164
First published on 4th September 2024
Abstract
The microbicidal activity of gramicidin D molecules (Gr) assembled as nanoparticles (NPs) against Staphylococcus aureus was found to be superior to that of other Gr formulations in bilayers. In combination with the antimicrobial polymer poly(diallyl dimethyl ammonium chloride) (PDDA), water dispersions and coatings on glass exhibited a remarkably broadened spectrum of activity, achieving complete killing of Gram-negative bacteria, Gram-positive bacteria and fungi at reduced Gr and PDDA doses. In this work, combinations of Gr NPs and polymers were cast on glass (hydrophilic) or polyethylene (hydrophobic) surfaces, modeling common surfaces on biomedical materials, to evaluate the effect of polymer positive charge on the antimicrobial activity. Decreasing positive charges of three different polymers, namely PDDA, chitosan (CH) and polyacrylamide (PA), reduced or abolished microbicidal activity both in the presence and absence of Gr NPs. At 4.7 μg Gr and 25 μg polymer, microbicidal activity increased from PA to CH to PDDA at pH 6.3. The results suggested that the Gr/polymer antimicrobial coatings can be used on both hydrophobic and hydrophilic biomedical materials, effectively imparting them with efficient defense against a broad spectrum of microbes.
Introduction
Colloids combined with oppositely charged polymers can create viscous liquids or soluble complexes,1–7 yield multilayered coatings,8–10 stabilize or flocculate particulates,11–14 lead to gelation,15–17 purify water,18–20 cause muco-adhesion,21,22 perform bio-separations,23,24 and flocculate bacteria.25,26 On the other hand, cationization has been used as a straightforward tool for delivering drugs, peptides, proteins, DNA, RNA, vaccines and other important pharmaceuticals in vivo. Most biomolecules are negatively charged, so they are electrostatically driven to cationic carriers, allowing innovative pharmaceutical applications for the derived assemblies.27 For example, nanoparticles (NPs) built from an antigen and an oppositely charged polyelectrolyte namely poly(diallyl dimethyl ammonium chloride) (PDDA) have recently been proved effective as immunoadjuvants for presenting the model antigen ovalbumin.28,29 Another example was the development of antimicrobial nanoparticles from cationic bilayer fragments surrounded by anionic carboxymethylcellulose and, finally, by a PDDA layer, which were used to deliver amphotericin B30 or simply to display effective microbicidal action by themselves.31,32Recently, we described the broad and potent antimicrobial activity of water dispersions of gramicidin (Gr) nanoparticles (NPs) and the water-soluble, hydrophilic polymer poly(diallyl dimethyl ammonium chloride) (PDDA).33 In particular, the self-assembly of gramicidin D as nanoparticles in aqueous solution was also described for the first time in 2022 by our group; scanning electron micrographs revealed nanometric spherical Gr NPs with a mean diameter of around 150 nm.33 In addition, we reported the construction of microbicidal transient coatings based on the co-deposition of cationic antimicrobial NPs14,30,31 and Gr NPs33 onto glass cover slips as substrates.34 The cationic antimicrobial NPs were formed layer-by-layer from cationic bilayer fragments (BF) of dioctadecyl dimethyl ammonium bromide (DODAB) consecutively surrounded by carboxy-methyl cellulose (CMC) and poly(diallyl dimethyl ammonium chloride) (PDDA) layers (DODAB BF/CMC/PDDA NPs) in water dispersions.14,30,31 Coatings obtained from casting and drying DODAB BF/CMC/PDDA NP and Gr NP dispersions in water onto glass coverslips effectively killed three different pathogens (Pseudomonas aeruginosa, Staphylococcus aureus and Candida albicans), resulting in complete loss of viability for all three strains tested at low doses of Gr and the cationic NPs.34
Here, we describe novel microbicidal coatings obtained from casting and drying water dispersions of Gr NPs and hydrophilic polymers onto materials of biomedical importance. The materials employed for casting and drying the Gr/polymer water dispersions were glass and polyethylene (PE). Glass is hydrophilic, whereas PE is hydrophobic. Thus, we model a variety of important biomedical materials that may eventually require antimicrobial coatings such as PE implants for total hip arthroplasty,35 acetabular cups around a metallic femoral head,36 oral implants,37 a variety of biomedical implants38 or orthopedic materials39 and other invasive medical devices such as catheters, prosthetic heart valves, stents, and intrauterine devices made of PE.40 In addition, some biomedical materials such as titanium dioxide exhibit special wettability, which can change with UV irradiation from hydrophobic to hydrophilic;41 thus, an antimicrobial coating able to act over a range of wettabilities such as the one described in this work would indeed be valuable.
Results and discussion
The effect of polymer nature and concentration on the physical properties of Gr NPs in water dispersion at the pH of water
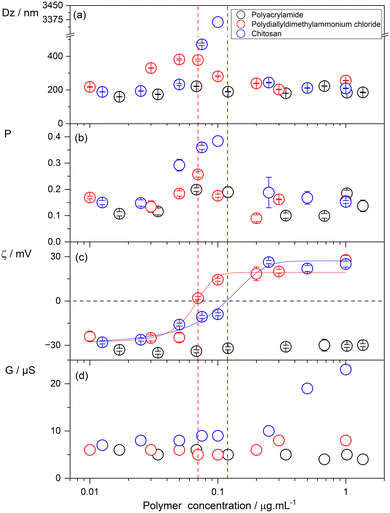
In this section, we describe the effect of polymer nature and concentration on the physical properties of Gr NPs for three different polymers at a fixed Gr concentration of 0.05 mM or 94.1 μg mL−1 (recalling that Gr molecular weight is 1882.3 g mol−1).Fig. 1 shows the effect of polymer concentration on the mean hydrodynamic diameter (Dz), polydispersity (P), zeta-potential (ζ) and conductance of the Gr NPs dispersed in pure water. For the positively charged polymers, over a range of low concentrations (0.01–1.00 μg mL−1), or chitosan (CH), Dz reached a maximum at 0.07 μg mL−1 PDDA (red circles) and 0.12 μg mL−1 CH (blue circles). At these concentrations, the zeta-potential was zero, showing that the absence of electrostatic repulsion between Gr NPs in dispersion promotes the aggregation of NPs and results in poor colloidal stability as often observed in the literature for similar systems. Increasing PDDA or CH concentration changed the zeta-potential from negative to positive values.
Consistently, the chitosan (CH) concentration that resulted in a zero zeta-potential for Gr NPs was about double the one determined for PDDA (Fig. 1). The chitosan polymer used in this work has a deacetylation degree of 49%, meaning that only 49% of the chitosan monomeric residues became charged by chitin deacetylation. Chitin is an N-acetyl-D-glucosamine polymer, which can be deacetylated to yield the cationic polyelectrolyte chitosan with pH-dependent cationic amino groups.
It is important to note that colloidal stability was recovered for polymer concentrations above those yielding maximal sizes and zero zeta-potentials (Fig. 1). In addition, the neutral polymer polyacrylamide (PA) did not affect the physical properties of the Gr NPs across the entire range of concentrations tested. The conductance of the dispersions (G) shown in Fig. 1(d) was rather low and mostly due to Gr NPs. Over the low range of polymer concentrations tested, conductance due to the polymers should be practically negligible. Increasing the concentration of the charged polymer in the Gr NP dispersions practically did not change G for PDDA but increased it for CH (Fig. 1). Previously, this same batch of CH at 100 μg mL−1 yielded a conductance of about 100 μS.42 In addition, the linear dependence of CH conductance on the CH concentration would yield G = 0.5 μS at 1 μg mL−1 CH. However, a conductance of approximately 23 μS was obtained for the Gr NP/CH dispersions at 1 μg mL−1 CH. This was probably due to the procedure of solubilizing CH in water. Solubilization required the addition of droplets of acetic acid, which possibly accounted for the additional conductance due to protons and acetate anions. Gr NPs’ conductance (G) in the presence of the non-conducting polymer polyacrylamide was 5–7 μS, consistent with previously reported G measurements for Gr NPs.33
Effects of coatings from gramicidin D and/or polymers on the surface wettability of glass or polyethylene
The water dispersions of Gr NPs and polymers can be cast and dried to create antimicrobial coatings on biomedical surfaces of interest. In this section, we evaluate whether both hydrophilic and hydrophobic surfaces can be coated by casting and drying the dispersions described.A simple procedure to evaluate the wettability of a surface is observing the shape of a water droplet on the surface. While glass is highly hydrophilic and the droplets are spread on its surface yielding contact angles of around 0°, polyethylene (PE) surfaces are hydrophobic with a reduced contact area between the droplet and the surface, yielding much higher contact angles. The images in Table 1 are reproducible and representative of more than three repeats for each sample of the same material. Furthermore, the functionality of the coatings was highly reproducible, as determined by their effects on the cell viability of several microbial cells reported in the next sub-section. This could also be well observed on the control surfaces (C+) examined in the absence of any coatings (Table 1). Photos of the droplets captured the changes in the hydrophobic/hydrophilic nature of the surfaces caused by the coatings. Gr barely affected the hydrophilicity of glass, possibly due to its nanoparticle (NP) nature and sparse NP distribution on the surface, as previously evidenced by scanning electron micrographs.33 In other words, the particulate nature of Gr dispersions would remain as such on the glass surface. However, on the polyethylene surface, the hydrophobic nature of Gr molecules would drive their interaction with the hydrophobic surface. Gr molecules would then leave the nanoparticles and interact with the polyethylene surface, becoming more homogeneously distributed on the surface and increasing wettability due to their terminal polar groups such as ethanolamine moieties. Cationic polymers, chitosan (CH) and PDDA, adsorb onto oppositely charged surfaces such as glass, reducing its hydrophilicity, as seen from the reduction in the spreading area of the water droplet (Table 1).
Curiously, polyacrylamide (PA) did not affect the hydrophilicity of glass, a result understandable from the hydrophilic nature of this neutral polymer, well known for its capability of forming hydrogels. On the other hand, PA and all other coatings increased the low wettability of the polyethylene surfaces. After washing out the coatings on polyethylene, surface hydrophobicity was recovered in all cases, with the exception of PA. It seems that the washing out procedure was not so effective at removing PA from the polyethylene surface. On glass, hydrophilicity was reduced by coatings with cationic polymers such as CH and PDDA but was not affected by PA. The washing out procedure apparently did not remove all CH from the surface since the reduction in the wettability of glass persisted afterwards. This was not the case for PDDA, which could be easily removed from the glass surface upon washing it out, allowing the usual hydrophilicity of glass to be completely recovered.
Microbicidal activity of Gr/polymer coatings deposited onto glass or polyethylene surfaces
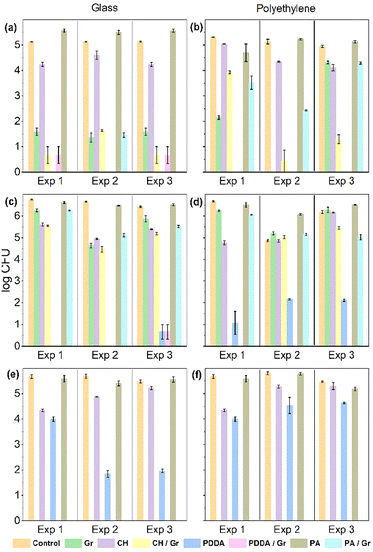
Fig. 2 compares the antimicrobial activities of Gr, CH, PA, PDDA, and their combinations. Gr and polymer doses tested were 4.71 and 25 μg, respectively. PA, a neutral polymer, did not display any antimicrobial activity in contrast to the cationic polymers.Among the cationic polymers, CH displayed a moderate activity on glass and a poor activity on PE. Gr activity against both bacteria tested was practically absent, in contrast with its remarkably high activity against C. albicans. Although PDDA was moderately effective against C. albicans, it was remarkably active against both bacterial strains tested when coatings were deposited on glass. On PE, PDDA activity against bacteria was reduced despite remaining the highest achieved among the three polymers tested. This result can be explained by PDDA's affinity for PE and its higher adsorption and permanence on the PE surface as compared to those on glass. PDDA's permanence on the PE surface could be inferred from the reduced surface hydrophobicity observed even after washing out the PDDA coating (Table 1). This would diminish the active PDDA dose in solution, reducing its bactericidal activity. In Fig. 2, only one of the coatings was able to completely reduce the cell viability of the three microbes tested: the coating PDDA/Gr deposited either on glass or on PE. This reconfirmed the synergistic action of PDDA and Gr on the coatings34 and opened new applications for the use of such coatings in biomedical materials displaying a range of hydrophobic/hydrophilic balances.
The antimicrobial effect was also evaluated for reproducibility when different coatings were independently prepared under the same conditions. Fig. 3 shows that the results were very similar for experiments 1, 2 and 3 performed against all microbial strains tested. In addition, we show in Fig. 3 the results for Escherichia coli, which were very similar to those obtained for Pseudomonas aeruginosa shown in Fig. 2. A major difference between Fig. 2 and 3 is the total amount of microbial cells employed for the experiments. In Fig. 2, one million cells were subjected to the coatings (see the control bar in Fig. 2), whereas in Fig. 3, the total number of cells tested was 10 times smaller, that is, one hundred thousand cells. The complete microbicidal effect achieved against about one million cells at the PDDA/Gr doses of 4.7 and 25 μg, respectively, was indeed remarkable.
Residual microbicidal activity of Gr/polymer coatings deposited onto glass or polyethylene surfaces after the washing out procedure
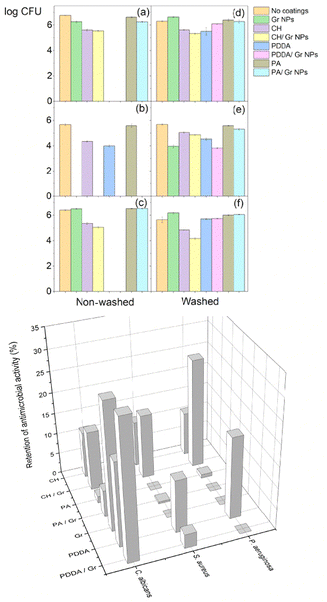
In order to test the stability of the coatings on the glass surfaces, their microbicidal activity was compared between non-washed and washed coatings (Fig. 3). In general, the washing out procedure substantially reduced the microbicidal activity of all coatings against the three microbial strains tested, as depicted in the comparisons between Fig. 4(a) and (d), (b) and (e), and (c) and (f). The retention of antimicrobial activity (%) was rarely higher than 20% of the full activity before washing out the coatings (Fig. 3, bottom). Since the self-assembly of Gr molecules yielded stable nanoparticles in water dispersions,33 the casting and drying of the Gr NPs on glass, an hydrophilic surface, did not disturb or disassemble the Gr NPs on the glass surface. Gr NPs were easily washed out from the glass surface, resulting in low retention of Gr's antimicrobial activity after washing out the coatings. The three hydrophilic polymers used in the coatings (CH, PA and PDDA) could also be easily removed from the glass surface by the washing-out procedure so their antimicrobial activity on glass was barely retained. The retention of antimicrobial activity for the coatings on polyethylene (PE) was also low, below 30% (Fig. 5). Coatings containing Gr exhibited the highest retention of activity, possibly because Gr NPs disassemble at the PE surface, driven by the hydrophobic interaction of individual Gr molecules with the PE hydrophobic surface. Consistently, the water droplet tests on the coated surfaces showed an increase in the hydrophilicity of the Gr coated PE surface as compared to the bare PE surface. This would be understandable because the polar terminals of the Gr molecules adsorbed on the PE surface increase surface hydrophilicity (Table 1).Inhibition of microbial growth on agar from polymers/Gr in filter paper disks
In order to ascertain the mobility of polymers and/or Gr on agar, filter paper disks containing the polymer, Gr or polymer/Gr dried from water dispersions were tested. The disks were placed on agar and used to determine the inhibition of growth. Inhibition halos were only observed for samples containing Gr tested against Candida albicans (Fig. 6). Polymers did not leave the paper disks to inhibit microbial growth (Fig. 6).For disks with polymer/Gr, Gr mobility on agar was not hampered by the polymer. Thus, once again the weak interaction between the polymer and Gr, previously reported for PDDA/Gr,33 was confirmed and extended to encompass the other two hydrophilic polymers tested, PA and CH. In agreement with the data in Fig. 2, which show the lack of microbicidal activity of Gr against S. aureus, no inhibition of growth was observed for Gr against S. aureus; no inhibition halos were observed (Fig. 6). Increasing Gr dose in the filter paper disk did not affect the area of the inhibition zone (Fig. 6). This showed that the inhibition of growth was related to the diffusion limit of the Gr molecule in the agar rather than to a low dose; all doses employed resulted in the same inhibition zone.
Materials and methods
Materials
Poly(diallyl dimethyl ammonium chloride) (PDDA), 35% w/v, low molecular weight (<100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), was obtained from Sigma (Steinheim, Germany). Chitosan (CH) (Fluka, 150
000 g mol−1), was obtained from Sigma (Steinheim, Germany). Chitosan (CH) (Fluka, 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), 49% acetylation degree, 4 g L−1, was prepared in dilute acetic acid, resulting in a final pH of 3.8. This stock solution was diluted in pure water (pH 6.3) and used to prepare diluted solutions. The proton dissociation constant (Ka) for the protonated amino groups of CH (100% deacetylated) has been considered as occurring at pH 6.5; thus, in our mixtures (pH 6.3–6.5), chitosan is indeed a polycation. Polyacrylamide (PA) was prepared through the polymerization of the acrylamide monomer using ammonium persulphate and N,N,N′,N′-tetramethylenediamine (TEMED) as initiators.43 30% acrylamide and 10% ammonium persulphate (APS) solutions were prepared in order to obtain a 10% polyacrylamide (PA) solution. All reagents and catalysts for polymerisation were obtained from Sigma (Steinheim, Germany). The peptide mixture named gramicidin D (peptides A, B and C, consisting mostly of Gr A), 2,2,2-trifluoroethanol (TFE), and Mueller–Hinton agar (MHA) were obtained from Sigma-Aldrich (St Louis, MO, U.S.A.). Stock solutions for gramicidin D, PDDA, CH and PA were prepared at 5 mM Gr in TFE, 20 mg mL−1 PDDA in pure water, 4 mg mL−1 CH in pure water at pH 3.5, and 30% PA in pure water.
000 g mol−1), 49% acetylation degree, 4 g L−1, was prepared in dilute acetic acid, resulting in a final pH of 3.8. This stock solution was diluted in pure water (pH 6.3) and used to prepare diluted solutions. The proton dissociation constant (Ka) for the protonated amino groups of CH (100% deacetylated) has been considered as occurring at pH 6.5; thus, in our mixtures (pH 6.3–6.5), chitosan is indeed a polycation. Polyacrylamide (PA) was prepared through the polymerization of the acrylamide monomer using ammonium persulphate and N,N,N′,N′-tetramethylenediamine (TEMED) as initiators.43 30% acrylamide and 10% ammonium persulphate (APS) solutions were prepared in order to obtain a 10% polyacrylamide (PA) solution. All reagents and catalysts for polymerisation were obtained from Sigma (Steinheim, Germany). The peptide mixture named gramicidin D (peptides A, B and C, consisting mostly of Gr A), 2,2,2-trifluoroethanol (TFE), and Mueller–Hinton agar (MHA) were obtained from Sigma-Aldrich (St Louis, MO, U.S.A.). Stock solutions for gramicidin D, PDDA, CH and PA were prepared at 5 mM Gr in TFE, 20 mg mL−1 PDDA in pure water, 4 mg mL−1 CH in pure water at pH 3.5, and 30% PA in pure water.
Software packages
All graphics were created using Origin 2022 software. References were formatted using Zotero software.Determination of the physical properties of gramicidin D dispersions in the presence of PDDA, CH or PA in pure water by dynamic light scattering (DLS)
Aliquots of a 5.0 mM gramicidin D stock solution in trifluoroethanol (TFE) once added to water dispersions yielded Gr nanoparticles (Gr NPs) over a range of Gr concentrations, while maintaining a final TFE concentration of 1%, as previously described.33,34 For interaction with polymers, water dispersions of Gr were prepared at 0.05 mM Gr by adding 0.020 mL of a 5.0 mM Gr solution in TFE to a final volume of 2.0 mL of polymer solution. Thereafter, dispersions were vortexed and their physical properties were evaluated by DLS over a range of polymer concentrations.Size, zeta-potential, polydispersity and conductance were determined by using a Zeta Plus Zeta-Potential Analyzer (Brookhaven Instruments Corporation, Holtsville, NY, U.S.A.) equipped with a 677 nm laser and dynamic light scattering at 90° for particle sizing.44 The zeta-average diameter is taken as the mean hydrodynamic diameter (Dz). Dz and the diffusion coefficient (D) for the particles in dispersion obey the Stokes–Einstein relationship, Dz = kT/(3πηD), where k is Boltzmann's constant, T is the temperature in Kelvin, and η is the medium viscosity. The algoritm in the apparatus was the non-negatively constrained least squares (NNLS) for multimodal size distributions.44 Polydispersities (P) were related to the width of Gaussian curves for size distributions. Zeta-potentials (ζ) were calculated by the apparatus software from the electrophoretic mobility μ and the equation of Smoluchowski, which is ζ = μη/ε, where η is the viscosity of the medium and ε is the medium dielectric constant. The electrodes in the apparatus also allowed determination of conductance for the dispersions (G, in μS).
Preparation of coatings from casting and drying the Gr/polymer dispersions on glass or polyethylene surfaces followed by determination of wettability from the shape of a 0.05 mL water droplet on the coatings
Substrates for depositing the coatings (glass coverslips and polyethylene sheets) were thoroughly cleaned with 70% ethanol under laminar flow and UV-Vis radiation before casting 0.050 mL of the Gr, polymer or Gr/polymer water dispersions and drying them overnight in a desiccator under vacuum. Final Gr and/or polymer masses in the dried coatings were fixed at 4.71 and/or 25 μg, respectively. The coated substrates were used for characterizing the wettability of the coatings by adding 0.050 mL water droplets and taking photos five minutes after depositing the droplets on the dried films. After applying the water droplet, bare glass or polyethylene surfaces were also photographed as controls. The wettability of the coatings was reproducible, and triplicate for each coating was represented by a single photo in Table 1. The coated substrates in triplicate were also used for the evaluation of their microbicidal activity as described below.Growth of microbes and determination of cell viability on Gr/polymer coatings by plating and colony-forming units (CFU) counting
Stocks of Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 29213) or Candida albicans (ATCC 90028) were kept at −20 °C in appropriate solutions. Pseudomonas aeruginosa PA14 was a kind gift from Dr Regina Baldini.45 All strains were reactivated for growing separately by streaking them on Mueller–Hinton agar (MHA) plates and incubating at 37 °C for 18–48 h. Thereafter, some colonies were added to a 0.264 M D-glucose isotonic solution and the turbidity of the suspensions at 625 nm was adjusted to a 0.5 McFarland scale. This turbidity was equivalent to 107–108 colony-forming units per mL, CFU mL−1. The 0.264 M D-glucose solution was used to avoid inactivation of cationic antimicrobials by the ionic strength or other negatively charged components of the culture medium (for example, amino acids or polysaccharides).For determining cell viability, 50 μL of the cell suspensions were added to the coatings prepared by casting and drying 50 μL of each water dispersion or solution (namely, Gr, PDDA, CH, PA, Gr/PDDA, Gr/CH, Gr/PA dispersions) onto glass coverslips or polyethylene sheets, as described in the previous section.
Coatings and microbes interacted for 1 h. Afterward, the coated coverslips with interacting microbes on top were placed into 10 mL of 0.264 M D-glucose isotonic solution. After stirring, three aliquots of 0.1 mL were withdrawn from the assay tube and were either directly plated onto MHA agar or diluted 10 to 106 times before plating 0.1 mL of each dilution and incubating (at 37 °C for 24 h) the plates for CFU determination. Thus, CFU counting was performed in triplicate in order to obtain a mean value and an error bar corresponding to the mean standard deviation. The logarithm of CFU was plotted against the types of coatings tested. CFU counting was taken as 1 for CFU counting equal to zero, so that log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU could be taken as zero. Three different coatings prepared under the same conditions gave reproducible results. The stability of the coatings under air was good and they could be used at any time after casting and drying. Under water however, the hydrophilic coatings would detach from the substrate, as evaluated from the washing-out experiments.
CFU could be taken as zero. Three different coatings prepared under the same conditions gave reproducible results. The stability of the coatings under air was good and they could be used at any time after casting and drying. Under water however, the hydrophilic coatings would detach from the substrate, as evaluated from the washing-out experiments.
Determination of the residual microbicidal activity of the coatings after the washing-out procedure in a water medium
The washing-out procedure involved immersing the coatings in a 0.264 M D-glucose solution for 1 h, followed by removing them from the solution, drying and assessing their interaction with the microbes, as described in the previous section. The residual activity of the coatings R(%) after the washing-out procedure was determined as follows:R = 100(log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFUcontrol – log CFUcontrol – log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFUwashed)/log CFUwashed)/log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFUcontrol. CFUcontrol. |
Determination of the inhibition of microbial growth on agar by Gr, PDDA, CH, PA and their combinations
In order to evaluate the mobility of Gr, polymers and their combinations on agar, inhibition halos were determined against S. aureus and C. albicans. Filter paper disks with a 3 mm diameter and 3 μm porosity were sterilized in a laminar flow cabinet before adding 0.01 mL of each dispersion prepared in 0.264 M D-glucose solution. As a control for disk and D-glucose solution sterility, 0.01 mL of 0.264 M D-glucose solution was routinely checked on filter paper disks. After depositing the water dispersions on the filter paper disks, they were dried overnight at room temperature and placed on MHA media that had been swabbed with the microbes. Polymer and/or Gr doses (in μg) numbered from 1 to 3 were as follows: (1) 0.5/0.941; (2) 0.25/0.47; and (3) 0.1/0.188, respectively.Conclusions
Nanoparticles of gramicidin D were first described and characterized in our lab in 2022.33 Gr formulations as NPs will possibly protect Gr molecules against proteases in vivo.46 There is a general trend in the literature to formulate antimicrobial peptides as assemblies to overcome their poor stability in vivo due to degradation. These assemblies can be self-assemblies or assemblies of modified peptides with added molecular moieties that are able to promote their assembly.47 Besides promoting protection against proteases, in certain instances, enhanced activity of the assembled peptides might be expected.33,46,47 The Gr NPs formulated in the present work showed a remarkable activity against Candida albicans and displayed a weak interaction with the three hydrophilic polymers PA, CH and PDDA. The microbicidal action of Gr was enhanced by the presence of PDDA but was unaffected by PA or CH, polymers that did not display a high antimicrobial action (Fig. 2). The cationic moieties in PDDA conferred a high microbicidal activity to the formulations against two other pathogens, S. aureus and P. aeruginosa. The Gr NP/PDDA coating deposited on either glass or PE was able to completely reduce the cell viability of the three microbes tested. This reconfirmed the synergistic action of PDDA and Gr on the coatings34 and opened new applications for the use of such coatings in biomedical materials displaying a variety of hydrophobic/hydrophilic balances. The mechanism of action is dual, with PDDA combining with biopolymers from the microbial cell wall facilitating Gr NPs’ interaction with the cell membrane and thus disrupting the cell ionic balance. Despite the transient character of the Gr/polymer coatings, which are easily detached from the surfaces upon washing out, they may still find utility for providing biomedical devices with timely protection over a limited period of time. Comparing the present combinations of gramicidin/polymers with a conventional antibiotic namely tetracycline (and its analogs) reveals important differences in their mechanisms of action. Tetracyclines also have a broad spectrum of activity against both Gram-positive and Gram-negative bacterial species. They bind to the 30S ribosomal subunit, which prevents the binding of aminoacyl-tRNA to the ribosome and consequently inhibits bacterial protein synthesis, a mechanism allowing the appearance of resistance via efflux pumps.48 In contrast, the gramicidin/polymer combinations act via the electrostatic interaction between the cationic polymer and the biopolymers of the cell wall, which causes cell wall disruption,31 thereby facilitating gramicidin access to the pathogen cell membrane.31–34,49–53 Therefore, the present coatings do not affect the pathogen metabolic pathways and kill the microbes by a dual and complementary mechanism: first, involving the interaction between the cationic polymer and the cell wall,31 and second, favoring the insertion of the gramicidin dimeric channel into the pathogen cell membranes. This synergistic mode of action not only kills resistant strains31 but also reduces the required doses of gramicidin and the polymer needed for complete microbicidal activity.33,34 In addition, the remarkable fungicidal activity imparted by gramicidin to the combination further broadened the microbicidal activity to include fungi as well. Therefore, this work points out that the complementary action of the self-assembled gramicidin nanoparticles and the hydrophilic, cationic polymer PDDA can be useful for developing antimicrobial coatings on a variety of important biomedical surfaces.Thanks to antimicrobial nanoscale materials, often built from self-assembly systems, recent advances against antimicrobial resistance have been achieved.54 Antimicrobial peptides (AMPs) are a novel class of alternatives that possess potent activity against a wide range of Gram-negative and positive bacteria with no ability to induce antimicrobial resistance. Novel developments involving AMPs often introduce chemical modifications to improve their bioavailability and protection against the in vivo environment.55 Gramicidin, a channel model of membrane proteins,56 self-assembles in aqueous solution to form useful nanoparticles.33 The coatings described in the present work may evolve to provide broad-spectrum antimicrobial coverage for a variety of biomedical surfaces, mainly in combination with the potent cationic and antimicrobial polymer PDDA.
Author contributions
Conceptualization, A. M. C.-R.; methodology, A. M. C.-R., L. C. S. C., B. R. B., R. T. R., G. M. Q., and A. C. B. M.; validation, A. M. C.-R., L. C. S. C., B. R. B., R. T. R., G. M. Q., and A. C. B. M.; formal analysis, A. M. C.-R., L. C. S. C., and R. T. R.; investigation, A. M. C.-R., L. C. S. C., B. R. B., R. T. R., G. M. Q., and A. C. B. M.; resources, A. M. C.-R.; data curation, A. M. C.-R.; writing – original draft preparation, A. M. C.-R.; writing – review and editing, A. M. C.-R.; visualization, A. M. C.-R.; supervision, A. M. C.-R.; project administration, A. M. C.-R.; and funding acquisition, A. M. C.-R. All authors have read and agreed to the published version of the manuscript.Data availability
The authors confirm that the data supporting the finding of this study are available within the article.Conflicts of interest
There are no conflicts of interest to declare. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.Acknowledgements
L. C. S. C., G. M. Q. and B. R. B. were undergraduate students receiving fellowships from Universidade de São Paulo, Programa Unificado de Bolsas (PUB). Regina Lúcia Baldini is gratefully acknowledged for the kind gift of P. aeruginosa PA14. A. M. C.-R. is the recipient of grants 304091/2023-5 and 302758/2019-4 from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).References
- J. Li, J.-F. Revol and R. H. Marchessault, J. Colloid Interface Sci., 1996, 183, 365–373 CrossRef CAS PubMed
.
-
S.-A. Jin and R. J. Spontak, in Handbook of Natural Polymers, ed. M. S. Sreekala, L. Ravindran, K. Goda and S. Thomas, Elsevier, 2023, vol. 1, pp. 203–249 Search PubMed
.
-
J. Desbrieres, S. Elkadaoui, A. Tolaimate, M. S. Sreekala, L. Ravindran, K. Goda and S. Thomas, in Handbook of Natural Polymers, Elsevier, 2023, vol. 1, pp. 123–151 Search PubMed
.
- R. C. W. Liu, Y. Morishima and F. M. Winnik, Macromolecules, 2001, 34, 9117–9124 CrossRef CAS
.
- H. J. Prado and M. C. Matulewicz, Eur. Polym. J., 2014, 52, 53–75 CrossRef CAS
.
- U. Choi, Colloids Surf., A, 1999, 157, 193–202 CrossRef CAS
.
- N. M. Kuznetsov, V. V. Kovaleva, S. I. Belousov and S. N. Chvalun, Mater. Today Chem., 2022, 26, 101066 CrossRef CAS
.
- Y. Lvov, K. Ariga, I. Ichinose and T. Kunitake, J. Am. Chem. Soc., 1995, 117, 6117–6123 CrossRef CAS
.
- K. Ariga, Y. Lvov and G. Decher, Phys. Chem. Chem. Phys., 2022, 24, 4097–4115 RSC
.
- J. J. Richardson, J. Cui, M. Björnmalm, J. A. Braunger, H. Ejima and F. Caruso, Chem. Rev., 2016, 116, 14828–14867 CrossRef CAS PubMed
.
- R. Divakaran and V. N. Sivasankara Pillai, Water Res., 2001, 35, 3904–3908 CrossRef CAS PubMed
.
-
A. Maiti, A. A. Mule, A. Kumar, A. Bhatnagar and P. Mondal, in Encyclopedia of Materials: Plastics and Polymers, ed. M. S. J. Hashmi, Elsevier, Oxford, 2022, pp. 392–410 Search PubMed
.
- M. Ashmore, J. Hearn and F. Karpowicz, Langmuir, 2001, 17, 1069–1073 CrossRef CAS
.
- L. D. Melo, E. M. Mamizuka and A. M. Carmona-Ribeiro, Langmuir, 2010, 26, 12300–12306 CrossRef CAS PubMed
.
- H. J. Watzke and C. Dieschbourg, Adv. Colloid Interface Sci., 1994, 50, 1–14 CrossRef CAS
.
- M. Hamidi, A. Azadi and P. Rafiei, Adv. Drug Delivery Rev., 2008, 60, 1638–1649 CrossRef CAS PubMed
.
- Y. Q. Almajidi, J. Gupta, F. S. Sheri, R. S. Zabibah, A. Faisal, A. Ruzibayev, M. Adil, M. J. Saadh, M. J. Jawad, F. Alsaikhan, A. Narmani and B. Farhood, Int. J. Biol. Macromol., 2023, 253, 127278 CrossRef CAS PubMed
.
-
O. N. Abiola, in Polymeric Materials for Clean Water, ed. R. Das, Springer International Publishing, Cham, 2019, pp. 77–92 Search PubMed
.
- S. Gulati, H. N. Lingan B, A. Baul, S. Kumar, R. Wadhwa, M. Trivedi, R. S. Varma and A. Amar, New J. Chem., 2022, 46, 17114–17139 RSC
.
- P. A. Alaba, N. A. Oladoja, Y. M. Sani, O. B. Ayodele, I. Y. Mohammed, S. F. Olupinla and W. M. W. Daud, J. Environ. Chem. Eng., 2018, 6, 1651–1672 CrossRef CAS
.
- K. A. Janes, P. Calvo and M. J. Alonso, Adv. Drug Delivery Rev., 2001, 47, 83–97 CrossRef CAS
.
- D. Luo, X. Ni, H. Yang, L. Feng, Z. Chen and L. Bai, Eur. J. Pharm. Sci., 2024, 192, 106630 CrossRef CAS PubMed
.
- E. Ruckenstein and X. Zeng, Biotechnol. Bioeng., 1997, 56, 610–617 CrossRef CAS
.
- B. Krajewska, Enzyme Microb. Technol., 2004, 35, 126–139 CrossRef CAS
.
- S. P. Strand, M. S. Vandvik, K. M. Vårum and K. Østgaard, Biomacromolecules, 2001, 2, 126–133 CrossRef CAS
.
- R. Yang, H. Li, M. Huang, H. Yang and A. Li, Water Res., 2016, 95, 59–89 CrossRef CAS PubMed
.
- A. M. Carmona-Ribeiro, Biomimetics, 2021, 7, 6 CrossRef PubMed
.
- Y. Pérez-Betancourt, B. d. C. L. F. Távora, M. Colombini, E. L. Faquim-Mauro and A. M. Carmona-Ribeiro, Vaccines, 2020, 8, 105 CrossRef PubMed
.
-
A. M. Carmona-Ribeiro, B. I. Mathiazzi and Y. Pérez-Betancourt, in Vaccine Design: Methods and Protocols, Volume 3. Resources for Vaccine Development, ed. S. Thomas, Springer US, New York, NY, 2022, pp. 233–245 Search PubMed
.
- D. B. Vieira and A. M. Carmona-Ribeiro, J. Nanobiotechnol., 2008, 6, 6 CrossRef PubMed
.
- L. D. de Melo Carrasco, J. L. M. Sampaio and A. M. Carmona-Ribeiro, Int. J. Mol. Sci., 2015, 16, 6337–6352 CrossRef PubMed
.
- L. M. Sanches, D. F. S. Petri, L. D. de Melo Carrasco and A. M. Carmona-Ribeiro, J. Nanobiotechnol., 2015, 13, 58 CrossRef PubMed
.
- Y. Pérez-Betancourt, R. Zaia, M. F. Evangelista, R. T. Ribeiro, B. M. Roncoleta, B. I. Mathiazzi and A. M. Carmona-Ribeiro, Pharmaceutics, 2022, 14, 2053 CrossRef PubMed
.
- R. Zaia, G. M. Quinto, L. C. S. Camargo, R. T. Ribeiro and A. M. Carmona-Ribeiro, Pharmaceuticals, 2023, 16, 816 CrossRef CAS PubMed
.
- H. Spece, R. V. Yarbrough and S. M. Kurtz, J. Arthroplasty, 2023, 38, 970–979 CrossRef
.
- A. Said, Khalij-Libya J Dent Med Res, 2022, 11–16 Search PubMed
.
- Z. Sun, L. Ma, X. Sun, A. J. Sloan, N. M. O'Brien-Simpson and W. Li, Aggregate, 2023, 4, e309 CrossRef CAS
.
- M. G. Drexelius and I. Neundorf, Int. J. Mater. Sci., 2021, 22, 13212 CAS
.
- B. Skerlavaj and G. Boix-Lemonche, Antibiotics, 2023, 12, 211 CrossRef CAS PubMed
.
- J. Chadha, N. Thakur, S. Chhibber and K. Harjai, Crit. Rev. Microbiol., 2023, 1–28 Search PubMed
.
- K. Liu, M. Cao, A. Fujishima and L. Jiang, Chem. Rev., 2014, 114, 10044–10094 CrossRef CAS PubMed
.
- D. B. Vieira, N. Lincopan, E. M. Mamizuka, D. F. S. Petri and A. M. Carmona-Ribeiro, Langmuir, 2003, 19, 924–932 CrossRef CAS
.
-
J. M. Walker, in Principles and tecniques of pratical biochemistry, ed. K. Wilson and J. M. Walker, Cambridge, 4th edn, 1994, pp. 425–461 Search PubMed
.
-
E. Grabowski and I. Morrison, in Measurement of suspended particles by quasi-elastic light-scattering, ed. B. Dahneke, Wiley Interscience, New York, NY, 1st edn, 1983, pp. 199–236 Search PubMed
.
- J. He, R. L. Baldini, E. Déziel, M. Saucier, Q. Zhang, N. T. Liberati, D. Lee, J. Urbach, H. M. Goodman and L. G. Rahme, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 2530–2535 CrossRef CAS PubMed
.
- A. M. Carmona-Ribeiro, Future Pharmacol., 2023, 3, 763–788 CrossRef
.
- L. Lombardi, Y. Shi, A. Falanga, E. Galdiero, E. de Alteriis, G. Franci, I. Chourpa, H. S. Azevedo and S. Galdiero, Biomacromolecules, 2019, 20, 1362–1374 CrossRef CAS PubMed
.
- F. Nguyen, A. L. Starosta, S. Arenz, D. Sohmen, A. Dönhöfer and D. N. Wilson, Biol. Chem., 2014, 395, 559–575 CrossRef CAS PubMed
.
- A. M. Carmona-Ribeiro and L. D. de Melo Carrasco, Int. J. Mol. Sci., 2013, 14, 9906–9946 CrossRef PubMed
.
- L. D. M. Carrasco, R. J. Bertolucci, R. T. Ribeiro, J. L. M. Sampaio and A. M. Carmona-Ribeiro, Front. Microbiol., 2016, 7, 1804 Search PubMed
.
- C. N. Galvão, L. M. Sanches, B. I. Mathiazzi, R. T. Ribeiro, D. F. S. Petri and A. M. Carmona-Ribeiro, Int. J. Mol. Sci., 2018, 19, 2965 CrossRef PubMed
.
- R. T. Ribeiro, C. N. Galvão, Y. P. Betancourt, B. I. Mathiazzi and A. M. Carmona-Ribeiro, Int. J. Mol. Sci., 2019, 20, 6150 CrossRef CAS PubMed
.
-
A. M. Carmona-Ribeiro, Biomimetic nanomaterials from the assembly of polymers, lipids and surfactants, ed. A. K. Dutta, IntechOpen, Rijka, Croatia, 2019, pp. 1–16 Search PubMed
.
- J. A. Doolan, G. T. Williams, K. L. F. Hilton, R. Chaudhari, J. S. Fossey, B. T. Goult and J. R. Hiscock, Chem. Soc. Rev., 2022, 51, 8696–8755 RSC
.
- W. Li, F. Separovic, N. M. O'Brien-Simpson and J. D. Wade, Chem. Soc. Rev., 2021, 50, 4932–4973 RSC
.
- J. Yoo and Q. Cui, Biophys. J., 2013, 104, 117–127 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2024 |